Abstract
Functional inactivation of the pRB pathway is a very frequent event in human cancer, resulting in deregulated activity of the E2F transcription factors. To understand the functional role of the E2Fs in cell proliferation, we have developed cell lines expressing E2F-1, E2F-2, and E2F-3 fused to the estrogen receptor ligand binding domain (ER). In this study, we demonstrated that activation of all three E2Fs could relieve the mitogen requirement for entry into S phase in Rat1 fibroblasts and that E2F activity leads to a shortening of the G0-G1 phase of the cell cycle by 6 to 7 h. In contrast to the current assumption that E2F-1 is the only E2F capable of inducing apoptosis, we showed that deregulated E2F-2 and E2F-3 activities also result in apoptosis. Using the ERE2F-expressing cell lines, we demonstrated that several genes containing E2F DNA binding sites are efficiently induced by the E2Fs in the absence of protein synthesis. Furthermore, CDC25A is defined as a novel E2F target whose expression can be directly regulated by E2F-1. Data showing that CDC25A is an essential target for E2F-1, since its activity is required for efficient induction of S phase by E2F-1, are provided. Finally, our results show that expression of two E2F target genes, namely CDC25A and cyclin E, is sufficient to induce entry into S phase in quiescent fibroblasts. Taken together, our results provide an important step in defining how E2F activity leads to deregulated proliferation.
Deregulation of cell cycle control mechanisms is a hallmark of human cancer. In particular, there is ample evidence for the deregulation of two control pathways containing the two prototypic tumor suppressor proteins, p53 and the retinoblastoma protein, pRB (88). p53 is believed to be a surveillance factor that can induce apoptosis or growth arrest under specific circumstances, such as DNA damage, hypoxia, or deregulated growth induced by oncogenes (for a review, see reference 55). The importance of p53 in the regulation of cell proliferation is illustrated by the frequent inactivation of the TP53 gene or mutations of the upstream regulators of p53 (e.g., MDM2 and p19ARF) in human tumors.
pRB occupies a central role in regulating the G1-S transition of the mammalian cell cycle, a very important moment of the cell cycle at which the cell decides whether it should proliferate, differentiate, or die (for reviews, see references 3 and 96). The importance of the pRB pathway to normal growth control is emphasized by the frequent inactivation of the RB-1 gene or mutation of upstream regulators of pRB (e.g., cyclin D1, CDK4, or p16INK4A) in human tumors. Of the numerous cellular proteins that interact with pRB, the best characterized are the E2F transcription factors, and it is widely believed that pRB, to a large extent, exerts its control of cell proliferation by binding to and inhibiting the activity of these transcription factors (see, e.g., references 57, 76, 99, and 101).
Mice with targeted disruptions of Rb have an increased number of cells in S phase in the central and peripheral nervous systems compared to wild-type mice, and the Rb−/− neuronal cells fail to undergo differentiation (8, 42, 51, 52, 58). Subsequently, the Rb−/− mice die between days 13.5 and 14.5 of gestation, exhibiting profound apoptotic cell death in the hemopoietic and nervous systems (8, 42, 51). Consistent with the E2Fs being key downstream targets of pRB, several similarities between the effects of E2F overexpression in tissue culture cells and the loss of Rb function in mice have been observed. For instance, ectopic expression of E2F-1, E2F-2, E2F-3, and, to a lesser extent, E2F-4 is sufficient to induce S phase in quiescent immortalized rat fibroblasts (13, 45, 57), whereas E2F-5 and E2F-6 are unable to do so (7, 13, 25, 57). Moreover, overexpression of E2F-1, but not other E2Fs, has been shown to induce apoptosis in tissue culture cells (13, 49, 77, 87, 98) and transgenic mice (27, 37). Recently, genetic evidence of E2F-1 being a critical downstream target for pRB in vivo was provided by two sets of data showing that Rb+/− E2f-1−/− mice live longer, that their incidence of pituitary tumors is reduced compared to that of Rb+/− mice, and that Rb−/− E2f-1−/− embryos survive longer than Rb−/− embryos (94, 99). Although the Rb+/− and Rb−/− mice survive longer in an E2f-1−/− genetic background, it is noteworthy that they still die, demonstrating (as expected) that E2F-1 is not the only target for pRB.
Thus, the E2Fs can be described as key downstream effectors in a pathway that is very frequently deregulated in human cancer and whose functional integrity is essential for normal cell proliferation. Therefore, it becomes important to understand how these transcription factors are regulated and to know which genes are regulated by the E2Fs (for reviews, see references 18, 30, and 91). The majority of E2F-regulated genes encode proteins that are involved in DNA replication and/or in cell cycle progression. These genes include those encoding DNA polymerase α (72), thymidine kinase (TK) (14), HsORC1 (66), dihydrofolate reductase (DHFR) (5, 60, 90), CDC6 (29, 68, 100), MCM2 to MCM7 (54), cyclin A (40, 85), and cyclin E (6, 26, 67), p107 (102), B-myb (50), c-myc (34, 92), CDC2 (11, 93), E2F-1 (38, 44, 64), and E2F-2 (86). Although the E2Fs have been reported to be essential for the proper cell cycle regulation of several of these genes, it is evident that deregulated E2F activity leads to only a marginal increase in the level of expression of most of these genes (12, 41). Moreover, since several of the proteins participating in DNA replication are very stable, it is not clear why the transcription of these genes needs to be cell cycle regulated. None of the known gene products whose expression is regulated by the E2Fs is able to induce S phase by itself, suggesting that combinations of two or more products are required or that the responsible and limiting targets which can regulate S-phase entry have not yet been identified.
In an effort to better understand how deregulation of the pRB pathway can result in hyperproliferation, we have generated cell lines in which is expressed E2F-1, E2F-2, or E2F-3 fused to a modified version of the estrogen receptor ligand binding domain (ER) (56). The use of ER fusion proteins allows the identification of primary genetic targets for these proteins, since the activation can occur in the absence of de novo protein synthesis. Moreover, the rapid activation of the ER fusion protein after addition of the ligand is another feature that should allow the identification of genes that are required for S-phase induction by the E2Fs. By using the ERE2F cell lines, we demonstrate that in the absence of de novo protein synthesis the E2Fs are sufficient to induce transcription of the genes encoding cyclin E and cyclin A, as well as cdc6, p107, E2f-1, and B-myb, whereas E2F activation has little effect on the transcription of TK, the thymidine synthase gene (TS), PCNA, DHFR, cdc2, or c-myc.
Finally, we have used the ERE2F cell lines to identify cdc25A as a novel E2F target gene. We show that the cell cycle-regulated expression of cdc25A is dependent on E2F and that CDC25A is required for E2F-induced S-phase entry. In addition, we show that CDC25A can cooperate with cyclin E, another target of the E2Fs, to induce S-phase entry in quiescent cells.
MATERIALS AND METHODS
Plasmids.
A BglII-XbaI 5′ fragment (172 bp) and a KpnI-BamHI 3′ fragment (199 bp) of the modified ER were generated by PCR, using pBSKER as a template (56). The two fragments were ligated to a 603-bp XbaI-KpnI fragment of the ER and cloned into the BamHI site of pBSKHA (32), thereby generating pBSKHAER. Sequencing of the resulting plasmid showed that no mutations were incorporated in the ER fragment. pBSKHAERE2F-1, pBSKHAERE2F-2, pBSKHAERE2F-3, and pBSKHAERE2F-4 were generated by cloning BamHI fragments containing the full open reading frames of the E2Fs into pBSKHAER. Coupled in vitro transcription-translation of the resulting plasmids resulted in 35S-labeled proteins of the expected size.
The ER and ERE2F fragments were subsequently cloned as blunt-end fragments into pBabepuro (62) which had been cut with BamHI and blunt ended by treatment with the Klenow fragment of DNA polymerase (1), resulting in pBabeHAER, pBabeHAERE2F-1, pBabeHAERE2F-2, pBabeHAERE2F-3, and pBabeHAERE2F-4. pBabeHAERE2F-1/VP-16 was generated by cloning a BamHI site-containing fragment of E2F-1 (encoding amino acids 1 to 368) fused to VP-16 (76) from pBSK111/VP16 (62a) into pBabeHAER. pCMVHAER was generated by cloning a blunt-end EcoRV-SacII fragment from pBSKHAER into BamHI-cut and blunt-ended pCMVneoBam (2). Subsequently, pCMVHAERE2F-1 and pCMVHAERE2F-1(E132) were generated by cloning the wild-type or the DNA binding mutant of E2F-1 (10), respectively, into the unique BamHI site of pCMVHAER.
The luciferase reporter plasmid CDC25A(−755/+423) luc was constructed by subcloning the 1,178-bp SacI fragment of the CDC25A promoter in pGL3 basic (Promega). Mutations in the E2F DNA binding sites were generated by PCR–site-directed mutagenesis. For the upstream site (m1), the first PCR was performed with primers A5 (5′-CTAGAGCTCCCAGGGGGCTAAG-3′) and A7 (5′-CCTAGTTGGCTTCAAACGGAATCC-3′), the second PCR was done with primers A6 (5′-CTAGAGCTCCCGCTCCTCTTCC-3′) and A8 (5′-GGATTCCGTTTGAAGCCAACTAGGAA-3′), and the final reaction was achieved with primers A5 and A6. For the downstream site (m2), the first PCR was performed with primers A5 and A9 (5′-CCGGCCTTTCAAGGTAATAGCGGC-3′), the second PCR was done with primers A6 and A10 (5′-GCTATTACCTTGAAAGGCCGGCCT-3′), and the final reaction was achieved with primers A5 and A6. The double mutant was generated with a three-fragment ligation, cloning in the SacI site of pGL3 basic the 730-bp SacI-PvuII fragment from m1 and the 430-bp PvuII-SacI fragment from m2. The 6× E2F luciferase construct used, pGL3 TATA basic 6× E2F, has been described previously (63) and was the kind gift of Ali Fattaey.
Generation of cell lines.
ERE2F-1 clones were generated by transfecting Rat1 cells with pCMVHAERE2F-1 constructs, using the calcium phosphate method (1). Cells were selected in Dulbecco’s modified Eagle medium (DMEM) containing 10% bovine calf serum (BCS) and supplemented with G418 (0.5 mg/ml) and then cloned by limiting dilution. ERE2F pools were generated by infecting Rat1 cells with retroviruses produced in Phoenix cells (kindly provided by Garry Nolan) transfected with pBabeHAERE2F constructs. Briefly, Phoenix cells were plated at a density of 2 million cells per 10-cm-diameter dish and 2 days later were transfected with 10 μg of DNA. Supernatants were collected after 2 days, filtered, and used to infect Rat1 cells. The viral supernatant was left on the cells for 3 h, and the procedure was repeated twice to increase the efficiency of infection. Two days after infection, the Rat1 cell cultures were split and puromycin-resistant cells were selected in medium supplemented with 2.5 μg of puromycin/ml. For starvation conditions, cells were grown in DMEM–0.1% BCS for 48 h and induced with fresh 10% serum or by the addition to starvation medium to which 4-hydroxytamoxifen (OHT) was added to a final concentration of 300 nM. Cycloheximide (CHX) was used at a final concentration of 10 μg/ml. The addition of CHX was shown to reduce protein synthesis, as measured by [35S]methionine incorporation, by more than 99%.
Immunofluorescence.
To stain for expression of the E2Fs, cells grown on coverslips were fixed and permeabilized in −20°C cold acetone-methanol (1:1) for 10 min. Coverslips were air dried and stained by two different procedures, depending on the level of protein expression. For transiently transfected U2OS cells, a two-step protocol that included a primary mouse monoclonal antibody (anti-E2F-1 [KH95], anti-E2F-2 [TFE22], or anti-E2F-3 [TFE31]) followed by a goat anti-mouse Cy3-conjugated antibody (Jackson ImmunoResearch Laboratories, Inc.) was used. Stable transfected Rat1 cells expressing a low level of protein were stained with the E2F-1-, E2F-2-, or E2F-3-specific antibodies as described above and developed by using a TSA-Direct kit (NEN Life Science Products) according to the manufacturer’s suggestions.
Transactivation assays.
U2OS cells were transfected by the calcium phosphate method (1) with 50 ng of pCMVE2F, 2 μg of reporter plasmid, and 0.5 μg of pCMVβ-gal per 60-mm-diameter dish. Cells were collected 40 h after addition of the precipitate, and lysates were tested for luciferase and β-galactosidase activities as described previously (7). Rat1 ERE2F cells were transfected with 2 μg of reporter plasmid and 0.5 μg of pCMVβ-gal per 60-mm-diameter dish. Fifteen hours after transfection, the cells were induced with OHT for 24 h and lysates were tested for luciferase and β-galactosidase activities. To determine the activity of the CDC25A promoter during the cell cycle, Rat1 cells were transfected with 5 μg of reporter plasmid and 1 μg of pCMVβ-gal per 10-cm-diameter dish. Twenty hours after transfection, the cells were starved for 40 h and subsequently induced with 10% serum. Samples were collected and tested for luciferase and β-galactosidase activities as well as protein content. The cell cycle profiles were analyzed with a Becton Dickinson FACScan flow cytometer.
RT-PCR.
Reverse transcription-PCR (RT-PCR) was performed on total RNA prepared by the guanidinium thiocyanate-acid phenol method (1). After DNase treatment, 1 μg of RNA was used for cDNA synthesis. In a 50-μl reaction volume were mixed RNA (denatured 1 min at 95°C), dithiothreitol (10 mM), deoxynucleoside triphosphates (0.25 mM each), RNasin (8 U), Superscript with the provided buffer (Life Technologies; 200 U), and random hexamers (25 μM; Pharmacia). Reaction mixtures were incubated at room temperature for 15 min, at 42°C for 45 min, and finally at 95°C for 5 min to inactivate the reverse transcriptase. PCR was performed on 2 μl of the 50-μl cDNA sample. In addition, a PCR sample contained deoxynucleoside triphosphates 50 μM each, buffer with MgCl2 (1.5 mM final concentration), primers (0.2 μM each), [32P]dCTP (0.1 μl of 3,000 Ci/mM; Amersham Life Science), Taq polymerase (1 U; Perkin-Elmer), and water (to a final volume of 30 μl). To perform semiquantitative PCR, for each couple of primers we determined on cDNA from cycling Rat1 RNA (at the best annealing temperature) how many cycles were required to detect a clear signal in the linear range. To do so, the reactions were done in triplicate and the samples were collected at the end of various cycles (i.e., 20, 24, or 28). Each sample was run on a 4% polyacrylamide gel in Howley buffer (40 mM Tris, 20 mM sodium acetate, 1 mM EDTA; pH 7.2), and the gel was dried and exposed to an autoradiographic film. Using a phosphorimager (Fuji Inc.), the intensities of the bands were evaluated, and the numbers were plotted to evaluate which conditions to use to be in a linear range. PCR were performed with a PTC-100 machine (MJ Research Inc.), dividing all of the primers into three groups corresponding to three different initial annealing temperatures, 54, 52, and 50°C. While designing the PCR program, we took advantage of the touchdown technique, decreasing the annealing temperature 0.5°C per cycle for the first 10 cycles. Below are indicated, respectively, the initial annealing temperature, the number of cycles, and the sequences of the upstream and downstream primers used for each of the tested genes: for the gene encoding cyclin E, 54°C, 23 cycles, using 5′-ACATTCTACTTGGCACAGGAC-3′ and 5′-TGAGACCTTCTGCATCAACTC-3′; for cdc6, 54°C, 23 cycles, using 5′-TTAAGCCGGATTCTGCAAGAC-3′ and 5′-TCTGGTATAAGGTGGGAAGTTC-3′; for the gene encoding cyclin A2, 54°C, 23 cycles, using 5′-ATGAGACCCTGCATTTGGCTG-3′ and 5′-TTGAGGTAGGTCTGGTGAAGG-3′; for B-myb, 50°C, 24 cycles, using 5′-TGAGGCAGTTTGGACAGC-3′ and 5′-TTGAGGTGGTTGTGCCAG-3′; for p107, 54°C, 23 cycles, using 5′-TCATTTGCACCTTCTACCC-3′ and 5′-AGTCTATGTGAGATCCTGG-3′; for E2f-1, 54°C, 23 cycles, using 5′-TCTTGGAGCTGCTGAGCC-3′ and 5′-TCTGCAGGGTCTGCAATGC-3′; for TK, 50°C, 23 cycles, using 5′-AGCTGATGAGGAGAGTAAG-3′ and 5′-ACAATCACTGTCTTGCCTG-3′; for TS, 50°C, 24 cycles, using 5′-TATGGATTCCAGTGGAGAC-3′ and 5′-TGCAATCATGTAGGTCAGC-3′; for DHFR, 50°C, 26 cycles, using 5′-TGGTTCTCCATTCCTGAG-3′ and 5′-AGGACGTACTAGGAACAG-3′; for cdc2, 54°C, 26 cycles, using 5′-ATATAGTCAGCCTGCAGGATG-3′ and 5′-AAGAGCTGGTCAATCTCTGAG-3′; for PCNA, 52°C, 24 cycles, using 5′-ACGTTGAGCAACTTGGAATCC-3′ and 5′-TGTTACTGTAGGAGACAGTGG-3′; for c-myc, 52°C, 24 cycles, using 5′-TAGTGCTGCATGAAGAGACAC-3′ and 5′-AGTCCAAGTTCTGTCAGAAGG-3′; for the gene encoding DNA polymerase α, 50°C, 28 cycles, using 5′-AGCTGATGGATGGTGAAG-3′ and 5′-AATACTCCTCTGCTGAGG-3′; for the gene encoding cyclin D1, 50°C, 24 cycles, using 5′-AGATGAAGGAGACCATTCC-3′ and 5′-ttcaatctgttcctggcag-3′; for the gene encoding cyclin D3, 50°C, 24 cycles, using 5′-TCATGCCATATCTGAAGCC-3′ and 5′-AGATCCAAATGCAGTGACC-3′; for cdc25A, 54°C, 23 cycles, using 5′-TCCAGTGAAGGCAGATGTTC-3′ and 5′-AGAACTCACAGTGGAACACG-3′; for cdc25B, 54°C, 24 cycles, using 5′-AGATGAAGCAGGCTACAGAG-3′ and 5′-ACCAGTGGAGCACTAATGAG-3′; and for the gene encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 54°C, 22 cycles, using 5′-ATCCGTTGTGGATCTGACATGC-3′ and 5′-TGTCATTGAGAGCAATGCCAGC-3′.
Immunoprecipitation and Western blotting.
Rat1 cells expressing ERE2F-1 were serum starved for 48 h in DMEM containing 0.1% BCS and subsequently stimulated for different lengths of time in medium containing 10% serum or in starvation medium to which 1 μM OHT was added. Cells were lysed in ELB (250 mM NaCl, 50 mM HEPES [pH 7.0], 0.1% Nonidet P-40) to which protease inhibitors and 1 mM dithiothreitol were added. After clearing of the lysate by centrifugation, the protein content in the lysate was determined by the method of Bradford. A 200-μg quantity of protein was used for each immunoprecipitation with a polyclonal rabbit antiserum to CDC25A (product no. 06-571; Upstate Biotechnology) by standard protocols (28). Subsequently, the immunoprecipitated proteins were separated on a sodium dodecyl sulfate–8% polyacrylamide gel and processed for Western blotting as described elsewhere (28). The blot was probed with a mouse monoclonal antibody to CDC25A (product no. sc-7389; Santa Cruz) and developed by using an enhanced chemiluminescence kit (ECL; Amersham).
Isolation of the human CDC25A promoter.
The CDC25A gene was cloned from a human placenta genomic library screened with a 166-bp probe obtained by PCR performed on human genomic DNA, using the following primers: an upstream primer, +70 bp from ATG of the published cDNA sequence (5′-TCGTGAAGGCGCTATTTGGC-3′); and a downstream primer, +236 bp from ATG (5′-ATCTGTTGACTCGGAGGAGC-3′) (22). Ten positive lambda clones were isolated from 106 plaques. A 32P-labeled oligonucleotide corresponding to the first 25 nucleotides of the published sequence (−459 to −434 of the ATG sequence) hybridized to a 1.2-kb SacI fragment in 9 of the 10 isolated phages. The 1.2-kb SacI fragment was cloned in pBluescript SK− (Stratagene) and sequenced. The sequence revealed a 1,178-bp SacI fragment starting 755 bp 5′ of the previously published start codon.
RNase protection assay.
RNase protection was performed as described elsewhere (1). A probe of 258 bp, containing the nucleotide sequence −558 to −315 relative to the ATG, was used. The probe was transcribed by using T7 RNA polymerase on BamHI-cut pBSK in which a PCR fragment (BamHI-XhoI) generated by PCR performed on the 5′ region of CDC25A with primers RNase up/BamHI (5′-ATCGGGATCCCGTAGCTGCCATTCGGTTGAG-3′) and RNase down/XhoI (5′-GGCGAGCTCGCAACGGCCCAGGCTCAC-3′) was cloned. The RNase protection assay was performed on RNA samples from proliferating HeLa cells and from U20S cells synchronized with a double thymidine block followed by a nocodazole block and released from the block for 14 h (79% of cells were in S phase).
Microinjection.
For DNA microinjection experiments, Rat1 cells were grown on glass coverslips to 30% confluency and then incubated in DMEM without serum for 48 h. At the time of microinjection, the cells had reached 60 to 80% confluency. Nuclear microinjections were performed with a Zeiss automated microinjection device connected to an Eppendorf injector, using the following settings: time of injection, 0.0 s, pressure, 50 to 150 hPa; angle, 45°; speed, 20. Glass capillaries (product no. GC120TF-10; Clark Electromedical Instruments) were pulled by using the P-87 puller from Sutter Instruments. The injection time per coverslip did not exceed 30 min. The injection medium was DMEM without serum. The DNA used for microinjection was diluted in filter-sterilized phosphate-buffered saline (PBS) to a final concentration of 100 ng/μl for pCMVEGFP (microinjection marker) or 20 ng/μl for pCMVE2F-1, pCMVCyclin E, and pCMVhCDC25A. After microinjection, the cells were allowed to recover for 3 h, and then bromodeoxyuridine (BrdU; 100 μM) was added to all dishes at the same time. Cells were fixed for immunostaining 16 h after the addition of BrdU. Cells were fixed in 4% paraformaldehyde for 5 min at room temperature, washed briefly with PBS, and then incubated for 30 s in 60 mM NaOH. After three washes in PBS, BrdU was detected with an anti-BrdU antibody from Becton Dickinson (BD347580) as a primary antibody and a Cy3-conjugated anti-mouse immunoglobulin G (IgG) from Jackson Laboratories as a secondary antibody. The DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI). Injected cells were identified by the green fluorescence emitted from enhanced green fluorescent protein. For each injection, between 100 and 150 injected cells were counted. The experiments were repeated three times with similar results.
For microinjection of antibodies, the antibodies were first dialyzed against PBS to remove NaN3 and then concentrated to 300 ng/μl by using Amicon G10 filters. Rabbit IgG was added as a microinjection marker at a concentration of 2 μg/μl. Rat1 ERE2F-1 cells were grown on glass coverslips to 50% confluency and then incubated in DMEM supplemented with 0.1% BCS for 48 h. Cytoplasmic microinjection was then performed in this medium, using the procedure described above. After a recovery period of several hours after the microinjection, BrdU (100 μM) and OHT (300 nM) were added to the medium. Twelve hours later, the cells were fixed as described above. Injected cells were identified with a goat-anti-rabbit IgG coupled to Cy3 (Jackson Laboratories), and BrdU staining was performed with the fluorescein isothiocyanate-labeled anti-BrdU antibody from Becton Dickinson (BD347583). DAPI was used for staining of DNA. The slides were analyzed under an Aristoplan fluorescence microscope (Leitz). For peptide competition experiments, the antibody was first incubated with a 10-fold molar excess of peptide, then dialyzed and concentrated as described above.
Nucleotide sequence accession number.
The nucleotide sequence of the human CDC25A promoter has been submitted to the DDJB-EMBL-GenBank databases under accession no. AJ242714.
RESULTS
Generation of cell lines expressing ERE2F-1 fusion proteins.
Previously, our laboratory has generated cell lines with tetracycline-regulated expression of the E2F transcription factors (57). Although the use of inducible expression of the E2F transcription factors or, alternatively, infection with adenoviruses containing the E2Fs is a useful technique for the understanding of the functional consequences of deregulated E2F expression (see, e.g., references 12, 13, 35, 57, and 87), neither technique allows the identification of primary target genes of the E2F transcription factors. As an alternative approach, the activities of several proteins have been made hormone dependent by fusion of these proteins to the hormone binding domain of the estrogen receptor (ER) (see, e.g., references 19, 75, 79, and 84). The generation of ER fusion proteins allows the identification of primary events, since the fusion proteins can be activated without de novo protein synthesis.
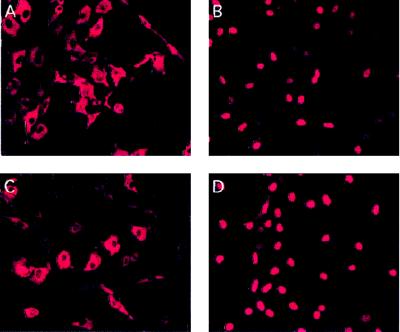
To test the feasibility of generating cell lines exhibiting hormone-dependent activation of the E2F transcription factors, we constructed expression plasmids in which a modified version of the ER was fused to the N terminus of E2F-1. This modified version of the ER does not respond to estrogen but rather to OHT (56). To test whether subsequent effects mediated by the ERE2F-1 fusion were a consequence of DNA binding, we also constructed plasmids expressing an ER fusion containing a DNA binding-deficient mutant of E2F-1 (E132) (10). The constructed expression plasmids were tested by transient transfection of Rat1 cells. Expression of ERE2F-1 was shown to lead to a 15-fold activation of a cotransfected reporter plasmid containing six E2F DNA binding sites after addition of OHT, while no transactivation was observed in cells expressing the E132 mutant (data not shown). Importantly, we did not observe a higher basal promoter activity of the reporter plasmid in the absence of OHT in cells expressing ERE2F-1 than in cells that were transfected with the reporter plasmid alone (data not shown), suggesting that the ERE2F-1 fusion protein was completely inactive in the absence of OHT. The subcellular localizations of the fusion proteins were tested by immunofluorescence analysis. U2OS cells were transiently transfected with pCMVHAERE2F-1 and stained for E2F-1 expression by using an anti-E2F-1 monoclonal antibody (Fig. 1). In the absence of OHT, the fusion protein is located in the cytoplasm, while after addition of OHT, the majority of the fusion protein is located in the nucleus (Fig. 1A and B). Identical results were obtained with cells expressing the ERE2F-1 E132 fusion protein (data not shown). The translocation of the fusion proteins from the cytosol to the nucleus strongly suggests that the activity of the ERE2F fusion proteins is regulated by their subcellular localization.
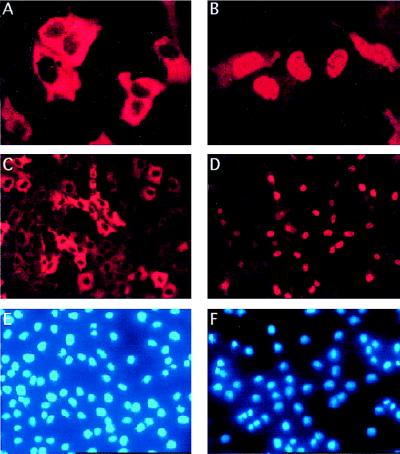
FIG. 1.
Subcellular localization of ERE2F-1. (A and B) Immunofluorescence of U2OS cells transiently transfected with pCMVHAERE2F-1, detected by using an anti-E2F-1 monoclonal antibody (KH95), in the absence (A) or in the presence (B) of OHT. (C to F) A selected pool of Rat1 clones expressing ERE2F-1, stained with an anti-E2F-1 monoclonal antibody, in the absence (C) or in the presence (D) of OHT and the corresponding DAPI staining (E and F).
To obtain cell lines that stably express ERE2F-1, Rat1 cells were transfected with pCMVHAERE2F-1 and pCMVHAERE2F-1(E132), and several clones were isolated and tested. For the cytomegalovirus-expressing clones, the data presented here are the results obtained with clone D (wild type) and clone Q (E132), which express comparable amounts of protein (data not shown). Similar results have, however, been obtained with several different clones. In addition, pools of cells were also selected after infection of Rat1 cells with retroviruses expressing ERE2F-1 or ERE2F-1/VP-16, in which the transactivation and pRB binding domain of E2F-1 is replaced by the VP-16 transactivation domain (76). Results for these pools of infected cells are also presented here. Since we do not have an anti-E2F-1 antibody that recognizes rat E2F-1 and human E2F-1 to the same extent, it is not possible to give the exact levels of E2F-1 overexpression in the clones. However, by using equal amounts of cell lysates from human U20S cells and Rat1 cells expressing the E2F-1 fusion proteins, we could estimate that the Rat 1 clones and the pools express 5- to 10-fold-higher levels of E2F-1 than do U2OS cells (data not shown).
Addition of OHT to the wild-type and E132 clones leads to nuclear translocation of the fusion proteins (data not shown), and as shown in Fig. 1C and D, a similar phenomenon is observed for the pool of infected cells expressing ERE2F-1. Our hypothesis is that the nuclear translocation is the consequence of OHT binding to the ER, the displacement of ER-associated cellular polypeptides, and the unmasking of the E2F-1 nuclear localization signal, located in the N terminus of E2F-1 (63). The pools (data not shown) and the clones (see Fig. 9D) were shown to transactivate a reporter plasmid containing six E2F DNA binding sites in an OHT-dependent manner. We also tested the capacity of the ERE2F-1 fusion protein to induce S phase in serum-deprived cells. The D and Q clones were kept in medium containing 0.1% serum for 48 h, after which either OHT was added to the starvation medium or fresh medium containing 10% serum was provided (Table 1). Addition of OHT to clone D (expressing wild-type E2F-1) but not to clone Q (expressing the E132 mutant) led to S-phase induction. The activation of E2F-1 by addition of OHT appears to shorten traversal of G0-G1 by approximately 6 to 7 h compared to serum stimulation (Table 1 and data not shown). Similarly, the activation of E2F-1 by addition of OHT to the E2F-1 pool (Table 1) and to the E2F-1/VP-16-expressing cells (data not shown) was sufficient to induce S phase in the treated cells. We could also detect a high rate of apoptosis in the D clone after OHT addition. Between 14 and 18 h, 30% of the cells were apoptotic, as indicated by the sub-G1 population detected by fluorescence-activated cell sorter (FACS) analysis (see Fig. 5C). There was no sign of apoptosis in the E132-expressing clones after addition of OHT, demonstrating the requirement of a functional DNA binding domain for cellular effects mediated by E2F-1 (10, 31, 39, 57, 74). In summary, the activated ERE2F-1 fusion protein shows all the expected properties of wild-type E2F-1, including DNA-dependent transactivation of E2F-dependent promoters and the capacity to induce S-phase entry and apoptosis in serum-starved fibroblasts. Moreover, since activated ERE2F-1/VP16 fusion protein elicits the same responses as ERE2F-1, these features are not due to inactivation of pRB by titration but rather are a genuine transactivation function of E2F-1. Finally, the facts that in the absence of OHT the fusion proteins are in the cytoplasm and that there is no detectable leakiness in the system make these inducible cell lines a powerful tool to study the functional effects of deregulated E2F-1 activity.
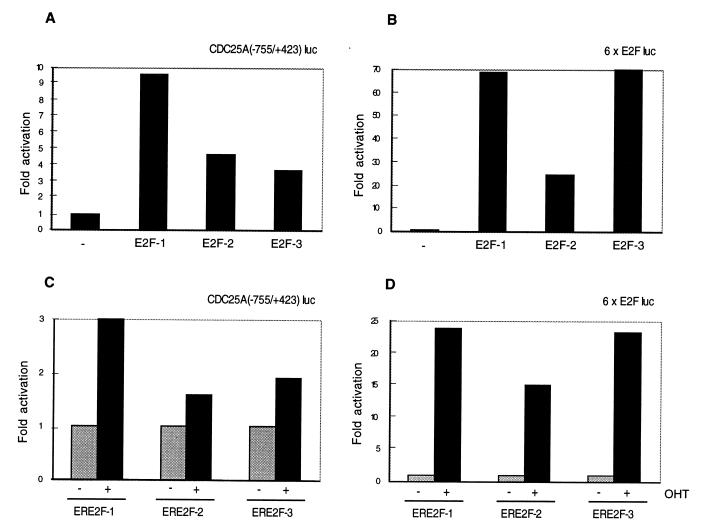
FIG. 9.
E2F transactivation of the CDC25A promoter. (A and B) Activation of the CDC25A promoter by transient transfection of pCMVE2F-1, pCMVE2F-2, and pCMVE2F-3. U2OS cells were transiently transfected with pCMVE2F-1, pCMVE2F-2, or pCMVE2F-3 together with CDC25A(−755/+423) luc (A) or with a synthetic E2F-responsive promoter, 6× E2Fluc (B). −, transfected with empty pcnv expression vector. (C and D) Activation of the CDC25A promoter in the ERE2F-expressing cell lines. Rat1 cells expressing ERE2F-1, ERE2F-2, or ERE2F-3 were transfected with CDC25A(−755/+423) luc (C) or with 6× E2Fluc (D). +, addition of OHT; −, without OHT. pCMVβ-gal was cotransfected in all experiments, and β-galactosidase activity served as a control for transfection efficiency. Numbers indicate fold induction relative to the sample transfected with empty pCMV (A and B) or fold induction after addition of OHT (C and D). The luciferase counts are normalized for β-galactosidase activity. The presented data are representative of at least three different experiments.
TABLE 1.
Activation of E2F-1 by OHT is sufficient for S-phase inductiona
| Cell line and treatment protocol | % of cells in cell cycle phase:
|
||
|---|---|---|---|
| G0-G1 | S | G-M | |
| ERE2F-1 clone D | |||
| 0 h | 91 | 5 | 4 |
| 8 h + OHT | 85 | 12 | 3 |
| 12 h + OHT | 2 | 98 | 0 |
| 18 h + OHT | 19 | 53 | 28 |
| 8 h + serum | 93 | 3 | 4 |
| 12 h + serum | 91 | 3 | 6 |
| 18 h + serum | 18 | 82 | 0 |
| ERE2F-1(E132) clone Q | |||
| 0 h | 89 | 5 | 6 |
| 12 h + OHT | 91 | 2 | 7 |
| 18 h + OHT | 92 | 2 | 6 |
| 18 h + serum | 54 | 46 | 0 |
| ERE2F-1 pool | |||
| 0 h | 92 | 6 | 2 |
| 13 h + OHT | 36 | 64 | 0 |
| 16 h + OHT | 28 | 72 | 0 |
| 19 h + OHT | 32 | 61 | 7 |
Cell cycle profiles of Rat1 cells expressing ERE2F-1 clone D, Rat1 cells expressing ERE2F-1(E132) clone Q, and a Rat1 ERE2F-1 pool were obtained. Cells were made quiescent by incubation in medium containing 0.1% serum for 48 h and subsequently incubated in the same medium to which OHT was added or, alternatively, stimulated with fresh medium containing 10% serum. Samples were taken for FACS analysis to determine the cell cycle profile of the cells at the indicated hours.
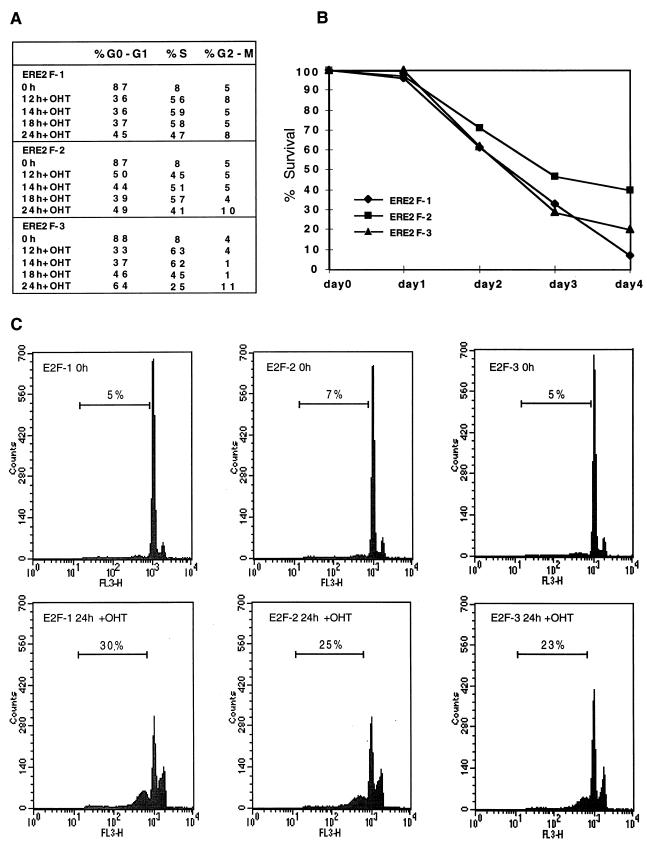
FIG. 5.
Activation of E2F-1, E2F-2, or E2F-3 results in S-phase entry and apoptosis. (A) E2F-1, E2F-2, or E2F-3 activation is sufficient for S-phase induction. Rat1 cells expressing ERE2F-1, ERE2F-2, or ERE2F-3 were grown in medium containing 0.1% serum for 48 h and subsequently induced with OHT. Cells were harvested for FACS analysis at the indicated times after addition of OHT, and the cell cycle profile was determined. (B) E2F-1, E2F-2, and E2F-3 induce cell death. Cells were serum starved for 48 h in medium with 0.1% serum, and OHT was subsequently added to the starvation medium. At the indicated times, cells were harvested and living cells were counted after being stained with Trypan blue. The percentage of surviving cells was calculated by using the number serum-starved cells in the absence of OHT as the reference point. (C) Activation of E2F-1, E2F-2, or E2F-3 induces apoptosis. FACS analysis of Rat1 cells expressing ERE2F-1, ERE2F-2, or ERE2F-3, showing the percentage of apoptotic cells (sub-G1 fraction) at 0 and 24 h after OHT addition. The cells were grown in medium with 0.1% serum for 48 h prior to addition of OHT.
Identification of genes as direct targets of E2F-1.
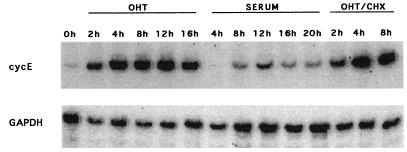
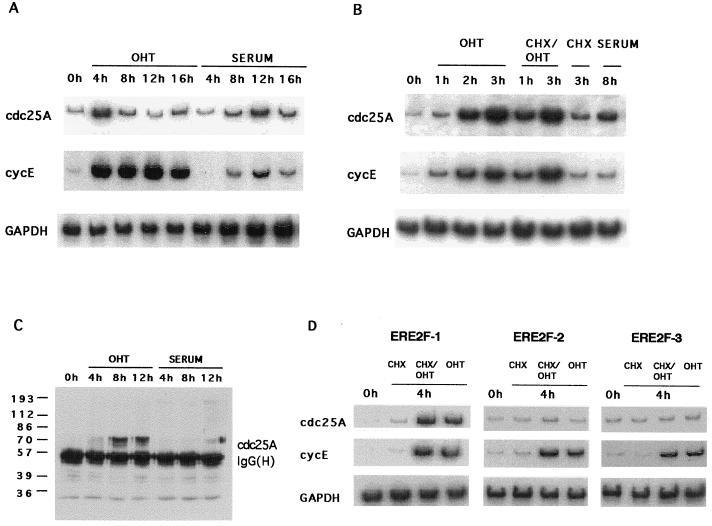
To identify the primary target genes that allow E2F-1 to induce S phase in serum-starved cells, Rat1 ERE2F-1 cells were serum starved for 48 h and then induced with serum or OHT for different lengths of time in the absence or presence of CHX. RNA was prepared, and RT-PCR was performed under linear reaction conditions as described in Materials and Methods. To test whether this method was feasible, we first analyzed the expression of the cyclin E gene (Fig. 2). As early as 2 h after addition of OHT, cyclin E mRNA levels were increased dramatically, and this increase was not abolished by simultaneous CHX treatment, demonstrating that E2F-1 activation alone is sufficient for increased transcription of the endogenous cyclin E gene (see also Fig. 7B). Consistent with the cyclin E gene being an important downstream target of E2F and the fact that cyclin E is limiting for progression through G0-G1 (69, 80), cyclin E transcription was activated 6 to 7 h earlier in cells treated with OHT than in serum-treated cells. This result is also in good agreement with the demonstration that activation of E2F-1 shortens the G1 phase of the cell cycle by approximately 6 to 7 h.
FIG. 2.
Cyclin E is a direct target of E2F-1. ERE2F-1 clone D cells were made quiescent by growing the cells in medium with 0.1% serum for 48 h. RT-PCR was performed on samples of total RNA from ERE2F-1 clone D that had been kept for 48 h in medium containing 0.1% serum and induced for different lengths of time by addition of OHT, serum, or OHT plus CHX. Primers specific for the amplification of cyclin E mRNA were used as described in Materials and Methods.
FIG. 7.
CDC25A is a direct target of E2F-1. (A and B) RT-PCR performed on RNA samples from ERE2F-1 clone D cells that were starved and induced with OHT or serum (A) or with OHT alone, CHX plus OHT, CHX alone, or serum alone (B). Primers for specific amplification of cdc25A, the cyclin E gene, and GAPDH were used as indicated and as described in Materials and Methods. (C) Activation of E2F-1 leads to accumulation of the cdc25A protein. Immunoprecipitation followed by Western blotting with cdc25A-specific antibodies was performed with 200-μg quantities of cell lysate at the indicated time points after addition of OHT or serum. The masses of molecular size markers are indicated to the left in kilodaltons. (D) Activation of E2F-2 and E2F-3 is not sufficient to induce transcription of CDC25A. RT-PCR was performed on RNA prepared from ERE2F-1, ERE2F-2, and ERE2F-3 pools as described above. Cyclin E served as an internal positive control. The cells were starved and induced for 4 h in the presence of CHX, CHX plus OHT, or OHT alone.
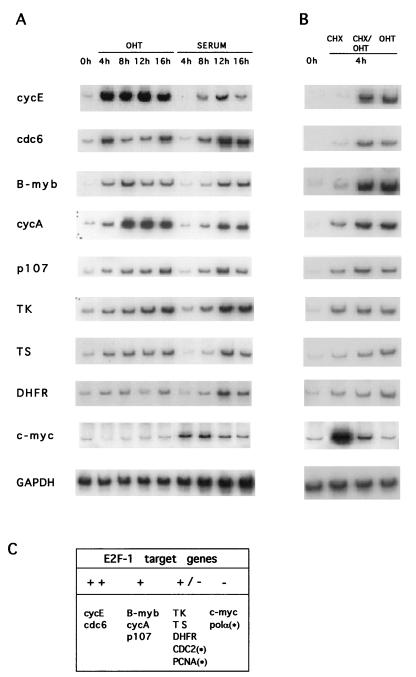
To analyze whether other genes containing E2F DNA binding sites in their promoters could be directly activated by E2F-1, RT-PCR was performed on RNA prepared from the ERE2F-1-expressing cells, using gene-specific primers. As shown in Fig. 3, activation of E2F-1 alone was sufficient to increase the transcription of cdc6, B-myb, the gene encoding cyclin A, and p107 in addition to the gene coding for cyclin E, whereas only minor activation was observed for TK, TS, DHFR, cdc2, and PCNA. We did not observe any significant increase in the level of c-myc or DNA polymerase α. None of the genes tested were upregulated by ERE2F-1(E132) when OHT was added to the cells (data not shown).
FIG. 3.
Genes activated by E2F-1. (A and B) RT-PCR was performed on RNA samples prepared from ERE2F-1 clone D (A) and from an ERE2F-1 pool (B). Cells were kept for 48 h in medium containing 0.1% serum and induced for different lengths of time with OHT or serum (A) or for 4 h in the presence of CHX, CHX plus OHT, or OHT alone (B). Primers specific for the cDNA amplification of the indicated genes were used as described in Materials and Methods. (C) Genes activated directly by E2F-1 expression. ++, upregulation over 10-fold; +, upregulation between 5- and 10-fold; +/−, upregulation between 1- and 5-fold; −, no upregulation observed; ∗, data not shown.
E2F-2 and E2F-3 induce S phase and apoptosis.
To extend the analyses of the regulation of E2F-dependent genes to the other members of the E2F family, we infected Rat1 cells with retroviruses expressing ERE2F-2, ERE2F-3, or ERE2F-4. Data concerning ERE2F-4 will not be presented here, since we were not able to show any transactivating potential of the fusion protein before or after addition of OHT. The lack of regulation of this protein may be due to the fact that the majority of ERE2F-4 remains in the cytoplasm after addition of OHT (data not shown). In contrast, the ERE2F-2 and ERE2F-3 proteins are localized in the cytoplasm in the absence of OHT, while nuclear translocation is observed after addition of OHT (Fig. 4). We noticed a slight nuclear staining in some of the ERE2F-2-expressing cells even in the absence of OHT, indicating that the activity of the ERE2F-2 fusion is not as tightly regulated as that of ERE2F-1 or ERE2F-3.
FIG. 4.
Subcellular localization of ERE2F-2 and ERE2F-3 in the presence or absence of OHT. (A and B) A pool of puromycin-resistant Rat1 cells infected with retroviruses expressing ERE2F-2, stained with an anti-E2F-2 monoclonal antibody (TFE22), in the absence (A) or in the presence (B) of OHT. (C and D) A pool of puromycin-resistant Rat1 cells infected with retroviruses expressing ERE2F-3, stained with an anti-E2F-3 monoclonal antibody (TFE31), in the absence (C) or in the presence (D) of OHT.
To test the functional properties of ERE2F-2 and ERE2F-3, we analyzed the abilities of the proteins to induce S-phase entry in low-serum medium and their abilities to transactivate a synthetic promoter. The data presented in Fig. 5A demonstrate that activation of ERE2F-2 or ERE2F-3 by OHT induce S-phase entry in serum-starved cells to a degree similar to activation of ERE2F-1. By transfection of a reporter plasmid containing six E2F DNA binding sites into ERE2F-2- or ERE2F-3-expressing cells, we also observed an OHT-dependent transactivation of this promoter construct (see Fig. 9D). These results are consistent with previously published results for transiently transfected or microinjected E2F-1, E2F-2, and E2F-3 (53, 57). To investigate the effects of constitutive activation of the E2Fs on cell proliferation, the ERE2F cell lines were grown in low-serum medium for 96 h in the absence or presence of OHT (Fig. 5B). While cells kept in medium without OHT proliferated slowly, there was a dramatic selection against proliferation of cells expressing active E2F-1, E2F-2, or E2F-3. In agreement with this, we observed a high level of cell death, as evidenced by floating and highly refractile cells (data not shown). No cell death was observed when the ERE2F-1(E132 cells) were grown in low-serum medium in the presence of OHT (data not shown). Since the observed phenotype is characteristic for cells undergoing apoptotic death, and it is known that ectopic expression of E2F-1 induces apoptosis (see the introduction), a flow-cytometric assay was used to test whether the cell death was due to apoptosis (65). As shown in Fig. 5C, activation of E2F-1, E2F-2, or E2F-3 induced apoptosis in ERE2F-expressing Rat1 cells grown in low-serum medium in the presence of OHT. In the absence of OHT, no sub-G1 peak was observed, nor did we observe a sub-G1 peak in ERE2F-1(E132)-expressing cells when they were grown in low-serum medium in the presence of OHT (data not shown). In agreement with these results, we have previously observed a strong selection against E2F-1, E2F-2, and E2F-3 expression in U2OS cells both with tetracycline-regulated expression and in colony formation assays (72a).
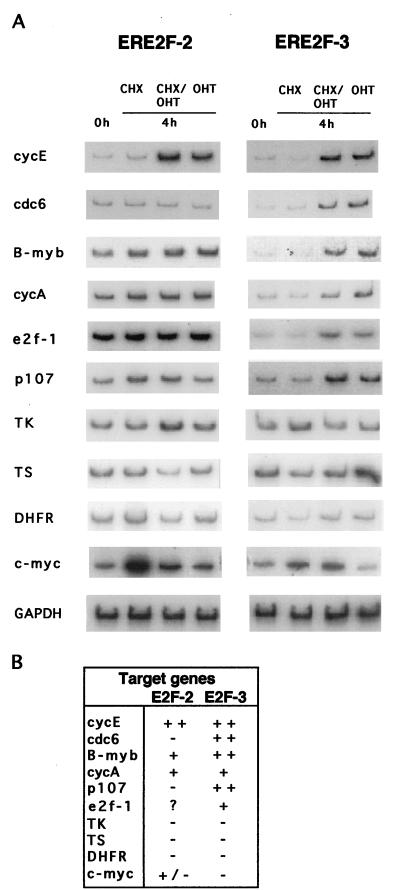
Since these data show that the selected pools of Rat1 cells expressing ERE2F-2 or ERE2F-3 contain regulated alleles of E2F-2 or E2F-3 with the expected properties, we performed RT-PCR as described for the ERE2F-1-expressing cells. ERE2F-2- or ERE2F-3-expressing cells were serum starved and subsequently grown in the presence of CHX alone, CHX plus OHT, or OHT alone for 4 h (Fig. 6A). As shown in Fig. 6A and summarized in Fig. 6B, cdc6, B-myb, p107, E2f-1, and the genes encoding cyclins E and A are all primary targets of E2F-3, as was previously shown for E2F-1. Surprisingly, E2F-2 was only able to upregulate the expression of the gene coding for cyclin E and, to some extent, the cyclin A gene and B-myb; however, a high basal level of the mRNA in uninduced cells made it difficult to estimate the degree of upregulation. In summary, the gene encoding cyclin E appears to be the best tested target of E2F-1, E2F-2, and E2F-3, followed by the cyclin A gene and B-myb. Comparison of the number of genes upregulated and the magnitudes of induction by the different E2Fs showed that E2F-1 appears to be a better transactivator than E2F-3, which in turn is a better transactivator than E2F-2. However, it is important to emphasize that this hierarchy may be different for as-yet-unknown, and therefore nontested, E2F target genes.
FIG. 6.
Genes activated by E2F-2 and E2F-3. (A) RT-PCR was performed on RNA samples prepared from ERE2F-2 and ERE2F-3. The cells were grown in medium containing 0.1% serum for 48 h, and OHT was subsequently added for 4 h in the presence of CHX, CHX plus OHT, or OHT alone. (B) Summary of genes activated directly by E2F-2 and E2F-3 expression. ++, upregulation over 10-fold; +, upregulation between 5- and 10-fold; +/−, upregulation between 1- and 5-fold; −, no upregulation observed; ?, increased levels in serum-starved cells.
CDC25A as a novel E2F target.
Since the use of ERE2F cell lines in combination with RT-PCR gave encouraging results when testing whether a gene can be a direct target of E2F regulation, we decided to use the system to identify other genes regulated by the E2Fs. For this purpose, we tested four genes (those encoding cyclins D1 and D3, as well as cdc25A and cdc25B) whose expression previously was shown to be cell cycle regulated (43, 46, 59) but with no indication that E2F activity participates in the regulation. Of the four tested genes, only cdc25A was shown to be affected by E2F expression (Fig. 7 and data not shown). As shown in Fig. 7A, a strong increase in the cdc25A mRNA level was observed 4 h after OHT addition, while serum stimulation of the same cell line led to the highest level of accumulation at 12 h. To confirm that the upregulation of cdc25A by E2F-1 was a primary effect, the level of cdc25A mRNA was measured shortly after OHT addition (Fig. 7B). An increase in cdc25A mRNA levels was observed as early as 1 h after OHT addition, and this increase became more pronounced at 2 to 3 h after stimulation. Moreover, cdc25A upregulation was sustained even in the presence of CHX, demonstrating that the transactivation of the cdc25A promoter by E2F-1 does not require de novo protein synthesis (Fig. 7B and D).
To investigate whether the abundance of the cdc25A protein increased in parallel with the mRNA level, cell extracts were prepared from Rat1 ERE2F-1 cells before and after stimulation with OHT or serum (Fig. 7C). A slight increase in the amount of cdc25A protein was observed as early as 4 h after OHT treatment, and the level was further increased at 8 and 12 h after OHT stimulation. In agreement with the slower kinetics of mRNA accumulation after serum stimulation, the level of the cdc25A protein did not increase until 12 h after addition of serum.
To analyze whether cdc25A is also a target for E2F-2 and E2F-3, RT-PCR analysis was performed on RNA samples from ERE2F-1, -2, and -3 cells that had been starved and then induced with OHT and CHX (Fig. 7D). In contrast to E2F-1, E2F-2 and E2F-3 were unable to stimulate the transcription of cdc25A within 4 h of OHT addition, whereas cyclin E mRNA levels were increased to similar extents by the three transcription factors. However, cdc25A mRNA levels were upregulated after treatment of ERE2F-2- or ERE2F-3-expressing cells with OHT for 12 h, suggesting that de novo synthesis of other proteins is required for the stimulation of cdc25A transcription by E2F-2 and E2F-3 (data not shown). In summary, our results suggest that the cell cycle regulation of cdc25A transcription is dependent on E2F.
E2F-dependent cell cycle regulation of CDC25A transcription.
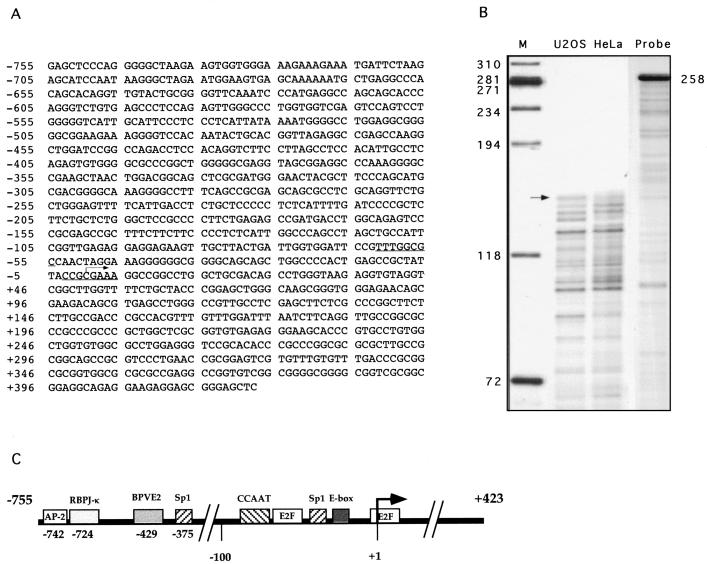
To understand the role played by E2Fs in the regulation of CDC25A transcription, we cloned the CDC25A gene from a human placenta genomic library. Ten positive lambda clones containing the 5′ end of the CDC25A cDNA were isolated, and hybridization with an oligonucleotide corresponding to the first 25 nucleotides of the published cDNA sequence (22) identified an approximately 1.2-kb SacI genomic fragment. This fragment was cloned and sequenced, and it was shown to contain an 1,178-bp insert (Fig. 8A). The 3′ SacI site is located 20 bp upstream of the initiation codon of the previously published cDNA sequence (22). To determine the position of transcription initiation in the CDC25A gene, RNase protection experiments were performed. As shown in Fig. 8B, an RNA probe of 258 nucleotides containing 237 nucleotides of the CDC25A gene yielded several protected fragments in RNAs prepared from both U2OS and HeLa cells. The longest of these protected fragments was approximately 120 nucleotides, corresponding to a position at 440 nucleotides 5′ of the CDC25A start codon. The 5′ end of the human CDC25A gene isolated by Galaktionov and Beach (22) corresponds to a position 442 nucleotides 5′ of the CDC25A start codon, and we have therefore decided to use this as the +1 reference point.
FIG. 8.
Sequence and schematic representation of the human CDC25A promoter. (A) Nucleotide sequence of the 1,178-bp CDC25A promoter SacI fragment containing 755 nucleotides upstream of the transcription initiation site and 423 nucleotides of the 5′ untranslated region of the human CDC25A cDNA. Two E2F DNA binding sites are underlined. The transcription initiation site, which coincides with the longest cDNA clone isolated, is indicated with an arrow. (B) RNase protection assay. RNA was prepared from HeLa and U2OS cells and processed for RNase protection, using a 258-nucleotide probe containing 237 nucleotides of the CDC25A gene surrounding the putative transcription initiation site. The longest protected fragment, corresponding to approximately 120 nucleotides, is indicated with an arrow. The length of the probe is indicated to the right, and the number of nucleotides in a double-stranded DNA molecular size marker is indicated to the left. RNA has a slower mobility than DNA, and it is estimated to be 5 to 10% different from DNA. (C) Schematic representation of transcription factor binding sites in the 1,178-bp human CDC25A promoter. The transcription start site is depicted with an arrow. The two E2F sites are indicated, as are DNA consensus binding sites for AP-2, RBPJ-κ, bovine papillomavirus E2 (BPVE2), CCAAT-box binding proteins, Sp-1, and bHLH (E-box).
An examination of the promoter region of the CDC25A gene revealed the presence of two putative consensus E2F DNA binding sites, as well as DNA binding sites for basic helix-loop-helix proteins (E box), Sp1, CCAAT box binding proteins, bovine papillomavirus E2, AP-2, and RBPJ-κ (Fig. 8C). As for most of the known genes containing E2F DNA binding sites, CDC25A has a TATA-less promoter.
To investigate whether the two putative E2F binding sites are responsible for the upregulation of CDC25A by E2F-1, the 1,178-bp SacI fragment was cloned in pGL3basic, resulting in CDC25A(−755/+423) luc. This construct was transfected into U2OS cells together with expression plasmids for E2F-1, -2, or -3 and cytomegalovirus β-galactosidase as a control for transfection efficiency. As shown in Fig. 9A, each of the three E2Fs was able to transactivate the CDC25A promoter, although E2F-1 was three times more efficient than E2F-2 or E2F-3. Similar data were obtained when the ERE2F-expressing cell lines were transfected and the E2Fs were activated by OHT (Fig. 9C). As a control for the transactivation activities of the E2Fs, we used a synthetic reporter construct containing six E2F DNA binding sites. As shown in Fig. 9B and D, E2F-1 and E2F-3 transactivated this construct to similar extents whereas E2F-2 was two- to threefold less efficient. In summary, our data show that the CDC25A promoter is efficiently transactivated by E2F-1, and to a lesser extent by E2F-2 and E2F-3, whereas we do not see any short-term effects of E2F-2 or E2F-3 on the induction of the endogenous CDC25A mRNA levels in the presence of CHX. Taken together, these data indicate that E2F-2 and E2F-3 are unable to transactivate the endogenous CDC25A promoter, perhaps because of site-specific preferences. The observed transactivation of the CDC25A luciferase construct by E2F-2 and E2F-3 may be indirect and could be a consequence of E2F-2- or E2F-3-induced cell cycle progression that leads to upregulation of other transcription factors, including E2F-1.
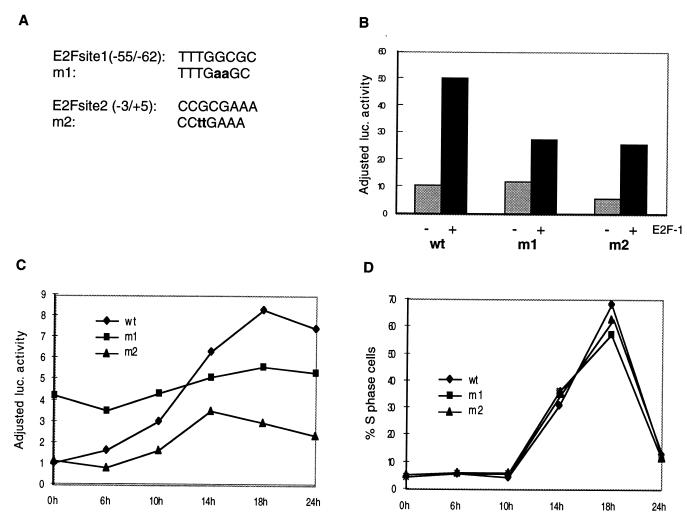
To test whether the transactivation of the CDC25A promoter by E2F-1 is dependent on the two identified putative E2F binding sites, these sites were mutated by PCR–site-directed mutagenesis (Fig. 10A). The different constructs were transfected into U2OS cells with and without an E2F-1 expression plasmid and pCMVβ-gal as a control for transfection efficiency. The transactivation of both mutants by E2F-1 was reduced compared to that of the wild-type promoter, suggesting that both sites can bind E2F-1 (Fig. 10B). In agreement with these results, gel retardation assays have shown that oligonucleotides containing either of the two sites can specifically bind E2F-containing complexes in vitro (data not shown). In contrast, the mutations introduced in m1 and m2 abolished the binding of E2F-containing complexes (data not shown). A construct with both E2F sites mutated was transfected into cells, and even though this construct does not contain any functional E2F DNA binding sites, its activity was slightly elevated after E2F-1 expression. These data suggest that there are other sites in the 1,178-bp construct which respond to cell cycle progression.
FIG. 10.
E2F-dependent cell cycle regulation of the CDC25A promoter. (A) Sequences of the putative E2F DNA binding sites in the CDC25A promoter. These sites were mutated to the indicated sequences by PCR–site-directed mutagenesis as described in Materials and Methods. (B) Both E2F DNA binding sites respond to E2F transactivation. Transfection of U2OS cells with CDC25A(−755/+423) luc (wild type) and mutants thereof, with (+) or without (−) pCMVE2F-1. The adjusted luciferase activities are the relative luciferase activities after normalization for the activity of cotransfected pCMVβ-gal. (C and D) The activity of the CDC25A promoter during the cell cycle. Rat1 cells transfected with CDC25A(−755/+422) luc wild type (wt), m1, or m2 were starved for 48 h and subsequently induced with serum. Samples were collected at different time points to evaluate the luciferase activity and the cell cycle profile as determined by FACS analysis. The adjusted luciferase activity in panel C indicates fold induction relative to the activity of the wild-type promoter at time 0 h, obtained by first normalizing the luciferase counts for the β-galactosidase activity of cotransfected pCMVβ-gal. In panel D are shown the percentages of S-phase cells for each time point. The data presented are representative of at least three different experiments.
Since we found that E2F can activate transcription from the CDC25A promoter, it was deemed interesting to determine the role of the E2F sites in the CDC25A promoter during the cell cycle. To this end, Rat1 cells were transiently transfected with the wild type or one of the two mutant constructs. Cotransfection was performed with a β-galactosidase-expressing plasmid. Next, cells were starved for 48 h in low-serum medium and subsequently stimulated with high-serum medium for different lengths of time before lysates were prepared for luciferase and β-galactosidase assays. Figure 10C shows that there was a significant upregulation of luciferase activity from the wild-type construct 10 to 15 h after serum stimulation (late G1-early S) (Fig. 10D), which corresponds to the time of CDC25A mRNA upregulation observed after serum stimulation (Fig. 7A) (43). A less-pronounced upregulation was detected for the two mutant constructs. Our results suggest that the 5′ E2F site plays a crucial role in the negative regulation of the CDC25A promoter in arrested cells while the 3′ E2F site is more important for the transactivation of the promoter. Based on these data, we conclude that the two E2F DNA binding sites participate in the cell cycle-regulated expression of the CDC25A promoter.
CDC25A cooperates with cyclin E to induce S phase.
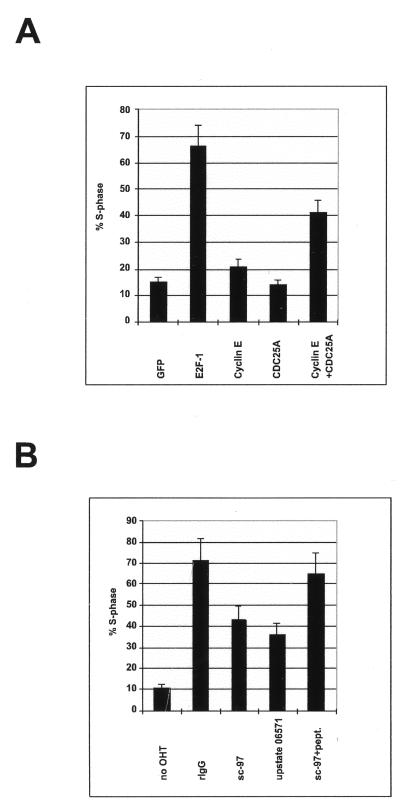
E2F activity can bypass the mitogen requirement for S-phase entry in rat fibroblasts. Since expression of none of the known E2F-regulated genes is sufficient to relieve the mitogen requirement for S-phase entry in serum-starved cells, it is not known how the E2Fs induce entry into S phase. Previous data have demonstrated that CDC25A is essential for entry into S phase (36, 43). CDC25A is a tyrosine phosphatase, and one of its activities is the removal of an inhibitory phosphate molecule from the G1 cyclin-dependent kinases (reviewed in reference 15). Studies of Drosophila genetics have demonstrated the requirement for cyclin E for E2F-induced S-phase entry (16, 17), and it has been shown that cyclin E is also required for entry into S phase (47, 70). Thus, it is very likely that cyclin E is also required for E2F-induced S-phase entry in mammalian cells. Previous publications have shown that cyclin E alone is not sufficient to induce S-phase entry in quiescent fibroblasts (69, 80). Cyclin E associates with CDK2, and the complex formed needs several modifications before it is active. Since CDK2 is present throughout the cell cycle, including quiescence (20, 83, 95), it is conceivable that the cyclin E-CDK2 complex is inactive as a result of CDC25A’s absence. To test whether CDC25A, alone or in combination with cyclin E, is sufficient to induce S-phase entry, serum-starved Rat1 fibroblasts were microinjected with plasmids expressing cyclin E or CDC25A, or with control plasmids (Fig. 11A). A plasmid expressing the green fluorescent protein was included in all microinjections to facilitate the identification of injected cells. After injection, the cells were cultured in medium containing BrdU, and cells that entered S phase were identified by using anti-BrdU antibodies. Injection of E2F-1 resulted in a significant number of cells undergoing DNA synthesis compared to control-injected cells. Expression of cyclin E or CDC25A alone was not sufficient to induce an increase in the number of S-phase cells compared to the control. However, when cyclin E and CDC25A were coexpressed, a significant increase in the number of S-phase cells was observed (Fig. 11A). This result is in agreement with recently published data demonstrating that performed active cyclin E-CDK2 or cyclin D1-CDK4 is sufficient to induce DNA synthesis when microinjected into quiescent cells (9). In contrast to the previously published data, our data demonstrate that two E2F targets, when coexpressed, are sufficient to initiate DNA synthesis in quiescent fibroblasts. Our data also demonstrate that coexpression of CDC25A and cyclin E is not as efficient at inducing S-phase entry as expression of E2F-1 alone, suggesting that other targets of E2F-1 are also limiting for progression through the G1 phase of the cell cycle.
FIG. 11.
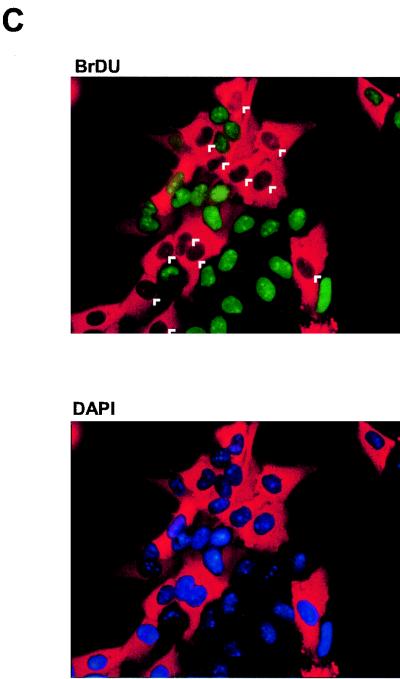
CDC25A cooperates with cyclin E in inducing S phase and is required for efficient E2F-1-induced S-phase entry. (A) Cyclin E and CDC25A cooperate to induce S phase in serum-starved Rat1 fibroblasts. E2F-1, cyclin E, CDC25A, or cyclin E plus CDC25A expression vectors (20 ng/μl each) were injected into serum-starved Rat1 fibroblasts. pCMVEGFP (100 ng/μl) was coinjected as an injection marker. At 16 h after microinjection, the percentage of BrdU-positive microinjected cells was determined. GFP, green fluorescent protein. (B) CDC25A is necessary for efficient E2F-1-induced S-phase entry. Rat1 ERE2F-1 cells were incubated in DMEM containing 0.1% serum for 48 h. The indicated antibodies were then microinjected (300 ng/ml) along with rabbit IgG as a microinjection marker (2 μg/μl). At 3 h after microinjection, OHT was added (300 nM) to induce E2F-1 activity. At 12 h after the induction of E2F-1, cells were harvested and processed for immunofluorescence analysis, and the percentage of BrdU in microinjected cells was determined. rIgG, rabbit IgG; pept., peptide. (C) An example of an antibody microinjection experiment. The percentage of BrdU in injected cells is lower in samples injected with antibodies against CDC25A.
CDC25A is required for efficient E2F-induced S-phase entry.
Since the results obtained so far suggest that CDC25A is a critical target for E2F-1 and that coexpression of CDC25A with another E2F target gene is sufficient to induce entry into S phase, we wanted to assess whether CDC25A is required for E2F-1-induced S-phase entry. Quiescent Rat1 cells expressing ERE2F-1 were microinjected with two affinity-purified polyclonal antibodies to CDC25A and the appropriate controls (Fig. 11B). Rabbit IgG was included in all injected samples to facilitate the identification of injected cells. After injection, cells were incubated in medium containing OHT and BrdU for 12 h. Cells undergoing DNA synthesis were identified with an anti-BrdU antibody (Fig. 11C). As shown in Fig. 11B, 72% of control-injected ERE2F-1-expressing cells entered S phase within the first 12 h after addition of OHT, whereas microinjection of the two affinity-purified antibodies to CDC25A significantly decreased (by 50%) the ability of E2F-1 to induce DNA synthesis. Moreover, preincubation of one of these antibodies with the antigenic peptide blocked the ability of the antibody to prevent E2F-1-induced S-phase entry. These data demonstrate that CDC25A is required for efficient induction of S-phase entry by E2F-1.
DISCUSSION
Elimination of cell cycle control mechanisms is one of the key features of human cancer. In particular, genes in the pRB pathway are very frequently found to be mutated. Several data have identified the E2F transcription factors as key downstream effectors in the pRB pathway. In agreement with this, ectopic expression of members of the E2F family recapitulates the phenotypes associated with the loss of function of the pRB pathway. To understand the underlying molecular mechanisms by which the E2Fs influence the cell phenotype, it is essential to identify the genes that are directly regulated by the E2Fs, i.e., what the primary targets are and how their deregulated expression results in hyperproliferation.
E2F target genes.
To identify the primary targets of the E2F transcription factors, we have developed cell lines expressing E2F-1, E2F-2, or E2F-3 fused to a modified version of the estrogen receptor ligand binding domain (ER). The activation of the E2Fs by OHT demonstrated that the ERE2F fusion proteins possess the expected biochemical features, including the abilities to transactive E2F-dependent synthetic promoters and to induce S-phase entry and apoptosis. Moreover, these changes were found to be dependent on the ability of E2F to bind to DNA but independent of pRB binding. Based on these results, we concluded that activation of the ERE2F fusion proteins by OHT is a reliable system to study the effects of deregulated E2F expression on gene activation and cell proliferation.
By employing a sensitive semiquantitative RT-PCR technique, we tested whether a series of genes containing E2F DNA binding sites in their promoters are transactivated by the E2Fs in the absence of de novo protein synthesis. Indeed, we found that several of the genes containing E2F DNA binding sites are very good direct targets for the E2F transcription factors. These genes encode proteins that are directly involved in regulating the initiation of DNA replication (CDC6), cell cycle control (cyclin E, cyclin A, and CDC25A), and growth control (B-myb, p107, and E2F-1). Several results indicate that these genes are direct targets of the E2Fs. First, all of these genes contain E2F DNA binding sites in their promoters, and transient-transfection experiments have shown that they respond to E2F overexpression (references 6, 26, 29, 40, 50, 67, 85, 100, and 102 and this report). Second, these genes are all induced in mid- to late G1 phase of the cell cycle (Fig. 3A), corresponding to the time at which pRB is phosphorylated (3, 88). Third, the kinetics of induction of these genes is very rapid after OHT addition (Fig. 3A and 7B). Fourth, the activation of these genes occurs in the absence of protein synthesis.
Interestingly, we also found that several genes that had been described as being regulated by the E2Fs were only mildly affected or not affected at all by (direct) E2F activity (5, 11, 14, 34, 60, 72, 90, 92, 93). There are numerous possible reasons for the lack of an effect of E2F on these genes, but one consideration is of particular interest. The experiments demonstrating the functional role of the E2F DNA binding site in these genes have all involved transient-expression assays that take the promoter out of its chromosomal context. Since the accessibility of DNA binding sites for transcription factors is to a large degree regulated by chromatin structure (see, e.g., reference 71), it is likely that the untimely expression of the E2Fs does not allow binding to a consensus E2F DNA binding element. Moreover, despite the presence of an E2F DNA binding consensus site, it may not be occupied due to interference by, for instance, other transcription factors. In agreement with such a notion is the finding that a previously identified E2F consensus site in the CDC2 promoter is never occupied during the mammalian cell cycle, as determined by in vivo genomic footprinting (11, 93).
Previously, two alternative strategies for analyzing the effects of deregulated E2F activity on genes containing E2F DNA binding sites have been described. One approach involved infecting serum-starved REF52 rat cells with recombinant adenoviruses expressing the various members of the E2F family (12, 13, 54). The expression of genes containing E2F DNA binding sites was analyzed 10 to 20 h after infection of the REF52 cells, and therefore it was not possible to analyze the primary effects of E2F expression on target genes with this assay. Thus, in agreement with results demonstrating that adenovirus-expressed E2Fs induce S-phase entry, it was observed that all cell cycle-regulated genes containing E2F DNA binding sites are indeed induced by E2F expression (see Fig. 2A in reference 13 and Fig. 1B in reference 54). In agreement with this result, we also found that addition of OHT to the ERE2F-expressing Rat1 cell lines induced upregulation of all cell cycle-regulated genes tested at later time points (Fig. 3A; compare 0 and 16 h).
In the other published approach, which more indirectly evaluates the effect of deregulated E2F expression, the cell cycle-regulated expression of genes containing E2F binding sites in mouse embryo fibroblasts (MEFs) derived from mice with targeted disruption of Rb or both p107 and p130 was analyzed (33, 41, 73). In accordance with our results, the loss of pRb in the established MEFs led to deregulated expression of the genes encoding cyclins E and A and of p107, whereas the loss of p107 and p130 led to deregulated expression of B-myb, cdc2, the cyclin A gene, E2f-1, and TS. Furthermore, in agreement with our results, no change in expression was observed for DHFR, TK, the DNA polymerase α gene, c-myc, PCNA, the cyclin D1 gene, or cdc25A. Although these results are surprisingly similar to ours, there are some unexpected differences. It is widely believed that E2F-1, E2F-2, and E2F-3 are specifically regulated by pRB (18, 53), and although our data show that B-myb is upregulated after expression of E2F-1, E2F-2, or E2F-3, such deregulation is not observed in Rb−/− MEFs. In addition, the loss of pRB is expected to lead to deregulation of E2F-1, E2F-2, and E2F-3 activities, which we and others have shown is sufficient to induce S-phase entry in quiescent cells (see the introduction). However, it is clear from the analyses of the Rb−/− MEFs that they can be made quiescent by serum starvation. We don’t know the reasons for these unexpected differences, but apart from the different cell lines used and E2F expression levels obtained, there are other possible explanations for the discrepancies. Many cell divisions are required for the establishment of MEFs, and the adaptive processes during early development may modulate the effects of any genetic mutation introduced into the embryo. Although it is clearly difficult to analyze for all possible genetic alterations and differences in gene expression profiles, it is evident that the Rb−/− MEFs express higher levels of p107 than wild-type MEFs. It is very likely that this higher level of p107 is sufficient to suppress the expression of B-myb (50), and it has been shown that p107 blocks E2F-1-induced S-phase entry (101).
Our results also demonstrate that E2F-1 is more active in inducing transcription of genes containing E2F DNA binding sites than is E2F-3, which is again more active than E2F-2. This hierarchy of activity was unexpected since E2F-1, E2F-2, and E2F-3 induce S-phase entry and apoptosis in Rat1 cells to similar extents and since E2F-1 and E2F-3 are equally good at inducing transcription from a synthetic promoter. Moreover, we have shown that the newly identified target for E2F-1, CDC25A (see further below), is not transactivated by E2F-2 or E2F-3 and that CDC6 is not a direct target of E2F-2. It is noteworthy, however, that CDC25A and CDC6 expression is increased in ERE2F-2 and ERE2F-3 OHT-treated cells before entry into S phase of the cell cycle, suggesting that the expression of other transcription factors as a consequence of E2F-2 or E2F-3 activation transactivates CDC25A and CDC6. A possible explanation for this observation is that the E2F-1 promoter contains two E2F DNA binding sites and is transactivated by E2F-1, E2F-2, and E2F-3 (38, 44, 64). Moreover, we have observed that the activation of E2F-3 is sufficient to transactivate the E2F-1 promoter in the absence of protein synthesis (Fig. 6) and that the activation of E2F-1, E2F-2, and E2F-3 by OHT leads to higher levels of the E2F-1 transcript shortly after addition of OHT (75a). Since CDC25A and CDC6 are both essential for entry into S phase (29, 36, 43, 100), these data may suggest that E2F-2- or E2F-3-induced S-phase entry is dependent on functional E2F-1. However, it is important to emphasize that at this stage we are only in the position to make conclusions about what happens in our Rat1 cell lines, and that the regulation of CDC25A by the different members of the E2F family should be studied in other cell lines and by other methods before firm conclusions are made. Moreover, there exists the formal possibility that CDC25A transcription is elevated due to derepression of the promoter, by a mechanism in which E2F-1 replaces a preexisting E2F-repressive complex on the promoter. In support of such a model is a recent report (published while this paper was undergoing review) that independently identified CDC25A as an E2F-responsive promoter and in which it was found that E2F-4/p130 is bound to the 5′ repressive element (41a) (Fig. 10A). However, we do not favor such a model, since E2F-2 and E2F-3 apparently do not lead to transactivation of the promoter and since we have not seen any measurable effects on cell cycle progression by using a protein in which a transactivation-deficient, but DNA-binding-capable, mutant of E2F-1 (amino acids 1 to 374) was fused to the estrogen receptor ligand binding domain (data not shown).
CDC25A is an E2F target.
To date, no systematic screen for E2F-regulated genes has been published. We believe that the ERE2F-expressing cell lines are excellent tools for performing such a screen. Meanwhile, however, we have used the ERE2F-expressing cell lines to test whether known cell cycle-regulated genes are induced by the E2Fs in the absence of protein synthesis. By performing such assays, we identified CDC25A as a direct target of E2F-1. The cloning and sequencing of the promoter identified two E2F DNA binding sites that can be transactivated by E2F-1 expression. Mutational analyses showed that the upstream-most E2F DNA binding site is essential for proper cell cycle regulation of the promoter whereas the downstream site appears to be important for the overall activity of the CDC25A promoter. To our surprise, our data suggest that CDC25A is a direct target of E2F-1 but not of E2F-2 or E2F-3. The reason for this apparent specificity is presently unknown. No data showing differences in the binding site specificities of E2F-1, E2F-2, and E2F-3 have so far been published. Moreover, our unpublished results suggest that the two E2F DNA binding sites can bind to E2F-1, E2F-2, and E2F-3 in vitro (95a), indicating that other factors implicated in regulating CDC25A may be important for the specificity of the transactivation. Future work will be required to investigate the molecular and cellular basis for the lack of CDC25A transactivation by E2F-2 and E2F-3 in our system.
The identification of CDC25A as an E2F target is interesting for several reasons. First, CDC25A is a tyrosine phosphatase whose activity is believed to be essential for the activation of the G1 cyclin-dependent kinases (15). Second, CDC25A is essential for entry into the S phase of the cell cycle (36, 43). Third, CDC25A has oncogenic properties, and it is overexpressed in human primary cancers (24, 97). Fourth, CDC25A has been described as an essential downstream target of c-Myc (23). In agreement with this proposed central role for CDC25A in the regulation of cell proliferation, we have shown that CDC25A activity is required for E2F-1-induced S-phase entry and that CDC25A can cooperate with cyclin E (another target of E2F-1) to induce entry into S phase.
In contrast to the modest CDC25A induction as a consequence of c-Myc activation (23), we observe a robust immediate transactivation by E2F-1. Indeed, a direct comparison of levels of CDC25A induction after c-Myc activation and after E2F-1 activation showed that CDC25A is only slightly elevated by c-Myc compared with E2F-1 (95a). Moreover, CDC25A expression is elevated in mid- to late G1 after serum stimulation, with kinetics that are in good agreement with it being a target gene for the E2F transcription factors (43) (Fig. 7A). In contrast, c-myc expression is elevated in early G1, and it is therefore expected that c-Myc target genes are induced in that period of the cell cycle. Finally, we show that the cell cycle-regulated expression of CDC25A is dependent on E2F, and we suggest that CDC25A is a bona fide E2F target gene which may, to a certain extent, also be regulated by c-Myc.
E2F-induced S-phase entry.
Deregulated expression of E2F-1, E2F-2, or E2F-3 can relieve the mitogen requirement for entry into S phase in rodent fibroblasts. The expression of the E2Fs shortens the traversal of the G0-G1 phases by 6 to 7 h compared to that of serum-stimulated Rat1 fibroblasts. In agreement with this result, we find that the direct targets of E2F transcription factors are induced 6 to 7 h earlier by E2F activation than by serum activation (Fig. 2 and 7B). Interestingly, previous results have shown that expression of cyclin E or cyclin A, two of the downstream targets for E2F, can accelerate the entry into S phase by 2 to 3 h (69, 80, 81), suggesting that these proteins are implicated in the biological effects mediated by the E2Fs. Although cyclins E and A have been shown to shorten the traversal of G1 by 2 to 3 h, they are unable to relieve the mitogen requirement for entry into S phase (69, 80, 81). However, it was recently demonstrated that the injection of physiological levels of active G1 kinases into serum-starved fibroblasts is sufficient to induce S-phase entry (9). In agreement with and as an extension of these data, we have shown that coexpression of two downstream targets of the E2Fs, cyclin E and CDC25A, is sufficient to induce S phase in quiescent fibroblasts, suggesting that both these targets are limiting for entry into the S phase of the cell cycle. It is important to note, however, that the coexpression of CDC25A and cyclin E is not as efficient at inducing S phase as is the expression of the E2Fs, suggesting that other direct targets of the E2Fs are limiting for the entry into S phase. Recent data from our laboratory have shown that another E2F target, CDC6, is limiting for fast progression through G1 (29). Unfortunately, it has not been possible to detect CDC6 after its microinjection into quiescent cells (63a). Therefore, we have been unable to test whether coexpression of CDC6 with cyclin E and CDC25A will relieve the mitogen requirement for entry into S phase as efficiently as the E2Fs.
E2F-induced apoptosis.
Several experiments have suggested a role for E2F-1 in apoptosis. Ectopic expression of E2F-1 has been shown to lead to p53-dependent and -independent apoptosis in tissue culture cells and transgenic mice (37, 39, 49, 74, 77, 87, 98). Lack of E2f-1 in the developing mouse leads to a decrease in thymocyte apoptosis (21). E2F-1 expression has been shown to lead to increased levels of p53 (35, 48), which may be a result of induced expression of p19ARF (4, 13, 82). In agreement with a role for p53 in E2F-1 induced apoptosis, recent results have shown that the apoptotic effect of E2F-1 can be overcome by overexpression of MDM2 (48). Rb−/− mice undergo p53-dependent apoptosis in the central nervous system and in the developing eye lens and p53-independent apoptosis in the peripheral nervous system (58, 61). The induction of apoptosis in pRb-deficient mice appears to be mediated by E2f-1, since Rb−/− mice survive longer in an E2f-1−/− genetic background than in a wild-type genetic background, most likely due to a decrease in apoptosis in the central nervous system and the developing lens (94).
In contrast to the current assumption that it is only E2F-1 and not the other members of the E2F family that can induce apoptosis (13, 48; see also Discussion in reference 94), we have demonstrated in this article that E2F-2 and E2F-3 are also efficient inducers of apoptosis. Consistent with these results, we observed a high level of cell death and selection against cell proliferation in cells expressing active E2F-1, E2F-2, or E2F-3. We have observed that even in high-serum medium, induced expression of E2F-1, E2F-2, or E2F-3 in clonal U2OS cells leads to cell death (72a). The reason for the discrepancy between our results and those of others (13, 48) is presently unknown. However, it may be ascribed to different technical aspects, such as the use of different cell lines and assays. It should be noted, however, that in the assays described by others, apoptosis as measured by a sub-G1 peak is not observed prior to 3 to 4 days after infection with recombinant adenoviruses expressing E2F-1 (13, 48). Therefore, it appears that our assay conditions are more sensitive than those previously described, since we detected an efficient apoptotic response less than 24 h after E2F activation. Moreover, E2F-1 may be a more efficient inducer of apoptosis in some cells, and E2F-2 and E2F-3 may in fact be dependent on E2F-1 function to induce apoptosis (as they may be dependent on E2F-1 to induce S-phase entry). Such a model may, in fact, also provide an explanation for the efficient suppression of apoptosis in pRb-deficient mice lacking E2f-1 (94).
Conclusions.
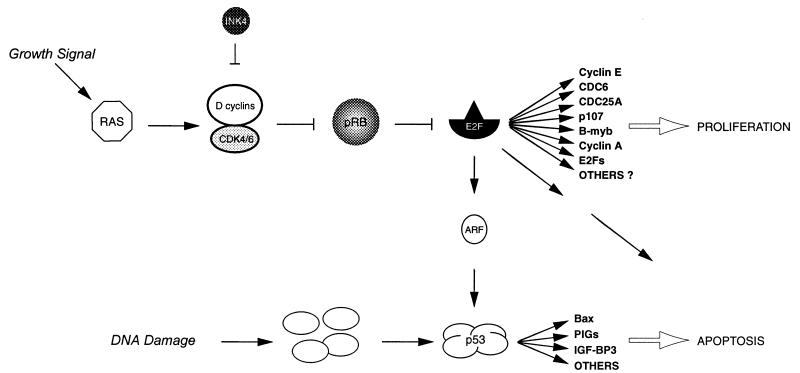
In Fig. 12, a model for the central role of the E2F transcription factors in regulating cell proliferation and apoptosis is presented. In this article we have identified several direct targets of the E2F transcription factors, including genes that are important for DNA replication, cell cycle control, and growth control. We have demonstrated that two of these genes (those encoding CDC25A and cyclin E) can cooperate in inducing S-phase entry in serum-starved cells. Furthermore, we have provided evidence that not only E2F-1 but also E2F-2 and E2F-3 expression leads to apoptosis. E2F-1 has been demonstrated to induce p53-dependent and -independent apoptosis, and it has been suggested that transactivation of ARF by the E2Fs provides the connection from the pRB pathway to the p53 pathway and the induction of apoptosis. The question, however, is what is mediating E2F-induced apoptosis. It is known that overexpression of ARF alone is not sufficient for the induction of apoptosis (78), suggesting that other targets of the E2F transcription factors are implicated in this event. Furthermore, several recent results have demonstrated that the activity of ARF is dependent on functional p53 (for a review, see reference 89), and it is therefore unclear how the E2Fs induce p53-independent apoptosis. The establishment of ERE2F-expressing cell lines provides an excellent tool for addressing these and other questions related to the role of the E2F transcription factors in cell cycle control.
FIG. 12.
Model for the central role of the E2F transcription factors in cell proliferation and apoptosis. The stimulation of resting cells with growth factors results in the induction of a signal cascade leading to the phosphorylation of the retinoblastoma protein (pRB) by D-type cyclins in association with CDK4 or CDK6, as well as the activation of the E2F transcription factors. The E2F transcription factors are essential for controlling several genes whose products are required for DNA replication, cell cycle progression, and/or growth. Deregulation of E2F activity as a result of upstream mutations in the pathway leads to premature entry into S phase of the cell cycle and—depending on the genetic status of the cell—to hyperproliferation or apoptosis. E2F-induced apoptosis occurs via p53-dependent and -independent pathways. One of the connections between the pRB and p53 pathways is provided by ARF, and the transactivation of ARF by the E2Fs may be involved in p53-dependent apoptosis. Presently we do not know which gene products are implicated in p53-independent E2F-induced apoptosis. PIGs, p53-induced genes; IGF-BP3, insulin growth factor-binding protein 3.
ACKNOWLEDGMENTS
We thank Karin Holm, Cristian Matteucci, Stefania Lupo, and Giuseppina Giardina for technical assistance in plasmid constructions, FACS analyses, and retroviral infections. We thank T. Littlewood for pBSKER, W. G. Kaelin for pSGE2F-1/VP16, and G. P. Nolan for Phoenix cells. We are grateful to the members of the Lattanzio family who donated the microinjection equipment. We thank Giulio Draetta and Nick Dyson for critical reading of the manuscript.
E.V. was supported by a fellowship from the Fondazione Italiana per la Ricerca sul Cancro, and H.M., G.H., and P.C. were supported in part by fellowships from the European Community’s TMR and Biomed 2 programs. This work was supported by grants from the Human Frontiers Science Program, the European Community’s TMR Programme, and the Associazione Italiana per la Ricerca sul Cancro (AIRC).
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1988. [Google Scholar]
- 2.Baker S J, Markowitz S, Fearon E R, Willson J K V, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 3.Bartek J, Bartkova J, Lukas J. The retinoblastoma protein pathway and the restriction point. Curr Opin Cell Biol. 1996;8:805–814. doi: 10.1016/s0955-0674(96)80081-0. [DOI] [PubMed] [Google Scholar]
- 4.Bates S, Phillips A C, Clark P A, Stott F, Peters G, Ludwig R L, Vousden K H. p14ARF links the tumour suppressors RB and p53. Nature. 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- 5.Blake M C, Azizkhan J C. Transcription factor E2F is required for efficient expression of the hamster dihydrofolate reductase gene in vitro and in vivo. Mol Cell Biol. 1989;9:4994–5002. doi: 10.1128/mcb.9.11.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Botz J, Zerfass-Thome K, Spitkovsky D, Delius H, Vogt B, Eilers M, Hatzigeorgiou A, Jansen-Dürr P. Cell cycle regulation of the murine cyclin E gene depends on an E2F binding site in the promoter. Mol Cell Biol. 1996;16:3401–3409. doi: 10.1128/mcb.16.7.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cartwright P, Müller H, Wagener C, Holm K, Helin K. E2F-6: a novel member of the E2F family is an inhibitor of E2F-dependent transcription. Oncogene. 1998;17:611–624. doi: 10.1038/sj.onc.1201975. [DOI] [PubMed] [Google Scholar]
- 8.Clarke A, Maandag E, van Roon M, van der Lugt N, van der Valk M, Hooper M, Berns A, te Riele H. Requirement for a functional Rb-1 gene in murine development. Nature. 1992;359:328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- 9.Connell-Crowley L, Elledge S J, Harper J W. G1 cyclin-dependent kinases are sufficient to initiate DNA synthesis in quiescent human fibroblasts. Curr Biol. 1998;8:65–68. doi: 10.1016/s0960-9822(98)70021-1. [DOI] [PubMed] [Google Scholar]
- 10.Cress W D, Johnson D G, Nevins J R. A genetic analysis of the E2F1 gene distinguishes regulation by Rb, p107, and adenovirus E4. Mol Cell Biol. 1993;13:6314–6325. doi: 10.1128/mcb.13.10.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalton S. Cell cycle regulation of the human cdc2 gene. EMBO J. 1992;11:1797–1804. doi: 10.1002/j.1460-2075.1992.tb05231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeGregori J, Kowalik T, Nevins J R. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeGregori J, Leone G, Miron A, Jakoi L, Nevins J R. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci USA. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dou Q-P, Markell P J, Pardee A B. Thymidine kinase transcription is regulated at G1/S phase by a complex that contains retinoblastoma-like protein and a cdc2 kinase. Proc Natl Acad Sci USA. 1992;89:3256–3260. doi: 10.1073/pnas.89.8.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Draetta G, Eckstein J. Cdc25 protein phosphatases in cell proliferation. Biochim Biophys Acta. 1997;1332:M53–M63. doi: 10.1016/s0304-419x(96)00049-2. [DOI] [PubMed] [Google Scholar]
- 16.Duronio R J, Brook A, Dyson N, O’Farrell P H. E2F-induced S phase requires cyclin E. Genes Dev. 1996;10:2505–2513. doi: 10.1101/gad.10.19.2505. [DOI] [PubMed] [Google Scholar]
- 17.Duronio R J, O’Farrell P H. Developmental control of the G1 to S transition in Drosophila: cyclin E is a limiting downstream target of E2F. Genes Dev. 1995;9:1456–1468. doi: 10.1101/gad.9.12.1456. [DOI] [PubMed] [Google Scholar]
- 18.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 19.Eilers M, Picard D, Yamamoto K R, Bishop J M. Chimaeras of myc oncoprotein and steroid receptors cause hormone-dependent transformation of cells. Nature. 1989;340:66–68. doi: 10.1038/340066a0. [DOI] [PubMed] [Google Scholar]
- 20.Elledge S J, Richman R, Hall F L, Williams R T, Lodgson N, Harper J W. CDK2 encodes a 33-kDa cyclin A-associated protein kinase and is expressed before CDC2 in the cell cycle. Proc Natl Acad Sci USA. 1992;89:2907–2911. doi: 10.1073/pnas.89.7.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Field S J, Tsai F Y, Kuo F, Zubiaga A M, Kaelin W G, Jr, Livingston D M, Orkin S H, Greenberg M E. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- 22.Galaktionov K, Beach D. Specific activation of cdc25 tyrosine phosphatases by B-type cyclins: evidence for multiple roles of mitotic cyclins. Cell. 1991;67:1181–1194. doi: 10.1016/0092-8674(91)90294-9. [DOI] [PubMed] [Google Scholar]
- 23.Galaktionov K, Chen X, Beach D. Cdc25 cell-cycle phosphatase as a target of c-myc. Nature. 1996;382:511–517. doi: 10.1038/382511a0. [DOI] [PubMed] [Google Scholar]
- 24.Galaktionov K, Lee A K, Eckstein J, Draetta G, Meckler J, Loda M, Beach D. Cdc25 phosphatases as potential human oncogenes. Science. 1995;269:1575–1577. doi: 10.1126/science.7667636. [DOI] [PubMed] [Google Scholar]
- 25.Gaubatz S, Wood J G, Livingston D M. Unusual proliferation arrest and transcriptional control properties of a newly discovered E2F family member, E2F-6. Proc Natl Acad Sci USA. 1998;95:9190–9195. doi: 10.1073/pnas.95.16.9190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geng Y, Eaton E N, Picón M, Roberts J M, Lundberg A S, Gifford A, Sardet C, Weinberg R A. Regulation of cyclin E transcription by E2Fs and retinoblastoma protein. Oncogene. 1996;12:1173–1180. [PubMed] [Google Scholar]
- 27.Guy C T, Zhou W, Kaufman S, Robinson M O. E2F-1 blocks terminal differentiation and causes proliferation in transgenic megakaryocytes. Mol Cell Biol. 1996;16:685–693. doi: 10.1128/mcb.16.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 29.Hateboer G, Wobst A, Petersen B O, Le Cam L, Vigo E, Sardet C, Helin K. Cell cycle-regulated expression of mammalian CDC6 is dependent on E2F. Mol Cell Biol. 1998;18:6679–6697. doi: 10.1128/mcb.18.11.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helin K. Regulation of cell proliferation by the E2F transcription factors. Curr Opin Genet Dev. 1998;8:28–35. doi: 10.1016/s0959-437x(98)80058-0. [DOI] [PubMed] [Google Scholar]
- 31.Helin K, Harlow E. Heterodimerization of the transcription factors E2F-1 and DP-1 is required for binding to the adenovirus E4 (ORF6/7) protein. J Virol. 1994;68:5027–5035. doi: 10.1128/jvi.68.8.5027-5035.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Helin K, Wu C-L, Fattaey A R, Lees J A, Dynlacht B D, Ngwu C, Harlow E. Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative transactivation. Genes Dev. 1993;7:1850–1861. doi: 10.1101/gad.7.10.1850. [DOI] [PubMed] [Google Scholar]
- 33.Herrera R E, Sah V P, Williams B O, Mäkelä T P, Weinberg R A, Jacks T. Altered cell cycle kinetics, gene expression, and G1 restriction point regulation in Rb-deficient fibroblasts. Mol Cell Biol. 1996;16:2403–2407. doi: 10.1128/mcb.16.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hiebert S W, Lipp M, Nevins J R. E1A-dependent trans-activation of the human MYC promoter is mediated by the E2F factor. Proc Natl Acad Sci USA. 1989;86:3594–3598. doi: 10.1073/pnas.86.10.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hiebert S W, Packham G, Strom D K, Haffner R, Oren M, Zambetti G, Cleveland J L. E2F-1:DP-1 induces p53 and overrides survival factors to trigger apoptosis. Mol Cell Biol. 1995;15:6864–6874. doi: 10.1128/mcb.15.12.6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffmann I, Draetta G, Karsenti E. Activation of the phosphatase activity of human cdc25A by a cdk2-cyclin E dependent phosphorylation at the G1/S transition. EMBO J. 1994;13:4302–4310. doi: 10.1002/j.1460-2075.1994.tb06750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holmberg C, Helin K, Sehested M, Karlström O. E2F-1 induced p53-independent apoptosis in transgenic mice. Oncogene. 1998;17:143–155. doi: 10.1038/sj.onc.1201915. [DOI] [PubMed] [Google Scholar]
- 38.Hsiao K-M, McMahon S L, Farnham P J. Multiple DNA elements are required for the growth regulation of the mouse E2F1 promoter. Genes Dev. 1994;8:1526–1537. doi: 10.1101/gad.8.13.1526. [DOI] [PubMed] [Google Scholar]
- 39.Hsieh J-K, Fredersdorf S, Kouzarides T, Martin K, Lu X. E2F1-induced apoptosis requires DNA binding but not transactivation and is inhibited by the retinoblastoma protein through direct interaction. Genes Dev. 1997;11:1840–1852. doi: 10.1101/gad.11.14.1840. [DOI] [PubMed] [Google Scholar]
- 40.Huet X, Rech J, Plet A, Vié A, Blanchard J M. Cyclin A expression is under negative transcriptional control during the cell cycle. Mol Cell Biol. 1996;16:3789–3798. doi: 10.1128/mcb.16.7.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hurford R K J, Cobrinik D, Lee M-H, Dyson N. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- 41a.Iavarone A, Massagué J. E2F and histone deacetylase mediate transforming growth factor β repression and cdc25A during keratinocyte cell cycle arrest. Mol Cell Biol. 1999;19:916–922. doi: 10.1128/mcb.19.1.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacks T, Fazeli A, Schmitt E, Bronson R, Goodell M, Weinberg R. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 43.Jinno S, Suto K, Nagata A, Igarashi M, Kanaoka Y, Nojima H, Okayama H. Cdc25A is a novel phosphatase functioning early in the cell cycle. EMBO J. 1994;13:1549–1556. doi: 10.1002/j.1460-2075.1994.tb06417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson D G, Ohtani K, Nevins J R. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 45.Johnson D G, Schwarz J K, Cress W D, Nevins J R. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 46.Kakizuka A, Sebastian B, Borgmeyer U, Hermans-Borgmeyer I, Bolado J, Hunter T, Hoekstra M F, Evans R M. A mouse cdc25 homolog is differentially and developmentally expressed. Genes Dev. 1992;6:578–590. doi: 10.1101/gad.6.4.578. [DOI] [PubMed] [Google Scholar]
- 47.Knoblich J A, Sauer K, Jones L, Richardson H, Saint R, Lehner C F. Cyclin E controls S phase progression and its downregulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell. 1994;77:107–120. doi: 10.1016/0092-8674(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 48.Kowalik T F, DeGregori J, Leone G, Jakoi L, Nevins J R. E2F1-specific induction of apoptosis and p53 accumulation which is blocked by MDM2. Cell Growth Differ. 1998;9:113–118. [PubMed] [Google Scholar]
- 49.Kowalik T F, DeGregori J, Schwarz J K, Nevins J R. E2F1 overexpression in quiescent fibroblasts leads to induction of cellular DNA synthesis and apoptosis. J Virol. 1995;69:2491–2500. doi: 10.1128/jvi.69.4.2491-2500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lam E W-F, Watson R J. An E2F-binding site mediates cell-cycle regulated repression of mouse B-myb transcription. EMBO J. 1993;12:2705–2713. doi: 10.1002/j.1460-2075.1993.tb05932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee E Y-H P, Chang C-Y, Hu N, Wang Y-C J, Lai C-C, Herrup K, Lee W-H, Bradley A. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- 52.Lee E Y-H P, Hu N, Yuan S-S F, Cox L A, Bradley A, Lee W-H, Herrup K. Dual roles of the retinoblastoma protein in cell cycle regulation and neuron differentiation. Genes Dev. 1994;8:2008–2021. doi: 10.1101/gad.8.17.2008. [DOI] [PubMed] [Google Scholar]
- 53.Lees J A, Saito M, Vidal M, Valentine M, Look T, Harlow E, Dyson N, Helin K. The retinoblastoma protein binds to a family of E2F transcription factors. Mol Cell Biol. 1993;13:7813–7825. doi: 10.1128/mcb.13.12.7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams R S, Nevins J R. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 1998;12:2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 56.Littlewood T D, Hancock D C, Daniellan P S, Parker M G, Evan G I. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lukas J, Petersen B O, Holm K, Bartek J, Helin K. Deregulated expression of E2F family members induces S-phase entry and overcomes p16INK4A-mediated growth suppression. Mol Cell Biol. 1996;16:1047–1057. doi: 10.1128/mcb.16.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Macleod K F, Hu Y, Jacks T. Loss of Rb activates both p53-dependent and independent cell death pathways in the developing mouse nervous system. EMBO J. 1996;15:6178–6188. [PMC free article] [PubMed] [Google Scholar]
- 59.Matsushime H, Roussel M F, Ashmun R A, Sherr C J. Colony-stimulating factor 1 regulates novel cyclins during the G1 phase of the cell cycle. Cell. 1991;65:701–713. doi: 10.1016/0092-8674(91)90101-4. [DOI] [PubMed] [Google Scholar]
- 60.Means A L, Slansky J E, McMahon S L, Knuth M W, Farnham P J. The HIP1 binding site is required for growth regulation of the dihydrofolate reductase gene promoter. Mol Cell Biol. 1992;12:1054–1063. doi: 10.1128/mcb.12.3.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morgenbesser S D, Williams B O, Jacks T, DePinho R A. p53-dependent apoptosis produced by pRb-deficiency in the developing mouse lens. Nature. 1994;371:72–74. doi: 10.1038/371072a0. [DOI] [PubMed] [Google Scholar]
- 62.Morgenstern J, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62a.Müller, H. Unpublished data.
- 63.Müller H, Moroni M C, Vigo E, Petersen B O, Bartek J, Helin K. Induction of S-phase entry by E2F transcription factors depends on their nuclear localization. Mol Cell Biol. 1997;17:5508–5520. doi: 10.1128/mcb.17.9.5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63a.Müller, H., B. O. Petersen, J. Lukas, and K. Helin. Unpublished data.
- 64.Neuman E, Flemington E K, Sellers W R, Kaelin W G., Jr Transcription of the E2F-1 gene is rendered cell cycle dependent by E2F DNA-binding sites within its promoter. Mol Cell Biol. 1994;14:6607–6615. doi: 10.1128/mcb.14.10.6607. . (Authors’ correction, 15:4660, 1995.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nicoletti I, Migliorati G, Pagliacci M C, Grignani F, Riccardi C. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J Immunol Methods. 1991;139:271–279. doi: 10.1016/0022-1759(91)90198-o. [DOI] [PubMed] [Google Scholar]
- 66.Ohtani K, DeGregori J, Leone G, Herendeen D R, Kelly T J, Nevins J R. Expression of the HsOrc1 gene, a human ORC1 homolog, is regulated by cell proliferation via the E2F transcription factor. Mol Cell Biol. 1996;16:6977–6984. doi: 10.1128/mcb.16.12.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ohtani K, DeGregori J, Nevins J R. Regulation of the cyclin E gene by transcription factor E2F1. Proc Natl Acad Sci USA. 1995;92:12146–12150. doi: 10.1073/pnas.92.26.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ohtani K, Tsujimoto A, Ikeda M, Nakamura M. Regulation of cell growth-dependent expression of mammalian CDC6 gene by the cell cycle transcription factor E2F. Oncogene. 1998;17:1777–1785. doi: 10.1038/sj.onc.1202105. [DOI] [PubMed] [Google Scholar]
- 69.Ohtsubo M, Roberts J M. Cyclin-dependent regulation of G1 in mammalian fibroblasts. Science. 1993;259:1908–1912. doi: 10.1126/science.8384376. [DOI] [PubMed] [Google Scholar]
- 70.Ohtsubo M, Theodoras A M, Schumacher J, Roberts J M, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol Cell Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pazin M J, Kadonaga J T. What’s up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 72.Pearson B E, Nasheuer H-P, Wang T S-F. Human DNA polymerase α gene: sequences controlling expression in cycling and serum-stimulated cells. Mol Cell Biol. 1991;11:2081–2095. doi: 10.1128/mcb.11.4.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72a.Petersen, B. O., and K. Helin. Unpublished results.
- 73.Philips A, Huet X, Plet A, Cam L L, Vié A, Blanchard J M. The retinoblastoma protein is essential for cyclin A repression in quiescent cells. Oncogene. 1998;16:1373–1381. doi: 10.1038/sj.onc.1201655. [DOI] [PubMed] [Google Scholar]
- 74.Phillips A C, Bates S, Ryan K M, Helin K, Vousden K. Induction of DNA synthesis and apoptosis are separable functions of E2F-1. Genes Dev. 1997;11:1853–1863. doi: 10.1101/gad.11.14.1853. [DOI] [PubMed] [Google Scholar]
- 75.Picard D, Salser S J, Yamamoto K R. A movable and regulable inactivation function within the steroid binding domain of the glucocorticoid receptor. Cell. 1988;54:1073–1080. doi: 10.1016/0092-8674(88)90122-5. [DOI] [PubMed] [Google Scholar]
- 75a.Prosperini, E., E. Vigo, and K. Helin. Unpublished results.
- 76.Qin X-Q, Livingston D M, Ewen M, Sellers W R, Arany Z, Kaelin W G., Jr The transcription factor E2F-1 is a downstream target for RB action. Mol Cell Biol. 1995;15:742–755. doi: 10.1128/mcb.15.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Qin X-Q, Livingston D M, Kaelin W G, Adams P. Deregulated E2F1 expression leads to S-phase entry and p53-mediated apoptosis. Proc Natl Acad Sci USA. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Quelle D E, Zindy F, Ashmun R A, Sherr C J. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 79.Reichmann E, Schwarz H, Deiner E M, Leitner I, Eilers M, Berger J, Busslinger M, Beug H. Activation of an inducible c-FosER fusion protein causes loss of epithelial polarity and triggers epithelial-fibroblastoid cell conversion. Cell. 1992;71:1103–1116. doi: 10.1016/s0092-8674(05)80060-1. [DOI] [PubMed] [Google Scholar]
- 80.Resnitzky D, Gossen M, Bujard H, Reed S I. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol Cell Biol. 1994;14:1669–1679. doi: 10.1128/mcb.14.3.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Resnitzky D, Hengst L, Reed S I. Cyclin A-associated kinase activity is rate limiting for entrance into S phase and is negatively regulated in G1 by p27Kip1. Mol Cell Biol. 1995;15:4347–4352. doi: 10.1128/mcb.15.8.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Robertson K D, Jones P A. The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and down-regulated by wild-type p53. Mol Cell Biol. 1998;18:6457–6473. doi: 10.1128/mcb.18.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rosenblatt J, Gu Y, Morgan D O. Human cyclin-dependent kinase 2 is activated during the S and G2 phases of the cell cycle and associate with cyclin A. Proc Natl Acad Sci USA. 1992;89:2824–2828. doi: 10.1073/pnas.89.7.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Samuels M L, Weber M J, Bishop J M, McMahon M. Conditional transformation of cells and rapid activation of the mitogen-activated protein kinase cascade by an estradiol-dependent human Raf-1 protein kinase. Mol Cell Biol. 1993;13:6241–6252. doi: 10.1128/mcb.13.10.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schulze A, Zerfass K, Spitkovsky D, Middendorp S, Berges J, Helin K, Jansen Dürr P, Henglein B. Cell cycle regulation of the cyclin A gene promoter is mediated by a variant E2F site. Proc Natl Acad Sci USA. 1995;92:11264–11268. doi: 10.1073/pnas.92.24.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sears R, Ohtani K, Nevins J R. Identification of positively and negatively acting elements regulating expression of the E2F2 gene in response to cell growth signals. Mol Cell Biol. 1997;17:5227–5235. doi: 10.1128/mcb.17.9.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shan B, Lee W-H. Deregulated expression of E2F-1 induces S-phase entry and leads to apoptosis. Mol Cell Biol. 1994;14:8166–8173. doi: 10.1128/mcb.14.12.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 89.Sherr C J. Tumor surveillance via ARF-p53 pathway. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 90.Slansky J E, Li Y, Kaelin W G, Farnham P J. A protein synthesis-dependent increase in E2F1 mRNA correlates with growth regulation of the dihydrofolate reductase promoter. Mol Cell Biol. 1993;13:1610–1618. doi: 10.1128/mcb.13.3.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Slansky J E, Farnham P J. Introduction to the E2F family: protein structure and gene regulation. Curr Top Microbiol Immunol. 1996;208:1–30. doi: 10.1007/978-3-642-79910-5_1. [DOI] [PubMed] [Google Scholar]
- 92.Thalmeier K, Synovzik H, Mertz R, Winnacker E-L, Lipp M. Nuclear factor E2F mediates basic transcription and trans-activation by E1a of the human MYC promoter. Genes Dev. 1989;3:527–536. doi: 10.1101/gad.3.4.527. [DOI] [PubMed] [Google Scholar]
- 93.Tommasi S, Pfeifer G P. In vivo structure of the human cdc2 promoter: release of a p130–E2F-4 complex from sequences immediately upstream of the transcription initiation site coincides with induction of cdc2 expression. Mol Cell Biol. 1995;15:6901–6913. doi: 10.1128/mcb.15.12.6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tsai K Y, Hu Y, Macleod K F, Crowley D, Yamasaki L, Jacks T. Mutation of E2F1 suppresses apoptosis and inappropriate S-phase entry and extends survival of Rb-deficient mouse embryos. Mol Cell. 1998;2:293–304. doi: 10.1016/s1097-2765(00)80274-9. [DOI] [PubMed] [Google Scholar]
- 95.Tsai L-H, Lees E, Faha B, Harlow E, Riabowol K. The cdk2 kinase is required for the G1-to-S transition in mammalian cells. Oncogene. 1993;8:1593–1602. [PubMed] [Google Scholar]
- 95a.Vigo, E., and K. Helin. Unpublished results.
- 96.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 97.Wu W, Fan Y-H, Kemp B L, Walsh G, Mao L. Overexpression of cdc25A and cdc25B is frequent in primary non-small cell lung cancer but is not associated with overexpression of c-myc. Cancer Res. 1998;58:4082–4085. [PubMed] [Google Scholar]
- 98.Wu X, Levine A J. p53 and E2F-1 cooperate to mediate apoptosis. Proc Natl Acad Sci USA. 1994;91:3602–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yamasaki L, Bronson R, Williams B O, Dyson N J, Harlow E, Jacks T. Loss of E2F-1 reduces tumorigenesis and extends the lifespan of Rb+/− mice. Nat Genet. 1998;18:360–364. doi: 10.1038/ng0498-360. [DOI] [PubMed] [Google Scholar]
- 100.Yan Z, DeGregori J, Shohet R, Leone G, Stillman B, Nevins J R, Williams R S. Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc Natl Acad Sci USA. 1998;95:3063–3068. doi: 10.1073/pnas.95.7.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhu L, van den Heuvel S, Helin K, Fattaey A, Ewen M, Livingston D, Dyson N, Harlow E. Inhibition of cell proliferation by p107, a relative of the retinoblastoma protein. Genes Dev. 1993;7:1111–1125. doi: 10.1101/gad.7.7a.1111. [DOI] [PubMed] [Google Scholar]
- 102.Zhu L, Zhu L, Xie E, Chang L-S. Differential roles of two tandem E2F sites in repression of the human p107 promoter by retinoblastoma and p107 proteins. Mol Cell Biol. 1995;15:3552–3562. doi: 10.1128/mcb.15.7.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]