Abstract
To study the molecular mechanisms of circadian gene expression, we have sought to identify genes whose expression in mouse liver is regulated by the transcription factor DBP (albumin D-site-binding protein). This PAR basic leucine zipper protein accumulates according to a robust circadian rhythm in nuclei of hepatocytes and other cell types. Here, we report that the Cyp2a4 gene, encoding the cytochrome P450 steroid 15α-hydroxylase, is a novel circadian expression gene. This enzyme catalyzes one of the hydroxylation reactions leading to further metabolism of the sex hormones testosterone and estradiol in the liver. Accumulation of CYP2A4 mRNA in mouse liver displays circadian kinetics indistinguishable from those of the highly related CYP2A5 gene. Proteins encoded by both the Cyp2a4 and Cyp2a5 genes also display daily variation in accumulation, though this is more dramatic for CYP2A4 than for CYP2A5. Biochemical evidence, including in vitro DNase I footprinting on the Cyp2a4 and Cyp2a5 promoters and cotransfection experiments with the human hepatoma cell line HepG2, suggests that the Cyp2a4 and Cyp2a5 genes are indeed regulated by DBP. These conclusions are corroborated by genetic studies, in which the circadian amplitude of CYP2A4 and CYP2A5 mRNAs and protein expression in the liver was significantly impaired in a mutant mouse strain homozygous for a dbp null allele. These experiments strongly suggest that DBP is a major factor controlling circadian expression of the Cyp2a4 and Cyp2a5 genes in the mouse liver.
Circadian rhythms in physiology and behavior are observed throughout the animal and plant kingdoms. They are believed to enable an organism to anticipate and adapt to rhythmic changes in its environment (35). These daily oscillations persist under constant conditions, that is, in the absence of external time cues such as changes in light intensities. In mammals, circadian rhythms influence many if not most aspects of physiology and behavior, including sleep-wake cycles, energy metabolism, heart beat, blood pressure, body temperature, renal plasma flow, and mating (for a review, see reference 18). Recently, genetic and biochemical approaches have identified genes which contribute to the generation and/or maintenance of circadian rhythms in mammals (37). However, little is known about the molecular mechanisms by which these genes ultimately generate overt rhythms in physiology and behavior.
Better understanding of these mechanisms will come from the identification and analysis of genes under circadian regulation in peripheral tissues. One example is the rodent CYP7 gene, encoding cholesterol 7α-hydroxylase, an enzyme of the cytochrome P450 superfamily which catalyzes the rate-limiting step in the metabolism of cholesterol to bile acids. In the rat liver, the CYP7 gene displays circadian variation in expression (28), which is regulated primarily at the level of CYP7 gene transcription (16). Recently, members of the PAR family of DNA-binding transcription factors albumin D-site-binding protein (DBP), thyroid embryonic factor (TEF), and hepatic leukemia factor (HLF), have been proposed to play a role in the circadian transcription of the CYP7 gene (4, 8, 16, 20). In fact, each of the PAR transcription factors displays circadian accumulation in the rodent liver as well as other tissues (17, 19), and mice homozygous for a targeted mutation of the dbp gene display altered circadian locomotor activity (24). Furthermore, the PAR transcription factors can bind specifically to a regulatory element in the CYP7 gene promoter and can increase expression of this promoter in cotransfection assays (4, 8, 16, 20). Although CYP7 mRNA accumulation still cycles in dbp−/− mice, it does so with a 4-h phase advance compared to wild-type animals (25). Hence, DBP appears to be involved in setting the precise timing of circadian CYP7 expression. Since the amplitude of daily variations in CYP7 mRNA accumulation is similar in dbp wild-type and mutant animals, it is conceivable that the other two PAR basic leucine zipper (bZip) proteins, TEF and HLF, can substitute for DBP in the regulation of circadian CYP7 transcription.
A broader approach to circadian target gene identification and study would employ subtractive cloning techniques to detect genes whose mRNA accumulation varies throughout the day. This approach has proven useful in the study of circadian expression genes in Neurospora crassa (26). Using SABRE (selective amplification via biotin-and restriction-mediated enrichment), a novel subtractive hybridization protocol, we have recently identified the mouse gene Cyp2a5, encoding the cytochrome P450 coumarin 7-hydroxylase, as a novel circadian target gene in the liver (15). Accumulation of CYP2A5 mRNA was found to vary with a high amplitude throughout the day, with peak accumulation detected in the evening (15) (see below). As coumarin is a mildly toxic compound found naturally in plants, circadian Cyp2a5 gene expression may serve to protect the night-feeding mouse from coumarin-induced toxicity via hydroxylation and metabolism of coumarin in the liver.
The mouse Cyp2a4 gene is a P450 superfamily member highly related to Cyp2a5 (>98% identity in amino acid sequences of their gene products [22]). Despite this high level of similarity, the Cyp2a4 gene product demonstrates a distinct enzymatic activity, namely, the 15α-hydroxylation of steroid hormones including testosterone and estrogens. This is one of several hydroxylation modifications of steroid hormones believed to lead to their further metabolism and/or excretion by the liver (42).
We report here that Cyp2a4, like Cyp2a5, displays circadian expression in mouse liver, with a circadian pattern of mRNA accumulation identical to that of CYP2A5. Furthermore, CYP2A4 and CYP2A5 proteins display circadian accumulation in mouse liver microsomes, as judged by isoelectric focusing (IEF) blot analysis. Circadian expression of both Cyp2a4 and Cyp2a5 genes appears to be regulated by the PAR family transcription factor DBP. This conclusion is supported by biochemical and genetic studies: the promoters of both the Cyp2a4 and Cyp2a5 genes contain high-affinity binding sites for DBP, and promoter sequences including these sites are necessary for their efficient activation by DBP in cotransfection assays. Moreover, in homozygous dbp knockout mice, circadian expression of CYP2A4 and CYP2A5 mRNAs and proteins is significantly reduced.
MATERIALS AND METHODS
Animals.
Wild-type mice used were isogenic 129/Ola strain mice. Mice homozygous for a disrupted allele of the dbp gene (dbp knockout [dbp−/−) mice [24]) were otherwise isogenic 129/Ola strain mice. Animals were housed under 12-h light/12-h dark conditions, with lights on at 7 a.m. (7 h; Zeitgeber time [ZT] 0) and lights off at 7 p.m. (19 h; ZT 12). For experiments, male mice between 10 and 16 weeks of age were kept in darkness for at least 3 days before sacrifice. Mice were removed from the darkened room with the aid of a red light and sacrificed within 10 min of removal. For these experiments, subjective times of sacrifice are expressed according to the same lighting schedule as above, on a 24-h scale, with 7 h equivalent to ZT 0 and 19 h equivalent to ZT 12.
Cloning of Cyp2a4 and Cyp2a5 gene-specific probes.
Cyp2a4- and Cyp2a5- specific cDNA probes were generated by reverse transcription-PCR) on poly(A)+ liver RNA from wild-type mice sacrificed at 20 h (ZT 13; 1 h after lights off). An antisense oligonucleotide complementary to bases 796 to 777 of the Cyp2a4 cDNA sequence was used to prime first-strand cDNA synthesis, using Superscript II RNase H-minus reverse transcriptase as recommended by the manufacturer (Life Technologies, Bethesda, Md.). PCR amplification was performed with a second, sense-strand oligonucleotide corresponding to positions 325 to 346 of the Cyp2a4 cDNA sequence (40). The PCR products were digested with restriction enzymes ApaI and ClaI, and the resulting 228-bp fragment (bases 471 to 698) was subcloned into a plasmid vector. Sequence analysis led to the identification of plasmids containing Cyp2a4- and Cyp2a5-specific cDNA fragments (pSH-CA and pCH-CA, respectively). However, the Cyp2a4-specific cDNA fragment was found to contain one mismatch to the published mouse Cyp2a4 sequence, a C-to-T conversion at position 501, which corresponds to the mouse Cyp2a5 cDNA sequence at this position. This mismatch presumably arose as an artifact of the PCR, as at the six other sequence positions differing between the Cyp2a4 and the Cyp2a5 cDNA sequences, the sequence corresponded to the Cyp2a4 cDNA sequence. Nonetheless, as a result of this mismatch, RNase protection experiments on mouse liver RNA with the Cyp2a4 probe leads to a partial digestion product of the Cyp2a4 probe of 197 bases in addition to the full-length protected product of 228 bases (Fig. 1B).
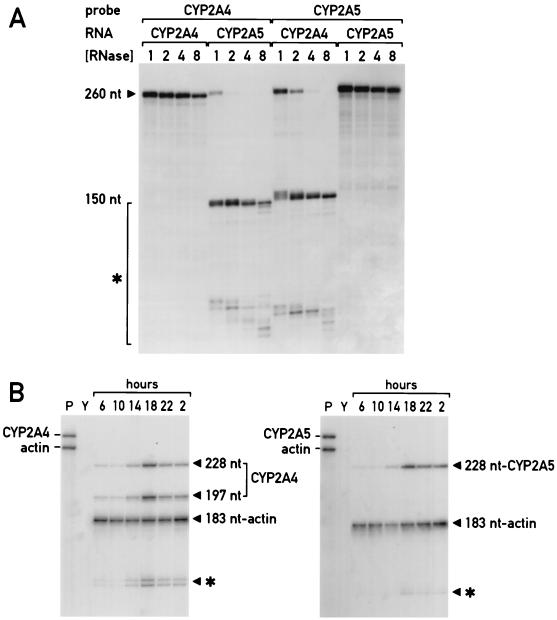
FIG. 1.
Circadian accumulation of CYP2A4 and CYP2A5 mRNA in total mouse liver RNA as detected by gene-specific RNase protection assays. (A) Titration of RNase digestion conditions to distinguish between the CYP2A4 and CYP2A5 mRNAs. Antisense riboprobes for either the mouse CYP2A4 or CYP2A5 genes as indicated were hybridized with sense-strand synthetic RNAs corresponding to either gene (RNA). Resulting hybrids were digested with increasing concentrations of RNase A and RNase T1 ([RNase]) as indicated, with “1” equal to RNase A at 10 μg/ml and RNase T1 at 0.35 U/ml (38). Full-length probe fragments of 260 nt detected by autoradiography of polyacrylamide gels are indicated (260 nt), as are partial degradation products of 150 nt and less, marked by brackets with an asterisk. (B) CYP2A4- and CYP2A5-specific RNase protection assays of mouse liver total RNA isolated around the clock. Ten micrograms of mouse total liver RNA isolated at the hours indicated (6 h [ZT 23] to 2 h [ZT 19]) were analyzed by RNase protection using CYP2A4 (left) or CYP2A5 (right) RNA probes and RNase concentration “8” from panel A. Full-length protected probe fragments for β-actin mRNA (183 nt) and CYP2A5 mRNA (228 nt) are indicated, as are the full-length (228-nt) and partially protected (197-nt) probe fragments corresponding to the CYP2A4 mRNA. Also indicated are partially protected probe fragments of approximately 150 nt and smaller (∗). Lanes P, undigested, full-length probes alone, as indicated; lanes Y, RNase protection on yeast RNA alone.
For use as an internal control, a mouse β-actin cDNA probe was generated by PCR amplification from oligo(dT)-primed double-stranded cDNA derived from 20 h (ZT 13) mouse liver mRNA (10), using primers to positions 622 to 644 (5′-CTGGCCGGGATCCGACAGACTAC-3′) and 823 to 801 (5′-ATGACCTGGCCTGCAGGCAG-CTC-3′). The upstream primer contained mutations (underlined) which inserted a BamHI restriction site at position 630, while the downstream primer contained mutations (underlined) of the β-actin sequence to insert a PstI site at position 812. These restriction sites were used to subclone the PCR fragments into plasmid vectors to generate plasmid pActin, which was sequenced to confirm its identity.
Cyp2a4 and Cyp2a5 genomic promoter DNA fragments were amplified from mouse 129/Ola strain genomic DNA by PCR. Primers used corresponded to positions 1 to 23 of the Cyp2a5 promoter (corresponding to positions −560 to −538 from the major transcription start site [GenBank accession no. M26204]; 5′-GGATCCTCTTCATTGAAAGACTC-3′) and 580 to 551 of this sequence (corresponding to positions +21 to −9 relative to the major transcription start site; 5′-TCAGCAAGCTTGCGGTGGGGTGATAGACAG-3′). hese sequences are identical in both Cyp2a4 and Cyp2a5 genes (22). The downstream PCR primer contained three base mismatches (underlined) to generate a HindIII restriction site downstream of the transcription start site, which was used to construct chloramphenicol acetyltransferase (CAT) reporter fusion genes for cotransfection experiments. The 580-bp PCR product was subcloned into a plasmid vector, and plasmids containing Cyp2a4 or Cyp2a5 gene promoter fragments were identified by restriction mapping and partial sequence analysis.
Mouse liver RNA isolation and RNase protection assays.
At hours of the day indicated, total RNA was isolated from mouse livers by the acid phenol-guanidine thiocyanate method as described previously (24). For each time point, three wild-type and three dbp−/− homozygous mice were sacrificed. Mouse liver samples were analyzed in RNase protection assays using 10 μg of total RNA, either pooled from the three individuals (Fig. 1b) or from each individual separately, for statistical analysis (see Fig. 6).
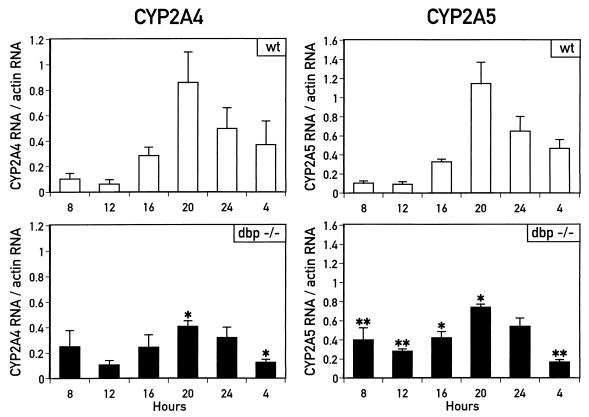
FIG. 6.
Circadian accumulation of CYP2A4 and CYP2A5 mRNAs is impaired in dbp−/− mice. Accumulation of CYP2A4 and CYP2A5 mRNAs in liver total RNA isolated from wild-type (wt) or dbp−/− mice sacrificed at the hours indicated (ZT 0 equal to 7 h; ZT 12 equal to 19 h) was determined by RNase protection assays. mRNA accumulation from each mouse was determined by phosphorimaging analysis (CYP2A5) or densitometry scanning (CYP2A4) and is expressed as the ratio of mRNA accumulation compared to accumulation of β-actin mRNA measured in the same RNA sample. Error bars indicate standard deviations for each group of individuals. Ratios of CYP2A4 and CYP2A5 mRNA to β-actin mRNA were compared between wild-type and dbp−/− mice for each time point by Student’s t test. For CYP2A4 mRNA, ratios were significantly different between wild-type and dbp−/− mice at 20 and 4 h (∗, P < 0.05); for CYP2A5 mRNA, ratios were significantly different between wild-type and dbp−/− mice at all times except 24 h, with confidence values as indicated (∗, P < 0.05; ∗∗, P < 0.01).
Accumulation of mouse CYP2A4, CYP2A5, and β-actin RNAs in mouse liver total RNA was determined by RNase protection assays as described previously (38). The antisense probe for the CYP2A4 mRNA was a 271-nucleotide (nt) riboprobe transcribed from EcoRI-linearized plasmid pSH-CA with T3 RNA polymerase in the presence of [α-32P]UTP. Likewise, the antisense probe for the CYP2A5 mRNA was a 271-nt riboprobe transcribed from EcoRI-linearized plasmid pCH-CA with T3 RNA polymerase in the presence of [α-32P]UTP. To determine the specificity of the probes used in the RNase protection assays, sense pseudo-mRNA molecules were transcribed from the plasmids described above. After linearization of the plasmids with Asp718, T7 RNA polymerase was used to generate a 324-nt nonradioactive RNA molecule containing the sense strand of the CYP2A4 or CYP2A5 mRNA, plus polylinker sequences. Hybridization of either of these 324-nt pseudo-mRNAs with their 271-nt homologous riboprobes and analysis by RNase protection results in a protected probe fragment of 260 nt, slightly longer than the gene-specific probe fragment, as a result of hybridized polylinker sequences. RNase protection analysis using conventional digestion conditions (RNase A, 10 μg/ml; RNase T1, 0.35 U/ml) of the CYP2A4 probe hybridized with the CYP2A5 pseudo-mRNA, as well as the CYP2A5 probe hybridized with the CYP2A4 pseudo-mRNA, were not sufficient to digest the CYP2A4-CYP2A5 hybrids because of their 97% sequence identity (Fig. 1A). Thus, twofold increments in concentrations of RNase A (up to 80 μg/ml) and RNase T1 (up to 2.8 U/ml) were tested. At the highest concentration, each probe gave rise to a 260-nt protected fragment when hybridized to its homologous pseudo-mRNA, while hybridization to the nonhomologous pseudo-mRNA gave rise to smaller (<150-nt) partially protected probe fragments (Fig. 1A).
These same RNase digestion conditions were used for analyzing mouse liver RNA for CYP2A4 or CYP2A5 RNA accumulation. With the CYP2A5 RNA probe, hybridization to the endogenous RNA gives rise to a protected probe fragment of 228 nt. With the CYP2A4 RNA probe, hybridization to the endogenous RNA leads to the production of two protected probe fragments: the full-length protected fragment of 228 nt, and a partially protected probe fragment of 197 nt as a result of the single base mismatch at nt 501 (see above). This partially protected fragment was not seen in the experiments using CYP2A4 RNA probe plus CYP2A4 pseudo-mRNA (Fig. 1A), as both the probe and the pseudo-mRNA contained the mutated residue at position 501. However, as no band of 197 nt was detected in RNase protection experiments with the CYP2A4 RNA probe and the CYP2A5 pseudo-mRNA (Fig. 1A), both the 229- nt and 197-nt bands can be assumed to be derived from the CYP2A4 mRNA.
In all RNase protection experiments, an internal control probe for mouse β-actin was included. This probe was generated by digesting plasmid pActin (see above) with BamHI, and transcribing with T3 RNA polymerase, in the presence of [α-32P]UTP. This results in the production of a 255-nt RNA probe molecule which when hybridized to cellular RNA and analyzed by RNase protection results produces a 183-nt protected fragment (Fig. 1B). This probe was included along with the CYP2A4 or CYP2A5 RNA probes to serve as an internal standard in the same reaction for each sample.
Following RNase digestion, reaction products were separated on polyacrylamide sequencing gels and detected by autoradiography. Radioactive signals were measured directly with a Bio-Rad GS-250 PhosphorImager system or, for circadian CYP2A4 RNA analysis, indirectly by densitometric scanning of autoradiographic films with a Shimadsu CS-9000 densitometric scanner. In experiments comparing RNA accumulation throughout the day, mouse liver β-actin mRNA levels were found to vary slightly, with greater (approximately twofold [7]) accumulation in the evening RNA samples than in the morning samples. To correct for possible variation in the RNA analysis, the levels of CYP2A4 and CYP2A5 RNA accumulation are expressed relative to levels of β-actin RNA accumulation. Statistical analysis of CYP2A4 and CYP2A5 RNA accumulation in liver RNA from wild-type or dbp−/− individual mice was performed by using the one-sided Student t test, with confidence level of P < 0.05 as the limit of significance.
IEF blot detection of CYP2A4 and CYP2A5 proteins in microsomal proteins of wild-type and dbp−/− mouse livers.
Livers were harvested every 4 h over a 24-h period from wild-type and dbp−/− mice kept in constant darkness. Livers were perfused with phosphate-buffered saline, frozen in dry ice, and stored at −80°C. Microsomes were prepared as described by van der Hoeven and Coon (41) and stored in 100-μl fractions at −80°C. Protein concentration was determined with the bicinchoninic assay reagent from Pierce Chemical Co. (Rockford, Ill.). The cytochrome P450 and cytochrome b5 concentrations were determined by differential spectrophotometry on a Uvikon 933 spectrophotometer. Sodium dithionite was used as reducing agent. The total P450 amount was estimated as ɛ450–490 = 91 mM−1 cm−1 for the reduced-CO minus reduced spectrum. In the case of cytochrome b5, we used ɛ424–409 = 185 mM−1 cm−1 for the reduced minus oxidized spectrum.
CYP2A4 and CYP2A5 amounts were measured on IEF blots as described previously (11). Protein IEF was performed in acrylamide vertical slab gels by the nonequilibrium pH gradient electrophoresis technique (33). The gels contained 1.6% Ampholines pH 7 to 9 and 0.4% Ampholines pH 3 to 10. Samples of microsomes (50 mg of protein) and of P450 2A4 and 2A5 standards were loaded at the anodic end of the focusing gel. After the run, the proteins were blotted on a polyvinylidene difluoride membrane (Immobilon P; Millipore, Bedford, Mass.), and the cytochrome P450 isozymes were visualized by double-antibody labeling in the same way as for a classical Western blotting. The first antibody was polyclonal anti-mouse P450tu, which cross-reacted both with 2A4 and 2A5 isozymes (11). Following reaction with the peroxidase-conjugated secondary antibody, the P450 2A isozymes were stained with diaminobenzidine. The intensity of the spots versus the concentration of cytochrome P450 2A was linear in the range of 20 to 200 ng. Quantitation of the spots by scanning the transfer membranes with an Apple scanner was followed by image intensity analysis using NIH Image software and statistical analysis using the one-sided Student t test, with the same limit of significance as specified above (P < 0.05).
DNase I footprinting on Cyp2a4, Cyp2a5, and CYP7 promoters with recombinant DBP.
Genomic DNA probes used for DBP binding studies were the rat CYP7 promoter fragment from −340 to −30 (16) and the mouse Cyp2a4 and Cyp2a5 promoter fragments from +10 to −560 (22). The CYP7 probe was end labeled at the HindIII site at −340 by filling in with [α-32P]dATP and Klenow enzyme (16), while the Cyp2a4 and Cyp2a5 probes were end labeled near the +10 end with [α-32P]dATP and Klenow enzyme, using a restriction site in the vector polylinker.
DNA binding and DNase I digestions were performed as described previously (16), using approximately 10 to 15 fmol of probe per reaction and serial dilutions of heparin-agarose-purified recombinant DBP (5). The concentration of functional DBP dimers had been determined previously by Scatchard analysis (5), and dilutions used for footprinting corresponded to final DBP dimer concentrations of 0.1 μM to 0.78 nM. The dissociation constant (KD) of DBP for a DNA binding site was determined as the concentration of total DBP protein ([proteintotal]) which demonstrates half-protection of that site from DNase I digestion. (This is because KD = [proteinfree] × [DNAfree]/[protein-DNA], where [proteinfree] equals the concentration of protein not bound to DNA, [DNAfree] equals the concentration of DNA not bound to protein, and [protein-DNA] equals the concentration of protein-DNA complexes. However, though [proteintotal] = [proteinfree] + [protein-DNA], we assume [proteintotal] ≅ [proteinfree], because [proteintotal] ≫ [DNAtotal]; at the protein concentration providing half-protection of the DNA, [DNAfree] ≅ [protein-DNA]; thus, KD ≅ [proteintotal] at half-protection of the DNA.) To permit direct comparison of KD values, reactions with the CYP7 and Cyp2a4 promoters, or the Cyp2a4 and Cyp2a5 promoters, were conducted in parallel using the same dilutions of recombinant DBP protein.
Transfection studies of Cyp2a4 and Cyp2a5 promoter reporter gene fusion constructions.
The Cyp2a4 and Cyp2a5 promoter fragments from −560 to +10 were cloned upstream of a bacterial CAT reporter gene (pCyp2a4-560CAT or pCyp2a5-560CAT [9]). Deletion mutants for both the Cyp2a4 and Cyp2a5 promoters were generated by PCR-based insertion of an XhoI restriction site at positions −275, −219, and −74 and subcloned into the CAT reporter plasmid. These constructs (pCyp2a4-275CAT, pCyp2a4-219CAT, pCyp2a4-74CAT, pCyp2a5-275CAT, pCyp2a5-219CAT, and pCyp2a5-74CAT) were designed to contain three, one, and none, respectively, of the recombinant DBP binding sites detected by DNase I footprint analysis (see Fig. 4).
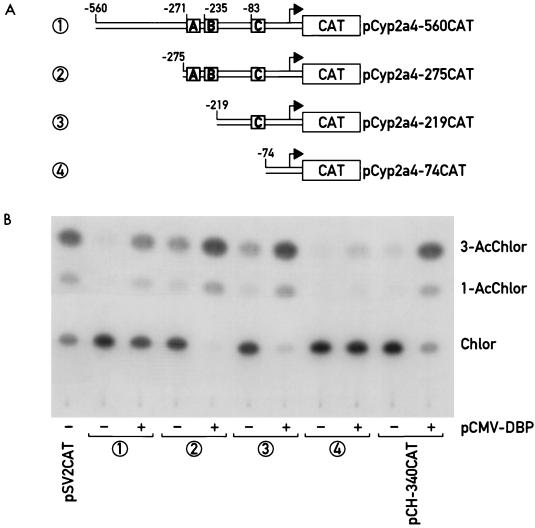
FIG. 4.
Efficient transactivation of the Cyp2a4 promoter by DBP requires sequences between positions −271 and −74. (A) CAT reporter genes (boxes labeled “CAT”) were fused to DNA elements containing various amounts of the Cyp2a4 promoter (from position +10 to positions between −560 and −74), to generate the plasmids indicated at right. Also indicated are the positions of recombinant DBP binding sites A, B, and C (shaded boxes) detected by DNase I footprinting experiments in each of the promoter elements. Arrow, major Cyp2a4 transcription start site (position +1). (B) CAT activity in HepG2 cell extracts transfected with the Cyp2a4 promoter CAT fusion plasmids. HepG2 cells were transfected with the Cyp2a4 promoter CAT fusion plasmid indicated by the circled number, corresponding to the same number in panel A, with (+) or without (−) the DBP expression vector pCMV-DBP (31). Plasmid pCH-340CAT, containing CYP7 promoter sequences to position −340, was also transfected with or without pCMV-DBP as a positive control for promoter activation by DBP (16). Plasmid pSV2CAT, containing the simian virus 40 late promoter (9), was included as a positive control for transfection efficiency. CAT activity in transfected cell lysates was determined by TLC to detect conversion of nonacetylated [14C]chloramphenicol (Chlor) to 1- and 3-acetyl-[14C]chloramphenicol (1-AcChlor and 3-AcChlor).
These promoter-reporter gene fusion plasmids were used in cotransfection experiments in human hepatoma HepG2 cells, with or without 5 μg of a DBP expression vector, pCMV-DBP (31). HepG2 cells were transfected by calcium phosphate precipitation as described previously (16), and CAT activity was measured by [14C]chloramphenicol conversion, as monitored by thin-layer chromatography (TLC) (9), and quantitated by using a Berthold Tracemaster 20 TLC linear analyzer and Chroma software. As a positive control for transfection, the vector containing the CAT gene driven by the simian virus 40 promoter (pSV2CAT [9]) was included in each series of transfections. In addition, plasmid pCH-340CAT was used as a positive control for DBP activation. This plasmid contains sequences of the rat CYP7 promoter to position −340 fused to the CAT gene. Cotransfection of this plasmid with the DBP expression vector leads to an increase in reporter gene activity of 7- to 10-fold (16). Experiments shown for all plasmid constructions are representative of results obtained reproducibly in at least three independent transfections.
EMSA with mouse liver nuclear proteins.
Nuclei from livers of wild-type or dbp−/− mice were prepared at different times of the day by homogenization and centrifugation through 2 M sucrose cushions (21). Soluble extracts were prepared from these purified nuclei by NaCl–urea–NP-40 extraction as described previously (24). Nuclear equivalent amounts of extracts (approximately 10 μg) were used for electrophoretic mobility shift analysis (EMSA) as previously described (24) except that salmon sperm DNA (0.4 μg/ml) was included as a nonspecific competitor. The probe used was an double-stranded DNA oligonucleotide containing the site C characterized by DNase I footprinting experiments (see above), corresponding to bases −82 to −57 from the major transcription start site of the Cyp2a4 promoter (22): 5′-GGTGAAATAGTTGCATAATCAAGACC-3′ 3′-CCACTTTATCAACGTATTAGTTCTGG-5′
The double-stranded oligonucleotide was radiolabeled near the 3′ termini, using [α-32P]dCTP and the Klenow enzyme. Following incubation on ice, probe-nuclear factor complexes were separated on a nondenaturing 0.25× Tris-borate-EDTA (TBE)–5% acrylamide gel and detected by autoradiography.
RESULTS
The CYP2A4 mRNA displays circadian accumulation in mouse liver.
We have reported that the mRNA encoding CYP2A5, a member of the cytochrome P450 gene superfamily, demonstrates circadian accumulation in mouse liver, with peak levels in the evening approximately 6- to 10-fold higher than in the morning (15) (see below). Another member of the cytochrome P450 gene superfamily, Cyp2a4, is over 98% identical to Cyp2a5 in both cDNA and protein sequences (22). To determine whether CYP2A4 mRNA also displayed a circadian accumulation pattern, RNase protection assay conditions using gene-specific probes which were stringent enough to distinguish between the mRNAs produced from these two highly related genes were developed. Probes specific for both mRNAs were generated by reverse transcription-PCR of the region from nt 471 to 698 of the Cyp2a4 and Cyp2a5 cDNAs, which contains 7 of 228 bases mismatched between the two cDNA sequences (40). These were used to generate uniformly labeled antisense RNA probes, as well as unlabeled, sense-strand CYP2A4- and CYP2A5-specific pseudo-mRNAs. Standard RNase protection assay conditions using the specific probes hybridized to either the CYP2A4 or CYP2A5 pseudo-mRNA were not sufficiently stringent to distinguish between the two synthetic RNAs (Fig. 1A). However, increasing the concentration of RNases A and T1 eightfold generated full-length protected bands for the homologous hybrids while digesting the CYP2A4-CYP2A5 cross-hybrids into shorter protected fragments (Fig. 1A). Thus, these conditions established with the synthetic RNAs were used to determine the accumulation of each mRNA in total mouse liver RNA throughout the day.
Total liver RNA isolated at 4-h intervals over 24 h from adult male mice was analyzed with both CYP2A4- and CYP2A5-specific probes to determine the accumulation of their mRNAs (Fig. 1B). In the experiment shown, two bands specific for the CYP2A4 mRNA are detected: a full-length protected fragment of 228 nt, and a partially protected fragment of 197 nt. These are distinct from the smaller, partially protected probe fragments attributed to the CYP2A5 mRNA (Fig. 1B). The CYP2A4 partially protected fragment arises due to RNase cleavage of a single-base mismatch in the probe-mRNA hybrid introduced by the PCR (see Materials and Methods). Nonetheless, the accumulation of CYP2A4-specific probe fragments demonstrates that as reported earlier for CYP2A5 mRNA (15), CYP2A4 mRNA displays circadian accumulation. Similar to CYP2A5 mRNA, peak accumulation of CYP2A4 mRNA is attained at approximately 22 h (ZT 15), with an amplitude of approximately 10-fold between peak and trough levels (Fig. 1B). Thus, CYP2A4 mRNA is a novel circadian expression mRNA, with kinetics of expression similar to those of CYP2A5 mRNA. Circadian CYP2A4 and CYP2A5 mRNA accumulation does not appear to be sex specific, as the accumulation of both mRNAs is greater in liver RNA from female BALB/c mice sacrificed at 20 h than in RNA from mice sacrificed at 8 h (14).
Circadian accumulation of CYP2A4 and CYP2A5 proteins in mouse liver microsomes.
To examine accumulation of the CYP2A4 and CYP2A5 proteins throughout the day, IEF blot analysis was used to measure CYP2A4 and CYP2A5 protein accumulation in liver microsomes isolated from mice sacrificed at different times during the day. IEF blot analysis (3) is required to distinguish between CYP2A4 and CYP2A5 proteins, as they migrate indistinguishably on sodium dodecyl sulfate-polyacrylamide gels, and antiserum directed against either protein recognizes both species equally well in conventional Western blotting. However, these proteins can be distinguished in nonfractionated rodent liver microsomes by taking advantage of the difference in isoelectric point for the two proteins (9.91 for CYP2A4 and 10.01 for CYP2A5). Samples are first separated on an IEF gel and then electrotransferred to a membrane which is treated with an antiserum recognizing both CYP2A4 and CYP2A5 proteins (11). As shown in Fig. 2A, species corresponding to CYP2A4 and CYP2A5 can be distinguished in nonfractionated mouse liver microsomes, here in samples harvested at 2 h and at 6 h, by comparison with purified CYP2A4 and CYP2A5. An additional species with an apparent pI slightly more basic than that of the CYP2A5 is detected in liver microsomes (Fig. 2A). Whether this species, which does not correspond to a species in either the purified CYP2A4 or CYP2A5 protein, is a related gene product or an unrelated antigen with which the antibody cross-reacts is unknown. However, its presence renders the quantitation of CYP2A5 accumulation by gel scanning more difficult.
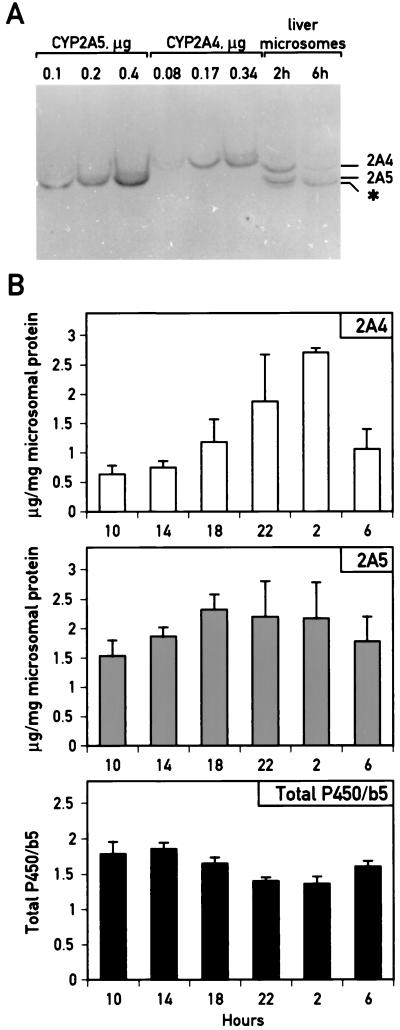
FIG. 2.
Circadian accumulation of CYP2A4 and CYP2A5 proteins in mouse liver microsomes. (A) IEF blot analysis of unfractionated liver microsomes from mice sacrificed at 2 h (ZT 19) and 6 h (ZT 23) to detect CYP2A4 and CYP2A5. Microsomal proteins isolated from mouse livers harvested at 2 or 6 h were separated on an IEF gel (top, acidic; bottom, basic), along with purified CYP2A4 and CYP2A5 as standards, and transferred to a hybridization membrane which was probed with an antiserum cross-reacting with both CYP2A4 and CYP2A5. The species in the unfractionated microsomes corresponding to these proteins are identified by comparison with the purified standards. In addition, a cross-reacting band migrating below CYP2A5 whose accumulation does not vary significantly throughout the day (2) is indicated with an asterisk. (B) Quantitation of CYP2A4 and CYP2A5 protein accumulation, and the ratio of total cytochromes P450 to cytochrome b5 in unfractionated mouse liver microsomes throughout the day (ZT 0 equal to 7 h; ZT 12 equal to 19 h). Values represent means ± standard deviation for two to four individuals per time point. Statistically significant differences in CYP2A5 accumulation were found between samples at 18 and 10 h and at 18 and 14 h (P < 0.05 for each). Statistically significant differences were found in CYP2A4 accumulation between 2 h and all other samples except 22 h (for instance, between 2 and 10 h, P < 0.00002). For total P450/b5 ratios, statistically significant differences were found between 14 and 22 h, between 10 and 22 h (P < 0.01 for both comparisons), between 14 and 2 h, and between 18 and 2 h (P < 0.05 for both comparisons).
To determine CYP2A4 and CYP2A5 protein accumulation throughout the day, liver microsomes were prepared every 4 h over a 24-h period from male mice kept in constant dark. Concentrations of total protein, cytochrome b5 and total cytochrome P450 were determined as indicated in Materials and Methods. The concentration of cytochrome b5 in liver microsomes was used as an internal control for isolation of microsomal proteins. In contrast to cytochrome P450, cytochrome b5 is not known to be inducible in animal liver by drug or hormonal treatment. In addition, this hemoprotein is less sensitive than cytochrome P450 to denaturation upon storage. We observed that its concentration did not vary significantly throughout the day (within 10% for all values [2]). However, total cytochrome P450 concentrations demonstrated slight yet significant circadian variations of approximately 1.3-fold (Fig. 2B; P < 0.01 for value at 22 h compared to value at 10 or 14 h; P < 0.05 for value at 2 h compared to value at 10 or 14 h). Peak cytochrome P450 concentrations were reached between 10 and 14 h, consistent with earlier reports on daily variations in total cytochrome P450 accumulation (13).
When equivalent amounts of total microsomal proteins were analyzed for CYP2A4 and CYP2A5 protein accumulation by IEF blotting, both proteins were found to demonstrate higher accumulation in the evening than in the morning (Fig. 2B). For CYP2A4, accumulation was greatest at 2 h, with a difference between peak and trough values of approximately fourfold (2 h versus 10 h). In contrast, CYP2A5 protein accumulation was maximal at 18 h, with a difference of only 1.5-fold. This less pronounced daily variation in CYP2A5 protein accumulation may reflect the difficulty in accurately measuring CYP2A5 protein accumulation due to the presence of the additional cross-reacting species (Fig. 2A); the abundance of this species does not vary throughout the day (2). Nonetheless, accumulation of CYP2A5 at 18 h differs significantly from CYP2A5 accumulation at either 10 or 14 h (P < 0.05 for either comparison). Peak accumulation of both CYP2A4 and CYP2A5 is reached in the subjective evening, distinct from the peak in total cytochromes P450 (Fig. 2B). This finding underscores the specific daily variations in CYP2A4 and CYP2A5 protein accumulation and argues against their being solely a reflection of global cytochrome P450 variations. Assays for the steroid 15α-hydroxylase and coumarin 7-hydroxylase activities, which are specific for the 2A4 and 2A5 isozymes, respectively, were consistent with the daily variations in CYP2A4 and CYP2A5 apoprotein as observed in IEF blots (2).
Detection of functional DBP binding sites in the Cyp2a4 and Cyp2a5 gene promoters.
In the rodent liver, each member of the PAR family of DNA-binding transcription factors, DBP, HLF, and TEF, demonstrates a circadian rhythm in its expression (17). These transcription factors therefore may contribute to the regulation of circadian expression genes in the liver. This has been proposed for another cytochrome P450 superfamily gene, the rat CYP7 gene encoding cholesterol 7α-hydroxylase. Biochemical analyses suggested that the circadian transcription of this gene might be regulated by DBP, as well as by other PAR bZip protein family members, through a high-affinity PAR bZip binding site in its promoter (16, 20).
To determine whether the Cyp2a4 and Cyp2a5 genes might also be regulated by the PAR family transcription factors, the promoter sequences of the two genes from positions +1 (the major transcription start site) to −560 (22) were examined for potential PAR factor binding motifs. These sequences are greater than 98% identical between the two genes, with only seven mismatches in the 560 bp. Three sequence motifs with at least 7-9-base identity to the consensus DNA binding sequence 5′-RTTAYGTAAY-3′ (R represents A or G; Y represents C or T) recognized by all PAR family members (5) were found in the promoters of both genes. The three sites were found within 300 bp upstream of the genes’ transcription start sites, commencing at −271 (motif A; 7 of 10 bases identical), −235 (B; 7 of 10 identical), and −83 (C; 9 of 10 identical), and were completely conserved between the two genes.
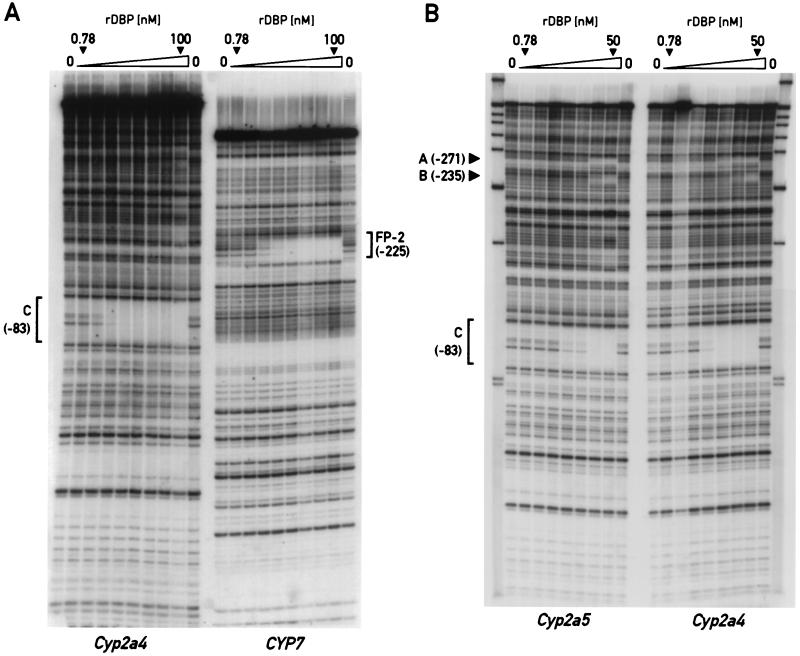
The ability of DBP to interact with these sites on the Cyp2a4 and Cyp2a5 promoters was determined by DNase I footprinting experiments using recombinant DBP and radiolabeled fragments of the Cyp2a4 and Cyp2a5 promoters from −560 to +10 relative to the transcription start site. In addition, DNase I footprinting was performed using the Cyp2a4 promoter fragment and a fragment of the rat CYP7 gene containing a previously characterized binding site for PAR bZip proteins (FP-2 [16]). Using known concentrations of recombinant DBP in excess over the probe DNA, the KD for the interaction of DBP with a particular binding site on the DNA fragment can be estimated as the protein concentration giving half-protection of the binding site (see Materials and Methods). As shown in Fig. 3A, DNase I footprinting reactions show half-protection of the site C on the Cyp2a4 promoter at a recombinant DBP dimer concentration of approximately 2 nM. Half-protection of the FP-2 site on the CYP7 promoter fragment with the same serial dilutions of recombinant DBP was achieved at approximately 3 nM DBP dimer. Thus, the KD for DBP binding at the Cyp2a4 promoter site C (≅2 nM) is at least as low as that for DBP binding to the CYP7 site FP-2 (≅3 nM). The FP-2 site has been demonstrated in cotransfection studies to be required for DBP-mediated transcription activation of the CYP7 promoter (16).
FIG. 3.
DNase I footprinting analysis of binding by recombinant DBP (rDBP) protein on the Cyp2a4, Cyp2a5, and CYP7 promoters. (A) Twofold-increasing concentrations of recombinant DBP (lowest and highest DBP dimer concentrations used are indicated) were used in DNase I footprinting assays with radiolabeled fragments of the mouse Cyp2a4 and rat CYP7 promoters. The major sites protected by DBP on these promoters are indicated: site C, between positions −83 and −74 on the Cyp2a4 promoter, and site FP-2, centered on position −225 on the CYP7 promoter. Half-protection of site C from DNase I digestion is estimated at between 1.56 and 3.12 nM DBP dimer, while half-protection of the FP-2 site is estimated at approximately 3 nM DBP dimer. (B) DNase I protection patterns of recombinant DBP on the mouse Cyp2a4 and Cyp2a5 promoters was determined by using twofold-increasing recombinant DBP dimer concentrations (lowest and highest recombinant DBP dimer concentrations used are indicated). Protection of each promoter from DNase I digestion by recombinant DBP is indicated at putative DBP binding sites A (between positions −271 and −262), B (between positions −235 and −226), and C (as in panel A). Half-protection at sites A and B on both the Cyp2a4 and Cyp2a5 promoters is observed at recombinant DBP dimer concentrations two to four times greater than that at site C.
A separate DNase I footprinting reaction was performed to visualize DBP binding at the other putative DBP binding sites, A and B, in addition to site C, on promoter fragments from both the Cyp2a4 and Cyp2a5 genes (Fig. 3B). In this experiment, half-protection of site C on both promoter fragments was achieved at a higher DBP dimer concentration than in the earlier experiment (approximately 6 nM), perhaps owing to a loss of DBP binding activity by freeze-thawing of protein stocks. However, the relative binding of DBP to sites A, B, and C on the Cyp2a4 and Cyp2a5 promoter fragments can still be evaluated. Half-protection of sites A and B was achieved at a higher DBP dimer concentration (12 to 25 nM) than that of site C (approximately 6 nM [Fig. 3B]). This is consistent with the greater divergence of the sequences of sites A and B from the DBP consensus DNA binding sequence (7 of 10 identical) than site C (9 of 10 identical).
The ability of DBP to influence expression of the Cyp2a4 and Cyp2a5 promoters was investigated in transient transfection assays. Chimeric reporter genes were constructed in which the promoters of the Cyp2a4 and Cyp2a5 genes were fused downstream of their cap sites to the bacterial CAT gene. Promoter constructs were generated so as to contain sequences from position +10 relative to the major transcription start site of either gene to −560 (pCyp2a4-560CAT and pCyp2a5-560CAT), −275 (pCyp2a4-275CAT and pCyp2a5-275CAT), −219 (pCyp2a4-219CAT and pCyp2a5-96CAT), and −74 (pCyp2a4-74CAT and pCyp2a5-74CAT). The deletion plasmids were designed such that the −560CAT and −275CAT constructs would both contain all three DBP binding sites (A, B, and C), −219CAT would contain only the promoter-proximal DBP binding site C, and −74CAT would contain none of the DBP binding sites detected by DNase I footprint analysis (Fig. 3 and 4). Transient cotransfection of these reporter constructs into the human hepatoma cell line HepG2 was performed either with or without an expression vector for DBP under the control of the cytomegalovirus major promoter. As a control for DBP-mediated transcription activation, a CAT reporter gene construct (CH-340CAT) which contains the promoter fragment of the rat CYP7 gene used in the DNase I footprinting experiments above was also cotransfected with or without the DBP expression vector. Previously, DBP was found to increase expression of this reporter gene construct in HepG2 cells (16).
When cotransfected with the DBP expression vector, expression of all of the Cyp2a4 promoter-CAT reporter gene constructs was increased over basal levels (Fig. 4). For CAT gene reporter plasmids containing Cyp2a4 promoter sequences to −560, −275, and −219, DBP induction of promoter expression was comparable to that of the CYP7 promoter-CAT reporter gene; for example, expression of the pCyp2a4-560CAT construct was induced more than sixfold (Fig. 4). Expression of reporter plasmid pCyp2a4-219CAT, which contains only site C of the DBP binding sites mapped by DNase I footprinting, was also efficiently activated by DBP. However, removal of Cyp2a4 promoter sequences to −74 severely reduced activation of reporter gene expression by DBP, though a slight (approximately twofold) activation was retained. Thus, the deletion of the DBP binding sites, and particularly site C, detected by DNase I footprinting experiments eliminates the majority of Cyp2a4 promoter activation by DBP in cotransfection assays. Nearly identical results were obtained in assays using the equivalent Cyp2a5 promoter constructs (14). In addition, the Cyp2a4 promoter construct pCyp2a4-560CAT was activated efficiently by another PAR transcription factor family member, TEF (14).
Circadian Cyp2a4 and Cyp2a5 gene expression is impaired in dbp−/− mice.
While the DNase I footprinting and cotransfection studies suggested that DBP may be a regulator of circadian expression of the Cyp2a4 and Cyp2a5 genes, a genetic approach was taken to determine whether DBP plays such a role in vivo, using a dbp−/− mouse strain generated by gene targeting techniques (24). Homozygous dbp−/− mice of this strain are viable and fertile but demonstrate a significant alteration in their endogenous circadian rhythms, with an average circadian period length approximately 30 min shorter than that of their wild-type counterparts in wheel-running assays (24).
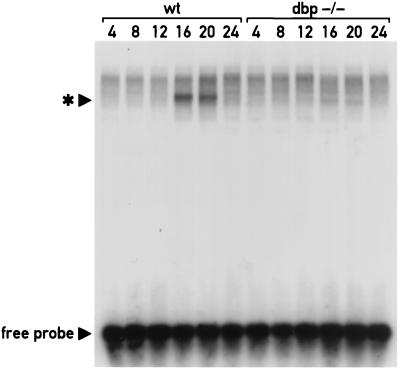
Sequences of the Cyp2a4 and Cyp2a5 promoters including the DBP binding site C were found to be important for the activation of these promoters by DBP in cotransfection studies. To determine whether the elimination of DBP expression in the dbp−/− mice led to a change in the population of liver nuclear factors capable of interacting with this site, EMSAs were performed with a radiolabeled oligonucleotide encompassing site C. This oligonucleotide probe was incubated with liver nuclear extracts isolated from wild-type or homozygous dbp−/− mice at different hours around the clock, in the presence of nonspecific competitor DNA. As shown in Fig. 5, the major complex detected with the site C oligonucleotide in wild-type mice demonstrated robust circadian expression, with the greatest accumulation detected in samples harvested at 16 and 20 h. This is consistent with the peak in accumulation of DBP and TEF proteins in mouse liver nuclei (23, 24) and also with the expected peak in circadian transcription rates of Cyp2a4 and Cyp2a5, as zenith levels of CYP2A4 and CYP2A5 mRNAs are observed at approximately 20 h (Fig. 1B). The site C oligonucleotide binding activity is absent in liver nuclear extracts prepared from dbp−/− mice (Fig. 5), indicating that the detection of this activity requires a functional dbp gene.
FIG. 5.
A circadian binding activity with a Cyp2a4 site C oligonucleotide in mouse liver nuclear extracts is reduced in dbp−/− mice. Approximately 10- μg aliquots of nuclear extracts from livers of wild-type (wt) or dbp−/− mice sacrificed at the hours indicated (ZT 0 equal to 7 h; ZT 12 equal to 19 h) were incubated with a 32P-radiolabeled double-stranded oligonucleotide corresponding to the Cyp2a4 promoter sequence from −82 to −57. These sequences comprise the DBP binding site C identified by DNase I footprinting studies. Complexes of the probe with nuclear factors were separated from free probe by electrophoresis on a 0.25× TBE-polyacrylamide gel and detected by autoradiography. Position of the free probe is indicated, as is the position of the major binding activity detected in liver nuclear extracts of wild-type mice (∗). This probe-nuclear factor complex is most abundant in samples harvested at 16 and 20 h but is nearly undetectable in samples harvested at other times during the day. The detection of this complex is greatly reduced in dbp−/− mice.
To determine whether the loss of DBP expression influences the circadian expression of the CYP2A4 and CYP2A5 mRNAs, total RNA was isolated from wild-type and dbp−/− mouse livers harvested every 4 h over 24 h. These RNAs were then examined by RNase protection assays for accumulation of CYP2A4 and CYP2A5 mRNAs. Liver RNA from each mouse was analyzed individually, and accumulation of the CYP2A4 and CYP2A5 mRNAs was determined by quantitation of the corresponding signals (see Materials and Methods). These mRNA signals were then corrected by using β-actin mRNA as an internal standard. Preliminary experiments demonstrated that β-actin mRNA accumulation remains largely constant throughout the day, though occasionally higher concentrations can be detected in RNA prepared in the evening (Fig. 1B). No significant difference was detected in β-actin mRNA between wild-type and dbp−/− individuals (7).
As shown in Fig. 6, circadian accumulation of the CYP2A4 and CYP2A5 mRNAs is significantly altered in dbp−/− mice. For both genes, the amplitude in circadian variation of their mRNA accumulation was decreased, from more than 12-fold in the wild type mice to approximately 4-fold in the dbp−/− mice. For the CYP2A5 mRNA, accumulation was significantly lower in the knockout mice than in the wild-type mice at 20 h (P < 0.05) and 4 h (P < 0.01) and significantly higher in the knockout mice than in the wild-type mice at 8, 12, and 16 h (P < 0.01 for 8 and 12 h; P < 0.05 for 16 h). CYP2A4 mRNA accumulation similarly differed between the knockout mice relative to the wild-type mice (Fig. 6). However, the standard deviations from the mean CYP2A4 mRNA accumulation levels were greater than those from the mean CYP2A5 mRNA accumulation levels. Nonetheless, significant differences were found between wild-type and dbp−/− mice in CYP2A4 mRNA accumulation at 20 h (P < 0.025) and at 4 h (P < 0.05).
Thus, RNase protection analyses of dbp−/− and wild-type mice indicate that DBP contributes to the circadian expression pattern of the CYP2A4 and CYP2A5 mRNAs. However, while circadian expression is significantly dampened, it is not completely eliminated in the absence of DBP. This may suggest that other circadian factors, perhaps the other PAR family transcription factors TEF and HLF, also contribute to circadian mRNA accumulation from the Cyp2a4 and Cyp2a5 genes.
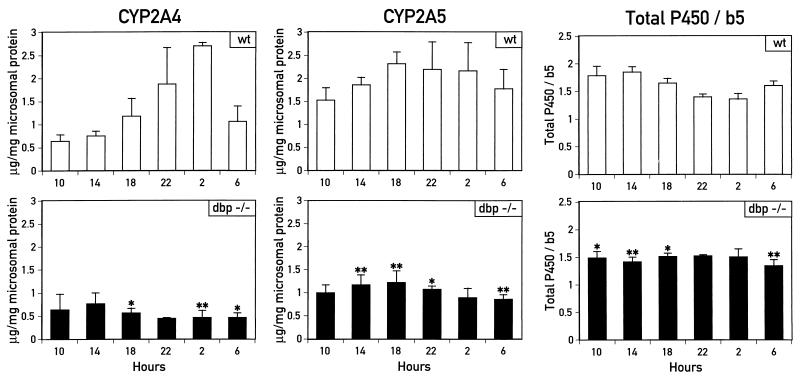
To determine whether the circadian accumulation of the CYP2A4 and CYP2A5 proteins detected in Fig. 2 was altered in dbp−/− mice, IEF blot analysis of liver microsomes from dbp−/− mice was performed as done previously for wild-type mouse liver microsomes (Fig. 2). Livers were harvested from dbp−/− mice at the same time as for wild-type mice (Fig. 2) and analyzed for content of non-P450 cytochrome b5, total cytochromes P450, and proteins CYP2A4 and CYP2A5. The results for the dbp−/− mice, along with the data for wild-type mice from Fig. 2, are shown in graphic form in Fig. 7.
FIG. 7.
Circadian accumulation of CYP2A4 and CYP2A5 proteins and total cytochromes P450 is also impaired in dbp−/− mice. Accumulation of CYP2A4 and CYP2A5 proteins and ratios of total cytochromes P450 to cytochrome b5 were measured in unfractionated liver microsomes isolated from dbp−/− mice sacrificed at the times indicated (ZT 0 equal to 7 h; ZT 12 equal to 19 h). Values for each sample are presented as the average of two to four individuals per time point ± standard deviation. Data for wild-type (wt) mice are taken from Fig. 2. Results were analyzed for statistical significance by Student’s t test. Values were found to be significantly different between wild-type and dbp−/− mice for CYP2A4 and CYP2A5 accumulation at times indicated (∗, P < 0.05; ∗∗, P < 0.01).
While levels of cytochrome b5 protein are similar between wild-type and knockout animals (2), the slight daily variation in total cytochrome P450 levels in liver microsomes from wild-type mice (Fig. 2 and 7) is no longer seen in the dbp−/− mice (Fig. 7). Thus, total cytochrome P450 levels are significantly lower in the dbp−/− mice than in wild-type mice in samples harvested at 6 h (P < 0.01), 10 h (P < 0.05), and 14 and 18 h (P < 0.01 for both).
More strikingly, CYP2A4 and CYP2A5 protein accumulation in the dbp−/− mice is significantly altered, with a near ablation of the circadian expression pattern detected in the wild-type mice (Fig. 7). Confidence values for significant differences in protein accumulation within samples from wild-type and dbp−/− mice are as follows: for CYP2A4, P < 0.05 at 6 and 18 h and P < 0.01 at 2 h; for CYP2A5, P < 0.05 at 10 h and P < 0.01 at 6, 14, and 18 h. Changes in circadian protein accumulation patterns are less striking in the case of CYP2A5, whose circadian accumulation was less robust than that of CYP2A4. However, as mentioned above, this may be due in part to the difficulty in measuring CYP2A5 protein accumulation distinct from that of the cross-reacting species migrating slightly faster than CYP2A5 on the IEF gels (Fig. 2). For CYP2A4, the more than fourfold amplitude in circadian expression is reduced in the dbp−/− mice to nearly constitutive expression at or slightly below minimal daily levels detected in wild-type mice. Thus, deletion of the dbp gene strongly impairs circadian expression of the CYP2A4 protein and, perhaps to a lesser degree, that of the CYP2A5 protein. Taken with the results for total P450 levels in dbp−/− mice, these results suggest that the Cyp2a4 and Cyp2a5 genes may represent a subclass of circadian cytochrome P450 genes regulated by DBP.
DISCUSSION
Previously, the CYP2A5 mRNA, encoding coumarin 7-hydroxylase, was found to display circadian accumulation (15). In this work, we report that mRNA accumulation from the highly related gene Cyp2a4, encoding steroid 15α-hydroxylase also displays circadian expression, as does the accumulation of proteins encoded by both of these genes. Furthermore, biochemical and genetic experiments indicate that the circadian expression of both of these genes is regulated by the PAR family transcription factor DBP. These results underscore the breadth with which circadian rhythms can influence physiology: in the liver alone, genes whose products are implicated in the metabolism of cholesterol, amino acids, xenobiotics, and androgens all display circadian expression patterns (15, 28, 34) (see above). Interestingly, the Cyp2a4, Cyp2a5, and CYP7 genes, encoding coumarin 7-hydroxylase, steroid hydroxylase, and cholesterol 7α-hydroxylase, respectively, are all members of the cytochrome P450 gene superfamily, whose gene products catalyze a wide range of mono-oxygen transferase reactions. Total cytochrome P450 protein levels vary slightly throughout the day in the liver (13) (see above), with highest levels near midday, different from peak accumulation times of the coumarin, steroid, and cholesterol hydroxylase proteins (Fig. 2 and reference 32). These daily variations in cytochrome P450 protein accumulation may reflect a cellular mechanism to restrict expression of some cytochromes P450, as their mono-oxygen transferase activity can produce damaging oxygen free radicals (1). Circadian regulation would thus restrict peak expression to the time of day when the function of the cytochrome P450 is most required. However, this does not appear to be true for all cytochromes P450: in rat liver, mRNA accumulation for the pentobarbital-inducible cytochrome P450 gene CYP2C6 remains constitutive throughout the day (16). Thus, slight daily variations in total cytochrome P450 accumulation may represent a subset of circadian cytochromes P450, such as Cyp2a4 and CYP7, superimposed on the bulk of constitutive cytochromes P450, such as CYP2C6. As total cytochrome P450 accumulation in livers of dbp−/− mice does not vary significantly throughout the day (Fig. 7), the dbp gene would appear to be an important regulator of circadian expression of this subset of circadian cytochromes P450.
Circadian expression of the Cyp2a4 gene, encoding steroid 15α-hydroxylase, is consistent with previous studies on daily fluctuations in serum levels of androgens, the presumed substrates of this enzyme. While studies on testosterone levels in male rats found a multiphasic circadian fluctuation in serum testosterone concentrations, testosterone levels consistently fell late in the subjective night, regardless of the lighting regimen or sampling method used (29). This would be consistent with the higher nighttime levels of hepatic CYP2A4 protein (Fig. 2) and steroid 15α-hydroxylase activity (2) detected in this study, leading to increased metabolism and clearance of testosterone from the serum. However, this may be only one component of a highly complex regulatory system, possibly involving additional endocrine, neural, and even behavioral inputs.
Despite considerable evidence for circadian variation of serum androgen concentrations, little is known about the physiological impact of such variations. Estrogens have been proposed to directly influence the circadian expression of other hormones, such as luteinizing hormone, and thus the female menstrual cycle (6, 30). Variations in testosterone levels may influence mating behavior, which normally occurs at the midpoint of the dark period, though the timing of serum testosterone peak levels in isolated male rats does not strictly coincide with mating time (29). An additional role for circadian androgen expression may be linked to the role of androgens as tumor promoters: the incidence of liver tumorigenesis is lower in female than in male mice, apparently as the result of a negative effect on hepatocarcinogenesis by ovarian hormones (36). Chronic testosterone administration increases tumor susceptibility in females, and male tumor susceptibility is reduced to female levels in a mutant mouse strain lacking high-affinity androgen receptors (12). Thus, as for circadian cytochrome P450 expression, circadian regulation of androgen serum concentrations may represent a mechanism to protect the organism from potentially damaging constitutive expression of androgens. Further studies with dbp−/− mice may determine whether altered circadian Cyp2a4 gene expression results in altered serum levels of steroid hormones and whether this in turn leads to alterations in sex hormone-associated physiology and behavior, such as mating, aggression, or tumor susceptibility.
In addition to 15α-hydroxylation, several other hydroxylation modifications of steroid hormones have been identified (42). Recent work has indicated that another cytochrome P450 steroid hydroxylase, the rat CYP2A1 gene, encoding testosterone 7α-hydroxylase, demonstrates circadian expression at the protein and enzyme level in the testes, but not the liver, of male rats (27, 39). In contrast, in female rats, caloric restriction uncovers a circadian rhythm in testosterone 7α-hydroxylase activity in the liver (27). Though distinct from circadian Cyp2a4 and Cyp2a5 gene expression presented here, the circadian expression of CYP2A1 underscores that regulation of steroid biogenesis is highly complex, demonstrating tissue-specific, sex-specific, and circadian regulation.
Biochemical and genetic experiments presented here indicate that the PAR transcription factor DBP can influence the circadian expression of the Cyp2a4 and Cyp2a5 genes. The strongest evidence for this derives from experiments with the dbp−/− mouse strain, in which circadian accumulation of their gene products in the liver is severely altered in the dbp−/− mice relative to wild-type mice. In mice, DBP mRNA is expressed with a dedicated circadian rhythm in the suprachiasmatic nucleus of the hypothalamus, an area of the brain believed to be important for circadian regulation (24). In addition to reduced circadian expression of the Cyp2a4 and Cyp2a5 genes reported here, dbp−/− mice display altered circadian locomotor activity, with changes in the precise timing and amplitude of wheel-running and infrared beam break activities (24). Thus, one and the same transcription factor appears to affect such completely different clock outputs as locomotor activity and liver metabolism. Interestingly, morning levels of CYP2A4 and CYP2A5 mRNA are generally higher in dbp−/− mice than in wild-type mice. As DBP is barely detectable in wild-type animals during the morning hours (43), the mechanisms through which it reduces the nadir levels of these transcripts are likely to be indirect, for example, by controlling the expression of a transcriptional repressor that accumulates in the morning.
While the circadian expression patterns of the CYP2A4 and CYP2A5 mRNAs are altered, they are not eliminated in dbp−/− mice. This implies that DBP, while an important player, is not the sole determinant of their circadian mRNA expression in the liver. Other members of the PAR transcription factor family, TEF and HLF, may contribute as well, given their overlapping expression patterns, transcription activation potentials, and DNA-binding-site preferences (4, 5, 8). In fact, given this high degree of similarity between the PAR family members, it is interesting that mutation of the DBP gene alone is sufficient to reduce circadian Cyp2a4 and Cyp2a5 gene expression to a significant degree. Perhaps multiple PAR factors represent different circadian regulatory pathways of common target genes, and thus elimination of any one family member reduces circadian expression. Alternatively, each PAR family member may have distinct target gene preferences, arising from specific protein-protein interactions or in vivo DNA-binding preferences more subtle than those observed in vitro. Further biochemical and genetic studies, including the generation of mutant mouse strains deficient in the other PAR factors and combinations thereof, will aid in determining whether the apparently highly similar PAR family members have overlapping or distinct target gene preferences in vivo.
ACKNOWLEDGMENTS
Luis Lopez-Molina and Daniel J. Lavery contributed equally to this study and should both be considered primary authors.
We are grateful to Nicolas Roggli for preparation of the artwork, to Frederic Bancel for purified P450 standards 2A4 and 2A5, and to Christian Larroque and Reinhard Lange for the anti-mouse P450 2A antibody.
This work was supported by the Swiss National Science Foundation, the State of Geneva, and Glaxo Wellcome Experimental Research, Geneva, Switzerland.
REFERENCES
- 1.Aust S D, Chignell C F, Bray T M, Kalyanaraman B, Mason R P. Free radicals in toxicology. Toxicol Appl Pharmacol. 1993;120:168–178. doi: 10.1006/taap.1993.1100. [DOI] [PubMed] [Google Scholar]
- 2.Bonfils, C. 1999. Unpublished results.
- 3.Bonfils C, Daujat M, Chevron M P, Dalet-Beluche I. Electrotransfer of microsomal cytochrome P450 after isoelectric focusing (IEF blots): resolution of human 2A and 3A isozymes. Anal Biochem. 1994;218:80–86. doi: 10.1006/abio.1994.1143. [DOI] [PubMed] [Google Scholar]
- 4.Falvey E, Fleury-Olela F, Schibler U. The rat hepatic leukemia factor (HLF) gene encodes two transcriptional activators with distinct circadian rhythms, tissue distributions and target preferences. EMBO J. 1995;14:4307–4317. doi: 10.1002/j.1460-2075.1995.tb00105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falvey E, Marcacci L, Schibler U. DNA-binding specificity of PAR and C/EBP leucine zipper proteins: a single amino acid substitution in the C/EBP DNA-binding domain confers PAR-like specificity to C/EBP. Biol Chem. 1996;377:797–809. [PubMed] [Google Scholar]
- 6.Fitzgerald K, Zucker I. Circadian organization of the estrous cycle of the golden hamster. Proc Natl Acad Sci USA. 1976;73:2923–2927. doi: 10.1073/pnas.73.8.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleury-Olela, F. 1999. Unpublished results.
- 8.Fonjallaz P, Ossipow V, Wanner G, Schibler U. The two PAR leucine zipper proteins, TEF and DBP, display similar circadian and tissue-specific expression, but have different target promoter preferences. EMBO J. 1996;15:351–362. [PMC free article] [PubMed] [Google Scholar]
- 9.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gubler U. A one tube reaction for the synthesis of blunt-ended double-stranded cDNA. Nucleic Acids Res. 1988;16:2726. doi: 10.1093/nar/16.6.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jounaidi Y, Bonfils C, Perin F, Negishi M, Lange R. Overexpression of a cytochrome P-450 of the 2a family (Cyp2a-5) in chemically induced hepatomas from female mice. Eur J Biochem. 1994;219:791–798. doi: 10.1111/j.1432-1033.1994.tb18559.x. [DOI] [PubMed] [Google Scholar]
- 12.Kemp C J, Leary C N, Drinkwater N R. Promotion of murine hepatocarcinogenesis by testosterone is androgen receptor-dependent but not cell autonomous. Proc Natl Acad Sci USA. 1989;86:7505–7509. doi: 10.1073/pnas.86.19.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Labrecque G, Belanger P M. Biological rhythms in the absorption, distribution, metabolism and excretion of drugs. Pharmacol Ther. 1991;52:95–107. doi: 10.1016/0163-7258(91)90088-4. [DOI] [PubMed] [Google Scholar]
- 14.Lavery, D. J. 1999. Unpublished results.
- 15.Lavery D J, Lopez-Molina L, Fleury-Olela F, Schibler U. Selective amplification via biotin- and restriction-mediated enrichment (SABRE), a novel selective amplification procedure for detection of differentially expressed mRNAs. Proc Natl Acad Sci USA. 1997;94:6831–6836. doi: 10.1073/pnas.94.13.6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lavery D J, Schibler U. Circadian transcription of the cholesterol 7 α hydroxylase gene may involve the liver-enriched bZIP protein DBP. Genes Dev. 1993;7:1871–1884. doi: 10.1101/gad.7.10.1871. [DOI] [PubMed] [Google Scholar]
- 17.Lavery D J, Schibler U. DBP and related transcription factors of the PAR family. In: Tronche F, Yaniv M, editors. Liver gene expression. New York, N.Y: Chapman & Hall; 1994. pp. 259–275. [Google Scholar]
- 18.Lavery, D. J., and U. Schibler. Circadian timing in animals. In E. Russo, D. Cove, L. Edgar, R. Jaenisch, and F. Salamini (ed.), Development: genetics, epigenetics and environmental regulation, in press. Springer-Verlag, Heidelberg, Germany.
- 19.Lavery D J, Schmidt E E, Schibler U. The PAR transcription factor family and circadian gene expression. In: Degli Agosti R, Bonzon M, Greppin H, editors. Foundations of biological rhythms. Geneva, Switzerland: University of Geneva; 1996. pp. 135–145. [Google Scholar]
- 20.Lee Y H, Alberta J A, Gonzalez F J, Waxman D J. Multiple, functional DBP sites on the promoter of the cholesterol 7 α-hydroxylase P450 gene, CYP7. Proposed role in diurnal regulation of liver gene expression. J Biol Chem. 1994;269:14681–14689. [PubMed] [Google Scholar]
- 21.Lichtsteiner S, Schibler U. A glycosylated liver-specific transcription factor stimulates transcription of the albumin gene. Cell. 1989;57:1179–1187. doi: 10.1016/0092-8674(89)90055-x. [DOI] [PubMed] [Google Scholar]
- 22.Lindberg R, Burkhart B, Ichikawa T, Negishi M. The structure and characterization of type I P-450(15) alpha gene as major steroid 15 α-hydroxylase and its comparison with type II P-450(15) alpha gene. J Biol Chem. 1989;264:6465–6471. [PubMed] [Google Scholar]
- 23.Lopez-Molina, L. 1999. Unpublished results.
- 24.Lopez-Molina L, Conquet F, Dubois-Dauphin M, Schibler U. The DBP gene is expressed according to a circadian rhythm in the suprachiasmatic nucleus and influences circadian behavior. EMBO J. 1997;16:6762–6771. doi: 10.1093/emboj/16.22.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez-Molina, L., D. Lavery, and U. Schibler. 1999. Unpublished results.
- 26.Loros J J, Denome S A, Dunlap J C. Molecular cloning of genes under control of the circadian clock in Neurospora. Science. 1989;243:385–388. doi: 10.1126/science.2563175. [DOI] [PubMed] [Google Scholar]
- 27.Manjgaladze M, Chen S, Frame L T, Seng J E, Duffy P H, Feuers R J, Hart R W, Leakey J E. Effects of caloric restriction on rodent drug and carcinogen metabolizing enzymes: implications for mutagenesis and cancer. Mutat Res. 1993;295:201–222. doi: 10.1016/0921-8734(93)90021-t. [DOI] [PubMed] [Google Scholar]
- 28.Mitropoulos K A, Balasubramaniam S, Gibbons G F, Reeves B E. Diurnal variation in the activity of cholesterol 7-hydroxylase in the livers of fed and fasted rats. FEBS Lett. 1972;27:203–206. doi: 10.1016/0014-5793(72)80620-3. [DOI] [PubMed] [Google Scholar]
- 29.Mock E J, Norton H W, Frankel A I. Daily rhythmicity of serum testosterone concentration in the male laboratory rat. Endocrinology. 1978;103:1111–1121. doi: 10.1210/endo-103-4-1111. [DOI] [PubMed] [Google Scholar]
- 30.Morin L P, Fitzgerald K M, Zucker I. Estradiol shortens the period of hamster circadian rhythms. Science. 1977;196:305–307. doi: 10.1126/science.557840. [DOI] [PubMed] [Google Scholar]
- 31.Mueller C R, Maire P, Schibler U. DBP, a liver-enriched transcriptional activator, is expressed late in ontogeny and its tissue specificity is determined posttranscriptionally. Cell. 1990;61:279–291. doi: 10.1016/0092-8674(90)90808-r. . (Erratum 65:following 914, 1991.) [DOI] [PubMed] [Google Scholar]
- 32.Noshiro M, Nishimoto M, Okuda K. Rat liver cholesterol 7 α-hydroxylase. Pretranslational regulation for circadian rhythm. J Biol Chem. 1990;265:10036–10041. [PubMed] [Google Scholar]
- 33.O’Farrell P Z, Goodman H M, O’Farrell P H. High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell. 1977;12:1133–1141. doi: 10.1016/0092-8674(77)90176-3. [DOI] [PubMed] [Google Scholar]
- 34.Ogawa H, Pitot H C, Fujioka M. Diurnal variation of the serine dehydratase mRNA level in rat liver. Arch Biochem Biophys. 1994;308:285–291. doi: 10.1006/abbi.1994.1040. [DOI] [PubMed] [Google Scholar]
- 35.Pittendrigh C S. Temporal organization: reflections of a Darwinian clock-watcher. Annu Rev Physiol. 1993;55:16–54. doi: 10.1146/annurev.ph.55.030193.000313. :16–54. [DOI] [PubMed] [Google Scholar]
- 36.Poole T M, Drinkwater N R. Two genes abrogate the inhibition of murine hepatocarcinogenesis by ovarian hormones. Proc Natl Acad Sci USA. 1996;93:5848–5853. doi: 10.1073/pnas.93.12.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schibler, U. 1998. Circadian rhythms. New cogwheels in the clockworks Nature 393:620–621. (News.) [DOI] [PubMed]
- 38.Schmidt E E, Schibler U. Cell size regulation, a mechanism that controls cellular RNA accumulation: consequences on regulation of the ubiquitous transcription factors Oct1 and NF-Y and the liver-enriched transcription factor DBP. J Cell Biol. 1995;128:467–483. doi: 10.1083/jcb.128.4.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seng J E, Gandy J, Turturro A, Lipman R, Bronson R T, Parkinson A, Johnson W, Hart R W, Leakey J E. Effects of caloric restriction on expression of testicular cytochrome P450 enzymes associated with the metabolic activation of carcinogens. Arch Biochem Biophys. 1996;335:42–52. doi: 10.1006/abbi.1996.0480. [DOI] [PubMed] [Google Scholar]
- 40.Squires E J, Negishi M. Reciprocal regulation of sex-dependent expression of testosterone 15 α-hydroxylase (P-450(15 alpha)) in liver and kidney of male mice by androgen. Evidence for a single gene. J Biol Chem. 1988;263:4166–4171. [PubMed] [Google Scholar]
- 41.van der Hoeven T A, Coon M J. Preparation and properties of partially purified cytochrome P-450 and reduced nicotinamide adenine dinucleotide phosphate-cytochrome P-450 reductase from rabbit liver microsomes. J Biol Chem. 1974;249:6302–6310. [PubMed] [Google Scholar]
- 42.Waxman D J, Ko A, Walsh C. Regioselectivity and stereoselectivity of androgen hydroxylations catalyzed by cytochrome P-450 isozymes purified from phenobarbital-induced rat liver. J Biol Chem. 1983;258:11937–11947. [PubMed] [Google Scholar]
- 43.Wuarin J, Schibler U. Expression of the liver-enriched transcriptional activator protein DBP follows a stringent circadian rhythm. Cell. 1990;63:1257–1266. doi: 10.1016/0092-8674(90)90421-a. [DOI] [PubMed] [Google Scholar]