Abstract
Background
African swine fever (ASF) is a hemorrhagic fever occurring in wild boars (Sus scrofa) and domestic pigs. The epidemic situation of ASF in South Korean wild boars has increased the risk of ASF in domestic pig farms. Although basic reproduction number (R0) can be applied for control policies, it is challenging to estimate the R0 for ASF in wild boars due to surveillance bias, lack of wild boar population data, and the effect of ASF-positive wild boar carcass on disease dynamics.
Objectives
This study was undertaken to estimate the R0 of ASF in wild boars in South Korea, and subsequently analyze the spatiotemporal heterogeneity.
Methods
We detected the local transmission clusters using the spatiotemporal clustering algorithm, which was modified to incorporate the effect of ASF-positive wild boar carcass. With the assumption of exponential growth, R0 was estimated for each cluster. The temporal change of the estimates and its association with the habitat suitability of wild boar were analyzed.
Results
Totally, 22 local transmission clusters were detected, showing seasonal patterns occurring in winter and spring. Mean value of R0 of each cluster was 1.54. The estimates showed a temporal increasing trend and positive association with habitat suitability of wild boar.
Conclusions
The disease dynamics among wild boars seems to have worsened over time. Thus, in areas with a high elevation and suitable for wild boars, practical methods need to be contrived to ratify the control policies for wild boars.
Keywords: African swine fever, wild boar, space-time clustering, basic reproduction number, South Korea
INTRODUCTION
The African swine fever virus (ASFV) induced African swine fever (ASF) is a catastrophic hemorrhagic fever disease affecting wild boar (Sus scrofa) and domestic pigs, and having a tremendously detrimental impact on the pig industry in Africa, Europe, and Asia [1]. Since the ASFV can spillover from wild boar to domestic pigs, this livestock-wildlife interaction has been highlighted and is considered one of the targets for formulating control policies of ASF [2,3]. It is important to study and understand the disease dynamics in wild boar [4,5,6].
Since the first case of ASF in wild boar in South Korea (October 2019), 1,416 cases have been reported until May 2021 [5]. Although the Korean government has implemented policies for ASF in wild boars, the epidemic situation has increased the risk of recurrence of ASF in domestic pig farms. In particular, recent cases in domestic pig farms are considered to be impacted by the ASF in wild boars [7].
Basic reproduction number (R0) is a key parameter of infectious disease epidemiology. It indicates the expected number of secondary cases directly infected from one primary case in a totally susceptible population [8,9]. This metric helps to understand the disease dynamics of infectious disease. Moreover, as the metric has characteristics of threshold centered on one, it helps determine whether the disease is spreading (R0 > 1), persisting (R0 = 1), or dying out (R0 < 1).
We anticipate that the R0 of ASF in wild boar can be applied to understand the disease dynamics and help develop control strategies. However, the lack of wild boar population data makes it hard to estimate R0. Even more, the spatiotemporally heterogeneous surveillance intensity for ASF in wild boar leads to artificial spatiotemporal disease patterns, which is called surveillance bias [5,10]. Those make it more challenging to estimate R0. Previous study that estimated the R0 of ASF in wild boars have tried to overcome the limitation [11]. For cases in the identified spatiotemporal clusters, the R0 was estimated by exponential growth assumption. This approach enabled the authors to estimate R0 without population data, which also relieve the effects of surveillance bias. Although the scan statistics, the clustering algorithm utilized in the study, have strength in that the algorithm can identify the clusters with statistical test, it is less likely to reflect transmission dynamics explicitly on the algorithm such as infectious period [12,13]. Moreover, the scan statistics has limitations to identify the true size and irregularly shaped clusters [14,15].
This study addresses the effects of ASF-positive wild boar carcass and the limitations of scan statistics by modifying the ST-DBSCAN, which is the density-based clustering algorithm, to incorporate the impacts of ASF-positive wild boar carcass on the disease dynamics. By applying this modified algorithm, we identified the spatiotemporally associated clusters [16]. For cases in each cluster, we estimated the R0 of ASF in wild boars, and analyzed the spatiotemporal heterogeneity and its association with the habitat suitability of wild boar.
MATERIALS AND METHODS
Data
The data for ASF-positive wild boar were retrieved from the database maintained by the Ministry of Environment, South Korea. The data include date of report, date of diagnosis, estimated death date, species (e.g., Sus scrofa), type of sample (i.e., carcass, capture, and hunt), and geographic location (i.e., latitude and longitude). The death date of carcass was estimated by researchers at the National Institute of Environmental Research. The death date of other sample types, including capture and hunt, was assumed to be the same as the date of report. Of the total 1,416 ASF-positive cases of wild boars, cases from carcasses, capture, and hunt comprised 1,354, 33, and 29 cases, respectively. In this study, we only included ASF-positive carcasses, due to the following reasons: 1) Due to the high severity and mortality (close to 100%) in wild boars, ASF-infected wild boars can transmit ASFV until 7 days after infection [17]. However, ASF-positive wild boar carcasses can also be a source of infection for weeks or months, due to the high environmental resistance of AFSV. Thus, ASF-positive wild boar carcasses are considered to be the main cause of transmission dynamics of ASFV in wild boars [12]. There is a need to further understand the impact of ASF-positive carcasses on the disease dynamics. 2) Wild boars are terrestrial mammals, and have a home range for roaming. Thus, the geographical locations of ASF-positive captured or hunted wild boars are affected by the locations of the traps and hunting activities, thereby leading to geographical bias. It is crucial to understand the impact of ASF-positive carcasses in ASF dynamics.
Estimation approach
Caution needs to be applied when analyzing the wild boar surveillance data for ASF. It is highly likely to be vulnerable to biases due to limited sensitivity of the surveillance system, and characteristics of the impact of ASF-positive wild boar carcasses on disease dynamics. Moreover, the lack of wild boar population data makes it hard to estimate the R0. To overcome these limitations, we applied a two-step approach: 1) spatiotemporal cluster analysis, and 2) statistical estimation of R0 in each cluster.
Local transmission clusters
Spatiotemporal cluster analysis was conducted to identify the local transmission clusters of ASF in wild boar. We defined the local transmission clusters as spatiotemporally associated cases of ASF in wild boar reflecting the disease dynamics of ASF and ecology of wild boars.
As the surveillance activities were focused on the previously reported regions [5], it can be assumed that the surveillance activities in local transmission clusters are relatively high and constant, thereby making it possible to alleviate biases from the imperfect detection sensitivity of the surveillance [11,18]. To identify the cluster, the transmission dynamics of ASFV need to be reflected by the clustering algorithm. Depending on the detection delay after death, ASF-positive boar carcasses can be sources of infection for different period of time, which leads to the fact that ASF-positive carcasses have varied infectious period (Fig. 1). In this study, we applied the ST-DBSCAN algorithm [16], which is based on the spatiotemporal density within a predefined temporal and geographical radius. This algorithm defines three parameters: spatial radius (eps), temporal radius (eps2), and minimal density (minpts). If the selected point has the neighborhood within the eps and eps2 distances, and the number of neighborhoods is higher than minpts, the point and its neighborhood are classified as a cluster. This process is serially conducted for all points, including points in the clusters. If a point in the cluster meets the criteria for cluster, its neighborhood will be included in the same cluster. Thus, this algorithm iteratively collects points into clusters. However, the algorithm has limitations in its inability to incorporate the characteristics of transmission dynamics of ASFV. Thus, the algorithm was modified to reflect the different infectious periods, depending on the lag of detection after the death of ASF-infected wild boar. Differences in the algorithm compared to the ST-DBSCAN were obtained for eps2. Thus, instead of the eps2, the modified algorithm selects the neighborhood points temporally located within the death time and detection time. Moreover, the buffers for death and detection date were included, which can construct a transmission network for cases that were infected at the initial stage of the infectious period, and at the time just before detection of the ASF-positive carcass, respectively (Fig. 1). To reflect the transmission viability of ASFV in ASF-positive carcasses, it was assumed that ASF-positive wild boar carcass continues to transmit ASFV until a certain period after death. To summarize, The modified algorithm of ST-DBSCAN needs to define the death date (eps_death) and detection date (eps_detect) instead of eps2, and also the buffers for death date (buf_death), detection date (buf_detect), transmission viability of ASFV (max_viab), spatial radius (eps), and minimal density (minpts). The ST-DBSCAN function in R software was retrieved from the GitHub of the developer of ST-DBSCAN (https://github.com/Kersauson/ST-DBSCAN), and the code for the modified algorithm can be found in the author's GitHub (https://github.com/borizook/ASF_ST_DBSCAN).
Fig. 1. Diagram illustrating the course of infection of ASF, and the parameters in the spatiotemporal clustering algorithm: E for the time of exposure to ASFV; I for the time to be infectious after the latent period from E; R for the time of death after infectious period from I; Duration of viable virus for the time period for transmission viability of ASFV from the R; D for the time of detection after detection delay from R; Buffer for death and Buffer for detection for the time periods for cases that were infected at the initial stage of the infectious period, and infected just before detection of the ASF-positive carcass, respectively.
ASF, African swine fever; ASFV, African swine fever virus.
In the current study, the eps parameter was defined as 3 km (the home range size), thus establishing the geographical boundaries for response strategies for ASF in the wild boars in South Korea [19]. Every eps_death and eps_detect for each carcass was assigned from date retrieved from the surveillance system (R and D in Fig. 1). As the ASF-positive wild boar starts shedding the ASFV between two and six days after infection (latent period in Fig. 1), and is dead within five to ten days after infection (infectious period in Fig. 1) [17,20], the buffers for the death and detection date were selected as three and ten days, respectively. For the max_viab parameter, given that the behavior of wild boars to their dead fellows usually include sniffing and poking on the carcass and not scavenging [13], the likelihood of infection from the totally skeletonized carcasses, whose ASFV can only be sampled from the bone marrow, was negligible. Thus, the max_viab was selected based on the experimental results of carcass decomposition of wild boars in various environmental conditions in South Korea (Duration of viable virus in Fig. 1) [21], which showed that the mean value of time to partial skeletonization was 45 days. Thus, the max_viab was selected as 45 days. When selecting the value of minpts, we considered the wild boar density in South Korea. According to the report “National Park Wild Boar Habitat Survey Research” by the Korea National Park Service [22], the minimum density of wild boars was 0.3 per square kilometer. We defined the high-risk cluster as spatiotemporally associated cases, with the number of ASF-positive wild boar carcasses exceeding the expected minimum number of wild boars within eps. Thus, the minpts was selected as 8.
Estimation of R0 in each cluster
Identified high-risk clusters were selected and the R0 was estimated for each cluster. It was assumed that the epidemic curves in each cluster grow at an exponential rate. The epidemic doubling time was estimated as follow [23]:
where yt is the number of observed cases at time t; k is the growth rate at which new cases occur; td is the epidemic doubling time. This formula was modified as follow [23]:
The modification was implemented on the premise that an infected case generates new cases of R0 during the infectious period (D), and the primary case eventually succumbs (i.e., carcass detection in this study). Thus, the relationship between R0 and k can be expressed as follow [23]:
where D is the infectious period. Combined together, the R0 can be estimated as follows [23]:
It is to be noted that the td remains constant at any time during the epidemic because of exponential growth assumption. td can be estimated from the observed epidemic curve in the identified clusters. However, the value of D need to be assumed. Since the ASF-positive carcass is a source of infection until detection, we considered the difference in value between death and detection date. As the ASF-infected wild boar continues to shed the ASFV for an average of 3.5 days [17,20], D was assumed as sum of the observed mean difference between death and detection date, and 3.5 days. Moreover, the maximum D value was restricted to the 45 days assigned to max_viab, which is the maximum period of transmission viability of ASFV in wild boar carcasses.
To identify the association between the basic reproduction number and environmental conditions of each cluster, we applied the habitat suitability of wild boar, extracted from the species distribution model (SDM) of wild boar developed by Kim and Pak [24]. SDM shows the quantitative continuous index for the environment suitability for wild boar on the base of environmental variables. The index ranges between 0 and 1. For the optimal habitat for wild boars, the index is scored 1; if the environment is unsuitable for wild boar, the index is scored 0. The median value of habitat suitability within a 3 km radius from the geographical location where the case was reported was extracted from SDM. Distribution of the median values of habitat suitability of cases in each cluster was then plotted. Cluster-level habitat suitability is defined as the median value of the extracted median values of habitat suitability. We applied Spearman's correlation analysis to assess the association between basic reproduction number and cluster-level habitat suitability. All clusters prior to April 2021 were analyzed. Clusters that ended in April 2021 were omitted for further analysis owing to the fact that these clusters may be incomplete due to cases reported after 6th May 2021, which was the end date of the retrieved surveillance data.
Statistical significance is defined as a p value < 0.05. All the data and statistical analyses were conducted using the R statistical software version 4.1.0 [25].
RESULTS
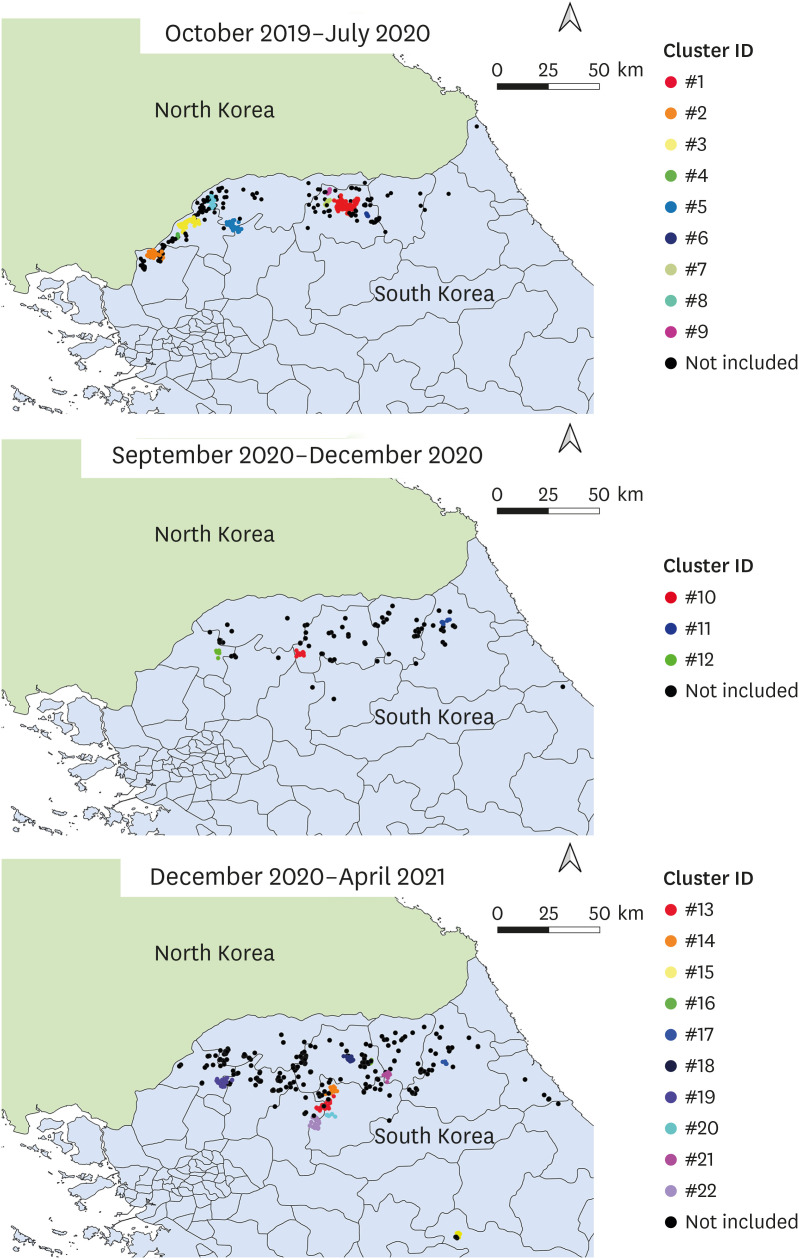
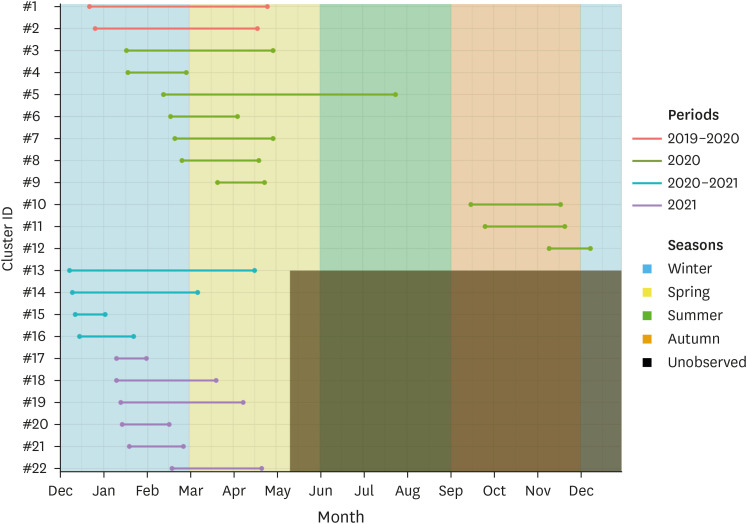
In all, 22 local transmission clusters were identified during the study period (Table 1). Of the total 1,416 cases, 753 cases (53.18%) were included in the clusters. Compared to the clusters identified during the period October 2019–July 2020, the identified clusters after September 2020 were located in the Eastern region of South Korea (Fig. 2). The number of cases included in each cluster ranged from 8 to 158, having a mean value of 34.22. The period of clusters revealed a seasonal pattern, with majority clusters identified during the winter to spring or autumn period (Fig. 3). The estimated doubling times ranged from 6.04 to 64.98 days, and the infectious period ranged from 7.5 to 23.5 days, which was higher in the recent clusters (Table 1). Mean value of the estimated basic reproduction number of each cluster was 1.54, ranging between 1.11 and 2.37 (Table 1). Cluster IDs 13, 19 and 22, whose end date was April 2020, were excluded for further analysis.
Table 1. Characteristics of the identified local transmission clusters, their reproduction numbers, and cluster-level habitat suitability, of African swine fever in wild boars in South Korea.
| Cluster ID | Number of cases | Period | Doubling time (95% CI) | Infectious period | Reproduction number (95% CI) | Habitat suitability (Min – Max) |
|---|---|---|---|---|---|---|
| 1 | 158 | 2019-12-22 – 2020-04-25 | 36.89 (35.77–38.08) | 10.5 | 1.20 (1.19–1.20) | 0.57 (0.51–0.64) |
| 2 | 53 | 2019-12-26 – 2020-04-18 | 58.65 (54.12–63.99) | 13.5 | 1.16 (1.15–1.17) | 0.39 (0.38–0.40) |
| 3 | 101 | 2020-01-17 – 2020-04-29 | 31.09 (29.77–32.52) | 8.5 | 1.19 (1.18–1.20) | 0.38 (0.35–0.40) |
| 4 | 10 | 2020-01-18 – 2020-02-28 | 47.48 (29.36–122.55) | 7.5 | 1.11 (1.04–1.18) | 0.36 (0.35–0.36) |
| 5 | 71 | 2020-02-12 – 2020-07-24 | 64.98 (61.84–68.45) | 13.5 | 1.14 (1.14–1.15) | 0.46 (0.40–0.51) |
| 6 | 15 | 2020-02-17 – 2020-04-04 | 17.44 (14.50–21.75) | 10.5 | 1.42 (1.33–1.50) | 0.63 (0.61–0.64) |
| 7 | 15 | 2020-02-20 – 2020-04-29 | 44.83 (36.05–59.10) | 15.5 | 1.24 (1.18–1.30) | 0.56 (0.56–0.56) |
| 8 | 15 | 2020-02-25 – 2020-04-19 | 26.77 (21.87–34.37) | 10.5 | 1.27 (1.21–1.33) | 0.40 (0.39–0.43) |
| 9 | 9 | 2020-03-21 – 2020-04-23 | 33.40 (20.37–91.03) | 23.5 | 1.49 (1.18–1.80) | 0.58 (0.57–0.59) |
| 10 | 16 | 2020-09-15 – 2020-11-17 | 17.37 (15.03–20.46) | 13.5 | 1.54 (1.46–1.62) | 0.53 (0.50–0.55) |
| 11 | 13 | 2020-09-25 – 2020-11-20 | 21.48 (17.85–26.81) | 9.5 | 1.31 (1.25–1.37) | 0.76 (0.75–0.76) |
| 12 | 19 | 2020-11-09 – 2020-12-08 | 6.04 (5.02–7.48) | 8.5 | 1.98 (1.79–2.17) | 0.34 (0.33–0.35) |
| 13 | 46 | 2020-12-08 – 2021-04-16 | 36.59 (34.63–38.76) | 15.5 | 1.29 (1.28–1.31) | 0.52 (0.46–0.56) |
| 14 | 34 | 2020-12-10 – 2021-03-07 | 25.89 (23.94–28.16) | 18.5 | 1.50 (1.46–1.54) | 0.51 (0.48–0.58) |
| 15 | 8 | 2020-12-12 – 2021-01-02 | 6.34 (4.68–9.51) | 10.5 | 2.15 (1.77–2.55) | 0.60 (0.59–0.61) |
| 16 | 8 | 2020-12-15 – 2021-01-22 | 16.45 (12.47–23.85) | 10.5 | 1.44 (1.31–1.58) | 0.59 (0.59–0.60) |
| 17 | 8 | 2021-01-10 – 2021-01-31 | 9.30 (6.43–16.35) | 17.0 | 2.27 (1.72–2.83) | 0.73 (0.72–0.74) |
| 18 | 25 | 2021-01-10 – 2021-03-20 | 14.81 (13.48–16.38) | 23.5 | 2.10 (1.99–2.21) | 0.55 (0.52–0.57) |
| 19 | 69 | 2021-01-13 – 2021-04-08 | 31.97 (30.01–34.18) | 18.5 | 1.40 (1.38–1.43) | 0.42 (0.36–0.47) |
| 20 | 14 | 2021-01-14 – 2021-02-16 | 12.76 (10.23–16.82) | 15.5 | 1.84 (1.64–2.05) | 0.48 (0.47–0.48) |
| 21 | 17 | 2021-01-19 – 2021-02-26 | 11.92 (9.95–14.75) | 23.5 | 2.37 (2.10–2.64) | 0.65 (0.63–0.66) |
| 22 | 29 | 2021-02-18 – 2021-04-21 | 23.61 (20.82–27.20) | 18.5 | 1.54 (1.47–1.62) | 0.46 (0.42–0.48) |
CI, confidence interval.
Fig. 2. Spatiotemporal distributions for the identified local transmission clusters in each corresponding period: October 2019 to July 2020 (Top); September 2020 to December 2020 (Middle); December 2020 to April 2021 (Bottom): Non included represents the cases which were not included in the identified clusters in each corresponding period.
Fig. 3. Plots showing the seasonality of the identified local transmission clusters. Colors are assigned with their corresponding period. Grey area is for the unobserved period after May 2021.
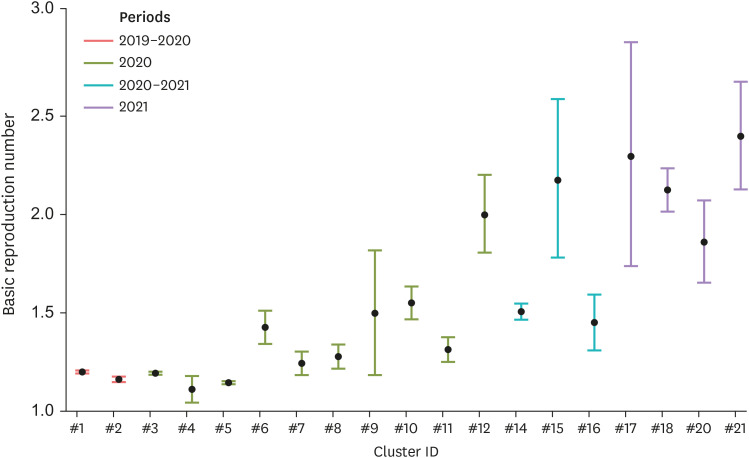
The estimated mean value of basic reproduction number of each cluster and its 95% confidence interval (95% CI) is plotted in Fig. 4. The results indicate that temporally recent clusters show a higher value of the basic reproduction numbers.
Fig. 4. Plot showing the estimated basic reproduction number (labeled as dot) and its 95% confidence interval for each cluster. The colors are assigned with their corresponding period.
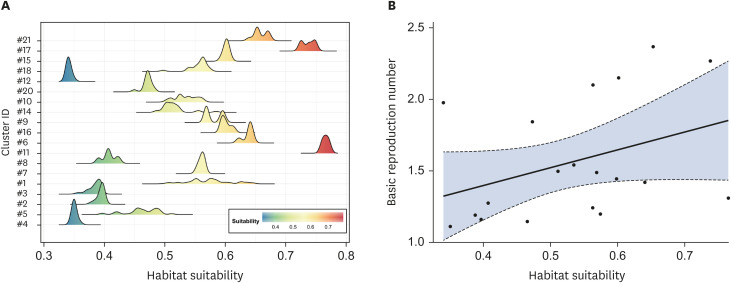
Median values of extracted habitat suitability ranged from 0.4 to 0.76, with mean value 0.52 (Table 1). Fig. 5A shows the distribution of median values of the habitat suitability of environments where a case was reported by each cluster. Cluster ID was ordered by the estimated basic reproduction number. The figure visually shows the positive association between habitat suitability and basic reproduction number. Results of the Spearman's correlation analysis shows a statistically significant positive association between the habitat suitability and basic reproduction number (rho = 0.47, p = 0.04). The scatter plot and its regression line showing the relationship between habitat suitability and basic reproduction number are plotted in Fig. 5B.
Fig. 5. Plots showing the association between habitat suitability of wild boar and basic reproduction number. (A) Ridge plots showing the distribution with habitat suitability of wild boar for each cluster. Cluster ID was ordered with its estimated basic reproduction number; (B) Scatter plot and regression line showing the association between the median habitat suitability of wild boars of each cluster and its basic reproduction number.
DISCUSSION
Basic reproduction number, as a key parameter for infectious disease epidemiology, would be helpful to understand the ASF dynamics and control ASF dynamics in wild boars in South Korea. To reduce bias from the imperfect detection sensitivity, we estimated R0 of the ASF in wild boar for the identified local transmission clusters incorporating the impact of ASF-positive wild boar carcasses on the disease dynamics. Totally, 22 identified clusters, mainly identified during the winter-spring season, showed R0 with a mean value of 1.54. Moreover, the estimated R0 had a temporal increasing trend, and were positively associated with the habitat suitability of wild boars.
Our results indicate that the clusters may show seasonal patterns, amassing during winter and spring. In particular, most of the identified clusters were initiated in December or January. This corresponds to the mating season of wild boars in South Korea. During the season, male wild boars tend to be non-solitary, travel long distances, and mate with five to ten females, which makes the contact network denser and larger. Moreover, the contact rate between wild boars also increases [26]. This results in higher risk of transmission events during the season than any other period. Our results confirmed previous reports for ASF [27] and other infectious diseases of wild boars [28]. Although the patterns should be studied further during 2021, it is highly expected that the seasonal pattern will be repeated, given the behavioral ecology of wild boars.
The estimated R0 ranged from 1.11 to 2.37 with a mean of 1.54, which was similar to the estimates from previous studies [11,29,30,31]. However, the current study revealed spatiotemporal variations in the values. Since R0 is the function of contact rate between hosts and infectious period, it is natural that R0 of a certain disease spatiotemporally varies under different environments [8,9]. However, it should be noted that R0 of ASF in the identified clusters in wild boars (1) is observed to increase with increasing time (Fig. 4), and (2) is higher in the regions where the environment is favorable to wild boars (Fig. 5). Given that the R0 is affected by the host density and time delay to detect the ASF-positive carcasses after death in the view of the transmission dynamics of ASFV in wild boars, the above results can be explained as follows. ASF in wild boar have been spread into the mountain ranges, whose environment is more suitable to wild boars (Fig. 2) [24]; thus, the higher density of wild boars results in increased contact rate between the animals (density-dependent contact rate in wild boars [32]), thereby resulting in the increased R0 (Fig. 5). Due to the high elevation of mountain ranges, the time required to detect ASF-positive wild boar carcass after death also increases, leading to a prolonged infectious period [5] (Table 1). As ASF in wild boars is expected to spread along the mountain ranges running from North to South in the Eastern regions of South Korea [5,33], it should be considered to contrive practical methods to impose the control policies for ASF in wild boars, to regions with high habitat suitability and high elevation.
The strength of association between R0 and the habitat suitability of wild boars (rho = 0.47) is considered to be the moderate according to the classification of strength of correlation [34]. This suggests that there are other factors such as age, sex, and the social network of wild boars demonstrating the R0 [26,35]. The reconstruction of transmission network would allow identifying individual level risk factors for spreading [36,37]. However, due to substantial imperfect detection of surveillance, it is expected to be challenging to reconstruct transmission network while disentangling the effects of surveillance activity and disease dynamics.
The modified algorithm of ST-DBSCAN with the parameters specified for ASF and ecology of wild boars enabled us to identify the clusters that can be considered as virtually linked transmission clusters. Moreover, the algorithm, as a modified version of ST-DBSCAN enables us to detect the irregularly shaped and closer to the true-sized cluster [15]. The cases not included in the clusters can be the ones that are not spatiotemporally related, in the context of ASF dynamics, to the cases in the clusters or the ones that did not meet the criterion for high-risk cluster (eps).
The results provided here should be interpreted with some cautions. First, there may be some errors in estimating the death date of wild boar carcass due to the interrelated factors, such as the decomposition of carcass [38]. However, the death date of all cases was estimated with the same logic applied by the National Institute of Environmental Research, which makes the temporal difference between the estimates and actual death date similar between the cases. Thus, we expect that the bias derived from the estimation error would be low. Secondly, there may be imperfect detection which can cause surveillance bias, although surveillance intensity within the regions of the identified clusters was supposed to be higher than other regions. However, since R0 is the ratio of the number of infected and reported individuals to the number of reported cases, the effect of surveillance bias on the estimates would disappear [9]. It has been known that other species such as scavengers and insects can spread ASF [39]. In this study, the estimation method for R0 did not disentangle the effect of the species on disease dynamics, thus the estimated R0 would be the averaged values of the transmission through direct and indirect contacts by wild boars and other species, respectively. Further studies for the contributions of other species to ASF dynamics are needed to separate R0 by each species. Nonetheless, as the other species are less likely to contribute to spreading ASF [40], the estimates can be considered to be derived from the direct contacts.
In this study, we estimated R0 of ASF in the identified local transmission clusters in wild boars. The estimated disease dynamics among wild boars seems to have worsened than before. However, the expected spreading and seasonal patterns of ASF in wild boars gives us clues to contain the epidemics. We believe that our results can be useful for designing and developing policies to eradicate ASF in wild boars, and potentially in domestic pig farms.
ACKNOWLEDGEMENTS
The authors would like to thank Jin A Kim (Korea Disease Control and Prevention Agency) for comments on the study.
Footnotes
Funding: This study was financially supported by the Institute of Planning and Evaluation for Technology in Food, Agriculture, and Forestry (IPET) grant funded by the Korea government (No. 321011-1).
Conflict of Interest: The authors declare no conflicts of interest.
- Conceptualization: Lim JS, Kim E, Ryu PD, Pak SI.
- Data curation: Lim JS, Pak SI.
- Formal analysis: Lim JS.
- Funding acquisition: Kim E.
- Methodology: Lim JS, Pak SI, Kim E.
- Writing - original draft: Lim JS.
- Writing - review & editing: Lim JS, Kim E, Ryu PD, Pak SI.
References
- 1.Dixon LK, Stahl K, Jori F, Vial L, Pfeiffer DU. African swine fever epidemiology and control. Annu Rev Anim Biosci. 2020;8(1):221–246. doi: 10.1146/annurev-animal-021419-083741. [DOI] [PubMed] [Google Scholar]
- 2.Andraud M, Bougeard S, Chesnoiu T, Rose N. Spatiotemporal clustering and Random Forest models to identify risk factors of African swine fever outbreak in Romania in 2018–2019. Sci Rep. 2021;11(1):2098. doi: 10.1038/s41598-021-81329-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chenais E, Ståhl K, Guberti V, Depner K. Identification of wild boar-habitat epidemiologic cycle in African swine fever epizootic. Emerg Infect Dis. 2018;24(4):810–812. doi: 10.3201/eid2404.172127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vergne T, Guinat C, Pfeiffer DU. Undetected circulation of African swine fever in wild boar, Asia. Emerg Infect Dis. 2020;26(10):2480–2482. doi: 10.3201/eid2610.200608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim JS, Vergne T, Pak SI, Kim E. Modelling the spatial distribution of ASF-positive wild boar carcasses in South Korea using 2019–2020 national surveillance data. Animals (Basel) 2021;11(5):1208. doi: 10.3390/ani11051208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schulz K, Staubach C, Blome S, Viltrop A, Nurmoja I, Conraths FJ, et al. Analysis of Estonian surveillance in wild boar suggests a decline in the incidence of African swine fever. Sci Rep. 2019;9(1):8490. doi: 10.1038/s41598-019-44890-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jo YS, Gortázar C. African swine fever in wild boar: assessing interventions in South Korea. Transbound Emerg Dis. 2021 doi: 10.1111/tbed.14106. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 8.Delamater PL, Street EJ, Leslie TF, Yang YT, Jacobsen KH. Complexity of the basic reproduction number (R0) Emerg Infect Dis. 2019;25(1):1–4. doi: 10.3201/eid2501.171901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim JS, Cho SI, Ryu S, Pak SI. Interpretation of the basic and effective reproduction number. J Prev Med Public Health. 2020;53(6):405–408. doi: 10.3961/jpmph.20.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haut ER, Pronovost PJ. Surveillance bias in outcomes reporting. JAMA. 2011;305(23):2462–2463. doi: 10.1001/jama.2011.822. [DOI] [PubMed] [Google Scholar]
- 11.Iglesias I, Muñoz MJ, Montes F, Perez A, Gogin A, Kolbasov D, et al. Reproductive ratio for the local spread of African swine fever in wild boars in the Russian Federation. Transbound Emerg Dis. 2016;63(6):e237–e245. doi: 10.1111/tbed.12337. [DOI] [PubMed] [Google Scholar]
- 12.Pepin KM, Golnar AJ, Abdo Z, Podgórski T. Ecological drivers of African swine fever virus persistence in wild boar populations: insight for control. Ecol Evol. 2020;10(6):2846–2859. doi: 10.1002/ece3.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Probst C, Globig A, Knoll B, Conraths FJ, Depner K. Behaviour of free ranging wild boar towards their dead fellows: potential implications for the transmission of African swine fever. R Soc Open Sci. 2017;4(5):170054. doi: 10.1098/rsos.170054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulldorff M, Heffernan R, Hartman J, Assunção R, Mostashari F. A space-time permutation scan statistic for disease outbreak detection. PLoS Med. 2005;2(3):e59. doi: 10.1371/journal.pmed.0020059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tango T. Spatial scan statistics can be dangerous. Stat Methods Med Res. 2021;30(1):75–86. doi: 10.1177/0962280220930562. [DOI] [PubMed] [Google Scholar]
- 16.Birant D, Kut A. ST-DBSCAN: an algorithm for clustering spatial–temporal data. Data Knowl Eng. 2007;60(1):208–221. [Google Scholar]
- 17.Gabriel C, Blome S, Malogolovkin A, Parilov S, Kolbasov D, Teifke JP, et al. Characterization of African swine fever virus Caucasus isolate in European wild boars. Emerg Infect Dis. 2011;17(12):2342–2345. doi: 10.3201/eid1712.110430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iglesias I, Perez AM, Sánchez-Vizcaíno JM, Muñoz MJ, Martínez M, de la Torre A. Reproductive ratio for the local spread of highly pathogenic avian influenza in wild bird populations of Europe, 2005–2008. Epidemiol Infect. 2011;139(1):99–104. doi: 10.1017/S0950268810001330. [DOI] [PubMed] [Google Scholar]
- 19.Ministry of Environment. Standard Operation Procedures for African Swine Fever in Wild Boar. Sejong: Ministry of Environment; 2019. [Google Scholar]
- 20.Blome S, Gabriel C, Dietze K, Breithaupt A, Beer M. High virulence of African swine fever virus Caucasus isolate in European wild boars of all ages. Emerg Infect Dis. 2012;18(4):708. doi: 10.3201/eid1804.111813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho HK, Kim ET, Jung BS, Pak SI. A preliminary investigation into the decomposition rate of wild boar carcasses in forest habitats. J Prev Vet Med. 2021;45(1):44–52. [Google Scholar]
- 22.Korea National Park Service. National Park Wild Boar Habitat Survey Research. Wonju: Korea National Park Service; 2019. [Google Scholar]
- 23.Anderson RM, May RM. Infectious Diseases of Humans: Dynamics and Control. Oxford: Oxford University Press; 1992. [Google Scholar]
- 24.Kim ET, Pak SI. Species distribution modeling for wild boar (Sus scropa) in the Republic of Korea using MODIS data. J Prev Vet Med. 2020;44(2):89–95. [Google Scholar]
- 25.R Core Team. R: a Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2021. [Google Scholar]
- 26.Morelle K, Podgórski T, Prévot C, Keuling O, Lehaire F, Lejeune P. Towards understanding wild boar Sus scrofa movement: a synthetic movement ecology approach. Mammal Rev. 2015;45(1):15–29. [Google Scholar]
- 27.O'Neill X, White A, Ruiz-Fons F, Gortázar C. Modelling the transmission and persistence of African swine fever in wild boar in contrasting European scenarios. Sci Rep. 2020;10(1):5895. doi: 10.1038/s41598-020-62736-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choe S, Cha RM, Yu DS, Kim KS, Song S, Choi SH, et al. Rapid spread of classical swine fever virus among South Korean wild boars in areas near the border with North Korea. Pathogens. 2020;9(4):244. doi: 10.3390/pathogens9040244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang A, Schlichting P, Wight B, Anderson WM, Chinn SM, Wilber MQ, et al. Effects of social structure and management on risk of disease establishment in wild pigs. J Anim Ecol. 2021;90(4):820–833. doi: 10.1111/1365-2656.13412. [DOI] [PubMed] [Google Scholar]
- 30.Loi F, Cappai S, Laddomada A, Feliziani F, Oggiano A, Franzoni G, et al. Mathematical approach to estimating the main epidemiological parameters of African swine fever in wild boar. Vaccines (Basel) 2020;8(3):8. doi: 10.3390/vaccines8030521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marcon A, Linden A, Satran P, Gervasi V, Licoppe A, Guberti V. R-0 estimation for the African swine fever epidemics in wild boar of Czech Republic and Belgium. Vet Sci. 2020;7(1):2. doi: 10.3390/vetsci7010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Podgórski T, Borowik T, Łyjak M, Woźniakowski G. Spatial epidemiology of African swine fever: host, landscape and anthropogenic drivers of disease occurrence in wild boar. Prev Vet Med. 2020;177:104691. doi: 10.1016/j.prevetmed.2019.104691. [DOI] [PubMed] [Google Scholar]
- 33.National Institute of Biological Resources. 2017 Wildlife Survey. Incheon: National Institute of Biological Resources; 2017. [Google Scholar]
- 34.Evans JD. Straightforward Statistics for the Behavioral Sciences. Belmont: Thomson Brooks/Cole Publishing Co.; 1996. [Google Scholar]
- 35.Pepin KM, Golnar A, Podgórski T. Social structure defines spatial transmission of African swine fever in wild boar. J R Soc Interface. 2021;18(174):20200761. doi: 10.1098/rsif.2020.0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robert A, Kucharski AJ, Gastañaduy PA, Paul P, Funk S. Probabilistic reconstruction of measles transmission clusters from routinely collected surveillance data. J R Soc Interface. 2020;17(168):20200084. doi: 10.1098/rsif.2020.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guzzetta G, Marques-Toledo CA, Rosà R, Teixeira M, Merler S. Quantifying the spatial spread of dengue in a non-endemic Brazilian metropolis via transmission chain reconstruction. Nat Commun. 2018;9(1):2837. doi: 10.1038/s41467-018-05230-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Probst C, Gethmann J, Amendt J, Lutz L, Teifke JP, Conraths FJ. Estimating the postmortem interval of wild boar carcasses. Vet Sci. 2020;7(1):6. doi: 10.3390/vetsci7010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vergne T, Andraud M, Bonnet S, De Regge N, Desquesnes M, Fite J, et al. Mechanical transmission of African swine fever virus by Stomoxys calcitrans: insights from a mechanistic model. Transbound Emerg Dis. 2021;68(3):1541–1549. doi: 10.1111/tbed.13824. [DOI] [PubMed] [Google Scholar]
- 40.Probst C, Gethmann J, Amler S, Globig A, Knoll B, Conraths FJ. The potential role of scavengers in spreading African swine fever among wild boar. Sci Rep. 2019;9(1):11450. doi: 10.1038/s41598-019-47623-5. [DOI] [PMC free article] [PubMed] [Google Scholar]