Abstract
Background
Recent studies have reported a dysfunctional gut microbiome in breastfed infants. Probiotics have been used in an attempt to restore the gut microbiome; however, colonization has been transient, inconsistent among individuals, or has not positively impacted the host’s gut.
Methods
This is a 2-year follow-up study to a randomized controlled trial wherein 7-day-old infants received 1.8 × 1010 colony-forming unit Bifidobacterium longum subsp. infantis (B. infantis) EVC001 (EVC) daily for 21 days or breast milk alone (unsupplemented (UNS)). In the follow-up study, mothers (n = 48) collected infant stool at 4, 6, 8, 10, and 12 months postnatal and completed the health-diet questionnaires.
Results
Fecal B. infantis was 2.5–3.5 log units higher at 6–12 months in the EVC group compared with the UNS group (P < 0.01) and this relationship strengthened with the exclusion of infants who consumed infant formula and antibiotics. Infants in the EVC group had significantly higher Bifidobacteriaceae and lower Bacteroidaceae and Lachnospiraceae (P < 0.05). There were no differences in any health conditions between the two groups.
Conclusions
Probiotic supplementation with B. infantis within the first month postnatal, in combination with breast milk, resulted in stable colonization that persisted until at least 1 year postnatal.
Impact
A dysfunctional gut microbiome in breastfed infants is common in resource-rich nations and associated with an increased risk of immune diseases.
Probiotics only transiently exist in the gut without persistent colonization or altering the gut microbiome.
This is the first study to show that early probiotic supplementation with B. infantis with breast milk results in stable colonization of B. infantis and improvements to the gut microbiome 1 year postnatal.
This study addresses a key gap in the literature whereby probiotics can restore the gut microbiome if biologically selected microorganisms are matched with their specific food in an open ecological niche.
Introduction
Human milk delivers a wide spectrum of biologically active molecules that aid in the development and maturation of the gut and the innate and adaptive immune systems and support the growth of protective intestinal microbiota. Specifically, human milk oligosaccharides (HMOs), the third most abundant component in human milk (~10–20 g/L),1,2 are a group of complex sugars that are nondigestible by the human infant and support the competitive growth of protective bifidobacterial strains within the intestine.3,4 In particular, the natural colonization of a protective subspecies of Bifidobacterium, Bifidobacterium longum subsp. infantis (B. infantis), unlike other bifidobacterial species in breastfed infants, is based on its genetic capabilities to bind, transport, and ferment HMOs into lactate and acetate.4,5 These fermentative products maintain a lower pH of the intestinal milieu, support the transport of these compounds into the intestinal epithelium for use by the host,6 create an undesirable environment for potential pathogens,7 and prevent the infiltration of toxic molecules produced by pathogenic bacteria by upregulating intestinal barrier function and inhibiting proinflammatory and apoptotic responses.8
Historically, the gut of the breastfed infant was dominated by a near monoculture of Bifidobacterium until the cessation of breastfeeding.9 However, findings from Henrick et al.10 reported a generational loss of Bifidobacterium in breastfed infants from resource-rich nations within the past 100 years accompanied by higher levels of enteropathogens and higher fecal pH.10 The reduction in Bifidobacterium and increase in potential pathogens in the infant gut microbiome are likely a result of the unintended consequences of antibiotic use, infant formula feeding,11 and cesarean section deliveries,12 all of which have been implicated in the increased risk for allergic and autoimmune diseases prevalent in resource-rich nations.13–15 Colonization of a dysfunctional gut microbiome in early infancy during the critical window of immune system development is reported to increase the risk for the development of immune disease later in life.16
We previously published findings from the IMPRINT Study in which healthy, term, breastfed infants supplemented with 1.8 × 1010 colony-forming units (CFUs) of B. infantis EVC001 per day for 21 consecutive days starting on day 7 postnatal demonstrated persistent colonization of fecal B. infantis 1 month post supplementation. Given the diversity among B. infantis strains,5,17 we selected B. infantis EVC001 because we knew this strain had the full cassette of genes needed to completely digest all HMOs from human milk. Supplementation with B. infantis EVC001 was well tolerated18 and increased fecal Bifidobacteriaceae by 79% and reduced enteropathogens by 80%, decreased fecal HMOs by 10-fold (consistent with increased HMO consumption by gut microbes), and increased fecal lactate and acetate by 2-fold, resulting in a decrease in fecal pH by 1 log unit.19 Intestinal colonization of B. infantis persisted 1 month post supplementation. These results are unprecedented as probiotics have only been found to transiently exist in the gut during supplementation in infants, without showing persistent colonization in most individuals or altering the gut microbiome composition in adults.20 In the follow-up study reported herein, infants who completed the IMPRINT Study at 2 months of age were followed up at 4, 6, 8, 10, 12, 18, and 24 months postnatal. The aims of this follow-up study were to determine if B. infantis colonization persisted up to 1 year postnatal and identify differences in reported health outcomes between B. infantis EVC001 supplemented and unsupplemented (UNS) infants.
Methods
Subjects and design
The details of the main 2-month-long IMPRINT Study are reported elsewhere.18 Briefly, mother–infant dyads were recruited in the Davis and Sacramento metropolitan region of Northern California. Mothers received either lactation support or lactation support and 1.8 × 1010 CFU of B. infantis EVC001 (ATCC SD-7035; manufactured by Evolve BioSystems, Inc.) to feed their infants daily from days 7 to 27 postnatal. Bifidobacterium infantis EVC001 was delivered as 156 mg of live bacteria (1.8 × 1010 CFU) diluted in 469 mg of lactose as an excipient. Mothers were trained by lactation consultants to mix the B. infantis EVC001 powder with 5 mL of expressed breast milk and feed the mixture to their infant using a feeding syringe. The probiotic was stored at −20 °C by the mothers during the study. Upon completing the parent trial when their infants were ~2 months of age, participants were offered the opportunity to enroll in two independent follow-up studies: follow-up #1, which was designed to determine if B. infantis persisted up to 1 year postnatal; and follow-up #2, which was designed to determine if B. infantis supplementation early in life was protective against the development of health conditions at 18 and 24 months postnatal. In the follow-up #1 study, mothers completed a paper questionnaire about their infants’ health and diet at 4, 6, 8, 10, and 12 months postnatal and collected one matching infant fecal sample at each time point. In the follow-up #2 study, mothers completed an online questionnaire about their infants’ health and diet at 18 and 24 months postnatal. The study and methods were approved by the UC Davis Institutional Review Board, and the study was registered at Clinicaltrials.gov (NCT02457338). All mothers provided written informed consent to participate in every aspect of the study.
Questionnaires
Follow-up studies
Mothers who completed the parent IMPRINT Study were invited to enroll in two different follow-up studies: follow-up #1 and follow-up #2. In follow-up #1, upon providing written informed consent, mothers completed up to five paper questionnaires that coincided with the collection of their infants’ stool at 4, 6, 8, 10, and 12 months postnatal (Supplementary File 1 (online)). In follow-up #2, upon providing email informed consent, mothers completed up to two questionnaires (without stool collection) at 18 and 24 months postnatal (Supplementary File 2 (online)). The questionnaires that were used in both follow-up studies prompted mothers to answer questions about their infants’ health and diet. Specifically, in follow-up #1, mothers were prompted to report on their infants’ health and diet over the past 2 months (either since completing the parent IMPRINT Study or since completing the previous questionnaire). In follow-up #2, mothers were prompted to report their infants’ health and diet over the past 6 months at 18 and 24 months postnatal. In both the follow-up #1 and #2 studies, mothers were asked questions about their infants’ dietary patterns (intake of breast milk, infant formula, and solid foods), use of medications, supplements and vitamins, illnesses, sick doctor visits, hospitalizations, antibiotic (oral/IV) usage, and probiotic intake.
In follow-up #1 only, mothers were asked if their infants were ever diagnosed with common infant health illnesses and conditions by a healthcare professional and infant age at diagnosis. The answer options were “diagnosed” or “not diagnosed.” Mothers were asked to report the frequency of common infant conditions and illnesses. The answer options were “never,” “sometimes,” “often,” “very often,” “unsure,” and “refuse.” When mothers answered “sometimes,” “often,” or “very often,” they were prompted to rate the severity of the gastrointestinal symptoms from 1 to 10, with 1 as the least severe and 10 as the most severe.
In follow-up #2, mothers were asked if their infants had experienced and were diagnosed with any allergies, wheezing, asthma, eczema, gastroesophageal reflux disease, and lactose intolerance. The answer options were “yes,” “no,” “unsure,” and “refuse.” When mothers answered “yes,” they were prompted to report the number of times their infants had experienced and if they had been diagnosed with common infant illnesses and conditions.
Samples
Follow-up #1
Fecal samples were collected at home at 4, 6, 8, 10, and 12 months postnatal. Fecal samples were stored in participants’ home freezers and transferred on dry ice to a −80 °C freezer for storage prior to DNA extraction. All individuals who processed and analyzed the samples were blinded to treatment allocation.
Molecular methods and analyses
As previously described,19 total DNA was extracted from ~100 mg of feces, using the Zymo Fecal DNA Miniprep Kit according to the manufacturer’s instructions (Zymo Research, Irvine, CA). Negative controls to detect kit contamination were included and failed to produce visible PCR bands in an agarose gel but were analyzed as quality controls. Samples were subjected to 16S ribosomal RNA (rRNA) gene sequencing as previously described.19 Quantification of the total B. infantis was performed by quantitative real-time PCR using Blon_2348 sialidase gene primers Inf2348F (5′-ATA CAG CAG AAC CTT GGC CT-3′), Inf2348_R (5′-GCG ATC ACA TGG ACG AGA AC-3′), and Inf2348_P (5′-/56-FAM/TTT CAC GGA /ZEN/TCA CCG GAC CAT ACG/3lABkFQ/-3′). The Blon_2348 gene is found in all B. infantis strains including EVC001. The primer and probe sequence specificity has been previously described.21 Each reaction contained 10 μL of 2× TaqMan Universal Master Mix II with UNG master mix (Thermo Fisher Scientific, Waltham, MA), 0.9 µM of each primer, 0.25 µM probe, and 5 μL of template DNA. Thermal cycling was performed on a QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA) and consisted of an initial UNG activation step of 2 min at 50 °C, followed by a 10-min denaturation at 95 °C, succeeded by 40 cycles of 15 s at 95 °C and 1 min at 60 °C. All samples were run in duplicate with a standard curve on each plate. Quantification of B. infantis was determined (CFU/g stool) using a standard curve of genomic DNA derived from a pure culture of B. infantis EVC001 using CFU counts and normalized for input stool wet weight.22 Standard curve genomic DNA was extracted from 1 mL aliquots of B. infantis EVC001 grown anaerobically at 37 °C for 16 h in deMann Rogosa Sharpe (MRS) medium (BD Biosciences, San Jose, CA) supplemented with 0.05% l-cysteine HCl. CFU counts of the 16-h B. infantis EVC001 culture were determined by serial dilution in 0.9% NaCl on MRS agar plates containing 0.05% l-cysteine HCl. Plates were incubated anaerobically at 37 °C for 48 h, then counted, and the CFU/mL value was calculated.
16S rRNA bioinformatics analysis
Sequences were analyzed using QIIME 1.9.1 (10.1038/nmeth.f.303). Open-reference operational taxonomical unit (OTU) picking was performed using UCLUST at 97% identity against the Greengenes database (v.13_8) (10.1128/AEM.03006-05), and chimera filtering was checked as part of the QIIME pipeline using USEARCH 6.1.23
A representative set of sequences was taken for each OTU and taxonomic classification was performed using UCLUST consensus taxonomy in QIIME. Representative sequences were then aligned using PyNAST (https://biocore.github.io/pynast/) to the Greengenes core reference alignment and a phylogenetic tree was built using FastTree.24 After quality filtering, a mean of 26,354 (±8830 [SD]) and a median of 27,646 reads were obtained per sample. Several multivariate linear modeling analyses (https://huttenhower.sph.harvard.edu/maaslin/) were computed to compare groups of samples at the family and genus levels, using the subject as a random effect to account for time and other clinical metadata, including treatment status, delivery mode, and feeding as fixed effects. Multivariate Association with Linear Models 2 (MaAsLin2) was run with a false-discovery rate (FDR) of 0.05, a minimum of 0.0001 for feature relative abundance filtering, and a minimum of 0.01 for feature prevalence filtering. Fixed effects used in the MaAsLin2 model include any use of the following by the infant: antibiotics, probiotics, probiotics containing B. infantis, infant formula, solid food, and breast milk. In addition, the model included delivery mode, supplementation allocation. Subject ID was used as a random effect and time was used as a continuous variable. P values were adjusted via FDR (Q values) and considered significant if Q value < 0.25. Raw data are accessible under the accession number PRJNA670448.
Diversity analysis
Rarefaction curves were computed to estimate the distribution of the identified OTUs at a depth of 1538 sequences/sample. Alpha-diversity was computed using the Shannon diversity index in QIIME. A nonparametric two-sample t test was used to compare alpha-diversity according to treatment status using Monte Carlo permutations (n = 999). Beta-diversity was computed using UniFrac distances and a dissimilarity matrix was constructed to estimate the global OTUs differences among samples and visualized via a principal coordinate analysis. A permutational multivariate analysis of variance using distance matrices (adonis) was used to assess OTU differences between treatments and the effect size (R2) of colonization by EVC001. P values for the principal coordinate analysis panel were computed using F tests based on sequential sums of squares from permutations of the raw data.
Statistics
The Mann–Whitney U test was used to compare mean ranks for fecal B. infantis between EVC and UNS groups at each time point during the first follow-up period (4, 6, 8, 10, and 12 months postnatal). The Mann–Whitney U test was performed on (1) all infants, (2) infants who had not used any infant formula, antibiotics, or probiotics since completing the parent IMPRINT Study (2 months postnatal), and (3) infants who used infant formula or antibiotics at 6 months postnatal (or time-point closest to 6 months). Because solid foods are commonly introduced to infants by 6 months of age, solid food consumption was not excluded in any of the analyses. Infant weight was measured at each study visit using a Pediatric Tanita digital scale and mean ranks for infant weight were compared between EVC and UNS groups using Mann–Whitney U test. The significance level for all Mann–Whitney U test analyses was set at an α 0.05 with a Bonferroni adjustment using the two-tailed exact test statistic, which is appropriate for small, unbalanced, or poorly distributed data. Mean ranks for frequency ordinal data and for severity continuous data were compared between EVC and UNS groups using Mann–Whitney U test. Categorical data that resulted in the answers “yes,” “no,” “unsure,” or “refuse” were analyzed using a two-sided Fisher’s exact test with an α 0.05 with a Bonferroni adjustment, whereby “unsure” and “refuse” responses were excluded from the analysis. SPSS version 25 was used for these analyses.
Results
Follow-up #1
Of the 68 mothers enrolled in the parent IMPRINT Study, 48 mothers enrolled in the follow-up #1 study. Of these 48 mother–infant dyads, n = 22 had received the UNS treatment and n = 26 had received the EVC treatment. There was a significantly higher number of primiparous women in the EVC group than in the UNS group (P < 0.01) (Table 1). There were no other differences in demographic, labor, delivery, and health history characteristics between the two groups. Infants enrolled in the EVC group were born at a younger gestational age than infants enrolled in the UNS group (P < 0.05) (Table 2); however, all infants were full term at birth. A detailed description of infants’ diet, intake of antibiotics and probiotics, and exposure to other infants via daycare are reported in Table 3. There were no differences in the number of infants who consumed breast milk; breast milk and infant formula; infant formula without breast milk; solid foods; used antibiotics or probiotics; or were enrolled in daycare at any time point (Table 3). There was no difference in weight between groups across time (Fig. 1).
Table 1.
Maternal demographics, labor, delivery, and health history.
| Characteristics | UNS (n = 22) | EVC (n = 26) | ||
|---|---|---|---|---|
| Mean | SD | Mean | SD | |
| Maternal age at enrollment (years) | 31.0 | 3.4 | 33 | 4.7 |
| Prepregnancy BMI | 24.5 | 3.1 | 26.2 | 3.5 |
| Pregnancy weight gain (kg) | 31.1 | 7.7 | 33.7 | 11.8 |
| Hours in labor | 22.0 | 26.0 | 11.3 | 12.6 |
| Ruptured membranes prior to birth (h) | 12.4 | 19.2 | 7.1 | 12.0 |
| Number of pregnancies | 2.0 | 1.5 | 2.8 | 1.7* |
| Number of live births | 1.5 | 1.0 | 2.2 | 1.1** |
| Parity, % (n) | ||||
| Primiparous | 77.3% (17) | 34.6% (9)** | ||
| Multiparous | 22.7% (5) | 65.4% (17) | ||
| Mode of delivery, % (n) | ||||
| Vaginal | 63.6% (14) | 69.2% (18) | ||
| Vaginal water birth | 18.2% (4) | 0% (0) | ||
| C-section, emergent | 13.6% (3) | 15.4% (4) | ||
| C-section, elective | 4.5% (1) | 15.4% (4) | ||
| Ethnicity, % (n) | ||||
| Not hispanic | 90.9% (20) | 76.9% (20) | ||
| Hispanic | 9.1% (2) | 23.1% (6) | ||
| Race, % (n) | ||||
| Asian | 4.5% (1) | 0% (0) | ||
| Black | 4.5% (1) | 0% (0) | ||
| White | 81.8% (18) | 73.1% (19) | ||
| Other | 0% (0) | 7.7% (2) | ||
| 2 or More races | 9.1% (2) | 19.2% (5) | ||
| Education, % (n) | ||||
| Some college, no degree, or AA degree | 13.6% (3) | 19.2% (5) | ||
| Bachelor’s degree (BA or BS) | 36.4% (8) | 34.6% (9) | ||
| Master’s, professional, or doctorate degree | 50% (11) | 46.2% (12) | ||
| Antibiotic use during labor, % (n) | ||||
| Yes | 18.2% (4) | 26.9% (7) | ||
| No | 81.8% (18) | 73.1% (19) | ||
| Gestational diabetes mellitus positive diagnosis, % (n) | ||||
| Yes | 9.1% (2) | 7.7% (2) | ||
| No | 90.9% (20) | 92.3% (24) | ||
| Group B Streptococcus (GBS) colonization positive diagnosis, % (n) | ||||
| Yes | 22.7% (5) | 30.8% (8) | ||
| No | 77.3% (17) | 69.2% (18) | ||
| Any allergy diagnosis in past 10 years, % (n) | ||||
| Yes | 36.4% (8) | 26.9% (7) | ||
| No | 63.6% (14) | 73.1% (19) | ||
| Asthma diagnosis in past 10 years, % (n) | ||||
| Yes | 22.7% (5) | 7.7% (2) | ||
| No | 77.3% (17) | 92.3% (24) | ||
| Hay fever diagnosis in past 10 years, % (n) | ||||
| Yes | 0% (0) | 7.7% (2) | ||
| No | 100% (22) | 92.3% (24) | ||
| Autoimmune disease diagnosis in past 10 years, % (n) | ||||
| Yes | 0% (0) | 15.4% (4) | ||
| No | 100% (22) | 84.6% (22) | ||
| Impaired glucose tolerance in past 10 years, % (n) | ||||
| Yes | 0% (0) | 0% (0) | ||
| No | 100% (22) | 100% (26) | ||
*P < 0.05.
**P < 0.01 for differences between treatment groups.
Table 2.
Infant characteristics.
| Infant Characteristics | UNS (n = 22) | EVC (n = 26) | ||
|---|---|---|---|---|
| Mean | SD | Mean | SD | |
| Gestational age at birth (weeks) | 40.2 | 1.0 | 39.5 | 1.3* |
| Birth weight (g) | 3669.2 | 587.8 | 3448.8 | 396.3 |
| Birth length (cm) | 51.1 | 2.4 | 50.5 | 2.2 |
| Gender, % (n) | ||||
| Male | 40.9% (9) | 65.4% (17) | ||
| Female | 59.1% (13) | 34.6% (9) | ||
*P < 0.05 for differences between treatment groups.
Table 3.
Infant diet and environmenta.
| Month | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Feeding and environment | 4 | 6 | 8 | 10 | 12 | |||||
| UNS | EVC | UNS | EVC | UNS | EVC | UNS | EVC | UNS | EVC | |
| Total (n) | 11 | 7 | 16 | 13 | 18 | 16 | 19 | 23 | 21 | 26 |
| Breast milka (n) | 8 | 6 | 12 | 9 | 10 | 10 | 9 | 13 | 10 | 12 |
| Breast milk and infant formulaa (n) | 2 | 1 | 1 | 4 | 3 | 4 | 2 | 5 | 4 | 5 |
| Infant formulaa (n) | 0 | 0 | 1 | 0 | 1 | 1 | 2 | 1 | 3 | 3 |
| Solids (n) | 1 | 0 | 14 | 9 | 18 | 16 | 19 | 23 | 21 | 26 |
| Antibiotics (n) | 0 | 0 | 1 | 0 | 2 | 1 | 4 | 4 | 3 | 4 |
| Probioticsb (n) | 1 | 0 | 1 | 0 | 3 | 1 | 4 | 2 | 1 | 3 |
| Daycare (n) | 27% | 0% | 50% | 23% | 50% | 25% | 47% | 35% | 57% | 50% |
aExcludes infants who took antibiotics and/or probiotics.
bn = 4 infants in the EVC and n = 5 in the UNS consumed five different probiotic supplement products at various times during the follow-up study. Participants in the study were able to recall the product names for four of the five probiotic supplements they fed to their infants. The four probiotic products recalled contained the following microorganisms: (1) Lactobacillus acidophilus and Lactobacillus helveticus (unspecified strains), (2) Bifidobacterium longum and Bifidobacterium infantis (unspecified strains), (3) proprietary probiotic blend containing five Lactobacillus and five Bifidobacterium species, and (4) B. infantis EVC001 (one participant enrolled in the study found one sachet of the study probiotic in her freezer and fed it to her infant at 11 months postnatal).
Fig. 1. Infant weight across time and between treatment groups for all infants.
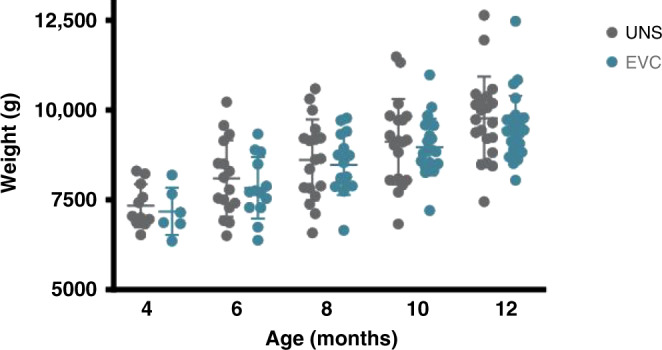
Sample size is not consistent with Table 3 due to missed weights for EVC: day 120, n = 6; day 300, n = 22; day 365, n = 25.
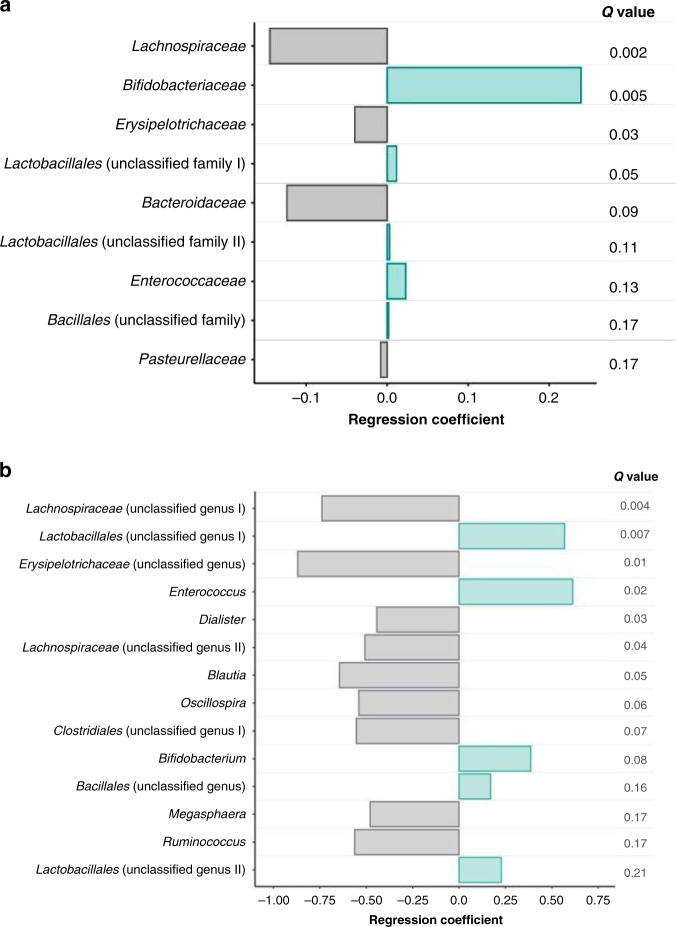
With the inclusion of all infants, fecal B. infantis was 2.5–3.5 logs higher at 6–12 months in the EVC group compared with the UNS group (P < 0.01) (Fig. 2a). In a subgroup of infants who did not receive infant formula, antibiotics, or probiotics, fecal B. infantis was 3.6–5.2 logs higher at 6–12 months in the EVC group compared with the UNS group (P < 0.001) (Fig. 2b). To further focus on these confounding variables, we conducted Mann–Whitney U testing on three subgroups: infants breast milk fed without intake of infant formula, antibiotics, or probiotics (BM); infants mixed fed with breast milk and infant formula without intake of antibiotics or probiotics (BM + FF); and infants exposed to antibiotics and/or additional probiotics (all feeding types) (ABX). We selected one time point, as close to 6 months as possible, when breast milk volume intake would be the highest and the introduction of solid foods would be minimal. For this analysis, for the BM subgroup, fecal B. infantis was 3.3 logs higher in infants in the EVC group compared to the UNS group (P < 0.0005). However, for both the BM + FF and ABX subgroups, fecal B. infantis was not different between EVC and UNS groups (Fig. 3). To further investigate how B. infantis supplementation influences the gut microbial composition across all time points, we used MaAsLin2, to determine if treatment altered gut microbial taxa. Infants in the EVC group had significantly higher Bifidobacteriaceae (R = 0.24, FDR-adjusted Q value < 0.01), Lactobacillales unclassified family I (R = 0.01, FDR-adjusted Q value = 0.05), Lactobacillales unclassified family II (R = 0.003, FDR-adjusted Q value = 0.12), Enterococcaceae (R = 0.02, FDR-adjusted Q value = 0.14), and Bacillales unclassified family (R = 0.002, FDR-adjusted Q value = 0.17) and significantly lower Lachnospiraceae (R = 0.14, FDR-adjusted Q value < 0.01), Erysipelotrichaceae (R = 0.04, FDR-adjusted Q value < 0.05), Bacteroidaceae (R = 0.12, FDR-adjusted Q value = 0.09), and Pasteurellaceae (R = 0.008, FDR-adjusted Q value = 0.17) compared with the UNS group (Fig. 4a) even after adjustments for infant formula, antibiotics, probiotics, delivery mode, postnatal age, and subject as a random variable. Of the taxa that were significantly different between treatments according to MaAsLin2, Mann–Whitney U test was used to compare differences for taxa between treatments for each time point and confirmed statistical differences for only fecal Bifidobacteriaceae and Lachnospiraceae at 6, 8, 10, and 12 months postnatal (P < 0.05) and Bacteroidaceae at 12 months postnatal (P < 0.01) (Table 4). The same MaAsLin2 modeling used on a family level showed higher correlation coefficients between supplementation and gut microbial composition on a genus level (Fig. 4b). The genera that were significantly different between treatments according to MaAsLin2 modeling were compared statistically at each time point using Mann–Whitney U test. Infants in the EVC group had significantly higher fecal Bifidobacterium at 6, 8, 10, and 12 months postnatal, and Enterococcus at 6 months postnatal and lower Lachnospiraceae (unclassified genus) at 6, 8, and 10 months postnatal; Ruminococcus at 8 months postnatal; and Erysipelotrichaceae (unclassified genus) at 6 and 8 months postnatal (Supplementary Table S1).
Fig. 2. Infant fecal B. infantis across time and between treatment groups.
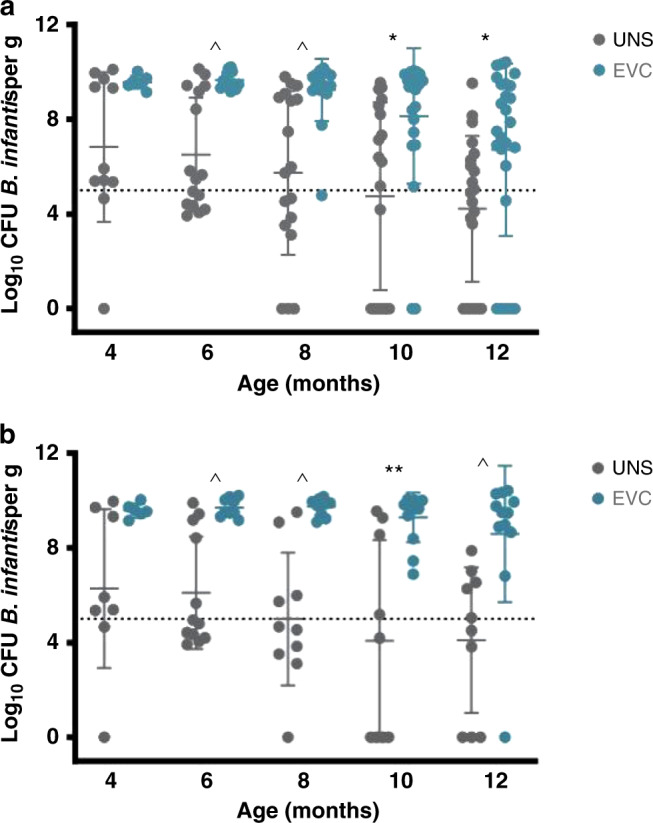
a Inclusion of all infants. b In a subgroup of infants who did not receive infant formula, antibiotics, or probiotics. *P < 0.01, **P < 0.001, and ^P < 0.0005 for differences between treatment groups.
Fig. 3. Infant fecal B. infantis at 6 months postnatal among three subgroups of infants based on diet and exposure to antibiotics.
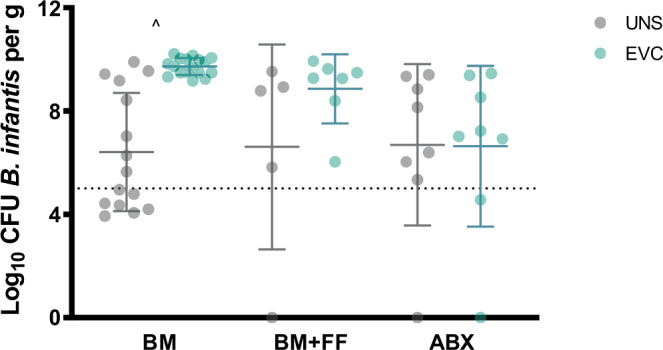
BM breast milk (including solids); BM + FF breast milk and formula-fed (including solids), ABX antibiotic use. ^P < 0.0005 for differences between treatment groups.
Fig. 4. Relationships between infant fecal microbial families and treatment groups based on MaAsLin2 for all infants.
a Family level. b Genus level. P values were adjusted via FDR (Q values) and considered significant if Q value < 0.25.
Table 4.
Infant fecal microbial families measured by 16s rRNA amplicon sequencing.
| Postnatal month | Family | % Mean relative abundance (SD) | |
|---|---|---|---|
| UNS | EVC | ||
| n = 11 | n = 7 | ||
| 4 | Bifidobacteriaceae | 59.8 (27.0) | 81.9 (9.72) |
| Coriobacteriaceae | 1.87 (5.62) | 0.248 (0.479) | |
| Bacteroidaceae | 7.19 (9.96) | 4.45 (5.05) | |
| Prevotellaceae | 0.001 (0.004) | 0 (0.001) | |
| Enterococcaceae | 0.476 (0.438) | 1.02 (1.05) | |
| Lactobacillaceae | 0.532 (0.711) | 0.061 (0.159) | |
| Clostridiaceae | 2.12 (5.77) | 1.30 (1.99) | |
| Lachnospiraceae | 8.67 (9.68) | 1.81 (3.21) | |
| Streptococcaceae | 1.19 (1.32) | 2.41 (2.53) | |
| Ruminococcaceae | 0.111 (0.257) | 0.008 (0.009) | |
| Veillonellaceae | 2.01 (3.50) | 0.251 (0.340) | |
| Erysipelotrichaceae | 3.72 (5.73) | 0.161 (0.423) | |
| Enterobacteriaceae | 8.31 (8.96) | 4.77 (4.64) | |
| Other bacteria | 4.03 (4.02) | 1.62 (1.04) | |
| n = 16 | n = 13 | ||
|---|---|---|---|
| 6 | Bifidobacteriaceae | 48.8 (26.9) | 73.4 (16.2)** |
| Coriobacteriaceae | 1.68 (4.05) | 0.232 (0.368) | |
| Bacteroidaceae | 10.8 (13.7) | 5.51 (6.65) | |
| Prevotellaceae | 0.387 (1.52) | 0.002 (0.003) | |
| Enterococcaceae | 0.494 (0.759) | 1.01 (0.867) | |
| Lactobacillaceae | 2.49 (8.49) | 0.693 (1.51) | |
| Clostridiaceae | 1.17 (1.59) | 1.26 (2.13) | |
| Lachnospiraceae | 9.98 (11.3) | 3.42 (7.95)* | |
| Streptococcaceae | 1.19 (1.41) | 1.79 (2.76) | |
| Ruminococcaceae | 0.342 (0.534) | 0.066 (0.169) | |
| Veillonellaceae | 5.65 (6.60) | 3.18 (5.30) | |
| Erysipelotrichaceae | 2.57 (3.41) | 0.248 (0.699) | |
| Enterobacteriaceae | 9.18 (8.56) | 6.34 (6.97) | |
| Other bacteria | 5.19 (8.88) | 2.81 (2.84) |
| n = 18 | n = 16 | ||
|---|---|---|---|
| 8 | Bifidobacteriaceae | 38.2 (24.6) | 61.2 (23.6)* |
| Coriobacteriaceae | 1.49 (4.78) | 0.349 (0.552) | |
| Bacteroidaceae | 17.4 (19.6) | 8.60 (10.8) | |
| Prevotellaceae | 0.965 (3.58) | 0.004 (0.008) | |
| Enterococcaceae | 0.678 (0.855) | 0.903 (0.938) | |
| Lactobacillaceae | 0.942 (1.60) | 1.09 (1.57) | |
| Clostridiaceae | 2.30 (2.66) | 2.62 (3.41) | |
| Lachnospiraceae | 14.7 (15.3) | 7.39 (13.2)* | |
| Streptococcaceae | 1.00 (1.58) | 1.20 (2.44) | |
| Ruminococcaceae | 1.19 (2.32) | 0.920 (3.00) | |
| Veillonellaceae | 6.88 (4.24) | 4.05 (4.27) | |
| Erysipelotrichaceae | 2.07 (2.87) | 0.872 (1.75) | |
| Enterobacteriaceae | 7.94 (8.37) | 7.21 (6.28) | |
| Other bacteria | 4.25 (5.90) | 3.64 (5.64) |
| n = 19 | n = 23 | ||
|---|---|---|---|
| 10 | Bifidobacteriaceae | 31.3 (20.7) | 48.2 (24.7)* |
| Coriobacteriaceae | 1.39 (2.61) | 0.733 (0.881) | |
| Bacteroidaceae | 15.2 (17.2) | 11.4 (12.5) | |
| Prevotellaceae | 2.06 (5.88) | 0.216 (0.744) | |
| Enterococcaceae | 0.483 (0.540) | 3.44 (13.1) | |
| Lactobacillaceae | 1.11 (1.95) | 0.656 (1.44) | |
| Clostridiaceae | 1.53 (1.65) | 1.62 (1.94) | |
| Lachnospiraceae | 19.5 (15.4) | 10.9 (13.5)* | |
| Streptococcaceae | 0.776 (1.05) | 1.09 (1.13) | |
| Ruminococcaceae | 3.75 (8.52) | 2.91 (5.50) | |
| Veillonellaceae | 7.49 (7.99) | 5.72 (6.28) | |
| Erysipelotrichaceae | 2.69 (4.17) | 0.699 (0.685) | |
| Enterobacteriaceae | 6.76 (8.24) | 4.58 (4.83) | |
| Other bacteria | 5.90 (8.97) | 7.80 (9.55) |
| n = 21 | n = 26 | ||
|---|---|---|---|
| 12 | Bifidobacteriaceae | 15.4 (14.0) | 33.1 (22.4)** |
| Coriobacteriaceae | 0.930 (2.06) | 0.779 (1.05) | |
| Bacteroidaceae | 28.2 (19.1) | 11.8 (11.1)** | |
| Prevotellaceae | 2.18 (8.73) | 2.16 (7.88) | |
| Enterococcaceae | 0.169 (0.479) | 0.657 (1.20) | |
| Lactobacillaceae | 0.405 (1.17) | 0.960 (2.04) | |
| Clostridiaceae | 0.951 (1.12) | 1.38 (1.31) | |
| Lachnospiraceae | 24.6 (11.5) | 17.4 (12.2)* | |
| Streptococcaceae | 1.64 (1.99) | 5.76 (14.7) | |
| Ruminococcaceae | 10.2 (10.1) | 7.49 (9.06) | |
| Veillonellaceae | 5.55 (9.60) | 7.15 (8.07) | |
| Erysipelotrichaceae | 1.13 (1.31) | 0.786 (1.08) | |
| Enterobacteriaceae | 3.34 (5.29) | 3.49 (4.40) | |
| Other bacteria | 5.33 (4.46) | 7.09 (5.92) |
*P < 0.05.
**P < 0.01 for differences between treatments groups.
To investigate if supplementation with B. infantis resulted in differences in gut-related symptoms, mothers were asked how often infants experienced symptoms (never = 0, sometimes = 1, often = 2, very often = 3, unsure = 4, and refuse = 5, whereby unsure and refuse responses were excluded from the statistical analysis) and to rate the severity of these symptoms. The mean frequencies for gastrointestinal symptoms were not statistically significant between treatments at any study time point. Reported severity for infant constipation was 83% higher in the UNS vs. EVC group (P < 0.001); however, neither value was considered severe (Supplementary Table S2). The frequency for illnesses, sick doctor visits, hospitalizations, ear infections, respiratory tract infections, other infections, thrush, allergy, wheezing, asthma, eczema, and other conditions were not significantly different between treatments across time (Supplementary Table S3). There was also no difference in reported use of antibiotics, antigas medication, gripe water, probiotics with or without B. infantis, prescribed medications, or over-the-counter medications (Supplementary Table S4).
Diversity analysis
Rarefaction curves were computed to assess differences in alpha-diversity composition as measured by the Shannon diversity index based on treatment status. No statistical difference was observed between groups (Supplementary Fig. 3a). Beta-diversity analysis was performed using UniFrac distances and the effect size of probiotic feeding was calculated, resulting in a significant (P = 0.001; adonis) although weak effect size (R2 = 0.05%; adonis) (Supplementary Fig. 3b).
Follow-up #2
Of the 68 mothers enrolled in the parent IMPRINT Study, 51 mothers enrolled in the follow-up #2 study. Of these participants, n = 19 in the UNS and n = 17 in the EVC group completed the 18-month health questionnaire and n = 21 in the UNS and n = 20 in the EVC group completed the 24-month health questionnaire. There were no treatment differences in the number of children who experienced or were diagnosed with any common infant conditions or experiences (Supplementary Table S5). There were no significant differences in the mean number of experiences or diagnoses of common infant conditions (Supplementary Table S6).
Discussion
The dominance of fecal Bifidobacterium and, specifically, B. infantis in the gut of breastfed infants has declined in recent decades in resource-rich nations, resulting in an increase in potential gut pathogens and immune dysfunction.10,25–29 Probiotic supplementation with B. infantis EVC001 in 7-day-old breastfed infants for 21 consecutive days resulted in a 7-log increase in fecal B. infantis, an increase in fecal Bifidobacteriaceae by 79%, a decrease in enteropathogens by 80%, an increase in fecal lactate and acetate by 2-fold, a decrease in fecal pH by 1-log,19 a decrease in antibiotic resistance genes, a sign of reduced enteropathogens known to harbor these genes,30 a reduction in mucin degradation,31 and reduced enteric inflammatory markers by several-fold27 during and 1 month post supplementation. These data demonstrate that the combination of breast milk and B. infantis EVC001 successfully restores the gut microbiome and biochemistry to historical norms observed a century ago.9
The infant gut microbiome is influenced by several maternal, dietary, and environmental factors, including delivery mode, feeding status (i.e., breast milk, infant formula, solid foods), and use of antibiotics. The current study showed that fecal B. infantis was 2.5–3.5 logs higher in infants in the EVC group compared with the UNS group at 6, 8, 10, and 12 months despite feeding status, and use of antibiotics or probiotics. The greatest difference in fecal B. infantis was observed in the earlier time points (6 and 8 months) when breast milk was the most abundant food source. The smallest difference in fecal B. infantis was observed at 12 months when infants’ diets were much more diverse and breast milk was less abundant. After excluding infants with confounding variables that impact the gut microbiome, such as infant formula, antibiotics, and probiotics, fecal B. infantis was 3.6–5.2 logs higher in infants in the EVC group compared with the UNS group at 6, 8, 10, and 12 months. We were unable to determine if probiotic intake during the 1-year follow-up period independently influenced fecal B. infantis abundance because six of the nine infants who consumed probiotics also received antibiotics. Taken together, these data suggest that a lack of HMOs, the preferred carbon source for B. infantis, and the use of antibiotics impact fecal B. infantis levels.
When infants were grouped by feeding type and exposures (breast milk fed without intake of infant formula, antibiotics or probiotics, mixed fed with breast milk and infant formula without intake of antibiotics or probiotics, and intake of antibiotics (all feeding types and probiotics), we found that fecal B. infantis was significantly higher in infants in the breast milk-fed group who were supplemented with B. infantis EVC001 compared with the UNS group. These findings further support the observation that breast milk is critical in supporting the colonization of B. infantis.
The UNS group had significantly higher Lachnospiraceae, including the genera, Ruminococcus, and Blautia, Bacteroidaceae, and lower Bifidobacteriaceae levels compared with the EVC group. These taxa differ in their preferences for carbohydrate substrates, metabolism of their preferred substrates into end products, and their consequent biochemical effects in the gut and on infant health. For example, members of the family Lachnospiraceae consist of spore-forming, anerobic bacteria that ferment complex plant polysaccharides into short-chain fatty acids (SCFAs), such as acetate, butyrate, and propionate.32 While gut microbes that produce SCFAs that lower luminal pH are considered beneficial, health outcomes associated with this family are mixed and likely vary with the genus or species and with host factors (e.g., infant vs. adult). For example, some members of this family that are commonly found in the human gut microbiome have been associated with a number of adverse health outcomes in adults (e.g., bloating, irritable bowel disease, metabolic disorders).33–35 In a prospective cohort study, the abundance of the family Lachnospiraceae at 3–4 months was higher in the gut of formula-fed infants compared to breastfed infants in a dose-dependent manner and associated with an 89% increase in the risk of overweight by 12 months.36 Emerging evidence suggests that the species, Ruminococcus gnavus, which belongs to the family Lachnospiraceae,37 may play a key role in allergy and immune development in infants38 and inflammation in the gut of adult patients with Crohn’s disease.39
In this study, we also found higher levels of the family Bacteroidaceae in the UNS group at 12 months postnatal. Bacteroidaceae is a family of Gram-negative, obligate anaerobic, nonsporulating bacilli that is commonly found in the healthy human adult colon. While most members of this family are considered commensals, some species, such as Bacteroides fragilis, include pathogenic strains.40 In addition, members of the Bacteroidaceae family contain an expanded set of genes encoded in polysaccharide utilization loci, allowing for the consumption of both dietary polysaccharides, as well as host-derived glycans.41 Specifically, Bacteroides thetaiotaomicron and B. fragilis, common members of the neonate gut, utilize a large set of mucin degradation polysaccharide utilization loci to catabolize HMOs.42 Previous studies in gnotobiotic mice have shown that downstream products derived from Bacteroides-driven HMO catabolism confer a growth advantage to potentially pathogenic Enterobacteriaceae, specifically Escherichia coli. This cross-feeding event was found to drive the E. coli bloom in a dextran sodium sulfate-induced colitis mouse model, thereby compounding the inflammatory response.43 On the other hand, the subspecies of B. longum, B. infantis, and specific strains of B. infantis5 such as EVC001 preferentially consume HMOs, which are fermented into acetate and lactate via the “bifid shunt.”4,5 These end products maintain a lower pH of the intestinal milieu, supporting the transport of these compounds into the intestinal epithelium for use by the host6 and creating an undesirable environment for potential pathogens.7 Acetate also blocks the infiltration of toxic molecules produced by pathogenic bacteria by enhancing intestinal barrier function and inhibiting proinflammatory and apoptotic responses.8 The clinical importance of infant fecal pH has been highlighted recently as a risk indicator for childhood stunting,44 and is also reflected in the updated reference range for infants provided by national diagnostic labs. The gut of infants enriched with the genus Bifidobacterium and low levels of potential pathogens decreases the risk of autoimmune diseases,15,16 supporting that supplementation with B. infantis EVC001 in early life may help protect infants from developing autoimmune diseases. Alpha-diversity was not different between groups; however, we found significant yet weak differences in beta-diversity between the two groups, suggesting that only a few OTUs were contributing to the overall beta-diversity in response to treatment status.
MaAsLin2 modeling also discovered that EVC supplementation was positively correlated with Enterococcaceae and Enterococcus. Confirmation of these data with statistical analyses at each time point found that Enterococcaceae was not different; however, Enterococcus was significantly higher by 0.5% in the EVC group compared with UNS at 6 months postnatal. Species that belong to the genus Enterococcus exert a range of functions in the gut as commensals to nosocomial pathogens that possess antibiotic resistance genes.45 In this study, Enterococcus represented a mean of 1% of the gut microbiome across both treatments and all time points, yet the variation was high ranging from 0.16% to 3.4% of the gut microbiome. For example, this genus represented 63% of the gut microbiome in one infant in the EVC group after the intake of antibiotics, but was reduced to 0% in this same infant 2 months later. We have previously reported that EVC supplementation reduced antibiotic resistance genes30 and that this taxon was not associated with enteric inflammation.27
In the follow-up #1 study, there was no difference in the frequency of illnesses, doctor visits, hospitalization, or health conditions between the EVC and UNS groups. Compared with the EVC group, participants in the UNS group reported a significantly higher score for the severity of their infants’ constipation (2.9 vs. 1.2), yet this value is not considered moderately or highly severe. In the follow-up #2 study, there were no differences between the EVC and UNS groups for the number of infants who were diagnosed with or experienced any common health conditions. While several larger studies have reported that probiotics can influence health conditions such as eczema,46,47 it is likely that the sample sizes in both follow-up #1 and follow-up #2 (n = 48 and n = 51, respectively) were too small to detect any significant differences in health outcomes or differences that may arise later in life.
One limitation of this study is that primers specific to the full genomic sequence for EVC001 were not used in this study. Based on the literature, B. infantis is an uncommon bifidobacterial subspecies found in infants who reside in Northern California.19,25 In the parent study published in Frese et al.,19 fecal B. infantis was on average 8 logs higher in infants supplemented with B. infantis EVC001 compared with UNS infants. Thus, we are confident that the several-fold difference in fecal B. infantis found in infants during the follow-up period is due to supplementation with EVC001 and not a random effect. Another limitation is that following completion of the parent study, factors that have confounding effects on the gut microbiome were not controlled. Although there were no significant differences in the number of infants among the different subgroups: breast milk; breast milk and infant formula; infant formula without breast milk; solid foods, or used antibiotics, probiotics, or were enrollees in daycare at any time point, given the small number in each subgroup it is possible that some of these factors had an impact on the gut microbiome. Second, different individuals participated in follow-up #1 and follow-up #2, limiting our ability to make direct comparisons between the gut microbiome results of follow-up #1 and the health outcomes measured in follow-up #2. Although follow-up #1 and #2 are independent of one another, both sets of participants stemmed from the parent study allowing us to make direct comparisons between treatment groups. Third, for both follow-up #1 and #2, not every participant provided a stool sample and questionnaire at every time point. As such, it was not possible to use paired data to compare the gut microbiome and health outcomes across time and, therefore, our statistical analyses were limited to the treatment group comparisons at each time point. Lastly, the parent study was originally designed to determine differences in the gut microbiome composition and fecal biochemistry at 1 month post-B. infantis EVC001 feeding and was not designed or powered to identify differences in health outcomes between treatment groups. Neither follow-up #1 nor #2 were designed or powered to detect differences in health outcomes between treatment groups. For example, previous longitudinal studies that have investigated the relationships between the early infant gut microbiome and atopic wheezing, and asthma have included both control and at-risk groups with sample sizes between 100 and 300 infants.48,49
Long-term colonization of a probiotic after cessation of its consumption has not been previously been demonstrated. These findings support the importance of matching a specific microorganism with a carbohydrate source that it selectively consumes thereby providing an open ecological niche for the microbe to occupy. We found that feeding breastfed infants a specific strain of B. infantis (EVC001) that efficiently utilizes all HMO structures in human milk for a brief period resulted in sustained colonization 1 year post supplementation. The gut microbiome in early infancy plays a critical role in immune system development and metabolic programming that has lifelong health impacts. Changes in the composition of the gut microbiome with lower protective microbes and higher potential pathogens associated with a Western lifestyle appear to increase the risks of developing allergic, inflammatory, and autoimmune diseases. Based on our findings, large clinical trials are warranted to determine whether B. infantis EVC001 supplementation early in life prevents the development of these diseases in child through adulthood.
Supplementary information
Acknowledgements
This work was supported by Evolve BioSystems, Inc. and, in part, by an NIEHS-funded predoctoral fellowship to C.E.O. (T32 ES007059) and its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS or NIH.
Author contributions
M.A.U. and J.T.S. conceived the study; J.T.S. designed the study; A.K.M., K.C., R.D.M., and S.A.F. acquired the data; G.C. and J.T.S. analyzed the data; C.E.O., G.C., S.A.F., B.M.H., M.A.U., and J.T.S. interpreted the data; C.E.O. and J.T.S. wrote the manuscript; A.K.M., K.C., R.D.M., G.C., S.A.F., B.M.H., and M.A.U. critically revised the manuscript for important intellectual content. All authors approved the final version of the manuscript for publication.
Competing interests
R.D.M., G.C., S.A.F., and B.M.H. are employees of Evolve BioSystems, Inc., a company focused on restoring the infant microbiome. Evolve BioSystems Inc. is committed to making data, materials, and analysis methods open and available upon request, where permitted. No other authors have conflicts of interest.
Statement of consent
All participants provided written informed consent to participate in the study described herein.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41390-020-01350-0) contains supplementary material, which is available to authorized users.
References
- 1.Totten SM, et al. Rapid-throughput glycomics applied to human milk oligosaccharide profiling for large human studies. Anal. Bioanal. Chem. 2014;406:7925–7935. doi: 10.1007/s00216-014-8261-2. [DOI] [PubMed] [Google Scholar]
- 2.Smilowitz JT, Lebrilla CB, Mills DA, German JB, Freeman SL. Breast milk oligosaccharides: Structure-function relationships in the neonate. Annu Rev Nutr. 2014;34:143–169. doi: 10.1146/annurev-nutr-071813-105721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LoCascio R, et al. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J. Agric. Food Chem. 2007;55:8914–8919. doi: 10.1021/jf0710480. [DOI] [PubMed] [Google Scholar]
- 4.Sela D, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl Acad. Sci. USA. 2008;105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LoCascio RG, Desai P, Sela DA, Weimer B, Mills DA. Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization. Appl. Environ. Microbiol. 2010;76:7373–7381. doi: 10.1128/AEM.00675-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.den Besten G, et al. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013;54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Limpt C, Crienen A, Vriesema A, Knol J. Effect of colonic short chain fatty acids, lactate and ph on the growth of common gut pathogens. Pediatr. Res. 2004;56:487–487. [Google Scholar]
- 8.Fukuda S, Toh H, Taylor TD, Ohno H, Hattori M. Acetate-producing bifidobacteria protect the host from enteropathogenic infection via carbohydrate transporters. Gut Microbes. 2012;3:449–454. doi: 10.4161/gmic.21214. [DOI] [PubMed] [Google Scholar]
- 9.Logan WR. The intestinal flora of infants and young children. J. Pathol. Bacteriol. 1913;18:527–551. [Google Scholar]
- 10.Henrick BM, et al. Elevated fecal pH indicates a profound change in the breastfed infant gut microbiome due to reduction of Bifidobacterium over the past century. mSphere. 2018;3:e00041–00018. doi: 10.1128/mSphere.00041-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azad MB, et al. Impact of cesarean section delivery and breastfeeding on infant gut microbiota at one year of age. Allergy Asthma Clin. Immunol. 2014;10:1–2. [Google Scholar]
- 12.Reyman M, et al. Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life. Nat. Commun. 2019;10:1–12. doi: 10.1038/s41467-019-13014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmadizar F, et al. Early‐life antibiotic exposure increases the risk of developing allergic symptoms later in life: a meta‐analysis. Allergy. 2018;73:971–986. doi: 10.1111/all.13332. [DOI] [PubMed] [Google Scholar]
- 14.Cardwell CR, et al. Caesarean section is associated with an increased risk of childhood-onset type 1 diabetes mellitus: a meta-analysis of observational studies. Diabetologia. 2008;51:726–735. doi: 10.1007/s00125-008-0941-z. [DOI] [PubMed] [Google Scholar]
- 15.Vatanen T, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165:842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olin A, et al. Stereotypic immune system development in newborn children. Cell. 2018;174:1277–1292.e1214. doi: 10.1016/j.cell.2018.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albert K, Rani A, Sela DA. Comparative pangenomics of the mammalian gut commensal Bifidobacterium longum. Microorganisms. 2020;8:7. doi: 10.3390/microorganisms8010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smilowitz JT, et al. Safety and tolerability of Bifidobacterium longum subspecies infantis EVC001 supplementation in healthy term breastfed infants: a phase I clinical trial. BMC Pediatr. 2017;17:133. doi: 10.1186/s12887-017-0886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frese SA, et al. Persistence of supplemented Bifidobacterium longum subsp. infantis EVC001 in breastfed infants. mSphere. 2017;2:e00501–e00517. doi: 10.1128/mSphere.00501-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maldonado-Gómez MX, et al. Stable engraftment of Bifidobacterium longum AH1206 in the human gut depends on individualized features of the resident microbiome. Cell Host Microbe. 2016;20:515–526. doi: 10.1016/j.chom.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Lawley B, et al. Differentiation of Bifidobacterium longum subspecies longum and infantis by quantitative PCR using functional gene targets. PeerJ. 2017;5:e3375. doi: 10.7717/peerj.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Penders J, et al. Quantification of Bifidobacterium spp., Escherichia coli and Clostridium difficile in faecal samples of breast-fed and formula-fed infants by real-time PCR. FEMS Microbiol. Lett. 2005;243:141–147. doi: 10.1016/j.femsle.2004.11.052. [DOI] [PubMed] [Google Scholar]
- 23.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 24.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis ZT, et al. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome. 2015;3:13. doi: 10.1186/s40168-015-0071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huda MN, et al. Stool microbiota and vaccine responses of infants. Pediatrics. 2014;134:e362–e372. doi: 10.1542/peds.2013-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henrick BM, et al. Colonization by B. infantis EVC001 modulates enteric inflammation in exclusively breastfed infants. Pediatr. Res. 2019;86:749–757. doi: 10.1038/s41390-019-0533-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fallani M, et al. Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J. Pediatr. Gastroenterol. Nutr. 2010;51:77–84. doi: 10.1097/MPG.0b013e3181d1b11e. [DOI] [PubMed] [Google Scholar]
- 29.Grzeskowiak L, et al. Distinct gut microbiota in southeastern African and northern European infants. J. Pediatr. Gastroenterol. Nutr. 2012;54:812–816. doi: 10.1097/MPG.0b013e318249039c. [DOI] [PubMed] [Google Scholar]
- 30.Casaburi G, et al. Early-life gut microbiome modulation reduces the abundance of antibiotic-resistant bacteria. Antimicrob. Resist. Infect. Control. 2019;8:131. doi: 10.1186/s13756-019-0583-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karav S, Casaburi G, Frese SA. Reduced colonic mucin degradation in breastfed infants colonized by Bifidobacterium longum subsp. infantis EVC001. FEBS Open Bio. 2018;8:1649–1657. doi: 10.1002/2211-5463.12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biddle A, Stewart L, Blanchard J, Leschine S. Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity. 2013;5:627–640. [Google Scholar]
- 33.Gosalbes MJ, et al. Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS ONE. 2011;6:1–9. doi: 10.1371/journal.pone.0017447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jalanka-Tuovinen J, et al. Intestinal microbiota in healthy adults: temporal analysis reveals individual and common core and relation to intestinal symptoms. PLoS ONE. 2011;6:1–13. doi: 10.1371/journal.pone.0023035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lippert K, et al. Gut microbiota dysbiosis associated with glucose metabolism disorders and the metabolic syndrome in older adults. Benef. Microbes. 2017;8:545–556. doi: 10.3920/BM2016.0184. [DOI] [PubMed] [Google Scholar]
- 36.Forbes JD, et al. Association of exposure to formula in the hospital and subsequent infant feeding practices with gut microbiota and risk of overweight in the first year of life. JAMA Pediatr. 2018;172:e181161–e181161. doi: 10.1001/jamapediatrics.2018.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ludwig, W., Schleifer, K.-H. & Whitman, W. B. in Bergey’s Manual® of Systematic Bacteriology (eds Garrity, V. P. et al.) 1–13 (Springer, New York, 2009).
- 38.Chua H-H, et al. Intestinal dysbiosis featuring abundance of Ruminococcus gnavus associates with allergic diseases in infants. Gastroenterology. 2018;154:154–167. doi: 10.1053/j.gastro.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 39.Henke MT, et al. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc. Natl Acad. Sci. USA. 2019;116:12672–12677. doi: 10.1073/pnas.1904099116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casterline BW, Hecht AL, Choi VM, Bubeck Wardenburg J. The Bacteroides fragilis pathogenicity island links virulence and strain competition. Gut Microbes. 2017;8:374–383. doi: 10.1080/19490976.2017.1290758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marcobal A, et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10:507–514. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang Y-L, Chassard C, Hausmann M, Von Itzstein M, Hennet T. Sialic acid catabolism drives intestinal inflammation and microbial dysbiosis in mice. Nat. Commun. 2015;6:1–11. doi: 10.1038/ncomms9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hossain, M. S. et al. Association of faecal pH with childhood stunting: results from a cross-sectional study. BMJ Paediatr. Open3, 1–6 (2019). [DOI] [PMC free article] [PubMed]
- 45.Dubin, K. & Pamer, E. G. Enterococci and their interactions with the intestinal microbiome. In Bugs as Drugs: Therapeutic Microbes for the Prevention and Treatment of Disease (eds Britton, R. A. & Cani, P. D.) 309–330 (American Society for Microbiology, Washington, 2018).
- 46.Kukkonen K, et al. Probiotics and prebiotic galacto-oligosaccharides in the prevention of allergic diseases: a randomized, double-blind, placebo-controlled trial. J. Allergy Clin. Immunol. 2007;119:192–198. doi: 10.1016/j.jaci.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 47.Cuello-Garcia CA, et al. Probiotics for the prevention of allergy: A systematic review and meta-analysis of randomized controlled trials. J. Allergy Clin. Immunol. 2015;136:952–961. doi: 10.1016/j.jaci.2015.04.031. [DOI] [PubMed] [Google Scholar]
- 48.Arrieta M-C, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015;7:307ra152–307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 49.Arrieta M-C, et al. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J. Allergy Clin. Immunol. 2018;142:424–434. e410. doi: 10.1016/j.jaci.2017.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.