Summary
Rggs are a group of transcriptional regulators with diverse roles in metabolism and virulence. Here, we present work on the Rgg1518/SHP1518 quorum sensing system of Streptococcus pneumoniae. The activity of Rgg1518 is induced by its cognate peptide, SHP1518. In vitro analysis showed that the Rgg1518 system is active in conditions rich in galactose and mannose, key nutrients during nasopharyngeal colonization. Rgg1518 expression is highly induced in the presence of these sugars and its isogenic mutant is attenuated in growth on galactose and mannose. When compared with other Rgg systems, Rgg1518 has the largest regulon on galactose. On galactose it controls up- or down regulation of a functionally diverse set of genes involved in galactose metabolism, capsule biosynthesis, iron metabolism, protein translation, as well as other metabolic functions, acting mainly as a repressor of gene expression. Rgg1518 is a repressor of capsule biosynthesis, and binds directly to the capsule regulatory region. Comparison with other Rggs revealed inter-regulatory interactions among Rggs. Finally, the rgg1518 mutant is attenuated in colonization and virulence in a mouse model of colonisation and pneumonia. We conclude that Rgg1518 is a virulence determinant that contributes to a regulatory network composed of multiple Rgg systems.
Keywords: Streptococcus pneumoniae, transcriptional regulation, Rgg, sugar metabolism, capsule synthesis, virulence
Graphical Abstract
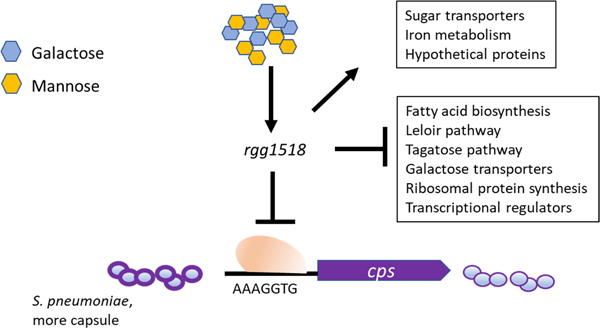
Abbreviated summary
The important human pathogen Streptococcus pneumoniae can sense the sugars present in the respiratory mucus including galactose and mannose by using a transcriptional regulator called Rgg1518. This regulator controls the expression of a large number of genes involved in vital metabolic processes and capsule, the most important pneumococcal virulence determinant. The absence of Rgg1518 adversely affects the microbe’s virulence and its survival in the host.
Introduction:
Streptococcus pneumoniae (pneumococcus) causes a diverse array of infections. It can exist as an asymptomatic colonizer of the upper respiratory tract or can spread to middle ear, lower respiratory tract, blood and central nervous system, causing diseases such as otitis media, pneumonia, septicaemia and meningitis, respectively (Weiser et al., 2018). S. pneumoniae is the leading agent of lower respiratory infection morbidity and mortality globally, causing more deaths than those caused by other major respiratory infectious agents combined in 2016 (Troeger et al., 2017). Unfortunately, there is a rising trend in the number of antibiotic resistant pneumococcal strains. The danger posed by the rising antibiotic resistance was recognised by the World Health Organisation, hence S. pneumoniae has been declared as a priority pathogen for which effective antibiotics are urgently needed.
The study of how the pneumococcus alters its phenotype in health and disease, and in different tissue sites, can reveal novel drug targets. S. pneumoniae encounters diverse environmental conditions in its human host, including fluctuations in oxygen and metal concentration, differences in concentration and type of nutrients, and changes in temperature (Sahin-Yilmaz and Naclerio, 2011; Shafeeq et al., 2011; Hajaj et al., 2017). This microbe has an innate capacity to sense and respond to changing environmental stimuli, employing various mechanisms including regulatory RNAs, standalone “one-component” regulators and two component regulatory systems (TCS) (Wilton et al., 2015; Cuevas et al., 2017; Kadam et al., 2017; Gómez-Mejia et al., 2018; Zhi et al., 2018). These mechanisms modulate an array of essential cellular functions, including acquisition and metabolism of nutrients, detoxification of reactive oxygen species, incorporation of genetic material, and expression of virulence factors.
Quorum sensing (QS) systems play a key role in sensing and responding to the environment. These multi-component regulatory mechanisms coordinate bacterial population responses to diverse stimuli (Kadam et al., 2017; Cuevas et al., 2017; Motib et al., 2019). The operation of these cell-cell communication systems relies on secretion and subsequent detection of signalling molecules by the producing and neighbouring cells. The response directs community- level synchronization of transcription, enabling microbes to adopt a phenotype consistent with the requirements of the specific host niche (Rutherford and Bassler, 2012). The pneumococcal QS systems can be divided broadly into two major categories: Autoinducer-2 (AI-2) and peptide mediated QS systems. AI-2 signaling molecule synthesis relies on the metabolic enzyme LuxS, which catalyses the conversion of S-ribosyl-homocysteine into homocysteine. It was shown by Trapetti et al that AI-2 QS signaling is mediated through a fructose-specific phosphoenolpyruvate-phosphotransferase system (FruA), and that AI-2 triggers upregulation of galactose catabolism through the Leloir pathway, ultimately enhancing capsule biosynthesis (Trappetti et al., 2017). In addition, the LuxS/AI-2 QS system plays an important role in biofilm synthesis, competence, and fratricide (Trappetti et al., 2017).
Peptide mediated pneumococcal QS systems are divided into three main groups based on peptide sequence features, transporters they utilize, and receptors; these are: (i) double glycine processing site peptides, (ii) peptides associated with the RRNPP (Rap, Rgg, NprR, PlcR, and PrgX) superfamily of quorum-sensing proteins, and (iii) lanthionine-containing peptides (Cook, LaSarre and Federle, 2013; Aggarwal et al., 2020).
Pneumococcal double glycine peptides, such as the competence stimulating peptide (CSP) and the bacteriocin inducing peptide (BIP), contain a conserved N-terminal leader sequence that terminates commonly in Gly-Gly residues. These peptides are exported after cleavage of the leader sequence via peptidase-containing ABC transporters. After secretion, some double-glycine processed peptides induce a response in the neighbouring cell by signalling via two component systems (TCS) (Håvarstein, et al., 1995; de Saizieu et al., 2000; Wang et al., 2020). The peptide interacts with the histidine kinase of the TCS, stimulates its autophosphorylation and transphosphorylation of the cognate response regulator, and initiate changes in the transcriptional composition of the cell.
Cyclic peptides, such as lanthionine-containing peptides, undergo post-translational modifications that result in the introduction of thioether amino acids lanthionine and methyllanthionine (Willey and van der Donk, 2007). Their distinctive structure is formed by LanBC or LanM enzymes, when serine or threonine residues in the pro-peptide are dehydrated and linked to cysteine thiols. The peptide is exported by dedicated LanT transporters (Sen et al., 1999; Koponen et al., 2002; Willey and van der Donk, 2007) and in various cases autoregulation via a two-component regulatory system also occurs, as exemplified by nisin autoinduction via NisRK in L. lactis (Kuipers et al., 1995).
Two RRNPP type QS systems have been studied in S. pneumoniae: Tpr/Phr and Rgg/SHP regulators (Hoover et al., 2017; Cuevas et al., 2017; Zhi et al., 2018; Motib et al., 2019). The Tpr (transcription factor regulated by Phr peptide) and Rgg (Regulator gene of glycosyltransferase) transcription factors are positively regulated by their cognate peptides Phr (phosphatase regulator) and SHP (Short Hydrophobic Peptides), respectively. The Tprs are part of the PlcR family, the best-characterized pair, TprA/PhrA, play a critical roles in pneumococcal colonization and virulence (Motib et al., 2019). Multiple Rgg regulators have been characterized in pneumococcus and other streptococcal species. The model that emerges is that SHPs are cleaved into active forms during export from precursor polypeptides that are typically shorter than 35 residues (Aggarwal et al., 2014; Zhi et al., 2018; Aggarwal et al., 2020). Multiple streptococcal species export SHPs through an ABC transporter, PptAB, whose homolog in Enterococcus faecalis exports sex pheromones (Varahan et al., 2014; Chang and Federle, 2016). The exact mechanism(s) directing SHPs to their transporter remain unclear. The mature peptides are re-imported into the cell via an oligopeptide permease system, where they interact with the cognate Rgg regulator, altering the cell’s transcriptional state (Chang et al., 2011; Fleuchot et al., 2011; Zhi et al., 2018; Aggarwal et al., 2020).
The pneumococcal pangenome encodes eight rgg homologs. In type 2 D39 strain there are 6 Rgg paralogues. Previously we characterised Rgg1952, Rgg144/SHP144 and Rgg939/SHP939 (Bortoni et al., 2009; Junges et al., 2017; Zhi et al., 2018). Rgg1952 and Rgg144 are part of the core genome. Rgg1952 is important in oxidative stress resistance. The Rgg144/SHP144 system represses capsule biosynthesis and induces VP1, a peptide that promotes biofilm development and virulence (Cuevas et al., 2017; Zhi et al., 2018). Its regulon contains genes involved in replication, recombination, translation, and nucleotide transport and metabolism (Zhi et al., 2018). It also controls multiple sugar transporters and its regulon is sugar-specific, with a broad response in mannose and a restricted response in galactose. Finally, Rgg939 is part of the accessory genome (Junges et al., 2017; Zhi et al., 2018). The diversity of genes regulated by Rgg939/SHP939 varies in response to environmental conditions, with an extensive regulon during growth on mannose and a limited regulon in galactose (Junges et al., 2017; Zhi et al., 2018). The regulon is predicted to function in cell division, iron transport, cell membrane biogenesis, and metabolism among others. Rgg939 also regulates capsule synthesis. While Zhi et al. (2018) suggested that Rgg939 can directly inhibit expression from the capsule locus, Junges et al. (2017), showed that Rgg939 expression positively regulates the expression of genes involved in production of compounds present in capsular polysaccharides of serotypes 12A and 12F. Rgg939/SHP939 signalling down regulated biofilm development on A549 epithelial cells (Junges et al., 2017). In murine models of disease, Rgg939/SHP939 was shown to be associated with a reduction in bacterial fitness in a pneumonia model (Junges et al., 2017), or alternatively, increased nasopharyngeal colonization and virulence in carriage and pneumonia models (Zhi et al., 2018).
In this study, we investigated the role of Rgg1518 (SPD_1518). This Rgg appears to be at the intersection of multiple RRNPP regulators, including Rgg144, Rgg939 and TprA. It is part of the accessory pneumococcal genome; present in approximately half of strains. We identified its peptide pheromone SHP1518, whose induction requires Rgg1518. Rgg1518 is highly induced on mannose and galactose, with an extensive regulon on galactose controlling the expression of genes involved in sugar transport, galactose metabolism and capsule synthesis. Finally, Rgg1518 promotes colonization and contributes to lung disease.
Results:
Rgg1518 promotes growth on galactose and mannose.
Previous studies on pneumococcal peptide mediated RRNPP family QS systems, namely Rgg144/SHP144, Rgg939/SHP939, and TprA/PhrA, indicated their role in host derived sugar metabolism (Zhi et al., 2018; Motib et al., 2019). Due to its homology to other Rggs, we hypothesised that Rgg1518 may also play a role in host-derived sugar metabolism, such as for galactose and mannose, which are plentiful in N- and O-linked glycans (King, 2010). To determine if Rgg1518 has a role in nutrient utilization, wild-type and Δrgg1518 strains were grown in BHI and CDM supplemented either with glucose, galactose or mannose. No difference in growth profiles was detected in BHI (SFigure 1) (p>0.05). Likewise, the strains grew equally well on CDM supplemented with glucose (Figure 1). In contrast, on galactose and mannose the growth rate (0.037±0.002 h−1, and 0.093±0.004 h−1 n=6) and yield of ∆rgg1518 was significantly lower than that of the wild type D39 (0.150±0.001 h−1, and 0.111±0.0028, h−1 n=6, respectively) and the complemented strain rescued the wild-type growth (0.14±0.001 h−1) (Figure 1). Notably, on galactose, the growth of the ∆rgg1518 displayed not only a lower max OD, but also an extended lag phase. We conclude that rgg1518 plays an important role in utilization of galactose and mannose.
Figure 1:
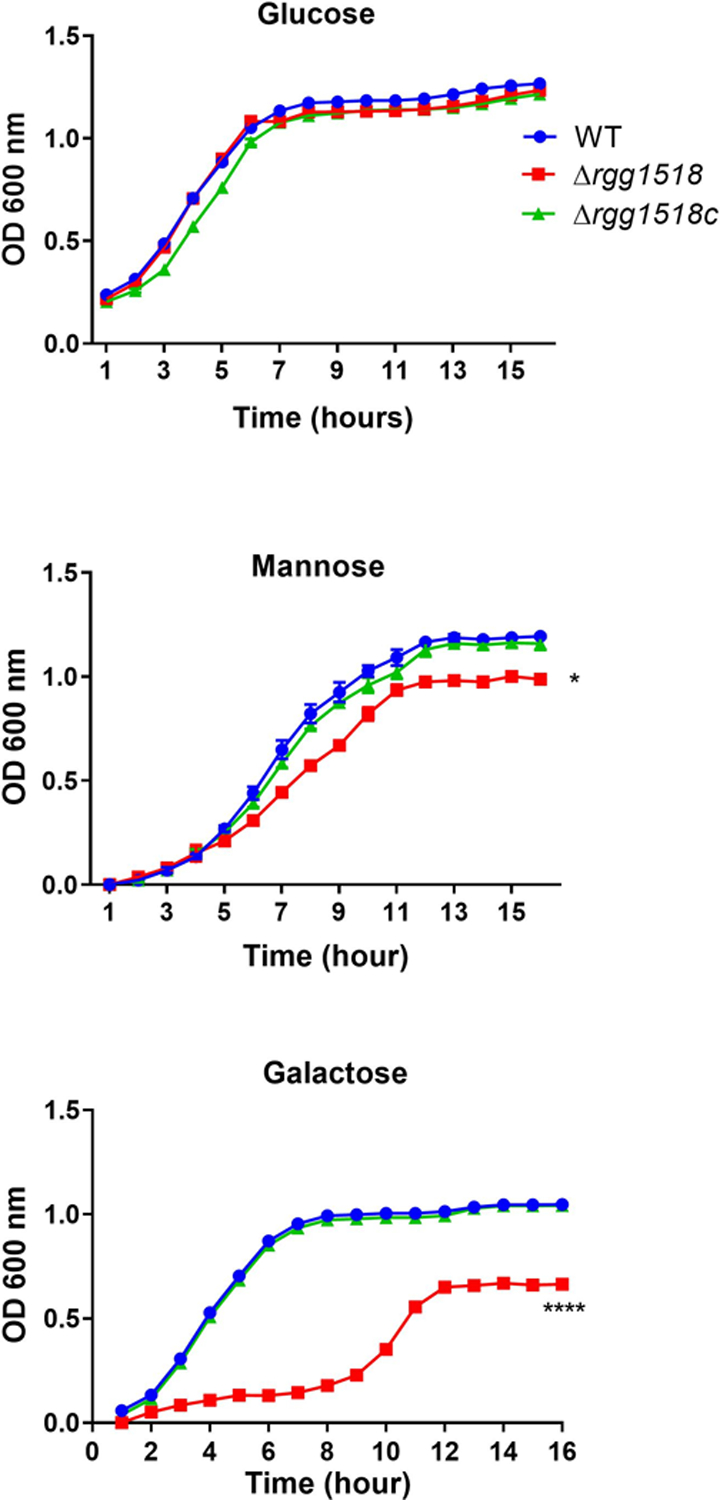
Growth profiles of pneumococcal strains in CDM supplemented with 55 mM glucose, galactose, or mannose. Experiment was repeated using 3 replicates of 6 independent biological samples; vertical bars indicate standard error of means (SEM). Significant differences were seen by comparing the growth rates of mutant strains to the wild type D39 using one-way ANOVA and Dunnett’s multiple comparisons test. (*p<0.05, ****p<0.0001).
The promoter for rgg1518 is induced by galactose and mannose.
The growth attenuation on galactose and mannose indicated a potential role for Rgg1518 on metabolism of these sugars. To investigate this further, we determined if the rgg1518 promoter (P) could be induced by galactose and mannose using LacZ reporter strains. Strains were grown in CDM-Glucose and then transferred to CDM with no sugar, or a single sugar for 1h. No significant induction of Prgg1518::lacZ-wild-type was observed on glucose (176±3.0 MU, n=3) relative to the no sugar control (172±2 MU, n=3) (p>0.05). In contrast, a significant promoter induction was detected on galactose (969±6 MU, n=3), and mannose (503±13 MU, n=3) (p<0.001 and p<0.01, for galactose and mannose) compared to no sugar control. These results show that the rgg1518 promoter is inducible by galactose and mannose (Figure 2).
Figure 2:
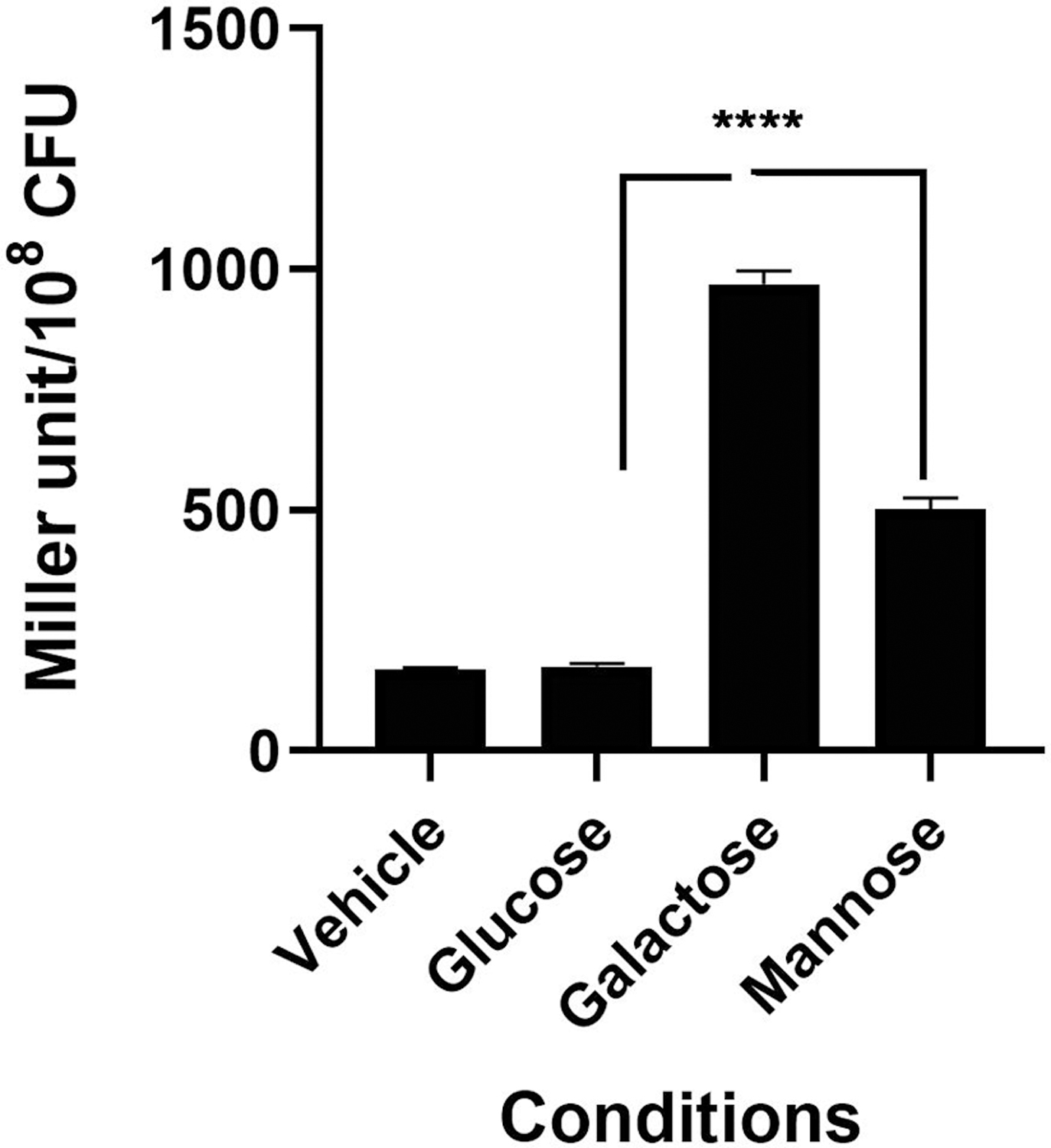
Induction analysis of rgg1518 promoter by different sugars using lacZ reporter assay. Strains were grown in CDM-Glucose and then transferred to CDM with no sugar, or a single sugar for 1h. Miller unit is expressed as nmol p-nitrophenol/min/ml. The values are the average of three independent experiments, each with three replicates, and vertical bars indicate standard error of means (SEM), (**p<0.01 and ***p<0.001 compared to expression on galactose).
Rgg1518 and SHP1518 form a quorum sensing system
Out of six Rgg homologous present in type 2 D39 pneumococcal strain, only Rgg144/SHP144 and Rgg939/SHP939 were demonstrated to be peptide mediated QS systems (Zhi et al., 2018). A recent study in D39 reported a putative shp1518 gene encoding for a 33 aa long peptide (MGFKKYLKNLPKNSGFLIWSWIQLIWFETWFWG) (Wang et al., 2020). The gene is located 93 nucleotides away from rgg1518 and inversely transcribed relative to rgg1518. We tested whether Rgg1518 and SHP1518 form a QS system. To this end, we tested the induction of Pshp1518::lacZ in wild type and Δrgg1518 backgrounds (Figure 3A). Induction of Pshp1518 required Rgg1518, as the deletion of rgg1518 led to a 3.75-fold decrease in Pshp1518 driven β-galactosidase activity relative to wild-type. Moreover, similar to Prgg1518, the Pshp1518 activity was significantly induced in galactose but not glucose. These results demonstrate that SHP1518 is induced by galactose and positively regulated by Rgg1518.
Figure 3:
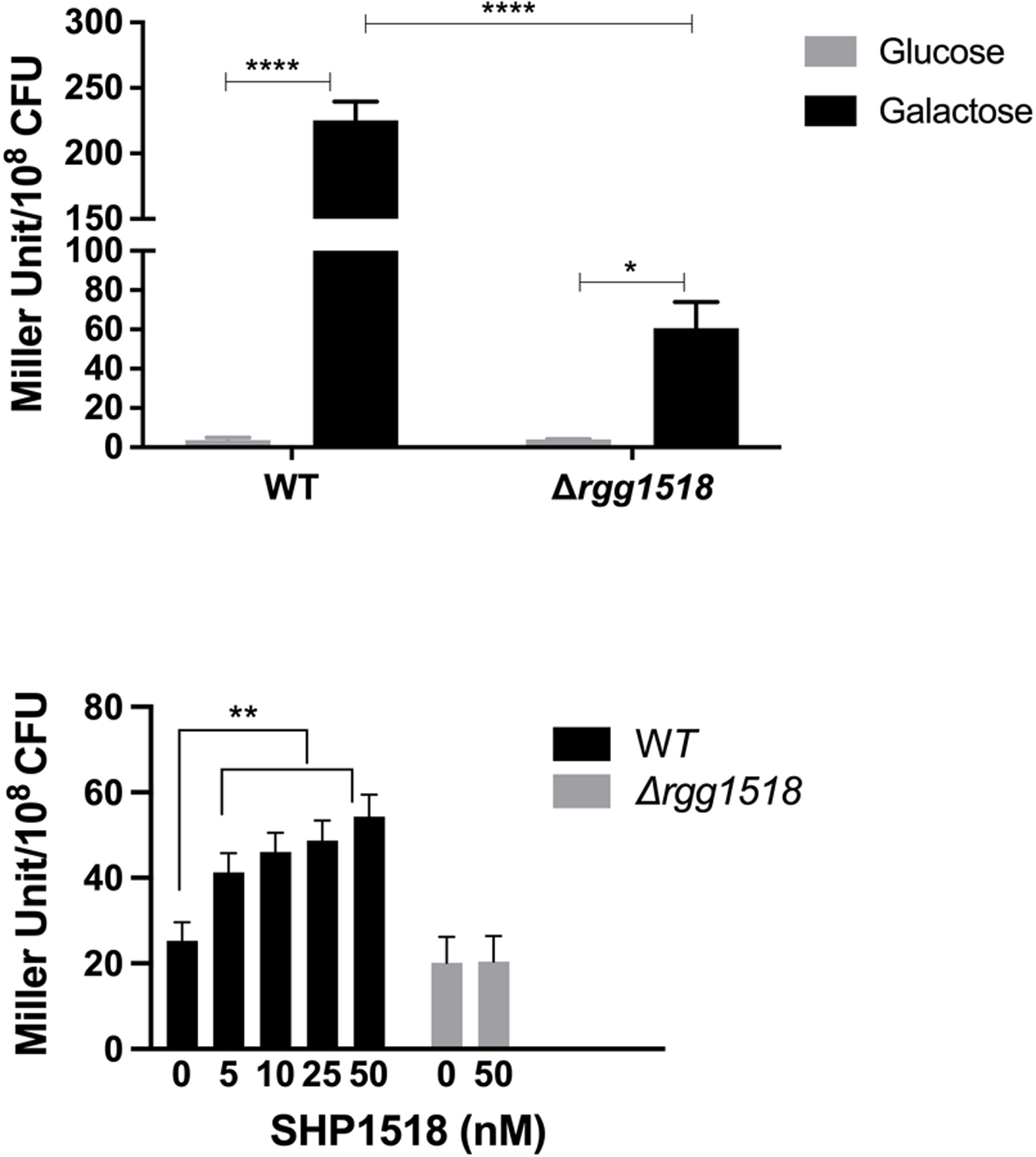
Expression levels (in Miller units) of pneumococcal transcriptional lacZ-fusion to the promoter region of shp1518 (Pshp1518:lacZ) in wild type (WT) and Δrgg1518 in glucose and galactose (A), and in the absence and presence of synthetic SHP1518 peptide in cultures grown in CDM supplemented with glucose (B).
Next, we identified an active form of SHP1518. We generated synthetic 12- (IQLIWFETWFWG) and 16- (IWSWIQLIWFETWFWG) residues C-terminal truncations of SHP1518. These peptides were evaluated for their ability to induce Pshp1518 in wild type and Δrgg1518. The reporter assay was performed on glucose, rather than galactose, to reduce induction of Pshp1518 by the native SHP1518. The 12 aa peptide did not induce Pshp1518 (data not shown). In contrast, the 16aa peptide showed a gradual dose-dependent induction of Pshp1518 relative to control. The difference in induction was significant and even at 5 nM and at 50 nM there was more than 2-fold increase relative to the no peptide control (p<0.01). Furthermore, the induction by the synthetic peptide required the presence of Rgg1518 (Figure 3B). We conclude that SHP1518 serves as the cognate peptide for Rgg1518.
SHPs have been defined and grouped based on (i.) the presence of lysine or arginine residues in their N-terminus and glutamate or aspartate residues at their C- terminus, and (ii.) encoded by a gene upstream of an rgg homolog and divergently transcribed. SHP1518 fits these criteria. SHPs are further classified into 3 groups based on the presence of aspartate (Group 1) or glutamate (Group 2) in the C-terminus (Ibrahim et al 2007a). Additionally, Group 3 peptides meet the criteria for SHP but are encoded by genes overlapping the end of the rgg genes in a convergent orientation (Fleuchot et al., 2011). Unlike Group 3, the shp1518 sequence does not overlap with rgg1518. Thus, SHP1518 can be classified as Group 2 based on its position on the locus and the glutamate residue in the C-terminus.
Role of Rgg1518 in regulation of other pneumococcal Rggs.
Previously, we reported the interaction between different Rgg/SHP systems, and a summary of regulon overlap among different Rggs on galactose and mannose have been graphically illustrated in Figure 4A and SFigure 3. Specifically, we found that the presence of non-cognate Rgg regulators is needed for maximal induction of SHP144 and SHP939 (Zhi et al. 2018). Further, in our previous work, both Rgg144 and Rgg939 were found to influence the transcription of adjacent to Rgg1518 (SPD_1513–1517); which here we show is also controlled by Rgg1518 (Zhi et al. 2018) (Table 1).
Figure 4:
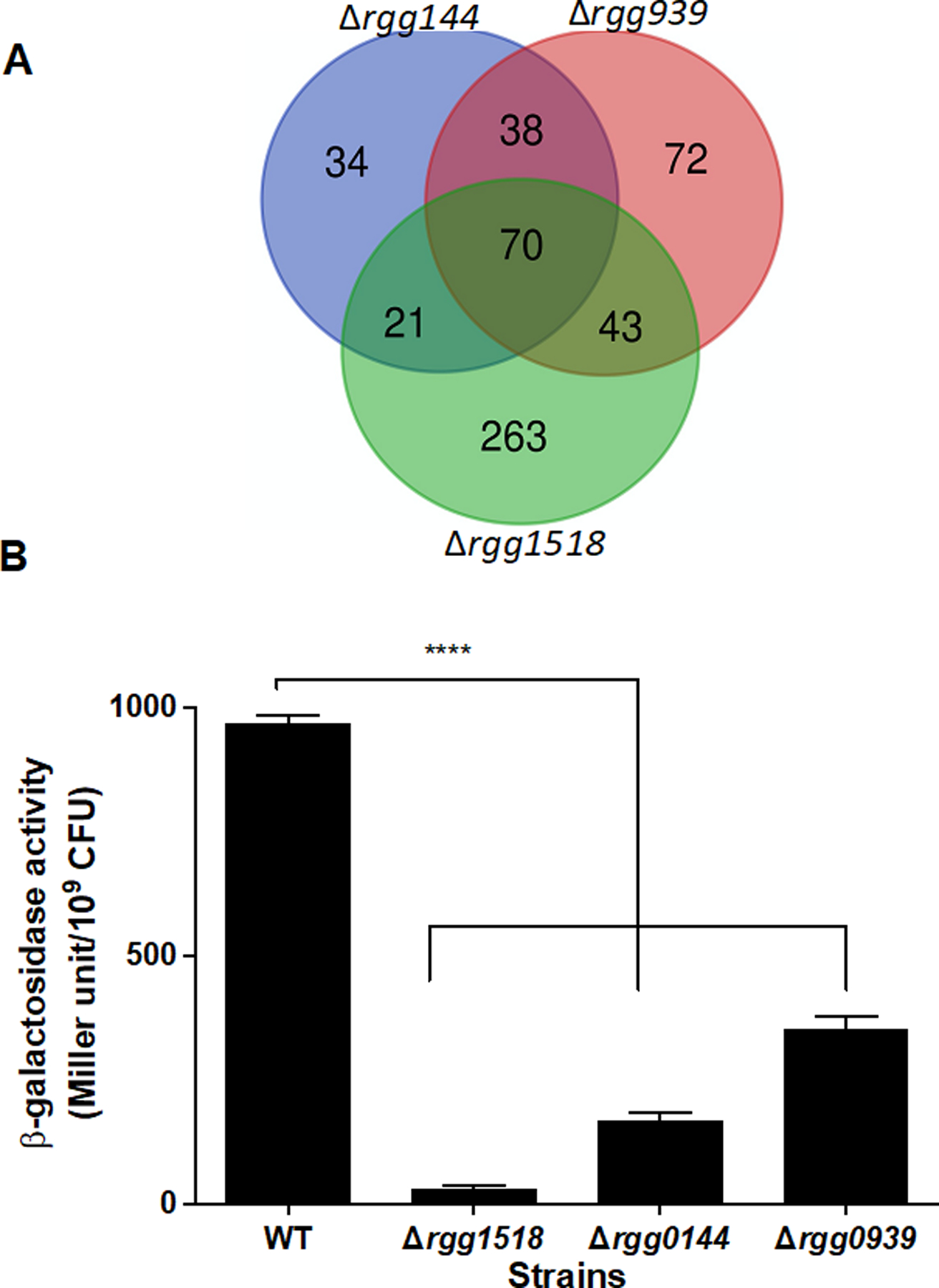
Analysis of regulatory interactions among Rggs. (A.) A Venn diagram showing the overlap between three pneumococcal Rggs. The diagram was produced by merging the differentially regulated genes on galactose and mannose for each Rgg by using VENNY (http://bioinfogp.cnb.csic.es/tools/venny/index.html). (B.) The activity of β-galactosidase, driven by rgg1518 putative promoter in different mutant strain backgrounds as measured in CDM-Gal, is expressed as nmol p-nitrophenol/min/ml. The values are the average of three independent experiments each with three replicates vertical bars indicate standard error of means (SEM), (****p<0.0001 compared to its expression in the wild type (WT) D39).
Table 1.
Some of the notable genomic loci that are found to be differentially expressed in Δrgg1518 relative to the wild type D39 grown in CDM supplemented with galactose. These were selected based on the number of effected genes in a given genomic locus. ‘-‘-‘ shows downregulation.
| Locus tag | Fold difference | Putative function |
|---|---|---|
| SPD_0113- SPD_0124 | 2.09–8.53 | Hypothetical protein |
| SPD_0145-SPD_0150 | 2.27–3.66 | CAAX amino terminal protease family protein |
| SPD_0179- SPD_0182 | 2.18–6.06 | Lipoprotein synthesis |
| SPD_0192-SPD_0219 | 2.66–8.71 | Ribosomal protein synthesis |
| SPD_0315- SPD_0334 | 2.28–6.36 | Capsule locus synthesis |
| SPD_0378- SPD_0390 | 2.16–7.19 | Fatty acid biosynthesis |
| SPD_0404- SPD_0408 | 2.08–3.09 | Acetolactate synthase synthesis |
| SPD_0705- SPD_0710 | 2.16–2.48 | Cell morphology/division |
| SPD_0750- SPD_0757 | 2.34–4.33 | Membrane protein synthesis |
| SPD_0760- SPD_0766 | 2.14–5.72 | FeS assembly |
| SPD_0905- SPD_0913 | 2.4–266 | Genetic information processing |
| SPD_0942- SPD_0951 | −2.06–2.27 | UDP-N-acetyl-D-mannosaminuronic acid dehydrogenase |
| SPD_0989- SPD_0991 | 3.62–3.83 | Protein synthesis |
| SPD_1050- SPD_1053 | 3.46–4.73 | Tagatose pathway, galactose catabolism |
| SPD_1344-SPD_1349 | 2.04–2.88 | acetyltransferase, GNAT family protein synthesis |
| SPD_1473- SPD_1480 | 2.63–5.48 | Cell division |
| SPD_1512- SPD_1517 | 2.64–9.48 | Membrane protein synthesis |
| SPD_1667- SPD_1671 | 2.38–7.63 | oligopeptide ABC transporter |
| SPD_1683- SPD_1698 | 2.02–5.53 | tRNA biosynthesis |
| SPD_1726- SPD_1730 | 2.1–4.81 | Pneumolysin/ hypothetical protein synthesis |
| SPD_1985- SPD_1989 | −2.1–2.14 | Mannose transport and metabolism |
| SPD_1942- SPD_1949 | 2.36–2.6 | Hypothetical protein |
| SPD_2066-SPD_2069 | 2.55–5.84 | DNA replication |
To further corroborate the interactions within the Rgg network, a Prgg1518-lacZ construct was introduced into Δrgg1518, Δrgg144, Δrgg939 and the wild type D39, generating the reporter strains Prgg1518::lacZ-∆rgg1518, Prgg1518::lacZ -∆rgg144, Prgg1518::lacZ-∆rgg939 and Prgg1518::lacZ-wt, respectively. β- galactosidase activity in the reporter strains were measured in CDM supplemented with galactose. The β- galactosidase activity was lower for Prgg1518::lacZ-∆rgg1518 (29.23±1.97 MU, n=3) compared to the activity in the wild type (970±16 MU, n=3), showing that Rgg1518 is required for its own expression (p<0.0001) (Figure 4). In addition, full induction of Prgg1518 required the presence of Rgg144 and Rgg939, as the induction of Prgg1518 was significantly lower in the ∆rgg144 and ∆rgg939 compared to the induction in the wild type (Prgg1518::lacZ-wt) (p<0.0001). We conclude that multiple Rggs form a regulatory network when pneumococci are metabolizing galactose.
Characterization of the Rgg1518 regulon.
In the respiratory tract, the pneumococcus uses both galactose and mannose from host glycans as a source of energy (King, 2010). Thus, we employed microarray analysis to compare gene expression between a wild-type and a strain with a deletion in rgg1518 (Δrgg1518). On galactose, we observed differential expression of 378 genes. Under these conditions 80 were down-regulated and 298 were up-regulated in the mutant (STable 1). This suggests that Rgg1518/SHP1518 acts mainly as a repressor, at least in the condition tested (CDM-galactose, mid-log phase). These genes are organized into 61 operons with an annotated role in the synthesis of capsule, fatty acid, hypothetical proteins, sugar and metal transporters, proteins involved in galactose metabolism as well as several transcriptional regulators. Moreover, among the highly regulated genes (4–9 fold difference) is the adjacent locus, encoding SPD_1513–1517. A selection of differentially expressed loci in Δrgg1518 relative to the wild type are shown in Table 1. On mannose, the size of the regulon (n=51) was smaller than that of galactose: 44 genes were downregulated and 7 were upregulated in the mutant, representing 6 operons (STable 2). Notably, the expression of genes adjacent to rgg1518 (SPD_1513-SPD_1517) were downregulated in the mutant relative to the wild type on mannose. It is very likely that the expression of some of these genes is affected indirectly by Rgg1518, hence additional studies are needed to determine genes directly controlled by Rgg1518.
Rgg1518 is a repressor of capsule synthesis on galactose.
Capsular polysaccharide (CPS) is an essential pneumococcal virulence determinant (Hyams et al., 2010). The microarray data revealed that Rgg1518 negatively regulates the capsule, as shown by the upregulation of the capsule synthesis locus (SPD_0315-SPD_0328) in Δrgg1518 on galactose (Table 1 and S1). The metabolism of galactose generates precursors for capsule synthesis (Carvalho et al., 2011). The type 2 capsule contains glucuronic acid (Kenne et al., 1975), therefore we quantified glucuronic acid in pneumococcal strains grown in CDM supplemented either with galactose or glucose (Figure 5). On galactose, the production of capsular polysaccharide was significantly higher in ∆rgg1518 (105±4 µg/109 CFU, n=6) than the wild type (63±3 µg/109 CFU, n=6), and the complemented mutant rescued wild type levels (63±4 µg/109 CFU, n=6) (p<0.001) (Figure 5A). On glucose, there was no significant difference in production of glucuronic acid among the wild type, ∆rgg1518 and ∆rgg1518c (60±6, 59±6, and 60±7 µg/109 CFU, n=6, respectively) (p>0.05) (Figure 5B). These results show that Rgg1518 is a repressor of capsule synthesis on galactose.
Figure 5:
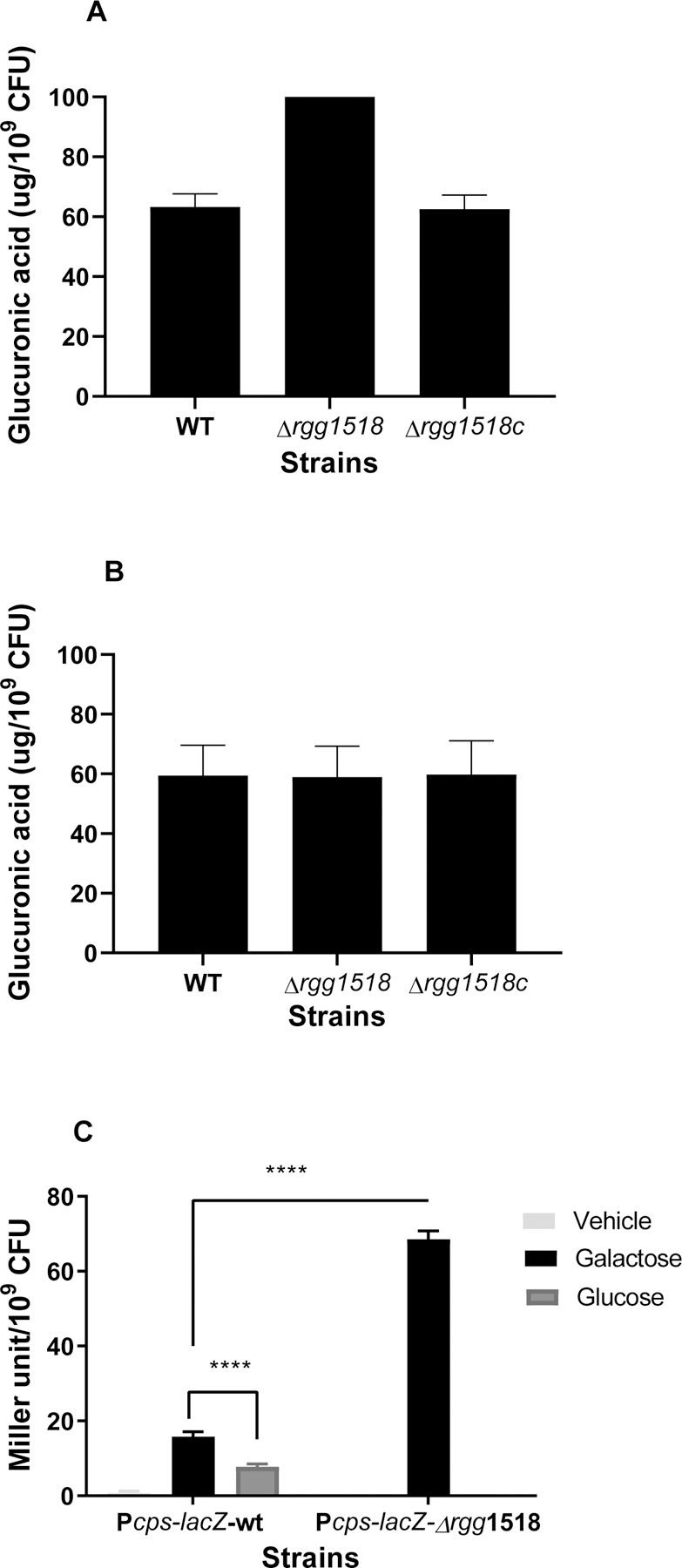
The quantification of glucuronic acid levels in pneumococcal strains grown in CDM supplemented either with galactose (A) or glucose (B). Influence of Rgg1518 on expression of the capsule locus (C) The activity of β-galactosidase, driven by cps putative promoter in different mutant strains, is expressed as nmol p-nitrophenol/min/ml. Vehicle refers to no sugar control. The activity was not measured in Δrgg1518 strain on glucose. The values are the average of at least three independent experiments each with three replicates vertical bars indicate standard error of means (SEM). ****p<0.0001.
Role of inactivated rgg1518 in capsule locus gene regulation.
Having established Rgg1518’s involvement in regulation of capsule synthesis, next we tested if the capsule locus is directly regulated by Rgg1518. To do this, we employed a LacZ reporter of the capsule promoter in wild type and ∆rgg1518 backgrounds. The β-galactosidase activities in the reporter strains were measured in strains grown in CDM supplemented with galactose or glucose. The activity in Pcps::lacZ-∆rgg1518 (69±2 MU/109 CFU, n=3) was more than four times that of the wild type (16±1 MU/109 CFU, n=3) (p<0.0001) (Figure 5C), supporting the view that Rgg1518 is a repressor of the cps locus.
In the next step, we employed electrophoretic mobility shift assay (EMSA) to investigate whether Rgg1518 directly interacts with the capsule locus. The results showed that the mobility pattern of DNA-protein complexes was different from that of DNA alone, showing the direct interaction of Rgg1518 with the promoter of the cps locus (Figure 6). There was no detectable change in mobility when Rgg1518 was incubated with gyrB, indicating that the Rgg1518-cps interaction is specific.
Figure 6:
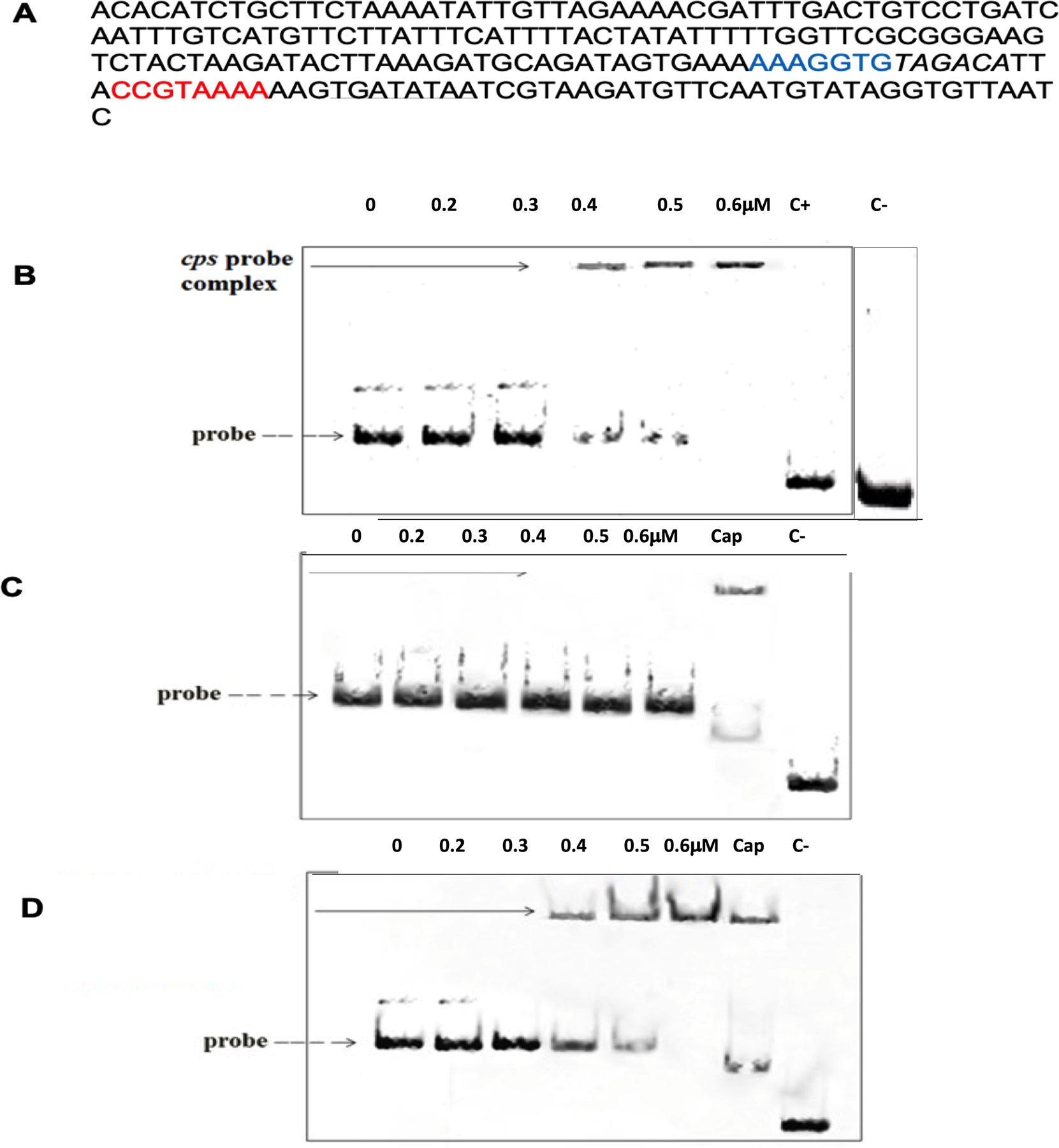
Direct interaction of Rgg1518 with the capsule (cps) locus. Concentrations stated at the top of the gel refer to the recombinant Rgg1518. (A) The putative promoter region of the cps locus is shown. The predicted Rgg binding motifs 1 and 2 are shown in blue and red, respectively. Italic sequence is the −35, and the underlined is the −10 sequence. (B) EMSA analysis showing the direct interaction of recombinant His-tagged Rgg1518 protein with the promoter of cps locus. (C) EMSA indicating the absence of the putative motif 1 abolishes the interaction of recombinant Rgg1518 with the probe. (D) EMSA indicating the absence of motif 2 did not affect the Rgg1518 binding to the probe. In each EMSA image, lane 1 contains only DNA probe as shown with a dotted line. In B, C and D ‘C+’ represents 10 nM of FAM-labeled gyrB amplicons incubated with 0.6 μM recombinant Rgg1518 while ‘C−’ shows gyrB amplicons without Rgg1518. In C and D, ‘Cap’ shows intact csp locus incubated with 0.6 mM Rgg1518. DNA-protein interaction is seen as an upward shift of DNA probe, represented by a black arrow.
To identify the Rgg1518 binding box, we hypothesized that the Rgg-box would be shared across multiple genes regulated in galatose. Thus, we selected 14 operons from our microarray analysis, all differentially expressed on galactose. We aligned the 225 to 260 bp upstream of the first gene in the operon and used MEME to identify conserved motifs (Bailey et al., 2015) (SFigure 2). We identified two putative binding motifs (motif 1: AAAGGTG and motif 2 CCGTAAAA), located near the core promoter elements as shown in Figure 6A. To investigate if either of these motifs was indeed the binding site for Rgg1518, DNA probes lacking either motif 1 or motif 2 were generated by splicing overlap extension, and Rgg1518 binding to these modified DNA probes was determined by EMSA. The absence of motif 2 did not effect Rgg1518 binding. In contrast, elimination of motif 1 abolished Rgg1518 binding. These data demonstrate a direct interaction between Rgg1518 and the cps promoter, and establish that motif 1 is required for binding of Rgg1518 to the Pcps promoter. In addition to the regulatory region of capsule locus, motif 1 has been identified in the regulatory regions of several genes represented in Rgg1518 regulon. The significance of this motif in the Rgg1518 regulon needs to be experimentally verified.
Contribution of Rgg1518 to pneumococcal colonization and virulence.
To explore the contribution of Rgg1518 to carriage we employed the murine CD1 pneumococcal colonisation model. One hour after infection, the bacterial load in the nasopharyngeal tissue was similar for all strains (p>0.05) (Figure 7A). However, at 3- and 7 days post-infection, the colony counts for ∆rgg1518 (Log10 1.45±0.09 CFU/mg and 0.95±0.104 CFU/mg, respectively n=5) was significantly lower than that of wild type (Log10 2.69±0.27 CFU/mg and Log10 2.98±0.17 CFU/mg n=4). Moreover, there was no significant difference in the counts of the complemented strain, ∆rgg1518c, and the wild type (p<0.001) at 3 (Log10 2.83±0.07 CFU/mg, n=5) and 7-days (Log10 2.85±0.087 CFU/mg n=5) post-infection (p>0.05). These results clearly prove the involvement of Rgg1518 in pneumococcal colonization of the murine nasopharynx.
Figure 7:
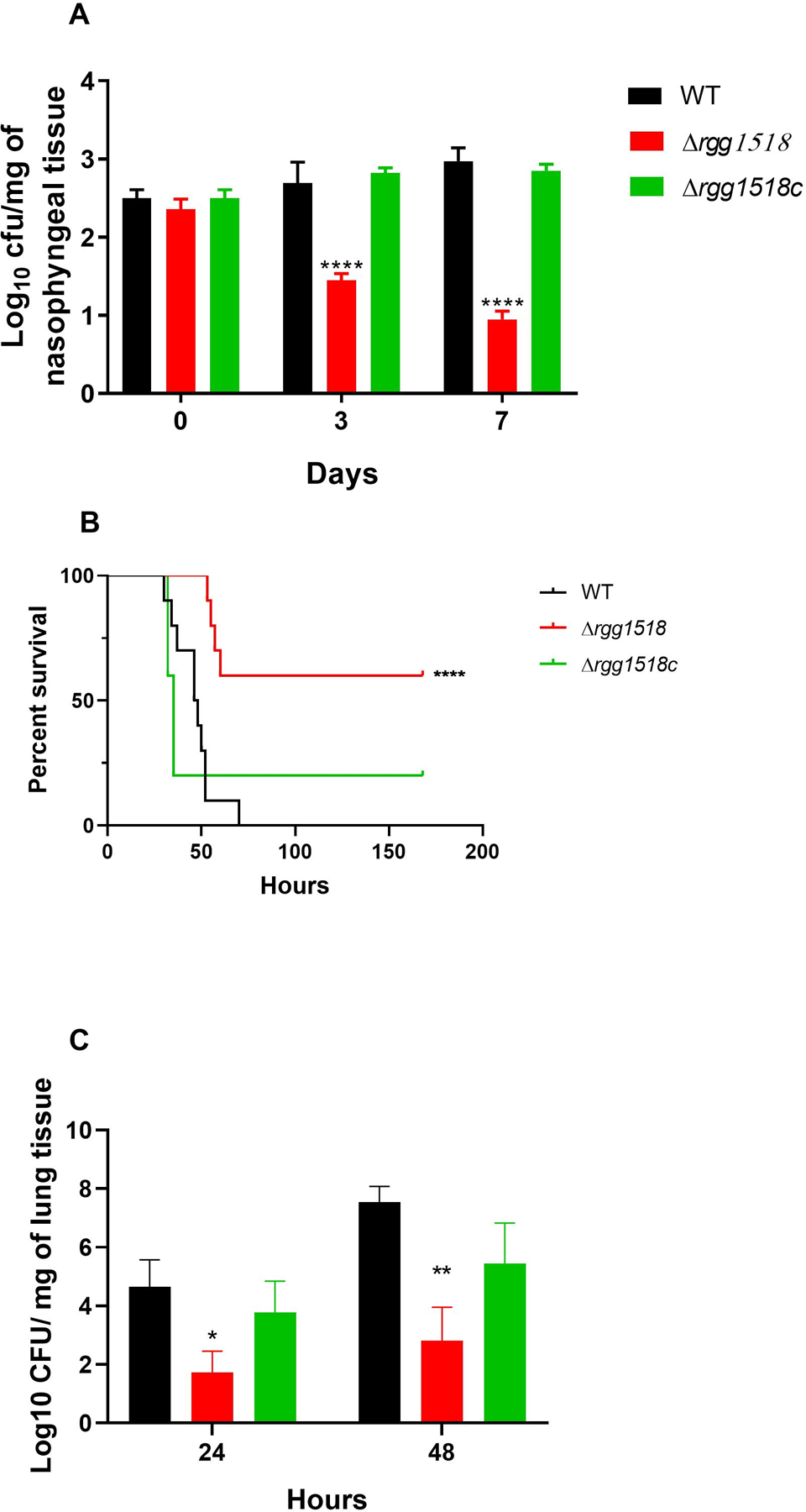
In vivo test of pneumococcal strains. (A) Δrgg1518 is impaired in nasopharyngeal colonization. Approximately 5 × 105 CFU of pneumococci was used to inoculate mice. Five mice were culled approximately 1 h after infection (day 0), day 3 and day 7, CFU/mg of pneumococci were defined by serial dilutions of homogenised nasopharyngeal. Each bar represents the average data collected from five mice. (B) The percent survival of mice infected intranasally with approximately 2×106 pneumococci/mouse. There are significant differences in percent survival of mice infected with the wild type, complemented mutant and mutant strains using Mantel-Cox test. (C) Development of bacteraemia after intranasal infection. Each column represents the average data for wild type D39 and mutant strains obtained from ten mice, while complemented strains were from five mice. Error bars illustrate the standard error of the mean. There are significant differences in bacterial counts between wild type and mutant strains using one-way ANOVA and Tukey’s multiple comparisons test, (*p<0.05 and **p<0.01, ***p<0.001, ****p<0.0001).
Colonization can extend beyond the nasopharynx causing pneumonia. To investigate the role of Rgg1518 in lung disease, we employed a murine model where pneumococci are inoculated in a high volume, such that they invade lung tissue. We analyzed mice inoculated with either wild type or the ∆rgg1518 strain for survival and bacterial load. The median survival time of mice infected intranasally with Δrgg1518 (123.3 h ± 18.2, n=10) was significantly longer than the wild type (47 h ± 3.1 n=10) and Δrgg1518c (60.4 h ± 26.9, n=5) infected groups (p<0.01) (Figure 7B). Further, there was no significant difference in survival time between the wild type and Δrgg1518c (p>0.05). Similarly, the bacterial counts for the ∆rgg1518 infected cohort were significantly lower at 24-and 48 h post-infection (Log10 1.62 ±0.74 CFU/ml, Log10 2.8±1.15 CFU/ml, respectively, n=10) in comparison with the wild type infected cohort at 24 (Log10 4.65±0.92 CFU/ml, n=10) and 48 h post-infection (Log10 7.55±0.53 CFU/ml, n=10) (p<0.05) (p<0.05, p<0.01) (Figure 7C). The progression of bacteraemia in cohort infected with the complemented strain Δrgg1518c at 24 h (Log10 3.78 ± 1.06 CFU/ml, n=5) and 48 h post-infection (Log10 5.44 ± 1.38 CFU/ml, n=5) was similar to that of wild type infected cohort (p>0.05). Together, these finding demonstrate that Rgg1518 promotes colonization and pathogenesis of the pneumococcus.
Discussion
In fluctuating host environments, community level synchronization of transcription enables S. pneumoniae to display a phenotype consistent with the requirements of the specific host niche. In this manner, peptide-mediated communication is critical to colonization and pathogenesis. In this study we present the transcriptional regulator Rgg1518 and its cognate peptide SHP1518. The Rgg1518 regulator is induced in galactose and mannose, required for galactose and mannose utilisation, and a negative regulator of capsule expression. It is critically positioned in a circuit of multiple Rgg communication systems implicated in the response to host carbohydrates. Finally, Rgg1518 promotes colonization and virulence.
This work suggests that Rgg1518 is part of an interconnected set of peptide-mediated regulators. Rgg144/SHP144 and Rgg939/SHP939 play an important role in sugar metabolism and capsule synthesis, and have a major impact on colonization and virulence (Junges et al., 2017; Zhi et al., 2018; Motib et al., 2019). In this study, we demonstrate that Rgg1518/SHP1518 is part of a larger network of Rgg regulators. First, Rgg144 and Rgg939 induce rgg1518 promoter activity. Second, there is overlap in the regulons for Rgg1518, Rgg144 and Rgg939 (SFigure 3 and Figure 4A), prominently they all include the locus immediately upstream of Rgg1518 (SPD1513-SPD1517), the VP1 operon and the capsular locus. While all these regulators play an important role in host-derived sugar metabolism, their roles differ in response to specific sugars. Rgg144 and Rgg939 are induced the highest by mannose and then galactose, while Rgg1518 is stimulated mainly by galactose (Junges et al., 2017; Zhi et al., 2018). These data suggest that these regulator-peptide systems are integrated, their relative roles are carbohydrate dependent, and their combined effects shape the bacterial transcriptional response to host nutrient availability.
Many questions remain regarding the connections and architecture of this multi-Rgg network. The systems have variable overlap not only in the sugars where the activity is most prominent, but also their distribution in the pangenome, and the nature and function of their regulons. While Rgg144 is part of the core genome, Rgg939 and Rgg1518 are part of the accessory genome, with distributions of approximately 54% and 38% of the strains in within sequenced pneumococcal genomes, respectively (Zhi et al., 2018). It is tempting to speculate that the distribution pattern of Rggs may be linked to fitness and pathogenic potential in diverse niches such as lungs, blood, middle ear and brain. The Rgg1518 regulon is larger than the regulon of Rgg144 or Rgg939, and is consistent with a master regulator. In galactose Rgg1518 controls, directly or indirectly, the expression of genes with diverse functions, notably those coding for carbohydrate metabolism and transport, capsule, fatty acid synthesis, iron transporters, and protein translation.
Regarding sugar metabolism, Rgg1518 appear to decrease overall galactose metabolism. The pneumococcus utilises galactose via the Leloir and tagatose catabolic pathways, and, in galactose, Rgg1518 represses genes in both of these pathways. Further, the utilisation of galactose leads to the generation of mixed acid products mainly via the activity of pyruvate formate lyase (Yesilkaya et al., 2009; Bidossi et al., 2012; Paixão et al., 2015). Rgg1518 also downregulates pyruvate formate lyase and transporters linked to galactose.
The connection between Rggs and capsule is clear for multiple Rgg systems. Here we show that Rgg1518 downregulates the capsule. In previous work, we demonstrated that the Rgg144 and Rgg939 systems also contributed to capsule synthesis repression in strain D39. The conditions where each Rgg is at play may vary. Rgg144 and Rgg939 repress capsule in mannose, but not on galactose (Zhi et al., 2018). In contrast, Rgg1518 inhibits the capsule in galactose. In mannose, only one gene from the capsule locus is downregulated by Rgg1518 (very likely due to the instability of the transcript on mannose). Thus, the role of Rgg1518 in capsule may be linked to galactose metabolism, and not mannose. In this study we demonstrated a direct interaction of recombinant Rgg1518 with the capsule locus promoter at the “AAAGGTG” sequence. While, we have previously shown the binding of Rgg144 and Rgg939 to the capsule locus promoter, their binding sites have not been identified. Thus, interesting questions remain regarding the overlap among Rgg-binding motifs on the capsule promoter, their role in diverse sugars, and whether there is a synergistic or antagonistic interaction among these Rggs for capsule locus expression.
The pneumococcal capsule has antiphagocytic and anticomplement activities (Janoff et al., 1999). Furthermore, by significantly preventing entrapment of S. pneumoniae in airway mucus, it decreases mucus-mediated clearance (Nelson et al., 2007). It has also been shown that capsule is essential for virulence and promotes colonization (Nelson et al., 2007; Geno et al., 2015). Contrary to the expectation, increased capsule synthesis in the mutant did not elevate the virulence, but rather we observed a decrease in the virulence potential of mutant as well as its colonisation in the nasopharynx. This apparent inconsistency of our results and the perceived function of capsule in colonization and virulence can be due to a number of different reasons. Firstly, previous studies reporting the importance of capsule in colonisation obtained their results using isogenic mutants devoid of capsule or the wild type strains. On the other hand, in our study we compared a capsule overexpressing mutant due to the rgg1518 mutation with the wild type. Therefore, the excess capsule production may hinder colonisation. Secondly, the in vivo conditions may be different than the in vitro assay conditions that indicated overexpression of capsule in the mutant. Finally, decreases in capsule have been linked to increase adhesion, likely via increase exposure of pneumococcal receptors on the bacterial surface (Hammerschmidt et al., 2005), which may in turn contribute to colonization and dissemination. We propose that the attenuation of virulence and colonisation in the Rgg1518 mutant is likely caused by the impact of Rgg1518 on multiple metabolic processes, where the capsule is one contributor. Specifically, in natural infections downregulation of the capsule may increase cell adhesion and in doing so contribute to increased colonization and virulence.
Experimental procedures
Bacterial strains and growth conditions:
The S. pneumoniae and Escherichia coli strains and plasmids constructed used in this study are listed in the STable 3. Brain Heart infusion (BHI) broth medium (Oxoid) or blood Agar Base (BAB) medium (Oxoid) supplemented with 5% (v/v) defibrinated horse blood were used to grow S. pneumoniae at 37ºC. When required, different antibiotics such as spectinomycin (100 μg/ml), kanamycin (200 μg/ml) and tetracycline (15 μg/ml) were used for supplementing the growth medium. Pneumococcal strains were grown anaerobically in chemically defined medium (CDM) supplemented with 55 mM of selected sugars (Motib et al., 2019).
Synthetic peptides:
Several synthetic peptides were used to determine the inducibility of Shp1518 (Table S4). The synthetic peptides were reconstituted as 6 mM stocks in dimethyl sulfoxide (DMSO) and stored at −80 °C.
Construction of genetically modified strains:
SOEing protocol (splicing by overlap extension) was used for mutagenesis using the primers listed in the STable 4 ( Al-Bayati et al., 2017). Briefly, a spectinomycin resistance cassette flanked by the DNA fragments of rgg1518 generated through two step PCR was transformed into S. pneumoniae type 2 D39 strain using competence stimulating peptide and the transformants were selected on BAB containing spectinomycin. The mutation was confirmed by PCR and sequencing. Genetic complementation of mutant strains were performed to eliminate the possibility polar effect of the mutation using a previously described protocol using pCEP (Guiral et al., 2006).
RNA extraction from bacterial cells, cDNA synthesis, and quantitative RT-PCR:
Pneumococcal RNA extraction was done using the Trizol method (Yesilkaya et al., 2009). SuperScript III reverse transcriptase (Invitrogen) was used to perform the first-strand cDNA synthesis according to the manufacturer’s instructions using DNase-treated RNA (~1μg) and random primers (Invitrogen), in 20 μl. qRT-PCR was done using SensiMixTM SYBR® Hi-ROX kit (Bioline, UK). The transcription level of selected genes was normalized to the expression level of house-keeping gyrB gene. The fold difference in expression was calculated by CT method (Livak and Schmittgen, 2001).
Microarray analysis:
Pneumococcal strains were grown to early exponential phase, OD600 approximately 0.3, in CDM supplemented with either 55 mM galactose or mannose. Three biological replicates were used for analysis. The MicroPrep software package was used to obtain the microarray data from the slides. CyberT implementation of a variant of t-test (http://bioinformatics.biol.rug.nl/cybert/index.shtml) was performed and false discovery rates (FDRs) were calculated (van Hijum et al., 2003). For differentially expressed genes, p < 0.001 and FDR < 0.05 were considered for significance threshold. For the determination of differentially expressed genes a Bayesian p-value of <0.001 and a fold-change cut-off of two was applied. All other procedures for the DNA microarray experiments and data analysis were performed as described before (Hajaj et al., 2017). Microarray data have been submitted to GEO (Gene Expression Omnibus) database under the accession number GSE154887
Construction of lacZ fusions and β-galactosidase assay:
The putative promoter region of rgg1518 was amplified and ligated into the pPP2 plasmid using the primers listed in STable 4 (Halfmann et al., 2007). The recombinant DNA molecule was transformed into E. coli TOP10 chemically competent cells for propagation in LA supplemented with ampicillin (100 μg/ml). After sequencing, the recombinant pPP2 was transformed into S. pneumoniae wild type D39 or selected mutants.
To evaluate promoter induction, strains were grown in CDM-Glucose and then transferred to CDM with no sugar, or a single sugar for 1h. The β-galactosidase activity was determined as previously described and the activity level was expressed for 109 CFU/ml.
Cloning and expression of recombinant proteins.
To obtain recombinant Rgg1518, rgg1518 was cloned into pLEICS-01 (Protex, University of Leicester). The construct was transformed into E. coli BL21 (DE3) pLysS for protein expression at 37°C. The expression was induced with 1 mM IPTG. The 10 prepacked gravity flow column His GraviTrap™ (GE Healthcare, UK) was used to purify protein. To confirm protein identity, the purified recombinant proteins were analysed by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (PNACL, University of Leicester).
Electrophoretic mobility shift assay (EMSA):
The presence of regulatory elements in the putative promoter regions of target genes was determined using the bacterial promoter prediction tool (BPROM) (Solovyev and Salamov, 2011) and the motif-sites were localized using (MEME) (Bailey and Elkan, 1994). For this analysis, we used the upstream regions of differentially regulated selected genes in Δrgg1518 predicted to be part of an operon including SPD_0315, SPD_1053, SPD_1634, SPD_1342, SPD_1461, SPD_1127, SPD_0113, SPD_0122, SPD_0193, SPD_0197, SPD_0208, SPD_0126, SPD_0341, SPD_0378, SPD_0404 and SPD_1512.
The genetic fragment containing the putative promoter sites of target genes were amplified using primers containing 5́-FAM fluorescent tags (STable 4), and the amplicons were purified using a commercial kit. Varying concentrations of recombinant Rgg1518 was incubated in 20 µL containing binding buffer (20 mM Tris-HCl pH 7.5, 30 mM KCl, 1 mM DTT, 1 mM EDTA pH 8.0 and 10% v/v glycerol), the DNA probe (50 nM), and peptide to a final concentration of 2 µM. The reaction mixture incubated at room temperature for 20 min. An aliquot (8 µL) of reaction mixture was loaded on 5% (v/v) polyacrylamide gel buffered with 20 mM potassium phosphate, pH 7.5, for 60 min at 200 V. All gel shifts were detected by fluorescence imaging using a Typhoon PhosphorImager.
Glucuronic acid assay:
Capsular polysaccharide (CPS) production was quantified by measuring the amount of glucuronic acid, which is found in the type 2 pneumococcal capsule (Zhi et al., 2018; Najmuldeen et al., 2019). Briefly, the pneumococcal cultures (500 μl) grown to late exponential phase in CDM supplemented with different sugars were mixed with 100 µl of 1% (v/v) Zwittergent 3–14 detergent (Sigma-Aldrich) in 100 mM citric acid (pH 2.0) and the mixture was kept at 50 °C for 20 min. The polysaccharides were precipitated with 1 ml of absolute ethanol. The pellet was dissolved in 200 µl distilled water, and added with 1200 µl 12.5 mM borax (Sigma) in H2SO4. The mixture was mixed thoroughly, boiled for 5 min. Then 20 µl 0.15% 3-hydroxydiphenol (Sigma) was added. The absorbance of the mixture at 520 nm was recorded, and the glucuronic acid content of the samples was quantified in comparison to a standard curve generated with the known concentrations of glucuronic acid (Sigma).
In vivo studies:
This study used 8–10 weeks old female CD1 mice (Charles River, UK). Standard inoculum was prepared as described previously (Kahya et al., 2017). To administer the dose, mice were lightly anaesthetised with 2.5% (v/v) Isoflurane (Isocare, UK) over oxygen (1.4 to 1.6 litres/min), in an anaesthetic box. To initiate infection, approximately 2 × 106 CFU of S. pneumoniae were administered equally dropwise into the nostrils. Mice were watched periodically for disease signs (hunched, piloerect, or lethargy) for 7 days. As soon they reached the lethargic stage, mice were sacrificed humanely. The time to reach lethargic stage was considered as survival time (Al-Bayati et al., 2017). The median survival times of mice were examined statistically by using the Mann-Whitney U test. To follow the progression of bacteraemia, 20 µl of venous blood from the tail vein was taken after 24- and 48 h after infection. The colonies were enumerated by plating the serial dilutions of the blood on blood agar plates. To evaluate Rgg1518’s contribution on colonisation, each mouse was given approximately 5×105 CFU of S. pneumoniae in 20 µl PBS. The bacterial counts in the nasopharynx were determined by plating the serial dilutions of tissue homogenates (Kahya et al., 2017).
Supplementary Material
Acknowledgement:
We are grateful for the support from the NIH (R01 AI139077–01A1 to NLH and R01 AI135060–01A1 to AU), also to the Eberly Family Trust to NLH. We also gratefully acknowledge the valuable support of the Preclinical Facility staff for the in vivo experiments in Leicester.
Footnotes
Ethics statement:In vivo studies were carried out under the UK Home Office approved project (60/4327) and personal licenses (I4142DBBE). The procedures were approved by the local ethic committee. The animals were placed in individually ventilated cages in a controlled environment. The mice were repeatedly monitored after infection in order to minimize suffering.
Conflict of interest: The authors declare no competing financial interests.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References:
- Aggarwal C, Jimenez JC, Nanavati D, and Federle MJ (2014) Multiple length peptide-pheromone variants produced by Streptococcus pyogenes directly bind Rgg proteins to confer transcriptional regulation. J Biol Chem 289: 22427–22436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal SD, Yesilkaya H, Dawid S, and Hiller NL (2020) The pneumococcal social network. PLoS Pathog 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bayati FAY, Kahya HFH, Damianou A, Shafeeq S, Kuipers OP, Andrew PW, and Yesilkaya H (2017) Pneumococcal galactose catabolism is controlled by multiple regulators acting on pyruvate formate lyase. Sci Rep 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, and Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36. [PubMed] [Google Scholar]
- Bidossi A, Mulas L, Decorosi F, Colomba L, Ricci S, Pozzi G, et al. (2012) A functional genomics approach to establish the complement of carbohydrate transporters in Streptococcus pneumoniae. PLoS One 7: e33320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortoni ME, Terra VS, Hinds J, Andrew PW, and Yesilkaya H (2009) The pneumococcal response to oxidative stress includes a role for Rgg. Microbiology 155: 4123–4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho SM, Kloosterman TG, Kuipers OP, and Neves AR (2011) CcpA ensures optimal metabolic fitness of Streptococcus pneumoniae. PLoS One 6:e26707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JC, and Federle MJ (2016) PptAB exports Rgg quorum-sensing peptides in Streptococcus. PLoS One 11:e0168461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JC, LaSarre B, Jimenez JC, Aggarwal C, and Federle MJ (2011) Two group a streptococcal peptide pheromones act through opposing rgg regulators to control biofilm development. PLoS Pathog 7:e1002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook LC, LaSarre B, and Federle MJ (2013) Interspecies communication among commensal and pathogenic streptococci. MBio 4:e00382–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas RA, Eutsey R, Kadam A, West-Roberts JA, Woolford CA, Mitchell AP, et al. (2017) A novel streptococcal cell-cell communication peptide promotes pneumococcal virulence and biofilm formation. Mol Microbiol 105: 554–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saizieu A. De, Gardes C, Flint N, Wagner C, Kamber M, Mitchell TJ, et al. (2000) Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J Bacteriol 182:4696–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleuchot B, Gitton C, Guillot A, Vidic J, Nicolas P, Besset C, et al. (2011) Rgg proteins associated with internalized small hydrophobic peptides: A new quorum-sensing mechanism in streptococci. Mol Microbiol 80:1102–1119. [DOI] [PubMed] [Google Scholar]
- Geno KA, Gilbert GL, Song JY, Skovsted IC, Klugman KP, Jones C, et al. (2015) Pneumococcal capsules and their types: Past, present, and future. Clin Microbiol Rev 34:e00320–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Mejia A, Gámez G, and Hammerschmidt S (2018) Streptococcus pneumoniae two-component regulatory systems: The interplay of the pneumococcus with its environment. Int J Med Microbiol 308:722–737. [DOI] [PubMed] [Google Scholar]
- Guiral S, Hénard V, Laaberki M-H, Granadel C, Prudhomme M, Martin B, and Claverys J-P (2006) Construction and evaluation of a chromosomal expression platform (CEP) for ectopic, maltose-driven gene expression in Streptococcus pneumoniae. Microbiology 152: 343–9. [DOI] [PubMed] [Google Scholar]
- Hajaj B, Yesilkaya H, Shafeeq S, Zhi X, Benisty R, Tchalah S, et al. (2017) CodY regulates thiol peroxidase expression as part of the pneumococcal defense mechanism against H2O2 stress. Front Cell Infect Microbiol 7:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann A, Hakenbeck R, and Brückner R (2007) A new integrative reporter plasmid for Streptococcus pneumoniae. FEMS Microbiol Lett 268: 217–24. [DOI] [PubMed] [Google Scholar]
- Håvarstein LS, Coomaraswamy G, and Morrison DA (1995) An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci U S A 92: 11140–11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hijum S.A.F.T. van, García de la Nava J, Trelles O, Kok J, and Kuipers OP (2003) MicroPreP: a cDNA microarray data pre-processing framework. Appl Bioinformatics 2:241–4. [PubMed] [Google Scholar]
- Ibrahim M, Guillot A, Wessner F, Algaron F, Besset C, Courtin P, et al. (2007) Control of the transcription of a short gene encoding a cyclic peptide in Streptococcus thermophilus: A new quorum-sensing system? J Bacteriol 189: 8845–8854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoff EN, Fasching C, Orenstein JM, Rubins JB, Opstad NL, and Dalmasso AP (1999) Killing of Streptococcus pneumoniae by capsular polysaccharide-specific polymeric IgA, complement, and phagocytes. J Clin Invest 104:1139–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junges R, Salvadori G, Shekhar S, Åmdal HA, Periselneris JN, Chen T, et al. (2017) A quorum-sensing system that regulates Streptococcus pneumoniae biofilm formation and surface polysaccharide production. mSphere 13:e00324–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam A, Eutsey RA, Rosch J, Miao X, Longwell M, Xu W, et al. (2017) Promiscuous signaling by a regulatory system unique to the pandemic PMEN1 pneumococcal lineage. PLoS Pathog 13: e1006339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahya HF, Andrew PW, and Yesilkaya H (2017) Deacetylation of sialic acid by esterases potentiates pneumococcal neuraminidase activity for mucin utilization, colonization and virulence. PLoS Pathog 13:e1006263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenne L, Lindberg B, and Svensson S (1975) The structure of capsular polysaccharide of the pneumococcus type II. Carbohydr Res 40:69–75. [DOI] [PubMed] [Google Scholar]
- King SJ (2010) Pneumococcal modification of host sugars: a major contributor to colonization of the human airway? Mol Oral Microbiol 25: 15–24. [DOI] [PubMed] [Google Scholar]
- Koponen O, Tolonen M, Qiao M, Wahlström G, Helin J, and Saris PEJ (2002) NisB is required for the dehydration and NisC for the lanthionine formation in the post-translational modification of nisin. Microbiology 148: 3561–3568. [DOI] [PubMed] [Google Scholar]
- Kuipers OP, Beerthuyzen MM, Ruyter P.G.G.A. De, Luesink EJ, and Vos W.M. De (1995) Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J Biol Chem 270:27299–304. [DOI] [PubMed] [Google Scholar]
- Livak KJ, and Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Motib A, Guerreiro A, Al-Bayati F, Piletska E, Manzoor I, Shafeeq S, et al. (2017) Modulation of quorum sensing in a gram-positive pathogen by linear molecularly imprinted polymers with anti-infective properties. Angew Chemie - Int Ed 56: 16555–16558. [DOI] [PubMed] [Google Scholar]
- Motib AS, Al-Bayati FAY, Manzoor I, Shafeeq S, Kadam A, Kuipers OP, et al. (2019) TprA/PhrA quorum sensing system has a major effect on pneumococcal survival in respiratory tract and blood, and its activity is controlled by CcpA and GlnR. Front Cell Infect Microbiol 9:326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najmuldeen H, Alghamdi R, Alghofaili F, and Yesilkaya H (2019) Functional assessment of microbial superoxide dismutase isozymes suggests a differential role for each isozyme. Free Radic Biol Med 134:215–228. [DOI] [PubMed] [Google Scholar]
- Nelson AL, Roche AM, Gould JM, Chim K, Ratner AJ, and Weiser JN (2007) Capsule enhances pneumococcal colonization by limiting mucus-mediated clearance. Infect Immun 75:83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paixão L, Oliveira J, Veríssimo A, Vinga S, Lourenço EC, Ventura MR, et al. (2015) Host glycan sugar-specific pathways in streptococcus pneumonia: Galactose as a key sugar in colonisation and infection. PLoS One 10:e0127483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST, and Bassler BL (2012) Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med 2:a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin-Yilmaz A, and Naclerio RM (2011) Anatomy and physiology of the upper airway. Proc Am Thorac Soc 8:31–39. [DOI] [PubMed] [Google Scholar]
- Sen AK, Narbad A, Horn N, Dodd HM, Parr AJ, Colquhoun I, and Gasson MJ (1999) Post-translational modification of nisin. Eur J Biochem 261: 524–532. [DOI] [PubMed] [Google Scholar]
- Shafeeq S, Yesilkaya H, Kloosterman TG, Narayanan G, Wandel M, Andrew PW, et al. (2011) The cop operon is required for copper homeostasis and contributes to virulence in Streptococcus pneumoniae. Mol Microbiol 81:1255–1270. [DOI] [PubMed] [Google Scholar]
- Solovyev V, and Salamov A (2011) Automatic Annotation of Microbial Genomes and Metagenomic Sequences. In Metagenomics and its Applications in Agriculture, Biomedicine and Environmental Studies. Li R. (ed.). Nova Science Publishers, pp. 61–78. [Google Scholar]
- Troeger C, Forouzanfar M, Rao PC, Khalil I, Brown A, Swartz S, et al. (2017) Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory tract infections in 195 countries: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis 17:1133–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trappetti C, McAllister LJ, Chen A, Wang H, Paton AW, Oggioni MR, et al. (2017) Autoinducer 2 signaling via the phosphotransferase FruA drives galactose utilization by Streptococcus pneumoniae, resulting in hypervirulence. MBio 8: e02269–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varahan S, Harms N, Gilmore MS, Tomich JM, and Hancock LE (2014) An ABC transporter is required for secretion of peptide sex pheromones in Enterococcus faecalis. MBio 5: e01726–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Medlin JS, Nguyen DR, Disbennett WM, and Dawid S (2020) Molecular determinants of substrate selectivity of a pneumococcal Rgg-regulated peptidase-containing ABC transporter. MBio 11: e02502–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser JN, Ferreira DM, and Paton JC (2018) Streptococcus pneumoniae: Transmission, colonization and invasion. Nat Rev Microbiol 16: 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey JM, and Donk W.A. van der (2007) Lantibiotics: Peptides of Diverse Structure and Function. Annu Rev Microbiol 61: 477–501. [DOI] [PubMed] [Google Scholar]
- Wilton J, Acebo P, Herranz C, Gómez A, and Amblar M (2015) Small regulatory RNAs in Streptococcus pneumoniae: Discovery and biological functions. Front Genet 6:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesilkaya H, Spissu F, Carvalho SM, Terra VS, Homer KA, Benisty R, et al. (2009) Pyruvate formate lyase is required for pneumococcal fermentative metabolism and virulence. Infect Immun 77: 5418–5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi X, Abdullah IT, Gazioglu O, Manzoor I, Shafeeq S, Kuipers OP, et al. (2018) Rgg-Shp regulators are important for pneumococcal colonization and invasion through their effect on mannose utilization and capsule synthesis. Sci Rep 8:6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


