Abstract
Glucocorticoids are potent anti-inflammatory agents widely used in the treatment of human disease. We have previously shown that the inflammatory cytokine monocyte chemoattractant protein 1 (MCP-1) is regulated posttranscriptionally by glucocorticoids in arterial smooth muscle cells (SMC). To elucidate the mechanism mediating this effect, in vitro-transcribed radiolabeled MCP-1 mRNA was incubated with cytoplasmic extracts from SMC and analyzed by gel electrophoresis. Extracts from SMC treated with platelet-derived growth factor (PDGF) did not degrade the transcripts for up to 3 h. In contrast, extracts from cells treated with 1 μM dexamethasone (Dex) alone or in combination with PDGF degraded the probe with a half-life of ≈15 min. Dex had maximal effect at concentrations above 0.01 μM and was effective on both rat and human MCP-1 transcripts. By deletion analysis, the Dex-sensitive region of the MCP-1 mRNA was localized to the initial 224 nucleotides (nt) at the 5′ end and did not involve an AU-rich sequence in the 3′ untranslated end. The 224-nt region conferred Dex sensitivity to heterologous mRNA. These studies provide new insights into the molecular mechanisms underlying the effect of glucocorticoids on gene expression.
Glucocorticoids are potent anti-inflammatory agents used to treat a wide variety of diseases. While considerable information is available on the cellular events mediating glucocorticoid activity (reviewed in references 4 and 8), the mechanisms involved in their anti-inflammatory effects remain to be fully elucidated. One component of the inflammatory response of particular importance in the arterial wall is the recruitment and adhesion of monocytes/macrophages to sites of inflammation and injury. Macrophages are the primary source of cholesterol-rich foam cells seen throughout the atherosclerotic plaque (20, 24, 28). Macrophages also produce a variety of cytokines and growth factors (15), which may stimulate a proliferative and migratory response at sites of injury. Macrophages are phagocytic and produce metalloproteinases, which degrade extracellular matrix proteins; these may play a role in destabilizing the atherosclerotic plaque or facilitating cellular migration (23, 30, 42). Finally, macrophages are an important source of tissue factor (32, 51, 63), the initiator of coagulation, and thus may play an important role in mediating plaque thrombosis.
Long-term glucocorticoid treatment has been reported to decrease macrophage accumulation and plaque size during the development of atherosclerosis (38). We have recently shown that dexamethasone (Dex) markedly inhibited the accumulation of macrophages and the development of intimal hyperplasia in the femoral arteries of cholesterol-fed, balloon-injured rabbits (47a). In addition, treatment of rat aortic smooth muscle cells (SMC) with Dex completely blocked the growth factor-mediated accumulation of monocyte chemotactic activity in the culture medium (48).
Monocyte chemoattractant protein 1 (MCP-1) is a low-molecular-weight cytokine secreted by endothelial cells (53), vascular SMC (65), monocytes/macrophages (70), and fibroblasts (59). MCP-1 is active as a chemoattractant at subnanomolar concentrations (22, 69). MCP-1 and its rodent analog, JE (16, 52), are not normally present in the arterial media or intima but have been found in human, primate and rabbit atherosclerotic plaques (16, 40, 60, 68, 71). In addition, MCP-1 mRNA is induced in the media within hours of experimental rodent balloon arterial injury (62). Recent data have demonstrated that despite the large number of agents capable of attracting monocytes, MCP-1 may be the only monocyte chemoattractant induced in cultured SMC by platelet-derived growth factor (PDGF) and minimally modified low-density lipoprotein (17, 48). MCP-1 may thus play a key role in attracting monocytes/macrophages to the developing atherosclerotic plaque and to sites of acute arterial injury. MCP-1 may also play a role in attracting monocytes/macrophages to other sites of inflammation, such as alveolar epithelial cells, inflammatory synovium, and meningioma (25, 45, 56).
Glucocorticoids have been shown to be inhibitors of MCP-1 synthesis in a variety of cell types (26, 27, 34, 37, 45, 48, 49). We previously showed that MCP-1 mRNA, protein, and activity were rapidly induced in cultured rat aortic SMC by PDGF and serum and in rat and rabbit aortas in response to balloon arterial injury (48, 62). The effect of PDGF and serum on MCP-1 mRNA levels was due to increases in both transcription and mRNA stability (7, 62). Dex, at doses as low as 0.01 μM, completely blocked the serum- or PDGF-induced accumulation of MCP-1 mRNA (49) and the secretion of monocyte chemotactic activity (48) by rat aortic SMC. This effect was seen with other glucocorticoids but not with mineralicorticoids, estrogen, progesterone, or testosterone. Reports from other laboratories have also demonstrated a profound effect of glucocorticoids on MCP-1 expression in human fibrosarcoma cells (37), 3T3 cells (27), alveolar epithelial cells (45), and human eosinophils (34). In addition, methyl prednisolone blocked the induction of MCP-1 expression in a rat model of renal ischemia (49).
The effect of Dex on MCP-1 mRNA accumulation in rat aortic SMC was due predominantly to a decrease in mRNA stability and not to changes in MCP-1 transcription (49). Thus, Dex reduced the half-life (t1/2) of MCP-1 mRNA in the presence of PDGF or serum from ≈3 h to ≈30 min (49). We have now used in vitro RNA decay assays and SMC transfections to elucidate further the mechanism underlying the effect of Dex on MCP-1 mRNA stability. We report the identification of a novel Dex-sensitive region on the 5′ end of the rat MCP-1 mRNA. This region also confers Dex sensitivity to heterologous mRNA. These studies provide new insights into the molecular mechanisms underlying the effect of glucocorticoids on gene expression.
MATERIALS AND METHODS
Cell culture.
Rat aortic SMC were isolated from the thoracic aortas of 200- to 300-g male Sprague-Dawley rats by enzymatic dissociation as previously described (49). Passages 5 to 17 were used for all experiments. Human pulmonary fibroblasts (catalog no. FHS 738) were obtained from the American Type Culture Collection (Baltimore, Md.). Cells were cultured at 37°C in 5% CO2 in Dulbecco modified Eagle medium (DMEM; Gibco Laboratories, Gaithersburg, Md.) supplemented with 10% heat-inactivated bovine serum. All experiments were performed at confluence (≈2 × 106 cells per 100-mm-diameter dish), 36 h after addition of fresh medium.
In vitro transcription.
MCP-1 constructs were generated by PCR using the full-length rat MCP-1 cDNA (GenBank accession no. AF058786) as the template. All constructs were verified by sequencing. The full-length human MCP-1 cDNA (29) cloned into the polylinker sites (HindIII and XbaI) of the pBluescript vector (Stratagene, La Jolla, Calif.) was the generous gift of Joan Berman (Albert Einstein School of Medicine, New York, N.Y.). Linearized templates were transcribed in vitro with T3 RNA polymerase (Boehringer Mannheim, Indianapolis, Ind.) in the presence of [α-32P]UTP (800 Ci/mmol; New England Nuclear, Boston, Mass.). Capped transcripts were generated by using a 4:1 ratio of m7G5′pppG (Boehringer Mannheim) to GTP. To generate polyadenylated MCP-1 RNA, synthetic poly(A-T) 30-mers were inserted at the 3′ end of the full-length MCP-1 cDNA. In vitro-transcribed tissue factor mRNA was generated from the full-length 1.6-kb rat cDNA (GenBank accession no. U07619). A 290-nucleotide (nt) fragment of pBluescript mRNA was in vitro transcribed from the T3 promoter (base 791) to the PvuI site (base 502) of linearized pBluescript KS (+/−) (Stratagene). Probes were purified on 4% polyacrylamide gels.
Preparation of cytosolic (S-100) extracts.
All procedures were performed at 4°C. Five 70% confluent plates of SMC were washed once with 5 ml of ice-cold phosphate-buffered saline (PBS) and scraped with a rubber policeman into 1.5 ml of PBS. The cells were spun at 500 × g for 10 min. The pellet was resuspended (5 × 107 cells per ml) in 10 mM Tris-Cl (pH 7.4)–10 mM KCl–1.5 mM MgCl2–0.5 mM dithiothreitol (DTT) and lysed with 20 strokes in a Dounce homogenizer (pestle B). The homogenate was adjusted to 100 mM KCl–1.5 mM MgCl2–10 mM Tris HCl (pH 7.4)–0.5 mM phenylmethylsulfonyl fluoride–0.5 mM DTT. The nuclei were separated by centrifugation at 14,000 × g for 2 min. The remaining cytoplasmic lysates (S-100 extracts) were centrifuged at 100,000 × g for 1 h at 4°C, and the supernatant stored at −80°C. Nuclear lysates were obtained by lysing the nuclei with 20 strokes in a Dounce homogenizer (pestle A). Protein concentrations of cytoplasmic or nuclear extracts were determined by the Bradford protein assay (Bio-Rad) (66). In general, cytoplasmic extracts had a final concentration of 250 to 500 ng/μl.
In vitro RNA decay assays.
Immediately before use, extracts were thawed and diluted to a final concentration of 0.2 μg/μl in 100 mM KCl–1.5 mM MgCl2–10 mM Tris HCl (pH 7.4)–0.5 mM phenylmethylsulfonyl fluoride–0.5 mM DTT. Duplicate 10-μl aliquots were incubated at 24°C with 1 μl of radiolabeled MCP-1 and pBluescript probes (≈5,000 cpm/μl). At the times indicated in the figures (30 min for most experiments), 0.5 μl of heparin (4 mg/ml, final concentration) and 1 μl of loading buffer (0.4% bromophenol blue, 0.4% xylene cyanol, and 50% glycerol in water) were added. The samples were loaded onto 4% native polyacrylamide gels and subjected to polyacrylamide gel electrophoresis (PAGE) for 2.5 h at 30 mA in 200 mM morpholinepropanesulfonic acid–100 mM Na acetate (pH 7.2). In some experiments, at the end of the incubation, samples were extracted with phenol, ethanol precipitated, and run on 4% denaturing gels containing 7 M urea. Gels were then exposed to X-Omat film. To quantify levels of RNA expression, autoradiograms were scanned in two dimensions with an Epson scanner. Densitometry, using 256 gray scales, was performed on a Macintosh 8500 computer using Image 2.0 software (developed by Wayne Rasband, National Institutes of Health). Densities are expressed as percentages of the density measured in the lane containing untreated probe (i.e., probe not exposed to extracts).
SMC transfections.
An ecdysone-inducible expression system (Invitrogen Corporation, San Diego, Calif.) was used to generate SMC-expressing wild-type and truncated MCP-1. This system has previously been shown to be insensitive to Dex (43). SMC were initially transfected with the pVgRXR vector, which encodes the receptor subunit for pronesterone. Stable lines were selected with Zeocin. Constructs were ligated into the pIND vector, which contains five modified ecdysone response elements upstream of a minimal heat shock promoter. A stable line of retinoid X receptor-expressing SMC was transfected with these constructs (2 μg) by using an Effectene transfection kit (Qiagen Inc., Valencia, Calif.). To normalize for transfection efficiency, cells were cotransfected with the pXP2 luciferase reporter construct (150 ng) driven by the Dex-insensitive cytomegalovirus promoter (44). Four hours after transfection, cells were washed and incubated for 24 h in fresh DMEM containing 10% serum and 10 μM pronesterone and then incubated overnight in serum-free DMEM and fresh pronesterone. Cells were subsequently treated with 1 μM Dex in DMEM and pronesterone for 3 h or left in DMEM and pronesterone without Dex. As controls for the efficacy of pronesterone treatment, some SMC were transfected for 4 h, incubated for 24 h in DMEM–10% serum without pronesterone, incubated overnight in DMEM alone, and then harvested. Expression of transcripts was analyzed by reverse transcriptase (RT) PCR (RT-PCR).
RT-PCR.
Total RNA was isolated from SMC grown on 100-mm-diameter plates, using an RNeasy Mini Kit (Qiagen). All RNA was treated for 15 min at 37°C with 200 U of RNase-free DNase I (Boehringer Mannheim) per ml. The integrity of the RNA was verified by electrophoresis on denaturing agarose gels. Prior to RT-PCR, all samples were tested for DNA contamination by performing PCR, in the absence of RT, with the primers indicated below. Only samples that were negative by PCR were used for the RT-PCR experiments. RT-PCR was performed by using the Superscript One-Step RT-PCR system (Life Technologies, Inc, Rockville, Md.) according to the manufacturer’s protocol. The 50-μl reaction mixture contained 0.5 μg of total RNA, 0.2 μM antisense primer, corresponding to a T7 site in the vector (5′ GTAATACGACTCACTCACTATAGGGC 3′), and 0.2 μM sense primer (5′ CCCAAGCTTGCAGAGACACAGACAGAGG 3′), containing a HindIII site and nt 1 to 19 of the rat JE mRNA (Genbank accession no. AF058786). These primer pairs amplified all MCP-1 constructs but did not amplify native MCP-1 mRNA. As an additional control for transfection efficiency, RT-PCR mixtures contained a second set of primers spanning a 501-bp segment of the neomycin resistance gene, corresponding to nt 1801 to 2302 of the pIND vector (sense, 5′ AATCGGCTGCTCTGATGCC; antisense, 5′ ATTCGGCAAGCAGGCATCG). The neomycin resistance gene is driven by the pronesterone-insensitive simian virus 40 promoter. The mixture was heated to 53°C for 30 min and denatured at 94°C for 2 min. This was followed by 35 cycles consisting of 30 s at 94°C, 1 min at 60°C, and 1 min at 72°C and 1 cycle of final extension at 72°C. RT-PCR products were examined on 2% agarose gels.
Luciferase assay.
SMC were washed twice at 25°C with PBS, lysed in luciferase cell culture lysis reagent (Promega, Madison, Wis.), and assayed for luciferase activity in a BioOrbit 1251 luminometer (Wallac, Gaithersburg, Md.), using luciferase assay reagent (Promega). For each construct tested, luciferase activity in pronesterone-stimulated cells (Dex treated or untreated) was normalized to the activity in unstimulated cells. Experiments were performed on duplicate plates and were repeated twice.
RESULTS
Cytoplasmic extracts from Dex-treated SMC reduce the t1/2 of MCP-1 mRNA.
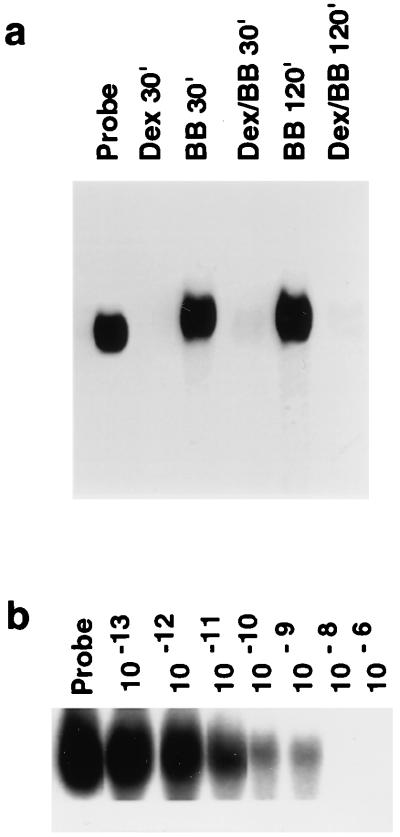
We have previously shown that Dex markedly decreases the t1/2 of MCP-1 mRNA in cultured rat SMC. To elucidate further the mechanism responsible for the effect of Dex on MCP-1 mRNA stability, in vitro-transcribed 32P-labeled mRNA was incubated with cytoplasmic extracts from SMC and analyzed on native 4% polyacrylamide gels. Extracts from SMC treated with PDGF BB (20 ng/ml) had no effect on the intensity of the radiolabeled band after 30 min of incubation (Fig. 1a). In contrast, no radiolabeled band was detected when the probe was incubated for 30 min with extracts from cells treated with 1 μM Dex alone or in combination with PDGF. Nuclear extracts had no effect on in vitro-transcribed MCP-1 mRNA when incubated with the probe for up to 3 h (data not shown). The effect of Dex treatment was maximal at 10−8 M and was absent at 10−11 M (Fig. 1b).
FIG. 1.
Dex mediated decay of MCP-1 mRNA in rat aortic SMC. (a) Cytoplasmic (S-100) extracts were isolated from SMC treated for 3 h with 1 μM Dex (Dex), 20 ng of PDGF BB (BB) per ml, or both (Dex/BB). 32P-labeled, in vitro-transcribed, full-length MCP-1 mRNA (Probe) was incubated with extracts for the times indicated and then analyzed by PAGE. (b) Cytoplasmic extracts were isolated from SMC treated for 3 h with various concentrations (from 10−13 to 10−6 M) of Dex. 32P-labeled, in vitro-transcribed, full-length MCP-1 mRNA (Probe) was incubated with extracts for 30 min and then analyzed by PAGE.
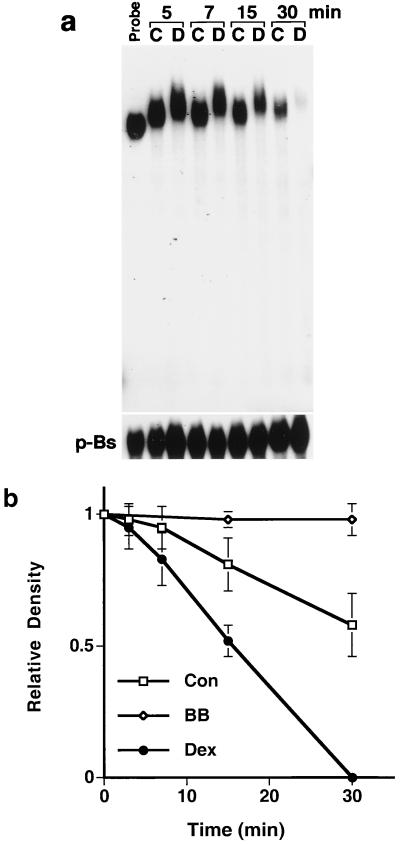
To better establish the rate of decay of the mRNA probe, assays were performed at time points ranging from 5 to 30 min. As shown in Fig. 2a (top panel), extracts from untreated SMC and SMC treated with Dex caused a shift in the mobility of the probe. Extracts from Dex-treated cells caused a greater shift than those from untreated SMC, suggesting that different proteins or different states of the same protein were bound under the two conditions. Neither extract caused a shift or decay when duplicate aliquots were incubated with in vitro-transcribed pBluescript mRNA (Fig. 2a, bottom panel). Figure 2b is a graphic representation of the t1/2 of the shifted complex derived from triplicate experiments. In the presence of extracts from quiescent SMC, the shifted complex had a t1/2 of ≈45 min. In contrast, the intensity of the shifted complex remained unchanged for 2 h in the presence of extracts from SMC treated with 20 ng of PDGF per ml. Most significantly, extracts from cells treated with 1 μM Dex alone or in combination with PDGF (not shown) reduced the t1/2 of the complex to ≈15 min.
FIG. 2.
Rate of in vitro MCP-1 mRNA decay in nondenaturing gels. (a) SMC were incubated for 36 h with DMEM–10% serum and then treated for 3 h with 1 μM Dex (D) or left untreated (C). Cytoplasmic extracts were incubated with the radiolabeled MCP-1 probe or with a radiolabeled p-Bluescript (p-Bs) mRNA fragment for the times indicated. (b) MCP-1 mRNA decay curves generated with cytoplasmic extracts from untreated SMC (Con), SMC treated with Dex (Dex), and SMC treated with PDGF BB (BB). PAGE was performed as shown in panel a. Gels were analyzed by densitometry. Curves represent the average ± standard error of the mean of triplicate experiments.
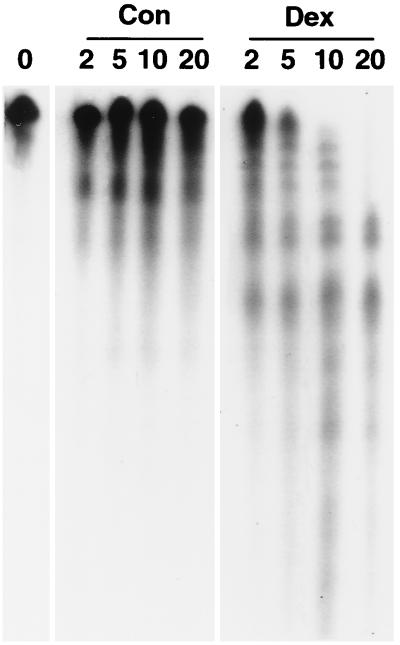
To verify that the loss of the radiolabeled band detected on gel shifts was due to differences in the decay of the in vitro-transcribed mRNA and not to Dex-mediated changes in protein binding, samples were also run on denaturing gels in the presence of 7 M urea. As shown in Fig. 3, Dex-treated extracts induced the rapid decay of radiolabeled MCP-1 mRNA. In contrast, MCP-1 mRNA decayed at a much lower rate when incubated with control extracts. The t1/2 of the Dex-mediated decay derived from duplicate experiments was ≈8 min. At 20 min, differences in the intensities of the principal band generated with control and Dex-treated extracts were similar on denaturing and nondenaturing gels.
FIG. 3.
In vitro MCP-1 mRNA decay in denaturing gels. Cytoplasmic extracts were isolated from untreated (Con) SMC (36 h after feeding with DMEM–10% serum) or from SMC treated for 3 h with Dex (Dex) and incubated with the radiolabeled MCP-1 probe for the times indicated (in minutes) and run on 4% denaturing polyacrylamide gels containing 6 M urea. The entire length of the gel is shown and is representative of three experiments.
Specificity of the Dex effect.
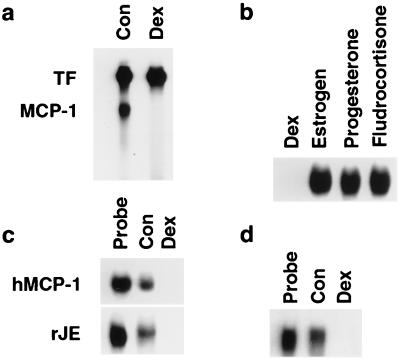
To examine the specificity of the RNA-destabilizing effect, extracts from Dex-treated SMC were incubated with in vitro-transcribed tissue factor mRNA. Like MCP-1, tissue factor acts as an immediate-early gene in rat aortic SMC (61). In contrast to their effect on MCP-1 mRNA, Dex-treated SMC extracts did not degrade tissue factor mRNA and may have stabilized the mRNA relative to extracts from untreated SMC (Fig. 4a). Cytoplasmic extracts from SMC treated with 10 μM estrogen, progesterone, or fludrocortisone had no effect on MCP-1 mRNA decay (Fig. 4b). This result supports previous findings in SMC culture demonstrating that the inhibition of MCP-1 mRNA accumulation was glucocorticoid specific (49). Cytoplasmic extracts from Dex-treated normal human pulmonary fibroblasts rapidly degraded transcribed human MCP-1 mRNA, suggesting that the Dex effect is not species specific and not limited to SMC (Fig. 4c). Extracts from Dex-treated human fibroblasts also efficiently degraded rat MCP-1 mRNA (Fig. 4c); in addition, those from rat SMC degraded human MCP-1 mRNA (Fig. 4d). This finding suggests that the degrading activity is not species specific and is conserved in evolution.
FIG. 4.
Specificity of Dex-mediated effects on mRNA stability. (a) In vitro-transcribed, radiolabeled tissue factor (TF) and MCP-1 mRNAs (Probe) were incubated together for 30 min with cytoplasmic extracts from untreated SMC (Con) and SMC treated for 3 h with 1 μM Dex and analyzed by nondenaturing PAGE as described for Fig. 3. (b) Radiolabeled MCP-1 mRNA was incubated for 30 min with extracts from SMC treated for 3 h with 1 μM Dex, estrogen, progesterone, or fludrocortisone. (c) Cytoplasmic extracts were isolated from untreated normal human pulmonary fibroblasts (Con) or fibroblasts treated for 3 h with 1 μM Dex (Dex). 32P-labeled, in vitro-transcribed, full-length human MCP-1 (hMCP-1) and rat JE (rJE) mRNAs were incubated with extracts for 30 min and then analyzed by nondenaturing PAGE. (d) Cytoplasmic extracts were isolated from untreated rat SMC (Con) or SMC treated for 3 h with 1 μM Dex (Dex). 32P-labeled, in vitro-transcribed human MCP-1 mRNA was incubated with extracts for 30 min and then analyzed by PAGE. All gels are representative of duplicate experiments.
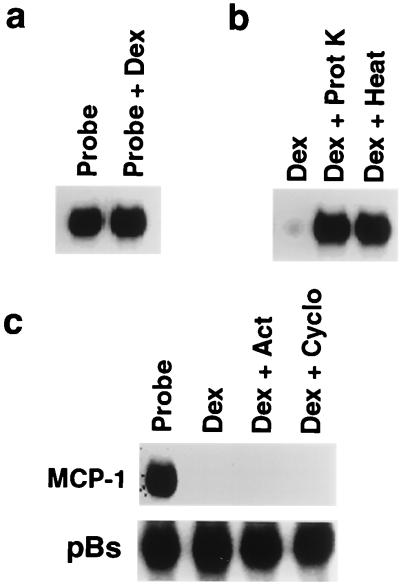
The effect of Dex on MCP-1 mRNA decay does not require de novo protein and RNA synthesis.
To determine whether the decrease in MCP-1 mRNA stability involved a direct effect of Dex on mRNA, decay assays were performed in which Dex (1 μM) was added directly to a reaction mixture containing in vitro-transcribed MCP-1 mRNA and extracts from PDGF-treated cells (Fig. 5a). The addition of Dex did not alter MCP-1 mRNA t1/2, suggesting that the destabilizing effect was secondary to the actions of Dex on SMC and likely due to the activation and/or synthesis of a trans-acting cytoplasmic factor. Dex also had no effect when added to in vitro-transcribed MCP-1 mRNA in the reaction buffer in the absence of cellular extracts (not shown), demonstrating that the preparation did not contain contaminating nucleases. Addition of proteinase K (2 mg/ml) to extracts from Dex-treated cells abolished the effect on MCP-1 mRNA stability, as did heating the extracts at 95°C for 10 min (Fig. 5b). These results suggest further that the effect of Dex on MCP-1 mRNA stability is mediated through a cytosolic trans-acting protein.
FIG. 5.
Dex-mediated effects on MCP-1 mRNA stability in SMC: cellular mechanisms. (a) Radiolabeled MCP-1 mRNA (Probe) was incubated with 1 μM Dex and cytoplasmic extract from PDGF-treated SMC. After 30 min, samples were analyzed by nondenaturing PAGE. (b) Radiolabeled MCP-1 mRNA was incubated for 3 h with cytoplasmic extracts from Dex-treated SMC (Dex). Aliquots of the same extract were treated for 30 min with 1 mg/ml proteinase K (Dex + Prot K) or heated at 95°C for 10 min (Dex + Heat) prior to incubation with the probe. (c) Radiolabeled MCP-1 or pBluescript (pBs) mRNA (Probe) was incubated with extracts from SMC treated for 3 h with Dex alone (Dex) or from SMC pretreated for 30 min with 10 μM actinomycin D (Act) or 10 μM cycloheximide (Cyclo) and then subsequently treated for 3 h with Dex in the presence of the same inhibitor. After 30 min, samples were analyzed by PAGE. All gels are representative of duplicate experiments.
To examine the effects of de novo protein and RNA synthesis on the destabilizing activity in SMC extracts, cells were incubated with actinomycin D (10 μM) or cycloheximide (10 μM) for 30 min prior to incubation with Dex (Fig. 5c). Neither inhibitor had a significant effect on the mRNA-destabilizing activity of Dex-treated SMC extracts, suggesting that the trans-acting factor is constitutively expressed and stable.
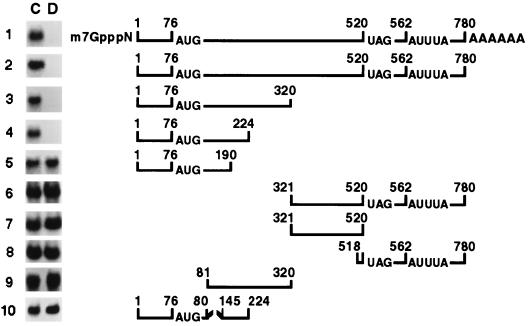
Localization of the Dex-sensitive region to the 5′ end of the MCP-1 mRNA.
The 5′ cap and poly(A) tail have been found to play an important role in regulating the stability of some mRNAs (54). No significant differences in Dex-mediated MCP-1 RNA decay were noted in the presence or absence of a 5′ cap or a 30-base poly(A) tail (Fig. 6). Only mRNAs containing the initial 224 nt from the 5′ end of the MCP-1 mRNA (Fig. 6, rows 1 to 4) retained sensitivity to Dex. mRNAs comprising 190 nt (row 5) or 80 nt (not shown) from the 5′ end failed to retain Dex sensitivity. In addition, deletion of 80 nt from the 5′ end of a Dex-sensitive 320-nt fragment (row 9) abolished its sensitivity to Dex-mediated RNA decay, as did deletion of 64 nt (bases 81 to 145) from the middle of the 224-nt fragment (row 10). These results suggest that the entire 224-nt fragment may be necessary for Dex-mediated decay. This may be due to the location of distinct binding and enzymatic sites at different ends of the 224-nt fragment. In addition, a minimum number of bases may be required for the initiation of RNase activity. All transcripts lacking the 5′ end of the MCP-1 mRNA (rows 6 to 8), including those containing putative AU-rich mRNA-destabilizing sequences in the 3′ untranslated region (UTR) (rows 6 and 8), were not responsive to Dex. This finding suggests that the AU-rich sequences are not involved in Dex-mediated decay of MCP-1 in vitro.
FIG. 6.
In vitro decay analysis of truncated MCP-1 transcripts. MCP-1 mRNA was 32P-labeled by in vitro transcription from full-length or truncated rat MCP-1 cDNA constructs generated by PCR. In row 1, transcripts were also 5′ capped (m7GpppN), and polyadenylated (AAAAAA). Cytoplasmic extracts from untreated SMC (C) or SMC treated with Dex (D) for 3 h were incubated with the labeled transcripts for 30 min and then examined by nondenaturing PAGE. The location of the 3′ AUUUA sequences, the AUG initiation codon, and the UAG stop codon are noted. The broken line in construct 10 indicates a deletion between nt 80 and 145. Gels are representative of experiments performed in triplicate. The numbering of nucleotides is based on the full-length rat cDNA (accession no. AF058786); nt 1 corresponds to nt 1040 of the rat JE genomic DNA (accession no. X71053).
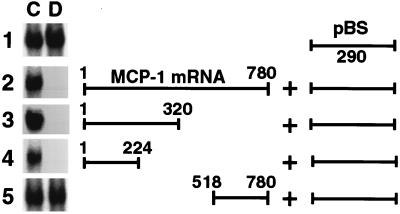
The Dex-sensitive region of the MCP-1 mRNA destabilizes heterologous mRNA.
To investigate further the properties of the Dex-sensitive region of the MCP-1 mRNA, transcripts were made from chimeric constructs containing MCP-1 cDNA ligated to a 290-nt fragment of pBluescript DNA (Fig. 7). Extracts from Dex-treated SMC had no effect on the t1/2 of the 290-nt pBluescript mRNA. However, chimeras of the 224-nt 5′ Dex-sensitive MCP-1 fragment and the pBluescript RNA rapidly degraded in the presence of extracts from Dex-treated SMC. The t1/2 of the chimeric mRNAs incubated with extracts from Dex-treated SMC was similar to that observed for the native MCP-1 mRNA. A chimeric transcript containing the 3′ UTR of the MCP-1 mRNA failed to respond to Dex. These data not only demonstrate the ability of the 224-nt 5′ region of the MCP-1 mRNA to convey Dex sensitivity to a heterologous mRNA but provide further evidence that the AU-rich region in the 3′ untranslated end does not mediate the in vitro Dex effect.
FIG. 7.
In vitro decay analysis of chimeric MCP-1 transcripts. Chimeric constructs were generated by ligating either full-length MCP-1 cDNA or MCP-1 cDNA fragments to a 290-bp fragment of the Dex-insensitive pBluescript DNA (pBS). In vitro-transcribed 32P-labeled chimeric transcripts were incubated for 30 min with cytoplasmic extracts from untreated SMC (C) or SMC treated for 3 h with Dex (D) and examined by nondenaturing PAGE. Gels are representative of triplicate experiments.
Analysis of Dex-sensitive mRNAs in transfected SMC.
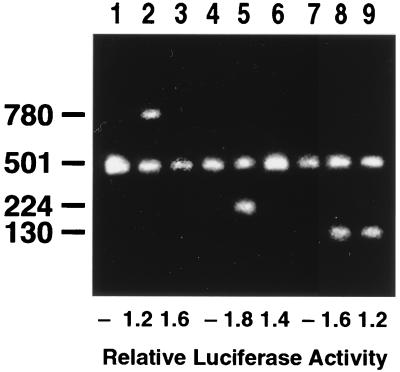
To ascertain whether the Dex-sensitive region of the MCP-1 mRNA would retain its sensitivity in vivo, wild-type and truncated MCP-1 constructs were transfected into SMC by using an ecdysone-inducible expression system. This system responds to ecdysone analogs, such as pronesterone, but is insensitive to Dex (43). Two methods were used to control for transfection efficiency. RT-PCR was performed with a second set of primers, corresponding to 501 bases of the neomycin resistance gene located in the pIND vector. The neomycin gene is driven by the pronesterone- and Dex-insensitive simian virus 40 promoter. In addition, cells were cotransfected with the pXP2 luciferase reporter construct, also driven by a pronesterone- and Dex-insensitive promoter. The relative luciferase activity (normalized to the lane not treated with pronesterone) for each construct used in the experiment is shown at the bottom of Fig. 8.
FIG. 8.
Analysis of MCP-1 transcripts in transfected SMC. DNA constructs containing nt 1 to 224 (lanes 1 to 3), nt 1 to 130 (lanes 4 to 6), or full-length rat MCP-1 (lanes 7 to 9) were ligated into the pIND vector and cotransfected with the pXP2 luciferase reporter plasmid into a retinoid X receptor-expressing SMC line. Cells were then incubated, as described in Materials and Methods, without pronesterone (lanes 1, 4, and 7), with pronesterone (lanes 2, 5, and 8), or with pronesterone plus 1 μM Dex for 3 h (lanes 3, 6, and 9). RNA was harvested and analyzed by RT-PCR using primers specific for each MCP-1 construct as well as primers corresponding to a 501-nt fragment of the neomycin resistance gene as an internal standard for transfection efficiency. For each construct, luciferase activity in pronesterone-treated cells (lanes 2, 3, 5, 6, 8, and 9) was normalized to that of cells not treated with pronesterone (lanes 1, 4, and 7). The gel is representative of duplicate experiments.
Pronesterone induced accumulation of exogenous MCP-1 mRNAs (Fig. 8, lanes 2, 5, and 8). Dex (1 μM) treatment of SMC transfected with the full-length MCP-1 construct resulted in the elimination of exogenous mRNA (lane 9). Similarly, Dex abolished the accumulation of mRNA in cells transfected with a construct containing the 224-nt 5′ Dex-sensitive region (lane 3). SMC transfected with a construct containing a 130-nt Dex-insensitive fragment derived from the 5′ end accumulated high levels of mRNA in the presence of Dex (lane 6).
DISCUSSION
This report describes the destabilization of MCP-1 mRNA by glucocorticoids. Glucocorticoids have been shown to repress the expression of a variety of genes, including those encoding prolactin (55), proliferin (35), pro-opiomelanocortin (12), members of the collagenase family (21), insulin (47), and collagen types I and IV (67). In many cases, this repression is transcriptional and involves binding of the ligand-glucocorticoid receptor complex to specific DNA sequences on the inhibited genes. The rat MCP-1 gene does not possess consensus sequences for either the classical glucocorticoid response element or a putative glucocorticoid inhibitory element (5, 19). Glucocorticoids have been shown to repress the expression of some genes, including those encoding interleukins 1, 2, 6, and 8, granulocyte-macrophage colony-stimulating factor, insulin, and collagen, in whole or in part, by posttranscriptional mechanisms, including changes in mRNA stability (reviewed in reference 1 and 54). Although some of these studies have shown an effect of glucocorticoids on mRNA t1/2, the RNA sequences involved have not been identified.
The Dex-sensitive region was localized to the 5′ end of the MCP-1 mRNA. Destabilizing elements on a variety of mRNAs have been found in the 3′ UTR and have been shown to involve AU-rich sequences (9–11, 13). One such AUUUA element (bases 563 to 567) is present in the MCP-1 3′ UTR. However, our in vitro data suggest that the region involved in the Dex-mediated decay of MCP-1 mRNA resides outside the 3′ UTR and does not involve the AUUUA element. Although additional in vivo data will be necessary to determine whether the 3′ AUUUA element plays any role in regulating MCP-1 mRNA stability, to our knowledge this is the first report identifying a glucocorticoid-sensitive region in the 5′ end of any eukaryotic mRNA. Despite the preponderance of data implicating 3′ UTR elements in regulating mRNA stability, mRNA-destabilizing sequences have been found in a 182-nt sequence in the c-myc coding region (6), in an amino-terminal 12-nt tetrapeptide sequence spanning the initiation codon of β-tubulin (3), and in a 56-nt purine-rich sequence in the coding region of c-fos (14). As determined with the MACAW program (57), the 224-nt Dex-sensitive region shares no significant homology with any of these regions.
Stem-loop structures have been shown to be important in regulating mRNA stability (54). Secondary structure analysis using the MFOLD program (72) revealed three potential stem-loop structures in the 224-nt region (bases 126 to 140, 151 to 186, and 187 to 212) with free energy levels of ≥−4 kcal/mol at 37°C; the sequences of these three stem-loops differ from those previously described as being involved in mRNA stability. Deletions involving the stem-loops at bases 126 to 140 (Fig. 6, row 5) and 187 to 212 (Fig. 6, row 10) abolished the response to Dex, raising the possibility that these structures are involved in mediating mRNA stability. One caveat, however, is that deletion of other regions, involving bases 1 to 40 (not shown) or 1 to 80 (Fig. 6, row 9), also eliminated Dex sensitivity. Unlike cis-acting transcriptional elements, which usually are comprised of short sequences, the cis-acting regions involved in mRNA stability may be considerably longer and encompass protein binding and catalytic sites. Therefore, multiple areas involving most of the 224-nt region may be required for Dex sensitivity. In addition, there may be a minimum size of mRNA required for catalytic activity.
The Dex-mediated decrease in MCP-1 mRNA stability appears to be secondary to its effect on the cell rather than a primary effect of Dex on the mRNA, in that Dex failed to alter mRNA stability when added directly to cytoplasmic extracts from control cells prior to performing the in vitro decay assays. The effect is at least in part mediated through a heat-labile protein(s), probably an RNase, because the activity was eliminated by proteinase K or heating. Of note, neither cycloheximide nor actinomycin D inhibited the Dex effect, suggesting that the protein(s) involved is constitutively expressed and that Dex mediates its activity via a posttranslational mechanism. This may include activation of a latent or relatively inactive RNase, via phosphorylation, glycosylation, or interference with a regulatory binding protein. Alternatively, binding of a Dex-responsive protein to the MCP-1 mRNA might alter its configuration, enhancing the binding or activity of the RNase. It should also be noted that although PDGF markedly increases the t1/2 of MCP-1 mRNA, the effect of Dex supersedes that of PDGF both in vitro and in vivo (49), reducing the t1/2 to similar levels. This suggests that the mechanism(s) underlying the stabilizing effects of PDGF may be different from those mediating the destabilizing effects of Dex and may involve different proteins and/or binding sites.
These results differ from those reported for other glucocorticoid-sensitive mRNAs, such as interleukins 1, 6, and 8, granulocyte-macrophage colony-stimulating factor, and c-myc, in which the effects of glucocorticoids are inhibited by cycloheximide (2, 64) or actinomycin D (33). The lack of response to cycloheximide or actinomycin D is similar to that reported for a polyribosome-associated histone mRNA exonuclease (46) and the estradiol-sensitive Xenopus albumin mRNA endonuclease (18, 36). Cytoplasmic extracts from SMC treated with estrogen had no effect on MCP-1 mRNA decay, suggesting that the properties of the Dex-sensitive protein is likely to be different from those of the estradiol-sensitive Xenopus endonuclease.
The effect of glucocorticoids on MCP-1 mRNA was not cell or species specific. Of particular note, extracts from rat SMC were effective on in vitro-transcribed MCP-1 mRNA from human and rat SMC. Likewise, extracts from human fibroblasts were effective on both species’ transcripts. This is in contrast to MCP-1 transcription, which is regulated differently in rat and human (reviewed by Bogdanov et al. [7]). These data suggest that the proteins mediating the Dex effect are conserved and that information obtained from the rat system may be applicable to human SMC and MCP-1. In addition, the efficacy of extracts from Dex-treated human pulmonary fibroblasts suggests that the effect is not limited to vascular SMC and may be relevant to nonvascular inflammatory processes.
The in vitro decay assay is a common approach to determining rates of mRNA decay (54). In general, mRNA degradation occurs more rapidly in intact cells than in in vitro assays (54). In this study, the rate of in vitro MCP-1 mRNA decay (t1/2 of ≈15 min on nondenaturing gels and ≈8 min on denaturing gels) was similar to, if not higher than, the rate previously determined (49) in cultured SMC (t1/2 ≈30 min). This finding suggests that the cytoplasmic extracts are capable of recapitulating the in vivo effect of Dex on MCP-1 mRNA stability. These extracts should therefore be useful for identification of the protein factor(s) responsible.
It is hard to assess the significance of the apparent higher decay rate in the in vitro system, given the inherent differences in the two assays. It is possible that it is the result of inactivation or loss of an inhibitor of the Dex-sensitive RNase. It should also be noted that virtually all of the experiments in this study were performed with uncapped, nonpolyadenylated transcripts. Although we did not see differences in the decay of uncapped, nonpolyadenylated and capped, polyadenylated transcripts after 30 min of incubation with cell extracts (Fig. 6), it is possible that the use of capped, polyadenylated transcripts would have generated a different t1/2. The higher decay rate calculated from denaturing gels, as opposed to nondenaturing gels, may be due in part to the ability of partially degraded radiolabeled MCP-1 mRNA fragments to remain attached to a protein complex for several minutes and thus appear as part of an intact complex on nondenaturing gels. These fragments may be represented by the lower-molecular-weight species seen at later time points on denaturing gels.
Glucocorticoids possess a wide variety of anti-inflammatory and antiproliferative properties. In SMC culture, Dex has been shown to inhibit cell cycle progression, mitogen-induced proliferation, protein synthesis, and PDGF A-chain expression (31, 39, 41, 50). Although the predominant concentration used for most studies has been 100 nM, maximum effects have been seen with as little as 10 nM (31). Inhibition of SMC growth has most often employed a 24-h pretreatment; however, recent studies have suggested that 6 to 8 h may be necessary to effectively inhibit vascular SMC growth (50) and as little as 1 h may suffice to inhibit airway SMC proliferation (58). In the present study, as well as in our previous study of cultured SMC (49), the maximum effect on MCP-1 mRNA was also seen with concentrations as low as 10 nM. As little as 1 h of exposure to Dex was necessary to obtain maximal inhibition of endogenous MCP-1 expression (49).
The complications of glucocorticoid treatment are protean and limit their long-term use to the most serious disease entities. The present study suggests that in contrast to many effects of glucocorticoids, which are based on stimulation or inhibition of transcription, there may be a smaller set of molecules, which are regulated by changes in mRNA stability. Identification of the Dex-sensitive protein(s) responsible for destabilizing MCP-1 mRNA may therefore provide novel approaches to emulating the anti-inflammatory properties of glucocorticoids without some of the deleterious side effects.
ACKNOWLEDGMENTS
This research was supported in part by National Institutes of Health grants HL43302 and HL61818 to M.B.T. M.P. is a recipient of a Clinician Scientist Award of the American Heart Association and an Arthur Ross Foundation Scholarship.
REFERENCES
- 1.Almawi W Y, Beyhum H N, Rahme A A, Rieder M J. Regulation of cytokine and cytokine receptor expression by glucocorticoids. J Leukoc Biol. 1996;60:563–572. doi: 10.1002/jlb.60.5.563. [DOI] [PubMed] [Google Scholar]
- 2.Amano Y, Lee S W, Allison A C. Inhibition by glucocorticoids of the formation of interleukin-1 alpha, interleukin-1 beta, and interleukin-6: mediation by decreased mRNA stability. Mol Pharmacol. 1993;43:176–182. [PubMed] [Google Scholar]
- 3.Bachurski C J, Theodorakis N G, Coulson R M, Cleveland D W. An amino-terminal tetrapeptide specifies cotranslational degradation of beta-tubulin but not alpha-tubulin mRNAs. Mol Cell Biol. 1994;14:4076–4086. doi: 10.1128/mcb.14.6.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes P J. Anti-inflammatory mechanisms of glucocorticoids. Biochem Soc Trans. 1995;23:940–945. doi: 10.1042/bst0230940. [DOI] [PubMed] [Google Scholar]
- 5.Beato M. Gene regulation by steroid hormones. Cell. 1989;56:335–344. doi: 10.1016/0092-8674(89)90237-7. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein P L, Herrick D J, Prokipcak R D, Ross J. Control of c-myc mRNA half-life in vitro by a protein capable of binding to a coding region stability determinant. Genes Dev. 1992;6:642–654. doi: 10.1101/gad.6.4.642. [DOI] [PubMed] [Google Scholar]
- 7.Bogdanov V Y, Poon M, Taubman M B. PDGF-specific regulation of the JE promoter in rat aortic smooth muscle cells. J Biol Chem. 1998;273:24932–24938. doi: 10.1074/jbc.273.38.24932. [DOI] [PubMed] [Google Scholar]
- 8.Boumpas D T, Chrousos G P, Wilder R L, Cupps T R, Balow J E. Glucocorticoid therapy for immune-mediated diseases: basic and clinical correlates. Ann Intern Med. 1993;119:1198–1208. doi: 10.7326/0003-4819-119-12-199312150-00007. [DOI] [PubMed] [Google Scholar]
- 9.Brewer G. An A+U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol Cell Biol. 1991;11:2460–2466. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brewer G, Ross J. Poly(A) shortening and degradation of the 3′ A+U-rich sequences of human c-myc mRNA in a cell-free system. Mol Cell Biol. 1988;8:1697–1708. doi: 10.1128/mcb.8.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci USA. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charron J, Drouin J. Glucocorticoid inhibition of transcription from episomal proopiomelanocortin gene promoter. Proc Natl Acad Sci USA. 1986;83:8903–8907. doi: 10.1073/pnas.83.23.8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C Y, Xu N, Shyu A B. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol Cell Biol. 1995;15:5777–5788. doi: 10.1128/mcb.15.10.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen C Y, You Y, Shyu A B. Two cellular proteins bind specifically to a purine-rich sequence necessary for the destabilization function of a c-fos protein-coding region determinant of mRNA instability. Mol Cell Biol. 1992;12:5748–5757. doi: 10.1128/mcb.12.12.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clinton S K, Libby P. Cytokines and growth factors in atherogenesis. Arch Pathol Lab Med. 1992;116:1292–1300. [PubMed] [Google Scholar]
- 16.Cochran H B, Reffel C A, Stiles D C. Molecular cloning of gene sequences regulated by platelet-derived growth factor. Cell. 1983;33:939–947. doi: 10.1016/0092-8674(83)90037-5. [DOI] [PubMed] [Google Scholar]
- 17.Cushing S D, Berliner J A, Valente A J, Territo M C, Navab M, Parhami F, Gerrity R, Schwartz C J, Fogelman A M. Minimally modified low density lipoprotein induces monocyte chemotactic protein 1 in human endothelial cells and smooth muscle cells. Proc Natl Acad Sci USA. 1990;87:5134–5138. doi: 10.1073/pnas.87.13.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dompenciel R E, Garnepudi V R, Schoenberg D R. Purification and characterization of an estrogen-regulated Xenopus liver polysomal nuclease involved in the selective destabilization of albumin mRNA. J Biol Chem. 1995;270:6108–6118. doi: 10.1074/jbc.270.11.6108. [DOI] [PubMed] [Google Scholar]
- 19.Evans R M. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faggiotto A, Ross R, Harker L. Studies of hypercholesterolemia in the nonhuman primate. I. Changes that lead to fatty streak formation. Arteriosclerosis. 1984;4:323–340. doi: 10.1161/01.atv.4.4.323. [DOI] [PubMed] [Google Scholar]
- 21.Frisch S M, Ruley H E. Transcription from the stromelysin promoter is induced by interleukin-1 and repressed by dexamethasone. J Biol Chem. 1987;262:16300–16304. [PubMed] [Google Scholar]
- 22.Furutani Y, Nomura H, Notake M, Oyamada Y, Toshikazu F, Yamada M, Larsen G C, Oppenheim J J, Matsushima K. Cloning and sequencing of the cDNA for human monocyte chemotactic and activating factor (MCAF) Biochem Biophys Res Commun. 1989;159:249–255. doi: 10.1016/0006-291x(89)92430-3. [DOI] [PubMed] [Google Scholar]
- 23.Galis Z S, Sukhova G K, Kranzhofer R, Clark S, Libby P. Macrophage foam cells from experimental atheroma constitutively produce matrix-degrading proteinases. Proc Natl Acad Sci USA. 1995;92:402–406. doi: 10.1073/pnas.92.2.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerrity R G. The role of the monocyte in atherogenesis. I. Transition of blood-borne monocytes into foam cells in fatty lesions. Am J Pathol. 1981;103:181–190. [PMC free article] [PubMed] [Google Scholar]
- 25.Harigai M, Hara M, Yoshimura T, Leonard E J, Inoue K, Kashiwazaki S. Monocyte chemoattractant protein-1 (MCP-1) in inflammatory joint diseases and its involvement in the cytokine network of rheumatoid synovium. Clin Immunol Immunopathol. 1993;69:83–91. doi: 10.1006/clin.1993.1153. [DOI] [PubMed] [Google Scholar]
- 26.Hartner A, Goppelt-Struebe M, Hocke G M, Sterzel R B. Differential regulation of chemokines by leukemia inhibitory factor, interleukin-6 and oncostatin M. Kidney Int. 1997;51:1754–1760. doi: 10.1038/ki.1997.241. [DOI] [PubMed] [Google Scholar]
- 27.Kawahara R S, Deng Z W, Deuel T F. Glucocorticoids inhibit the transcriptional induction of JE, a platelet-derived growth factor-inducible gene. J Biol Chem. 1991;266:13261–13266. [PubMed] [Google Scholar]
- 28.Klurfeld D M. Identification of foam cells in human atherosclerotic lesions as macrophages using monoclonal antibodies. Arch Pathol Lab Med. 1985;109:445–449. [PubMed] [Google Scholar]
- 29.Leonard E J, Yoshimura T. Human monocyte chemoattractant protein-1 (MCP-1) Immunol Today. 1990;11:97–101. doi: 10.1016/0167-5699(90)90035-8. [DOI] [PubMed] [Google Scholar]
- 30.Libby P, Sukhova G, Lee R T, Galis Z S. Cytokines regulate vascular functions related to stability of the atherosclerotic plaque. J Cardiovasc Pharmacol. 1995;25(Suppl 2):S9–S12. doi: 10.1097/00005344-199500252-00003. [DOI] [PubMed] [Google Scholar]
- 31.Longenecker J P, Kilty L A, Johnson L K. Glucocorticoid inhibition of vascular smooth muscle cell proliferation: influence of homologous extracellular matrix and serum mitogens. J Cell Biol. 1984;98:534–540. doi: 10.1083/jcb.98.2.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackman N. Regulation of the tissue factor gene. FASEB J. 1995;9:883–889. doi: 10.1096/fasebj.9.10.7615158. [DOI] [PubMed] [Google Scholar]
- 33.Maroder M, Martinotti S, Vacca A, Screpanti I, Petrangeli E, Frati L, Gulino A. Post-transcriptional control of c-myc proto-oncogene expression by glucocorticoid hormones in human T lymphoblastic leukemic cells. Nucleic Acids Res. 1990;18:1153–1157. doi: 10.1093/nar/18.5.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyamasu M, Misaki Y, Izumi S, Takaishi T, Morita Y, Nakamura H, Matsushima K, Kasahara T, Hirai K. Glucocorticoids inhibit chemokine generation by human eosinophils. J Allergy Clin Immunol. 1998;101:75–83. doi: 10.1016/S0091-6749(98)70196-4. [DOI] [PubMed] [Google Scholar]
- 35.Mordacq J C, Linzer D I. Co-localization of elements required for phorbol ester stimulation and glucocorticoid repression of proliferin gene expression. Genes Dev. 1989;3:760–769. doi: 10.1101/gad.3.6.760. [DOI] [PubMed] [Google Scholar]
- 36.Moskaitis J E, Buzek S W, Pastori R L, Schoenberg D R. The estrogen-regulated destabilization of Xenopus albumin mRNA is independent of translation. Biochem Biophys Res Commun. 1991;174:825–830. doi: 10.1016/0006-291x(91)91492-u. [DOI] [PubMed] [Google Scholar]
- 37.Mukaida N, Zachariae C C, Gusella G L, Matsushima K. Dexamethasone inhibits the induction of monocyte chemotactic-activating factor production by IL-1 or tumor necrosis factor. J Immunol. 1991;146:1212–1215. [PubMed] [Google Scholar]
- 38.Naito M, Yasue M, Asai K, Yamada K, Hayashi T, Kuzuya M, Funaki C, Yoshimine N, Kuzuya F. Effects of dexamethasone on experimental atherosclerosis in cholesterol-fed rabbits. J Nutr Sci Vitaminol (Tokyo) 1992;38:255–264. doi: 10.3177/jnsv.38.255. [DOI] [PubMed] [Google Scholar]
- 39.Nakano T, Raines E W, Abraham J A, Wenzel F G T, Higashiyama S, Klagsbrun M, Ross R. Glucocorticoid inhibits thrombin-induced expression of platelet-derived growth factor A-chain and heparin-binding epidermal growth factor-like growth factor in human aortic smooth muscle cells. J Biol Chem. 1993;268:22941–22947. [PubMed] [Google Scholar]
- 40.Nelken N A, Coughlin S R, Gordon D, Wilcox J N. Monocyte chemoattractant protein-1 in human atheromatous plaques. J Clin Investig. 1991;88:1121–1127. doi: 10.1172/JCI115411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nichols N R, Olsson C A, Funder J W. Steroid effects on protein synthesis in cultured smooth muscle cells from rat aorta. Endocrinology. 1983;113:1096–1101. doi: 10.1210/endo-113-3-1096. [DOI] [PubMed] [Google Scholar]
- 42.Nikkari S T, O’Brien K D, Ferguson M, Hatsukami T, Welgus H G, Alpers C E, Clowes A W. Interstitial collagenase (MMP-1) expression in human carotid atherosclerosis. Circulation. 1995;92:1393–1398. doi: 10.1161/01.cir.92.6.1393. [DOI] [PubMed] [Google Scholar]
- 43.No D, Yao T P, Evans R M. Ecdysone-inducible gene expression in mammalian cells and transgenic mice. Proc Natl Acad Sci USA. 1996;93:3346–3351. doi: 10.1073/pnas.93.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nordeen S K. Luciferase reporter gene vectors for analysis of promoters and enhancers. BioTechniques. 1988;6:454–458. [PubMed] [Google Scholar]
- 45.Paine R D, Rolfe M W, Standiford T J, Burdick M D, Rollins B J, Strieter R M. MCP-1 expression by rat type II alveolar epithelial cells in primary culture. J Immunol. 1993;150:4561–4570. [PubMed] [Google Scholar]
- 46.Peltz S W, Brewer G, Groppi V, Ross J. Exonuclease activity that degrades histone mRNA is stable when DNA or protein synthesis is inhibited. Mol Biol Med. 1989;6:227–238. [PubMed] [Google Scholar]
- 47.Philippe J, Missotten M. Dexamethasone inhibits insulin biosynthesis by destabilizing insulin messenger ribonucleic acid in hamster insulinoma cells. Endocrinology. 1990;127:1640–1645. doi: 10.1210/endo-127-4-1640. [DOI] [PubMed] [Google Scholar]
- 47a.Poon, M., S. D. Gertz, J. T. Fallon, P. Wiegman, J. W. Berman, I. J. Sarembock, and M. B. Taubman. Submitted for publication. [DOI] [PubMed]
- 48.Poon M, Hsu W C, Bogadanov V Y, Taubman M B. Secretion of monocyte chemotactic activity by cultured rat aortic smooth muscle cells in response to PDGF is due predominantly to the induction of JE/MCP-1. Am J Pathol. 1996;149:307–317. [PMC free article] [PubMed] [Google Scholar]
- 49.Poon M, Megyesi J, Green R S, Zhang H, Rollins B J, Safirstein R, Taubman M B. In vivo and in vitro inhibition of JE gene expression by glucocorticoids. J Biol Chem. 1991;266:22375–22379. [PubMed] [Google Scholar]
- 50.Reil T D, Sarkar R, Kashyap V S, Sarkar M, Gelabert H A. Dexamethasone suppresses vascular smooth muscle cell proliferation. J Surg Res. 1999;85:109–114. doi: 10.1006/jsre.1999.5665. [DOI] [PubMed] [Google Scholar]
- 51.Rivers A P R, Hathaway E W, Weston L W. The endotoxin-induced coagulant activity of human monocytes. Br J Haematol. 1975;30:311–316. doi: 10.1111/j.1365-2141.1975.tb00547.x. [DOI] [PubMed] [Google Scholar]
- 52.Rollins B J, Stier P, Ernst T, Wong G G. The human homolog of the JE gene encodes a monocyte secretory protein. Mol Cell Biol. 1989;9:4687–4695. doi: 10.1128/mcb.9.11.4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rollins B J, Yoshimura T, Leonard E J, Pober J S. Cytokine-activated human endothelial cells synthesize and secrete a monocyte chemoattractant, MCP-1/JE. Am J Pathol. 1990;136:1229–1233. [PMC free article] [PubMed] [Google Scholar]
- 54.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sakai D D, Helms S, Carlstedt-Duke J, Gustafsson J A, Rottman F M, Yamamoto K R. Hormone-mediated repression: a negative glucocorticoid response element from the bovine prolactin gene. Genes Dev. 1988;2:1144–1154. doi: 10.1101/gad.2.9.1144. [DOI] [PubMed] [Google Scholar]
- 56.Sato K, Kuratsu J, Takeshima H, Yoshimura T, Ushio Y. Expression of monocyte chemoattractant protein-1 in meningioma. J Neurosurg. 1995;82:874–878. doi: 10.3171/jns.1995.82.5.0874. [DOI] [PubMed] [Google Scholar]
- 57.Schuler G D, Altschul S F, Lipman D J. A workbench for multiple alignment construction and analysis. Proteins. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- 58.Stewart A G, Fernandes D, Tomlinson P R. The effect of glucocorticoids on proliferation of human cultured airway smooth muscle. Br J Pharmacol. 1995;116:3219–3226. doi: 10.1111/j.1476-5381.1995.tb15127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strieter R M, Wiggins R, Phan S H, Wharram B L, Showell H J, Remick D G, Chensue S W, Kunkel S L. Monocyte chemotactic protein gene expression by cytokine-treated human fibroblasts and endothelial cells. Biochem Biophys Res Commun. 1989;162:694–700. doi: 10.1016/0006-291x(89)92366-8. [DOI] [PubMed] [Google Scholar]
- 60.Takeya M, Yoshimura T, Leonard E J, Takahashi K. Detection of monocyte chemoattractant protein-1 in human atherosclerotic lesions by an anti-monocyte chemoattractant protein-1 monoclonal antibody. Hum Pathol. 1993;24:534–539. doi: 10.1016/0046-8177(93)90166-e. [DOI] [PubMed] [Google Scholar]
- 61.Taubman M B, Marmur J D, Rosenfield C L, Guha A, Nichtberger S, Nemerson Y. Agonist-mediated tissue factor expression in cultured vascular smooth muscle cells. Role of Ca2+ mobilization and protein kinase C activation. J Clin Investig. 1993;91:547–552. doi: 10.1172/JCI116234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taubman M B, Rollins B J, Poon M, Marmur J, Green R S, Berk B C, Nadal-Ginard B. JE mRNA accumulates rapidly in aortic injury and in platelet-derived growth factor-stimulated vascular smooth muscle cells. Circ Res. 1992;70:314–325. doi: 10.1161/01.res.70.2.314. [DOI] [PubMed] [Google Scholar]
- 63.Thiruvikraman S V, Guha A, Roboz J, Taubman M B, Nemerson Y, Fallon J T. In situ localization of tissue factor in human atherosclerotic plaques by binding of digoxigenin-labeled factors VIIa and X. Lab Investig. 1996;75:451–461. [PubMed] [Google Scholar]
- 64.Tobler A, Meier R, Seitz M, Dewald B, Baggiolini M, Fey M F. Glucocorticoids downregulate gene expression of GM-CSF, NAP-1/IL-8, and IL-6, but not of M-CSF in human fibroblasts. Blood. 1992;79:45–51. [PubMed] [Google Scholar]
- 65.Valente A J, Graves D T, Vialle-Valentin C E, Delgado R, Schwartz C J. Purification of a monocyte chemotactic factor secreted by nonhuman primate vascular cells in culture. Biochemistry. 1988;27:4162–4168. doi: 10.1021/bi00411a039. [DOI] [PubMed] [Google Scholar]
- 66.Wang X, Kiledjian M, Weiss I M, Liebhaber S A. Detection and characterization of a 3′ untranslated region ribonucleoprotein complex associated with human alpha-globin mRNA stability. Mol Cell Biol. 1995;15:1769–1777. doi: 10.1128/mcb.15.3.1769. . (Erratum, 15:2331.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weiner F R, Czaja M J, Jefferson D M, Giambrone M A, Tur-Kaspa R, Reid L M, Zern M A. The effects of dexamethasone on in vitro collagen gene expression. J Biol Chem. 1987;262:6955–6958. [PubMed] [Google Scholar]
- 68.Yla-Herttuala S, Lipton B A, Rosenfeld M E, Sarkioja T, Yoshimura T, Leonard E J, Witztum J L, Steinberg D. Expression of monocyte chemoattractant protein 1 in macrophage-rich areas of human and rabbit atherosclerotic lesions. Proc Natl Acad Sci USA. 1991;88:5252–5256. doi: 10.1073/pnas.88.12.5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yoshimura T, Robinson E A, Tanaka S, Appella E, Kuratsu J, Leonard E J. Purification and amino acid analysis of two human glioma-derived monocyte chemoattractants. J Exp Med. 1989;169:1449–1459. doi: 10.1084/jem.169.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yoshimura T, Yuhki N, Moore S K, Appella E, Lerman M I, Leonard E J. Human monocyte chemoattractant protein-1 (MCP-1). Full-length cDNA cloning, expression in mitogen-stimulated blood mononuclear leukocytes, and sequence similarity to mouse competence gene JE. FEBS Lett. 1989;244:487–493. doi: 10.1016/0014-5793(89)80590-3. [DOI] [PubMed] [Google Scholar]
- 71.Yu X, Dluz S, Graves D T, Zhang L, Antoniades H N, Hollander W, Prusty S, Valente A J, Schwartz C J, Sonenshein G E. Elevated expression of monocyte chemoattractant protein 1 by vascular smooth muscle cells in hypercholesterolemic primates. Proc Natl Acad Sci USA. 1992;89:6953–6957. doi: 10.1073/pnas.89.15.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]