Abstract
Defined as the dysfunction and/or cell death caused by toxic lipids accumulation in hepatocytes, hepatic lipotoxicity plays a pathological role in nonalcoholic fatty liver disease. The cellular and molecular mechanisms underlying lipotoxicity remain to be elucidated. In this study, using AML12 cells, a nontransformed murine hepatocyte cell line, exposed to palmitate (a 16-C saturated fatty acid) as an experimental model, we investigated the role and mechanisms of nicotinamide N-methyltransferase (NNMT), a methyltransferase catalyzing nicotinamide methylation and degradation, in hepatic lipotoxicity. We initially identified activating transcription factor 4 (ATF4) as a major transcription factor for hepatic NNMT expression. Here, we demonstrated that palmitate upregulates NNMT expression via activating ATF4 in a mechanistic target of rapamycin complex 1 (mTORC1)-dependent mechanism in that mTORC1 inhibition by both Torin1 and rapamycin attenuated ATF4 activation and NNMT upregulation. We further demonstrated that the mTORC1-dependent ATF4 activation is an integral signaling event of unfolded protein response (UPR) as both ATF4 activation and NNMT upregulation by tunicamycin, a well-documented endoplasmic reticulum (ER) stress inducer, are blunted when hepatocytes were pretreated with Torin1. Importantly, our data uncovered that NNMT upregulation contributes to palmitate-induced hepatotoxicity as NNMT inhibition, via either pharmacological (NNMT inhibitors) or genetic approach (siRNA transfection), provided protection against palmitate lipotoxicity. Our further mechanistic exploration identified protein kinase A (PKA) activation to contribute, at least, partially to the protective effect of NNMT inhibition against lipotoxicity. Collectively, our data demonstrated that NNMT upregulation by the mTORC1-ATF4 pathway activation contributes to the development of lipotoxicity in hepatocytes.
Keywords: ATF4, lipotoxicity, mTORC1, NNMT, palmitate, PKA
INTRODUCTION
Nonalcoholic fatty liver disease (NAFLD) is considered as the hepatic manifestation of metabolic syndrome whose global increase is parallel to the increase in diabetes and obesity. The pathological progression of the disease ranges from hepatic steatosis (fatty liver) to steatohepatitis, which is characterized by hepatocyte cell death and immune cells infiltration, to late-stage fibrosis/cirrhosis and hepatocellular carcinoma. Despite the intensive research in last several decades, the molecular mechanisms behind its initiation and progression remain elusive and there is currently no FDA-approved treatments for this disease (1).
NAFLD initiation and progression are closely linked to obesity, where one of the major hallmarks is the elevated levels of circulatory free fatty acids (FFAs) caused by uncontrolled adipose tissue lipolysis derived from peripheral insulin resistance (2). Of all FFAs, it is the saturated fatty acids (SFAs) that are toxic to a wide range of cells, including hepatocytes, resulting in a pathological environment defined as lipotoxicity (3, 4), a term which is used to describe the cellular toxicity, including cellular dysfunction and cell death, derived from ectopic lipid accumulation in nonadipose tissues (5, 6) Palmitate is a 16-carbon SFA that represents the most abundant SFA found in human circulation (7). Elevated plasma concentrations of palmitate have been implicated in NAFLD initiation and development (4, 8). However, the exact mechanisms underlying palmitate induced-hepatotoxicity remains to be clearly defined.
Nicotinamide N-methyltransferase (NNMT) is an intracellular methyltransferase that uses S-adenosylmethionine as a methyl donor to catalyze the methylation/degradation of nicotinamide, the primary endogenous precursor for NAD+ biosynthesis via the salvage pathway (9). Abundantly expressed in the metabolically active tissues, such as the liver, NNMT has been increasingly considered to be an important regulator for cellular metabolism and energy homeostasis. Whereas the whole body NNMT overexpression worsened hepatic steatosis upon chronic high-fat diet feeding (9, 10), NNMT knockdown protected mice from diet-induced obesity (11) and NNMT deficiency mice had showed improved insulin sensitivity in a diet-induced obesity model (12). We were the first to document that endoplasmic reticulum (ER) stress upregulates NNMT expression in hepatocytes via ATF4 activation, which has been shown to play a pathological role in the development of alcoholic liver disease (13). Currently, it remains completely unknown whether and how NNMT contributes to palmitate-induced lipotoxicity in hepatocytes.
ER stress, a term coined to describe the perturbed ER integrity of ER due to the accumulation of unfolded or misfolded proteins inside ER lumen, activates a signaling network termed the unfolded protein response (UPR). UPR consists of three canonical signaling pathways: inositol-requiring enzyme 1-α (IRE1α), RNA-dependent protein kinase-like ER kinase (PERK)/activating transcription factor 4 (ATF4), and activating transcription factor 6 (ATF6) (14). Mammalian target of rapamycin (mTOR) is an evolutionarily conserved protein serine/threonine kinase with two structurally and functionally distinct kinase complexes, mTOR complex 1 (mTORC1) and mTORC2, that are formed using mTOR as a core component, which regulate distinct cellular processes. mTORC1 activation integrates a wide range of extracellular and intracellular signals to regulate cell growth, anabolism, and protein synthesis, depending on growth factors stimulation and availability of nutrients (15). ER stress has been well-documented as a central mechanism implicated in palmitate-induced lipotoxicity in various cell types, including hepatocytes (16–18). The mechanistic involvement of mTORC1 activation in palmitate-induced lipotoxicity has also been reported in several types of cells (19, 20). Recently, we reported that mTORC1 activation contributes to palmitate-induced cell death and lipid secretion in hepatocytes via activating IRE1α (21), one of three canonical signaling pathways activated upon ER stress, suggesting that mTORC1 and ER stress may coconspire to contribute to palmitate hepatotoxicity.
In this study, employing both in vitro cell culture studies with AML12 cells, a nontransformed mouse hepatocyte cell line, and in vivo investigations with male C57 BL/6 mice, we demonstrate that palmitate exposure upregulates NNMT expression through activating mTORC1-ATF4 pathway in hepatocytes. Furthermore, we demonstrate that NNMT upregulation contributes to palmitate-induced hepatocyte cell death.
MATERIALS AND METHODS
Reagents
AML12 cells, a nontransformed mouse hepatocyte cell line was purchased from American Type Culture Collection (ATCC, Manassas). Cell culture and transfection reagents were purchased from Invitrogen (Carlsbad, CA). Palmitate, GSK2006414, and tunicamycin were purchased from Sigma-Aldrich (St. Louis, MO). Torin1 and rapamycin were purchased from APExBIO Technology LLC (Houston, TX). H 89 dihydrochloride and SQ 22536 were from TOCRIS (Minneapolis, MN). JBSNF000088 was purchased from MCE (Monmouth Junction, NJ) and II399 provided by Dr. Rong Huang from Purdue University.
For Western blotting, antibodies for pS6 (S235/236) (Product No.: 4858 s, dilution 1:1,000), S6 (Product No.: 2317 s, dilution 1:1,000), ATF4 (Product No.: 11815, dilution 1:1,000), and PKA substrates (Production No.: 9624 s, dilution 1:1,000) were purchased from Cell Signalling Technology (Beverly, CA). Anti-NNMT antibody (Product No.: ab11978, dilution 1:1,000) was purchased from Abcam (Cambridge, MA) and anti-β-actin antibody (Product No.: AC026, dilution 1:10,000) from ABclonal Science Inc. (Woburn, MA). Antibodies were validated by either siRNA knockdown (ATF4) or pharmacological inhibition of target proteins (pS6 and PKA substrates).
Animal Studies
All animal studies were approved by the Institutional Animal Care and Use Committee at the University of Illinois at Chicago (Chicago, IL) and consistent with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Ten-week-old male C57BL/6N mice (Charles River Lab, Chicago, IL) were injected intraperitoneally with either tunicamycin (2 mg/kg body wt), a widely-used ER stress inducer, or isovolumic vehicle control (150 mM dextrose) as previously described (13). Mice were anesthetized with inhalational isoflurane and liver samples harvested after 16 h.
Cell Culture
AML12 hepatocytes were cultured as monolayers using Dulbecco’s modified Eagle’s medium (DMEM)/F12 supplemented with 10% fetal bovine serum, 100 units/mL penicillin, 0.1 mg/mL streptomycin, 40 ng/mL dexamethasone, and ITS, containing 5 mg/L insulin, 5 mg/L transferrin, and 5 μg/L selenium. Cells were grown in 75 cm2 flasks and maintained at 37°C in a humidified atmosphere of air and 5% CO2. Cells were grown at 80% confluence before the exposure of treatments in various experiments.
siRNA Transfection
Cultured AML12 hepatocytes were transfected with mouse ATF4 siRNA (Santa Cruz, sc-35113). Cells in exponential growth phase were plated in 6-well or 24-well plate and allowed to adhere for 24 h before transfection with siRNA. Transfection was performed according to Invitrogen’s Lip 2000 protocol. In brief, cells were cultured to 60%–70% confluence and transfected with 2 μL siRNA (20 mM) using 2 μL Lipo 2000. As a control, cells were transfected with scramble RNA under the same conditions. The results of transfection were identified by RT-PCR following a 12-h siRNA transfection.
Cell Lysates and Western Blotting
Total protein from AML12 hepatocytes and liver tissues was obtained using lysis buffer from Thermo Fisher Scientific Inc. (Rockford, ). The samples were chilled on ice with frequent vortex for 15 min and centrifuged for 10 min at 12,000 g. Protein concentrations of samples were determined using Enhanced BCA Protein Assay Kit from Thermo Fisher Scientific Inc. (Rockford, IL) according to the manufacturer’s instructions. Equals amounts of protein (30 μg) were loaded onto 10% or 8% SDS-PAGE depending on the molecular weight of the desired proteins and transferred to a nitrocellulose transfer membrane (Pall Corporation, New Port Richey, FL). Following transfer, membranes were cut based on molecular weight of target proteins and blocked in 5% (wt/vol) nonfat dry milk in PBS-0.1% Tween 20 and probed with specific antibodies. Following incubation of primary antibodies, membranes were washed with PBST, then incubated with fluorescein-conjugated secondary antibody (Cat No.: 102673-330 from VWR) at 1:10,000 dilution in blocking buffer at room temperature for 1 h, followed by washing with PBST. Immunoreactive bands of predicted molecular mass were visualized using LI-COR Odyssey CLx system and quantified with Image Studio v. 4.0.
LDH Release Assay
AML12 cells were seeded at 1 × 105 in 24-well plates overnight. After the indicated treatments, culture medium was collected and detected using CytoSelect LDH Cytotoxicity Assay Kit (Cell Biolabs, Inc., San Diego, CA) according to the manufacturer’s instructions. The absorption at OD450 was measured using a microplate spectrophotometer (SPECTRAmax340PC; Molecular Devices Corp., Sunnyvale, CA). The relative LDH release levels were scaled as folds of the control untreated groups.
Total RNA Isolation and Quantitative Real-Time PCR
Total RNA from AMl12 hepatocytes were isolated via phenol-chloroform extraction. For each sample, 1.0 μg total RNA was reverse transcribed using a high-capacity cDNA reverse transcription kit (Applied Biosystem, Vilnius, Lithuania). The cDNA was then amplified in MicroAmp Optical 96-well reaction plates with an SYBR Green PCR Master Mix (Applied Biosystems, Warrington, UK) on a Life Technologies ABI 7500 FAST sequence detection system. Relative gene expression was calculated after normalization by a house-keeping gene (Gapdh and 18srna).
Primers.
Gapdh: 5′- ATGCCAGTGAGCTTCCCGTTCAG-3′ and 3′- CATCACTGCCACCCAGAAGACTG-5′; 18srna: 5′- GTAACCCGTTGAACCCCATT-3′ and 3′- CCATCCAATCGGTAGTAGCG-5′; Nnmt: 5′- CTTTGGGTCCAGACACTGTGCA-3′ and 3′- CCAGAGCCAATGTCAATCAGGAG-5′; Atf-4: 5′- AACCTCATGGGTTCTCCAGCGA-3′ and 3′- CTCCAACATCCAATCTGTCCCG-5′; Xbp1: 5′- TAGACCTCTGGGAGTTCCTCCA-3′ and 3′- TGGACTCTGACACTGTGCCTC-5′; Xbp1s: 5′- GAGGCAACAGTGTCAGAGTCC-3′ and 3′- TGCTGAGTCCGCAGCAGGTG-5′.
Intracellular Total NAD Assay
The intracellular total NAD content in AML12 cells was measured following the instruction of NAD/NADH Quantitation Colorimetric Kit (BioVision K337-100, Milpitas, CA), as described by the manufacturer. Briefly, AML12 cells on six-well culture plates were washed with ice-cold PBS, lysed with 400 μL of extraction buffer, and centrifuged at 20,000 g for 5 min at 4°C. Total NAD concentrations were measured using supernatants and expressed relative to cell numbers for each independent experiment.
Statistical Analysis
All data sets are expressed as means ± SD. Statistical analysis was performed using a Student’s t test or one-way ANOVA for comparison of two groups and three or more groups, respectively. GraphPad Prism software (v. 9). Statistical tests were used as described in the figure legends and statistical significance was indicated as follows: *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant.
RESULTS
Palmitate Upregulates NNMT Expression in AML12 Hepatocytes
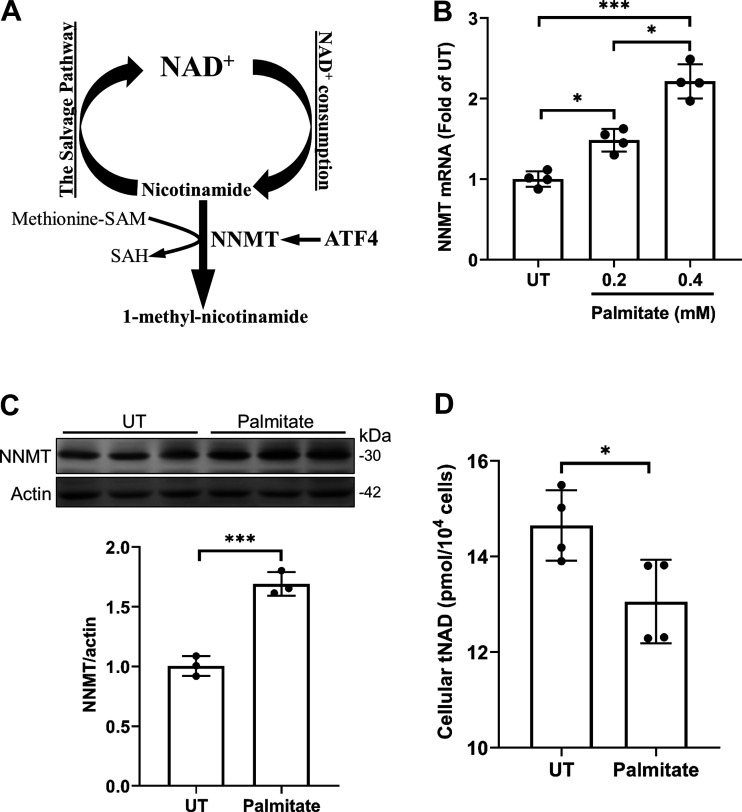
NNMT is a methyltransferase that catalyzes SAM-dependent degradation of nicotinamide, the primary endogenous precursor for cellular NAD+ biosynthesis via a salvage pathway (Fig. 1A). The effect of palmitate on NNMT expression in hepatocytes was directly examined in AML12 cells, a non-transformed mouse hepatocyte cell line. AML12 cells were treated with albumin-bound palmitate at 0, 0.2, and 0.4 mM for 16 h and NNMT expression, at both mRNA and protein levels, was determined by real-time qPCR and Western blotting, respectively. As shown in Fig. 1B, palmitate exposure increased NNMT gene expression in a dose-dependent manner. An increase in NNMT protein abundance was observed upon 0.4 mM palmitate exposure for 16 h (Fig. 1C). Correspondingly, intracellular total NAD concentrations were significantly reduced upon palmitate (0.4 mM) exposure for 16 h, suggesting that palmitate exposure decreased NNMT enzymatic activity.
Figure 1.
Palmitate upregulates NNMT expression in hepatocytes. A: NNMT catalyzes SAM-dependent methylation/degradation of nicotinamide, which serves as a predominant endogenous precursor for NAD+ biosynthesis via the salvage pathway. B: AML12 cells were treated with the indicated concentrations of palmitate for 16 h. NNMT gene expression was quantified using real time-qPCR. Data are expressed as means ± SD, n = 4 separate experiments. Differences between the 2 groups were determined using Student’s t test (*P < 0.05; ***P < 0.001 vs. control). C: AML12 cells were treated with the 0.4 mM palmitate for 16 h. Protein abundance of NNMT was detected by Western blotting. The signal of NNMT protein band was measured by densitometry and then divided by the signal of its corresponding actin abundance in the same sample. Data are expressed as means ± SD, n = 3 separate experiments. Student’s t test was used for statistical evaluation (***P < 0.001 vs. control). D: AML12 cells were treated with the 0.4 mM palmitate for 16 h. Intracellular total NAD (tNAD) concentrations were measured. Data are expressed as means ± SD, n = 3 separate experiments. Student’s t test was used for statistical evaluation (*P < 0.05 vs. control). NNMT, nicotinamide N-methyltransferase.
ATF4 Activation Contributes to Palmitate-Induced NNMT Upregulation
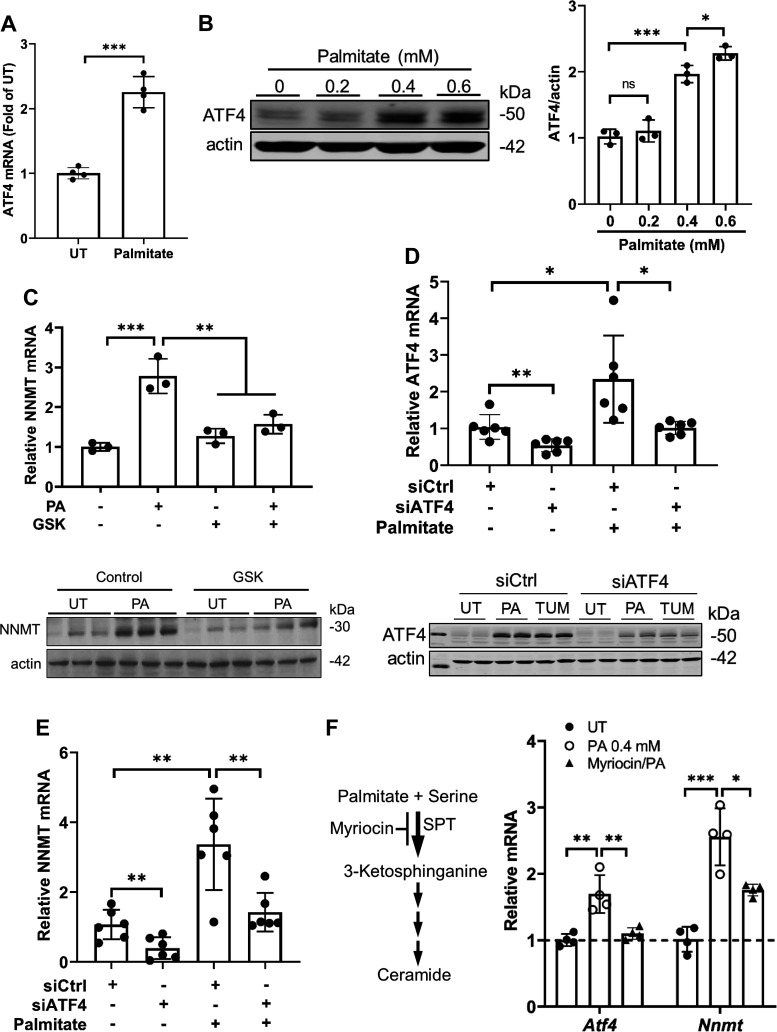
We have previously reported that the transcription factor ATF4 mediates NNMT expression in hepatocytes and the liver, contributing to the pathogenesis of alcoholic liver disease (13). To determine whether ATF4 activation contributes to palmitate-elicited NNMT upregulation, in this study, we first examined the effect of palmitate exposure on ATF4 activation in AML12 cells. As shown in Fig. 2, A and B, palmitate exposure upregulates ATF4 expression at both mRNA (Fig. 2A) and protein levels (Fig. 2B). To determine the mechanistic role of ATF4 activation in palmitate-induced NNMT upregulation, we then employed both pharmacological (GSK2006414, a specific inhibitor of PERK, an upstream kinase for ATF4 activation) and genetic (ATF4 siRNA transfection) approaches to inhibit ATF4 before palmitate exposure. As expected, ATF4 inhibition via either approach blunted palmitate-induced NNMT upregulation (Fig. 2, C–E), suggesting that ATF4 activation plays a mechanistic role in NNMT overexpression in response to palmitate challenge. Palmitate is a precursor for ceramide biosynthesis via the de novo pathway with serine palmitoyl transferase (SPT) being the rate-limiting enzyme. The pretreatment of AML12 cells with myriocin, a specific inhibitor of SPT, alleviated palmitate-induced upregulation of ATF4 and NNMT (Fig. 2G), suggesting that ceramide generation contributes, at least partially, to palmitate-elicited ATF4 activation.
Figure 2.
ATF4 activation contributes to palmitate induced NNMT upregulation. A: AML12 were treated with palmitate (0.4 mM) for 16 h. Total RNA was extracted, and ATF4 gene expression quantified using real time-qPCR. Data are expressed as means ± SD, n = 4 separate experiments. Differences between the two groups were determined using Student’s t test (***P < 0.001 vs. control). B: AML12 cells were treated with the indicated concentrations of palmitate for 16 h, ATF4 protein abundance was determined by Western blotting. The signal of ATF4 protein band was measured by densitometry and then divided by the signal of its corresponding actin abundance in the same sample. Data are expressed as means ± SD, n = 3 separate experiments. Student’s t test was used for statistical evaluation (*P < 0.05; ***P < 0.001 vs. control). C: AML12 cells were pretreated with GSK2006414 (20 nM), a selective inhibitor of PERK, for 2 h before 0.4 mM palmitate exposure for 16 h. NNMT expression was quantified by real time-qPCR for mRNA and Western blotting for protein, respectively. Data are expressed as means ± SD, n = 3 separate experiments. Differences between the two groups were determined using Student’s t test (**P < 0.01; ***P < 0.001 vs. control). D and E: AML12 cells were transfected with either scramble siRNA (siCtrl) or ATF4 siRNA (siATF4) for 24 h and treated with palmitate (PA at 0.4 mM) or tunicamycin (TUM at 5 µM) for 16 h. ATF4 (D) and NNMT (E) expression was quantified and data were expressed as means ± SD, n = 6 separate experiments. Differences between the two groups were determined using Student’s t test (*P < 0.05; **P < 0.01 vs. corresponding control cells). F: AML12 cells were pretreated with myriocin (20 µM), a specific inhibitor of serine palmitoyltransferase (SPT), for 2 h before 0.4 mM palmitate exposure for 16 h. ATF4 and NNMT gene expression was quantified by real time-qPCR. Data are expressed as means ± SD, n = 3 separate experiments. Differences between the two groups were determined using Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001 vs. control). ATF, activating transcription factor; NNMT, nicotinamide N-methyltransferase.
mTORC1 Activation upon Palmitate Exposure Contributes to NNMT Upregulation
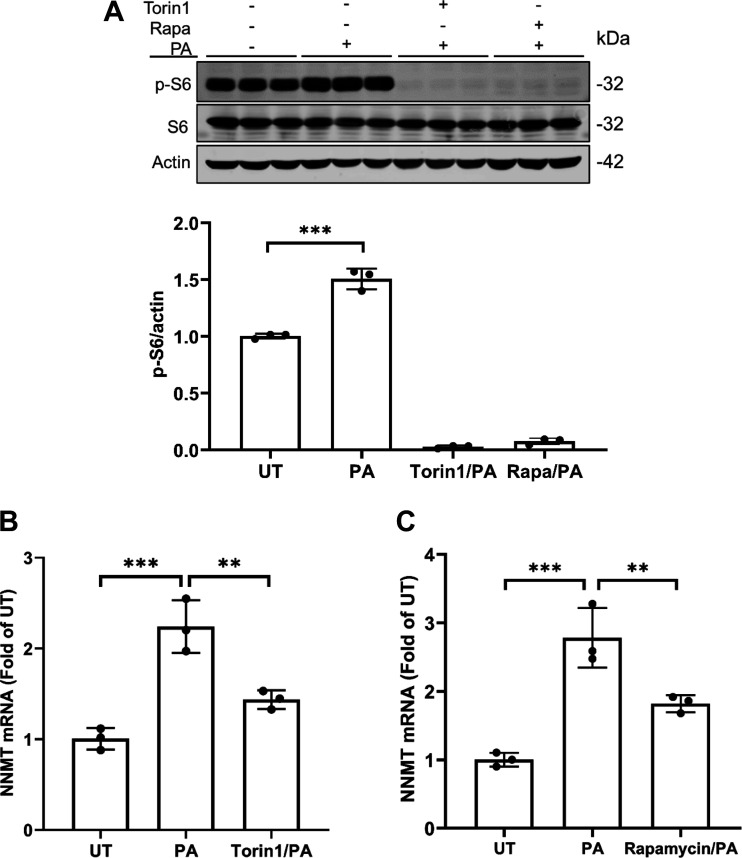
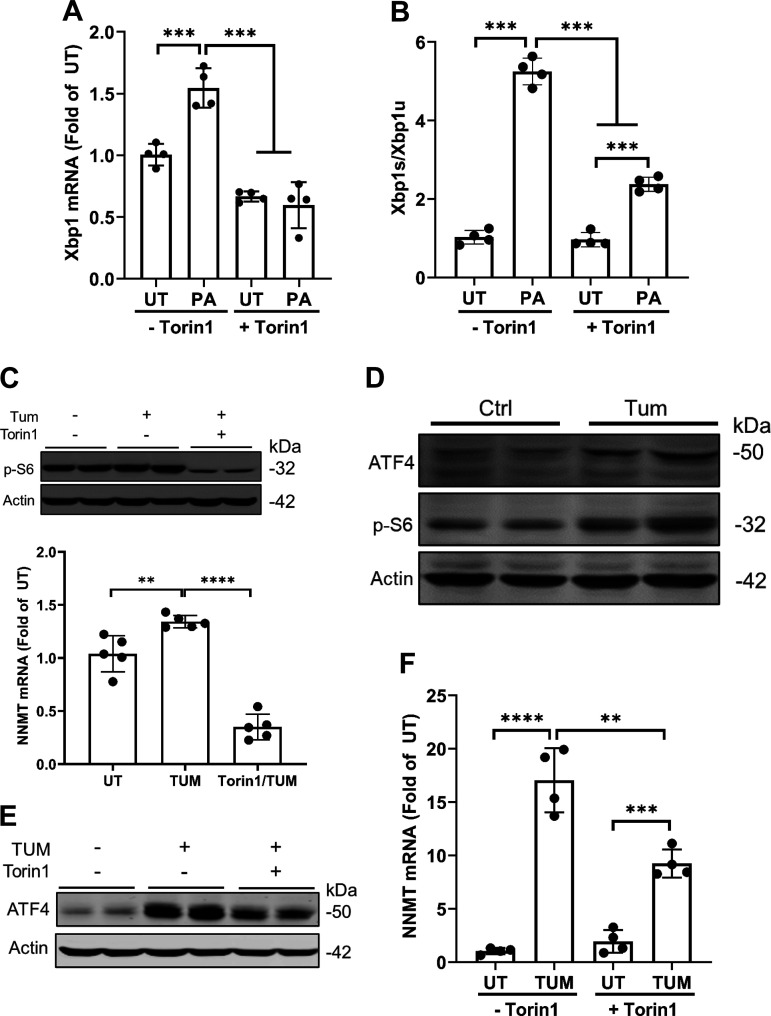
In a recent study, we demonstrated that mTORC1 activation contributes to palmitate-induced hepatotoxicity (21). In line with our previous study, here we showed that palmitate exposure activated mTORC1 in AML12 cells, evidenced by increased p-S6 protein abundance upon palmitate challenge (Fig. 3A). To determine whether the mTORC1 activation is mechanistically involved in palmitate-induced NNMT upregulation, we pretreated AML12 cells with two mechanistically different mTORC1 inhibitors, Torin1 and rapamycin, respectively, for 2 h before palmitate exposure for 16 h. NNMT gene expression was determined by real time-qPCR. As shown in Fig. 3, B and C, mTORC1 inhibitors attenuated NNMT mRNA increment in response to palmitate exposure, indicating that mTORC1 activation plays a mechanistic role in palmitate-induced NNMT upregulation in hepatocytes.
Figure 3.
mTORC1 activation upon palmitate exposure contributes to NNMT upregulation. A: AML12 cells were pretreated with or without Torin1 (0.25 µM) or rapamycin (Rapa at 50 nM) for 2 h before palmitate (0.4 mM) exposure for 16 h. Total protein was extracted. Protein abundance of phosphorylated (p-)S6, total-S6, and actin were detected by Western blotting. The signal of p-S6 protein band was measured by densitometry and then divided by the signal of its corresponding actin abundance in the same sample. Data are expressed as means ± SD, n = 3 separate experiments. Student’s t test was used for statistical evaluation (***P < 0.001 vs. control). B and C: AML12 cells were pretreated with or without Torin1 (0.25 µM) (B) or rapamycin (Rapa at 50 nM) (C) for 2 h before palmitate (0.4 mM) exposure for 16 h. Total RNA was extracted. NNMT gene expressions were detected by real time-qPCR. Data are expressed as means ± SD, n = 3 separate experiments. Differences between the two groups were determined using Student’s t test (**P < 0.01; ***P < 0.001 vs. control). NNMT, nicotinamide N-methyltransferase.
mTORC1 Activation is an Upstream Signaling Event in Palmitate-Induced ATF4 Activation
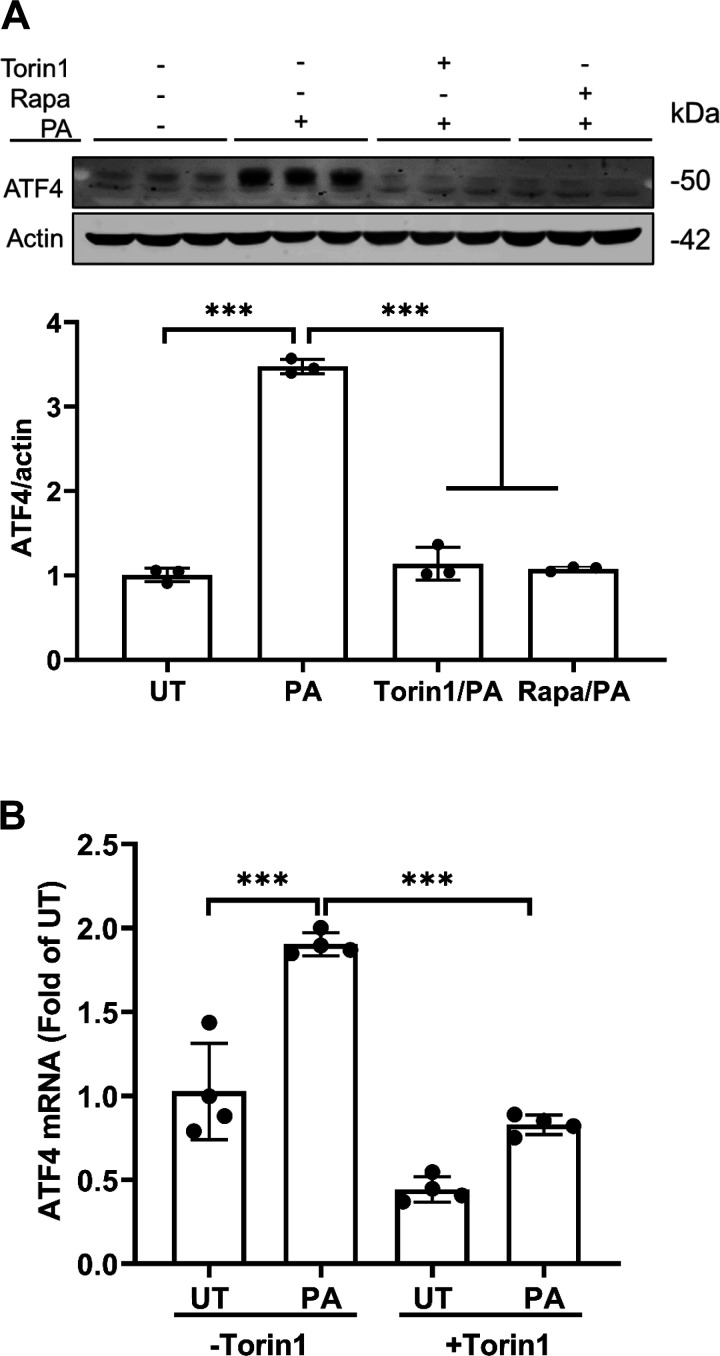
The critical involvement of mTORC1 activation in palmitate-induced NNMT upregulation prompted us to posit that mTORC1 is an upstream signaling event of ATF4 activation in response to palmitate exposure. To test this, we pretreated AML12 cells with or without mTORC1 inhibitors, Torin1 or rapamycin, for 2 h before palmitate exposure for 16 h. ATF4 expression at both mRNA and protein levels was determined. As shown in Fig. 4, A and B, mTORC1 inhibition ameliorated palmitate-induced ATF4 upregulation, suggesting that mTORC1 activation is an upstream signaling event in palmitate-induced ATF4 activation.
Figure 4.
mTORC1 activation is an upstream event in palmitate induced ATF4 activation. A: AML12 cells were pretreated with or without Torin1 (0.25 µM) or rapamycin (Rapa at 50 nM) for 2 h before palmitate (0.4 mM) exposure for 16 h. Total protein was extracted. Protein abundance of ATF4 and actin were detected by Western blotting. The signal of ATF4 protein band was measured by densitometry and then divided by the signal of its corresponding actin abundance in the same sample. Data are expressed as means ± SD, n = 3 separate experiments. Student’s t test was used for statistical evaluation (***P < 0.001 vs. control). B: AML12 were pretreated with Torin1 (0.25 µM) for 2 h before a 16-h palmitate (0.4 mM) exposure. Total RNA was extracted. ATF4 mRNA levels were detected by real time-qPCR. Data are expressed as means ± SD, n = 4 separated experiments. Differences between the two groups were determined using Student’s t test (***P < 0.001 vs. control). ATF, activating transcription factor.
mTORC1 Activation Contributes to Palmitate-Induced ER Stress and Downstream ATF4 Activation
Palmitate exposure induces ER stress in a variety of cell types, including hepatocytes (16, 18, 22, 23). This notion was further validated by our observation that a 16-h palmitate exposure of AML12 cells significantly increased the ratio between Xbp1s (spliced Xbp1) and Xbp1u (un-spliced Xbp1) (Xbp1s/Xbp1u), an indicator of activation of IRE1α pathway, the most conserved arm of three canonical pathways activated upon ER stress (Fig. 5, A and B). Intriguingly, Torin1 pretreatment compromised palmitate-induced elevation of Xbp1s/Xbp1u (Fig. 5, A and B), indicating that mTORC1 activation was required for palmitate-induced ER stress elicitation. To test the generality of this notion, we conducted both in vitro cell culture experiments in AML12 cells and in vivo animal studies with male C57 BL/6 mice using tunicamycin, a well-established ER stress inducer. In AML12 cells, tunicamycin activated mTORC1 (Fig. 5C), which was attenuated by Torin1 pretreatment. In mice, hepatic ER stress induction by tunicamycin administration through IP injection, which was evidenced by ATF4 upregulation, was concomitant with hepatic mTORC1 activation, evidenced by increased p-S6 protein abundance (Fig. 5D). Furthermore, both ATF4 activation and NNMT upregulation upon tunicamycin treatment in AML12 cells were blunted by mTORC1 inhibition (Fig. 5, E and F).
Figure 5.
mTORC1 activation contributes to palmitate-induced ER stress and NNMT upregulation. A and B: AML12 cells were pretreated with Torin1 (0.25 µM) for 2 h before the palmitate (0.4 mM) exposure for 16 h. Total RNA was extracted. The gene expressions of Xbp1, Xbp1s, and Xbp1u were quantified by real time-qPCR and Xbp1s/Xbp1u ratio calculated. Data are expressed as means ± SD, n = 4 different experiments. Differences between the two groups were determined using Student’s t test (***P < 0.001vs. control). C: AML12 cells were pretreated with Tornin1 for 2 h before tunicamycin (10 μm) treatment for 16 h. Protein abundance of p-S6 and actin was detected by Western blotting. The signal of p-S6 protein band was measured by densitometry and then divided by the signal of its corresponding actin abundance in the same sample. Data are expressed as means ± SD, n = 5 separate experiments. Student’s t test was used for statistical evaluation (**P < 0.01; ****P < 0.0001 vs. control). D: Ten-week-old male C57BL/6N mice were injected with tunicamycin (2 mg/kg body wt ip) or isovolumic vehicle (150 mM dextrose) and 16 h later livers were harvested. Protein abundance of ATF4, p-S6 and actin was detected by Western blotting. E: AML12 cells were pretreated with Torin1 (0.25 µM) for 2 h before tunicamycin (10 μm) treatment for 16 h. Protein abundance of ATF4 was detected by Western blotting. F: AML12 cells were pretreated with Torin1 for 2 h before tunicamycin (10 μm) treatment for 16 h. Total RNA was extracted and NNMT gene expression quantified by real time-qPCR. All data were expressed as means ± SD, n = 4 separated experiments. Differences between the two groups were determined using Student’s t test (**P < 0.01; ***P < 0.001; ****P < 0.0001 vs. control). NNMT, nicotinamide N-methyltransferase; XBP1, X-box binding protein 1.
NNMT Upregulation Contributes to Palmitate-Induced Cell Death
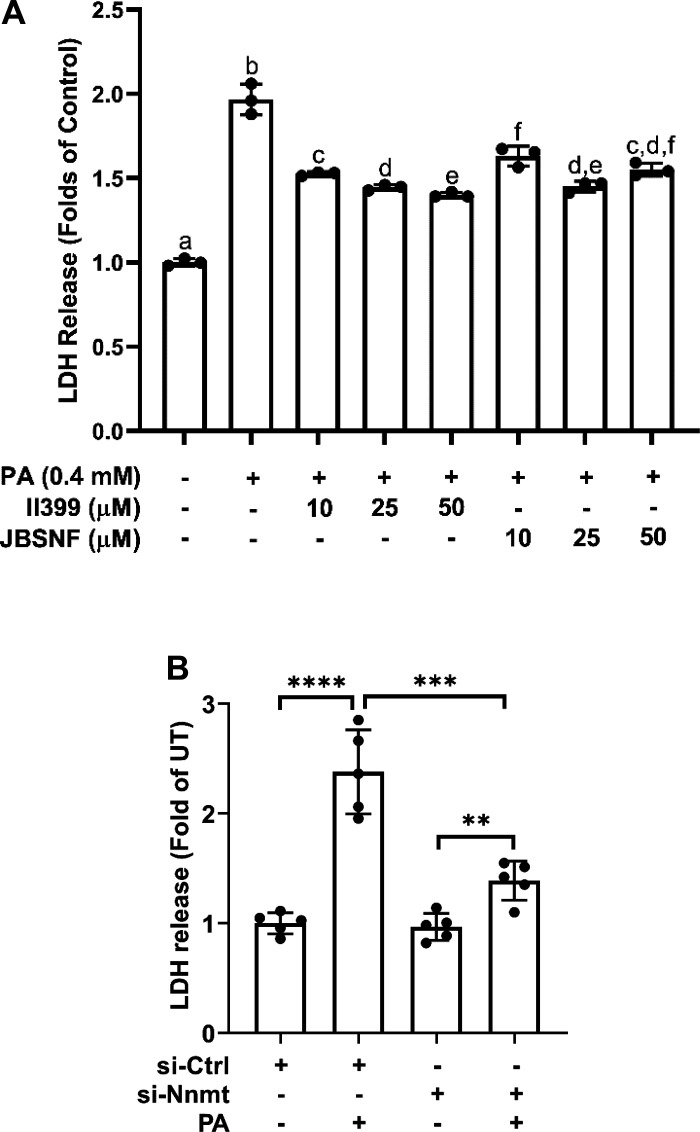
To determine whether NNMT upregulation contributes to palmitate-induced lipotoxicity in hepatocytes, NNMT suppression was achieved through both pharmacological and genetic approaches. As shown in Fig. 6A, both chemical inhibitors for NNMT, JBSNF-000088 and II390 (24), alleviated palmitate-induced cell death in AML12 cells. Similarly, siRNA knockdown of NNMT led to a significant reduction of LDH release upon a 16-h palmitate exposure (Fig. 6B).
Figure 6.
NNMT inhibition protects against palmitate-induced cell death. A: AML12 cells were pretreated with either JBSNF-000088 (20 µM) or II399 (20 µM) at the indicated concentrations for 4 h before palmitate (0.4 mM) exposure for 16 h. Cell viability was determined by LDH release measurement. Data are expressed as mean ± SD, n = 3 separated experiments. Bars with different character differ significantly (P < 0.05). B: AML12 cells were transfected with either scramble siRNA or NNMT siRNA for 24 h and treated with palmitate at 0.4 mM for 16 h. Cell death was determined by LDH release measurement. Data are expressed as means ± SD, n = 3 separated experiments. Differences between the two groups were determined using Student’s t test (**P < 0.01; ***P < 0.001; ***P < 0.001 vs. control). NNMT, nicotinamide N-methyltransferase.
Protein Kinase A Activation is Attributable to the Protective Effect of NNMT Inhibition against Palmitate-Induced Cell Death
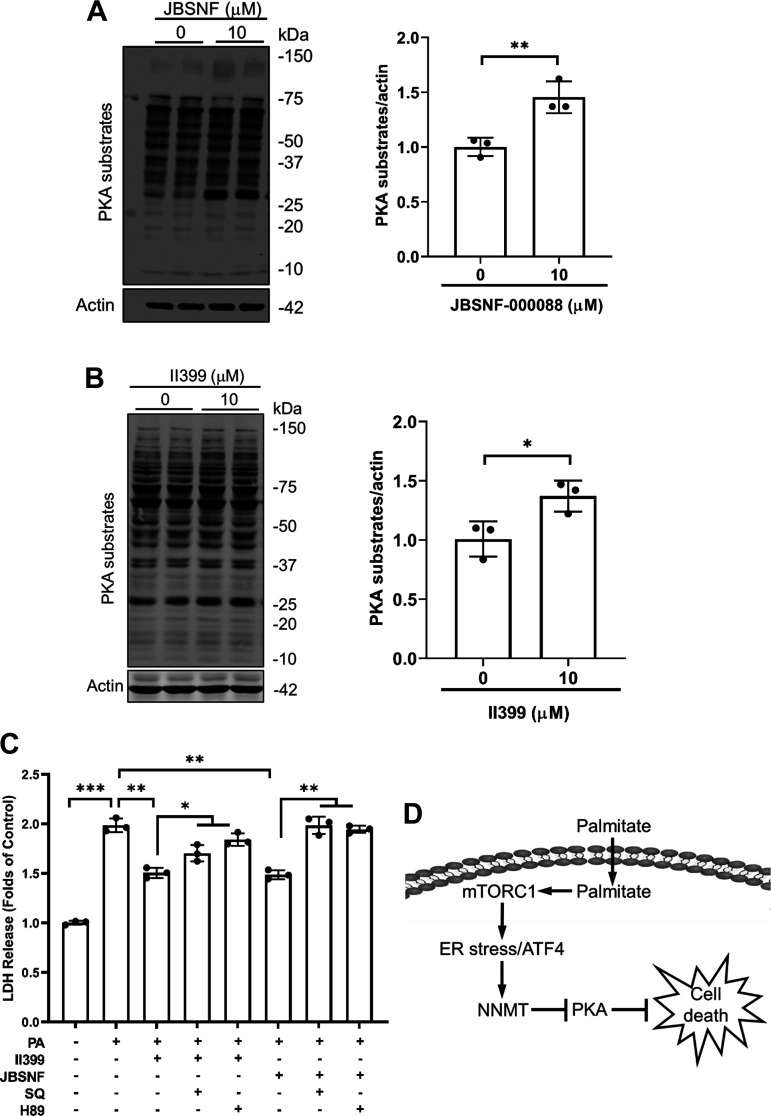
In search of the mechanism(s) underlying the protective effect of NNMT inhibition against palmitate lipotoxicity, a small-scale inhibitor screening was performed, and we identified PKA activation to be mechanistically involved in the process. We uncovered that NNMT inhibitors, JBSNF-000088 and II390, activated PKA in AML12 cells (Fig. 7, A and B). Importantly, the protective effects of NNMT inhibitors were compromised when PKA was inhibited via a 2-h pretreatment with either SQ22536, a specific inhibitor of adenylyl cyclase for intracellular cAMP formation, or H89, a specific inhibitor for PKA (Fig. 7C).
Figure 7.
Protein kinase A (PKA) inhibition compromises the protective effect of NNMT inhibition against palmitate-induced cell death. A and B: AML12 cells were treated with either JBSNF-000088 (25 µM) or II399 (25 µM) for 6 h. Total proteins were isolated and PKA substrates detected by Western blotting. The signal of PKS substrates was measured by densitometry and then divided by the signal of its corresponding actin abundance in the same sample. Data are expressed as means ± SD, n = 3 separate experiments. Student’s t test was used for statistical evaluation (*P < 0.05; ***P < 0.01 vs. untreated cells). C: AML12 cells were pretreated with either JBSNF-000088 (25 µM) or II399 (25 µM) at the presence/absence of PKA inhibitor, either SQ22536 (200 µM) or H89 (10 µM) for 4 h before palmitate exposure for 16 h. Cell death was determined by LDH release. All data are expressed as means ± SD, n = 3 separated experiments. Differences between the two groups were determined using Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001 vs control). D: schematic illustration of the role and mechanism of NNMT upregulation in palmitate-induced hepatocyte lipotoxicity. The mTORC1-ATF4 pathway activation contributes to palmitate-elicited NNMT upregulation and protein kinase A (PKA) activation contributes to NNMT inhibition-conferred protection against hepatolipotoxicity. NNMT, nicotinamide N-methyltransferase.
DISCUSSION
We are the first to identify ATF4 as a transcription factor mediating NNMT expression in the liver (13). In a very recent study, we reported that mTORC1 activation is mechanistically involved in palmitate-induced cell death in AML12 hepatocytes (25). In this present study, we demonstrated that palmitate exposure activates ATF4 in a mTORC1-dependent mechanism and the mTORC1-ATF4 pathway activation contributes to palmitate-elicited NNMT upregulation. Importantly, we have provided initial evidence that NNMT upregulation contributes to palmitate lipotoxicity in hepatocytes. Specifically, NNMT inhibition through both pharmacological and genetic approach bestows protection against palmitate-induced cell death in hepatocytes. Interestingly, we uncovered that NNMT inhibition is associated with PKA activation and PKA inhibition offsets the protective effect of NNMT inhibitors, suggesting that PKA activation contributes to NNMT inhibition-mediated amelioration of palmitate-induced lipotoxic effects in hepatocytes.
The PERK-eIF2α-ATF4 pathway is one of the three canonical pathways that is activated in response to ER stress (14, 22, 23). Upon ER stress, PERK activation selectively activates ATF4 via phosphorylating (activating) the eukaryotic initiation factor 2 (eIF2α) at serine 51 (26). In addition to suppressing global protein synthesis, eIF2α activation paradoxically enhances the ATF4 translation. ER stress plays a mechanistic role in palmitate-elicited lipotoxicity in various cell types, including hepatocyte(16–18). In this study, our data clearly demonstrate that ATF4 activation contributes to palmitate-induced NNMT upregulation in hepatocytes. This notion is supported by the evidence that palmitate activates ATF4 and the genetic knockdown of ATF4 prevents palmitate-triggered NNMT upregulation in AML12 cells.
Other than the translational upregulation by the PERK-eIF2α pathway in response to ER stress, the expression of ATF4, as a key effector of the integrated stress response, is also mediated both transcriptionally and translationally by several other kinases (27), among which mTORC1 has been emerging as an important regulator for ATF4 expression and activation. In the human lung fibroblasts, transforming growth factor-β1 (TGF-β1) stimulation increased ATF4 protein production via mTORC1 activation to promote glycine biosynthesis for collagen production (28). In HEK293T cells, mTORC1 activation upregulates ATF4 expression both transcriptionally and translationally to regulate amino acids homeostasis (29). In contrast, it has been recently reported that mTORC1 activation is associated with ATF4 inhibition, which elicits prooxidant and pro-death effects in MEFs (30). We recently reported that mTORC1 activation upon palmitate exposure contributes to palmitate-induced cellular dysfunction and cell death in hepatocytes (25). The concomitant occurrence of mTORC1 and ATF4 activation in response to palmitate exposure prompted us to investigate whether a causal relationship exists between these two events in this study. As expected, both mTORC1 inhibitors, Torin1 and rapamycin, blunted palmitate-elicited ATF4 activation and NNMT upregulation upon palmitate exposure, indicating that mTORC1 activation is an upstream signaling event for palmitate-induced ATF4 activation. Considering the ER stress-inducing property of palmitate, we further tested the generality of this signaling event for ER stress. Interestingly, tunicamycin, a well-characterized ER stress inducer, similarly activates mTORC1 in AML12 cells and furthermore in mice livers. Importantly, Torin1 pretreatment prevents tunicamycin-induced ATF4 activation and NNMT upregulation, suggesting that mTORC1 activation is an integral signaling event of UPR in response to ER stress induction. Indeed, in line with several other reports using different cells (19, 20, 30), we previously reported that palmitate-induced IRE1α activation is mTORC1-dependent in hepatocytes (21), suggesting the existence of a cross talk between mTORC1 and ER stress/UPR. Further investigations are warranted to examine how mTORC1 cross talk with the PERK-eIF2α-AFT4 pathway of UPR.
NNMT is a methyltransferase which catalyzes the reaction for nicotinamide degradation (methylation) using S-adenosylmethionine (SAM) as a methyl donor to produce 1-methylnicotinamide (1-MNA) and S-adenosylhomocysteine (SAH) (Fig. 1A). Nicotinamide is a predominant endogenous precursor for NAD+ biosynthesis via the salvage pathway. As NNMT-catalyzed transmethylation reaction is a major route for nicotinamide degradation, NNMT competes with the NAD+ salvage pathway for nicotinamide availability. As a result, dysregulated NNMT expression can exert significant influence on cellular SAM and NAD+ homeostasis (31). Emerging evidence supports that NNMT, which is abundantly expressed in the liver, plays a critical role in regulating liver functions. Whole body NNMT overexpression leads to reduced hepatic NAD+ and SAM levels in mice (32). Importantly, the NNMT-overexpressing mice manifest worsened hepatic steatosis in the setting of chronic high-fat diet feeding, which is associated with increased expression of the genes involved in fatty acid uptake and decreased VLDL secretion (32). We previously reported that hepatic NNMT upregulation is mediated by transcription factor ATF4 and contributes to the pathogenesis of alcoholic fatty disease development (13). Our observation that palmitate exposure upregulates NNMT expression in hepatocytes encouraged us to further explore the potential beneficial role of NNMT inhibition on NAFLD. In this study, we set out to examine if NNMT upregulation contributes to palmitate lipotoxicity in hepatocytes and our data uncovered that both chemical inhibitors of NNMT and NNMT siRNA transfection are protective against palmitate hepatotoxicity, indicative of a mechanistic implication of NNMT upregulation in hepatic lipotoxicity.
NNMT plays a central role in the regulation of cellular SAM and NAD+ metabolism via catalyzing SAM-dependent methylation of nicotinamide, a main endogenous precursor for NAD+ biosynthesis. As SAM is a predominant methyl donor for most intracellular transmethylation reactions and NAD+ has a potential to regulate (de)acetylation reactions via regulating SIRT1 activation, the mechanisms behind the protective effects of NNMT inhibition against lipotoxicity can be complex and multifactorial. We previously reported that exogenous nicotinamide supplementation confers protection against palmitate-induced cell death in hepatocytes via upregulating SIRT1 expression in a cAMP/PKA pathway-dependent mechanism (33). In search of mechanism(s) that underlie the hepatoprotective effect of NNMT inhibition, we identified PKA as a target for NNMT inhibition through a small-scale inhibitor screening. Our observations that NNMT inhibition activates PKA and PKA inhibition offsets the protective effect of NNMT inhibitors on palmitate-induced lipotoxicity suggest that PKA activation contributes, at least partially, to the hepatoprotective effect of NNMT inhibition upon palmitate exposure.
In conclusion, this study provides evidence that the mTORC1-ATF4 pathway activation in hepatocytes contributes to palmitate-elicited NNMT upregulation. Importantly, we demonstrate for the first time that NNMT inhibition provides protection against hepatic lipotoxicity. The mechanistic involvement of NNMT upregulation in palmitate-induced hepatocyte cell death suggests the clinical beneficial potential of NNMT inhibitors in the treatment of a variety of metabolic disorders including NAFLD. Further investigations are necessary to clarify the underpinning mechanisms behind the hepatoprotective effects of NNMT inhibition and to explore its clinical relevance.
GRANTS
This work was in part funded by US National Institutes of Health (NIH) Grants NIAAA R01AA026603 (to Z.S.).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Z.S. conceived and designed research; A.G., J.W., Q.S., I.D.I., L.L., and J.P. performed experiments; A.G., J.W., Q.S., I.D.I., L.L., J.P., and R.H. analyzed data; A.G., J.W., and Y.J. interpreted results of experiments; A.G., J.W. and Q.S. prepared figures; A.G. and J.W. drafted manuscript; Y.J., R.H., and Z.S. edited and revised manuscript; Z.S. approved final version of manuscript.
REFERENCES
- 1.Raza S, Rajak S, Upadhyay A, Tewari A, Anthony Sinha R. Current treatment paradigms and emerging therapies for NAFLD/NASH. Front Biosci (Landmark Ed) 26: 206–237, 2021. doi: 10.2741/4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Godoy-Matos AF, Silva Júnior WS, Valerio CM. NAFLD as a continuum: from obesity to metabolic syndrome and diabetes. Diabetol Metab Syndr 12: 60, 2020. doi: 10.1186/s13098-020-00570-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Csak T, Ganz M, Pespisa J, Kodys K, Dolganiuc A, Szabo G. Fatty acid and endotoxin activate inflammasomes in mouse hepatocytes that release danger signals to stimulate immune cells. Hepatology 54: 133–144, 2011. doi: 10.1002/hep.24341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ogawa Y, Imajo K, Honda Y, Kessoku T, Tomeno W, Kato S, Fujita K, Yoneda M, Saito S, Saigusa Y, Hyogo H, Sumida Y, Itoh Y, Eguchi K, Yamanaka T, Wada K, Nakajima A. Palmitate-induced lipotoxicity is crucial for the pathogenesis of nonalcoholic fatty liver disease in cooperation with gut-derived endotoxin. Sci Rep 8: 11365, 2018. doi: 10.1038/s41598-018-29735-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asrih M, Jornayvaz FR. Metabolic syndrome and nonalcoholic fatty liver disease: Is insulin resistance the link? Mol Cell Endocrinol 418: 55–65, 2015. doi: 10.1016/j.mce.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 6.Estadella D, da Penha Oller do Nascimento CM, Oyama LM, Ribeiro EB, Dâmaso AR, de Piano A. Lipotoxicity: effects of dietary saturated and transfatty acids. Mediators Inflamm 2013: 137579, 2013. doi: 10.1155/2013/137579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Almeida IT, Cortez-Pinto H, Fidalgo G, Rodrigues D, Camilo ME. Plasma total and free fatty acids composition in human non-alcoholic steatohepatitis. Clin Nutr 21: 219–223, 2002. doi: 10.1054/clnu.2001.0529. [DOI] [PubMed] [Google Scholar]
- 8.Rada P, González-Rodríguez Á, García-Monzón C, Valverde ÁM. Understanding lipotoxicity in NAFLD pathogenesis: is CD36 a key driver? Cell Death Dis 11: 802, 2020. doi: 10.1038/s41419-020-03003-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pissios P. Nicotinamide N-methyltransferase: more than a vitamin B3 clearance enzyme. Trends Endocrinol Metab 28: 340–353, 2017. doi: 10.1016/j.tem.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberti A, Fernández AF, Fraga MF. Nicotinamide N-methyltransferase: at the crossroads between cellular metabolism and epigenetic regulation. Mol Metab 45: 101165, 2021. doi: 10.1016/j.molmet.2021.101165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kraus D, Yang Q, Kong D, Banks AS, Zhang L, Rodgers JT, Pirinen E, Pulinilkunnil TC, Gong F, Wang Y-C, Cen Y, Sauve AA, Asara JM, Peroni OD, Monia BP, Bhanot S, Alhonen L, Puigserver P, Kahn BB. Nicotinamide N-methyltransferase knockdown protects against diet-induced obesity. Nature 508: 258–262, 2014. doi: 10.1038/nature13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brachs S, Polack J, Brachs M, Jahn-Hofmann K, Elvert R, Pfenninger A, Bärenz F, Margerie D, Mai K, Spranger J, Kannt A. Genetic nicotinamide N-methyltransferase (Nnmt) deficiency in male mice improves insulin sensitivity in diet-induced obesity but does not affect glucose tolerance. Diabetes 68: 527–542, 2019. doi: 10.2337/db18-0780. [DOI] [PubMed] [Google Scholar]
- 13.Song Q, Chen Y, Wang J, Hao L, Huang C, Griffiths A, Sun Z, Zhou Z, Song Z. ER stress-induced upregulation of NNMT contributes to alcohol-related fatty liver development. J Hepatol 73: 783–793, 2020. doi: 10.1016/j.jhep.2020.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8: 519–529, 2007. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 15.Sabatini DM. Twenty-five years of mTOR: uncovering the link from nutrients to growth. Proc Natl Acad Sci USA 114: 11818, 2017. doi: 10.1073/pnas.1716173114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karaskov E, Scott C, Zhang L, Teodoro T, Ravazzola M, Volchuk A. Chronic palmitate but not oleate exposure induces endoplasmic reticulum stress, which may contribute to INS-1 pancreatic beta-cell apoptosis. Endocrinology 147: 3398–3407, 2006. doi: 10.1210/en.2005-1494. [DOI] [PubMed] [Google Scholar]
- 17.Perry BD, Rahnert JA, Xie Y, Zheng B, Woodworth-Hobbs ME, Price SR. Palmitate-induced ER stress and inhibition of protein synthesis in cultured myotubes does not require Toll-like receptor 4. PLoS One 13: e0191313, 2018. doi: 10.1371/journal.pone.0191313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu S, Nam SM, Kim JH, Das R, Choi SK, Nguyen TT, Quan X, Choi SJ, Chung CH, Lee EY, Lee IK, Wiederkehr A, Wollheim CB, Cha SK, Park KS. Palmitate induces ER calcium depletion and apoptosis in mouse podocytes subsequent to mitochondrial oxidative stress. Cell Death Dis 6: e1976, 2015. doi: 10.1038/cddis.2015.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He L, Weber KJ, Diwan A, Schilling JD. Inhibition of mTOR reduces lipotoxic cell death in primary macrophages through an autophagy-independent mechanism. J Leukoc Biol 100: 1113–1124, 2016. doi: 10.1189/jlb.3A1015-463R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marafie SK, Al-Shawaf EM, Abubaker J, Arefanian H. Palmitic acid-induced lipotoxicity promotes a novel interplay between Akt-mTOR, IRS-1, and FFAR1 signaling in pancreatic β-cells. Biol Res 52: 44, 2019. doi: 10.1186/s40659-019-0253-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, Griffiths A, Wang J, Zhang T, Song Q, Song Z. Inositol-requiring enzyme 1α links palmitate-induced mTOR activation and lipotoxicity in hepatocytes. Am J Physiol Cell Physiol 319: C1130–C1140, 2020. doi: 10.1152/ajpcell.00165.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Achard CS, Laybutt DR. Lipid-induced endoplasmic reticulum stress in liver cells results in two distinct outcomes: adaptation with enhanced insulin signaling or insulin resistance. Endocrinology 153: 2164–2177, 2012. doi: 10.1210/en.2011-1881. [DOI] [PubMed] [Google Scholar]
- 23.Guo W, Wong S, Xie W, Lei T, Luo Z. Palmitate modulates intracellular signaling, induces endoplasmic reticulum stress, and causes apoptosis in mouse 3T3-L1 and rat primary preadipocytes. Am J Physiol Endocrinol Metab 293: E576–E586, 2007. doi: 10.1152/ajpendo.00523.2006. [DOI] [PubMed] [Google Scholar]
- 24.Chen D, Li L, Diaz K, Iyamu ID, Yadav R, Noinaj N, Huang R. Novel propargyl-linked bisubstrate analogues as tight-binding inhibitors for nicotinamide N-methyltransferase. J Med Chem 62: 10783–10797, 2019. doi: 10.1021/acs.jmedchem.9b01255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Chen Y, Song Q, Griffiths A, Song Z. mTORC1-IRE1α pathway activation contributes to palmitate-elicited triglyceride secretion and cell death in hepatocytes. Exp Biol Med (Maywood) 245: 1268–1279, 2020. doi: 10.1177/1535370220928276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rozpedek W, Pytel D, Mucha B, Leszczynska H, Diehl JA, Majsterek I. The role of the PERK/eIF2α/ATF4/CHOP signaling pathway in tumor progression during endoplasmic reticulum stress. Curr Mol Med 16: 533–544, 2016. doi: 10.2174/1566524016666160523143937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luhr M, Torgersen ML, Szalai P, Hashim A, Brech A, Staerk J, Engedal N. The kinase PERK and the transcription factor ATF4 play distinct and essential roles in autophagy resulting from tunicamycin-induced ER stress. J Biol Chem 294: 8197–8217, 2019. doi: 10.1074/jbc.RA118.002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selvarajah B, Azuelos I, Platé M, Guillotin D, Forty EJ, Contento G, Woodcock HV, Redding M, Taylor A, Brunori G, Durrenberger PF, Ronzoni R, Blanchard AD, Mercer PF, Anastasiou D, Chambers RC. mTORC1 amplifies the ATF4-dependent de novo serine-glycine pathway to supply glycine during TGF-β(1)-induced collagen biosynthesis. Sci Signal 12: eaav3048, 2019. doi: 10.1126/scisignal.aav3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park Y, Reyna-Neyra A, Philippe L, Thoreen CC. mTORC1 balances cellular amino acid supply with demand for protein synthesis through post-transcriptional control of ATF4. Cell Rep 19: 1083–1090, 2017. doi: 10.1016/j.celrep.2017.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torrence ME, MacArthur MR, Hosios AM, Valvezan AJ, Asara JM, Mitchell JR, Manning BD. The mTORC1-mediated activation of ATF4 promotes protein and glutathione synthesis downstream of growth signals. ELife 10: e63326, 2021. doi: 10.7554/eLife.63326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loring HS, Thompson PR. Kinetic Mechanism of Nicotinamide N-Methyltransferase. Biochemistry 57: 5524–5532, 2018. doi: 10.1021/acs.biochem.8b00775. [DOI] [PubMed] [Google Scholar]
- 32.Komatsu M, Kanda T, Urai H, Kurokochi A, Kitahama R, Shigaki S, Ono T, Yukioka H, Hasegawa K, Tokuyama H, Kawabe H, Wakino S, Itoh H. NNMT activation can contribute to the development of fatty liver disease by modulating the NAD+metabolism. Sci Rep 8: 8637, 2018. doi: 10.1038/s41598-018-26882-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, Dou X, Li S, Zhang X, Zeng Y, Song Z. Nicotinamide ameliorates palmitate-induced ER stress in hepatocytes via cAMP/PKA/CREB pathway-dependent Sirt1 upregulation. Biochim Biophys Acta 1853: 2929–2936, 2015. doi: 10.1016/j.bbamcr.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]