Abstract
This study aims at assessing the usefulness of deep learning to enhance the diagnostic ability of oximetry in the context of automated detection of pediatric obstructive sleep apnea (OSA). A total of 3196 blood oxygen saturation (SpO2) signals from children were used for this purpose. A convolutional neural network (CNN) architecture was trained using 20-min SpO2 segments from the training set (859 subjects) to estimate the number of apneic events. CNN hyperparameters were tuned using Bayesian optimization in the validation set (1402 subjects). This model was applied to three test sets composed of 312, 392, and 231 subjects from three independent databases, in which the apnea-hypopnea index (AHI) estimated for each subject (AHICNN) was obtained by aggregating the output of the CNN for each 20-min SpO2 segment. AHICNN outperformed the 3% oxygen desaturation index (ODI3), a clinical approach, as well as the AHI estimated by a conventional feature-engineering approach based on multi-layer perceptron (AHIMLP). Specifically, AHICNN reached higher four-class Cohen’s kappa in the three test databases than ODI3 (0.515 vs 0.417, 0.422 vs 0.372, and 0.423 vs 0.369) and AHIMLP (0.515 vs 0.377, 0.422 vs 0.381, and 0.423 vs 0.306). In addition, our proposal outperformed state-of-the-art studies, particularly for the AHI severity cutoffs of 5 e/h and 10 e/h. This suggests that the information automatically learned from the SpO2 signal by deep-learning techniques helps to enhance the diagnostic ability of oximetry in the context of pediatric OSA.
Index Terms—: Oximetry, deep learning, convolutional neural networks (CNN), apnea–hypopnea index (AHI), pediatric obstructive sleep apnea (OSA)
I. Introduction
OBSTRUCTIVE sleep apnea (OSA) is a highly prevalent condition among the pediatric population (1%−5%) [1]. Pediatric OSA is characterized by recurrent respiratory pauses (apneas) and airflow reductions (hypopneas), which leads to oxygen desaturations and arousals that cause restless sleep [1], [2]. Untreated OSA is associated to metabolic and cardiovascular malfunctioning, as well as neurobehavioral abnormalities that diminish children’s health and quality of life, [1], [3].
The gold standard diagnosis test is polysomnography (PSG) [1]. PSG requires children to spend the night in a specialized sleep unit while being recorded up to 32 biomedical signals [3], [4]. These recordings are used to score apneas and hypopneas in order to obtain the apnea-hypopnea index (AHI), which is the clinical variable used to diagnose OSA [2]. Despite its effectiveness, several limitations of PSG have been pointed out [5], [6], including its complexity, cost, high intrusiveness, and limited availability. This results in a delay in the diagnosis and treatment of OSA of the affected children [7].
In order to overcome these limitations, the scientific community has explored the use of simplified tests that increase the accessibility and effectiveness of pediatric OSA diagnosis. In this respect, the blood oxygen saturation signal (SpO2) from nocturnal oximetry has been frequently proposed as a clinically valuable tool for the screening of OSA in children due to its simplicity, reliability and suitability [8], [9]. The SpO2 signal measures the oxygen content in the hemoglobin of the blood [10], thus containing information of the oxygen desaturations associated to apneic events from OSA [2].
Promising results have been obtained in previous studies from the automated analysis of the SpO2 signal following a feature-engineering methodology [11]–[18]. First, a set of hand-crafted features from oximetry was obtained using different signal processing algorithms: conventional oximetric indices, statistical parameters, nonlinear methods, and frequency domain techniques [11]–[18]. Then, thresholding rules [11], [12] and machine-learning algorithms [13]–[18] were used with these features to determine the presence and severity of pediatric OSA. Nonetheless, these conventional feature-engineering approaches require considerable knowledge in order to identify, a priori, a set of relevant features to extract from the data [19]. In addition, the level of abstraction that classical methods provide is low, which limits their ability to identify complex patterns in the data [19]. This may result in missing relevant information from the SpO2 signals linked to apneic events.
These issues can be solved by using deep-learning algorithms, which automatically learn complex patterns for detection or classification tasks from raw data using architectures with multiple levels of representation [19]. These algorithms have beaten conventional approaches in many fields, such as image recognition, language processing, and time series analysis [19]. In the OSA context, recent studies have focused on the application of deep-learning techniques to detect sleep stages [20], apneic events [21], and/or estimate AHI in adult OSA patients [21]. Their findings suggest that deep-learning algorithms are appropriate to analyze different physiological signals from PSG, such as electrocardiogram, electroencephalogram, airflow, or oximetry [20], [21].
Specifically, the majority of these studies employed deep-learning architectures based on convolutional neural networks (CNN) [21], which are the most widely-used deep-learning algorithm [19]. Despite being originally inspired for image analysis, CNNs have proven its suitability for time series classification in a big variety of domains [22], including biomedical signal analysis [23]–[26]. CNN have a multi-layer architecture, with shared weights, sparse connections, and pooling operations, which allows them to identify both short- and long-term patterns occurring in different parts of the time series [25], while reducing the computational cost of other deep-learning algorithms [19]. This CNN property may be useful to identify desaturations in the SpO2 signal associated to apneic events that may occur at different times. In addition, CNNs provide higher levels of representation that allow to learn more complex features [19], which may be useful to detect complex patterns in long segments of the SpO2 signal, such as clusters of desaturations [27].
The novelty of this research is the use of a new deep-learning model based on CNN that allows to accurately diagnose pediatric OSA with a high generalization ability from the raw oximetry signal. We hypothesize that deep-learning approaches could help to automatically extract the relevant information of the oximetry signal in the context of pediatric OSA diagnosis. Consequently, the main goal of this study is to evaluate the usefulness of deep-learning to estimate the AHI from ovenight oximetry in children with suspected OSA. To achieve this goal, a CNN architecture is trained to estimate the number of apneic events from 20-min SpO2 segments, which is a novel approach in the context of pediatric OSA. The output of the CNN for each segment is then aggregated to estimate the AHI in pediatric OSA patients using a large cohort of 3196 SpO2 recordings from three different datasets.
One related conference paper developed by our own group has already been published showing preliminary results [28]. Despite the fact that our previous work also applied CNNs to analyze SpO2 recordings, there are some essential differences with this research. Our main contribution is that our deep-learning based methodology allows to diagnose pediatric OSA using the oximetry signal. In this sense, our previous work showed promising results in detecting apneic events (event-based approach) from the oximetry signal using a CNN [28]. In the current study, we have investigated whether those indications may be extended to obtain a new deep-learning model based on CNN that allows to directly estimate the AHI, thus being able to conduct a complete automatic diagnosis (subject-based), including the assessment of the pediatric OSA severity degrees. Instead of training a CNN to detect individual apneic events (binary output), in this research we have trained a CNN to regress the number of apneic events in SpO2 segments (continuous output), which allows to accurately analyze SpO2 segments with several apneic events. A two-step aggregation procedure (averaging plus linear regression) has been also included to accurately estimate the AHI of each subject from the outputs of the CNN. We have also incorporated novel elements to improve the training and optimization process of the deep-learning model (Huber loss, batch shuffling, learning rate scheduler, early stopping, and Bayesian optimization). Additionally, in the present study, we have designed and prospectively assessed a new model using three independent datasets, leading to a sample size seven times larger than in our preliminary work (3196 vs. 453). This contributes to increase the generalization ability of our current proposal. Finally, another contribution of our work is that we have performed a thorough comparison with two conventional approaches to properly assess the validity of our proposal. Particularly, we have compared the results of the proposed approach with the 3% oxygen desaturation index (ODI3), a conventional clinical approach commonly used for comparison purposes [11], [13]–[18], as well as with the AHI estimated by a classical feature-engineering approach.
II. Subjects And Signals Under Study
A total of 3196 sleep studies of children ranging from 0 to 18 years of age composed the population under study. Three large datasets were used: (i) the Childhood Adenotonsillectomy Trial (CHAT) dataset, a public multicenter database composed of 1638 sleep studies (clinical trial identifier: NCT00560859) [29], [30]; (ii) the University of Chicago (UofC) dataset, a private database composed of 980 pediatric subjects; and (iii) the Burgos University Hospital (BUH) dataset, a private database composed of 578 subjects. All subjects from the three datasets were referred to overnight PSG due to clinical suspicion of OSA. An informed consent was obtained from all legal caretakers of the children and the Ethics Committee of the different sleep centers involved in the study approved the protocols.
SpO2 recordings were acquired during PSG using sampling rates ranging from 1 to 512 Hz. The guidelines of the AASM were used to quantify sleep and score apneas and hypopneas by pediatric sleep specialists from the different centers [2], [31]. The AHI, obtained as the number of apneas and hypopneas per hour of sleep, was used to diagnose pediatric OSA [2]. Common clinically used AHI cutoffs of 1, 5, and 10 events per hour (e/h) were used in this study to classify children into four OSA severity degrees: no-OSA (AHI<1 e/h), mild OSA (1≤AHI<5 e/h), moderate OSA (5≤AHI<10 e/h), and severe OSA (AHI≥10 e/h) [3], [32], [33].
Data was divided into three sets: training set, employed to train the deep-learning algorithms; validation set, used for hyperparameters optimization; and test set, employed to evaluate the diagnostic performance of the deep-learning methods. Only the CHAT database contains annotations of time location of apnea and hypopnea events, which are needed in the deep-learning models as the output labels for training. Accordingly, the training set was composed of 859 SpO2 recordings from the baseline (453 subjects) and follow-up groups (406 subjects) of the CHAT database [30]. The subjects from the remaining group of the CHAT dataset, nonrandomized (779 subjects), as well as the subjects of the UofC and BUH sets, were randomly divided into a validation set (60%) and a test set (40%), being 60%−40% a common proportion used in previous studies for validation and test purposes [14], [17].
In this way, the validation set was composed of 1402 SpO2 recordings from the CHAT (467, 60% of the 779 subjects from the nonrandomized group), UofC (588, 60% of the 980 subjects), and BUH (347, 60% of the 578 subjects) databases, whereas the test set was composed of 312 subjects from the CHAT dataset (40% of the nonrandomized group), 392 subjects from the UofC dataset (40%), and 231 subjects from the BUH dataset (40%). Table I shows clinical and demographic data from the subjects under study.
TABLE I.
Demographic and clinical data from children under study
| All | Training set | Validation set | CHAT Test set |
UofC Test set | BUH Test set |
|
|---|---|---|---|---|---|---|
| SpO 2 recordings (n) | 3196 | 859 | 1402 | 312 | 392 | 231 |
| Age (years) | 6 [5–8] | 7 [6–8] | 6 [4–8] | 7 [6–8] | 6 [3–9] | 5 [4–7] |
| Males (n) | 1735 (54.6%) | 417 (48.5%) | 740 (52.8%) | 143 (45.8%) | 254 (64.8%) | 160 (69.3%) |
| BMI (kg/m 2 ) | 17.2 [15.4–21.1] | 17.3 [15.5–22.3] | 16.9 [15.2–20.7] | 17.1 [15.4–19.9] | 18.1 [15.8–21.7] | 16.0 [14.7–18.0] |
| AHI (e/h) | 2.1 [0.7–6.3] | 3.1 [1.4–6.9] | 1.7 [0.6–5.9] | 0.8 [0.4–1.7] | 3.3 [1.4–7.8] | 2.3 [0.9–6.4] |
| AHI<1 (n) | 1015 (31.8%) | 173 (20.1%) | 516 (36.8%) | 187 (59.9%) | 77 (19.6%) | 62 (26.8%) |
| 1≤AHI<5 (n) | 1230 (38.5%) | 395 (46.0%) | 493 (35.2%) | 76 (24.4%) | 169 (43.1%) | 97 (42.0%) |
| 5≤AHI<10 (n) | 447 (14.0%) | 170 (19.8%) | 164 (11.7%) | 18 (7.8%) | 63 (16.1%) | 32 (13.9%) |
| AHI≥10 (n) | 504 (15.8%) | 121 (14.1%) | 229 (16.3%) | 31 (9.9%) | 83 (21.2%) | 40 (17.3%) |
SpO2: blood oxygen saturation signal; BMI: Body Mass Index; AHI: Apnea-Hypopnea Index, CHAT = Childhood Adenotonsillectomy Trial, UofC = University of Chicago, BUH = Burgos University Hospital. Data are presented as median [interquartile range] or n (%)
III. Methodology
A. Proposed CNN model
The proposed solution, depicted in Fig.1, consists of three steps: (1) signals segmentation; (2) CNN architecture; and (3) AHI estimation.
Fig. 1.
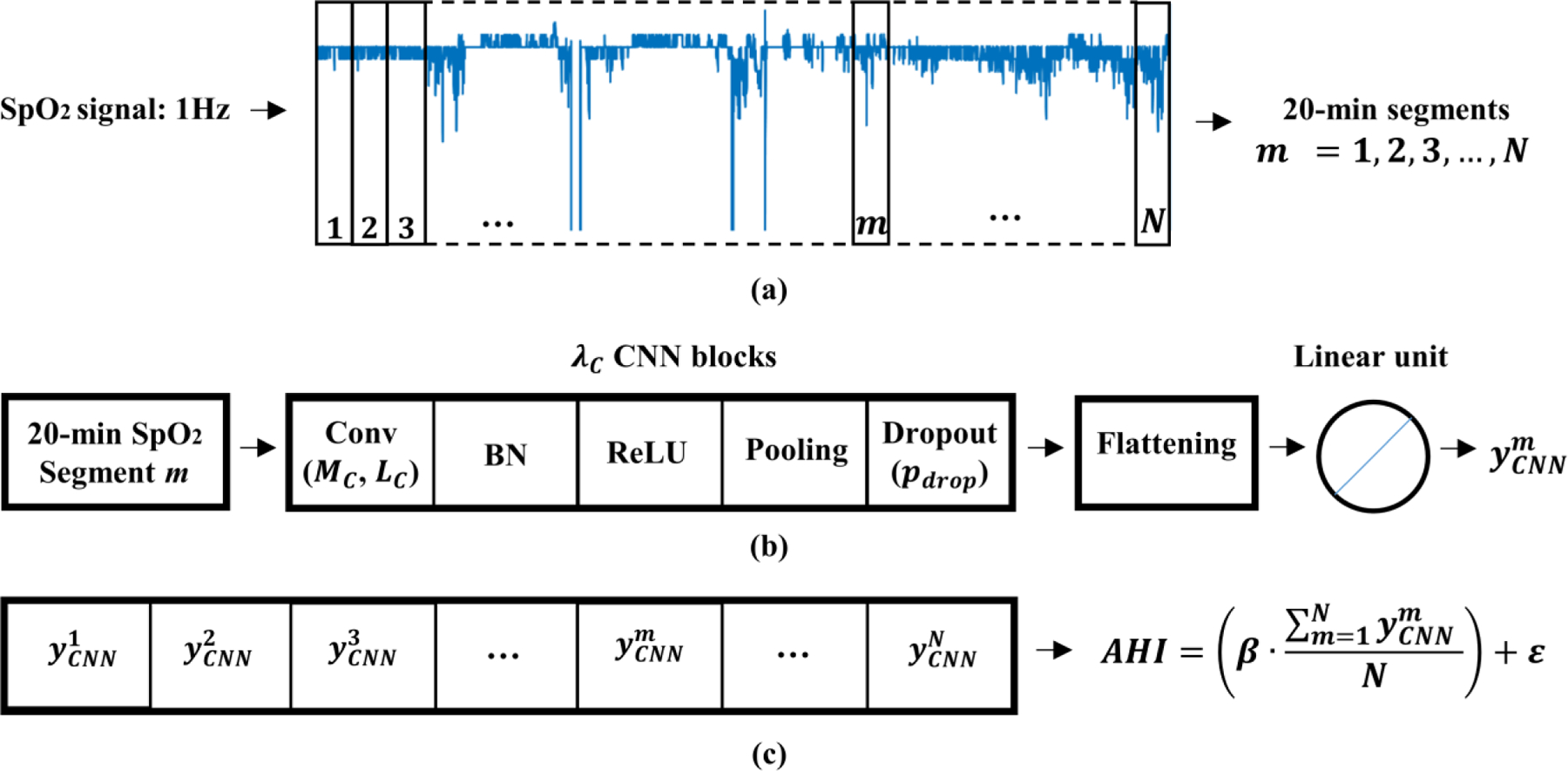
Overview of the proposed methodology. (a) Signals segmentation, (b) CNN architecture, and (c) AHI estimation.
1). Signals segmentation
First, SpO2 recordings were down-sampled to a sample rate of 1 Hz in order to homogenize the frequency. SpO2 signals from each subject were then divided into 20-min segments (1200 samples), as shown in Fig. 1 (a). This segment size (20-min) allows to detect clusters of desaturations, which have a minimum duration of 10-min [27]. Finally, each 20-min SpO2 segment in the training set is labelled with the annotations provided by sleep technicians [30]. The output label for each segment was obtained as the number of apnea and hypopnea events associated to 3% oxygen desaturations occurring in these 20 minutes, according to the annotation event files of the CHAT dataset [30].
2). CNN architecture
CNN are the most popular deep-learning technique to process multidimensional arrays, such as 1D signals or 2D images. In this study, CNN were used to process raw oximetry data. Figure 1 (b) shows the architecture of the proposed CNN. The input of the CNN architecture is the 20-min SpO2 segment. The CNN architecture processes the input by the use of λC stacked convolutional blocks, each one composed of: convolutional layer, batch normalization, activation, pooling, and dropout [19].
The convolutional layer extracts feature maps from the input data a[n] using convolutional filters (kernels) [19]:
| (1) |
where is the lth feature map (l= 1, …, MC, being MC the number of filters) in the convolutional block = 1, …, λC, and are the weights and biases of each convolutional kernel in the convolutional block i, and LC is the kernel size.
After the convolution, batch normalization is applied to normalize the feature maps [19]. Then, a non-linear function is used to decide which feature maps are activated, depending on a rule or a threshold [19]. In this study, a rectified linear unit (ReLU) activation function, which is the standard choice for deep-learning [19], was used:
| (2) |
The output of the ReLU is fed into a max-pooling layer, which applies a maximum operation with a pool factor K=2, which is a widely used value, in order to reduce dimensionality as well as computational cost [19]. Finally, dropout operation was included in the training phase in order to avoid overfitting [19]. Dropout randomly removes some units with a probability pdrop at each batch of a training epoch [19].
After λC convolutional blocks, a flattening layer is used to transform the 2-D feature maps into a 1-D series [19]. Then, a linear activation unit is used to obtain the output of the network, which accounts for the apneic events associated to desaturations for the corresponding input 20-min SpO2 segment.
3). AHI estimation
Based on the output of the CNN for each 20-min SpO2 segment m = 1, 2, 3, …, N, the AHI of each patient can be estimated. First, the average of the output of the CNN obtained for each SpO2 segment is computed:
| (3) |
where N is the number of 20-min SpO2 segments of the oximetry signal. This step is necessary as the number of 20-min SpO2 segments is different for each patient. Then, the AHI is obtained using the following expression, as shown in Fig. 1 (c):
| (4) |
where β and ε are the intercept and disturbance term of a linear regression model, which was fitted using the validation set. This linear regression corrects the trend of the CNN to underestimate the AHI, which is caused by [34]: (i) the AHI estimated by the CNN is obtained using the total recording time, while the AHI from PSG uses total sleep time; (ii) there are apneic events that are not associated to oxygen desaturations, so they cannot be detected by the CNN.
B. CNN training and optimization process
The training data were fed into the CNN in batches of 100 during 500 epochs. He-normal method was used to initialize the weights and biases of each layer [35]. Then, the adaptive moment estimation (Adam) algorithm was used with an initial learning rate of 0.001 to update the weights and biases in each training batch [36]. Huber loss [37] was the function used to minimize Adam algorithm in the validation set. This loss function has a tunable hyperparameter, delta (δ), that allows to control the importance of outliers [37]:
| (5) |
where ym is the target variable and is the output of the CNN for a segment m. At each training epoch, training data were shuffled in order to improve the convergence of the optimization algorithm [19], so the batches were different. In addition, the learning rate was decreased by a factor of 2 after 10 epochs of non-improvement in the loss function value of the validation set, which helps to obtain a converged stable set of final weights [19]. Finally, early stopping [19] was applied to stop training after 30 epochs of non-improvement in order to reduce the training time, restoring weights to those that achieved the best performance in the validation set.
The hyperparameters of the CNN architecture to optimize were the number of filters in each convolutional layer (MC), the kernel size of each convolutional layer (LC), the number of CNN blocks (λC), the dropout probability (pdrop), and the delta parameter of the Huber loss (δ). Bayesian optimization with tree-structured Parzen estimator (BO-TPE) [38] implemented in Hyperopt library [39] was used to obtain the optimum values of these hyperparameters. BO-TPE is considered more efficient than grid search or random search for hyperparameters optimization, since it uses past evaluation results to form a probabilistic model that attempts to optimize the objective function in an iterative way [40].
Keras framework with Tensorflow backend was used to implement the CNN-based architecture [41]. CNNs were trained on a NVIDIA GeForce RTX 2080 GPU in a Windows 10 environment.
C. Comparison with conventional approaches
The following conventional methods have been applied in order to compare the diagnostic performance of the proposed deep-learning model:
1). Clinical approach:
ODI3. ODI3 was estimated as the number of desaturations of at least 3% per hour of recording [42]. This parameter has shown its usefulness in the clinical OSA context, and is usually employed for comparison purposes [11], [13]–[18].
2). Classical feature-engineering approach:
multilayer perceptron (MLP) neural network trained using features extracted from the 20-min SpO2 segments. This approach is divided into the following common four steps: (i) signal preprocessing, where artifacts were removed from SpO2 recordings following the methodology employed in previous studies [14], [15], [17]; (ii) feature extraction, where up to 23 features were extracted from each 20-min SpO2 segment, the same features as in the previous study by Hornero et al. [14]; (iii) segment-based AHI estimation; where a MLP model was trained with the set of 23 SpO2 features to estimate the number of apneic events associated to desaturations in each segment; (iv)subject-based AHI estimation, where the AHI of each subject is obtained from the output of the MLP for each 20-min SpO2 segment using the procedure described in (3) and (4).
D. Statistical Analysis
The agreement between the estimated AHI by the CNN architecture (AHICNN) and the actual AHI from PSG (AHIPSG) was assessed by means of scatter and error distribution plots, as well as the intra-class correlation coefficient (ICC) and root mean square error (RMSE). The overall agreement of AHICNN to estimate the severity of OSA was assessed by means of the confusion matrices, as well as Cohen’s kappa index (kappa) and 4-class accuracy. ICC, RMSE, kappa, and 4-class accuracy were also obtained for ODI3 and the AHI estimated by the MLP (AHIMLP). Additionally, the diagnostic ability of AHICNN was assessed for each of the AHI cutoffs that define the OSA severity degrees (1, 5, and 10 e/h) by means of sensitivity (Se, percentage of OSA positive patients rightly classified), specificity (Sp, percentage of OSA negative children rightly classified), positive predictive value (PPV, proportion of positive test results that are true positives), negative predictive value (NPV, proportion of negative test results that are true negatives), positive likelihood ratio (LR+, Se/(1-Sp)), negative likelihood ratio (LR-, (1-Se)/Sp), and accuracy (Acc, percentage of subjects correctly classified).
IV. Results
A. Training and validation sets
Training and validation sets were used to optimize the CNN architecture. BO-TPE was used to find the optimum values of the hyperparameters of the CNN architecture: MC, LC, λC, pdrop, and δ. The search space of the BO-TPE is shown in Table II. The training set was used to train the CNN models at each iteration of the BO-TPE procedure, whereas kappa was obtained in the validation set as the objective function to optimize. The training of most of the CNNs was finished by early stopping criterion after 80–120 epochs, thus contributing to reduce the training time. The results of the BO-TPE algorithm are shown in Figure 2. For each hyperparameter, the values of kappa in the validation set are given. These values are represented in a boxplot. It can be seen that there is not a high dependence of kappa on the hyperparameter values. Slightly higher overall kappa values are obtained when λC = 6 and LC = 5, as well as with increasing values of MC and decreasing values of δ, whereas pdrop had little effect on the value of kappa. Finally, MC = 64, LC = 5, λC = 6, pdrop = 0.1, and δ = 1.5 were obtained as the optimum values for the hyperparameters, since this combination reached the highest kappa, as shown in Table II.
TABLE II.
Search space of BO-TPE for the CNN hyperparameters
| Hyperparameter | Search space | Optimum value |
|---|---|---|
| M C | 8, 16, 32, 64 | 64 |
| L C | 3, 5, 7 | 5 |
| λ C | 4, 5, 6, 7, 8 | 6 |
| P drop | 0:0.025:0.3 | 0.1 |
| δ | 0.5:0.5:6 | 1.5 |
BO-TPE= Bayesian optimization with tree-structured Parzen estimator; CNN = Convolutional neural network; MC = number of filters; LC = kernel size; λC = number of convolutional blocks; Pdrop = dropout probability; δ =delta value of the Huber loss
Fig. 2.
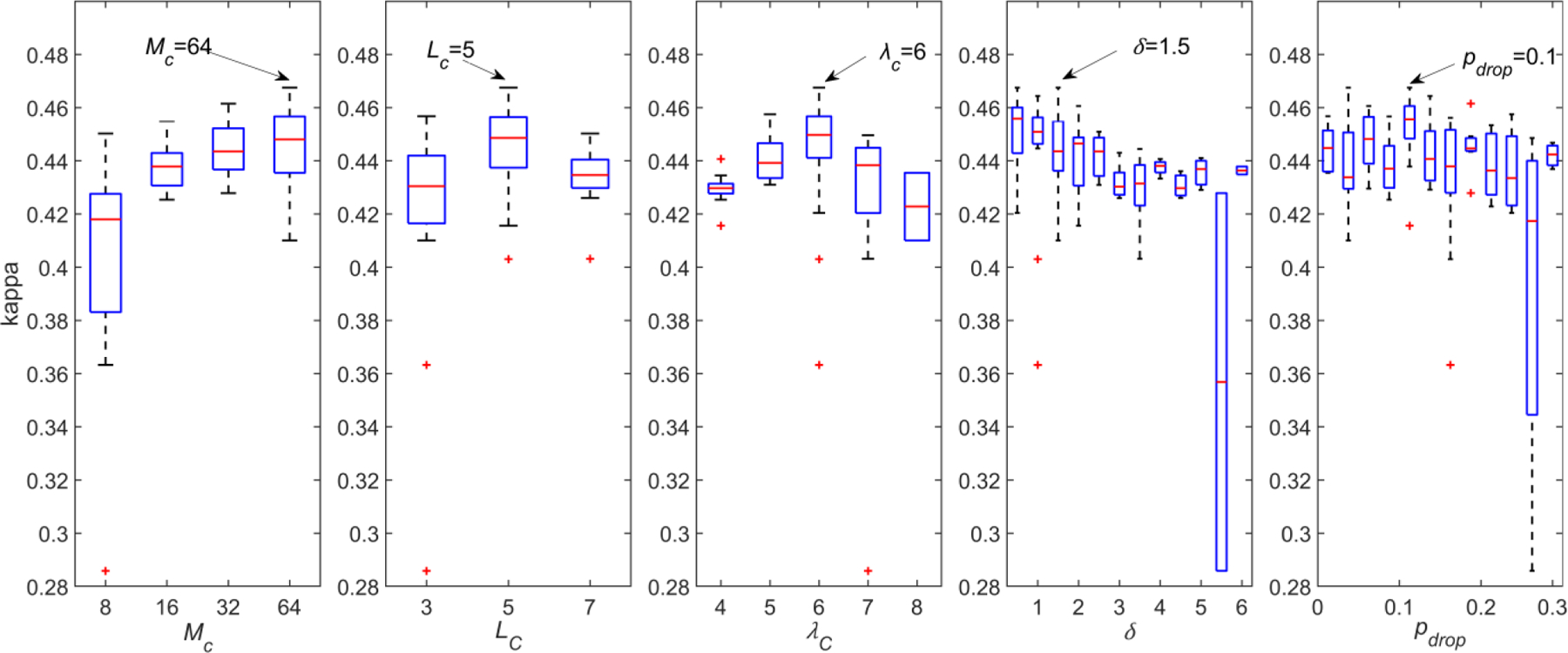
Results of the BO-TPE for every hyperparameter in the validation set.
B. Test set
1). Diagnostic performance of the CNN model.
Figure 3 shows the scatter plots of AHICNN compared to AHIPSG in the CHAT, UofC and BUH test sets, respectively. ICC and RMSE are also shown. Points of the scatter plot of AHICNN in the CHAT test set are more concentrated near the diagonal line, which is reflected in a higher agreement (ICC=0.960 and RMSE=2.89) than in the UofC (ICC=0.917 and RMSE=5.45) and BUH test sets (ICC=0.583 and RMSE=10.44). Figure 4 shows the error distribution plots of AHICNN in the three test sets. Mean error was low in the three test sets. Nonetheless, 95% confidence intervals of AHICNN were higher in the UofC (21.69 e/h) and BUH (28.84 e/h) test sets than in the CHAT test set (12.80 e/h). In addition, there are some outliers in AHICNN that can be observed in the UofC and BUH sets, as reported by the maximum error.
Fig. 3.
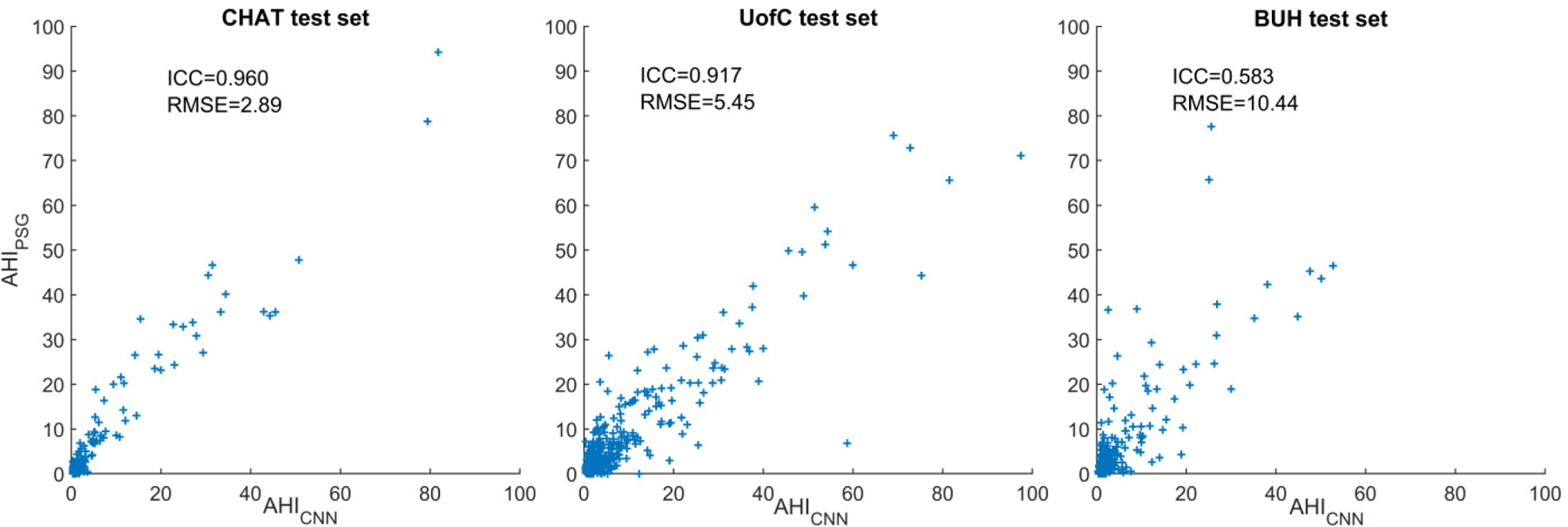
Scatter plots comparing AHICNN with AHIPSG in the CHAT, UofC, and BUH test databases.
Fig. 4.
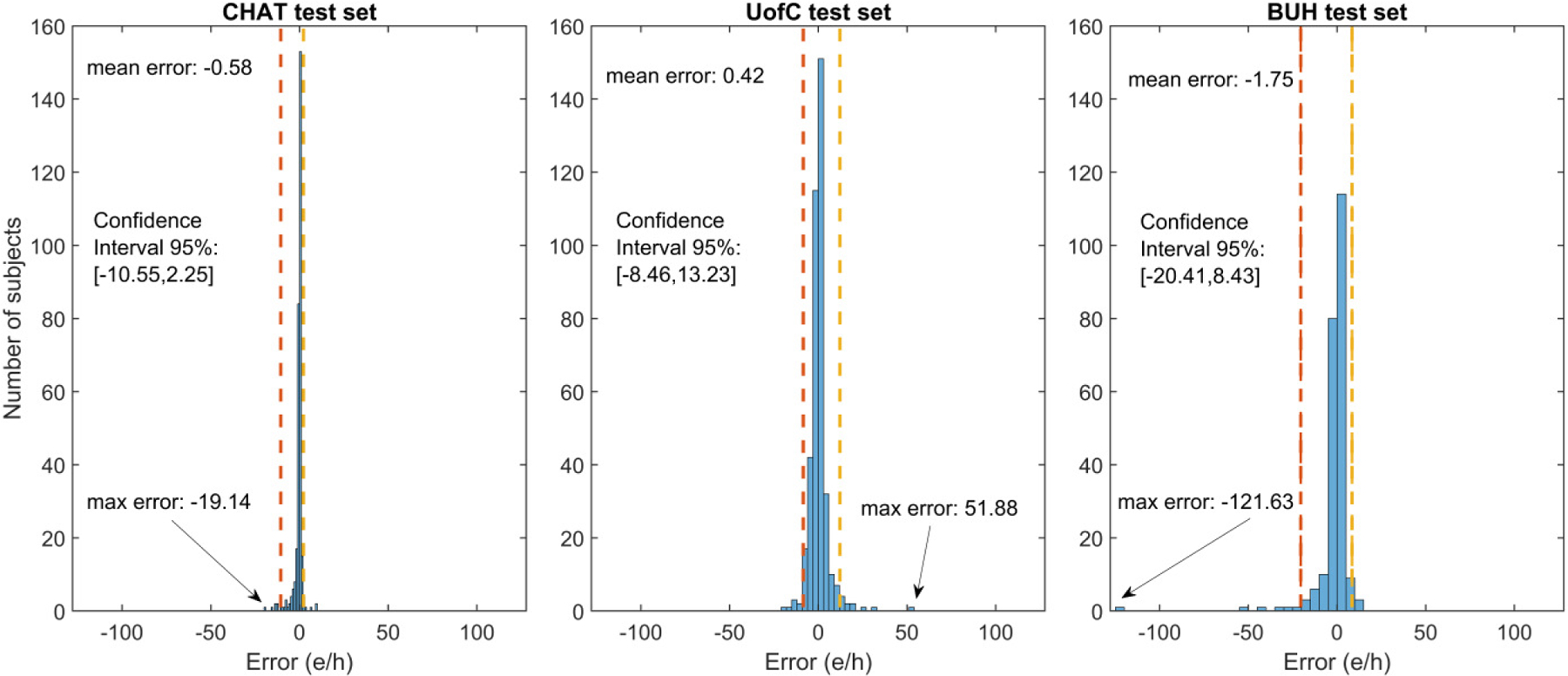
Error distribution of AHICNN in the CHAT, UofC, and BUH test databases.
Fig. 5 shows the confusion matrices of AHICNN, evaluated in the three test sets. AHICNN rightly assigned 72.8% (227/312), 60.2% (236/392), and 61.0% (141/231) of subjects in the three test sets to their actual OSA severity group. Table III shows diagnostic ability statistics of AHICNN for the AHI severity cutoffs of 1, 5, and 10 e/h, which are derived from the confusion matrix. Notice that AHICNN reached a higher kappa in the CHAT test set (0.515) than in the UofC (0.422) and BUH test sets (0.423). Higher performance metrics were obtained in the CHAT test set for the three AHI cutoffs, especially for the AHI cutoffs of 5 and 10 e/h.
Fig. 5.
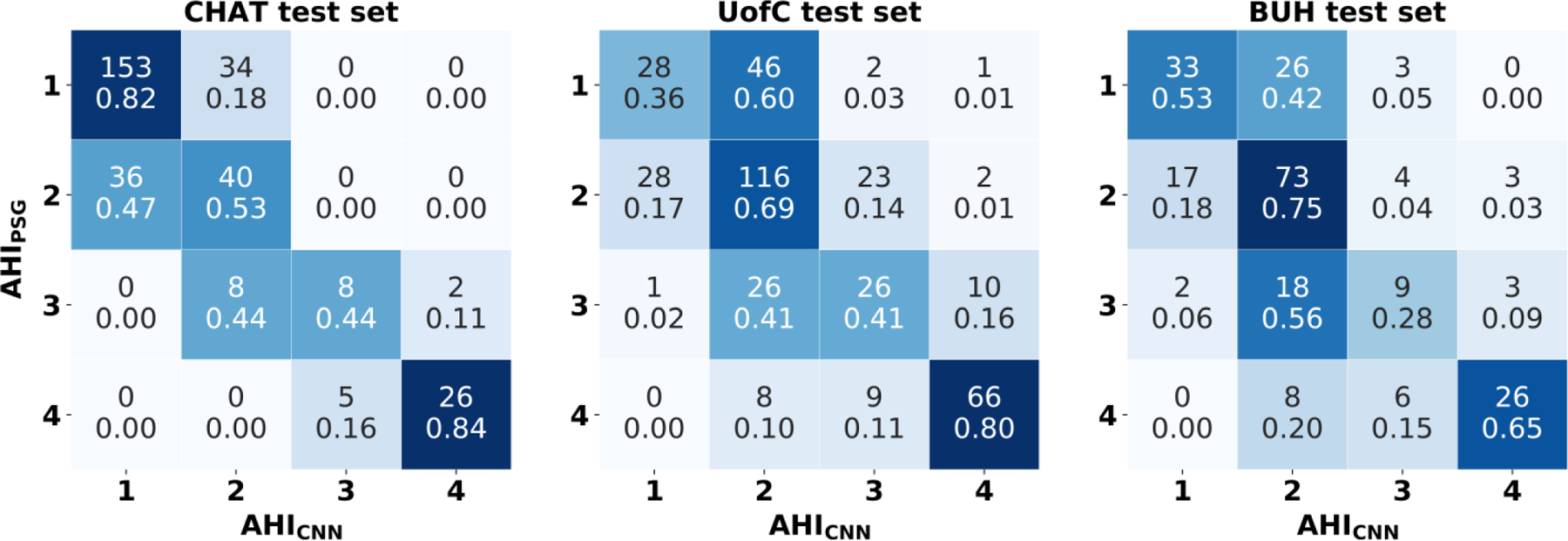
Confusion matrices of AHICNN in the CHAT, UoFC, and BUH test databases. 1: No OSA (AHI < 1 e/h); 2: Mild OSA (1 ≤ AHI < 5 e/h); 3: Moderate OSA (5 ≤ AHI < 10 e/h); 4: Severe OSA (AHI≥10 e/h).
TABLE III.
diagnostic ability of AHICNN for the ahi cutoffs= 1e/h, 5 e/h, and 10 e/h in the CHAT, UofC and BUH test databases
| Estimated AHI |
CHAT test set | UofC test set | BUH test set | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| AHI = 1 e/h |
AHI =5 e/h |
AHI = 10 e/h |
AHI = 1 e/h |
AHI =5 e/h |
AHI = 10 e/h |
AHI = 1 e/h |
AHI =5 e/h |
AHI = 10 e/h |
|
| Se (%) | 71.2 | 83.7 | 83.9 | 90.8 | 76.0 | 79.5 | 88.8 | 61.1 | 65.0 |
| Sp (%) | 81.8 | 100 | 99.3 | 36.4 | 88.6 | 95.8 | 53.2 | 93.7 | 96.9 |
| PPV (%) | 72.4 | 100 | 92.9 | 85.4 | 79.8 | 83.5 | 83.8 | 81.5 | 81.3 |
| NPV (%) | 81.0 | 97.0 | 98.2 | 49.1 | 86.2 | 94.6 | 63.5 | 84.2 | 93.0 |
| LR+ | 3.92 | N.D | 117.84 | 1.43 | 6.68 | 18.90 | 1.90 | 9.72 | 20.69 |
| LR− | 0.35 | 0.16 | 0.16 | 0.25 | 0.27 | 0.21 | 0.21 | 0.42 | 0.36 |
| Acc (%) | 77.6 | 97.4 | 97.8 | 80.1 | 83.9 | 92.3 | 79.2 | 83.5 | 91.3 |
| kappa | 0.515 | 0.422 | 0.423 | ||||||
CNN = Convolutional neural network, AHI = apnea-hypopnea index, Se = sensitivity, Sp = specificity, PPV = positive predictive value, NPV = negative predictive value, LR+ = positive likelihood ratio, LR- = negative likelihood ratio, Acc = accuracy, kappa = Cohen’s kappa index, N.D = not defined, CHAT = Childhood Adenotonsillectomy Trial, UofC = University of Chicago, BUH = Burgos University Hospital.
2). Comparison with conventional approaches
Table IV shows the comparison of AHICNN with ODI3 and AHIMLP in the three test sets. It can be seen that AHICNN showed a higher diagnostic capability than ODI3 and AHIMLP in the CHAT, UofC, and BUH test sets, as derived from the values of ICC, RMSE, kappa, and 4-class accuracy.
TABLE IV.
Diagnostic performance of AHICNN vs. ODI3 and AHIMLP in the CHAT, UofC and BUH test databases
| Test set | ICC | RMSE | 4-class kappa | 4-class Accuracy (%) | |
|---|---|---|---|---|---|
| CHAT | AHICNN | 0.960 | 2.89 | 0.515 | 72.8 |
| ODI3 | 0.871 | 4.63 | 0.417 | 65.1 | |
| AHIMLP | 0.832 | 5.51 | 0.377 | 63.3 | |
|
| |||||
| UofC | AHICNN | 0.917 | 5.45 | 0.422 | 60.2 |
| ODI3 | 0.861 | 6.21 | 0.372 | 56.6 | |
| AHIMLP | 0.890 | 6.02 | 0.381 | 56.9 | |
|
| |||||
| BUH | AHICNN | 0.583 | 10.44 | 0.423 | 61.0 |
| ODI3 | 0.520 | 10.64 | 0.369 | 57.6 | |
| AHIMLP | 0.500 | 11.05 | 0.306 | 52.4 | |
AHICNN = apnea-hypopnea index (AHI) estimated by our convolutional neural network architecture, ODI3 = 3% oxygen desaturation index, AHIMLP = AHI estimated by the multi-layer perceptron neural network trained with features from the blood oxygen saturation (SpO2) signal, ICC = intra-class correlation coefficient, RMSE = root mean squared error, kappa = Cohen’s kappa index, CHAT = Childhood Adenotonsillectomy Trial, UofC = University of Chicago, BUH = Burgos University Hospital.
Table V summarizes the comparison of the performance of our proposal with state-of-the-art studies aimed at simplifying the detection of pediatric OSA and its severity using the SpO2 signal [11]–[18]. Notice that none of the studies that employed a validation approach reported a higher accuracy for the AHI cutoffs of 5 e/h and 10 e/h than the proposed CNN-based architecture in the CHAT, UofC, and BUH datasets.
TABLE V.
Summary of the state-of-the-art studies in the context of pediatric OSA detection using spo2 recordings.
| Studies | Number of subjects (Total dataset/test set) | AHI (e/h) | Methods (Feature/ classification) | Validation | Se (%) | Sp (%) | Acc (%) |
|---|---|---|---|---|---|---|---|
| Tsai et al. [11] | 148/148 | 1 | ODI4/ Thresholding | No | 77.7 | 88.9 | 79.0 |
| 5 | 83.8 | 86.5 | 85.1 | ||||
| 10 | 89.1 | 86.0 | 87.1 | ||||
|
| |||||||
| Pia-Villa et al. [12] | 268/268 | 1 | Clusters of desaturations and clinical history/ Thresholding | Direct validation* | 91.6 | 40.6 | 85.8 |
| 5 | 40.6 | 97.9 | 69.4 | ||||
|
| |||||||
| Álvarez et al. [13] | 50/50 | 1 | Statistical moments, spectral, nonlinear features, and classical indices /LR | Bootstrapping | 89.6 | 71.5 | 85.5 |
| 3 | 82.9 | 84.4 | 83.4 | ||||
| 5 | 82.2 | 83.6 | 82.8 | ||||
|
| |||||||
| Hornero et al. [14] | 4191/3602 | 1 | Statistical, spectral, nonlinear features, and ODI3 / Regression MLP | Training-test | 84.0 | 53.2 | 75.2 |
| 5 | 68.2 | 87.2 | 81.7 | ||||
| 10 | 68.7 | 94.1 | 90.2 | ||||
|
| |||||||
| Vaquerizo-Villar et al. [15] | 298/75 | 5 | Bispectrum, spectral features, ODI3,and anthropometric variables / Multiclass MLP | Feature optimization-training-test | 61.8 | 97.6 | 81.3 |
| 10 | 60.0 | 94.5 | 85.3 | ||||
|
| |||||||
| Crespo et al. [16] | 176/176 | 1 | Statistical moments, spectral, nonlinear features, and classical indices/ LDA, QDA, and LR | Bootstrapping | 93.9 | 37.8 | 84.3 |
| 3 | 74.6 | 81.8 | 77.7 | ||||
| 5 | 70.0 | 91.4 | 82.7 | ||||
|
| |||||||
| Vaquerizo-Villar et al. [17] | 981/392 | 1 | Nonlinear features and ODI3 / Regression MLP | Training-test | 97.1 | 23.3 | 82.7 |
| 5 | 78.8 | 83.7 | 81.9 | ||||
| 10 | 77.1 | 94.8 | 91.1 | ||||
|
| |||||||
| Xu et al. [18] | 432/432 | 1 | Statistical, spectral, nonlinear features, and ODI3 / Regression MLP | Training-test | 95.3 | 19.1 | 79.6 |
| 5 | 77.8 | 80.5 | 79.4 | ||||
| 10 | 73.5 | 92.7 | 88.2 | ||||
|
| |||||||
| Our proposal: CHAT set | 3196/312 | 1 | CNN architecture | Training-validation-test | 71.2 | 81.8 | 77.6 |
| 5 | 83.7 | 100 | 97.4 | ||||
| 10 | 83.9 | 99.3 | 97.8 | ||||
|
| |||||||
| Our proposal: UofC set | 3196/392 | 1 | CNN architecture | Training-validation-test | 90.8 | 36.4 | 80.1 |
| 5 | 76.0 | 88.1 | 83.9 | ||||
| 10 | 79.5 | 95.8 | 92.3 | ||||
|
| |||||||
| Our proposal: BUH set | 3196/231 | 1 | CNN architecture | Training-validation-test | 88.8 | 53.2 | 79.2 |
| 5 | 61.1 | 93.7 | 83.5 | ||||
| 10 | 65.0 | 96.9 | 91.3 | ||||
Direct validation of a scoring criteria against AHI from polysomnography. CNN = Convolutional Neural Networks, AHI = apnea-hypopnea index, Se = sensitivity, Sp = specificity, Acc = accuracy, ODI3 = 3% oxygen desaturation index, ODI4 = 4% oxygen desaturation index, LR = Logistic Regression, MLP = Multi-layer perceptron, LDA = Linear Discriminant analysis, QDA = Quadratic discriminant analysis, CHAT = Childhood Adenotonsillectomy Trial, UofC = University of Chicago, BUH = Burgos University Hospital.
V. Discussion
In the present study, we assessed the potential usefulness of a new CNN architecture to enhance the diagnostic ability of the oximetry signal in the context of pediatric OSA. A CNN-based deep-learning model was built and trained to estimate pediatric OSA severity using raw SpO2 data. This model was validated in a database of 3196 SpO2 recordings from three different datasets. The proposed CNN model showed a high diagnostic ability, improving the diagnostic performance of ODI3 and AHIMLP.
A. CNN architecture
To the best of our knowledge, this is the first study that provides a deep-learning model able to automatically detect pediatric OSA and its severity from the oximetry signal. Our results showed that that the proposed CNN-based architecture is able to discern patterns linked with apneic events present in the oximetry signal of children with OSA. Recent studies have also shown the usefulness of deep-learning to analyze different physiological signals from PSG in adult OSA patients [21]. In this regard, the studies developed by Biswal et al. [43], Choi et al. [44], Van Steenkiste et al. [45], and Nikkonen et al. [46] reached accuracies in the range 57%−91% to classify subjects into the four adult OSA severity degrees (AHI<5, 5≤AHI<15, 15≤AHI<30, and AHI≥30 e/h). Despite some of these studies reported higher accuracies, they focus on adult patients, whereas our study applies a CNN to the context of pediatric OSA. In this respect, scoring rules for apnea and hypopnea events are more restrictive in children than in adults [31]. In addition, AHI cutoffs for mild, moderate and severe OSA are lower in children (1, 5, and 10 e/h in contrast to 5, 15, and 30 e/h), which changes the diagnosis and treatment of these patients [3], [32], [47]. Due to these remarkable differences, automated diagnosis of OSA is more challenging in children and thus higher performances are commonly reached in adult patients.
These aforementioned studies in the context of adult OSA applied different deep-learning architectures to raw PSG signals: recurrent neural networks (RNN) [43], [45], multi-layer perceptron (MLP) [46], and CNN [44]. From these architectures, CNN hold advantage over RNN and MLP in terms of computational cost, since they do not include recurrent and/or fully-connected layers. This facilitates the integration of the proposed architecture in wearable and portable devices. In order to corroborate the suitability of CNNs for our problem, we also applied the RNN architecture proposed by Van Steenkiste et al. [45]. This RNN architecture did not obtain a better performance than our CNN model (AHICNN outperformed the RNN architecture in terms of ICC: 0.960 vs 921, 0.917 vs 0.812, and 0.583 vs 0.480 in the CHAT, UofC, and BUH test sets), while having a higher computational cost. This agrees with a recent review of deep learning for time series classification (TSC), where CNN-based architectures achieved the highest performance for TSC in an experiment where more than 8000 deep-learning models were trained and assessed on 97 different time series datasets [22].
With respect to the hyperparameters of the CNN architecture, Fig. 2 shows the low dependence of kappa from the validation set on the optimum hyperparameter values, which highlights the reliability of the proposed solution to automatically learn OSA-related features from the oximetry signal. We also assessed the effect of varying the segment size and the overlap between segments. Different values of the segment size (5 min, 10 min, 30 min, and 60 min), and overlap (50%, and 90%) were tested. Regarding segment size, none of the tested values achieved higher kappa in the validation set than the segment size of our optimum CNN model (20 min), which is appropriate to detect clusters of desaturations [27]. Changing the overlap between segments did not result in a better performance while significantly increased training and validation process.
B. Diagnostic performance
As aforementioned, the AHI estimated by our proposed optimum CNN architecture (AHICNN) outperformed a conventional clinical index ODI3 as well as a classical feature-engineering approach (AHIMLP) in the three test sets. Our AHICNN achieved a higher overall agreement with AHIPSG, as well as a higher diagnostic capability to predict pediatric OSA severity. In contrast to traditional clinical (ODI3) and feature-engineering (AHIMLP) approaches, AHICNN automatically learns features from the SpO2 recordings associated to apneic events through a multi-layer architecture that provides a high level of abstraction. According to our results, CNNs can detect additional information on the OSA-related changes occurring in the SpO2 signal that helps to enhance its diagnostic ability.
Looking at the confusion matrices of Fig. 5, it can be seen that 95.2% (BUH), 96.1% (UofC), and 100% (CHAT) of class 1 (no-OSA) patients have an estimated AHICNN < 5 e/h (class 1 or class 2). In addition, 94.4% (BUH), 97.8% (UofC), and 100% (CHAT) of subjects with an AHICNN≥5 e/h actually show an AHIPSG ≥1 e/h, whereas 90.6% (BUH), 96.2% (UofC), and 100% (CHAT) predicted as severe OSA (AHICNN ≥10 e/h) are at least moderate OSA patients. Hence, a possible screening protocol can be derived to show the clinical usefulness of our proposal as follows: i) if AHICNN <1 e/h, discard the presence of OSA because most of these patients (96.2% in BUH, 98.2% in UofC, and 100% in CHAT) will have an AHIPSG <5 e/h. If symptoms persist, these children may be eventually referred to PSG, as recommended by Alonso-Álvarez et al [3]; ii) if 1≤AHICNN<5 e/h, suggest PSG, since doubts arise about the actual diagnosis of the patients; iii) if 5≤AHICNN<10 e/h, consider treatment, since most probably (86.4% in BUH, 96.7% in UofC, and 100% in CHAT) these subjects have at least a mild degree of OSA; iv) if AHICNN≥10 e/h, suggest treatment, since most of these children (90.6% in BUH, 96.2% in UofC, and 100% in CHAT) have an AHIPSG ≥5 e/h, and also consider a further observation of these patients, since they are prone to have residual OSA after PSG [1]. This screening protocol would avoid the need for 45.9% (BUH), 50.0% (UofC), and 73.7% (CHAT) of complete PSGs, thus contributing to a reduction in the waiting lists and medical costs associated with the diagnosis of OSA, as well as to provide a more suitable diagnostic procedure for children.
Comparing the results of the proposed approach in the three test sets, it is important to highlight the high diagnostic performance obtained by AHICNN in CHAT, where there is a higher increase in the performance of AHICNN with respect to ODI3 and AHIMLP in terms of overall accuracy, kappa, RMSE, and ICC. The proposed CNN model also performed well in the UofC and BUH datasets. Despite not being as remarkable as in the CHAT dataset, AHICNN also outperformed ODI3 and AHIMLP in most of the performance metrics. As it can be seen in the scatter plots (Fig. 3), error distribution plots (Fig. 4), and confusion matrices (Fig. 5), AHICNN performed better in the CHAT dataset than in the UofC and BUH datasets. However, the results are still remarkable considering that the optimum CNN model was trained in the CHAT dataset. In this sense, Collop et al. [48] state that there is a high variability in the scoring of polysomnographies among different sleep technologists, which may affect the external assessment of our proposed deep-learning methodology in two independent databases. In the current work, we tried to minimize this variability by using a validation set composed of subjects from the three datasets to optimize the hyperparameters of the CNN architecture.
The varying diagnostic performance could also be due to some differences in the clinical characteristics among datasets. As observed in the scatter plots (Fig. 3), AHI from PSG has a different distribution in each dataset. The mean values of AHI are 4.2 e/h, 9.3 e/h, and 5.9 e/h in the CHAT, UofC and BUH test sets. In addition, interquartile range are also different: 0.4–1.7 in the CHAT dataset, 1.5–9.3 in the UofC dataset, and 0.6–5.3 in the BUH dataset. The age of children is also different in each dataset. CHAT is composed of children ranging 5 to 10 years of age, whereas UofC dataset is composed of children from 0 to 13 years of age and children in the BUH dataset range from 0 up to 18 years of age. Sampling rate values of SpO2 recordings also vary among datasets: (i) 1, 2, 10, 12, 16, 200, 256, and 512 Hz in the CHAT dataset; (ii) 25, 200, and 500 Hz in the UofC dataset; (iii) 200 Hz in the BUH dataset. Finally, the population groups of CHAT and UofC datasets are children from the United States of America (USA), whereas BUH dataset is composed of Spanish patients. In this respect, there are differences in race and obesity prevalence between these countries. Health system is also different: mostly public in Spain vs. private in USA. This influences the socioeconomic level of the patients, thus having a considerable impact on the health condition. Consequently, these differences in sampling rate values, age range, AHI distribution, and patient characteristics among countries may have resulted in a lower diagnostic performance in the UofC and BUH datasets. This agrees with previous studies that also reported differences in the diagnostic performance among sleep datasets with different clinical characteristics [43], [46], [49].
C. Comparison with state-of-the-art studies
Table V shows the details of previous studies focused on the analysis of the SpO2 signal in the automated detection of pediatric OSA and its severity [11]–[18]. The first studies focused on the use of conventional oximetric indices [11], [12]. Nonetheless, these studies did not employ a hold-out approach to further assess their methodological approaches.
Recent studies focused on the use of automated signal processing and machine learning methods to enhance the diagnostic ability of the oximetry signal [13]–[18]. These studies followed a three-stage feature-engineering methodology to detect pediatric OSA and its severity [13]–[18]. The diagnostic accuracies reported in these studies ranged between 75.2% and 85.5% Acc for an AHI cutoff of 1 e/h, 79.4%−82.8% Acc using an AHI cutoff of 5 e/h, and 85.3%−91.1% using an AHI cutoff of 10 e/h. From these studies, only Hornero et al. [14], Xu et al. [18], and Vaquerizo-Villar et al. [15] evaluated the diagnostic performance of an AHI estimation model for the common AHI cutoffs of 1, 5, and 10 e/h. As aforementioned, our optimum CNN model showed a higher diagnostic performance in the CHAT, UofC, and BUH datasets than state-of-the-art studies for the AHI cutoffs of 5 and 10 e/h. In addition, a higher Sp for the AHI cutoff of 1 e/h was obtained in the CHAT, UofC, and BUH datasets than the reported by Xu et al. [18] and Vaquerizo-Villar et al. [15], which is useful to discard the presence of OSA in pediatric patients. Beyond the superior performance of our CNN model, it uses raw data, i.e., does not require neither prior pre-processing nor human-driven assumptions regarding the SpO2 information needed.
D. Limitations
In spite of the promising results of our proposal, some limitations should be considered. First, the CNN model training procedures were conducted using only the CHAT database, since the other two datasets do not contain the annotation files with the time locations of apneic events. This, together with the differences in sampling rate values, age range, AHI distribution, and patient characteristics among countries, may have resulted in a lower diagnostic performance in the UofC and BUH datasets. Nonetheless, our proposed approach showed a higher diagnostic ability than a conventional clinical index, ODI3, as well as a classical feature-engineering approach, AHIMLP, in all the datasets. Another limitation is that different optimization runs could result in different values of the hyperparameters, as shown in Fig. 2. However, preliminary analysis on our data showed that kappa values in the validation set were similar among different runs, which highlights the reliability of our CNN architecture. Regarding the explanation of the features extracted by the CNN, a new limitation arises. In this regard, the application of methods for explainable deep-learning models would help to further understand the perturbations in oximetry dynamics caused by apneic events, as well as the influence of the different elements of the CNN architecture. Future research may also focus on the use of pretrained deep-learning architectures especially suited for the time series classification field, which might increase the diagnostic performance of traditional architectures based on CNN and RNN, analogous to the pretrained deep-learning networks existing in the image processing field [50]. Another limitation is that we used the AHI without including central sleep apnea (CSA) events, as originally conducted in the study that designed the CHAT database [30]. In this respect, our proposal could also be used to estimate other physiological parameters, such as the apnea index, obstructive apnea index, central apnea index, and/or ODI. Additionally, the use of SpO2 together with other physiological signals from PSG may help to improve the diagnostic ability of our proposal at the cost of higher complexity in the test, since some physiological perturbation of apneic events may not be detected by the oximetry signal alone [1]. Finally, another future goal would be further validation of our proposed methodology in a database of oximetry signals recorded at home.
VI. Conclusion
In summary, we investigated the ability of a novel deep-learning model based on CNN to automatically detect pediatric OSA and its severity from the raw oximetry signal. Our results suggest that deep learning is an appropriate tool to automatically learn discriminative features from oximetry dynamics associated to apneic events. The proposed CNN architecture reached a high diagnostic performance, outperforming the ODI3, a clinical approach, as well as the AHIMLP from a conventional feature-engineering approach. In addition, we achieved higher performance than the reported by previous studies, particularly for moderate-to-severely affected children. The extensive validation of our proposal in three independent datasets as well as the design of a screening protocol highlight the applicability of our results. Therefore, we conclude that deep-learning techniques could be potentially used to enhance the diagnostic ability of the oximetry signal in the context of pediatric OSA.
ACKNOWLEDGMENT
In the loving memory of Dr. Joaquín Terán-Santos and Dra. María. L. Alonso-Álvarez, for all their help and encouragement, and for providing us with the BUH database.
Funding
This work was supported by ‘Ministerio de Ciencia, Innovación y Universidades - Agencia Estatal de Investigación’ and ‘European Regional Development Fund (FEDER)’ under projects DPI2017-84280-R and RTC-2017-6516-1, by “European Commission” and “FEDER” under project ‘Análisis y correlación entre la epigenética y la actividad cerebral para evaluar el riesgo de migraña crónica y episódica en mujeres’ (‘Cooperation Programme Interreg V-A Spain-Portugal POCTEP 2014-2020’), by Sociedad Española de Neumología y Cirugía Torácica (SEPAR) under project 649/2018, by Sociedad Española de Sueño (SES) under project “Beca de Investigación SES 2019”, and by ‘Centro de Investigación Biomédica en Red en Bioingeniería, Biomateriales y Nanomedicina, (CIBER-BBN), Spain’ through ‘Instituto de Salud Carlos III’ co-funded with FEDER funds. The Childhood Adenotonsillectomy Trial (CHAT) was supported by National Institutes of Health (NIH) grants HL083075, HL083129, UL1-RR-024134, and UL1 RR024989. The National Sleep Research Resource was supported by the National Heart, Lung, and Blood Institute (R24 HL114473, 75N92019R002).
F. Vaquerizo-Villar was in receipt of a ‘Ayuda para contratos predoctorales para la Formacin de Profesorado Universitario (FPU)’ grant from the Ministerio de Educacin, Cultura y Deporte (FPU16/02938). D. lvarez is supported by a “Ramn y Cajal” grant (RYC2019-028566-I) from the ‘Ministerio de Ciencia e Innovacin - Agencia Estatal de Investigacin’ co-funded by the European Social Fund (ESF). V. Barroso-Garca and E. Santamara-Vazquez were in a receipt of a ‘Ayuda para financiar la contratacin predoctoral de personal investigador’ grant from the Consejera de Educacin de la Junta de Castilla y Len and the ESF. L. Kheirandish-Gozal and D. Gozal were supported by NIH grants HL130984, HL140548, and AG061824.
Contributor Information
Fernando Vaquerizo-Villar, Biomedical Engineering Group, Universidad de Valladolid, Paseo de Belen 15, 47011, Valladolid, Spain and CIBER-BBN, Spain..
Daniel Álvarez, Biomedical Engineering Group, Universidad de Valladolid, Paseo de Belen 15, 47011, Valladolid, Spain; Hospital Universitario Ro Hortega of Valladolid, Spain and CIBER-BBN, Spain..
Leila Kheirandish-Gozal, Department of Child Health, The University of Missouri School of Medicine, Columbia, Missouri, USA..
Gonzalo C. Gutiérrez-Tobal, Biomedical Engineering Group, Universidad de Valladolid, Paseo de Belen 15, 47011, Valladolid, Spain and CIBER-BBN, Spain..
Verónica Barroso-García, Biomedical Engineering Group, Universidad de Valladolid, Paseo de Belen 15, 47011, Valladolid, Spain and CIBER-BBN, Spain..
Eduardo Santamaría-Vázquez, Biomedical Engineering Group, Universidad de Valladolid, Paseo de Belen 15, 47011, Valladolid, Spain; and CIBER-BBN, Spain..
Félix del Campo, Biomedical Engineering Group, Universidad de Valladolid, Paseo de Belen 15, 47011, Valladolid, Spain; Hospital Universitario Ro Hortega of Valladolid, Spain and CIBER-BBN, Spain..
David Gozal, Department of Child Health, The University of Missouri School of Medicine, Columbia, Missouri, USA..
Roberto Hornero, Biomedical Engineering Group, Universidad de Valladolid, Paseo de Belen 15, 47011, Valladolid, Spain and CIBER-BBN, Spain..
REFERENCES
- [1].Marcus CL et al. , “Diagnosis and Management of Childhood Obstructive Sleep Apnea Syndrome,” Pediatrics, vol. 130, no. 3, pp. e714–e755, 2012. [DOI] [PubMed] [Google Scholar]
- [2].Berry RB et al. , “Rules for scoring respiratory events in sleep: update of the 2007 AASM manual for the scoring of sleep and associated events: deliberations of the sleep apnea definitions task force of the American Academy of Sleep Medicine,” J. Clin. sleep Med, vol. 8, no. 5, p. 597, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Alonso-Álvarez ML et al. , “Documento de consenso del síndrome de apneas-hipopneas durante el sueño en niños,” Arch Bronconeumol, vol. 47, no. Supl 5, pp. 2–18, 2011. [Google Scholar]
- [4].Kaditis A, Kheirandish-Gozal L, and Gozal D, “Pediatric OSAS : Oximetry can provide answers when polysomnography is not available,” Sleep Med. Rev, vol. 27, pp. 96–105, 2016. [DOI] [PubMed] [Google Scholar]
- [5].Kheirandish-Gozal L, “What is ‘Abnormal’ in pediatric sleep?,” Respir. Care, vol. 55, no. 10, pp. 1366–1374, 2010. [PubMed] [Google Scholar]
- [6].Katz ES, Mitchell RB, and Ambrosio CMD, “Obstructive Sleep Apnea in Infants,” Am. J. Respir. Crit. Care Med, vol. 185, no. 8, pp. 805–816, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nixon GM, Kermack AS, Davis GM, Manoukian JJ, Brown A, and Brouillette RT, “Planning adenotonsillectomy in children with obstructive sleep apnea: the role of overnight oximetry,” Pediatrics, vol. 113, no. 1, pp. e19–e25, 2004. [DOI] [PubMed] [Google Scholar]
- [8].del Campo F, Crespo A, Cerezo-Hernández A, Gutiérrez-Tobal GC, Hornero R, and Álvarez D, “Oximetry use in obstructive sleep apnea,” Expert Rev. Respir. Med, vol. 12, no. 8, pp. 665–681, 2018. [DOI] [PubMed] [Google Scholar]
- [9].Netzar N, Eliasson AH, Netzar C, and Kristo DA, “Overnight Pulse Oximetry for Sleep- Disordered Breathing in Adults,” CHEST J, vol. 120, no. 2, pp. 625–633, 2001. [DOI] [PubMed] [Google Scholar]
- [10].McClatchey KD, Clinical laboratory medicine Lippincott Williams & Wilkins, 2002. [Google Scholar]
- [11].Tsai C et al. , “Usefulness of desaturation index for the assessment of obstructive sleep apnea syndrome in children,” Int. J. Pediatr. Otorhinolaryngol, vol. 77, no. 8, pp. 1286–1290, 2013. [DOI] [PubMed] [Google Scholar]
- [12].Villa MP et al. , “Diagnosis of Pediatric Obstructive Sleep Apnea Syndrome in Settings With Limited Resources,” JAMA Otolaryngol. Neck Surg, vol. 141, no. 11, pp. 990–996, 2015. [DOI] [PubMed] [Google Scholar]
- [13].Álvarez D et al. , “Automated Screening of Children With Obstructive Sleep Apnea Using Nocturnal Oximetry : An Alternative to Respiratory Polygraphy in Unattended Settings,” J Clin Sleep Med, vol. 13, no. 5, pp. 7–11, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hornero R et al. , “Nocturnal Oximetry-based Evaluation of Habitually Snoring Children.,” Am. J. Respir. Crit. Care Med, vol. 196, no. 12, pp. 1591–1598, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Vaquerizo-Villar F et al. , “Utility of bispectrum in the screening of pediatric sleep apnea-hypopnea syndrome using oximetry recordings,” Comput. Methods Programs Biomed, vol. 156, pp. 141–149, 2018. [DOI] [PubMed] [Google Scholar]
- [16].Crespo A et al. , “Assessment of oximetry-based statistical classifiers as simplified screening tools in the management of childhood obstructive sleep apnea,” Sleep Breath, pp. 1–11, 2018. [DOI] [PubMed]
- [17].Vaquerizo-Villar F et al. , “Detrended fluctuation analysis of the oximetry signal to assist in paediatric sleep apnoea-hypopnoea syndrome diagnosis,” Physiol. Meas, vol. 39, no. 11, p. 114006, 2018. [DOI] [PubMed] [Google Scholar]
- [18].Xu Z et al. , “Cloud algorithm-driven oximetry-based diagnosis of obstructive sleep apnoea in symptomatic habitually snoring children,” Eur. Respir. J, vol. 53, no. 2, 2019. [DOI] [PubMed] [Google Scholar]
- [19].Goodfellow I, Bengio Y, and Courville A, Deep Learning MIT Press, 2016. [Google Scholar]
- [20].Faust O, Razaghi H, Barika R, Ciaccio EJ, and Acharya UR, “A review of automated sleep stage scoring based on physiological signals for the new millennia,” Comput. Methods Programs Biomed, vol. 176, pp. 81–91, 2019. [DOI] [PubMed] [Google Scholar]
- [21].Mostafa SS, Mendonça F, Ravelo-García AG, and Morgado-Dias F, “A systematic review of detecting sleep apnea using deep learning,” Sensors (Switzerland), vol. 19, no. 22, pp. 1–26, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ismail Fawaz H, Forestier G, Weber J, Idoumghar L, and Muller PA, “Deep learning for time series classification: a review,” Data Min. Knowl. Discov, vol. 33, no. 4, pp. 917–963, 2019. [Google Scholar]
- [23].Faust O, Hagiwara Y, Hong TJ, Lih OS, and Acharya UR, “Deep learning for healthcare applications based on physiological signals: A review,” Comput. Methods Programs Biomed, vol. 161, pp. 1–13, 2018. [DOI] [PubMed] [Google Scholar]
- [24].Roy Y, Banville H, Albuquerque I, Gramfort A, Falk TH, and Faubert J, “Deep learning-based electroencephalography analysis: A systematic review,” J. Neural Eng, vol. 16, no. 5, 2019. [DOI] [PubMed] [Google Scholar]
- [25].Ebrahimi Z, Loni M, Daneshtalab M, and Gharehbaghi A, “A review on deep learning methods for ECG arrhythmia classification,” Expert Syst. with Appl. X, vol. 7, p. 100033, 2020. [Google Scholar]
- [26].Murat F, Yildirim O, Talo M, Baloglu UB, Demir Y, and Acharya UR, “Application of deep learning techniques for heartbeats detection using ECG signals-analysis and review,” Comput. Biol. Med, vol. 120, no. April, p. 103726, 2020. [DOI] [PubMed] [Google Scholar]
- [27].Brouillette RT, Morielli A, Leimanis A, Waters KA, Luciano R, and Ducharme FM, “Nocturnal pulse oximetry as an abbreviated testing modality for pediatric obstructive sleep apnea,” Pediatrics, vol. 105, no. 2, pp. 405–412, 2000. [DOI] [PubMed] [Google Scholar]
- [28].Vaquerizo-Villar F et al. , “Convolutional Neural Networks to Detect Pediatric Apnea- Hypopnea Events from Oximetry,” in 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society, 2019, pp. 3555–3558. [DOI] [PubMed] [Google Scholar]
- [29].Redline S et al. , “The Childhood Adenotonsillectomy Trial (CHAT): Rationale, Design, and Challenges of a Randomized Controlled Trial Evaluating a Standard Surgical Procedure in a Pediatric Population,” Sleep, vol. 34, no. 11, pp. 1509–1517, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Marcus CL et al. , “A Randomized Trial of Adenotonsillectomy for Childhood Sleep Apnea,” N. Engl. J. Med, vol. 368, no. 25, pp. 2366–2376, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Iber C, Ancoli-Israel S, Chesson A, and Quan SF, “The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology and Technical Specification,” J. Clin. Sleep Med, vol. 3, no. 7, p. 752, 2007. [PMC free article] [PubMed] [Google Scholar]
- [32].Church GD, “The Role of Polysomnography in Diagnosing and Treating Obstructive Sleep Apnea in Pediatric Patients,” Curr. Probl. Pediatr. Adolesc. Health Care, vol. 42, no. 1, pp. 22–25, 2012. [DOI] [PubMed] [Google Scholar]
- [33].Tan H-L, Gozal D, Ramirez HM, Bandla HPR, and Kheirandish-Gozal L, “Overnight polysomnography versus respiratory polygraphy in the diagnosis of pediatric obstructive sleep apnea,” Sleep, vol. 37, no. 2, pp. 255–260, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Deviaene M, Testelmans D, Buyse B, Borzée P, Van Huffel S, and Varon C, “Automatic Screening of Sleep Apnea Patients Based on the SpO 2 Signal,” IEEE J. Biomed. Heal. Informatics, vol. 23, no. 2, pp. 607–617, 2019. [DOI] [PubMed] [Google Scholar]
- [35].He K, Zhang X, Ren S, and Sun J, “Delving deep into rectifiers: Surpassing human-level performance on imagenet classification,” in Proceedings of the IEEE international conference on computer vision, 2014, pp. 1026–1034. [Google Scholar]
- [36].Kingma DP and Ba JL, “Adam: A method for stochastic optimization,” 3rd Int. Conf. Learn. Represent. ICLR 2015 - Conf. Track Proc., pp. 1–15, 2015. [Google Scholar]
- [37].Huber PJ, “Robust Estimation of a Location Parameter,” Ann. Math. Stat, vol. 35, no. 1, pp. 73–101, 1964. [Google Scholar]
- [38].Bergstra J, Bardenet R, Bengio Y, and Kégl B, “Algorithms for hyper-parameter optimization,” in Advances in Neural Information Processing Systems 24: 25th Annual Conference on Neural Information Processing Systems 2011, NIPS 2011, 2011, pp. 1–9. [Google Scholar]
- [39].Bergstra J, Komer B, Eliasmith C, Yamins D, and Cox DD, “Hyperopt: A Python library for model selection and hyperparameter optimization,” Comput. Sci. Discov, vol. 8, no. 1, 2015. [Google Scholar]
- [40].Snoek J, Larochelle H, and Adams RP, “Practical Bayesian Optimization of Machine Learning Algorithms,” Adv. Neural Inf. Process. Syst, pp. 2951–2959, 2012.
- [41].Chollet F, “Keras,” 2015. [Online]. Available: https://github.com/fchollet/keras.
- [42].Taha BH et al. , “Automated detection and classification of sleep-disordered breathing from conventional polysomnography data,” Sleep, vol. 20, no. 11, pp. 991–1001, 1997. [DOI] [PubMed] [Google Scholar]
- [43].Biswal S, Sun H, Goparaju B, Brandon Westover M, Sun J, and Bianchi MT, “Expert-level sleep scoring with deep neural networks,” J. Am. Med. Informatics Assoc, vol. 25, no. 12, pp. 1643–1650, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Choi SH et al. , “Real-time apnea-hypopnea event detection during sleep by convolutional neural networks,” Comput. Biol. Med, vol. 100, no. February, pp. 123–131, 2018. [DOI] [PubMed] [Google Scholar]
- [45].Van Steenkiste T, Groenendaal W, Deschrijver D, and Dhaene T, “Automated Sleep Apnea Detection in Raw Respiratory Signals using Long Short-Term Memory Neural Networks,” IEEE J. Biomed. Heal. Informatics, vol. PP, no. c, p. 1, 2018. [DOI] [PubMed] [Google Scholar]
- [46].Nikkonen S, Afara IO, Leppänen T, and Töyräs J, “Artificial neural network analysis of the oxygen saturation signal enables accurate diagnostics of sleep apnea,” Sci. Rep, vol. 9, no. 1, p. 13200, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Epstein LJ et al. , “Clinical guideline for the evaluation, management and long-term care of obstructive sleep apnea in adults.,” J. Clin. Sleep Med, vol. 5, no. 3, pp. 263–76, 2009. [PMC free article] [PubMed] [Google Scholar]
- [48].Collop NA, “Scoring variability between polysomnography technologists in different sleep laboratories,” Sleep Med, vol. 3, no. 1, pp. 43–47, 2002. [DOI] [PubMed] [Google Scholar]
- [49].Álvarez D et al. , “Assessment of Feature Selection and Classification Approaches To Enhance Information From Overnight Oximetry in the Context of Apnea Diagnosis,” Int. J. Neural Syst, vol. 23, no. 05, p. 1350020, 2013. [DOI] [PubMed] [Google Scholar]
- [50].Canziani A, Culurciello E, and Paszke A, “An analysis of deep neural network models for practical applications,” pp. 1–7, 2016. [Google Scholar]


