Abstract
We report that the recently introduced commercial strain of Vibrio natriegens (Vmax X2) supports robust unnatural amino acid mutagenesis, generating exceptional yields of soluble protein containing up to 5 noncanonical α-amino acids (ncAA). The isolated yields of ncAA-containing superfolder green fluorescent protein (sfGFP) expressed in Vmax X2 are up to 25-fold higher than those achieved using commercial expression strains (Top10 and BL21) and more than 10-fold higher than those achieved using two different genomically recodedEscherichia colistrains that lack endogenous UAG stop codons and release factor 1 and have been optimized for improved fitness and preferred growth temperature (C321.ΔA.opt and C321.ΔA.exp). In addition to higher yields of soluble protein, Vmax X2 cells also generate proteins with significantly lower levels of misincorporated natural α-amino acids at the UAG-programmed position, especially in cases where the ncAA is a moderate substrate for the chosen orthogonal aminoacyl tRNA synthetase (aaRS). This increase in fidelity implies that the use of Vmax X2 cells as the expression host can obviate the need for time-consuming directed evolution experiments to improve the selectivity of an aaRS toward highly desired but suboptimal ncAA substrates.
Short abstract
The engineered strain Vmax X2 is an excellent vehicle for genetic code expansion, generating high titers of proteins containing multiple, diverse, and unnatural amino acids with very high fidelity.
Introduction
Noncanonical amino acid mutagenesis, often referred to as genetic code expansion (GCE), is a powerful tool for introducing unique chemical functionality or reactivity into an expressed protein.1−3 Literally hundreds of diverse noncanonical α-amino acids (ncAA), including those that support protein labeling or conjugation, can be introduced into proteins3−6 biosynthesized in laboratory Escherichia coli strains, pathogenic7 and soil bacteria,8 yeast,9 mammalian cells,9−11 and whole organisms.12−14 Genetic code expansion in vivo demands the coexpression of one or more aminoacyl-tRNA synthetase (aaRS)/tRNA pairs that are orthogonal in the expression host. Commonly used pairs include variants of pyrrolysyl aminoacyl-tRNA synthetase (PylRS)/pylT from Methanosarcina,15−19 tyrosyl aminoacyl-tRNA synthetase (TyrRS)/tRNATyr from Methanococcus jannaschii,20−23 and others;24−26 newly identified orthogonal pairs include those from Ilumatobacter nonamiensis, Sorangium cellulosum, and Archaeoglobus fulgidus.24
In addition to its utility in basic research, GCE has significant and growing importance in the pharmaceutical and biotechnological industries. The ability to efficiently introduce reactive bioorthogonal functionality into a therapeutic antibody provides a streamlined route to homogeneous antibody–drug conjugates with high (∼95%) conjugation efficiency.27−29 The incorporation of noncanonical α-amino acids can be leveraged to confer favorable therapeutic properties, such as increased circulation half-life and improved bioactivity.30−32 Finally, GCE expands the chemical space that is accessible for the development of novel macrocycles and therapeutic peptides.33−35
For almost all genetic code expansion applications, yield and purity are paramount.36 One factor that can limit the yield of a protein carrying one or more ncAAs is competition between the acylated suppressor tRNA and release factor 1 (RF1), both of which recognize the amber UAG stop codon. Recognition of the acylated suppressor tRNA leads to incorporation of the ncAA, whereas recognition by RF1 triggers early translation termination.37,38 Another factor that can limit the yield of the desired ncAA-containing product is that the orthogonality of an aaRS/tRNA pair is rarely absolute, resulting in suppressor tRNAs that are acylated incorrectly with one or more α-amino acids. To circumvent this issue, researchers have developed E. coli strains that lack RF1, including those that have been genomically recoded to eliminate all or some of the 321 endogenous UAG stop codons in E. coli.39−41 While these strains can improve expression yields, genomically recoded organisms that lack RF1 suffer from fitness defects as well as higher levels of misincorporation events when utilizing suboptimal aaRS/tRNA pairs.42−45
Cell-free translation systems offer the opportunity to omit RF1 as well as tune the individual levels of near-cognate tRNAs to potentially increase incorporation fidelity, but these systems are significantly more costly than cellular bioproduction.46,47 Nevertheless, cell-free translation is used widely to substantially reprogram the genetic code and incorporate multiple, distinct ncAAs (even non-α-amino acid and 1,3-dicarbonyl monomers) into a single protein chain, especially on a small scale.33,48−52 Further challenges arise when high-yield expression of the target protein in E. coli demands low temperatures. Many orthogonal synthetases are derived from thermophilic organisms and exhibit minimal activity at temperatures below 25 °C.53,54 The requirement for higher expression temperatures can lower the yield of target proteins that are unstable and/or insoluble under these conditions. Consequently, there is a need for novel avenues to improve the expression yield of proteins containing ncAAs.
Vibrio natriegens, a bacteria isolated originally from saltmarsh mud, is the fastest growing organism on record and expresses many recombinant proteins in exceptional yields and at a variety of temperatures (20–37 °C).55−61 Previous research has revealed considerable compatibility between E. coli and V. natriegens in terms of commonly used genetic elements (i.e., promoters, ribosome binding sites, etc.) and plasmids.57,62 This compatibility facilitates the use of numerous extensively optimized E. coli GCE plasmid systems with little or no plasmid modification. Vmax X2 cells also possess advantages for the production of protein destined for use in animals, where endotoxin contamination remains a persistent concern, reportedly generating endotoxin titers akin to those seen in cells like ClearColi cells.63,64 Moreover, it was recently reported that V. natriegens is amenable to GCE via the use of MmPylRS.65
Here, we demonstrate that the recently introduced commercial strain of V. natriegens (Vmax X2) supports the robust incorporation of noncanonical α-amino acids into proteins, generating exceptional yields of soluble protein containing up to 5 ncAAs.65 Yields are especially high when ncAAs are introduced using the M. jannaschii tyrosyl aminoacyl-tRNA synthetase (TyrRS)/tRNATyr variant pCNFRS.20,21 The isolated yields of ncAA-containing superfolder green fluorescent protein (sfGFP) expressed in Vmax X2 are up to 25-fold higher than those achieved using commercial expression strains (Top10 and BL21) and more than 10-fold higher than those achieved using two different genomically recoded E. coli strains that lack endogenous UAG stop codons and have been optimized for improved fitness and preferred growth temperature (C321.ΔA.opt and C321.ΔA.exp, Addgene strains 87359 and 49018).39,40 The rapid doubling time of Vmax X2 (∼10–14 min)57 also translates into a highly convenient 3 day workflow for protein expression, as opposed to the traditional 4 day workflow for protein expression using traditional E. coli strains. In addition to high yields, Vmax X2 cells also generate proteins with significantly lower levels of misincorporated natural α-amino acids at the UAG-programmed position, especially in cases where the ncAA is a subpar substrate for the chosen orthogonal aminoacyl tRNA synthetase (aaRS). Thus, the use of Vmax X2 can obviate the need for time-consuming directed evolution experiments to improve the specific activity of highly desirable but nonpreferred aaRS substrates.
Results
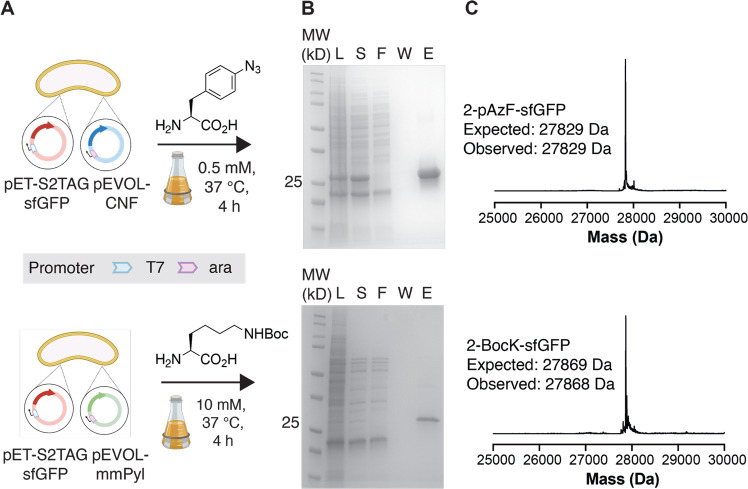
To evaluate Vmax X2 as a host organism for genetic code expansion, we first asked if it would support the incorporation of a single ncAA into sfGFP using two orthogonal translation systems (OTSs) that are used commonly in E. coli. The first is the p-cyano-l-phenylalanyl aminoacyl-tRNA synthetase (pCNFRS)-tRNACUATyr pair20,21 derived from M. jannaschii, while the second is the pyrrolysyl-tRNA synthetase (PylRS)-tRNACUAPyl pair15 derived from Methanosarcina mazei. Both of these aaRS/tRNA pairs support the incorporation of chemically diverse ncAA into proteins in E. coli, but only the activity of the PylRS-tRNACUAPyl pair15 has been tested in Vibrio natriegens.65 Specifically, we asked whether either of these aaRS/tRNA pairs would support the incorporation of p-azido-l-phenylalanine (using pCNFRS) or Boc-l-lysine (using PylRS) at position 2 of sfGFP (Figure 1A). Vmax X2 cells were transformed with either pEVOL-mmPyl or pEVOL-CNF66 (along with pET-S2TAG sfGFP67) and grown in Vmax-optimized media supplemented with either 0.5 mM 4-azido-l-phenylalanine (pAzF) or 10 mM Boc-l-lysine (BocK)68 at a temperature of 37 °C. Vmax X2 cells grew quickly under these conditions, reaching saturation after approximately 4 h (Figure 2A and Figure S1).
Figure 1.
Genetic code expansion in Vmax X2. (A) Vmax X2 cells were transformed with pET-S2TAGsfGFP67 and either pEVOL-CNF66 or pEVOL-mmPyl to induce the expression of sfGFP bearing an ncAA at the second position of sfGFP. Cells were induced and incubated for 4 h at 37 °C in the presence of 0.5 mM p-azido-l-phenylalanine (pAzF) (pCNFRS) or 10 mM l-Boc-lysine (BocK) (MmPylRS). (B) SDS-PAGE gels illustrate proteins produced in Vmax X2 cells transformed with pET-S2TAGsfGFP and either pEVOL-CNF or pEVOL-mmPyl. L = lysate; S = supernatant; F = flow-through; W = wash; E = elution. (C) Intact protein mass spectra of 2TAG sfGFP variants purified from Vmax X2 cells coexpressing pCNFRS (top) or MmPylRS (bottom) in the presence of pAzF or BocK, respectively.
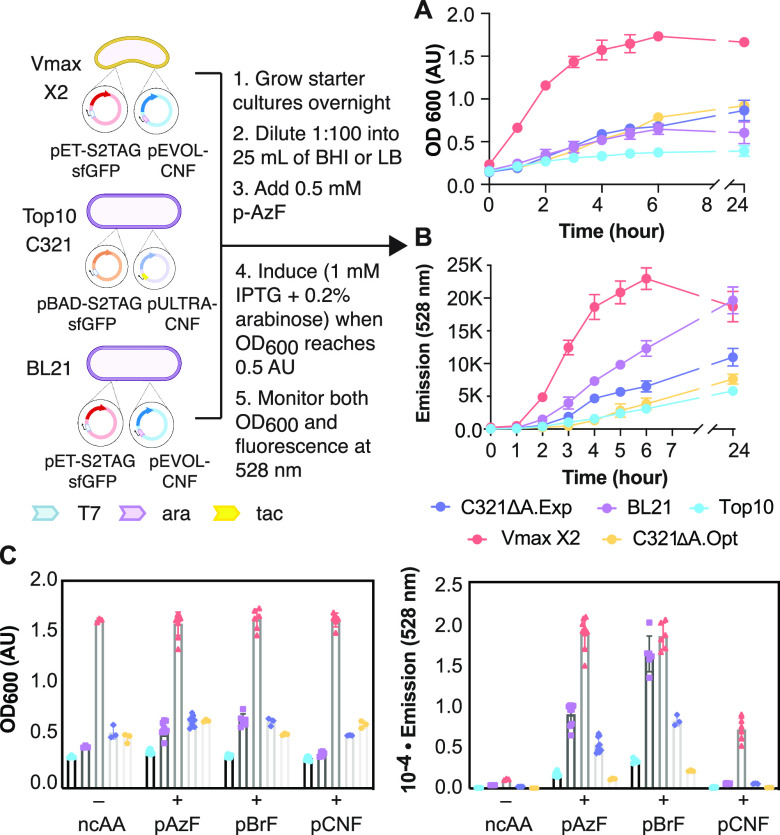
Figure 2.
Growth and sfGFP expression in Vmax X2 versus traditional (Top10, BL21) and genomically recoded (C321)39,40E. coli strains. (A) Plot of the OD600 of each cell growth as a function of time. Vmax X2 and BL21 cells were transformed with pET-S2TAGsfGFP and pEVOL-CNF, whereas Top10 and C321 cells were transformed with pBAD-S2TAGsfGFP and pULTRA-CNF (Top10, C321) to induce expression of sfGFP bearing an ncAA at the second position of sfGFP. After induction, cells were grown for 24 h at 37 °C (Vmax X2, BL21, Top10, C321.ΔA.exp) or 34 °C (C321.ΔA.opt) in the presence of 0.5 mM pAzF. (B) Plot of the emission of each cell growth at 528 nm as a function of time. (C) Plots comparing the OD600 and 528 nm fluorescence of each growth at the 4 h time point in the presence or absence of pAzF, p-bromo-l-phenylalanine (pBrF), or p-cyano-l-phenylalanine (pCNF).
The 2TAG sfGFP variants expressed in Vmax X2 during a 4 h incubation were isolated using immobilized metal affinity chromatography (IMAC) and their purity and identity assessed using SDS-PAGE and mass spectrometry (Figure 1B,C). In each case, an SDS-PAGE evaluation of the isolated protein products revealed a prominent band just above the 25 kDa marker as expected, whose intact mass spectrum was consistent with the incorporation of a single copy of either BocK or pAzF. To verify that the noncanonical α-amino acid was introduced into the expected (second) position, the proteins produced in Vmax X2 cells were digested with the endoproteinase Glu-C and analyzed further by LC-MS/MS (Figure S2). Sequence matching of the digested peptides confirmed the incorporation of BocK and pAzF at position 2 of sfGFP. These data indicate that the orthogonal synthetases pCNFRS and PylRS are expressed and active in Vmax X2 cells and acylate their cognate tRNAs with the provided ncAA and that the charged tRNA is utilized by the Vmax X2 translational machinery. The yield of sfGFP containing an ncAA at position 2 was 8.8 mg (BocK) and 387.2 mg (pAzF) per liter of culture. The yield of sfGFP containing BocK at position 2 is more than 8-fold higher than that observed previously (∼1 mg/L), likely due to the use of a stronger promoter for the sfGFP transcript, as well as ncAA-dependent effects.65
Next, we set out to evaluate how the yield of ncAA-containing sfGFP produced in Vmax X2 cells compared to those obtained in several E. coli strains used for protein expression and genetic code expansion. The strains evaluated included Top10 (a broad utility strain related to DH10B), BL21 (optimized for protein expression from T7 promoters), as well as two genomically recoded strains, C321.ΔA.exp39 and C321.ΔA.opt,40 in which the 321 endogenous UAG stop codons are replaced by the alternative stop codon UAA. Both GROs are derived from the original C321.ΔA; the former has been engineered to lower the rate of spontaneous mutagenesis and can be grown at 37 °C, and the latter carries additional mutations that improve doubling time. Because C321 strains lack T7 RNA polymerase,39 sfGFP expression in these strains is under the control of the commonly used pBAD promoter. All strains were grown under their own optimized conditions and in the presence of 0.5 mM pAzF; both OD600 and fluorescence at 528 nm (λmax for sfGFP) were monitored as a function of time (Figure 2A).
As expected, the Vmax X2 cultures grew faster than all others and reached saturation at an OD600 of 1.5 approximately 4 h after induction. All other strains required more than 24 h to reach an OD600 of 1.0. As judged by the emission value at 528 nm, sfGFP expression in Vmax X2 reached a maximal value after 6 h of expression and then decreased slightly. In the case of all other strains, the signal at 528 nm increased linearly over time over the entire course of the experiment (24 h). The greatest difference in OD600 and fluorescence occurred at the 4 h time point (Figure 2A).
Examination of the OD600 and emission at 528 nm after 4 h of incubation in the presence of three different pCNFRS substrates (pAzF, pBrF, and pCNF) reveals several trends (Figure 2C). First, as expected, the Vmax X2 growth rate exceeded that of any other strain in the absence of an ncAA or in the presence of 0.5 mM pAzF, pBrF, or pCNF. The presence or the identity of the ncAA had little or no effect on the growth rate of any strain examined. The changes in fluorescence at 528 nm show more significant changes and greater dependence on ncAA identity. For example, although the values for 528 nm emission in the presence of pAzF mirrored the OD600 values across all strains, the values in the presence of pBrF were higher than expected in BL21. The values for 528 nm emission in the presence of pCNF were low in all strains other than Vmax X2. Despite these differences, two overarching trends emerge: in all cases, the 528 nm emission values suggest that higher levels of sfGFP are produced in Vmax X2 than in any other strain tested and that the yields in BL21 cells exceed those obtained in either C321.ΔA.exp39 or C321.ΔA.opt.40 A plot of 528 nm fluorescence/OD600 (Figure S1C) shows that Vmax X2 cells often express less sfGFP per cell, except in the case where pCNF was incorporated, compared to BL21 and C321.ΔA.exp.
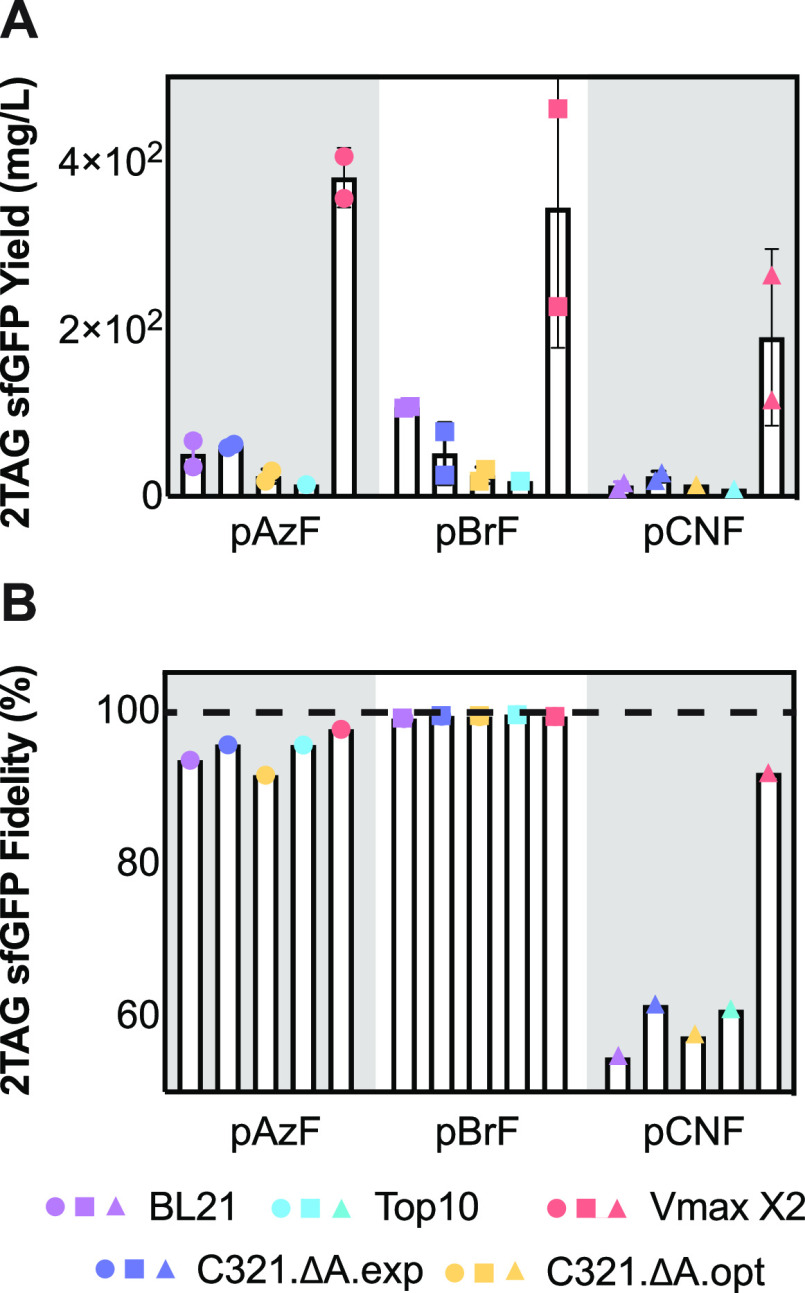
Although fluorescence emission at 528 nm is often taken as a measure of sfGFP expression, for most applications it is the isolated, purified protein yield that matters more, and isolated yields can be affected negatively if aggregation occurs at a high protein concentration.69 To evaluate whether the isolated yields would parallel fluorescence at 528 nm, we isolated sfGFP from each strain after a 4 h growth (Figure 3A and Table S1). The unoptimized yield of isolated sfGFP produced in Vmax X2 cells was at least 2-fold and in some cases as much as 25-fold higher than the yield obtained in any E. coli strain tested. In the case of pAzF, the yield of sfGFP produced in Vmax was 25-fold higher than the yield obtained in either Top10 or C321.ΔA.opt cells. In fact, although protein yields can be affected negatively by high expression titers, for Vmax X2, the opposite trend is observed: significantly more soluble protein is obtained per unit 528 nm absorbance in Vmax X2 than in any other strain, across all ncAA examined (Figure S3). As discussed more extensively below, these experiments necessitated the use of different plasmid/promoter systems for the expression of sfGFP and the aaRS, as Top10 and C321 strains do not encode T7 RNA polymerase (T7 RNAP), and the use of different promoters may also affect protein yields. Integration of T7 RNAP into the C321 strains would allow the use of the T7 promoter in these cells and would likely improve protein yields.70 Notwithstanding, these experiments highlight the ability of Vmax X2 cells to produce significant amounts of ncAA-containing protein at short time periods.
Figure 3.
Yield and purity of sfGFP containing a single ncAA when expressed in Vmax X2 versus E. coli strains. (A) Isolated yield (mg/L) and (B) fidelity (%) of sfGFP containing the indicated ncAA at position 2 when expressed in the indicated strain. Cells were grown for 4 h and lysed via sonication, and sfGFP was isolated using IMAC. Yields were determined using the Pierce 660 nm protein assay (Thermo Scientific) and a BSA standard curve. Fidelity was determined from LC-MS/MS data as the fraction of the Glu-C-generated N-terminal peptide MXKGEE containing the desired ncAA (X) at position 2 relative to all other detectable amino acids at that position. The dashed line indicates 100% fidelity. Additional LC-MS/MS data are found in Figures S4 and S5.
One strain-dependent complication of GCE is misincorporation of an endogenous α-amino acid in place of the desired ncAA.42−44 Many factors can influence the misincorporation rate, including inefficient transport of the ncAA into cells, imperfect specificity of the aaRS enzyme for the ncAA over natural α-amino acids, differences in EF-Tu-mediated delivery of the misacylated tRNA to the ribosome, and slower rates of accommodation, elongation, or translocation of misacylated tRNAs.43,71,72 Misincorporation at an amber stop codon can also arise from near-cognate suppression, in which natural (and correctly acylated) tRNAs pair imperfectly with a UAG codon and direct the incorporation of one or more proteinogenic α-amino acids in place of the ncAA. The frequency of near-cognate suppression events depends on many factors, including the concentration of charged suppressor tRNA relative to near-cognate tRNAs and the presence of RF1, with a higher occurrence of near-cognate suppression when the level of charged tRNA is low, and RF1 is absent.42 It has been found that G/U mismatch as well as the third-base wobble mismatch are the most common errors during codon recognition, followed by some other single-base mismatches (A/C, U/U, and U/C).73,74 It follows that the aminoacyl-tRNA of tyrosine (both codons), glutamine (CAG codon), tryptophan, lysine (AAG codon), and glutamic acid (GAG codon) are likely near-cognate suppressors of UAG, in agreement with observations here and elsewhere.75 Additionally, phenylalanine and tyrosine misincorporations are also expected due to tRNA mischarging for their structural similarities to the ncAAs (pAzF, pBrF, and pCNF).
We used high-resolution LC-MS/MS to evaluate and compare the strain-dependent fidelity of ncAA incorporation at position 2 of sfGFP using pCNFRS (Figure 3B and Figure S6). In each case, sfGFP samples were isolated from each strain after 4 h using IMAC, denatured, reduced and alkylated with iodoacetamide, and digested with Glu-C. The peptide fragments so-generated were analyzed on an LC-MS/MS system composed of an Agilent 1290-II HPLC and a Thermo Fisher Q Exactive Biopharma mass spectrometer. The mass spectrometry data were searched against the sfGFP sequence using MassAnalyzer (an in-house developed program, available in Biopharma Finder from Thermo Fisher) for potential amino acid substitutions.73
The LC-MS/MS data revealed that the yield of sfGFP containing the desired ncAA at position 2 was both strain- and ncAA-dependent (Figure S6). As expected from the reported substrate preferences of pCNFRS, the highest and lowest fidelity were observed with pBrF and pCNF, respectively.21 The yield of sfGFP containing pBrF at position 2 ranged from a high of 99.7% (in Top10 cells) to a low of 99.4% (BL21 cells). With pAzF, the range was slightly greater, from a high of 97.6% (Vmax X2) to a low of 91.7% (C321.ΔA.opt). The lowest fidelity was observed for the incorporation of pCNF. Here, the fidelity was low (between 54.5% and 61.6%, in line with previously reported values76) in all strains except Vmax X2, where the fidelity was 91.8%. It is notable that the most significant impurities, regardless of strain, contained Phe in place of the desired ncAA, although in certain cases a significant amount of Tyr was also detected (Figure S6). The improved fidelity of sfGFP containing an ncAA at position 2 when produced in Vmax X2 could be due to many factors, including differences in ncAA permeability, aaRS activity and expression levels, tolerance of Vmax X2 EF-Tu to misacylated tRNAs, and even the relative activity and expression of Vmax X2 RF1.77,78 Another potential factor is degradation of the ncAA due to cellular metabolic processes, which may differ betweenE. coliand V. natriegens.79,80 Although ncAA side chains can also degrade during MS analysis or sample workup, as all samples were prepared identically, we would expect that such occurrences would be fairly constant. Nevertheless, if the goal is a homogeneous and uniquely modified ncAA-containing protein in high yield and purity, Vmax X2 outperformed other strains, even those that funnel GCE through a single UAG channel. This statement is especially true in the case of ncAA possessing moderate to low specific activity, which could obviate the need for directed evolution to improve specific activity further.
As noted above, the plasmids used to express the aaRS/tRNA pair and sfGFP in Vmax X2 cells were identical with the plasmids used in BL21, but those used in C321 and Top10 cells were different. In the case of Vmax X2 and BL21, the aaRS/tRNA pair was under the control of an arabinose-dependent promoter, and sfGFP expression was under the control of the T7 promoter. However, as C321.ΔA.exp, C321.ΔA.opt, and Top10 cells lack T7 RNA polymerase, in this case, aaRS/tRNA expression was under the control of a tac promoter, and sfGFP was under the control of an arabinose promoter. We wondered about the extent to which these differences in promoter/inducer identity affected the relative yield of sfGFP in each cell line (Figure S7). To evaluate this question, we made use of a plasmid in which the expression of sfGFP and the aaRS/tRNA pair expression were under the control of T5 promoter and arabinose promoters, respectively, both of which are compatible with all cell types examined here.45 In this case, the plasmid encoding sfGFP carried a UAG codon at position 151. When under the control of a T5 promoter and in the presence of pBrF, although the OD600 of Vmax X2 cells was highest, the value of 528 nm emission at the 4 h time point was highest for BL21 and C321.ΔA.exp cells, lowest for C321.ΔA.opt and Top10 cells, and intermediate for Vmax X2 cells (Figure S7A). The isolated yield of 151TAG sfGFP was comparable whether the expression was performed in BL21, C321.ΔA.exp, or Vmax X2—approximately 100 mg/L of culture. These yields were at least 2-fold lower than those obtained in Vmax X2 cells when sfGFP expression was under the control of a T7 promoter (Figure S7B,C). Given that the T5 promoter is recognized by endogenous RNAPs, the lower yields using this promoter in Vmax X2 may arise due to differences in the relative affinity of V. natriegens andE. coliRNAP for the T5 promoter or differences in the kinetics of V. natriegens andE. coliRNAP-mediated transcription. Thus, the use of the T7 promoter is recommended for achieving optimal yields. It is also important to note that these experiments have focused on the expression of a single protein (sfGFP). Given the differences betweenE. coli and V. natriegens in terms of metabolic processes, translation, and cellular milieu, it is likely that there are a subset of proteins that are better expressed in V. natriegens and vice versa.81
For many ncAA applications relating to fundamental research or the preparation of homogeneous antibody-drug conjugates (ADCs), one ncAA per polypeptide chain is generally sufficient. For others, such as the design of sequence-defined protein materials, multiple copies of one (or more) ncAA may be desired.82 These applications push the limits of genetic code expansion, as the isolated yields of such materials from standardE. colistrains can be low due to the increased frequency of RF1-mediated termination events.45 Strains that lack RF1, such as B95 and C321 derivatives, have been reported to support greatly improved yields of model proteins containing multiple copies of a single ncAA such as pAzF.39,41,45 Likewise, cell-free expression systems derived from these strains have been utilized.60,70,83 These systems are able to produce appreciable amounts of material containing up to 40 noncanonical α-amino acids,70 but fidelity is evaluated only rarely.
We wondered whether Vmax X2 would also provide advantages in yield or purity for the expression of proteins whose coding sequences contained multiple UAG codons. Thus, we examined the strain-dependent yield and purity of sfGFP containing pBrF at five positions with sfGFP (S2, D36, K101, E132, and D190). All of these positions have been shown previously to accept one or more ncAAs.83,84 As before, all strains were grown under their own optimized conditions and in the presence of 0.5 mM pBrF; both OD600 and fluorescence at 528 nm were monitored as a function of time (Figure 4A,B). As observed previously, Vmax X2 cells grew the most quickly (Figure 4A), and there was no direct correlation between the 528 nm emission value at 4 h and the isolated yield. Although the average 528 nm emission of C321.ΔA.exp cells was higher than that of the Vmax X2 growth (Figure 4D), the isolated yield of sfGFP from Vmax X2 was significantly higher (Figure 4E). The fidelity of sfGFP containing five copies of pBrF was also higher in Vmax X2 (>98.7%) than in C321.ΔA.exp (Figure S8). Other detected amino acids include phenylalanine, glutamine (C321 strain only), tyrosine, lysine, leucine/isoleucine, and proline (Figure S8). None of the otherE. colistrains generated sufficient material for a detailed LC-MS/MS analysis.
Figure 4.
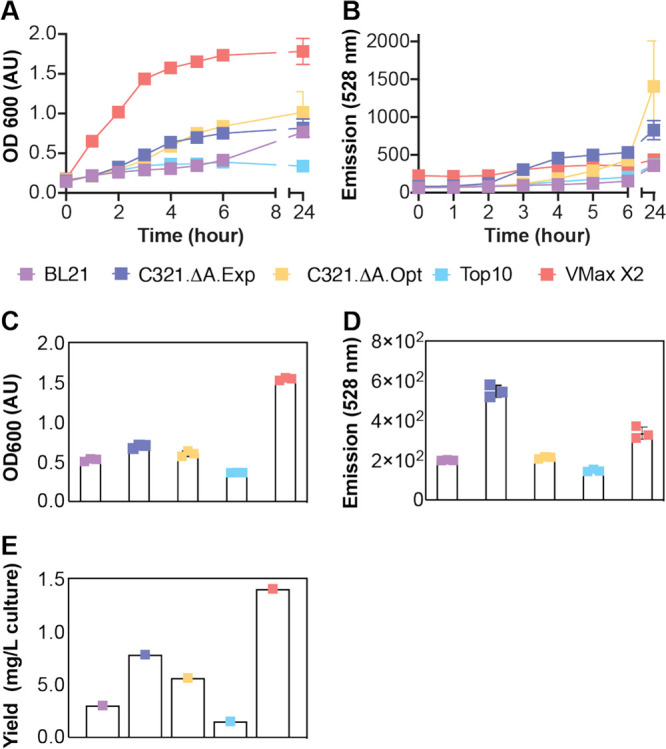
Yield of sfGFP containing five copies of pBrF ncAA when expressed in Vmax X2 versusE. colistrains. (A) Time-dependent growth curves; (B) time-dependent increase in 528 nm fluorescence; (C) OD600; (D) 528 nm fluorescence; and (E) isolated yields after 4 h. Vmax X2 and BL21 cells were transformed with pET-5XTAG-sfGFP and pEVOL-CNF, whereas Top10 and C321 cells were transformed with pET-5XTAG-sfGFP and pULTRA-CNF (Top10, C321), to induce expression of sfGFP bearing an ncAA at five positions with sfGFP (S2, D36, K101, E132, and D190). After induction, cells were grown for 4 h at 37 °C (Vmax X2, BL21, Top10, C321.ΔA.exp) or 34 °C (C321.ΔA.opt) in the presence of 0.5 mM pBrF.
In summary, here, we show that Vmax X2 is capable of producing exceptional yields of soluble protein containing up to 5 ncAAs. The yields realized with Vmax X2 and plasmids under the control of a T7 promoter are up to 25-fold higher than those achieved using commercial expression strains (Top10 and BL21) and more than 10-fold higher than those achieved using two different genomically recodedE. colistrains that lack endogenous UAG stop codons and have been optimized for improved fitness and preferred growth temperature (C321.ΔA.opt and C321.ΔA.exp, Addgene strains 87359 and 49018).39,40 In addition to high yields, Vmax X2 cells also generate proteins with significantly lower levels of misincorporated natural α-amino acids at the UAG-programmed position, especially in cases in which the ncAA is only a moderate substrate for the chosen aaRS. Thus, the use of Vmax X2 could obviate the need for time-consuming directed evolution experiments to improve the selectivity of a synthetase for subpar substrates.
Acknowledgments
This work was supported by the Center for Genetically Encoded Materials (C-GEM), an NSF Center for Chemical Innovation (NSF 2002182). O.A. was supported in part by Agilent Technologies as an Agilent Fellow. S.S.G. was supported by an NSF Predoctoral Fellowship (NSF 1752814) and an NIH Chemistry-Biology Interface Training Grant (T32GM067543). Figures were created with Biorender.com.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.1c00499.
Methods, protein sequences, and additional data and figures including LC-MS/MS analyses, yield data, MS/MS identification, amino acid distributions, and a GCE comparison (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Chin J. W. Expanding and Reprogramming the Genetic Code. Nature 2017, 550 (7674), 53–60. 10.1038/nature24031. [DOI] [PubMed] [Google Scholar]
- Young D. D.; Schultz P. G. Playing with the Molecules of Life. ACS Chem. Biol. 2018, 13 (4), 854–870. 10.1021/acschembio.7b00974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T.; Lajoie M. J.; Englert M.; Soll D. Rewriting the Genetic Code. Annu. Rev. Microbiol. 2017, 71, 557–577. 10.1146/annurev-micro-090816-093247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D. R.; Magliery T. J.; Pastrnak M.; Schultz P. G. Engineering a TRNA and Aminoacyl-TRNA Synthetase for the Site-Specific Incorporation of Unnatural Amino Acids into Proteins in Vivo. Proc. Natl. Acad. Sci. U. S. A. 1997, 94 (19), 10092–10097. 10.1073/pnas.94.19.10092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Magliery T. J.; Liu D. R.; Schultz P. G. A New Functional Suppressor TRNA/Aminoacyl-tRNA Synthetase Pair for the in Vivo Incorporation of Unnatural Amino Acids into Proteins. J. Am. Chem. Soc. 2000, 122 (20), 5010–5011. 10.1021/ja000595y. [DOI] [Google Scholar]
- Chin J. W. Expanding and Reprogramming the Genetic Code of Cells and Animals. Annu. Rev. Biochem. 2014, 83 (1), 379–408. 10.1146/annurev-biochem-060713-035737. [DOI] [PubMed] [Google Scholar]
- Gan Q.; Lehman B. P.; Bobik T. A.; Fan C. Expanding the Genetic Code of Salmonella with Non-Canonical Amino Acids. Sci. Rep. 2016, 6, 39920. 10.1038/srep39920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopatniuk M.; Myronovskyi M.; Luzhetskyy A. Streptomyces Albus: A New Cell Factory for Non-Canonical Amino Acids Incorporation into Ribosomally Synthesized Natural Products. ACS Chem. Biol. 2017, 12 (9), 2362–2370. 10.1021/acschembio.7b00359. [DOI] [PubMed] [Google Scholar]
- Hancock S. M.; Uprety R.; Deiters A.; Chin J. W. Expanding the Genetic Code of Yeast for Incorporation of Diverse Unnatural Amino Acids via a Pyrrolysyl-TRNA Synthetase/TRNA Pair. J. Am. Chem. Soc. 2010, 132 (42), 14819–14824. 10.1021/ja104609m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K. A.; Deiters A. Genetic Code Expansion of Mammalian Cells with Unnatural Amino Acids. Curr. Protoc Chem. Biol. 2015, 7 (3), 187–199. 10.1002/9780470559277.ch150038. [DOI] [PubMed] [Google Scholar]
- Beranek V.; Reinkemeier C. D.; Zhang M. S.; Liang A. D.; Kym G.; Chin J. W. Genetically Encoded Protein Phosphorylation in Mammalian Cells. Cell Chem. Biol. 2018, 25 (9), 1067–1074. 10.1016/j.chembiol.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiss S.; Chin J. W. Expanding the Genetic Code of an Animal. J. Am. Chem. Soc. 2011, 133 (36), 14196–14199. 10.1021/ja2054034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst R. J.; Krogager T. P.; Maywood E. S.; Zanchi R.; Beránek V.; Elliott T. S.; Barry N. P.; Hastings M. H.; Chin J. W. Genetic Code Expansion in the Mouse Brain. Nat. Chem. Biol. 2016, 12 (10), 776–778. 10.1038/nchembio.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W.; Liu J.; Deiters A. Genetic Code Expansion in Animals. ACS Chem. Biol. 2018, 13 (9), 2375–2386. 10.1021/acschembio.8b00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan W.; Tharp J. M.; Liu W. R. Pyrrolysyl-TRNA Synthetase: An Ordinary Enzyme but an Outstanding Genetic Code Expansion Tool. Biochim. Biophys. Acta, Proteins Proteomics 2014, 1844 (6), 1059–1070. 10.1016/j.bbapap.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan G.; James C. M.; Krzycki J. A. Pyrrolysine Encoded by UAG in Archaea: Charging of a UAG-Decoding Specialized TRNA. Science 2002, 296 (5572), 1459–1462. 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- Willis J. C. W.; Chin J. W. Mutually Orthogonal Pyrrolysyl-TRNA Synthetase/TRNA Pairs. Nat. Chem. 2018, 10 (8), 831–837. 10.1038/s41557-018-0052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T.; Miller C.; Guo L.-T.; Ho J. M. L.; Bryson D. I.; Wang Y.-S.; Liu D. R.; Söll D. Crystal Structures Reveal an Elusive Functional Domain of Pyrrolysyl-TRNA Synthetase. Nat. Chem. Biol. 2017, 13 (12), 1261–1266. 10.1038/nchembio.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H.; Peak-Chew S. Y.; Chin J. W. Genetically Encoding Nε-Acetyllysine in Recombinant Proteins. Nat. Chem. Biol. 2008, 4 (4), 232–234. 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- Wang L.; Brock A.; Herberich B.; Schultz P. G. Expanding the Genetic Code of Escherichia Coli. Science 2001, 292 (5516), 498–500. 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- Young D. D.; Young T. S.; Jahnz M.; Ahmad I.; Spraggon G.; Schultz P. G. An Evolved Aminoacyl-TRNA Synthetase with Atypical Polysubstrate Specificity. Biochemistry 2011, 50 (11), 1894–1900. 10.1021/bi101929e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahn N.; Tharp J. M.; Crnković A.; Söll D.. Engineering Aminoacyl-TRNA Synthetases for Use in Synthetic Biology. In The Enzymes; Elsevier, 2020; Vol. 48, pp 351–395. 10.1016/bs.enz.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes A. L.; Miyake-Stoner S. J.; Peeler J. C.; Nguyen D. P.; Hammer R. P.; Mehl R. A. Enhancing the Utility of Unnatural Amino Acid Synthetases by Manipulating Broad Substrate Specificity. Mol. BioSyst. 2009, 5 (9), 1032. 10.1039/b904032c. [DOI] [PubMed] [Google Scholar]
- Cervettini D.; Tang S.; Fried S. D.; Willis J. C. W.; Funke L. F. H.; Colwell L. J.; Chin J. W. Rapid Discovery and Evolution of Orthogonal Aminoacyl-TRNA Synthetase-TRNA Pairs. Nat. Biotechnol. 2020, 38 (8), 989–999. 10.1038/s41587-020-0479-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A.; Xiao H.; Yang P.-Y.; Soundararajan G.; Schultz P. G. A Tryptophanyl-TRNA Synthetase/TRNA Pair for Unnatural Amino Acid Mutagenesis in E. Coli. Angew. Chem., Int. Ed. 2013, 52 (19), 5106–5109. 10.1002/anie.201301094. [DOI] [PubMed] [Google Scholar]
- Italia J. S.; Addy P. S.; Wrobel C. J. J.; Crawford L. A.; Lajoie M. J.; Zheng Y.; Chatterjee A. An Orthogonalized Platform for Genetic Code Expansion in Both Bacteria and Eukaryotes. Nat. Chem. Biol. 2017, 13 (4), 446–450. 10.1038/nchembio.2312. [DOI] [PubMed] [Google Scholar]
- Axup J. Y.; Bajjuri K. M.; Ritland M.; Hutchins B. M.; Kim C. H.; Kazane S. A.; Halder R.; Forsyth J. S.; Santidrian A. F.; Stafin K.; et al. Synthesis of Site-Specific Antibody-Drug Conjugates Using Unnatural Amino Acids. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (40), 16101–16106. 10.1073/pnas.1211023109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam T. J.; Wold E.; Wahl A.; Smider V. V. Antibody Conjugates with Unnatural Amino Acids. Mol. Pharmaceutics 2015, 12 (6), 1848–1862. 10.1021/acs.molpharmaceut.5b00082. [DOI] [PubMed] [Google Scholar]
- Beck A.; Goetsch L.; Dumontet C.; Corvaïa N. Strategies and Challenges for the next Generation of Antibody-Drug Conjugates. Nat. Rev. Drug Discovery 2017, 16 (5), 315–337. 10.1038/nrd.2016.268. [DOI] [PubMed] [Google Scholar]
- Cho H.; Daniel T.; Buechler Y. J.; Litzinger D. C.; Maio Z.; Putnam A.-M. H.; Kraynov V. S.; Sim B.-C.; Bussell S.; Javahishvili T.; et al. Optimized Clinical Performance of Growth Hormone with an Expanded Genetic Code. Proc. Natl. Acad. Sci. U. S. A. 2011, 108 (22), 9060–9065. 10.1073/pnas.1100387108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieblich S. A.; Fang K. Y.; Cahn J. K. B.; Rawson J.; LeBon J.; Ku H. T.; Tirrell D. A. 4 S -Hydroxylation of Insulin at ProB28 Accelerates Hexamer Dissociation and Delays Fibrillation. J. Am. Chem. Soc. 2017, 139 (25), 8384–8387. 10.1021/jacs.7b00794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezhdo A.; Islam M.; Huang M.; Van Deventer J. A. Future Prospects for Noncanonical Amino Acids in Biological Therapeutics. Curr. Opin. Biotechnol. 2019, 60, 168–178. 10.1016/j.copbio.2019.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T.; Sengoku T.; Hirata K.; Ogata K.; Suga H. Ribosomal Synthesis and de Novo Discovery of Bioactive Foldamer Peptides Containing Cyclic β-Amino Acids. Nat. Chem. 2020, 12 (11), 1081–1088. 10.1038/s41557-020-0525-1. [DOI] [PubMed] [Google Scholar]
- Zambaldo C.; Luo X.; Mehta A. P.; Schultz P. G. Recombinant Macrocyclic Lanthipeptides Incorporating Non-Canonical Amino Acids. J. Am. Chem. Soc. 2017, 139 (34), 11646–11649. 10.1021/jacs.7b04159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost J. R.; Jacob N. T.; Papa L. J.; Owens A. E.; Fasan R. Ribosomal Synthesis of Macrocyclic Peptides in Vitro and in Vivo Mediated by Genetically Encoded Aminothiol Unnatural Amino Acids. ACS Chem. Biol. 2015, 10 (8), 1805–1816. 10.1021/acschembio.5b00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.; Liu T. Therapeutic Applications of Genetic Code Expansion. Synth. Syst. Biotechnol. 2018, 3 (3), 150–158. 10.1016/j.synbio.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.; Korostelev A.; Lancaster L.; Noller H. F. Crystal Structures of 70S Ribosomes Bound to Release Factors RF1, RF2 and RF3. Curr. Opin. Struct. Biol. 2012, 22 (6), 733–742. 10.1016/j.sbi.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi M. R. Polypeptide Chain Termination in Vitro: Isolation of a Release Factor. Proc. Natl. Acad. Sci. U. S. A. 1967, 58 (3), 1144–1151. 10.1073/pnas.58.3.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie M. J.; Rovner A. J.; Goodman D. B.; Aerni H. R.; Haimovich A. D.; Kuznetsov G.; Mercer J. A.; Wang H. H.; Carr P. A.; Mosberg J. A.; et al. Genomically Recoded Organisms Expand Biological Functions. Science 2013, 342 (6156), 357–360. 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov G.; Goodman D. B.; Filsinger G. T.; Landon M.; Rohland N.; Aach J.; Lajoie M. J.; Church G. M. Optimizing Complex Phenotypes through Model-Guided Multiplex Genome Engineering. Genome Biol. 2017, 18 (1), 100. 10.1186/s13059-017-1217-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T.; Hoshi H.; Ohtake K.; Takahashi M.; Yamaguchi A.; Hayashi A.; Yokoyama S.; Sakamoto K. Highly Reproductive Escherichia Coli Cells with No Specific Assignment to the UAG Codon. Sci. Rep. 2015, 5 (1), 9699. 10.1038/srep09699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer J. N.; Hosseinzadeh P.; Gottfried-Lee I.; Van Fossen E. M.; Zhu P.; Bednar R. M.; Karplus P. A.; Mehl R. A.; Cooley R. B. Overcoming Near-Cognate Suppression in a Release Factor 1-Deficient Host with an Improved Nitro-Tyrosine TRNA Synthetase. J. Mol. Biol. 2020, 432 (16), 4690–4704. 10.1016/j.jmb.2020.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aerni H. R.; Shifman M. A.; Rogulina S.; O’Donoghue P.; Rinehart J. Revealing the Amino Acid Composition of Proteins within an Expanded Genetic Code. Nucleic Acids Res. 2015, 43 (2), e8 10.1093/nar/gku1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George S.; Aguirre J. D.; Spratt D. E.; Bi Y.; Jeffery M.; Shaw G. S.; O’Donoghue P. Generation of Phospho-Ubiquitin Variants by Orthogonal Translation Reveals Codon Skipping. FEBS Lett. 2016, 590 (10), 1530–1542. 10.1002/1873-3468.12182. [DOI] [PubMed] [Google Scholar]
- Zheng Y.; Lajoie M. J.; Italia J. S.; Chin M. A.; Church G. M.; Chatterjee A. Performance of Optimized Noncanonical Amino Acid Mutagenesis Systems in the Absence of Release Factor 1. Mol. BioSyst. 2016, 12 (6), 1746–1749. 10.1039/C6MB00070C. [DOI] [PubMed] [Google Scholar]
- Gan Q.; Fan C. Increasing the Fidelity of Noncanonical Amino Acid Incorporation in Cell-Free Protein Synthesis. Biochim. Biophys. Acta, Gen. Subj. 2017, 1861 (11), 3047–3052. 10.1016/j.bbagen.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claassens N. J.; Burgener S.; Vögeli B.; Erb T. J.; Bar-Even A. A Critical Comparison of Cellular and Cell-Free Bioproduction Systems. Curr. Opin. Biotechnol. 2019, 60, 221–229. 10.1016/j.copbio.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T.; Suga H. Ribosomal Elongation of Cyclic γ-Amino Acids Using a Reprogrammed Genetic Code. J. Am. Chem. Soc. 2020, 142 (11), 4965–4969. 10.1021/jacs.9b12280. [DOI] [PubMed] [Google Scholar]
- Tsiamantas C.; Rogers J. M.; Suga H. Initiating Ribosomal Peptide Synthesis with Exotic Building Blocks. Chem. Commun. 2020, 56 (31), 4265–4272. 10.1039/D0CC01291B. [DOI] [PubMed] [Google Scholar]
- Lee J.; Schwarz K. J.; Kim D. S.; Moore J. S.; Jewett M. C. Ribosome-Mediated Polymerization of Long Chain Carbon and Cyclic Amino Acids into Peptides in Vitro. Nat. Commun. 2020, 11 (1), 4304. 10.1038/s41467-020-18001-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ad O.; Hoffman K. S.; Cairns A. G.; Featherston A. L.; Miller S. J.; Söll D.; Schepartz A. Translation of Diverse Aramid- and 1,3-Dicarbonyl-Peptides by Wild Type Ribosomes in Vitro. ACS Cent. Sci. 2019, 5 (7), 1289–1294. 10.1021/acscentsci.9b00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharp J. M.; Walker J. A.; Söll D.; Schepartz A.. Initiating Protein Synthesis with Noncanonical Monomers in Vitro and in Vivo. In Methods in Enzymology; Elsevier, 2021; Vol. 656, pp 495–519. 10.1016/bs.mie.2021.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieille C.; Zeikus G. J. Hyperthermophilic Enzymes: Sources, Uses, and Molecular Mechanisms for Thermostability. Microbiol. Mol. Biol. Rev. 2001, 65 (1), 1–43. 10.1128/MMBR.65.1.1-43.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L.; Qin X.; Huang Y.; Cao W.; Wang C.; Wang Y.; Ling X.; Chen H.; Wu D.; Lin Y.; et al. Thermophilic Pyrrolysyl-TRNA Synthetase Mutants for Enhanced Mammalian Genetic Code Expansion. ACS Synth. Biol. 2020, 9 (10), 2723–2736. 10.1021/acssynbio.0c00257. [DOI] [PubMed] [Google Scholar]
- Vmax X2 Chemically Competent Cells; SGI-DNA: La Jolla, CA, 2020.
- Lee H. H.; Ostrov N.; Wong B. G.; Gold M. A.; Khalil A. S.; Church G. M. Functional Genomics of the Rapidly Replicating Bacterium Vibrio Natriegens by CRISPRi. Nat. Microbiol 2019, 4 (7), 1105–1113. 10.1038/s41564-019-0423-8. [DOI] [PubMed] [Google Scholar]
- Weinstock M. T.; Hesek E. D.; Wilson C. M.; Gibson D. G. Vibrio Natriegens as a Fast-Growing Host for Molecular Biology. Nat. Methods 2016, 13 (10), 849–851. 10.1038/nmeth.3970. [DOI] [PubMed] [Google Scholar]
- Becker W.; Wimberger F.; Zangger K. Vibrio Natriegens: An Alternative Expression System for the High-Yield Production of Isotopically Labeled Proteins. Biochemistry 2019, 58 (25), 2799–2803. 10.1021/acs.biochem.9b00403. [DOI] [PubMed] [Google Scholar]
- Schleicher L.; Muras V.; Claussen B.; Pfannstiel J.; Blombach B.; Dibrov P.; Fritz G.; Steuber J. Vibrio Natriegens as Host for Expression of Multisubunit Membrane Protein Complexes. Front. Microbiol. 2018, 9, 2537. 10.3389/fmicb.2018.02537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Soye B. J.; Davidson S. R.; Weinstock M. T.; Gibson D. G.; Jewett M. C. Establishing a High-Yielding Cell-Free Protein Synthesis Platform Derived from Vibrio Natriegens. ACS Synth. Biol. 2018, 7 (9), 2245–2255. 10.1021/acssynbio.8b00252. [DOI] [PubMed] [Google Scholar]
- Wiegand D. J.; Lee H. H.; Ostrov N.; Church G. M. Establishing a Cell-Free Vibrio Natriegens Expression System. ACS Synth. Biol. 2018, 7 (10), 2475–2479. 10.1021/acssynbio.8b00222. [DOI] [PubMed] [Google Scholar]
- Tschirhart T.; Shukla V.; Kelly E. E.; Schultzhaus Z.; NewRingeisen E.; Erickson J. S.; Wang Z.; Garcia W.; Curl E.; Egbert R. G.; et al. Synthetic Biology Tools for the Fast-Growing Marine Bacterium. ACS Synth. Biol. 2019, 8 (9), 2069–2079. 10.1021/acssynbio.9b00176. [DOI] [PubMed] [Google Scholar]
- CODEX DNA. Vmax X2 Cells.
- Mamat U.; Woodard R. W.; Wilke K.; Souvignier C.; Mead D.; Steinmetz E.; Terry K.; Kovacich C.; Zegers A.; Knox C. Endotoxin-Free Protein Production—ClearColiTM Technology. Nat. Methods 2013, 10 (9), 916–916. 10.1038/nmeth.f.367. [DOI] [Google Scholar]
- Ozer E.; Alfonta L. Genetic Code Expansion of Vibrio Natriegens. Front. Bioeng. Biotechnol. 2021, 9, 95. 10.3389/fbioe.2021.594429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young T. S.; Ahmad I.; Yin J. A.; Schultz P. G. An Enhanced System for Unnatural Amino Acid Mutagenesis in E. Coli. J. Mol. Biol. 2010, 395 (2), 361–374. 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- Kwok H. S.; Vargas-Rodriguez O.; Melnikov S. V.; Söll D. Engineered Aminoacyl-TRNA Synthetases with Improved Selectivity toward Noncanonical Amino Acids. ACS Chem. Biol. 2019, 14 (4), 603–612. 10.1021/acschembio.9b00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkelmann D. L.; Willis J. C. W.; Beattie A. T.; Chin J. W. Engineered Triply Orthogonal Pyrrolysyl-TRNA Synthetase/TRNA Pairs Enable the Genetic Encoding of Three Distinct Non-Canonical Amino Acids. Nat. Chem. 2020, 12 (6), 535–544. 10.1038/s41557-020-0472-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebendiker M.; Danieli T. Production of Prone-to-Aggregate Proteins. FEBS Lett. 2014, 588 (2), 236–246. 10.1016/j.febslet.2013.10.044. [DOI] [PubMed] [Google Scholar]
- Des Soye B. J.; Gerbasi V. R.; Thomas P. M.; Kelleher N. L.; Jewett M. C. A Highly Productive, One-Pot Cell-Free Protein Synthesis Platform Based on Genomically Recoded Escherichia Coli. Cell Chem. Biol. 2019, 26 (12), 1743–1754. 10.1016/j.chembiol.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruna K.; Alkazemi M. H.; Liu Y.; Söll D.; Englert M. Engineering the Elongation Factor Tu for Efficient Selenoprotein Synthesis. Nucleic Acids Res. 2014, 42 (15), 9976–9983. 10.1093/nar/gku691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonczak A. K.; Morris J.; Tippmann E. M. Advances in the Mechanism and Understanding of Site-Selective Noncanonical Amino Acid Incorporation. Curr. Opin. Struct. Biol. 2011, 21 (4), 481–487. 10.1016/j.sbi.2011.04.004. [DOI] [PubMed] [Google Scholar]
- Zhang Z.; Shah B.; Bondarenko P. V. G/U and Certain Wobble Position Mismatches as Possible Main Causes of Amino Acid Misincorporations. Biochemistry 2013, 52 (45), 8165–8176. 10.1021/bi401002c. [DOI] [PubMed] [Google Scholar]
- Wong H. E.; Huang C.-J.; Zhang Z. Amino Acid Misincorporation Propensities Revealed through Systematic Amino Acid Starvation. Biochemistry 2018, 57 (49), 6767–6779. 10.1021/acs.biochem.8b00976. [DOI] [PubMed] [Google Scholar]
- Roy B.; Leszyk J. D.; Mangus D. A.; Jacobson A. Nonsense Suppression by Near-Cognate TRNAs Employs Alternative Base Pairing at Codon Positions 1 and 3. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (10), 3038–3043. 10.1073/pnas.1424127112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horness R. E.; Basom E. J.; Thielges M. C. Site-Selective Characterization of Src Homology 3 Domain Molecular Recognition with Cyanophenylalanine Infrared Probes. Anal. Methods 2015, 7 (17), 7234–7241. 10.1039/C5AY00523J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalanne J.-B.; Taggart J. C.; Guo M. S.; Herzel L.; Schieler A.; Li G.-W. Evolutionary Convergence of Pathway-Specific Enzyme Expression Stoichiometry. Cell 2018, 173 (3), 749–761. 10.1016/j.cell.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalanne J.-B.; Li G.-W.. First-Principles Model of Optimal Translation Factors Stoichiometry. bioRxiv preprint, 2021. 10.1101/2021.04.02.438287. [DOI] [PMC free article] [PubMed]
- Long C. P.; Gonzalez J. E.; Cipolla R. M.; Antoniewicz M. R. Metabolism of the Fast-Growing Bacterium Vibrio Natriegens Elucidated by 13C Metabolic Flux Analysis. Metab. Eng. 2017, 44, 191–197. 10.1016/j.ymben.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F.; Blombach B. Metabolic Engineering of Vibrio Natriegens. Essays Biochem. 2021, 65 (2), 381–392. 10.1042/EBC20200135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.; Dong F.; Wu M.; Tao R.; Yang J.; Wu M.; Jiang Y.; Yang S.; Yang L. Vibrio Natriegens as a PET-Compatible Expression Host Complementary to Escherichia Coli. Front. Microbiol. 2021, 12, 627181. 10.3389/fmicb.2021.627181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israeli B.; Vaserman L.; Amiram M. Multi-Site Incorporation of Nonstandard Amino Acids into Protein-Based Biomaterials. Isr. J. Chem. 2020, 60 (12), 1118–1128. 10.1002/ijch.201900043. [DOI] [Google Scholar]
- Hong S. H.; Ntai I.; Haimovich A. D.; Kelleher N. L.; Isaacs F. J.; Jewett M. C. Cell-Free Protein Synthesis from a Release Factor 1 Deficient Escherichia Coli Activates Efficient and Multiple Site-Specific Nonstandard Amino Acid Incorporation. ACS Synth. Biol. 2014, 3 (6), 398–409. 10.1021/sb400140t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. B. F.; Xu J.; Shen Z.; Takimoto J. K.; Schultz M. D.; Schmitz R. J.; Xiang Z.; Ecker J. R.; Briggs S. P.; Wang L. RF1 Knockout Allows Ribosomal Incorporation of Unnatural Amino Acids at Multiple Sites. Nat. Chem. Biol. 2011, 7 (11), 779–786. 10.1038/nchembio.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.