Abstract
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) seroprevalence studies bridge the gap left from case detection, to estimate the true burden of the COVID-19 pandemic. While multiple anti-SARS-CoV-2 immunoassays are available, no gold standard exists.
Methods
This serial cross-sectional study was conducted using plasma samples from 8999 healthy blood donors between April-September 2020. Each sample was tested by four assays: Abbott SARS-Cov-2 IgG assay, targeting nucleocapsid (Abbott-NP) and three in-house IgG ELISA assays (targeting spike glycoprotein, receptor binding domain, and nucleocapsid). Seroprevalence rates were compared using multiple composite reference standards and by a series of Bayesian Latent Class Models.
Result
We found 13 unique diagnostic phenotypes; only 32 samples (0.4%) were positive by all assays. None of the individual assays resulted in seroprevalence increasing monotonically over time. In contrast, by using the results from all assays, the Bayesian Latent Class Model with informative priors predicted seroprevalence increased from 0.7% (95% credible interval (95% CrI); 0.4, 1.0%) in April/May to 0.7% (95% CrI 0.5, 1.1%) in June/July to 0.9% (95% CrI 0.5, 1.3) in August/September. Assay characteristics varied over time. Overall Spike had the highest sensitivity (93.5% (95% CrI 88.7, 97.3%), while the sensitivity of the Abbott-NP assay waned from 77.3% (95% CrI 58.7, 92.5%) in April/May to 64.4% (95% CrI 45.6, 83.0) by August/September.
Discussion
Our results confirmed very low seroprevalence after the first wave in Canada. Given the dynamic nature of this pandemic, Bayesian Latent Class Models can be used to correct for imperfect test characteristics and waning IgG antibody signals.
Introduction
Worldwide, more than 159 million people have been diagnosed with coronavirus disease 2019 (COVID-19), as of May 13, 2021 [1]. Yet, this is likely an underestimation of the true burden of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) given testing is primarily used to confirm suspected infection as opposed to broad surveillance. For example, in Canada, testing was only accessible early in the pandemic to people who had symptoms, were known contacts of a case or had a relevant travel history [2]. This meant community transmission by asymptomatic or mildly symptomatic individuals was likely underestimated. Determining the proportion of individuals with evidence of an immune response to SARS-CoV-2 can provide a more comprehensive assessment of prevalence to assist public health officials in making policy decisions. This prompted an urgent need for seroprevalence studies and accurate anti-SARS-CoV-2 immunoassays to estimate the true burden of disease.
While multiple commercial and in-house immunoassays to detect anti-SARS-CoV-2 antibodies are available, to date no gold standard exists [3]. Furthermore, laboratorians have described multiple examples of discordance between assays [4]. Some of this variability is in part due to the assays which vary significantly by the isotype (i.e. IgA, IgM, IgG), viral antigens (i.e. spike or nucleocapsid protein and whether full-length or partial), and test performance (i.e. sensitivity/specificity). It is also known that anti-SARS-COV-2 antibodies wane over time which can further affect the sensitivity and specificity of the assays [5–7]. Additionally, biological differences between individuals can lead to different antibody profiles. Given these overlapping challenges of estimating seroprevalence, relying on a single assay (regardless which assay this may be) may bias results.
In the absence of a gold standard, using results from multiple assays may improve accuracy. However, which methods are appropriate for estimating SARS-CoV-2 seroprevalence has not been defined. One method is to use a composite reference standard (CRS); a traditional approach used in clinical settings based on prespecified rules based on results from multiple assays [8]. More recently, Bayesian Latent Class Analysis (BLCA) has become more mainstream in diagnostic studies [9]. In contrast to CRS which classifies individuals as either positive or negative, BLCA uses a likelihood-based approach from multiple imperfect assays to estimate test characteristics and prevalence. Given the uncertainty of the assay performance, we evaluated multiple methodological approaches to estimate SARS-COV-2 seroprevalence during the first COVID-19 wave in Canada using four unique assays.
Methods
Study design and population sampling
We conducted a serial cross-sectional study among blood donors in Canada between April and September 2020 (prior to COVID-19 vaccine availability). Canadian Blood Services (CBS) collects approximately 850,000 blood donations per year from a combination of fixed and mobile sites in all larger cities and most urban areas from all provinces in Canada except Quebec [10]. Blood donors (>17 years old) must meet numerous selection criteria to ensure that they are in good health and at low risk of infectious disease. Beginning in March 2020, donors were deferred for two weeks if they were diagnosed with SARS-CoV-2 infection or if they were in contact with a known case. Each month 1500 deidentified samples were randomly selected by collection site by region, age and sex to be reflective of the donor population across Canada. Data on the collection site, birth year, sex and Forward Sortation Area (FSA) of the residential postal code for each donor were extracted. The Research Ethics Board of the Canadian Blood Services and Lunenfeld-Tanenbaum Research Institute (LTRI) (REB study #20-0194-E) approved this study and exempted study-specific consent.
SARS-CoV-2 antibody testing
Retention EDTA plasma samples were aliquoted and frozen at -20°C at the CBS laboratory in Ottawa. Each sample was tested for SARS-CoV-2 IgG antibodies using four assays. The Abbott Architect SARS-Cov-2 IgG assay which targets the nucleocapsid antigen (Abbott-NP), (Abbott, Chicago IL) and three in-house IgG ELISA chemiluminescent assays recognizing distinct recombinant viral antigens: full length spike glycoprotein (Spike), spike glycoprotein receptor binding domain (RBD), and nucleocapsid (NP), were tested at the CBS laboratory in Ottawa and the Gingras laboratory [11, 12] at the LTRI in Toronto, respectively. Table 1 summarizes each antibody assay by: platform, antigen targets and how reactivity was determined.
Table 1. Assay characteristics.
| Assay | Assay platform | Capture Antigen (IgG) | Manufacture | Cut-offs (positive) | Cut-off reference |
|---|---|---|---|---|---|
| Abbott-NP | Chemiluminescent microparticle immunoassay | Nucleocapsid | Abbott | ≥1.40 | Manufacture |
| Spike | Chemiluminescent ELISA | spike | Gingras Lab | ≥0.190 | 3 SD + negative meana |
| RBD | Chemiluminescent ELISA | RBD | Gingras Lab | ≥0.186 | 3 SD + negative meana |
| NP | Chemiluminescent ELISA | Nucleocapsid | Gingras Lab | ≥0.396 | 3 SD + negative meana |
Abbott-NP, Abbott Architect SARS-Cov-2 IgG assay targeting nucleocapsid antigen; Spike, full length spike glycoprotein; RBD, spike glycoprotein receptor binding domain; NP, nucleocapsid.
a3 Standard deviations (SD) + negative mean is a standard approach to choosing cut-off thresholds for ELISA based assays. Briefly, the relative ratio values of the negative controls from 20–22 different tests were used. Ratios are transformed on the log10-scale and then mean 3 SD determines the cutoff. Values are then exponentiated to identify cut off listed in the table.
Analysis
We evaluated the correlation between the individual assays by kappa statistics. In the absence of a gold standard, we examined multiple approaches to estimate seroprevalence. First, seroprevalence was estimated by individual assays based on pre-defined thresholds (Table 1). Then we used a series of composite reference standards to identify a “true” positive if a sample was reactive by a combination of two or more assays. Finally, we estimated seroprevalence using Bayesian Latent Class Models (BLCM).
Bayesian latent class analysis
In this study the “latent” unobservable target was evidence of SARS-CoV-2 infection (based on IgG positivity). Instead of relying on one imperfect assay, this iterative model leverages the data from multiple imperfect assays to estimate the “true” prevalence and test characteristics [13, 14]. Given any one of four assays could assign an individual to be positive or negative, there was a maximum of 16 (24), possible diagnostic phenotypes. We assumed each assay was independent of the others, conditional on the individual’s unknown antibody status. This means that the probability of obtaining a given diagnostic phenotype depended on the probability that an individual had been truly infected with SARS-CoV-2 and on the outcome of each assay given the underlying exposure status. Briefly, we estimated parameters in a Bayesian framework using a Gibbs sampler to produce Markov chain Monte Carlo (MCMC) simulations. We ran 50,000 iterations, with the first 5000 steps discarded as burn-in [15]. Given the uncertainty of assay performance in this donor population, we compared goodness of fit parameters using informative, weakly informative and non-informative priors. Informative priors were based on the manufactures assumed sensitivity assumed specificity (S1 Table). Expert opinion defined weakly informative priors (sensitivity ranging from 60%-100% and specificity from 90%-100%, for each assay). We assumed an uniformed distribution for the model using non-informative priors. We verified convergence of all MCMC chains. We reported posterior means and 95% credible intervals (CrI) for all estimated parameters overall and by two-month intervals using SAS (version 9.1, SAS Institute, Cary, NC). For more additional details on Bayesian Latent Class analysis please refer to Cheung et al, 2021 [16].
Results
Between April and September 2020, a total of 8999 healthy blood samples were assessed for SARS-CoV-2 antibodies by four distinct assays. Most donors (96%) were between 20–69 years old, there were slightly more male donors (52%) compared to female donors (48%) and there was representation from all provinces across Canada except Quebec. Donor characteristics remained consistent over the study period (S2 Table).
Individual assays
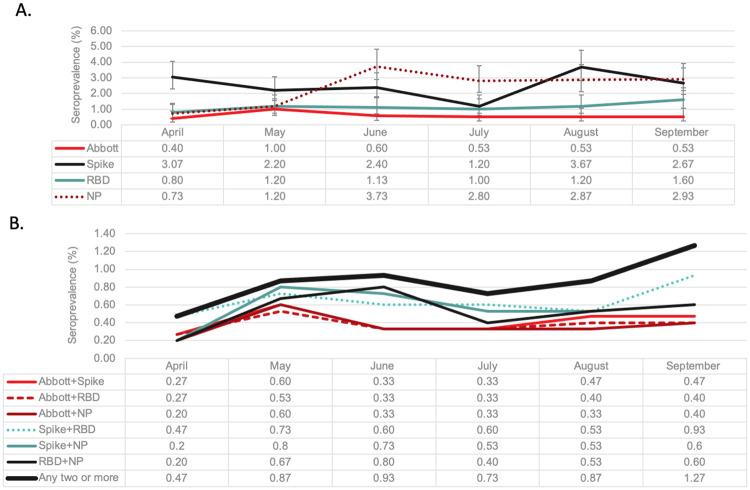
We evaluated seroprevalence rates over time by the individual assays (Fig 1a). Overall, there was significant variability by the assays over time. The Abbott-NP assay consistently remained lower than the ELISA-based assays. Seroprevalence based on the spike assay was 3.1% in May, dropped to 1.2% in July and then plateaued around 3%. Rates were lower and more stable by RBD that started at 0.8% and increased to 1.6% by September. In contrast the NP assay increased significantly from May (1.2%) until June (3.7%). The signal to cut off ratios remained relatively stable over time for all assays (S1 Fig). Overall, the correlation between the assays was low (kappa score, 0.28 (95% CI 0.21, 0.34)). Given concurrent negative results the percent agreement was highest between Abbott-NP and RBD (kappa 0.43 (95% CI 0.33, 0.51).
Fig 1.
A. Seroprevalence by month over the first COVID-19 wave in Canada by individual assays. Each line represents seroprevalence rates (summarized in table below) monthly between April and September 2020 (during the first COVID-19 Wave) based thresholds for each assay. Abbott Architect SARS-Cov-2 IgG assay (Abbott-NP) and three in-house IgG ELISA assays recognizing distinct recombinant viral antigens: full length spike glycoprotein (Spike), spike glycoprotein receptor binding domain (RBD), and nucleocapsid (NP). B. Seroprevalence by month over the first COVID-19 wave in Canada by various composite reference standards (results from four anti-SARS-CoV-2 immunoassays). Each line represents seroprevalence rates based on predefined definitions. CRS based on a combination of reactive samples using Abbott-NP, Spike, RBD and NP. Positivity based on “any two or more” was determined by a reactive sample from two or more assays. Since we are not comparing CRS, we did not include 95% CI for each data point (all overlapping).
Composite reference standards
Given screening occurred in a low prevalence setting, to minimize false positive results, we assumed a true positive was more likely when two or more assays were positive. A priori, relying on two pre-specified assays resulted in a range of seroprevalence estimates that ranged from 0.2% to 0.5% in April to 0.4% to 1.4% in September (Fig 1b). Any two assays (from four) resulted in a seroprevalence that increased significantly over time from 0.5% (95% CI 0.3%, 1.1%) in April to 1.3% (95% CI 0.8, 2.0) in September (p = 0.02) (Fig 1b).
Bayesian latent class model
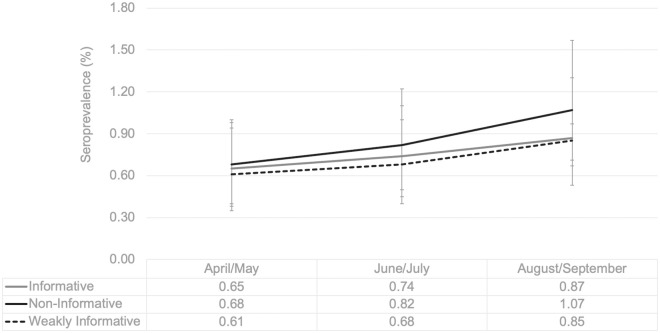
From 16 possible diagnostic phenotypes, 13 were observed among the 8999 sampled (eight phenotypes had at least 10 observations). The most frequent profile was “all negative” (95.0% (95% CI 94.6, 95.5) followed by only positive by the individual ELISA-based assays (Spike only (1.8%, 95% CI 1.5, 2.0) and NP only (1.7, 95% CI 1.5, 2.1). Only 32 samples were positive for all four assays (0.4, 95% CI 0.3, 0.5) (Table 2). Overall seroprevalence was estimated to be 0.8% (95% CrI 0.6, 1.0%); 0.8% (95% CrI 0.6, 1.0%); 0.8% (95% CrI 0.7, 1.0%) using informative, weakly informative and non-informative priors, respectively. Fig 2 illustrates temporal trends in seroprevalence by BLCA comparing the various models. The model with the non-informative prior consistently was higher than the other two models, but the difference was not statistically significant. Given the uncertainty of test characteristics, we compared the observed vs predicted values of the three BLCM and found the informative model had the best model fit and identified the “all negative” phenotype most accurately (S3 Table).
Table 2. Diagnostic phenotypes of anti-SARS-CoV-2 immunoassay results from 8999 samples tested.
| Commercial | In-house ELISA | Observed | ||||
|---|---|---|---|---|---|---|
| Abbott-NP | Spike | RBD | NP | Number | % (95% CI) | |
| (n = 54) | (n = 228) | (n = 104) | (n = 214) | |||
| NONE | - | - | - | - | 8552 | 95.0 (94.6, 95.5) |
| ALL | + | + | + | + | 32 | 0.4 (0.3, 0.5) |
| All—Abbott-NP | - | + | + | + | 9 | 0.1 (0.1, 0.2) |
| NP Only | - | - | - | + | 156 | 1.7 (1.5, 2.1) |
| RBD Only | - | - | + | - | 39 | 0.4 (0.3, 0.6) |
| Spike Only | - | + | - | - | 158 | 1.8 (1.5, 2.0) |
| Spike + RBD | - | + | + | - | 15 | 0.2 (0.1, 0.3) |
| RBD + NP | - | - | + | + | 7 | 0.1 (0.0, 0.3) |
| Abbott—NP Only | + | - | - | - | 17 | 0.2 (0.1, 0.3) |
| Spike + Abbott-NP | + | + | - | - | 2 | 0.0 (0.0, 0.1) |
| Spike + NP | - | + | - | + | 9 | 0.1 (0.1, 0.2) |
| All–NP | + | + | + | - | 2 | 0.0 (0.0, 0.1) |
| All–RBD | + | + | - | + | 1 | 0.0 (0.0, 0.1) |
Abbott-NP, Abbott Architect SARS-Cov-2 IgG assay targeting nucleocapsid antigen; Spike, full length spike glycoprotein; RBD, spike glycoprotein receptor binding domain; NP, nucleocapsid.
Fig 2. Seroprevalence estimates by different BLCM (informative, weakly informative and non-informative priors).
Each line represents seroprevalence rates (summarized in table below) derived from posterior means of three BLCMs (comparing informative, weakly informative and non-informative priors) bi-monthly between April and September 2020 (during the first COVID-19 Wave). Error bars represent 95% Credible Intervals.
The test characteristics (sensitivity and specificity) varied significantly by the different assays (Table 3). Overall, the ELISA based assays had higher sensitivity than the Abbott-NP. Abbott-NP had a sensitivity of 58.5% (95% CrI 46.3, 70.6%) and a specificity of 99.8% (95% CrI 99.7, 99.9%). RBD had the highest specificity (99.5% (95% CrI 99.3, 99.7%)) and NP had the lowest specificity (98.2% (95% CrI 97.9, 98.4%)). Negative predictive values of all assays were very high (ranging from 99.4% to 99.8%). The Abbott-NP had the highest positive predictive value at 87.5% (95% CrI 81.3, 93.8%) while the ELISA based assays that ranged from 21.0 to 59.8%. The ELISA-based assays did not significantly wane over the first six months of the pandemic, the test characteristics of the Abbott assay varied more (Table 3). Sensitivity Abbott-NP assay waned the most from 77.3% (95%CrI 58.7, 92.5) in April/May to 64.4% (95% CrI 45.6, 83.0) in August/September. Similar trends were observed using weakly informative and non-informative priors (S4 Table).
Table 3. Seroprevalence and assay characteristics overall and bi-monthly based on the Bayesian latent class analysis with informative priors.
| Overall | April/May | June/July | August/September | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Seroprevalence | 0.76% (95% CrI 0.58, 0.97) | 0.65% (95% CrI 0.38, 0.98) | 0.74% (95% CrI 0.45, 1.11) | 0.87% (95% CrI 0.53, 1.29%) | ||||||
| Sensitivity | PPV | Specificity | NPV | Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | |
| Spike | 93.5% | 27.7% | 98.1% | 99.8% | 95.0% | 98.0% | 93.6% | 98.8% | 93.6% | 97.6% |
| (88.7, 97.3) | (23.2, 33.7) | (97.9, 98.4) | (99.6, 100.0) | (90.1, 98.2) | (97.4, 98.4) | (88.0, 97.6) | (98.4, 99.2) | (88.0, 97.6) | (97.0, 98.1) | |
| RBD | 89.1% | 59.8% | 99.5% | 99.8% | 89.3% | 99.5% | 89.2% | 99.6% | 88.7% | 99.3% |
| (84.1, 93.5) | (50.0, 71.4) | (99.3, 99.7) | (99.6, 99.9) | (83.7, 93.8) | (99.3, 99.8) | (98.4, 99.2) | (83.6, 93.9) | (83.0, 93.4) | (99.0, 99.6) | |
| NP | 78.8% | 21.0% | 98.2% | 99.6% | 79.9% | 99.5% | 80.5% | 97.3% | 78.5% | 97.6% |
| (74.1, 83.2) | (17.2, 25.3) | (97.9, 98.4) | (99.3, 99.7) | (74.9, 84.5) | (99.2, 99.7) | (75.6, 84.9) | (96.7, 97.9) | (73.5, 83.3) | (97.1, 98.1) | |
| Abbott-NP | 58.5% | 87.5% | 99.8% | 99.4% | 77.3% | 99.7% | 60.2% | 99.8% | 64.4% | 99.9% |
| (46.3, 70.6) | (81.3, 93.8) | (99.7, 99.9) | (99.1, 99.6) | (58.7, 92.5) | (99.5, 99.9) | (41.2, 78.5) | (99.5, 99.9) | (45.6, 83.0) | (99.8, 100.0) | |
CrI, Creditable Interval; PPV, Positive Predictive Value; NPV, Negative Predictive Value; Abbott-NP, Abbott Architect SARS-Cov-2 IgG assay targeting nucleocapsid antigen; Spike, full length spike glycoprotein; RBD, spike glycoprotein receptor binding domain; NP, nucleocapsid.
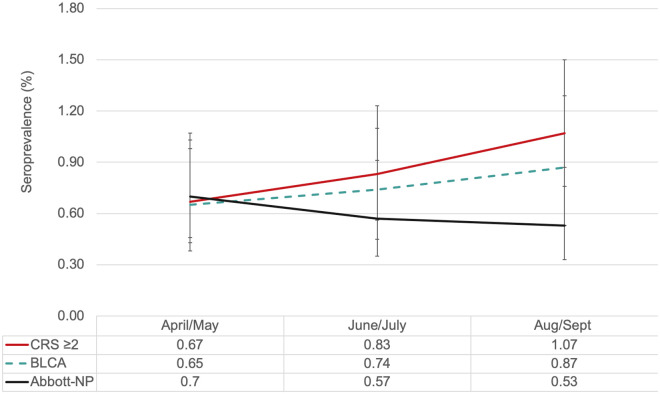
Overall the latent class model and CRS (using the rule > = 2 reactive assays out of four) yielded similar results and there was no evidence of waning seroprevalence rates over time (Fig 3). While the Abbott-NP (a common commercial assay) did wane over time.
Fig 3. Summary comparison of seroprevalence rates by analytical methods.
Each line represents seroprevalence rates (summarized in table below) derived from four analytical methods bi-monthly between April and September 2020 (during the first COVID-19 Wave). > = 2 proteins (positivity was determined by a reactive sample from two or more assays), BLCA-Bayesian latent class analysis with informative priors, results are posterior means and error bars are 95% CrI and Abbott-NP is a single commercial assay.
Discussion
In the absence of a gold standard, we evaluated multiple assays and methodological approaches to estimate SARS-CoV-2 seroprevalence in healthy Canadian blood donors. None of the individual assays resulted in seroprevalence increasing monotonically over time. Seroprevalence estimates were similar by either BLCM or a composite reference standard when at least two positive assays (out of four) were used to determine a “true” result. However, by using the BLCM, we were able to derive time-updated test characteristics that could be used to adjust for waning antibody signals.
Gaps in laboratory testing during the first wave, the significant proportion of asymptomatic or pauci-symptomatic infections, as well as a continuing pandemic have prompted public health authorities in Canada to continue to invest in serological surveys to evaluate the true burden of SARS-CoV-2. Yet unique biological and epidemiological challenges exist when estimating seroprevalence, particularly in low prevalence settings. We recently conducted a scoping review and identified 33 seroprevalence studies among blood donors worldwide. From the 33 studies, 27 unique assay combinations were identified, more than half of studies used a single assay to determine prevalence and less than a third accounted for imperfect test performance [17]. Results from this study suggest relying on a single assay to determine prevalence in a low prevalence setting may significantly bias results.
The variability in the number of diagnostic phenotypes may be associated with the interindividual variability of the immune response. SARS-CoV-2 infects cells using a spike glycoprotein to bind to human angiotensin-converting enzyme 2 (ACE2) [18–20]. The receptor binding domain attached to spike mediates both viral binding and fusion events and all proteins are targets for neutralizing monoclonal antibodies [21, 22]. Biologically, it is not clear why a person may differentially express antibodies against SARS-CoV-2, but among our sample, we found 13 distinct diagnostic phenotypes. The discordance between assays may also be a product of imperfect test characteristics. In this study, we used one commercial assay for which the manufacture originally reported a sensitivity of 95.9% and specificity of 99.6%. Later real-world reports suggested the sensitivity was as low as 92.7% [23–28]. Results from this study suggest significantly lower sensitivity. While it is customary to assume that assay performance remains static, amid this dynamic pandemic, waning antibody signals may compromise correct classification of prior SARS-CoV-2 exposure. We have previously shown in a longitudinal study that the NP signal in the ELISA-based assay wanes faster than spike or RBD [12]. Consistent with previous reports, we found the nucleocapsid signal from the Abbott assay also wanes faster than spike or RBD [29, 30]. This suggests that NP-based assays may be identifying more recent exposures.
It should be noted that waning antibody signals do not necessarily mean waning cellular mediated immunity. Indeed, recent studies suggest in the absence of detectable antibody signals there is evidence of neutralization associated with longer lasting immunity [31, 32]. Therefore, without adjusting for waning antibody signals we may be underestimating SARS-CoV-2 seroprevalence. At this point in time, it remains unknown what the true measures or correlates of immunity are in the Canadian population. The data presented here does not address whether some blood donors may have mounted a cellular immune response with an antibody response that waned by the time of serologic testing. We also note that the presence of antibodies does not imply that those antibodies are neutralizing; although we have assessed for spike and RBD antibodies, we have not attempted to understand the neutralizing capacity of these donor specimens against wild type strains of SARS-CoV-2 or emerging variants in Canada. In the next steps of our analysis we will be undertaking studies to understand the neutralizing capacity of these donor specimens to SARS-CoV-2.
Our study has several strengths. First this study is nested within a large national seroprevalence survey which to date has tested >179,000 samples using the Abbott-NP assay since the beginning of the pandemic in Canada. While we tested only a fraction of the samples, the sample demographic and seroprevalence rates (based on Abbott-NP) were very similar, illustrating the generalizability of our results nationally [33].
Given the uncertainty around the assay characteristics specifically among a donor population, we used multiple methodological approaches to estimate seroprevalence and report all findings. One of the strengths of the BLCA is the ability to estimate assay performance in the absence of a gold standard. Given limited resources, it may not be feasible to evaluate seroprevalence using four unique assays. However, smaller nested studies with more comprehensive antibody data within larger surveys can be used to correct for measurement errors. For example, we used the sensitivity and specificity of the BCLA from this study to adjust for national seroprevalence estimates recently published between May-July 2020 [33]. We reported seroprevalence was 0.74% (95%CI 0.68, 0.80), but after reanalyzing the data with updated sensitivity/specificity, based on the BLCM with informative priors, we found that the corrected seroprevalence was 27% higher at 0.94% (95% CI 0.83, 1.05%). As the pandemic continues, the proportion of recent and older infections will continue to vary over time and having an ability to correct these time varying assay characteristics will become even more important.
Our study also has weaknesses. This study was conducted among blood donors, based on selection criteria to be allowed to donate blood donors may be healthier than the general population [34]. However, a recent study compared seroprevalence estimates from European blood donors to household surveys targeting the general population and found seroprevalence rates to be very similar [35]. In this analysis we assumed the assays were conditionally independent, meaning there were no jointly false-positive or false-negative results between the assays. We evaluated this assumption by assessing changes in sensitivity and specificity of each assay after leaving one assay out. Although we found insignificant changes in test characteristics (S5 Table), it is possible this assumption does not hold, potentially biasing results. Future studies are planned to explore different correlation structures. Assay performance is based on predefined thresholds. For the Abbott assay we used the manufacturer’s ≥1.4 cut off, but recent reports do suggest reducing the threshold to >0.8 to increase sensitivity and to account for waning antibody signals. However, the sensitivity and specificity was not provided by the manufacture to evaluate this alternative threshold. All four assays only probed for IgG meaning that we did not measure IgM and IgA, which may provide some neutralizing capacity in some individuals as anti-SARS-CoV-2 IgG titers begin to rise. In other donors, different profiles of anti-SARS-COV-2 IgM, IgA and IgG may also provide different profiles of humoral protection. Finally, we did not assess for the SARS-CoV-2 neutralizing capacity of donor specimens nor the avidity of the IgG antibody responses in those donors.
Conclusions
We used multiple analytical methods and assays to confirm very low seroprevalence (~1%) among a healthy population of Canadian blood donors after the first COVID-19 wave [36]. We also found antibody signals by all the assays waned over time and this impacted seroprevalence rates. These findings suggest significant limitations to using a single assay to estimate SARS-CoV-2 seroprevalence in a low prevalence setting. We recommend that seroprevalence studies use multiple assays on either their entire sample or a representative subset to estimate seroprevalence more accurately in the future. As seroprevalence studies enter a new era of tracking natural and vaccine induced humoral immunity, highly sensitive methods will continue to be needed to adjust for waning antibodies and imperfect test characteristics.
Supporting information
Red lines represent thresholds. Abbott-NP (1.4) (n = 54 positive) based on the manufacture’s recommendations. Spike (0.190) (n = 228 positive); RBD (0.186) (n = 104 positive); and NP (0.396) (n = 214 positive). Abbott-NP, Abbott Architect SARS-Cov-2 IgG assay targeting nucleocapsid antigen; Spike, full length spike glycoprotein; RBD, spike glycoprotein receptor binding domain; NP, nucleocapsid.
(DOCX)
1Sensitivity and Specificity are based on manufactures, given the uncertainty the range was based on expert opinion. Abbott-NP, Abbott Architect SARS-Cov-2 IgG assay targeting nucleocapsid antigen; Spike, full length spike glycoprotein; RBD, spike glycoprotein receptor binding domain; NP, nucleocapsid.
(DOCX)
(DOCX)
Abbott-NP, Abbott Architect SARS-Cov-2 IgG assay targeting nucleocapsid antigen; Spike, full length spike glycoprotein; RBD, spike glycoprotein receptor binding domain; NP, nucleocapsid.
(DOCX)
A. Assay Characteristics Overall and Bi-monthly based on the Bayesian Latent Class Analysis with Non-Informative Priors. PPV, positive predictive value; NPV, negative predictive value; Abbott-NP, Abbott Architect SARS-Cov-2 IgG assay targeting nucleocapsid antigen; Spike, full length spike glycoprotein; RBD, spike glycoprotein receptor binding domain; NP, nucleocapsid. B. Assay Characteristics Overall and Bi-monthly based on the Bayesian Latent Class Analysis with Weakly-Informative Priors. PPV, positive predictive value; NPV, negative predictive value; Abbott-NP, Abbott Architect SARS-Cov-2 IgG assay targeting nucleocapsid antigen; Spike, full length spike glycoprotein; RBD, spike glycoprotein receptor binding domain; NP, nucleocapsid.
(DOCX)
Abbott-NP, Abbott Architect SARS-Cov-2 IgG assay targeting nucleocapsid antigen; Spike, full length spike glycoprotein; RBD, spike glycoprotein receptor binding domain; NP, nucleocapsid.
(DOCX)
(XLS)
(DOCX)
Acknowledgments
We would like to thank Craig Jenkins, Valerie Conrod and Canadian Blood Services operations staff for their assistance with this project.
Data Availability
All relevant data are within the manuscript and its Supporting information files.
Funding Statement
SJD received funding through the Canadian Institutes of Health Research (VR2-172723) and Alberta Innovates (G2020000360 Drews), ACG received funding through the Krembil Foundation to the Sinai Health System Foundation. The robotics equipment used for the ELISA assays is housed in the Network Biology Collaborative Centre at the Lunenfeld-Tanenbaum Research Institute (ACG), a facility supported by Canada Foundation for Innovation funding, by the Ontarian Government and by Genome Canada and Ontario Genomics (OGI-139). Commercial Abbott Architect SARS-Cov-2 IgG assay kit costs were partially supported by Abbott Laboratories, Abbott Park, Illinois. Abbott analyzers used at Canadian Blood Services were provided by the COVID-19 Immunity task Force (CITF). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.World Health Organization. Coronavirus (COVID-19) Dashboard | WHO Coronavirus (COVID-19) Dashboard with Vaccination Data. [Cited 2021 May 7] https://covid19.who.int.
- 2.Detsky AS, Bogoch II. COVID-19 in Canada: Experience and Response. JAMA. 2020;324(8):743–744. doi: 10.1001/jama.2020.14033 [DOI] [PubMed] [Google Scholar]
- 3.Lisboa Bastos M, Tavaziva G, Abidi SK, Campbell JR, Haraoui L, Johnston JC, et al. Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ. 2020; 370:m2516.doi: 10.1136/bmj.m2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ward MD, Mullins KE, Pickett E, Merrill V, Ruiz M, Rebuck H, et al. Performance of four automated SARS-CoV-2 serology assay platforms in a large cohort including susceptible COVID-19 negative and COVID-19 positive patients. J Appl Lab Med. 2021; 6(4) 942–952. doi: 10.1093/jalm/jfab014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shioda K, Lau MSY, Kraay ANM, Nelson KN, Siegler AJ, Sullivan PS, et al. Estimating the cumulative incidence of SARS-CoV-2 infection and the infection fatality ratio in light of waning antibodies. Epidemiology. 2021; 32(4)518–524 doi: 10.1097/EDE.0000000000001361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Favresse J, Eucher C, Elsen M, Gillot C, Van Eeckhoudt S, Dogné JM, et al. Persistence of Anti-SARS-CoV-2 Antibodies Depends on the Analytical Kit: A Report for Up to 10 Months after Infection. Microorganisms. 2021;9(3):556. doi: 10.3390/microorganisms9030556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chia WN, Zhu F, Ong SWX, Young BE, Fong SW, LeBet N, et al. Dynamics of SARS-CoV-2 neutralising antibody responses and duration of immunity: a longitudinal study. Lancet Microbe. 2021; 2(6)E240–249. 10.1016/S2666-5247(21)00025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pepe M. Incomplete data and imperfect reference tests. The Statistical Evaluation of Medical Tests for Classification and Prediction. Oxford University Press; 2003: pp.168–213. [Google Scholar]
- 9.van Smeden M, Naaktgeboren CA, Reitsma JB, Moons KJM, de Groot JAH. Latent Class Models in Diagnostic Studies When There is No Reference Standard—A Systematic Review. Am J Epidemiol, 2014;179(4):423–31. doi: 10.1093/aje/kwt286 [DOI] [PubMed] [Google Scholar]
- 10.Canadian Blood Services. [Cited 2021 May 7] https://www.blood.ca/en.
- 11.Abe KT, Li Z, Samson R, Samavarchi-Tehrani P, Valcourt EJ, Wood H, et al. A simple protein-based surrogate neutralization assay for SARS-CoV-2. JCI Insight 2020; 5(19):e142362. 10.1172/jci.insight.142362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isho B, Abe KT, Zuo M, Jamal AJ, Rathod B, Wang JH, et al. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci Immunol 2020; 5(52):eabe5511. doi: 10.1126/sciimmunol.abe5511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis FI, Torgerson PR. A tutorial in estimating the prevalence of disease in humans and animals in the absence of a gold standard diagnostic. Emerg Themes Epidemiol 2012; 9(1):9. 10.1186/1742-7622-9-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisman DN, Greer AL, Brouhanski G, Drews SJ. Of gastro and the gold standard: evaluation and policy implications of norovirus test performance for outbreak detection. J Transl Med 2009; 26;7:23. doi: 10.1186/1479-5876-7-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joseph L, Gyorkos TW, Coupal L. Bayesian estimation of disease prevalence and the parameters of diagnostic tests in the absence of a gold standard. Am J of epidemiol.1995;141(3), 263–272. doi: 10.1093/oxfordjournals.aje.a117428 [DOI] [PubMed] [Google Scholar]
- 16.Cheung A, Dufour S, Jones G, Kostoulas P, Stevenson MA, Singanallur NB, et al. Bayesian latent class analysis when the reference test is imperfect. Rev Sci Tech. 2021Jun;40(1):271–286. doi: 10.20506/rst.40.1.3224 [DOI] [PubMed] [Google Scholar]
- 17.Saeed S, Uzicanin S, Lewin A, Lieshout-Krikke R, Faddy H, Erikstrup C, et al. Current Challenges of SARS-CoV-2 seroprevalence studies among blood donors: A scoping review. medRxiv 2021.05.13.21257177[Preprint]; 2021[cited 2021 May 7]. 10.1101/2021.05.13.21257177. [DOI]
- 18.Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 2020; 11:1620. 10.1038/s41467-020-15562-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARSCoV-2 by full-length human ACE2. Science 2020; 367:1444–1448. doi: 10.1126/science.abb2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020; 181:271–280. doi: 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coughlin M, Lou G, Martinez O, Masterman SK, Olsen OA, Moksa AA, et al. Generation and characterization of human monoclonal neutralizing antibodies with distinct binding and sequence features against SARS coronavirus using XenoMouse. Virology 2007; 361:93–102. doi: 10.1016/j.virol.2006.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coughlin M, Babcook J, Prabhakar BS. Human monoclonal antibodies to SARS coronavirus inhibit infection by different mechanisms. Virology 2009; 394:39–46. doi: 10.1016/j.virol.2009.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di GS, Ciccullo A, Posteraro B, Lombardi F, Borghetti A, Sanguinetti M. Still much to learn about the diagnostic role of SARS-CoV-2 antibody detection. Clin Infect Dis 2020; 71(16):2299–2300. doi: 10.1093/cid/ciaa532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bryan A, Pepper G, Wenner MH, Fink S, Morishima C, Chaudhary A, et al. Performance Characteristics of the Abbott Architect SARS-CoV-2 IgG Assay and Seroprevalence Testing in Idaho. J Clin Microbiol 2020; 58(8):e00941–20. doi: 10.1128/JCM.00941-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hachim A, Kavian N, Cohen CA, Chin AWH, Chu DK, Mok CK, et al. ORF8 and ORF3b antibodies are accurate serological markers for early and late SARS-CoV-2 infection. Nat Immunol 2020; 21(10):1293–1301. 10.1038/s41590-020-0773-7 [DOI] [PubMed] [Google Scholar]
- 26.Abbott. SARS-COV-2 Immunoassays. [Cited 2021 May 7] https://www.corelaboratory.abbott/us/en/offerings/segments/infectious-disease/sars-cov-2.
- 27.National SARS-CoV-2 Serology Assay Evaluation Group. Performance characteristics of five immunoassays for SARS-CoV-2: a head-to-head benchmark comparison. Lancet Infect Dis 2020; 20(12):1390–4000. doi: 10.1016/S1473-3099(20)30634-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan SS, Saw S, Chew KL, Huak CY, Khoo C, Pajarillaga A, et al. Head-to-head evaluation on diagnostic accuracies of six SARS-CoV-2 serological assays. Pathology 2020; 52(7):770–777. doi: 10.1016/j.pathol.2020.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolotin S, Tran V, Osman S, Brown KA, Buchan SA, Joh E, et al. SARS-CoV-2 Seroprevalence Survey Estimates Are Affected by Anti-Nucleocapsid Antibody Decline. J Infect Dis 2021; 223(8):1334–1338. doi: 10.1093/infdis/jiaa796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buss LF, Prete CA, Abrahim CMM, Mendrone A, Salomon T, Almeida-Neto C, et al. Three-quarters attack rate of SARS-CoV-2 in the Brazilian Amazon during a largely unmitigated epidemic. Science 2021; 371(6526): 288–292. doi: 10.1126/science.abe9728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cox RJ, Brokstad KA. Not just antibodies: B cells and T cells mediate immunity to COVID-19. Nat Rev Immunol 2020; 20, 581–582. doi: 10.1038/s41577-020-00436-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen KW, Linderman SL, Moodie Z, Czartoski J, Lai L, Mantus G, et al. Longitudinal analysis shows durable and broad immune memory after SARS-CoV-2 infection with persisting antibody responses and memory B and T cells. medRxiv 2021.04.19.21255739 [Preprint] 10.1101/2021.04.19.21255739 [DOI] [PMC free article] [PubMed]
- 33.Saeed S, Drews SJ, Pambrun C, Yi Q- L, Osmond L, O’Brien SF. SARS-CoV-2 seroprevalence among blood donors after the first COVID-19 wave in Canada. Transfusion 2021; 61(3):862–872. doi: 10.1111/trf.16296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atsma F, de Vegt F. The healthy donor effect: a matter of selection bias and confounding. Transfusion 2011; 51(9):1883–5. doi: 10.1111/j.1537-2995.2011.03270.x [DOI] [PubMed] [Google Scholar]
- 35.Grant R, Dub T, Andrianou X, Nohynek H, Wilder-Smith A, Pezzotti P, et al. SARS-CoV-2 population-based seroprevalence studies in Europe: a scoping review. BMJ Open 2021; 11(4):e045425. doi: 10.1136/bmjopen-2020-045425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Government of Canada. COVID-19 daily epidemiology update. [Cited 2021 May 7] https://health-infobase.canada.ca/covid-19/epidemiological-summary-covid-19-cases.html?measure=tested&stat=num&measure=tested&stat=num
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Red lines represent thresholds. Abbott-NP (1.4) (n = 54 positive) based on the manufacture’s recommendations. Spike (0.190) (n = 228 positive); RBD (0.186) (n = 104 positive); and NP (0.396) (n = 214 positive). Abbott-NP, Abbott Architect SARS-Cov-2 IgG assay targeting nucleocapsid antigen; Spike, full length spike glycoprotein; RBD, spike glycoprotein receptor binding domain; NP, nucleocapsid.
(DOCX)
1Sensitivity and Specificity are based on manufactures, given the uncertainty the range was based on expert opinion. Abbott-NP, Abbott Architect SARS-Cov-2 IgG assay targeting nucleocapsid antigen; Spike, full length spike glycoprotein; RBD, spike glycoprotein receptor binding domain; NP, nucleocapsid.
(DOCX)
(DOCX)
Abbott-NP, Abbott Architect SARS-Cov-2 IgG assay targeting nucleocapsid antigen; Spike, full length spike glycoprotein; RBD, spike glycoprotein receptor binding domain; NP, nucleocapsid.
(DOCX)
A. Assay Characteristics Overall and Bi-monthly based on the Bayesian Latent Class Analysis with Non-Informative Priors. PPV, positive predictive value; NPV, negative predictive value; Abbott-NP, Abbott Architect SARS-Cov-2 IgG assay targeting nucleocapsid antigen; Spike, full length spike glycoprotein; RBD, spike glycoprotein receptor binding domain; NP, nucleocapsid. B. Assay Characteristics Overall and Bi-monthly based on the Bayesian Latent Class Analysis with Weakly-Informative Priors. PPV, positive predictive value; NPV, negative predictive value; Abbott-NP, Abbott Architect SARS-Cov-2 IgG assay targeting nucleocapsid antigen; Spike, full length spike glycoprotein; RBD, spike glycoprotein receptor binding domain; NP, nucleocapsid.
(DOCX)
Abbott-NP, Abbott Architect SARS-Cov-2 IgG assay targeting nucleocapsid antigen; Spike, full length spike glycoprotein; RBD, spike glycoprotein receptor binding domain; NP, nucleocapsid.
(DOCX)
(XLS)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting information files.