Abstract
Background
Bovine group A rotavirus (BoRVA) is a major cause of severe gastroenteritis in newborn dairy calves. Only one study has investigated the G and P genotypes among dairy calves in a few regions of China, which were G6 and P[5]. Therefore, data on the prevalence and molecular characteristics of BoRVA in dairy calves in China remains limited.
Objectives
The purpose of this study was to investigate the prevalence and molecular characteristics of BoRVA in dairy calves in China.
Methods
269 dairy calves diarrheic samples from 23 farms in six provinces in China were collected to detect BoRVA using reverse transcription polymerase chain reaction.
Results
71% of samples were determined to be BoRVA-positive. Two G genotypes (G6, G10) and two P genotypes (P[1], P[5]) were identified, and G6P[1] BoRVA was the predominant strain. Moreover, the VP7 and VP4 gene sequences of these dairy calf BoRVA strains revealed abundant genetic diversity. Interestingly, eight out of 17 complete G6 VP7 sequences were clustered into G6 lineage VI and analysis showed the strains were closely related to Chinese yak BoRVA strains.
Conclusions
The results of this study show that BoRVA circulates widely among dairy calves in China, and the dominant genotype in circulation is G6P[1], first report on molecular characteristics of complete P[5] VP4 genes in chinese dairy calves. These results will help us to further understand the prevalence and genetic evolution of BoRVA among dairy calves in China and, thus, prevent the disease more effectively.
Keywords: Dairy calf, BoRVA, prevalence, molecular characteristics
INTRODUCTION
Rotavirus A (RVA) is an important pathogen causing acute diarrhea in children and the young of other animal species worldwide [1,2]. Bovine rotavirus A (BoRVA) is considered to be the most common cause of severe gastroenteritis in cattle, and the significant morbidity and mortality it leads to, in newborn calves in particular, causes huge economic losses [3,4]. Diarrhea is a common symptom in newborn calves and results in a major loss of capital to the livestock industry. To date, bovine coronavirus, bovine viral diarrhea virus, and nebovirus have been identified as important diarrhea-causing viruses circulating in Chinese calves [5,6]. Moreover, previous studies have provided clear evidence that BoRVA can be transmitted to humans, either directly or through multiple reassortment events during the evolution of the strains [7,8]; therefore, control of the virus has significance to public health.
VP7 is the outer capsid protein of rotaviruses and participates in both a membrane-displacing assembly step and a membrane-disrupting entry step [9]. Moreover, VP7 is highly immunogenic and induces neutralizing antibodies [9]. The neutralizing epitopes on the surface of the VP7 protein are aggregated around two regions, 7-1 (7-1a, 7-1b) and 7-2 [10], and mutation at these epitopes leads to viral evasion of monoclonal antibody neutralization [11]. Previous studies have suggested that amino acid changes of strains may facilitate escape from vaccine-induced immunity because the specific epitopes cannot be recognized [12].
VP4, located on the surface of mature virus particles, independently elicits neutralizing antibodies and induces protective immunity [9]. Through proteolysis, VP4 is cleaved into two fragments, termed VP5 (amino acid site 1-231) and VP8 (amino acid site 248-776). VP8 forms the head of the VP4 spike, and it interacts with receptors on host cells that are necessary for virion attachment and enhances RV infection [9]. VP5 can promote cell membrane penetration and mediate heterotypic protective immunity [13,14]. To date, the neutralizing epitopes of VP5 have been identified in five regions, 5-1 to 5-5 [11], and those of VP8 are clustered into four regions: 8-1 to 8-4 [15]. Changes in these epitopes may lead to changes in the antigenicity of VP4 proteins, which, in turn, can escape the recognition of neutralizing antibodies and impact the effectiveness of existing vaccines [11].
VP7 and VP4 respectively define the G and P genotypes of RVA strains. Thus far, at least 36 G genotypes and 51 P genotypes have been described for strains from humans and animals worldwide (https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification/rcwg). Notably, the genotypes of circulating strains can change over time and with the geographical area of sample collection [16]. Vaccination is an effective means to protect against predominant RVA strains, but there is no efficacious cross-protection to different genotypes [9,17]. Therefore, the surveillance of rotavirus G and P genotypes is important for understanding the epidemiology of rotaviruses and to construct an effective vaccine. G6, G8, and G10, in conjunction with P[1], P[5], P[7] and P[11], are considered the common bovine genotypes [3] and phenotypes, and G6P[5], G6P[11], and G10P[11] are the most common combinations of G genotypes and P genotypes [18,19,20,21]. Moreover, six (G6 I to G6 VI) and ten (G10 I to G6 X) lineages can be distinguished within the G6 and G10 genotypes, respectively [22,23,24]. In China, several G genotypes (G6, G8, and G10) and P genotypes (P[1], P[5], P[7], and P[11]) have been detected in the bovine population [25,26,27]. The only relevant study previously conducted showed G6 and P[5] to be common G and P genotypes among dairy calves in some regions of China, and the dominant genotype combination was G6P[5] [25,27]. Information on the prevalence and molecular characteristics of BoRVA is, therefore, still limited. The purpose of this study was to investigate the prevalence and molecular characteristics of BoRVA in dairy calves in China.
MATERIALS AND METHODS
Specimen collection
A total of 269 diarrheic samples were collected from dairy calves (< 2 mon old, large-scale breeding) from January to December 2018. There were 11, 67, 27, 86, 40, and 38 diarrheic samples collected from Shandong Province (three farms), Liaoning Province (four farms), Henan Province (three farms), Xinjiang Province (seven farms), Shanxi Province (five farms), and Jilin Province (one farm), respectively. All samples were shipped on ice and stored at −80°C in our laboratory.
RNA extraction and cDNA synthesis
The fecal samples were fully resuspended in phosphate-buffered saline (1:5 w/v) and centrifuged at 11,000 × g for 10 min. Viral RNA was extracted from samples using RNAiso Plus (TaKaRa Bio Inc., Japan) and reverse transcribed using the PrimeScript RT Reagent Kit (TaKaRa Bio Inc.) in accordance with the manufacturer's instructions. The resulting cDNA was stored at −20°C for use in further analysis.
Detection of BoRVA and co-infection with other diarrhea-causing viruses
BoRVA was detected using a previously reported reverse transcription polymerase chain reaction (RT-PCR) assay that was established and validated in our laboratory [26,28]. The primer sequences were as follows: BoRVA-F, 5′-CCACCAGGTATGAATTGGAC-3′, and BoRVA-R, 5′-GAGTAATCACTCAGATGGCG-3′ (targeting the VP6 gene; fragment length 231 bp). To investigate co-infection with other major diarrhea-causing viruses, the specific RT-PCR assays, established and validated in our laboratory previously, were used to screen for neboviruses (NeVs), norovirus (NoV), and bovine coronavirus (BCoV) [28,29], and a specific RT-PCR assay was used to screen for bovine viral diarrhea virus (BVDV) [5].
Determination of G/P genotypes of BoRVA
Samples detected as BoRVA-positive were subjected to G and P genotyping according to the methods described in previous reports for a VP7 gene fragment length of 887 bp and a VP4 gene fragment length of 664 bp [30,31]. All the amplification products were purified using the Omega Gel kit (Omega), cloned into the pMD19-T simple vector (TaKaRa Bio Inc.), and sequenced (Sangon Biotech) in both directions. Genotype assignments were carried out using the RotaC v2.0 online tool according to the genotyping recommendations of the RCWG.
Complete VP7 and VP4 gene amplification of dairy calf BoRVA strains
To investigate the molecular characteristics of VP7 and VP4 genes of dairy calf BoRVA strains prevalent in China, the complete VP7 and VP4 gene sequences were amplified from each sample with successfully identified genotypes. Briefly, four primer pairs were designed to amplify the complete VP7 gene, and five primer pairs were designed to the complete VP4 gene. Primer information and PCR conditions were as shown in Supplementary Table 1. All PCR products were purified using the Omega Gel Kit (Omega) following the manufacturer's instructions, after which they were ligated to the pMD19-T simple vector prior to sequencing.
Sequence, phylogeny, and recombination analyses
The sequences were assembled using SeqMan software (version 7.0; DNASTAR Inc., USA) and compared with other sequences available in Genbank using BLAST. The homologies of the nucleotide and deduced amino acid sequences were determined using the MegAlign program of DNASTAR 7.0 software (DNASTAR Inc.). Phylogenetic trees based on nucleotide sequences were constructed using the maximum likelihood method and the Kimura two-parameter model in MEGA 7.0. Bootstrap analyses were performed on the basis of 1000 replicates. Recombination analysis was performed using SimPlot software (version 3.5.1) and the Recombination Detection Program 4.0 (RDP 4.0, version 4.96) with the RDP, GeneConv, Chimaera, MaxChi, BootScan, SiScan, and 3Seq methods.
Ethics approval and consent to participate
This study was approved by the Animal Ethics Committee of Southwest Minzu University. Experimental protocols for obtaining bovine clinical samples used in this study were carried out in strict accordance with the Animal Ethics Procedures and Guidelines of China. This study did not involve animal experiments besides the fecal sampling of diarrhea calves that visited farm for clinical treatment. The sample collection work was approved with signed informed consent from the cattle farm owner.
RESULTS
Detection and of BoRVA and co-infections in dairy calves
Among the 269 diarrheic samples from dairy calves, 191 (71.00%) were found to be BoRVA-positive by RT-PCR, and the BoRVA-positive rates were 92.4% (62/67), 90.9% (10/11), 68.4% (26/38), 62.9% (17/27), 60.4% (52/86), and 60.0% (24/40) in Liaoning, Shandong, Jilin, Henan, Xinjiang, and Shanxi provinces, respectively. Notably, BoRVA was distributed in all farms across the six provinces. Moreover, 145 out of the 191 BoRVA-positive diarrheic samples were co-infected with BCoV, NoV, NeVs, and/or BVDV: detailed information is shown in Table 1.
Table 1. The co-infection of rotavirus A positive samples with other diarrhoea-causing viruses.
| Viruses | Positive rate (%) (positive/number tested) | 95% CI |
|---|---|---|
| BoRVA (only) | 21.47 (41/191) | 15.6 to 27.4 |
| BoRVA+BNoV | 23.04 (44/191) | 17.2 to 28.9 |
| BoRVA+BVDV | 8.38 (16/191) | 4.5 to 12.3 |
| BoRVA+NeVs | 13.61 (26/191) | 9.7 to 17.5 |
| BoRVA+BCoV | 5.76 (11/191) | 1.8 to 9.7 |
| BoRVA+BCoV+NeVs | 8.90 (17/191) | 5 to 12.8 |
| BoRVA+BVDV+NeVs | 2.62 (5/191) | 0.7 to 4.6 |
| BoRVA+BVDV+BCoV | 1.05 (2/191) | −0.9 to 3 |
| BoRVA+BVDV+BCoV+NeVs | 5.24 (10/191) | 1.3 to 9.2 |
| BoRVA+BNoV+NeVs | 0.05 (1/191) | −1.5 to 2.5 |
| BoRVA+BNoV+ BCoV | 4.19 (8/191) | 2.2 to 6.2 |
| BoRVA+BVDV+BCoV+BNoV | 1.57 (3/191) | −0.4 to 3.5 |
| BoRVA+NeVs+BCoV+BNoV | 3.66 (7/191) | 1.7 to 5.6 |
| BoRVA+NeVs+BVDV+BNoV | 2.62 (5/191) | 0.7 to 4.6 |
| BoRVA+BVDV+BCoV+BNoV+NeVs | 8.38 (16/191) | 4.5 to 12.3 |
CI, confidence interval; BoRVA, bovine rotavirus A; BNoV, bovine norovirus; BVDV, bovine viral diarrhea virus; NeVs, neboviruses; BCoV, bovine coronavirus.
G/P genotyping of BoRVA in dairy calves
We successfully genotyped 55 samples from the 191-BoRVA positive samples. The G genotypes of 54 samples were determined, leading to 49 samples being identified as G6 (9 in Shandong, 36 in Liaoning, and 4 in Xinjiang), and the remaining 5 samples were determined to be G10 (5 in Xinjiang). Meanwhile, the P genotypes of the 36 samples were determined: 21 samples were P[1] (9 in Shandong, 11 in Liaoning, and 1 in Xinjiang), and the remaining 15 samples were P[5] (15 in Liaoning). Altogether, the G and P genotypes were successfully identified for 35 of the samples, of which 20 samples were G6P[1] (9 from Shandong, 10 from Liaoning, and 1 from Xinjiang) and 15 from Liaoning Province were G6P[5]. The detailed genotype information for each farm is shown in Supplementary Table 2.
Molecular characteristics of complete G6 VP7 genes in dairy calves
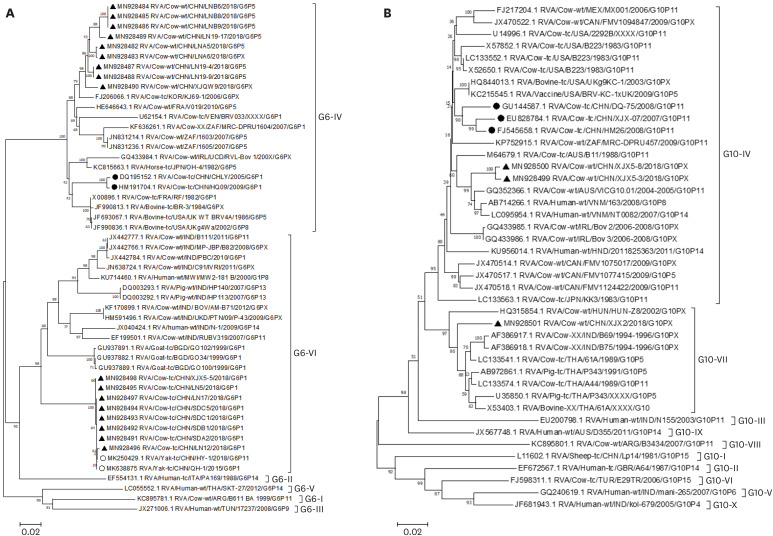
In total, 17 complete VP7 genes of G6 were obtained from three provinces (Shandong, Liaoning, and Xinjiang). The complete VP7 gene of 17 G6 dairy calf BoRVA strains was 981 bp, encoding 326 amino acids. A phylogenetic tree was constructed on the basis of all complete VP7 genes of G6 lineages IV and VI, the representative strains of other G6 lineages, and all Chinese BoRVA G6 VP7 sequences. The eight strains from this study (four from Shandong, three from Liaoning, and one from Xinjiang) clustered into G6 lineage VI, and the remaining nine strains (eight from Liaoning, one from Xinjiang) clustered into G6 lineage IV (Fig. 1A). Interestingly, G6 lineage VI and IV strains coexisted in Liaoning and Xinjiang provinces. Eight strains of lineage VI in this study had the highest identity with two Chinese yak BoRVA strains (GenBank accession no. MK250429 and MK638875). And the eight strains shared nucleotide identity of 99.7%–100% and amino acid identity of 99.4%–100% with each other. When the sequences were compared with the other G6 lineage VI strains available in GenBank, they shared 88.3%–90.7% nucleic acid and 94.8%–97.9% amino acid identities. Compared with all the G6 lineage VI strains available in GenBank, the eight strains of lineage VI in this study with the two yak BoRVA strains shared 24 nucleotide changes (G57A, T63A, A69G, T96C, A126G, A150G, T156C, A180T, T207C, G216A, A237G, C247T, G259A, A330G, T363C, C453T, C471T, T522C, A528G, C537T, A648G, A762G, T795C, C949T), resulting in one amino acid change at the neutralizing epitope 7-1a (V87I) region. These changes resulted in the sequences being placed in a unique branch in the phylogenetic tree. Compared with the only two G6P[1] BoRVA strains characterized from China, the VP7 genes of the eight lineage VI strains from this study and two yak strains had 25 identical amino acid changes, resulting in seven amino acid changes at the neutralizing epitope 7-1a (V87I, D100N, E130D), 7-1b (N211D, D213N, A242T, N238D), and 7-2 (Q148L) regions.
Fig. 1. Phylogenetic tree based on G6 (A) and G10 (B) VP7 complete nucleotide sequences. Sequence alignments and clustering were performed using ClustalW in MEGA 7.0 software. The tree was constructed with the maximum likelihood method, with bootstrap values calculated for 1000 replicates. ▴ represents the VP7 gene of dairy calf BoRVA strains from this study (GenBank no. MN928482-MN928498; GenBank no. MN937499-MN937501); ∘ represents the VP7 gene of two yak G6 BoRVA strains; • represents the VP7 gene of BoRVA strains in China.
Nine strains of lineage IV in this study shared 97.2%–100% nucleotide identity and 99.1%–100% amino acid identity with each other and 90.9%–97.4% nucleotide identity and 94.8%–99.4% amino acid identity with the G6 lineage IV RVA strains available in GenBank. In addition, they shared 92.8%–93.7% nucleotide identity and 96.3%–97.5% amino acid identity with the only two G6P[1] BoRVA strains from China in GenBank. Nine strains of lineage IV identified in this study were closely related to BoRVA strain KJ69-1. Compared with the two G6P[1] BoRVA strains from China, the VP7 genes of all nine lineage IV sequences had eight amino acid changes (M22I, I29M, V42T, D69N, L118F, T187M, I268V, and F317L). The amino acid changes in the G6 VP7 gene are shown in Supplementary Fig. 1. No recombination events occurred in the G6 VP7 genes.
Molecular characteristics of complete G10 VP7 genes in dairy calves
Three complete VP7 genes of G10 were obtained from Xinjiang Province. The complete VP7 gene found in three G10 dairy calf BoRVA strains was 981 bp long and encoded 326 amino acids. We constructed a phylogenetic tree based on all complete VP7 genes of G10 lineages IV and VII, representative strains of the other G10 lineages, and the Chinese BoRVA G10 VP7 sequences. The results showed that the two strains from this study were clustered into G10 lineage IV, and the remaining one strain was in G10 lineage VII (Fig. 1B). Two strains of lineage IV VP7 sequences shared 99.4% nucleotide identity and 100% amino acid identity with each other, 87.9%–97.3% nucleotide and 93.3%–98.8% amino acid identity with other G10 lineage IV strains available in GenBank. The two G10 lineage IV strains were closely related to Human strain NT0082 (97.1%–97.3% nucleotide and 98.8% amino acid identities). Compared with the other G10 lineage IV BoRVA strains in GenBank, the VP7 genes of the two sequences from this study had one amino acid change (V47I). Moreover, compared with the three G10 BoRVA strains from China, the two sequences had three and two amino acid changes at the neutralizing epitopes 7-1a (S100N) and 7-2 (L148I) regions, respectively.
The remaining one G10 VP7 sequence was located in lineage VII and shared 94.2–97.0% nucleotide identity and 96.6%–99.7% amino acid identity with all G10 lineage VII RVA strains available in GenBank. This strain was closely related to dairy calf BoRVA strain 61A (97.0% nucleotide identity and 99.7% amino acid identity). Compared with other G10 lineage VII BoRVA strains, there was one amino acid change located at neutralizing epitopes 7-2 (L146S). Moreover, compared with the three G10 BoRVA strains from China, this strain had four amino acid variations (L19F, T44A, S146L, N278A). No recombination events had occurred in the G10 VP7 genes.
Molecular characteristics of complete P[1] VP4 genes in dairy calves
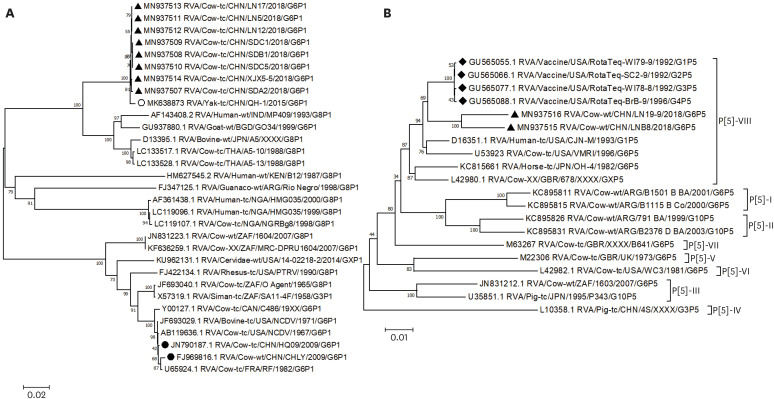
We obtained eight complete VP4 genes of P[1] from three provinces (four from Shandong, three from Liaoning, and one from Xinjiang) containing 2331 nucleotides encoding 776 amino acids. Eight P[1] strains in this study and the Chinese yak QH-1 strain (GenBank accession no. MK638873) showed the highest similarity, with 99.4%–99.5% nucleotide identity and 98.0%–99.1% amino acid identity between them. The eight P[1] strains also shared 80.5%–92.8% nucleotide identity and 90.6%–98.1% amino acid identity with other P[1] strains from GenBank.
A phylogenetic tree was then created using all complete VP4 genes of the P[1] genotype available in GenBank, which showed the VP4 genes of the eight P[1] strains were clustered together with Chinese yak strain QH-1 on a unique branch that was obviously genetically distinct from the Chinese G6P[1] strains (Fig. 2A). Compared with the other complete VP4 sequences of the P[1] RVA strains obtained from GenBank, the eight P[1] strains shared six nucleotide changes (G190A, T307C, C641A, A1275 C, A1374G, and A1512G) with the QH-1 strain, leading to two amino acid changes (V64I, A214E) at the VP8 region. These shared changes meant the strains were located on a unique branch. Moreover, compared with the two G6P[1] BoRVA strains from China, the VP4 gene of the P[1] RVA strains contained 56 amino acid changes, of which four were located at the neutralizing epitopes 8-1 (F194Y, T196S), 8-3 (Q114S), and 5-1 (L440M) regions. The amino acid changes in the P[1] VP4 gene are shown in Supplementary Fig. 2. No recombination events occurred in P[1] VP4 genes.
Fig. 2. Phylogenetic tree based on P[1] (A) and P[5] (B) VP4 complete nucleotide sequences. Sequence alignments and clustering were performed with ClustalW in MEGA 7.0 software. The tree was constructed using the maximum likelihood method, with bootstrap values calculated for 1000 replicates. ▴ represents the VP4 gene dairy calf BoRVA strains from this study (GenBank no. MN937507-MN937514; GenBank no. MN937515-MN937516); ∘ represents the VP4 gene of one yak P[1] BoRVA strain; • represents the VP4 gene of BoRVA strains in China.
Molecular characteristics of complete P[5] VP4 genes in dairy calves
There were two complete VP4 genes of P[5] obtained from Liaoning province, and the complete VP4 genes of these two strains had 2331 nucleotides and encoded 776 amino acids. The two P[5] VP4 gene sequences shared 96.8% nucleotide identity and 97.9% amino acid identity with each other, and they shared 89.8%–96.3% nucleotide and 95.2%–98.0% amino acid sequence identities with other P[5] strains recorded in GenBank. These two P[5] strains were closely related to the four RVA vaccine strains of RotaTeq.
When we constructed a phylogenetic tree based on all complete VP4 genes of P[5] lineage VIII and representative strains of other P[5] lineages, the VP4 genes of the two P[5] strains were clustered into P[5] lineage VIII and appeared to be closely related to four RVA vaccine strains of RotaTeq (Fig. 2B). Compared with the other complete VP4 sequences from P[5] RVA strains in GenBank, the two P[5] strains from this study had two unique amino acids (D151E and Q266R). Moreover, compared with the four RotaTeq RVA vaccine strains, the VP4 gene of the two P[5] strains had 48 unique nucleotides, resulting in eight amino acid changes, of which six were within the VP8 region (V44G, I148V, D151E, M167I, K172E, R188G) and two were in the VP5 region (Q266R, V391A). The amino acid changes in the P[5] VP4 gene are shown in Supplementary Fig. 3. No recombination events occurred in P[5] VP4 genes.
DISCUSSION
Prevalence of BoRVA in dairy calves in China
BoRVA is considered to be an important pathogen of cattle because of its conspicuous economic impact on the cattle industry [3,4]. However, data on the prevalence and molecular characteristics of BoRVA in China are incomplete. In this study, all dairy calves were less than 2-mon-old, the observed BoRVA prevalence rate was 71.0%. Positive samples were distributed across six provinces that are the major dairy calf production areas in China, and the geographical distance between the two farthest farms was more than 2,000 km. The results showed that BoRVA is circulating widely among young age of dairy calves in China, and this information is useful for efforts on the diagnosis and control of diarrhea in dairy calves. Furthermore, a high rate of co-infection with NeVs, NoV, BCoV, and BVDV was found in this study, and similar data have been reported previously in the USA [32]. This may lead to an increase in clinical severity and the difficulty in diagnosing and controlling dairy calf diarrhea.
In this study, only 55 samples were successfully genotyped from 191 RVA-positive samples, the low genotyping rate may be related to the low virus amounts or/and the changes in viral nucleotide sequences in clinical samples. The G6 genotypes and P[1] genotypes were the most frequently identified. Notably, the G6 strains in this study were clustered into lineages VI and IV, and G6 VI lineage with P[1], which was the predominant strain combination, was found in three provinces. The G6 IV lineage, which was mainly combined with P[5] and occasionally with P[1], was only observed in Liaoning Province. G6 Lineage IV is typical of BoRVA strains [16], while G6 lineage VI strains also infect various other hosts, such as humans, goats, yak, and pigs. In this study, the wide distribution of a lineage VI strain in Chinese dairy calves warrants the continuous co-surveillance of samples from the above host sources. To date, only one previous study has reported the G and P genotypes among dairy calves in China, of which the predominant strain was G6P[5] [25]. This is the first to report G6P[1] BoRVA among dairy calves in China, which was discovered to be the predominant strain in these animals. The differences in the genotypes of dairy calf BoRVA strains may be associated with the geographical sampling area and the period of sample collection [16,33]. It is well known that vaccination is an effective means to prevent predominant RVA strains; however, cross-protection between different genotypes is not known to occur [11]. The results may be useful for understanding the epidemiology of BoRVA and in constructing an effective vaccine to control diarrhea among dairy calves in China. The findings support the idea that research into BoRVA is continually needed in China.
Molecular characteristics of VP7 and VP4 genes in dairy calves
In this study, eight G6 lineage VI strains with two Chinese yak BoRVA strains were clustered within an independent branch, these strains have emerged and spread in the Chinese bovine population. Notably, compared with the four RVA vaccine strains of RotaTeq, the VP4 gene of the two P[5] strains had amino acid variation in the neutralizing antigen epitope (one site in 8-1) and sialic acid binding site (R188G). The residues that contact sialic acid are conserved in sialic acid-dependent rotavirus strains [15]. Therefore, this phenomenon may affect receptor-binding capacity, leading to changes in the virulence of the RVA strains, and the biological significance of this variant warrants further investigation. To the best of our knowledge, until now, there were no complete VP4 sequences of P[5] BoRVA from China in GenBank, thus this is the first report on the molecular characteristics of a VP4 gene of domestic P[5] BoRVA strains. Additionally, the P[5]-VIII lineage has been described for the first time in RVA strains in Chinese cattle herds. Previous studies have suggested that the amino acid changes of RVA strains may facilitate viral escape from vaccine-induced immunity because the specific epitopes cannot be recognized [12,34]; therefore, the biological significance of this variant needs further investigation. Whether amino acid changes in these strains affect the antigenicity of VP7 and VP4 also needs to be determined. The results of this study have a particular reference to the development of a BoRVA vaccine, as there are still no commercial BoRVA vaccines available in China. The results will contribute to the prevention of dairy calf diarrhea and our understanding of the genetic evolution of BoRVA in China.
In conclusion, the results of our study have shown that BoRVA is widely transmitted among dairy calves in China, and G6P[1] is the dominant genotype in circulation. First report on molecular characteristics of complete P[5] VP4 genes in Chinese dairy calves. We believe these results will contribute to a better understanding of the molecular epidemiology of rotaviruses and the development of vaccines.
ACKNOWLEDGEMENTS
We thank all the dairy calves farms for submitting the samples to our laboratory.
Footnotes
Funding: This work was funded by the innovation team for emerging animal diseases, Southwest Minzu University (grant number 2020NTD02); and Leading Talents Support Program of State Ethnic Affairs Committee. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest: The authors declare that they have no conflicts of interest.
- Conceptualization: Liu X, Yan N.
- Data curation: Liu X, Yan N.
- Formal analysis: Wang Y, Zhang B.
- Funding acquisition: Yue H, Tang C.
- Investigation: Liu X, Yan N.
- Methodology: Liu X, Yan N.
- Software: Wang Y, Zhang B.
- Writing - original draft: Liu X, Yan N.
- Writing - review & editing: Yue H, Tang C.
SUPPLEMENTARY MATERIALS
Oligonucleotide primers used for complete VP7 and VP4 gene
BRVA genotyping in each farm
Amino acid variants of the complete VP7 genes of all 8 G6 lineage VI strains in this study. Green indicates the unique aa variants sites in all 8 strains in this study and 2 Yak strains compared with the other G6 lineage VI strains in GenBank; Blue indicates the unique aa variants sites in all 8 strains in this study and 2 Yak strains compared with the only 2 G6P[1] BRVA strains in China. The neutralization epitopes are shown in red numbers.
Amino acid variants of the complete VP4 genes of all 8 P[1] strains in this study. Green indicates the unique aa variants sites in all 8 P[1] strains in this study and Yak strain QH-1 compared with the other P[1] strains in GenBank; Blue indicates the unique aa variants sites in all 8 P[1] strains in this study and Yak strain QH-1 compared with the only 2 G6P[1] BRVA strains in China. The neutralization epitopes are shown in red numbers. Orange indicates sialic acid binding site.
Amino acid variants of the complete VP4 genes of all 2 P[5] strains in this study. Green indicates the unique aa variants sites in all 2 P[5] strains in this study compared with the other P[5] strains in GenBank; Blue indicates the unique aa variants sites in all 2 P[5] strains in this study compared with the 4 vaccine strains. The neutralization epitopes are shown in red numbers. Orange indicates sialic acid binding site.
References
- 1.Clark A, Black R, Tate J, Roose A, Kotloff K, Lam D, et al. Estimating global, regional and national rotavirus deaths in children aged <5 years: Current approaches, new analyses and proposed improvements. PLoS One. 2017;12(9):e0183392. doi: 10.1371/journal.pone.0183392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barros BC, Chagas EN, Bezerra LW, Ribeiro LG, Duarte Júnior JW, Pereira D, et al. Correction: rotavirus A in wild and domestic animals from areas with environmental degradation in the Brazilian Amazon. PLoS One. 2019;14(1):e0211311. doi: 10.1371/journal.pone.0211311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papp H, László B, Jakab F, Ganesh B, De Grazia S, Matthijnssens J, et al. Review of group A rotavirus strains reported in swine and cattle. Vet Microbiol. 2013;165(3-4):190–199. doi: 10.1016/j.vetmic.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al Mawly J, Grinberg A, Prattley D, Moffat J, Marshall J, French N. Risk factors for neonatal calf diarrhoea and enteropathogen shedding in New Zealand dairy farms. Vet J. 2015;203(2):155–160. doi: 10.1016/j.tvjl.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gong X, Liu L, Zheng F, Chen Q, Li Z, Cao X, et al. Molecular investigation of bovine viral diarrhea virus infection in yaks (Bos gruniens) from Qinghai, China. Virol J. 2014;11(1):29. doi: 10.1186/1743-422X-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He Q, Guo Z, Zhang B, Yue H, Tang C. First detection of bovine coronavirus in Yak (Bos grunniens) and a bovine coronavirus genome with a recombinant HE gene. J Gen Virol. 2019;100(5):793–803. doi: 10.1099/jgv.0.001254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komoto S, Adah MI, Ide T, Yoshikawa T, Taniguchi K. Whole genomic analysis of human and bovine G8P[1] rotavirus strains isolated in Nigeria provides evidence for direct bovine-to-human interspecies transmission. Infect Genet Evol. 2016;43:424–433. doi: 10.1016/j.meegid.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 8.Quaye O, McDonald S, Esona MD, Lyde FC, Mijatovic-Rustempasic S, Roy S, et al. Rotavirus G9P[4] in 3 countries in Latin America, 2009–2010. Emerg Infect Dis. 2013;19(8):1332–1333. doi: 10.3201/eid1908.130288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Estes MK, Greenberg HB. In: Fields Virology. 6th ed. Knipe DM, Howley PM, editors. Philadelphia: Lippincott Williams & Wilkins; 2013. Rotaviruses; pp. 1347–1401. [Google Scholar]
- 10.Aoki ST, Settembre EC, Trask SD, Greenberg HB, Harrison SC, Dormitzer PR. Structure of rotavirus outer-layer protein VP7 bound with a neutralizing Fab. Science. 2009;324(5933):1444–1447. doi: 10.1126/science.1170481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dormitzer PR, Nason EB, Prasad BV, Harrison SC. Structural rearrangements in the membrane penetration protein of a non-enveloped virus. Nature. 2004;430(7003):1053–1058. doi: 10.1038/nature02836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeller M, Heylen E, De Coster S, Van Ranst M, Matthijnssens J. Full genome characterization of a porcine-like human G9P[6] rotavirus strain isolated from an infant in Belgium. Infect Genet Evol. 2012;12(7):1492–1500. doi: 10.1016/j.meegid.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Rodríguez JM, Chichón FJ, Martín-Forero E, González-Camacho F, Carrascosa JL, Castón JR, et al. New insights into rotavirus entry machinery: stabilization of rotavirus spike conformation is independent of trypsin cleavage. PLoS Pathog. 2014;10(5):e1004157. doi: 10.1371/journal.ppat.1004157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nair N, Feng N, Blum LK, Sanyal M, Ding S, Jiang B, et al. VP4- and VP7-specific antibodies mediate heterotypic immunity to rotavirus in humans. Sci Transl Med. 2017;9(395):eaam5434. doi: 10.1126/scitranslmed.aam5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dormitzer PR, Sun ZY, Wagner G, Harrison SC. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 2002;21(5):885–897. doi: 10.1093/emboj/21.5.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Badaracco A, Garaicoechea L, Matthijnssens J, Louge Uriarte E, Odeón A, Bilbao G, et al. Phylogenetic analyses of typical bovine rotavirus genotypes G6, G10, P[5] and P[11] circulating in Argentinean beef and dairy herds. Infect Genet Evol. 2013;18:18–30. doi: 10.1016/j.meegid.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 17.Ruiz V, Mozgovoj MV, Dus Santos MJ, Wigdorovitz A. Plant-produced viral bovine vaccines: what happened during the last 10 years? Plant Biotechnol J. 2015;13(8):1071–1077. doi: 10.1111/pbi.12440. [DOI] [PubMed] [Google Scholar]
- 18.Badaracco A, Garaicoechea L, Rodríguez D, Uriarte EL, Odeón A, Bilbao G, et al. Bovine rotavirus strains circulating in beef and dairy herds in Argentina from 2004 to 2010. Vet Microbiol. 2012;158(3-4):394–399. doi: 10.1016/j.vetmic.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 19.Collins PJ, Mulherin E, Cashman O, Lennon G, Gunn L, O'Shea H, et al. Detection and characterisation of bovine rotavirus in Ireland from 2006–2008. Ir Vet J. 2014;67(1):13. doi: 10.1186/2046-0481-67-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pourasgari F, Kaplon J, Karimi-Naghlani S, Fremy C, Otarod V, Ambert-Balay K, et al. The molecular epidemiology of bovine rotaviruses circulating in Iran: a two-year study. Arch Virol. 2016;161(12):3483–3494. doi: 10.1007/s00705-016-3051-0. [DOI] [PubMed] [Google Scholar]
- 21.Mohamed FF, Mansour SM, El-Araby IE, Mor SK, Goyal SM. Molecular detection of enteric viruses from diarrheic calves in Egypt. Arch Virol. 2017;162(1):129–137. doi: 10.1007/s00705-016-3088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cowley D, Donato CM, Roczo-Farkas S, Kirkwood CD. Novel G10P[14] rotavirus strain, northern territory, Australia. Emerg Infect Dis. 2013;19(8):1324–1327. doi: 10.3201/eid.1908.121653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fritzen JT, Oliveira MV, Lorenzetti E, Miyabe FM, Viziack MP, Rodrigues CA, et al. Longitudinal surveillance of rotavirus A genotypes circulating in a high milk yield dairy cattle herd after the introduction of a rotavirus vaccine. Vet Microbiol. 2019;230:260–264. doi: 10.1016/j.vetmic.2019.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jamnikar-Ciglenecki U, Kuhar U, Sturm S, Kirbis A, Racki N, Steyer A. The first detection and whole genome characterization of the G6P[15] group A rotavirus strain from roe deer. Vet Microbiol. 2016;191:52–59. doi: 10.1016/j.vetmic.2016.05.019. [DOI] [PubMed] [Google Scholar]
- 25.Wei S, Gong Z, Che T, Guli A, Tian F. Genotyping of calves rotavirus in China by reverse transcription polymerase chain reaction. J Virol Methods. 2013;189(1):36–40. doi: 10.1016/j.jviromet.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Yan N, Li R, Wang Y, Zhang B, Yue H, Tang C. High prevalence and genomic characteristics of G6P[1] bovine rotavirus A in yak in China. J Gen Virol. 2020;101(7):701–711. doi: 10.1099/jgv.0.001426. [DOI] [PubMed] [Google Scholar]
- 27.Elkady G, Zhu J, Peng Q, Chen M, Liu X, Chen Y, et al. Isolation and whole protein characterization of species A and B bovine rotaviruses from Chinese calves. Infect Genet Evol. 2021;89:104715. doi: 10.1016/j.meegid.2021.104715. [DOI] [PubMed] [Google Scholar]
- 28.Guo Z, He Q, Zhang B, Yue H, Tang C. Detection and molecular characteristics of neboviruses in dairy cows in China. J Gen Virol. 2019;100(1):35–45. doi: 10.1099/jgv.0.001172. [DOI] [PubMed] [Google Scholar]
- 29.Guo Z, He Q, Yue H, Zhang B, Tang C. First detection of nebovirus and Norovirus from cattle in China. Arch Virol. 2018;163(2):475–478. doi: 10.1007/s00705-017-3616-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gómara MI, Cubitt D, Desselberger U, Gray J. Amino acid substitution within the VP7 protein of G2 rotavirus strains associated with failure to serotype. J Clin Microbiol. 2001;39(10):3796–3798. doi: 10.1128/JCM.39.10.3796-3798.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simmonds MK, Armah G, Asmah R, Banerjee I, Damanka S, Esona M, et al. New oligonucleotide primers for P-typing of rotavirus strains: strategies for typing previously untypeable strains. J Clin Virol. 2008;42(4):368–373. doi: 10.1016/j.jcv.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 32.Cho YI, Han JI, Wang C, Cooper V, Schwartz K, Engelken T, et al. Case-control study of microbiological etiology associated with calf diarrhea. Vet Microbiol. 2013;166(3-4):375–385. doi: 10.1016/j.vetmic.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.da Silva Medeiros TN, Lorenzetti E, Alfieri AF, Alfieri AA. Phylogenetic analysis of a G6P[5] bovine rotavirus strain isolated in a neonatal diarrhea outbreak in a beef cattle herd vaccinated with G6P[1] and G10P[11] genotypes. Arch Virol. 2015;160(2):447–451. doi: 10.1007/s00705-014-2271-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDonald SM, Matthijnssens J, McAllen JK, Hine E, Overton L, Wang S, et al. Evolutionary dynamics of human rotaviruses: balanceng reassortment with preferred genome constellations. PLoS Pathog. 2009;5(10):e1000634. doi: 10.1371/journal.ppat.1000634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligonucleotide primers used for complete VP7 and VP4 gene
BRVA genotyping in each farm
Amino acid variants of the complete VP7 genes of all 8 G6 lineage VI strains in this study. Green indicates the unique aa variants sites in all 8 strains in this study and 2 Yak strains compared with the other G6 lineage VI strains in GenBank; Blue indicates the unique aa variants sites in all 8 strains in this study and 2 Yak strains compared with the only 2 G6P[1] BRVA strains in China. The neutralization epitopes are shown in red numbers.
Amino acid variants of the complete VP4 genes of all 8 P[1] strains in this study. Green indicates the unique aa variants sites in all 8 P[1] strains in this study and Yak strain QH-1 compared with the other P[1] strains in GenBank; Blue indicates the unique aa variants sites in all 8 P[1] strains in this study and Yak strain QH-1 compared with the only 2 G6P[1] BRVA strains in China. The neutralization epitopes are shown in red numbers. Orange indicates sialic acid binding site.
Amino acid variants of the complete VP4 genes of all 2 P[5] strains in this study. Green indicates the unique aa variants sites in all 2 P[5] strains in this study compared with the other P[5] strains in GenBank; Blue indicates the unique aa variants sites in all 2 P[5] strains in this study compared with the 4 vaccine strains. The neutralization epitopes are shown in red numbers. Orange indicates sialic acid binding site.