Abstract
Purpose
To evaluate the association between a previously published 313 variant–based breast cancer (BC) polygenic risk score (PRS313) and contralateral breast cancer (CBC) risk, in BRCA1 and BRCA2 pathogenic variant heterozygotes.
Methods
We included women of European ancestry with a prevalent first primary invasive BC (BRCA1 = 6,591 with 1,402 prevalent CBC cases; BRCA2 = 4,208 with 647 prevalent CBC cases) from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA), a large international retrospective series. Cox regression analysis was performed to assess the association between overall and ER-specific PRS313 and CBC risk.
Results
For BRCA1 heterozygotes the estrogen receptor (ER)-negative PRS313 showed the largest association with CBC risk, hazard ratio (HR) per SD = 1.12, 95% confidence interval (CI) (1.06–1.18), C-index = 0.53; for BRCA2 heterozygotes, this was the ER-positive PRS313, HR = 1.15, 95% CI (1.07–1.25), C-index = 0.57. Adjusting for family history, age at diagnosis, treatment, or pathological characteristics for the first BC did not change association effect sizes. For women developing first BC < age 40 years, the cumulative PRS313 5th and 95th percentile 10-year CBC risks were 22% and 32% for BRCA1 and 13% and 23% for BRCA2 heterozygotes, respectively.
Conclusion
The PRS313 can be used to refine individual CBC risks for BRCA1/2 heterozygotes of European ancestry, however the PRS313 needs to be considered in the context of a multifactorial risk model to evaluate whether it might influence clinical decision-making.
INTRODUCTION
Heterozygotes of germline pathogenic variants in BRCA1 or BRCA2 (henceforth BRCA1/2 heterozygotes) have a higher risk of developing contralateral breast cancer than nonheterozygotes.1 The estimated cumulative 10-year contralateral breast cancer risk varies across studies between 18.5% and 34.2% for BRCA1 heterozygotes and between 10.8% and 29.2% for BRCA2 heterozygotes,1–6 compared to 4–6% in the population.7,8 Whether or not to undergo a risk-reducing contralateral mastectomy, which is an invasive intervention and associated with side effects such as postoperative surgical complications, inability to breast feed in the future, and psychosocial burden,9 is an important and difficult decision for BRCA1/2 heterozygotes who have been just confronted with their first breast cancer diagnosis. Precise individualized risk estimates could facilitate decision making for these women.
Two important factors influencing contralateral breast cancer risk in BRCA1/2 heterozygotes are the age at diagnosis of the first breast tumor and a family history of breast cancer.2,4,5,10 The effect of family history on contralateral breast cancer risk suggests a role for other genetic factors. In the last decade, more than 180 common low risk variants have been associated with breast cancer risk in genome-wide association studies (GWAS).11–13 Individually, these variants are associated with small increases in risk, but when combined as polygenic risk scores (PRS) they may improve disease-related risk stratification for women of European and Asian ancestry in the population.14–16 A limited number of studies have shown that variants associated with the risk of a first primary breast cancer are also associated with the risk of contralateral breast cancer.17–19 Furthermore, the PRS derived from the general population has also been shown to be associated with breast cancer risk in BRCA1/2 heterozygotes.20–24
The most predictive, well validated PRS for breast cancer in the general population is based on 313 breast cancer–associated variants (PRS313); it showed an association with breast cancer in ten prospective studies with an odds ratio (OR) per standard deviation (SD) of 1.61 and an area under the receiver–operator characteristic curve of 0.630.14 Among BRCA2 heterozygotes, this same PRS313 was also associated with breast cancer risk, hazard ratio (HR) per SD = 1.31, 95% confidence interval (CI) (1.27–1.36).24 Among BRCA1 heterozygotes, the largest association with breast cancer risk was found using the estrogen receptor (ER)-negative PRS313 (which uses the same variants but with weights adapted to provide better prediction for ER-negative disease), HR = 1.29, 95% CI (1.25–1.33).24 Although these effect sizes were smaller than those for the general population, the 313 variant–based PRS could have a substantial impact on the high absolute risks24 associated with BRCA1/2 pathogenic variants.25 Whether variants associated with breast cancer are associated with contralateral breast cancer risk for BRCA1/2 heterozygotes as well, individually or combined in a PRS, has not been investigated previously. If so, the PRS may be useful to guide choices for risk management, especially regarding invasive risk-reducing contralateral mastectomy. In this study, we investigated whether the 313 variant–based PRS for breast cancer is associated with contralateral breast cancer risk among women of European ancestry with pathogenic variants in BRCA1/2 and explored the implications for contralateral breast cancer risk prediction for these women.
MATERIALS AND METHODS
Study participants
We used retrospective cohort data from heterozygotes participating in the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA).26 Briefly, CIMBA participants are heterozygotes of pathogenic variants in BRCA1 or BRCA2 who are 18 years or older at the time of inclusion and have phenotypic data available.26 CIMBA includes 81 individual studies of which the majority of the participants were ascertained through cancer genetics clinics.26 Although studies in CIMBA include individuals of non-European ancestry, our analyses were, due to power considerations (small numbers available for analyses and expected lower estimates for the PRS313 in Asian ancestry based on results of women in the general breast cancer population19), restricted to women of European ancestry with available array genotyping data (31,195 women of 67 studies).
Women were eligible for this retrospective analysis if they developed an invasive primary breast tumor without metastatic disease at least 1 year before the baseline age. Women without information about metastatic disease were assumed to have no metastatic disease (n = 9,242 of whom 2,140 had a known negative lymph node status). Baseline age was defined as the age at local ascertainment (97%), or when this was not known, age at genetic testing (2%) or age at last follow-up (1%). Women were excluded if no information was available about the age at baseline or if they had developed synchronous contralateral breast cancer. Synchronous contralateral breast cancer was defined as contralateral breast cancer within one year after the first primary breast cancer, which was based on the exact date of cancer diagnosis or, if this was not available, on the age at diagnosis. A schematic overview of the selection is shown in Fig. S1. In total, 6,591 women with BRCA1 and 4,208 women with BRCA2 pathogenic variants were included in this study, among whom 1,402 BRCA1 heterozygotes and 647 BRCA2 heterozygotes have had contralateral breast cancer. The diagnosis of primary and contralateral breast cancer was confirmed by pathology records, tumor registry data, or medical records by the individual studies. Available phenotypic information for all participants is shown in Table 1, including the number of participants for whom the information was not available for each of the variables. Information about the ER status of the first primary breast cancer compared to the contralateral breast cancer is shown in Table S1.
Table 1.
Characteristics of the participants.
| BRCA1 heterozygotes | BRCA2 heterozygotes | ||||
|---|---|---|---|---|---|
| UBC, n (%) | CBC, n (%) | UBC, n (%) | CBC, n (%) | ||
| N | 5,189 | 1,402 | 3,561 | 647 | |
| Genotyping array | iCOGS | 895 (17) | 200 (14) | 383 (11) | 80 (12) |
| OncoArray | 4,294 (83) | 1,202 (86) | 3,178 (89) | 567 (88) | |
| Birth cohort | <1920 | 25 (0.5) | 8 (0.6) | 23 (0.6) | 9 (1) |
| 1920–1929 | 143 (3) | 46 (3) | 121 (3) | 30 (5) | |
| 1930–1939 | 392 (8) | 130 (9) | 341 (10) | 99 (15) | |
| 1940–1949 | 1,060 (20) | 386 (28) | 793 (22) | 172 (27) | |
| 1950–1959 | 1,540 (30) | 452 (32) | 1,104 (31) | 202 (31) | |
| 1960–1969 | 1,354 (26) | 298 (21) | 822 (23) | 115 (18) | |
| ≥1970 | 675 (13) | 82 (6) | 357 (10) | 20 (3) | |
| Variant classa | I | 3,354 (65) | 904 (64) | 3,207 (90) | 570 (88) |
| II | 1,345 (26) | 374 (27) | 125 (4) | 25 (4) | |
| III | 490 (9) | 124 (9) | 229 (6) | 52 (8) | |
| BRRM | 160 (3) | 0 | 101 (3) | 0 | |
| Deceased | N | 44 (0.8) | 12 (0.9) | 19 (0.5) | 2 (0.3) |
| Family historyb | No BC | 583 (11) | 175 (12) | 289 (8) | 78 (12) |
| 1 BC | 906 (17) | 270 (19) | 760 (21) | 127 (20) | |
| ≥ 2 BC | 1,250 (24) | 363 (26) | 1,120 (31) | 210 (32) | |
| Unknown | 2,450 (47) | 594 (42) | 1,392 (39) | 232 (36) | |
| Characteristics of first BC | |||||
| Age at diagnosis | Mean | 41.8 | 38.5 | 44.5 | 41.8 |
| Range | 19–82 | 19–68 | 18–85 | 21–75 | |
| ER status | Positive | 570 (11) | 92 (7) | 1,302 (37) | 182 (28) |
| Negative | 1,738 (33) | 402 (29) | 424 (12) | 61 (9) | |
| Unknown | 2,881 (56) | 908 (65) | 1,835 (52) | 404 (62) | |
| Node status | Positive | 797 (15) | 182 (13) | 781 (22) | 119 (18) |
| Negative | 1,544 (30) | 441 (31) | 877 (25) | 151 (23) | |
| Unknown | 2,848 (55) | 779 56) | 1,903 (53) | 377 (58) | |
| Tumor sizec | T1 | 1,261 (24) | 314 (22) | 842 (24) | 136 (21) |
| T2 | 771 (15) | 211 (15) | 553 (16) | 87 (13) | |
| T3 | 67 (13) | 12 (0.9) | 78 (2) | 8 (1) | |
| T4 | 16 (0.5) | 2 (0.1) | 22 (0.6) | 2 (0.3) | |
| Unknown | 3,074 (59) | 863 (62) | 2,066 (58) | 414 (64) | |
| Chemotherapyd | Yes | 1,099 (21) | 236 (17) | 821 (23) | 123 (19) |
| No | 576 (11) | 212 (15) | 503 (14) | 129 (20) | |
| Unknown | 3,514 (68) | 954 (68) | 2,237 (63) | 395 (61) | |
| Adjuvant hormone therapy | Yes | 493 (10) | 125 (9) | 795 (22) | 111 (17) |
| No | 1,103 (21) | 288 (21) | 474 (13) | 135 (21) | |
| Unknown | 3,593 (69) | 989 (71) | 2,292 (64) | 401 (62) | |
| Adjuvant trastuzumab therapy | Yes | 11 (0.2) | 1 (0.1) | 20 (0.6) | 0 (0) |
| No | 1,161 (22) | 351 (25) | 983 (28) | 218 (34) | |
| Unknown | 4,017 (77) | 1,050 (75) | 2,558 (72) | 429 (66) | |
| Radiotherapy | Yes | 1,090 (21) | 277 (20) | 797 (22) | 158 (24) |
| No | 535 (10) | 141 (10) | 420 (12) | 84 (13) | |
| Unknown | 3,564 (69) | 984 (70) | 2,344 (66) | 405 (63) | |
| Characteristics of CBC | |||||
| Age at diagnosis | Mean | – | 47.3 | – | 51.24 |
| Range | – | 26–80.5 | – | 23.8–86 | |
| Invasiveness | Invasive | – | 1,267 (90) | – | 545 (84) |
| Noninvasive | – | 135 (10) | – | 102 (16) | |
| ER status | Positive | – | 101 (7) | – | 197 (30) |
| Negative | – | 446 (32) | – | 50 (8) | |
| Unknown | – | 855 (61) | – | 400 (62) | |
| PRS313 | |||||
| Standardized PRS313 mean (SD) | Overall BC | 0.08 (1.01) | 0.13 (1.01) | 0.09 (1.02) | 0.27 (1.04) |
| ER-positive BC | 0.07 (1.01) | 0.09 (1.01) | 0.08 (1.01) | 0.27 (1.03) | |
| ER-negative BC | 0.09 (1.00) | 0.23 (0.99) | 0.07 (1.02) | 0.23 (1.07) | |
BC breast cancer, BRRM bilateral risk-reducing mastectomy, CBC contralateral breast cancer, ER status estrogen receptor status of the tumor, PRS polygenic risk score, SD standard deviation, UBC unilateral breast cancer.
aVariant class: I = unstable or no protein, II = stable mutant protein, III = consequence unknown.
bFamily history was defined as the number of first- or second-degree relatives affected with BC, ranging from 0 to ≥2.
cTumor size: T1 = ≤ 2 cm (≤0.79 inches), T2 = > 2cm-5cm (>0.79–1.97 inches), T3 = > 5 cm (>1.97 inches), T4 = any size, with direct extension to the chest wall or skin.
dIncluding neoadjuvant and adjuvant chemotherapy.
Genotyping and polygenic risk score calculation
For most of the participants, genotyping was performed with the Illumina OncoArray.27 The remaining participants were genotyped with the Illumina iCOGS array.11 Details about the quality control procedures and correlation between the arrays have been described previously.19,24,28–31 European ancestry was determined using genetic data and multidimensional scaling. More detailed information about the genotyping and PRS calculation is provided in the Supplementary methods.
We used the 313 variant–based PRS for breast cancer developed in an independent study using data from the general population as described previously;14 correlation between PRS based on the two genotyping arrays was high.19 The PRS for overall breast cancer (PRS313) and two ER-specific PRS, the ER-positive PRS313 and ER-negative PRS313 were calculated. The variants and their corresponding weights used in the PRS as published previously14 and the imputation quality are listed in Table S2. The three PRS were standardized to the mean from all CIMBA participants, including both unaffected and affected women, and to the SD in Breast Cancer Association Consortium (BCAC) population controls that were included in the validation data set.14 Using these SDs, the HR estimates for the associations of the standardized PRS313 in our study are directly comparable with the OR estimates reported in the BCAC population-based study14 and the HR estimates reported for primary breast cancer in BRCA1 and BRCA2 heterozygotes.24
Statistical analysis
To assess the associations between the three PRS and contralateral breast cancer risk in BRCA1/2 heterozygotes, Cox regression analyses were performed. The time at risk was started one year after the first breast cancer diagnosis based on the exact date, or if not available, on the age of developing the first breast tumor. Time at risk of participants was censored at age at baseline, i.e., end of follow-up in these analyses, prophylactic contralateral mastectomy, or death, whichever was earlier (Fig. S2). Incidence of a metachronous contralateral breast cancer, invasive or in situ, before baseline was considered as an event in the main analyses. The proportional hazard assumption was evaluated by using Schoenfeld residuals against the transformed time. A sensitivity analysis was performed considering invasive contralateral breast cancer only as an event. Women who developed an in situ contralateral breast cancer were censored at the age at diagnosis of the in situ contralateral breast cancer. Furthermore, a sensitivity analysis was performed including information about distant relapse, which was available for 1,725 BRCA1 and 1,450 BRCA2 heterozygotes. In total 55 BRCA1 heterozygotes and 101 BRCA2 heterozygotes were censored at the age of distant relapse of which 13 and 11 women were excluded from the analyses, respectively, because they developed distant relapse in the year before the baseline age.
Analyses were stratified by country (Table S3), adjusted for birth cohort (quartiles of the observed distribution), and clustered on family membership using a unique family identifier to account for the inclusion of related individuals. For BRCA1 and BRCA2 respectively, there were 5,923 and 3,752 clusters, of which 554 and 362 clusters had more than one participant. The main analyses assessed the association with the PRS as a continuous covariate. We evaluated the linearity of the association using restricted cubic splines with three knots, which showed no evidence for violation of the linearity assumption. The discriminatory ability of the best-performing PRS was evaluated by Harrell’s C-index.32 C-indexes were calculated stratified by country and clustered on family membership.
The influence of possible confounding variables on the observed associations was assessed using the PRS exhibiting the largest associations. Possible confounding variables included breast cancer family history, age at diagnosis of the first breast cancer, pathological characteristics, and treatment of the first breast cancer. Each variable was added to the model one by one and in addition, a full model that included all possible confounders together was fitted. If the addition of a variable resulted in a change of more than 10% in the log HR, the variable was retained as a covariate in the final Cox regression model. To avoid excluding many participants with missing data for one of these included variables (Table 1), missing data were imputed using multiple imputation by chained equations (MICE).33 Imputation was started with the least missing variable and progressed in order of increased amount of missing data. Using this method, ten complete data sets for analyses were created and mean parameter estimates were derived.
Secondary analyses were performed for ER-positive and ER-negative cases only, based on the ER status of the contralateral breast cancer, after imputation as described above. The average number of ER-positive and ER-negative cases in the ten imputed data sets is shown in Table S4. In these analyses the event of interest was either ER-positive or ER-negative contralateral breast cancer. Contralateral breast cancer cases with the alternative ER status were censored at the age of contralateral breast cancer.
The interaction between the PRS with the age at first breast cancer diagnosis was tested in the final model, treating the PRS as a continuous variable. Furthermore, the effect size of the PRS was evaluated for groups based on the age at first primary breast cancer diagnosis (<40 years; 40 to 50 years; ≥50 years).1,20 The association of the PRS and contralateral breast cancer risk was tested separately for heterozygotes of pathogenic variants that lead to unstable or no protein (class I) and heterozygotes of pathogenic variants that lead to mutant stable protein (class II). Finally, analyses were performed to test the association between a categorized PRS and contralateral breast cancer risk to establish whether the results were consistent with those under a continuous PRS model. The categories were defined on the basis of the distribution of the PRS in unilateral breast cancer cases, using PRS percentiles (0–5th, 5th−10th, 10th−20th, 20th−40th, 40th−60th [reference], 60th−80th, 80th−90th, 90th−95th, 95th−100th).
Cumulative risks
Absolute contralateral breast cancer risks were calculated at percentiles of the best-performing continuous PRS for both BRCA1 and BRCA2 heterozygotes, using the log HR per SD and including an interaction term with the continuous age at first breast cancer diagnosis (at age 35, 45, and 55 for the corresponding age groups as described below). For this purpose, we constrained the incidence of contralateral breast cancer, by age at first breast cancer and in years after the first breast cancer, and averaged over all PRS categories to agree with external contralateral breast cancer incidence estimates, as described previously.23 These external incidence estimates were based on prospective cohort data from three consortia on heterozygotes of pathogenic BRCA1 and BRCA2 variants,1 the International BRCA1/2 Carrier Cohort Study (IBCCS), the Breast Cancer Family Registry (BCFR), and the Kathleen Cunningham Foundation Consortium for Research Into Familial Breast Cancer (kConFab). Because the contralateral breast cancer incidences vary with the age of first breast cancer diagnosis, incidences were calculated for three different groups based on the age of the first breast cancer diagnosis (<40 years, 40 to 50 years, ≥50 years).1
All statistical tests were performed with R version 3.5.0.34 Statistical significance was defined as a two-sided p value <0.05.
RESULTS
In the analyses, 6,591 BRCA1 and 4,208 BRCA2 heterozygotes of European ancestry who had developed an invasive first primary breast cancer before entry in CIMBA were identified. The median follow-up time was 6.0 and 5.4 years for BRCA1 and BRCA2 heterozygotes, respectively. In total, 1,402 BRCA1 and 647 BRCA2 heterozygotes were diagnosed with a metachronous contralateral breast cancer before enrollment in CIMBA. The cumulative 10-year risk of developing contralateral breast cancer in this cohort was 25%, 95% CI (23.5–26.4%) and 18.8%, 95% CI (17.1–20.5%) for BRCA1 and BRCA2 heterozygotes, respectively (Fig. S3). Patient and tumor characteristics as well as the PRS distributions are shown in Table 1 and Fig. S4.
PRS and contralateral breast cancer risk
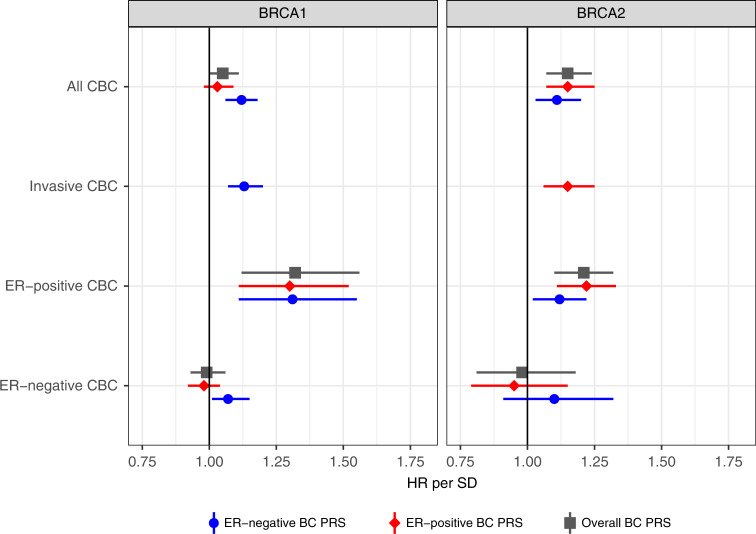
Results of the association analyses between the PRS and contralateral breast cancer risk are shown in Table 2, Table S4, and Fig. 1.
Table 2.
Results of association analyses between the PRS313 and contralateral breast cancer risk.
| BRCA1 heterozygotes (ER-negative PRS313) | BRCA2 heterozygotes (ER-positive PRS313) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| UBC cases, n | CBC cases, n | HRa | 95% CI | P | UBC cases, n | CBC cases, n | HRa | 95% CI | P | ||
| PRS continuous | All CBC | 5,189 | 1,402 | 1.12 | 1.06–1.18 | 5.98×10-5 | 3,561 | 647 | 1.15 | 1.07–1.25 | 1.94×10-4 |
| Invasive CBC | 5,324 | 1,267 | 1.13 | 1.07–1.20 | 3.15×10-5 | 3,663 | 545 | 1.15 | 1.06–1.25 | 6.02×10-4 | |
| Categorical PRS percentiles | 0–5 | 260 | 48 | 0.81 | 0.59–1.11 | 0.188 | 166 | 28 | 1.06 | 0.71–1.58 | 0.782 |
| 5–10 | 259 | 54 | 0.77 | 0.57–1.03 | 0.082 | 198 | 26 | 0.68 | 0.44–1.04 | 0.074 | |
| 10–20 | 519 | 131 | 0.94 | 0.76–1.15 | 0.544 | 355 | 51 | 0.91 | 0.66–1.25 | 0.554 | |
| 20–40 | 1,038 | 230 | 0.83 | 0.70–0.98 | 0.031 | 697 | 108 | 0.87 | 0.68–1.13 | 0.295 | |
| 40–60 (reference) | 1,037 | 282 | 1.00 | 695 | 123 | 1.00 | |||||
| 60–80 | 1,038 | 313 | 1.04 | 0.88–1.22 | 0.664 | 734 | 128 | 0.96 | 0.75–1.23 | 0.748 | |
| 80–90 | 519 | 170 | 1.11 | 0.92–1.34 | 0.255 | 358 | 90 | 1.35 | 1.03–1.77 | 0.030 | |
| 90–95 | 259 | 82 | 1.18 | 0.92–1.51 | 0.185 | 178 | 46 | 1.35 | 0.96–1.90 | 0.082 | |
| 95–100 | 260 | 92 | 1.24 | 0.98–1.56 | 0.074 | 180 | 47 | 1.31 | 0.94–1.82 | 0.116 | |
| PRS*age BC1 continuous | Main effect | 5,189 | 1,402 | 1.48 | 1.15–1.89 | 2.03×10-3 | 3,561 | 647 | 1.53 | 1.11–2.12 | 0.010 |
| Interaction effect | 0.99 | 0.99–1.00 | 0.025 | 0.99 | 0.99–1.00 | 0.089 | |||||
| PRS effect per age group | <40 | 2,339 | 815 | 1.22 | 1.14–1.31 | 4.79×10-8 | 1,238 | 268 | 1.23 | 1.09–1.38 | 5.78×10-4 |
| 40–50 | 1,821 | 456 | 0.99 | 0.90–1.09 | 0.785 | 1,306 | 261 | 1.19 | 1.05–1.34 | 6.91×10-3 | |
| ≥50 | 1,029 | 131 | 1.03 | 0.86–1.24 | 0.715 | 1,017 | 118 | 0.97 | 0.81–1.15 | 0.698 | |
| Variant classb | Class I | 3,354 | 904 | 1.11 | 1.03–1.18 | 4.32×10-3 | 3,207 | 570 | 1.16 | 1.07–1.26 | 1.99×10-4 |
| Class II | 1,345 | 374 | 1.15 | 1.04–1.28 | 4.75×10-3 | 125 | 25 | 0.91 | 0.65–1.28 | 0.594 | |
BC1 first primary breast cancer, CBC contralateral breast cancer, CI confidence interval, HR hazard ratio, PRS polygenic risk score, UBC unilateral breast cancer.
aHRs for association with breast cancer and the continuous PRS313 are reported per standard deviation of the PRS in population-based controls.
bClass I pathogenic variants result in an unstable or no protein. Class II pathogenic variants yield stable mutant proteins.
Fig. 1. Association between the PRS and contralateral breast cancer risk for BRCA1 and BRCA2 heterozygotes.
Effect size of the association between contralateral breast cancer and the three different PRS313 after testing for covariates for the following selections: all contralateral breast cancer, invasive contralateral breast cancer only, ER-negative contralateral breast cancer, and ER-positive contralateral breast cancer. The numbers of unilateral and contralateral breast cancer cases and effect sizes are shown in Table 2 and Table S4. CBC contralateral breast cancer, ER estrogen receptor, HR hazard ratio, PRS polygenic risk score, SD standard deviation.
BRCA1 heterozygotes
For BRCA1 heterozygotes the ER-negative PRS313 showed the largest association with all contralateral breast cancer, HR per SD = 1.12, 95% CI (1.06–1.18), p value = 6.0×10−5, C-index 0.53, 95% CI (0.51–0.55). There was no evidence of violation of the proportional hazard assumption, p value = 0.840.
Neither sequential inclusion of possible confounders nor including all these confounders in one model changed the log HR estimate for the ER-negative PRS313 association more than 10% when compared with the model with no confounders (Table S5).
Considering only invasive contralateral breast cancer as the event of interest resulted in a similar association with the ER-negative PRS313, HR per SD = 1.13, 95% CI (1.07–1.20), p value = 3.2×10−5.
Censoring at distant metastasis relapse, if applicable, did not change the effect size of the ER-negative PRS313, HR per SD = 1.12, 95% CI (1.06–1.18), p value = 4.9×10-5.
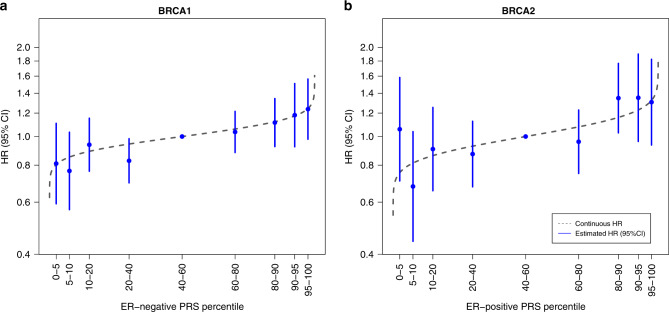
The HR estimates for association with contralateral breast cancer for different quantiles of the ER-negative PRS313, were consistent with the predicted HRs from the model using the continuous ER-negative PRS313 (Table 2 and Fig. 2).
Fig. 2. Association between categories of the PRS and contralateral breast cancer risk for BRCA1 and BRCA2 heterozygotes.
HRs and 95% CI for percentiles of the ER-negative PRS313 for BRCA1 heterozygotes and the ER-positive PRS313 for BRCA2 heterozygotes, relative to the middle quintile. The PRS percentile groups were 0–5%, 5–10%, 10–20%, 20–40%, 40–60% (reference), 60–80%, 80–90%, 90–95%, and 95–100% based on the distribution in unilateral breast cancer cases. The numbers and corresponding effect sizes are shown in Table 2. The gray line represents the distribution based on the HR of the continuous ER-negative PRS313 and ER-positive PRS313 and the distribution in unilateral breast cancer cases of BRCA1 and BRCA2 heterozygotes respectively. CI confidence interval, ER estrogen receptor, HR hazard ratio, PRS polygenic risk score.
For ER-positive contralateral breast cancer as event, the PRS313 showed the largest association, HR per SD = 1.32, 95% CI (1.12–1.56), p value = 0.002. For ER-negative contralateral breast cancer as event, only the ER-negative PRS313 showed a significant association, HR per SD = 1.07, 95% CI (1.01–1.15), p value = 0.036 (Table S4).
BRCA2 heterozygotes
For BRCA2 heterozygotes the largest association was seen with the ER-positive PRS313, HR per SD = 1.15, 95% CI (1.07–1.25), p value = 1.9×10−4, C-index 0.57, 95% CI (0.54–0.59). There was no evidence of violation of the proportional hazard assumption, p value = 0.300.
Neither sequential inclusion of possible confounders, nor including all these confounders in one model, changed the log HR estimate for the ER-positive PRS313 association more than 10% when compared with the model with no confounders (Table S5).
Considering only invasive contralateral breast cancer as the event of interest resulted in a similar association, HR per SD for the ER-positive PRS313 = 1.15, 95% CI (1.06–1.25), p value = 6.0×10−4.
Censoring at distant metastasis relapse, if applicable, did not change the effect size of the ER-positive PRS313, HR per SD = 1.15, 95% CI (1.07–1.24), p value = 2.1×10-4.
The HR estimates for association with contralateral breast cancer for different quantiles of the ER-positive PRS313, were consistent with the predicted estimates using the continuous PRS313 (Table 2 and Fig. 2).
The ER-positive PRS313 showed the largest association with ER-positive contralateral breast cancer for BRCA2 heterozygotes, HR per SD = 1.22, 95% CI (1.11–1.33), p value = 2.2×10−5 (Table S4). None of the PRS showed significant associations with ER-negative contralateral breast cancer for BRCA2 heterozygotes, but the ER-negative PRS313 exhibited the largest HR estimate, HR per SD = 1.10, 95% CI (0.91–1.32), p value = 0.346.
Interaction with age at first breast cancer diagnosis
A significant interaction between the age at first breast cancer diagnosis and the ER-negative PRS313 was found for BRCA1 heterozygotes: HR per year = 0.99, 95% CI (0.99–1.00), p value = 0.025. For BRCA2 heterozygotes a similar magnitude of interaction was observed with the ER-positive PRS313, although the interaction was not significant, HR per year = 0.99, 95% CI (0.99–1.00), p value = 0.09.
Categorizing age at first breast cancer diagnosis for BRCA1 heterozygotes resulted in HRs per SD of the ER-negative PRS313 of 1.22, 95% CI (1.14–1.31); 0.99, 95% CI (0.90–1.09);, and 1.03, 95% CI (0.86–1.24) for ages <40 years, 40–50 years, and ≥50 year respectively. For BRCA2 heterozygotes the corresponding estimates for ER-positive PRS313 were 1.23, 95% CI (1.09–1.38); 1.19, 95% CI (1.05–1.34); and 0.97, 95% CI (0.81–1.15) respectively (Table 2).
Analyses by predicted variant effect on protein expression
For BRCA1 heterozygotes, the HRs for association between the ER-negative PRS313 and contralateral breast cancer risk were similar for heterozygotes of pathogenic variants, which lead to a stable mutant protein (class II) compared with those leading to no protein or an unstable protein (class I). For BRCA2 heterozygotes, the ER-positive PRS313 effect size for the association with contralateral breast cancer risk was nonsignificantly smaller among heterozygotes of a pathogenic variant that lead to a stable mutant protein, although statistical power to detect these associations was low and the confidence intervals overlap with the overall estimate (Table 2).
Cumulative risks
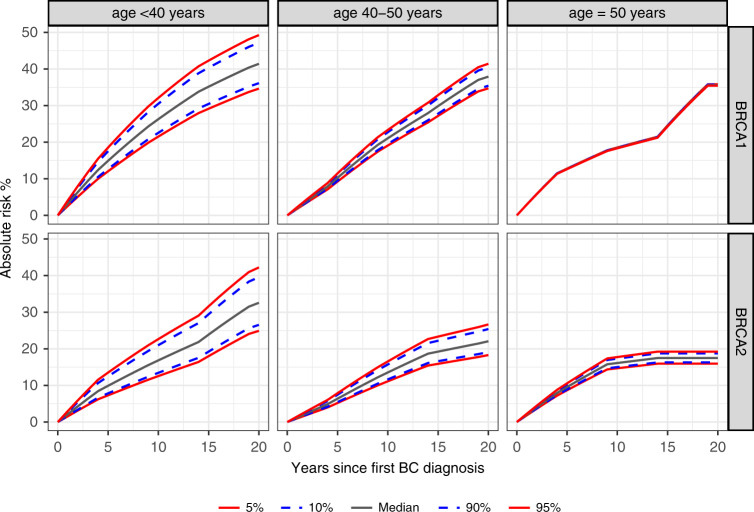
Estimate cumulative contralateral breast cancer risks, by categories of age at diagnosis of the first breast cancer are shown in Fig. 3. The largest risk difference was seen for women with a first breast cancer diagnosis before the age of 40, with BRCA1 heterozygotes at the 5th percentile of the ER-negative PRS313 having a 10- and 20-year risk of 22% and 35% compared with 32% and 49% at the 95th percentile, respectively. For BRCA2 heterozygotes, the 10- and 20-year risks in this category were 13% and 25% at the 5th percentile of the ER-positive PRS313 compared with 23% and 42% for women at the 95th percentile.
Fig. 3. Absolute contralateral breast cancer risk by PRS percentiles per age category of the first breast cancer diagnosis for BRCA1 and BRCA2 heterozygotes.
Predicted absolute contralateral breast cancer risks by percentile of the continuous ER-negative PRS313 for BRCA1 heterozygotes and ER-positive PRS313 for BRCA2 heterozygotes. The assumed contralateral breast cancer incidences were from a study that estimated breast cancer incidence in a large prospective cohort of BRCA1 and BRCA2 heterozygotes.20 The age categories were based on the age at diagnosis of the first primary breast tumor. Risks were calculated including the interaction between the PRS and the continuous age of first breast cancer diagnosis. The lines for different percentiles of the PRS are overlapping for the age category ≥50 year for BRCA1 heterozygotes. BC breast cancer, CBC contralateral breast cancer, PRS polygenic risk score.
DISCUSSION
In this study we investigated the associations between an established PRS based on 313 variants for primary first breast cancer and contralateral breast cancer risks among BRCA1 and BRCA2 heterozygotes of European ancestry enrolled in the large international retrospective CIMBA cohort. We showed significant albeit modest associations among both BRCA1 and BRCA2 heterozygotes between the PRS and contralateral breast cancer risk. For BRCA1 heterozygotes, the largest association was seen with the ER-negative PRS313, while for BRCA2 heterozygotes, both the PRS313 and ER-positive PRS313 showed similar associations with contralateral breast cancer risk that were somewhat larger than the ER-negative PRS313 association. These findings are consistent with previous studies on the effects of disease-specific PRS on the first breast cancers in BRCA1 and BRCA2 heterozygotes20,24 and with the higher relative prevalence of ER-negative and ER-positive contralateral breast cancers respectively, in this cohort.
For both BRCA1 and BRCA2 heterozygotes, the strength of the association was greater for ER-positive contralateral breast cancers compared with ER-negative contralateral breast cancers (in the case of BRCA1, even if the ER-negative PRS was used), although most of the confidence intervals overlapped. The effect sizes for the PRS are also larger for ER-positive disease in the general population, perhaps because ER-positive disease is commoner and the power to identify genetic variants has been greater for ER-positive disease. With larger data sets, it should be possible to develop better subtype specific PRS for contralateral breast cancer.
Although we found clear associations between the PRS and contralateral breast cancer risk, the magnitude of these associations (expressed in terms of HRs) were smaller than previously reported for the first breast cancers. For BRCA1 heterozygotes, the HR per SD for the association between the ER-negative PRS313 and breast cancer was 1.29, 95% CI (1.25–1.33),24 compared with 1.12, 95% CI (1.06–1.18) for contralateral breast cancer in this study. For BRCA2 heterozygotes, the HR per SD for the association between the ER-positive PRS313 and breast cancer was 1.31, 95% CI (1.26–1.36),24 compared with 1.15, 95% CI (1.07–1.24) for contralateral breast cancer in this study. This lower relative risk is consistent with a general pattern of a lower relative risk in a higher risk population, as seen in the lower relative risk for contralateral breast cancer than first breast cancer in the general population,19 and the lower relative risk for the first cancer in BRCA1/2 heterozygotes than in the general population.24 The attenuated estimate might be explained by several factors, some of which are speculative. BRCA1/2 pathogenic variant heterozygotes in this study were selected based on having a first breast cancer; these women will have on average a higher PRS, but also higher frequencies of other genetic and nongenetic risk factors than women who do not develop breast cancer at all. This can lead to a weaker association with the PRS as women with the largest PRS may have lower risks due to other factors, a phenomenon related to index event bias.35 There could also be negative interactions between the PRS effect and other risk factors (for example, treatment factors). However, in this study, we have shown that adjustment for the known contralateral breast cancer risk factors did not change the effect size of the PRS, which was also shown in population-based studies.17,19 Finally, although we tried to exclude potential early metastases misdiagnosed as second primaries by excluding women who developed a contralateral breast cancer the first year after the primary diagnosis, it is possible that a small percentage of contralateral breast cancers were metastases.36
A limitation of this study is that participants were recruited through clinical genetic centers, resulting in ascertainment bias, as individuals are more likely to have a strong family of breast cancer and/or be affected at a young age to be referred for testing. This was a historical cohort in which follow-up was prior to entry into CIMBA, so that all cases are prevalent. Therefore, the breast cancer patients included in the analyses are likely to be at higher contralateral breast cancer risk when compared with the general BRCA1/2 heterozygote breast cancer population. Indeed, the estimated 20-year risks of developing contralateral breast cancer in this study were higher compared to a previously published study with a prospective design:1 47% versus 40% for BRCA1 heterozygotes and 40% versus 26% for BRCA2 heterozygotes, respectively. While this is unlikely to introduce a significant bias in the relative risk estimates, a prospective cohort would clearly be preferably, although this will take several years to achieve. Finally, the PRS was developed using data sets of women of European ancestry, since our data set included insufficient samples of women of other ancestries, and our results were exclusively based on women of European ancestry. Therefore, caution is required when applying this to non-European ancestry populations. However, a population study found clear associations between the PRS, based on the same 313 variants or a subset of these variants, and (contralateral) breast cancer also in women of Asian ancestry. The effect size of these associations were slightly weaker, possibly reflecting the fact that this PRS was developed in a cohort of women of European ancestry.16,19 These results suggest that there might be an association with the PRS as well in BRCA1/2 heterozygotes of Asian ancestry. Future studies including a sufficient number of individuals of Asian ancestry are needed to confirm this statement.
Although the relative risks of the PRS for contralateral breast cancer were modest, differences in the PRS may still have an important effect on the absolute risk, which is high. BRCA1 and BRCA2 heterozygotes under age 40 at first breast cancer, at the 5th and 95th percentile of the PRS, differed by 10% in 10-year contralateral breast cancer risk. These absolute risk differences are modest, but might be of relevance for the choices regarding preventive surgery if incorporated into a multifactorial model that includes other predictive factors, such as family history and adjuvant systemic treatment of the first breast cancer.37,38 In the context of such a comprehensive model, further research is needed to investigate whether the PRS would contribute to the choices that women make for follow-up or preventive surgery.
To summarize, we have investigated the associations between PRS based on 313 variants with contralateral breast cancer risk in a large international series of BRCA1/2 heterozygotes. We found that the PRS is associated with contralateral breast cancer risk in both BRCA1 and BRCA2 heterozygotes of European ancestry and that PRS can be used to refine estimates of contralateral breast cancer risks in these women. However, for women with a first breast cancer after the age of 50, PRS may be of less value in the prediction of the contralateral breast cancer risk. Incorporating risk factors other than PRS and including ER-specific estimates may further improve contralateral breast cancer risk prediction. Before implementation in a diagnostic setting, our results should be validated in a prospective cohort of BRCA1 and BRCA2 heterozygotes.
Supplementary information
Acknowledgements
We acknowledge all the families, clinicians, family doctors, researchers, research nurses, research assistants, and technicians who contribute to the individual studies from which we used the data for this research and paper.
Author contributions
Conceptualization: A.C.C., M.R., M.K.S. Formal analysis: I.M.M.L., A.J.v.d.B., J.A.M.V., D.R.B. Resources: All authors. Supervision: A.C.C., M.R., M.K.S. Writing—original draft: I.M.M.L., A.C.C., M.R., M.K.S. Writing—review & editing: All authors.
Funding
This work was supported by the Alpe d’HuZes/Dutch Cancer Society (KWF Kankerbestrijding) project 6253 and Dutch Cancer Society (KWF Kankerbestrijding) project UL2014-7473. CIMBA: The CIMBA data management and data analysis were supported by Cancer Research–UK grants C12292/A20861, C12292/A11174. G.C.T. and A.B.S. are NHMRC Research Fellows. iCOGS: the European Community’s Seventh Framework Programme under grant agreement number 223175 (HEALTH-F2-2009-223175) (COGS), Cancer Research UK (C1287/A10118, C1287/A 10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692, C8197/A16565), the National Institutes of Health (CA128978) and Post-Cancer GWAS initiative (1U19 CA148537, 1U19 CA148065 and 1U19 CA148112–the GAME-ON initiative), the Department of Defence (W81XWH-10-1-0341), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer (CRN-87521), and the Ministry of Economic Development, Innovation and Export Trade (PSR-SIIRI-701), Komen Foundation for the Cure, the Breast Cancer Research Foundation, and the Ovarian Cancer Research Fund. OncoArray: the PERSPECTIVE and PERSPECTIVE I&I projects funded by the Government of Canada through Genome Canada and the Canadian Institutes of Health Research, the Ministère de l’Économie, de la Science et de l’Innovation du Québec through Genome Québec, and the Quebec Breast Cancer Foundation; the NCI Genetic Associations and Mechanisms in Oncology (GAME-ON) initiative and Discovery, Biology and Risk of Inherited Variants in Breast Cancer (DRIVE) project (NIH grants U19 CA148065 and X01HG007492); and Cancer Research UK (C1287/A10118 and C1287/A16563). BCFR: UM1 CA164920 from the National Cancer Institute. The content of this paper does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the BCFR. BFBOCC: Lithuania (BFBOCC-LT): Research Council of Lithuania grant SEN-18/2015. BIDMC: Breast Cancer Research Foundation. BMBSA: Cancer Association of South Africa (PI Elizabeth J. van Rensburg). BRI-COH: S.L.N. is partially supported by the Morris and Horowitz Families Professorship. CNIO: Spanish Ministry of Health PI16/00440 supported by FEDER funds, the Spanish Ministry of Economy and Competitiveness (MINECO) SAF2014-57680-R and the Spanish Research Network on Rare diseases (CIBERER). COH-CCGCRN: Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under grant number R25CA112486, and RC4CA153828 (PI: J. Weitzel) from the National Cancer Institute and the Office of the Director, National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. CONSIT TEAM: Associazione Italiana Ricerca sul Cancro (AIRC; IG2015 number 16732) to P. Peterlongo. DEMOKRITOS: European Union (European Social Fund–ESF) and Greek national funds through the Operational Program “Education and Lifelong Learning” of the National Strategic Reference Framework (NSRF)–Research Funding Program of the General Secretariat for Research & Technology: SYN11_10_19 NBCA. Investing in knowledge society through the European Social Fund. DFKZ: German Cancer Research Center. EMBRACE: Cancer Research UK Grants C1287/A10118 and C1287/A11990. D.G.E. and F.L. are supported by an NIHR grant to the Biomedical Research Centre, Manchester. The Investigators at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust are supported by an NIHR grant to the Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust. R.E. and E.B. are supported by Cancer Research UK Grant C5047/A8385. R.E. is also supported by NIHR support to the Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust. FCCC: A.K.G. was in part funded by the NCI (R01 CA214545), The University of Kansas Cancer Center Support Grant (P30 CA168524), The Kansas Institute for Precision Medicine (P20 GM130423), and the Kansas Bioscience Authority Eminent Scholar Program. A.K.G. is the Chancellors Distinguished Chair in Biomedical Sciences Professorship. FPGMX: A. Vega is supported by the Spanish Health Research Foundation, Instituto de Salud Carlos III (ISCIII), partially supported by FEDER funds through Research Activity Intensification Program (contract grant numbers: INT15/00070, INT16/00154, INT17/00133), and through Centro de Investigación Biomédica en Red de Enferemdades Raras CIBERER (ACCI 2016: ER17P1AC7112/2018); Autonomous Government of Galicia (Consolidation and structuring program: IN607B), and by the Fundación Mutua Madrileña. The German Consortium for Hereditary Breast and Ovarian Cancer (GC-HBOC) is funded by the German Cancer Aid (110837, 70111850, coordinator: Rita K. Schmutzler, Cologne) and the Ministry for Innovation, Science and Research of the State of North Rhine-Westphalia (#323-8.0302.16.02-132142). GEMO: initially funded by the French National Institute of Cancer (INCa, PHRC Ile de France, grant AOR 01 082, 2001-2003, grant 2013-1-BCB-01-ICH-1), the Association “Le cancer du sein, parlons-en!” Award (2004), the Association for International Cancer Research (2008-2010), and the Foundation ARC pour la recherche sur le cancer (grant PJA 20151203365). It also received support from the Canadian Institute of Health Research for the “CIHR Team in Familial Risks of Breast Cancer” program (2008–2013), and the European commission FP7, Project «Collaborative Ovarian, breast and prostate Gene-environment Study (COGS), Large-scale integrating project» (2009–2013). GEMO is currently supported by the INCa grant SHS-E-SP 18-015. GEORGETOWN: The Survey, Recruitment, and Biospecimen Collection Shared Resource at Georgetown University (NIH/NCI grant P30-CA051008), the Fisher Center for Hereditary Cancer and Clinical Genomics Research, and the Nina Hyde Center for Breast Cancer Research. G-FAST: Bruce Poppe is a senior clinical investigator of FWO. Mattias Van Heetvelde obtained funding from IWT. HCSC: Spanish Ministry of Health PI15/00059, PI16/01292, and CB-161200301 CIBERONC from ISCIII (Spain), partially supported by European Regional Development FEDER funds. HEBCS: Helsinki University Hospital Research Fund, the Finnish Cancer Society and the Sigrid Juselius Foundation. The HEBON study is supported by the Dutch Cancer Society grants NKI1998-1854, NKI2004-3088, NKI2007-3756, the Netherlands Organisation of Scientific Research grant NWO 91109024, the Pink Ribbon grants 110005 and 2014-187.WO76, the BBMRI grant NWO 184.021.007/CP46 and the Transcan grant JTC 2012 Cancer 12-054. HRBCP: Hong Kong Sanatorium and Hospital, Dr Ellen Li Charitable Foundation, The Kerry Group Kuok Foundation, National Institute of Health1R 03CA130065, and North California Cancer Center. HUNBOCS: Hungarian Research Grants KTIA-OTKA CK-80745, NKFI_OTKA K-112228 and TUDFO/51757/2019-ITM. ICO: Contract grant sponsor: Supported by the Carlos III National Health Institute funded by FEDER funds–a way to build Europe–(PI16/00563, PI19/00553 and CIBERONC); the Government of Catalonia (Pla estratègic de recerca i innovació en salut (PERIS) Project MedPerCan, 2017SGR1282 and 2017SGR496); and CERCA program.IHCC: supported by grant PBZ_KBN_122/P05/2004 and the program of the Minister of Science and Higher Education under the name “Regional Initiative of Excellence” in 2019–2022 project number 002/RID/2018/19 amount of financing 12 000 000 PLN. ILUH: Icelandic Association “Walking for Breast Cancer Research” and by the Landspitali University Hospital Research Fund. INHERIT: Canadian Institutes of Health Research for the “CIHR Team in Familial Risks of Breast Cancer” program–grant CRN-87521 and the Ministry of Economic Development, Innovation and Export Trade–grant # PSR-SIIRI-701. IOVHBOCS: Ministero della Salute and “5×1000” Istituto Oncologico Veneto grant. IPOBCS: Liga Portuguesa Contra o Cancro. kConFab: The National Breast Cancer Foundation, and previously by the National Health and Medical Research Council (NHMRC), the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia. KOHBRA: the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), and the National R&D Program for Cancer Control, Ministry of Health & Welfare, Republic of Korea (HI16C1127; 1020350; 1420190). KUMC: NIGMS P20 GM130423 (to A.K.G.). MAYO: NIH grants CA116167, CA192393 and CA176785, an NCI Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA116201), and a grant from the Breast Cancer Research Foundation. MCGILL: Jewish General Hospital Weekend to End Breast Cancer, Quebec Ministry of Economic Development, Innovation and Export Trade. Marc Tischkowitz is supported by the funded by the European Union Seventh Framework Program (2007Y2013)/European Research Council (Grant No. 310018). MODSQUAD: MH CZ–DRO (MMCI, 00209805) and LM2018125, MEYS–NPS I–LO1413 to LF, and by Charles University in Prague project UNCE204024 (MZ). MSKCC: the Breast Cancer Research Foundation, the Robert and Kate Niehaus Clinical Cancer Genetics Initiative, the Andrew Sabin Research Fund and a Cancer Center Support Grant/Core Grant (P30 CA008748). NAROD: 1R01 CA149429-01. NCI: the Intramural Research Program of the US National Cancer Institute, NIH, and by support services contracts NO2-CP-11019-50, N02-CP-21013-63 and N02-CP-65504 with Westat, Inc, Rockville, MD. NICCC: Clalit Health Services in Israel, the Israel Cancer Association and the Breast Cancer Research Foundation (BCRF), NY. NNPIO: the Russian Foundation for Basic Research (grants 17-00-00171, 18-515-45012 and 19-515-25001). NRG Oncology: U10 CA180868, NRG SDMC grant U10 CA180822, NRG Administrative Office and the NRG Tissue Bank (CA 27469), the NRG Statistical and Data Center (CA 37517) and the Intramural Research Program, NCI. OSUCCG: Ohio State University Comprehensive Cancer Center. PBCS: supported by the “Fondazione Pisa per la Scienza, project nr. 127/2016. Maria A Caligo was supported by the grant: “n. 127/16 Caratterizzazione delle varianti missenso nei geni BRCA1/2 per la valutazione del rischio di tumore al seno” by Fondazione Pisa, Pisa, Italy; SEABASS: Ministry of Science, Technology and Innovation, Ministry of Higher Education (UM.C/HlR/MOHE/06) and Cancer Research Initiatives Foundation. SMC: the Israeli Cancer Association. SWE-BRCA: the Swedish Cancer Society. UCHICAGO: NCI Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA125183), R01 CA142996, 1U01CA161032 and by the Ralph and Marion Falk Medical Research Trust, the Entertainment Industry Fund National Women’s Cancer Research Alliance and the Breast Cancer research Foundation. O.I.O. is an ACS Clinical Research Professor. UCLA: Jonsson Comprehensive Cancer Center Foundation; Breast Cancer Research Foundation. UCSF: UCSF Cancer Risk Program and Helen Diller Family Comprehensive Cancer Center. UKFOCR: Cancer Research h UK. UPENN: Breast Cancer Research Foundation; Susan G. Komen Foundation for the cure, Basser Research Center for BRCA. UPITT/MWH: Hackers for Hope Pittsburgh. VFCTG: Victorian Cancer Agency, Cancer Australia, National Breast Cancer Foundation. WCP: B.Y.K. is funded by the American Cancer Society Early Detection Professorship (SIOP-06-258-01-COUN) and the National Center for Advancing Translational Sciences (NCATS), grant UL1TR000124.
Data availability
CIMBA data is available on request. To receive access to the data, a concept form must be submitted, which will then be reviewed by the CIMBA Data Access Coordination Committee (DACC). Please contact Lesley McGuffog (email: ljm26@medschl.cam.ac.uk) to get access to these concept forms (http://cimba.ccge.medschl.cam.ac.uk/contact/).
Ethics declaration
All participants were recruited by the host institutions under protocols approved by local ethics review boards and provided written informed consent.24
Competing interests
C. Isaacs is consultant to Astra Zeneca, Novartis, Pfizer, Genentech, PUMA, Seattle Genetics, and received research support from Tesaro. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Inge M. M. Lakeman, Alexandra J. van den Broek.
Shared last authors: Antonis C. Antoniou, Mark Robson, Marjanka K. Schmidt.
Lists of authors and their affiliations appears at the end of the paper.
Contributor Information
Marjanka K. Schmidt, Email: mk.schmidt@nki.nl
GEMO Study Collaborators:
Emmanuelle Barouk-Simonet, Muriel Belotti, Pascaline Berthet, Yves-Jean Bignon, Valérie Bonadona, Brigitte Bressac-de Paillerets, Bruno Buecher, Sandrine Caputo, Olivier Caron, Laurent Castera, Virginie Caux-Moncoutier, Chrystelle Colas, Marie-Agnès Collonge-Rame, Isabelle Coupier, Antoine de Pauw, Capucine Delnatte, Camille Elan, Laurence Faivre, Sandra Fert Ferrer, Marion Gauthier-Villars, Paul Gesta, Sophie Giraud, Lisa Golmard, Claude Houdayer, Christine Lasset, Maïté Laurent, Dominique Leroux, Michel Longy, Véronique Mari, Sylvie Mazoyer, Noura Mebirouk, Isabelle Mortemousque, Fabienne Prieur, Pascal Pujol, Claire Saule, Helene Schuster, Nicolas Sevenet, Hagay Sobol, Johanna Sokolowska, and Laurence Venat-Bouvet
EMBRACE Collaborators:
Munaza Ahmed, Julian Barwell, Angela Brady, Paul Brennan, Carole Brewer, Jackie Cook, Rosemarie Davidson, Alan Donaldson, Alison M. Dunning, Jacqueline Eason, Diana M. Eccles, Helen Gregory, Helen Hanson, Patricia A. Harrington, Alex Henderson, Shirley Hodgson, M. John Kennedy, Fiona Lalloo, Clare Miller, Patrick J. Morrison, Kai-ren Ong, Aoife O’Shaughnessy-Kirwan, Jo Perkins, Mary E. Porteous, Mark T. Rogers, Lucy E. Side, Katie Snape, and Lisa Walker
OCGN Investigators:
Gord Glendon and Anna Marie Mulligan
HEBON Investigators:
Christi J. van Asperen, Cora M. Aalfs, Muriel A. Adank, Margreet G. E. M. Ausems, Marinus J. Blok, Encarna B. Gómez Garcia, Bernadette A. M. Heemskerk-Gerritsen, Antoinette Hollestelle, Agnes Jager, Linetta B. Koppert, Marco Koudijs, Mieke Kriege, Hanne E. J. Meijers-Heijboer, Arjen R. Mensenkamp, Thea M. Mooij, Jan C. Oosterwijk, Ans M. W. van den Ouweland, Frederieke H. van der Baan, Annemieke H. van der Hout, Lizet E. van der Kolk, Rob B. van der Luijt, Carolien H. M. van Deurzen, Helena C. van Doorn, Klaartje van Engelen, Liselotte P. van Hest, Theo A. M. van Os, Senno Verhoef, Maartje J. Vogel, and Juul T. Wijnen
KconFab Investigators:
Jonathan Beesley, Stephen Fox, Helene Holland, Kelly-Anne Phillips, and Amanda B. Spurdle
Supplementary information
The online version contains supplementary material available at 10.1038/s41436-021-01198-7.
References
- 1.Kuchenbaecker KB, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017;317:2402–2416. doi: 10.1001/jama.2017.7112. [DOI] [PubMed] [Google Scholar]
- 2.Graeser MK, et al. Contralateral breast cancer risk in BRCA1 and BRCA2 mutation carriers. J. Clin. Oncol. 2009;27:5887–5892. doi: 10.1200/JCO.2008.19.9430. [DOI] [PubMed] [Google Scholar]
- 3.Mavaddat N, et al. Cancer risks for BRCA1 and BRCA2 mutation carriers: results from prospective analysis of EMBRACE. J. Natl. Cancer Inst. 2013;105:812–822. doi: 10.1093/jnci/djt095. [DOI] [PubMed] [Google Scholar]
- 4.Metcalfe K, et al. Predictors of contralateral breast cancer in BRCA1 and BRCA2 mutation carriers. Br. J. Cancer. 2011;104:1384–1392. doi: 10.1038/bjc.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhiem K, et al. The risk of contralateral breast cancer in patients from BRCA1/2 negative high risk families as compared to patients from BRCA1 or BRCA2 positive families: a retrospective cohort study. Breast Cancer Res. 2012;14:R156. doi: 10.1186/bcr3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Kolk DM, et al. Penetrance of breast cancer, ovarian cancer and contralateral breast cancer in BRCA1 and BRCA2 families: high cancer incidence at older age. Breast Cancer Res. Treat. 2010;124:643–651. doi: 10.1007/s10549-010-0805-3. [DOI] [PubMed] [Google Scholar]
- 7.Lizarraga IM, Sugg SL, Weigel RJ, Scott-Conner CE. Review of risk factors for the development of contralateral breast cancer. Am. J. Surg. 2013;206:704–708. doi: 10.1016/j.amjsurg.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Kramer I, et al. The influence of adjuvant systemic regimens on contralateral breast cancer risk and receptor subtype. J. Natl. Cancer Inst. 2019;111:709–718. doi: 10.1093/jnci/djz010. [DOI] [PubMed] [Google Scholar]
- 9.Carbine NE, Lostumbo L, Wallace J, Ko H. Risk-reducing mastectomy for the prevention of primary breast cancer. Cochrane Database Syst. Rev. 2018;4:Cd002748. doi: 10.1002/14651858.CD002748.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van den Broek AJ, et al. Impact of age at primary breast cancer on contralateral breast cancer risk in BRCA1/2 mutation carriers. J. Clin. Oncol. 2016;34:409–418. doi: 10.1200/JCO.2015.62.3942. [DOI] [PubMed] [Google Scholar]
- 11.Michailidou K, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 2013;45:353–352. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michailidou, K. et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 551, 92–94 (2017). [DOI] [PMC free article] [PubMed]
- 13.Lilyquist, J., Ruddy, K. J., Vachon, C. M. & Couch, F. J. Common genetic variation and breast cancer risk—past, present, and future. Cancer Epidemiol. Biomarkers Prev. 27, 380–394 (2018). [DOI] [PMC free article] [PubMed]
- 14.Mavaddat N, et al. Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am. J. Hum. Genet. 2019;104:21–34. doi: 10.1016/j.ajhg.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mavaddat, N. et al. Prediction of breast cancer risk based on profiling with common genetic variants. J. Natl. Cancer Inst. 107, djv036 (2015). [DOI] [PMC free article] [PubMed]
- 16.Ho WK, et al. European polygenic risk score for prediction of breast cancer shows similar performance in Asian women. Nat. Commun. 2020;11:3833. doi: 10.1038/s41467-020-17680-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robson, M. E. et al. Association of common genetic variants with contralateral breast cancer risk in the WECARE Study. J. Natl. Cancer Inst.109, djx051 (2017). [DOI] [PMC free article] [PubMed]
- 18.Sawyer S, et al. A role for common genomic variants in the assessment of familial breast cancer. J. Clin. Oncol. 2012;30:4330–4336. doi: 10.1200/JCO.2012.41.7469. [DOI] [PubMed] [Google Scholar]
- 19.Kramer I, et al. Breast cancer polygenic risk score and contralateral breast cancer risk. Am. J. Hum. Genet. 2020;107:837–848. doi: 10.1016/j.ajhg.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuchenbaecker, K. B. et al. Evaluation of polygenic risk scores for breast and ovarian cancer risk prediction in BRCA1 and BRCA2 mutation carriers. J. Natl. Cancer Inst.109, djw302 (2017). [DOI] [PMC free article] [PubMed]
- 21.Antoniou AC, et al. Common variants in LSP1, 2q35 and 8q24 and breast cancer risk for BRCA1 and BRCA2 mutation carriers. Hum. Mol. Genet. 2009;18:4442–4456. doi: 10.1093/hmg/ddp372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antoniou AC, et al. Common breast cancer-predisposition alleles are associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. Am. J. Hum. Genet. 2008;82:937–948. doi: 10.1016/j.ajhg.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antoniou AC, et al. Common breast cancer susceptibility alleles and the risk of breast cancer for BRCA1 and BRCA2 mutation carriers: implications for risk prediction. Cancer Res. 2010;70:9742–9754. doi: 10.1158/0008-5472.CAN-10-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barnes DR, et al. Polygenic risk scores and breast and epithelial ovarian cancer risks for carriers of BRCA1 and BRCA2 pathogenic variants. Genet. Med. 2020;22:1653–1666. doi: 10.1038/s41436-020-0862-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gail MH, Pfeiffer RM. Breast cancer risk model requirements for counseling, prevention, and screening. J. Natl. Cancer Inst. 2018;110:994–1002. doi: 10.1093/jnci/djy013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chenevix-Trench G, et al. An international initiative to identify genetic modifiers of cancer risk in BRCA1 and BRCA2 mutation carriers: the Consortium of Investigators of Modifiers of BRCA1 and BRCA2 (CIMBA) Breast Cancer Res. 2007;9:104. doi: 10.1186/bcr1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amos CI, et al. The OncoArray Consortium: a network for understanding the genetic architecture of common cancers. Cancer Epidemiol. Biomarkers Prev. 2017;26:126–135. doi: 10.1158/1055-9965.EPI-16-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaudet MM, et al. Identification of a BRCA2-specific modifier locus at 6p24 related to breast cancer risk. PLoS Genet. 2013;9:e1003173. doi: 10.1371/journal.pgen.1003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Couch FJ, et al. Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genet. 2013;9:e1003212. doi: 10.1371/journal.pgen.1003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuchenbaecker KB, et al. Associations of common breast cancer susceptibility alleles with risk of breast cancer subtypes in BRCA1 and BRCA2 mutation carriers. Breast Cancer Res. 2014;16:3416. doi: 10.1186/s13058-014-0492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milne RL, et al. Identification of ten variants associated with risk of estrogen-receptor-negative breast cancer. Nat. Genet. 2017;49:1767–1778. doi: 10.1038/ng.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrell FE, Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat. Med. 1996;15:361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 33.Azur MJ, Stuart EA, Frangakis C, Leaf PJ. Multiple imputation by chained equations: what is it and how does it work? Int. J. Methods Psychiatr. Res. 2011;20:40–49. doi: 10.1002/mpr.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria (2019).
- 35.Dahabreh IJ, Kent DM. Index event bias as an explanation for the paradoxes of recurrence risk research. JAMA. 2011;305:822–823. doi: 10.1001/jama.2011.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Begg CB, et al. Contralateral breast cancers: Independent cancers or metastases? Int. J. Cancer. 2018;142:347–356. doi: 10.1002/ijc.31051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akdeniz D, et al. Risk factors for metachronous contralateral breast cancer: a systematic review and meta-analysis. Breast. 2019;44:1–14. doi: 10.1016/j.breast.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 38.Giardiello D, et al. Prediction and clinical utility of a contralateral breast cancer risk model. Breast Cancer Res. 2019;21:144. doi: 10.1186/s13058-019-1221-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CIMBA data is available on request. To receive access to the data, a concept form must be submitted, which will then be reviewed by the CIMBA Data Access Coordination Committee (DACC). Please contact Lesley McGuffog (email: ljm26@medschl.cam.ac.uk) to get access to these concept forms (http://cimba.ccge.medschl.cam.ac.uk/contact/).