Abstract
Trithorax (TRX) and ASH1 belong to the trithorax group (trxG) of transcriptional activator proteins, which maintains homeotic gene expression during Drosophila development. TRX and ASH1 are localized on chromosomes and share several homologous domains with other chromatin-associated proteins, including a highly conserved SET domain and PHD fingers. Based on genetic interactions between trx and ash1 and our previous observation that association of the TRX protein with polytene chromosomes is ash1 dependent, we investigated the possibility of a physical linkage between the two proteins. We found that the endogenous TRX and ASH1 proteins coimmunoprecipitate from embryonic extracts and colocalize on salivary gland polytene chromosomes. Furthermore, we demonstrated that TRX and ASH1 bind in vivo to a relatively small (4 kb) bxd subregion of the homeotic gene Ultrabithorax (Ubx), which contains several trx response elements. Analysis of the effects of ash1 mutations on the activity of this regulatory region indicates that it also contains ash1 response element(s). This suggests that ASH1 and TRX act on Ubx in relatively close proximity to each other. Finally, TRX and ASH1 appear to interact directly through their conserved SET domains, based on binding assays in vitro and in yeast and on coimmunoprecipitation assays with embryo extracts. Collectively, these results suggest that TRX and ASH1 are components that interact either within trxG protein complexes or between complexes that act in close proximity on regulatory DNA to maintain Ubx transcription.
The control of body segment identity in many organisms is achieved in large part by the activities of homeotic genes. In Drosophila, the combined activities of the transiently expressed segmentation genes initiate the pattern of expression of the homeotic genes at the blastoderm stage, and additional factors and mechanisms are required to preserve these patterns at later stages of development. Two groups of genes, the trithorax group (trxG) (reviewed in reference 12) and the Polycomb group (PcG) (reviewed in references 2, 18, and 24) play major roles in the maintenance of the active and the repressed state, respectively. It is thought that trxG and PcG proteins are assembled into multiprotein complexes, which function by maintaining either open or closed domains of chromatin structure. This chromatin association has been established for several trxG proteins. The yeast and human homologues of the Drosophila trxG proteins BRAHMA, SNR1, and MOIRA have been shown to be components of a 2-MDa yeast and human SWI-SNF chromatin remodeling complex (7, 17, 32), and a similar Drosophila complex was recently characterized (16). GAGA factor, a DNA binding protein encoded by the trxG gene Trithorax-like (10), is required for the function of another Drosophila chromatin remodeling complex, the NURF complex (30). These protein complexes possess ATP-dependent activities that facilitate the binding of other transcription factors. While the SWI-SNF complex has not been shown to be stably associated with chromatin, other trxG and PcG proteins are localized to multiple sites on salivary gland polytene chromosomes (1, 6, 13, 19, 28, 31). It has been proposed that the SWI-SNF complex is recruited to DNA by interaction with TRX-containing protein complexes formed at trxG response elements (TREs). This possibility was suggested by the finding that TRX interacts with SNR1, a trxG protein and a component of the SWI-SNF complex (20).
trxG genes encode proteins with several conserved domains. TRX, ASH1, and ASH2 each contain PHD fingers, which are Cys-rich Zn finger-like motifs implicated in protein-protein interactions (1, 15, 28). In addition, TRX and ASH1 contain a conserved SET domain (15, 28), an approximately 130-amino-acid (aa) region found in a number of other chromatin-associated proteins, including the PcG protein E(Z) (11) and the modifier of position effect variegation Su(Var)3-9 (29). The SET domains of TRX and ALL-1/HRX have been shown to interact with several other proteins (8, 20).
Based on analyses of genetic interactions between trx and two other trxG genes, ash1 and ash2, it was proposed that these proteins function in multimeric protein complexes (23). This was corroborated by the finding that TRX binding to polytene chromosomes was decreased at nonpermissive temperatures in larvae homozygous for a temperature-sensitive ash1 allele (13). Furthermore, some of the ASH1 binding sites on polytene chromosomes were found to be similar (28) to those reported for TRX (13), suggesting that these proteins may regulate a similar set of genes. However, with the exception of the interaction between TRX and the SNR1 component of the SWI-SNF complex (20), no biochemical evidence is available concerning direct interactions between TRX and other trxG proteins or the existence of a protein complex containing TRX.
The existing data suggest that TREs and PcG response elements (PREs) tend to be clustered together in the regulatory regions of the bithorax complex (BX-C) (4, 5, 27). In the best-characterized TRE-PRE-containing module of the bxd regulatory region of Ubx, a TRE was mapped to a 90-bp DNA element (27). It is not known whether other trxG proteins are required for the activity of this element. Here we provide evidence that TRX and ASH1 act together through the same bxd subregion. Mutations in each gene affect the activity of this region. Both proteins colocalize at most of their binding sites on polytene chromosomes, as well as at the insertion sites of transgenes containing this region. Furthermore, they are associated with each other in embryos and larvae. This interaction appears to be mediated through their SET domains, since these domains are capable of interacting both in vivo and in vitro, and the interaction requires the entire SET domain of TRX. Thus, these proteins may be components of the same or interacting protein complexes.
MATERIALS AND METHODS
Immunoprecipitation from Drosophila embryonic nuclear extracts.
Four- to 18-h-old embryos were collected and nuclear protein extracts were prepared as previously described (9, 25, 27). One hundred microliters of protein extract (4 mg/ml) in a mixture of 50 mM Tris-HCl (pH 7.4), 250 mM NaCl, 5 mM EDTA, 0.1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 1-mg/ml leupeptin, 1-mg/ml aprotinin, and 1-mg/ml pepstatin was kept on ice for 30 min. After centrifugation at 14,000 rpm for 2 min, the supernatant was collected and diluted by adding 400 μl of the same buffer, but lacking NaCl. The solution was precleared by incubation with protein A-Sepharose beads for 40 min, followed by centrifugation at 14,000 rpm for 5 min. The supernatant was incubated with either anti-TRX or anti-ASH1 antibodies or preimmune serum for 2 h, and then 20 μl of protein A-Sepharose beads was added, and incubation was continued for 1 h. Protein A-Sepharose beads were centrifuged at 6,000 rpm for 5 min and washed six times with 0.5 ml of binding buffer. Proteins were recovered by adding to the beads 50 μl of the sodium dodecyl sulfate-containing sample buffer and boiling for 5 min. ASH1 and TRX proteins in the immunoprecipitated pellet were analyzed by immunoblotting.
In situ hybridization and immunostaining of polytene chromosomes.
Drosophila polytene chromosome spreads were prepared from salivary glands of third-instar larvae and processed as described previously (27). The DNA of the pCaSpeR vector containing the mini-white marker gene was labeled with digoxigenin-11-dUTP (Boehringer Mannheim) by using a random-priming DNA labeling kit (Boehringer Mannheim) and utilized as a probe for in situ hybridization in conjunction with anti-digoxigenin-fluorescein antibody (Boehringer Mannheim). Fluorescent double labeling of proteins (TRX and ASH1) on polytene chromosomes was carried out as described previously (20), by using newly generated rat anti-TRX polyclonal antibody (N1 domain) (20) at a 1:20 dilution and anti-ASH1 rabbit polyclonal antibody (28) at a 1:40 dilution. Goat Texas red-conjugated anti-rabbit immunoglobulin G (IgG) and fluorescein-conjugated anti-rat IgG (Jackson Immunoresearch Labs) were used as secondary antibodies at a 1:200 dilution. DNA was counterstained with Hoechst 33258 (Sigma). The slides were mounted in Vectashield mounting medium for fluorescence visualization (Vector). Image files of labeled chromosomes were acquired with a Zeiss microscope equipped with a digital camera and processed with the Adobe Photoshop program.
Generation and analysis of transgenic lines.
The 4-kb N constructs were made by inserting a BamHI-KpnI fragment from the bxd-pbx region of Ubx (nucleotides 216,487 to 220,533 [GenBank accession no. U31961]) into a pCaSpeR3 vector (27). Injections into a homozygous yw;+/+;+/+ strain were performed by standard procedures (26). To test the effect of the ash1 mutations, ash1B1, ash112, and ash122, on white gene expression in the N lines, transformants from each tested line were crossed to flies from balancer stocks containing mutant loci. For all comparisons, flies of the same sex and age were compared, and to avoid the potential effects from balancer chromosomes on eye color, comparisons were made with unbalanced heterozygotes for each transgenic line.
Yeast two-hybrid assays.
cDNA inserts were cloned by PCR into the pGBT9-DBD or pACTII-AD vector (Clontech). Association between the proteins in question was determined by synthesis of β-galactosidase in SFY526 and HF7c yeast reporter strains and assayed according to the Matchmaker two-hybrid protocol (Clontech). Point mutants were generated by a PCR-based strategy using oligonucleotide-directed mutagenesis. To examine the synthesis of the mutated proteins in yeast, the inserts were recloned into the yeast vector pAS1-CYH2; following transformation, the resultant proteins were detected by Western blotting.
In vitro binding assays.
ASH1 and TRX SET-spanning polypeptides were expressed as glutathione S-transferase (GST) fusion proteins in Escherichia coli, purified, and immobilized on affinity matrix beads according to standard methodology. 35S-labeled TRX and ASH1 polypeptides were prepared by utilizing the TNT T7 coupled transcription-translation reticulocyte lysate system (Promega) according to the manufacturer’s instructions. The radiolabeled polypeptides were diluted in binding buffer (20 mM Tris [pH 8.0], 0.2% Triton X-100, 2 mM EDTA, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 1-mg/ml chymostatin, 2-mg/ml antipain, 2-mg/ml pepstatin A, 2-mg/ml aprotinin, 5-mg/ml leupeptin) and incubated at 4°C for 2 h with beads containing equal amounts of immobilized appropriate GST fusion protein or GST alone. The beads were washed three times with 1 ml of binding buffer and boiled in 2× sample buffer, and the eluted proteins were resolved on SDS–10% polyacrylamide gel and visualized by autoradiography.
In vitro immunoprecipitation.
T7-tagged ASH1 or TRX polypeptides, two unrelated Drosophila proteins (T7-tagged p62 nuclear porin-related protein and T7-tagged Dsup35 protein cloned by us in the context of other experiments), and 35S-labeled TRX or ASH1 SET-spanning polypeptide were synthesized in a coupled transcription-translation system (Promega). Immunoprecipitation was performed by incubation (2 h at 4°C) of equal amounts of T7-tagged proteins with radiolabeled 35S-labeled polypeptides followed by incubation (2 h at 4°C) with 5 mg of anti-T7 monoclonal antibody (MAb) (Novagen) in 0.5 ml of binding buffer containing 50 mM Tris (pH 7.4), 150 mM NaCl, 5 mM EDTA, 0.1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 1-mg/ml chymostatin, 2-mg/ml antipain, 2-mg/ml pepstatin A, 2-mg/ml aprotinin, and 5-mg/ml leupeptin. Thirty microliters of protein G-Sepharose beads was added, and incubation was continued for 1 h at 4°C. Protein G-Sepharose beads were centrifuged and washed three times with 1 ml of binding buffer. Proteins were recovered by boiling in 30 ml of 2× sample buffer, and bound radiolabeled proteins were analyzed by electrophoresis and autoradiography. Amounts of the T7-tagged proteins were determined by Western blot analysis.
RESULTS
TRX and ASH1 are associated in embryos and colocalize on larval polytene chromosomes.
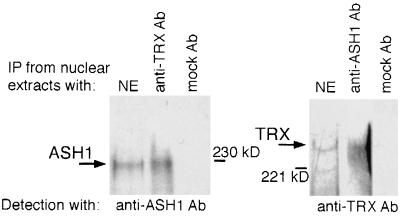
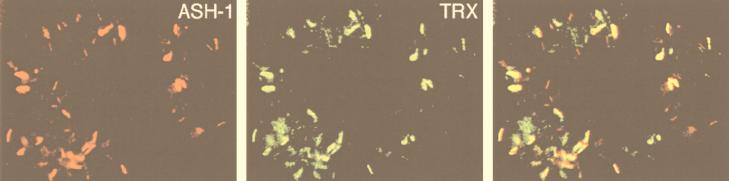
To explore the possibility that endogenous TRX and ASH1 are capable of oligomerization, we examined whether the two proteins are associated in embryonic nuclear extracts. Following immunoprecipitation of proteins from nuclear extracts with anti-TRX antibody, we detected ASH1 in the immunoprecipitated material (Fig. 1), indicating that TRX and ASH1 may be associated in embryos. Similar results were obtained in a reciprocal experiment (Fig. 1). To address whether TRX and ASH1 colocalize on salivary gland chromosomes, we examined simultaneously the binding sites of ASH1 and TRX in double immunostaining experiments, using rabbit anti-ASH1 antibody and rat anti-TRX antibody. ASH1 binds to ∼100 sites on polytene chromosomes, with very strong signals associated with ∼20 of these sites (28), while TRX has been localized to 16 strong sites and to a number of weaker binding sites (6, 13). The results of our immunostaining experiments showed that almost all of the strong binding sites and at least one-half of the weak binding sites for TRX colocalize with ASH1 binding sites (Fig. 2).
FIG. 1.
Endogenous ASH1 and TRX coimmunoprecipitate (IP) in 0- to 20-h Drosophila embryonic nuclear extracts. Anti-TRX Ab, N1 antibody (13); mock Ab, unrelated PHIT antibody; NE, nuclear extract. Anti-ASH1 antibody is described in reference 28. The positions of molecular size markers are indicated.
FIG. 2.
Endogenous TRX and ASH1 proteins colocalize on salivary gland polytene chromosomes. Merging of green and red signals representing TRX and ASH1, respectively, identifies the sites (yellow bands on the right panel) where the two proteins colocalize.
Both TRX and ASH1 bind to a Ubx regulatory region.
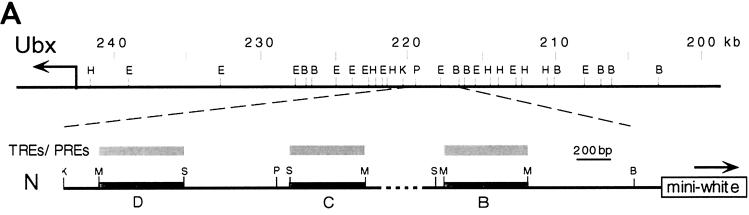
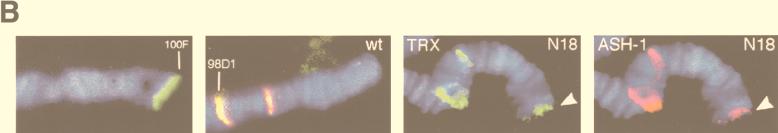
TRX and ASH1 are required for maintenance of expression of several homeotic genes, including Ubx (3, 5, 14, 15, 22, 23). In the bxd regulatory region of Ubx, TREs regulated by TRX were analyzed in detail and were localized to three neighboring 400-bp DNA fragments (5, 27). These three elements are included in the 4-kb N transgene (Fig. 3A). This transgene is a site of binding of TRX on salivary gland polytene chromosomes (Fig. 3A) (27). In double immunostaining experiments using rat anti-TRX and rabbit anti-ASH1 antibodies, we examined whether the ASH1 protein also binds to the site of insertion of the N transgene. Figure 3B shows that in the N18 transgenic line, the insert is located at the tip of chromosome 3R at polytene band 100F (Fig. 3B, panel 1). Analysis of polytene chromosomes of wild-type larvae showed no strong binding of either of the endogenous TRX or ASH1 proteins in this region (Fig. 3B, panel 2). However, we observed a new signal on polytene chromosomes of the N18 line for each protein at the site of insertion of the transgene (Fig. 3B, panels 3 and 4). The results of this experiment suggest that both proteins bind to the 4-kb bxd regulatory element, confirming the conclusion that these proteins directly associate in vivo on the same regulatory region.
FIG. 3.
TRX and ASH1 bind in vivo to a 4-kb subregion of the bxd regulatory region of Ubx. (A [Top]) Partial map of the BX-C upstream of the Ubx promoter. (Bottom) Map of previously analyzed TRE-PRE-containing expression maintenance modules in the bxd region of Ubx (27). TRX and specific PcG genes that were previously shown to interact genetically with the central C module (27) are indicated above the map. N indicates the mini-white reporter gene-containing construct used to detect ash1-dependent TRE activity in transgenic flies and to show colocalization of TRX and ASH1 on polytene chromosomes. K, KpnI; M, MspI; S, Sau3A; P, PstI; B, BamHI. (B) Colocalization of the endogenous TRX (green) and ASH1 (red) proteins on salivary gland polytene chromosomes at the site of insertion of the N18 transgene. (First panel) In situ hybridization of pCaSpeR-βgal DNA to polytene chromosomes of the N18 line. (Second panel) TRX- and ASH1-specific antibody staining showing colocalization to the distal portion of the wild-type 3R chromosome. TRX and ASH1 colocalize at 98D1, the cytological localization of the endogenous fkh gene. (Third and fourth panels) TRX and ASH1 antibody staining, respectively, each showing binding to the site of insertion of the N18 construct at 100F (arrowheads).
Response elements for both trx and ash1 are contained within the same regulatory region of Ubx.
The experiments described above suggest that ASH1 binds in vivo to the same bxd subregion that contains both binding sites for TRX and three functionally important TREs (27). We sought, therefore, to address the question of whether ash1 is required for the activity of this region. This TRE-PRE-containing regulatory region is capable of regulating mini-white gene expression in the eyes of transgenic flies in a trxG- and PcG-dependent manner (27). We therefore tested whether ash1 mutations affected the eye color in these flies, which is entirely dependent on mini-white transgene expression. A number of transgenic N lines containing a mini-white reporter gene (Fig. 3A) were examined in the background of three ash1 mutant alleles, ash1B1, ash112, and ash122. As Fig. 4 shows, the eye color is strongly reduced relative to that of the wild type in heterozygotes for the strong allele, ash122. Similarly, in most of the other N lines, we observed a clear decrease in the eye color in flies heterozygous for either of the three ash1 alleles tested as follows. The ratios of numbers of lines showing decreased white gene expression in response to heterozygous ash1 alleles versus number of transgenic lines tested were 7/7, 5/7, and 5/7 for ash1B1, ash112, and ash122, respectively. This suggests that the activity of this TRE-containing region depends strongly on the level of ASH1. These results are very similar to those obtained with the same transgenic lines and the null trxB11 allele (27). Although the sensitivity of the mini-white reporter depends on the site of insertion of the transgene, our results suggest that the 4-kb N construct contains one or more ash1 response elements, in addition to the three trx-dependent response elements identified previously (27). This further strengthens our hypothesis that TRX and ASH1 interact in vivo, since they exert their function through the same bxd DNA. Although such a possibility was anticipated from the genetic analysis, this is the first demonstration that the same regulatory subregion contains response elements for two physically interacting trxG proteins.
FIG. 4.
Effect of the ash122 mutation in heterozygotes on expression of the mini-white gene. Eye color due to expression of the mini-white transgene in the N18-15 heterozygous line (left) is decreased in ash122 heterozygotes (right).
Physical interaction domains of TRX and ASH1 span the SET domain.
By applying yeast two-hybrid assays as well as other methodologies, we recently found that the SET domains of both TRX and ASH1 proteins can self-associate (21b). The self-associating TRX fragment (aa 3540 to 3759) spans the ∼130-aa SET domain and includes an additional ∼90 aa of upstream sequence. The self-interacting ASH1 region includes the entire SET domain (residues 1318 to 1448) in addition to upstream sequence (aa 1160 to 1317). An alternative self-associating region of ASH1 (aa 1245 to 1525) also includes the entire SET domain. Mutations within the SET domain of both TRX and ASH1 prevent self-association. We examined whether those TRX and ASH1 regions can also undergo hetero-oligomerization. Indeed, the two polypeptides interacted strongly in yeast, as evidenced by activation of both the HIS and lacZ reporters (not shown). To confirm this result, we applied GST pull-down methodology as well as coimmunoprecipitation analysis. A C-terminal TRX polypeptide (TRX SET) was synthesized and radiolabeled in a coupled transcription-translation system and tested for binding to the relevant ASH1 polypeptide (ASH1 SET) linked to GST. As can be seen in Fig. 5A (left), this ASH1-linked resin bound 10- to 20-fold more TRX SET than did GST resin alone. The reciprocal experiment is shown in Fig. 5A (right panel). For in vitro coimmunoprecipitation analysis, the same TRX polypeptide was radiolabeled and mixed with unlabeled epitope-tagged (T7) ASH1 SET. The labeled TRX SET coimmunoprecipitated with the T7-ASH1 SET but not with two unrelated T7-tagged proteins (Fig. 5B, left). Similar results were obtained in a reciprocal experiment. Finally, plasmids encoding the T7-tagged ASH1 SET and HA-tagged TRX SET were transiently cotransfected into COS cells. The epitope-tagged polypeptides produced in vivo were also found to coimmunoprecipitate (Fig. 5C).
FIG. 5.
ASH1 and TRX SET domains interact in vitro. Interaction was analyzed by GST pull-down assays (A), coimmunoprecipitation (IP) of in vitro-produced polypeptides (B), and coimmunoprecipitation of polypeptides expressed in transfected cells (C). The radiolabeled TRX SET polypeptide in panels A and B and the epitope-tagged polypeptides in panel B were made by coupled transcription-translation. GST-ASH1 SET and GST polypeptides in panel A were produced in bacteria. In panel B, p62 related and Dsup35 are Drosophila proteins, which served in this experiment as controls for specificity. HA (hemagglutinin) and T7 are epitope tags. The positions of the molecular size markers are indicated on the left.
To address the biological significance of this hetero-oligomerization, we mutagenized conserved residues within the SET domain and tested their effects on interaction in yeast. Thirteen different mutations at either single amino acids or nearby pairs of amino acids were constructed, 10 at highly conserved residues and 3 controls at nonconserved residues within TRX SET. Each of the alterations of conserved amino acids resulted in the loss of most or all of the capacity of TRX SET to interact with ASH1 SET in yeast (Fig. 6). In contrast, the three alterations of nonconserved residues, located within the SET domain or immediately upstream of it, did not affect the interaction (Fig. 6). In all cases, we confirmed synthesis of the mutated polypeptides in yeast (not shown). Finally, a more limited mutagenesis analysis of conserved residues within the ASH1 SET domain showed that conversion of GRG (residues 1310 to 1321) to VRV, PN (1391 and 1392) to AY, I (1414) to A, or DY (1423 and 1424) to AA resulted in the loss of most or all of the interaction in yeast (not shown). These results argue for the functional significance of the TRX SET-ASH1 SET interactions seen in yeast and in vitro and suggest that the association in embryos between full-length TRX and ASH1 is direct and involves binding between their SET domains.
FIG. 6.
Effect of point mutations in TRX SET on its interaction with ASH1 SET as determined in yeast two-hybrid assays. The association between the polypeptides was determined by synthesis of β-galactosidase in the SFY526 reporter strain, as assayed on filters. “Strong” and “very weak” interactions indicate that color developed in less than 1 h or in more than 12 h, respectively.
DISCUSSION
Several criteria are used to define trxG genes. (i) Mutations in members of this group have synergistic effects, i.e., mutations in one member enhance the mutant phenotypes of other group members; (ii) trxG mutants suppress the phenotypes of PcG mutants; and (iii) trxG genes are required to maintain homeotic gene expression. Although all of these criteria apply to most of the genes in this family, it is apparent that the trxG gene encodes a variety of proteins with different biochemical properties. Some of the genes encode proteins that are components of chromatin remodeling complexes, while others, such as trx and ash1, encode proteins that are associated with chromosomes but have no known biochemical activities. Previous studies have shown that trx and ash1 interact genetically (23). Here we provide evidence that the TRX and ASH1 proteins colocalize on polytene chromosomes, that they can exert their function through the same DNA region, and that they coimmunoprecipitate from embryonic extracts and appear to physically interact through their conserved SET domains.
The endogenous TRX and ASH1 proteins are localized to many of the same sites on salivary gland polytene chromosomes (Fig. 2). This suggests that they may coregulate many of the same target genes. Prompted by TRX binding sites on polytene chromosomes, we previously showed that TRX regulates the region-specific homeotic gene fork head (fkh) (13). The cytological location of fkh is 98D1. ASH1 also binds to this cytological region (28), and we showed that the proteins colocalize at this site (Fig. 3B), suggesting that ASH1 is also required for regulation of the fkh gene. It is likely that the true number of TRX-ASH1-regulated genes is substantially larger than the number of strong sites detected on salivary gland polytene chromosomes (∼20), since some of their target genes are probably not expressed in this tissue and, therefore, are not bound by TRX and ASH1.
Direct evidence for a physical interaction between endogenous TRX and ASH1 is provided by their coimmunoprecipitation from embryonic extracts (Fig. 1). In principle, physical interaction between TRX and ASH1 might occur within the same complex or between two separate complexes, perhaps in association with neighboring DNA sequences. It has been shown recently that ASH1 is a component of a large-molecular-weight complex or complexes (16). Our preliminary data on the distributions of TRX and ASH1 in glycerol gradients also suggest that they are components of several protein complexes of various sizes. It is possible that different TRX-ASH1 complexes are present at different stages of embryonic development. Indeed, in extracts from very early embryos (0 to 4 h after egg laying), TRX and ASH1 comigrate in fractions that contain complexes of about 1.5 MDa, while in older embryos (4 to 8 h), they are both found in fractions containing larger complexes of several megadaltons (22a). It is not yet known whether these correspond to complexes formed at the bxd TREs that we have identified (27).
Through application of several methodologies, we have shown physical interaction between the SET domains of TRX and ASH1. Perhaps the best evidence for the biological significance of this interaction is the mutagenesis analysis (Fig. 6). Interestingly, the TRX SET domain is apparently a multifunctional domain, because the interaction between TRX and SNR1 (21) also requires the entire SET domain; that is, the same point mutations in conserved amino acids of TRX SET abolish its interaction with both SNR1 and ASH1 in yeast (21). In principle, interactions of TRX SET with ASH1 and/or SNR1 may be mutually exclusive or may involve all three proteins. Currently, we favor the first alternative, because three-hybrid analysis of yeast (21a) indicates that homo-oligomerization of TRX SET may inhibit its interaction with SNR1.
Since genetic experiments suggest that both trx and ash1 are involved in regulation of homeotic gene expression (23), we were particularly interested in determining whether binding of the proteins to polytene chromosomes and/or genetic responsiveness is conferred by the same DNA sequences. To test this, we first analyzed whether both proteins bind in vivo to a well-characterized TRE-PRE-containing bxd regulatory module located 25 kb upstream of the Ubx promoter (4, 5, 27). Indeed, on salivary gland polytene chromosomes, both proteins are found at the site of insertion of a transgene containing this 4-kb bxd subregion (Fig. 3B). This indicates that TRX and ASH1 DNA binding elements may be close to each other. In addition, we have shown that ASH1 is required for full function of the same regulatory region in vivo (Fig. 4). Since this 4-kb region contains three trx-responsive TREs, this leaves open the possibility that TRX and ASH1 may function through the same DNA elements. Experiments aimed at fine mapping of the ash1 response element(s) within this region of Ubx are currently in progress. Nonetheless, our results suggest that TRX and ASH1 may act in concert on one or more bxd TREs. Two interesting possibilities are that both TRX and ASH1 are components of the same protein complex or that they are interacting components of two separate protein complexes that form on closely situated TREs. The physical association between TRX and ASH1 (probably through interaction of their SET domains) is apparently required for TRX binding to chromosomes, since we previously showed that TRX is only weakly associated with chromosomes in ash1 mutant larvae (13). These close physical and functional associations on Ubx regulatory DNA provide a biochemical rationale for the genetic interactions between trx and ash1 mutants.
While genetic data indicate that the trxG and PcG mutations suppress each other’s effects, and it has been shown that in cultured cells Pc protein can prevent TRX-induced transactivation of the Ubx promoter (5), there is little biochemical evidence that the protein products of these groups directly interact in regulation of their target genes. The fine mapping of a TRE-PRE-containing module of the bxd regulatory region, described above (Fig. 3A), identified a TRE closely juxtaposed with, but separable from, two PREs (27). These results argue against earlier models of a direct competition between trxG and PcG complexes for binding sites. However, the close proximity of the elements suggests that protein complexes formed at these sites might be involved in direct physical interactions. Such interactions might allow functional competition between nearby trxG and PcG complexes. Further illumination of this issue can be provided by analyses of direct interactions between trxG and PcG proteins and determination of whether such interactions are essential for their function on particular TREs and PREs. It will therefore be important to identify additional trxG proteins involved in the regulation of particular TREs, so as to obtain a more detailed picture of the interactions required for their function.
ACKNOWLEDGMENTS
We thank J. B. Jaynes for critical comments on the manuscript.
This work was supported by grant CA 50507-05 from the National Cancer Institute and grant GM 53058, as well as grants from DKFZ, the Minerva Foundation, and the Israeli Academy of Sciences.
S. Tillib, S. Smith, and Y. Sedkov contributed equally to this work.
REFERENCES
- 1.Adamson A L, Shearn A. Molecular genetic analysis of Drosophila ash2, a member of the trithorax group required for imaginal disc pattern formation. Genetics. 1996;144:621–633. doi: 10.1093/genetics/144.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bienz M, Müller J. Transcriptional silencing of homeotic genes in Drosophila. BioEssays. 1995;17:775–784. doi: 10.1002/bies.950170907. [DOI] [PubMed] [Google Scholar]
- 3.Breen T R, Harte P J. trithorax regulates multiple homeotic genes in the bithorax and Antennapedia complexes and exerts different tissue-specific, parasegment-specific and promoter-specific effects on each. Development. 1993;117:119–134. doi: 10.1242/dev.117.1.119. [DOI] [PubMed] [Google Scholar]
- 4.Chan C-S, Rastelli L, Pirrotta V A. Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 1994;13:2553–2564. doi: 10.1002/j.1460-2075.1994.tb06545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang Y-L, King B O, O’Connor M, Mazo A, Huang D-H. Functional reconstruction of trans regulation of the Ultrabithorax promoter by the products of two antagonistic genes, trithorax and Polycomb. Mol Cell Biol. 1995;15:6601–6612. doi: 10.1128/mcb.15.12.6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chinwalla V, Jane E P, Harte P J. The Drosophila trithorax protein binds to specific chromosomal sites and is co-localized with Polycomb at many sites. EMBO J. 1995;14:2056–2065. doi: 10.1002/j.1460-2075.1995.tb07197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crosby M A, Miller C, Alon T, Watson K L, Verrijzer C P, Goldman-Levi R, Zak N B. The trithorax group gene moira encodes a Brahma-associated putative chromatin-remodeling factor in Drosophila melanogaster. Mol Cell Biol. 1999;19:1159–1170. doi: 10.1128/mcb.19.2.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui X, De Vivo I, Slany R, Miyamoto A, Firestein R, Cleary M L. Association of SET domain and myotubularin-related proteins modulates growth control. Nat Gen. 1998;18:331–337. doi: 10.1038/ng0498-331. [DOI] [PubMed] [Google Scholar]
- 9.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Karch F. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature. 1994;371:806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- 11.Jones R S, Gelbart W M. The Drosophila Polycomb-group gene Enhancer of zeste contains a region with sequence similarity to trithorax. Mol Cell Biol. 1993;13:6357–6366. doi: 10.1128/mcb.13.10.6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kennison J A. The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu Rev Genet. 1995;29:289–303. doi: 10.1146/annurev.ge.29.120195.001445. [DOI] [PubMed] [Google Scholar]
- 13.Kuzin B, Tillib S, Sedkov Y, Mizrokhi L, Mazo A. The Drosophila trithorax gene encodes a chromosomal protein and directly regulates the region specific homeotic gene fork head. Genes Dev. 1994;8:2478–2490. doi: 10.1101/gad.8.20.2478. [DOI] [PubMed] [Google Scholar]
- 14.LaJeunesse D, Shearn A. Trans-regulation of thoracic homeotic selector genes of the Antennapedia and bithorax complexes by the trithorax group genes: absent, small, and homeotic discs 1 and 2. Mech Dev. 1995;53:123–139. doi: 10.1016/0925-4773(95)00430-0. [DOI] [PubMed] [Google Scholar]
- 15.Mazo A M, Huang D H, Mozer B A, Dawid I B. The trithorax gene, a trans-acting regulator of the bithorax complex in Drosophila, encodes a protein with zinc-binding domains. Proc Natl Acad Sci USA. 1990;87:2112–2116. doi: 10.1073/pnas.87.6.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papoulas O, Beek S J, Moseley S L, McCallum C M, Sarte M, Shearn A, Tamkun J W. The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development. 1998;125:3955–3966. doi: 10.1242/dev.125.20.3955. [DOI] [PubMed] [Google Scholar]
- 17.Peterson C L, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 18.Pirrotta V. PcG complexes and chromatin silencing. Curr Opin Genet Dev. 1997;7:249–258. doi: 10.1016/s0959-437x(97)80135-9. [DOI] [PubMed] [Google Scholar]
- 19.Rastelli L, Chan C S, Pirotta V. Related chromosome binding sites for zeste, suppressors of zeste and Polycomb group proteins in Drosophila and their dependence on Enhancers of zeste function. EMBO J. 1993;12:1513–1522. doi: 10.1002/j.1460-2075.1993.tb05795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rozenblatt-Rosen O, Rozovskaia T, Buracov D, Sedkov Y, Tillib S, Blechman J, Croce C, Mazo A, Canaani E. The C-terminal SET domains of ALL-1 and Trithorax interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc Natl Acad Sci USA. 1998;95:4152–4157. doi: 10.1073/pnas.95.8.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rozovskaia, T. Unpublished observations.
- 21a.Rozovskaia, T. Unpublished observations.
- 21b.Rozovskaia, T., et al. Unpublished data.
- 22.Sedkov Y, Tillib S, Mizrokhi L, Mazo A. The bithorax complex is regulated by trithorax earlier during Drosophila embryogenesis than is the Antennapedia complex, correlating with a bithorax-like expression pattern of distinct early trithorax transcripts. Development. 1994;120:1907–1917. doi: 10.1242/dev.120.7.1907. [DOI] [PubMed] [Google Scholar]
- 22a.Sedkov, Y., and T. Nakamura. Unpublished observations.
- 23.Shearn A. The ash-1, ash-2 and trithorax genes of Drosophila melanogaster are functionally related. Genetics. 1989;121:517–525. doi: 10.1093/genetics/121.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simon J. Locking in stable states of gene expression: transcriptional control during Drosophila development. Curr Opin Cell Biol. 1995;7:376–385. doi: 10.1016/0955-0674(95)80093-x. [DOI] [PubMed] [Google Scholar]
- 25.Soeller W C, Poole S J, Kornberg T. In vitro transcription of the Drosophila engrailed gene. Genes Dev. 1988;2:68–81. doi: 10.1101/gad.2.1.68. [DOI] [PubMed] [Google Scholar]
- 26.Spradling A C. P element-mediated transformation. In: Roberts D B, editor. Drosophila, a practical approach. Washington, D.C: IRL Press; 1986. [Google Scholar]
- 27.Tillib S, Petruk S, Sedkov Y, Kuzin A, Fujioka M, Goto T, Mazo A. Trithorax- and Polycomb-group response elements within an Ultrabithorax transcription maintenance unit consist of closely situated but separable sequences. Mol Cell Biol. 1999;19:5189–5202. doi: 10.1128/mcb.19.7.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tripoulas N, LaJeunesse D, Gildea J, Shearn A. The Drosophila ash1 gene product, which is localized at specific sites on chromosomes, contains a SET domain and a PHD finger. Genetics. 1996;143:913–928. doi: 10.1093/genetics/143.2.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tschiersch B, Hofmann A, Krauss V, Dorn R, Korge G, Reuter G. The protein encoded by the Drosophila position-effect variegation gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 1994;13:3822–3831. doi: 10.1002/j.1460-2075.1994.tb06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsukiyama T, Daniel C, Tamkun J, Wu C. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 KDa subunit of the nucleosome remodeling factor. Cell. 1995;83:1021–1026. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- 31.Tsukiyama T, Wu C. Chromatin remodeling and transcription. Curr Opin Genet Dev. 1997;7:182–191. doi: 10.1016/s0959-437x(97)80127-x. [DOI] [PubMed] [Google Scholar]
- 32.Wang W, Côte J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kalpana G V, Gaff S P, Yaniv M, Workman J L, Crabtree G R. Purification to biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5380. [PMC free article] [PubMed] [Google Scholar]