Abstract
Sinomenine is a pure alkaloid that can be isolated from the root of Sinomenium acutum and has been found to exert anti-inflammatory and immunosuppressive effects. The present study investigated the effects of sinomenine hydrochloride (SIN) on inflammation and the gut microbiota composition in the colon of mouse models of dextran sulfate sodium (DSS)-induced colitis. DSS-induced mice colitis was established by treating the mice with drinking water containing 3% (w/v) DSS for 7 days. The disease activity index of each mouse was calculated on a daily basis. All mice were sacrificed on day 11, then the weight of their spleen and length of their colons were measured. The histological analysis was measured by hematoxylin-eosin staining. Oral administration of SIN (100 mg/kg/day) attenuated the DSS-induced increases in the disease activity indices and spleen indices, DSS-induced shortening of the colon length and histological damage. In addition, reverse transcription-quantitative PCR data showed that SIN treatment effectively regulated the expression of inflammatory mediators, specifically by suppressing the expression of proinflammatory gene (TNF-α, IL-6 and inducible nitric oxide synthase) whilst increasing those associated with inhibiting inflammation (IL-10 and arginine 1). Gut microbiota analysis was conducted using 16S ribosomal DNA sequencing. The results revealed that SIN improved bacterial community homeostasis and diversity, which were damaged by DSS. Furthermore, western blotting showed that the activation of the NOD-, LRR- and pyrin domain-containing protein 3 (NLRP3) inflammasome was markedly suppressed by SIN treatment. In conclusion, these results indicated that SIN may ameliorate experimental colitis by modulating the gut microbiota composition and suppressing the activation of the NLRP3 inflammasome in mice. Overall, these findings suggested a broad protective effect of SIN in treating inflammatory gut diseases, including ulcerative colitis.
Keywords: Sinomenine hydrochloride, ulcerative colitis, dextran sulfate sodium-induced colitis, gut microbiota composition, NOD-, LRR- and pyrin domain-containing protein 3 inflammasome
Introduction
Ulcerative colitis (UC) is a complex, chronic and non-specific inflammatory condition of the gastrointestinal tract (1). It is one of the most common subtypes of inflammatory bowel disease (2). The etiology of UC remains unclear, although it is considered to involve genetic, environmental and immunological factors, all of which contribute to the disease at least in part by interacting with the gut microbiota (3,4). In particular, the composition and function of the gut microbiota have been increasingly reported to influence the immune system and physiological functions in the colon (5). In fact, treatment options that are currently available for UC primarily focus on preventing colonic inflammation by restoring the composition of the gut microbiota (6,7).
The NOD-, LRR- and pyrin domain-containing protein 3 (NLRP3) inflammasome is a multi-protein complex comprising NLRP3, PYD and CARD domains that contain pro-caspase-1, which has been found to serve a role in UC (8). The NLRP3 inflammasome is stimulated by a diverse series of agonists (such as ATP and UV radiation, and pattern-associated molecular patterns derived from bacteria, virus, fungus, and protozoa) (9), before activates caspase-1, which results in the maturation of the proinflammatory cytokine IL-1β (10). It was previously reported that the activation of NLRP3 inflammasome served an inflammatory role in dextran sulfate sodium (DSS)-induced colitis (11).
Sinomenine (CAS: 115-53-7) is a pure alkaloid that was originally extracted from the Chinese herb, Sinomenium acutum (Rehder and Wilson) of the Menispermaceae family (12). Sinomenine hydrochloride (SIN) is a hydrochloride form of sinomenine that has attracted the interest of researchers due to its reported safety and potent anti-inflammatory and immunoregulatory effects both in vitro and in vivo (13,14). It has been reported that SIN demonstrated notable therapeutic efficacy for treating collagen-induced arthritis in a rat model (15). However, little is currently known of the potential effects of SIN on human UC. To date, several sinomenine derivatives or Sinomenium acutum extracts have been applied to treat rheumatoid arthritis in the clinic, such as Sinomenone hydrochloride enteric-coated tablets (16). In addition, several pharmacokinetic studies of sinomenine have been performed, which suggested that sinomenine can be metabolized in humans and rats in vivo (17,18). In addition, sinomenine can be metabolized by human microsomal proteins in vitro (17,18). Liu et al (19) previously found that oral administration of sinomenine may be applicable for future clinical studies, because at a dosage of 90 mg/kg in rats, it achieved ~80% in terms of bioavailability. A previous study also demonstrated that SIN alleviated dextran sulfate sodium (DSS)-induced colitis in mice, by decreasing the levels of oxidative stress in thecolon (20). However, to the best of our knowledge, whether SIN exerts effects on the composition of the gut microbiota remains to be investigated.
UC has been reported to be partially caused by disruptions in the balance between the immune system and the gut microbiota (21). Consequently, the present study aimed to assess the effects of SIN on colonic inflammation and the composition of gut microbiota in a mouse model of DSS-induced colitis. The present results may provide novel insights into the possible application of SIN as a therapeutic agent for regulating microbial homeostasis in UC.
Materials and methods
Reagents
SIN (cat. no. S105956) was purchased from Shanghai Aladdin Biochemical Technology Co., Ltd. and was dissolved in 0.9% NaCl. DSS (cat. no. 0216011080; molecular weight, 36,000-50,000 Da) was purchased from MP Biomedicals, LLC.
Animals and study design
In total, 15 female C57BL/6 mice (age, 6-8 weeks; body weight, 18-22 g) were purchased from Cavens Laboratory Animal Co., Ltd. (Changzhou, China). All animal experiments were approved by the Ethics Committee of Changzhou No. 2 People's Hospital [approval no. SCXK (SU) 2016-0010; Changzhou, China] and animal protocols were strictly conducted in accordance with the Animal Research: Reporting of in vivo Experiments guidelines (22). All mice were housed in pathogen-free animal facilities under a standard 12-h light/dark cycle at 25˚C and 40-50% humidity. All mice were group-housed in a standard rodent unit with free access to food and water.
The 15 animals were randomized into the following three groups (5 mice per group): i) Control; ii) DSS model; and iii) DSS + SIN (100 mg/kg) groups. Colitis was not induced in the control group and these mice were fed with standard food and water during the whole experiment. Mice in the DSS and DSS + SIN groups were treated with 3% DSS, which was supplemented in drinking water, for 7 days and then provided with normal drinking water for 3 days for recovery (23). By contrast, mice in the DSS + SIN group were provided with 100 mg/kg SIN by oral gavage from days 1 to 10 daily. On day 11, all the mice were sacrificed by CO2 suffocation. The air replacement rate of CO2 was 10-30% of the container volume per min, which lasted for ~5 min.
During the experiment, body weight, stool consistency and fecal bleeding scores of each animal were observed to calculate the disease activity index (DAI) (15,24) every day. The scoring system for the DAI is presented in Table I. Following sacrifice, the spleens were obtained from each mice and weighed. Spleen index (%)=Spleen weight (mg)/body weight on day 11 (g) x10. In addition, after the mice were sacrificed, the colons were removed from each mouse and the colon length was measured. The dissected colon tissues were then washed with cold PBS before one section of the distal colon was stored at -80˚C for subsequent biochemical examination. The remaining tissue was fixed in 4% paraformaldehyde at room temperature for 24 h for further histopathological analysis.
Table I.
Disease activity index scoring systema.
| Score | Weight loss (%)b | Stool consistency | Blood in stool |
|---|---|---|---|
| 0 | None | Well-formed stools | Negative |
| 1 | 1-5% | Well-formed stools | Negative |
| 2 | 6-10% | Pasty stools that did not adhere to the anus | Hemoccult positive |
| 3 | 11-15% | Pasty stools that did not adhere to the anus | Hemoccult positive |
| 4 | >15% | Liquid stools that adhered to the anus | Gross bleeding |
aThe disease activity index score=the sum of the scores for weight loss, stool consistency and blood in stool/3.
bBody weight change (%)=[(Weight change at day X)-(Weight at day 1)]/(Weight at day 1) x100.
Histological analysis
The fixed colon sections were dehydrated with an ascending ethanol gradient (70-100%) and embedded in paraffin before 4-µm colon sections were obtained and stained with the Hematoxylin-Eosin Staining kit (cat. no. G1120; Beijing Solarbio Science & Technology Co., Ltd.) according to the manufacturer's protocol (25). Briefly, the sections were deparaffinized in xylene at room temperature, rehydrated with a decreasing concentration of ethanol (100-70%) and washed with distilled water. The sections were incubated with hematoxylin solution for 10 min and eosin solution for 2 min, both at room temperature before being washed with water. The stained sections were dehydrated in an ascending ethanol gradient (90-100%), cleared with xylene and observed using a light microscope (magnifications, x40, x100 and x200; Olympus BX51; Olympus Corporation).
Measurement of colonic inflammatory cytokines
The mRNA expression levels of TNF-α, IL-6, IL-10, inducible nitric oxide synthase (iNOS) and arginase 1 (Arg1) in the colon tissues of mice were determined using reverse transcription-quantitative PCR (RT-qPCR). Total RNA was extracted from colon tissues using a RNAsimple Total RNA kit (cat. no. DP419; Tiangen Biotech Co., Ltd.) according to manufacturer's protocol. Total RNA was reverse transcribed into cDNA using a PrimeScript™ RT reagent kit with gDNA Eraser (Perfect Real-time; cat. no. RR047A; Takara Bio, Inc.) at 37˚C for 5 min and 85˚C for 5 sec before the samples were cooled to 4˚C. qPCR was subsequently performed using an AceQ® qPCR SYBR® Green Master mix (Low ROX Premixed; cat. no. Q131-02; Vazyme Biotech Co., Ltd.) on an ABI 7500 Real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The following thermocycling conditions were used for the qPCR: Initial denaturation at 95˚C for 5 min; followed by 40 cycles at 95˚C for 10 sec and 60˚C for 30 sec. The fold changes in the expression levels of each gene were calculated using the 2-ΔΔCq method (26). The mRNA expression levels for each target gene were normalized to the expression levels of GAPDH. The primers used for the qPCR are listed in Table II.
Table II.
Primer sequences for reverse transcription-PCR.
| Gene | Sequences (5'-3') |
|---|---|
| TNF-α | F: CATCTTCTCAAAATTCGAGTGAC |
| R: TGGGAGTAGACAAGGTACAACCC | |
| IL-6 | F: GCTGGTGACAACCACGGCCT |
| R: AGCCTCGACTTGTGAAGTGGT | |
| IL-10 | F: AGATGATGACCCTTTTGGCTC |
| R: AGATGATGACCCTTTTGGCTC | |
| iNOS | F: CCAACCTGCAGGTCTTCGATG |
| R: GTCGATGCACAACTGGGTGAAC | |
| Arg 1 | F: GAACCCAACTCTTGGGAAGAC |
| R: GGAGAAGGCGTTTGCTTAGTT | |
| GAPDH | F: AAGGTCGGAGTCAACGGATTT |
| R: AGATGATGACCCTTTTGGCTC |
iNOS, inducible nitric oxide synthase; Arg1, arginine 1.
Gut microbiota 16S ribosomal (r)DNA analysis
Murine fecal samples were collected between days 8 and 11 and then stored at -80˚C for use in subsequent experiments. Bacterial DNA was isolated from the samples using an E.Z.N.A.™ stool DNA kit (cat. no. D5625-01; Omega Bio-Tek, Inc.) according to the manufacturer's protocol. Determination of DNA quality and 16S rDNA analysis were conducted by Guangzhou RiboBio Co., Ltd. DNA integrity was detected by 1% agarose gel electrophoresis, and was amplified using NEBNext® Ultra™ II Q5 Master Mix (cat. no. M0544L 250rxn; New England Biolabs, Inc.) for 15 cycles with a specific primer set containing the RiboBio® barcode (Guangzhou RiboBio Co., Ltd.). The products were verified using an Agilent 2200 Bioanalyzer (Agilent Technologies, Inc.). Bacterial 16S rRNA gene sequences (V3-V4 region) were performed using a MiSeq Reagent Kit v3 (600 cycle) (cat. no. MS-102-3003; Illumina, Inc.) on a Illumina HiSeq 2500 platform (Illumina Inc.) and 250 bp paired-end reads were obtained. The DNA loading concentration was 10 pmol, which measured using Qubit 2.0. The paired-end reads were merged using Fast Length Adjustment of SHort reads (27) and then assigned to each sample based on their unique barcodes conducted by QIIME software (version 1.7; QIIME development team; https://qiime.org/). Within-sample microbiota diversity (α-diversity) was evaluated using Chao1 and Shannon (28), which measured gut microbiota richness and evenness. Additional α-diversity assessment was performed with PD Whole Tree, which measure species phylogeny (29). All α-diversity indices were calculated with QIIME (version 1.7; QIIME development team). Principal component analysis (PCA) and principal coordinate analysis (PCoA) was measured at the operational taxonomic units (OTU) level and hierarchical clustering tree on Genus level to compare the differences among microbial communities (30). The community structure was based on the weighted UniFrac distance (31). In this study, the OTUs were clustered with a similarity cutoff value of 97% using UPARSE software (version 7.0) (32).
Western blotting
Total protein was extracted from colon samples using Western & IP cell lysis buffer (cat. no. P0013; Beyotime Institute of Biotechnology) supplemented with the PMSF protease inhibitor (Beyotime Institute of Biotechnology) according to the manufacturer's protocol. Protein concentration was determined using an Enhanced BCA Protein Assay kit (cat. no. P0010; Beyotime Institute of Biotechnology). Protein extracts were stored at -70˚C until required for further experimentation. Proteins (40 µg/lane) were separated via 12% SDS-PAGE, transferred onto PVDF membranes (cat. no. IPVH00010; EMD Millipore) and blocked using blocking buffer (cat. no. P0023B-100 ml; Beyotime Institute of Biotechnology) for 2 h at 37˚C. The membranes were then incubated with the following primary antibodies overnight at 4˚C: Anti-NLRP3 (1:1,000; cat. no. ab210491; rabbit monoclonal; Abcam), anti-caspase-1 (1:1,000; cat. no. ab207802; rabbit monoclonal; Abcam) and anti-β-actin (1:1,000; cat. no. 4970T; rabbit monoclonal; Cell Signaling Technology, Inc.). Following primary antibody incubation, the membranes were washed with TBS-Tween 20 (0.05%) and incubated with an HRP-conjugated Affini-pure goat anti-rabbit IgG (H + L) secondary antibody (1:7,500; cat. no. BA1054; Boster Biological Technology) for 1.5 h at room temperature. All antibodies were diluted in the primary antibody dilution buffer (cat. no. P0023A-100 ml; Beyotime Institute of Biotechnology). Protein bands were visualized using SuperSignal West Pico Chemiluminescent substrate (cat. no. 34077; Thermo Fisher Scientific, Inc.) and immunoblots were quantified using ImageJ version 1.8 software (National Institutes of Health).
Statistical analysis
All data are presented as the mean ± SEM. Statistical differences among groups were determined using Kruskal-Wallis followed by Dunn's test, one-way ANOVA and Sidak's multiple comparison test with GraphPad Prism 5 software (GraphPad Software, Inc.). P≤0.05 was considered to indicate a statistically significant difference.
Results
SIN alleviates DSS-induced colitis in mice
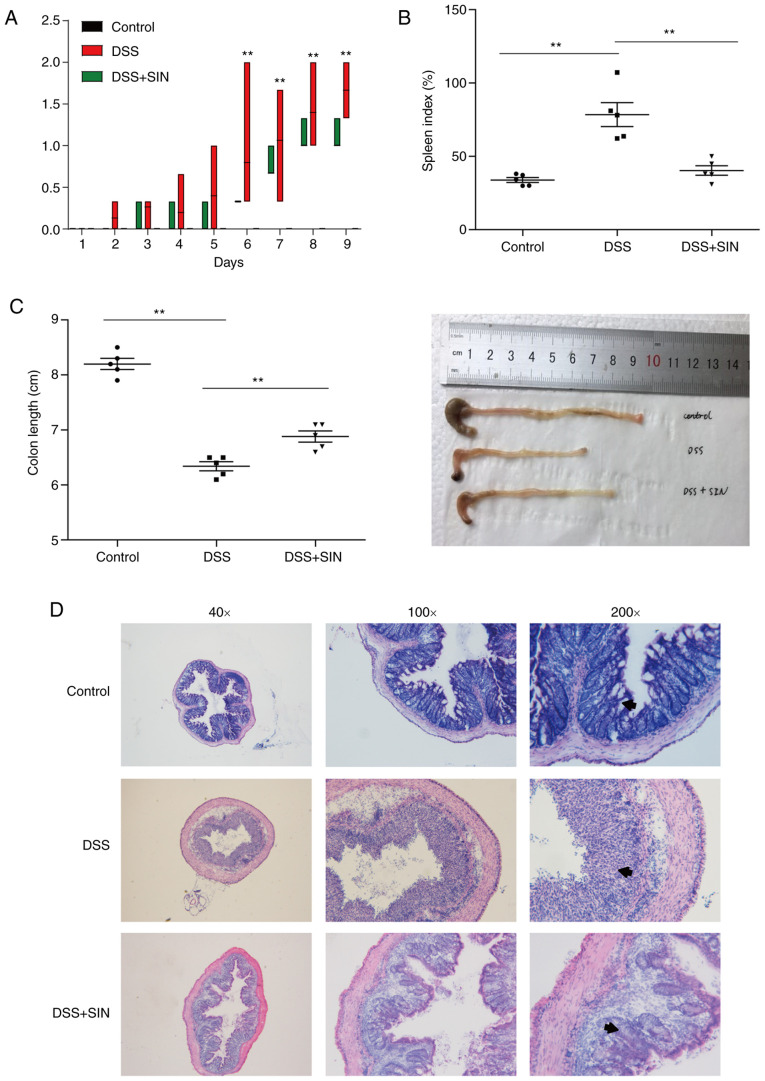
Mice treated with 3% DSS developed severe colitis, which exhibited significantly higher DAI scores compared with those in control mice from day 6 (P<0.01; Fig. 1A). Notably, the DAI score of DSS + SIN group was decreased compared with that in the DSS group from day 4, though there was no statistical difference (Fig. 1A). SIN supplementation significantly reduced the DAI scores spleen indices compared with those in the untreated DSS-induced mice (Fig. 1B). DSS-induced colonic shortening was also significantly improved by SIN administration (Fig. 1C). Pathologically, the colons of mice in the control group had an intact structure (Fig. 1D). As the arrows indicated, gland defects, mucosal ulcerations and inflammatory cell infiltration were observed in the colons of DSS-induced mice, whilst SIN supplementation attenuated this histological damage (Fig. 1D).
Figure 1.
SIN alleviates DSS-induced colitis in mice. (A) Disease activity index. **P<0.01, DSS vs. control. (B) Spleen index and (C) colon length. The data are presented as the mean ± SEM. **P<0.01 as indicated. (D) Hematoxylin and eosin staining. DAI, disease activity index; DSS, dextran sulfate sodium; SIN, sinomenine hydrochloride.
SIN regulates the expression levels of inflammatory cytokines in the colon
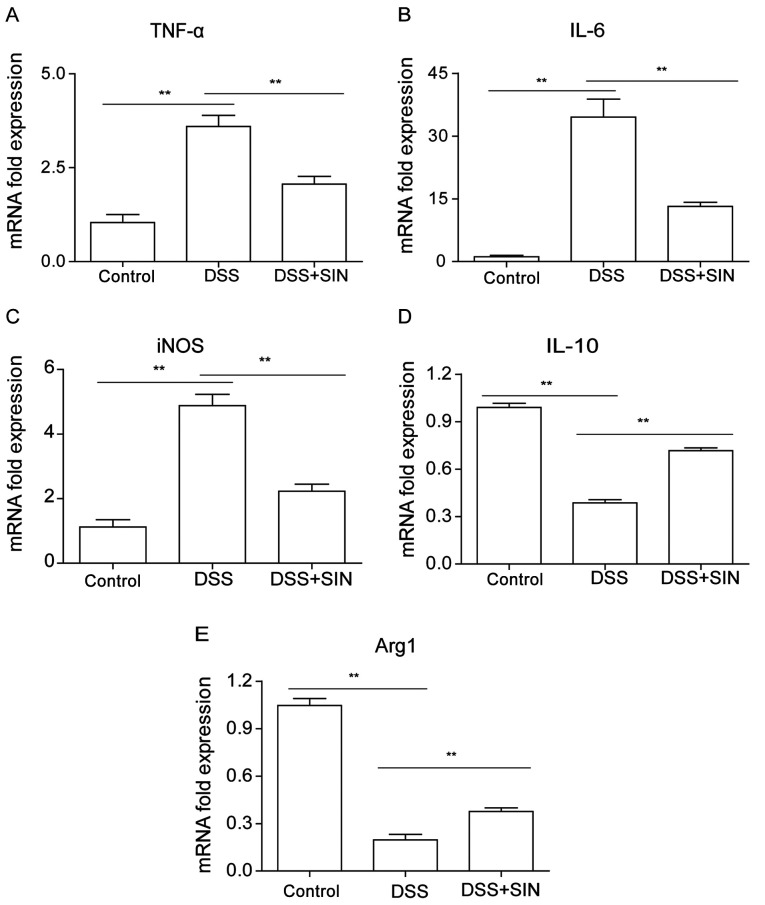
As presented in Fig. 2A-C, the mRNA expression levels of TNF-α, IL-6 and iNOS were found to be significantly upregulated in the colon tissues of mice treated with DSS, whilst the combined treatment of DSS with SIN significantly suppressed the increased expression. DSS treatment significantly downregulated IL-10 and Arg1 mRNA expression levels in the colon compared with those in the control group, which were significantly reversed by SIN treatment (Fig. 2D and E).
Figure 2.
Effect of SIN on the mRNA expression of inflammatory cytokines in mice with DSS-induced colitis. Reverse transcription-quantitative PCR was performed to analyze the expression levels of (A) TNF-α, (B) IL-6, (C) iNOS, (D) IL-10 and (E) Arg1 in colon tissues. Data are presented as the mean ± SEM. **P<0.01 as indicated. Arg1, arginine 1; DSS, dextran sulfate sodium; iNOS, inducible nitric oxide synthase; SIN, sinomenine hydrochloride.
SIN moderates the gut microbiota composition in the colons of mice
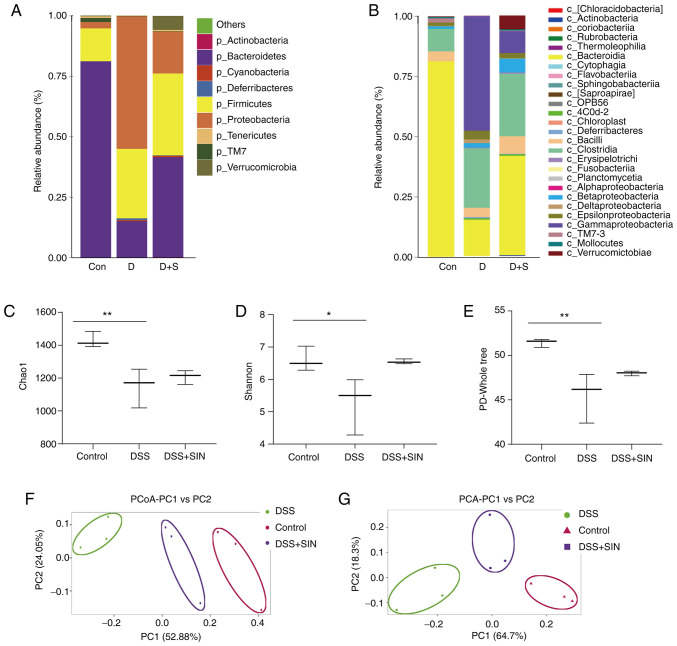
The species abundance and community composition of the gut microbiota in mice from the different treatment groups were analyzed using 16S rDNA sequencing. On the phylum level, the relative abundance of Bacteroidetes in mice was markedly reduced following DSS treatment, which was partially reversed following SIN treatment (Fig. 3A). Conversely, the relative abundance of Firmicutes and Proteobacteria was increased in DSS group compared with that in the control group (Fig. 3A). In the DSS + SIN group, the abundance of Proteobacteria was markedly decreased compared with that in the DSS-only group (Fig. 3A). On a class level, Bacteroidia, Clostridia and γ-proteobacteria were the dominant species present (Fig. 3B). Following DSS treatment, the abundance of Bacteroidia was decreased, whereas the abundance of Clostridia and γ-proteobacteria was increased compared with that in the control group (Fig. 3B). SIN treatment markedly reduced the abundance of γ-proteobacteria whilst increasing the abundance of Bacteroidia compared with that in the DSS-only group. However, no differences were observed in the abundance of Clostridia between the DSS and DSS + SIN groups (Fig. 3B).
Figure 3.
Analysis of gut microbiota composition and diversity. (A) Phylum level microbial changes. (B) Class level microbial changes. Microbiome diversity differences based on the (C) Chao1, (D) Shannon and (E) PD-Whole tree. (F) PCoA and (G) PCA of gut microbial communities. *P<0.05 and **P<0.01 as indicated. PCoA, principal coordinate analysis; PCA, principal component analysis; DSS, dextran sulfate sodium; SIN, sinomenine hydrochloride; PC, principal component.
Subsequently, comparisons of the α-diversity of different groups were performed using the Chao1(29), Shannon (33) and PD-Whole tree indices (30). DSS treatment significantly decreased the α-diversity of gut microbiota community compared with that in the control group (Fig. 3C-E). Although those in the DSS + SIN group exhibited markedly increased Shannon and PD-Whole tree indices, the differences were not significant (Fig. 3D and E). A marked difference was identified in the microbial β-diversity based on principal coordinate and principal component analyzes between the DSS and control groups, whilst the microbial β-diversity in the DSS + SIN group appeared to be closer to that of the control group (Fig. 3F and G).
SIN inhibits the activation of the NLRP3 inflammasome
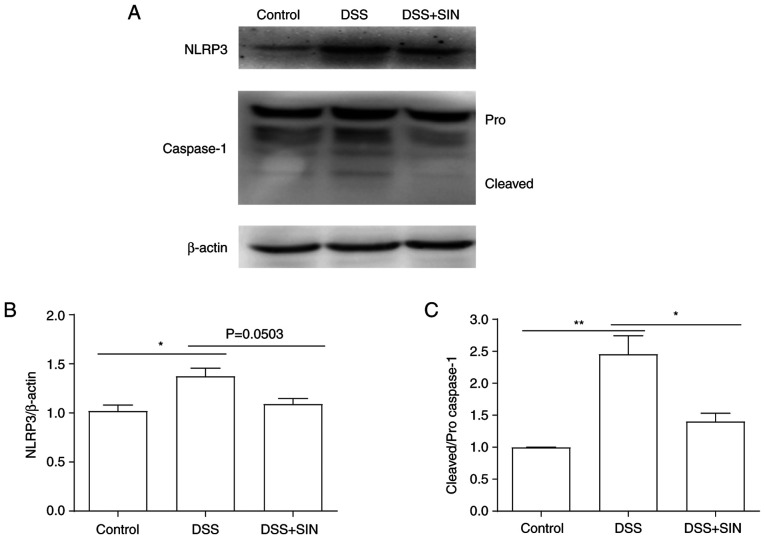
The results in Fig. 4 revealed that activation of the NLRP3 inflammasome and cleaved caspase-1 expression levels were significantly increased following DSS treatment, whilst treatment with SIN markedly decreased both the activation of the NLRP3 inflammasome and downregulated the expression levels of cleaved caspase-1. However, no significance was observed in NLRP3 expression between the DSS and DSS + SIN groups. The expression levels of pro-caspase-1 remained unchanged following the different treatments (Fig. 4).
Figure 4.
SIN inhibits DSS-induced NLRP3 inflammasome activation. (A) Expression levels of NLRP3 and caspase-1 in colon tissues were analyzed using western blotting. The quantified protein ratios of NLRP3:β-actin and cleaved caspase-1:pro-caspase-1 were analyzed and presented in (B) and (C), respectively. *P<0.05 and **P<0.01 as indicated. DSS, dextran sulfate sodium; NLRP3, NOD-, LRR- and pyrin domain-containing protein 3; SIN, sinomenine hydrochloride.
Discussion
Anti-inflammatory and immunosuppressive drugs are commonly used for treating UC in the clinic, with examples including glucocorticoids, azathioprine, biological agents, janus kinase inhibitors (TNF antibodies, ustekinumab, vedolizumab, and tofacitinib) and calcineurin inhibitors (34). However, these drugs can cause numerous side effects, such as Sweet's syndrome (35). Traditional Chinese herbal medicine has attracted interest due to its reported effects in preventing and controlling digestive diseases (36,37). Park et al (36) reported that pristimerin attenuated tumorigenesis in a mouse model of experimental colitis-associated colon cancer. In another study, Zheng et al (37) revealed that silibinin administration ameliorated colitis and inhibited colitis-associated tumorigenesis in a mouse model. Sinomenine can be extracted from the herb Sinomenium acutum, such that SIN is its form of hydrochloride (12). Given that there were few reports about the effects of SIN on diseases in the digestive system (20,38), the present study aimed to determine the possible effects and underlying mechanism of SIN on a mouse model of DSS-induced colitis, which has been previously found to successfully mimic the clinical symptoms of UC (39). Consistent with the findings of previous studie (20,38), the results of the present study revealed that the DAI and spleen indices were markedly increased, and colon length was shortened in DSS-treated mice compared with those in the control group. Notably, SIN treatment significantly attenuated the DSS-induced effects on the DAI, spleen index and colon length. Histological analysis revealed that SIN prevented crypt destruction and inflammatory cell infiltration caused by DSS treatment. These findings suggest that SIN may exert protective effects against UC in the colon of mice.
Recent studies suggested that inflammatory cytokines and mediators serve important roles in facilitating mucosal immune responses (40,41). High levels of inflammatory cytokines, including TNF, IFN-γ and IL-6 can cause intestinal dysfunction, which is associated with the severity of UC (42). Macrophages are abundant within the intestine, where its polarization occurs during the development of UC (43). M1 macrophages are characterized by the high expression levels of iNOS, which produce a wide range of proinflammatory cytokines, including TNF-α and IL-6, to aggravate UC (32,33). By contrast, M2 macrophages release anti-inflammatory mediators, including Arg1 and IL-10, to reduce UC symptoms (44,45). The present results revealed that SIN treatment significantly downregulated the DSS-induced upregulation of TNF-α, IL-6 and iNOS expression. Moreover, SIN treatment increased the production of IL-10 and Arg1, which were initially decreased by DSS.
Changes in the gut microbiota profile is considered to be one of the key factors associated with UC (46). The colonic immune system can be activated by gut microbiota (47). Loss of diversity in the species of flora in the gut can affect the host's gut immune responses, leading to increased risk of inflammation and pathological damage (48-50). The results of the present study demonstrated that SIN may exert an anti-colitis effect by modulating the gut microbiota composition. The loss of phylum Bacteroidetes (class Bacteroidia) species diversity in patients with UC was previously found, which was in accordance with the endoscopic index of severity (51,52). In the present study, DSS treatment decreased the phylum Bacteroidetes (class Bacteroidia) abundance, whilst SIN treatment increased the abundance compared with that in the DSS-only group. During gut inflammation, Enterobacteriaceae family (phylum Proteobacteria) has previously exhibited a dysbiotic expansion (53). The present study also observed that the phylum Proteobacteria and class γ-proteobacteria were expanded after DSS treatment, whilst SIN administration suppressed their abundance, compared with that in the DSS-only group. However, the mechanism through which SIN regulated the composition of the gut microbiota in this mouse model of colitis requires further investigation.
The NLRP3 inflammasome is a large multimeric protein complex (8). Activation of the NLRP3 inflammasome has been found to promote colon inflammation and DSS-induced UC by activating caspase-1(54). In the present study, the expression levels of NLRP3 and caspase-1 in colonic tissues were analyzed, and the results demonstrated that SIN treatment inhibited the activation of the NLRP3 inflammasome in DSS-induced colitis.
In conclusion, the present results demonstrated that SIN treatment alleviated DSS-induced colitis not only by regulating the gut microbiota composition, but also by inhibiting the activation of the NLRP3 inflammasome. These beneficial effects may provide novel preclinical evidence for the potential application of SIN for treating UC.
Acknowledgements
Not applicable.
Funding Statement
Funding: The present study was supported by grants from the National Natural Science Foundation of China (grant no. 81803498), China Postdoctoral Science Foundation (grant no. 2020M670012ZX), Jiangsu Natural Science Foundation (grant no. BK20181155), Jiangsu Planned Projects for Postdoctoral Research Funds (grant no. 2019K157), Changzhou Sci&Tech Program (grant no. CJ20200090), Young Science & Technology Project of Changzhou Health Commission (grant no. QN202032) and Funding from Young Talent Development Plan of Changzhou Health Commission (grant nos. CZQM2020071 and CZQM2020063).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the Sequence Read Archive repository; https://www.ncbi.nlm.nih.gov/bioproject/739479.
Authors' contributions
YZ designed the study and wrote the manuscript. YZ, SC, XS and WG conducted the experiments and performed the statistical analysis. YZ, LW and LT supervised the experiments, analyzed the data and revised the manuscript. YZ and LW confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent for participation
Research involving animals was approved by the Ethics Committee of the Affiliated Changzhou No. 2 People's Hospital of Nanjing Medical University (Changzhou, China). All animal protocols performed in this study were conducted strictly based on the guidelines of the Jiangsu Committee on Animal Care.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Null KD, Xu Y, Pasquale MK, Su C, Marren A, Harnett J, Mardekian J, Manuchehri A, Healey P. Ulcerative colitis treatment patterns and cost of care. Value Health. 2017;20:752–761. doi: 10.1016/j.jval.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 2.van den Brink G, van Gaalen MAC, de Ridder L, van der Woude CJ, Escher JC. Health care transition outcomes in inflammatory bowel disease: A multinational delphi study. J Crohns Colitis. 2019;13:1163–1172. doi: 10.1093/ecco-jcc/jjz044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munyaka PM, Rabbi MF, Khafipour E, Ghia JE. Acute dextran sulfate sodium (DSS)-induced colitis promotes gut microbial dysbiosis in mice. J Basic Microbiol. 2016;56:986–998. doi: 10.1002/jobm.201500726. [DOI] [PubMed] [Google Scholar]
- 4.Alavala S, Sangaraju R, Nalban N, Sahu BD, Jerald MK, Kilari EK, Sistla R. Stevioside, a diterpenoid glycoside, shows anti-inflammatory property against dextran sulphate sodium-induced ulcerative colitis in mice. Eur J Pharmacol. 2019;855:192–201. doi: 10.1016/j.ejphar.2019.05.015. [DOI] [PubMed] [Google Scholar]
- 5.Klimesova K, Kverka M, Zakostelska Z, Hudcovic T, Hrncir T, Stepankova R, Rossmann P, Ridl J, Kostovcik M, Mrazek J, et al. Altered gut microbiota promotes colitis-associated cancer in IL-1 receptor-associated kinase M-deficient mice. Inflamm Bowel Dis. 2013;19:1266–1277. doi: 10.1097/MIB.0b013e318281330a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang K, Jin X, You M, Tian W, Le Leu RK, Topping DL, Conlon MA, Wu L, Hu F. Dietary propolis ameliorates dextran sulfate sodium-induced colitis and modulates the gut microbiota in rats fed a western diet. Nutrients. 2017;9(875) doi: 10.3390/nu9080875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeom Y, Kim BS, Kim SJ, Kim Y. Sasa quelpaertensis leaf extract regulates microbial dysbiosis by modulating the composition and diversity of the microbiota in dextran sulfate sodium-induced colitis mice. BMC Complement Altern Med. 2016;16(481) doi: 10.1186/s12906-016-1456-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He R, Li Y, Han C, Lin R, Qian W, Hou X. L-Fucose ameliorates DSS-induced acute colitis via inhibiting macrophage M1 polarization and inhibiting NLRP3 inflammasome and NF-kB activation. Int Immunopharmacol. 2019;73:379–388. doi: 10.1016/j.intimp.2019.05.013. [DOI] [PubMed] [Google Scholar]
- 9.Davis BK, Wen H, Ting JP. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu Rev Immunol. 2011;29:707–735. doi: 10.1146/annurev-immunol-031210-101405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lian D, Lai J, Wu Y, Wang L, Chen Y, Zhang Y, Boini KM, Huang Y, Chen Y. Cathepsin B-mediated NLRP3 inflammasome formation and activation in angiotensin II-induced hypertensive mice: Role of macrophage digestion dysfunction. Cell Physiol Biochem. 2018;50:1585–1600. doi: 10.1159/000494656. [DOI] [PubMed] [Google Scholar]
- 11.He X, Wei Z, Wang J, Kou J, Liu W, Fu Y, Yang Z. Alpinetin attenuates inflammatory responses by suppressing TLR4 and NLRP3 signaling pathways in DSS-induced acute colitis. Sci Rep. 2016;6(28370) doi: 10.1038/srep28370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan K, Liu ZQ, Jiang ZH, Zhou H, Wong YF, Xu HX, Liu L. The effects of sinomenine on intestinal absorption of paeoniflorin by the everted rat gut sac model. J Ethnopharmacol. 2006;103:425–432. doi: 10.1016/j.jep.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 13.Chen H, Wang Y, Jiao FZ, Yang F, Li X, Wang LW. Sinomenine attenuates acetaminophen-induced acute liver injury by decreasing oxidative stress and inflammatory response via regulating TGF-β/smad pathway in vitro and in vivo. Drug Des Devel Ther. 2020;14:2393–2403. doi: 10.2147/DDDT.S248823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Wang K, Ren Y, Zhang L, Tang XJ, Zhang HM, Zhao CQ, Liu PJ, Zhang JM, He JJ. MAPK signaling mediates sinomenine hydrochloride-induced human breast cancer cell death via both reactive oxygen species-dependent and -independent pathways: An in vitro and in vivo study. Cell Death Dis. 2014;5(e1356) doi: 10.1038/cddis.2014.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tong B, Yu J, Wang T, Dou Y, Wu X, Kong L, Dai Y, Xia Y. Sinomenine suppresses collagen-induced arthritis by reciprocal modulation of regulatory T cells and Th17 cells in gut-associated lymphoid tissues. Mol Immunol. 2015;65:94–103. doi: 10.1016/j.molimm.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 16.Huang RY, Pan HD, Wu JQ, Zhou H, Li ZG, Qiu P, Zhou YY, Chen XM, Xie ZX, Xiao Y, et al. Comparison of combination therapy with methotrexate and sinomenine or leflunomide for active rheumatoid arthritis: A randomized controlled clinical trial. Phytomedicine. 2019;57:403–410. doi: 10.1016/j.phymed.2018.12.030. [DOI] [PubMed] [Google Scholar]
- 17.Yao YM, Tan ZR, Hu ZY, Guo X, Cheng ZN, Wang LS, Zhou HH. Determination of sinomenine in human plasma by HPLC/ESI/ion trap mass spectrum. Clin Chim Acta. 2005;356:212–217. doi: 10.1016/j.cccn.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 18.Long LH, Wu PF, Chen XL, Zhang Z, Chen Y, Li YY, Jin Y, Chen JG, Wang F. HPLC and LC-MS analysis of sinomenine and its application in pharmacokinetic studies in rats. Acta Pharmacol Sin. 2010;31:1508–1514. doi: 10.1038/aps.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu ZQ, Chan K, Zhou H, Jiang ZH, Wong YF, Xu HX, Liu L. The pharmacokinetics and tissue distribution of sinomenine in rats and its protein binding ability in vitro. Life Sci. 2005;77:3197–3209. doi: 10.1016/j.lfs.2005.05.054. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Y, Liu H, Song J, Cao L, Tang L, Qi C. Sinomenine alleviates dextran sulfate sodium-nduced colitis via the Nrf2/NQO-1 signaling pathway. Mol Med Rep. 2018;18:3691–3698. doi: 10.3892/mmr.2018.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gkouskou KK, Deligianni C, Tsatsanis C, Eliopoulos AG. The gut microbiota in mouse models of inflammatory bowel disease. Front Cell Infect Microbiol. 2014;4(28) doi: 10.3389/fcimb.2014.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: The ARRIVE guidelines for reporting animal research. J Pharmacol Pharmacother. 2010;1:94–99. doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bang B, Lichtenberger LM. Methods of inducing inflammatory bowel disease in mice. Curr Protoc Pharmacol. 2016;72:5.58.1–5.58.42. doi: 10.1002/0471141755.ph0558s72. [DOI] [PubMed] [Google Scholar]
- 24.Vong LB, Tomita T, Yoshitomi T, Matsui H, Nagasaki Y. An orally administered redox nanoparticle that accumulates in the colonic mucosa and reduces colitis in mice. Gastroenterology. 2012;143:1027–1036.e3. doi: 10.1053/j.gastro.2012.06.043. [DOI] [PubMed] [Google Scholar]
- 25.Feldman AT, Wolfe D. Tissue processing and hematoxylin and eosin staining. Methods Mol Biol. 2014;1180:31–43. doi: 10.1007/978-1-4939-1050-2_3. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Magoč T, Salzberg SL. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang FG, Bai RX, Yan WM, Yan M, Dong LY, Song MM. Differential composition of gut microbiota among healthy volunteers, morbidly obese patients and post-bariatric surgery patients. Exp Ther Med. 2019;17:2268–2278. doi: 10.3892/etm.2019.7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter SJ, Hunter GR, Blackston JW, Liu N, Lefkowitz EJ, Van Der Pol WJ, Morrow CD, Paulsen JA, Rogers LQ. Gut microbiota diversity is associated with cardiorespiratory fitness in post-primary treatment breast cancer survivors. Exp Physiol. 2019;104:529–539. doi: 10.1113/EP087404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lozupone C, Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011;5:169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar RC. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 33.Dong T, Feng Q, Liu F, Chang LK, Zhou X, Han M, Tian X, Zhong N, Liu S. Alteration of stomach microbiota compositions in the progression of gastritis induces nitric oxide in gastric cell. Exp Ther Med. 2017;13:2793–2800. doi: 10.3892/etm.2017.4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kucharzik T, Koletzko S, Kannengiesser K, Dignass A. Ulcerative colitis-diagnostic and therapeutic algorithms. Dtsch Arztebl Int. 2020;117:564–574. doi: 10.3238/arztebl.2020.0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moreno Márquez C, Maldonado Pérez B, Castro Laria L. Infliximab as rescue treatment in Sweet's syndrome related to corticodependent ulcerative colitis. J Crohns Colitis. 2018;12:755–756. doi: 10.1093/ecco-jcc/jjy018. [DOI] [PubMed] [Google Scholar]
- 36.Park JH, Kim JK. Pristimerin, a naturally occurring triterpenoid, attenuates tumorigenesis in experimental colitis-associated colon cancer. Phytomedicine. 2018;42:164–171. doi: 10.1016/j.phymed.2018.03.033. [DOI] [PubMed] [Google Scholar]
- 37.Zheng R, Ma J, Wang D, Dong W, Wang S, Liu T, Xie R, Liu L, Wang B, Cao H. Chemopreventive effects of silibinin on colitis-associated tumorigenesis by inhibiting IL-6/STAT3 signaling pathway. Mediators Inflamm. 2018;2018(1562010) doi: 10.1155/2018/1562010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiong H, Tian L, Zhao Z, Chen S, Zhao Q, Hong J, Xie Y, Zhou N, Fu Y. The sinomenine enteric-coated microspheres suppressed the TLR/NF-κB signaling in DSS-induced experimental colitis. Int Immunopharmacol. 2017;50:251–262. doi: 10.1016/j.intimp.2017.06.033. [DOI] [PubMed] [Google Scholar]
- 39.Wang K, Jin X, Li Q, Sawaya ACHF, Le Leu RK, Conlon MA, Wu L, Hu F. Propolis from different geographic origins decreases intestinal inflammation and bacteroides spp. populations in a model of DSS-induced colitis. Mol Nutr Food Res. 2018;62(e1800080) doi: 10.1002/mnfr.201800080. [DOI] [PubMed] [Google Scholar]
- 40.Wang W, Li X, Zheng D, Zhang D, Peng X, Zhang X, Ai F, Wang X, Ma J, Xiong W, et al. Dynamic changes and functions of macrophages and M1/M2 subpopulations during ulcerative colitis-associated carcinogenesis in an AOM/DSS mouse model. Mol Med Rep. 2015;11:2397–2406. doi: 10.3892/mmr.2014.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao D, Dong M, Dai C, Wu S. Inflammation and inflammatory cytokine contribute to the initiation and development of ulcerative colitis and its associated cancer. Inflamm Bowel Dis. 2019;25:1595–1602. doi: 10.1093/ibd/izz149. [DOI] [PubMed] [Google Scholar]
- 42.Guan Q, Zhang J. Recent advances: The imbalance of cytokines in the pathogenesis of inflammatory bowel disease. Mediators Inflamm. 2017;2017(4810258) doi: 10.1155/2017/4810258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou X, Li W, Wang S, Zhang P, Wang Q, Xiao J, Zhang C, Zheng X, Xu X, Xue S, et al. YAP aggravates inflammatory bowel disease by regulating M1/M2 macrophage polarization and gut microbial homeostasis. Cell Rep. 2019;27:1176–1189.e5. doi: 10.1016/j.celrep.2019.03.028. [DOI] [PubMed] [Google Scholar]
- 44.Aass HCD, Hellum M, Trøseid AS, Brandtzaeg P, Berg JP, Øvstebø R, Henriksson CE. Whole-blood incubation with the Neisseria meningitidis lpxL1 mutant induces less pro-inflammatory cytokines than the wild type, and IL-10 reduces the MyD88-dependent cytokines. Innate Immun. 2018;24:101–111. doi: 10.1177/1753425917749299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu L, Liang L, Liang H, Wang M, Lu B, Xue M, Deng J, Chen Y. Fusobacterium nucleatum aggravates the progression of colitis by regulating M1 macrophage polarization via AKT2 pathway. Front Immunol. 2019;10(1324) doi: 10.3389/fimmu.2019.01324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cui H, Cai Y, Wang L, Jia B, Li J, Zhao S, Chu X, Lin J, Zhang X, Bian Y, Zhuang P. Berberine regulates Treg/Th17 balance to treat ulcerative colitis through modulating the gut microbiota in the colon. Front Pharmacol. 2018;9(571) doi: 10.3389/fphar.2018.00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ilott NE, Bollrath J, Danne C, Schiering C, Shale M, Adelmann K, Krausgruber T, Heger A, Sims D, Powrie F. Defining the microbial transcriptional response to colitis through integrated host and microbiome profiling. ISME J. 2016;10:2389–2404. doi: 10.1038/ismej.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Forbes JD, Van Domselaar G, Bernstein CN. The gut microbiota in immune-mediated inflammatory diseases. Front Microbiol. 2016;7(1081) doi: 10.3389/fmicb.2016.01081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao X, Cao Q, Cheng Y, Zhao D, Wang Z, Yang H, Wu Q, You L, Wang Y, Lin Y, et al. Chronic stress promotes colitis by disturbing the gut microbiota and triggering immune system response. Proc Natl Acad Sci USA. 2018;115:E2960–E2969. doi: 10.1073/pnas.1720696115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang M, Sun K, Wu Y, Yang Y, Tso P, Wu Z. Interactions between intestinal microbiota and host immune response in inflammatory bowel disease. Front Immunol. 2017;8(942) doi: 10.3389/fimmu.2017.00942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishikawa D, Sasaki T, Osada T, Kuwahara-Arai K, Haga K, Shibuya T, Hiramatsu K, Watanabe S. Changes in intestinal microbiota following combination therapy with fecal microbial transplantation and antibiotics for ulcerative colitis. Inflamm Bowel Dis. 2017;23:116–125. doi: 10.1097/MIB.0000000000000975. [DOI] [PubMed] [Google Scholar]
- 52.Ishikawa D, Sasaki T, Takahashi M, Kuwahara-Arai K, Haga K, Ito S, Okahara K, Nakajima A, Shibuya T, Osada T, et al. The microbial composition of bacteroidetes species in ulcerative colitis is effectively improved by combination therapy with fecal microbiota transplantation and antibiotics. Inflamm Bowel Dis. 2018;24:2590–2598. doi: 10.1093/ibd/izy266. [DOI] [PubMed] [Google Scholar]
- 53.Zhu W, Winter MG, Byndloss MX, Spiga L, Duerkop BA, Hughes ER, Büttner L, de Lima Romão E, Behrendt CL, Lopez CA, et al. Precision editing of the gut microbiota ameliorates colitis. Nature. 2018;553:208–211. doi: 10.1038/nature25172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rathinam VAK, Chan FK. Inflammasome, inflammation, and tissue homeostasis. Trends Mol Med. 2018;24:304–318. doi: 10.1016/j.molmed.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the Sequence Read Archive repository; https://www.ncbi.nlm.nih.gov/bioproject/739479.