ABSTRACT
Orientia tsutsugamushi is an obligate intracellular bacterium that causes scrub typhus, a potentially fatal rickettsiosis, and for which no genetic tools exist. Critical to addressing this technical gap is to identify promoters for driving expression of antibiotic resistance and fluorescence reporter genes in O. tsutsugamushi. Such promoters would need to be highly conserved among strains, expressed throughout infection, and exhibit strong activity. We examined the untranslated regions upstream of O. tsutsugamushi genes encoding outer membrane protein A (ompA), 22-kDa type-specific antigen (tsa22) and tsa56. The bacterium transcribed all three during infection of monocytic, endothelial and epithelial cells. Examination of the upstream noncoding regions revealed putative ribosome binding sites, one set of predicted −10 and −35 sequences for ompA and two sets of −10 and −35 sequences for tsa22 and tsa56. Comparison of these regions among geographically diverse O. tsutsugamushi patient isolates revealed nucleotide identities ranging from 84.8 to 100.0%. Upon examination of the candidates for the ability to drive green fluorescence protein expression in Escherichia coli, varying activities were observed with one of the tsa22 promoters being the strongest. Identification and validation of O. tsutsugamushi promoters is an initial key step toward genetically manipulating this important pathogen.
Keywords: Orientia tsutsugamushi, scrub typhus, Rickettsia, promoter, Rickettsiales
This study identifies and functionally validates promoter elements in Orientia tsutsugamushi.
INTRODUCTION
Obligate intracellular bacterial pathogens have significantly impacted human health throughout history and continue to pose substantial public health threats. Their evolutionarily reliance on eukaryotic cells makes them excellent models for studying host–pathogen interactions and for uncovering novel aspects of host cell biology (McClure et al. 2017). Among all species in the six genera that infect humans (Anaplasma, Chlamydia, Coxiella, Ehrlichia, Orientia and Rickettsia), Orientia tsutsugamushi is the deadliest (Xu et al. 2017). Trombiculid mites are the primary reservoir for the bacterium and transmit it to vertebrate animals and humans. Orientiatsutsugamushi infection in humans results in scrub typhus, a disease long known to be endemic to the Asia-Pacific (reviewed in Xu et al. 2017; Luce-Fedrow et al. 2018). Notably, recent reports of non-travel related cases of scrub typhus in Chile, Peru and the United Arab Emirates indicate that the disease is endemic to South America and the Middle East (Izzard et al. 2010; Weitzel et al. 2016, 2019; Kocher et al. 2017; Abarca et al. 2020). Phylogenetic analyses of isolates recovered from patients in Chile and Dubai identified two novel species, Candidatus Orientia chiloensis and Candidatus Orientia chuto, respectively (Izzard et al. 2010; Abarca et al. 2020). The reservoirs and vectors of these two new species are unknown. Orientia tsutsugamushi invades dendritic cells, monocytes and macrophages at the mite bite site, after which it is presumed that the infected leukocytes disseminate via the lymphatics (Paris et al. 2012). The bacterium then egresses to infect endothelial cells of the skin and other major organs. Scrub typhus clinical manifestations include nonspecific flu-like symptoms, fever, rash, eschar at the mite bite site, headache, myalgia, cough, lymphadenopathy, nausea, vomiting and abdominal pain. Severe complications can occur and include multiorgan failure, acute respiratory distress syndrome, interstitial pneumonia, myocarditis, pericarditis, meningoencephalitis and disseminated vascular coagulation (Luce-Fedrow et al. 2018). In the absence of antibiotic therapy or when it is delayed, the scrub typhus median mortality rate of 6% can be as high as 70% (Xu et al. 2017). Pharmacologic treatment options are limited, immunity is short-lived and there is no preventative vaccine (Xu et al. 2017; Luce-Fedrow et al. 2018; Wongsantichon et al. 2020).
Scrub typhus molecular pathogenesis is poorly understood. Conspicuously, mutational approaches for nonessential genes have been developed for species of all obligate intracellular genera except Orientia (reviewed in McClure et al. 2017). Hence, there is a major technical gap in the ability to study this global health threat. Gene disruption by plasmid-based allelic exchange or transposon-mediated insertion of antibiotic resistance and fluorescence reporter cassette would allow for selection of mutants and identification/recovery of host cells containing the mutants. These feats have been successfully performed for Rickettsia, Anaplasma, Ehrlichia, Coxiella and Chlamydia spp. (Baldridge et al. 2005; Felsheim et al. 2006; Liu et al. 2007; Binet and Maurelli 2009; Driskell et al. 2009; Baldridge et al. 2010; Felsheim et al. 2010; Clark et al. 2011; Beare 2012; Beare et al. 2012; Chen et al. 2012; Cheng et al. 2013; Beare and Heinzen 2014; Crosby et al. 2014, 2015, 2020; Kokes et al. 2015; Noriea, Clark and Hackstadt 2015; Oliva Chávez et al. 2015; Mueller, Wolf and Fields 2016; McKuen et al. 2017; Mueller, Wolf and Fields 2017; Wang et al. 2017; Keb, Hayman and Fields 2018; LaBrie et al. 2019; Wang et al. 2019; Wang et al. 2020; Arroyave et al. 2021; O'Conor et al. 2021). For optimal expression, in addition to codon-optimizing the antibiotic resistance and reporter genes for the high AT content of the O. tsutsugamushi chromosome (Nakayama et al. 2008), their expression would have to be driven from an O. tsutsugamushi promoter. In this study, we sought to identify suitable O. tsutsugamushi promoters of genes that are expressed throughout infection, are conserved among clinically relevant isolates that are geographically diverse and/or commonly used in laboratory studies, and exhibit strong activity. The findings presented herein are a first step toward developing genetic tools for this understudied pathogen.
METHODS
Cultivation of uninfected and O. tsutsugamushi infected cells
HeLa 229 human cervical epithelial cells (CCL-2; American Type Culture Collection [ATCC], Manassas, VA) and THP-1 cells (TIB-202; ATCC) were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented 2 mM l-glutamine (ThermoFisher Scientific, Waltham, MA) and 10% fetal bovine serum (FBS; Gemini Bio-Products, Sacramento, CA, USA) at 35°C in a humidified incubator with 5% CO2. RF/6A monkey choroidal endothelial cells (CRL-1780; ATCC) were cultured in Dulbecco's Modified Eagle's Medium (DMEM; ThermoFisher Scientific) supplemented with 10% FBS, 2 mM l-glutamine, 19 MEM nonessential amino acids (Invitrogen) and 15 mM HEPES. Orientia tsutsugamushi str. Ikeda was maintained in HeLa cells as described for uninfected cells except that it had 1× Anti-Anti (ThermoFisher Scientific) and the FBS concentration was 1%, the latter of which slowed host cell growth to allow for a high percentage (>90%) of infection to be achieved by 3–4 days. To obtain O. tsutsugamushi for experimental use, infected HeLa cells were mechanically disrupted by adding glass beads and shaking the culture flasks followed by differential centrifugation at 250 × g for 5 min to remove intact cells and cellular debris. The resulting supernatant was centrifuged at 2739 × g for 10 min to recover O. tsutsugamushi for use in infection studies. In infection experiments, O. tsutsugamushi infected and uninfected control cells were maintained in the appropriate media containing 1% FBS.
Synchronous infection, RNA isolation and reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR)
HeLa, THP-1 and RF/6A cells were synchronously infected with O. tsutsugamushi at a multiplicity of infection (MOI) of 10 as verified by assessing duplicate coverslips using antiserum targeting the bacterium's TSA56 (56-kDa type-specific antigen) (Beyer et al. 2017) and immunofluorescence microscopy. The cells, which had been seeded onto coverslips and infected, were washed with PBS and then fixed and permeabilized with –20°C methanol at 2 h postinfection. Blocking in 5% (vol/vol) bovine serum albumin (BSA) in PBS was performed followed by successive incubations with rabbit anti-TSA56 (1:1000) and Alexa Fluor 488-conjugated goat anti-rabbit (Invitrogen, Carlsbad, CA, 1:1000) in 5% BSA. Blocking and antibody incubations were performed for 1 h at room temperature with three PBS washes between each step. Samples were incubated with 0.1 µg ml–1 4′6-diamidino-2-phenylindole (DAPI; Invitrogen) in PBS for 1 min, washed three times with PBS and mounted with ProLong Gold Antifade mounting media (Invitrogen). Coverslips were viewed with an Olympus BX51 spinning disc confocal microscope (Olympus, Shinjuku City, Tokyo, Japan) for enumeration of the mean number of O. tsutsugamushi per cells. Cells were scored for immunosignal subcellular localization by counting 100 cells per coverslip. Triplicate infected cultures in 25-cm2 flasks were washed at 2 h to remove unbound bacteria. Fresh media was added and the cells were placed in a humidified incubator with 5% CO2 and set to 35°C. Total RNA was isolated at 24, 48 and 72 h using the RNeasy Mini Kit (Qiagen, Germantown, MD). One microgram RNA was treated with amplification grade DNase (Invitrogen). cDNA was generated using iScript Reverse Transcription Supermix according to the manufacturer's protocol (Bio-Rad, Hercules, CA). To verify successful removal of genomic DNA, parallel reactions performed in the absence of reverse transcriptase were used as template for PCR with human GAPDH-specific primers (Rodino et al. 2019) and MyTaq polymerase (Bioline, Taunton, MA). After an initial denaturing step at 95°C for 1 min, thermal cycling conditions were 35 cycles of 95°C for 15 s, 55°C for 15 s and 72°C for 10 s, followed by a final extension at 72°C for 30 s. Amplicons were analyzed with 2.0% agarose gels in 40 mM Tris-acetate-2 mM EDTA (pH 8). qPCR using cDNA from synchronously infected cells as template was performed with SsoFast EvaGreen supermix (Bio-Rad) and primers targeting O. tsutsugamushi 16S rDNA (ott16S) nucleotides 911–1096 (VieBrock et al. 2014), ompA nucleotides 57–260 (Evans et al. 2018a), tsa22 nucleotides 339–509 (Table 1) and tsa56 nucleotides 21–176 (Table 1). Orientia tsutsugamushi gene-specific primers were designed according to the annotated Ikeda str. genome (GenBank accession NC_010793.1) (Nakayama et al. 2008). Thermal cycling conditions used were 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 55°C for 5 s. Relative expression was determined using the 2−ΔΔCT method (Livak and Schmittgen 2001) as part of the CFX Maestro for Mac 1.0 software package (Bio-Rad).
Table 1.
Oligonucleotides utilized in this study.
| Designationa | Sequence (5′–3′) |
|---|---|
| tsa22-339F | TGTAGCTGCAGGCATACAAAC |
| tsa22-509R | GCGCTAGCAGCTTGAAGTTTAG |
| tsa56-21F | TGCTAGTGCAATGTCTGCGT |
| tsa56-176R | GCATCAGCTGAATCCAAGCG |
| PompA-fl BamHI Fb | GACTGCGGATCCATCTAGTTTAAAGGCGATTTTAAAAAAAATTG |
| PompA-fl EcoRI Rb | GACTGCGAATTCACTTGCTTATCCATCAAAATATTTAAAATG |
| Ptsa22-fl BamHI Fb | GACTGCGGATCCAGCCTGTTTTAATATTTAGCTAAC |
| Ptsa22-fl EcoRI Rb | GACTGCGAATTCAAAGATAATTCCTTATAATATTAGTAATACTTAAAAATATAC |
| Ptsa22-down BamHI Fb | GACTGCGGATCCCATGATATATGATAGAGGATCAATAC |
| Ptsa22-up BamHI, EcoRI Fb | GACTGCGGATCCAGCCTGTTTTAATATTTAGCTAACTAAAAGAAATATTTTGTAAGATAATACAGTATATGT ATAATGAATTCGCAGTC |
| Ptsa22-up EcoRI, BamHI Rb | GACTGCGAATCCATTATACATATACTGTATTATCTTACAAAATATTTCTTTTAGTTAGCTAAATATT AAAACAGGCTGGATCCGCAGTC |
| Ptsa56-fl BamHI Fb | GACTGCGGATCCTTTTTATGTGGGCTAATTTTAG |
| Ptsa56-fl EcoRI Rb | GACTGCGAATTCTCTAATCTCCTTAAAAGAATTAAATTTATTTTTTAG |
| Ptsa56-down BamHI Fb | GACTGCGGATCCAAATAAAAATAAATTTTACAATGGATAAAAC |
| Ptsa56-up EcoRI Rb | GACTGCGAATTCTTTATATATAACTTAAAGACAACATTCAATAG |
| pPROBEseqF | TAAACTGCCAGGAATTGGGGA |
| pPROBEseqR | CACCCTCTCCACTGACAGAAAA |
F and R refer to primers that bind to the sense and antisense strands, respectively. The terms down and up refer to each of the two putative promoter elements that are closest and furthest, respectively, from the start codon. P, putative promoter; fl, full length.
Boldface text indicates extra nucleotides upstream of restriction sites. Restriction sites are underlined.
In silico analyses
Noncoding regions upstream of the ompA, tsa22 and tsa56 initiation codons were assessed for the presence of sequences exhibiting similarity to the consensus Escherichia coli −10 and −35 RNA polymerase binding sites using BPROM (http://www.softberry.com/berry.phtml?topic=bprom&group=programs&subgroup=gfindb) (Solovyev and Salamov 2011). The upstream regions were defined as the nucleotides beginning at the −1 position relative to the initiation codon and extending upstream until the next open reading frame of each annotated chromosome. CLUSTAL Ω (Madeira et al. 2019) was used to generate alignments of and calculate % nucleotide identities among the ompA, tsa22 and tsa56 upstream regions of O. tsutsugamushi strains Boryong (NC_009488.1), Gilliam (NZ_LS398551.1), Karp (NZ_LS398548.1), Kato (NZ_LS398550.1), TA686 (NZ_LS398549.1), UT76 (NZ_LS398552.1), UT176 (NZ_LS398547.1) and Wuj/2014 (NZ_CP044031.1) relative to Ikeda.
Promoter constructs
pPROBE-NT was a gift from Dr Steven Lindow (Addgene plasmid # 37818; http://n2t.net/addgene:37818; RRID:Addgene 37818). The 5′ untranslated regions (UTRs) upstream of the O. tsutsugamushi str. Ikeda ompA, tsa22 and tsa56 start codons were PCR amplified using primers listed in Table 1, Platinum Taq DNA Polymerase High Fidelity (ThermoFisher, Waltham, MA), and DNA that had been isolated from O. tsutsugamushi str. Ikeda infected HeLa cells using the DNeasy Blood and Tissue kit (Qiagen, Valenica, CA) according to the manufacturer's protocol. The primers utilized contained BamHI or EcoRI sites for the purpose of cloning into pPROBE-NT. After an initial denaturing step at 94°C for 2 min, thermal cycling conditions were 30 cycles of 94°C for 15 s, 53°C for 30 s and 68°C for 30 s, followed by a final extension at 68°C for 5 min. The resulting amplicons were purified using the NucleoSpin Gel and PCR Clean-up Kit (Machery-Nagel, Bethlehem, PA), digested with BamHI and EcoRI (New England Biolabs, Ipswich, MA), and subsequently cloned using T4 DNA ligase (New England Biolabs) into pPROBE-NT that had been digested with BamHI and EcoRI. The recombinant plasmids were transformed into chemically competent Stellar E. coli HST08 cells (Takara, Mountain View, CA) followed by the addition of SOC (super optimal broth with catabolite repression) medium (0.5% yeast extract, 2% tryptone, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, 20 mM glucose) (Takara) and incubation at 37°C with agitation at 250 RPM for 1 h. Aliquots of each culture were plated onto Luria–Bertani (LB) agar plates containing 50 μg ml–1 kanamycin and incubated at 37°C overnight. Colony PCR using vector-specific primers pPROBEseqF and pPROBEseqR (Table 1) was performed to identify colonies that harbored plasmids containing inserts of the expected size. Plasmids were isolated from PCR-positive colonies using the QIAprep Spin Miniprep Kit (Qiagen). Recombinant plasmid insert integrity was confirmed by sequence analysis (Genewiz, South Plainfield, NJ).
Promoter activity assays
To qualitatively assess for the abilities of selected putative promoter O. tsutsugamushi sequences to drive green fluorescence protein (GFP) expression, E. coli HST08 (Takara Bio, Mountain View, CA) transformed with pPROBE-NT constructs were streaked onto LB agar containing 50 μg ml–1 kanamycin. After overnight incubation at 37°C, the plates were imaged under blue light illumination using the ChemiDoc Touch Imaging System (Bio-Rad, Hercules, CA). To quantitatively measure GFP expression, single colonies of E. coli HST08 transformants containing the recombinant pPROBE-NT plasmids or empty vector were inoculated into LB broth containing 50 μg ml–1 kanamycin and grown aerobically under constant agitation at 37°C to an optical density of 0.4 at 600 nm. Five lakh cells were pelleted, washed three times with phosphate buffered saline (PBS; 1.05 mM KH2PO4, 155 mM NaCl, 2.96 mM Na2HPO4, pH 7.4) and lysed with radioimmunoprecipitation assay buffer (50 mM Tris–HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA [pH 8]) containing Halt Protease and Phosphatase Inhibitor Cocktail (ThermoFisher Scientific, Waltham, MA). Lysates were resolved by SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) in 4–15% TGX polyacrylamide gels (Bio-Rad), transferred onto nitrocellulose membrane and screened by western blot analysis as described (VieBrock et al. 2014). Antibodies used were rabbit anti-GFP (Invitrogen, Carlsbad, CA; catalog #A6455) and horseradish peroxidase-conjugated goat anti-rabbit IgG (Cell Signaling, Danvers, MA; #7074S) at concentrations of 1:1000 and 1:10 000, respectively. Blots were imaged using the ChemiDoc Touch Imaging System. Anti-GFP immunosignal intensities were quantified using the Image Lab 6.0 software package (Bio-Rad). Duplicate gels that were stained with Coomassie Brilliant Blue R250 (Bio-Rad) were also imaged for total protein. The GFP immunosignal was normalized to total protein signal per sample.
Statistical analyses
Statistical analyses were performed using the Prism 7.0 software package (GraphPad, San Diego, CA). Two-way analysis of variance with Tukey's post-hoc test was used to test for significant differences among the groups. P-values < 0.05 were considered statistically significant.
RESULTS
Orientia tsutsugamushi expresses ompA, tsa22 and tsa56 throughout infection of mammalian host cells
An O. tsutsugamushi promoter being considered for expressing antibiotic resistance and fluorescence reporter genes should first be confirmed to drive expression of its endogenous gene throughout infection. Genes encoding OmpA (outer membrane protein A), TSA22 (22-kDa type-specific antigen) and TSA56 were selected as candidates for promoter activity analysis because O. tsutsugamushi expresses all three proteins during infection of tissue culture cells, laboratory mice and/or scrub typhus patients (Lee et al. 2008; Chao et al. 2009; Cho et al. 2010; Lin et al. 2012; Beyer et al. 2017; Ha et al. 2017). The tsa22 gene was also chosen because its 5′ untranslated region was shown to be highly conserved among seven strains and prior to this study was the only putative O. tsutsugamushi promoter sequence that had been assessed for conservation (Ge et al. 2005). Another reason for our inclusion of ompA was its demonstrated 93.6–100.0% nucleotide identity among 51 isolates (Evans et al. 2018a). Orientia tsutsugamushi str. Ikeda, which was isolated from a patient in Japan, causes severe disease and has a fully annotated genome (Tamura et al. 1984; Nakayama et al. 2008), was used for these studies. Total RNA that had been isolated from synchronously infected HeLa (epithelial), RF/6A (endothelial) and THP-1 (monocytic) cells at 24, 48 and 72 h was subjected to RT-qPCR analyses using gene-specific primers. All three were expressed at all time points examined in each cell line (Fig. 1), indicating that their promoters are active throughout O. tsutsugamushi infection of mammalian host cells.
Figure 1.
Orientia tsutsugamushi transcriptionally expresses ompA, tsa22 and tsa56 during infection of mammalian host cells. HeLa, RF/6A or THP-1 cells were synchronously infected with O. tsutsugamushi (Ot) followed by collection of total RNA at 24, 48 or 72 h. RT-qPCR was performed using gene-specific primers. Relative ompA-, tsa22- and tsa56-to-Ot 16S rRNA gene (ott16S) expression was determined using the 2−ΔΔCT method. Data are mean values ± SD from three experiments performed in triplicate.
Predicted ompA, tsa22 and tsa56 promoters are conserved among O. tsutsugamushi isolates
The noncoding regions upstream of O. tsutsugamushi str. Ikeda ompA, tsa22 and tsa56 were examined to identify sequences similar to E. coli −10 and −35 RNA polymerase binding sites and potential ribosome binding site (RBS) sequences. The bacterial σ70 promoter prediction program, BPROM (Solovyev and Salamov 2011), identified one set of −10 and −35 sequences beginning at 53 nucleotides upstream of the ompA start codon (Fig. 2A). BPROM identified two pairs of promoter elements beginning at 36 and 111 nucleotides upstream of the tsa22 translational start site and 45 and 214 bases upstream of the tsa56 initiation codon (Fig. 2B and C). Putative RBS sequences (Steitz and Jakes 1975) were detected seven to nine nucleotides upstream of the ompA, tsa22 and tsa56 start codons (Fig. 2). To differentiate between the respective tsa22 and tsa56 promoter candidates (Ptsa22 and Ptsa56), the elements closest to the start codon were assigned the designation ‘down’ (Ptsa22-down and Ptsa56-down), and those further upstream were given the designation ‘up’ (Ptsa22-up and Ptsa56-up) (Table 2). Among the five promoter candidates, the −10 regions differed by 1 to 5 nucleotides and the −35 regions differed by 2 to 4 nucleotides relative to each other (Fig. 3). The distance between the −10 and −35 sequences was identical for the Ptsa22-down, Ptsa56-down and Ptsa56-up, but was one nucleotide shorter for Ptsa22-up and five nucleotides longer for PompA. The five O. tsutsugamushi −10 and −35 sequences were similar to those that had been identified in other Rickettsiales species and validated for activity in E. coli (Policastro and Hackstadt 1994; Barbet et al. 2005; Peddireddi, Cheng and Ganta 2009), although the similarity was more discernible with the Rickettsia rickettsii sequences than with those for Anaplasma phagocytophilum or Ehrlichia chaffeensis. The predicted promoter regions for O. tsutsugamushi str. Ikeda were compared with corresponding sequences for the Boryong (South Korea), Gilliam (Burma), Kato (Japan) and Karp (New Guinea) strains along with Karp isolates UT76, UT176 (both from northeastern Thailand) and the Wuj/2014 isolate (China), each of which were recovered from scrub typhus patients (Bengtson 1945; Rights and Smadel 1948; Shishido et al. 1958; Chang et al. 1990; Blacksell et al. 2008; James et al. 2016; Yao et al. 2020). Nucleotide identities ranging from 84.79 to 100.00% were observed (Table 3).
Figure 2.
The UTR of ompA, tsa22 and tsa56 each contains a predicted RBS and −10 and −35 sequences. Upstream sequences of ompA(A), tsa22(B) and tsa56(C) were evaluated for the presence of RBS and −10 and −35 sequences. Initiation codons are indicated by white boldface text and purple highlighting. Putative RBS sequences are underlined. (A) The ompA UTR −10 and −35 sequences are denoted by boldface text. The two sets of −10 and −35 sequences in the UTRs of tsa22 (B) and tsa56 (C), which are designated ‘down’ and ‘up’ based on proximity to the start codon, are indicated by blue and red boldface text, respectively. Nucleotides cloned into pPROBE-NT for assessing promoter activity of tsa22-down and tsa56-down are highlighted gray, while those for evaluating promoter activity of tsa22-up and tsa56-up are highlighted yellow.
Table 2.
Putative O. tsutsugamushi Ikeda str. promoter regions assessed in this study.
| Promoter designation | Nucleotide rangea | Length | Genome coordinates |
|---|---|---|---|
| PompA | –1 to –213 | 213 | 1338189 to 1338401 |
| Ptsa22-down | –1 to –110 | 110 | 1578808 to 1578917 |
| Ptsa22-up | –111 to –175 | 65 | 1578743 to 1578807 |
| Ptsa56-down | –1 to –213 | 213 | 988304 to 988516 |
| Ptsa56-up | –214 to –544 | 331 | 987973 to 988303 |
Nucleotide range is relative to the initiation codon of each gene.
Figure 3.
Comparison of −10 and −35 regions of O. tsutsugamushi ompA, tsa22 and tsa56 with those of other Rickettsiales species and E. coli. The −10 and −35 sequences are indicated by boldface text. Dashes were introduced to enable alignment with the portion of the O. tsutsugamushi ompA UTR sequence depicted.
Table 3.
Nucleotide identities of putative promoters among O. tsutsugamushi strains.
| Strain | Genome coordinates | Length | Percent identitya |
|---|---|---|---|
| PompA | |||
| Boryong | 1771158 to 177364 | 207 | 84.79 |
| Gilliam | 442789 to 442967 | 179 | 91.67 |
| Karp | 261035 to 261245 | 211 | 97.66 |
| Kato | 315245 to 315458 | 214 | 99.53 |
| TA686 | 1656491 to 1656693 | 203 | 88.26 |
| UT76 | 469266 to 469480 | 215 | 94.50 |
| UT176 | 141889 to 142103 | 216 | 94.50 |
| Wuj/2014 | 1335472 to 1335684 | 213 | 95.37 |
| Ptsa22-down | |||
| Boryong | 24947 to 25056 | 110 | 99.09 |
| Gilliam | 22852 to 22961 | 110 | 100.00 |
| Karp | 19481 to 19590 | 110 | 96.36 |
| Kato | 72271 to 72380 | 110 | 100.00 |
| TA686 | 2061082 to 2061191 | 110 | 99.09 |
| UT76 | 21335 to 21445 | 111 | 99.10 |
| UT176 | 347754 to 347863 | 110 | 100.00 |
| Wuj/2014 | 965889 to 965999 | 111 | 99.10 |
| Ptsa22-up | |||
| Boryong | 24883 to 24946 | 64 | 93.85 |
| Gilliam | 22787 to 22851 | 65 | 96.92 |
| Karp | 19417 to 19480 | 64 | 90.77 |
| Kato | 72381 to 72445 | 65 | 100.00 |
| TA686 | 2062017 to 2061081 | 65 | 98.46 |
| UT76 | 21446 to 21510 | 65 | 100.00 |
| UT176 | 348864 to 347928 | 65 | 100.00 |
| Wuj/2014 | 965824 to 965888 | 65 | 100.00 |
| Ptsa56-down | |||
| Boryong | 581099 to 581311 | 213 | 100.00 |
| Gilliam | 2036332 to 2036544 | 213 | 99.06 |
| Karp | 1873572 to 1873785 | 214 | 97.66 |
| Kato | 860782 to 860994 | 213 | 100.00 |
| TA686 | 2031496 to 2031708 | 213 | 99.06 |
| UT76 | 1676193 to 1676405 | 213 | 100.00 |
| UT176 | 840850 to 841062 | 213 | 97.65 |
| Wuj/2014 | 463969 to 464181 | 213 | 100.00 |
| Ptsa56-up | |||
| Boryong | 581312 to 581645 | 334 | 98.20 |
| Gilliam | 2036002 to 2036331 | 330 | 97.89 |
| Karp | 1873238 to 1873571 | 334 | 92.84 |
| Kato | 860995 to 861325 | 331 | 100.00 |
| TA686 | 2031707 to 2032041 | 335 | 91.45 |
| UT76 | 1675864 to 1676192 | 329 | 98.19 |
| UT176 | 841063 to 841394 | 332 | 97.60 |
| Wuj/2014 | 463640 to 463968 | 329 | 98.19 |
Identity relative to O. tsutsugamushi str. Ikeda.
Evaluation of promoter activities of the sequences upstream of the ompA, tsa22 and tsa56 coding regions
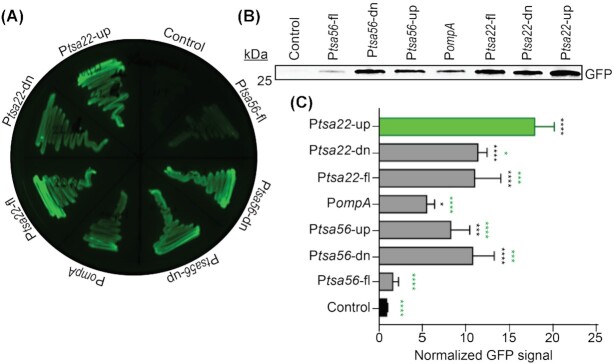
Functional activity of the candidate promoter regions was evaluated using an approach similar to those used to validate R. rickettsii, E. chaffeensis and Anaplasma spp. promoters (Policastro and Hackstadt 1994; Barbet et al. 2005; Peddireddi, Cheng and Ganta 2009). PompA, Ptsa22-down, Ptsa22-up, Ptsa56-down and Ptsa56-up were examined. The entire noncoding sequences upstream of tsa22 and tsa56 containing both the ‘down’ and ‘up’ promoter elements, referred to as Ptsa22-fl (full length) and Ptsa56-fl, respectively, were also tested. Each sequence was inserted upstream of the green fluorescent protein (gfp) open reading frame in pPROBE-NT followed by examination of GFP expression by recombinant E. coli transformed with each construct. Qualitative assessment of LB agar plates streaked with bacteria under blue light illumination revealed that all of the promoter regions drove relatively strong GFP expression with the exception of Ptsa56-fl, which exhibited GFP signal that was comparable to the background level observed for E. coli transformed with empty vector (Fig. 4A). To quantitatively measure promoter activity, GFP antibody was used to probe Western-blotted whole cell lysates. GFP signal was normalized to total protein signal of duplicate Coomassie brilliant blue-stained samples. Consistent with the qualitative assessment, the Ptsa56-fl-driven GFP level was barely above the background of empty pPROBE-NT (Fig. 4B). The six other promoters exhibited activities that were significantly greater than the vector control, thereby confirming their functional activity. Of these six, PompA exhibited the weakest activity. Notably, Ptsa22-up displayed the most robust promoter activity, as the GFP level for this transformant significantly exceeded those of the other five including Ptsa22-fl. These results functionally validate promoter activities of noncoding regions upstream of three O. tsutsugamushi genes and signify Ptsa22-up as the strongest.
Figure 4.
Promoter activity evaluation of ompA, tsa22 and tsa56 upstream noncoding regions. Putative promoter regions of tsa56 (Ptsa56-fl [full length], Ptsa56-down [dn], Ptsa56-up, ompA [PompA] and tsa22 [Ptsa22-fl, Ptsa22-down and Ptsa22-up]) were cloned upstream of promoter-less gfp in pPROBE-NT. The recombinant plasmids or empty pPROBE-NT (control) were transformed into E. coli. (A) LB agar plate streaked with E. coli transformed with the indicated constructs imaged under blue light illumination to detect GFP fluorescence. (B) Western-blotted whole cell lysates of E. coli transformed with the indicated vector were screened with GFP antibody. (C) Mean ± GFP signal intensity of triplicate samples each normalized to total protein signal of duplicate Coomassie brilliant blue stained gels. Data in panels (B) and (C) are representative of three experiments with similar results. Statistically significant (*P < 0.05; ***P < 0.003; ****P < 0.0001) values are indicated. Black asterisks denote GFP signal values that are statistically significant versus control, whereas green asterisks indicate statistical significance of Ptsa22-up GFP signal compared with those produced from other O. tsutsugamushi promoter sequences.
DISCUSSION
Orientia tsutsugamushi and other Orientia species are the etiologic agents of scrub typhus, an infection that can be debilitating, has a high fatality rate and is an emerging global health threat (Xu et al. 2017; Luce-Fedrow et al. 2018). Their obligate intracellular lifestyle makes them challenging to study, as deleting or disrupting essential genes would prevent recovery of the mutants. Clever approaches have been applied to studying O. tsutsugamushi that circumvent its genetic intractability including using fluorescent probes to label and track live bacteria during infection of tissue culture cells, validating the ability of adhesin candidates to mediate binding to/infection of host cells when heterologously expressed on the E. coli surface, and ectopically expressing oriential effectors and mutated versions thereof in host cells to determine if they phenocopy aspects of infection, identify eukaryotic interaction partners and discern their functional roles (Ha et al. 2011; VieBrock et al. 2014; Atwal et al. 2016; Beyer et al. 2017; Evans et al.2018b; Nguyen et al. 2021). Still, the development of genetic tools for Anaplasma, Chlamydia, Coxiella, Ehrlichia and Rickettsia spp. has arguably enabled greater strides in understanding the molecular pathogenesis of these organisms. For instance, random transposon insertional mutagenesis and allelic exchange, some cases of which were followed up by genetic complementation, facilitated discovery of new virulence factors and immunopathological molecules, genes required for optimal fitness or survival in host cells and mammals, and novel vaccine targets (Binet and Maurelli 2009; Driskell et al. 2009; Chen et al. 2012; Cheng et al. 2013; Crosby et al. 2015, 2020; Oliva Chávez et al. 2015; Mueller, Wolf and Fields 2016; McClure et al. 2017; McKuen et al. 2017; LaBrie et al. 2019; Wang et al. 2020; Arroyave et al. 2021; O'Conor et al. 2021). These approaches involve chromosomal insertion of a cassette that expresses an antibiotic resistance gene that permits mutant selection and often a fluorescence reporter that enables visualization of mutants and recovery of host cells harboring them. The essential first step in constructing such cassettes is to identify promoters that will drive selection and reporter gene expression in the transformed bacterium. The driving rationale for this study was to identify promoters in O. tsutsugamushi that can ultimately be used in constructs for genetically manipulating this important pathogen.
Orientia tsutsugamushi infects leukocytes and endothelial cells during mammalian infection and a variety of cell lines in vitro, including HeLa cells, which are commonly used to study host interactions with the pathogen (VieBrock et al. 2014; Giengkam et al. 2015; Beyer et al. 2017; Evans et al. 2018a; Rodino et al. 2019). Any promoter used for selection and reporter marker expression would need to be active throughout infection of host cells. All three candidates assessed—ompA, tsa22 and tsa56—met this criterion as they were expressed at throughout infection of HeLa, THP-1 and RF/6A cells. Examination of the UTRs upstream of each revealed that they are highly conserved among geographically diverse O. tsutsugamushi isolates. This is important because it suggests that cassettes/constructs harboring these promoters could be used to drive gene of interest expression in multiple O. tsutsugamushi strains. Examination of the ompA, tsa22 and tsa56 noncoding regions revealed the presence of a putative RBS in each and a total of five potential sets of −10 and −35 sequences that were similar to the consensus E. coli sequences and to several that had been previously confirmed in other Rickettsiales members (Policastro & Hackstadt 1994; Barbet et al. 2005; Peddireddi, Cheng and Ganta 2009).
In the absence of being able to directly assess functionality of the candidate promoter regions in O. tsutsugamushi, each was examined for the ability to drive promoter-less GFP expression in E. coli. Notably, this and similar approaches using E. coli as a heterologous host validated promoters in Anaplasma, Ehrlichia and Rickettsia that were able to be subsequently used to drive selection marker and fluorescence reporter gene expression when transpositionally inserted into the chromosomes of these bacteria (Policastro and Hackstadt 1994; Baldridge et al. 2005; Barbet et al. 2005; Felsheim et al. 2006; Peddireddi, Cheng and Ganta 2009; Baldridge et al. 2010; Felsheim et al. 2010; Clark et al. 2011; Chen et al. 2012; Cheng et al. 2013; Crosby et al. 2015, 2020; Oliva Chávez et al. 2015; Wang et al. 2017, 2020; O'Conor et al. 2021). This precedent highlights that promoters identified herein should be functional if inserted into the O. tsutsugamushi chromosome. Ptsa22-up drove GFP expression significantly greater than all other candidates, denoting it as the top choice for inclusion in constructs for future genetic manipulation studies. Ptsa22-down, Ptsa22-fl, Ptsa56-up and Ptsa56-down also exhibited strong promoter activities and therefore also remain in contention for inclusion in future studies. As Ptsa56-fl exhibited no promoter activity, it has been eliminated from further consideration. Likewise, PompA is no longer being considered because, in addition to exhibiting relative weak promoter activity, its nucleotide identity among the isolate sequences examined ranged from 84.79 to 95.47% relative to Ikeda, whereas the identities of the Ptsa22 and Ptsa56 promoters ranged from 90.77 to 100%.
In closing, this study identifies a total of five O. tsutsugamushi promoter elements that exhibit strong activity when transformed into E. coli, drive relatively consistent gene expression throughout infection of three different mammalian host cell types and exhibit high degrees of nucleotide identity among diverse strains. By satisfying each of these criteria, they represent excellent candidates for driving selection marker and reporter expression when inserted into the O. tsutsugamushi chromosome and are a foundation on which genetic manipulation approaches for this understudied pathogen can be built.
ACKNOWLEDGMENTS
We thank Drs Hwan Kim (Stony Brook University), Sean P. Riley (University of Maryland) and Juan J. Martinez (Louisiana State University) for helpful discussions and Haley Adcox for critical review of this manuscript.
Contributor Information
Jason R Hunt, Department of Microbiology and Immunology, Virginia Commonwealth University (VCU) Medical Center, School of Medicine, VCU, Richmond, VA 23298, USA.
Jason A Carlyon, Department of Microbiology and Immunology, Virginia Commonwealth University (VCU) Medical Center, School of Medicine, VCU, Richmond, VA 23298, USA.
FUNDING
This work was supported by the National Institutes of Health—National Institute of Allergy and Infectious Diseases grants R01 AI123346 and R21 AI152513 and the Virginia Commonwealth University Presidential Research Quest Fund (to JAC).
Conflict of Interest
None declared.
REFERENCES
- Abarca K, Martínez-Valdebenito C, Angulo Jet al. . Molecular description of a novel Orientia species causing scrub typhus in Chile. Emerg Infect Dis. 2020;26:2148–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyave E, Hyseni I, Burkhardt Net al. . Rickettsia parkeri with a genetically disrupted phage integrase gene exhibits attenuated virulence and induces protective immunity against fatal rickettsioses in mice. Pathogens (Basel, Switzerland). 2021;10:819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwal S, Giengkam S, VanNieuwenhze Met al. . Live imaging of the genetically intractable obligate intracellular bacteria Orientia tsutsugamushi using a panel of fluorescent dyes. J Microbiol Methods. 2016;130:169–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge GD, Burkhardt N, Herron MJet al. . Analysis of fluorescent protein expression in transformants of Rickettsia monacensis, an obligate intracellular tick symbiont. Appl Environ Microbiol. 2005;71:2095–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge GD, Burkhardt NY, Oliva ASet al. . Rickettsial ompB promoter regulated expression of GFPuv in transformed Rickettsia montanensis. PLoS One. 2010;5:e8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet AF, Agnes JT, Moreland ALet al. . Identification of functional promoters in the msp2 expression loci of Anaplasma marginale and Anaplasma phagocytophilum. Gene. 2005;353:89–97. [DOI] [PubMed] [Google Scholar]
- Beare PA, Heinzen RA. Gene inactivation in Coxiella burnetii. Methods Mol Biol. 2014;1197:329–45. [DOI] [PubMed] [Google Scholar]
- Beare PA, Larson CL, Gilk SDet al. . Two systems for targeted gene deletion in Coxiella burnetii. Appl Environ Microbiol. 2012;78:4580–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beare PA. Genetic manipulation of Coxiella burnetii. Adv Exp Med Biol. 2012;984:249–71. [DOI] [PubMed] [Google Scholar]
- Bengtson IA. Apparent serological heterogeneity among strains of Tsutsugamushi disease (scrub typhus). Public Health Rep (1896–1970). 1945;60:1483–8. [PubMed] [Google Scholar]
- Beyer AR, Rodino KG, VieBrock Let al. . Orientia tsutsugamushi Ank9 is a multifunctional effector that utilizes a novel GRIP-like Golgi localization domain for Golgi-to-endoplasmic reticulum trafficking and interacts with host COPB2. Cell Microbiol. 2017;19:cmi.12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binet R, Maurelli AT. Transformation and isolation of allelic exchange mutants of Chlamydia psittaci using recombinant DNA introduced by electroporation. Proc Natl Acad Sci USA. 2009;106:292–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacksell SD, Luksameetanasan R, Kalambaheti Tet al. . Genetic typing of the 56-kDa type-specific antigen gene of contemporary Orientia tsutsugamushi isolates causing human scrub typhus at two sites in north-eastern and western Thailand. FEMS Immunol Med Microbiol. 2008;52:335–42. [DOI] [PubMed] [Google Scholar]
- Chang WH, Kang JS, Lee WKet al. . Serological classification by monoclonal antibodies of Rickettsia tsutsugamushi isolated in Korea. J Clin Microbiol. 1990;28:685–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao CC, Garland DL, Dasch GAet al. . Comparative proteomic analysis of antibiotic-sensitive and insensitive isolates of Orientia tsutsugamushi. Ann N Y Acad Sci. 2009;1166:27–37. [DOI] [PubMed] [Google Scholar]
- Chen G, Severo MS, Sakhon OSet al. . Anaplasma phagocytophilum dihydrolipoamide dehydrogenase 1 affects host-derived immunopathology during microbial colonization. Infect Immun. 2012;80:3194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Nair AD, Indukuri VVet al. . Targeted and random mutagenesis of Ehrlichia chaffeensis for the identification of genes required for in vivo infection. PLoS Pathog. 2013;9:e1003171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho BA, Cho NH, Seong SYet al. . Intracellular invasion by Orientia tsutsugamushi is mediated by integrin signaling and actin cytoskeleton rearrangements. Infect Immun. 2010;78:1915–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark TR, Lackey AM, Kleba Bet al. . Transformation frequency of a mariner-based transposon in Rickettsia rickettsii. J Bacteriol. 2011;193:4993–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby FL, Brayton KA, Magunda Fet al. . Reduced Infectivity in cattle for an outer membrane protein mutant of Anaplasma marginale. Appl Environ Microbiol. 2015;81:2206–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby FL, Munderloh UG, Nelson CMet al. . Disruption of VirB6 paralogs in Anaplasma phagocytophilum attenuates its growth. J Bacteriol. 2020;202:e00301–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby FL, Wamsley HL, Pate MGet al. . Knockout of an outer membrane protein operon of Anaplasma marginale by transposon mutagenesis. BMC Genomics. 2014;15:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell LO, Yu XJ, Zhang Let al. . Directed mutagenesis of the Rickettsia prowazekii pld gene encoding phospholipase D. Infect Immun. 2009;77:3244–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SM, Adcox HE, VieBrock Let al. . Outer membrane protein A conservation among Orientia tsutsugamushi isolates suggests its potential as a protective antigen and diagnostic target. Trop Med Infect Dis. 2018a;3:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SM, Rodino KG, Adcox HEet al. . Orientia tsutsugamushi uses two Ank effectors to modulate NF-κB p65 nuclear transport and inhibit NF-κB transcriptional activation. PLoS Pathog. 2018b;14:e1007023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsheim RF, Chávez AS, Palmer GHet al. . Transformation of Anaplasma marginale. Vet Parasitol. 2010;167:167–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsheim RF, Herron MJ, Nelson CMet al. . Transformation of Anaplasma phagocytophilum. BMC Biotech. 2006;6:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Tong M, Li Aet al. . Cloning and sequence analysis of the 22-kDa antigen genes of Orientia tsutsugamushi strains Kato, TA763, AFSC 7, 18-032460; TH1814, and MAK 119. Ann N Y Acad Sci. 2005;1063:231–8. [DOI] [PubMed] [Google Scholar]
- Giengkam S, Blakes A, Utsahajit Pet al. . Improved quantification, propagation, purification and storage of the obligate intracellular human pathogen Orientia tsutsugamushi. PLoS Negl Trop Dis. 2015;9:e0004009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha NY, Cho NH, Kim YSet al. . An autotransporter protein from Orientia tsutsugamushi mediates adherence to nonphagocytic host cells. Infect Immun. 2011;79:1718–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha NY, Kim Y, Min CKet al. . Longevity of antibody and T-cell responses against outer membrane antigens of Orientia tsutsugamushi in scrub typhus patients. Emerg Microbes Infect. 2017;6:e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzard L, Fuller A, Blacksell SDet al. . Isolation of a novel Orientia species (O. chuto sp. nov.) from a patient infected in Dubai. J Clin Microbiol. 2010;48:4404–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James SL, Blacksell SD, Nawtaisong Pet al. . Antigenic relationships among human pathogenic Orientia tsutsugamushi isolates from Thailand. PLoS Negl Trop Dis. 2016;10:e0004723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keb G, Hayman R, Fields KA. Floxed-cassette allelic exchange mutagenesis enables markerless gene deletion in Chlamydia trachomatis and can reverse cassette-induced polar effects. J Bacteriol. 2018;200:e00479–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher C, Jiang J, Morrison ACet al. . Serologic evidence of scrub typhus in the Peruvian Amazon. Emerg Infect Dis. 2017;23:1389–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokes M, Dunn JD, Granek JAet al. . Integrating chemical mutagenesis and whole-genome sequencing as a platform for forward and reverse genetic analysis of Chlamydia. Cell Host Microbe. 2015;17:716–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBrie SD, Dimond ZE, Harrison KSet al. . Transposon mutagenesis in Chlamydia trachomatis identifies CT339 as a ComEC homolog important for DNA uptake and lateral gene transfer. mBio. 2019;10:e01343–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Cho NH, Kim SYet al. . Fibronectin facilitates the invasion of Orientia tsutsugamushi into host cells through interaction with a 56-kDa type-specific antigen. J Infect Dis. 2008;198:250–7. [DOI] [PubMed] [Google Scholar]
- Lin CC, Chou CH, Lin TCet al. . Molecular characterization of three major outer membrane proteins, TSA56, TSA47 and TSA22, in Orientia tsutsugamushi. Int J Mol Med. 2012;30:75–84. [DOI] [PubMed] [Google Scholar]
- Liu ZM, Tucker AM, Driskell LOet al. . Mariner-based transposon mutagenesis of Rickettsia prowazekii. Appl Environ Microbiol. 2007;73:6644–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- Luce-Fedrow A, Lehman ML, Kelly DJet al. . A review of scrub typhus (Orientia tsutsugamushi and related organisms): then, now, and tomorrow. Trop Med Infect Dis. 2018;3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F, Park YM, Lee Jet al. . The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure EE, Chavez ASO, Shaw DKet al. . Engineering of obligate intracellular bacteria: progress, challenges and paradigms. Nat Rev Microbiol. 2017;15:544–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKuen MJ, Mueller KE, Bae YSet al. . Fluorescence-reported allelic exchange mutagenesis reveals a role for Chlamydia trachomatis TmeA in invasion that is independent of host AHNAK. Infect Immun. 2017;85:e00640–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller KE, Wolf K, Fields KA. Chlamydia trachomatis Transformation and Allelic Exchange Mutagenesis. Curr Protoc Microbiol. 2017;45:11A.3.1–3.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller KE, Wolf K, Fields KA. Gene deletion by fluorescence-reported allelic exchange mutagenesis in Chlamydia trachomatis. mBio. 2016;7:e01817–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Yamashita A, Kurokawa Ket al. . The Whole-genome sequencing of the obligate intracellular bacterium Orientia tsutsugamushi revealed massive gene amplification during reductive genome evolution. DNA Res. 2008;15:185–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen YTH, Kim C, Kim Yet al. . The Orientia tsutsugamushi ScaB autotransporter protein is required for adhesion and invasion of mammalian cells. Front Microbiol. 2021;12:626298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noriea NF, Clark TR, Hackstadt T. Targeted knockout of the Rickettsia rickettsii OmpA surface antigen does not diminish virulence in a mammalian model system. mBio. 2015;6:e00323–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Conor MC, Herron MJ, Nelson CMet al. . Biostatistical prediction of genes essential for growth of Anaplasma phagocytophilum in a human promyelocytic cell line using a random transposon mutant library. Pathog Dis. 2021;79:ftab029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva Chávez AS, Fairman JW, Felsheim RFet al. . An O-methyltransferase is required for infection of tick cells by Anaplasma phagocytophilum. PLoS Pathog. 2015;11:e1005248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris DH, Phetsouvanh R, Tanganuchitcharnchai Aet al. . Orientia tsutsugamushi in human scrub typhus eschars shows tropism for dendritic cells and monocytes rather than endothelium. PLoS Negl Trop Dis. 2012;6:e1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peddireddi L, Cheng C, Ganta RR. Promoter analysis of macrophage- and tick cell-specific differentially expressed Ehrlichia chaffeensis p28-Omp genes. BMC Microbiol. 2009;9:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Policastro PF, Hackstadt T. Differential activity of Rickettsia rickettsii opmA and ompB promoter regions in a heterologous reporter gene system. Microbiology. 1994;140:2941–9. [DOI] [PubMed] [Google Scholar]
- Rights FL, Smadel JE. Studies on scrib typhus; tsutsugamushi disease; heterogenicity of strains of R. tsutsugamushi as demonstrated by cross-vaccination studies. J Exp Med. 1948;87:339–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodino KG, Adcox HE, Martin RKet al. . The obligate intracellular bacterium Orientia tsutsugamushi targets NLRC5 to modulate the major histocompatibility complex class I pathway. Infect Immun. 2019;87:813–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shishido A, Ohtawara M, Tateno Set al. . The nature of immunity against scrub typhus in mice. I. The resistance of mice, surviving subcutaneous infection of scrub typhus Rickettsia, to intraperitoneal reinfection of the same agent. Jap J Med Sci Biol. 1958;11:383–99. [Google Scholar]
- Solovyev V, Salamov A. Automatic annotation of microbial genomes and metagenomic sequences. In: Li RW (ed). Metagenomics and Its Applications in Agriculture, Biomedicine and Environmental Studies, Nova Science, 2011, 61–78. [Google Scholar]
- Steitz JA, Jakes K. How ribosomes select initiator regions in mRNA: base pair formation between the 3' terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc Natl Acad Sci USA. 1975;72:4734–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura A, Takahashi K, Tsuruhara Tet al. . Isolation of Rickettsia tsutsugamushi antigenically different from Kato, Karp, and Gilliam strains from patients. Microbiol Immunol. 1984;28:873–82. [DOI] [PubMed] [Google Scholar]
- VieBrock L, Evans SM, Beyer ARet al. . Orientia tsutsugamushi ankyrin repeat-containing protein family members are Type 1 secretion system substrates that traffic to the host cell endoplasmic reticulum. Front Cell Infect Microbiol. 2014;4:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, LaBrie SD, Carrell SJet al. . Development of transposon mutagenesis for Chlamydia muridarum. J Bacteriol. 2019;201:e00366–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Nair ADS, Alhassan Aet al. . Multiple Ehrlichia chaffeensis genes critical for its persistent infection in a vertebrate host are identified by random mutagenesis coupled with in vivo infection assessment. Infect Immun. 2020;88:e00316–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wei L, Liu Het al. . A genetic system for targeted mutations to disrupt and restore genes in the obligate bacterium, Ehrlichia chaffeensis. Sci Rep. 2017;7:15801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzel T, Dittrich S, López Jet al. . Endemic scrub typhus in South America. N Engl J Med. 2016;375:954–61. [DOI] [PubMed] [Google Scholar]
- Weitzel T, Martínez-Valdebenito C, Acosta-Jamett Get al. . Scrub typhus in continental Chile, 2016–2018. Emerg Infect Dis. 2019;25:1214–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongsantichon J, Jaiyen Y, Dittrich Set al. . Orientia tsutsugamushi. Trends Microbiol. 2020;28:780–1. [DOI] [PubMed] [Google Scholar]
- Xu G, Walker DH, Jupiter Det al. . A review of the global epidemiology of scrub typhus. PLoS Negl Trop Dis. 2017;11:e0006062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L, Lu Q, Ruan Wet al. . Gene genotypes and variation of 5 cases with Orientia tsutsugamushi infection. Chin J Exp Clin Infect Dis (Electronic Edition). 2020;14:110–6. [Google Scholar]