Highlight
-
•
This study conducted a battery of bioinformation analysis to calculate novel prognostic markers based on metabolism-related DEGs and verified the new prognostic index on the basis of the prognostic models.
-
•
We use a series of algorithms and analyses to develop prognostic index based on MRGs analysis in Ewing's sarcoma, and ultimately identified 74 differentially expressed MRGs and 25 differentially expressed MRGs affecting prognosis.
-
•
We found the 18-MRGs markers can help judge the prognosis of Ewing's sarcoma clinically, thereby helping to individualize the treatment of patients in different groups, and provide a solid foundation for the treatment and prognosis of Ewing's sarcoma.
Keywords: Metabolism-related genes, Ewing's sarcoma, Prognosis, Bioinformatics
Abstract
Objective
By combining the expression profiles of metabolism-related genes (MRGS) with clinical information, the expression quantities of MRGS and the influence on development and prognosis were systematically analyzed, so as to provide a theoretical basis for the clinical study on the prognosis of Ewing's sarcoma.
Methods
MRGs expression profiles of 64 patients with Ewing's sarcoma were obtained from GEO dataset. Univariate Cox regression analysis was used to identify metabolization-related differentially expressed genes (DEGs) related with prognosis in Ewing's sarcoma patients. Then, multivariate Cox analysis was used to calculate novel prognostic markers based on metabolism-related DEGs. Besides, We validate the model using ICGC datasets. Finally, the new prognostic index was verified on the basis of the prognostic models.
Results
Multivariate Cox regression analysis identified 74 metabolization-related DEGs, 25 of which were associated with Ewing's sarcoma patients' overall survival. Subsequently, we used 25 DEGs to construct metabolism-related prognostic signature for patients with Ewing's sarcoma. Based on the 18 DEGs regression coefficient, we propose the formula of each patient's risk score, and then divided the patients into high-risk group and low-risk group. The results indicated that the survival rate and survival time were higher in the low-risk group and lower in the high-risk group. Multivariate Cox analysis showed that risk score index was an independent prognostic factor for Ewing's sarcoma.
Conclusion
The experimental results suggest that the 18 metabolism-related DEGs marker may be effective in predicting the prognosis of Ewing's sarcoma to some extent, helping to individualize treatment of patients at different risks.
Introduction
The incidence rate of Ewing's sarcoma is extremely high, ranking second in cancer, and is fairly common in children and adolescents with bone tumors [1]. This is a highly malignant tumor of unknown origin, with no clear etiology and pathogenesis at present [2]. Among childhood cancers, the incidence of Ewing's sarcoma is 1.4% [3]. Since scientists first proposed the concept and nature of Ewing's sarcoma in the 1920s, its clinical treatment has made great progress. According to the common treatment of cancer, the current treatment methods for Ewing's sarcoma mainly include surgery, chemotherapy and radiotherapy, etc. After long-term exploration and efforts, although the prognosis of patients with localized tumors is good, the survival rate of patients with tumors with strong metastatic ability is very low [4]. Clinically, there are many factors used to evaluate the prognosis of Ewing's sarcoma, including tumor stage, classification and subtype, etc. Although these clinicopathological features can evaluate tumor prognosis to a certain extent, they cannot provide accurate and detailed prognostic information. Therefore, this may affect the judgment of clinical prognosis of patients. Some high-risk patients may face tumor metastasis and spread due to untimely or inadequate treatment. At the same time, those low-risk patients may damage certain functions of the body due to excessive treatment. This will greatly affect the treatment and rehabilitation of patients. Then, we desperately need to find some innovative molecular markers related to the development and prognosis of Ewing's sarcoma to predict the prognosis accurately and effectively, so as to treat patients favorably and improve and perfect the prognosis of patients.
Table 1.
Multivariate Cox regression analysis of identified metabolism-related DEGs.
| Coef | HR | HR.95L | HR.95H | P value | |
|---|---|---|---|---|---|
| ACSL1 | 17.55421 | 42,043,297 | 176,623.2 | 1E+10 | 3.24E-10 |
| ACADM | −51.0836 | 6.53E-23 | 1.97E-30 | 2.16E-15 | 7.35E-09 |
| NME4 | −9.4952 | 7.52E-05 | 7.49E-10 | 7.551533 | 0.106116 |
| PSAT1 | 8.027627 | 3064.461 | 1.377943 | 6,815,178 | 0.041202 |
| PAFAH1B3 | 22.68579 | 7.12E+09 | 2,368,232 | 2.14E+13 | 2.82E-08 |
| UAP1 | −66.2224 | 1.74E-29 | 8.46E-39 | 3.57E-20 | 1.42E-09 |
| ALDH18A1 | 9.21071 | 10,003.7 | 0.100493 | 9.96E+08 | 0.116729 |
| IMPDH2 | 160.2667 | 4.01E+69 | 4.89E+45 | 3.28E+93 | 1.17E-08 |
| CYP26B1 | −12.8493 | 2.63E-06 | 1.91E-08 | 0.000362 | 3.16E-07 |
| ENPP1 | 13.89245 | 1,079,974 | 5342.295 | 2.18E+08 | 2.92E-07 |
| TYMS | −7.26825 | 0.000697 | 2.89E-06 | 0.167972 | 0.00939 |
| NME1 | −76.8606 | 4.17E-34 | 4.19E-47 | 4.15E-21 | 4.82E-07 |
| LPCAT1 | 52.36241 | 5.50E+22 | 2.06E+14 | 1.47E+31 | 1.22E-07 |
| ALDH7A1 | −40.7763 | 1.95E-18 | 3.79E-25 | 1.01E-11 | 2.33E-07 |
| DNMT1 | −18.5259 | 9.00E-09 | 7.04E-16 | 0.115043 | 0.026488 |
| MIF | −20.4121 | 1.37E-09 | 5.69E-15 | 0.000327 | 0.00124 |
| RRM2 | 58.41263 | 2.33E+25 | 2.97E+14 | 1.84E+36 | 5.04E-06 |
| KDSR | −14.8962 | 3.39E-07 | 3.90E-10 | 0.000295 | 1.61E-05 |
Table 2.
Univariate and multivariate Cox regression analysis of prognosis for ES in GEO dataset.
| Term | HR | HR.95L | HR.95H | P value | |
|---|---|---|---|---|---|
| uniCox | gender | 0.790139 | 0.410652 | 1.520314 | 0.480558 |
| age | 0.99665 | 0.950605 | 1.044925 | 0.889417 | |
| PRS type | 1.241552 | 0.827521 | 1.862733 | 0.295882 | |
| Risk | 34.56558 | 11.19088 | 106.7636 | 7.40E-10 | |
| multiCox | gender | 1.69212 | 0.803458 | 3.563684 | 0.166323 |
| age | 1.024969 | 0.974313 | 1.078258 | 0.340251 | |
| PRS type | 0.820787 | 0.513314 | 1.312437 | 0.409566 | |
| Risk | 46.84929 | 14.29588 | 153.5307 | 2.12E-10 |
Table 3.
Univariate and multivariate Cox regression analysis of prognosis for ES in ICGC dataset.
| Term | HR | HR.95L | HR.95H | P value | |
|---|---|---|---|---|---|
| uniCox | gender | 1.257858 | 0.537467 | 2.943821 | 0.596945 |
| age | 0.963 | 0.910988 | 1.017982 | 0.183235 | |
| PRS type | 2.089491 | 0.846148 | 5.15982 | 0.110098 | |
| Risk | 5.930699 | 2.160648 | 16.27901 | 0.00055 | |
| multiCox | gender | 0.967994 | 0.387212 | 2.419895 | 0.944525 |
| age | 0.947195 | 0.876128 | 1.024027 | 0.172788 | |
| PRS type | 1.723881 | 0.594937 | 4.995096 | 0.315732 | |
| Risk | 5.274356 | 1.858176 | 14.97104 | 0.001784 |
At present, studies have found that metabolism-related genes are closely related to the occurrence, development and prognosis of tumors. Cancerous cells undergo a gradual process of adaptation to metabolism, which allows tumor cells to grow and proliferate rapidly, thus supporting the occurrence and development of tumors [5]. That is to say, During the above adaptation process, the tumor will rewrite its own nutritional metabolism program to meet the energy metabolism and biosynthesis of cancerous cells [6]. In addition, scientists have put forward a term "Warburg effect" for the metabolism of tumor cells, that is, even under the conditions of sufficient oxygen supply, the glycolysis process of tumor cells will be stronger than that of the original cells [7]. One study proposed that the occurrence and prognosis of esophageal cancer are closely related to the methylation level of folate metabolism-related genes, and that low serum and cell folic acid levels are factors that promote the occurrence of esophageal cancer [8]. A study found and identified 13 differentially expressed MRGs related to the prognosis of gastric cancer, and established a metabolic model that can be used to judge the prognosis of gastric cancer patients [9].
At present, many studies have confirmed that metabolism is closely related to Ewing's sarcoma. For example, Tadashi Kondo [10] said that through proteomic analysis, they proposed a new mechanism for the occurrence and development of Ewing's sarcoma, that is, the EWS-FLI-1 transition, and pointed out that EWS-FLI-1 has a certain regulatory effect on IL-6 secretion, thereby promoting tumor growth and metastasis. The lack of EWS-FLI-1 can lead to the excessive secretion of IL-6 and other soluble factors, which activate STAT signaling in bystander cells that maintain EWS-FLI-1 expression, and these factors can prevent the spontaneous apoptosis of tumor cells [11]. However, most studies focused on the relationship between one or a few MRGs and the occurrence, development, or prognosis of Ewing's sarcoma. There is a lack of studies using MRGs expression profiles containing dozens or even hundreds of MRGs to screen and identify molecular markers related to the prognosis of Ewing's sarcoma.
In our study, we integrated the MRGS expression profile of patients with Ewing's sarcoma, and combined with clinical data to obtain prognostic-related metabolic genes, so as to construct a prognostic model of MRGs and calculate the patient's risk value. In addition, it was verified by using the sample data of Ewing's sarcoma in the ICGC dataset. So we can systematically analyze the differences in the expression of MRGs and their influence on tumor prognosis. Therefore, the use of metabolic-related DEGs markers can effectively predict the prognosis of Ewing's sarcoma, thereby helping to individualize the treatment of patients in different risk states.
Material and methods
Metabolism-related genes and GEO data acquisition
851 MRGs were obtained from GSEA database (http://www.gsea-msigdb.org/gsea/index.jsp). The expression levels and clinical correlation of MRGs were obtained from the Gene Expression Omnibus (GEO) dataset (https://www.ncbi.nlm.nih.gov/gds) [12] (GSE17679), including 64 Ewing's samples and 18 normal samples [13], and ICGC dataset (https://dcc.icgc.org/), including 43 Ewing's samples.
Identification of differentially expressed genes
Identify differentially expressed genes (DEG) related to metabolism between tumor samples and non-tumor samples according to the following criteria: false discovery rate (FDR) <0.05 and [log2 (fold change)]> 2 [14]. Univariate Cox regression analysis was used to determine the metabolic-related DEG related to patient survival. Correlogram of prognostic MRGs were plotted using “corrgram” package in R (v. 4.0.0), including heat map, volcano map and box map [15].
Functional enrichment
The DAVID database Online Enrichment Tool (https://david.ncifcrf.gov/tools.jsp/) and Metascape online analysis tools (https://metascape.org/gp/index.html#/main/step1) was used for Gene Ontology (GO) and the Kyoto Gene and Genomic Encyclopedia (KEGG) pathway enrichment. After GO and KEGG analysis, we can understand the functions and pathways behind DEGs related to metabolism, that is, the biological significance of MRGs.
Construction of the MPI model
Multivariate Cox regression analysis combined with clinical data was used to identify metabolic genes related to prognosis [16] (https://CRAN.R-project.org/package=survival). The expression of each autophagy gene related to prognosis is weighted by regression coefficient, and then combined. After that, a risk score formula was established for each patient based on the combined results, and the patient's risk value was calculated. According to the risk score formula and the risk value, with the median value as the demarcation point, all patients were divided into high-risk groups and low-risk groups. The dataset is then validated for model validation
Statistical analysis
The Kaplan-Meier survival curve was utilized to analyze and evaluate the survival difference between the two groups, and any differences in survival were evaluated with a log rank test. The risk curve was used to compare the patient's risk score and survival time. ROC curve was used to test the accuracy of model prediction. Conduct univariate and multivariate independent prognostic analysis and clinical correlation analysis, including the influence of age and gender on prognosis [16] (https://CRAN.R-project.org/package=survival). All statistical tests were bilateral, and P value<0.05 was regarded statistically significant.
Results
Identification of differentially expressed MRGs
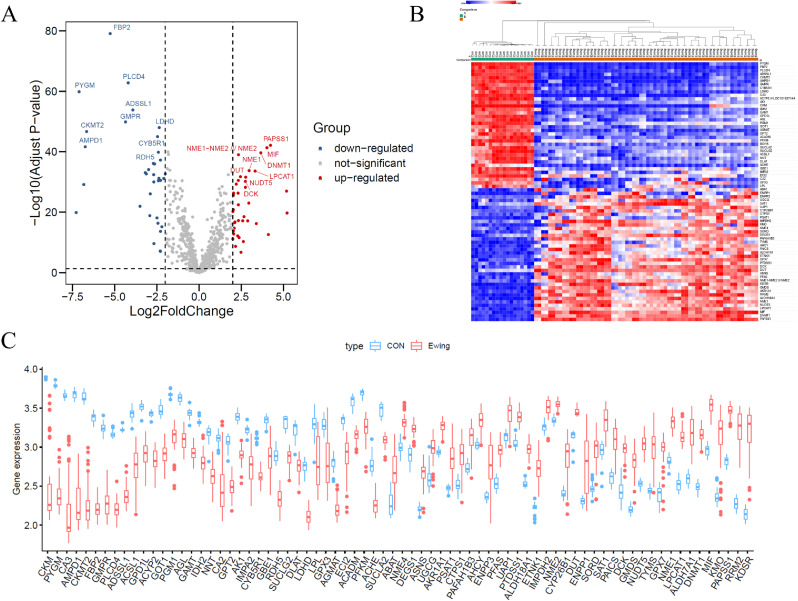
We finally obtained 74 differentially expressed MRGs, including 38 up-regulated and 36 down-regulated MRGs (Fig. 1A and 1B). In addition, a boxplot was visualized to show the expression pattern of 74 differentially expressed MRGs between Ewing's sarcoma and non-tumor tissue (Fig. 1C).
Fig. 1.
Metabolism-related differentially expressed genes (DEGs). Volcanic map (A) and Heat map (B) showed DEGs between tumor samples and non-tumor samples. Red dots represented significantly up-regulated genes, blue dots represented significantly down-regulated genes, while grey dots indicated no differences. (C)Expression patterns of metabolism-related DEGs in tumor and non-tumor samples. The red block represented tumor samples and the blue block represented non-tumor samples. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
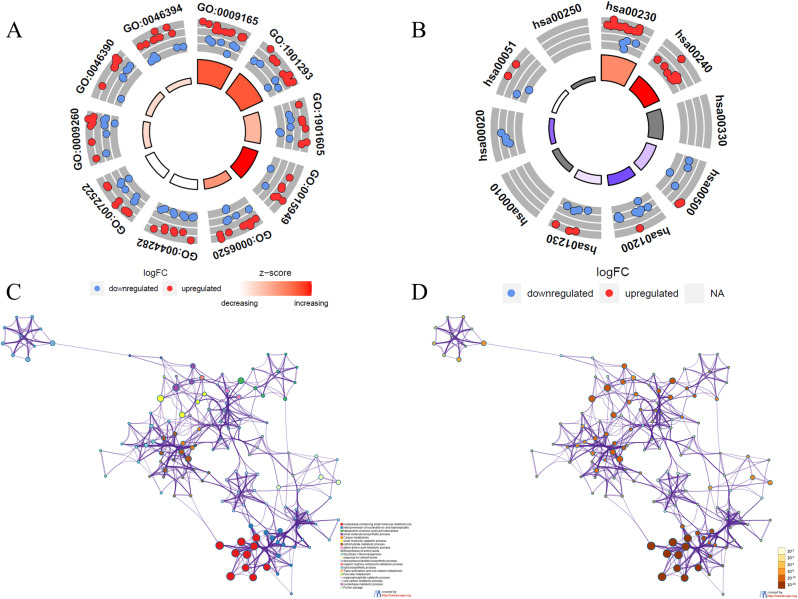
Functional enrichment of the differentially expressed MRGs
Functional enrichment analysis of 74 differentially expressed MRGs provides a biological understanding of these genes. GO enrichment shows that differential genes are mainly involve in nucleotide biosynthetic process, nucleotide phosphate biosynthetic process and small molecule catabolic process (Fig. 2A and Supplementary Figure 1A). KEGG enrichment shows that pathways of differentially expressed MRGs mainly involve pathways in purine metabolism, arginine and proline metabolism, starch and sucrose metabolism and glycolysis and gluconeogenesis (Fig. 2B and Supplementary Figure 1B). Metascape results show that pathways of differential genes are mainly include in small molecule biosynthetic process, deoxyribonucleotide biosynthetic process and biosynthesis of amino acids (Fig. 2C, D and Supplementary Figure 1C).
Fig. 2.
The circle diagram showed the GO functional enrichment of metabolism-related DEGs (A), and KEGG pathway enrichment of metabolism-related DEGs(B). (C) colored by cluster ID, where nodes that share the same cluster ID are typically close to each other in DEGs. (D) colored by p-value, where terms containing more genes tend to have a more significant P-value in DEGs.
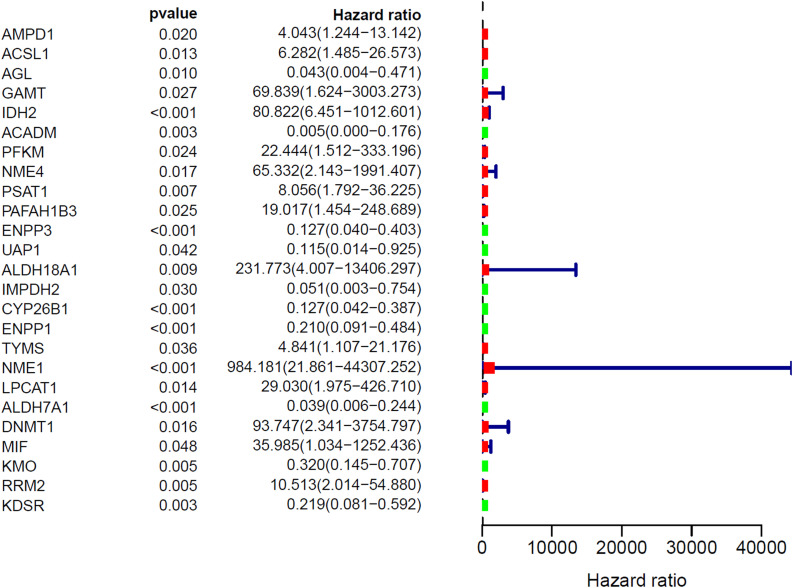
Identification of prognostic MRGs
The forest map of hazard ratios showed the hazard ratios of 25 MRGs (Fig. 3). In the forest map of hazard ratios, green indicates that the genes are positively associated with favorable prognosis in Ewing's sarcoma, including 10 genes (AGL, ACADM, ENPP3, UAP1, IMPDH2, CYP26B1, ENPP1, ALDH7A1, KMO, KDSR), and red indicates that the genes are positively associated with poor prognosis in Ewing's sarcoma, including 15 genes (AMPD1, ACSL1, GAMT, IDH2, PFKM, NME4, PSAT1, PAFAH1B3, ALDH18A1, TYMS, NME1, LPCAT1, DNMT1, MIF, RRM2). Correlogram showed prognostic MRGs intercorrelations (Supplementary Fig. 2). Among them, ENPP1 is strongly positively correlated with NME4, TYMS, NME1 and RRM2. ALDH18A1 is strongly positively correlated with ENPP1, CYP26B1, KDSR, IMPDH2 and UAP1.
Fig. 3.
The Hazard ratio forest map showed the significant prognostic value of the genes.
Construction of metabolic prognostic index
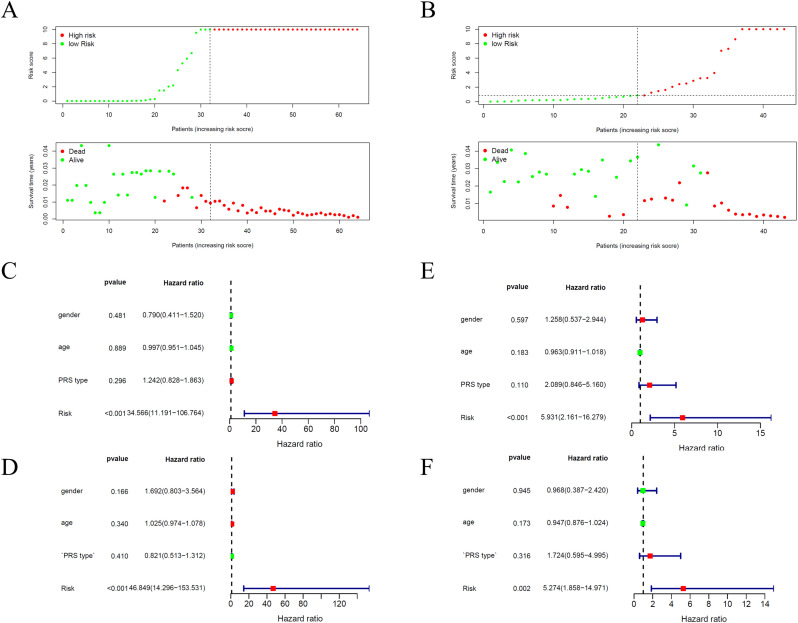
We pooled prognostic MRGs for multivariate Cox regression analysis and constructed Metabolic prognostic index (MPI). Patients were divided into two groups with risk score, which could be calculated based on the MPI. Fig. 4A showed distribution of risk score, survival status and survival rate of patients of the two groups.
Fig. 4.
Survival status of patients in the high Risk Score and low Risk Score groups and Expression spectrum of identified genes in high Risk Score and low Risk Score groups for GEO dataset(A) and ICGC dataset(B). (C)The forest map of univariate Cox regression analysis and (D) multivariate Cox regression analysis in ES patients for GEO dataset. (E)The forest map of univariate Cox regression analysis and (F) multivariate Cox regression analysis in ES patients for ICGC dataset.
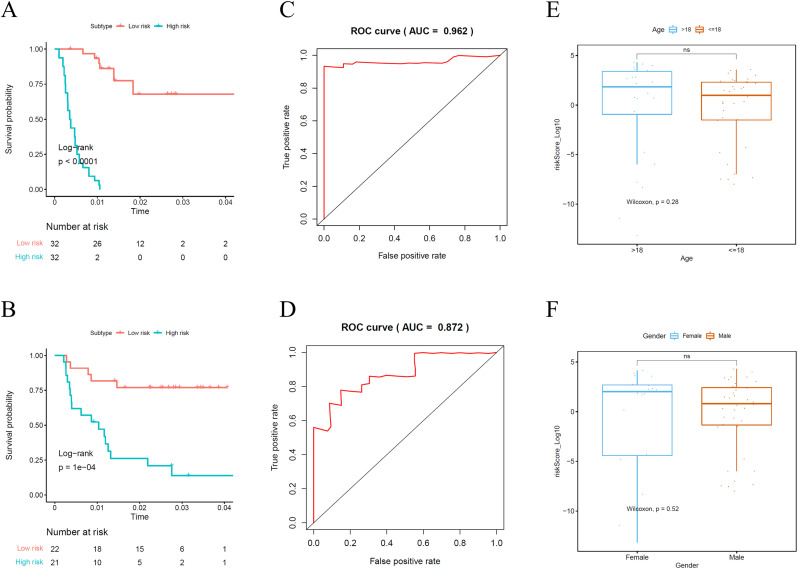
In order to determine the role of the MPI in predicting clinical outcomes in Ewing's sarcoma patients, K-M survival curves were plotted to analyze different survival times between high-risk and low-risk groups. K-M analysis showed that the survival rate of patients in the high-risk group was significantly lower than that in the low-risk group (Fig. 5A). Univariate analysis showed that MPI was significantly associated with patient prognosis (Fig. 4C). In addition, after adjusting for clinicopathological features such as gender, age and PRS type, MPI remained an independent prognostic indicator for Ewing's sarcoma patients in multivariate analysis (Fig. 4D). The area under the curve of the corresponding receiver operating characteristic (ROC) curve for survival is 0.962 (Fig. 5C). This indicated that the prognostic index based on MRGs has a certain role and potential in the prediction of prognosis.
Fig. 5.
Kaplan-Meier survival curve analysis of survival differences between high RiskScore and low RiskScore groups for GEO dataset(A) and ICGC dataset(B). Receiver operating characteristic (ROC) curve for GEO dataset(C) and ICGC dataset(D) validated the prognostic significance of the established prognostic model. (E) (F) Clinicopathological significance of the prognostic index of ES.
In addition, this prognostic model was validated by ICGC data. Fig. 4B showed distribution of risk score, survival status and survival rate of patients of the two groups. Fig. 5B shows the K-M survival curve, which shows that the survival rate of patients in the high-risk group is low. Univariate and multivariate analysis also indicated that MPI was obvious correlated with prognosis (Fig. 4E and F). The area under the curve of the corresponding receiver operating characteristic (ROC) curve for survival is 0.872 (Fig. 5D). This validation more fully demonstrates that the prognostic model is not stochastic.
Clinical correlation analysis and differential expression of MRGs
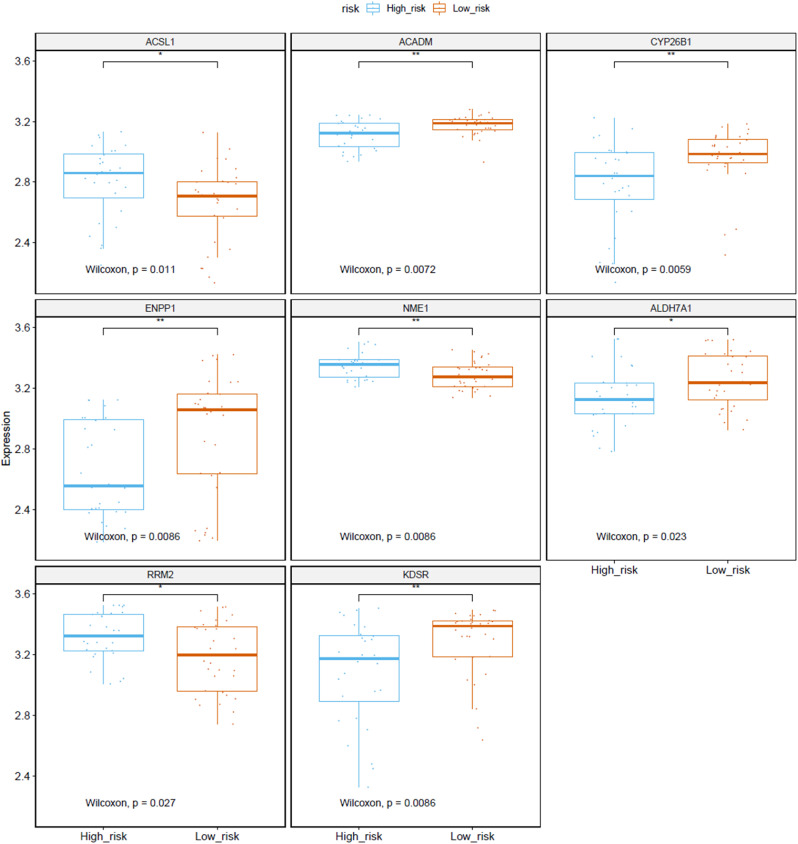
Clinical correlation analyses compared risk scores for different ages and genders. The results showed that the risk score of ≤ 18 age group was lower than that of >18 age group (P > 0.05) (Fig. 5E). And female risk scores were relatively higher than male risk scores (P > 0.05) (Fig. 5F) Fig. 6 show that the expression levels of ACSL1, ACADM, CYP26B1, ENPP1, NME1, ALDH7A1, RRM2 and KDSR in high-risk and low-risk groups are obvious and significant difference.
Fig. 6.
Genes of Metabolic prognostic index expressed in high Risk Score and low Risk Score groups. (statistical significance).
Discussion
Ewing's sarcoma is a highly aggressive and metastatic tumor with a high incidence in children and young adults. The occurrence of Ewing's sarcoma requires multiple steps, which are related to the genetic and epigenetic changes of intracellular proto-oncogenes and tumor suppressor genes [17]. There are age and gender differences in Ewing's sarcoma. It mainly occurs in men aged 5–25, and 80% of them occur in men under 20 years of age [18]. The current treatment of Ewing's sarcoma mainly includes chemotherapy, radiotherapy and surgical resection to control its occurrence [19]. Although a large number of studies have shown that metabolism is involved in the occurrence and development of Ewing's sarcoma, MRGS has not been comprehensively analyzed to explore its clinical application and significance.
To metabolically analyze Ewing's sarcoma prognosis-related genes, we screened and identified 76 differentially expressed MRGs and 25 prognostic MRGs. Our results suggested that a prognostic model based on 18 MRGs can be used to classify and stratify prognosis in patients with Ewing's sarcoma, thus facilitating the individualization of treatment plans based on patient risk. According to GO and KEGG analysis, we discovered that these MRGs influenced the prognosis of Ewing's sarcoma mainly through the following functions and pathways, including small molecule catabolic process, nucleotide phosphate biosynthetic process, arginine and proline metabolism and glycolysis and gluconeogensis. The role of the regulation of small-molecule metabolic processes, amino acid, proteasome and ribosome biosynthesis in eukaryotes in Ewing's sarcoma development and prognosis has been identified and validated by studies [20]. Cancer metabolism is characterized by high consumption of glucose and its incomplete breakdown to lactate. Pentose phosphate pathway (PPP) plays an important role in cancer metabolic reprogramming, mainly through glucose-6-phosphate dehydrogenase (G6PD) [21].
We screened and obtained valuable MRGs that can predict the prognosis of Ewing's sarcoma. Many previous studies have confirmed that some of these MRGs are closely related to the prognosis of Ewing's sarcoma or other malignant tumors. Fatty acid metabolism plays an important role in the occurrence and development of cancer, but it needs an initial step known as fatty acid activation. Long-chain acyl-CoA synthetases (ACSLs), which are responsible for activation of the most abundant long-chain fatty acids, are commonly deregulated in cancer. Among the five family isoforms, ACSL1 and ACSL4 are able to promote ungoverned cell growth, facilitate tumor invasion and evade programmed cell death [22]. ACADM can promote the epithelial-mesenchymal transition process of breast cancer cells and improve the migration and invasion ability. ACADM is an oncogene in breast cancer, so it is speculated that it may also play a role in Ewing's sarcoma [23]. OX40L transgenic Ewing sarcoma cells showed retained the expression of certain Ewing sarcoma-associated (anti)gens including lipase member I, CCND1 and CYP26B1. OX40L-expressing tumor cells showed a trend for enhanced immune stimulation against Ewing sarcoma cells. This suggests that CYP26B1 takes part in Ewing's sarcoma [24]. ENPP1 has been found to be related to the stemness of tumor cells in cancers such as breast cancer and glioblastoma, and is involved in tumor occurrence, nausea growth and metastasis. Therefore, ENPP1 can also be used as a biomarker for the diagnosis of malignant tumors and used to predict the prognosis of patients [25]. NME1 is a tumor metastasis suppressor gene, mapped to chromosomes 17q21.3 [26]. Studies have shown that EcoR1 (rs34214448) polymorphism of NME1 gene revealed significant association with increased risk of breast cancer development [27]. Therefore, it can be speculated that NME1 is related to Ewing's sarcoma. More and more evidence showed that ALDH7A1, which is one of the superfamily members of ALDH, can degrade and detoxify acetaldehyde produced by cell metabolism, and thus affect the occurrence and prognosis of many cancers [28]. Targeting the ATR, CHK1, and WEE1 kinases in Ewing sarcoma cells activates CDK2 and reduce the ability of DNA replication by promoting the proteasome-mediated degradation of RRM2 [29]. In addition, the translation inhibitor 4E-BP1 can regulate the expression level of RRM2, thereby reducing its expression, inhibiting the occurrence and development of tumors, and improving the prognosis of Ewing's sarcoma [30]. Both indicated an association between RRM2 and Ewing's sarcoma. Alterations of sphingolipid metabolism and associated signaling pathways are a potential pathway for cancer development. In particular, ceramides are involved in the regulation of cellular proliferation, differentiation, senescence, and apoptosis and are known to function as important regulators of malignant transformation as well as tumor progression [31]. KDSR is the key enzyme in the de novo sphingolipid synthesis [32], so it may have an important effect on Ewing's sarcoma.
The above research still has limitations to a certain extent. First of all, our research is retrospective and needs to use existing data to conduct experiments, so statistical bias is unavoidable. Besides, there are few samples of Ewing's sarcoma in various databases. This study uses two datasets of GEO and ICGC, but the number of samples is still small. Therefore, the prognostic model has some limitations, and the potential mechanism of MRG's influence on prognosis needs further exploration and reveal.
In summary, this study comprehensively analyzed the expression profile and clinical data of MRGs, and determined the prognostic-related MRGs of Ewing's sarcoma. The metabolism-related DEGs marker may be effective in predicting the prognosis of Ewing's sarcoma to some extent, and helping to individualize treatment of patients at different risks.
Conclusion
In this study, we used a series of algorithms and analyses to develop prognostic index based on MRGs analysis in Ewing's sarcoma, and ultimately identified 74 differentially expressed MRGs and 25 differentially expressed MRGs affecting prognosis. The 18-MRGs markers can help judge the prognosis of Ewing's sarcoma clinically, thereby helping to individualize the treatment of patients in different groups, and provide a solid foundation for the treatment and prognosis of Ewing's sarcoma.
Declaration of Competing Interest
These is no conflict of interests.
Acknowledgments
Author contribution
Zhaoyu Fu and Bo Wu designed the study and collected data. Bo Yu drafted the manuscript. Yuanyuan Hou, Hongyu Wang and Yuting Jiang contributed to the writing. Zhaoyu Fu, Mingxi Liu and Dong Zhu provided critical feedback and contributed to the review of the manuscript. All authors contributed to the article approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (Grant no. 12072129).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101225.
Contributor Information
Zhaoyu Fu, Email: fuzy19@mails.jlu.edu.cn.
Bo Yu, Email: yubo7018@jlu.edu.cn.
Mingxi Liu, Email: liumingxi@jlu.edu.cn.
Bo Wu, Email: wubo20@mails.jlu.edu.cn.
Yuanyuan Hou, Email: houyy7018@mails.jlu.edu.cn.
Hongyu Wang, Email: wanghy7018@mails.jlu.edu.cn.
Yuting Jiang, Email: jiangyt7018@jlu.edu.cn.
Dong Zhu, Email: zhu_dong@mail.jlu.edu.cn.
Appendix. Supplementary materials
References
- 1.Balamuth N.J., Womer R.B. Ewing's sarcoma. Lancet Oncol. 2010;11(2):184–192. doi: 10.1016/S1470-2045(09)70286-4. PMID: 20152770. [DOI] [PubMed] [Google Scholar]
- 2.Eggli K.D., Quiogue T., Moser R.P., Jr. Ewing’s sarcoma. Radiol. Clin. N. Am. 1993;31(2):325–337. PMID:8446752. [PubMed] [Google Scholar]
- 3.Ottaviani G., Jaffe N. The epidemiology of osteosarcoma. Cancer Treat. Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. PMID: 20213383. [DOI] [PubMed] [Google Scholar]
- 4.Karosas A.O. Ewing's sarcoma. Am. J. Health Syst. Pharm. 2010;67(19):1599–1605. doi: 10.2146/ajhp090526. PMID: 20852160. [DOI] [PubMed] [Google Scholar]
- 5.Vander Heiden M.G., DeBerardinis R.J. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168(4):657–669. doi: 10.1016/j.cell.2016.12.039. PMID:28187287PMCID: PMC5329766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeBerardinis R.J., Chandel N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016;2(5) doi: 10.1126/sciadv.1600200. PMID:27386546PMCID: PMC4928883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z., Zhang H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell. Mol. Life Sci. 2016;73(2):377–392. doi: 10.1007/s00018-015-2070-4. Epub 2015 Oct 23PMID: 26499846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong Y., Yi X., Yujie Z., Huixia Z., Yan C. Relationship between the methylation of folic acid metabolism-related genes and the incidence and prognosis of esophageal cancer among ethnic Kazakhs. J. Cancer. 2018;9(16):2865–2875. doi: 10.7150/jca.25270. PMID: 30123355PMCID: PMC6096358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wen F., Huang J., Lu X., Huang W., Wang Y., Bai Y., Ruan S., Gu S., Chen X., Shu P. Identification and prognostic value of metabolism-related genes in gastric cancer. Aging. 2020;12(17):17647–17661. doi: 10.18632/aging.103838. Epub ahead of printPMID:32920549PMCID: PMC7521523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kondo T. Current status of proteomics in Ewing's sarcoma. Proteomics Clin Appl. 2019;13(3) doi: 10.1002/prca.201700130. Epub 2018 Aug 23PMID: 29992772. [DOI] [PubMed] [Google Scholar]
- 11.Anderson J.L., Titz B., Akiyama R., Komisopoulou E., Park A., Tap W.D., Graeber T.G., Denny C.T. Phosphoproteomic profiling reveals IL6-mediated paracrine signaling within the Ewing sarcoma family of tumors. Mol. Cancer Res. 2014;12(12):1740–1754. doi: 10.1158/1541-7786.MCR-14-0159. Epub 2014 Aug 4PMID: 25092916. [DOI] [PubMed] [Google Scholar]
- 12.Savola S., Klami A., Myllykangas S., Manara C., Scotlandi K., Picci P., Knuutila S., Vakkila J. High Expression of Complement Component 5 (C5) at tumor site associates with superior survival in Ewing’s sarcoma family of tumour patients. ISRN Oncol. 2011;2011 doi: 10.5402/2011/168712. Epub 2011 Oct 2PMID:22084725PMCID: PMC3196920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Savola S., Klami A., Myllykangas S., Manara C. High expression of complement component 5 (C5) at tumor site associates with superior survival in Ewing’s sarcoma family of tumour patients. ISRN Oncol. 2011;2011 doi: 10.5402/2011/168712. PMID:22084725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. Epub 2015 Jan 20PMID:25605792PMCID: PMC4402510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wright K. Corrgram: plot a correlogram. 2017.
- 16.Therneau T. 2021. A Package for Survival Analysis in R. R Package Version 3.2-9. [Google Scholar]
- 17.Li Z., Yu X., Shen J., Wu W.K., Chan M.T. MicroRNA expression and its clinical implications in Ewing's sarcoma. Cell Prolif. 2015;48(1):1–6. doi: 10.1111/cpr.12160. Epub 2014 Dec 22PMID: 25530497PMCID: PMC6496123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Astekar M., Saxena S., Murari A., Manjunatha B.S. Ewing’s sarcoma in maxilla. BMJ Case Rep. 2019;12(2) doi: 10.1136/bcr-2018-227819. PMID:30796079PMCID: PMC6388892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore D.D., Haydon R.C. Ewing's sarcoma of bone. Cancer Treat. Res. 2014;162:93–115. doi: 10.1007/978-3-319-07323-1_5. PMID: 25070232. [DOI] [PubMed] [Google Scholar]
- 20.Deng Y., Xie Q., Zhang G., Li S., Wu Z., Ma Z., He X., Gao Y., Wang Y., Kang X., Wang J. Slow skeletal muscle troponin T, titin and myosin light chain 3 are candidate prognostic biomarkers for Ewing's sarcoma. Oncol. Lett. 2019;18(6):6431–6442. doi: 10.3892/ol.2019.11044. Epub 2019 Nov 4PMID: 31807166PMCID: PMC6876326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cossu V., Bonanomi M., Bauckneht M., Ravera S., Righi N., Miceli A., Morbelli S., Orengo A.M., Piccioli P., Bruno S., Gaglio D., Sambuceti G., Marini C. Two high-rate pentose-phosphate pathways in cancer cells. Sci. Rep. 2020;10(1):22111. doi: 10.1038/s41598-020-79185-2. PMID:33335166PMCID: PMC7746718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang Y., Zhou J., Hooi S.C., Jiang Y.M., Lu G.D. Fatty acid activation in carcinogenesis and cancer development: essential roles of long-chain acyl-CoA synthetases. Oncol. Lett. 2018;16(2):1390–1396. doi: 10.3892/ol.2018.8843. Epub 2018 May 30PMID:30008815PMCID: PMC6036470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu Y., Zhao L., Li R. [Medium-chain acyl-CoA dehydrogenase enhances invasion and metastasis ability of breast cancer cells] Nan Fang Yi Ke Da Xue Xue Bao. 2019;39(6):650–656. doi: 10.12122/j.issn.1673-4254.2019.06.04. ChinesePMID: 31270042PMCID: PMC6743927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reuter D., Staege M.S., Kühnöl C.D., Föll J. Immunostimulation by OX40 ligand transgenic ewing sarcoma cells. Front. Oncol. 2015;5:242. doi: 10.3389/fonc.2015.00242. PMID: 26579494PMCID: PMC4621427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawaguchi M., Han X., Hisada T., Nishikawa S., Kano K., Ieda N., Aoki J., Toyama T., Nakagawa H. Development of an ENPP1 fluorescence probe for inhibitor screening, cellular imaging, and prognostic assessment of malignant breast cancer. J. Med. Chem. 2019;62(20):9254–9269. doi: 10.1021/acs.jmedchem.9b01213. Epub 2019 Oct 3PMID: 31536342. [DOI] [PubMed] [Google Scholar]
- 26.Antar S., Mokhtar N., Abd Elghaffar M.A., Seleem A.K. Association of polymorphisms in metastasis suppressor genes NME1 and KISS1 with breast cancer development and metastasis. J. Egypt. Natl. Cancer Inst. 2020;32(1):24. doi: 10.1186/s43046-020-00037-1. PMID: 32462300. [DOI] [PubMed] [Google Scholar]
- 27.Rajagopal T., Seshachalam A., Akshaya R.L., Rathnam K.K., Talluri S., Jothi A., Dunna N.R. Association of HOTAIR (rs920778 and rs1899663) and NME1 (rs16949649 and rs2302254) gene polymorphisms with breast cancer risk in India. Gene. 2020;762 doi: 10.1016/j.gene.2020.145033. Epub 2020 Aug 8PMID:32781191. [DOI] [PubMed] [Google Scholar]
- 28.Wang H., Tong L., Wei J., Pan W., Li L., Ge Y., Zhou L., Yuan Q., Zhou C., Yang M. The ALDH7A1 genetic polymorphisms contribute to development of esophageal squamous cell carcinoma. Tumour Biol. 2014;35(12):12665–12670. doi: 10.1007/s13277-014-2590-9. Epub 2014 Sep 12PMID: 25213698. [DOI] [PubMed] [Google Scholar]
- 29.Koppenhafer S.L., Goss K.L., Terry W.W., Gordon D.J. Inhibition of the ATR-CHK1 pathway in ewing sarcoma cells causes DNA damage and apoptosis via the CDK2-mediated degradation of RRM2. Mol. Cancer Res. 2020;18(1):91–104. doi: 10.1158/1541-7786.MCR-19-0585. Epub 2019 Oct 24PMID: 31649026PMCID: PMC6942212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goss K.L., Koppenhafer S.L., Waters T., Terry W.W., Wen K.K., Wu M., Ostergaard J., Gordon P.M., Gordon D.J. The translational repressor 4E-BP1 regulates RRM2 levels and functions as a tumor suppressor in Ewing sarcoma tumors. Oncogene. 2021;40(3):564–577. doi: 10.1038/s41388-020-01552-0. Epub 2020 Nov 15PMID:33191406PMCID: PMC7856031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barron K.A., Jeffries K.A., Krupenko N.I. Sphingolipids and the link between alcohol and cancer. Chem. Biol. Interact. 2020;322 doi: 10.1016/j.cbi.2020.109058. Epub 2020 Mar 11PMID: 32171848PMCID: PMC8371698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park K.H., Ye Z.W., Zhang J., Hammad S.M., Townsend D.M., Rockey D.C., Kim S.H. Author correction: 3-ketodihydrosphingosine reductase mutation induces steatosis and hepatic injury in zebrafish. Sci. Rep. 2020;10(1):11971. doi: 10.1038/s41598-020-67912-8. Erratum for: Sci Rep. 2019 Feb 4;9(1):1138PMID: 32665573PMCID: PMC7360750. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.