Abstract
Nutrients, drugs, and hormones influence transcription during differentiation and metabolism by binding to high-affinity nuclear receptors. In the absence of ligand, some but not all nuclear receptors repress transcription as a heterodimer with retinoid X receptor (RXR). Here we define a novel role for helix 12 (H12) in sterically masking the corepressor (CoR) binding site in apo-RXR. Removing H12 converts RXR to a potent transcriptional repressor. The length but not the specific sequence of H12 is critical for masking RXR’s intrinsic repression function. This contrasts with the amphipathic character required for mediating ligand-dependent activation and coactivator recruitment. Physiologically, we show that heterodimerization of RXR with apo-thyroid hormone receptor (TR) unmasks the CoR binding site in RXR and allows the TR-RXR heterodimer to repress. A molecular mechanism that involves sequence-specific interaction between RXR H12 and the coactivator-binding surface of the nuclear receptor is proposed for this heterodimerization-mediated unmasking. Peroxisome proliferator-activated receptor γ does not interact as well with RXR H12, thus explaining its inability to repress transcription as an RXR heterodimer. The requirement to unmask RXR’s latent repression function explains why only certain RXR partners repress transcription.
Nuclear hormone receptors (NHRs) allow cells to regulate gene transcription in response to nutritional, metabolic, and hormonal signals. Many NHRs repress transcription in the unliganded state (3, 6, 13, 19, 39). This silences target genes and amplifies the ligand signal. Repression is a function of the ligand-binding domain (LBD) of unliganded (apo-) NHR, which recruits corepressor (CoR) molecules to target genes. The main NHR CoRs are N-CoR (for nuclear receptor corepressor) (24, 47, 63) and SMRT (for silencing mediator for retinoid and thyroid hormone receptors) (8, 43). The CoRs are components of multiprotein repression complexes that also include mSin3 and histone deacetylase (2, 23, 37), the latter providing an enzymatic link to the nucleosome (22, 26). The biological importance of repression by NHRs is increasingly clear from studies of human diseases, including thyroid hormone resistance (51) and acute promyelocytic leukemia (32).
Ligand binding to the NHR leads to dissociation of the CoR complex. This depression step accounts for a portion of the increase in gene activity associated with hormone binding. In addition, the hormone-bound (holo-) NHR LBD recruits a coactivator (CoA) complex containing multiple histone acetyltransferases (HATs), including CREB-binding protein (CBP), p300/CBP-associated factor, and a member of the steroid receptor coactivator class of CoAs (reviewed in references 18 and 50). The localized HAT activity is thought to counteract the repressive effects of deacetylated histone and thus lead to enhanced transcription of the hormonally responsive gene.
Remarkably, the stability of the huge CoR and CoA complexes with NHR on DNA is dependent upon the absence or presence of a small lipophilic ligand. NHR apo- and holoreceptor conformations are similar overall, with 12 highly ordered α helices (H1 to H12) (reviewed in reference 61). Numerous differences in the position of these helices between the apo- and holoreceptors have been inferred by comparing the apo-retinoid X receptor (RXR) (5) with holo-retinoic acid receptor (RAR) (42) and holo-thyroid hormone receptor (TR) (56) structures. The most striking difference is in the positioning of H12, which is rotated back toward the ligand-binding pocket in the holoreceptor structure. The change in position of H12 is much less marked in apo- versus holo-peroxisome proliferator-activated receptor γ (PPARγ) (38, 54). H12 contains an amphipathic region whose sequence is critical to the ligand-dependent recruitment of the CoA complex (14, 21). This amphipathic helix also plays a less-well-defined role in CoR release by NHRs (31, 66).
The structures of apo- and holoreceptors have provided important insights into the molecular underpinnings of hormone action. However, information is limited because NHRs have not been cocrystallized as heterodimers. Indeed, the majority of NHRs, including the TR, RAR, and PPAR, require heterodimerization with RXR for high-affinity binding to hormonally responsive elements (HREs) in target genes (reviewed in reference 34). The RXR-NHR interface also determines target gene specificity by restricting the spacing between directly repeated half-sites that constitute the HRE, thus contributing to the precision of hormone action (40, 53). RXR-TR and RXR-RAR heterodimers interact with corepressors off and on DNA, in vitro as well as in vivo (9, 28, 60, 64). The observation that two repression domains are required for CoR complex formation (25, 64) suggests an active role of RXR. Consistent with this, TR mutants that cannot heterodimerize with RXR are defective in repression (4, 39), and interaction with RXR via a heterologous dimerization interface rescues this defect (66).
RXR by itself weakly interacts with CoRs (48) and only weakly represses transcription (36, 44, 66). Here we show that this is because H12 of RXR sterically prevents CoR binding. Deletion of H12 reveals the potential of RXR to bind CoR and repress transcription. Remarkably, unlike other functions of H12, such as CoA recruitment (7, 21, 55), it is the length and not the sequence of H12 that is critical to masking this intrinsic repression function. Moreover, heterodimerization with receptors that reposition H12, such as TR, unmasks RXR’s latent repressive potential. Not all RXR-heterodimerizing NHRs are capable of the latter interaction, thereby providing an explanation of variations in their repression function.
MATERIALS AND METHODS
Receptor and CoR expression plasmids.
Most plasmids have been described previously (66). Others were created by using PCR, endonuclease restriction digestion, or a combination of these techniques followed by ligation. Gal4-PPARγ contains amino acids 204 to 505 of mouse PPARγ2 cDNA. Details of other constructs are described in the text and/or figure legends. All constructs were directly sequenced.
Interaction assays.
Mammalian two-hybrid and glutathione S-transferase (GST) pull-down assays were performed as previously described (66). Gel shift assays have been previously described (64).
Transcription assays.
Transient transfection of 293T cells and luciferase reporter assays were performed as previously described (66). Cells were transfected either with calcium phosphate as previously described (66) or by Lipofectamine reagent (Gibco/BRL) in accordance with the manufacturer’s instructions. In each case, β-galactosidase expression vector was cotransfected as a control for transfection efficiency. Results shown are normalized to β-galactosidase expression. Fold activation in the mammalian two-hybrid assay was normalized to luciferase activity in the absence of the VP16-prey vector. For fold repression, results were normalized to the Gal4 DNA-binding domain (DBD) or receptor alone and the reciprocal was plotted. In every case, experiments were repeated two to six times, and duplicates and the range of the results of a representative experiment are shown.
Limited proteolytic digestion assays.
Protease digestions and analyses were performed as previously described (49). Trypsin concentrations and times of incubation are provided in the figure legends.
Loop building.
The Ω loop of RXRΔ449 was built using the coordinates of apo-RXRα (5) and Swiss-Pdb Viewer software (20). Free energies (force field scores) were calculated for all conformations during loop building, and the lowest energy conformation without amino acid clashes is shown in Fig. 2G. All structures were displayed using MOLMOL software (27).
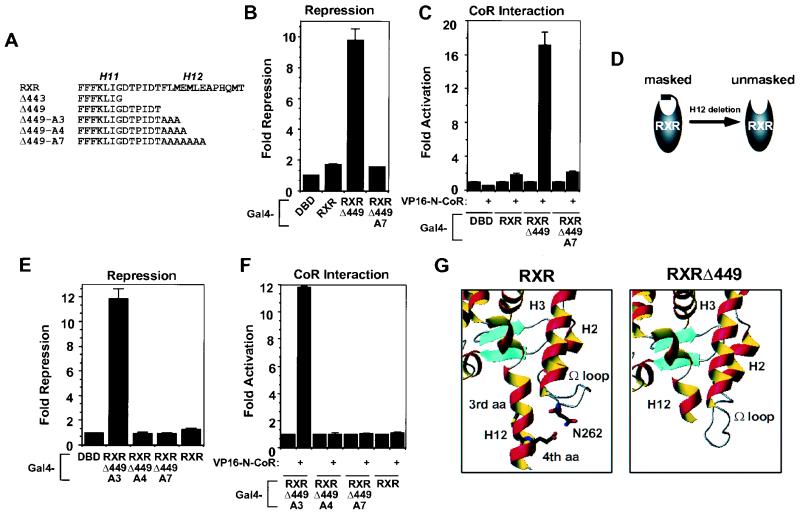
FIG. 2.
Length is the critical determinant of the RXR H12 mask. (A) RXRα mutants used in the study. Sequences shown begin at amino acid 437. The positions of H11 and H12 are indicated. (B and C) Seven alanine residues in place of H12 are sufficient for interference with repression (B) and CoR interaction (C) with RXR. Fold repression values for Gal4-RXRΔ449 or Gal4-RXRΔ449-A7 are shown. (D) Schematic of conclusion from panels B and C. (E and F) Four but not three alanines after amino acid 443 are sufficient to block repression and CoR interaction. Three, four, and seven alanine residues were attached to the Gal4-RXRΔ449 (designated RXRΔ449-A3, -A4, and -A7), and these were assayed for repression (E) and CoR interaction activities (F). (G) Potential structural basis of the H12 mask. The apo-RXR structure is from Bourguet et al. (5). Note that the Ω loop flips outward, and the side chain of the fourth but not the third amino acid after 449 contacts the Ω loop, especially N262. The RXRΔ449 structure was modeled using Swiss-Pdb Viewer software. The lowest energy (force field score) conformation without amino acid clashes is shown. Note that the Ω loop is flipped down.
RESULTS
RXR H12 masks the intrinsic repression function of apo-RXR by preventing CoR interaction.
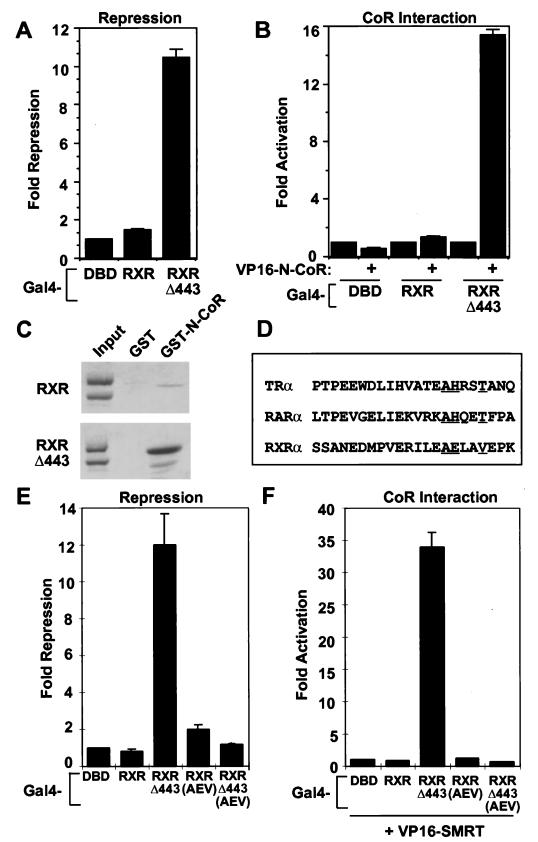
Unlike TR, the RXR LBD only minimally represses transcription when fused to the Gal4 DBD (Fig. 1A and references 32 and 62). Deletion of the RXR C terminus at amino acid 443 converts RXR LBD into a potent repression domain (Fig. 1A), consistent with the results of others (30, 44). Here, we show that the increased repression by the C-terminal deletion correlates with a dramatic increase in the ability of RXR to interact with N-CoR in vivo (Fig. 1B) and in vitro (Fig. 1C). Figure 1D aligns H1 of RXR with the “CoR box” that is required for CoR interaction with TR and RAR; repression by TR is abolished by mutation of the underlined A, H, and T residues to G, G, and A, respectively (24). Mutation of the homologous amino acids (AEV to GGA) in RXR in the context of RXRΔ443 abrogated repression (Fig. 1E) as well as CoR interaction with RXR (Fig. 1F). This result indicated that the CoR interaction surface in RXRΔ443 is similar to that in TR and RAR, although in the case of RXR it is obscured by H12.
FIG. 1.
RXR H12 masks an intrinsic repression function. (A) Deletion of the RXR C terminus converts RXR LBD into a potent repression domain. Full-length human RXRα LBD or RXR LBDΔ443 was fused to Gal4 DBD, and 1 μg was transfected into 293T cells along with a (Gal4 × 5)-simian virus 40-luciferase reporter gene. (B) Enhanced interaction between RXRΔ443 and N-CoR in vivo. Mammalian two-hybrid assay with Gal4-RXR or Gal4-RXRΔ443 as bait and VP16-N-CoR as prey. (C) Enhanced interaction between RXRΔ443 and N-CoR in vitro: GST pull-down of RXR or RXRΔ443 with GST alone or GST-N-CoR interaction domain. The input lane shows 10% of input. (D) CoR box sequence comparison. Rat TRα sequence is shown from amino acid 160. Human RARα sequence is shown from amino acid 180. Human RXRα sequence is shown from amino acid 224. Amino acids critical for interaction of CoR with TR and RAR are underlined, as are the homologous amino acids in RXR. (E and F) CoR box is required for repression (E) and interaction (F) between RXRΔ443 and CoR: mammalian two-hybrid assay with Gal4-RXRΔ443 or Gal4-RXRΔ443(AEV) as bait and VP16-SMRT as prey.
Length is the critical determinant of the H12 mask.
We next tested the role of RXR H12 using the C-terminal mutants summarized in Fig. 2A. Deletion at amino acid 449 was sufficient to unmask RXR repression (Fig. 2B). The core amphipathic region of RXR H12 is comprised of amino acids 450 to 456. Remarkably, complete replacement of these amino acids with alanines was sufficient to mask the repression function of RXR (Fig. 2B) as well as the ability of RXR to interact with N-CoR (Fig. 2C). Our results suggest that the presence of H12, rather than its primary amino acid sequence or amphipathicity, was critical for masking repression (Fig. 2D). To further test this hypothesis, one to seven alanine residues were attached to the C terminus of RXRΔ449 (designated RXRΔ449-A1 through -A7). An extension of one to three alanine residues did not block potent repression by RXRΔ449 (Fig. 2E and data not shown). Strikingly, substitution of a single additional alanine (four total), as well as a total of five to seven alanines, was sufficient to block repression (Fig. 2E and data not shown). N-CoR interaction was similarly permitted by the tri-alanine extension but prevented by addition of a fourth alanine residue (Fig. 2F). These results show that the H12 mask is steric and that the length of H12 is the critical factor in RXR’s inability to interact with CoR and repress transcription.
In the solved apo-RXR crystal structure, H12 makes multiple tight contacts with the Ω loop between H1 and H3 (5). Since the CoR box is located in H1, we reasoned that unmasking of the CoR interaction surface by deletion of H12 might be due to a conformational change in the Ω loop, which is flipped outward in apo-RXR (Fig. 2G, left). As previously noted (5), this position of the Ω loop is constrained by amino acid clashes between the Ω loop and H12, especially the fourth amino acid, whose side chain sticks out toward the Ω loop and makes van der Waals and hydrogen bonding contacts with N262 (d = 2.91 Å). By contrast, modeling studies predict that the Ω loop would be flipped down in the conformation of lowest free energy for RXRΔ449 (Fig. 2G, right). This analysis, although containing numerous assumptions, is based upon the solved apo-RXR structure and provides a plausible explanation for why RXRΔ449A3 but not RXRΔ449A4 is able to interact with CoR.
Apo-RXR exists as an intermediate between CoR- and CoA-interacting conformations.
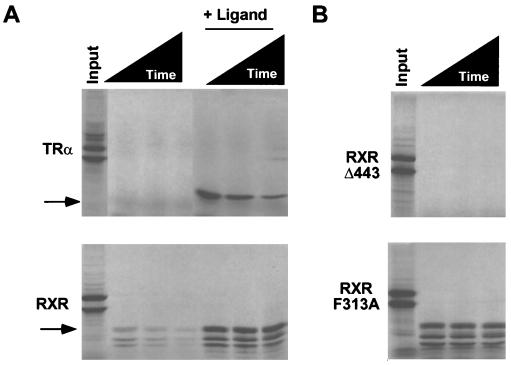
The ability of RXR H12 to sterically prevent corepressor interaction contrasts with apo-TR and apo-RAR, where H12 does not have this function. This suggests that the conformation of apo-RXR is fundamentally different from that of other NHRs, perhaps more akin to the liganded conformation that excludes CoR binding. The conformation of NHR LBDs can be probed by limited protease digestion, wherein the C terminus of the aporeceptor is more prone to proteolytic cleavage than that of the holo-NHR (1). As predicted by the functional studies, RXR LBD was partially protected under conditions that led to complete proteolysis of apo-TR LBD (Fig. 3A). Like apo-TR, apo-RXRΔ443 was completely proteolyzed (Fig. 3B). Unlike RXR, however, deletion of H12 from apo-TR did not affect its proteolytic stability (data not shown). Although the conformation of apo-RXR was different from that of the H12 deletion, it was also distinct from that of holo-RXR, which was protected from proteolysis to a far greater extent (Fig. 3A). This protection was due to the holoreceptor conformation, and not interference by ligand, because a constitutively active mutant (F313A) (55) was similarly protected from proteolytic digestion even in the absence of ligand (Fig. 3B).
FIG. 3.
Apo-RXR exists as an intermediate between CoR- and CoA-interacting conformations. (A) Apo-RXR and apo-TR are differentially sensitive to proteolysis. Arrows point to protected fragments. Proteins were translated in reticulocyte lysate and treated with trypsin (300 μg/ml) for various times (5, 10, and 20 min). The T3 concentration was 1 μM and SR11237 was used at a concentration of 50 μM. (B) Differential protease sensitivity of apo-RXRΔ443 and RXR-F313A.
The CoR interaction surface of RXR is critical for repression by the RXR-TR heterodimer.
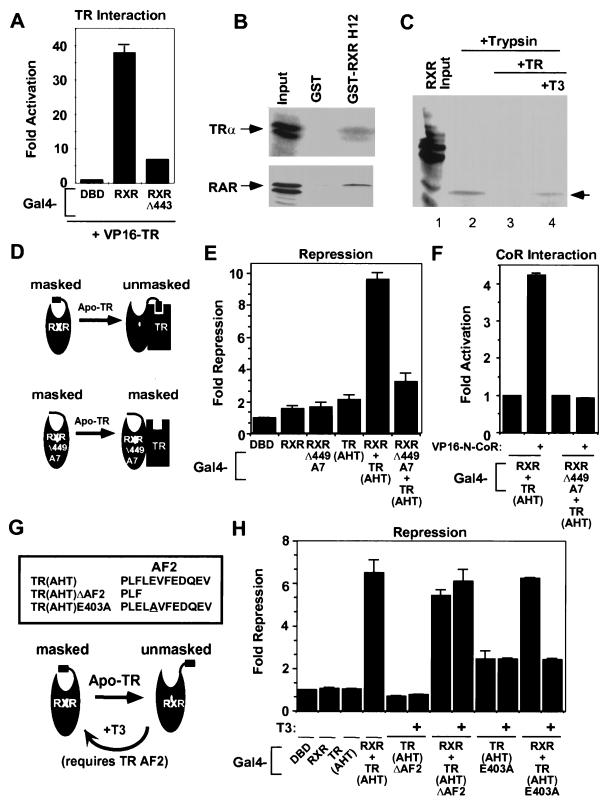
We next explored the ability of RXR to rescue repression-defective TR. The TR CoR box mutant, Gal4-TR(AHT), is inactive in repression assays (references 24 and 66) (Fig. 5). As previously shown (66), Gal4-RXR rescued the repression-defective Gal4-TR(AHT) by restoring repression (Fig. 4A) as well as CoR interaction (Fig. 4B and C). The contribution of the CoR interaction surface of RXR to repression by the TR(AHT)-RXR heterodimer was confirmed by using the Gal4-RXR(AEV) CoR box mutant, which was unable to rescue the TR CoR box mutant in terms of either repression (Fig. 4A) or CoR interaction (Fig. 4B and C).
FIG. 5.
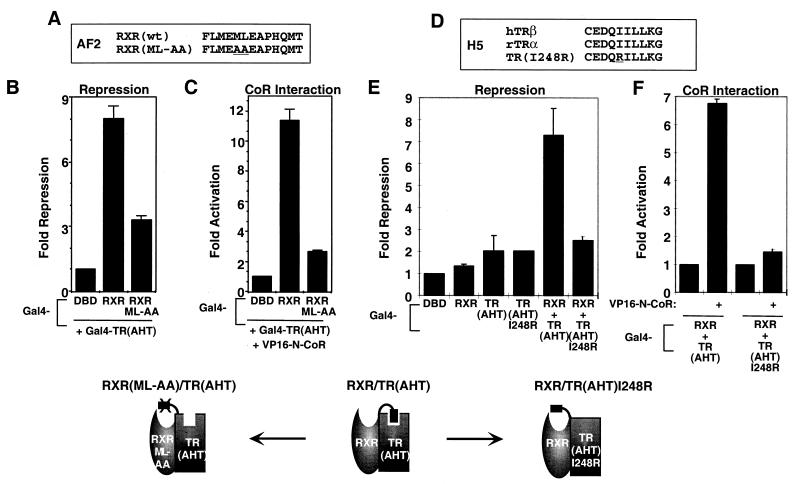
Unmasking the CoR interaction domain of RXR by “H12 docking” with a heterodimer partner. (A) RXR H12 contributes to the affinity of RXR for TR: mammalian two-hybrid assay with Gal4-RXR or Gal4-RXRΔ449 and VP16-N-CoR. (B) RXR H12 interacts specifically with apo-TR and apo-RAR in the GST pull-down assay. (C) Heterodimerization with TR alters protease sensitivity of RXR. Labeled RXR was incubated with a fivefold excess of unlabeled TR (in the presence and absence of T3 [12.5 μM]), then exposed to trypsin (1 mg/ml) for 3 min. (D) Model of conclusions from panels A, B, and C. (E and F) Gal4-RXRΔ449A7 cannot rescue Gal4-TR(AHT) for repression (E) and CoR interaction (F). (G) H12-AF2 mutants of TR used in panel H. (H) Ligand binding by TR abolishes complementation of repression between Gal4-RXR and Gal4-TR(AHT)E403A but not Gal4-TR(AHT)ΔAF2. Gal4-TR(AHT)ΔAF2, Gal4-TR(AHT)E403A, and Gal4-RXR were transfected separately or together and assayed for repression of (Gal4 × 5)-simian virus 40-luciferase reporter in the presence and absence of T3 (1 μM).
FIG. 4.
RXR provides a CoR interaction surface in the RXR-TR heterodimer. (A) Gal4-RXR, but not the RXR CoR box mutant, rescues Gal4-TR(AHT) for repression. (B and C) Gal4-RXR, but not the RXR CoR box mutant, rescues Gal4-TR(AHT) for interaction with VP16-SMRT (B) and VP16-N-CoR (C) in the mammalian two-hybrid assay.
Interaction of RXR H12 with TR unmasks the CoR interaction domain of RXR.
The requirement for the RXR CoR box indicated that the masking effect of H12 is relieved by heterodimerization with TR. We hypothesized that H12 of RXR interacts with apo-TR, thereby repositioning H12 and unmasking the CoR interaction surface of RXR. The major RXR heterodimer interface involves H10 and H11 and, as previously shown (35), RXR H12 is not required for interaction with TR (Fig. 5A). However, mammalian two-hybrid experiments showed that while it is not required for heterodimerization, RXR H12 contributes to the interaction of RXR with apo-TR (Fig. 5A). The ability of RXR H12 to interact with TR was directly tested in a GST pull-down experiment, comparing GST alone with GST fused to the 20 C-terminal amino acids (443 to 462) that are deleted in RXRΔ443. Indeed, Fig. 5B shows that RXR H12 interacted specifically with apo-TR. TR(AHT) also interacted with RXR H12 (data not shown). The interaction between RXR H12 and TR was relatively weak but reproducible, similar to that between RAR and RXR H12 (Fig. 5B and reference 58).
Since the protease sensitivities of apo-RXR with and without H12 are distinguishable, we next used the protease assay to probe the conformational change in RXR induced by TR heterodimerization. The presence of apo-TR reduced the proteolytic stability of the C-terminal RXR fragment characteristic of the aporeceptor conformation that does not bind CoR (Fig. 5C; compare lanes 2 and 3). This result indicates that interaction with TR repositions RXR H12 to unmask the CoR interaction surface (Fig. 5D). This predicts that the ability of TR to reposition H12 should be sequence specific. Thus, we tested for complementation of TR repression by RXRΔ449-A7. This H12 sequence mutant differed from wild-type RXR in that it failed to rescue TR(AHT) either for repression (Fig. 5E) or for CoR interaction (Fig. 5F). Thus, the specific sequence of H12 is an important determinant of CoR binding to the TR-RXR heterodimer.
T3 binding releases CoR from TR-RXR heterodimers. The strong TR-RXR interaction mediated by H10 and H11 (the “ninth heptad”) is ligand independent, but we hypothesized that the weak interaction of RXR H12 with TR might be sensitive to ligand-induced changes in the conformation of TR. This was tested in the protease sensitivity assay. Indeed, the ability of TR heterodimerization to allosterically promote the “unmasked” conformation of RXR was reversed by T3 (Fig. 5C, compare lanes 3 and 4), indicating that H12 does not interact with holo-TR. We next used an H12 deletion mutant (ΔAF2, missing the last nine amino acids) and an H12 point (E403A) mutant of Gal4-TR(AHT) to test the hypothesis that repositioning of TR H12 in the presence of T3 is involved in the undocking of RXR H12 and thus the release of CoR from the TR-RXR heterodimer (Fig. 5G). T3 does not activate either TR mutant because of the alterations in H12 that prevent CoA binding. Because of their CoR box mutations, these TR mutants only weakly repress transcription. However, Gal4-RXR can rescue this defect (Fig. 5H). Addition of T3 to the Gal4-RXR–Gal4-TR(AHT)ΔAF2 dimer has no effect on repression. The concentration of T3 used (1 μM) was well above the Kd of the mutant Gal4-TRα1(AHT)ΔAF2 (the measured Kd was 27 nM). By contrast, addition of T3 to the Gal4-RXR–Gal4-TR(AHT)E403A dimer relieves repression. This suggests that the T3-dependent switch in position of TR H12 (which is deleted in the ΔAF2 mutant) prevents RXR H12 from interacting with TR, thereby remasking the RXR repression function. Thus, RXR H12 differentially recognizes the liganded and unliganded conformations of TR and this switch regulates the repression of the TR-RXR heterodimer.
The sequence specificity of the interaction between apo-TR and RXR H12 is similar to that of receptor-CoA interactions.
Recent crystallographic data have indicated that the AF2 helix of apo-PPARγ interacts with the CoA binding pocket of another PPARγ molecule (38). A similar interaction has been observed in the estrogen receptor crystal structure (52). This led us to hypothesize that RXR AF2 mutations that prevented CoA binding would similarly fail to rescue repression by TR(AHT). To test this idea, two amino acids in the RXR AF2 amphipathic helix were mutated to alanine (ML454AA) (Fig. 6A). This mutation has previously been shown to abolish ligand-dependent activation and CoA recruitment by RXR (55). In the TR(AHT) complementation assay, this mutant displayed markedly reduced repression (Fig. 6B) as well as interaction with CoR (Fig. 6C).
FIG. 6.
Sequences in TR and RXR required for the unmasking of repression. (A) Sequences of RXR AF2 and the ML-AA mutant. (B and C) The ML-AA mutant of RXR fails to rescue TR(AHT) for repression (B) or N-CoR interaction (C). (D) Sequences of H5 of TRα and the I248R mutant. (E and F) The I248R mutant, which does not interact with coactivators, cannot functionally interact with RXR for repression (E) and N-CoR interaction (F).
RXR H12 contains a sequence similar to the LXXLL motif (LXXML in RXR) that is important for CoA interaction with a hydrophobic cleft, comprised of key amino acids in H3, H4, H5, and H12 in TR (15). We therefore reasoned that this surface might also be the TR interaction site of RXR H12. To test this, we created a mutation in H5 [TR(AHT)I248R; Fig. 6D], corresponding to the I302R mutation in TRβ that has been shown to bind T3 normally but to be defective in CoA binding and activation (16). As predicted, wild-type RXR could not rescue this mutant in either repression (Fig. 6E) or CoR interaction (Fig. 6F). These data indicate that a sequence-specific interaction between H12 of RXR and the hydrophobic cleft of apo-TR is responsible for unmasking the CoR interaction domain of RXR and thus allows the TR-RXR heterodimer to repress transcription.
PPARγ is unable to interact with RXR H12 and unmask the RXR CoR interaction surface.
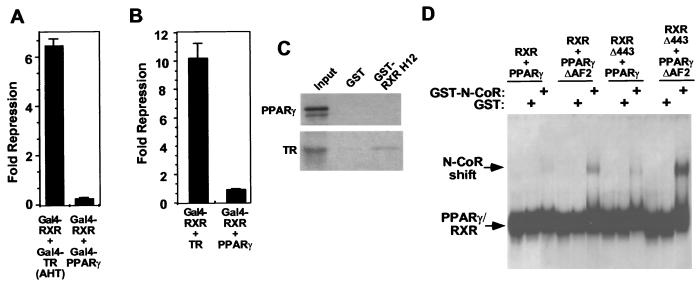
Although PPARγ binds to N-CoR and SMRT in GST pull-down assays, the PPARγ-RXR heterodimer fails to interact with CoRs on DNA and, thus, full-length PPARγ does not repress transcription (64). Gal4-PPARγ is a weak repressor, consistent with the detectable but weak ability of PPAR to bind N-CoR and SMRT in vitro (41, 64). However, unlike its effects on Gal4-TR(AHT), Gal4-RXR was unable to complement Gal4-PPARγ for repression (Fig. 7A). Furthermore, full-length TR but not full-length PPARγ complemented Gal4-RXR for repression (Fig. 7B). This suggested that RXR H12 would not dock with PPARγ. Indeed, this was confirmed by GST pull-down assay (Fig. 7C and reference 58).
FIG. 7.
PPARγ does not repress transcription because it is unable to dock with RXR H12 and unmask the RXR CoR interaction surface. (A) Gal4-RXR does not complement Gal4-PPARγ in repression. (B) Wild-type TR but not wild-type PPARγ complements Gal4-RXR in repression. (C) PPARγ does not interact with the GST-RXR H12 in the GST pull-down assay. GST was fused to amino acids 443 to 462 of hRXRα. (D) PPARγ-RXRΔ443 heterodimer interacts with N-CoR on DNA: gel shift assay using in vitro-translated PPARγ and RXR proteins and bacterially expressed GST or GST-N-CoR interaction domain, binding to PPRE from the acyl- CoA oxidase gene.
The PPARγ-RXRΔ443 heterodimer, unlike wild-type PPARγ-RXR heterodimers, was able to interact with N-CoR on a PPAR response element (PPRE) (Fig. 7D). Interestingly, deleting AF2 from both PPARγ and RXR increased CoR binding on DNA even further. Consistent with this, deletion of PPARγ H12 increased repression, especially in the context of Gal4 fusion proteins (data not shown). This is not surprising in light of the apo-PPARγ structure, in which the position of H12 appears to be similar to that in the holoreceptor.
DISCUSSION
Apo-RXR has a conformation distinct from repressive and active states.
We have shown that apo-RXR exists in a neutral conformation intermediate between that of CoR and CoA binding conformations. This conformation is characterized by a novel protease sensitivity and by the inability to interact with CoA or CoR. In this conformation, H12 restricts CoR interaction as a function of the length and not the specific sequence of H12. This is in sharp contrast to H12 of other receptors, such as TR and RAR, where H12 does not restrict CoR interaction (8). Thus, not all apo-NHR conformations are the same. Moreover, the crystal structure of apo-RXR cannot be used to reliably predict the structure of other apo-NHRs.
Deletion of H12 unmasks RXR’s repression function.
The repression function of RXR is similar to that of other NHRs, in that an intact CoR box is required. Deletion of H12 alters the conformation of the receptor in such a way as to provide access of CoRs to the appropriate interaction surfaces in apo-RXR. Unlike other functions of H12, which depend upon the sequence of the amphipathic α helix, the ability of RXR H12 to mask the repression surface is primarily determined by the length of H12. Furthermore, introducing RXR H12 into TR does not block repression (24a). These data clearly indicate that regulation of repression by H12 is primarily steric, rather than allosteric. This argues against the allosteric model suggested by Schulman et al. (44), in which the activation function of RXR H12 neutralizes its repression function. The modeling analysis shown in Fig. 2G provides a plausible explanation for the steric basis of the H12 mask.
Molecular mechanism for heterodimerization-mediated unmasking of RXR’s repression function.
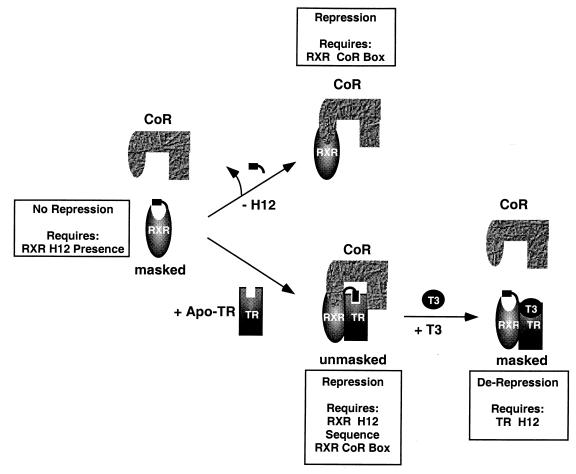
The more physiological mechanism that we have identified for repositioning H12 in RXR is heterodimerization with TR. Interaction of RXR H12 with the CoA binding surface of TR unmasks the CoR interaction surface of RXR by removing steric hindrance (Fig. 8). This mechanism requires the sequence of the RXR H12 to be specifically recognized by the heterodimer partner. It is likely that RAR behaves similarly, since biochemical studies and molecular modeling suggest that H12 of RXR can interact with the CoA binding pocket of RAR (58). The interaction between RXR H12 and TR (or RAR) is weaker than that between the main heterodimerization interfaces. This may allow reversible interaction between RXR H12 and TR to occur without disrupting the DNA binding of the heterodimer, which mediates activation as well as repression. The concept that RXR differentially interacts with unliganded and liganded TR is supported by the observation that the TR CoR box is required for RXR interaction in the absence but not in the presence of T3 (12, 66). Here we have shown that T3 binding releases RXR H12 by a TR H12-dependent mechanism, thereby contributing to the release of CoR from the heterodimer (Fig. 8).
FIG. 8.
Model of the role of RXR H12 in masking and unmasking CoR interaction. H12 of RXR sterically masks the CoR interaction domain. The intrinsic affinity of RXR for N-CoR and SMRT can be unmasked by deletion of RXR H12 (top) or by heterodimerization with TR (bottom). T3 binding to TR-RXR heterodimer alters the conformation of TR in a manner that interferes with H12 docking, thereby contributing to CoR dissociation and derepression. For simplicity, the CoR is depicted in isolation but is most likely in a complex including some combination of proteins known to associate with N-CoR or SMRT, including Sin3 (2, 23, 37), HDAC (2, 23, 37), SUN-CoR (62), and ETO (17, 33, 57).
Regulation of repression by RXR.
PPARγ interacts with both N-CoR and SMRT in solution, albeit weakly (41). This is consistent with the observation that apo-PPARγ itself is in an intermediate conformation, as suggested by structural studies (38, 54) as well as protease assays (49). PPARγ-RXR heterodimers bind CoRs extremely weakly when bound to a PPRE (64), and here we show that deletion of H12 from RXR (or both RXR and PPARγ) dramatically enhances the CoR binding affinity of the PPARγ-RXR heterodimer. This correlates with the lack of detectable interaction between RXR H12 and PPARγ (Fig. 7 and reference 58). The failure of RXR H12 to interact with PPARγ explains the inability of the wild-type PPAR-RXR heterodimer to recruit CoRs to the PPRE. Consistent with this, the PPARγ-RXR heterodimer does not repress transcription from a PPRE (64). A recent report suggests that microinjection of antibody to SMRT reverses the inhibitory effect of activated Ras on ligand-dependent PPARγ activation (29). However, the effect of apo-PPARγ on basal expression of a PPRE-containing reporter gene was not analyzed in that study. Indeed, to our knowledge, there is no published report of PPARγ significantly repressing basal transcription when bound to a heterodimer binding site. The present results suggest that PPARγ’s lack of repressive function is due to the inability of PPARγ to interact with RXR H12, coupled with the reduced inherent CoR binding affinity relative to TR and RAR.
The complexity of NHR function allows ligand, receptor, and target gene specificity while retaining generally successful strategies for DNA-binding and transcriptional regulation. Numerous NHRs utilize RXR heterodimerization as a mechanism for enhancing DNA binding affinity. Here we have shown that repression by RXR heterodimers is restricted to partners like TR and RAR that can dock with RXR H12 and unmask a latent repression function in apo-RXR. Thus, RXR governs partner-specific repression, in addition to target gene recognition (40, 53, 65) and the capacity of the partner’s ligand to synergize with RXR ligands (11, 45, 46, 58, 59). The ability of RXR to dictate receptor-specific functions has enabled NHRs to share a successful strategy for target gene recognition while permitting diverse ligands to have quantitatively and qualitatively different effects on gene transcription.
ACKNOWLEDGMENTS
We thank Clarice Chen and Myles Brown for reading the manuscript and for valuable discussions.
This work was supported by NIH grants DK43806 and DK45586 (to M.A.L.). DNA sequencing was performed by the University of Pennsylvania sequencing facility, supported in part by the University of Pennsylvania Center for the Molecular Study of Digestive Diseases (P30 DK50306.)
J.Z. and X.H. contributed equally to this work.
REFERENCES
- 1.Allan G F, Leng X, Tsai S Y, Weigel N L, Edwards D P, Tsai M J, O’Malley B W. Hormone and antihormone induce distinct conformational changes which are central to steroid receptor activation. J Biol Chem. 1992;267:19513–19520. [PubMed] [Google Scholar]
- 2.Alland L, Muhle R, Hou H, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 3.Baniahmad A, Kohne A C, Renkawitz R. A transferable silencing domain is present in the thyroid hormone receptor, in the v-erbA onco-gene product and in the retinoic acid receptor. EMBO J. 1992;11:1015–1023. doi: 10.1002/j.1460-2075.1992.tb05140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baniahmad A, Leng X, Burris T P, Tsai S Y, Tsai M-J, O’Malley B W. The τ4 activation domain of the thyroid hormone receptor is required for release of a putative corepressor(s) necessary for transcriptional silencing. Mol Cell Biol. 1995;15:76–86. doi: 10.1128/mcb.15.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-alpha. Nature. 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 6.Brent G A, Dunn M K, Harney J W, Gulick T, Larsen P R, Moore D D. Thyroid hormone aporeceptor represses T3-inducible promoters and blocks activity of the retinoic acid receptor. New Biol. 1989;1:329–336. [PubMed] [Google Scholar]
- 7.Cavailles V, Dauvois S, Danielian P S, Parker M G. Interaction of proteins with transcriptionally active estrogen receptors. Proc Natl Acad Sci USA. 1994;91:10009–10013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J D, Evans R M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 9.Chen J D, Umesono K, Evans R M. SMRT isoforms mediate repression and anti-repression of nuclear receptor heterodimers. Proc Natl Acad Sci USA. 1996;93:7567–7571. doi: 10.1073/pnas.93.15.7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J-Y, Penco S, Ostrowski J, Balaguer P, Pons M, Starrett J E, Reczek P, Chambon P, Gronmeyer H. RAR-specific agonist/antagonists which dissociate transactivation and AP1 transrepression inhibit anchorage-independent cell proliferation. EMBO J. 1995;14:1187–1197. doi: 10.1002/j.1460-2075.1995.tb07102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J Y, Clifford J, Zusi C, Starrett J, Tortolani D, Ostrowski J, Reczek P R, Chambon P, Gronemeyer H. Two distinct actions of retinoid-receptor ligands. Nature. 1996;382:819–822. doi: 10.1038/382819a0. [DOI] [PubMed] [Google Scholar]
- 12.Collingwood T N, Butler A, Tone Y, Clifton-Bligh R J, Parker M G, Chatterjee V K. Thyroid hormone-mediated enhancement of heterodimer formation between thyroid hormone receptor β and retinoid X receptor. J Biol Chem. 1997;272:13060–13065. doi: 10.1074/jbc.272.20.13060. [DOI] [PubMed] [Google Scholar]
- 13.Damm K, Thompson C C, Evans R M. Protein encoded by v-erbA functions as a thyroid-hormone receptor antagonist. Nature. 1989;339:593–597. doi: 10.1038/339593a0. [DOI] [PubMed] [Google Scholar]
- 14.Danielian P S, White R, Lees J A, Parker M G. Identification of a conserved region required for hormone dependent transcriptional activation by steroid hormone receptors. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter J D, Fletterick R J, Yamamoto K R. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng W, Ribeiro R C J, Wagner R L, Nguyen H, Apriletti J W, Fletterick R J, Baxter J D, Kushner P J, West B L. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science. 1998;280:1747–1749. doi: 10.1126/science.280.5370.1747. [DOI] [PubMed] [Google Scholar]
- 17.Gelmetti V, Zhang J, Fanelli M, Minucci S, Pelicci P G, Lazar M A. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol Cell Biol. 1998;18:7185–7191. doi: 10.1128/mcb.18.12.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glass C K, Rose D W, Rosenfeld M G. Nuclear receptor coactivators. Curr Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 19.Graupner G, Wills K N, Tzukerman M, Zhang X-K, Pfahl M. Dual regulatory role for thyroid-hormone receptors allows control of retinoic-acid receptor activity. Nature. 1989;340:653–656. doi: 10.1038/340653a0. [DOI] [PubMed] [Google Scholar]
- 20.Guex N, Peitsch M C. SWISS-MODEL and the Swiss-Pdb viewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 21.Halachmi S, Marden E, Martin G, MacKay H, Abbondanza C, Brown M. Estrogen receptor-associated proteins: possible mediators of hormone-induced transcription. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- 22.Hassig C A, Schreiber S L. Nuclear histone acetylases and deacetylases and transcriptional regulation: HATs off to HDACs. Curr Opin Chem Biol. 1998;1:300–308. doi: 10.1016/s1367-5931(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 23.Heinzel T, Lavinsky R M, Mullen T-M, Soderstrom M, Laherty C D, Torchia J, Yuang W-M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 24.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass C K, Rosenfeld M G. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 24a.Hu, X., and M. A. Lazar. Unpublished results.
- 25.Jeannin E, Robyr D, Desvergne B. Transcriptional regulatory patterns of the myelin basic protein and malic enzyme genes by the thyroid hormone receptors α1 and β1. J Biol Chem. 1998;273:24239–24248. doi: 10.1074/jbc.273.37.24239. [DOI] [PubMed] [Google Scholar]
- 26.Kadosh D, Struhl K. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 28.Kurokawa R, Soderstrom M, Horlein A, Halachmi S, Brown M, Rosenfeld M G, Glass C K. Polarity-specific activities of retinoic acid receptors determined by a co-repressor. Nature. 1995;377:451–454. doi: 10.1038/377451a0. [DOI] [PubMed] [Google Scholar]
- 29.Lavinsky R M, Kristen J, Thorsten H, Torchia J, Mullen T-M, Schiff R, Del-Rio A L, Ricote M, Ngo S, Gemsch J, Hilsenbeck S G, Osborne C K, Glass C K, Rosenfeld M G. Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complexes. Proc Natl Acad Sci USA. 1998;95:2920–2925. doi: 10.1073/pnas.95.6.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leng X, Blanco J, Tsai S Y, Ozato K, O’Malley B W, Tsai M-J. Mouse retinoid X receptor contains a separable ligand-binding and transactivation domain in its E region. Mol Cell Biol. 1995;15:255–263. doi: 10.1128/mcb.15.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin B C, Hong S H, Krig S, Yoh S M, Privalsky M L. A conformational switch in nuclear hormone receptors is involved in coupling hormone binding to corepressor release. Mol Cell Biol. 1997;17:6131–6138. doi: 10.1128/mcb.17.10.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin R J, Nagy L, Inoue S, Shao W, Miller W H, Evans R M. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 33.Lutterbach B, Westendorf J J, Linggi B, Patten A, Moniwa M, Davie J R, Huynh K D, Bardwell V J, Lavinsky R M, Rosenfeld M G, Glass C, Seto E, Hiebert S W. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol Cell Biol. 1998;18:7176–7184. doi: 10.1128/mcb.18.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mangelsdorf D J, Evans R M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 35.Marks M S, Hallenback P L, Nagata T, Segars J H, Appella E, Nikodem V M, Ozato K. H-2RIIBP (RXRβ) dimerization provides a mechanism for combinatorial diversity in the regulation of retinoic acid and thyroid hormone responsive genes. EMBO J. 1992;11:1419–1435. doi: 10.1002/j.1460-2075.1992.tb05187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin B, Renkawitz R, Muller M. Two silencing sub-domains of v-erbA synergize with each other, but not with RXR. Nucleic Acids Res. 1994;22:4899–4905. doi: 10.1093/nar/22.23.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagy L, Kao H-Y, Chakvarkti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 38.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 39.Qi J-S, Desai-Yajnik V, Greene M E, Raaka B M, Samuels H H. The ligand-binding domains of the thyroid hormone/retinoid receptor gene subfamily function in vivo to mediate heterodimerization, gene silencing, and transactivation. Mol Cell Biol. 1995;15:1817–1825. doi: 10.1128/mcb.15.3.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rastinejad F, Perlmann T, Evans R M, Sigler P B. Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature. 1995;375:203–211. doi: 10.1038/375203a0. [DOI] [PubMed] [Google Scholar]
- 41.Reginato M J, Bailey S T, Krakow S L, Minami C, Ishii S, Takaka H. A potent antidiabetic thiazolidinedione with unusual PPARγ-activating properties. J Biol Chem. 1998;273:32679–32684. doi: 10.1074/jbc.273.49.32679. [DOI] [PubMed] [Google Scholar]
- 42.Renaud J-P, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Crystal structure of the RARγ ligand-binding domain bound to all-trans retinoic acid. Nature. 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 43.Sande S, Privalsky M L. Identification of TRACs, a family of co-factors that associate with and modulate the activity of nuclear hormone receptors. Mol Endocrinol. 1996;10:813–825. doi: 10.1210/mend.10.7.8813722. [DOI] [PubMed] [Google Scholar]
- 44.Schulman I G, Juguilon H, Evans R M. Activation and repression by nuclear hormone receptors: hormone modulates an equilibrium between active and repressive states. Mol Cell Biol. 1996;16:3807–3813. doi: 10.1128/mcb.16.7.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schulman I G, Li C, Schwabe J W R, Evans R M. The phantom ligand effect: allosteric control of transcription by the retinoid X receptor. Genes Dev. 1997;11:299–308. doi: 10.1101/gad.11.3.299. [DOI] [PubMed] [Google Scholar]
- 46.Schulman I G, Shao G, Heyman R A. Transactivation by retinoid X receptor-peroxisome proliferator-activated receptor γ (PPARγ) heterodimers: intermolecular synergy requires only the PPARγ hormone-dependent activation function. Mol Cell Biol. 1998;18:3483–3494. doi: 10.1128/mcb.18.6.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seol W, Choi H S, Moore D D. Isolation of proteins that interact specifically with the retinoid X receptor: two novel orphan receptors. Mol Endocrinol. 1995;9:72–85. doi: 10.1210/mend.9.1.7760852. [DOI] [PubMed] [Google Scholar]
- 48.Seol W, Mahon M J, Lee Y-K, Moore D D. Two receptor interacting domains in the nuclear hormone receptor corepressor RIP13/N-CoR. Mol Endocrinol. 1996;10:1646–1655. doi: 10.1210/mend.10.12.8961273. [DOI] [PubMed] [Google Scholar]
- 49.Shao D, Rangwala S M, Bailey S T, Krakow S L, Reginato M J, Lazar M A. Interdomain communication regulating PPARγ ligand binding. Nature. 1998;396:377–380. doi: 10.1038/24634. [DOI] [PubMed] [Google Scholar]
- 50.Shibata H, Spencer T E, Onate S A, Jenster G, Tsai S Y, Tsai M J, O’Malley B W. Role of co-activators and co-repressors in the mechanism of steroid/thyroid receptor action. Rec Prog Horm Res. 1997;52:141–164. [PubMed] [Google Scholar]
- 51.Tagami T, Jameson J L. Nuclear corepressors enhance the dominant negative activity of mutant receptors that cause resistance to thyroid hormone. Endocrinology. 1998;139:640–650. doi: 10.1210/endo.139.2.5742. [DOI] [PubMed] [Google Scholar]
- 52.Tanenbaum D M, Wang Y, Williams S P, Sigler P B. Crystallographic comparison of the estrogen and progesterone receptor’s ligand binding domains. Proc Natl Acad Sci USA. 1998;95:5998–6003. doi: 10.1073/pnas.95.11.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Umesono K, Murakami K K, Thompson C C, Evans R M. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uppenberg J, Svensson S, Jaki M, Bertilsson G, Jendeberg L, Berkenstam A. Crystal structure of the ligand binding domain of the human nuclear receptor PPARγ. J Biol Chem. 1998;273:31108–31112. doi: 10.1074/jbc.273.47.31108. [DOI] [PubMed] [Google Scholar]
- 55.Vivat V, Zechel C, Wurtz J M, Bourguet W, Kagechika H, Umemiya H, Shudo K, Moras D, Gronemeyer H, Chambon P. A mutation mimicking ligand-induced conformational change yields a constitutive RXR that senses allosteric effects in heterodimers. EMBO J. 1997;16:5697–5709. doi: 10.1093/emboj/16.18.5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wagner R L, Apriletti J W, McGrath M E, West B L, Baxter J D, Fletterick R J. A structural role for hormone in the thyroid hormone receptor. Nature. 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 57.Wang J, Hoshino T, Redner R L, Kajigaya S, Liu J M. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc Natl Acad Sci USA. 1998;95:10860–10865. doi: 10.1073/pnas.95.18.10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Westin S, Kurokawa R, Nolte R T, Wisely G B, McInerney E M, Rose D W, Milburn M V, Rosenfeld M G, Glass C K. Interactions controlling the assembly of nuclear receptor heterodimers and co-activators. Nature. 1998;395:199–202. doi: 10.1038/26040. [DOI] [PubMed] [Google Scholar]
- 59.Willy P J, Umesono K, Ong E S, Evans R M, Heyman R A, Mangelsdorf D J. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes Dev. 1995;9:1033–1045. doi: 10.1101/gad.9.9.1033. [DOI] [PubMed] [Google Scholar]
- 60.Wong C-W, Privalsky M L. Transcriptional silencing is defined by isoform- and heterodimer-specific interactions between nuclear hormone receptors and corepressors. Mol Cell Biol. 1998;18:5724–5733. doi: 10.1128/mcb.18.10.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wurtz J M, Bourguet W, Renaud J P, Vivat V, Chambon P, Moras D, Gronemeyer H. A canonical structure for the ligand binding domain of nuclear receptors. Nat Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- 62.Zamir I, Dawson J, Lavinsky R M, Glass C K, Rosenfeld M G, Lazar M A. Cloning and characterization of a corepressor and potential component of the nuclear hormone receptor repression complex. Proc Natl Acad Sci USA. 1997;94:14400–14405. doi: 10.1073/pnas.94.26.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zamir I, Harding H P, Atkins G B, Horlein A, Glass C K, Rosenfeld M G, Lazar M A. A nuclear hormone receptor corepressor mediates transcriptional silencing by receptors with different repression domains. Mol Cell Biol. 1996;16:5458–5465. doi: 10.1128/mcb.16.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zamir I, Zhang J, Lazar M A. Stoichiometric and steric principles governing repression by nuclear hormone receptors. Genes Dev. 1997;11:835–846. doi: 10.1101/gad.11.7.835. [DOI] [PubMed] [Google Scholar]
- 65.Zechel C, Shen X Q, Chambon P, Gronemeyer H. Dimerization interfaces formed between the DNA binding domains determine the cooperative binding of RXR/RAR and RXR/TR heterodimers to DR5 and DR4 elements. EMBO J. 1994;13:1414–1424. doi: 10.1002/j.1460-2075.1994.tb06395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang J, Zamir I, Lazar M A. Differential recognition of liganded and unliganded thyroid hormone receptor by retinoid X receptor regulates transcriptional repression. Mol Cell Biol. 1997;17:6887–6897. doi: 10.1128/mcb.17.12.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]