SUMMARY
The Piwi-interacting RNA (piRNA) pathway suppresses transposable elements and promotes fertility in diverse organisms. Maturation of piRNAs involves pre-piRNA trimming followed by 2′-O-methylation at their 3′ termini. Here, we report that the 3′ termini of Caenorhabditis elegans piRNAs are subject to nontemplated nucleotide addition, and piRNAs with 3′ addition exhibit extensive base-pairing interaction with their target RNAs. Animals deficient for PARN-1 (pre-piRNA trimmer) and HENN-1 (2′-O-methyltransferase) accumulate piRNAs with 3′ nontemplated nucleotides. In henn-1 mutants, piRNAs are shortened prior to 3′ addition, whereas long isoforms of untrimmed piRNAs are preferentially modified in parn-1 mutant animals. Loss of either PARN-1 or HENN-1 results in modest reduction in steady-state levels of piRNAs. Deletion of both enzymes leads to depletion of piRNAs, desilenced piRNA targets, and impaired fecundity. Together, our findings suggest that pre-piRNA trimming and 2′-O-methylation act collaboratively to protect piRNAs from tailing and degradation.
Graphical abstract
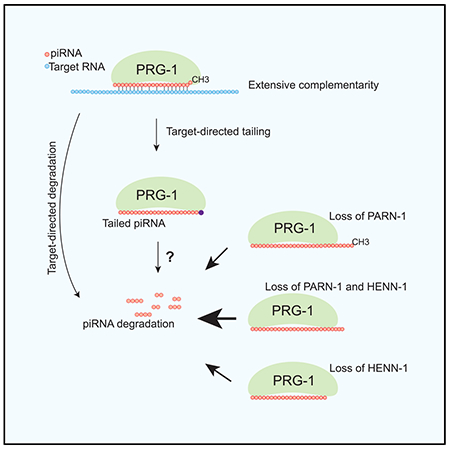
In brief
Pastore et al. show that, in C. elegans, target RNAs trigger piRNA 3′ tailing and degradation, and 3′-end processing of piRNAs protects piRNAs from tailing and degradation.
INTRODUCTION
Argonaute (AGO) proteins and their small noncoding RNAs play a vital role in regulating gene expression (Ghildiyal and Zamore, 2009). The Piwi protein, a germline-enriched AGO, and Piwi-interacting RNAs (piRNAs) are essential for gametogenesis and germline maintenance (Carmell et al., 2007; Cox et al., 1998; Goh et al., 2015; Gou et al., 2014; Houwing et al., 2007; Lin and Spradling, 1997). The length, sequence, and genomic origin of piRNAs vary between species. However, their biogenesis pathway is similar (Luteijn and Ketting, 2013; Ozata et al., 2019; Weick and Miska, 2014). In flies and mice, piRNAs are derived from long single-stranded transcripts (Aravin et al., 2006; Brennecke et al., 2007; Li et al., 2013; Malone et al., 2009). Piwi proteins initiate cleavage of piRNA precursors and generate 5′-monophosphorylated (5′-monoP) intermediates that are loaded onto another Piwi protein (Gainetdinov et al., 2018; Han et al., 2015; Mohn et al., 2015). An endonuclease (Zucchini in flies; PLD6 in mice) cleaves 3′ to the footprint of the Piwi protein, which releases Piwi-bound pre-piRNA and establishes 5′-monoP intermediates for Piwi loading (Ipsaro et al., 2012; Nishimasu et al., 2012; Olivieri et al., 2010; Pane et al., 2007). This stepwise fragmentation of long precursors produces tail-to-head strings of phased pre-piRNAs (Gainetdinov et al., 2018; Han et al., 2015; Homolka et al., 2015; Mohn et al., 2015). The 3′ ends of pre-piRNA must be further trimmed to an optimal length to be accommodated by Piwi proteins (Kawaoka et al., 2011; Saxe et al., 2013; Vourekas et al., 2012). This trimming process requires 3′-to-5′ exonucleases (Ding et al., 2017; Izumi et al., 2016; Nishimura et al., 2018; Tang et al., 2016; Zhang et al., 2017) and Tudor-domain protein Papi/Tdrkh (Honda et al., 2013; Liu et al., 2011; Saxe et al., 2013). The last step of piRNA maturation is the 2′-O-methylation at their 3′ termini, a reaction catalyzed by a conserved methyltransferase, Hen1 (Horwich et al., 2007; Kirino and Mourelatos, 2007; Saito et al., 2007).
In Caenorhabditis elegans, piRNAs are expressed from thousands of genomic loci, of which most are localized at two large genomic clusters on chromosome IV (Batista et al., 2008; Das et al., 2008; Gu et al., 2012; Ruby et al., 2006). Mature piRNAs are loaded into Piwi protein PRG-1 and referred to as 21U-RNAs because of their strong propensity for a 5′-monoP uridine residue and length of 21 nucleotides (nt) (Batista et al., 2008; Das et al., 2008; Gu et al., 2012; Ruby et al., 2006). 21U-RNA precursors (pre-piRNAs) are produced from 25- to 29-nt capped small RNA precursors (csRNAs) that are transcribed by RNA polymerase II and initiate precisely two nucleotides upstream of the 5′ end of mature piRNAs (Cecere et al., 2012; Gu et al., 2012). csRNAs are stabilized by proteins containing 5′ cap and 5′ phosphate RNA-binding domains (Cordeiro Rodrigues et al., 2019; Goh et al., 2015). To generate 21U-RNAs, the 5′ cap and first two nucleotides of csRNAs are removed, and extra nucleotides are trimmed from their 3′ end. The 3′-end processing involves 3′-to-5′ trimming mediated by the exonuclease PARN-1 followed by 2′-O-methylation catalyzed by the methyltransferase HENN-1 (Billi et al., 2012; Kamminga et al., 2012; Montgomery et al., 2012; Tang et al., 2016).
Although much is known about piRNA biogenesis, little is known about its degradation. This stands in contrast to the significant body of work on microRNA (miRNA) decay (Bartel, 2018; Sanei and Chen, 2015). For example, extensive base complementarity between the miRNA and its target RNA results in degradation of the bound miRNAs in a process known as target-directed miRNA degradation (TDMD) (Ameres et al., 2010; Baccarini et al., 2011; Bitetti et al., 2018; de la Mata et al., 2015; Fuchs Wightman et al., 2018). In TDMD, the base-paring interaction facilitates dislocation of the miRNA 3′ terminus, making it susceptible to enzymatic attacks (Sheu-Gruttadauria et al., 2019). An array of terminal nucleotidyltransferases (TENTs) add nontemplated A/U nucleotides to 3′ ends of miRNAs, a process known as 3′ tailing (Yang et al., 2020; Yu and Kim, 2020; Zhao et al., 2012). The adenylation and uridylation of miRNAs are associated with trimming in a 3′-to-5′ direction and turnover of specific miRNAs (Faehnle et al., 2014; Haas et al., 2016; Katoh et al., 2015). In a tailing- and trimming-independent manner, highly complementary RNA targets can trigger TDMD through proteolysis of the ARGO protein (Han et al., 2020; Kleaveland et al., 2018; Shi et al., 2020). So far, the mechanism(s) that drive piRNA decay are largely unknown.
Here, we report a quality-control mechanism for piRNAs that is mediated by 3′ tailing. Extensive base-pair interactions between piRNAs and their target RNAs are associated with piRNA 3′ nontemplated nucleotide addition. Loss of either parn-1 or henn-1 leads to the increased frequency of piRNA tailing. In the adult germline in which piRNAs are actively produced, 3′ addition has limited effects on steady-state piRNA levels. However, in early embryos in which the transcription of piRNA genes is inactive, 3′ modifications lead to a decrease in piRNA levels. Simultaneous deletion of both enzymes results in collapse of piRNAs, desilencing of piRNA targets, and a strong decline in fertility. Together, our findings suggest that highly complementary target RNAs can induce piRNA tailing and demonstrate that pre-piRNA trimming and 2′-O-methylation act independently to protect piRNAs from tailing-mediated degradation.
RESULTS
PARN-1 and HENN-1 are required for 21U-RNA accumulation
In C. elegans, 5′ processing of csRNAs occurs prior to or independently of 3′ processing (Luteijn and Ketting, 2013; Ozata et al., 2019; Weick and Miska, 2014). The 3′ termini of mature piRNAs are generated by trimming followed by 2′-O-methylation catalyzed by PARN-1 and HENN-1 (Billi et al., 2012; Kamminga et al., 2012; Montgomery et al., 2012; Tang et al., 2016). To examine the interplay between these two key processes on piRNA 3′ termini, we cloned and deep sequenced small RNAs isolated from wild-type C. elegans and henn-1, parn-1, and henn-1; parn-1 mutant young adults and embryos. To capture piRNA 3′ extensions and modifications, we generated a reference genome containing a 10-nt 3′ extension of annotated piRNA genes. Sequencing reads that mapped to the reference with no mismatches were obtained, analyzed, and plotted (Figures 1A–1D; Table S1). In all genetic backgrounds and developmental stages tested, piRNA abundance determined from two biological replicates was well correlated (Pearson’s ρ = 0.94–1) (Figures S1A–S1H).
Figure 1. Loss of henn-1, parn-1, and henn-1; parn-1 destabilizes piRNAs.
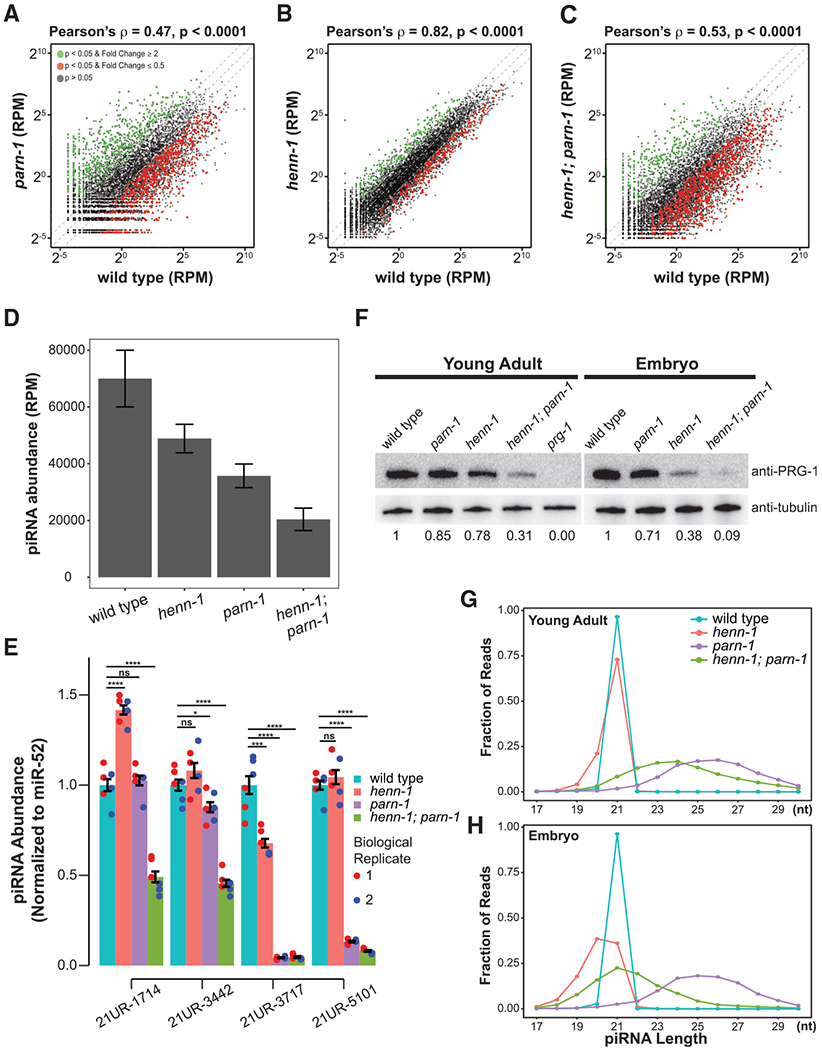
(A–C) Scatterplots showing the expression of piRNAs in parn-1 (A), henn-1 (B), and henn-1; parn-1 mutant young adults (C) relative to wild-type control (n = 10,870). Green and red dots represent piRNA expression in mutants that significantly changed (p < 0.05) with a fold change greater than or equal to 2 or less than or equal to 0.5, respectively. The central diagonal line represents a fold change of 1, the top diagonal line represents a fold change of 2, and the bottom diagonal line represents a fold change of 0.5. The Welch two-sample t test was used to calculate p values; correlation statistics were calculated using the Pearson’s correlation test (for a perfect correlation, the correlation coefficient ρ = 1 or −1; and for no correlation, ρ = 0).
(D) Barplot showing the total normalized reads per million (RPM) mapping to piRNA loci (n = 10,870) in wild type and henn-1, parn-1, and henn-1; parn-1 mutant animals. Error bars represent standard deviation in the total RPM mapping to piRNA loci in two biological replicates.
(E) Barplots showing the expression of 21UR-1714, 21UR-3442, 21UR-3717, and 21UR-5101 in wild-type and henn-1, parn-1, and henn-1; parn-1 mutant young adults measured using qRT-PCR. piRNA cycle threshold (Ct) values were normalized to miRNA miR-52 Ct values. The error bars represent the standard deviation. Blue and red dots represent technical triplicates from biologically distinct samples. A two-sample t test was used to statistically compare mutant expression to that of wild type. Significance indicators are as follows: ns (no significance), *p ≥ 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
(F) Western blotting analysis quantifying the abundance of the PRG-1 protein in wild-type and henn-1, parn-1, and henn-1; parn-1 mutant young adults and embryos. A prg-1 negative control was included in the young adult sample. A β-tubulin control served as the loading control. Relative signals with respect to wild-type PRG-1 blotting are indicated below each blot.
(G and H) Line graphs showing the length distribution of piRNAs in wild-type (blue), henn-1 (red), parn-1 (purple), and henn-1; parn-1 (green) young adult (G) and embryo (H). The y axis represents the fraction of reads at each length indicated on the x axis.
C. elegans deficient for PARN-1 accumulates untrimmed piRNAs that appear to be 2′-O-methylated at their 3′ termini (Tang et al., 2016). piRNA levels in parn-1 mutant young adults generally correlated with their corresponding wild-type counterparts (Pearson’s ρ = 0.47) (Figure 1A; Tang et al., 2016). Yet, a subset of piRNAs was significantly differentially expressed. Among them, 628 were downregulated in parn-1 mutant young adults compared to 577 with increased expression (Figure 1A). Overall, piRNA levels decreased to 51.0% of the wild type (Figure 1D).
It is challenging to compare levels of wild-type piRNAs with those in henn-1 mutants because 2′-O-methylation inhibits 3′ adaptor ligation during high-throughput sequencing library preparation as described (Munafó and Robb, 2010; Svendsen et al., 2019). To minimize such a bias, we attempted to clone 2′-O-methyl RNA species by adding polyethylene glycol during the adaptor ligation step as described (Dard-Dascot et al., 2018; Munafó and Robb, 2010). To quantify the difference in ligation efficiency, we included synthetic single-strand RNA spike-ins with either 2′-hydroxyl or 2′-O-methyl 3′ ends in our library preparation protocol. We found that 2′-O-methyl RNA was at least 5-fold less efficiently detected than 2′-hydroxyl RNA. We thus further normalized piRNA-matching reads in henn-1 and henn-1; parn-1 mutant young adults and embryos by reducing their read counts by 5-fold. After such normalization, piRNA levels in henn-1 mutants were reduced to 69.8% as compared to the wild type (Figures 1B and 1D). In henn-1; parn-1 mutant young adult animals, remarkably, levels of 846 piRNAs were significantly decreased (Figure 1C). Steady-state piRNA levels were further reduced to 29.2% relative to the wild type (Figure 1D).
Because HENN-1 is required for piRNA accumulation in embryos (Billi et al., 2012; Montgomery et al., 2012; Svendsen et al., 2019), we next profiled small RNAs from wild-type and mutant early embryos. When analyzing the reads mapping piRNA loci, we found that the abundance of piRNAs in henn-1 and parn-1 mutant embryos was reduced to 87.5% and 60.9%, respectively, as compared to that of the wild type (Figure S1I). The synergistic phenotype was observed in henn-1; parn-1 mutant embryos in which piRNA abundance was further reduced to 18.9% of the wild type (Figure S1I). To corroborate these observations, we used qRT-PCR to quantify the levels of four randomly selected piRNAs in young adults. Because qRT-PCR does not require adaptor ligation, this approach should be less sensitive to the 2′-O-methyl modification. Although there was variation between individual piRNAs, their expression levels tended to be lower in either parn-1 or henn-1 single-mutant animals, with the strongest reduction in henn-1; parn-1 double-mutant strains (Figure 1E).
The steady-state levels of piRNAs and the PRG-1 protein are co-dependent (Batista et al., 2008; Das et al., 2008; Weick et al., 2014). Consistent with the change in piRNA levels, levels of the PRG-1 protein were lower in henn-1 and parn-1 single mutants when assayed by western blot. Deletion of both henn-1 and parn-1 reduced the PRG-1 level to ~30% in young adults and ~10% in the embryos relative to wild type (Figure 1F). We therefore concluded that both 3′ trimming and 2′-O-methylation are required for accumulation of piRNA and the Piwi protein during both germline development and embryogenesis.
In wild-type worms, piRNAs have a uniform length of 21 nt (Batista et al., 2008; Das et al., 2008; Ruby et al., 2006). parn-1 mutant animals accumulate piRNAs that are several nucleotides longer (Figure 1G; Tang et al., 2016). In agreement with previous findings, henn-1 mutants produced piRNAs that were mainly 21 nt, with a small fraction showing 1- to 2-nt truncations (Figure 1G; Kamminga et al., 2012; Svendsen et al., 2019). In henn-1; parn-1 young adults in which piRNAs failed to be trimmed and methylated, piRNAs had 3′ extensions but became 1 to 2 nt shorter than their counterparts in parn-1 mutants (Figure 1G). Strikingly, such 3′ shortening became more prevailing in embryos. For example, in henn-1 mutant embryos, the length of piRNAs was 18~21 nt and peaked at 20 nt (Figures 1H; Svendsen et al., 2019). In henn-1; parn-1 double mutants, piRNA size showed a broad range, with the peak shifting from 24 nt in young adults to 21 nt in embryos (Figure 1H). These observations suggest that piRNA 3′ ends must be properly trimmed and methylated to prevent 3′ shortening and degradation.
Activities of PARN-1 and HENN-1 suppress piRNA tailing
To explore the relationship between 3′ shortening and 3′ addition of piRNA, we developed a computational pipeline to define nontemplated nucleotides (see STAR Methods). Briefly, we removed one nucleotide from the 3′ end of sequencing reads that fail to align to the genome reference and re-aligned these trimmed reads to the reference. We conducted three iterations, which allowed us to detect up to three occurrences of nontemplated additions from individual reads (Figures 2A and 2B).
Figure 2. Nontemplated nucleotide addition occurs at piRNA 3′ termini.
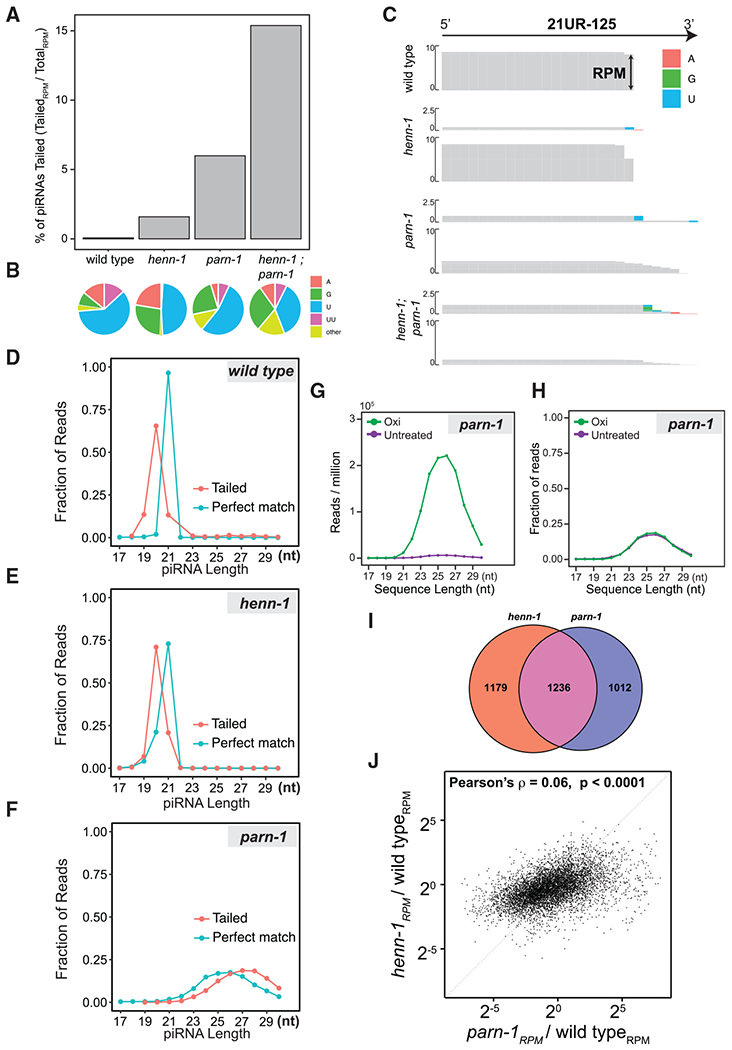
(A) Barplot showing the ratio of tailed piRNA reads to total reads mapping to piRNA loci in wild-type, henn-1, parn-1, and henn-1; parn-1 young adults.
(B) Pie charts showing the composition of nontemplated nucleotides in wild-type, henn-1, parn-1, and henn-1; parn-1 young adults.
(C) Genome browser view of sequencing reads mapped to 21UR-125. The height of each bar is representative of the RPM. Nontemplated nucleotides are indicated by the colored boxes. For each mutant, reads with nontemplated nucleotide additions are shown above perfectly matching reads. The scale of perfect match reads and tailed reads was 0–10 and 0–2.5, respectively.
(D–F) Line plots showing the distribution of tailed (red line) and perfect match reads (blue line) in wild-type(D), henn-1 (E), and parn-1 (F) young adults. The y axis represents the fraction of reads at each length indicated on the x axis. Lengths of tailed reads are reported after removing nontemplated nucleotides.
(G and H) Line plots showing the length distribution of perfectly matching reads mapping to piRNA loci in parn-1 mutants under untreated (purple) or oxidized (Oxi) conditions (green). Reads were grouped into bins according to their length, and the total normalized RPM (G) and the fraction of reads (H) of each group were plotted.
(I) Venn diagram showing the overlap of tailed piRNAs in parn-1 and henn-1 young adults. The number of tailed piRNAs specific to each genotype and the numbers of tailed piRNAs in both genotypes are indicated.
(J) Scatterplot showing the relationship between the fold change (mutant versus wild type) of piRNAs in henn-1 and that of parn-1 mutant young adults. The correlation coefficient was generated using Pearson’s correlation test. The diagonal line represents no difference in fold change.
We found that 3′ modifications added to piRNAs in both young adults and early embryos were mainly mononucleotides (Figures S2A–S2H). In wild-type young adult animals, the frequency of nontemplated addition was 0.07%, and 272 piRNAs contained detectable 3′ additions (Figure 2A; Table S2). The 3′ modifications were mainly mono-uridylation (60.6%) followed by mono-adenylation (14.0%) (Figure 2B). In henn-1 mutant young adults, the frequency of nontemplated additions increased ~23-fold and 2,415 piRNAs contained 3′ additions. In parn-1 mutant young adults, nontemplated additions increased ~85-fold, and in total 2,248 piRNAs had detectable 3′ additions. Loss of henn-1 and parn-1 led to an even higher level of tailing with frequency reaching 15.4% (~219-fold increase when compared to wild type). In all genetic backgrounds, mono-uridylation was the most abundant modification. Although guanylation was uncommon in the wild type, 23.7% of piRNAs in parn-1, 27.0% in henn-1, and 29.0% in henn-1; parn-1 mutant animals contained nontemplated guanine, indicating that different TENTs act on unmethylated and untrimmed piRNAs (Figure 2B). A closer inspection of randomly selected piRNA loci—21UR-125 and 21UR-205—confirmed the presence of nontemplated nucleotides in mutants (Figures 2C and S2I). Bioinformatic analyses of embryo samples revealed elevated 3′ tailing in the mutants. The frequency of nontemplated nucleotides was 0.07%, 5.21%, 4.54%, and 10.1% in wild type, henn-1, parn-1, and henn-1; parn-1 mutants, respectively (Figure S2J).
To gain more insight into the sequential order of 3′ shortening and tailing, we compared the length of piRNA perfectly matching reads to the length of tailed reads after removal of nontemplated nucleotides. If 3′ shortening occurs prior to tailing, tailed piRNAs with the removal of 3′ additions are expected to be shorter than untailed piRNA species. On the contrary, if tailing precedes 3′ shortening, we expect to find a portion of tailed reads equal to or longer than perfect match reads. In wild-type and henn-1 animals, tailed reads were 1 to 2 nt shorter than perfect match reads, indicating 21U-RNAs are truncated prior to 3′ tailing (Figures 2D and 2E; Svendsen et al., 2019). In these scenarios, the 3′ truncation may be required to expose piRNA 3′ termini to TENT(s).
In contrast to henn-1 mutants, tailed reads in parn-1 mutants were several nucleotides longer than perfect match reads (Figure 2F). One explanation for this finding is that these long piRNA isoforms are not fully methylated and thus are more susceptible to 3′ tailing. To examine the methylation status of untrimmed piRNAs, we conducted sodium-periodate-mediated oxidation experiments and sequenced oxidation-resistant small RNAs. The vicinal diol at the 3′ end of unmodified small RNAs is oxidized by sodium meta-periodate to a dialdehyde, rendering them poor substrates for small RNA cloning, whereas RNAs with 2′-O-methylation at their 3′ termini are resistant to oxidation. Consistent with previous studies, untrimmed piRNAs in parn-1 mutants were indeed enriched by oxidation when normalized to the total number of reads (Figure 2G; Tang et al., 2016). However, untreated and oxidation-resistant piRNA showed a very similar length distribution, suggesting untrimmed piRNA isoforms are efficiently 2′-O-methylated regardless of length (Figure 2H). It is thus unlikely that the absence of 2′-O-methylation is responsible for 3′ tailing of untrimmed piRNAs in parn-1 mutants.
Untrimmed piRNAs in parn-1 mutants are loaded onto and stabilized by PRG-1 (Tang et al., 2016). A speculative but intriguing possibility is that the 3′ ends of longer piRNA isoforms could not be properly accommodated by the PAZ domain of PRG-1 and therefore were more susceptible to catalysis of TENT(s). Future structural analysis of the PRG-1 complex is required to determine the interaction between the PAZ domain and 3′ termini of piRNA of different lengths.
Consistent with the idea that different mechanisms may drive the tailing of unmethylated piRNAs versus untrimmed piRNAs, overlapping yet distinct piRNA subsets exhibited 3′ additions in henn-1 and parn-1 mutants (Figure 2I). We compared the change in unmethylated piRNA abundance to the change in corresponding untrimmed piRNA abundance. We did not observe a general correlation between piRNA fold change in henn-1 mutants and that in parn-1 mutants (Pearson’s ρ = 0.06; Figure 2J). These findings suggest that distinct piRNA populations were differentially expressed in the absence of 2′-O-methylation or pre-piRNA trimming.
piRNA tailing may be linked to instability
Previous studies showed that 3′ tailing is associated with miRNA turnover (Ameres et al., 2010; Faehnle et al., 2014; Haas et al., 2016; Katoh et al., 2015). To explore the relationship between piRNA tailing and its degradation, we calculated the change of piRNA steady-state levels in the mutant strains relative to wild type and asked if the change in piRNA abundance correlated with tailing frequency. To our surprise, we found poor correlation in henn-1 mutants (Spearman’s ρ = 0.047 for young adult; Spearman’s ρ = 0.054 for embryos; Figures S3A and S3B) and in parn-1 mutants (Spearman’s ρ = 0.086 for young adult; Spearman’s ρ = −0.013 for embryos; Figures S3C and S3D), as well as in henn-1; parn-1 mutants (Spearman’s ρ = 0.12 for young adult; Spearman’s ρ = 0.024 for embryos; Figures S3E and S3F). These findings suggest at least two models, as follows: (1) degradation of piRNAs is independent of 3′ tailing; and (2) degradation of piRNAs correlates with 3′ tailing, but the synthesis of piRNAs obscures the correlation (Figure 3A).
Figure 3. piRNA tailing may be linked to instability.
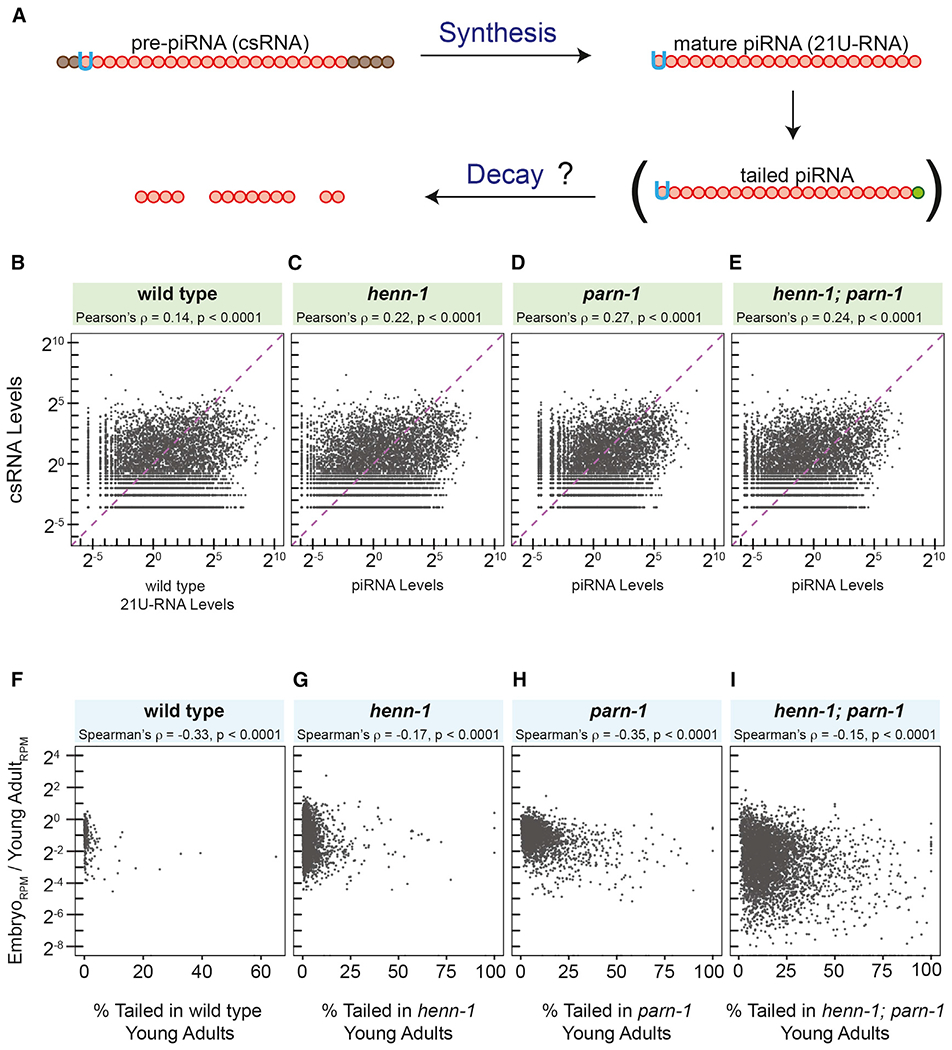
(A) Model illustrating determinants of steady-state levels of piRNA as well as the role of 3′ tailing in piRNA turnover.
(B–E) Scatterplots displaying the correlation between wild-type csRNA abundance and mature piRNA abundance in wild type (B) and henn-1 (C), parn-1 (D), and henn-1; parn-1 (E) mutant animals. Pearson’s correlation test was used to calculate ρ.
(F–I) Scatterplots showing the correlation between the fold change of tailed piRNAs (embryo versus young adult) compared to the tailing frequency of piRNAs in young adults, in wild type (F) and henn-1 (G), parn-1 (H), and henn-1; parn-1 (I) mutants. Lowly abundant piRNA reads were filtered by removing reads with a standardized RPM of less than −1.5. The Spearman’s correlation test was used to calculate ρ.
These two conflicting models make distinct predictions. For example, the latter model predicts that, with increased decay because of 3′ tailing, levels of untrimmed and unmethylated piRNAs should better correlate with their corresponding csRNAs. We compared steady-state levels of piRNAs to corresponding csRNAs that were previously characterized by Mello and colleagues (Gu et al., 2012). In wild-type strains, levels of csRNAs and mature piRNA were modestly correlated (Pearson’s ρ = 0.14; Figure 3B). In support of the second model, there was a better correlation between csRNA and unmethylated piRNA levels in the henn-1 mutant strains (Pearson’s ρ = 0.22; Figure 3C). Similarly, we found a stronger association between csRNA and untrimmed piRNA levels in parn-1 (Pearson’s ρ = 0.27; Figure 3D) and henn-1; parn-1 mutant animals (Pearson’s ρ = 0.24; Figure 3E).
By inhibiting the synthesis of csRNAs, the steady-state piRNA level should be largely determined by the turnover rates. In this scenario, the second model predicts that 3′ tailing frequency would be associated with piRNA turnover. C. elegans piRNAs are actively transcribed in the adult germ cells and passed on to the embryos (Goh et al., 2014; Kasper et al., 2014; Weick et al., 2014; Weng et al., 2019). Germ cells in the early embryos, however, are transcriptionally quiescent (Seydoux et al., 1996). We thus calculated the change in piRNA levels in embryos versus young adults and examined the relationship between the change of abundance and 3’ tailing frequency in young adults. Consistent with the notion that 3′ tailing is involved in piRNA decay, we detected a moderate but statistically significant inverse correlation between the tailing frequency and piRNA reduction in the wild-type strain (Spearman’s ρ = −0.33), as well as in henn-1 (Spearman’s ρ = −0.17), parn-1 (Spearman’s ρ = −0.35), and henn-1; parn-1 mutant animals (Spearman’s ρ = −0.15) (Figures 3F–3I). Taken together, our findings imply that 3′ tailing may play a role in piRNA turnover during the maternal-to-zygotic transition. Identification of the TENT(s) that act on piRNA will be essential to determine whether tailed piRNAs are degradation intermediates or side products formed prior to or during degradation. In addition, kinetic analyses, such as direct measurement of decay rates of tailed piRNAs, will be necessary to determine the role of 3′ tailing in piRNA decay.
Loss of parn-1 and henn-1 results in decreased WAGO-22G-RNAs and fecundity
Upon targeting, the PRG-1/piRNA complex recruits the RNA-dependent RNA polymerase (RdRP). RdRP uses RNA targets as a template to synthesize secondary small RNAs, referred to as 22G-RNAs based on their propensity for a 5′ G residue and a length of 22 nt (Gu et al., 2009; Maniar and Fire, 2011; Smardon et al., 2000). These 22G-RNAs are loaded onto an expanded group of worm AGOs (WAGOs) that function downstream of piRNAs to maintain and propagate epigenetic silencing of many endogenous genes (Ashe et al., 2012; Bagijn et al., 2012; Buckley et al., 2012; Lee et al., 2012; Shen et al., 2018; Shirayama et al., 2012). RdRP is also responsible for generation of 22G-RNAs that engage the CSR-1 AGO protein. However, biogenesis of CSR-1-22G-RNAs is independent of piRNA activity (Claycomb et al., 2009; Gu et al., 2009; Lee et al., 2012).
We next tested whether deletion of parn-1 and henn-1 affects the synthesis of 22G-RNAs. In total, 1,125 WAGO 22G-RNA loci and 3,207 CSR-1 22G-RNA loci were examined (Table S3; Claycomb et al., 2009; Gu et al., 2009; Svendsen et al., 2019). CSR-1 22G-RNA production was generally not affected upon deletion of henn-1 and parn-1 (Figures 4A–4C). In contrast, levels of WAGO 22G-RNAs exhibited significant changes in the mutant animals (Figures 4A–4D; Svendsen et al., 2019; Tang et al., 2016). Specifically, the median abundance of WAGO 22G-RNAs in parn-1 and henn-1 was reduced to 97.5% and 83.4% of wild type, respectively. Consistent with a stronger depletion of piRNAs in henn-1; parn-1 animals, the median abundance of WAGO 22G-RNAs was reduced to 56.5% of the wild-type strain (Figure 4D).
Figure 4. Expression of WAGO-class 22G RNAs and fertility are reduced in henn-1, parn-1, and henn-1; parn-1 mutants.
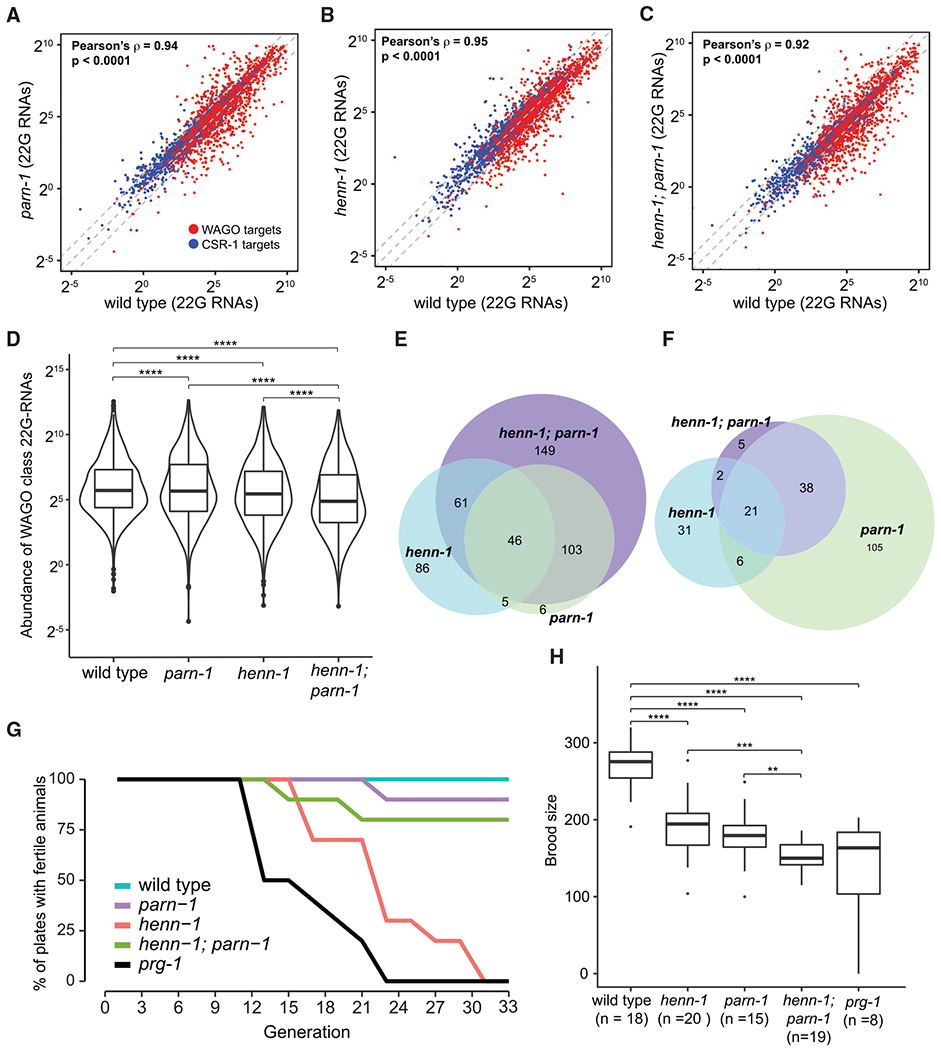
(A–C) Scatterplots visualizing the expression of 22G-RNAs mapped to C. elegans genes in parn-1 (A), henn-1 (B), and henn-1; parn-1 (C) mutants compared to that of the wild type. WAGO-class 22G (n = 1,125) RNAs are highlighted in red, and CSR-class 22G RNAs (n = 3,207) are shown in blue. The central diagonal line represents a fold change of 1, the top diagonal line represents a fold change of 2, and the bottom diagonal line represents a fold change of 0.5. Pearson’s correlation test was used to calculate ρ.
(D) Boxplots quantifying the overall expression of WAGO-class 22G RNAs (n = 1,125) in wild type and parn-1, henn-1, and henn-1; parn-1 mutants. The paired Wilcoxon rank-sum test was used to determine statistical differences between the sample groups. Significance indicators are as follows: ns (no significance), p ≥ 0.05; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
(E and F) The overlaps of PRG-1 targets that are downregulated (E) or upregulated (F) in henn-1 (blue), parn-1 (green), and henn-1; parn-1 (purple) young adults compared to wild-type. PRG-1 targets were considered down- or upregulated if the log2 fold change was less than or equal to −1 or greater than or equal to 1, respectively, compared to wild-type.
(G) Germline mortality assay in wild type and prg-1, henn-1, parn-1, and henn-1; parn-1 mutants. Percentages of 10 independent lines of animals that remained fertile at every 2 generations 25°C were plotted.
(H) Brood size assay showing the fertility of wild type (n = 18) and parn-1 (n = 24), henn-1 (n = 20), henn-1; parn-1 (n = 19), and prg-1 (n = 8) mutant animals. The two-sample t test was used to compare wild-type brood size to that of henn-1 and parn-1 mutants, as well as to compare parn-1 mutants with henn-1; parn-1 mutants. The Welch two-sample t test was used to compare wild-type and henn-1; parn-1 mutant brood size as well as to compare henn-1 and henn-1; parn-1 mutants. The Wilcoxon rank-sum test was used to compare wild-type brood size with that of henn-1; parn-1 mutants. Significance indicators are as follows, ns(no significance), p ≥ 0.05; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
We further examined 890 genes whose 22G-RNA accumulation depends on PRG-1/piRNA activity (Reed et al., 2020; Tang et al., 2016). A total of 198 genes exhibited a significant depletion of 22G-RNAs in henn-1 mutant animals, whereas 160 genes with depleted 22G-RNAs were found in the parn-1 mutant strain (Figure 4E). Consistent with the notion that both parn-1 and henn-1 function in stabilizing piRNAs, deletion of both henn-1 and parn-1 led to a reduction of 22G-RNAs mapped to 359 genes (Figure 4E).
We were surprised to find that among 890 PRG-1/piRNA targets, 60 in henn-1, 170 in parn-1, and 66 in the double-mutant animals displayed elevated 22G-RNA levels (Figure 4F). By inspecting some PRG-1/piRNA targets showing increased 22G-RNAs upon loss of parn-1, we found an intriguing genetic interaction between parn-1 and henn-1 in regulating 22G-RNA biogenesis (Figures 4F and S4A–S4C). For example, at clec-238 and fbxc-39 genomic loci, depletion of parn-1 appeared to restore 22G-RNA production of henn-1 mutants (Figures S4A and S4B). At the pdx-1 locus, 22G-RNAs mapping to specific regions–exons 4-6 and 3′ UTR–were increased in parn-1 and henn-1; parn-1 mutants as compared to henn-1 and wild-type strains. One speculative explanation is that by extending the potential base pairing, the untrimmed piRNAs in parn-1 and henn-1; parn-1 animals may stabilize the interactions with certain target sequences and thus promote the production of 22G-RNAs. Together, these data suggest that HENN-1 and PARN-1 act in the piRNA pathway to regulate the production of WAGO 22G-RNAs.
Disruption of the piRNA pathway causes infertility in diverse animals (Carmell et al., 2007; Cox et al., 1998; Goh et al., 2015; Gou et al., 2014; Houwing et al., 2007). In C. elegans prg-1 mutants, this sterility is progressive, and fertility declines over generations, known as a mortal germline phenotype (Batista et al., 2008; Das et al., 2008; Simon et al., 2014; Wang and Reinke, 2008). We performed two assays to assess the fertility of mutant animals. In the first assay, we outcrossed mutants with wild-type strains, tracked 10 prg-1, henn-1, parn-1, and double-mutant lines, and scored whether or not each line produced progenies every 2 generations. Wild-type animals remained fertile in the course of the experiment (~33 generations) (Figure 4G). As expected, propagation of prg-1 and henn-1 animals at 25°C resulted in progressive sterility (Figure 4G; Simon et al., 2014; Svendsen et al., 2019). Animals deficient for PARN-1 displayed moderate decline in fertility over generations (Figure 4G). Surprisingly, henn-1; parn-1 double mutants became sterile more slowly than henn-1 single mutants, suggesting that the loss of parn-1 suppresses progressive sterility of henn-1 mutants.
As a second assay for fertility, we measured the brood sizes of outcrossed strains at approximately generation 8 when animals were relatively fertile. Under such conditions, the wild-type strain produced ~276 progeny/animal (Figure 4H). Consistent with findings from previous studies (Svendsen et al., 2019; Tang et al., 2016), we found that as compared to the wild type, parn-1 and henn-1 animals displayed a decrease in brood size, producing ~180 and ~194 progeny/animals, respectively (Figure 4H). Strikingly, henn-1; parn-1 double mutants exhibited an additive fertility deficit and produced ~150 progeny/animal, which is comparable to that of prg-1 mutants (Figure 4H). These findings imply that HENN-1 genetically interacts with PARN-1 to modulate germline immortality and promote fertility.
Tailed piRNAs show an extensive base-paring interaction with their target RNAs
Previous studies showed that a 3′ addition of miRNAs is triggered through binding to target RNAs with extensive complementarity (Ameres et al., 2010; de la Mata et al., 2015; Xie et al., 2012). In wild-type animals, 3′ nontemplated additions were found on 272 piRNAs (Table S2). We thus wondered if extensive base-pairing interactions between piRNAs and their targets are associated with piRNA tailing.
We examined the piRNA-RNA interactome, as measured by a method combining crosslinking, ligation, and sequencing of hybrids (CLASH) (Helwak et al., 2013; Shen et al., 2018). The CLASH experiment included an RNase treatment step prior to hybrid ligation and the 3′-end sequences of piRNA were not preserved. We thus cross-referenced tailed piRNAs defined by this study to base-pairing interactions revealed by CLASH reads obtained by Mello and colleagues (Shen et al., 2018). In silico folding for each base-pairing interaction was conducted, and Gibbs free energy (ΔG) of the most energetically favorable interactions was calculated as described (Figure 5A; Lorenz et al., 2011; Shen et al., 2018). To examine weak and stable interactions, we split chimeric reads into two bins of decreasing ΔG (−15 < ΔG <0, and ΔG<−15 kCal/mol). As a control, we considered a set of untailed piRNAs whose expression levels were comparable to those of tailed piRNAs (Figure S5A). We detected no sequence bias between tailed piRNAs and the control group (Figures S5B and S5C). We next computed the abundance of CLASH chimeric reads for each piRNA species in tailed group and control (Figure 5B). In the group representing weak interactions (−15 < ΔG < 0 kCal/mol), we detected no significant differences in CLASH reads between tailed piRNAs and the control (Figure 5B). In contrast, when examining the bin showing stable base-pair interactions, we found more piRNA/target RNA interactions for tailed piRNAs than those for the control (1.9-fold increase in ΔG < −15 kCal/mol hybrids) (Figure 5B).
Figure 5. Tailed piRNAs share a high degree of base pairing with their target RNAs.
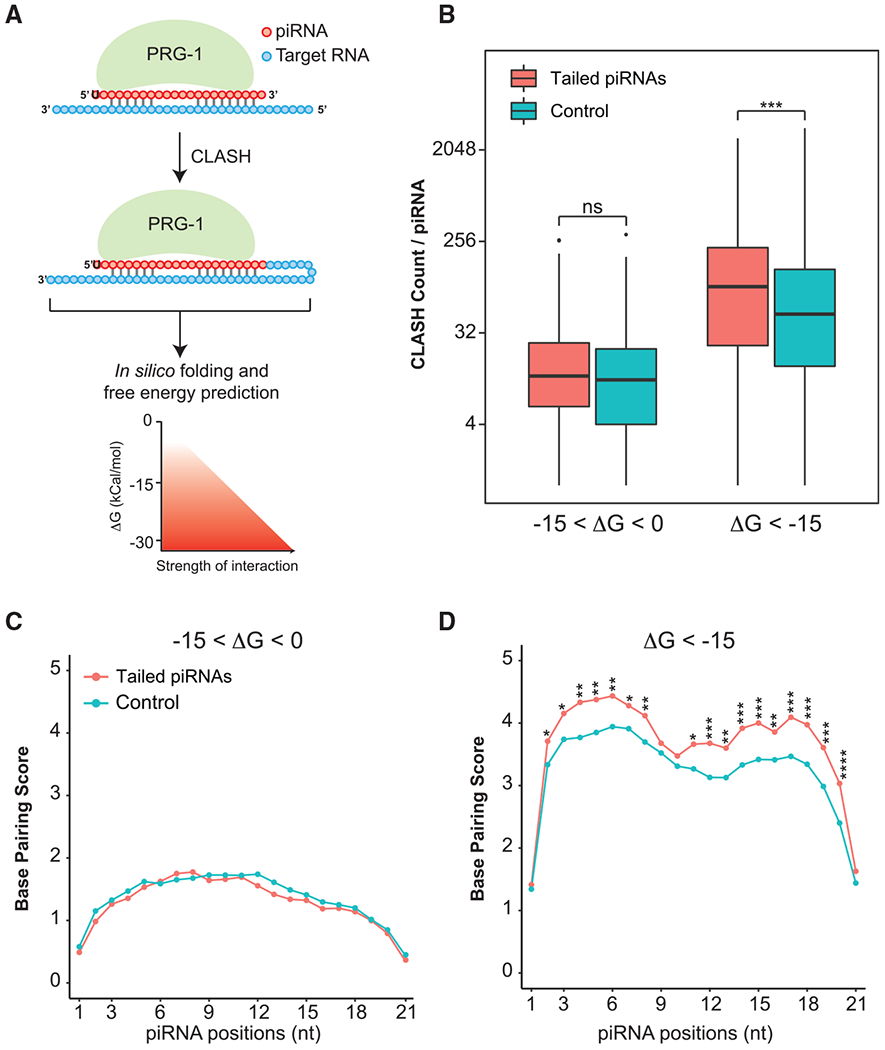
(A) Illustration displaying PRG-1 crosslinking, ligation, and sequencing of hybrids (CLASH) as described (Shen et al., 2018). Following sequencing, the CLASH reads were subject to in silico folding and free energy predictions. A schematic is provided at the bottom of the figure to illustrate the relationship between Gibbs free energy (kCal/mol) and the strength of the piRNA-target interaction.
(B) Boxplots showing the number of CLASH hybrid counts in tailed piRNA and control set. Hybrids with different binding energies (−15 < ΔG < 0, ΔG < −15 kCal/mol) are plotted. The Wilcoxon rank-sum test was used to derive p values to check for statistical difference between the groups. Significance indicators are as follows: ns (no significance), p ≥ 0.05; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
(C and D) Perfect Watson-Crick base pairing between piRNAs and CLASH-defined targets. Line graphs show the mean CLASH counts at each position of the piRNA for hybrids with −15 < ΔG < 0 (C) and ΔG < −15 (D). The Welch two-sample t test was used to calculate p values. Significance indicators are as follows: ns (no significance), p ≥ 0.05; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
To determine the extent of complementarity required to induce tailing, we conducted in silico folding to examine the Watson-Crick base pairing between piRNAs and their targets revealed by CLASH reads. For hybrids with −15 < ΔG < 0 kCal/mol, base-pairing patterns of tailed piRNAs were indistinguishable from those of the control (Figure 5C). When examining stable interactions, we found that tailed piRNAs exhibited increased base-pairing frequencies at several positions of piRNAs (Figure 5D). There were preferred interactions within the seed region, particularly at positions 4 to 6. Additionally, more 3′ supplementary pairing between tailed piRNAs and their targets was detected (Figure 5D).
To directly test the causal relationship between base-pairing interactions and piRNA 3′ tailing and/or degradation, we mutated piRNA target sites in endogenous transcripts and examined the effect on piRNA expression. We focused on the abundant piRNAs with detectable 3′ addition (Table S2) and searched for their target sites defined by the CLASH experiment (hybrid reads with ΔG < −15 kCal/mol and read count of >100). In addition, we manually selected target sites showing extensive complementarity to the 5′ and 3′ ends of piRNAs and mismatches in the central region (Figure 5D). Among tailed piRNAs, 21UR-4864 was mono-uridylated and interacted with a set of transcripts as defined by CLASH (Tables S2 and S4). The base-pairing interaction between 21UR-4864 and w03h9.2 stood out; the CLASH read count was 131, ΔG was −29.08 kCal/mol, the target site could base pair with 5′ and 3′ ends of 21UR-4864 with a central 3-nt mismatch (Figure 6A), and the w03h9.2 transcript was highly enriched in the C. elegans germline (Li et al., 2014; Ortiz et al., 2014).
Figure 6. 21UR-4864 tailing and degradation are target directed.
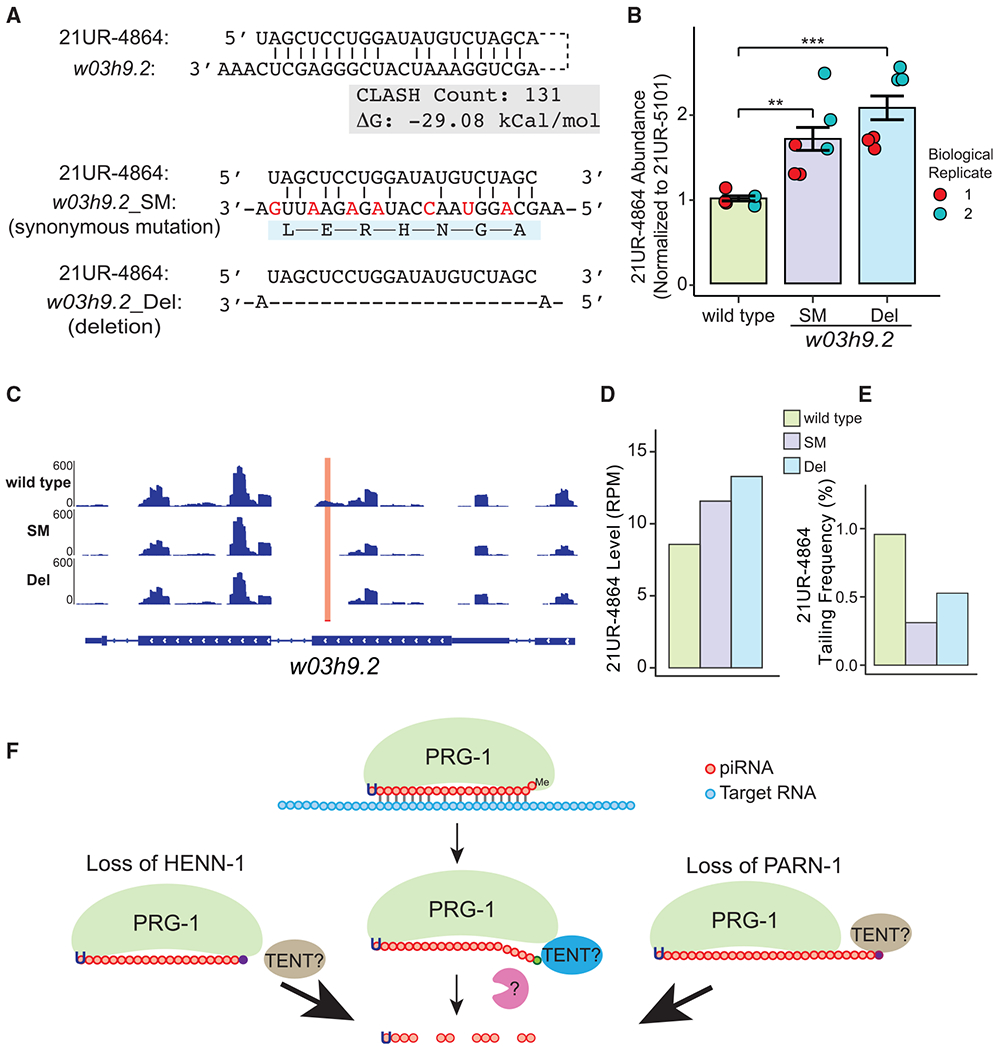
(A) The top schematic illustrates the base-paring interaction between 21UR-4864 and w03h9.2, CLASH read count, and binding energy of this interaction. The middle schematic illustrates synonymous mutations (SMs) that disrupt base pairing between 21UR-4864 and w03h9.2. Mutated nucleotides are shown in red, and amino acid are shown below the mRNA sequence. The bottom schematic illustrates complete deletion of the 21UR-4864 target site at w03h9.2. Potential A:U, G:C base pairs or G:U wobble base pairs are marked.
(B) Barplot showing the abundance of 21UR-4864 in wild type (green) and strains containing SM (purple) and deletion (blue) of the 21UR-4864 target site at w03h9.2 in two independent biological replicates as measured by qRT-PCR. Technical triplicates are shown as dots in blue and red. 21UR-4864 Ct values were normalized to 21UR-5101 Ct values. A Student’s two-sample t test was used to derive p values. Significance indicators are as follows: **p < 0.01; ***p < 0.001.
(C) The normalized coverage of antisense reads mapping to w03h9.2 in wild type and strains containing SMs and deletion of the 21UR-4864 target site at w03h9.2 are visualized using the IGV Genome Browser. A red bar and highlighted region mark the 21UR-4864 piRNA target site.
(D) Barplot showing the abundance of 21UR-4864 in wild type (green) and strains containing SMs (purple) and deletion (blue) of the 21UR-4864 target site at w03h9.2 as measured by small RNA sequencing. The normalized RPM of 21UR-4864 is shown on the y axis.
(E) Barplot showing tailing frequency of 21UR-4864 in wild type (green), and strains containing SMs (purple) and deletion (blue) of the 21UR-4864 target site at w03h9.2 as measured by RNA sequencing.
(F) Model illustrating the mechanism of target-directed piRNA tailing and the role of pre-piRNA trimming and 2′-O-methylation in protecting piRNA from 3′ tailing and degradation. TENT, terminal nucleotidyltransferase.
We reasoned if 3′ tailing and degradation of piRNAs are induced by base-pairing interactions, disrupting the binding between 21UR-4864 and w03h9.2 should have an effect on 21UR-4864 stability. CRISPR-Cas9 genome editing was used to mutate 21UR-4864 target sequences in the w03h9.2 transcript. We introduced synonymous mutations (SMs) that maintain the w03h9.2 open reading frame but the disrupt 21UR-4864 target site (Figure 6A). Additionally, we generated a deletion (Del) that completely removes 21UR-4864 target sequences and deletes 7 amino acids on the W03H9.2 protein (Figure 6A). We quantified the abundance of 21UR-4864 with qRT-PCR and found that the 21UR-4864 level increased approximately 2-fold in both SM and Del mutants (Figure 6B). To assess nontemplated nucleotide addition on piRNAs, we cloned and deep sequenced small RNAs isolated from wild-type and SM and Del mutant animals. 22G-RNA signals near the 21UR-4864 target site were reduced in SM and Del mutant worms, suggesting an intact piRNA target site is required for the production of 22G-RNAs (Figures 6A and 6C; Ashe et al., 2012; Lee et al., 2012). Furthermore, small RNA sequencing confirmed the elevated level of 21UR-4864 in SM and Del mutant worms compared to the wild type (Figure 6D). Importantly, the frequency of 21UR-4864 with a 3′ nontemplated nucleotide (mono-uridylation) is reduced in both piRNA target site mutants (Figure 6E). Taken together, our analyses revealed that similar to miRNAs, extensive complementarity between piRNAs and mRNA can trigger 3′ tailing and degradation of piRNAs in C. elegans.
DISCUSSION
In contrast to piRNA biogenesis, the molecular principles for piRNA degradation are poorly understood (Huang et al., 2017; Ozata et al., 2019; Weick and Miska, 2014). In this study, we characterized tailing, trimming, and decay of C. elegans piRNAs (Figure 6). Our analyses revealed that in wild-type animals a subset of piRNAs possess detectable 3′ nontemplated nucleotides. C. elegans piRNAs interact with thousands of germline transcripts and generally permit mismatches at various positions of piRNAs (Shen et al., 2018). When compared to piRNAs with no detectable 3′ additions, piRNAs showing 3′ additions exhibit extensive base pairing to their targets, raising the possibility that complementary target RNAs elicit piRNA tailing and degradation. The target sites that trigger piRNA decay may differ from the sites that cause target silencing in that they base pair extensively to both 5′ ends and 3′ ends of piRNAs. Indeed, a structural analysis of the AGO complex revealed that 3′-end complementarity facilitates conformational changes that expose miRNA 3′ termini to nucleotidyltransferases and/or nucleases (Sheu-Gruttadauria et al., 2019). It is possible that mismatches in the central region may also be essential to induce piRNA tailing and degradation. These mismatches could conceivably inhibit or slow down the cleavage and/or release of target RNAs. Although pairing rules for piRNA target recognition were well characterized, pairing patterns that drive for piRNA degradation are unclear and require further investigation (Shen et al., 2018; Svendsen and Montgomery, 2018; Zhang et al., 2018). Our findings, together with studies in miRNAs and small interfering RNAs (siRNAs) (Ameres et al., 2010; Baccarini et al., 2011; Bitetti et al., 2018; de la Mata et al., 2015), suggest that complementarity is a general mechanism for 3′ nontemplated nucleotide addition of small RNAs.
We report that both pre-piRNA trimming and 2′-O-methylation protect piRNAs from 3′ addition (Figure 6). Previous work showed that C. elegans deficient for HENN-1 accumulates piRNAs with 1- to 2-nt truncations (Kamminga et al., 2012; Svendsen et al., 2019). Our analyses revealed that short isoforms are subject to nontemplated nucleotide addition, indicating 3′ truncation occurs prior to tailing in henn-1 animals. In contrast, long isoforms of piRNAs in parn-1 mutants are preferentially modified, indicating tailing of untrimmed piRNAs occurs prior to or independently of 3′ truncation. Therefore HENN-1 and PARN-1 protect piRNA against nontemplated nucleotide addition through distinct mechanisms. Indeed, simultaneous deletion of both enzymes results in a higher frequency of 3′ tailing and stronger depletion of piRNAs than those of henn-1 or parn-1 single mutants. A recent pre-print revealed that both PNLDC1 (mouse homolog of PARN-1) and HENMT1 (mouse homolog of HENN-1) are required for the accumulation of piRNAs (Gainetdinov et al., 2020). Together, these findings suggest that despite many differences in piRNA lengths and sequences in nematodes and mammals, the quality-control mechanism for piRNAs is nevertheless conserved.
We show that the composition of piRNA 3′ nontemplated nucleotides are distinct between wild-type and parn-1 or henn-1 mutant strains. For example, uridylation is the major form of 3′ modifications in wild-type animals (Figure 2B). Additionally, 3′ guanylation and a few other modifications of piRNAs are found in parn-1 as well as in henn-1 mutant animals. These data suggest that different TENTs act on 21U-RNAs and untrimmed and unmethylated piRNAs (Figure 6). Members of the TENT family act in small RNA pathways in a variety of organisms (Yu and Kim, 2020). In C. elegans, the nucleotidyltransferases CDE-1 is required for the uridylation of CSR-1-22G-RNAs (van Wolfswinkel et al., 2009). RDE-3, another TENT required for WAGO-22G-RNA accumulation but dispensable for CSR-1-22G-RNA production, adds alternating stretches of polyuridine-guanine tails to the RNA template for RdRP amplification (Chen et al., 2005; Shukla et al., 2020). An in cellulo tethering assay identified several C. elegans TENTs with uridylation and/or guanylation activities (Preston et al., 2019), making them good candidates for modifying piRNA. Together with these studies, our work lays the foundation for future studies to identify enzymatic factors responsible for nontemplated nucleotide addition of piRNAs.
Although piRNA 3′ tailing was detected in wild-type animals and became more prevailing in henn-1 and parn-1 mutant animals, we do not know if 3′ tailing causes piRNA degradation. There are several challenges to establishing a causal relationship between these two processes. First, it is difficult to identify and quantify tailed piRNA species. Although there are a large number of piRNAs, the abundance of individual piRNAs is low in C. elegans. The median abundance of piRNAs in C. elegans is 0.43 reads per million (RPM) (n = 15,366), as compared to 32.2 RPM for miRNAs (n = 223). Our bioinformatic analyses revealed 272 tailed piRNAs from small RNA sequencing runs with over 50 million reads (Tables S1 and S2). A higher sequencing depth will facilitate the identification of more piRNAs containing 3′ nontemplated nucleotides. Second, we found that a significant portion of piRNAs in parn-1 and henn-1 mutant animals possess more than one type of nontemplated nucleotides. Different 3′ modifications have various or even opposite effects on small RNA stability. For example, 3′-mono-adenylation mediated by the noncanonical poly(A) polymerase GLD-2 stabilizes mouse miR-122, whereas 3′-oligo-adenylation by the same enzyme promotes degradation of miR-122 (Katoh et al., 2009, 2015). Third, we measured the steady-state abundance of piRNAs; the production and degradation dynamics of tailed species remain largely obscure. A pulse-chase approach and metabolic incorporation strategies will be required for elucidating the kinetics of 3′ additions and their effect on piRNA turnover (Kingston and Bartel, 2019; Marzi et al., 2016; Reichholf et al., 2019). Finally, it is worth noting that 3’ tailing and trimming can be uncoupled from nucleolytic degradation of small RNAs. Cyrano, a mammalian long noncoding RNA, has been shown to induce 3′ tailing and decay of miRNA-7 through extensive complementarity (Kleaveland et al., 2018). However, degradation of miRNA-7 is independent of 3′ tailing and trimming. Instead, the highly complementary target triggers proteolysis of the ARGO protein through the ubiquitin proteasome pathway (Han et al., 2020; Kleaveland et al., 2018; Shi et al., 2020). The interplay between ribonuclease-mediated small RNA decay and ubiquitination-mediated AGO proteolysis remains to be determined.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests concerning resources and reagents can be directed to and fulfilled by the Lead Contact Wen Tang (tang.542@osu.edu).
Materials availability
Strains used in this study are available for order from the Caenorhabditis Genetics Center and are available upon request.
Data and code availability
WAGO-1 and HRDE-1/WAGO-9 immunoprecipitation data are available fromGene Expression Omnibus (GEO) under the accession number GEO: GSE137734. CSR-1 immunoprecipitation data are available under GEO: GSE18165. Small RNA sequencing data from wild-type and rde-3 strains are available under GEO: GSE18215. CIP-TAP sequencing data for csRNAs are available under GEO: GSE40053. PRG-1 CLASH sequencing data are available from Sequence Read Archive (SRA) under SRA: SRP131397. Sequencing data used to define PRG-1 targets are available under GEO: GSE141243 and SRA: SRS1021265. Small RNA sequencing data generated for use in this study are available under SRA: PRJNA683039.
Custom R, python and shell scripts used in this analysis are freely available upon request.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Caenorhabditis elegans strains
The Bristol strain N2 was designated as the wild-type C. elegans strain (Brenner, 1974). Other strains used in this study are listed in Key resources table. All strains were maintained with an E. coli OP50 diet on Nematode Growth Media (NGM). All animals were maintained at 20 °C unless otherwise indicated.
KEY RESOURCES TABLE
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Anti-PRG-1 | Batista et al., 2008 | PMC2570341 |
| Rat polyclonal anti-alpha-Tubulin | Bio-Rad | Cat# MCA77G; RRID: AB_325003 |
| Goat Anti-Rabbit IgG H&L (HRP) | Abcam | Cat# ab6721; RRID: AB_955447 |
| Goat Anti-Rat IgG H&L (HRP) | Abcam | Cat# ab97057; RRID: AB_10680316 |
| Bacterial and virus strains | ||
| Bacteria: OP50 | Caenorhabditis Genetics Center | https://cgc.umn.edu/strain/OP50 |
| Chemicals, peptides, and recombinant proteins | ||
| TRI Reagent | Thermo Fisher Scientific | Cat# AM9738 |
| PIR-1 | Li et al., 2020 | PMC6961543 |
| T4 RNA Ligase 1 | New England Biolabs | Cat# M0437 |
| T4 RNA Ligase 2, truncated | New England Biolabs | Cat# M0242 |
| SUPERaseIn | Thermo Fisher Scientific | Cat# AM2694 |
| Q5 High-Fidelity DNA Polymerase | New England Biolabs | Cat# M0491 |
| Ex TaqDNA Polymerase | TaKaRa | Cat# RR001C |
| dNTPs | Roche | Cat# 3622614001 |
| Sodium Hypochlorite Solution | Thermo Fisher Scientific | Cat# SS2901 |
| Sodium meta-Periodate | Thermo Fisher Scientific | Cat# S398-100 |
| Sodium Tetraborate Decahydrate | Thermo Fisher Scientific | Cat# S246-500 |
| Boric acid | Sigma Aldrich | Cat# B0394-100G |
| Glycerol | Thermo Fisher Scientific | Cat# AC327255000 |
| PVDF membrane | Bio-Rad | Cat# 1620177 |
| Alt-R S.p. Cas9 Nuclease V3 | Integrated DNA Technologies | Cat# 1081058 |
| Critical commercial assays | ||
| mirVana miRNA Isolation Kit | Thermo Fisher Scientific | Cat# AM1560 |
| PowerUp SYBR Green Master Mix | Thermo Fisher Scientific | Cat# A25742 |
| SuperScript III Reverse Transcriptase | Thermo Fisher Scientific | Cat# 18080093 |
| Clarity Max ECL Western Blotting Substrate | Bio-Rad | Cat# 1705062 |
| Zero Blunt TOPO PCR Cloning Kit | Thermo Fisher Scientific | Cat# 450245 |
| Deposited data | ||
| Small RNA sequencing data from WAGO-1 and HRDE-1/WAGO-9 IP | Svendsen et al., 2019 | GEO: GSE137734 |
| Small RNA sequencing data from CSR-1 IP | Claycomb et al., 2009 | GEO: GSE18165 |
| Small RNA sequencing data from rde-3 mutants | Gu et al., 2009 | GEO: GSE18215 |
| CIP-TAP cloning data | Gu et al., 2012 | GEO: GSE40053 |
| Small RNA sequencing from prg-1 mutants | Tang et al., 2016 and Reed et al., 2020 | SRA: SRS1021265; GEO: GSE141243 |
| CLASH data | Shen et al., 2018 | SRA: SRP131397 |
| Small RNA sequencing data from N2, and henn-1, parn-1, henn-1; parn-1, N2-Oxidized, parn-1-Oxidized, and 21ur-4864 target site mutants | This study | SRA: PRJNA683039 |
| Experimental models: Organisms/strains | ||
| C. elegans: N2 Bristol (wild type) | Caenorhabditis Genetics Center | https://cgc.umn.edu/strain/N2 |
| C. elegans: henn-1 (tm4477) | Billi et al., 2012 | PMC3330095 |
| C. elegans: parn-1 (tm869) | Tang et al., 2016 | PMC4785802 |
| C. elegans: prg-1 (tm872) | Batista et al., 2008 | PMC2570341 |
| C. elegans: henn-1 (tm4477); parn-1 (tm869) | This study | |
| C. elegans: 21ur-4864 target site mutation on w03h9.2 | This study | |
| C. elegans: 21ur-4864 target site deletion on w03h9.2 | This study | |
| Oligonucleotides | ||
| See Table S5 | N/A | N/A |
| Recombinant DNA | ||
| pRF4 injection marker, rol-6(su1006) | Mello et al., 1991 | PMC453137 |
| Software and algorithms | ||
| Bowtie version 1.2.3 | Langmead et al., 2009 | http://bowtie-bio.sourceforge.net/manual.shtml |
| BEDtools version 2.26.2 | Quinlan and Hall, 2010 | https://bedtools.readthedocs.io/en/latest/ |
| Trim Galore version 0.6.4 | The Babraham Institute | https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
| FASTX-Toolkit version 0.0.14 | Hannon lab | http://hannonlab.cshl.edu/fastx_toolkit/ |
| BEDOPS version 2.4.39 | Neph et al., 2012 | https://bedops.readthedocs.io/en/latest/ |
| R version 4.0.3 | The R Project for Statistical Computing | https://www.r-project.org/ |
| ggplot2 version 3.3.2 | tidyverse | https://ggplot2.tidyverse.org/ |
| Integrative Genomics Viewer 2.8.12 | Broad Institute | https://software.broadinstitute.org/software/igv/ |
METHOD DETAILS
CRISPR/Cas9 genome editing
CRISPR lines were generated via Cas9 editing using short single-stranded oligo donors (ssdonors) as previously reported (Ghanta and Mello, 2020). In brief, two ssdonors were used to introduce synonymous mutations and deletion at target site of 21UR-4864 of w03h9.2. 20 μl injection mix contained pre-assembled Cas9 ribonucleoprotein complex (5 μg Cas9, 2 μg guide RNA, 1 μg tracrRNA) and 1.1 μg ssdonors (Integrated DNA Technologies). The vector pRF4 was used as a co-injection marker (Mello et al., 1991). Sequences of guide RNA and ssdonors used in this study are listed in Table S5.
Western blotting analysis
40 synchronized wild-type, henn-1, parn-1, and henn-1/parn-1 L1s were grown at 20°C and harvested before egg-laying for young adult protein lysate preparation. 850 embryos per strain were isolated from gravid adults grown at 20°C for embryo protein lysate preparation. Samples were separated on precast denaturing polyacrylamide gels, transfer onto PVDF membrane (Bio-Rad), and probed with anti-PRG-1 antibody (Batista et al., 2008) and rat polyclonal anti-alpha tubulin (Bio-Rad, MCA77G). HRP goat anti-rabbit IgG H&L (Abcam, ab6734) or rabbit anti-rat IgG H&L (Abcam, ab6721) secondary antibodies were used for detection using Clarity Max ECL Western Blotting Substrate (Bio-Rad).
Brood size assay
Wild-type, outcrossed henn-1, parn-1, henn-1/parn-1, and prg-1 animals are passed approximately 8 generations. Newly hatched L1 larvae were placed singly on plates. Halfway through egg-laying, animals were transferred to fresh plates until egg-laying stopped. The brood size for each animal was calculated by adding the progeny on the original and transferred plates.
Germline mortality assay
Prior to starting the assay, animals were freshly outcrossed. 10 lines of N2, henn-1, parn-1, henn-1; parn-1 and prg-1 respectively, were grown continuously at 25°C on OP50. Four L1s for each line were transferred to fresh plates seeded with OP50 bacteria every two generations (approximately every 4-5 days). Lines were recorded as fertile until animals no longer produce viable progeny.
RNA Extraction and Small RNA enrichment
Approximately 10,000 synchronized wild-type and mutant young adults were harvested before egg-laying for young adult samples. Embryos were obtained by hypochlorite treatment (Fisher Scientific, SS2901) of gravid adult hermaphrodites. Total RNAs were isolated from young adult and embryos using TRI Reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. Small RNAs were enriched using MirVana miRNA Isolation Kit (Thermo Fisher Scientific), according to the manufacturer’s instructions.
Oxidation of RNA with sodium periodate
5 μg small RNA enriched by the MirVana Kit (Thermo Fisher Scientific) was oxidized 25 mM sodium periodate (Fisher Scientific) in borax/boric acid buffer (0.06 M borax, 0.06 M boric acid, pH 8.6) in the dark at room temperature for 30 min. 20 μL of 50% glycerol was added and incubated for additional 15 min to quench sodium periodate. RNA was recovered by ethanol precipitation, dissolved in nuclease free water, and subjected to small RNA cloning.
Real-time quantitative RT-PCR
1.5 μg of total RNA isolated from two biologically distinct young adult samples was reverse transcribed in technical triplicates using Superscript III Reverse Transcriptase (Thermo Fisher Scientific) with gene-specific P1-P6 antisense primers that are listed Table S5. Quantitative PCR was performed in 20 ul reactions using cDNA with PowerUp SYBR Green Master Mix (Thermo Fisher Scientific), gene-specific primers P7-P12 and P13 universal primer in a CFX Connect Real-Time PCR System (Bio-Rad). Ct values were determined using CFX Manager v3.1 software (Bio-Rad) and the relative abundances of individual piRNAs were calculated using the ΔΔCt method. Error bars in the graph indicate the standard deviation (SD) in all statistical analysis.
Small RNA-sequencing library preparation
Small RNA samples from wild-type and mutants were pretreated with a recombinant 5′ polyphosphatase PIR-1 that removes the γ and β phosphates from 5′-triphosphorylated RNA (Li et al., 2020). The resulting monophosphorylated small RNAs were ligated to a 3′ adaptor (5′rAppAGATCGGAAGAGCACACGTCTGAACTCCAGTCA/3ddC/3′, IDT) using T4 RNA ligase 2 in the presence of 25% PEG8000 (NEB) at 15°C overnight. The 5′ adaptor (rArCrArCrUrCrUrUrUrCrCrCrUrArCrArCrGrArCrGrCrUrCrUrUrCrCrGrArUrCrU, IDT) was then ligated to the product using T4 RNA ligase 1 (NEB) at 15°C for 4 h. The ligated products were converted to cDNA using SuperScript III Reverse Transcriptase (Thermo Fisher Scientific). The cDNAs were amplified by PCR, and the libraries were sequenced on an Illumina Novaseq platform (SP 2 X 50 bp) at the OSU Comprehensive Cancer Center genomics core.
Analysis of small RNA sequencing datasets
Raw small RNA sequencing reads were first processed to remove low quality reads and adapters using TrimGalore (v0.6.4). Following adaptor removal and quality control, reads were aligned to a genome reference (Wormbase release WS230) or modified references containing synonymous mutations and deletion at target site of 21UR-4864 of w03h9.2 using Bowtie (v1.2.3) (Langmead et al., 2009), then and converted to bed files using BEDOPS (v2.4.39) (Neph et al., 2012). Following alignment to our genomic reference reads were also aligned to a reference containing annotated exon-exon junctions. To account for variation in sequencing depth between samples, each read was normalized to the total number of mapped reads. To account for difference in ligation efficiency between unmethylated and 2′-O-methyl RNAs, piRNA-matching reads in henn-1 and henn-1; parn-1 mutants were further normalized by reducing their read counts by 5-fold. WAGO 22G-RNA loci (n = 1125) were defined as genes (1) with small RNAs enriched in WAGO-1 or WAGO-9/HRDE-1 immunoprecipitation and (2) depleted of small RNAs in rde-3 mutants (Gu et al., 2009; Svendsen et al., 2019). CSR-1 22G-RNA loci (n = 3207) were defined as genes with small RNAs enriched in CSR-1 immunoprecipitation experiments (Claycomb et al., 2009). PRG-1/piRNA targets (n = 890) were defined as genes depleted of small RNAs in prg-1 mutants (Reed et al., 2020; Tang et al., 2016).
Bioinformatic analysis of nontemplated nucleotide addition
A piRNA reference was generated by extending 3′ ends of annotated piRNA loci by 10 nucleotides (Wormbase release WS230) using BEDtools (v2.26.2) (Quinlan and Hall, 2010). Nontemplated nucleotide addition was detected a custom computational pipeline. Briefly, sequencing reads mapped to the piRNA reference using the Bowtie command bowtie–best–strata -a–no-unal -m 1 -un -f -v 0, were denoted as reads without mismatches. One nucleotide from the 3′ end of unmapped reads was removed. And these trimmed reads were aligned to the reference. Reads aligning to the piRNA reference following removal of 3’ nucleotides were denoted as tailed reads. Unmapped reads were then re-submitted into the pipeline. At each round of alignment, sequencing reads were aligned to the piRNA reference allowing N-1 mismatches, where N is the pipeline iteration. This step allowed us to exclude multinucleotide tails where the number of mismatches is not equal to the length of the tail. The cycle of nucleotide removal followed by alignment was repeated a total of three times, allowing for a maximum of a trinucleotide tail. All read counts were then normalized to the total number of mapped reads of each sample.
Untailed and tailed reads were further processed to exclude sequencing errors and misalignments. First, we preformed size selection on our aligned reads, requiring read length to be within 17 to 30 nucleotides long. Second, aligned reads must start with 5′ T and align to position 1, 2, or 3 of annotated piRNA genes. Third, tailed piRNA reads containing stretches of the same nucleotide preceding the tail were removed. Finally, tailed reads must be present in both biological replicates.
Capped small RNA sequencing dataset analysis
Abundance of piRNA precursors (csRNAs) was based on CIP-TAP sequencing data which is available under GSE40053 (Gu et al., 2012). To detect piRNA precursors, a reference was constructed to have a two-nucleotide extension at the 5′ end of annotated piRNA loci and a ten-nucleotide addition to the 3′ ends of annotated piRNA loci (Wormbase release WS230). Processed sequencing reads were aligned to the piRNA reference using Bowtie. Following alignment, the abundance of csRNA and piRNA reads was analyzed using a custom R script.
CLASH dataset analysis
Publicly available Crosslinking, ligation and sequencing of hybrids (CLASH) data were downloaded from NCBI under ascension available under GSE38723 (Shen et al., 2018). CLASH chimeras were mapped and analyzed as described (Shen et al., 2018). Gibbs free energy (ΔG) of piRNA-target hybrids were calculated using RNAfold in Vienna RNA Package (version 2.3.5) (Lorenz et al., 2011; Shen et al., 2018). To conduct CLASH dataset analysis, we randomly selected a control set composed of 272 untailed piRNAs whose expression levels are comparable to tailed piRNAs. Following generation of a suitable control, CLASH reads for each piRNA in the control and tailed population were examined. CLASH reads were placed into bins according to the predicted binding energy of the piRNA-target interaction. Using custom R scripts, we analyzed the CLASH counts per piRNA as well as the Watson-Crick base-pairing at each piRNA position for piRNA-target chimeras.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analysis was preformed using custom R (v4.0.3) scripts and R packages dplyr (v1.0.2) and tidyr (v1.1.2). Plotting was conducted using ggplot2 (v3.3.2). Specific statistical tests used in our analyses are indicated in the figure legends. Data were evaluated using homoscedasticity assumption tests before performing parametric statistical tests.
Supplementary Material
Highlights.
Nontemplated nucleotides are added at the 3′ end of piRNAs
Base-pairing interaction with target RNAs induces degradation of piRNAs
Activities of PARN-1 and HENN-1 protect piRNAs from 3′ tailing
Deletion of parn-1 and henn-1 leads to reduction in WAGO-22G-RNAs and low fecundity
ACKNOWLEDGMENTS
We thank D. Schoenberg and I. Gainetdinov for discussion and critical comments, S. Tu and H. Chen for providing curated CLASH data, W. Gu for providing csRNA data, P. Yan and Genomics Shared Resource for the support in Illumina sequencing (R50 CA211524 and P30 CA016058), and the Ohio Supercomputer Center for computing resources. Some of the C. elegans and bacterial strains were provided by the Caenorhabditis Genetics Center, which is supported by the NIH (P40 OD010440). This work was supported by the NIH Pathway to Independence Award (R00 GM124460) and Maximizing Investigators’ Research Award (R35 GM142580) to W.T.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109640.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, and Zamore PD (2010). Target RNA-directed trimming and tailing of small silencing RNAs. Science 328, 1534–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, lovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. (2006). A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442, 203–207. [DOI] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, et al. (2012). piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150, 88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarini A, Chauhan H, Gardner TJ, Jayaprakash AD, Sachidanandam R, and Brown BD (2011). Kinetic analysis reveals the fate of a micro-RNA following target regulation in mammalian cells. Curr. Biol 21, 369–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagijn MP, Goldstein LD, Sapetschnig A, Weick EM, Bouasker S, Lehrbach NJ, Simard MJ, and Miska EA (2012). Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science 337, 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2018). Metazoan MicroRNAs. Cell 173, 20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. (2008). PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell 31, 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billi AC, Alessi AF, Khivansara V, Han T, Freeberg M, Mitani S, and Kim JK (2012). The Caenorhabditis elegans HEN1 ortholog, HENN-1, methylates and stabilizes select subclasses of germline small RNAs. PLoS Genet. 8, e1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitetti A, Mallory AC, Golini E, Carrieri C, Carreño Gutiérrez H, Perlas E, Pérez-Rico YA, Tocchini-Valentini GP, Enright AJ, Norton WHJ, et al. (2018). MicroRNA degradation by a conserved target RNA regulates animal behavior. Nat. Struct. Mol. Biol 25, 244–251. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, and Hannon GJ (2007). Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103. [DOI] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, and Kennedy S (2012). A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489, 447–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, and Hannon GJ (2007). MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 12, 503–514. [DOI] [PubMed] [Google Scholar]
- Cecere G, Zheng GX, Mansisidor AR, Klymko KE, and Grishok A (2012). Promoters recognized by forkhead proteins exist for individual 21U-RNAs. Mol. Cell 47, 734–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Simard MJ, Tabara H, Brownell DR, McCollough JA, and Mello CC (2005). A member of the polymerase beta nucleotidyltransferase superfamily is required for RNA interference in C. elegans. Curr. Biol 15, 378–383. [DOI] [PubMed] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. (2009). The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139, 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeiro Rodrigues RJ, de Jesus Domingues AM, Hellmann S, Dietz S, de Albuquerque BFM, Renz C, Ulrich HD, Sarkies P, Butter F, and Ketting RF (2019). PETISCO is a novel protein complex required for 21U RNA biogenesis and embryonic viability. Genes Dev. 33, 857–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, and Lin H (1998). A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12, 3715–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dard-Dascot C, Naquin D, d’Aubenton-Carafa Y, Alix K, Thermes C, and van Dijk E (2018). Systematic comparison of small RNA library preparation protocols for next-generation sequencing. BMC Genomics 19, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, et al. (2008). Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol. Cell 31,79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata M, Gaidatzis D, Vitanescu M, Stadler MB, Wentzel C, Scheiffele P, Filipowicz W, and Großhans H (2015). Potent degradation of neuronal miRNAs induced by highly complementary targets. EMBO Rep. 16, 500–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D, Liu J, Dong K, Midic U, Hess RA, Xie H, Demireva EY, and Chen C (2017). PNLDC1 is essential for piRNA 3′ end trimming and transposon silencing during spermatogenesis in mice. Nat. Commun 8, 819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faehnle CR, Walleshauser J, and Joshua-Tor L (2014). Mechanism of Dis3l2 substrate recognition in the Lin28-let-7 pathway. Nature 514, 252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs Wightman F, Giono LE, Fededa JP, and de la Mata M (2018). Target RNAs Strike Back on MicroRNAs. Front. Genet 9, 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gainetdinov I, Colpan C, Arif A, Cecchini K, and Zamore PD (2018). A Single Mechanism of Biogenesis, Initiated and Directed by PIWI Proteins, Explains piRNA Production in Most Animals. Mol. Cell 71, 775–790.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gainetdinov I, Colpan C, Cecchini K, Albosta P, Jouravleva K, Vega-Badillo J, Lee Y, Özata DM, and Zamore PD (2020). Terminal Modification, Sequence, and Length Determine Small RNA Stability in Animals. bioRxiv. 10.1101/2020.09.08.287979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanta KS, and Mello CC (2020). Melting dsDNA Donor Molecules Greatly Improves Precision Genome Editing in Caenorhabditis elegans. Genetics 216, 643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, and Zamore PD (2009). Small silencing RNAs: an expanding universe. Nat. Rev. Genet 10, 94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh WS, Seah JW, Harrison EJ, Chen C, Hammell CM, and Hannon GJ (2014).A genome-wide RNAi screen identifies factors required for distinct stages of C. elegans piRNA biogenesis. Genes Dev. 28, 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh WSS, Falciatori I, Tam OH, Burgess R, Meikar O, Kotaja N, Hammell M, and Hannon GJ (2015). piRNA-directed cleavage of meiotic transcripts regulates spermatogenesis. Genes Dev. 29, 1032–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou L-T, Dai P, Yang J-H, Xue Y, Hu Y-P, Zhou Y, Kang J-Y, Wang X, Li H, Hua M-M, et al. (2014). Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 24, 680–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D Jr., Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. (2009). Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell 36, 231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Lee HC, Chaves D, Youngman EM, Pazour GJ, Conte D Jr., and Mello CC (2012). CapSeq and CIP-TAP identify Pol II start sites and reveal capped small RNAs as C. elegans piRNA precursors. Cell 151, 1488–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas G, Cetin S, Messmer M, Chane-Woon-Ming B, Terenzi O, Chicher J, Kuhn L, Hammann P, and Pfeffer S (2016). Identification of factors involved in target RNA-directed microRNA degradation. Nucleic Acids Res. 44,2873–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BW, Wang W, Li C, Weng Z, and Zamore PD (2015). Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 348, 817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, LaVigne CA, Jones BT, Zhang H, Gillett F, and Mendell JT (2020). A ubiquitin ligase mediates target-directed microRNA decay independently of tailing and trimming. Science 370, eabc9546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helwak A, Kudla G, Dudnakova T, and Tollervey D (2013). Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 153, 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homolka D, Pandey RR, Goriaux C, Brasset E, Vaury C, Sachidanandam R, Fauvarque M-O, and Pillai RS (2015). PIWI Slicing and RNA Elements in Precursors Instruct Directional Primary piRNA Biogenesis. Cell Rep. 12, 418–428. [DOI] [PubMed] [Google Scholar]
- Honda S, Kirino Y, Maragkakis M, Alexiou P, Ohtaki A, Murali R, Mourelatos Z, and Kirino Y (2013). Mitochondrial protein BmPAPI modulates the length of mature piRNAs. RNA 19, 1405–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, and Zamore PD (2007). The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 17, 1265–1272. [DOI] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. (2007). A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129, 69–82. [DOI] [PubMed] [Google Scholar]
- Huang X, Fejes Tóth K, and Aravin AA (2017). piRNA Biogenesis in Drosophila melanogaster. Trends Genet. 33, 882–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, and Hannon GJ (2012). The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature 491, 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi N, Shoji K, Sakaguchi Y, Honda S, Kirino Y, Suzuki T, Katsuma S, and Tomari Y (2016). Identification and Functional Analysis of the Pre-piRNA 3′ Trimmer in Silkworms. Cell 164, 962–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamminga LM, van Wolfswinkel JC, Luteijn MJ, Kaaij LJ, Bagijn MP, Sapetschnig A, Miska EA, Berezikov E, and Ketting RF (2012). Differential impact of the HEN1 homolog HENN-1 on 21U and 26G RNAs in the germline of Caenorhabditis elegans. PLoS Genet. 8, e1002702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper DM,Wang G, Gardner KE, Johnstone TG, and Reinke V (2014). The C. elegans SNAPc component SNPC-4 coats piRNA domains and is globally required for piRNA abundance. Dev. Cell 31, 145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T, and Suzuki T (2009). Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 23, 433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T, Hojo H, and Suzuki T (2015). Destabilization of microRNAs in human cells by 3′ deadenylation mediated by PARN and CUGBP1. Nucleic Acids Res. 43, 7521–7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka S, Izumi N, Katsuma S, and Tomari Y (2011). 3′ end formation of PIWI-interacting RNAs in vitro. Mol. Cell 43, 1015–1022. [DOI] [PubMed] [Google Scholar]
- Kingston ER, and Bartel DP (2019). Global analyses of the dynamics of mammalian microRNA metabolism. Genome Res. 29, 1777–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, and Mourelatos Z (2007). Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat. Struct. Mol. Biol 14, 347–348. [DOI] [PubMed] [Google Scholar]
- Kleaveland B, Shi CY, Stefano J, and Bartel DP (2018). A Network of Noncoding Regulatory RNAs Acts in the Mammalian Brain. Cell 174, 350–362.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Gu W, Shirayama M, Youngman E, Conte D Jr., and Mello CC (2012). C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell 150, 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XZ, Roy CK, Dong X, Bolcun-Filas E, Wang J, Han BW, Xu J, Moore MJ, Schimenti JC, Weng Z, and Zamore PD (2013). An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol. Cell 50, 67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JJ, Huang H, Bickel PJ, and Brenner SE (2014). Comparison of D. melanogaster and C. elegans developmental stages, tissues, and cells by modENCODE RNA-seq data. Genome Res. 24, 1086–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Dai H, Nguyen AP, and Gu W (2020). A convenient strategy to clone small RNA and mRNA for high-throughput sequencing. RNA 26, 218–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, and Spradling AC (1997). A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 724, 2463–2476. [DOI] [PubMed] [Google Scholar]
- Liu L, Qi H, Wang J, and Lin H (2011). PAPI, a novel TUDOR-domain protein, complexes with AGO3, ME31B and TRAL in the nuage to silence transposition. Development 138, 1863–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, and Hofacker IL (2011). ViennaRNA Package 2.0. Algorithms Mol. Biol 6, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn MJ, and Ketting RF (2013). PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat. Rev. Genet 14, 523–534. [DOI] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, and Hannon GJ (2009). Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137, 522–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniar JM, and Fire AZ (2011). EGO-1, a C. elegans RdRP, modulates gene expression via production of mRNA-templated short antisense RNAs. Curr. Biol 21,449–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzi MJ, Ghini F, Cerruti B, de Pretis S, Bonetti P, Giacomelli C, Gorski MM, Kress T, Pelizzola M, Muller H, et al. (2016). Degradation dynamics of microRNAs revealed by a novel pulse-chase approach. Genome Res. 26, 554–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, and Ambros V (1991). Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Handler D, and Brennecke J (2015). Noncoding RNA. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 348, 812–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery TA, Rim YS, Zhang C, Dowen RH, Phillips CM, Fischer SE, and Ruvkun G (2012). PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 ortholog henn-1. PLoS Genet. 8, e1002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munafó DB, and Robb GB (2010). Optimization of enzymatic reaction conditions for generating representative pools of cDNA from small RNA. RNA 16, 2537–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Kuehn MS, Reynolds AP, Haugen E, Thurman RE, Johnson AK, Rynes E, Maurano MT,Vierstra J, Thomas S, et al. (2012). BEDOPS: high-performance genomic feature operations. Bioinformatics 28,1919–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L, Matsumoto N, Nishizawa T, Nakanaga K, Aoki J, et al. (2012). Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature 491, 284–287. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Nagamori I, Nakatani T, Izumi N, Tomari Y, Kuramochi-Miyagawa S, and Nakano T (2018). PNLDC1, mouse pre-piRNA Trimmer, is required for meiotic and post-meiotic male germ cell development. EMBO Rep. 19, e44957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, and Brennecke J (2010). An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 29, 3301–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz MA, Noble D, Sorokin EP, and Kimble J (2014). A new dataset of spermatogenic vs. oogenic transcriptomes in the nematode Caenorhabditis elegans. G3 (Bethesda) 4, 1765–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozata DM, Gainetdinov I, Zoch A, O’Carroll D, and Zamore PD (2019). PIWI-interacting RNAs: small RNAs with big functions. Nat. Rev. Genet 20, 89–108. [DOI] [PubMed] [Google Scholar]
- Pane A, Wehr K, and Schüpbach T (2007). zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev. Cell 12, 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston MA, Porter DF, Chen F, Buter N, Lapointe CP, Keles S, Kimble J, and Wickens M (2019). Unbiased screen of RNA tailing activities reveals a poly(UG) polymerase. Nat. Methods 16, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed KJ, Svendsen JM, Brown KC, Montgomery BE, Marks TN, Vijayasarathy T, Parker DM, Nishimura EO, Updike DL, and Montgomery TA (2020). Widespread roles for piRNAs and WAGO-class siRNAs in shaping the germline transcriptome of Caenorhabditis elegans. Nucleic Acids Res. 48, 1811–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichholf B, Herzog VA, Fasching N, Manzenreither RA, Sowemimo I, and Ameres SL (2019). Time-Resolved Small RNA Sequencing Unravels the Molecular Principles of MicroRNA Homeostasis. Mol. Cell 75, 756–768.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, and Bartel DP (2006). Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127, 1193–1207. [DOI] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, and Siomi MC (2007). Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 21, 1603–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanei M, and Chen X (2015). Mechanisms of microRNA turnover. Curr. Opin. Plant Biol 27, 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxe JP, Chen M, Zhao H, and Lin H (2013). Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. EMBO J. 32, 1869–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G, Mello CC, Pettitt J, Wood WB, Priess JR, and Fire A (1996). Repression of gene expression in the embryonic germ lineage of C. elegans. Nature 382, 713–716. [DOI] [PubMed] [Google Scholar]
- Shen EZ, Chen H, Ozturk AR, Tu S, Shirayama M, Tang W, Ding YH, Dai SY, Weng Z, and Mello CC (2018). Identification of piRNA Binding Sites Reveals the Argonaute Regulatory Landscape of the C. elegans Germline. Cell 172, 937–951.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu-Gruttadauria J, Pawlica P, Klum SM, Wang S, Yario TA, Schirle Oakdale NT, Steitz JA, and MacRae IJ (2019). Structural Basis for Target-Directed MicroRNA Degradation. Mol. Cell 75, 1243–1255.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CY, Kingston ER, Kleaveland B, Lin DH, Stubna MW, and Bartel DP (2020). The ZSWIM8 ubiquitin ligase mediates target-directed microRNA degradation. Science 370, eabc9359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D Jr., and Mello CC (2012). piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150, 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Yan J, Pagano DJ, Dodson AE, Fei Y, Gorham J, Seidman JG, Wickens M, and Kennedy S (2020). poly(UG)-tailed RNAs in genome protection and epigenetic inheritance. Nature 582, 283–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M, Sarkies P, Ikegami K, Doebley AL, Goldstein LD, Mitchell J, Sakaguchi A, Miska EA, and Ahmed S (2014). Reduced insulin/IGF-1 signaling restores germ cell immortality to Caenorhabditis elegans Piwi mutants. Cell Rep. 7, 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, and Maine EM (2000). EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol 10, 169–178. [DOI] [PubMed] [Google Scholar]
- Svendsen JM, and Montgomery TA (2018). piRNA Rules of Engagement. Dev. Cell 44, 657–658. [DOI] [PubMed] [Google Scholar]
- Svendsen JM, Reed KJ, Vijayasarathy T, Montgomery BE, Tucci RM, Brown KC, Marks TN, Nguyen DAH, Phillips CM, and Montgomery TA (2019). henn-1/HEN1 Promotes Germline Immortality in Caenorhabditis elegans. Cell Rep. 29, 3187–3199.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Tu S, Lee HC, Weng Z, and Mello CC (2016). The RNase PARN-1 Trims piRNA 3′ Ends to Promote Transcriptome Surveillance in C. elegans. Cell 164, 974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wolfswinkel JC, Claycomb JM, Batista PJ, Mello CC, Berezikov E, and Ketting RF (2009). CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell 139, 135–148. [DOI] [PubMed] [Google Scholar]
- Vourekas A, Zheng Q, Alexiou P, Maragkakis M, Kirino Y, Gregory BD, and Mourelatos Z (2012). Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat. Struct. Mol. Biol 19, 773–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, and Reinke V (2008). A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr. Biol 18, 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weick EM, and Miska EA (2014). piRNAs: from biogenesis to function. Development 141, 3458–3471. [DOI] [PubMed] [Google Scholar]
- Weick E-M, Sarkies P, Silva N, Chen RA, Moss SMM, Cording AC, Ahringer J, Martinez-Perez E, and Miska EA (2014). PRDE-1 is a nuclear factor essential for the biogenesis of Ruby motif-dependent piRNAs in C. elegans. Genes Dev. 28, 783–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng C, Kosalka J, Berkyurek AC, Stempor P, Feng X, Mao H, Zeng C, Li WJ, Yan YH, Dong MQ, et al. (2019). The USTC co-opts an ancient machinery to drive piRNA transcription in C. elegans. Genes Dev. 33, 90–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Ameres SL, Friedline R, Hung JH, Zhang Y, Xie Q, Zhong L, Su Q, He R, Li M, et al. (2012). Long-term, efficient inhibition of microRNA function in mice using rAAV vectors. Nat. Methods 9, 403–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Shao TJ, Bofill-De Ros X, Lian C, Villanueva P, Dai L, and Gu S (2020). AGO-bound mature miRNAs are oligouridylated by TUTs and subsequently degraded by DIS3L2. Nat. Commun 11, 2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, and Kim VN (2020). A tale of non-canonical tails: gene regulation by post-transcriptional RNA tailing. Nat. Rev. Mol. Cell Biol 21, 542–556. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Guo R, Cui Y, Zhu Z, Zhang Y, Wu H, Zheng B, Yue Q, Bai S, Zeng W, et al. (2017). An essential role for PNLDC1 in piRNA 3′ end trimming and male fertility in mice. Cell Res. 27, 1392–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Tu S, Stubna M, Wu WS, Huang WC, Weng Z, and Lee HC (2018). The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science 359, 587–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yu Y, Zhai J, Ramachandran V, Dinh TT, Meyers BC, Mo B, and Chen X (2012). The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr. Biol 22, 689–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
WAGO-1 and HRDE-1/WAGO-9 immunoprecipitation data are available fromGene Expression Omnibus (GEO) under the accession number GEO: GSE137734. CSR-1 immunoprecipitation data are available under GEO: GSE18165. Small RNA sequencing data from wild-type and rde-3 strains are available under GEO: GSE18215. CIP-TAP sequencing data for csRNAs are available under GEO: GSE40053. PRG-1 CLASH sequencing data are available from Sequence Read Archive (SRA) under SRA: SRP131397. Sequencing data used to define PRG-1 targets are available under GEO: GSE141243 and SRA: SRS1021265. Small RNA sequencing data generated for use in this study are available under SRA: PRJNA683039.
Custom R, python and shell scripts used in this analysis are freely available upon request.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


