Abstract
The bioactive peptides that can inhibit angiotensin-I converting enzyme (ACE, EC. 3. 4.15.1) are considered as possible cures of hypertension. Food-derived angiotensin-I converting enzyme inhibitory (ACEi) peptides have gained more attention because of their reduced side effects. In this study, we reported the method for purifying ACEi peptides from the lees of traditional Chinese rice wine and evaluated the product's biochemical properties. After three steps of reversed-phase high-performance liquid chromatography (RP-HPLC), for the first time, we isolated, purified, and identified two novel peptides: LIIPQH and LIIPEH, both of which showed strong ACEi activity (IC50-values of 120.10 ± 9.31 and 60.49±5.78 μg/ml, respectively). They were further categorized as mixed-type ACE inhibitors and were stable against both ACE and gastrointestinal enzymes during in vitro digestion. Together, these results suggest that the rice wine lees that produced as a by-product during rice wine production can be utilized in various fields related to functional foods and antihypertensive medicine.
Keywords: hypertension, angiotensin-I converting enzyme, rice wine lees, ACE-inhibitory peptides, structure-activity
Introduction
Raised blood pressure is a major cardiovascular risk factor (1). As one of the leading risk factors for global mortality, uncontrolled hypertension causes stroke, myocardial infarction, cardiac failure, dementia, and other diseases (2). According to the World Health Organization (WHO), hypertension causes an estimated 9.4 million deaths yearly and represents 7% of the global disease burden (3). Once hypertension develops, lifelong pharmaceutical treatment is typically required. Several types of antihypertensive drugs have been developed based on different therapeutic targets, including angiotensin-I converting enzyme (ACE) inhibitors (4). Angiotensin-I converting enzyme (dipeptidyl carboxypeptidase, EC 3.4.15.1) is a zinc metallopeptidase that is found in male genital, vascular endothelial, neuro-epithelial, and absorptive epithelial cells (5). Angiotensin-I converting enzyme plays dual roles in the renin-angiotensin system (RAS) to regulate blood pressure by degrading angiotensin-I (Ang I) into angiotensin-II (Ang II), a strong vasopressor. Moreover, in the kallikrein-kinin system (KKS), ACE triggers inactivation of bradykinin, an important vasodilator (6, 7). Currently, ACE inhibitors are considered to be the first-line treatment for hypertension (8). Compared to other regimens, treatment regimens based on ACE inhibitors have been shown to reduce risk of coronary artery disease, cardiovascular death, and heart failure, and these treatments are frequently used for this reason (7, 9). However, the long-term administration of several synthetic ACE-inhibition drugs, such as captopril, enalapril, and lisinopril, is still associated with undesirable side effects, including headache, dry cough, and edema (9–11). Furthermore, drug treatment can be costly, and the high prevalence of hypertension makes it a challenge for resource-constrained settings (3). Therefore, the development of safe, cost-effective, and efficient treatment approaches is urgently needed. Angiotensin-I converting enzyme inhibitory peptides derived from food sources represent an effective way to treat hypertension without incurring unacceptable side effects (12). Hence, much interest has been focused on the extraction of ACE inhibitors from natural food sources as a replacement for synthetic drugs (13).
Numerous protein-based foods, including milk (14, 15), eggs (8), fish (16, 17), and rice (18–20), have been used to prepare angiotensin-I converting enzyme inhibitory (ACEi) peptides through enzymatic hydrolysis. Rice (Oryzae sativa, Family: Oryzeae) is a major cereal crop worldwide (21). It is a dietary staple for about half of the world's population and is particularly common in Asian countries, where it serves as a substantial source of dietary protein for millions of people (21–23). Compared to those of other cereals, rice proteins have a preferable amino acid structure due to their essential amino acid composition (24). In addition, the high availability of processing by-products has led rice to attract even much attention as a potential source of biologically active hydrolysates and peptides (25). Wine lees is the main by-product of rice wine production, and more than 400,000 tons are generated annually. Wine lees are composed of solid and liquid fractions whose compositions depend on their regions of origin and the associated agronomic and edaphoclimatic characteristics (26). The protein content of wine lees is approximately 28.0% (dry basis), making it a good protein resource.
Six novel ACEi peptides, AVQ, YPQ, NQL, AYLQ, VLPVLS, and VLPSLN, have successfully been identified from solid fractions of wine lees (27). However, due to the high moisture and organic acid contents, the liquid fraction of wine less has received comparatively less attention. As a product of rice fermentation, the liquid phase of lees contains considerable amounts of peptides and proteins. Therefore, it is urgent to development the comprehensive utilization of the liquid phase of rice wine lees, as it has the potential to provide new ideas for the treatment of rice wine lees to reduce environmental pressure.
In this study, we purified and identified ACEi peptides from rice wine waste using RP-HPLC and MOLDI-TOF-MASS. The IC50-values of the products were determined, and their inhibition types were kinetically characterized using a Lineweaver–Burk plot. Additionally, we investigated the stability of these compounds against digestive proteases and ACE. In doing so, this work contributes to the comprehensive utilization of rice wine waste and provides a theoretical basis for the development of effective antihypertensive peptides.
Materials and Methods
Materials and Chemicals
Rice wine lees was obtained from Guangdong Shunde Winery Co., Ltd., Shunde, China. Angiotensin-I converting enzyme (from rabbit lung) and N-Hippuryl-His-Leu hydrate (HHL) were purchased from Sigma Co., USA. Other chemicals and reagents were of analytical grade.
Physicochemical Characterization of Rice Wine Lees Extract
To prepare Rice Wine Lees Extract (RWLE) for characterization, large particles were removed by centrifuging at 4,000 rpm for 10 min, after which the supernatant was concentrated and lyophilized. The molecular weight distribution of RWLE was determined as previously described (28) with some modifications. Specifically, the molecular weight distribution was determined via gel permeation chromatography (GPC) with a TSK gel filtration column, G2000 SWXL (300 × 7.8 mm; Tosoh, Tokyo, Japan). The mobile phase, water/acetonitrile/trifluoroacetic acid (80/20/0.1, v/v/v), was delivered at a flow rate of 0.5 ml/min. The determination was carried out at room temperature, and the UV detector was operated at 220 nm. A calibration curve was obtained using the following standards: insulin (5,808 Da), bacitracin (1422 Da), tetrapeptide GGYR (451 Da), and tripeptide GGG (189 Da). Then, a molecular weight calibration curve (Y = −0.336X + 9.043, R2 =0.993) was obtained. In addition, fractions from different molecular weights were collected, concentrated, and tested using the ACE inhibition assay described below.
Measurement of ACEi Activity
Angiotensin-I converting enzyme inhibitory activity was measured using the method described in Wu et al. (29) with some modifications. For each reaction, 10 μl of 0.25 U/ ml ACE solution (prepared in 0.01 M potassium phosphate buffer containing 0.5 M NaCl, pH 7.0) was mixed with 10 μl of inhibitor solution and incubated for 5 min at 37°C. Subsequently, 30 μl of HHL solution (5 mM in 0.1 M sodium borate buffer containing 0.3 M NaCl, pH 8.3) was added, followed by incubation at 37°C for 1 h. The reaction was terminated with the addition of 70 μl 1 M HCl before subsequent assays were performed. For the control group, 10 μl ACE solution and 80 μl 1 M HCl were simultaneously added to the mixture. A blank sample was prepared by replacing the inhibitor solution with 10 μl borate buffer. Samples were then separated using a C18 column (4.6 × 150 mm, 5 μm) at a flow rate of 1.0 ml/min. The mobile phase was water/acetonitrile/trifluoroacetic acid (75/25/0.1, v/v/v) with an isocratic gradient of 25% B in 15 min. Absorbance was monitored at 228 nm. Angiotensin-I converting enzyme activity (%) was calculated according to the following equation:
Where A1, A2, and B are peak areas corresponding to Hippuric Acid in the control group, the blank and the sample, respectively. The IC50-value was defined as the concentration of inhibitor required to inhibit 50% of enzyme activity and was calculated by performing regression analysis of ACE inhibition vs. sample concentration.
Purification of ACEi Peptides
Angiotensin-I converting enzyme inhibitory peptides were first isolated by preparative high-performance liquid chromatography (LC-8, Shimadzu, Japan) using a reversed-phase (RP) column [Shimadzu PRC-ODS(K) 30 × 250 mm, 15 μm] of C18. Solvent A was 0.1% trifluoroacetic acid in double-distilled water, and solvent B was 0.1% trifluoroacetic acid in 100% acetonitrile. A linear gradient of solvent B was applied at a flow rate of 10 ml/min for 55 min as follows: 10–34% solvent B for 30 min and 30–45% solvent B for 45 min. Fractions were collected, concentrated, and tested using an ACE-inhibition assay. The fraction with the highest ACEi activity was further separated using an additional C18 column (ECOSIL 300 × 20 mm,10 μm, Germany) and the same mobile phases. A linear gradient of solvent B was applied at a flow rate of 10 ml/min for 45 min as follows: 15–19% solvent B for 5 min and 19–23% solvent B for 45 min. To further purify the ACE inhibitor, active fractions were analyzed using HPLC to identify monomeric peptides with increased ACEi activity. The active fraction solution collected previously was filtered through a 0.45 μm membrane filter and separated using an ECOSIL C18 column (260 × 4.6 mm, 5 μm). Mobile phases were solvent A (double-distilled water with 0.1% trifluoroacetic acid) and solvent B (acetonitrile with 0.1% trifluoroacetic acid). Elution was performed using water/acetonitrile/trifluoroacetic acid (80/20/0.1, v/v/v) with 5–15% solvent B (0–10 min) and 15–20% solvent B (10–45 min) at a flow rate of 1 ml/min. Main peaks were collected, concentrated, and lyophilized. All elution steps were monitored at 214 nm. The ACEi activity of the eluted peaks was tested.
Amino Acid Sequencing of Purified Peptides
The amino acid sequences of active peptides displaying ACEi activity were analyzed using a matrix-assisted laser desorption ionization time-of-flight mass spectrophotometer (MALDI-TOF-MASS, Bruker Daltonik GmbH, Bremen, Germany).
Peptide recognition was performed using an automated protein/peptide sequencer (model PPSQ-53A, Shimadzu, Tokyo, Japan). The purified peptide was dissolved in deionized water and transferred onto a polyvinylidene fluoride membrane. After fixation with polybrene, the amino acid sequence of the peptide was characterized.
Synthesis of Identified Peptides
The sequences of the identified peptides were submitted to Synpeptide Co., Ltd. (Shanghai, China) for peptides synthesis using solid-phase methods. Purity and molecular weights of synthesized peptides were determined using HPLC and LC-MS.
Stability of Purified Peptides During In Vitro Digestion by Gastrointestinal Enzymes
Peptide stability was measured according to the methods described in Chen et al. (30) with some modifications. For this assay, 2% (w/w) pepsin was dissolved in a KCl–HCl buffer (0.1 mM) adjusted to pH 2.0, and 2% (w/w) chymotrypsin was dissolved in a KCl–NaOH (0.1 mM) buffer adjusted to pH 7.0. Peptides were dissolved at 1 mg/ml in pepsin or chymotrypsin solutions and incubated at 37°C for 4 h. Reactions were terminated by boiling for 15 min. After centrifugation at 4,000 rpm for 15 min, the supernatant was collected, pH was adjusted to 8.0, and samples were assayed for ACEi activity. The supernatant was also analyzed using the previously described RP-HPLC conditions to evaluate changes to the peptides after in vitro digestion.
Control groups were prepared by incubating inactivated pepsin and/or chymotrypsin (boiled for 15 min) with the sample peptides. After incubation, the mixture was processed and analyzed as described.
Stability of Peptides Against ACE
The stability of peptides against ACE was assayed according to the methods of Deng (31), with some modifications. For each reaction, 30 μl of peptides (2 mg/ml) was combined with 30 μl 0.25 U/ml ACE solution and incubated at 37°C for 30 min. Angiotensin-I converting enzyme was then inactivated by boiling the solution for 10 min. As described in section Measurement of ACEi Activity, 20 μl of each peptide sample was used for the detection of ACEi activity.
Kinetic Parameters of ACEi Peptides
Angiotensin-I converting enzyme inhibition of purified peptides was determined using Lineweaver–Burk plots according to reported methods (32). Samples were added at two fixed concentrations to each reaction mixture, and enzyme activity was measured for different concentrations of the substrate HHL according to the ACE inhibition assay. The curves were described by 1/area and 1/[S], and the kinetics of ACE inhibitors were estimated by comparing the resulting curves.
Design of ACEi Peptides According to Source Proteins
As protein structure has a significant impact on ACE-inhibition (33–35), we chose to prune or add peptides according to UNIPROT (https://sparql.uniprot.org/) BLAST results. The newly designed peptides were synthesized and further assessed for ACE-inhibition capacity as described above.
Statistical Analysis
Analysis of variance was performed with SPSS 13.0 software (Armonk, NY, USA). Linear regression was performed using Origin (Microsoft Corp., Seattle, WA, USA).
All experiments were repeated at least three times, and mean values were used. Data were subjected to analysis of variance, and Duncan values were calculated with a confidence interval of 95% to compare means.
Results and Discussion
Physicochemical Properties and ACEi Activity of Rice Wine Lees Extract
Rice Wine Lees Extract was prepared as described and found to exhibit potent ACEi activity, with an IC50-value of 806.67 ± 58.97 μg/ml. Previously, wine and wine lees have been subjected to recovery or transformation of components into high value-added compounds (26).
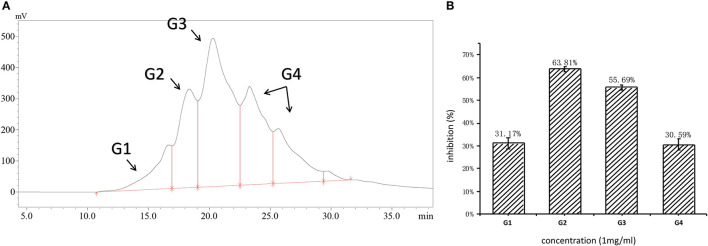
The molecular weight distribution of RWLE was broad, ranging from 50 to 25,000 Da, indicating that it contained proteins and free amino acids (Table 1). Accordingly, RWLE was further divided into four components, G1–G4 (Figure 1A). As shown by the associated ratios, the 50–500 Da molecular weight segment accounted for a significant proportion of RWLE (37.89%).
Table 1.
Molecular distribution of RWLE.
| MW (Da) | Distribution (%) |
|---|---|
| 3k−25k | 10.19 |
| 500–3k | 18.68 |
| 50–500 | 37.89 |
| <50 | 33.24 |
Figure 1.
(A) Molecular distribution of RWLE; (B) inhibition of different molecular fractions (G1–G4).
Fractions G1–G4 were collected individually, and their ACEi activities were further determined. As shown in Figure 1B, G2 possessed the highest ACEi capacity at a concentration of 1 mg/ml. Therefore, we predicted that the molecular weights of ACEi peptides would fall in the range of 500–3,000 Da. This prediction is consistent with previous reports that antihypertensive peptides are typically short, with molecular weights >2,000 Da (36). Larger peptides are not capable to contact the ACE active site easily, resulting in reduced inhibitory capacity.
ACEi Peptide Purification
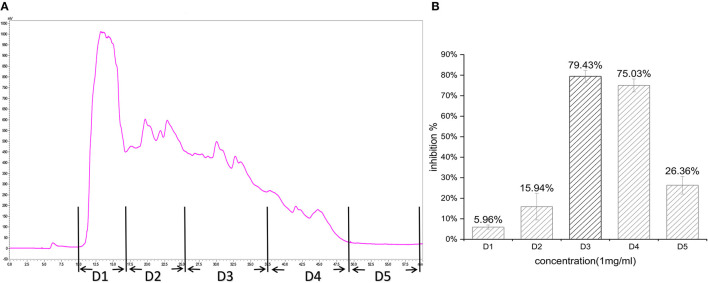
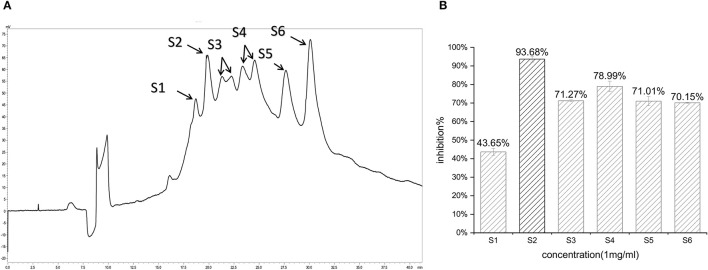
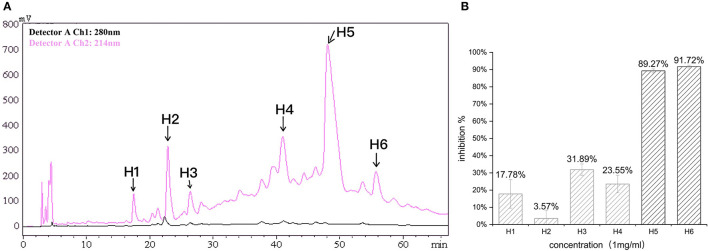
In the first step, fractions between 20 and 55 min in the RP-HPLC chromatogram were collected and termed D1–D5 (Figure 2A). Fraction D3 showed the lowest IC50-value (643.33 ± 17.64 μg/ml) and was further separated into different fractions (Figure 2B). The seven main fractions were designated as S1–S7 (Figure 3A). Each fraction was subsequently collected. All of them exhibited ACEi activity at a concentration of 1 mg/ml, and the inhibition rate was in the range of 43.65–93.68% (Figure 3B). Importantly, activity analysis showed that Fraction S2 had the lowest IC50 (479.00 ± 59.36 μg/ml). Thus, this fraction was further purified by RP-HPLC using an analytical C18 column. As shown in Figure 4A, S2 was subsequently separated into six peaks (H1–H6). Among them, H5 and H6 were the two most potent ones in ACE inhibition with IC50-value of 120.10 ± 9.31 and 60.49 ± 5.78 μg/ml, respectively (Figure 4B). Accordingly, they were subjected to MOLDI-TOF-MASS to identify their amino acid sequence.
Figure 2.
(A) Reverse-phase HPLC chromatogram of first-step purification of RWLE peptides; (B) inhibitory activity of five pooled fractions (D1–D5).
Figure 3.
(A) Reverse-phase HPLC chromatogram of second-step purification of the G3 fraction; (B) inhibitory activity of six pooled fractions (S1–S6).
Figure 4.
(A) Reverse-phase HPLC chromatogram of third-step purification of the S2 fraction; (B) inhibitory activity of six fractions (H1–H6).
Identification of Amino Acid Sequence of Purified ACEi Peptides
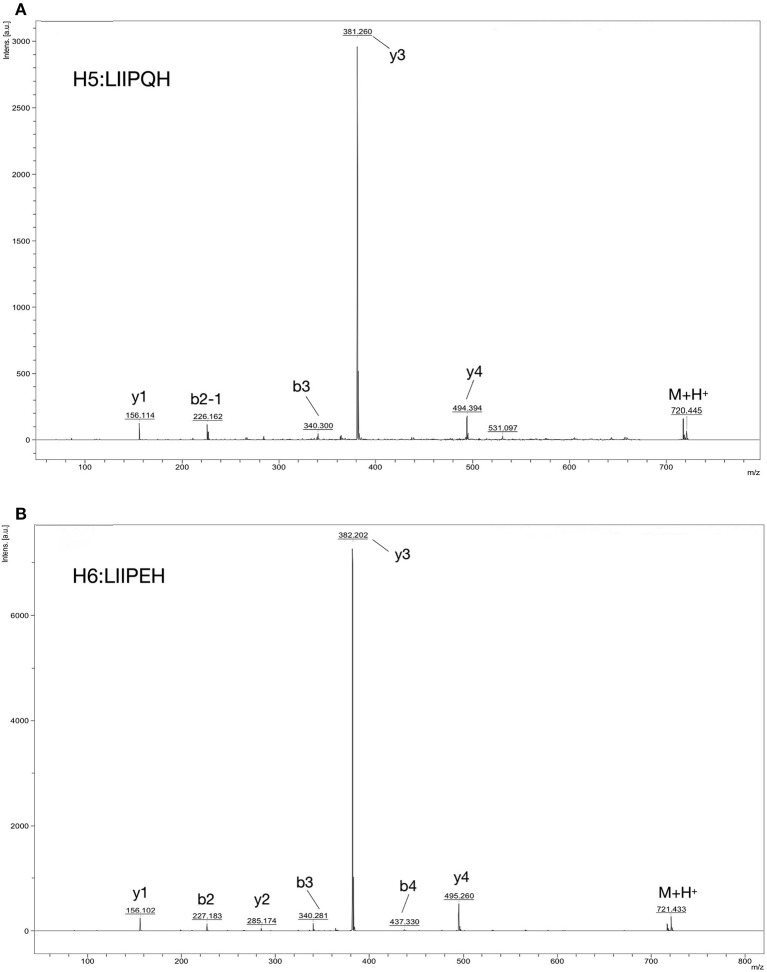
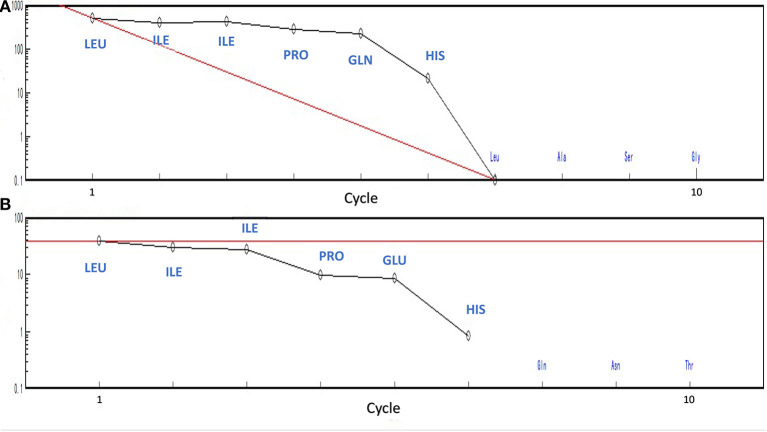
MOLDI-TOF-MS/MS was used to determine the amino acid sequence of the peptides (Figure 5). H5 was found to have a molecular mass of 719.85 Da, and H6 had a molecular mass of 720.85 Da. Because Leu and Ile have the same molecular weight, we used an automated protein/peptide sequencer to further validate the peptides sequence. The results showed that H5 had a sequence of Ile-Leu-Leu-Pro-Gln-His, while H6 had a sequence of Ile-Leu-Leu-Pro-Glu-His (Figure 6). Markedly, it is the first time to isolate and identify these two novel peptides from food proteins. Additionally, their molecular weights correspond to the aforementioned results that the most active ACEi peptide was found to fall within the molecular weight range of <2,000 Da.
Figure 5.
Identification of ACE-inhibition peptides from RWLE using MOLDI-TOF-MS. (A) Molecular mass and amino acid sequence of peptide H5; (B) molecular mass and amino acid sequence of peptide H6.
Figure 6.
Identification of peptide's sequence using PPSQ (A) peptide H5 and (B) peptide H6.
Interestingly, these two novel peptides are all oligopeptides with six amino acid residues and shared the sequence LIIP. It has been previously reported that most ACEi peptides are short, containing 2–12 amino acids (37). Peptides with hydrophobic amino acids, especially those with aliphatic chains such as Gly, Ile, and Leu at the N-terminus, have very high ACEi activity (38–40). Moreover, the presence of Leu at the final position has been claimed to enhance the ACE-inhibiting effects of an amino acid sequence (41, 42).
The C-domain of ACE is responsible for generating Ang II from Ang I under physiological conditions (43). Of note, the C-domain catalytic site of ACE consists of three subsites—S1, S1′, and S2′–which accommodate the three hydrophobic C-terminal residues of its natural substrate, angiotensin I (44). Hence, preferred ACE inhibitors contain hydrophobic (aromatic or branched side chains) amino acid residues at each of the three C-terminal positions (39). Based on studies further exploring ACEi penta- and hexapeptides and their structure-activity relationships, some rules have been concluded. These peptides contain, at minimum, two highly hydrophobic amino acid residues (8). Notably, the sequences of RWLE-derived ACE-inhibition peptides are consistent with these relationships between the primary structures of ACEi peptides and their inhibitory effects.
However, comparing the primary structures, the second N-terminal amino acids of H5 and H6 are different. Specifically, H5 contains Gln, an amino acid with a negative charge, while H6 contains Glu, a neutral amino acid. Although the role of positive charge at an amino acid remains unknown, it has been suggested that the presence of positively charged amino acids in the C-terminal region of the peptide could contribute to ACE-I activity and substantially increase inhibitory potency (45). This phenomenon might be explained by the presence of negatively charged Glu403 and Glu162 residues at the S2 and S1′ subsites of the sACE C-domain, respectively (43). The preference for positive charges at these two subsites might enhance selectivity to the sACE C-domain over the N-domain, therefor weakening ACE's ability to produce ANG II (46, 47). In theory, glutamic acid could act an important contributor to ACE inhibition by chelating zinc at the ACE active center and through hydrophobic interactions with ACE (48), which might be another reason for the differing IC50-values. Furthermore, the binding affinity of long peptides to ACE is influenced by the structure adopted in the specific environment of the binding site (49). Therefore, conformational changes in the peptide backbone can be induced by the polarity of these two residues and influence ACEi capacity (50).
In this work, the IC50-values of synthetic H5, H6, and the products in each purification step are summarized in Table 2. We found that synthetic peptides have no significant differences in comparison to natural peptides. Since limited amounts of natural peptides could be isolated from RWLE, synthetic peptides were used to further study the inhibition patterns and assess biological stability against ACE and gastrointestinal enzymes.
Table 2.
IC50-values of the products in each purification step.
| Inhibitor | IC50 (μg/ml) |
|---|---|
| RWLE | 806.67 ± 58.97 |
| G3 | 643.33 ± 17.64 |
| S2 | 479.00 ± 59.36 |
| H5 | 120.10 ± 9.31 |
| H6 | 60.49 ± 5.78 |
| H5 (synthesis) | 143.33 ± 5.77 |
| H6 (synthesis) | 56.67 ± 12.02 |
Stability of the Purified Peptides Against Gastrointestinal Enzymes
In contrast to chemically synthesized low-molecular-weight compounds, peptides are easily inactivated by gastrointestinal enzymatic digestion (51). In addition to demonstrating high inhibitory activity, purified peptides must remain sufficiently bioavailability during hydrolysis by gastrointestinal proteases after oral administration to bind their target sites and exhibit an antihypertensive activity in vivo (52).
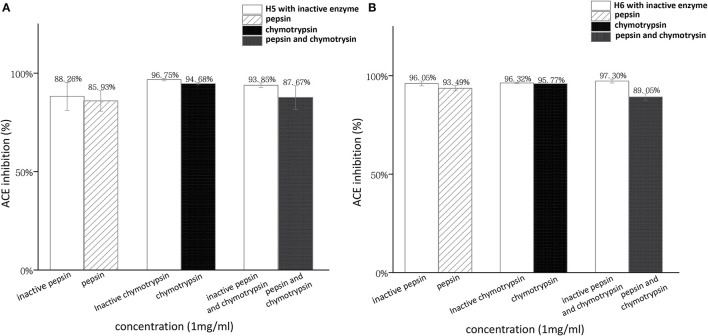
In this work, LIIPQH and LIIPEH were subjected to in vitro digestion by pepsin and chymotrypsin (Figure 7). Although ACEi capacity of H5 and H6 was decreased by the combination of two gastrointestinal enzymes, the differences were not statistically significant (p > 0.05), suggesting that the peptides were stable within the gastrointestinal environment and could maintain their activities after being absorbed. Proline- and hydroxyproline-containing peptides are generally resistant to degradation by digestive enzymes (41, 53), so the great stability of these two peptides might be a result of a proline residue found in the third C-terminal position.
Figure 7.
Stability of RWLE peptides against gastrointestinal enzymes (A) peptide H5 and (B) peptide H6.
Enzymatic Studies of ACEi Peptides
Due to the broad substrate specificity of ACE, some ACEi peptides may become ACE substrates and be subsequently cleaved into smaller fragments (54). Accordingly, ACEi peptides can be classified into three categories based on the results of ACE-ACEi peptides preincubation testing. The first type is true inhibitors, the second type is real substrates, and the third type is the pro-drug type (7). The stability of ACEi peptides we purified against ACE were summarized in Table 3, showing that both LIIPQH and LIIPEH had lower IC50-values compared to the control (co-incubation with inactive ACE). Therefore, these peptides could be categorized as pro-drug ACE-inhibitors, which showed improved ACEi activity due to the formation of a more active fragment after hydrolysis by ACE.
Table 3.
Stability of RWLE peptides toward ACE.
| Peptide | IC50 with | IC50 without |
|---|---|---|
| pre-incubation (mg/ml) | pre-incubation (mg/ml) | |
| LIIPQH | 82.0 ± 19.92 | 65.88 ± 18.42 |
| LIIPEH | 51.0 ± 7.51 | 36.98 ± 21.08 |
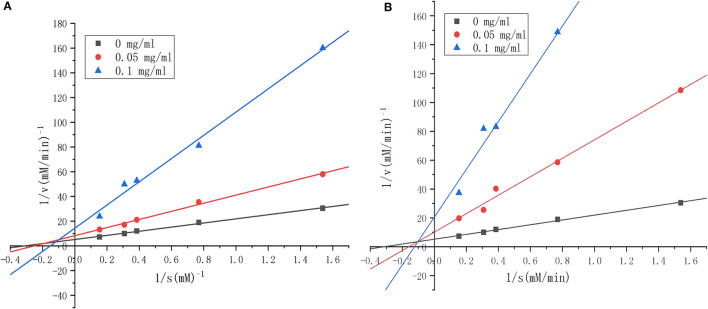
To clarify the kinetics of inhibition, Lineweaver–Burk plots were generated. Various concentrations of substrate were incubated with ACE solution in the absence or presence of RWLE peptides at 0.05 or 0.1 mg/ml. The results were illustrated in Figure 8A (H5) and Figure 8B (H6). We found that all lines intersected at similar x-intercepts, but had different slopes and y-intercepts, suggesting that H5 and H6 acted as non-competitive inhibitors (mostly mixed-type) toward ACE. Thus, H5 and H6 could bind to both free enzymes and the enzyme-substrate complex to reduce catalysis (55).
Figure 8.
Lineweaver–Burk plots of angiotensin I-converting enzyme (ACE) inhibition by different concentrations of synthesized RWLE peptides across varying substrate concentrations (0–0.1 mg/ml). (A) Lineweaver–Burk plots of peptide H5 and (B) Lineweaver–Burk plots of peptide H6. V = initial rate of reaction [ΔA228 (nm)/min].
As Km and Vm are important parameters of enzymatic reactions, we further determined these values for the novel ACEi peptides. Vmax for the uninhibited ACE reaction was 0.191 min−1.Though Vm values decreased as peptide concentration increased for both H5 and H6, H6 appeared to be more effective (Table 4). The finding of a lower Vmax in the presence of H6 was consistent with its lower IC50-value compared to H5. Together, these results suggested that the activation energy of the catalytic reaction increased in the presence of peptides because of the decreased enzyme reaction rate. We hypothesized that the peptides blocked the substrate from binding to the ACE active site (56), resulting in the reduction of enzymatic efficiency. Based on a linear regression fit of the kinetic data, the Km-value of ACE activity in the absence of inhibiting peptide was estimated as 3.199 (Table 4). The Km-values increased when ACEi peptide was added to the reaction, suggesting that a higher concentration of substrate was required for the ACE catalytic reaction. The catalytic efficiency (CE) of the uninhibited ACE-catalyzed reaction was 0.060. In the presence of H5 or H6, it was decreased in a dose-dependent manner. The reduction of CE was directly related to the lower Vmax and IC50-values, suggesting that the peptides had binding affinity toward the target enzyme (7).
Table 4.
Kinetic constants of ACE-catalyzed reactions at different peptide concentrations.
| Catalytic parameters | Control | H5 | H6 | ||
|---|---|---|---|---|---|
| Concentration (mg/ml) | 0 | 0.05 | 0.1 | 0.05 | 0.1 |
| Vm (ΔA/min) | 0.191 | 0.120 | 0.070 | 0.099 | 0.049 |
| Km (mg) | 3.199 | 3.948 | 6.552 | 6.308 | 8.054 |
| CE | 0.060 | 0.030 | 0.011 | 0.016 | 0.006 |
Km is the Michaelis constant, Vmax is maximum reaction velocity, CE is the catalytic efficiency (Vmax/Km).
The classification of “mixed-type inhibition” indicates that the peptide binds to ACE at both active and non-active sites, consequently reducing ACE's catalytic activity (57). As the space within the ACE active site is limited, which is hard for large peptides to bind. Therefore, peptides H5 and H6 may preferentially bind to the non-active site of ACE (57). Some research has explored mixed-competitive ACE inhibitors, including NMAINPSKENLCSTFCK and EKVNELSK isolated from casein (56, 57) and the synthesized peptides WG and PRY (55).
Structure-Activity Relationship of ACEi Peptides According to Source Proteins
The BLAST program was used for homology searches between manually obtained sequences and sequences in the Uniprot database. Analysis result indicated that a total of 27 rice proteins contained the H5 peptide sequence, while H6 had no identifiable source protein. H6 and H5 showed highly similar structure, differing in only one amino acid. Therefore, we speculated that H6 was a modified peptide generated during fermentation, while H5 corresponded to the source protein. To explore the structure-functional relationships, only H5 peptide that conformed to the rice protein sequence was selected to conduct further analysis.
Based on BLAST results, we determined that all source proteins belonged to Oryza sativa subsp. japonica, and most of them were glutelin. In addition, we found that the first amino acid linked to the N-terminus of H5 was Leu, and the amino acids connected to the C-terminus were His or Tyr. Thus, we synthesized three new peptides with increased terminal amino acid content.
Additionally, Pro, Trp, and Lys seem to be the most effective ones in increasing a peptide's ACEi potential (39). Between H5 and H6, the common sequence was identified as LIIP. Accordingly, new peptides with decreased terminal amino acids and exposed proline residues were also synthesized, and their IC50-values were measured (Table 5).
Table 5.
IC50-values of synthetic peptides.
| Peptide | IC50 (μg/ml) |
|---|---|
| LIIPQHH | 1,206.67 ± 36.83 |
| LIIPQHY | 1,082.00 ± 38.85 |
| LLIIPQH | 829.00 ± 14.57 |
| IIPQH | 1,082.67 ± 36.83 |
| LIIPQ | 1,390.67 ± 21.40 |
| LIIP | 928.67 ± 24.01 |
| H5 (LIIPQH) | 143.33 ± 5.77 |
| H6 (LIIPEH) | 56.67 ± 12.02 |
Among LIIPQHH, LIIPQHY, and LLIIPQH, we found LLIIPQH possessed the highest ACEi capacity. This might be due to the fact that, among the added amino acids, Leu was the only hydrophobic residue, further emphasizing the importance of terminal hydrophobic amino acids for ACE inhibition. It has been suggested that amino acids with positive charge can promote ACE inhibition. However, despite containing two positively charged amino acids, LIIPQHH did not show the lowest IC50-value. In conclusion, the inclusion of positively charged amino acids improves inhibition rate only under certain conditions, rather than universally. The different IC50-values of LIIPQHH and LIIPQHY could attribute to the differences in the polarity of C-terminal residues, which might induce conformational changes in the peptide backbone that influence ACEi capacity.
For pruned peptides, IIPQH had a lower IC50-value than LIIPQ. which might be due to the positive charge of His that enhanced ACE inhibition. This is supported by a separated research, in which three of five ACE inhibitor peptides obtained from cherry seeds had a positively charge amino acid (H) in their C-terminal positions (58).
However, the activities of IIPQH and LIIPQ were not as high as that of LIIP. These differences might be a result of the C-terminal proline residues. C-terminal proline residues were found in lisinopril and enalapril peptides, suggesting that the presence of proline residues at the C-terminus was a specific feature associated with high ACEi activity and played a key role in ACE binding (38, 59). In addition, the imidazole ring of proline might easily interact with amino acid residues in the active center of ACE (60).
After the addition or subtraction of amino acids, the peptide sequence with the best activity was identified as LLIIPQH, followed by LIIP. However, all newly synthesized peptides exhibited lower ACEi activity than purified peptides obtained by us. Similar results had been found that some peptides showed high homology with ACEi peptides. Although these peptides shared the subsequence VTSTAV, not all of them exhibited significant ACEi effects (41).
Conclusion
In this study, we obtained antihypertensive peptides from RWLE without enzymatic steps. Two novel peptides were purified and identified: LIIPQH and LIIPEH. They both showed strong ACEi activity and were classified as mixed-type inhibitors. Markedly, these two peptides showed great stability to ACE and digestive enzymes in vitro. Furthermore, we studied the structure-activity relationship of ACEi peptides by synthesizing similar peptides. Together, this study suggested that novel peptides with ACEi activity can be derived from rice wine lees and utilized to develop functional foods or antihypertensive medicine.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author Contributions
ZH, GL, YC, and MS designed the experiment. ZH, GL, and ZQ conducted the activity evaluation experiments and collected and analyzed the data. ZH, GL, YC, and MS wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by Guangdong Provincial Key Laboratory of Nutraceuticals and Functional Foods (2018B030322010), the Program for Guangdong Introducing Innovative and Entrepreneurial Teams (2019ZT08N291) China Postdoctoral Science Foundation (2020M672651), and Guangdong Province Modern Agricultural Industrial Technology System Innovation Team Project (2021KJ117).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1.Christopher P., Cannon M. Cardiovascular disease and modifiable cardiometabolic risk factors. Clin Cornerstone. (2008) 9:24–38; discussion 39–41. 10.1016/S1098-3597(09)62037-8 [DOI] [PubMed] [Google Scholar]
- 2.Das UN. Is angiotensin-II an endogenous pro-inflammatory molecule? Med Sci Monit. (2005) 11:RA155–162. [PubMed] [Google Scholar]
- 3.Organization WH . Overview. (2014). Available online at: https://www.who.int/publications/i/item/9789241564854 (accessed October 26, 2014).
- 4.McManus RJ, Caulfield M, Williams B. NICE hypertension guideline 2011: evidence based evolution. BMJ. (2012) 344:e181. 10.1136/bmj.e181 [DOI] [PubMed] [Google Scholar]
- 5.Chamata Y, Watson KA, Jauregi P. Whey-derived peptides interactions with ACE by molecular docking as a potential predictive tool of natural ACE inhibitors. Int J Mol Sci. (2020) 21:864. 10.3390/ijms21030864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riordan JF. Angiotensin-I-converting enzyme and its relatives. Genome Biol. (2003) 4:225. 10.1186/gb-2003-4-8-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shih YH, Chen FA, Wang LF, Hsu JL. Discovery and study of novel antihypertensive peptides derived from Cassia obtusifolia seeds. J Agric Food Chem. (2019) 67:7810–20. 10.1021/acs.jafc.9b01922 [DOI] [PubMed] [Google Scholar]
- 8.Fan H, Wang J, Liao W, Jiang X, Wu J. Identification and characterization of gastrointestinal-resistant angiotensin-converting enzyme inhibitory peptides from egg white proteins. J Agric Food Chem. (2019) 67:7147–56. 10.1021/acs.jafc.9b01071 [DOI] [PubMed] [Google Scholar]
- 9.Turnbull F, Neal B, Algert C, Chalmers J, Woodward M, MacMahon S, et al. Effects of different blood-pressure-lowering regimens on major cardiovascular events: results of prospectively-designed overviews of randomised trials. Lancet. (2003) 362:1527–35. 10.1016/S0140-6736(03)14739-3 [DOI] [PubMed] [Google Scholar]
- 10.Gavras H, Gavras I. Angiotensin converting enzyme inhibitors. Properties and side effects. Hypertension. (1988) 11:II37–41. 10.1161/01.HYP.11.3_Pt_2.II37 [DOI] [PubMed] [Google Scholar]
- 11.Sweeting MJ, Thompson SG, Brown LC, Greenhalgh RM, Powell JT. Use of angiotensin converting enzyme inhibitors is associated with increased growth rate of abdominal aortic aneurysms. J Vasc Surg. (2010) 52:1–4. 10.1016/j.jvs.2010.02.264 [DOI] [PubMed] [Google Scholar]
- 12.Himaya SWA, Ngo D-H, Ryu B, Kim S-K. An active peptide purified from gastrointestinal enzyme hydrolysate of Pacific cod skin gelatin attenuates angiotensin-1 converting enzyme (ACE) activity and cellular oxidative stress. Food Chem. (2012) 132:1872–82. 10.1016/j.foodchem.2011.12.020 [DOI] [Google Scholar]
- 13.Rui X, Boye JI, Simpson BK, Prasher SO. Purification and characterization of angiotensin I-converting enzyme inhibitory peptides of small red bean (Phaseolus vulgaris) hydrolysates. J Funct Foods. (2013) 5:1116–24. 10.1016/j.jff.2013.03.008 [DOI] [Google Scholar]
- 14.Sakurai N, Nishio S, Akiyama Y, Miyata S, Oshima K, Nadano D, et al. Apical-to-basolateral transepithelial transport of cow's milk caseins by intestinal Caco-2 cell monolayers: MS-based quantitation of cellularly degraded alpha- and beta-casein fragments. J Biochem. (2018) 164:113–25. 10.1093/jb/mvy034 [DOI] [PubMed] [Google Scholar]
- 15.Clare DA, Swaisgood HE. Bioactive milk peptides: a prospectus. J Dairy Sci. (2000) 83:1187–95. 10.3168/jds.S0022-0302(00)74983-6 [DOI] [PubMed] [Google Scholar]
- 16.Balti R, Nedjar-Arroume N, Bougatef A, Guillochon D, Nasri M. Three novel angiotensin I-converting enzyme (ACE) inhibitory peptides from cuttlefish (Sepia officinalis) using digestive proteases. Food Res Int. (2010) 43:1136–43. 10.1016/j.foodres.2010.02.013 [DOI] [Google Scholar]
- 17.Neves AC, Harnedy PA, O'Keeffe MB, FitzGerald RJ. Bioactive peptides from Atlantic salmon (Salmo salar) with angiotensin converting enzyme and dipeptidyl peptidase IV inhibitory, and antioxidant activities. Food Chem. (2017) 218:396–405. 10.1016/j.foodchem.2016.09.053 [DOI] [PubMed] [Google Scholar]
- 18.Kuba M, Tanaka K, Sesoko M, Inoue F, Yasuda M. Angiotensin I-converting enzyme inhibitory peptides in red-mold rice made by Monascus purpureus. Proc Biochem. (2009) 44:1139–43. 10.1016/j.procbio.2009.06.007 [DOI] [Google Scholar]
- 19.Chen J, Liu S, Ye R, Cai G, Ji B, Wu Y. Angiotensin-I converting enzyme (ACE) inhibitory tripeptides from rice protein hydrolysate: purification and characterization. J Funct Foods. (2013) 5:1684–92. 10.1016/j.jff.2013.07.013 [DOI] [Google Scholar]
- 20.Wang X, Chen H, Fu X, Li S, Wei J A. novel antioxidant and ACE inhibitory peptide from rice bran protein: biochemical characterization and molecular docking study. Lwt. (2017) 75:93–9. 10.1016/j.lwt.2016.08.047 [DOI] [Google Scholar]
- 21.Sohail M, Rakha A, Butt MS, Iqbal MJ, Rashid S. Rice bran nutraceutics: a comprehensive review. Crit Rev Food Sci Nutr. (2017) 57:3771–80. 10.1080/10408398.2016.1164120 [DOI] [PubMed] [Google Scholar]
- 22.Muthayya S, Sugimoto JD, Montgomery S, Maberly GF. An overview of global rice production, supply, trade, and consumption. Ann N Y Acad Sci. (2014) 1324:7–14. 10.1111/nyas.12540 [DOI] [PubMed] [Google Scholar]
- 23.Amagliani L, O'Regan J, Kelly AL, O'Mahony JA. The composition, extraction, functionality and applications of rice proteins: a review. Trends Food Sci Tech. (2017) 64:1–12. 10.1016/j.tifs.2017.01.008 [DOI] [Google Scholar]
- 24.Al Doury MKW, Hettiarachchy NS, Horax R. Rice-endosperm and rice-bran proteins: a review. J Amer Oil Chem Soc. (2018) 95:943–56. 10.1002/aocs.12110 [DOI] [Google Scholar]
- 25.Rani S, Pooja K, Pal GK. Exploration of rice protein hydrolysates and peptides with special reference to antioxidant potential: computational derived approaches for bio-activity determination. Trends Food Sci Tech. (2018) 80:61–70. 10.1016/j.tifs.2018.07.013 [DOI] [Google Scholar]
- 26.Perez-Bibbins B, Torrado-Agrasar A, Salgado JM, de Souza Oliveira RP, Dominguez JM. Potential of lees from wine, beer and cider manufacturing as a source of economic nutrients: an overview. Waste Manage. (2015) 40:72–81. 10.1016/j.wasman.2015.03.009 [DOI] [PubMed] [Google Scholar]
- 27.Wei D, Fan W, Xu Y. In vitro production and identification of Angiotensin Converting Enzyme (ACE) inhibitory peptides derived from distilled spent grain prolamin isolate. Foods. (2019) 8:390. 10.3390/foods8090390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.G B I and C S . High-performance gel permeation chromatography of proteins and peptides on columns of TSK-G2000-SW and TSK-G3000-SW: a volatile solvent giving separation based on charge and size of polypeptides. Anal Biochem. (1986) 155:141–8. 10.1016/0003-2697(86)90239-3 [DOI] [PubMed] [Google Scholar]
- 29.Wu S, Feng X, Lu Y, Lu Y, Liu S, Tian Y. Purification of angiotensin I-converting enzyme (ACE) inhibitory peptides from casein hydrolysate by IMAC-Ni(2). Amino Acids. (2017) 49:1787–91. 10.1007/s00726-017-2475-5 [DOI] [PubMed] [Google Scholar]
- 30.Chen J, Wang Y, Zhong Q, Wu Y, Xia W. Purification and characterization of a novel angiotensin-I converting enzyme (ACE) inhibitory peptide derived from enzymatic hydrolysate of grass carp protein. Peptides. (2012) 33:52–8. 10.1016/j.peptides.2011.11.006 [DOI] [PubMed] [Google Scholar]
- 31.Deng Z, Liu Y, Wang J, Wu S, Geng L, Sui Z, et al. Antihypertensive effects of two novel Angiotensin I-Converting Enzyme (ACE) inhibitory peptides from Gracilariopsis lemaneiformis (Rhodophyta) in Spontaneously Hypertensive Rats (SHRs). Mar Drugs. (2018) 16:299. 10.3390/md16090299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng Y, Li Y, Zhang Y, Ruan X, Zhang R. Purification, characterization, synthesis, in vitro ACE inhibition and in vivo antihypertensive activity of bioactive peptides derived from oil palm kernel glutelin-2 hydrolysates. J Funct Foods. (2017) 28:48–58. 10.1016/j.jff.2016.11.021 [DOI] [Google Scholar]
- 33.Tondo AR, Caputo L, Mangiatordi GF, Monaci L, Lentini G, Logrieco AF, et al. Structure-based identification and design of angiotensin converting enzyme-inhibitory peptides from whey proteins. J Agric Food Chem. (2020) 68:541–8. 10.1021/acs.jafc.9b06237 [DOI] [PubMed] [Google Scholar]
- 34.Qian B, Tian C, Huo J, Ding Z, Xu R, Zhu J, et al. Design and evaluation of four novel tripeptides as potent angiotensin converting enzyme (ACE) inhibitors with anti-hypertension activity. Peptides. (2019) 122:170171. 10.1016/j.peptides.2019.170171 [DOI] [PubMed] [Google Scholar]
- 35.De Oliveira TV, Guimaraes AP, Bressan GC, Maia ER, Coimbra J, Poleto MD, et al. Structural and molecular bases of angiotensin-converting enzyme inhibition by bovine casein-derived peptides: an in silico molecular dynamics approach. J Biomol Struct Dyn. (2021) 39:1386–403. 10.1080/07391102.2020.1730243 [DOI] [PubMed] [Google Scholar]
- 36.Iwaniak A, Minkiewicz P, Darewicz M. Food-originating ACE inhibitors, including antihypertensive peptides, as preventive food components in blood pressure reduction. Compr Rev Food Sci Food Safety. (2014) 13:114–34. 10.1111/1541-4337.12051 [DOI] [PubMed] [Google Scholar]
- 37.Hernandez-Ledesma B. del Mar Contreras M, Recio I. Antihypertensive peptides: production, bioavailability and incorporation into foods. Adv Colloid Interface Sci. (2011) 165:23–35. 10.1016/j.cis.2010.11.001 [DOI] [PubMed] [Google Scholar]
- 38.Rao S-Q, Liu S, Ju T, Xu W-Q, Mei G-M, Xu Y-S, et al. Design of substrate-type ACE inhibitory pentapeptides with an antepenultimate C-terminal proline for efficient release of inhibitory activity. Biochem Eng J. (2012) 60:50–5. 10.1016/j.bej.2011.09.018 [DOI] [Google Scholar]
- 39.Murray BA, FitzGerald RJ. Angiotensin converting enzyme inhibitory peptides derived from food proteins: biochemistry, bioactivity and production. Curr Pharm Des. (2007) 13:773–91. 10.2174/138161207780363068 [DOI] [PubMed] [Google Scholar]
- 40.Fitzgerald RJ, Murray BA, Walsh DJ. Hypotensive peptides from milk proteins. J Nutr. (2004) 134:980S−8S. 10.1093/jn/134.4.980S [DOI] [PubMed] [Google Scholar]
- 41.Tavares T, Contreras Mdel M, Amorim M, Pintado M, Recio I, Malcata FX. Novel whey-derived peptides with inhibitory effect against angiotensin-converting enzyme: in vitro effect and stability to gastrointestinal enzymes. Peptides. (2011) 32:1013–9. 10.1016/j.peptides.2011.02.005 [DOI] [PubMed] [Google Scholar]
- 42.Kim S-K, Byun H-G, Park P-J, Shahidi F. Angiotensin I converting enzyme inhibitory peptides purified from bovine skin gelatin hydrolysate. J Agric Food Chem. (2001) 49:2992–7. 10.1021/jf001119u [DOI] [PubMed] [Google Scholar]
- 43.Fan HB, Liao W, Wu JP. Molecular interactions, bioavailability, and cellular mechanisms of angiotensin-converting enzyme inhibitory peptides. J Food Biochem. (2019) 43:e12572. 10.1111/jfbc.12572 [DOI] [PubMed] [Google Scholar]
- 44.Brew K. Structure of human ACE gives new insights into inhibitor binding and design. Trends Pharmacol Sci. (2003) 24:391–4. 10.1016/s0165-6147(03)00196-2 [DOI] [PubMed] [Google Scholar]
- 45.Soleymanzadeh N, Mirdamadi S, Mirzaei M, Kianirad M. Novel β-casein derived antioxidant and ACE-inhibitory active peptide from camel milk fermented by Leuconostoc lactis PTCC1899: identification and molecular docking. Int Dairy J. (2019) 97:201–8. 10.1016/j.idairyj.2019.05.012 [DOI] [Google Scholar]
- 46.Masuyer G, Schwager SLU, Sturrock ED, Isaac RE, Acharya KR. Molecular recognition and regulation of human angiotensin-I converting enzyme (ACE) activity by natural inhibitory peptides. Sci Rep. (2012) 2:717. 10.1038/srep00717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Corradi HR, Chitapi I, Sewell BT, Georgiadis D, Dive V, Sturrock ED, et al. The structure of testis angiotensin-converting enzyme in complex with the C domain-specific inhibitor RXPA380. Biochemistry. (2007) 46:5473–8. 10.1021/bi700275e [DOI] [PubMed] [Google Scholar]
- 48.Aluko RE. Structure and function of plant protein-derived antihypertensive peptides. Curr Opin Food Sci. (2015) 4:44–50. 10.1016/j.cofs.2015.05.002 [DOI] [Google Scholar]
- 49.FitzGerald RJ, Meisel H. Milk protein-derived peptide inhibitors of angiotensin-I-converting enzyme. Brit J Nutr. (2000) 84:S33–7. 10.1017/S0007114500002221 [DOI] [PubMed] [Google Scholar]
- 50.Alemán A, Giménez B, Pérez-Santin E, Gómez-Guillén MC, Montero P. Contribution of Leu and Hyp residues to antioxidant and ACE-inhibitory activities of peptide sequences isolated from squid gelatin hydrolysate. Food Chem. (2011) 125:334–41. 10.1016/j.foodchem.2010.08.058 [DOI] [Google Scholar]
- 51.Dang Y, Zhou T, Hao L, Cao J, Sun Y, Pan D. In vitro and in vivo studies on the angiotensin-converting enzyme inhibitory activity peptides isolated from broccoli protein hydrolysate. J Agric Food Chem. (2019) 67:6757–64. 10.1021/acs.jafc.9b01137 [DOI] [PubMed] [Google Scholar]
- 52.Yoshikawa M, Fujita H, Matoba N, Takenaka Y, Yamamoto T, Yamauchi R, et al. Bioactive peptides derived from food proteins preventing lifestyle-related diseases. Biofactors. (2000) 12:143–6. 10.1002/biof.5520120122 [DOI] [PubMed] [Google Scholar]
- 53.Vermeirssen V, Van Camp J, Verstraete W. Bioavailability of angiotensin I converting enzyme inhibitory peptides. Br J Nutr. (2004) 92:357–66. 10.1079/bjn20041189 [DOI] [PubMed] [Google Scholar]
- 54.Tagliazucchi D, Martini S, Shamsia S, Helal A, Conte A. Biological activities and peptidomic profile of in vitro-digested cow, camel, goat and sheep milk. International Dairy Journal. (2018) 81:19–27. 10.1016/j.idairyj.2018.01.014 [DOI] [Google Scholar]
- 55.Fu Y, Alashi AM, Young JF, Therkildsen M, Aluko RE. Enzyme inhibition kinetics and molecular interactions of patatin peptides with angiotensin I-converting enzyme and renin. Int J Biol Macromol. (2017) 101:207–13. 10.1016/j.ijbiomac.2017.03.054 [DOI] [PubMed] [Google Scholar]
- 56.Tu M, Liu H, Zhang R, Chen H, Mao F, Cheng S, et al. Analysis and evaluation of the inhibitory mechanism of a novel angiotensin-i-converting enzyme inhibitory peptide derived from casein hydrolysate. J Agric Food Chem. (2018) 66:4139–44. 10.1021/acs.jafc.8b00732 [DOI] [PubMed] [Google Scholar]
- 57.Tu M, Wang C, Chen C, Zhang R, Liu H, Lu W, et al. Identification of a novel ACE-inhibitory peptide from casein and evaluation of the inhibitory mechanisms. Food Chem. (2018) 256:98–104. 10.1016/j.foodchem.2018.02.107 [DOI] [PubMed] [Google Scholar]
- 58.Garcia MC, Endermann J, Gonzalez-Garcia E, Marina ML. HPLC-Q-TOF-MS identification of antioxidant and antihypertensive peptides recovered from cherry (Prunus cerasus L) subproducts. J Agric Food Chem. (2015) 63:1514–20. 10.1021/jf505037p [DOI] [PubMed] [Google Scholar]
- 59.Li P, Jia J, Fang M, Zhang L, Guo M, Xie J, et al. In vitro and in vivo ACE inhibitory of pistachio hydrolysates and in silico mechanism of identified peptide binding with ACE. Process Biochem. (2014) 49:898–904. 10.1016/j.procbio.2014.02.007 [DOI] [Google Scholar]
- 60.Terashima M, Oe M, Ogura K, Matsumura S. Inhibition strength of short peptides derived from an ACE inhibitory peptide. J Agric Food Chem. (2011) 59:11234–7. 10.1021/jf202902r [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.