Abstract
DNA methylation of promoter-associated CpG islands is involved in the transcriptional repression of vertebrate genes. To investigate the mechanisms underlying gene inactivation by DNA methylation, we characterized a human MBD1 protein, one of the components of MeCP1, which possesses a methyl-CpG binding domain (MBD) and cysteine-rich (CXXC) domains. Four novel MBD1 isoforms (MBD1v1, MBD1v2, MBD1v3, and MBD1v4) were identified by the reverse transcription-PCR method. We found that these transcripts were alternatively spliced in the region of CXXC domains and the C terminus. Green fluorescent protein-fused MBD1 was localized to multiple foci on the human genome, mostly in the euchromatin regions, and particularly concentrated in the pericentromeric region of chromosome 1. Both the MBD sequence and genome methylation were required for proper localization of the MBD1 protein. We further investigated whether MBD1 isoforms are responsible for transcriptional repression of human genes. A bacterially expressed MBD1 protein bound preferentially to methylated DNA fragments containing CpG islands from the tumor suppressor genes p16, VHL, and E-cadherin and from an imprinted SNRPN gene. All MBD1 isoforms inhibited promoter activities of these genes via methylation. Interestingly, MBD1 isoforms v1 and v2 containing three CXXC domains also suppressed unmethylated promoter activities in mammalian cells. These effects were further manifested in Drosophila melanogaster cells, which lack genome methylation. Sp1-activated transcription of methylated p16 and SNRPN promoters was inhibited by all of the MBD1 isoforms, whereas the isoforms v1 and v2 reduced Sp1-activated transcription from unmethylated promoters as well. These findings suggested that the MBD1 isoforms have different roles in methylation-mediated transcriptional silencing in euchromatin.
DNA methylation at position 5 of cytosine within CpG dinucleotides is the major epigenetic modification of mammalian genomes and is involved in a wide range of biological phenomena, including genomic imprinting (3, 33), X chromosome inactivation (2), and tissue-specific gene expression (3). Lack of a gene encoding the DNA methyltransferase is also reported to be lethal to mice at midgestation (34). In addition, aberrant DNA methylation patterns have been linked to altered gene expression in certain genetic diseases and tumors (4, 16). These observations are based upon the fact that genome methylation regulates gene transcription and higher-order chromatin structure (29). There are two possible mechanisms by which methylation can suppress gene expression (37). One is a direct mechanism: some transcription factors, such as E2F1, cannot bind their recognition sequences when they are methylated (5, 24). In contrast, transcription factors, such as Sp1, are not sensitive to the methylation status of the recognition sites (23, 30). In such cases, methylation may repress transcription by additional mechanisms. The other is an indirect mechanism: repressor molecules which bind specifically to methylated DNA and induce subsequent chromatin assembly cause the gene inactivation. This is supported by observations that methylation-dependent silencing of genes occurs only after chromatin is assembled (13) and that methylation has additional repressive effects on transcription conferred by nucleosomes alone (29). Thus, trans-acting factors targeted for methylated sequences are likely to play a role in gene repression and chromatin structure.
The proteins, designated methyl-CpG binding proteins (MeCPs), either independently or together with other components of chromatin, contribute to the regulation of gene transcription via methylation (5, 9, 29). They have no DNA sequence specificity except for the requirement for CpG methylation, suggesting that these proteins affect the genome globally. Two MeCPs, MeCP1 (9, 37) and MeCP2 (32, 41), have been initially determined to bind specifically to methylated DNA. In mammalian genomes, approximately 60 to 90% of CpG dinucleotides are methylated, and most of them are dispersed on the vast chromosomes. However, the CpG islands, a cluster of CpG sequences, are almost unmethylated in the promoter regions of actively transcribed genes (1). Thus, the distribution of methylated cytosines on the genome is disproportionate. MeCP1 was reported to bind preferentially to DNA containing a cluster of symmetrically methylated CpGs (37), while MeCP2 can bind to a single methylated CpG pair (42). MeCP2 is more abundant than MeCP1 and is thought to associate mainly with dispersed methylated CpGs to prevent inappropriate transcription (32). MeCP2 provides a transcriptional noise reduction system on the genome (8), and this may be consistent with a recent report that Mar-binding protein, which is involved in the loop domain organization of chromatin, is homologous to MeCP2 (51).
A human protein, MBD1 (formerly PCM1), was reported to be a component of the MeCP1 complex and to repress transcription from a methylated adenovirus promoter in vitro (15). Recently, three additional human and mouse proteins, named MBD2, MBD3, and MBD4, have been identified based on a sequence similarity to the methyl-CpG binding domain (MBD) of MeCP2 and MBD1 (20). MBD1, MBD2, and MBD4 bound specifically to oligonucleotide probes with methylated CpG. The expression of MBD1 and MBD2 was detected in somatic tissues but not in embryonic stem cells or germ cells, which are deficient in MeCP1 activity. Among these MBD-containing proteins, only MBD1 was demonstrated to have sequences similar to a cysteine-rich CXXC domain, which was originally found in DNA methyltransferase (7). The conserved presence of a CXXC domain represents the first link between MeCPs and DNA methyltransferase, although the significance of the CXXC domain is poorly understood. The lines of evidence suggest that MBD1 is involved in methylation-mediated repression of genes whose promoters carry CpG islands. However, the involvement of MBD1 in gene transcription and chromatin structure in mammalian genomes remains to be elucidated. In the present study, therefore, we focused on the characterization of MBD1, and our data have demonstrated that four novel MBD1 isoforms which are alternatively spliced in the region of CXXC domains play multiple roles in methylation-mediated gene silencing in the euchromatin of the human genome.
MATERIALS AND METHODS
Cloning of human MBD1 isoforms.
The method for PCR-based, full-length cDNA cloning with the sequence information of human expressed sequence tags (EST) in databases was reported previously (36). Briefly, our tblastn search of the GenBank database with the amino acid sequences of the MBD of human MeCP2 (accession no. P51608) revealed that several EST clones, including H85883, had sequence similarity. Amplification of the corresponding cDNA clone was carried out by a nested PCR procedure with a human fetal brain cDNA library as a template and specific primers designed from the EST sequences. The cDNA and deduced amino acid sequences were similar but not identical to those of MBD1 (accession no. Y10746), according to Cross et al. (15). A set of primers corresponding to the 5′ and 3′ untranslated regions of MBD1 cDNA, as described below, amplified four alternatively spliced MBD1 isoforms from primary cultured human fibroblasts.
Methylation-specific PCR assay.
Genomic DNA was extracted from the lung cancer cell line NCI-H1299. The DNA (1 μg) was treated with sodium bisulfite, and the bisulfite-modified DNA was amplified with p16 gene-specific primers as described previously (21). The sets of primers were p16-W-sense (5′-CAGAGGGTGGGGCGGACCGC-3′) and p16-W-antisense (5′-CGGGCCGCGGCCGTGG-3′) for the unmodified sequence, p16-M-sense (5′-TTATTAGAGGGTGGGGCGGATCGC-3′) and p16-M-antisense (5′-GACCCCGAACCGCGACCGTAA-3′) for the methylated sequence, and p16-U-sense (5′-TTATTAGAGGGTGGGGTGGATTGT-3′) and p16-U-antisense (5′-CAACCCCAAACCACAACCATAA-3′) for the unmethylated sequence. The sets of primers p16-M and p16-U were used for the amplification of bisulfite-modified DNA. PCR was performed in a 25-μl reaction volume for 40 cycles with AmpliTaq Gold DNA polymerase (Perkin-Elmer, Branchburg, N.J.).
Construction of GST-fused MBD1 protein and band shift assay.
The MBD1v1 cDNA fragment (amino acids 1 to 421) was subcloned into a pGEX-2TH bacterial expression vector. Expression and purification of the glutathione S-transferase (GST)-fused MBD1 protein were performed as described previously (36). DNA fragments containing the promoter regions of human p16, E-cadherin, VHL, and SNRPN genes (accession no. X94154, L34545, U19763, and U41384, respectively) were amplified from human genomic DNA. The sets of primers used were as follows: p16-sense (5′-CACGCTAGCACAGCGTCCCCTTGCCTGGAA-3′) and p16-antisense (5′-CCGAAGCTTCCATGCTGCTCCCCGCCGC-3′), E-cadherin-sense (5′-AGCGCTAGCAGGCTAGAGGGTCACCGCGT-3′) and E-cadherin-antisense (5′-CCGAAGCTTCACAGGTGCTTTGCAGTTCCG-3′), VHL-sense (5′-TAGGCTAGCTACAGTAACGAGTTGGCCTAGC-3′) and VHL-antisense (5′-CAGAAGCTTCGGGCCGGACGCCGCGG-3′) and SNRPN-sense (5′-TTGGCTAGCGTAGCATGCTTTTAGAGTTAGG-3′) and SNRPN-antisense (5′-CGCAAGCTTGACAGATGCGTCAGGCATCTC-3′), containing NheI and HindIII sites (underlined). As competitor DNA, a portion of the SNRPN CpG island was also amplified by the primers SNRPN-CF (5′-CCCTCTCATTGCAACAGTGCTGT-3′) and SNRPN-DR (5′-CGAGGGTTCCTAAAGGGTACGC-3′). A cycling reaction was performed in a 25-μl reaction volume for 35 cycles with a cloned Pfu DNA polymerase (Stratagene, La Jolla, Calif.). These PCR products were methylated by SssI methyltransferase as indicated by the manufacturer (New England Biolabs, Beverly, Mass.). For a band shift assay, the unmethylated or methylated DNA fragments (0.2 μg each) were incubated with the GST-MBD1 protein or GST (0.3 μg each) in a binding buffer containing 20 mM HEPES (pH 7.4), 1 mM EDTA, 3 mM MgCl2, 10 mM 2-mercaptoethanol, 0.4% glycerol, and 0.1% Triton X-100 on ice for 30 min. Competitor DNA (0.4 μg each), either unmethylated or methylated, was added to the reaction. The DNA-protein complexes were then electrophoresed on 1 or 3% agarose gels and were stained with ethidium bromide.
In vitro transcription analysis.
The PCR products amplified from the promoter regions of p16, E-cadherin, VHL, and SNRPN genes were double digested with NheI and HindIII and ligated upstream of a luciferase cDNA in a pGL3-Basic vector (Promega, Madison, Wis.). The template DNAs for in vitro transcription were amplified from the promoter-inserted pGL3 constructs by primers DF (5′-TGCAGGTGCCAGAACATTTCTCT-3′) and ER (5′-CTCTATGCATTTATTATTAGCTATTTATCGTTTCATAGCTTCTGC-3′) (the 3′ untranslated sequence of human SNRPN mRNA [underlined] was added to ensure the stability of the transcript). These PCR products were methylated with SssI methyltransferase. GST-MBD1 protein was incubated on ice for 60 min with unmethylated or methylated DNA fragments (0.1 μg) in the binding buffer. The transcription was then initiated by adding nucleoside triphosphate NTP mix (0.4 mM [each] ATP, CTP, GTP, and UTP) and 8 U of HeLa nuclear extract (Promega), and the samples were incubated for 60 min at 30°C. After extraction once with phenol-chloroform and precipitation in ethanol, RNA-containing samples were dissolved in nuclease-free water.
Primer extension assay.
The primer ER was labeled with T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (3,000 Ci/mmol) (Amersham, Buckinghamshire, England). The radiolabeled primer was mixed with the above-mentioned RNA-containing samples in a 20-μl reaction buffer containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, dNTP mix (1 mM [each] dATP, dGTP, dCTP, and dTTP), and 200 U of SuperScript II reverse transcriptase (Gibco BRL, Rockville, Md.). After incubation for 1 h at 42°C, the samples were extracted with phenol-chloroform and precipitated in ethanol, electrophoresed on an 8% polyacrylamide gel, and exposed to X-ray film.
Construction of expression plasmids.
The full-length cDNAs for MBD1v1 to MBD1v4 isoforms were PCR amplified with cDNA fragments from primary cultured human fibroblasts and specific primers. These are a forward primer (5′-CTGCTTGTTACCTCTAGAATGGCTGAGGACTGGCTGGAC-3′) and a reverse primer (5′-AGTTTTTCTAGAAAGCTTTTCATACAATCTCTGTACTTGCTG-3′), each containing an XbaI site (underlined). Cycling reactions were performed in a 25-μl reaction volume for 30 cycles with a cloned Pfu DNA polymerase. Each cycle included denaturation at 96°C for 1 min, annealing at 55°C for 2 min, and extension at 72°C for 5 min 30 s. The PCR fragments were digested with XbaI and ligated into pCGN mammalian expression vectors (termed pCGN-MBD1v1 to pCGN-MBD1v4) and into pAc5.1/V5-His Drosophila melanogaster expression vectors (termed pAc5.1-MBD1v1 to pAc5.1-MBD1v4) (Invitrogen, Carlsbad, Calif.). To express epitope-tagged MBD1, the MBD1 vectors were transfected into COS-7, HeLa, NCI-H1299, and CHO cells by the liposome-mediated gene transfer method. The pAc5.1-MBD1 vectors were transfected into Drosophila Schneider cell line 2 (SL2) derived from Drosophila embryos (provided by R. M. Evans) by the calcium phosphate method. To express enhanced green fluorescent protein (EGFP) fusion proteins, we ligated cDNA for full-length MBD1 isoforms into the eukaryotic expression vector pEGFP-C1 (Clontech, Palo Alto, Calif.). In addition, the cDNA fragments encoding amino acids 1 to 112 (which include both MBD and nuclear localization signal [NLS]), amino acids 1 to 75 (which include MBD alone), and amino acids 70 to 112 (which include NLS alone) of MBD1 were also subcloned into the pEGFP-C1. These constructs were named pEGFP-MBD1(MBD+NLS), pEGFP-MBD1(MBD), and pEGFP-MBD1(NLS). The full-length cDNA for MeCP2 was amplified with cDNA fragments from U251 glioma cells. The specific primers are a forward primer (5′-CAGCTCTCTAGAGGATCCATGGTAGCTGGGATGTTAGGG-3′) and a reverse primer (5′-AATCCGTCTAGAGGATCCTCAGCTAACTCTCTCGGTCAC-3′), each containing an XbaI site (underlined). The PCR fragments were digested with XbaI and ligated into a pEGFP-C1 vector (termed pEGFP-MeCP2). The full-length cDNA for E2F1 was amplified from an expression plasmid pDCE2F (26) (provided by K. Ohtani) with a forward primer (5′-GCGCGGGGATCCGATATCATGGCCTTGGCCGGGGCCC-3′) and a reverse primer (5′-TGGTCCTCTAGAGAAATCCAGGGGGGTGAGGTC-3′) containing an EcoRV and an XbaI site (underlined), respectively, and it was digested with EcoRV and XbaI and ligated into a pAc5.1/V5-His vector (termed pAc5.1-E2F1).
Cell cultures and preparation of cell lysates.
COS-7, NCI-H1299, and HeLa cells were cultured in a 1:1 mixture of Dulbecco’s modified Eagle’s minimum essential medium and Ham’s F12 nutrient medium (Gibco BRL) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (Bio-Whittaker, Walkersville, Md.). CHO cells were grown in the above-mentioned medium supplemented with 0.1 mM minimum essential medium nonessential amino acid solution (Gibco BRL). 5-Aza-2′-deoxycytidine (Sigma, St. Louis, Mo.) was used for the cell treatment. SL2 cells were cultured in Schneider’s Drosophila medium (Gibco BRL) with 10% (vol/vol) heat-inactivated fetal bovine serum (Gibco BRL) and 2 mM glutamine.
For preparing cell lysates, at 48 h after the transfection of pCGN-MBD1, the cells on plates were washed with phosphate-buffered saline (PBS) and lysed with a lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.5% Triton X-100, 50 μg of aprotinin/ml, 100 μg of phenylmethanesulfonyl fluoride/ml, 10 μg of leupeptin/ml, 10 μg of pepstatin/ml, and 180 μg of sodium orthovanadate/ml) on ice for 5 min. After centrifugation at 3,000 rpm for 5 min, the pellet and the supernatant were separately solubilized by a Laemmli sample buffer (2% sodium dodecyl sulfate, 100 mM dithiothreitol, 60 mM Tris-HCl [pH 6.8], and 0.001% bromphenol blue) as nuclear and cytosolic fractions, respectively.
Western blot analysis.
Samples containing equal amounts of protein (15 μg) from the cell lysates were separated by sodium dodecyl sulfate–8% polyacrylamide gel electrophoresis and then transferred to a nitrocellulose membrane with a constant current of 140 mA for 2 h. The membrane was blocked for 1 h at 4°C with PBS containing 10% nonfat dry milk and then incubated with an anti-hemagglutinin (HA) monoclonal antibody (12CA5; Boehringer Mannheim, Mannheim, Germany) or rabbit anti-MBD1 polyclonal antibody, which we had prepared with the GST-fused MBD1 protein, in PBS containing 0.03% Tween 20 for 1 h. After being washed with PBS containing 0.3% Tween 20, the membrane was incubated with species-appropriate horseradish peroxidase-conjugated secondary antibodies for 40 min. After the membrane was washed with PBS containing 0.3% Tween 20, visualization was performed with an enhanced-chemiluminescence detection system (Amersham).
Luciferase assay.
The promoter-inserted pGL3 vectors were either unmethylated or methylated by SssI methyltransferase. At 48 h after the cotransfection of the promoter-inserted pGL3, pCGN-MBD1, and pRL-SV40, which was used for monitoring the transfection efficiency into CHO cells, the cells were collected and lysed by a lysis buffer offered by the manufacturer (Promega). For Drosophila cells, we cotransfected the promoter-inserted pGL3, pAc5.1-MBD1, and either an Sp1 expression plasmid, pPacSp1 (14) (provided by J. T. Kadonaga), or an E2F1 expression plasmid, pAc5.1-E2F1. The insertless pCGN, actin 5C (A5C) (provided by R. M. Evans), or the pAc5.1/V5-His vector was used as a mock transfection. The luciferase activities were determined by the Dual-luciferase reporter assay system (Promega) and a luminometer (Niti-on, Funabashi, Japan).
CLSM analysis.
The COS-7, NCI-H1299, and HeLa cells were incubated for 24 h at 37°C after the transfection of EGFP-fused MBD1 expression vectors. After being washed two times with PBS, the cells were fixed with 4% paraformaldehyde in PBS for 10 min and then treated with 0.2% Triton X-100 for 5 min. After being washed with PBS, the cells were incubated with anti-kinetochore autoantibody from a patient with calcinosis, Reynaud’s phenomenon, esophageal motility disorders, sclerodactyly, and telangiectasia (CREST) syndrome (40) (provided by H. Nakakuma) in PBS containing 0.3% bovine serum albumin for 60 min at room temperature. After being washed with PBS, the cells were incubated with a fluorolink Cy3-labeled goat anti-human immunoglobulin G (Amersham) for 60 min. After being washed with PBS, the samples were mounted in 80% glycerol. The cells were counterstained with propidium iodide (Sigma) at a final concentration of 1 μg/ml. The cells were visualized with a confocal laser scanning microscope (CLSM; Olympus, Tokyo, Japan).
Deconvolution system.
After being washed two times with PBS, HeLa cells were similarly fixed with 4% paraformaldehyde in PBS for 10 min and then treated with 0.2% Triton X-100 for 5 min. After washing the cells with PBS, we incubated them with anti-GFP polyclonal antibodies (Clontech) in PBS containing 0.3% bovine serum albumin for 55 min at room temperature, and DAPI (4′,6-diamidino-2-phenylindole) (Sigma) was added to make a final concentration of 0.2 μg/ml for nuclear staining for 5 min. The following deconvolution system was used to show the localization of MBD1 and DAPI: a Zeiss Axioplan2 MOT epifluorescence microscope equipped with a filter wheel system and a cooled charge-coupled camera (PentaMax-1317K1; Princeton Instruments, Inc.) controlled by a Power Macintosh 9600/200MP computer running the software program IPLab (Signal Analystics Co.). Several images for each cell captured at different stage positions and then deconvoluted by the software HazeBuster (VayTek, Inc.) are shown in the figures in the same focal plane.
Nucleotide sequence accession numbers.
The accession numbers for MBD1v1, -v2, -v3, and -v4 are AF078830, AF078831, AF078832, and AF078833, respectively.
RESULTS
Identification of four MBD1 splice isoforms.
We cloned an MBD1 cDNA by a reverse transcription-PCR method from primary cultured human fibroblasts. Four kinds of full-length cDNAs appeared to be expressed equally in the cells. The deduced amino acid sequences were very similar but not identical to that of MBD1 (Y10746) (15). We did not isolate a cDNA encoding MBD1 (Y10746) from fibroblasts. The term MBD1 (Y10746) is used throughout the text to distinguish this isoform from the general name of the protein. The four novel MBD1 isoforms were confirmed to be alternatively spliced, and these splice variants were designated MBD1v1, MBD1v2, MBD1v3, and MBD1v4 (Fig. 1A). There were one MBD (amino acids 7 to 61) and one putative NLS (amino acid sequence, K84KRKK88) in the common N-terminal region. MBD1v3 and MBD1v4 had CXXC domains 1 and 2, while MBD1 (Y10746) had CXXC domains 2 and 3. In contrast, MBD1v1 and MBD1v2 possessed CXXC domains 1, 2, and 3. In addition, MBD1v2 had a different C-terminal sequence with a downstream stop codon, due to additional alternative splicing events. From searches of the human database, we found the presence of only one CXXC domain in DNA methyltransferase (7) and ALL-1/HRX (35), which is related to translocation breakpoints in leukemia and has amino acid sequence identity with trithorax proteins of Drosophila (Fig. 1B).
FIG. 1.
Protein structure of MBD1 splice isoforms and sequence alignment of the CXXC domains. (A) Diagrams of MBD1 isoforms. These include MBD1v1, MBD1v2, MBD1v3, and MBD1v4 and a previously described MBD1 (formerly PCM1 [Y10746]). They show one methyl-CpG binding domain (MBD), one putative NLS (KKRKK), and two or three cysteine-rich (CXXC) domains, due to alternative splicing events, designated as CXXC1 to CXXC3. The numbers below the diagrams indicate the positions of amino acids (a.a.) from the N terminus. (B) Sequence alignment of the CXXC domains between MBD1 isoforms, ALL1/HRX, and DNA (cytosine-5)-methyltransferase (MTase). Conserved cysteine residues are indicated by boldface type. The accession numbers of the sequences are Q03164 (ALL1/HRX) and X63692 (MTase).
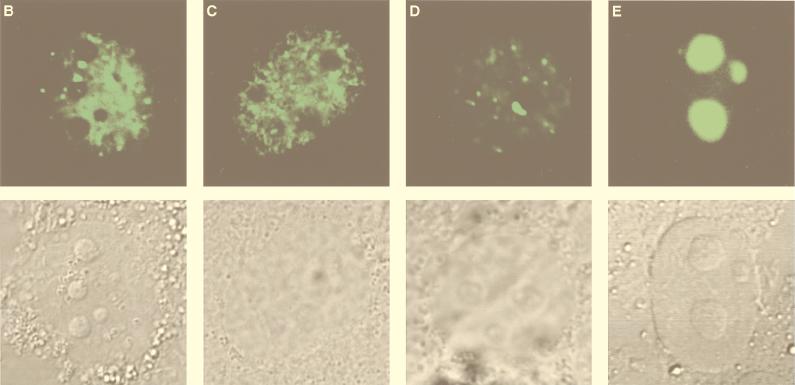
Localization of MBD1 isoforms in the nuclei of mammalian cells.
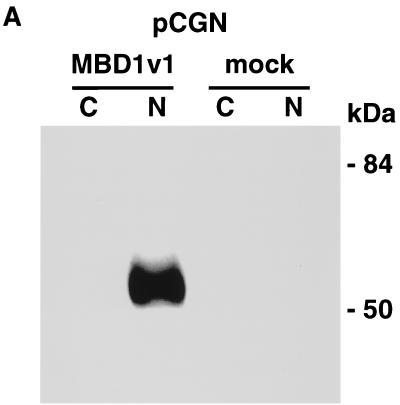
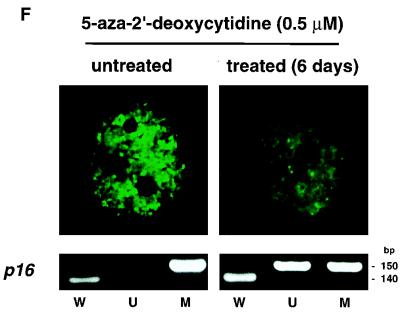
To characterize the subcellular localization of MBD1, a Western blot analysis of COS-7 cells which had been transfected with pCGN (mock transfection) or pCGN-MBD1v1 (amino acids 1 to 421) was performed with an anti-HA epitope monoclonal antibody (12CA5). HA-tagged MBD1 was found not in a cytosolic fraction but in a nuclear fraction from lysed cells (Fig. 2A). A very similar result was obtained by expressing full-length MBD1 isoforms (data not shown). In order to visualize the intranuclear localization of MBD1 isoforms in intact cells, EGFP-MBD1 fusion proteins were transiently expressed in COS-7 cells and observed by CLSM (Fig. 2B to E). The EGFP-MBD1 isoforms showed punctate distribution in the interphase nuclei except for the nucleolus. Multiple foci of different sizes with intense staining were seen in the nuclei of transfected cells. The result obtained with EGFP-fused full-length MBD1v1 is shown in Fig. 2B. We then constructed three additional vectors—pEGFP-MBD1(MBD+NLS), expressing an EGFP fused to both MBD and putative NLS; pEGFP-MBD1(MBD), expressing an EGFP fused to MBD alone; and pEGFP-MBD1(NLS), expressing an EGFP fused to putative NLS alone—and they were expressed in COS-7 cells to clarify the roles of these domains. EGFP-MBD1(MBD+NLS) presented punctate labeling of the nuclei (Fig. 2C) similar to that of full-length EGFP-MBD1v1. EGFP-MBD1(MBD) still tended to locate in the nucleus and form multiple foci (Fig. 2D), and a portion of EGFP-MBD1(MBD) was retained in the cytoplasm of some transfected cells. In contrast, EGFP-MBD1(NLS) lacking MBD was localized uniformly in the nucleolus (Fig. 2E). To demonstrate that the interaction of MBD1 with chromosomal DNA is sensitive to genome methylation, we utilized a lung cancer cell line, NCI-H1299, which had demonstrated the presence of hypermethylation in the promoter region of the p16 gene. EGFP-MBD1(MBD+NLS) was expressed in the cells which were untreated or treated with 5-aza-2′-deoxycytidine (0.5 μM), a cytidine analog to inhibit DNA methylation, for 6 days (Fig. 2F). EGFP-MBD1(MBD+NLS) revealed a reduced punctate formation and was distributed nonspecifically throughout the nuclei of the treated cells. The methylation-specific PCR assay with bisulfite modification of genomic DNAs from these cells demonstrated that the treatment with 5-aza-2′-deoxycytidine induced the altered levels of genomic demethylation. Both M and U primers, corresponding to methylated and unmethylated p16 promoter sequences, respectively, allowed the amplification of expected fragments in the treated cells, while the use of M primers, but not U primers, gave an amplified product in the untreated cells. These findings indicate that both MBD and genome methylation are required for the precise localization of MBD1 protein.
FIG. 2.
Subcellular localization of MBD1 protein. (A) Western blot analysis. pCGN-MBD1v1 (amino acids 1 to 421) was transfected into COS-7 cells. HA-tagged MBD1 was present in the nuclear fraction (N) but not in the cytosolic fraction (C). (B to F) EGFP-fused MBD1 was transiently expressed in COS-7 and NCI-H1299 cells and observed with a CLSM. (B to E) The lower row shows a transmitted view (Nomarski) of the upper. (B) The result of the full-length MBD1v1 is shown in the upper row. Three vectors were constructed: pEGFP-MBD1(MBD+NLS), expressing an EGFP fused to both MBD and putative NLS (C); pEGFP-MBD1(MBD), expressing an EGFP fused to MBD alone (D); and pEGFP-MBD1(NLS), expressing an EGFP fused to putative NLS alone (E). (F) Localization of EGFP-MBD1(MBD+NLS) in the nuclei of NCI-H1299 cells which were treated with 5-aza-2′-deoxycytidine (0.5 μM) for 6 days. The methylation-specific PCR with bisulfite modification of DNA (lower panels) demonstrated the demethylation of the promoter region of human p16 gene after the treatment. W, U and M indicate specifically amplified fragments corresponding to unmodified, unmethylated, and methylated sequences, respectively.
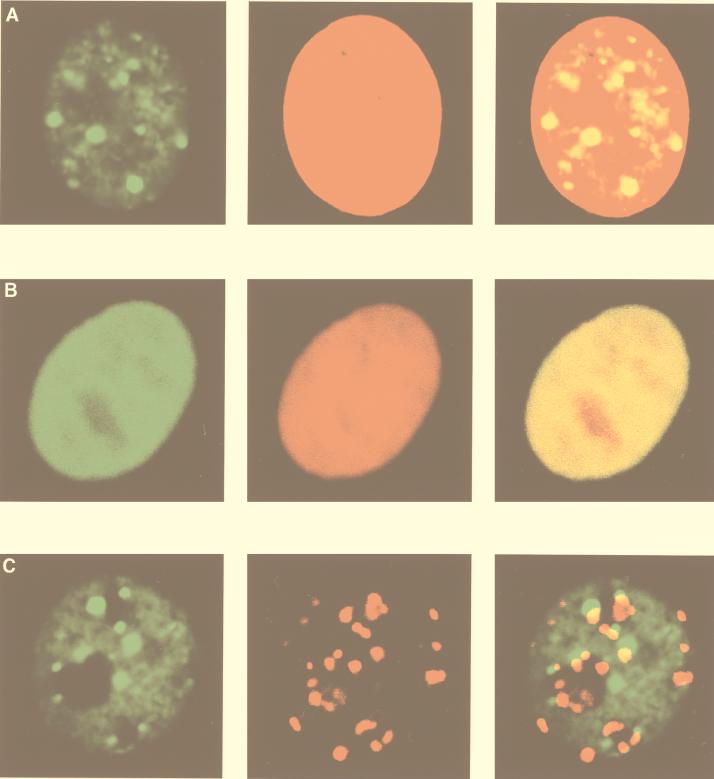
Intranuclear territories of MBD1 and other chromosomal proteins.
We further observed the localization of MBD1 and other chromosomal proteins in human HeLa cells. CLSM showed that EGFP-MBD1(MBD+NLS) produced a punctate staining pattern in HeLa cells (Fig. 3A), while EGFP-MeCP2 was observed diffused throughout the nucleus (Fig. 3B). Chromosomal DNA was counterstained with propidium iodide. The merged images in the right-hand panels reveal that EGFP-MBD1 localized not to the nuclear periphery but to the interior of the nucleus. Thus, these two MeCPs showed very different localization patterns for the same assay, indicating that MBD1 is distinct from MeCP2. Next, interphase kinetochores were labeled with autoantibodies from sera of a patient with the CREST syndrome of scleroderma (Fig. 3C), as described previously (40). The foci of CREST antigens in kinetochores did not appear to colocalize with those of MBD1, suggesting that MBD1 is not associated with kinetochores.
FIG. 3.
Intranuclear territories of MBD1 and other chromosomal proteins in human HeLa cells. EGFP-MBD1(MBD+NLS) (green) transfected into HeLa cells produced a punctate staining pattern in the nucleus (A and C), while EGFP-MeCP2 (green) was distributed throughout the nucleus (B). Chromosomal DNAs were counterstained with propidium iodide (red) (A and B). MBD1(MBD+NLS) did not colocalize with kinetochores, which were labeled with autoantibodies from a patient with the CREST syndrome of scleroderma (red) (C). The right-hand panels show merges of the left-hand and the middle panels.
Localization of MBD1 in euchromatin regions and the pericentromeric region of human chromosome 1.
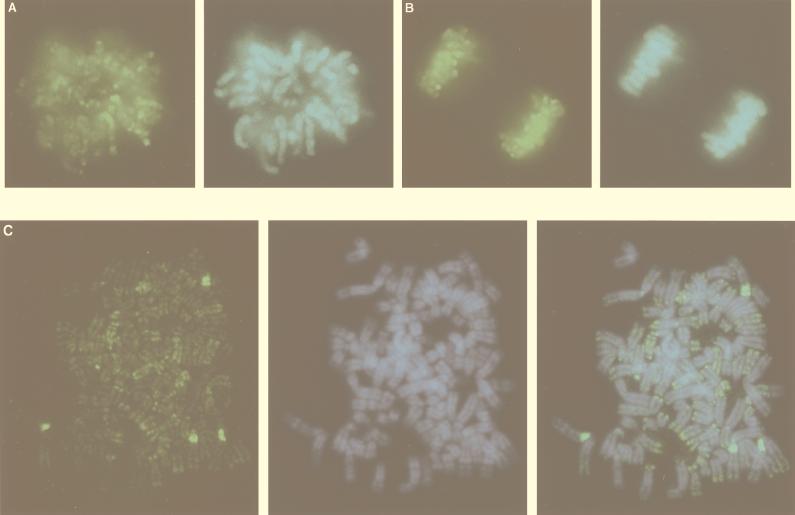
A deconvolution system with a highly sensitive charge-coupled camera was utilized to detect the localization of MBD1 in the nucleus. The chromosomal DNAs, especially in the heterochromatin regions, were counterstained with DAPI (39) (Fig. 4A to C). In HeLa cells, EGFP-MBD1(MBD+NLS) localized throughout all the chromosomes (Fig. 4A shows the metaphase image) and was reorganized and inherited in cell division (Fig. 4B). EGFP-MBD1 tended to be found mostly in negatively DAPI-stained euchromatin regions on the human chromosomes. To clarify whether MBD1 associates with specific chromosomal regions, we analyzed the localization of EGFP-MBD1 in metaphase chromosome spreads of the transfected cells (Fig. 4C). The EGFP-MBD1 was localized predominantly in DAPI-negative euchromatic regions on the whole chromosomes, and the merged image of EGFP-MBD1 and DAPI demonstrated their complementary staining pattern. This is the first observation that MBD1 targets the sites in euchromatin regions, which are rich in transcribed genes on the genome. The telomeric region of several chromosomes also appeared to be labeled by MBD1. In addition, four regions of intense staining were found in the pericentromeric regions of human chromosome 1, as determined by fluorescence in situ hybridization with chromosome 1-specific probes (data not shown). The HeLa cells exhibited tetraploidy of chromosome 1, but we did not find any structural abnormalities of this chromosome. This may be consistent with the existence of specifically methylated repeat structures in the pericentromeric region of chromosome 1 (47).
FIG. 4.
Localization of MBD1 on human chromosomes. EGFP-MBD1(MBD+NLS) (green) was found in HeLa cells on the metaphase chromosomes (A), on the chromosomes during mitosis (B), and on the metaphase chromosome spreads (C), using a deconvolution system with a highly sensitive charge-coupled camera. Chromosomal DNAs, especially in the heterochromatin regions, were counterstained with DAPI (blue) (A to C). (C) MBD1 localized mostly in negatively DAPI-stained euchromatic regions on human chromosomes and to pericentromeric regions of chromosome 1. The right-hand panel shows a merge of left-hand and middle panels.
Band shift analysis and in vitro transcription assay.
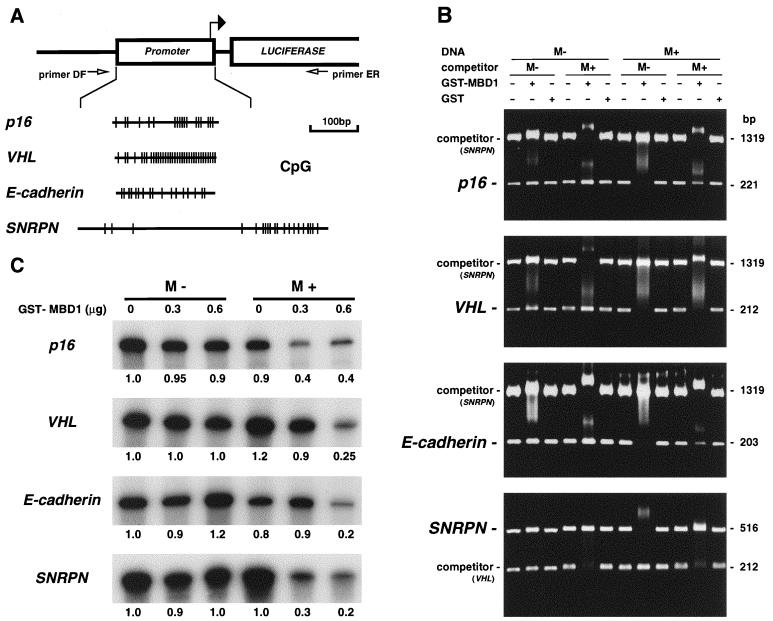
The fact that MBD1 exists largely in euchromatin regions prompted us to investigate whether MBD1 associates with human genes via DNA methylation. We PCR amplified genomic DNAs containing CpG islands from the promoter regions of four genes and then cloned these fragments upstream of a luciferase cDNA in a pGL3-Basic vector (Fig. 5A). The different types of CpG-rich gene promoters were analyzed to demonstrate convincing data that MBD1 can generally target many genes on the genome. Tumor suppressor genes, including p16, VHL, and E-cadherin, have been shown to have aberrantly hypermethylated CpG islands in cancer cells (17, 22, 38), resulting in the transcriptional repression advantageous to tumor progression. The CpG island of an imprinted SNRPN gene is normally unmethylated and heavily methylated on the active paternal and inactive maternal alleles, respectively, and monoallelic expression is required for mammalian development (48). The nucleotide component of each cloned DNA was as follows: p16 (221 bp long; G+C content, 70.1%; CpG/GpC = 0.69), VHL (212 bp long; G+C content, 69.4%; CpG/GpC = 1.20), E-cadherin (203 bp long; G+C content, 70.4%; CpG/GpC = 0.65), and SNRPN (516 bp long; G+C content, 52.0%; CpG/GpC = 0.43). The MBD1v1 protein (containing one MBD and three CXXC domains) fused to GST was expressed in Escherichia coli. First, the GST-MBD1 protein was incubated with the PCR fragments containing a CpG island which had been either unmethylated (M−) or methylated (M+) by SssI methyltransferase, which methylates all cytosine residues within the CpG dinucleotide. A band shift analysis was performed by agarose gel electrophoresis (Fig. 5B). The DNA-MBD1 complex was found to be shifted to a slow-migrating, higher-molecular-mass band or to disappear into the upper part. The recombinant MBD1 bound preferentially to all of the methylated DNAs, while MBD1 was only slightly associated with the unmethylated versions. The methylated DNA-MBD1 complexes were unaffected by the presence of an excess of unmethylated competitor DNA (containing a portion of the SNRPN CpG island (1,319 bp long; G+C content, 61.3%; CpG/GpC = 0.68) or a VHL CpG island [212 bp long, same as above]), but the complex formations were abolished when the methylated competitor DNA was added to the reaction. The MBD1 binding to the methylated fragment containing a VHL CpG island, which had the highest content of CpG dinucleotides in this study, was not inhibited by the methylated SNRPN competitor, indicating that MBD1 binds preferentially to a high density of methylated CpGs. GST alone bound to neither methylated nor unmethylated DNAs, and purified histone proteins bound strongly to the DNAs regardless of the methylation status (data not shown). Next, to test whether MBD1 affects transcription from these promoter sequences, the GST-MBD1 was added to in vitro-transcription reaction mixtures containing HeLa nuclear extract and either unmethylated or methylated DNA fragments which were PCR amplified from the promoter-inserted pGL3 vectors with primers DF and ER (Fig. 5A). A primer extension analysis with a radiolabeled ER primer was then performed to detect a specific transcript driven by the promoter (Fig. 5C). The in vitro-transcription products were electrophoresed on an 8% polyacrylamide gel, and the radioactivities of the bands were quantitatively elucidated by the bioimaging analyzer MacBAS version 2.51 (Fuji Film, Tokyo, Japan). The unmethylated and methylated promoters had similar transcriptional activities in the absence of GST-MBD1, suggesting that DNA methylation alone does not complete the gene repression. The addition of GST-MBD1 (0.6 μg) repressed transcription from the methylated promoters by two- (p16), four- (E-cadherin), and five-fold (VHL and SNRPN) compared with the results in the absence of MBD1. The unmethylated promoters, however, were little affected by the presence of MBD1. The lack of DNA template indicated that there was no transcription product in the reaction, and GST protein alone did not affect the result (data not shown).
FIG. 5.
Effect of MBD1 on promoter-associated CpG islands of p16, VHL, E-cadherin, and SNRPN genes. (A) PCR-amplified DNA fragments from the gene promoters were subcloned upstream of a luciferase cDNA in a pGL3-Basic vector. The position of the transcription start site and the oligonucleotide primers DF and ER to amplify DNA fragments utilized for the in vitro transcription assay are shown by solid and open arrows, respectively. The presence of CpG dinucleotides within the inserted promoter sequences of four genes is indicated by vertical lines. (B) Band shift of methylated DNAs complexed with MBD1. Unmethylated (M−) and methylated (M+) DNA fragments containing CpG islands from these genes were incubated with GST-fused MBD1v1 or GST alone. The in vitro methylation of CpG sequences was performed with SssI methyltransferase. +, present; −, absent. (C) Transcriptional repression by MBD1 in a methylation-dependent manner. GST-MBD1 was incubated with either unmethylated (M−) or methylated (M+) DNA fragments which were PCR amplified from the promoter-inserted pGL3 vectors with primers DF and ER. Transcripts from the promoters were synthesized in HeLa nuclear extract and detected by the primer extension method with a radiolabeled ER primer. The amount of a predicted cDNA product was measured by a Bioimaging analyzer, MacBAS version 2.51, and the relative amount of transcript compared with that of the unmethylated promoter without GST-MBD1 is indicated below each panel.
Transcriptional repression by MBD1 isoforms in mammalian cells.
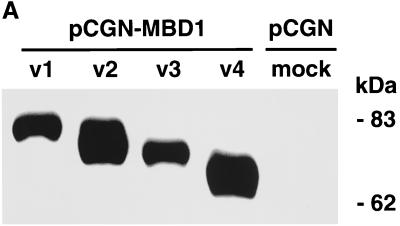
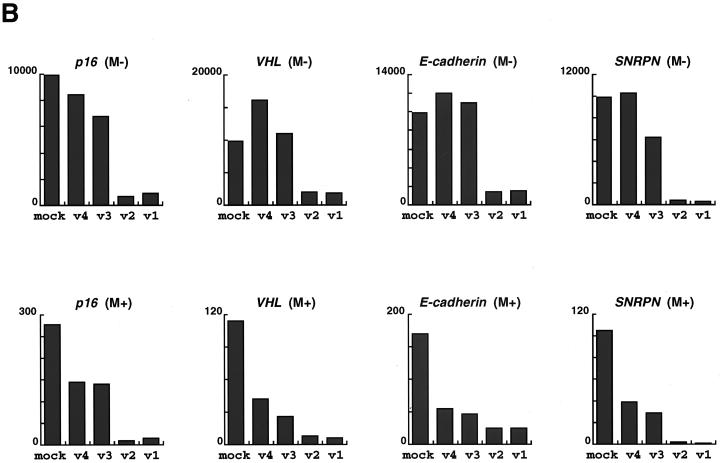
To determine whether MBD1 isoforms are involved in the regulation of gene activities in the cell, we constructed four HA-tagged full-length MBD1 expression vectors (pCGN-MBD1v1 to pCGN-MBD1v4) and transfected each of them into CHO cells. A Western blot analysis was performed with an anti-HA epitope monoclonal antibody, and MBD1 isoforms were found to be approximately 70 to 80 kDa in molecular mass (Fig. 6A). In addition, the promoter-inserted pGL3 vectors (pGL3-p16, pGL3-VHL, pGL3-E-cadherin, and pGL3-SNRPN) were utilized to express a Photinus pyralis luciferase under the promoter-associated CpG islands of the genes (Fig. 5A). Both the promoter-inserted pGL3 and pCGN-MBD1 vectors in the appropriate combinations were cotransfected into CHO cells, and the level of luciferase activity was measured with a luminometer. The pRL-SV40 vector expressing Renilla reniformis luciferase was simultaneously used as an internal control for correcting the transfection efficiency. The average data from three or four independent experiments are shown throughout this report. In the mock transfections shown in the left bar of each panel (Fig. 6B), the relative luciferase activity of the methylated construct (M+) was repressed by 35- (p16), 90- (VHL), 60- (E-cadherin) and 90-(SNRPN) fold compared with that of the unmethylated version (M−), probably due to the involvement of endogenous cellular factors (9, 41, 46). The expression of the MBD1 isoforms repressed transcription from all of the methylated constructs. MBD1v1 and MBD1v2, which contain three CXXC domains, inhibited the luciferase activities more than did MBD1v3 and MBD1v4 (both have two CXXC domains) in the methylated promoters tested. On the other hand, MBD1v1 and MBD1v2 could repress transcription from the unmethylated promoters whereas MBD1v3 and MBD1v4 had a subtle influence on the unmethylated promoter activity. This suggested that MBD1 isoforms v1 and v2 associate with unmethylated as well as methylated promoters, possibly through interaction with certain cellular factors affecting transcription in the cell.
FIG. 6.
Transcriptional repression by MBD1 isoforms in mammalian cells. (A) Expression of HA-tagged MBD1 isoforms. Four vectors expressing MBD1 isoforms, pCGN-MBD1v1 to pCGN-MBD1v4, were transfected into CHO cells, and a Western blot analysis with an anti-HA epitope monoclonal antibody was performed. (B) Inhibition of p16, VHL, E-cadherin, and SNRPN gene promoter activities by MBD1 isoforms. Both promoter-inserted pGL3 and pCGN-MBD1 vectors in the appropriate combinations were cotransfected into CHO cells. The level of luciferase activity in the cotransfection of unmethylated promoter-inserted pGL3 and pCGN (mock) was normalized to 10,000 in each gene promoter. The average of relative activities from four independent experiments is indicated by each bar. Unmethylated (M−) and methylated (M+) promoter-inserted pGL3 vectors are shown in the upper and lower rows, respectively.
Transcriptional regulation by MBD1 isoforms in Drosophila cells.
The above-mentioned results in mammalian cells suggest that endogenous MeCPs participate in constitutive repression of transcription from the methylated constructs. In addition, we did not exclude the possibility that unmethylated constructs might be methylated de novo to some degree after being transfected into the cells (7). Thus, the levels of factors influencing gene transcription appear to be high in mammalian cells. Finally, in order to determine whether MBD1 isoforms are truly involved in the repression of these tumor suppressor and imprinted genes via DNA methylation, we employed D. melanogaster cells, which lack genome methylation (50), as host cells. Drosophila cells possess a general transcription machinery homologous to that of mammalian cells (18, 19), but the methylation-insensitive transcription factor Sp1 and endogenous MeCPs are known to be deficient or present at very low levels (14, 30). First, we tested the effect of cytosine methylation of the promoter-inserted pGL3 vectors in SL2 cells. Since the p16 and SNRPN promoter sequences which were cloned into the pGL3 vectors had three putative Sp1 binding motifs (GGCTGG, GGGTGG, and CGGCGG) and one putative Sp1 site (GGGAGG), respectively, we utilized pGL3-p16 and pGL3-SNRPN in combination with the Sp1 expression vector pPacSp1 for a transient expression assay. As shown in Fig. 7A, both the unmethylated and methylated promoters of these genes could confer extremely weak transcriptional activities without cotransfection of pPacSp1. Cotransfection of pPacSp1 led to approximately 10- (p16) and 50- (SNRPN) fold increases in the promoter activity from methylated as well as unmethylated constructs. The relative luciferase activity of the methylated construct was found to be almost equal to that of the unmethylated version in Drosophila cells, indicating that Sp1 can transactivate both p16 and SNRPN promoters in Drosophila SL2 cells even when these promoters are methylated. To clarify whether these constructs might alter their methylation status in the Drosophila cells, we utilized methylation-sensitive transcription factor E2F1 instead of Sp1. The effect of cytosine methylation on transcriptional activation by E2F1 was investigated with the pGL3-VHL and pGL3-SNRPN vectors in combination with the E2F1 expression vector pAc5.1-E2F1. The VHL and SNRPN promoters contained one putative E2F binding motif (TTCGCGC) and three putative E2F sites (CTGGCGC, TTGCCGC, and ATGGCGC), respectively. Neither the unmethylated nor the methylated promoters of these genes showed transcriptional activities in the absence of coexpression of E2F1. Cotransfection of pAc5.1-E2F1 and unmethylated pGL3 constructs led to approximately 4- (VHL) and 25-(SNRPN) fold increases in luciferase activity, while transcription from methylated constructs could not be stimulated any more. Thus, methylation alone could inhibit the transcriptional activity of E2F1 but not of Sp1. Therefore, we confirmed that the constructs used here did not change their methylation status in Drosophila SL2 cells. Next, we examined whether MBD1 can independently down-regulate p16 and SNRPN promoters activated by Sp1 and whether MBD1 isoforms with two or three CXXC domains have different effects on these methylated and unmethylated promoters. The full-length cDNAs encoding MBD1 isoforms were subcloned into a pAc5.1/V5-His vector, which can induce a high expression in Drosophila cells. pAc5.1-MBD1v1 to pAc5.1-MBD1v4 vectors were each transfected into Drosophila SL2 cells, and a Western blot analysis was performed with an anti-MBD1 polyclonal antibody to confirm their expression (data not shown). The effect of MBD1 isoforms on the Sp1-activated transcription from methylated and unmethylated promoters was examined by coexpressing Sp1 and one of the MBD1 isoforms (Fig. 7B). The MBD1 isoforms intensely repressed transcription from both p16 and SNRPN promoters when the promoters were methylated. Interestingly, MBD1v1 and -v2 also inhibited the activities of unmethylated promoters, although the repression levels in unmethylated promoters were lower than those in the methylated versions. In contrast, MBD1v3 and -v4 somewhat increased transcription activities from unmethylated constructs, compared with the results of insertless mock transfections. MBD1 isoforms had a very similar effect on E2F1-activated transcription from unmethylated promoters of VHL and SNRPN genes (data not shown). Thus, MBD1 isoforms can regulate transcription from an unmethylated promoter which is activated by the distinct transcription factor Sp1 or E2F1. Taken together, our independent transcription analyses in the both mammalian and Drosophila cells consistently demonstrated that MBD1 isoforms display multiple effects on transcription from both methylated and unmethylated promoters, depending upon the alternatively spliced CXXC domains.
FIG. 7.
Transcriptional regulation by MBD1 isoforms in Drosophila SL2 cells. (A) Unmethylated (M−) or methylated (M+) pGL3 construct (pGL3-p16 and pGL3-SNRPN or pGL3-VHL and pGL3-SNRPN) was cotransfected in combination with mock A5C vector (solid bars), pPacSp1 expressing transcription factor Sp1 (hatched bars on the left), or pAc5.1-E2F1 expressing transcription factor E2F1 (hatched bars on the right) into SL2 cells. The level of luciferase activity in the presence of unmethylated pGL3 and mock vectors was normalized to 1 for each gene promoter. (B) Methylated (solid bars) or unmethylated (hatched bars) promoter-inserted pGL3 vector was cotransfected with pPacSp1 and MBD1-expressing plasmids (pAc5.1-MBD1v1 to pAc5.1-MBD1v4) or insertless plasmid (mock). The luciferase activity of unmethylated pGL3 in the combination of pPacSp1 and mock was normalized to 100 for each gene promoter, and the relative luciferase activities are presented.
DISCUSSION
In this report, we have presented evidence that MBD1 isoforms can provide an important gene regulation system in mammals, according to the methylation status of many genes in euchromatin regions. These MBD1 isoforms repress transcription preferentially from methylated gene promoters, and MBD1v1 and -v2 also affect unmethylated promoter activities.
The patterns of genome methylation are propagated through mitosis in order to stably maintain the DNA structure and transcriptional state of the genome. There are other types of heritable transcriptional regulation systems in organisms that seem to lack genome methylation. The best-known example is homeotic gene regulation in Drosophila (45). On the basis of reciprocal homeotic phenotypes in several mutants, two classes of genes have been identified: the trithorax group (trxG), which is responsible for sustaining the active state of homeotic gene expression, and the Polycomb group (PcG), which encodes a stable repressor. A direct linkage between DNA methylation and the PcG-trxG system is supported by the recent identification of the cysteine-rich CXXC domain shown in MBD1, DNA methyltransferase, and ALL-1/HRX. The CXXC region in DNA methyltransferase is reported to be a regulatory sequence which can give Zn-binding activity and sense the methylation status of unmethylated and hemimethylated CpG sites (7). It has been found that the amino-terminal region containing the CXXC domain in DNA methyltransferase inhibits its de novo methylation activity (6). However, certain genes in organisms whose genomes do not contain detectable methylated cytosines encode sequences highly homologous to the CXXC domain (45), suggesting the presence of undiscovered roles of the CXXC domain. Our transcription assays imply that an alternatively spliced CXXC region causes MBD1 isoforms to respond differently to specific sites on the genome which are densely or sparsely methylated and unmethylated. Thus, the CXXC domain is thought to be a regulatory element of MBD1.
The localization of EGFP-tagged MBD1 was observed in intact mammalian cells. Full-length MBD1 formed multiple foci in the nucleus, which may indicate chromatin complex formation, reported as MeCP1 complex (31, 37). There was no significant difference in subnuclear localization among MBD1 isoforms, although we did not determine whether these isoforms assemble in the same or different sites in the nucleus. Both the MBD sequence and genome methylation were demonstrated to be indispensable for normal localization of the protein. The localization pattern of MBD1 was quite distinct from that of MeCP2, which exists throughout the nucleus. Thus, these two MBD-containing proteins have different roles in interaction with methylated DNA in the cell. In addition, the increase of the acetylated core histones H3 and H4 in the presence of sodium butyrate and histone deacetylase inhibitor had much less effect on the MBD1-containing focus complexes (data not shown). A link between DNA methylation and histone deacetylation was recently reported (27, 43). Our data, however, emphasized that the localization of MBD1 is dependent on genome methylation rather than on histone acetylation.
A deconvolution system with a highly sensitive charge-coupled camera revealed that MBD1 is present mostly in the euchromatin regions of the human genome. MBD1-containing complexes are likely to target methylated genes for their repression and to construct local heterochromatin regions within the euchromatin region. On the other hand, MBD1 concentrated in the pericentromeric regions of chromosome 1q12, which is known to contain the longest region of heterochromatin adjacent to the centromere, known as juxtacentromeric heterochromatin (47). This heterochromatin region consists of a classical satellite 2 sequence that is normally methylated. Of note, the pericentromere of chromosome 1 is related to the occurrence of hypomethylation and recombination in a rare human genetic disease, immunodeficiency, centromeric region instability, and facial anomalies (25), and many kinds of cancer (47). We further determined the chromosomal location of the MBD1 gene at 6.29cR from WI-6206 on chromosome 18q21 (data not shown), which is frequently mutated in cancers (49). Thus, MBD1 may be involved in chromosome structure and genome stability. Recently, it was reported that three human PcG proteins, BMI1, RING1, and hPc2, are tightly associated with the heterochromatin regions in 1q12 (44). Taken together, these data indicate that MBD1 may be involved in local heterochromatin production within the euchromatin region and in the construction of pericentromeric heterochromatin on chromosome 1, either independently or together with other proteins.
We next demonstrated gene repression by MBD1 in a methylation-dependent manner by using CpG-rich promoters from tumor suppressor and imprinted genes. Recombinant MBD1 could preferentially bind methylated DNAs of the gene promoters and inhibited transcription from methylated rather than unmethylated promoters. We further focused on the significance of MBD1 isoforms, using a transient transfection system in mammalian cells. The cytosine methylation of promoter-inserted constructs gave very low transcriptional activities, even in the absence of MBD1 coexpression. Thus, mammalian cells appear to have high gene repression activity, probably due to the involvement of endogenous MeCPs (9, 41, 46) or de novo methylation (7). Under such conditions, MBD1 isoforms more strongly repressed transcription from methylated promoters. Interestingly, the MBD1 isoforms v1 and v2 containing three CXXC domains could also reduce transcription from unmethylated promoters. Finally, in order to demonstrate the precise mechanism of gene regulation by MBD1, we investigated the effect of these isoforms on Sp1- or E2F1-activated transcription in methylation-deficient Drosophila SL2 cells. Sp1 and E2F1 are known to be methylation-insensitive and methylation-sensitive transcription factors, respectively. In Sp1-activated transcription, MBD1v1 and -v2 repressed gene expression from both methylated and unmethylated promoters, while MBD1v3 and -v4 inhibited the transcriptional activities only when the promoter was methylated. In addition, MBD1v1 and -v2, but not MBD1v3 and -v4, similarly repressed transcription from unmethylated promoters that were transactivated by E2F1 (data not shown). Accordingly, we concluded that MBD1 can affect both methylated and unmethylated promoters, depending upon the presence of alternatively spliced CXXC3 sequence. Thus, all of the MBD1 isoforms could inhibit transcription from methylated genes by DNA contact through the MBD sequence, and MBD1v1 and -v2 might acquire the ability to repress unmethylated promoters by interaction between the CXXC3 and the general transcriptional machineries, some chromosomal proteins, or DNA itself. Another protein that can bind to methylated DNA is MDBP-2-H1, which belongs to the histone H1 family (28). The phosphorylation of MDBP-2-H1 is required for the binding and suppressive activities of the methylated promoter (11, 12). The regulation of MBD1 by the RNA-splicing events exemplifies a new molecular mechanism in methylation-mediated transcriptional regulation. Boyes and Bird reported that MeCP1 can repress transcription from sparsely methylated promoters and that the inhibition was fully overcome by the presence of enhancer (10). In contrast, densely methylated genes could not be reactivated by the enhancer. Sparsely methylated genes seem to form an unstable complex with MeCP1. Recently, Hendrich and Bird pointed out that MBD1 binds to DNA with low levels of methylation in DNA methyltransferase-deficient mouse cells (20). These observations may be explained by the content of MBD1 isoforms in the MeCP1 complex, since these isoforms can have different effects on DNAs due to their methylation status. Possibly, these MBD1 isoforms may allow methylated genes to be transcribed or unmethylated genes to be repressed, which is known as methylation-mediated chromatin remodeling. Therefore, MBD1 isoforms are thought to play an important role in the establishment and maintenance of local chromatin states to regulate gene activities.
ACKNOWLEDGMENTS
We thank Y. Kimura, N. Mugita, T. Kino, and H. Takeshima (Kumamoto University School of Medicine) for their technical assistance; J. T. Kadonaga for providing the pPacSp1 plasmid; R. M. Evans for providing the A5C vector; K. Ohtani for providing the pDCE2F plasmid; H. Nakakuma for providing sera from a patient with CREST syndrome; and T. Arino for secretarial assistance.
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science and Culture, Japan (M.N.), and from the Science and Technology Agency, Japan (K.O.).
REFERENCES
- 1.Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci USA. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballabio A, Willard H F. Mammalian X-chromosome inactivation and the XIST gene. Curr Opin Genet Dev. 1992;2:439–447. doi: 10.1016/s0959-437x(05)80155-8. [DOI] [PubMed] [Google Scholar]
- 3.Bartolomei M S, Tilghman S M. Genomic imprinting in mammals. Annu Rev Genet. 1997;31:493–525. doi: 10.1146/annurev.genet.31.1.493. [DOI] [PubMed] [Google Scholar]
- 4.Baylin S B, Herman J G, Graff J R, Vertino P M, Issa J P. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 5.Bergman Y, Mostoslavsky R. DNA demethylation: turning genes on. Biol Chem. 1998;379:401–407. doi: 10.1515/bchm.1998.379.4-5.401. [DOI] [PubMed] [Google Scholar]
- 6.Bestor T H. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J. 1992;11:2611–2617. doi: 10.1002/j.1460-2075.1992.tb05326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bestor T H, Verdine G L. DNA methyltransferases. Curr Opin Cell Biol. 1994;6:380–389. doi: 10.1016/0955-0674(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 8.Bird A P. Gene number, noise reduction and biological complexity. Trends Genet. 1995;11:94–100. doi: 10.1016/S0168-9525(00)89009-5. [DOI] [PubMed] [Google Scholar]
- 9.Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64:1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- 10.Boyes J, Bird A. Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J. 1992;11:327–333. doi: 10.1002/j.1460-2075.1992.tb05055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruhat A, Jost J P. In vivo estradiol-dependent dephosphorylation of the repressor MDBP-2-H1 correlates with the loss of in vitro preferential binding to methylated DNA. Proc Natl Acad Sci USA. 1995;92:3678–3682. doi: 10.1073/pnas.92.9.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruhat A, Jost J P. Phosphorylation/dephosphorylation of the repressor MDBP-2-H1 selectively affects the level of transcription from a methylated promoter in vitro. Nucleic Acids Res. 1996;24:1816–1821. doi: 10.1093/nar/24.10.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buschhausen G, Wittig B, Graessmann M, Graessmann A. Chromatin structure is required to block transcription of the methylated herpes simplex virus thymidine kinase gene. Proc Natl Acad Sci USA. 1987;84:1177–1181. doi: 10.1073/pnas.84.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Courey A J, Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 15.Cross S H, Meehan R R, Nan X, Bird A. A component of the transcriptional repressor MeCP1 shares a motif with DNA methyltransferase and HRX proteins. Nat Genet. 1997;16:256–259. doi: 10.1038/ng0797-256. [DOI] [PubMed] [Google Scholar]
- 16.Feinberg A P, Rainier S, DeBaun M R. Genomic imprinting, DNA methylation, and cancer. J Natl Cancer Inst Monogr. 1995;1995:21–26. [PubMed] [Google Scholar]
- 17.Graff J R, Herman J G, Lapidus R G, Chopra H, Xu R, Jarrard D F, Isaacs W B, Pitha P M, Davidson N E, Baylin S B. E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res. 1995;55:5195–5199. [PubMed] [Google Scholar]
- 18.Heberlein U, England B, Tjian R. Characterization of Drosophila transcription factors that activate the tandem promoters of the alcohol dehydrogenase gene. Cell. 1985;41:965–977. doi: 10.1016/s0092-8674(85)80077-5. [DOI] [PubMed] [Google Scholar]
- 19.Heiermann R, Pongs O. In vitro transcription with extracts of nuclei of Drosophila embryos. Nucleic Acids Res. 1985;13:2709–2730. doi: 10.1093/nar/13.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herman J G, Graff J R, Myohanen S, Nelkin B D, Baylin S B. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herman J G, Latif F, Weng Y, Lerman M I, Zbar B, Liu S, Samid D, Duan D S, Gnarra J R, Linehan W M, Baylin S B. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci USA. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holler M, Westin G, Jiricny J, Schaffner W. Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes Dev. 1988;2:1127–1135. doi: 10.1101/gad.2.9.1127. [DOI] [PubMed] [Google Scholar]
- 24.Iguchi-Ariga S M M, Schaffner W. CpG methylation of the cAMP responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- 25.Jeanpierre M, Turleau C, Aurias A, Prieur M, Ledeist F, Fischer A, Viegas P E. An embryonic-like methylation pattern of classical satellite DNA is observed in ICF syndrome. Hum Mol Genet. 1993;2:731–735. doi: 10.1093/hmg/2.6.731. [DOI] [PubMed] [Google Scholar]
- 26.Johnson D G, Ohtani K, Nevins J R. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 27.Jones P L, Veenstra G J, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 28.Jost J P, Hofsteenge J. The repressor MDBP-2 is a member of the histone H1 family that binds preferentially in vitro and in vivo to methylated nonspecific DNA sequences. Proc Natl Acad Sci USA. 1992;89:9499–9503. doi: 10.1073/pnas.89.20.9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kass S U, Pruss D, Wolffe A P. How does DNA methylation repress transcription? Trends Genet. 1997;13:444–449. doi: 10.1016/s0168-9525(97)01268-7. [DOI] [PubMed] [Google Scholar]
- 30.Kudo S. Methyl-CpG-binding protein MeCP2 represses Sp1-activated transcription of the human leukosialin gene when the promoter is methylated. Mol Cell Biol. 1998;18:5492–5499. doi: 10.1128/mcb.18.9.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamond A I, Earnshaw W C. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 32.Lewis J D, Meehan R R, Henzel W J, Maurer F I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 33.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 34.Li E, Bestor T H, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 35.Ma Q, Alder H, Nelson K K, Chatterjee D, Gu Y, Nakamura T, Canaani E, Croce C M, Siracusa L D, Buchberg A M. Analysis of the murine All-1 gene reveals conserved domains with human ALL-1 and identifies a motif shared with DNA methyltransferases. Proc Natl Acad Sci USA. 1993;90:6350–6354. doi: 10.1073/pnas.90.13.6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Makino K, Kuwahara H, Masuko N, Nishiyama Y, Morisaki T, Sasaki J, Nakao M, Kuwano A, Nakata M, Ushio Y, Saya H. Cloning and characterization of NE-dlg: a novel human homolog of the Drosophila discs large (dlg) tumor suppressor protein interacts with the APC protein. Oncogene. 1997;14:2425–2433. doi: 10.1038/sj.onc.1201087. [DOI] [PubMed] [Google Scholar]
- 37.Meehan R R, Lewis J D, McKay S, Kleiner E L, Bird A P. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 38.Merlo A, Herman J G, Mao L, Lee D J, Gabrielson E, Burger P C, Baylin S B, Sidransky D. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 39.Miller O J, Schnedl W, Allen J, Erlanger B F. 5-Methylcytosine localised in mammalian constitutive heterochromatin. Nature. 1974;251:636–637. doi: 10.1038/251636a0. [DOI] [PubMed] [Google Scholar]
- 40.Moroi Y, Peebles C, Fritzler M J, Steigerwald J, Tan E M. Autoantibody to centromere (kinetochore) in scleroderma sera. Proc Natl Acad Sci USA. 1980;77:1627–1631. doi: 10.1073/pnas.77.3.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nan X, Campoy F J, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 42.Nan X, Meehan R R, Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 44.Saurin A J, Shiels C, Williamson J, Satijn D P, Otte A P, Sheer D, Freemont P S. The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J Cell Biol. 1998;142:887–898. doi: 10.1083/jcb.142.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schumacher A, Magnuson T. Murine Polycomb- and trithorax-group genes regulate homeotic pathways and beyond. Trends Genet. 1997;13:167–170. [PubMed] [Google Scholar]
- 46.Stein R, Razin A, Cedar H. In vitro methylation of the hamster adenine phosphoribosyltransferase gene inhibits its expression in mouse L cells. Proc Natl Acad Sci USA. 1982;79:3418–3422. doi: 10.1073/pnas.79.11.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Surralles J, Puerto S, Ramirez M J, Creus A, Marcos R, Mullenders L H, Natarajan A T. Links between chromatin structure, DNA repair and chromosome fragility. Mutat Res. 1998;404:39–44. doi: 10.1016/s0027-5107(98)00093-1. [DOI] [PubMed] [Google Scholar]
- 48.Sutcliffe J S, Nakao M, Christian S, Orstavik K H, Tommerup N, Ledbetter D H, Beaudet A L. Deletions of a differentially methylated CpG island at the SNRPN gene define a putative imprinting control region. Nat Genet. 1994;8:52–58. doi: 10.1038/ng0994-52. [DOI] [PubMed] [Google Scholar]
- 49.Thiagalingam S, Lengauer C, Leach F S, Schutte M, Hahn S A, Overhauser J, Willson J K, Markowitz S, Hamilton S R, Kern S E, Kinzler K W, Vogelstein B. Evaluation of candidate tumour suppressor genes on chromosome 18 in colorectal cancers. Nat Genet. 1996;13:343–346. doi: 10.1038/ng0796-343. [DOI] [PubMed] [Google Scholar]
- 50.Urieli S S, Gruenbaum Y, Sedat J, Razin A. The absence of detectable methylated bases in Drosophila melanogaster DNA. FEBS Lett. 1982;146:148–152. doi: 10.1016/0014-5793(82)80723-0. [DOI] [PubMed] [Google Scholar]
- 51.Weitzel J M, Buhrmester H, Stratling W H. Chicken MAR-binding protein ARBP is homologous to rat methyl-CpG-binding protein MeCP2. Mol Cell Biol. 1997;17:5656–5666. doi: 10.1128/mcb.17.9.5656. [DOI] [PMC free article] [PubMed] [Google Scholar]