Abstract
Activating signal cointegrator 1 (ASC-1) harbors an autonomous transactivation domain that contains a putative zinc finger motif which provides binding sites for basal transcription factors TBP and TFIIA, transcription integrators steroid receptor coactivator 1 (SRC-1) and CBP-p300, and nuclear receptors, as demonstrated by the glutathione S-transferase pull-down assays and the yeast two-hybrid tests. The ASC-1 binding sites involve the hinge domain but not the C-terminal AF2 core domain of nuclear receptors. Nonetheless, ASC-1 appears to require the AF2-dependent factors to function (i.e., CBP-p300 and SRC-1), as suggested by the ability of ASC-1 to coactivate nuclear receptors, either alone or in cooperation with SRC-1 and p300, as well as its inability to coactivate a mutant receptor lacking the AF2 core domain. By using indirect immunofluorescence, we further show that ASC-1, a nuclear protein, is localized to the cytoplasm under conditions of serum deprivation but is retained in the nucleus when it is serum starved in the presence of ligand or coexpressed CBP or SRC-1. These results suggest that ASC-1 is a novel coactivator molecule of nuclear receptors which functions in conjunction with CBP-p300 and SRC-1 and may play an important role in establishing distinct coactivator complexes under different cellular conditions.
The nuclear receptor superfamily is a group of ligand-dependent transcriptional regulatory proteins that function by binding to specific DNA sequences named hormone response elements in the promoters of target genes (reviewed in reference 36). The superfamily includes receptors for a variety of small hydrophobic ligands, such as steroids, T3, and retinoids, as well as a large number of related proteins that do not have known ligands, referred to as orphan nuclear receptors. Functional analysis of nuclear receptors has shown that there are two major activation domains. The N-terminal domain (AF1) contains a ligand-independent activation function, whereas the extreme C-terminal helix of the ligand binding domain (LBD) serves as an integral component of the ligand-dependent transactivation function AF2, which also includes several essential helixes in broad parts of the LBD. This C-terminal AF2 core region, which is relatively highly conserved among nuclear receptors, undergoes an allosteric change upon ligand binding, and deletion or point mutations in this region often impair transcriptional activation without changing ligand and DNA binding affinities (36).
Transcriptional activation of nuclear receptors appears to involve at least two separate processes: derepression and activation. Repression is mediated in part by the interaction of unliganded receptors with corepressors such as the nuclear receptor corepressor (N-CoR) (8) and SMRT (20). However, ligand binding triggers the dissociation of these corepressors and the concomitant recruitment of coactivators. These putative receptor-interacting coactivators include RIP-140 and RIP-160 (5, 6), ERAP-140 and ERAP-160 (17), TIF1 (28), TRIP1 (30), ARA70 (55), CBP and its functional homolog p300 (16), steroid receptor coactivator 1 (SRC-1) (42, 54), xSRC-3 (25), AIB1 (1), TIF2 (50), RAC3 (34), ACTR (9), TRAM-1 (47), and p/CIP (48). In particular, the last eight proteins are highly related to each other and can enhance transcriptional activation by several nuclear receptors. These coactivators are postulated to function in transmitting the signal of ligand-induced conformational change to the basal transcription machinery. As expected, many coactivators fail to interact with nuclear receptors mutated for the C-terminal AF2 core region (6, 28, 50). Recently, chromatin remodeling by cofactors was also suggested to contribute, through histone acetylation-deacetylation, to receptor-mediated transcriptional regulation (49, 52, 56). Accordingly, SRC-1 (46) and its homologue ACTR (9), along with CBP-p300 (3, 41), were shown to contain histone acetyltransferase activities and form a complex with histone acetyltransferase protein P/CAF (53). In contrast, it was shown that SMRT (20) and N-CoR (8), nuclear receptor corepressors, form complexes with Sin3 and histone deacetylase proteins (19, 40).
CBP and p300 define a distinct class of coactivator molecules, functionally interacting with many different transcription factors, including nuclear receptors, CREB, NF-κB, bHLH factors, p53, STATs, AP-1, and SRF-TCF (reviewed in reference 16). In particular, CBP and p300 have been found to interact directly with nuclear receptors in a ligand- and AF2-dependent manner (7, 18, 23, 54). Interestingly, CBP-p300 was recently found to interact with SRC-1 (54) and form a complex with a series of cellular proteins with molecular masses ranging from 44 to 270 kDa (11). SRC-1 and p/CIP were recently shown to coactivate multiple transcription factors, including CREB and STATs (48), NF-κB (39), AP-1 (32), and SRF (26). Based on this broad spectrum of action, these proteins (i.e., CBP-p300 and SRC-1) were renamed transcription integrators.
Herein, we describe the initial characterizations of a novel protein, ASC-1, originally isolated based on its association with nuclear receptors (31). ASC-1 is a transcription coactivator of nuclear receptors, associating with specific components of the RNA polymerase II complex, the hinge domain of nuclear receptors, and transcription integrators SRC-1 and CBP-p300. Strikingly, ASC-1, a nuclear protein, localizes to the cytoplasm under conditions of serum starvation but is retained in the nucleus when serum starved in the presence of ligand or coexpressed CBP or SRC-1, raising an interesting possibility that ASC-1 may play an important role in establishing distinct coactivator complexes under different cellular conditions.
MATERIALS AND METHODS
Antibodies and plasmids.
A monoclonal antibody against hemagglutinin (HA) epitope and fluorescein isothiocyanate (FITC)-rhodamine-conjugated antibodies as well as rabbit polyclonal antibodies against retinoic acid receptor alpha (RARα) were purchased from Boehringer (Mannheim, Germany). A rabbit polyclonal antibody was raised and affinity purified against the ASC-1 zinc finger region (the ASC-1 residues 125 to 237). A PCR-amplified fragment encoding a full-length ASC-1 was cloned into EcoRI and XhoI restriction sites of pJ3H (kind gift of J. K. Chung at KAIST, Korea) to express HA-tagged ASC-1 in mammalian cells. ASCdn, a mutant ASC-1 deleted for the zinc finger region (the ASC-1 residues 165 to 216), was constructed by using two-step PCR procedures (2). LexA, B42, T7, glutathione S-transferase (GST), and Gal4 fusion vectors to express ASC-1, ASCΔN, ASCΔNΔC, p300-N, p300-C, and CBP-C were constructed by inserting appropriate PCR fragments into EcoRI and XhoI/SalI sites of p202PL (2), pJG4-5 (2), pcDNA3 (Invitrogen, San Diego, Calif.), pGEX4T-1 (Amersham Pharmacia Biotech), and pCMX-GAL4-N (kind gift of Ron Evans at the Salk Institute), respectively. PCR fragments encoding the TRβ1 residues 164 to 265 and 260 to 444 were inserted into EcoRI and SalI sites of p202PL to express LexA–TR-D and LexA–TR-E, respectively. LexA vectors to express B42, retinoid X receptor (RXR)-DEF, thyroid hormone receptor (TR)-DEF, TR-DEF459, SRC-C and SRC-D as well as T7 vectors to express SMRT, SRC-1, SRC-C, and SRC-D were as previously described (2, 20, 29, 32, 33, 39). GST-vectors encoding RXR, RXRΔAF2, RAR, TR, estrogen receptor alpha (ERα), SRC-1, CBP-1, CBP-5, TBP, and TFIIA, the mammalian expression vectors for Gal4-VP16, p300, SRC-1, TR, RAR, ERα, and RXR, the transfection indicator construct pRSV–β-gal as well as the reporter constructs TREpal-LUC, ERE-LUC, DR5-LUC, and Gal4-TK-LUC were as previously described (29, 32, 39, 57).
Northern and Western blot analyses.
An RNA blot (Clontech, Palo Alto, Calif.) containing 2 μg of poly(A)+ (polyadenylated) RNA from various human tissues was hybridized with a random-primed 32P-labeled DNA probe (encompassing amino acids 125 to 237 of ASC-1) and exposed on X-ray film, as described (2). Western analyses were done as previously described (2).
GST pull-down assays.
Approximately 2 to 4 μg of the GST fusions or GST alone expressed in Escherichia coli was bound to glutathione-Sepharose-4B beads (Pharmacia, Piscataway, N.J.), and incubated with labeled proteins expressed by in vitro translation by using the TNT-coupled transcription-translation system, with conditions as recommended by the manufacturer (Promega, Madison, Wis.). Specifically bound proteins were eluted from beads with 40 mM reduced glutathione in 50 mM Tris (pH 8.0) and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography as described (2). Where total nuclear extracts were used, Western analysis was exploited to confirm specific bindings as indicated (see Fig. 4B).
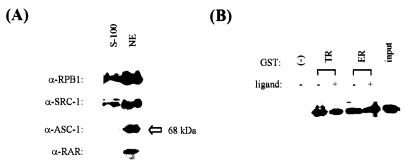
FIG. 4.
Expression of endogenous ASC-1 in HeLa cells. (A) HeLa cells were fractionated into nuclear (NE) and cytosolic (S-100) fractions as described (2) and were probed with indicated antibodies by Western analysis. α-RPB1 and α-SRC-1 are monoclonal antibodies against the RNA polymerase II largest subunit (8WG16) and SRC-1, respectively. (B) HeLa nuclear extracts were incubated with bacterially expressed GST-fusions to TR and ER or GST alone either in the presence or absence of ligand, as indicated. Specifically bound proteins were released by glutathione, resolved by SDS-PAGE, and probed with a rabbit polyclonal antibody against ASC-1 by Western analysis. Approximately 20% of the nuclear extracts used in the binding reactions were loaded as an input.
Yeast two-hybrid test.
For the yeast two-hybrid tests, plasmids encoding LexA fusions and B42 fusions were cotransformed into Saccharomyces cerevisiae EGY48 containing the lacZ reporter plasmid, SH/18-34 (2). Plate and liquid assays of β-gal expression were carried out as described (29). Similar results were obtained in more than two similar experiments.
Immunofluorescence.
Rat-1 fibroblast cells were microinjected with HA-tagged ASC-1 plasmid (25 μg/ml) and fixed at indicated times after serum starvation, followed by indirect immunostaining with anti-HA antibody and FITC-rhodamine-conjugated antibodies as previously described (21). The image was photographed with a Zeiss AxioplanII camera equipped with PIXERA.
Cell culture and transfections.
HeLa or CV-1 cells were grown in 24-well plates with medium supplemented with 10% fetal calf serum for 24 h and were transfected with 100 ng of lacZ expression vector pRSV-β-gal and 100 ng of a luciferase reporter gene, along with various expression vectors. Total amounts of expression vectors were kept constant by adding appropriate amounts of pcDNA3. These cells were incubated, either in the presence or absence of 0.1 μM of ligand, with medium containing 10% fetal calf serum for 36 h. Cells were then harvested, luciferase activity was assayed as described (2), and the results were normalized to the lacZ expression. Similar results were obtained in more than two similar experiments.
RESULTS
Isolation of a full-length segment of cDNA encoding ASC-1.
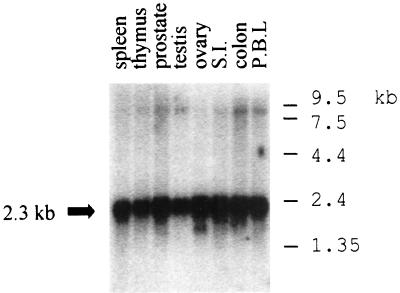
TRIP4 was isolated as a partial clone of a protein that specifically interacts with a series of nuclear receptors including TR and RXR (31). On the basis of its ability to functionally interact with nuclear receptors and other signal-dependent transcription factors (unpublished data), we have renamed this protein ASC-1. ASC-1 is encoded by an approximately 2,300-nucleotide mRNA which is expressed in all the human and mammalian tissues examined (Fig. 1 and results not shown). Extensive screening of a few human cDNA libraries produced a number of full-length cDNA clones in which the initiator methionine is preceded by a few in-frame stop codons. Overall, ASC-1 shows no significant homology to any gene or protein in current databases. However, ASC-1 contains a putative zinc finger motif with an arrangement of metal binding residues similar to those of E1A (14) and a putative regulatory factor VAC1 (51) (i.e., CX2CX12–13CX2CX4C) (Fig. 2A).
FIG. 1.
Expressions of ASC-1. An RNA blot (Clontech) containing 2 μg of poly(A)+ mRNA from the indicated human tissues was hybridized with an ASC-1 probe under standard conditions (2). Equivalent loading was verified by hybridization with an actin cDNA probe (results not shown). S.I., small intestine; P.B.L., peripheral blood leukocyte. Size markers are as indicated.
FIG. 2.
(A) ASC-1 amino acid sequence. Two potential nuclear localization signals are underlined. Cysteine and histidine residues are indicated in boldface (those conserved with E1A [14] and VAC1 [51] are underlined). The positions of the deletions in ASCΔN and ASCΔNΔC are indicated by arrows. (B) Schematic diagram of ASC-1 and its deletion mutants. The putative zinc finger domain is as indicated. ASCΔN consists of the ASC-1 residues 125 to 581, and ASCΔNΔC includes only the putative zinc finger domain (residues 125 to 237). The zinc finger region is specifically deleted in ASCdn (i.e., the ASC-1 residues 165 to 216).
ASC-1 contains a distinct autonomous transactivation domain.
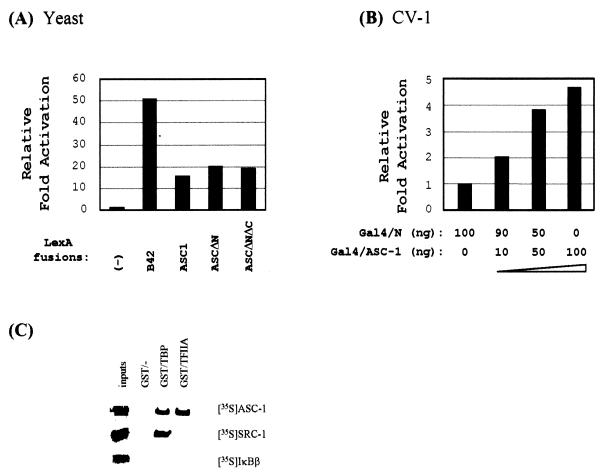
Previous results in yeast demonstrated that the originally isolated TRIP4 clone, referred to here as ASCΔN (Fig. 2B), was able to activate transcription when fused to LexA (31). Since a similar transactivation was also observed with LexA fusions to a full-length ASC-1 and ASCΔNΔC, which consists only of the putative zinc finger domain (residues 125 to 237), this transactivation function maps to the zinc finger region (Fig. 3A). Similarly, GAL4 fusions to a full-length ASC-1 and ASCΔNΔC stimulated, in a dose-dependent manner, expression of the GAL4-LUC reporter construct (13) in mammalian cells (Fig. 3B and results not shown). This autonomous transactivation function associated with ASC-1, particularly in yeast, where nuclear receptors do not exist, led us to examine if ASC-1 directly associates with basal transcription factors TBP and TFIIA (reviewed in reference 4). Indeed, ASC-1 readily interacted with both proteins in vitro, whereas SRC-1 interacted only with TBP (Fig. 3C). In contrast, IκBβ did not interact with either protein. Similar results were obtained with ASCΔNΔC (results not shown), indicating that the zinc finger region is sufficient for these interactions.
FIG. 3.
Autonomous transactivation function of ASC-1. (A) The yeast one-hybrid results. The indicated LexA-plasmids were transformed into a yeast strain containing an appropriate lacZ reporter gene, as described (29). The results are expressed as induction fold (n-fold) over the value obtained with LexA/−, which was given an arbitrary value of 1. The data are representative of at least two similar experiments and the standard deviations were less than 5%. (B) CV-1 cells were transfected with lacZ expression vector and increasing amounts of Gal4–ASC-1-expression vector, along with a reporter construct Gal4-TK-LUC, as described. Similar results were also obtained with HeLa cells and Gal4-ASCΔNΔC (results not shown). The results are expressed as n-fold over the value obtained with 100 ng of Gal4/N, which was given an arbitrary value of 1. The data are representative of at least two similar experiments, and the standard deviations were less than 5%. (C) In vitro interaction of ASC-1 with basal factors. Bacterially produced GST-TBP, GST-TFIIA, or GST alone was bound to a glutathione-agarose column and incubated with equivalent amounts of the indicated 35S-labeled ASC-1, SRC-1, or IκBβ produced by in vitro translation, as described (2). Specifically bound proteins were released by glutathione and resolved by SDS-PAGE. Approximately 20% of the labeled proteins used in the binding reactions were loaded as inputs.
ASC-1 binds the hinge domain of nuclear receptors.
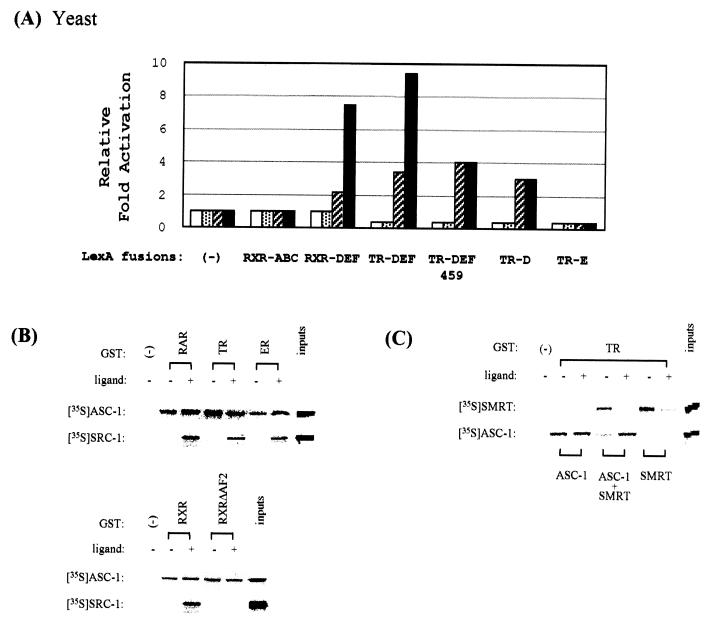
Western analysis revealed that endogenous ASC-1 in HeLa cells is, as expected, a predominantly nuclear protein of approximately 68 kDa (Fig. 4A). In addition, HeLa nuclear extracts specifically retained GST fusions to TR, and ERα contained ASC-1, either in the presence or absence of ligand (Fig. 4B). As previously reported (31), coexpression of the B42-ASCΔN or B42-ASCΔNΔC fusion protein and a LexA fusion to either the TR-DEF or the RXR-DEF domains stimulated LacZ expression in yeast, which became further enhanced in the presence of their ligands, T3 and 9-cis-RA (results not shown). Similar results were also obtained with a B42 fusion to the full-length ASC-1 (Fig. 5A). These results suggested either ligand-dependent interaction between ASC-1 and receptors or ASC-1-mediated transcriptional coactivation of ligand-dependent transactivation in yeast. Since the C-terminal AF2 core helix of the receptors is the target for other ligand-dependent coactivators, we examined whether the zinc finger region of ASC-1 contacts this AF2 core region. However, all of the ASC-1 constructs interacted in a T3-independent manner with LexA–TR-DEF459, a previously described mutant TR in the AF2 core region (29). In addition, ASC-1 didn’t show any interaction with the N-terminal ABC domains of RXR (RXR-ABC) and all of the ASC-1 fusions to B42 specifically bound a LexA fusion including only the hinge region of TR (LexA–TR-D), but did not bind a LexA fusion to the TR-LBD (LexA–TR-E). Overall, these results suggest that ligand-independent contacts between the ASC-1 zinc finger domain and the receptor hinge domain are a major component of their interaction.
FIG. 5.
ASC-1 interacts with nuclear receptors. (A) The indicated B42 and LexA plasmids were transformed into a yeast strain containing an appropriate lacZ reporter gene, as described (29). TR-D and TR-E include the rat TRβ1 residues 164 to 265 (the hinge domain) and 260 to 444 (the LBD), respectively. Open and stippled boxes indicate the presence of B42 alone, whereas hatched and closed boxes indicate the presence of B42–ASC-1. Stippled and closed boxes include 1 μM T3 or 9-cis-RA. The results are expressed as induction fold (n-fold) over the value obtained with LexA/− and B42 alone, which was given an arbitrary value of 1. The data are representative of at least two similar experiments and the standard deviations were less than 5%. Similar results were also obtained with ASCΔN and ASCΔNΔC. (B) In vitro interaction of ASC-1 with nuclear receptors. Bacterially produced GST alone or GST fusions to RARα, TRβ, ERα, RXR, and RXRΔAF2 (57) were bound to a glutathione-agarose column and incubated with equivalent amounts of the indicated 35S-labeled ASC-1 or SRC-1 produced by in vitro translation, as described (2). +, presence of 0.1 μM of each cognate ligand. Approximately 20% of the labeled proteins used in the binding reactions were loaded as inputs. (C) Ligand-dependent interaction of ASC-1 with TR in the presence of SMRT (20). GST alone or GST-TR bound to a glutathione-agarose column was incubated with ASC-1 or SMRT labeled by in vitro translation. +, presence of 0.1 μM T3. Approximately 20% of the labeled proteins used in the binding reactions were loaded as inputs.
These results were further confirmed by direct in vitro binding. As shown in Fig. 5B, ASC-1 interacted in a ligand-independent manner with GST fusions to RXR, RAR, TR, and ERα, but did not bind luciferase and other control proteins (results not shown). Similar to the yeast results with LexA–TR-DEF459, ASC-1 interacted with RXRΔAF2, a recently described AF2 mutant (57), in a 9-cis-RA-independent fashion. Similar results were also obtained with ASCΔNΔC (results not shown). However, SRC-1 bound only the wild-type RXR as well as RAR, TR, and ERα in a ligand-dependent manner, as expected (42). N-CoR (8) and SMRT (20) bind to the hinge regions of unliganded receptors that appear to overlap the binding site for ASC-1. Indeed, ASC-1 interacted with TR in a ligand-dependent manner when SMRT was added in the in vitro binding reactions (Fig. 5C), probably due to competitive bindings between ASC-1 and SMRT, since SMRT interacted only with unliganded TR as previously described (20). These results strongly suggest that the receptor–ASC-1 interactions are ligand-dependent in vivo, as these corepressor molecules are ubiquitously expressed in most tissues (8, 20).
ASC-1 binds to integrators SRC-1 and CBP-p300.
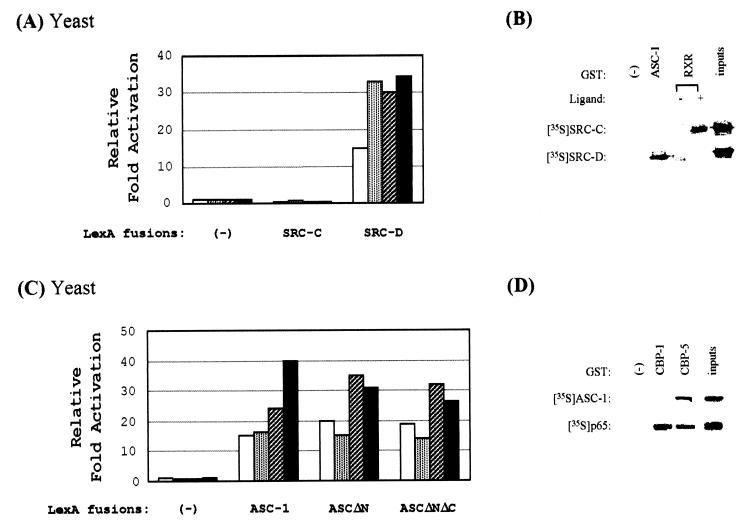
Surprisingly, ASC-1 was also found to associate with transcription integrator SRC-1 (26, 32, 39, 48) in yeast (Fig. 6A). In particular, the interaction was mapped to a subregion of SRC-1 (i.e., SRC-D, residues 759 to 1141), encompassing the previously defined CBP-p300 binding domain (18, 23, 42, 54). Consistent with these yeast results, ASC-1 also interacted with SRC-D (residues 759 to 1141) but not with SRC-C (residues 568 to 779) in vitro, whereas liganded RXR interacted only with SRC-C (Fig. 6B), as previously described (42). In addition, ASC-1 associated with transcription integrator CBP-p300 (16) in yeast (Fig. 6C). ASC-1 interacted with p300-C (the p300 residues 2041 to 2157) and CBP-C (the CBP residues 1868 to 2441) but not with p300-N (the p300 residues 1 to 117), localizing the interaction domain to a subregion of CBP-p300 that includes the previously defined SRC-1 binding sites (i.e., the p300 residues 2041 to 2157) (18, 23, 42, 54). These yeast results were also confirmed by direct in vitro bindings, in which ASC-1 interacted with CBP-5 (residues 1891 to 2441) but not with CBP-1 (residues 1 to 450), whereas p65 interacted with both (Fig. 6D), as previously reported (15). Similar results were also obtained with ASCΔN and ASCΔNΔC (results not shown), indicating that the zinc finger region is sufficient for all of these interactions.
FIG. 6.
ASC-1 interacts with SRC-1–CBP-p300. (A) The indicated B42 and LexA plasmids were transformed into a yeast strain containing an appropriate lacZ reporter gene, as described (29). SRC-C and SRC-D include the SRC-1 residues 568 to 779 and 759 to 1141, respectively. Open boxes indicate B42 alone, whereas stippled, hatched, and closed boxes indicate the presence of B42 fusions to ASC-1, ASCΔN, and ASCΔNΔC, respectively. The results are expressed as induction fold (n-fold) over the value obtained with LexA/− and B42 alone, which was given an arbitrary value of 1. The data are representative of at least two similar experiments and the standard deviations were less than 5%. (B) GST alone, GST-ASC-1, or GST-RXR bound to a glutathione-agarose column was incubated with SRC-C or SRC-D labeled by in vitro translation. +, presence of 0.1 μM 9-cis-RA. Approximately 20% of the labeled proteins used in the binding reactions were loaded as inputs. (C) The indicated B42 and LexA plasmids were transformed into a yeast strain containing an appropriate lacZ reporter gene, as described (29). CBP-C includes the CBP residues 1868 to 2441, whereas p300-N and p300-C include the p300 residues 1 to 117 and 2041 to 2157, respectively. Open boxes indicate B42 alone, whereas stippled, hatched, and closed boxes indicate the presence of B42 fusions to p300-N, p300-C, and CBP-C, respectively. The results are expressed as n-fold over the value obtained with LexA/− and B42 alone, which was given an arbitrary value of 1. The data are representative of at least two similar experiments and the standard deviations were less than 5%. (D) GST alone, GST-CBP-1, or GST-CBP-5 bound to a glutathione-agarose column was incubated with ASC-1 or p65 labeled by in vitro translation. Approximately 20% of the labeled proteins used in the binding reactions were loaded as inputs.
The zinc finger region of ASC-1 forms a ternary complex with CBP, SRC-1, and RXR.
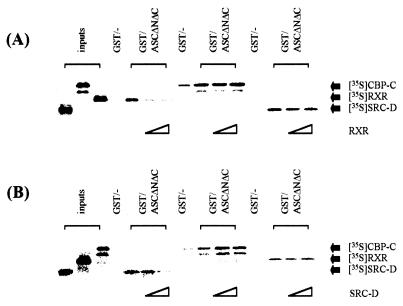
It was surprising that a relatively small region of the ASC-1 (residues 125 to 237) encompassing the putative zinc finger motif was found to be responsible for interactions with receptors, SRC-1, CBP-p300, and basal factors. In addition, ASC-1 interacted with a region in SRC-1 that encompasses the CBP-p300-interaction domain (54). Similarly, ASC-1 interacted with a region in CBP-p300 that was previously shown to contain the SRC-1 binding site (54). Thus, we examined whether the zinc finger region of ASC-1 is able to form a ternary complex with these proteins. CBP-C (the CBP residues 1868 to 2441) contains the SRC-1 binding domain, whereas SRC-D (the SRC-1 residues 759 to 1141) includes the CBP-p300 binding domain. However, CBP-C and SRC-D do not include the domains interacting with nuclear receptors. As shown in Fig. 7, radiolabeled CBP-C, a full-length RXR, and SRC-D efficiently bound to a GST fusion to ASCΔNΔC but not GST alone in vitro. Inclusion of increasing amounts of unlabeled RXR disrupted bindings of radiolabeled RXR and ASCΔNΔC, as expected, but not the ASCΔNΔC-CBP-C and ASCΔNΔC-SRC-D interactions (Fig. 7A). These results demonstrate that the zinc finger region of ASC-1 (i.e., ASCΔNΔC) and RXR can form a ternary complex with either CBP-C or SRC-D. Similarly, ASCΔNΔC and SRC-D were found to form a ternary complex with CBP-C or RXR (Fig. 7B). In particular, it was notable that the CBP-C-ASCΔNΔC interactions became further stimulated in the presence of SRC-D. From these results, we concluded that the zinc finger region of ASC-1 is capable of forming a ternary complex with CBP, SRC-1, and RXR.
FIG. 7.
ASC-1 forms a ternary complex with RXR, CBP, and SRC-1. GST alone or GST-ASCΔNΔC bound to a glutathione-agarose column was incubated with CBP-C, RXR, or SRC-D labeled by in vitro translation. Increasing amounts of RXR (A) or SRC-D (B), in vitro translated in the absence of [35S]methionine, was added to the binding reaction as a competitor, as indicated. Approximately 20% of the labeled proteins used in the binding reactions were loaded as inputs.
ASC-1 relocates between nucleus and cytosol, depending on different cellular conditions.
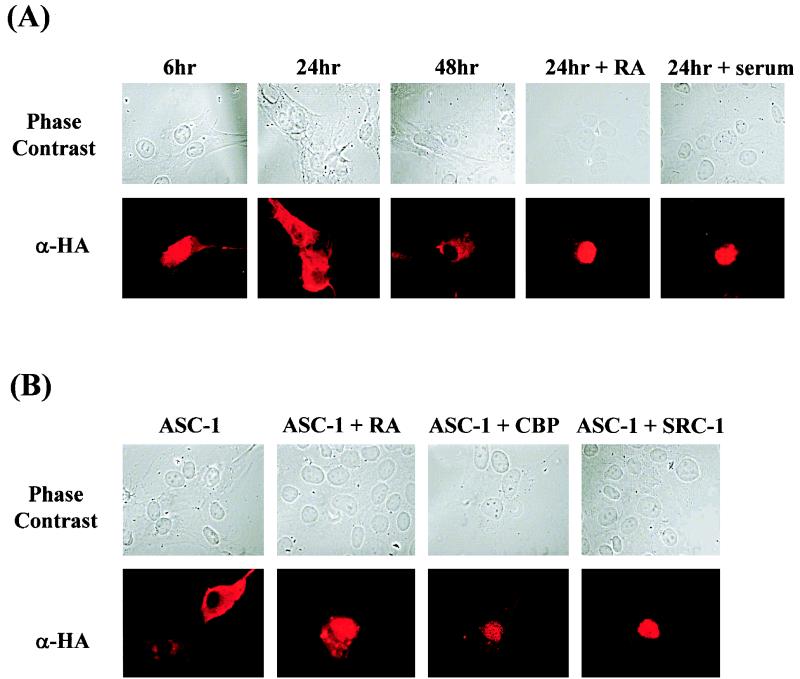
The HA-tagged, overexpressed ASC-1 was normally found exclusively in the nucleus, as expected (results not shown). In contrast, it accumulated in the cytoplasm in a time-dependent manner when deprived of serum (Fig. 8A). ASC-1 was found throughout the cytoplasm and nucleus after 6 h of serum starvation, whereas it was found mostly in the cytoplasm after 24 to 48 h of starvation. Interestingly, when 9-cis-RA or serum was added after 24 h of serum starvation, ASC-1 was found exclusively in nucleus 24 h later (Fig. 8A). Similarly, the cytoplasmic accumulation was not observed when cells were serum starved for 24 h in the presence of ligand or coexpressed CBP or SRC-1, resulting in nuclear localization of ASC-1 (Fig. 8B). Similar results were also obtained with charcoal-stripped serum (results not shown). Taken together, these results indicate that localization of ASC-1 is tightly regulated under different cellular conditions, and ASC-1 can directly associate with CBP, SRC-1, and liganded receptors in vivo.
FIG. 8.
Association of ASC-1 with SRC-1, CBP, and liganded receptor in vivo. (A) Time-dependent accumulation of ASC-1 to cytoplasm in serum-starved cells. Rat-1 fibroblast cells were microinjected with HA-tagged ASC-1 plasmid (25 μg/ml) and fixed at indicated times during serum starvation, followed by indirect immunostaining with anti-HA antibody and FITC-rhodamine-conjugated antibodies as previously described (21). Serum (10%) or 0.1 μM 9-cis-RA was added to the media following 24 h of serum starvation and was observed 24 h later by indirect immunostaining, as indicated. The image was photographed with a Zeiss AxioplanII camera equipped with PIXERA. The picture is representative of approximately 50 injected cells which gave similar results. Similar results were also obtained with HeLa cells (results not shown). (B) Inhibition of cytoplasmic accumulation of ASC-1 by ligand, SRC-1, or CBP in serum-starved cells. Rat-1 fibroblast cells were microinjected with either HA-tagged ASC-1 (25 μg/ml) alone, HA-tagged ASC-1 plus SRC-1 (25 μg/ml each), or HA-tagged ASC-1 plus CBP (25 μg/ml each), were serum starved for 24 h either in the presence or absence of 0.1 μM 9-cis-RA as indicated, and were processed for immunostaining. Similar results were also obtained with HeLa cells (results not shown).
ASC-1 as a novel transcription coactivator of nuclear receptors.
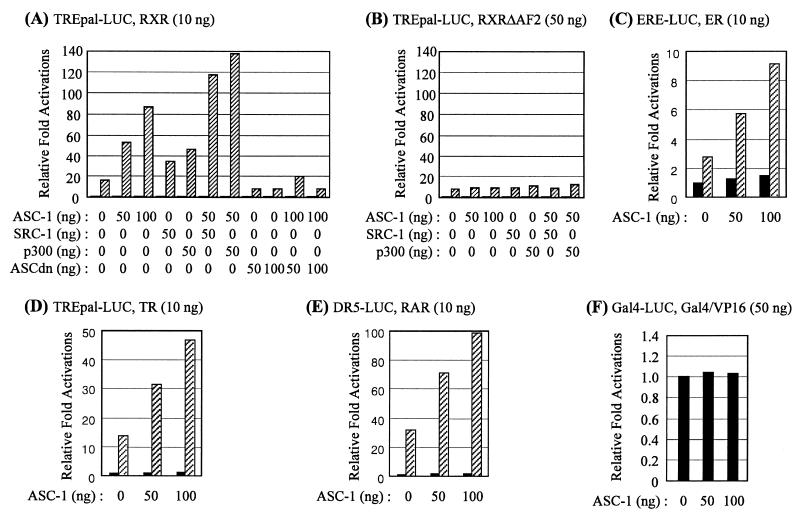
To assess the functional consequences of these interactions, ASC-1 was cotransfected into CV-1 cells along with a reporter construct, TREpal-LUC. Increasing amounts of cotransfected ASC-1 enhanced the 9-cis-RA-dependent activation of this reporter in a dose-dependent manner, with cotransfection of 100 ng of ASC-1 increasing the activation approximately fivefold. Consistent with the specific interactions of ASC-1 with SRC-1 and CBP, the ASC-1-enhanced level of the reporter gene expression was further stimulated by cotransfected SRC-1 or by the CBP homologue p300 (Fig. 9A). A mutant ASC-1 deleted for the zinc finger domain (ASCdn) showed a strong dominant-negative phenotype; coexpression of ASCdn impaired the 9-cis-RA-dependent activation of the reporter, either in the presence or absence of additional wild-type ASC-1. Similar dominant-negative phenotypes were also observed with ASCΔNΔC (results not shown). These results strongly attest to the importance of ASC-1 in the nuclear receptor transactivation function in vivo. Interestingly, ASC-1 was not able to coactivate the RXRΔAF-driven transactivations (Fig. 9B), indicating that ASC-1 has to function in conjunction with AF2-dependent coactivators, such as CBP and SRC-1. Similar results were also obtained with a series of different reporter constructs responsive to ERα, TR, RAR, or RXR in various cell types (Fig. 9C, D, and E and results not shown). In contrast, the basal expression of the reporter in the absence of ligand remained relatively constant with increasing amount of cotransfected ASC-1. Similarly, ASC-1 did not affect expression of the control plasmids pRSV–β-gal or TK-LUC (results not shown), or GAL4-VP16-mediated transactivation of the GAL4-LUC reporter construct (13) (Fig. 9F). These results demonstrated that ASC-1 is a bona fide transcription coactivator molecule of nuclear receptors.
FIG. 9.
ASC-1 potentiates gene expressions mediated by nuclear receptors. HeLa cells were transfected with β-gal expression vector and increasing amounts of ASC-1-expression vector, either in the absence or presence of SRC-1 or p300 expression vector, along with indicated reporter constructs, as described. For specific activation of each reporter construct, 10 ng of RXR (A), 50 ng of RXRΔAF2 (B), 10 ng of ERα (C), 10 ng of TRβ (D), 10 ng of RARα (E), or 50 ng of Gal4-VP16 (F) was also added. Hatched boxes indicate the presence of 0.1 μM each cognate ligand. The results are expressed as induction fold (n-fold) over the value obtained in the absence of ligand and ASC-1, which was given an arbitrary value of 1. The data are representative of at least two similar experiments, and the standard deviations were less than 5%. Similar results were also obtained with CV-1 cells (results not shown).
DISCUSSION
Transcription coactivators (reviewed in reference 45) can function not only to transmit the signal of ligand-induced conformational change to the basal transcription machinery but also to modulate chromatin structure. In this report, we have described the initial characterization of ASC-1, which shows various properties consistent with its role as a novel transcription coactivator molecule of nuclear receptors. These include associations with nuclear receptors (Fig. 4 and 5), the basal transcription machinery (Fig. 3), and transcription integrators CBP and SRC-1 (Fig. 6), an autonomous transactivation function (Fig. 3), and coactivation of transactivation mediated by nuclear receptors (Fig. 9).
The interactions of ASC-1 with nuclear receptors are ligand independent in vitro (Fig. 5B), whereas inclusion of SMRT (20) in the binding reactions resulted in ligand-dependent interactions of ASC-1 with receptors (Fig. 5C). The hinge domain of nuclear receptor is a major determinant in interactions with ASC-1 (Fig. 5A) as well as SMRT (20). Since SMRT is released from liganded nuclear receptors and ASC-1 is still able to bind to liganded receptors, the receptor–ASC-1 bindings should become ligand dependent in vivo, where SMRT is ubiquitously expressed. The indirect immunofluorescence results are also consistent with this notion (Fig. 8). However, it was intriguing that the relatively strong basal interactions of ASC-1 with receptors were further stimulated by ligand in yeast, whereas the interactions of ASC-1 with TR-DEF459, a previously described AF2 mutant TR (29), were not. These results may have been caused simply by ligand-induced stabilization of receptor conformation to facilitate the ASC-1–receptor interactions in yeast. Alternatively, proteins functionally homologous to SMRT, an AF2-dependent coactivator such as SRC-1–CBP-p300, or both may exist in yeast, and ASC-1 may cooperate with these putative yeast proteins to mediate the ligand- and AF2-dependent coactivation of nuclear receptors in yeast. In this regard, it is notable that our database search revealed the highly conserved ASC-1 homologues in the higher eukaryotes Caenorhabditis elegans and Schizosaccharomyces pombe as well as a relatively distant ASC-1 homologue in S. cerevisiae (results not shown).
Recent biochemical studies suggest that transcription coactivators function as a complex with other related proteins (45). Thus far, several groups of such distinct macromolecular complexes have been described. The TAF components of TFIID (reviewed in reference 4) and the SRB-MED components bound to polymerase II (reviewed in reference 38) comprise those that are ultimately associated with the general transcription machinery. B-cell-specific OCA-B (35), a group of distinct nuclear proteins named thyroid hormone receptor associated proteins (TRAPs) (12), and a transcriptionally active nuclear complex that interacts only with liganded vitamin D receptor (VDR) (DRIPs) (43) define a group of coactivator complexes with rather specific functions. TRAPs purified from HeLa cells grown in the presence of thyroid hormone (T3) were found to markedly activate transcription by liganded TR in vitro, whereas DRIPs consisting of a complex of at least 10 different proteins ranging from 65 to 250 kDa were found to coactivate the VDR-dependent transactivation. The DRIPs were distinct from the CBP-p300 complex, although like these coactivators, their interaction also required the AF2 transactivation motif of VDR (43). Finally, the CBP-p300 coactivator complex defines a distinct coactivator complex that directly binds and coactivates a wide spectrum of different transcription factors. In particular, CBP-p300 was found to be complexed with SRC-1 (18, 23, 42, 54) and a series of cellular proteins with molecular masses ranging from 44 to 270 kDa (11). Purification and analysis of various proteins in this group revealed that they are components of the human SWI-SNF complex and that p270 is an integral member of this complex. In addition, different classes of mammalian transcription factors—nuclear receptors, CREB, and STATs—were recently shown to functionally require distinct components of the CBP-p300 coactivator complex, based on their platform or assembly properties (27). RAR, CREB, and STATs were further demonstrated to require different histone acetyltransferase activities within the CBP-p300 complex to activate transcription. Recently, p300 and CBP, despite their similarities, have been shown to have distinct functions during retinoid-induced differentiation of embryonic carcinoma F9 cells (24). Overall, these results suggest that distinct coactivator complexes appear to exist among CBP-p300-containing coactivator complexes in the cell, which should be responsive to distinct activating signals and differentially integrate various signaling pathways.
We have presented a few pieces of experimental evidence that support direct associations of ASC-1 with SRC-1 and CBP-p300. First, ASC-1 was shown to physically bind CBP-p300 and SRC-1, as demonstrated by the yeast two-hybrid tests and the GST pull-down assays (Fig. 6). Second, ASC-1 was found to colocalize, at least under serum-starved conditions, with CBP and SRC-1 in vivo, as demonstrated by the indirect immunofluorescence of Rat-1 fibroblast cells (Fig. 8). Finally, while the ASC-1 interaction domain does not include the AF-2 domain of nuclear receptors (Fig. 5), ASC-1 was shown to cooperate with SRC-1 and p300 in coactivating transactivation by nuclear receptors (Fig. 9A). In addition, ASC-1 was not able to coactivate the mutant RXR that lacks the AF2 domain (i.e., RXRΔAF2) (Fig. 9B), clearly indicating that ASC-1 has to function in conjunction with AF2-dependent factors such as CBP-p300 and SRC-1. These results suggest that ASC-1 may represent an active member of the CBP-p300 complexes, along with SRC-1 and related proteins. Alternatively, it may represent a constituent of distinct coactivator complexes that, in turn, functionally interact with the CBP–SRC-1 complexes. Consistent with the latter possibility, it was recently shown that CBP, p300, and SRC-1 exist as distinct steady-state coactivator complexes in vivo (37). In addition, we have also isolated a novel complex of proteins from HeLa nuclei based on their tight association with ASC-1 which exhibited a fractionation profile distinct from that of CBP, p300, or SRC-1 and didn’t appear to contain any of these proteins (unpublished results).
One of the most striking features of ASC-1 was its interesting relocation property (Fig. 8). When deprived of serum, ASC-1 accumulated in cytoplasm, while it was exclusively nuclear in the presence of ligand or coexpressed CBP or SRC-1. However, it is not known whether this cytoplasmic accumulation under conditions of serum deprivation is due to the inability of newly synthesized ASC-1 to enter the nucleus or active relocation of the existing nuclear ASC-1 to the cytoplasm. Nonetheless, these relocation properties raise an interesting possibility that ASC-1 may play a critical role in establishing distinct coactivator complexes under different cellular conditions (as summarized in Fig. 10). When deprived of serum, for instance, coactivator complexes devoid of ASC-1 may predominate within the nucleus (due to the cytoplasmic accumulation of ASC-1), which may preferentially transactivate a set of transcription factors activated by serum starvation while shutting down general transcription activities. Candidate genes potentially regulated by these non-ASC-1-containing coactivator complexes include those specifically expressed at growth arrest (10, 22, 44). It is also notable that ASC-1 is likely to function with transcription factors other than nuclear receptors, based on its associations with SRC-1 and CBP, which in turn functionally interact with transcription factors in diverse signaling pathways. Indeed, we have found that ASC-1 is required for transactivation by multiple transcription factors, including AP-1 and NF-κB (unpublished data).
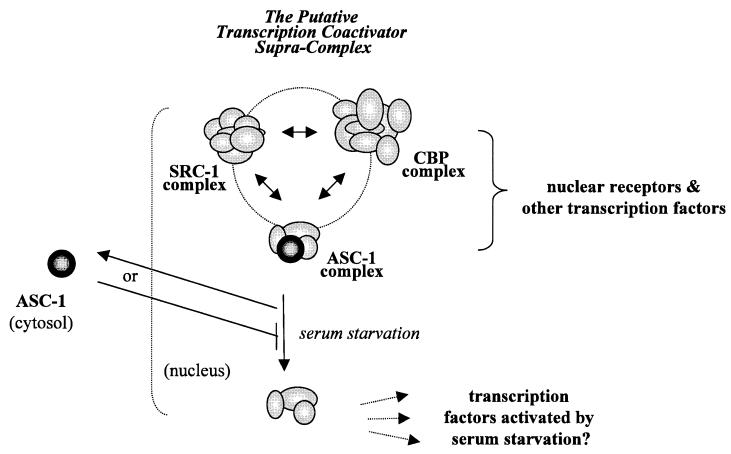
FIG. 10.
Model of ASC-1 actions. Distinct putative coactivator complexes, each containing ASC-1, SRC-1, or CBP, exist in vivo (37 and unpublished results) and mediate transactivations by a number of distinct transcription factors. Each of these complexes recognizes a discrete part or subunit of target transcription factors and may cooperate with each other for efficient coactivation. Alternatively, these distinct coactivator complexes may form a larger supracomplex, as suggested by direct bindings among SRC-1, CBP, and ASC-1. Serum starvation may lead to enrichment of non-ASC-1-containing coactivator complexes within the nucleus, which may preferentially regulate genes turned on under serum-deprived conditions.
In conclusion, we have described a novel coactivator molecule capable of associating with liganded receptors, SRC-1, and CBP in vivo and which may play a critical role in integrating different cellular conditions into the transcription machinery. Surprisingly, all of the functions of ASC-1 described in this report appear to require only the putative zinc-binding domain, whereas the actions of the much larger CBP, p300, and SRC-1 proteins are dependent on distinct interaction domains for their various targets. However, it should be noted that only some of these interactions are likely to exist within the ASC-1-containing complex. Finally, it’s notable that ASC-1 contains multiple phosphorylation sites, potentially responsive to various signals, suggesting that ASC-1 may directly respond to various cellular regulatory signals. Overall, studies of this coactivator protein should provide important insights into the multifactorial control of biological processes under regulation of multiple signal transduction pathways in vivo.
ACKNOWLEDGMENTS
We thank Ming Tsai, J. K. Chung, David Livingston, and Ronald Evans for plasmids and antibodies. We also thank Soo-Kyung Lee for the plasmids encoding LexA–TR-D and LexA–TR-E.
This work was supported in part by grants from the NIH to D.D.M. and the Korean Ministry of Health and Welfare to B.H.J. Y.C.L. and J.W.L. are supported by the National Creative Research Initiatives of the Korean Ministry of Science and Technology.
REFERENCES
- 1.Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X Y, Sauter G, Kallioniemi O P, Trent J M, Meltzer P S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Associates; 1995. [Google Scholar]
- 3.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 4.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 5.Cavailles V, Dauvois S, L’Horset F, Lopez G, Hoare S, Kushner P J, Parker M G. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavailles V, Dauvois S, Danielian P S, Parker M G. Interaction of proteins with transcriptionally active estrogen receptors. Proc Natl Acad Sci USA. 1994;91:10009–10013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Role of CBP/p300 in nuclear receptor signaling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 8.Chen J D, Evans R M. A transcriptional corepressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 9.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 10.Coccia E M, Cicala C, Charlesworth A, Ciccarelli C, Rossi G B, Philipson L, Sorrentino V. Regulation and expression of a growth arrest-specific gene (gas5) during growth, differentiation, and development. Mol Cell Biol. 1992;12:3514–3521. doi: 10.1128/mcb.12.8.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dallas P B, Cheney I W, Liao D W, Bowrin V, Byam W, Pacchione S, Kobayashi R, Yaciuk P, Moran E. p300/CREB binding protein-related protein p270 is a component of mammalian SWI/SNF complexes. Mol Cell Biol. 1998;18:3596–3603. doi: 10.1128/mcb.18.6.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fondell J D, Ge H, Roeder R G. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forman B M, Umesono K, Chen J, Evans R M. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell. 1995;81:541–550. doi: 10.1016/0092-8674(95)90075-6. [DOI] [PubMed] [Google Scholar]
- 14.Geisberg J V, Lee W S, Berk A J, Ricciardi R P. The zinc finger region of the adenovirus E1A transactivating domain complexes with the TATA box binding protein. Proc Natl Acad Sci USA. 1994;91:2488–2492. doi: 10.1073/pnas.91.7.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerritsen M E, Williams A J, Neish A S, Moore S, Shi Y, Collins T. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc Natl Acad Sci USA. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldman P S, Tran V K, Goodman R H. The multifunctional role of the co-activator CBP in transcriptional regulation. Recent Prog Horm Res. 1997;52:103–119. [PubMed] [Google Scholar]
- 17.Halachmi S, Marden E, Martin G, Mackay H, Abbondanza C, Brown M. Estrogen receptor-associated proteins: possible mediators of hormone-induced transcription. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- 18.Hanstein B, Eckner R, DiRenzo J, Halachmi S, Liu H, Searcy B, Kurokawa R, Brown M. p300 is a component of an estrogen receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 20.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass C K, Rosenfeld M G. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor corepressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 21.Jhun B H, Rose D W, Seely B L, Rameh L, Cantley L, Saltiel A R, Olefsky J M. Microinjection of the SH2 domain of the 85-kilodalton subunit of phosphatidylinositol 3-kinase inhibits insulin-induced DNA synthesis and c-fos expression. Mol Cell Biol. 1994;14:7466–7475. doi: 10.1128/mcb.14.11.7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ju Y T, Chang A C Y, She B R, Tsaur M L, Hwang H M, Chao C C K, Cohen S N, Lin-Chao S. gas7: a gene expressed preferentially in growth-arrested fibroblasts and terminally differentiated purkinje neurons affects neurite formation. Proc Natl Acad Sci USA. 1998;95:11423–11428. doi: 10.1073/pnas.95.19.11423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 24.Kawasaki H, Eckner R, Yao T P, Taira K, Chiu R, Livingston D M, Yokoyama K K. Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9-cell differentiation. Nature. 1998;393:284–289. doi: 10.1038/30538. [DOI] [PubMed] [Google Scholar]
- 25.Kim H-J, Lee S K, Na S-Y, Choi H-S, Lee J W. Molecular cloning of xSRC-3, a novel transcription coactivator from Xenopus, that is related to AIB1, p/CIP, and TIF2. Mol Endocrinol. 1998;12:1038–1047. doi: 10.1210/mend.12.7.0139. [DOI] [PubMed] [Google Scholar]
- 26.Kim H-J, Kim J H, Lee J W. Steroid receptor coactivator-1 interacts with serum response factor and coactivates serum response element-mediated transactivations. J Biol Chem. 1998;273:28564–28567. doi: 10.1074/jbc.273.44.28564. [DOI] [PubMed] [Google Scholar]
- 27.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T M, Glass C K, Rosenfeld M G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 28.Le Douarin B, Zechel C, Garnier J M, Lutz Y, Tora L, Pierrat B, Heery D, Gronemeyer H, Chambon P, Losson R. The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee J W, Moore D D, Heyman R A. A chimeric thyroid hormone receptor constitutively bound to DNA requires retinoid X receptor for hormone-dependent transcriptional activation in yeast. Mol Endocrinol. 1994;8:1245–1252. doi: 10.1210/mend.8.9.7838157. [DOI] [PubMed] [Google Scholar]
- 30.Lee J W, Ryan F, Swaffield J, Johnston S, Moore D D. Interaction of thyroid hormone receptor with a conserved transcriptional mediator. Nature. 1995;373:91–94. doi: 10.1038/374091a0. [DOI] [PubMed] [Google Scholar]
- 31.Lee J W, Choi H S, Gyuris J, Brent R, Moore D D. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol Endocrinol. 1995;9:243–254. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- 32.Lee S-K, Kim H-J, Na S-Y, Kim T-S, Choi H-S, Im S-Y, Lee J W. Steroid receptor coactivator-1 coactivates AP-1-mediated transactivations through interactions with the c-Jun and c-Fos subunits. J Biol Chem. 1998;273:16651–16654. doi: 10.1074/jbc.273.27.16651. [DOI] [PubMed] [Google Scholar]
- 33.Lee S-K, Choi H-S, Song M R, Lee M O, Lee J W. Estrogen receptor, a common interaction partner for a subset of nuclear receptors. Mol Endocrinol. 1998;12:1184–1192. doi: 10.1210/mend.12.8.0146. [DOI] [PubMed] [Google Scholar]
- 34.Li H, Gomes P J, Chen J D. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc Natl Acad Sci USA. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo Y, Fujii H, Gerster T, Roeder R G. A novel B cell-derived coactivator potentiates the activation of immunoglobulin promoters by octamer-binding transcription factors. Cell. 1992;71:231–241. doi: 10.1016/0092-8674(92)90352-d. [DOI] [PubMed] [Google Scholar]
- 36.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKenna N J, Nawaz Z, Tsai S Y, Tsai M-J, O’Malley B W. Distinct steady-state nuclear receptor coregulator complexes exist in vivo. Proc Natl Acad Sci USA. 1998;95:11697–11702. doi: 10.1073/pnas.95.20.11697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myers L C, Gustafsson C M, Bushnell D A, Lui M, Erdjument-Bromage H, Tempst P, Kornberg R D. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 1998;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Na S-Y, Lee S-K, Han S-J, Choi H-S, Im S-Y, Lee J W. Steroid receptor coactivator-1 interacts with the p50 subunit and coactivates NFκB-mediated transactivations. J Biol Chem. 1998;273:10831–10834. doi: 10.1074/jbc.273.18.10831. [DOI] [PubMed] [Google Scholar]
- 40.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 41.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 42.Onate S A, Tsai S Y, Tsai M J, O’Malley B M. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 43.Rachez C, Suldan Z, Ward J, Chang C P, Burakov D, Erdjument-Bromage H, Tempst P, Freedman L P. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 1998;12:1787–1800. doi: 10.1101/gad.12.12.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider C, King R M, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54:787–793. doi: 10.1016/s0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- 45.Shibata H, Spencer T E, Onate S A, Jenster G, Tsai S Y, Tsai M J, O’Malley B W. Role of co-activators and co-repressors in the mechanism of steroid/thyroid receptor action. Recent Prog Horm Res. 1997;52:141–164. [PubMed] [Google Scholar]
- 46.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O’Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 47.Takeshita A, Cardona G R, Koibuchi N, Suen C S, Chin W W. TRAM-1, a novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J Biol Chem. 1997;272:27629–27634. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- 48.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional coactivator p/CIP binds CBP and mediates nuclear receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 49.Truss M, Bartsch J, Schelbert A, Hache R J G, Beato M. Hormone induces binding of receptors and transcription factors to a rearranged nucleosome on the MMTV promoter in vivo. EMBO J. 1995;14:1737–1751. doi: 10.1002/j.1460-2075.1995.tb07163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voegel J J, Heine M J, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 51.Weisman L S, Wickner W. Molecular characterization of VAC1, a gene required for vacuole inheritance and vacuole protein sorting. J Biol Chem. 1992;267:618–623. [PubMed] [Google Scholar]
- 52.Wong J, Shi Y-B, Wolffe A P. A role for nucleosome assembly in both silencing and activation of the Xenopus TRbA gene by the thyroid hormone receptor. Genes Dev. 1995;9:2696–2711. doi: 10.1101/gad.9.21.2696. [DOI] [PubMed] [Google Scholar]
- 53.Yang X-J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 54.Yao T P, Ku G, Zhou N, Scully R, Livingston D M. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc Natl Acad Sci USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeh S, Chang C. Cloning and characterization of a specific coactivator, ARA70, for the androgen receptor in human prostate cells. Proc Natl Acad Sci USA. 1996;93:5517–5521. doi: 10.1073/pnas.93.11.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zaret K S, Yamamoto K R. Reversible and persistent changes in chromatin structure accompany activation of a glucocorticoid dependent enhancer element. Cell. 1984;38:29–38. doi: 10.1016/0092-8674(84)90523-3. [DOI] [PubMed] [Google Scholar]
- 57.Zavacki A M, Lehmann J M, Seol W, Willson T M, Kliewer S A, Moore D D. Activation of the orphan receptor RIP14 by retinoids. Proc Natl Acad Sci USA. 1997;94:7909–7914. doi: 10.1073/pnas.94.15.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]