Abstract
Background
Mast cells are skin immune sentinels located in the upper dermis, where wheal formation and sensory nerve stimulation take place. Skin inflammation is occasionally accompanied by mast cell-driven responses with wheals, angioedema, or both. Immunoglobulin E (IgE) antibodies are regarded as typical stimuli to drive mast cell activation. However, various causative factors, including microbial infections, can drive IgE-independent mast cell response. When infected, the innate immunity orchestrates an immune response by activating receptor signaling via Toll-like receptors (TLRs).
Objective
In this study, we determined the effect of TLR7 stimulation on mast cells to investigate the possible mechanism of IgE-independent inflammatory response.
Methods
Human mast cell (HMC) line, HMC-1 cells were treated with TLR7 agonist and the morphologic alteration was observed in transmission electron microscopy. Further, TLR7 agonist treated HMC-1 cells were conducted to RNA sequencing to compare transcriptomic features.
Results
HMC-1 cells treated with TLR7 agonist reveals increase of intracellular vesicles, lipid droplets, and ribosomes. Also, genes involved in pro-inflammatory responses such as angiogenesis are highly expressed, and Il12rb2 was the most highly upregulated gene.
Conclusion
Our data suggest that TLR7 signaling on mast cells might be a potential therapeutic target for mast cell-driven, IgE-independent skin inflammation.
Keywords: Mast cell, Skin inflammation, Toll-like receptor 7
INTRODUCTION
Mast cells play a crucial role in skin immune response by expressing important stimulatory receptors on the cell surface. Classical mast cell activation occurs through the high affinity immunoglobulin E (IgE) receptor, FcεRI. Activation occurs when adjacent receptors, occupied by receptor-bound IgE, are crosslinked by re-exposure to the original or a crossreactive bivalent or multivalent antigen1. Many inflammatory skin diseases are mediated by such IgE-dependent mast activation, including atopic dermatitis and acute urticaria2. Besides classical IgE-mediated activation, it is emerging that the novel mast cell activation that are not only independent of IgE crosslinking but also express unique cytokine secretion profiles3. The diverse stimulatory receptors expressed on mast cells implicate multiple roles of mast cells in the pathogenesis underlying skin inflammation.
Importantly, mast cells respond to a wide range of pathological and environmental stimuli owing to their expression of Toll-like receptors (TLRs). TLRs are pattern-recognition receptors that initiate innate immune responses via the recognition of pathogen-associated molecular patterns (PAMPs)4. TLRs can also sense endogenous molecules that are released after cellular stress or tissue injury, known as danger-associated molecular patterns (DAMPs). Activation of TLRs in immune cells leads to the synthesis of various pro-inflammatory cytokines and chemokines via transcriptional regulation5. Also, TLRs are considered cellular sensors for detecting exogenous and endogenous ligands in primary sensory neurons to initiate itchy sensation (pruritus) that is associated with skin infections and tissue injuries. For example, TLR7 is expressed by a subset of primary sensory neurons that co-express itch-signaling components such as the transient receptor potential vanilloid subtype 1, which and play an important role in itchy sensation6. Other studies show that TLR7 agonists effectively mimic psoriatic skin inflammation in mouse with increase of dermal mast cells7. Thus, TLR-7 stimulus could be a mediator in the development of skin inflammation. Considering that mast cells predominantly reside in skin as a skin immune sentinel, we hypothesized that mast cells may act as a key player in the pathogenesis of infectious skin inflammation via TLR7 activation. In this study, we investigated transcriptome portrait of mast cells in stimulation with TLR7 and provided possible therapeutic targets in skin inflammatory disease.
MATERIALS AND METHODS
All procedures were approved by the Ewha Womans University College of Medicine Animal Care and Use Committee (EUM 20-024).
Cell culture
The human mast cell (HMC) line, HMC-1, was grown in Iscove's Modified Dulbecco's Medium (IMDM; GIBCO, Grand Island, NY, USA) containing 10% FBS, streptomycin (100 µg/ml), and penicillin (100 U/ml). The cells were stimulated with 2 µg/ml imiquimod (R837; Invivogen, San Diego, CA, USA) for 24 hours to examine the effects of TLR7 ligand on HMC-1 cells. The cultured HMC-1 cells were visualized by toluidine blue staining.
Reverse transcription polymerase chain reaction (RT-PCR)
For the analysis of TLR7 gene expression, HMC-1 cells cultured in the presence or absence of imiquimod for 24 hours were harvested and homogenized in TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA). Total RNA (1 µg) was transcribed into complementary DNA using a reverse transcription reagent (ELPIS-Biotech Inc., Daejeon, Korea) according to the manufacturer's instructions. TLR7 (201 bp) and internal control gene GAPDH (192 bp) were amplified using the primers listed in Table 1. The band pixel densities of TLR7 were divided by the pixel densities of the corresponding GAPDH bands for quantitation using UN-SCAN-IT-gel 6.1 software (Silk Scientific Inc., UT, USA).
Table 1. Primers used for reverse transcription polymerase chain reaction (RT-PCR) and quantitative RT-PCR.
| Primer | Sequence | Product size (bp) |
|---|---|---|
| Tlr7 | F: 5’-CCTTGAGGCCAACAACATCT-3’ | 201 |
| R: 5’-GTAGGGACGGCTGTGACATT-3’ | ||
| Muc22 | F: 5’-GGCCACTGACGTTTCTATCCA-3’ | 121 |
| R: 5’-GGCCGTGAAGTCCATTCCAG-3’ | ||
| Ern2 | F: 5’-TCGAAGGACCAATGTACGTCA-3’ | 117 |
| R: 5’-GGATGGTGAATGGCAGTTTCAT-3’ | ||
| Rps17 | F: 5’-GTTCGCACCAAAACCGTGAAG-3’ | 339 |
| R: 5’-GTTGGACAGACTGCCGAAGT-3’ | ||
| Epha1 | F: 5’-GTGGCCTCTACCTCGCTTTC-3’ | 124 |
| R: 5’-CCAGGCAGAGTGTCTGGGA-3’ | ||
| Kcnt1 | F: 5’-GACCCGTCCTTCCAGAACG-3’ | 173 |
| R: 5’-ACGCGCACAATGTAGAGCA-3’ | ||
| Nlrp3 | F: 5’-CGTGAGTCCCATTAAGATGGAGT-3’ | 191 |
| R: 5’-CGTGAGTCCCATTAAGATGGAGT-3’ | ||
| Ccl4 | F: 5’-CTGTGCTGATCCCAGTGAATC-3’ | 279 |
| R: 5’-TCAGTTCAGTTCCAGGTCATACA-3’ | ||
| Il12rb2 | F: 5’-AAAATAGATGCGTGCAAGAGAGG-3’ | 211 |
| R: 5’-GGGGAAGACCTGTGACTTGAG-3’ | ||
| Gapdh | F: 5’-GGTAAAGTGGATATTGTTGCCATCAATG-3’ | 192 |
| R: 5’-GGAGGGATCTCGCTCCTGGAAGATGGTG-3’ |
F: Forward, R: Reverse.
Transmission electron microscopy (TEM)
HMC-1 cells were cultured in the presence or absence of imiquimod (2 µg/ml) for 24 hours at a concentration with 106 cells/ml in 3 ml media. After centrifugation, the cell pellets were fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4). Specimens were washed in 0.1 M phosphate buffer and post-fixed with 1% osmium tetroxide in 0.1 M phosphate buffer (pH 7.4) for 1 hour, dehydrated with ethanol, and embedded in epoxy resin. Ultrathin sections, approximately 60~70 nm thick, were cut by an EM UC7 ultramicrotome (Leica, Wetzlar, Germany) using a diamond knife. Sections were contrasted with uranyl acetate followed by lead citrate and observed with H-7650 TEM (Hitachi, Tokyo, Japan) at an accelerating voltage of 80 kV.
RNA extraction, library construction, and sequencing
RNA was extracted from control HMC-1 cells and imiquimod-treated HMC-1 cells (Qiagen, Hilden, Germany) and subsequently column purified with RNeasy mini kit (Qiagen). Purified RNA was treated with DNase I (New England Biolabs, Ipswich, MA, USA) to remove genomic DNA. RNA quantity and quality were examined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) with an RNA integrity number ≥8. cDNA libraries were prepared with 1 µg of starting total RNA using a TruSeq RNA library Prep kit v2 (Illumina, San Diego, CA, USA), and transcriptome sequencing was performed using a NovaSeq S4 Reagent kit and Illumina NovaSeq with 151 bp paired-end reads per sample (Macrogen, Seoul, Korea).
Sequence annotation and identification of differentially expressed genes
FASTQ-formatted sequencing data were de-multiplexed to assign reads to the originating sample. The raw sequences were quality checked using FastQC v.0.11.7 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), the low-quality bases with quality scores <30 and the adaptor contamination were removed by Trimmomatic v.0.38 (https://www.usadellab.org/cms/?page=trimmomatic) using the parameters ‘LEADING:3 TRAILING:3 SLIDINGWINDOW:4:20 MINLEN:36’. Trimmed reads were mapped to the human genome reference (UCSC hg19) using TopHat v2.0.13 (https://ccb.jhu.edu/software/tophat/index.shtml). The total mapped read numbers for each transcript were determined and normalized to detect fragments per kilobase of exon per million fragments mapped (FPKMs) using Cufflinks v2.2.1 (https://cole-trapnell-lab.github.io/cufflinks/releases/v2.2.1/). Genes with more than one zero FPKM value out of the analyzed samples were excluded to filter potentially significant gene expressions. For differentially expressed gene (DEG) analysis, the values of log2 (FPKM+1) were calculated, and then normalized by quantile. Transcripts with absolute fold-change values larger than 2 with a p-value ≤0.05 were included in the analysis as DEG. Hierarchical clustering analysis was performed using complete linkage and Euclidean distance as a measure of similarity to display the DEG expression patterns. All DEG data analysis was conducted using R 3.2.2 (https://www.r-project.org).
Gene ontology and enrichment analysis
Functional groups and pathways encompassing the DEGs were identified based on the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway analysis using the Database for Annotation, Visualization, and Integrated Discovery (DAVID v.6.8; https://david.ncifcrf.gov/) software. The threshold was set as modified Fisher Exact p-value (EASE score) ≤0.05.
Quantitative reverse transcription PCR (qRT-PCR)
For validation of DEG, HMC-1 cells were cultured in the presence or absence of imiquimod for 24 hours followed by extraction of RNA using TRIzol (Thermo Fisher Scientific). Complementary DNA was synthesized using reverse transcription reagent (ELPIS-Biotech) according to the manufacturer's instructions. Real-time PCR analysis was performed on a StepOnePlus instrument (Applied Biosystems, Foster City, CA, USA) using a SensiFAST SYBR Hi-ROX kit (Bioline, London, UK). All gene expression values (Muc22, Ern2, Rps17, Epha1, Kcnt1, Nlrp3, Ccl4, and Il12rb2) were normalized to the GAPDH reference gene using the primers listed in Table 1.
Statistical analysis
Data are presented as mean±standard error of the mean (SEM). The statistical significance was analyzed by Student t-test using GraphPad PRISM 7 software (GraphPad Software Inc., San Diego, CA, USA). For all analyses, p<0.05 was considered statistically significant.
RESULTS
Imiquimod directly activates mast cells in vitro
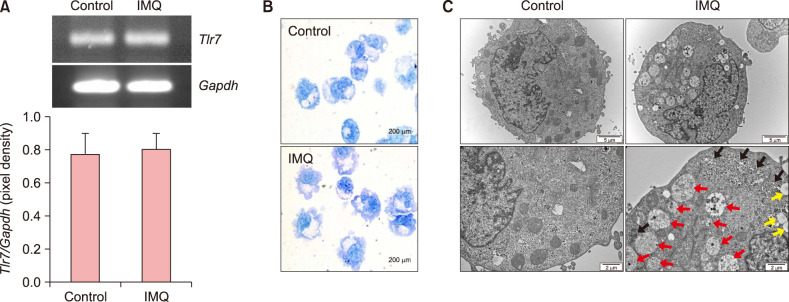
We treated the human mast cell line, HMC-1, with imiquimod, an agonist for TLR7, to investigate the effect of TLR7 stimulation. When we detected the TLR7 transcripts in HMC-1 cells, TLR7 mRNA expression was stable and not increased by the imiquimod treatment (Fig. 1A). To confirm the morphological characteristics under TLR7 stimulation, HMC-1 cells cultured with or without imiquimod were visualized by toluidine blue staining. Imiquimod-treated HMC-1 cells were larger and had increased villi surrounding the cells (Fig. 1B). These results prompted us to investigate other phenotypic alterations. As shown in Fig. 1C, TEM showed apparent differences with or without TLR7 stimulation. Imiquimod-treated HMC-1 cells showed an increase in vesicles, lipid droplets, and ribosomes compared to control HMC-1 cells. Vesicles indicate an activated state of HMC-1 cells, though most were already de-granulated. In particular, the increase of ribosomes is significant when HMC-1 cells were stimulated with imiquimod (Fig. 1C).
Fig. 1. Human mast cell (HMC)-1 cells show morphologic alterations in response to Toll-like receptor 7 (TLR7) stimulation. (A) Reverse transcription polymerase chain reaction was used to determine TLR7 expression in HMC-1 cells cultured with or without imiquimod (IMQ) for 24 h. The GAPDH gene was the internal control. The band pixel density of the TLR7 amplicon was divided by the pixel density of the corresponding GAPDH band. (B) HMC-1 cells cultured with or without IMQ were visualized by toluidine blue stain (original magnification, ×1,000). (C) Transmission electron microscope images of control HMC-1 cells (left) or IMQ-treated HMC-1 cells (right). The red arrows point to vesicles containing one or more smaller vesicles. Yellow arrows indicate lipid droplets. Black arrows point to ribosomes next to the rough endoplasmic reticulum. Original magnification, ×10,000 (top) and ×20,000 (bottom).
Transcriptome sequencing differentiates Toll-like receptor7-stimulated mast cells
By RNA sequencing, an average of 82 million 151-base long reads was processed, and approximately 98% of reads were mapped to the human genomic DNA reference. Mapped reads were assembled into known transcripts and condensed into FPKM expression values.
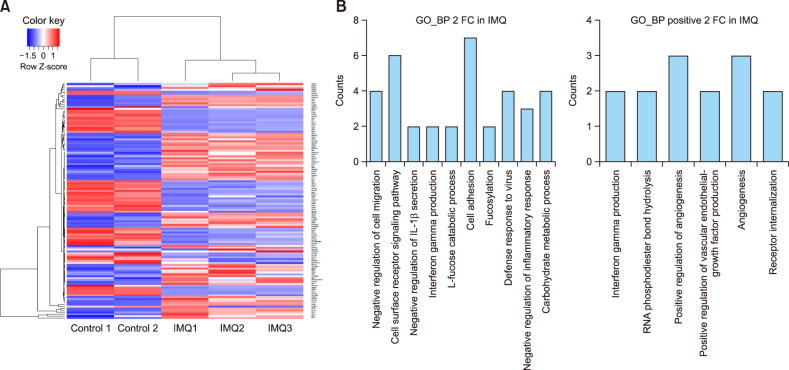
To visualize differential transcription between untreated HMC-1 cells and imiquimod-treated HMC-1 cells, a heatmap was generated showing |log2 (fold-change)|>2 (Fig. 2A). Twenty-five transcripts upregulated in imiquimod-treated HMC-1 with a log2 base fold-change > 2 are listed in Table 2.
Fig. 2. Transcription differs between control human mast cell (HMC)-1 cells and imiquimod-treated HMC-1 cells. (A) A heatmap of hierarchical clustering indicates differentially expressed gene (rows) between control HMC-1 cells and imiquimod-treated HMC-1 cells (fold-change>2, p<0.05). Red indicates upregulation and blue indicates downregulation. (B) Functional enrichment analysis of highly regulated genes in control HMC-1 cells and imiquimod-treated HMC-1 cells were annotated in the GO category of biological process (left). Distribution of positively regulated genes was annotated in the GO category of biological process (right).
Table 2. Twenty-five most upregulatedtranscripts in imiquimod-treated human mast cell (HMC)-1 cells compared to untreated cells.
| Gene_ID | Gene_symbol | Gene_description | Fold-change |
|---|---|---|---|
| 3595 | IL12RB2 | Interleukin 12 receptor, subunit beta 2 | 316 |
| 26824 | RNU11 | RNA, U11 small nuclear | 5 |
| 8339 | H2BC8 | H2B clustered histone 8 | 4.28 |
| 6218 | RPS17 | Ribosomal protein S17 | 3.96 |
| 8875 | VNN2 | Vanin 2 | 3.79 |
| 100507679 | MUC22 | Mucin 22 | 2.96 |
| 57582 | KCNT1 | Potassium sodium-activated channel subfamily T member 1 | 2.87 |
| 103344718 | HOTS | H19 opposite tumor suppresso | 2.84 |
| 150946 | GAREM2 | GRB2-associated regulator of MAPK1 subtype 2 | 2.81 |
| 146227 | BEAN1 | Brain-expressed associated with NEDD4 1 | 2.67 |
| 2041 | EPHA1 | EPH receptor A1 | 2.67 |
| 728113 | ANXA8L1 | Annexin A8 like 1 | 2.64 |
| 103091864 | SNHG22 | Small nucleolar RNA host gene 22 | 2.57 |
| 6351 | CCL4 | C-C motif chemokine ligand 4 | 2.55 |
| 23632 | CA14 | Carbonic anhydrase 14 | 2.54 |
| 9478 | CABP1 | Calcium-binding protein 1 | 2.49 |
| 1545 | CYP1B1 | Cytochrome P450 family 1 subfamily B member 1 | 2.47 |
| 83643 | CCDC3 | Coiled-coil domain containing 3 | 2.43 |
| 2626 | GATA4 | GATA binding protein 4 | 2.43 |
| 10595 | ERN2 | Endoplasmic reticulum to nucleus signaling 2 | 2.39 |
| 2117 | ETV3 | ETS variant transcription factor 3 | 2.38 |
| 9945 | GFPT2 | Glutamine-fructose-6-phosphate transaminase 2 | 2.38 |
| 1829 | DSG2 | Desmoglein 2 | 2.37 |
| 10859 | LILRB1 | Leukocyte immunoglobulin-like receptor B1 | 2.37 |
| 114548 | NLRP3 | NLR family pyrin domain containing 3 | 2.34 |
We categorized the 218 highly differentially regulated genes between control HMC-1 and imiquimod-treated HMC-1 into enriched GO categories of biological processes. Genes that were negatively regulated for cell migration, signaling pathways for cell surface receptors, cell adhesion, defense response to viruses, and carbohydrate metabolic processes were prevalent in the biological processes (Fig. 2B, left). Because we focused on the TLR7-mediated activation response in mast cells, we chose to analyze positively expressed genes in the biological process category. HMC-1 cells stimulated with imiquimod highly expressed genes involved in angiogenesis-half of the six categories were positive regulation of angiogenesis, angiogenesis, and vascular endothelial growth factor production (Fig. 2B, right).
RT-qPCR results validate RNA sequencing
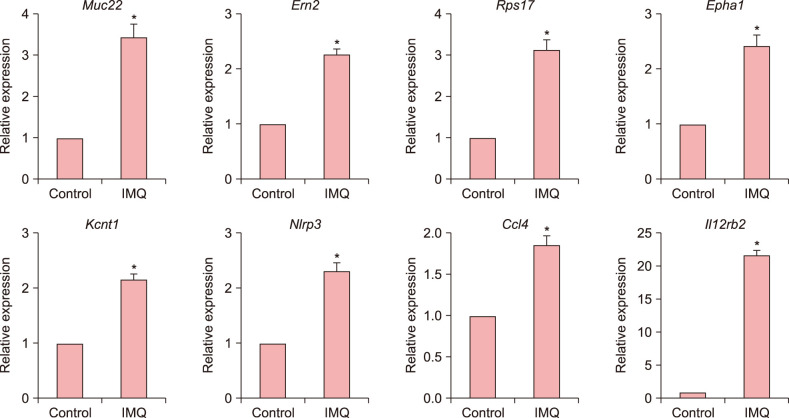
We selected eight genes involved in various inflammatory process such as angiogenesis, cell adhesion, migration, and vascular permeability for validation. Expression of Muc22, Ern2, Rps17, Epha1, Kcnt1, Nlrp3, Ccl4, and Il12rb2 was confirmed in both control HMC-1 and imiquimod-treated HMC-1 cells using RT-qPCR. We found that expression of all eight genes increased in imiquimod-treated HMC-1 cells, confirming the results of the RNA sequencing analysis. Although the fold-changes of upregulated genes did not appear substantial by RNA sequencing, RT-qPCR validated the fold-changes. In particular, Il12rb2, the most highly expressed gene when analyzed by RNA sequencing, was also most upregulated by quantitative PCR (Fig. 3).
Fig. 3. Validation of genes upregulated in response to imiquimod. The mRNA expression of Muc22, Ern2, Rps17, Epha1, Kcnt1, Nlrp3, Ccl4, and Il12rb2 in control human mast cell (HMC)-1 cells and imiquimod-treated HMC-1 cells was analyzed by quantitative reverse transcription polymerase chain reaction. Values are presented as the mean±standard error of the mean (*p<0.05).
DISCUSSION
In this study, we demonstrated that TLR7 activation stimulates mast cells, leading to the upregulation of genes involved in the inflammatory process. Specifically, we showed that mast cells activate in response to TLR7 stimulation with imiquimod, phenotypically presented by the increases in vesicles, lipid droplets, and ribosomes. Furthermore, transcriptome sequencing indicates that TLR7 activation increases the expression of genes involved in pro-inflammatory responses such as angiogenesis.
Mast cells can sense a plethora of agents by expressing various receptors including TLRs. TLRs are divided into two groups according to their subcellular localization and respective PAMPs: (i) cell surface TLRs (TLR1, 2, 4, 5, 6, and 10) that recognize microbial membrane components such as lipids, lipoproteins, and protein; and (ii) intracellular TLRs (TLR3, 7/8, and 9) that recognize viral or endogenous nucleic acids4. Infections by microorganisms such as bacteria or viruses could be an underlying cause in inflammatory skin disease8,9. Besides, some viral infection occur acute urticaria together with the onset of viral symptoms10,11. Thus, TLR signals might be an important mediator of infectious inflammatory skin disease.
TLR7 was originally identified as recognizing imidazoquinoline derivatives such as imiquimod and resiquimod, and guanine analogs such as loxoribine12. Intradermal injection of imiquimod, R848, and loxoribine induced scratching in wild-type mice, which was reduced in Tlr7-/-mice6, indicating that TLR7 is an itch mediator and a potential therapeutic target for anti-itch treatment in skin diseases. On the other hand, imiquimod-treated mice showed psoriasis-like skin inflammation with increase of dermal mast cells, suggesting that a TLR7 agonist could be direct stimulator of mast cells in psoriatic skin inflammation7.
Our results strongly support that TLR7 activation on mast cells can induce an inflammatory response. The obvious increase of intracellular components such as vesicles, lipid droplets, and ribosomes in imiquimod-treated HMC-1 cells indicate that active biological processes respond to TLR7 stimulation. Transcription analysis showed upregulation of pro-inflammatory genes when TLR7 activated mast cells, particularly in angiogenesis. Importantly, most skin inflammatory diseases are characterized by excessive angiogenesis13. Among 25 top-upregulated genes, we particularly validated eight genes involved in various inflammatory process that possibly contribute to angiogenic immunopathogenesis. Mucins (MUCs) are family of glycoproteins that are present in the mucus coating of epithelial surface and are used as ligands for cell adhesion and also promote angiogenesis. MUC gene expression has been found to be altered in inflammatory states14. TLR7 stimulated mast cells showed upregulation of one of MUCs coding gene, Muc22 and positive regulator of mucin gene expression, Ern2. The upregulation of Rps17 gene which provides instructions for making ribosomal proteins support outstanding increase of ribosomes in TLR7-stimulated mast cells. Epha1 gene is involved in cell migration, adhesion, and angiogenesis15,16. In addition, upregulated Kcnt1, Nlrp3, and Ccl4 are expected to contribute to promote inflammatory process such as immune cell activation and migration. In particular, Nlrp3 is expected to increase vascular permeability by producing active form of interleukin (IL)-1β. Of note, the most highly upregulated gene, Il12rb2, implied that TLR7 signaling may enhance IL-12 response in mast cells. Upregulation of the Il12rb2 gene is associated with several diseases including leprosy17, and is thought to contribute to the inflammatory response and host defense mechanism. In fact, IL-12 is key player in the regulation of Th1 response which are orchestrated mainly by macrophage and dendritic cells in viral infection. Our result indicate that mast cells also could be direct responder in addition to T cells in IL-12 abundant microenvironment. Enhanced expression of receptor for IL-12 could activates downstream pathway such as NF-κB that ultimately induce pro-inflammatory cytokines. Interestingly, several mast cell-derived pro-inflammatory cytokines have been implicated in Th17 cell-skewing inflammation, including IL-1β and IL-618. Therefore, mast cells could be responsible for producing pro-inflammatory cytokines as the consequence of increased IL-12 response when TLR7 stimulated. The specific outcome of IL-12 effect on mast cells should be handled within our next expanded research. Also, our findings should be broaden into early and rate response in actual mast cell degranulation. Considering that other chronic inflammatory autoimmune disease such as psoriasis can be initiated by excessive activation of endosomal TLRs, particularly TLR7, TLR8, and TLR919, TLR7 signaling in mast cells is able to expand our point of view on immune network between inflammatory skin disease.
In summary, we demonstrated that TLR7 directly mediate mast cell activation and the TLR7 or IL-12 signaling could be candidates in the search for therapeutic targets for skin inflammatory disease.
Footnotes
CONFLICTS OF INTEREST: The authors have nothing to disclose.
FUNDING SOURCE: This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea Government (MSIT) (No. NRF-2019R1H1A1035603 and NRF-2020 R1C1C1012769) and RP-Grant 2019 of Ewha Womans University. The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
DATA SHARING STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Galli SJ, Tsai M. IgE and mast cells in allergic disease. Nat Med. 2012;18:693–704. doi: 10.1038/nm.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang TB, Kim BS. Pruritus in allergy and immunology. J Allergy Clin Immunol. 2019;144:353–360. doi: 10.1016/j.jaci.2019.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lyons DO, Pullen NA. Beyond IgE: alternative mast cell activation across different disease states. Int J Mol Sci. 2020;21:1498. doi: 10.3390/ijms21041498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 5.West AP, Koblansky AA, Ghosh S. Recognition and signaling by toll-like receptors. Annu Rev Cell Dev Biol. 2006;22:409–437. doi: 10.1146/annurev.cellbio.21.122303.115827. [DOI] [PubMed] [Google Scholar]
- 6.Liu T, Xu ZZ, Park CK, Berta T, Ji RR. Toll-like receptor 7 mediates pruritus. Nat Neurosci. 2010;13:1460–1462. doi: 10.1038/nn.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho KA, Park M, Kim YH, Woo SY. Th17 cell-mediated immune responses promote mast cell proliferation by triggering stem cell factor in keratinocytes. Biochem Biophys Res Commun. 2017;487:856–861. doi: 10.1016/j.bbrc.2017.04.141. [DOI] [PubMed] [Google Scholar]
- 8.Yanase Y, Takahagi S, Hide M. Chronic spontaneous urticaria and the extrinsic coagulation system. Allergol Int. 2018;67:191–194. doi: 10.1016/j.alit.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Griffin PM, Kevat DA, McCarthy JS, Woods ML. Chronic urticaria following acute hepatitis A. BMJ Case Rep. 2012;2012:bcr2012006479. doi: 10.1136/bcr-2012-006479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mareri A, Adler SP, Nigro G. Herpesvirus-associated acute urticaria: an age matched case-control study. PLoS One. 2013;8:e85378. doi: 10.1371/journal.pone.0085378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chuamanochan M, Chiewchanvit S, Tovanabutra N, Rujiwetpongstorn R, Laosakul K, Maurer M. A case of dengue fever presenting with acute urticaria. Asian Pac J Allergy Immunol. 2019 doi: 10.12932/AP-150419-0539. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 12.Hemmi H, Kaisho T, Takeuchi O, Sato S, Sanjo H, Hoshino K, et al. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol. 2002;3:196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 13.Richarz NA, Boada A, Carrascosa JM. Angiogenesis in dermatology - insights of molecular mechanisms and latest developments. Actas Dermosifiliogr. 2017;108:515–523. doi: 10.1016/j.ad.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Volin MV, Shahrara S, Haines GK, 3rd, Woods JM, Koch AE. Expression of mucin 3 and mucin 5AC in arthritic synovial tissue. Arthritis Rheum. 2008;58:46–52. doi: 10.1002/art.23174. [DOI] [PubMed] [Google Scholar]
- 15.Pasquale EB. Eph receptor signalling casts a wide net on cell behaviour. Nat Rev Mol Cell Biol. 2005;6:462–475. doi: 10.1038/nrm1662. [DOI] [PubMed] [Google Scholar]
- 16.Kuijper S, Turner CJ, Adams RH. Regulation of angiogenesis by Eph-ephrin interactions. Trends Cardiovasc Med. 2007;17:145–151. doi: 10.1016/j.tcm.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Kim J, Uyemura K, Van Dyke MK, Legaspi AJ, Rea TH, Shuai K, et al. A role for IL-12 receptor expression and signal transduction in host defense in leprosy. J Immunol. 2001;167:779–786. doi: 10.4049/jimmunol.167.2.779. [DOI] [PubMed] [Google Scholar]
- 18.Cho KA, Suh JW, Sohn JH, Park JW, Lee H, Kang JL, et al. IL-33 induces Th17-mediated airway inflammation via mast cells in ovalbumin-challenged mice. Am J Physiol Lung Cell Mol Physiol. 2012;302:L429–L440. doi: 10.1152/ajplung.00252.2011. [DOI] [PubMed] [Google Scholar]
- 19.Lai CY, Su YW, Lin KI, Hsu LC, Chuang TH. Natural modulators of endosomal toll-like receptor-mediated psoriatic skin inflammation. J Immunol Res. 2017;2017:7807313. doi: 10.1155/2017/7807313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.