Abstract
Activation of the complement system is mediated by the interaction between pathogens and pattern recognition molecules (PRMs); mannose-binding lectin (MBL), ficolins, and collectin-10/-11 from the lectin pathway and C1q from the classical pathway. Lectin pathway activation specifically depends on proteases named MBL-associated serine proteases (MASPs) that are found in complexes with PRMs. In this study, we hypothesize that MASPs can recognize selected pathogens independently of PRMs. Using different clinical strains of opportunistic fungi, we have observed that MASPs directly recognize certain fungal pathogens in a way that can facilitate complement activation. Among these were Aspergillus fumigatus − a dangerous pathogen, especially for immunocompromised patients. In flow cytometry and fluorescence microscopy, we found that MASP-1 and −3 bound to all A. fumigatus growth stages (conidia, germ tubes, and hyphae), whereas rMASP-2 and the nonproteolytic rMAP-1 did not. Bound rMASPs could recruit rMBL and rficolin-3 to A. fumigatus conidia in a nonclassical manner and activate complement via rMASP-2. In experiments using recombinant and purified components, rMASP-1 increased the neutrophilic phagocytosis of conidia. In serum where known complement activation pathways were blocked, phagocytosis could be mediated by rMASP-3. We have encountered an unknown pathway for complement activation and found that MASP-1 and MASP-3 have dual functions as enzymes and as PRMs.
Keywords: Lectin pathway, Mannose-binding lectin-associated serine protease, Pattern recognition molecules, Fungi, Aspergillosis
Introduction
Host defense against pathogens is an important feature of the complement system. Through recognition of pathogen-associated molecular patterns (PAMPs) on, for example, fungi or bacteria, an immediate response is initiated to eliminate the microorganism. In the complement system, recognition of PAMPs is executed by collagenous pattern recognition molecules (PRMs) belonging to the two pathways called the lectin and the classical pathway. The lectin pathway PRMs, mannose-binding lectin (MBL), ficolin-1, −2, and −3, and collectin-10 and −11, bind directly to their respective PAMP ligand [1]. In contrast, the PRM in the classical pathway, C1q, in general binds to immune complexes composed of either IgG or IgM and antigen.
The complement system consists of proteolytic cascades that amplify the number of effector molecules around the activation site. The proteases that initiate the lectin pathway are three so-called MBL/ficolin/CL-associated serine proteases (MASP-1, −2, and −3). MASPs form complexes with PRMs, which put the MASPs in position to initiate the cascade on the surface of the pathogen [2]. MASP-1 cleaves complement component 2 (C2) while MASP-2 cleaves both C2 and C4. This results in the generation of the C3 convertase C4b2b (formerly known as C4bC2a) [3], which cleaves C3 into the important opsonin C3b and the anaphylatoxin C3a [4]. MASP-3 has recently been shown to be involved in activation of the third complement pathway, the alternative pathway, which is an amplification loop that boosts C3b formation [5]. MASP-1 and −3 are splicing variants of the MASP1 gene and have identical heavy chains but different protease domains. MASP1 also encodes a third variant called MBL/ficolin-associated protein-1 (MAP-1). MAP-1 lacks a protease domain and is therefore not involved in C3 convertase formation [6, 7]. MASP-2 is a product of a different gene, MASP2, but has the same overall domain organization as MASP-1 and −3 [8].
A. fumigatus is an opportunistic fungus that infects the lungs of immunocompromised patients via airborne conidia (spores). From the lungs, the fungus can spread to other organs by forming hyphae structures, which are outgrowths that can penetrate the epithelia [9]. The disease is called invasive aspergillosis and is associated with high mortality rates. Even though better prophylactic treatment has led to decreased mortality rates during recent years, the numbers are still high with a 30–50% mortality rate for infected acute myeloid leukemia patients and 20% for infected stem cell transplanted patients [10, 11]. Furthermore, A. fumigatus drug resistance challenges the continued use of drugs currently applied to treat infections in the clinic [12]. Thus, the development of new types of therapeutics is needed and an increased understanding of the interplay between the pathogen and the immune system is therefore valuable. In terms of complement, in vitro studies have deciphered the paths to activation [13, 14] and in vivo studies have shown that complement is important for fungal clearance [15, 16]. Additionally, complement works in close collaboration with PTX3, a nonredundant innate immune protection molecule against A. fumigatus [17, 18]. Based on preliminary findings, we hypothesized that MASP-1 and MASP-3 from the lectin pathway bind directly to A. fumigatus, and our objective was to explore whether this interaction could lead to complement activation and phagocytosis of the fungal pathogen.
Materials and Methods
Fungi
A. fumigatus strain 6871 was obtained from a fatal case of invasive aspergillosis and was a kind gift from Professor Luigina Romani from the Infectious Diseases Institute at the University of Perugia, Italy. A. terreus, A. niger, A. flavus, L. corymbifera, R. arrhizus, M. circinelloides, and C. albicans are clinical isolates collected at the Division of Hygiene and Medical Microbiology at the Medical University of Innsbruck, Austria. A. fumigatus strain Af293 is used in many basic research laboratories and we obtained the strain at the Division of Hygiene and Medical Microbiology at the Medical University of Innsbruck. Fungi were grown on Sabouraud glucose agar with chloramphenicol (89579; Sigma-Aldrich, Copenhagen, Denmark) for 4 days at 37°C except M. circinelloides that incubated at 30°C. C. albicans was afterward cultured in YPD liquid media to sporulate. Conidia were harvested in PBS/0.1% Tween 20, filtered, and washed thoroughly. Finally, conidia used for flow cytometry were heat-inactivated for 15 min at 121°C and aliquots were stored at −80°C. Living conidia of strain Af293 were kept at 4°C until it was used for maturation into different growth stages.
Primary Antibodies
In-house produced antibodies (Abs): mouse anti-MASP-1/-3/MAP-1 mAb 8B3 [19], mouse anti-ficolin-1/-2 mAb FCN106 [20], mouse anti-ficolin-3 mAb FCN313 [19], and mouse anti-CL-11 mAb Hyb15 [21]. Commercial Abs: anti-MASP-2 mAb 8B5 (HM2190; Hycult Biotech, Copenhagen, Denmark), anti-MASP-3 mAb 38:12-3 (HM2216; Hycult Biotech), mouse anti-MBL mAbs (HYB 131-1 and −11; Bioporto Diagnostics, Gentofte, Denmark), biotinylated rabbit anti-C4c and FITC-conjugated a-C3c pAb (0369 and F0201; Dako, Agilent Technologies, Glostrup, Denmark).
Secondary Antibodies
HRP-conjugated streptavidin (RPN1051; GE Healthcare, Broendby, Denmark), HRP-conjugated rabbit anti-rat (P0450; Dako), FITC-conjugated Streptavidin (F0422; Dako, Agilent Technologies), APC-conjugated streptavidin (SA1005; Thermo Fisher Scientific, Slangerup, Denmark), PE-conjugated goat anti-mouse (P9287; Sigma-Aldrich), Alexa flour 488-coupled goat anti-mouse (A10667; Thermo Fisher Scientific), and FITC-conjugated goat anti-mouse Ab (F0479; Dako, Agilent Technologies).
Inhibitory Antibodies
Anti-MBL mAb clone 3F8 makes a conformational change in MBL that inhibits ligand binding [22]. The related anti-MBL mAb clone 1C10 was used as a noninhibitory control [23]. Anti-C1q mAb clone CLB/C1q85 isotype IgG1 that binds to the globular head region of C1q (MW1828; Sanquin, Amsterdam, The Netherlands).
Recombinant and Serum Purified Proteins
Recombinant proteins were expressed and purified essentially as previously described [24, 25]. In short, rMBL, rficolin-1, rficolin-2, rficolin-3, rMASP-1, −2, and −3, and rMAP-1 were expressed in CHO-DG44 cells, whereas rMASP-1/-3 heavy chain was expressed in flip-in CHO cells. rficolin-1, −2, and −3 were cultivated in RPMI 1640 medium (Sigma-Aldrich) supplemented with 10% FCS, 100 U/mL penicillin, 0.1 mg/mL streptomycin, 2 mM L-glutamine, and 200 nM methotrexate. rMASP-1, −2, and −3, rMAP-1, and rMASP-1/-3 heavy chain was cultivated in Power CHO serum-free media (Lonza, Vallensbaek, Denmark) supplemented with 20 U/mL penicillin, 20 μg/mL streptomycin, and 2 mM L-glutamine. Miscellaneous MASPs were purified from a pool of fresh human serum from 3 healthy donors by indirect affinity chromatography using anti-ficolin-3 mAb FCN334 followed by elution with 10 mM EDTA. Purified complement components were purchased from Comptech, Tyler, TX: C2 (A112), C3 (A113), and C4 (A105).
Western Blotting
The content of MASPs and PRMs after MASP purification from serum was tested by Western blotting. The sample was run on a 4–12% bis-tris polyacrylamide gel under nonreducing conditions (Life Technologies); 0.5 μg of the following controls were included: rficolin-1, rficolin-2, rficolin-3, rMBL, and rCL-11. Proteins were blotted onto nitrocellulose membranes (GE Healthcare) and probed with anti-MASP-1/-3/MAP-1 mAb 8B3*, anti-MASP-3 mAb 38:12-3, anti-ficolin-1/-2 mAb FCN106*, anti-ficolin-3 mAb FCN334*, anti-MBL HYB131-11*, and anti-CL-11 mAb HYB15*. As secondary detection, HRP-conjugated streptavidin was used for the biotinylated Abs (*) and HRP-conjugated rabbit anti-rat was used for 38:12-3. Membranes were developed with Super Signal West Femto Chemiluminescent Substrate (Thermo Fisher Scientific) (*biotinylated).
MASP Binding to A. fumigatus Resting Conidia − Flow Cytometry
A. fumigatus conidia strain 6871 (1 × 107 cells/mL) were incubated with the following MASP variants for 30 min at 37°C in barbital (5 mM barbital sodium, 145 mM NaCl, 2 mM CaCl2, 1 mM MgCl2 [pH 7.4])/0.5% BSA buffer: 5 μg/mL rMASP-1 and −3 and serum purified MASPs; 2.5 μg/mL rMAP-1; 3.6 μg/mL rMASP-1/-3 heavy chain (equal molar amounts of 55.6 nM). The binding was measured with 10 μg/mL anti-MASP-1/-3/MAP-1 mAb 8B3 and 10 μg/mL mAb anti-MASP-2 mAb 8B5 followed by FITC-conjugated goat anti-mouse. Barbital/0.5% BSA buffer was used in all steps. Binding was measured as mean fluorescence intensity by gating on the conidia in a forward scatter/side scatter dot plot using flow cytometry (Gallios; Beckman Coulter). The same gating strategy was used for all experiments in which fluorescence was measured on the surface of conidia. Data were analyzed with Kaluza soſtware (Beckman Coulter).
MASP Binding to 4 Growth Stages of A. fumigatus − Microscopy
A. fumigatus conidia strain Af293 were grown in time intervals of 0, 4, 8, and 16 h at 37°C in RPMI 1640 medium to get different growth stages of the fungus (a different strain was used to test for strain specificity). For the 16 h experiments, A. fumigatus (1 × 105 cells/mL) was grown and stained directly on microscopy glass slides. For the 0, 4, and 8 h, A fumigatus (1 × 106 cells/mL) was grown and stained in facs tubes and finally transferred to glass slides by cytospin. The following MASP variants were applied for 30 min at 37°C in barbital/0.5% BSA buffer: 5 μg/mL rMASP-1 and −3 and serum purified MASPs; 2.5 μg/mL rMAP-1; and 3.6 μg/mL rMASP-1/-3 heavy chain (equal molar amounts of 55.6 nM). 10 μg/mL mAb 8B3 followed by Alexa flour 488-coupled goat anti-mouse was used to detect the binding. Images were obtained on a Zeiss Axio Observer using a Plan-Apochromat 63x/1.40 Oil DIC M27 objective and Zen pro software (Zeiss). Image processing was performed with the Zen Blue pro software (Zeiss).
MASP Binding to Other Fungi
Binding of rMASP-1 and −3 and rMAP-1 to various opportunistic fungi was tested using the following species: A. terreus, A. niger, A. flavus, L. corymbifera, R. arrhizus, M. circinelloides, and C. albicans. Binding was tested on heat-inactivated conidia and measured in flow cytometry as previously described.
PRM Recruitment
We tested if rMASPs bound to A. fumigatus conidia strain 6871 were able to recruit PRMs to the fungal surface. We used 5 μg/mL rMBL and rficolin-3 to use a binding and a nonbinding PRM. rMBL was allowed to associate with an MBL inhibitor (3F8) and a related mock inhibitor (1C10) and mannose before incubation with MASP-bound A. fumigatus (see “MASP binding to A. fumigatus − flow cytometry”). Binding of rMBL and rficolin-3 was detected with anti-MBL mAb HYB131-1* and anti-ficolin-3 mAb FCN334* followed by FITC-conjugated streptavidin. Measurements were done by flow cytometry.
Binding of MASP/PRM Complexes
5 μg/mL of rficolin-3 and rMASP-1 or −3 were preincubated for 30 min at room temperature before addition to A. fumigatus conidia strain 6871 conidia. Binding of rficolin-3 was detected with anti-ficolin-3 mAb FCN334 followed by FITC-conjugated goat anti-mouse. Measurements were done by flow cytometry.
PRM Recruitment and Complement Activation
Binding between MASPs and A. fumigatus conidia strain 6871 was established as previously described. Next, 5 μg/mL rMBL and rficolin-3 were combined with activated rMASP-2 to make rMBL/rMASP-2 and rficolin-3/rMASP-2 complexes, which were added to the conidia for 30 min at 37°C. After washing, purified complement components C2, C3, and C4 (2, 20, and 10 μg/mL) were added for 30 min at 37°C. C4b and C3b deposition were measured in flow cytometry using rabbit anti-C4c*/APC-conjugated streptavidin and FITC-conjugated a-C3c pAb.
Complement Activation in MBL-Defect Serum with C1q Inhibition
MASP-bound A. fumigatus conidia strain 6871 were incubated with MBL-defect serum combined with a C1q inhibitory mAb (CLB/C1q85) for 30 min at 37°C. C4b and C3b were measured in flow cytometry using rabbit anti-C4c*/APC-conjugated streptavidin and FITC-conjugated a-C3c pAb. Serum was obtained from an MBL-defect healthy individual (homozygous for the MBL2 D/D variant [rs5030737]), resulting in low MBL serum concentration and defect MBL.
Phagocytosis
PRM recruitment and complement activation (as described above) were performed with FITC-conjugated A. fumigatus conidia strain 6871. In parallel, neutrophils were isolated from EDTA plasma from a healthy donor using Polymorphprep (Axis-shield, Oslo, Norway). Neutrophils and treated FITC-conjugated conidia were mixed in a ratio of 1 neu:5 con (2 × 106:1 × 107 cells/mL) and incubated 20 min at 37°C. The level of neutrophilic phagocytosis was based on the level of FITC-positive neutrophils. Gating was set on granulocytes in an forward scatter/side scatter dot plot, and the level of fluorescence was measured in flow cytometry.
Statistics
Statistical analyses were performed with GraphPad Prism 6 (GraphPad Soſtware, San Diego, CA, USA). The results represent the means ± SD of 3 independent experiments. We used 1-way ANOVA, and paired samples with planned comparisons shown with brackets in the graphs. Bonferroni's correction was used to adjust for multiple comparisons. p values: ns p > 0.05; *p ≤ 0.05; **p ≤ 0.01.
Ethical Approval
Blood samples were obtained with consent from voluntary donors and approved by the Local Ethical Committee of the Capital Region of Denmark.
Results
Recombinant and Purified MASPs Bind to A. fumigatus
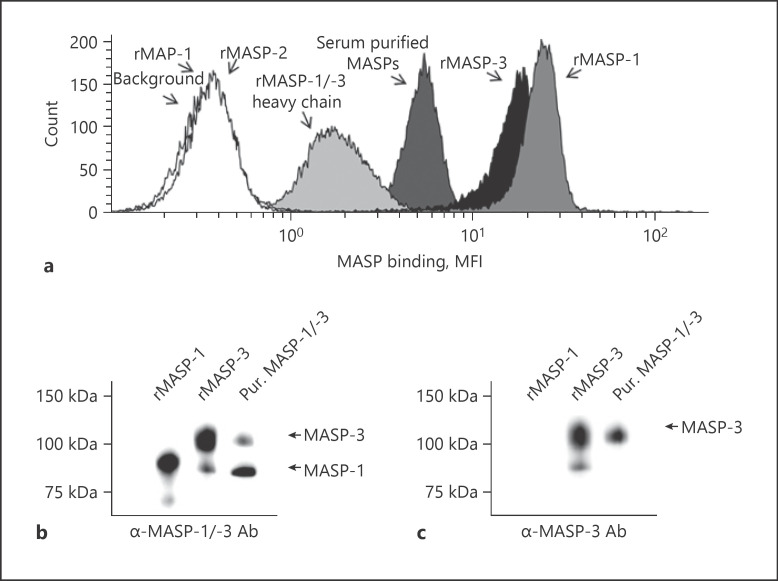
MASPs are a central initiating part of the complement system and observations of unexpected binding patterns made us investigate a possible interaction between these proteins and the fungus A. fumigatus. We measured the binding of various recombinant and serum-purified MASPs to heat-inactivated A. fumigatus conidia strain 6871 by flow cytometry. We have previously seen that heat inactivation does not compromise the binding of complement components [13]. The results showed that both rMASP-1 and rMASP-3, and to a minor degree rMASP-1/-3 heavy chain, bound to A. fumigatus. No binding was detected for rMAP-1 and rMASP-2 (Fig. 1a). Furthermore, we showed that serum-purified MASP-1 and −3 could also bind to A. fumigatus (Fig. 1a). Western blots of the purified MASPs were done to verify the content of MASP-1 and −3 (Fig. 1b, c). Additionally, we confirmed that the purified product did not contain detectable amounts of the PRMs ficolins, MBL, and CL-11 (see online suppl. Fig. 1; see www.karger.com/doi/10.1159/000514546 for all online suppl. material).
Fig. 1.
Recombinant and purified MASP-1 and −3 bind to a. fumigatus. a Histogram showing binding of the following MASP variants to A. fumigatus strain 6871 resting conidia: rMASP-1, −2, and −3, rMAP-1, rMASP-1/-3 heavy chain, and purified MASP-1/-3 from serum. b, c The purification was evaluated in Western blots detecting MASP-1 and −3 (b) and MASP-3 (c). Lane 1: rMASP-1. Lane 2: rMASP-3. Lane 3: purified MASP-1/-3 mix. The shown histogram is representative of 3 independent experiments (see also online suppl. Fig. 1 for further evaluation of the serum-purified MASPs). MASP, mannose-binding lectin-associated serine protease; MFI, mean fluorescence intensity.
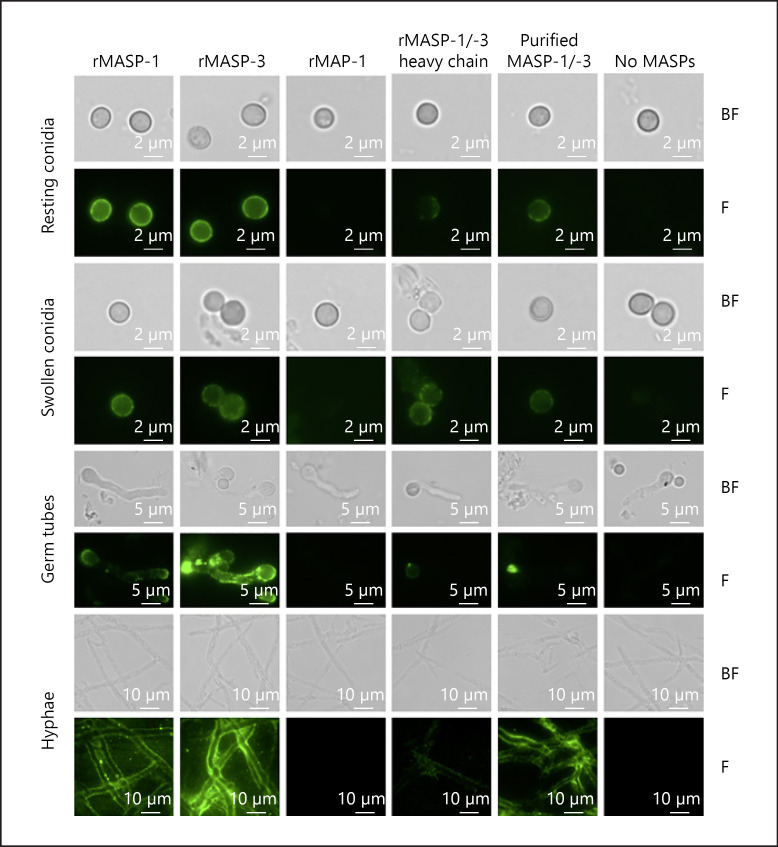
Next, we tested the binding of MASPs to different growth stages of living A. fumigatus using fluorescence microscopy. Results are visualized in Figure 2, which illustrates bright-field (B) and fluorescent (F) images of A. fumigatus conidia, swollen conidia, germ tubes, and hyphae of the strain Af293. These data showed that rMASP-1, MASP-3, and the serum-purified MASP-1/-3 bound to all growth stages. rMASP-1/-3 heavy chain showed a vague interaction and rMAP-1 did not bind to any of the 4 growth stages.
Fig. 2.
MASP-1 and MASP-3 bind to 4 growth stages of A. fumigatus. Resting conidia, swollen conidia, germ tubes, and hyphae from A. fumigatus strain Af293 (rows) were incubated with different MASP variants (columns). We applied a primary mAb (8B3) that recognizes a common epitope on all the included MASP variants. Binding was detected in fluorescence microscopy using an Alexa flour 488-conjugated secondary pAb. Both BF and F images are shown. Images were obtained on a Zeiss Axio Observer using a Plan-Apochromat. ×63/1.40 Oil DIC M27. Exposure time and image editing (brightness/background) were done equally between the samples and the negative controls (no MASPs). Shown images are representative of 2 independent experiments. MASP, mannose-binding lectin-associated serine protease; BF, bright-field; F, fluorescent.
MASP Binding to Different Opportunistic Fungi
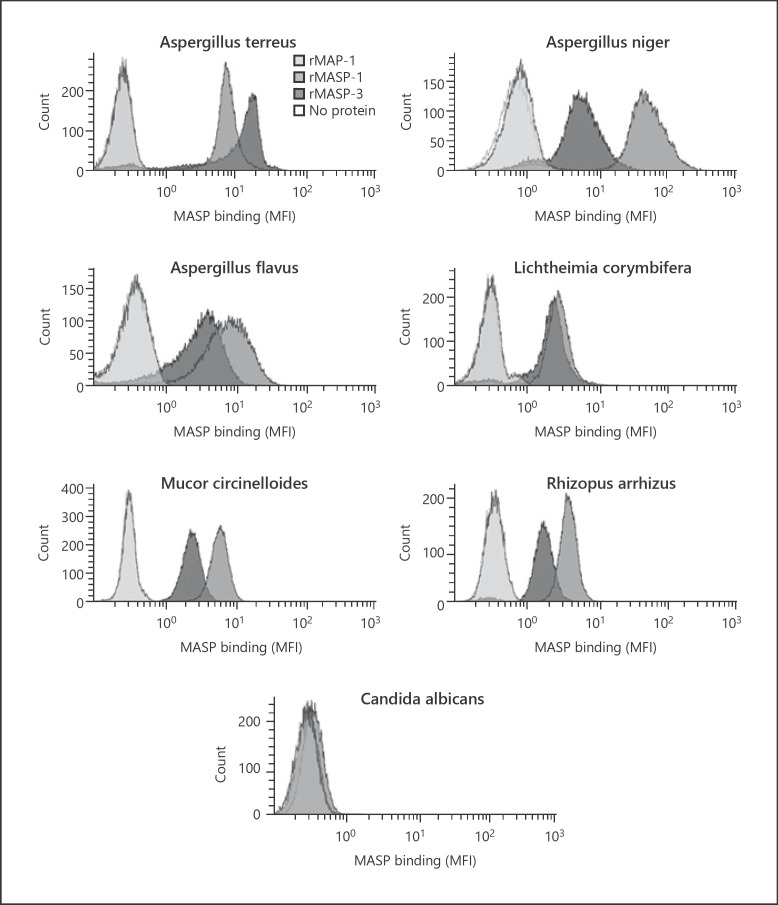
MASP binding was tested on different clinical isolates of the genus Aspergillus, different isolates from the order Mucorales, and the yeast C. albicans. rMASP-1 and −3 bound to all tested Aspergillus fungi (A. terreus, A. niger, and A. flavus) and Mucorales fungi (L. corymbifera, R. arrhizus, and M. circinelloides). However, no binding was observed on C. albicans. rMAP-1 did not bind to any of the fungi tested (Fig. 3).
Fig. 3.
MASP binding to different opportunistic fungi. Histograms showing binding of rMASP-1, rMASP-3, and rMAP-1 to conidia from various opportunistic fungi; A. terreus, A. niger, A. flavus, L. corymbifera, R. arrhizus, M. circinelloides, and C. albicans. Binding was measured with anti-MASP-1/-3/MAP-1 mAb 8B3 using flow cytometry. Histograms are representative of 2 independent experiments. MASP, mannose-binding lectin-associated serine protease; MFI, mean fluorescence intensity.
MASPs Recruit PRMs to the Fungal Surface
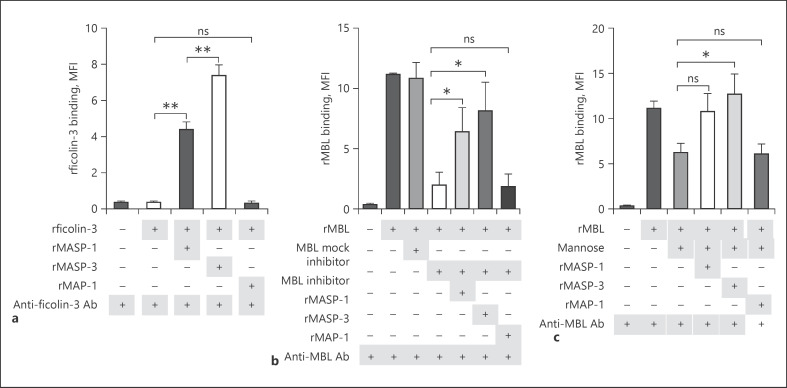
To evaluate the ability of MASPs to recruit PRMs to the fungal surface, we included recombinant MBL and ficolin-3. These served as two differently behaving PRMs concerning A. fumigatus, a PRM known to bind (MBL) and one that does not bind A. fumigatus (ficolin-3). As expected, rficolin-3 did not independently bind, but we found that rMASP-1 and −3 could recruit rficolin-3 to the fungus (Fig. 4a). rMBL bound to A. fumigatus independently; however, upon inhibition of MBL binding by either an inhibitory Ab (Fig. 4b) or mannose (Fig. 4c), it appeared that MBL interaction with the fungi was reestablished by the MASPs. The presence of rMAP-1 did not affect the binding of rficolin-3 and rMBL.
Fig. 4.
MASPs recruit PRMs to the fungal surface. A. fumigatus strain 6871 resting conidia were incubated with rMASP-1, rMASP-3, and rMAP-1 to test the recruitment of rficolin-3 (a), rMBL + MBL inhibitor (b), and rMBL + mannose (c). Results represent the means of 3 independent experiments ±SD. *p ≤ 0.05, **p ≤ 0.01, 1-way ANOVA, Bonferroni's correction. MASP, mannose-binding lectin-associated serine protease; PRMs, pattern recognition molecules; MBL, mannose-binding lectin; MFI, mean fluorescence intensity.
Preformed Complexes of Ficolin-3 and MASPs Bind to A. fumigatus
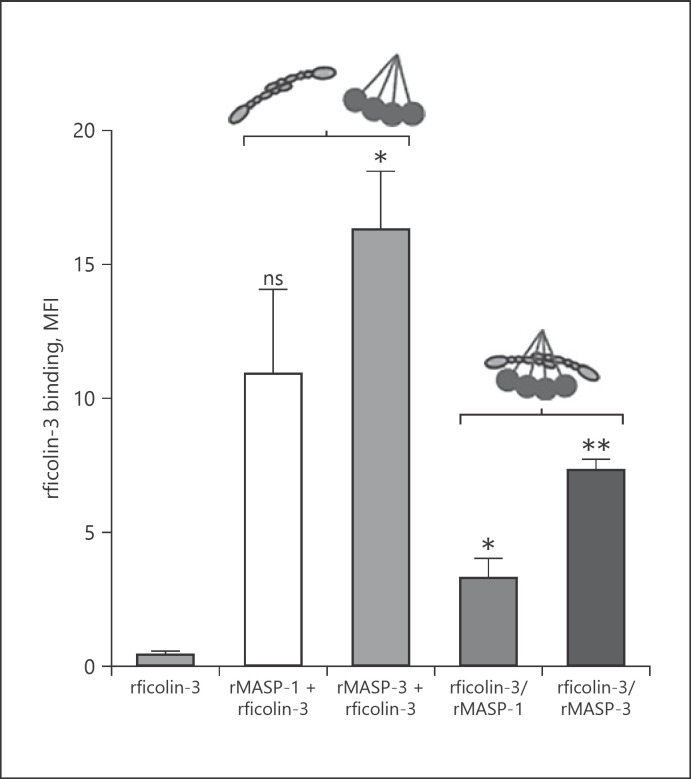
The presence of free MASPs (not associated with PRMs) in the blood/tissues is debatable and thus we tested whether complexes between MASPs and ficolin-3 could also bind to A. fumigatus. As Figure 5 shows, both rMASP-1/rficolin-3 and rMASP-3/rficolin-3 could bind to the conidia, whereas rficolin-3 alone could not. Though it appears that binding of complexes is less pronounced than stepwise binding (first rMASPs then rficolin-3), it is difficult to make a direct comparison as the concentrations of formed rficolin-3/rMASP complexes versus free proteins cannot be accurately determined and normalized. The observation that the level of rficolin-3 binding seems to be higher via rMASP-3 compared to rMASP-1 is perhaps not surprising as it has previously been suggested that MASP-3 has a preference for ficolin-3 as its associated PRM [19].
Fig. 5.
Preformed complexes of rficolin-3 and rMASPs bind to A. fumigatus. Preformed complexes of rficolin-3 and rMASP-1 or rMASP-3 were incubated with A. fumigatus strain 6871 resting conidia and afterward binding of rficolin-3 was measured using flow cytometry. Stepwise incubations as shown in Fig. 4 were also included. Results represent the means of 3 independent experiments ±SD. *p ≤ 0.05, **p ≤ 0.01, 1-way ANOVA, Bonferroni's correction. MASP, mannose-binding lectin-associated serine protease; MFI, mean fluorescence intensity.
MASP-Recruited PRM/MASP-2 Complexes Activate Complement
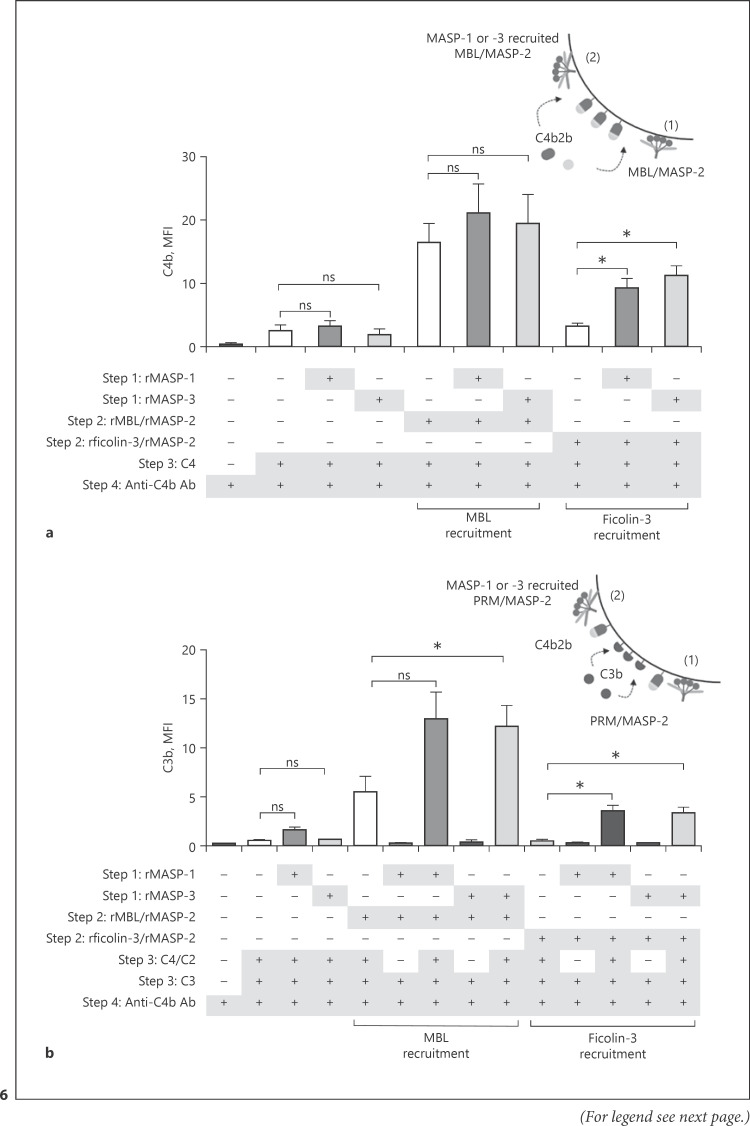
Next, we addressed whether MASP-recruited PRMs retained their function in complement activation. Purified complement components C2, C4, and C3 were applied after rMASP-1 and rMASP-3-recruitment of rficolin-3/rMASP-2 and rMBL/rMASP-2 complexes (rMASP-2 was in a preactivated enzyme state). MASP recruitment of rficolin-3/rMASP-2 complexes significantly increased both C4b and C3b (Fig. 6a, b). Recruitment of rMBL/rMASP-2 slightly increased C4b and more clearly elevated C3b deposition, though only significantly for rMASP-3 (Fig. 6a, b). As shown in Figure 6b, there was no C3b deposition without the presence of C2 and C4, implying that C3 convertase (C4b2b) formation was required for C3 cleavage. The recruitment/activation mechanism is illustrated in Figure 10.
Fig. 6.
MASP-recruited PRM/MASP-2 complexes activate complement. A. fumigatus strain 6871 resting conidia were incubated with rMASP-1 or rMASP-3 to recruit PRM/MASP-2 complexes before the addition of purified C2, C4, and C3 and deposition of C4b (a), C3b (b) on the cell surface was measured by flow cytometry. When using rMBL/rMASP-2 complexes, both situation 1 and 2 in the top right drawing is possible. For rficolin-3/rMASP-2 complexes, only rMASP-recruitment, situation 2, is possible as ficolin-3 does not bind independently. Results represent the means of 3 independent experiments ±SD. *p ≤ 0.05, **p ≤ 0.01, 1-way ANOVA, Bonferroni's correction. MASP, mannose-binding lectin-associated serine protease; PRM, pattern recognition molecule; MBL, mannose-binding lectin; MFI, mean fluorescence intensity.
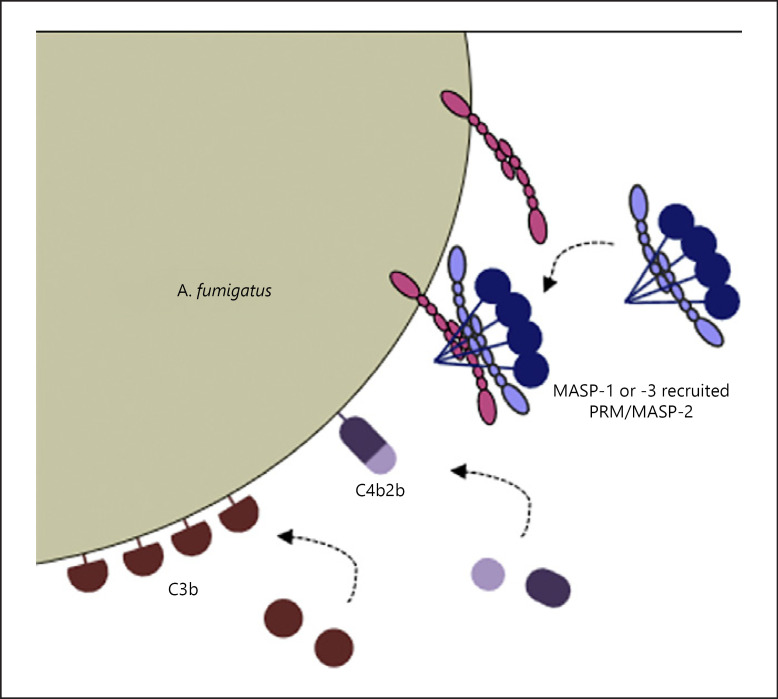
Fig. 10.
Illustration of the mechanism behind MASP recruitment of PRMs and subsequent complement activation. MASP-1 and MASP-3 can bind directly to the opportunistic pathogenic fungus A. fumigatus in a manner possibly implicating the CCP2 and protease domains of the MASPs. In theory, MASPs can bind with either 1 or 2 “arms” of the dimer − here the latter is depicted. The bound MASPs possibly recruit PRMs and thereby facilitate complement activation leading to C3b deposition on A. fumigatus. MASP, mannose-binding lectin-associated serine protease; PRM, pattern recognition molecule.
MASP-1 Facilitates Phagocytosis of A. fumigatus in a Pure System
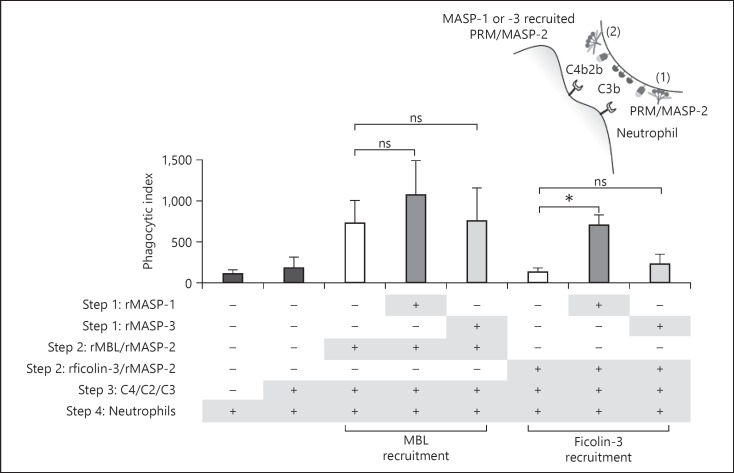
We speculated if the MASP-generated C3b facilitated phagocytosis of A. fumigatus conidia, as C3b is the main opsonizing molecule in the complement system. To test this, we used purified components in the same manner as in the previous experiment and added isolated neutrophils as the final step. The phagocytic index was measured by the percentage of phagocytic neutrophils × mean fluorescence intensity and the results are shown in Figure 7. The recruitment of rficolin-3/rMASP-2 complexes by rMASP-1 resulted in significantly increased phagocytosis, whereas rMASP-3 did not have any significant effect. Hence, the MASP-3-driven increase in C3b deposition seen in Figure 6b did not translate into increased phagocytosis. Additionally, increased C3b via rMBL/rMASP-2 facilitated by either of the rMASPs did not generate significantly increased phagocytosis (Fig. 7).
Fig. 7.
MASP-1 facilitates phagocytosis of A. fumigatus in a pure system. A. fumigatus strain 6871 FITC-conjugated resting conidia were incubated with rMASP-1 or rMASP-3 to recruit rPRM/rMASP-2 complexes. Next, purified C4, C2, and C3 were added and subsequently neutrophils isolated from human blood. Phagocytosis was measured by flow cytometry and based on that the phagocytic index was calculated as the percentage of phagocytizing neutrophils × MFI. Situation 1 and 2 in the top right drawing represent the action of rMBL/rMASP-2 complexes, whereas only situation 2 occurs for rficolin-3/rMASP-2. Graphs show the phagocytic index after rMASP-recruitment of rMBL/rMASP-2 complexes or rficolin-3/rMASP-2 complexes. Results represent the means of 3 independent experiments ±SD. *p ≤ 0.05, **p ≤ 0.01, 1-way ANOVA, Bonferroni's correction. MASP, mannose-binding lectin-associated serine protease; MFI, mean fluorescence intensity; PRMs, pattern recognition molecules; MBL, mannose-binding lectin.
A. fumigatus-Bound MASPs Activate Complement in Serum
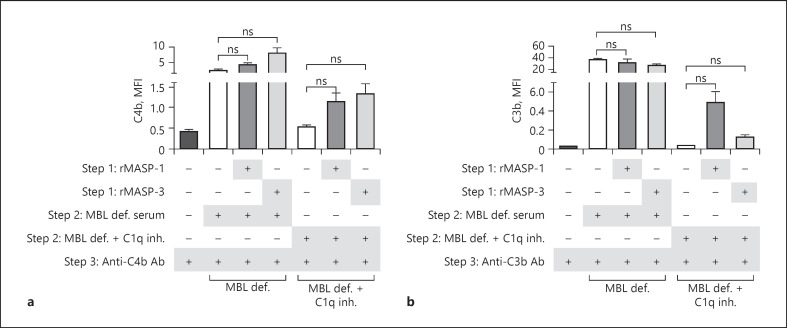
To further investigate the consequence of MASPs binding to the surface of A. fumigatus, we tested the effects in human serum. More specifically, in MBL-defect serum combined with a C1q inhibitory mAb to exclude the contribution from previously identified classical and lectin pathway activation on A. fumigatus [13]. Figure 8a and b shows a tendency that prebound rMASP-1 and MASP-3 increased the C4b deposition and that rMASP-1 was able to induce a higher C3b deposition than rMASP-3. The experiment with MBL-defect serum without C1q inhibition shows that classical pathway activation, via C1q, exceeds the MASP-driven activation.
Fig. 8.
A. fumigatus-bound MASPs activate complement in serum. A. fumigatus strain 6871 resting conidia were incubated with rMASP-1 or rMASP-3 and then added to MBL-defect serum ± C1q inhibitory mAb. Deposition of C4b, C3b was detected by flow cytometry. Results represent the means of 3 independent experiments ±SD. *p ≤ 0.05, 1-way ANOVA, Bonferroni's correction. MASP, mannose-binding lectin-associated serine protease; MBL, mannose-binding lectin.
MASP-3 Facilitates Phagocytosis of A. fumigatus in Serum
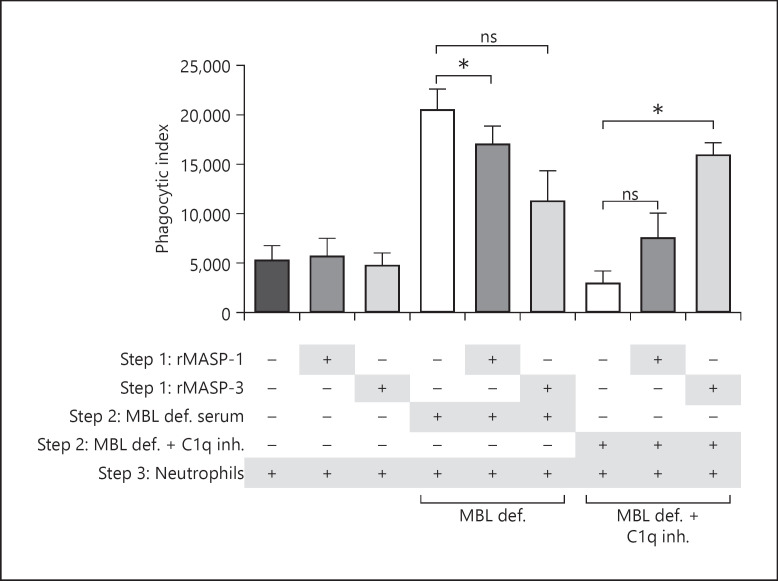
Finally, we did the follow-up phagocytosis assay in connection with the previous complement activation experiments in serum. In this setup, opsonization using rMASP-3 and MBL-defect serum + C1q inhibitory mAb significantly increased the level of phagocytosis. Though rMASP-1 was best at mounting a C3b response on A. fumigatus, rMASP-1 did not promote phagocytosis as potently as rMASP-3. In MBL-defect serum, it seemed that the MASPs lowered the level of phagocytosis (Fig. 9). To be noted, the negative control values differ from the controls in the previous phagocytosis experiment, which is most likely due to neutrophil donor differences.
Fig. 9.
MASP-3 facilitates phagocytosis of A. fumigatus in a serum system. A. fumigatus strain 6871 FITC-conjugated resting conidia were incubated with rMASP-1 or rMASP-3 before adding MBL-defect serum ± C1q inhibitory mAb and subsequently neutrophils isolated from human plasma. Phagocytosis was measured by flow cytometry and based on that the phagocytic index was calculated as the percentage of phagocytizing neutrophils × MFI. Results represent the means of 3 independent experiments ±SD. *p ≤ 0.05, 1-way ANOVA, Bonferroni's correction. MASP, mannose-binding lectin-associated serine protease; MFI, mean fluorescence intensity; MBL, mannose-binding lectin.
Discussion/Conclusion
In our investigations of complement responses to A. fumigatus, we found that recombinant and purified MASP-1 and −3 bound directly to the fungus. Besides A. fumigatus, we also observed binding to other opportunistic fungal species. This was surprising since MASPs have not previously been found to have pattern recognition traits. MASPs are known to associate with PRMs in the lectin pathway where they function as initiators of the proteolytic cascade. PRMs can bind various molecular patterns on pathogens and altered self-surface antigens via their C-terminal recognition domains. The binding domains are bundled up via collagen-like domains toward the N-terminal end and oligomerization of the polypeptide chains gives a high avidity toward the ligand [26]. MASPs have a completely different structure and tend to form head-to-tail dimers [27]. MASPs have a light chain containing a protease domain (SP) and a heavy chain (CUB1-EGF-CUB2-CCP1-CCP2) through which they dimerize and bind to PRMs [28, 29].
By studying the binding patterns of the different MASP variants, we can pinpoint the A. fumigatus-interacting binding site to the CCP2-SP domain area, as MASP-1, MASP-3, and their common heavy chain bind to the fungi whereas MAP-1 does not. The heavy chain of MAP-1 differs from MASP-1 and −3 by having only the first CCP1 domain and 17 unique residues as the C-terminal [27]. This indicates that the CCP2 domain might have a central function in the interaction with the fungal cell surface. Moreover, the protease domain may stabilize the binding since rMASP-1 and −3 had a better interaction with A. fumigatus than the heavy chain itself. We found that rMASP-1 and −3 bound to all the different growth stages of A. fumigatus, from resting conidia that initiate the infection to hyphae structures that extend the fungal territory and disseminate the bloodstream. Thus, despite cell wall changes in the developmental phases of the fungus [30], rMASP-1 and rMASP-3 retained their binding ability. The binding did not appear to be strain specific and conidia of other Aspergillus species, as well as different Mucorales fungi, also bound rMASP-1 and −3. Interestingly C. albicans did not bind and it could be speculated whether the morphology is playing a role as C. albicans is a yeast, whereas the other tested fungi are filamentous.
The functional part of our experiments showed that A. fumigatus-bound rMASP-1 and rMASP-3 were able to activate complement via recruited rPRM/rMASP-2 complexes (rMBL/rMASP-2 and rficolin-3/rMASP-2) (illustrated in Fig. 10). The premise for complement activation in this setup was that the PRMs were able to bind two MASP dimers simultaneously, namely, the A. fumigatus-bound MASP-1 or MASP-3 and the PRM-associated MASP-2. Gringas et al. [29] have proposed that MASP dimers bind two collagen-like stalks; thus, in theory four stalks are the minimum amount to create the basis for having two MASP dimers bound to one PRM. We have previously speculated if this is a feasible PRM-MASP interaction model for activating complement [31] and Degn et al. [32] have shown that a pentameric molecule is likely the minimum level of PRM oligomerization, which is able to contain two MASP dimers. The rficolin-3 and rMBL used in our experiments are highly oligomerized, which means that these proteins are likely able to bind 2 MASP dimers. In serum, MBL mostly forms trimers and tetramers of the trimeric units [33], whereas ficolin-3 may primarily exist in hexamers [34], which altogether might explain the complexity and heterogenicity of the activation systems. Complement activation was shown in a controlled setting using recombinant and purified serum proteins. Thus, binding and/or activation were not mediated indirectly by unknown components present in the system, thereby providing a clear basic understanding of the mechanisms. However, it has the disadvantage of oversimplifying the system. Hence, we also tested the effects in serum by eliminating the two PRM-activation sources on A. fumigatus − C1q/classical and MBL-driven lectin pathway [13]. Both rMASP-1 and −3 increased the deposition of C4b, but interestingly only rMASP-1 increased C3b (Fig. 8). According to the literature, MASP-1 might be able to directly cleave C3 [4, 35], whereas a similar mechanism has not been described for MASP-3, which could explain the discrepancy. However, our experiments using purified components do not indicate that surface-bound rMASP-1 cleaves C3 directly. Differences in substrate specificity between MASP-1 and MASP-3 could be an alternative explanation; it is known that MASP-3 cannot cleave C2, whereas surface-bound rMASP-1 could contribute to cleavage of endogenous C2 to induce more C3 convertase (C4b2b) and thereby more C3b. This could indeed be reflected in the serum setting where all downstream components are present.
The MASP-1 driven C3b increase was only detectable if both the classical and MBL-driven lectin pathway were blocked (Fig. 8b). Thus, MASP binding could represent an additional part of complement activation and be more important in extravascular situations where the amount of other PRMs might be low. On the other hand, MASP-1 and MASP-3 have different tasks in the immune system that could also be relevant concerning fungal-binding. Previous studies have shown that MASP-3 cleaves pro-factor D to active factor D, a pivotal molecule of the alternative pathway that aids C3b amplification on resting conidia of A. fumigatus [5, 14]. MASP-3 has also been shown to some degree to inhibit lectin pathway activation [19, 36], but the true physiological relevance of these observations remains to be established. Regarding MASP-1, there are multiple functions to consider; it can activate the coagulation [37] and kallikrein-kinin systems [38] and activate endothelial cells [39]. The purpose of the A. fumigatus-MASP interaction found in our study could therefore also be related to some of these functions.
Furthermore, we speculate if neutrophilic phagocytosis is directly influenced by MASP-1 and MASP-3. In the pure system using recombinant and purified components, binding of rMASP-1 and not rMASP-3 induced phagocytosis (Fig. 7). Surprisingly, rMASP-3 was the main facilitator of phagocytosis in a serum situation where classical and lectin PRM interference was abolished. The discrepancy between the pure and serum phagocytosis experiments suggest that the effect of MASP-3 may be dependent on a cofactor in serum. And regarding MASP-1, C1-inhibitor may sterically hinder the MASP-1-driven phagocytosis in serum, whereas MASP-3 (i.e., not influenced by C1-inhibitor) is free to interact with a hypothetical receptor.
MASP-1 and MASP-3 from the lectin pathway seem to work as direct recognition molecules toward A. fumigatus, which implicate more broad functions of innate effector enzymes than previously anticipated. Moreover, this study expands our current view on the complement system in general and the lectin pathway in particular.
Statement of Ethics
Blood samples were obtained with consent from voluntary donors and approved by the Local Ethical Committee of the Capital Region of Denmark.
Conflict of Interest Statement
The authors have no conflict of interest to declare.
Funding Sources
The work was carried out as a part of the BRIDGE − Translational Excellence Programme (bridge.ku.dk) at the Faculty of Health and Medical Sciences, University of Copenhagen, funded by the Novo Nordisk Foundation. Grant agreement No. NNF18SA0034956. The work was also supported by grants from the Danish Research Foundation of Independent Research (DFF-6110-00489), the Sven Andersen Research Foundation, Novo Nordisk Research Foundation, Rigshospitalet and the Austrian Science Funds (FWF) doctoral program “Host response in opportunistic infections” (HOROS, W1253).
Author Contributions
A.R.: study design, experimental work, data interpretation, drafting the article, and final approval. R.W.: critical revision of the article and final approval. P.G.: data interpretation, critical revision of the article, and final approval. M.-O.S.: study design, data interpretation, critical revision of the article, and final approval.
Supplementary Material
Supplementary data
Acknowledgements
The authors thank Jytte Bryde Clausen for technical assistance regarding the production of recombinant proteins and Katrine Pilely for assisting with the microscopy.
References
- 1.Garred P, Genster N, Pilely K, Bayarri-Olmos R, Rosbjerg A, Ma YJ, et al. A journey through the lectin pathway of complement-MBL and beyond. Immunol Rev. 2016;274((1)):74–97. doi: 10.1111/imr.12468. [DOI] [PubMed] [Google Scholar]
- 2.Wallis R. Interactions between mannose-binding lectin and MASPs during complement activation by the lectin pathway. Immunobiology. 2007;212((4–5)):289–99. doi: 10.1016/j.imbio.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bohlson SS, Garred P, Kemper C, Tenner AJ. Complement nomenclature-deconvoluted. Front Immunol. 2019;10:1–6. doi: 10.3389/fimmu.2019.01308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsushita M, Thiel S, Jensenius JC, Terai I, Fujita T. Proteolytic activities of two types of mannose-binding lectin-associated serine protease. J Immunol. 2000;165((5)):2637–42. doi: 10.4049/jimmunol.165.5.2637. [DOI] [PubMed] [Google Scholar]
- 5.Dobó J, Szakács D, Oroszlán G, Kortvely E, Kiss B, Boros E, et al. MASP-3 is the exclusive pro-factor D activator in resting blood: the lectin and the alternative complement pathways are fundamentally linked. Sci Rep. 2016;6:31877. doi: 10.1038/srep31877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skjoedt MO, Hummelshoj T, Palarasah Y, Honore C, Koch C, Skjodt K, et al. A novel mannose-binding lectin/ficolin-associated protein is highly expressed in heart and skeletal muscle tissues and inhibits complement activation. J Biol Chem. 2010;285((11)):8234–43. doi: 10.1074/jbc.M109.065805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Degn SE, Hansen AG, Steffensen R, Jacobsen C, Jensenius JC, Thiel S. MAp44, a human protein associated with pattern recognition molecules of the complement system and regulating the lectin pathway of complement activation. J Immunol. 2009;183((11)):7371–8. doi: 10.4049/jimmunol.0902388. [DOI] [PubMed] [Google Scholar]
- 8.Gál P, Dobó J, Závodszky P, Sim RB. Early complement proteases: C1r, C1s and MASPs. A structural insight into activation and functions. Mol Immunol. 2009;46((14)):2745–52. doi: 10.1016/j.molimm.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 9.Dagenais TR, Keller NP. Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin Microbiol Rev. 2009;22((3)):447–65. doi: 10.1128/CMR.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dragonetti G, Criscuolo M, Fianchi L, Pagano L. Invasive aspergillosis in acute myeloid leukemia: are we making progress in reducing mortality? Med Mycol. 2017;55((1)):82–6. doi: 10.1093/mmy/myw114. [DOI] [PubMed] [Google Scholar]
- 11.Neofytos D, Horn D, Anaissie E, Steinbach W, Olyaei A, Fishman J, et al. Epidemiology and outcome of invasive fungal infection in adult hematopoietic stem cell transplant recipients: analysis of multicenter prospective antifungal therapy (PATH) alliance registry. Clin Infect Dis. 2009;48((3)):265–73. doi: 10.1086/595846. [DOI] [PubMed] [Google Scholar]
- 12.Vermeulen E, Lagrou K, Verweij PE. Azole resistance in Aspergillus fumigatus: a growing public health concern. Curr Opin Infect Dis. 2013 Dec;26((6)):493–500. doi: 10.1097/QCO.0000000000000005. [DOI] [PubMed] [Google Scholar]
- 13.Rosbjerg A, Genster N, Pilely K, Skjoedt MO, Stahl GL, Garred P. Complementary roles of the classical and lectin complement pathways in the defense against Aspergillus fumigatus. Front Immunol. 2016;7:473. doi: 10.3389/fimmu.2016.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braem SG, Rooijakkers SH, van Kessel KP, de Cock H, Wösten HA, van Strijp JA, et al. Effective neutrophil phagocytosis of Aspergillus fumigatus is mediated by classical pathway complement activation. J Innate Immun. 2015;7((4)):364–74. doi: 10.1159/000369493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsoni SV, Kerrigan AM, Marakalala MJ, Srinivasan N, Duffield M, Taylor PR, et al. Complement C3 plays an essential role in the control of opportunistic fungal infections. Infect Immun. 2009;77((9)):3679–85. doi: 10.1128/IAI.00233-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaur S, Gupta VK, Thiel S, Sarma PU, Madan T. Protective role of mannan-binding lectin in a murine model of invasive pulmonary aspergillosis. Clin Exp Immunol. 2007;148((2)):382–9. doi: 10.1111/j.1365-2249.2007.03351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garlanda C, Hirsch E, Bozza S, Salustri A, De Acetis M, Nota R, et al. Non-redundant role of the long pentraxin PTX3 in anti-fungal innate immune response. Nature. 2002 Nov;420((6912)):182–6. doi: 10.1038/nature01195. [DOI] [PubMed] [Google Scholar]
- 18.Inforzato A, Doni A, Barajon I, Leone R, Garlanda C, Bottazzi B, et al. PTX3 as a paradigm for the interaction of pentraxins with the complement system. Semin Immunol. 2013;25((1)):79–85. doi: 10.1016/j.smim.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Skjoedt MO, Palarasah Y, Munthe-Fog L, Jie Ma Y, Weiss G, Skjodt K, et al. MBL-associated serine protease-3 circulates in high serum concentrations predominantly in complex with Ficolin-3 and regulates Ficolin-3 mediated complement activation. Immunobiology. 2010;215((11)):921–31. doi: 10.1016/j.imbio.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Honoré C, Rørvig S, Munthe-Fog L, Hummelshøj T, Madsen HO, Borregaard N, et al. The innate pattern recognition molecule Ficolin-1 is secreted by monocytes/macrophages and is circulating in human plasma. Mol Immunol. 2008;45((10)):2782–9. doi: 10.1016/j.molimm.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 21.Bayarri-Olmos R, Kirketerp-Moller N, Pérez-Alós L, Skjodt K, Skjoedt MO, Garred P. Development of a quantitative assay for the characterization of human collectin-11 (CL-11, CL-K1) Front Immunol. 2018;9((September)):2238. doi: 10.3389/fimmu.2018.02238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao HUI, Wakamiya N, Suzuki Y, Hamonko MT, Stahl GL. Identification of human mannose binding lectin (MBL) recognition sites for novel inhibitory antibodies. Hybrid Hybridomics. 2002 Feb;21((1)):25–36. doi: 10.1089/15368590252917610. [DOI] [PubMed] [Google Scholar]
- 23.Collard CD, Väkevä A, Morrissey MA, Agah A, Rollins SA, Reenstra WR, et al. Complement activation after oxidative stress: role of the lectin complement pathway. Am J Pathol. 2000;156((5)):1549–56. doi: 10.1016/S0002-9440(10)65026-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsen F, Madsen HO, Sim RB, Koch C, Garred P. Disease-associated mutations in human mannose-binding lectin compromise oligomerization and activity of the final protein. J Biol Chem. 2004 May;279((20)):21302–11. doi: 10.1074/jbc.M400520200. [DOI] [PubMed] [Google Scholar]
- 25.Hummelshoj T, Thielens NM, Madsen HO, Arlaud GJ, Sim RB, Garred P. Molecular organization of human Ficolin-2. Mol Immunol. 2007;44((4)):401–11. doi: 10.1016/j.molimm.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 26.Kjaer TR, Jensen L, Hansen A, Dani R, Jensenius JC, Dobó J, et al. Oligomerization of mannan-binding lectin dictates binding properties and complement activation. Scand J Immunol. 2016;84((1)):12–9. doi: 10.1111/sji.12441. [DOI] [PubMed] [Google Scholar]
- 27.Skjoedt MO, Roversi P, Hummelshøj T, Palarasah Y, Rosbjerg A, Johnson S, et al. Crystal structure and functional characterization of the complement regulator mannose-binding lectin (MBL)/ficolin-associated protein-1 (MAP-1) J Biol Chem. 2012;287((39)):32913–21. doi: 10.1074/jbc.M112.386680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teillet F, Gaboriaud C, Lacroix M, Martin L, Arlaud GJ, Thielens NM. Crystal structure of the CUB1-EGF-CUB2 domain of human MASP-1/3 and identification of its interaction sites with mannan-binding lectin and ficolins. J Biol Chem. 2008;283((37)):25715–24. doi: 10.1074/jbc.M803551200. [DOI] [PubMed] [Google Scholar]
- 29.Gingras AR, Girija UV, Keeble AH, Panchal R, Mitchell DA, Moody PC, et al. Structural basis of mannan-binding lectin recognition by its associated serine protease MASP-1: implications for complement activation. Structure. 2011 Nov;19((11)):1635–43. doi: 10.1016/j.str.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 30.Latgé J-P, Beauvais A, Chamilos G. The cell wall of the human fungal pathogen Aspergillus fumigatus: biosynthesis, organization, immune response, and virulence. Annu Rev Microbiol. 2017;71((1)):99–116. doi: 10.1146/annurev-micro-030117-020406. [DOI] [PubMed] [Google Scholar]
- 31.Rosbjerg A, Munthe-Fog L, Garred P, Skjoedt MO. Heterocomplex Formation between MBL/Ficolin/CL-11-associated serine protease-1 and −3 and MBL/Ficolin/CL-11-associated protein-1. J Immunol. 2014 Mar;192((9)):4352–60. doi: 10.4049/jimmunol.1303263. [DOI] [PubMed] [Google Scholar]
- 32.Degn SE, Kjaer TR, Kidmose RT, Jensen L, Hansen AG, Tekin M, et al. Complement activation by ligand-driven juxtaposition of discrete pattern recognition complexes. Proc Natl Acad Sci U S A. 2014;111((37)):13445–50. doi: 10.1073/pnas.1406849111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teillet F, Dublet B, Andrieu JP, Gaboriaud C, Arlaud GJ, Thielens NM. The two major oligomeric forms of human mannan-binding lectin: chemical characterization, carbohydrate-binding properties, and interaction with MBL-associated serine proteases. J Immunol. 2005;174((5)):2870–7. doi: 10.4049/jimmunol.174.5.2870. [DOI] [PubMed] [Google Scholar]
- 34.Hummelshoj T, Fog LM, Madsen HO, Sim RB, Garred P. Comparative study of the human ficolins reveals unique features of Ficolin-3 (Hakata antigen) Mol Immunol. 2008 Mar;45((6)):1623–32. doi: 10.1016/j.molimm.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 35.Matsushita M, Fujita T. Cleavage of the third component of complement (C3) by mannose-binding protein-associated serine protease (MASP) with subsequent complement activation. Immunobiology. 1995;194((4–5)):443–8. doi: 10.1016/S0171-2985(11)80110-5. [DOI] [PubMed] [Google Scholar]
- 36.Møller-Kristensen M, Thiel S, Sjöholm A, Matsushita M, Jensenius JC. Cooperation between MASP-1 and MASP-2 in the generation of C3 convertase through the MBL pathway. Int Immunol. 2007;19((2)):141–9. doi: 10.1093/intimm/dxl131. [DOI] [PubMed] [Google Scholar]
- 37.Dobó J, Schroeder V, Jenny L, Cervenak L, Závodszky P, Gál P. Multiple roles of complement MASP-1 at the interface of innate immune response and coagulation. Mol Immunol. 2014 Oct;61((2)):69–78. doi: 10.1016/j.molimm.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 38.Dobó J, Major B, Kékesi KA, Szabó I, Megyeri M, Hajela K, et al. Cleavage of kininogen and subsequent bradykinin release by the complement component: mannose-binding lectin-associated serine protease (MASP)-1. PLoS One. 2011;6((5)):e20036. doi: 10.1371/journal.pone.0020036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Megyeri M, Makó V, Beinrohr L, Doleschall Z, Prohászka Z, Cervenak L, et al. Complement protease MASP-1 activates human endothelial cells: PAR4 activation is a link between complement and endothelial function. J Immunol. 2009;183((5)):3409–16. doi: 10.4049/jimmunol.0900879. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data