Abstract
Purpose
To investigate the effect of PLXNA1 variants on the phenotype of patients with autosomal dominant and recessive inheritance patterns and to functionally characterize the zebrafish homologs plxna1a and plxna1b during development.
Methods
We assembled ten patients from seven families with biallelic or de novo PLXNA1 variants. We describe genotype–phenotype correlations, investigated the variants by structural modeling, and used Morpholino knockdown experiments in zebrafish to characterize the embryonic role of plxna1a and plxna1b.
Results
Shared phenotypic features among patients include global developmental delay (9/10), brain anomalies (6/10), and eye anomalies (7/10). Notably, seizures were predominantly reported in patients with monoallelic variants. Structural modeling of missense variants in PLXNA1 suggests distortion in the native protein. Our zebrafish studies enforce an embryonic role of plxna1a and plxna1b in the development of the central nervous system and the eye.
Conclusion
We propose that different biallelic and monoallelic variants in PLXNA1 result in a novel neurodevelopmental syndrome mainly comprising developmental delay, brain, and eye anomalies. We hypothesize that biallelic variants in the extracellular Plexin-A1 domains lead to impaired dimerization or lack of receptor molecules, whereas monoallelic variants in the intracellular Plexin-A1 domains might impair downstream signaling through a dominant-negative effect.
INTRODUCTION
Plexins are a large family of cell surface receptors for the axon guidance molecules semaphorins. Plexin-A1 and its co-receptor Neuropilin-1 (NRP1) bind different classes of semaphorins.1–4 The Plexin cytoplasmic domain contains two segments (C1 and C2) that have sequence similarity to GTPase-activating protein (GAP) and form a functional GAP domain.5 Before semaphorin binding, plexin is an inactive monomer or dimer in which the RapGAP activity is autoinhibited.6 Semaphorin-induced dimerization of the plexin extracellular region promotes formation of the activating dimer of the cytoplasmic region, which converts the GAP domain to the active state through an allosteric mechanism.6–8 The Plexin-A1 GAP domains show dual specificity for Rac and Rap GTPases.9 Plxna1 null mice exhibit different axonal abnormalities (e.g., abnormal proprioceptive neuronal and oligodendrocyte morphology, slight defasciculation of optic chiasm, aberrant crossing of commissural axons, agenesis of corpus callosum [CC], and defects in the olfactory and neuroendocrine reproductive systems).10–13 Additionally, Plxna1 null mice exhibit neuronal abnormalities with rarefied interneurons in developing cortex and a decreased cortical thickness.14 Recently, van der Klaauw et al. implicated rare monoallelic variants in plexins and semaphorins in the expression of severe obesity.15 They found 40 rare variants in 13 plexin and semaphorin genes. Notably, nine variants were associated with neurodevelopmental phenotypes in the respective patients. Previously, three studies reported monoallelic de novo variants in PLXNA1 to be associated with infantile-onset epilepsy, intellectual disability with autism spectrum disorder (ASD), epileptic encephalopathy, or schizophrenia in the respective patients.16–19
Here, we describe four families with rare biallelic and three families with rare/novel monoallelic de novo variants in PLXNA1. The observed clinical phenotypes establish a range of neurological disease associated with presumably pathogenic variant alleles at this locus. Shared phenotypic features comprise global developmental delay, brain and eye anomalies. Seizures were predominantly reported in patients with monoallelic variants. Morpholino knockdown of the zebrafish homologs plxna1a and plxna1b in zebrafish larvae causes anomalies of the central nervous system and the eye as observed in our patients.
MATERIALS AND METHODS
Exome sequencing
Exome sequencing and subsequent analysis was performed by established procedures (see Supplementary information). GeneMatcher20,21 and matchbox22 facilitated the identification of additional patients with biallelic and monoallelic pathogenic variants in PLXNA1.
3D modeling of protein structure
The 3D protein structural models were built using I-Tasser.23 Sequences were trimmed from the N-terminal (1,020 amino acids) for prediction of the respective Plexin-A1 amino acid changes. Structural comparison of variants were done in Chimera after superimposing the structure of mutant onto the wild-type structure using SuperPose (superpose.wishartlab.com). Amino acid conservation was obtained from the Consurf server.24
Zebrafish husbandry and embryo maintenance
Zebrafish were maintained and raised according to national law and recommendations by Westerfield25 in our fish facility in Bonn, Germany. Zebrafish larvae (zfl) of wild-type AB/TL strain and transgenic Tg(-3.1ngn1:GFP)26 were obtained by natural spawning and raised at 28 °C on a 14-hour light–10-hour dark cycle.
Knockdown with morpholino oligonucleotides microinjections and mRNA rescue
The human PLXNA1 gene has two zebrafish orthologs (plxna1a, ENSDARG00000105452; and plxna1b, ENSDARG00000114823).27 Knockdown was performed using specific Morpholino® oligonucleotides (MO) synthetized by GeneTools, LLC. We designed one MO targeting a splice site (splice blocking [SB]) and one MO targeting the AUG translational start site (translational blocking [TB]) for each of the orthologs. In one-cell or two-cell embryos 2.2 ng (1.7 nL/embryo) of plxna1a SB MO (5’-AAGGAGATGCAGATACTTACACACT-’3), 2.9 ng plxna1a TB MO (5’-CCCCTACCATACGGCAGCATTTTTC-’3), 4.4 ng plxna1b SB MO (5’-AGCAGATAATTCTCTTACCGAGATC-’3), 1.5 ng plxna1b TB MO (5’-GCCACATATCTGCACTGGTCCTTGA-’3), or 4.4 ng of standard control MO (5’-CCTCTTACCTCAGTTACAATTTATA-‘3) was injected into the yolk. plxna1b SB MO and plxna1b TB MO were established previously in a model for wound healing but not early embryonic development.28,29
For messenger RNA (mRNA) rescue experiments, 150 pg of in vitro transcribed human PLXNA1 mRNA was co-injected into the yolk of one-cell embryos together with plxna1a SB MO. PLXNA1 mRNA was transcribed from complementary DNA (cDNA) clone HsCD00863277 (Harvard Medical School) containing NM_032242.3 using the mMESSAGE mMACHINE T7 Ultra Kit (Thermo Fisher Scientific). Prior to transcription, the orf of clone HsCD00863277 was changed into the stop codon of NM_032242.3 using the mutagenesis In-Fusion HD Cloning kit (Takara).
RNA isolation and reverse transcription polymerase chain reaction
To test splice-blocking effect of the designed plxna1a SB MO, total RNA was extracted from pools of 20 larvae with TRIzol reagent (Thermo Fisher Scientific). Then, 1 µg of RNA was used for cDNA synthesis with iScript™ Reverse Transcription Supermix (Bio-Rad). Polymerase chain reaction (PCR) was performed with plxna1a forward primer (5’- GATGAAGAAGATCTTGGTGAACT-‘3) and intron-spanning plxna1a reverse primer (5’- AAGAACCAGCTGGACTTCAG-‘3); for control eef1α1 was used as housekeeping gene.30
Imaging and phenotyping
Zfl were phenotyped at 2 days postfertilization (dpf) using a ZEISS Stemi508 for brightfield imaging. The timepoint of 2 dpf was chosen since the phenotype was most prominent. The phenotype category is defined by the presence of at least two of the following features: hydrocephalus, general hypopigmentation, reduced head or eye size. Diameter of head and eyes was measured with NIS-Element Viewer software. To account for variation and growth effects, eye size was calculated as diameter normalized to head.31 Zfl were anesthetized at 2 dpf with 0.03% tricaine (Sigma-Aldrich), fixed in 1.25% low-melting agarose for fluorescence imaging with a ZEISS Axio Zoom.V16 stereo microscope. Phenotypic differences and dorsal root ganglions (DRG) in Tg(-3.1ngn1:GFP) were analyzed with the ZEN 2.3 software. To account for variation in embryo size, DRG were counted in somites cranial of the yolk sac and thereby normalized.
Statistical analyses
Two-tailed Student’s t-test, Mantel–Cox, and two-way analysis of variance (ANOVA) were used for analysis using GraphPad Prism version 6. Survival was analyzed using Kaplan–Meier survival curves.
RESULTS
Biallelic and monoallelic PLXNA1 variants
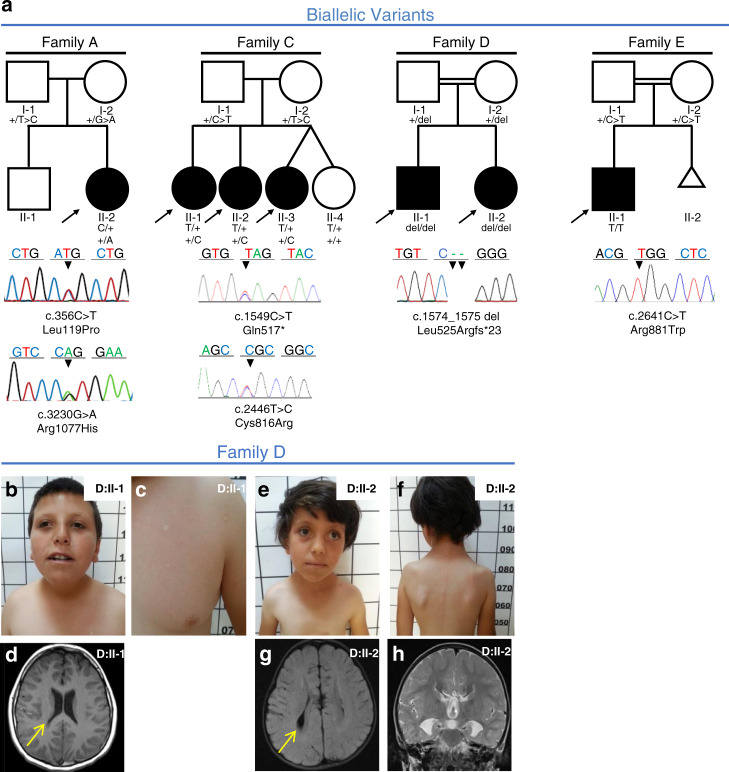
In four families, we identified seven patients with biallelic variants in PLXNA1 segregating with the disease (Fig. 1a). Clinical findings are summarized in Table 1; detailed case reports can be found in the Supplementary information. Six patients showed global developmental delay (6/7) whereas one patient had isolated language regression (1/7). Three patients had cerebral anomalies (3/7). Brain magnetic resonance image (MRI) studies of patient D:II-1 showed dysmorphic ventricular system and prominent Virchow–Robins (perivascular) spaces at the level of the semiovale in both hemispheres. MRI studies of his sister (D:II-2) showed agenesis of the CC and colpocephaly (Fig. 1d, g, h). While the affected patients II-1 and II-3 in family C had unremarkable cerebral MRI studies, their affected sister (C:II-2) showed a dysplastic “mega CC.” Three patients had ASD (3/7), and four had eye anomalies (4/7) comprising optic disc hypoplasia without visual deficits, strabismus, and ptosis in (D:II-1, D:II-2); ptosis in (C:II-2); and nystagmus in (C:II-3). Three patients showed craniofacial dysmorphisms (3/7). Only one patient presented with seizures (1/7); patient A:II-1 had 15 episodes of febrile and nonfebrile seizures between 15 months and 4.5 years of age.
Fig. 1. Families with biallelic PLXNA1 variants.
a Pedigrees and Sanger sequencing results of four families with biallelic variants in PLXNA1. b–h Photographs and brain magnetic resonance image (MRI) of affected siblings of family D. b, c D:II-1 showing mild microphthalmia, depressed nasal bridge, short neck, and hypopigmented stains that were absent in both parents. d T1 axial MRI of D:II-1 showing dysmorphic ventricular system most prominent in posterior horns (arrow). e, f D:II-2 showing strabismus, sparse lateral eyebrows, flattened nasal bridge, large earlobes, and hypopigmented lesions on the torso measuring less than 1 cm. g, h Axial T2 FLAIR MRI of D:II-2 showing dysmorphic ventricular system (arrow in g), and frontal steer horn sign typically seen in corpus callosum (CC) agenesis in the T2 coronal (h).
Table 1.
Clinical features of ten patients with rare and novel biallelic and monoallelic variants in PLXNA1.
| Family ID | Family A | Family C | Family D | Family E | Family F | Family G | Family H | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Patient | II-2 | II-1 | II-2 | II-3 | II-1 | II-2 | II-1 | II-1 | II-1 | II-1 |
| Zygosity | Compound heterozygous | Compound heterozygous | Homozygous | Homozygous | De novo | De novo | De novo | |||
| c.356T>C, p.(Leu119Pro) c.3230G>A, p.(Arg1077His) |
c.1549C>T, p.(Gln517*) c.2446T>C, p.(Cys816Arg) |
c.1574_1575del, p.(Leu525Argfs*23) | c.2641C>T, p.(Arg881Trp) | c.3554G>A, p.(Arg1185Gln) | c.4483C>T, p.(Arg1495Trp) | c.5242C>T, p.(Arg1748Cys) | ||||
| gnomAD MAF% | NR; 0.003% (hom=0) | 0.001% (hom=0); NR | NR | 0.0004% (hom=0) | 0.001% (hom=0) | NR | NR | |||
| GDD | Yes | Yes | Yes | Yes | Yes | Yes | Noa | Yes | Yes | Yes |
| Seizures | 15 episodes of febrile and nonfebrile | - | - | - | - | - | - | Neonatal-onset absence-like | Generalized tonic–clonic | Neonatal-onset atonic |
| Craniofacial anomalies | - | - | Cupped ears, small teeth, microcephaly | - | Sparse lateral eyebrows, depressed nasal bridge, large earlobes, short neck | - | High forehead, hypertelorism, posteriorly rotated ears, smooth philtrum | Unilateral facial palsy, dysmorphic right auricle | Macrocephaly, mild face hypotrophy | |
| Eye anomalies | - | - | Ptosis | Nystagmus | Optic disc hypoplasia, strabismus, mild ptosis | - | Mild ptosis | Optic disc hypoplasia, impaired vision | Enophthalmia | |
| Cerebral anomalies | - | - | Dysplastic “mega” CC | - | Dysmorphic VS, prominent Virchow–Robins spaces | Agenesis of CC, colpocephaly | Several bilateral high signal foci in the subcortical white matter | Normal brain MRI | Enlarged VS, thin CC, brainstem hypoplasia, agenesis of the posterior pituitary | Periventricular leukoencephalopathy, basal ganglia calcifications, subtentorial atrophy |
| Neurologic findings | - | - | Unitateral sensorineural hearing loss, ASD | - | ADHD, ASD | Bilateral peripheral axonal neuropathy | Lower limb hyper-reflexia, ASD | Muscular hypotonia | Bilateral sensorineural hearing loss, vestibule-cochlear nerve agenesis, muscular hypotonia | Spastic paraparesis, pyramidal signs |
| Other | Hypoplastic right kidney | VUR | - | Hypopigmented skin lesions | Elevated CK, hypo- and hyperpigmented skin | IUGR, DCM, joint hypermobility, hand bone and rib anomalies | Dextrocardia, SVC, esophageal atresia | - | ||
ADHD attention deficit hyperactivity disorder, ASD autism spectrum disorder, CC corpus callosum, CK creatine kinase, CSF cerebrospinal fluid, DCM dilated cardiomyopathy, GDD global developmental delay, IUGR intrauterine growth restriction, MAF minor allele frequency, MRI magnetic resonance image, NR not reported, SVC left-sided superior vena cava, VS ventricular system, VUR vesicoureteral reflux.
aIsolated language regression.
In three further patients, we identified monoallelic de novo variants in PLXNA1 (Fig. 2a). Clinical findings are summarized in Table 1; detailed case reports can be found in the Supplementary information. All three patients showed global developmental delay (3/3) and craniofacial dysmorphisms (3/3). Two had hypotonia (2/3) and two had cerebral anomalies (2/3). MRI studies of patient H:II-1 showed periventricular leukoencephalopathy, basal ganglia calcifications, and infratentorial atrophy. MRI studies of patient G:II-1 showed enlarged ventricular system, mild thinning of the CC, delayed myelination, hypoplasia of the brainstem, and agenesis of the posterior pituitary (Fig. 2c, d). All three had eye anomalies (3/3) characterized by enophthalmia (H:II-1), optic disc hypoplasia with impaired vision (G:II-1), and ptosis (F:II-1). Notably, all three patients presented with seizures (3/3) including neonatal-onset atonic seizures (H:II-1), childhood-onset generalized tonic–clonic seizures (G:II-1), and neonatal-onset absence-like seizures (F:II-1).
Fig. 2. Families with monoallelic PLXNA1 variants.
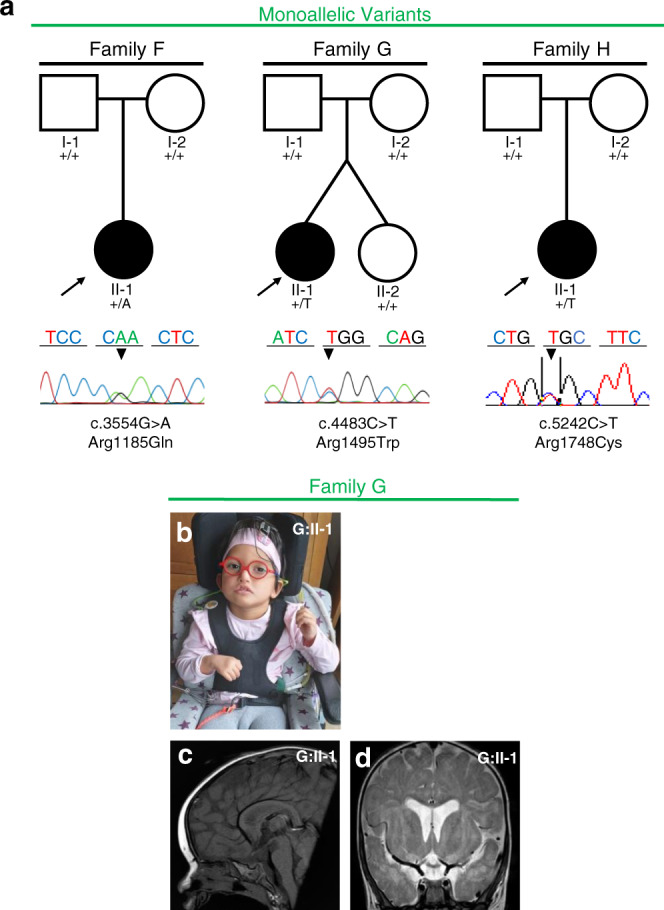
a Pedigrees and Sanger sequencing results of three families with monoallelic de novo variants in PLXNA1. b–d Photographs and brain magnetic resonance image (MRI) of affected patient of family G. b G:II-1 showing unilateral facial palsy, dysmorphic right auricle and bilateral sensorineural hearing loss due to agenesis of vestibulocochlear nerves requiring cochlear implants. c Midsagittal T1 MRI of G:II-1 showing absent posterior pituitary and mild hypoplasia of brainstem. d Coronal T2 MRI of G:II-1 showing mild dilatation of the ventricular system, delayed myelination including the periventricular region.
Besides the above reported patients, we identified four additional patients with biallelic and five additional patients with monoallelic variants in PLXNA1. In all patients, the clinical significance of the identified variants remains uncertain. In one of the patients with biallelic variants (family J) and in three of the patients with monoallelic variants (families M, N, and P), either no parent or only one parent was available for segregation analysis. Patient L:II-3, carrying a rare de novo variant, showed severe muscular hypotonia during the neonatal period prompting exome analysis; however, his hypotonia had resolved spontaneously at six months of age and the family was lost to follow up. Analogous family O was lost to follow up and the outcome of pregnancy of O:II-1 remains unknown. In family B we identified compound heterozygous missense variants; although the inheritance pattern appears plausible, both variants are fairly common with a minor allele frequency (MAF) of 0.1%. Additionally, the phenotype in this patient appears exceptional with the presence of inflammatory changes in the cerebrospinal fluid (CSF) that are not otherwise observed in other subjects herein. For these reasons, these variants have been classified as variants of unknown significance (VUS). A detailed description of all additional patients can be found in the Supplementary information, Figures S1, S2.
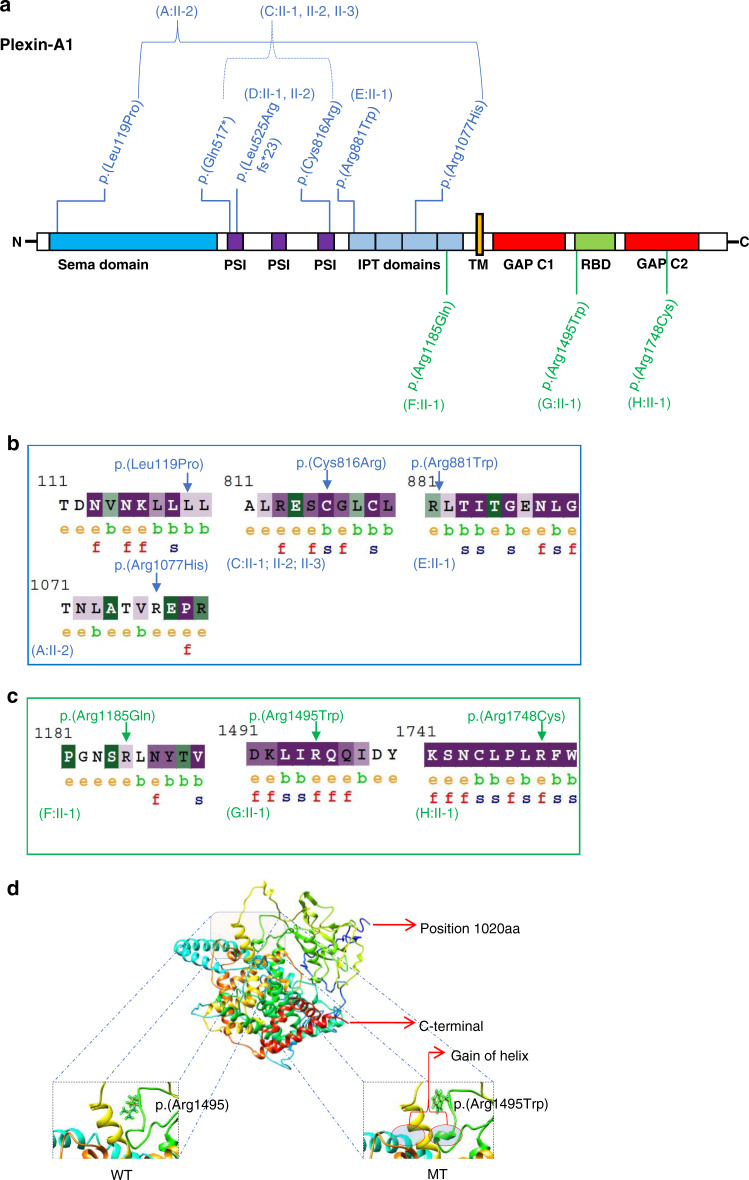
Structural modeling and in silico analysis of Plexin-A1 protein variants
The observed distribution of both biallelic variants and monoallelic variants over the Plexin-A1 domains appears to be nonrandom (Fig. 3). Structural models showed a sequence identity of 83%, coverage of 64% and normalized Z-score of 3.09. Z-score values >1 are considered indicative of correctly folded and good modeled structures and a close approximation of the native structure.32 From the structural modeling of mutated Plexin-A1, we observed that all modeled (n = 4) biallelic and all modeled (n = 3) monoallelic variants likely cause a distortion in the native protein (Figure S3). Superimposition of mutant p.(Arg1495Trp) onto the wild-type structure showed a gain of helix in the mutant protein in close proximity to the variant location (Fig. 3d). The UniProt protein database reports ten putative disulfide bonds in the Plexin-A1 protein. A truncated protein resulting from the p.(Leu525Argfs*23) or the p.(Gln517*) may therefore lack five of those disulfide bonds (515–532; 521–563; 524–541; 535–547; 598–617). In silico analyses using SIFT, CADD, and PolyPhen-2 predicted all monoallelic de novo variants and most biallelic variants to be damaging. ConSurf analysis predicted residues at all positions of the three monoallelic missense variants (p.[Arg1185], p.[Arg1495], and p.[Arg1748]) to be exposed, suggesting that they have a functional role (Fig. 3c). The intracellular residues p.(Arg1495) and p.(Arg1748) are evolutionarily conserved based on their ConSurf analysis suggesting that these residues react highly sensitively if altered.
Fig. 3. Plexin-A1 variant distribution, conservation, and modeling.
a Schematic protein domain structure adapted from St. Clair et al.5 and localization of Plexin-A1 variants. Note the nonrandom concentration of the biallelic variants (blue) in the extracellular domains and the monoallelic variants (green) toward the intracellular domains. GAP GTPase-activating protein, IPT Ig domain shared by plexins and transcription factors, PSI plexin-semaphorin-integrin domain, RBD Rho GTPase-binding domain, TM transmembrane region. b, c ConSurf analysis of the biallelic (b) and monoallelic (c) missense variants. The monoallelic variants (p.[Arg1185], p.[Arg1495], and p.[Arg1748]) are exposed suggesting functional residues in the protein. The intracellular residues p.(Arg1495) and p.(Arg1748) are evolutionarily conserved based on their ConSurf analysis suggesting that these residues react highly sensitively if altered. The biallelic missense variants (p.[Leu119Pro], p.[Cys816Arg], p.[Arg881Trp], p.[Arg1077His]) are noticed to be less conserved compared to the monoallelic variants with the p.(Cys816Arg) variant representing an exemption of this observation. e exposed, b buried, f functional, s structural. d 3D protein structure prediction shows replacement of the arginine long side chain by a flat aromatic ring of tryptophan (p.[Arg1495Trp]). Superimposition of the p.(Arg1495Trp) variant onto the wild-type structure showed that there is a gain of helix in the altered protein in close proximity to the site of variant. MT mutated, WT wild type.
Knockdown of PLXNA1 homologs plxna1a and plxna1b leads to anomalies of zebrafish central nervous system development
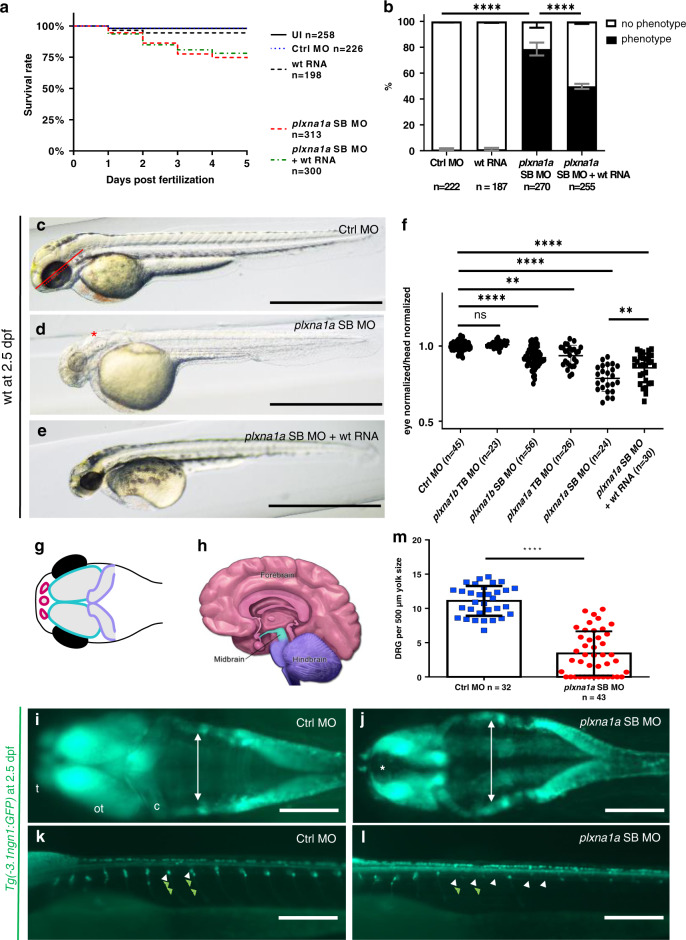
The zebrafish protein Plexin-A1a has a slightly higher amino acid sequence homology with the human Plexin-A1 protein compared to Plexin-A1b (82% vs. 73%, calculated with CLUSTAL_omega from EMBL-EBI). Here, knockdown of plxna1a and plxna1b in developing zebrafish larvae (zfl) with a splice blocking (SB MO) and a translational blocking morpholino (TB MO) for each of the two paralogs resulted in an overlapping phenotype. Since Plexin-A1a shows the higher homology to human Plexin-A1 and since the knockdown with plxna1a SB MO resulted in the most intense phenotype with an only mildly increased mortality (Fig. 4a), we focused on this MO for further analysis. Following the plxna1a knockdown, we observed hydrocephalus in midbrain and hindbrain ventricles, generalized hypopigmentation, reduced head size (Fig. 4b, d), reduced eye diameter (Fig. 4d, f), and slightly increased mortality. This phenotype was observed in approximately 80% of plxna1a SB MO morphants (n = 270) but only in 1% of control MO-injected zfl (n = 222, p < 0.0001 [two-way ANOVA]) at 2 dpf (Fig. 4b). The measured eye size was normalized to head length to account for variation of embryo size.31 This ratio was significantly lower in plxna1a SB MO morphants compared to controls (0.31 ± 0.007 vs. 0.48 ± 0.02, p < 0.0001 [two-way ANOVA] N = 3) (Fig. 4f). Although we observed a reduced head length following the knockdown of plxna1a compared to controls, reduction of eye size was still significant after normalization. Efficiency of knockdown with the plxna1a SB MO was demonstrated by reverse transcription polymerase chain reaction (RT-PCR) with a decrease of wild-type plxna1a expression and presence of an alternative band without exon 5, but no change in eef1α1 expression as control (Figure S5). Similarly, RT-PCR for the plxna1b SB MO confirmed its efficiency (data not shown) as previously demonstrated.29
Fig. 4. Knockdown of plxna1a leads to cerebral anomalies and eye anomalies in zebrafish larvae.
a Quantification of survival (N = 3), zfl injected with plxna1a splice blocking Morpholino (SB MO) show a slight but significant reduction (71% with a p value <0.0001, two-way analysis of variance [ANOVA]) of survival rate at 5 dpf compared to Ctrl MO (95%) and uninjected zfl (UI, 98%). Survival of plxna1a SB MO is not significantly rescued by co-injection of wt plxna1 RNA (p value <0.0001, Mantel–Cox test). b The graph shows 100% of surviving zfl at 2 dpf. 79% of plxna1a SB MO-injected zfl show a central nervous system (CNS) phenotype as hydrocephalus, smaller head and eye size (p value <0.0001, two-way ANOVA, N = 3) compared to 0% of UI and 1% of Ctrl MO-injected. The phenotype of plxna1a SB MO-injected zfl is significantly rescued by co-injection of wild-type (wt) PLXNA1 RNA (50% vs. 79%) (p value <0.0001, two-way ANOVA). Data are presented as means with standard error of the mean (SEM). c–e Brightfield images of zfl injected with Ctrl MO, plxna1a SB MO, or plxna1a SB MO + wt human RNA. Hydrocephalus (asterisk), hypopigmentation, smaller head and eye size are visible. The phenotype of plxna1a SB MO-injected zfl (d) is partially rescued by co-injection of wt PLXNA1 RNA (e). f Eye–head ratio of injected zebrafish larvae at 2 dpf. Measurement of the eye (dotted line) and head (distance between anterior tip up to the otic vesicle) (continuous line) was performed as visualized (c). Injection of plxna1b SB MO, plxna1a TB MO and plxna1a SB MO significantly reduced eye–head ratio (**p value 0.0024 or ****p value <0.0001; ordinary one-way ANOVA, N = 3), while wt RNA injection in plxna1a SB MO-injected zfl significantly rescues the phenotypic effect (p value 0.016). Data are presented as means with standard error of the mean (SEM). g Schematic of the CNS visible in dorsally mounted Tg(-3,1ngn1:GFP) zfl at 2 dpf. Pink: forebrain, turquoise: midbrain, purple: cerebellum (part of hindbrain), black: eyes. h Schematic of adult human brain (adapted from Midbrain. Blausen Medical. Retrieved on 29 February 2016. http://blausen.com/?Topic=9703). Pink: forebrain, turquoise: midbrain, purple: hindbrain. i, j Tg(-3.1ngn1:GFP) zfl are mounted ventral and imaged from dorsal, the anterior to the left. The white arrows mark lateral borders of the hindbrain ventricle and asterisk mark dilatation of the forebrain ventricle (j). plxna1a SB MO-injected zfl show a dilatation of the ventricle at 2 dpf corresponding to the hydrocephalus seen in brightfield images. (j) Note the hypoplasia of telencephalon, mesencephalon, and cerebellum compared to the control (i). c cerebellum, ot optic tectum, t telencephalon. k, l Tg(-3.1ngn1:GFP) zfl are mounted lateral, anterior to the left. plxna1a SB MO-injected zfl have a reduced number of dorsal root ganglions (DRG) (white arrowheads) and corresponding somites lack outgrowing axons (green arrowheads). m Quantification of DRG, normalized to yolk size (Figure S6). In plxna1a SB MO-injected zfl, the number of DRG is significantly reduced (mean of 3.44 ± 0.49 DRG/500 µm, Ctrl MO 11.1 ± 0.38 DRG/500 µm. P value <0.0001, unpaired t-test, N = 3). White scale bars in all figures: 200 µm. Black scale bars 1,000 µm. **p value <0.01 ****p value <0.0001.
The observed phenotypic spectrum follows the previously published spatiotemporal expression pattern from plxna1a/plxna1b in situ hybridization studies.27 We confirmed the same expression pattern performing immunohistochemistry with an antibody that detects both Plexin-A1a and Plexin-A1b due to their high similarity (Figure S4). Co-injection of human PLXNA1 RNA together with the plxna1a SB MO did not result in a reduction of mortality, but it could significantly rescue the morphologic phenotype (Fig. 4e, b, f).
To assess the impact of the plxna1a knockdown on the central nervous system and axonal outgrowth, we used the transgenic Tg(-3.1ngn1:GFP) reporter line, showing GFP expression in pineal gland, dorsal midbrain, hindbrain, Rohon-Beard sensory neurons, and DRG.26 Following the plxna1a SB MO injection into Tg(-3.1ngn1:GFP) zfl, morphants displayed hypoplasia of the telencephalon, mesencephalon, and cerebellum (Fig. 4g, h) as well as dilatation of the ventricles (Fig. 4i, j). Additionally, morphants showed a decrease of migrated DRG cells in the spinal cord above the yolk. The respective somites lack axon outgrowth compared to controls (Fig. 4k–m). To account for variation in embryo size we normalized the DRG count to the yolk sac diameter (Figure S6). Following the plxna1a knockdown the number of DRG at 2 dpf was significantly reduced to 3.4 DRG/500 µm (n = 32) compared to 11.1 DRG/500 µm in controls (n = 43) (p < 0.0001 [unpaired t-test]) (Fig. 4m, Figure S6).
DISCUSSION
Neurodevelopmental disorders (NDDs) display extensive genetic and phenotypic heterogeneity.33 With the implementation of exome sequencing and family-based rare variant analyses, examples of gene-first/genotype-driven approaches to characterize associated phenotypic spectrums have been illustrated for NDDs.34,35 Here, we describe ten patients with NDD ranging from 1.9 years to 42 years from four families with biallelic variants and three families with monoallelic de novo variants in PLXNA1. Biallelic and monoallelic variants lead to a phenotypic spectrum primarily affecting the central nervous system. Shared phenotypic features comprise global developmental delay (9/10), congenital anomalies of the brain (6/10), and eye anomalies (7/10). All three patients with monoallelic de novo variants displayed seizures (3/3), but only one of seven patients with biallelic variants (1/7).
The phenotypes observed in plxna1a and plxna1b zebrafish morphants resemble the phenotypic spectrum we observed in the herein reported patients. plxna1a SB MO morphants show a reduced eye diameter when compared to controls (Fig. 4c–f), indicating that plxna1a and plxna1b are important for eye development analogous to the observed eye phenotypes in our patients. Eye development in zebrafish appears to involve other Plexins as well, since plxna2 knockdown also leads to reduced relative eye diameter.5 Notably, we observed reduced pigmentation in plxna1a SB MO morphants compared to controls, which may correlate with the skin abnormalities observed in patients D:II-1, D:II-2, and E:II-1 respectively, presenting with hypo- and hyperpigmented skin anomalies. Another phenotypic feature of the plxna1a SB morphants is hydrocephalus (Fig. 4d, j). Here, we report three patients (D:II-1, D:II-2, G:II-1) with an abnormal dilatation of the ventricular system. Additionally, the patient reported by Park et al. showed prominence of ventricles.16 plxna1a SB MO morphants showed hypoplasia of forebrain, midbrain, and hindbrain obtained in the transgenic reporter fish Tg(-3.1ngn1:GFP) (Fig. 4j). Accordingly, six of our ten patients presented with structural cerebral anomalies (C:II-2, D:II-1, D:II-2, E:II-1, G:II-1, H:II-1) (Table 1, Fig. 4g, h). The temporal and spatial expression pattern of Plexins in zfl has been studied in detail by Emerson et al., suggesting a dynamic role in neuronal development.27 plxna1a and plxna1b are expressed in the optic vesicle, neural retina, optic tectum, optic chiasm, hypothalamus, medulla oblongata, forebrain, and ventricle of zfl. Here we confirmed this expression pattern (Figure S4). The phenotypic spectrum of plxna1a SB MO morphants follows the expression pattern in early development of zfl, suggesting a specific observation. Finally, we detected a decrease of migrated DRG cells in the spinal cord. Consecutively, the axon outgrowth is missing in the respective somites supporting the role of PLXNA1 as a mediator of axon guidance.10 Accordingly, a recent report demonstrated impaired midline crossing of axons in the CC in Plxna1 knockout mice at E17.5 and agenesis of the CC in newborn mice (P0.5).11 Here, we report three patients (C:II-2, D:II-2, G:II-1) with CC anomalies. Analogously, Belyk et al. suggested that polymorphisms in PLXNA1 are associated with altered developmental trajectory of the CC microstructure.36 Additionally, three patients had either signs of congenital cranial neuropathies including sensorineural hearing loss with or without agenesis of vestibulocochlear nerves (C:II-2, G:II-1), unilateral facial palsy (G:II-1), and peripheral axonal neuropathy (D:II-2). These features may be indicative of axonal dysfunction and reflective of the role of PLXNA1 in axonal guidance. Since the publication of the only two existing Plxna1 knockout mouse models in 2006 numerous articles describe histopathological abnormalities affecting axonal and neuronal phenotypes. While there are numerous links between the patients reported here and our zebrafish model, others remain without correspondence: for example, it remains unclear whether the Plxna1-null mice are developmentally delayed or develop seizures.
Hence, the biallelic and monoallelic variants in PLXNA1 reported here lead to a distinct overlapping phenotypic spectrum. So far over 30 loci have been linked to disease genes presenting with both recessive (biallelic) and dominant (monoallelic) inheritance patterns.37 Investigations of allelic series suggested that allelic heterogeneity may be explained in part by the functional consequences of pathogenic variants, i.e., loss-of-function (LoF), gain-of-function, or dominant-negative mechanisms.35 Recently, Harel et al. reviewed 13 of these genes with allelic heterogeneity.38 For some of these genes, the gnomAD constraint metric (probability of loss of function intolerance [pLI] score) for loss of function is 0, basically indicating complete tolerance for heterozygous LoF alleles. For example, individuals harboring a heterozygous deletion in ATAD3A are unaffected suggesting a dominant-negative pathogenic mechanism or a gain-of-function mechanism for de novo missense variants rather than haploinsufficiency.39 However, other genes with reported allelic heterogeneity, e.g., KIF1A, COL6A1, ROR2, and here PLXNA1 have a pLI score of 1, despite the fact that some healthy parents are heterozygous with LoF variants and affected patients carry monoallelic de novo missense variants. Hence, functional consequences of potentially pathogenic variant alleles alone cannot explain allelic heterogeneity. Interestingly, for PLXNA1, gnomAD reports in total 20 individuals with homozygous extracellular and only two individuals with homozygous intracellular missense variants (https://gnomad.broadinstitute.org/). Since the extracellular Plexin-A1 domains comprise 1,245 amino acids and the intracellular domains comprise only 629 amino acids (33%), random distribution of homozygous missense variants should have led to the observation of more intracellular homozygous missense variants. This imbalance might suggest that intracellular missense variants are less tolerated compared to extracellular missense variants. Accordingly, we observed only one patient with a homozygous intracellular missense variant of uncertain clinical significance (K:II-3).
We hypothesize that the here reported biallelic LoF might lead to nonsense-mediated decay (NMD) and the extracellular missense variants lead to impaired dimerization of the Plexin-A1 receptor. Both mechanisms would require two affected alleles in order to affect downstream signaling, whereas the monoallelic intracellular (de novo) missense variants might impair signaling through a dominant-negative effect. Extracellular receptor dimerization and ligand binding may be correct in the intracellular monoallelic situation; however, these variants may harm the dimerized Plexin-A1 receptor macromolecule through a dominant-negative effect in the intracellular domains (Figures S7, S8). However, this concept does not implicate the action of the Plexin-A1 co-receptor NRP1 and other protein–protein interactions of the receptor. Furthermore, two of the previously published monoallelic de novo missense variants reside in extracellular domains of Plexin-A116,17 analogous to the de novo missense variant reported here in patient F:II-1. Finally, we report one de novo LoF variant in a neonate with severe neonatal hypotonia (L:II-3), which resides also in the extracellular domains of Plexin-A1. While the hypotonia resolved spontaneously in this patient, indicating that the identified Plexin-A1 variant might be clinically insignificant, the family was lost to follow up and the outcome remains unknown.
As outlined earlier, binding of semaphorins activates the cytoplasmic GAP domain of Plexin-A1 and alterations of conserved arginine residues in the GAP domain have been shown to diminish this activity.9 Previously, Rohm et al. altered three arginine residues—p.(Arg1429), p.(Arg1430), and p.(Arg1746)—of the murine protein in cultured cells.9 The murine p.(Arg1746) is the orthologous amino acid residue of the human p.(Arg1748). The data of Rohm et al. suggest that the novel de novo p.(Arg1748Cys) change of patient H:II-1 is functionally abolishing downstream signaling of Plexin-A1, supporting our hypothesis on the pathogenic mechanism of intracellular de novo missense variants in PLXNA1 (Figure S7). Interestingly, we identified an additional missense variant altering an arginine residue in close proximity to the Plexin-A1 GAP domains (Fig. 3) in patient G:II-1. Hence, the same mechanism described by Rohm et al. for the p.(Arg1748Cys) change may also apply to the additional missense variant identified here. Remarkably, the sequence of the Plexin-A1 GAP domain is highly similar to SYNGAP1, a Ras/Rap GTPase-activating protein that is one of the most frequently mutated genes in pediatric patients with intellectual disability and seizures.40 In these children, the disease-causing genetic mechanism is dominant de novo (MIM 612621) with the majority of pathogenic variants in SYNGAP1 being LoF alleles.40 Hence, in support of the above-proposed concept, we might see a dominant-negative effect in patients with monoallelic de novo missense variants leading to impaired downstream GTPase-activating function of the SYNGAP1 related protein Plexin-A1. Notably, all monoallelic variants observed cluster toward the C-terminal domains of Plexin-A1 harboring the two GAP domains (Fig. 3). Analogously, seizures were observed in all three patients with monoallelic de novo variants and only in one patient with biallelic variants.
In conclusion, our study provides evidence that biallelic and monoallelic variants in PLXNA1 result in a novel neurodevelopmental syndrome mainly comprising developmental delay and brain and eye anomalies.
Supplementary information
Acknowledgements
We thank the families for their collaboration and participation in this study. This work was supported in part by the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG) (RE 1723/5–1, BO102/1–3 and TH 1327/2–1) to H.R. and H.T. and a DFG equipment grant (INST 1172/37–1 FUGG) for a multiphoton microscope setup. G.C.D. is supported by BONFOR grant O-120.0001 and by the Herbert-Reeck foundation (2019). J.C.K. is supported by BonnNi grant Q614.0754. Further we are thankful for the support of the Zebrafish Core Facility (Bonn Medical Faculty). This work was supported in part by the National Institutes of Health, National Institute of Neurologic Disease and Stroke [R35 NS105078] and the Muscular Dystrophy Association [512848] to J.R.L.; and a jointly funded National Human Genome Research Institute (NHGRI), and National Heart, Lung, and Blood Institute (NHLBI) grant to the Baylor-Hopkins Center for Mendelian Genomics [UM1 HG006542]. J.E.P. is supported by NHGRI [K08 HG008986]. T.M. is supported by the Uehara Memorial Foundation. D.P. is supported by Clinical Research Training Scholarship in Neuromuscular Disease partnered by the American Academy of Neurology (AAN), American Brain Foundation (ABF) and Muscle Study Group (MSG), and International Rett Syndrome Foundation (IRSF grant 3701–1). Two families were collected as part of the SYNaPS Study Group collaboration funded by The Wellcome Trust and strategic award (Synaptopathies) funding (WT093205 MA and WT104033AIA). This research was conducted as part of the Queen Square Genomics group at University College London, supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre. H.H. is funded by The MRC (MR/S01165X/1, MR/S005021/1, G0601943), The National Institute for Health Research University College London Hospitals Biomedical Research Centre, Rosetree Trust, Ataxia UK, MSA Trust, Brain Research UK, Sparks GOSH Charity, Muscular Dystrophy UK, Muscular Dystrophy Association. Sequencing and analysis were provided by the Broad Institute of MIT and Harvard Center for Mendelian Genomics and was funded by the NHGRI, the National Eye Institute, and the National Heart, Lung and Blood Institute grant UM1 HG008900 and in part by NHGRI grant R01 HG009141. The study was supported by the DESIRE project, EU Seventh Framework Programme (grant agreement number 602531 to R.G.). The work was supported by SFARI and Simons VIP foundations. This study was supported by the National Health and Medical Research Council of Australia (APP1048816 and APP1136197 S.T.C., 1080587 S.T.C., D.G.M.). E.C.O. was supported by NHMRC ECR fellowship GNT1090428. The Djavad Mowafaghian Foundation generously enabled this molecular research to take place at the Djavad Mowafaghian Centre for Brain Health. This work was also supported by the Canada Excellence Research Chair, Leading Edge Endowment Fund, Rare Disease Foundation, Grocholski Foundation, and Alva Foundation. This study makes use of data generated by the DECIPHER community. A full list of centers who contributed to the generation of the data is available from http://decipher.sanger.ac.uk and via email from decipher@sanger.ac.uk. Funding for the project was provided by Wellcome.
Author contributions
Conceptualization: G.C.D., J.E.P., J.R.L., H.R. Data curation: J.P., H.T., H.R. Formal analysis: G.C.D., J.C.K., E.M. Investigation: G.C.D., J.P., J.C.K., E.M., H.R., H.B.E., Z.C.A., E.K., T.M., D. Marafi, J.M.F., S.N.J., J.V.H., T.C.D., B.D., O.D., H.S.A., F.S.A., R.M., H.H., S.E., N.D., V.S., T.S., S.H., F.B., J.H., K.M.R., M.W., I.G., M.D., B.K., J.B., P.C., C.N., D.H., S.H., E.V., L.B.W., K.J.J., E.C.O., S.T.C., D. MacArthur, S.S., A.Z., K.P., V.O., W.K.C., S.A.O., R.A., S.F., P.R.M., R.G., A.V., B.H., R.E.S., G.E.H., J.E.B., M.M., T.L., Ö.Y., D.P. Methodology: G.C.D., B.O. Resources: B.O., J.R.L., H.R. Visualization: J.P., J.C.K. Writing—original draft: G.C.D., J.P., T.M., D. Marafi, D.P., J.E.P., J.R.L., H.R. Writing—review & editing: G.C.D., J.R.L., H.R.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
All variants have been deposited into ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/)under Lupski Lab, Baylor-Hopkins CMG, Baylor College of Medicine, including VCV000867235 through VCV000867245.
Ethics Declaration
The research project was approved by the Ethics Committee of the Medical Faculty of the University of Bonn (number 208/08). All families provided consent according to the respective research protocols including patient photographs, approved by each of the institutional review boards (IRBs), the respective IRB protocols are named in the Supplementary information. Molecular findings and clinical descriptions are provided in Table 1 and in the Supplementary information. Husbandry and experimental procedures were in accordance to the European Legislation for the Protection of Animals used for Scientific Purposes (Directive 2010/62/EU). All zebrafish experiments were performed at ≤5 dpf before independent feeding and by national law no ethical approval is necessary for this time period.
Competing interests
J.R.L. has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals and Novartis and is a member of the Scientific Advisory Board of Baylor Genetics, and is a co-inventor on multiple US and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis (CMA) and clinical exome sequencing offered at Baylor Genetics. S.T.C. is director of Frontier Genomics Pty Ltd (Australia). Frontier Genomics has not traded (as of December 2019). Frontier Genomics Pty Ltd (Australia) has no existing financial relationships that will benefit from publication of these data. R.E.S. is an employee of GeneDx, Inc. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Gabriel C. Dworschak, Jaya Punetha.
Supplementary information
The online version contains supplementary material available at 10.1038/s41436-021-01196-9.
References
- 1.Tamagnone L, et al. Plexins are a large family of receptors for transmembrane, secreted, and GPI-anchored semaphorins in vertebrates. Cell. 1999;99:71–80. doi: 10.1016/S0092-8674(00)80063-X. [DOI] [PubMed] [Google Scholar]
- 2.Winberg ML, et al. Plexin A is a neuronal semaphorin receptor that controls axon guidance. Cell. 1998;95:903–916. doi: 10.1016/s0092-8674(00)81715-8. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi T, et al. Plexin-neuropilin-1 complexes form functional semaphorin-3A receptors. Cell. 1999;99:59–69. doi: 10.1016/S0092-8674(00)80062-8. [DOI] [PubMed] [Google Scholar]
- 4.Wang, N., Dhumale, P., Chiang, J. & Püschel, A. W. The Sema3A receptor Plexin-A1 suppresses supernumerary axons through Rap1 GTPases. Sci Rep. 10.1038/s41598-018-34092-5 (2018). [DOI] [PMC free article] [PubMed]
- 5.St. Clair RM, et al. Fyn-dependent phosphorylation of PlexinA1 and PlexinA2 at conserved tyrosines is essential for zebrafish eye development. FEBS J. 2018;285:72–86. doi: 10.1111/febs.14313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, et al. Plexins are GTPase-activating proteins for Rap and are activated by induced dimerization. Sci. Signal. 2012;5:ra6. doi: 10.1126/scisignal.2002636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kong Y, et al. Structural basis for plexin activation and regulation. Neuron. 2016;91:548–60. doi: 10.1016/j.neuron.2016.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang, Y., Pascoe, H. G., Brautigam, C. A., He, H. & Zhang, X. Structural basis for activation and non-canonical catalysis of the Rap GTPase activating protein domain of plexin. Elife. 10.7554/eLife.01279 (2013). [DOI] [PMC free article] [PubMed]
- 9.Rohm B, Rahim B, Kleiber B, Hovatta I, Püschel AW. The semaphorin 3A receptor may directly regulate the activity of small GTPases. FEBS Lett. 2000;486:68–72. doi: 10.1016/s0014-5793(00)02240-7. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida Y, Han B, Mendelsohn M, Jessell TM. PlexinA1 signaling directs the segregation of proprioceptive sensory axons in the developing spinal cord. Neuron. 2006;52:775–88. doi: 10.1016/j.neuron.2006.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hossain M, Tsuzuki T, Sakakibara K, Imaizumi F. PlexinA1 is crucial for the midline crossing of callosal axons during corpus callosum development in BALB / cAJ mice. PLoS One. 2019;5:1–23. doi: 10.1371/journal.pone.0221440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marcos S, et al. Defective signaling through plexin-A1 compromises the development of the peripheral olfactory system and neuroendocrine reproductive axis in mice. Hum. Mol. Genet. 2017;26:2006–17. doi: 10.1093/hmg/ddx080. [DOI] [PubMed] [Google Scholar]
- 13.Kuwajima T, et al. Optic chiasm presentation of Semaphorin6D in the context of Plexin-A1 and Nr-CAM promotes retinal axon midline crossing. Neuron. 2012;74:676–90. doi: 10.1016/j.neuron.2012.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrews WD, Davidson K, Tamamaki N, Ruhrberg C, Parnavelas JG. Altered proliferative ability of neuronal progenitors in PlexinA1 mutant mice. J. Comp. Neurol. 2016;524:518–34. doi: 10.1002/cne.23806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Klaauw AA, et al. Human semaphorin 3 variants link melanocortin circuit development and energy balance. Cell. 2019;176:729–.e18. doi: 10.1016/j.cell.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park K, Seltzer LE, Tuttle E, Mirzaa GM, Paciorkowski AR. PLXNA1 developmental encephalopathy with syndromic features: a case report and review of the literature. Am. J. Med. Genet. A. 2017;173:1951–4. doi: 10.1002/ajmg.a.38236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Epi4K Consortium, Epilepsy Phenome/Genome Project. Allen AS, et al. De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–21. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliver, K. L., Lukic, V., Freytag, S., Scheffer, I. E., Berkovic, S. F. & Bahlo, M. In silico prioritization based on coexpression can aid epileptic encephalopathy gene discovery. Neurol. Genet. 10.1212/NXG.0000000000000051 (2016). [DOI] [PMC free article] [PubMed]
- 19.Fromer M, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–84. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–30. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sobreira NLM, et al. Matchmaker Exchange. Curr Protoc Hum Genet. 2017;95:9.31.1–9.31.15. doi: 10.1002/cphg.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arachchi H, et al. matchbox: An open-source tool for patient matching via the Matchmaker Exchange. Hum. Mutat. 2018;39:1827–34. doi: 10.1002/humu.23655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010;5:725–38. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westerfield, M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish. https://www.Zfinorg/Zf_info/Zfbook/Zfbk.html (2007).
- 26.Blader P, Plessy C, Strähle U. Multiple regulatory elements with spatially and temporally distinct activities control neurogenin1 expression in primary neurons of the zebrafish embryo. Mech. Dev. 2003;120:211–8. doi: 10.1016/S0925-4773(02)00413-6. [DOI] [PubMed] [Google Scholar]
- 27.Emerson SE, Light SE, Ebert AM. Neuronal expression patterns of the PlexinA family during zebrafish development. Gene Expr. Patterns. 2018;27:56–66. doi: 10.1016/j.gep.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 28.Ton QV, Kathryn Iovine M. Semaphorin3d mediates Cx43-dependent phenotypes during fin regeneration. Dev. Biol. 2012;366:195–203. doi: 10.1016/j.ydbio.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoo, S. K. et al. Plexins function in epithelial repair in both Drosophila and zebrafish. Nat. Commun. 10.1038/ncomms12282 (2016). [DOI] [PMC free article] [PubMed]
- 30.Teng, Y. et al. Loss of zebrafish lgi1b leads to hydrocephalus and sensitization to pentylenetetrazol induced seizure-like behavior. PLoS One. 10.1371/journal.pone.0024596 (2011). [DOI] [PMC free article] [PubMed]
- 31.Emerson SEST, et al. Identification of target genes downstream of semaphorin6A/PlexinA2 signaling in zebrafish. Dev. Dyn. 2017;246:539–49. doi: 10.1002/dvdy.24512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dakal TC, Kumar R, Ramotar D. Structural modeling of human organic cation transporters. Comput. Biol. Chem. 2017;68:153–63. doi: 10.1016/j.compbiolchem.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 33.Hu WF, Chahrour MH, Walsh CA. The diverse genetic landscape of neurodevelopmental disorders. Annu. Rev. Genomics Hum. Genet. 2014;15:195–213. doi: 10.1146/annurev-genom-090413-025600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilissen C, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–7. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 35.White J, et al. POGZ truncating alleles cause syndromic intellectual disability. Genome Med. 2016;8:3. doi: 10.1186/s13073-015-0253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belyk, M., Kraft, S. J., Brown, S. & Neurocognition and Genetics Study. PlexinA polymorphisms mediate the developmental trajectory of human corpus callosum microstructure. J. Hum. Genet. 60, 147–50, 10.1038/jhg.2014.107 (2015). [DOI] [PMC free article] [PubMed]
- 37.Posey JE, et al. Insights into genetics, human biology and disease gleaned from family based genomic studies. Genet. Med. 2019;21:798–812. doi: 10.1038/s41436-018-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harel T, et al. Monoallelic and biallelic variants in EMC1 identified in individuals with global developmental delay, hypotonia, scoliosis, and cerebellar atrophy. Am J. Hum. Genet. 2016;98:562–70. doi: 10.1016/j.ajhg.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harel T, et al. Recurrent de novo and biallelic variation of ATAD3A, encoding a mitochondrial membrane protein, results in distinct neurological syndromes. Am J. Hum. Genet. 2016;99:831–45. doi: 10.1016/j.ajhg.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holder, J. L., Hamdan, F. F. & Michaud, J. L. in GeneReviews (eds Adam, M. P. et al.) SYNGAP1-related intellectual disability. (University of Washington, Seattle, 1993).https://www.ncbi.nlm.nih.gov/books/NBK537721/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All variants have been deposited into ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/)under Lupski Lab, Baylor-Hopkins CMG, Baylor College of Medicine, including VCV000867235 through VCV000867245.