Abstract
The recent re-emergence of multidrug-resistant pathogens has exacerbated their threat to worldwide public health. The evolution of the genomics era has led to the generation of huge volumes of sequencing data at an unprecedented rate due to the ever-reducing costs of whole-genome sequencing (WGS). We have developed the Rapid Microbial Analysis Pipeline (rMAP), a user-friendly pipeline capable of profiling the resistomes of ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter species) using WGS data generated from Illumina’s sequencing platforms. rMAP is designed for individuals with little bioinformatics expertise, and automates the steps required for WGS analysis directly from the raw genomic sequence data, including adapter and low-quality sequence read trimming, de novo genome assembly, genome annotation, single-nucleotide polymorphism (SNP) variant calling, phylogenetic inference by maximum likelihood, antimicrobial resistance (AMR) profiling, plasmid profiling, virulence factor determination, multi-locus sequence typing (MLST), pangenome analysis and insertion sequence characterization (IS). Once the analysis is finished, rMAP generates an interactive web-like html report. rMAP installation is very simple, it can be run using very simple commands. It represents a rapid and easy way to perform comprehensive bacterial WGS analysis using a personal laptop in low-income settings where high-performance computing infrastructure is limited.
Keywords: command line, ESKAPE, pipeline, rapid microbial analysis, rMAP, whole-genome sequencing
Data Summary
The source code for single-nucleotide polymorphism (SNP) sites is available from GitHub under GNU GPL v3; (https://github.com/GunzIvan28/rMAP)
The authors confirm that all supporting data, code and protocols have been provided within the article. All sequencing reads from the exemplary data sets are publicly stored in the SRA database; accession IDs are provided.
Impact Statement.
The evolution of the genomics era has led to the generation of massive chunks of sequencing data and different bioinformatics tools have been developed to analyse these data. The ever-reducing costs of whole-genome sequencing (WGS) have led to diagnostic and research laboratories obtaining genome sequencing technologies. The considerable bioinformatics skills needed to analyse the large volume of genomic data from these platforms and the complex format in which results are presented offer two important impediments in the implementation of WGS. To the best of our knowledge, there is currently no published all-in-one bioinformatics tool that successfully provides: genome-assembly statistics; single-nucleotide polymorphism (SNP) variant calling; phylogenetic analysis; antimicrobial resistance, plasmid and virulence factor profiling; multi-locus sequence typing; pangenome analysis; and insertion sequence characterization (IS) for ESKAPE pathogens. Therefore, we introduce rMAP (https://github.com/GunzIvan28/rMAP), a rapid microbial analysis pipeline for comprehensive analysis of bacterial WGS data. This is an open-source, user-friendly, command-line and scalable pipeline for conducting WGS analysis of Illumina sequencing reads. It represents a rapid and easy way to perform comprehensive bacterial WGS analysis using personal laptops, especially in low-income settings where high-performance computing infrastructure is limited. rMAP generates a web-like html interactive report (https://gunzivan28.github.io/rMAP/) that can be shared and interpreted by microbiologists.
Introduction
The recent re-emergence of multidrug-resistant pathogens through persistent misuse of antibiotics has exacerbated their threat to worldwide human public health and well-being. Such organisms, consisting of Staphylococcus aureus, Pseudomonas aeruginosa and Klebsiella species belonging to the ESKAPE pathogen group, have been flagged among the most notorious micro-organisms expressing tremendously high levels of antimicrobial resistance by the World Health Organization (WHO), and have been reported by many studies to contribute to the high frequency of nosocomial infections which have led to high morbidity and mortality rates all over the world [1–3].
In the same spirit, rapid advances in diagnostic science and personalized medicine have seen the emergence of high-throughput next-generation sequencing technologies to replace conventional microbiology laboratories, and this has greatly reduced diagnostic costs and turnaround times for results for infectious pathogens as a way of keeping pace with emerging multidrug-resistant varieties. Next-generation processes generally involve parallel sequencing, producing vast quantities of genomic data, and extensive modern computation infrastructure is required to make sense of the sequencing data in downstream analysis. Furthermore, another bottleneck in the deployment of high-throughput sequencing (HTS) technologies is the ability to analyse the increasing amount of data produced in a fit-for-purpose manner [4]. The field of microbial bioinformatics is thriving and quickly adapting to technological changes, which creates difficulties for clinical microbiologists with little or no bioinformatics background in following the complexity and increasingly obscure jargon of this field [4].
The routine application of whole-genome sequencing (WGS) requires cheap, user-friendly techniques that can be used on-site by personnel who have not specialized in big data management [5, 6]. The ability of bioinformaticists to analyse, compare, interpret and visualize the vast increase in bacterial genomes is valiantly trying to keep up with these developments [7]. Many biologists are drowning in too much data, and in desperate need of a tool capable of deciphering this complex information, and it is predicted that these trends will continue in the foreseeable future as the generation of genome data becomes cheaper and abundant [7].
Therefore, we introduce the Rapid Microbial Analysis Pipeline (rMAP), a one-stop toolbox that uses WGS illumina data to characterize the resistomes of bacteria of ESKAPE origin. This is an open-source, user-friendly, command-line, automated and scalable pipeline for conducting analysis of HTS data produced by Illumina platforms. rMAP takes raw sequencing data as input and performs bacterial bioinformatic analysis steps, including: adapter and low-quality sequence trimming, de novo genome assembly, genome annotation, SNP variant calling, phylogenetic inference by maximum likelihood, antimicrobial resistance profiling, plasmid profiling, virulence factor determination, multi-locus sequence typing (MLST), pangenome analysis and insertion sequence (IS) characterization.
Methods
Pipeline architecture
rMAP is a tool implemented in four programming languages, namely Shell script, Python, Perl and R. It was precompiled and supports the Linux 64-bit architecture and macOS version 10.14.6 (Mojave) and above. It was originally built using WSL Ubuntu 20.04.1 LTS (Focal Fossa) and Ubuntu 18.04.4 LTS (Bionic Beaver) and the binaries are compatible with noarch–Unix-style operating systems.
rMAP was built using a collection of published reputable tools such as FASTQC [8], MultiQC [9], Trimmomatic [10], Shovill, Megahit [11], Prokka [12], Freebayes, SnpEff [13], IQtree [14], BWA [15], Samtools [16], Roary [17] and ISMapper [18], just to mention a few. All of the tools and third-party dependences required by rMAP are resolved and containerized within a conda environment as a single package so as not to interfere with already existing programs. The programs in the conda environment are built on top of Python version 3.7.8 [19] and are compatible with R statistical package version 4.0.2 [20]. A full list of the packages used by rMAP is provided in Table 1.
Table 1.
Comprehensive list of third-party tools and algorithms used in rMAP
|
Software |
Version |
Summary |
|---|---|---|
|
Abricate |
1.0.1 |
Detection of antimicrobial resistance genes, plasmids and virulence factors |
|
AMRfinder |
3.8.4 |
Detection of antimicrobial resistance genes from assembled contigs |
|
Any2fasta |
0.4.2 |
Converts any genomic data format to fasta format |
|
Assembly-stats |
1.0.1 |
Summarizes quality assembly metrics from contigs |
|
Biopython.convert |
1.0.3 |
Conversion and manipulation of different genomic data formats |
|
BMGE |
1.12 |
Block mapping and gathering with entropy for removal of ambiguously aligned reads from multiple sequence alignments |
|
BWA |
0.7.17 |
Burrow–Wheeler algorithm for fast alignment of short sequence reads |
|
Cairosvg |
2.4.2 |
Converts SVG to PDF and PNG formats |
|
Fastqc |
0.11.9 |
Quality control and visualization of HTS data |
|
Fasttree |
2.1.10 |
Ultra-fast inference of phylogeny using the maximum-likelihood method |
|
Freebayes |
1.3.2 |
Bayesian-based haplotype prediction of nucleotide variants |
|
ISMapper |
2.0.1 |
Detection of insertion sequences within genomes |
|
IQtree |
2.0.3 |
Inference of phylogeny using the maximum-likelihood method |
|
Kleborate |
1.0.0 |
Screening for AMR genes and MLSTs from genome assemblies |
|
Lxml |
4.5.2 |
Parsing of XML and HTML using Python |
|
Mafft |
7.471 |
Algorithm for performing multiple sequence alignments |
|
Multiqc |
1.9 |
Aggregates numerous HTML quality reports into a single file |
|
Megahit |
1.2.9 |
Ultra-fast genome assembly algorithm |
|
Mlst |
2.19.0 |
Characterization and detection of clones within a population of pathogenic isolates |
|
Nextflow |
20.07.1 |
Portable next-generation workflow language that enables reproducibility and development of pipelines |
|
Parallel |
20200722 |
Executes jobs in parallel |
|
Prinseq |
0.20.4 |
Trims, filters and reformats genomic sequence data |
|
Prodigal |
2.6.3 |
Prediction of protein-coding genes in prokaryotic genomes |
|
Prokka |
1.14.6 |
Fast and efficient annotation of prokaryotic assembled genomes |
|
Quast |
5.0.2 |
Quality assembly assessment tool |
|
Roary |
3.13.0 |
Large-scale pangenome analysis |
|
R-base |
4.0.2 |
Statistical data computing and graphical software |
|
Samclip |
0.4.0 |
Filters SAM file for soft and hard clipped alignments |
|
Samtools |
1.9 |
Tools for manipulation of next-generation sequence data |
|
Shovill |
1.0.9 |
Illumina short-read assembler for bacterial genomes |
|
Snippy |
4.3.6 |
Rapid haploid bacterial variant caller |
|
Snpeff |
4.5covid19 |
Functional effect and variant predictor suite |
|
SRA-tools |
2.10.8 |
Toolbox for acquisition and manipulation of sequences from the NCBI |
|
Trimmomatic |
0.39 |
Illumina short-read adapter trimming algorithm |
|
Unicycler |
0.4.8 |
A hybrid assembly pipeline for Illumina and long-read sequence data |
|
Vt |
2015.11.10 |
A tool for normalizing variants in genomic sequence data |
Overview of rMAP workflow
rMAP can be used with an unlimited number of samples of different species and origins. However, it was built to target pathogens of public health concern exhibiting high levels of antimicrobial resistance (AMR) and nosocomial infections. It can be applied to isolates of human and animal origin to give insights into the transmission dynamics of AMR genes at the human–animal interface.
Benchmarking datasets
The pipeline was tested on numerous bacterial pathogens from the ESKAPE group isolated from different origins (clinical, faecal, animal and sewage), sequenced on Illumina platforms and obtained from the publicly available repositories the Sequence Read Archive (SRA) and the European Nucleotide Archive (ENA) under the following accessions: Enterococcus species (SRR8948878, SRR8948879, SRR8948880, SRR8948881, SRR8948882, SRR8948883, SRR8948884, SRR8948885, SRR8948886, SRR8948887, SRR8948888, SRR8948889, SRR8948890, SRR8948891), Acinetobacter baumannii (ERR1989084, ERR1989100, ERR1989115, ERR3197698, SRR3666962, SRR5739056, SRR6037664, SRR8289559, SRR8291681), Klebsiella species (SRR8753739, SRR8753737, SRR8291573, SRR11816972, SRR9703249, SRR9029107, SRR9029108, SRR8610335, SRR8610353, SRR8610357, SRR8610351, SRR8610354, SRR9964283, SRR9044171, SRR5687278, SRR5514226, SRR5514224, SRR5514223) and Staphylococcus aureus (ERR1794900, ERR1794901, ERR1794902, ERR1794903, ERR1794904, ERR1794905, ERR1794906, ERR1794907, ERR1794908, ERR1794909, ERR1794910, ERR1794911, ERR1794912, ERR1794913, ERR1794914). The GenBank references used include A. baumannii strain 36–1512, accession CP059386.1, GI 1880620189; Enterococcus faecalis strain KB1, accession CP015410.2, GI 1173533644; and S. aureus subsp. aureus strain MRSA252, accession BX571856.1, GI 49240382.
Core pipeline features
rMAP requires three mandatory parameters; the input directory that contains sequence reads in either fastq or fastq.gz formats, an output user-defined directory and a reference genome in either GenBank or fasta format. A full GenBank reference genome file is recommended for the --reference option to obtain an annotated VCF files. The raw fastq files are directly submitted to rMAP, with no prior bioinformatics treatment, as follows:
rMAP -t 8 --reference --input dir_name--output dir_name --quality --assembly megahit --amr --varcall --phylogeny --pangenome --gen-ele
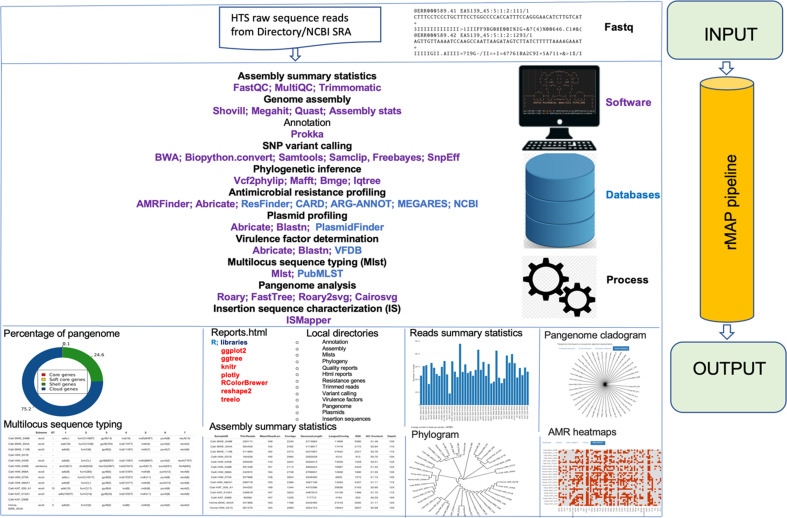
The pipeline’s features can be summarized in the order of: SRA sequence download, quality control, adapter trimming, de novo assembly, resistome profiling, variant calling, phylogenetic inference, pangenome analysis, insertion sequence mapping and report generation, as shown in Fig. 1.
Fig. 1.
Schematic graphical representation of rMAP pipeline workflow and associated tools.
Sequence read archive download
rMAP is able to retrieve sequences from the NCBI’s SRA using fastq-dump [21]. A user simply creates a list containing the sample accession numbers to be downloaded saved at the home directory. The downloaded sequences are saved in a default directory called SRA-READS created by rMAP.
Quality assessment and filtering
The pipeline autodetects any non-zipped fastq reads and parses them to the fastq.gz format for optimization purposes during downstream analysis. Fastqc [8] generates sequence quality reports and statistics from each individual sample, which are then aggregated into a single graphically interactive html report using MultiQC [9].
Adapter and low sequence read trimming
Trimmomatic [10] is used to trim off adapters using a set of pre-defined Illumina library preparation adapters saved in fasta format and low sequence regions from the raw input sequence reads. The pipeline’s default parameters for quality and minimum sequence length are set at a phred quality score of 27 and 80 base pairs, respectively, to accommodate sequencing data that may not be of the very high recommended quality (i.e. 33).
De novo assembly and annotation
Two assemblers are selected for this purpose for a user to choose from – Shovill [22] and Megahit [11] – each demonstrating an advantage over the other. Both algorithms take the trimmed reads as their input and perform k-mer-based assembly to produce contigs. Megahit exhibited very fast computational speeds, almost half those of its counterpart, but with slightly lower quality assembly metrics. Assembly with Shovill involves guided mapping of the contigs to a reference and numerous rounds of genome polishing using pilon to remove gaps,and takes more time but produces good quality assembly metrics (N50, L50, genome length). Prodigal [23] is used to predict open reading frames from the assembled contigs, which are then functionally annotated using Prokka [12].
Variant calling
The trimmed reads are aligned against a an indexed reference in the fasta format using the Burrows–Wheeler aligner [15] to produce SAM files. Soft and hard clipped alignments are removed from the sequence alignment map (SAM) files using Samclip (https://github.com/tseemann/samclip). Samtools [16] then sorts, marks duplicates and indexes the resultant binary alignment map (BAM) files. Freebayes [24] calls variants using Bayesian models to produce variant call format (VCF) files containing single-nucleotide polymorphism (SNP) information, which is filtered using bcftools (https://github.com/samtools/bcftools) and normalized of biallelic regions using Vt [25]. The filtered VCF files are annotated using snpEff [13]. Raw, tab-separated, annotated and filtered VCF files are available for the users to manipulate.
Resistome profiling
The conceptualization of rMAP was aimed at exhaustively exploiting the resistome of pathogenic bacteria. AMRfinder plus [26] predicts resistance genes using its database. Mass screening for antimicrobial resistance genes is performed using the CARD [27], ARG-ANNOT [28], NCBI, ResFinder and MEGARES [29] databases. Plasmids and virulence factors are typed from the assembled genomes using PlasmidFinder [30] and the Virulence Factor Database (VFDB) [31], respectively, using Abricate (https://github.com/tseemann/abricate). Multi-locus sequence typing is performed using Mlst (https://github.com/tseemann/mlst).
Phylogenetic inference
Because of the computationally demanding requirements of algorithms in terms of RAM and core threads during phylogenetic analysis, rMAP incorporates the use of SNP-based analysis, which has been proven to be faster than using sequencing data to infer phylogeny. A single VCF file containing all the samples and their SNPs is generated towards the end the variant calling stage, which is transposed by vcf2phylip [32] into a multi-alignment fasta file. Multi-sequence alignment is performed using Mafft [33], with the removal of ambiguously aligned reads and the selection of informative regions to infer phylogeny using BMGE [34]. IQtree [14] tests various substitution models and constructs trees from the alignments using the maximum-likelihood method with 1000 bootstraps. The resulting trees are visualized in rectangular (phylogram), circular (phylogram) and circular (cladogram) forms.
Pangenome analysis
Roary [17] is employed by rMAP to perform core and accessory pangenome analysis across the input samples using general feature format (.gff) files generated from the annotation step. Fasttree is used to convert the core genome alignment to the newick format. The scalable vector graphic (SVG) file obtained from the pangenome analysis is converted to a portable network graphic (PNG) file format by cairosvg (https://cairosvg.org/). The resulting trees are visualized in rectangular (phylogram), circular (phylogram) and circular (cladogram) forms.
Insertion sequence (IS) analysis
rMAP interrogates for the presence of mobile genetic elements, in particular insertion sequences, using ISMapper [18], which basically spans the lengths of the entire genome of a sequence searching for homology against a set of well-known insertion sequence families commonly found in ESKAPE isolates [35] and the ISfinder database (https://www-is.biotoul.fr/index.php), as shown in Table 2.
Table 2.
ESKAPE group insertion sequence families (both Gram-positive and Gram-negative) used by rMAP
|
Sequence name |
Determinant genes |
Conferred resistance |
|---|---|---|
|
IS903 |
aphA1 |
Kanamycin |
|
ISApl1 |
mcr-1 |
Colistin |
|
ISEc69 |
mcr-2 |
Colistin |
|
ISAba14 |
aphA6 |
Kanamycin |
|
ISAba1 |
blaOXA-23 |
Carbapenems, beta-lactams |
|
IS16 |
VanB1 |
Vancomycin |
|
IS256 |
cfr |
Phenicols, lincosamides, oxazolidinones, pleuromutilins, streptogramin A |
|
IS257-2 |
aadD, ble, fosB5, fusB, tetL, tetK, aacA-aphD, vatA, dfrK |
Kanamycin, bleomycin, fosfomycin, fusidic acid, tetracycline, gentamicin, streptogramin A, trimethoprim |
|
IS1182 |
aadE, aphaA-3, sat4 |
Streptomycin, kanamycin, neomycin, streptothricin |
|
IS1216 |
cfr, str |
Phenicols, lincosamides, oxazolidinones, pleuromutilins, streptomycin, streptogramin A |
|
IS1272 |
mecR |
Methicillin |
|
IS1182 |
aphA-3, aadE |
Aminoglycoside |
|
ISEnfa4 |
cfr |
Phenicols, lincosamides, oxazolidinones, pleuromutilins, streptogramin A |
|
ISEcp1 |
CTX-M |
Cefotaxime, ceftriaxone, aztreonam |
|
ISSau1 |
SCCmec |
Methicillin |
|
ISKpn23 |
blaBKC-1 |
Carbapenems, cephalosporins, monobactams |
Results
Reporting and visualization of the reports
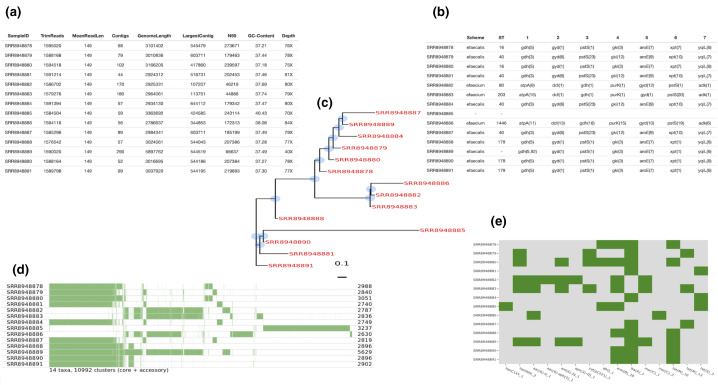
rMAP stores and formats reports from each stage of the pipeline under one directory called ‘reports’ and uses R-base [20] with a set of R packages, including ggtree [36], RcolorBrewer, ggplot2 [37], knitr [38], rmarkdown [39], plotly [40], reshape2 [41] and treeio [42], to generate a web-like html interactive report with explanations at every stage of analysis that can easily be shared and interpreted by inexperienced bioinformatics individuals. An example of such a report can be accessed via https://gunzivan28.github.io/rMAP/. The reporting format for rMAP was mainly adapted from the Tormes [6] pipeline. The results from a successful run can be found under the user-defined output directory and consist of files from assembly, annotation, insertion sequences, mlsts, pangenomes, phylogeny, plasmids, quality reports, quast assembly stats, reports, resistance genes, trimmed reads, variant calling and virulence factors for further analysis. rMAP retains all of the intermediate files generated after a successful run to be interrogated further by experienced bioinformatics users. The contents extracted from the intermediate files and summarized in the html report with a short description are summarized in Table 3. Examples of visuals generated by the pipeline are illustrated in Fig. 2.
Table 3.
Summary of some stages of intermediate files generated from rMAP
|
Analysis |
Metrics |
Description |
|---|---|---|
|
Assembly |
Genome length, average genome length, N50, GC content and sequencing depth |
|
|
Phylogeny |
|
|
|
Variant calling |
SNPs |
|
|
Antimicrobial resistance profiling |
Contig, gene, identity, product |
|
|
Pangenome analysis |
Core genes, soft core genes, shell genes, cloud genes |
|
Fig. 2.
Selected interactive Enterococcus species HTML reports. (a) Genome assembly summary statistics for the different Enterococcus species isolates. These include common genome analysis key metrics for checking assembly quality. (b) Table of multi-locus sequence typing (MLST) distribution. (c) SNP-based approximately maximum-likelihood phylogenetic tree. Three different formats are available, i.e circular (phylogram), circular (cladogram) and rectangular (phylogram). An approximately maximum-likelihood phylogenetic tree is computed based on SNPs detected via read mapping against a reference genome and stored in a standard Newick file format. (d) Pangenome analysis including a schematic representation of gene presence (colour) or absence (blank) between samples. (e) Antibiotic resistance profile. Presence/absence of antibiotic resistance genes (coverage and identity >90 %) for each sample. An antibiotic resistance profile is computed based on Resfinder, CARD, ARG-ANNOT, NCBI and MEGARES annotations for each isolate and transformed into an overview that allows a rapid resistome comparison of all analysed isolates.
Computational infrastructure and benchmarking
The original philosophy of creating rMAP was to create a tool that can be easily installed and run on a descent personal computer. The pipeline was successfully compiled on two personal computers with the following specifications: Dell Inspiron 5570 8th Gen Intel Core i7-8550U CPU @1.80 GHz (8 CPUs), ~2.0 GHz with 12 GB of RAM and 1 TB of hard disk space running Windows subsystem Linux (WSL) Ubuntu 20.04.1 LTS (Focal Fossa) and Ubuntu 18.04.4 LTS (Bionic Beaver) and a MacBook pro Intel Core i7 CPU @3.0 GHz, 16 GB of RAM and 2 TB of SSD space running on macOS Mojave. When provided with the same samples, the MacBook performed better because of its hardware compared to Ubuntu. Depending on the number of samples provided in the input, rMAP generates intermediate files ranging between 10 and 30 GB. The wall clock runtimes and benchmarking statistics for each bacterial species on different platforms are summarized in Table 4.
Table 4.
rMAP’s wall clock runtimes for different bacterial species across different operating system platforms
|
Genomes |
Genome size |
Ubuntu |
macOS Mojave |
|---|---|---|---|
|
~2.9 Mbp |
22 h |
18 h |
|
|
~3.9 Mbp |
22 h |
19 h |
|
|
14 Enterococcus spp. |
~2.9 Mbp |
21 h |
17 h |
Discussion
Although other pipelines developed under the same philosophy and functionality as rMAP, such as Tormes [6], ASA3P [43] and the recently published Bactopia [44], exist, we noticed that each of these had a shortcoming that we aimed to address. In terms of usability, Tormes [6] was the most friendly pipeline, with one major drawback, where it could never be launched without a tab-separated metadata file complying with a set criteria. It was also more oriented to bacterial species-specific analyses, namely Escherichia coli and Salmonella species. ASA3P [43] and Bactopia [44] required a bioinformatics-competent user for operation, since they are written in complex languages, namely, Groovy and Nextflow, respectively. Other similar pipelines, such as Nullarbor (https://github.com/tseemann/nullarbor), were extremely difficult to compile and use compared to their counterparts, requiring a metadata file conforming with set criteria. In cases where metadata files are required, the different software flagged errors or halted task executions as the correct conforming metadata files were essential for the downstream analyses.
rMAP, on the other hand, comes with features aimed at overcoming the limitations of its counterparts. It requires no prior preprocessing of the sequences or metadata files. The user only provides three essential requirements, namely, an input directory, an output directory and a reference genome to run the pipeline. The pipeline is written in basic programming languages that do not require advanced expertise or troubleshooting to be launched. rMAP is highly portable and capable of operating on decent personal computers running on either Ubuntu or macOS. Installation is quite easy and straightforward from the GitHub repository (https://github.com/GunzIvan28/rMAP), with the binaries and dependences built within conda environment packages. Most of all, rMAP shows a high sensitivity towards analysis and is not limited to the ESKAPE group pathogens, but also applies to other Enterobacteria, such as E. coli and Salmonella species.
As a significant limitation, rMAP is coded exclusively in Bash and is not implemented within a modern workflow language manager, such as Snakemake or Nextflow. The ultimate consequence of this is that a user will have to either restart the whole run or manually check which steps have completed successfully and resume the run by only selecting options that were not performed while excluding the computed steps from the main command script. Implementation of the pipeline within a modern workflow language will feature in the next release of the software.
Conclusion
rMAP is a robust, scalable, user-friendly, automated bioinformatics analysis workflow for Illumina WGS reads that has demonstrated efficiency in the analysis of public health-significant pathogens. Therefore, we recommend it as a tool for continuous monitoring and surveillance that is suitable for assessing antimicrobial resistance gene trends, especially in low-income countries with limited computational bioinformatics infrastructure.
Availability and future directions
The source code is available on GitHub under a GPL3 licence at https://github.com/GunzIvan28/rMAP. Questions and issues can be sent to ivangunz23@gmail.com and bug reports can be filed as GitHub issues. Although rMAP itself is published and distributed under a GPL3 licence, some of its dependences bundled within the rMAP volume are published under different licence models.
Funding information
This work was supported through the Grand Challenges Africa programme (GCA/AMR/rnd2/058). Grand Challenges Africa is a programme of the African Academy of Sciences (AAS) implemented through the Alliance for Accelerating Excellence in Science in Africa (AESA) platform, an initiative of the AAS and the African Union Development Agency (AUDA-NEPAD). GC Africa is supported by the Bill and Melinda Gates Foundation (BMGF) and The African Academy of Sciences and partners. The views expressed herein are those of the authors and not necessarily those of the AAS and its partners. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgements
I. S. is a current trainee of the Makerere University/Uganda Virus Research Institute (UVRI) Centre of Excellence for Infection and Immunity Research and Training (MUII) programme. He is also supported by MUII. MUII is supported through the DELTAS Africa Initiative (grant no. 107743). The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS), Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency), with funding from the Wellcome Trust (grant no. 107743) and the UK Government. We are equally grateful for the support of NIH Common Fund, through the OD/Office of Strategic Coordination (OSC) and the Fogarty International Center (FIC), NIH award number U2R TW 010672. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the supporting offices. We also acknowledge all the different authors for the various open source code for inspiring the conception and design of this pipeline. Special thanks go to Narciso Quijada, Torsten Seeman, Oliver Schwengers and Robert Pettit. We express our gratitude to the NCBI and ENA for providing the dataset resources used in development of this tool.
Author contributions
Both authors contributed equally in conceptualization; formal analysis; funding acquisition; methodology; resources; software; validation; visualization; writing – original draft; writing – review and editing.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
No ethics permission was required for work. All data used were anonymized by the submitting authors/institutions.
Footnotes
Abbreviations: AMR, antimicrobial resistance; ESKAPE, Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species; IS, insertion sequence; MLST, multi-locus sequence type; rMAP, rapid microbial analysis pipeline; SNP, single nucleotide polymorphism; VCF, variant call format; WGS, whole-genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files.
References
- 1.Sserwadda I, Lukenge M, Mwambi B, Mboowa G, Walusimbi A, et al. Microbial contaminants isolated from items and work surfaces in the post- operative ward at Kawolo General Hospital, Uganda. BMC Infect Dis. 2018;18:68. doi: 10.1186/s12879-018-2980-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mulani MS, Kamble EE, Kumkar SN, Tawre MS, Pardesi KR. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: a review. Front Microbiol. 2019;10 doi: 10.3389/fmicb.2019.00539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma Y-X, Wang C-Y, Li Y-Y, Li J, Wan Q-Q, Chen J-H, et al. Considerations and caveats in combating ESKAPE pathogens against nosocomial infections. Adv Sci. 2020;7:1901872. doi: 10.1002/advs.201901872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carriço JA, Rossi M, Moran-Gilad J, Van Domselaar G, Ramirez M. A primer on microbial bioinformatics for nonbioinformaticians. Clin Microbiol Infect. 2018;24:342–349. doi: 10.1016/j.cmi.2017.12.015. [DOI] [PubMed] [Google Scholar]
- 5.Hyeon J-Y, Li S, Mann DA, Zhang S, Li Z, et al. Quasimetagenomics-based and real-time-sequencing-aided detection and subtyping of Salmonella enterica from food samples. Appl Environ Microbiol. 2018;84:e02340–17. doi: 10.1128/AEM.02340-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quijada NM, Rodríguez-Lázaro D, Eiros JM, Hernández M. TORMES: an automated pipeline for whole bacterial genome analysis. Bioinformatics. 2019;35:4207–4212. doi: 10.1093/bioinformatics/btz220. [DOI] [PubMed] [Google Scholar]
- 7.Land M, Hauser L, Jun S-R, Nookaew I, Leuze MR, et al. Insights from 20 years of bacterial genome sequencing. Funct Integr Genomics. 2015;15:141–161. doi: 10.1007/s10142-015-0433-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrews S. FastQC: a Quality Control Tool for High Throughput Sequence Data. Cambridge, UK: Babraham Bioinformatics, Babraham Institute; 2010. [Google Scholar]
- 9.Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 12.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 13.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. The sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31::3691–:3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawkey J, Hamidian M, Wick RR, Edwards DJ, Billman-Jacobe H, et al. ISMapper: identifying transposase insertion sites in bacterial genomes from short read sequence data. BMC Genomics. 2015;16:667. doi: 10.1186/s12864-015-1860-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossum G. Python reference manual. 1995.
- 20.Ihaka R, RJJoc G. Statistics G. R: a language for data analysis and graphics. 1996;5:299–314. [Google Scholar]
- 21.ncbi/sra-tools NCBI - National Center for Biotechnology Information/NLM/NIH. 2020.
- 22.Seemann T. Tseemann/shovill. 2020 https://github.com/tseemann/shovill
- 23.Hyatt D, Chen G-. L, LoCascio PF, Land ML, Larimer FW, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv:12073907 [q-bio] 2012 [Google Scholar]
- 25.Tan A, Abecasis GR, Kang H. Unified representation of genetic variants. Bioinformatics. 2015;31:2202–2204. doi: 10.1093/bioinformatics/btv112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feldgarden M, Brover V, Haft DH, Prasad AB, Slotta DJ, et al. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother. 2019;63:e00483–19. doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother. 2014;58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doster E, Lakin SM, Dean CJ, Wolfe C, Young JG, et al. MEGARes 2.0: a database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic Acids Res. 2019;48::D561–D9. doi: 10.1093/nar/gkz1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carattoli A, Zankari E, García-Fernández A, Larsen MV, Lund O, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu B, Zheng D, Jin Q, Chen L, Yang JJ. Nar. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019;47 doi: 10.1093/nar/gky1080. D687–D692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ortiz E. vcf2phylip V1. 5: convert a VCF matrix into several matrix formats for phylogenetic analysis. Zenodo. 2018 [Google Scholar]
- 33.Katoh K, Standley DMJMb, evolution MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Criscuolo A, Gribaldo S. BMGE (block mapping and Gathering with entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Partridge SR, Kwong SM, Firth N, Jensen SO, SOJCmr J. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev. 2018;31 doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 2017;8:28–36. [Google Scholar]
- 37.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer; 2016. p. 266. p. [Google Scholar]
- 38.Xie Y. Dynamic documents with R and knitr. CRC Press; 2015. [Google Scholar]
- 39.Grolemund G, Allaire JJ, Xie Y. R Markdown: the definitive guide. 2018.
- 40.Plotly Plotly R Graphing library. https://plotly.com/r/
- 41.Wickham H. Reshaping data with the reshape package. J Stat Softw. 2007;21:1–20. doi: 10.18637/jss.v021.i12. [DOI] [Google Scholar]
- 42.Yu G, Smith DK, Zhu H, Guan Y, Lam TTY. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 2017;8:28–36. [Google Scholar]
- 43.Schwengers O, Hoek A, Fritzenwanker M, Falgenhauer L, Hain T, et al. ASA3P: an automatic and scalable pipeline for the assembly, annotation and higher-level analysis of closely related bacterial isolates. PLoS Comput Biol. 2020;16:e1007134. doi: 10.1371/journal.pcbi.1007134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petit RA, Read TD. Bactopia: a flexible pipeline for complete analysis of bacterial genomes. bioRxiv. 2020:2020.02.28.969394 doi: 10.1128/mSystems.00190-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Plotly Plotly R Graphing library. https://plotly.com/r/