Abstract
N6-methyladenosine (m6A) is a prevalent internal modification in eukaryotic RNAs regulated by the so-called “writers”, “erasers”, and “readers”. m6A has been demonstrated to exert critical molecular functions in modulating RNA maturation, localization, translation and metabolism, thus playing an essential role in cellular, developmental, and disease processes. Circular RNAs (circRNAs) are a class of non-coding RNAs with covalently closed single-stranded structures generated by back-splicing. CircRNAs also participate in physiological and pathological processes through unique mechanisms. Despite their discovery several years ago, m6A and circRNAs has drawn increased research interest due to advances in molecular biology techniques these years. Recently, several scholars have investigated the crosstalk between m6A and circRNAs. In this review, we provide an overview of the current knowledge of m6A and circRNAs, as well as summarize the crosstalk between these molecules based on existing research. In addition, we present some suggestions for future research perspectives.
Keywords: N6-methyladenosine, Circular RNA, Crosstalk
Background
RNA modifications (e.g., N6-methyladenosine [m6A], 5-methylcytosine, pseudouridine, N4-acetylcytidine, ribose methylations, and N1-methylguanosine), have recently emerged as vital post-transcriptional epigenetic modulators of gene expression in eukaryotes [1, 2]. Among these RNA modifications, m6A represents the most common and well-studied to date. m6A is a reversible modification that methylated adenosine at the N6 position of almost every type of RNA molecule, including mRNAs, small nuclear RNAs, ribosomal RNAs, and non-coding RNAs [1–3]. m6A was first discovered in the 1970s and developed rapidly during the past few years due to the advances in high-throughput m6A sequencing and methylated RNA m6A immunoprecipitation [4]. Moreover, m6A has been demonstrated to exert critical molecular functions in modulating RNA maturation, localization, translation, and metabolism. m6A dynamically exists and is involved in a variety of physiological and pathological processes, including growth, development, aging and diseases [4–6].
Circular RNAs (circRNAs) are a class of endogenous RNAs with covalently closed single-stranded structures also present in eukaryotes [7, 8]. Most circRNAs are non-coding RNAs while a proportion of cytoplasmic circRNAs have the coding potential to be translated into peptides [9, 10]. These molecules were also discovered several years ago, but has recently attracted the attention of researchers due to the advances in high-throughput RNA sequencing and bioinformatics [11]. Similar to other types of RNAs, circRNAs are involved in the maintenance of the normal physiological function of the human body, as well as the occurrence and development of a variety of human diseases [11–13]. While distinct from other RNA molecules, circRNAs possess unique biogenesis, biology, and characterization. Therefore, they may present peculiarities in response to RNA modifications.
Recently, some scholars have combined these two recent hot topics to investigate the crosstalk between them. In this review, we provide an overview of the current knowledge of m6A as well as circRNAs, and summarize the crosstalk between m6A modification and circular RNAs based on existing research. In addition, we have found that many questions still remain unanswered in this area and present some suggestions for future research perspectives.
RNA m6A modification
Similar to DNA methylation, RNA m6A methylation is catalyzed and recognized by corresponding enzymes, methyltransferases- “writers”, demethylases- “erasers” and “readers”. Subsequently, these modified RNAs will present with a different fate in maturation, localization, translation and metabolism, thereby influencing various molecular cellular processes. The specific details are described below and a summary is presented in Fig. 1.
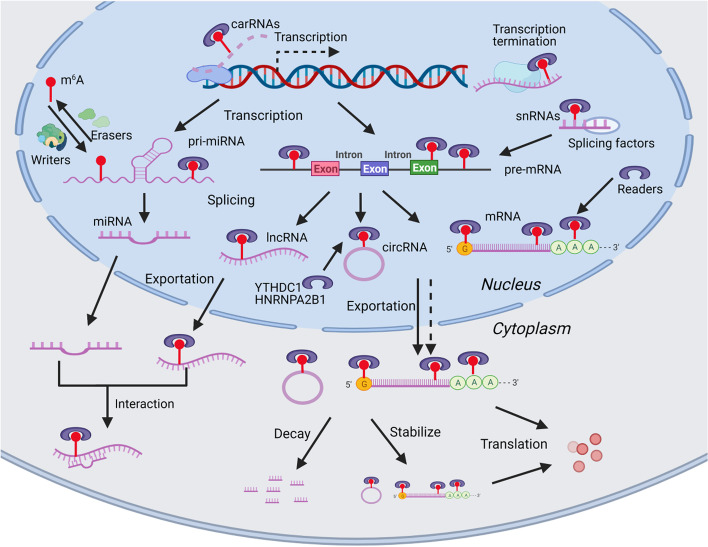
Fig. 1.
Overview of m6A modification. m6A modification is installed by the multicomponent m6A methyltransferases complex (writers) and removed by demethylases (erasers). The m6A modification is then identified by m6A readers which determine the fate of these RNAs and involved in various cellular processes. In the nucleus, m6A are identified by nuclear readers and modulates RNA transcription (transcription activation and termination), splicing (mRNAs, miRNAs, lncRNAs and circRNAs maturing) and structure (influence readers binding and splicing). Mature RNAs modified by m6A in the nucleus are recognized by readers, which subsequently mediate subcellular localization. In the cytoplasm, m6A are identified by cytoplasmic readers and modulates RNA stability (enhance stability or facilitate degradation), translation (promote translation via multiple mechanisms), and binding capacity (RNA-RBP interaction and RNA-RNA interaction)
Participants of m6A modification: writers, erasers, and readers
m6A writers
The m6A is installed by the multicomponent m6A methyltransferases complex (MTC), known as “writers”. The currently reported writers include methyltransferase-like 3 (METTL3), methyltransferase-like 14 (METTL14), methyltransferase-like 5 (METTL5), methyltransferase-like 16 (METTL16), Cbl proto-oncogene-like 1 (HAKAI), Wilms’ tumor 1-associating protein (WTAP), Vir Like M6A Methyltransferase Associated (VIRMA), RNA Binding Motif Protein 15/15B (RBM15/15B), Zinc Finger CCCH-Type Containing 4 (ZCCHC4), and Zinc Finger CCCH-Type Containing 13 (ZC3H13). These enzymes perform their respective duties and jointly complete the “writing” task. According to current knowledge, METTL3, METTL5, and METTL16 function as catalytic cores in the complex which catalyze m6A modification via methyltransferase domains [3–5, 14–16]. Other components typically play auxiliary roles, such as structural stabilization, reorganization of special RNA sites, and directing MTC location [3–5, 14–16].
m6A erasers
The m6A installed by writers can be removed by demethylases, or so-called “erasers”, which include fat mass and obesity-associated protein (FTO) and AlkB homolog 5 (ALKBH5) [14–16]. These two demethylases are localized in nuclear speckles and can oxidatively eliminate both DNA and RNA methylation, with a particularity for m6A methylation [15–17]. The identification of m6A demethylation provides evidence for the possible reversibility of the m6A modification.
m6A readers
To exert their biological functions, the m6A modifications determined by m6A writers and erasers must be identified by m6A readers. The currently reported readers include the YT521-B homology (YTH) domain family proteins (YTHDF1, YTHDF2, and YTHDF3), YTH domain containing proteins (YTHDC1 and YTHDC2), heterogeneous nuclear ribonucleoprotein (HNRNPC, HNRNPG, and HNRNPA2B1), insulin-like growth factor 2 mRNA-binding proteins (IGF2BP1, IGF2BP2, IGF2BP3), eukaryotic translation initiation factor 3 (EIF3), proline rich coiled-coil 2A (PRRC2A), and staphylococcal nuclease and tudor domain containing 1 (SND1). These RNA binding proteins (RBPs) have conserved m6A-binding domains that can specifically recognize m6A modifications. RBPs bind to m6A methylated RNAs and determine the fate of these RNAs, thus regulating various cellular processes such as transcription, splicing and maturing, exportation, translation, decay and others [3–5, 14–16]. Therefore, the m6A readers represent intermediaries for RNA m6A modification and different RNA fates.
A more comprehensive summary than previous reviews on the classification and functions of m6A writers, erasers, and readers is presented in Table 1. It appears that compared with a simple m6A installation and elimination function of writers and erasers, the roles of readers are more complicated and diverse, which is an area of keen research interest.
Table 1.
Writers, erasers and readers of RNA m6A modification
| Category | Factors | Roles | Refs |
|---|---|---|---|
| Writers | METTL3 | m6A catalytic subunit | [14, 15] |
| METTL14 | Forms heterodimer with METTL3 to stabilize METTL3 and assist recognizing the subtract | [14, 18] | |
| METTL16 | m6A catalytic subunit | [19] | |
| METTL5 | Ribosome 18S m6A methyltransferase | [20, 21] | |
| TRMT112 | Forms heterodimeric complex with METTL5 as a methyltransferase activator to stabilize METTL5 | [21] | |
| ZCCHC4 | Ribosome 28S m6A methyltransferase | [22, 23] | |
| HAKAI | Essential member of the MTC | [24] | |
| WTAP | Promotse m6A methyltransferase activity and localization in nuclear speckles | [25] | |
| VIRMA | Binds the MTC and recruit it to specific RNA region | [26] | |
| RBM15/15B | Binds the MTC and recruit it to specific RNA site | [27] | |
| ZC3H13 | Promotes nuclear localization of MTC to modulate m6A in the nucleus | [28] | |
| Erasers | FTO | Eliminates m6A by oxidation | [14–18] |
| ALKBH5 | Eliminates m6A by oxidation | [14–18] | |
| Readers | YTHDF1 | Facilitates the ribosome assembly of m6A-mRNAs and interacts with the initiation factor to promote translation; cooperates with YTHDF2 and YTHDF3 to mediate degradation of m6A-mRNAs | [29, 30] |
| YTHDF2 | Reduces m6A-mRNAs stability; stabilize m6A-mRNAs specifically in cancer stem cells; cooperates with YTHDF1 and YTHDF3 to mediate degradation of m6A-mRNAs | [29–31] | |
| YTHDF3 | Cooperates with YTHDF1 and YTHDF2 to mediate degradation of m6A-mRNAs | [30, 32] | |
| YTHDC1 | Promotes RNA splicing and translocation; facilitates the decay of m6A-modified chromosome-associated regulatory RNAs; together with its target m6A-RNAs to regulate chromatin modification and retrotransposon repression; regulate histone methylation | [33–38] | |
| YTHDC2 | Facilitates the translation and decrease the abundance of m6A-RNAs; has 3′-5′ RNA helicase activity and decrease the stability of m6A-mRNAs | [39–41] | |
| HNRNPC/G | Responsible for pre-mRNA processing and affect the alternative splicing of target m6A-mRNAs | [42–44] | |
| HNRNPA2B1 | Binds to m6A-containing pri-miRNAs to promote pri-miRNA processing; may regulate mRNA splicing by binding to m6A-containing pre-mRNAs; facilitates m6A modification and nucleocytoplasmic trafficking of mRNAs | [45, 46] | |
| IGF2BP1/2/3 | Regulates m6A-RNAs stability, subcellular localization and translation | [47–49] | |
| EIF3 | Facilitates translation of m6A-mRNAs by recruiting the 43S complex | [50, 51] | |
| PRRC2A | Enhances m6A-mRNAs stability | [52] | |
| SND1 | Enhances m6A-RNAs stability | [53] |
Biological functions of m6A modification
The modulation of m6A methylation on RNAs begins during transcription and is largely dependent on the subcellular localization of writers, erasers, and readers. The writers are primarily localized in the nucleus, so the writing processes predominantly occur during the nuclear phase [14–18]. The eraser ALKBH5 mainly exists and functions as a demethylase in the nucleus, and the eraser FTO exerts demethylase activity both in nucleus and cytoplasm [14–18, 54]. Thus, the erasing processes may occur in the nucleus and cytoplasm. Some readers are localized and “read” m6A in the nucleus, which may influence nuclear processes, such as transcription and RNA splicing. In addition, some readers are able to assist with m6A-RNAs export from the nucleus to the cytoplasm. Readers in the cytoplasm may regulate cytosolic processes, such as translation and degradation.
m6A modulates RNA transcription, splicing, and structure
RNA m6A modification is a post-transcriptional regulation which appears not to be related to transcription; however, a recent study demonstrated that m6A modification on chromosome-associated regulatory RNAs (carRNAs), including promoter-associated RNAs, enhancer RNAs, and repeat RNAs, can induce carRNA decay by YTHDC1 and impact the open chromatin state and downstream transcription [36]. Moreover, RNA m6A modification play a critical role in transcription termination by facilitating the formation of co-transcriptional R-loops to decrease the readthrough activity of Pol II [55]. Reports have confirmed that m6A modification on primary miRNAs (pri-miRNAs) promotes the recognition and processing by the microRNA microprocessor complex protein, DGCR8, thereby enhancing miRNA maturation [45, 56]. The regulation of m6A on pre-mRNA splicing has been validated in Drosophila [57], whereas the precise regulation pattern remains largely unknown in mammals. Nevertheless, some efforts have been made to consummate the pathways through which m6A modulates pre-mRNA splicing in mammals. For example, the m6A reader, HNRNPG, may use Arg-Gly-Gly motifs to co-transcriptionally interact with RNA polymerase II and m6A-modified nascent pre-mRNA to modulate alternative splicing [44]. Additionally, YTHDC1 can recruit and promote pre-mRNA splicing factors to enter the binding regions of targeted mRNAs to modulate mRNA splicing [33]. In addition, the m6A modification on U2 and U6 snRNAs may influence the splicing of specific pre-mRNA transcripts [58, 59]. Evidence also shows that m6A can alter RNA structures to affect RNA-protein interactions in cells. For instance, m6A alters the local structure in mRNA and lncRNA and thereby influences the binding of HNRNPC to mediate pre-mRNA processing [42]. m6A located near splice sites in nascent pre-mRNA modulates HNRNPG binding, which influences RNAPII occupancy patterns and promotes HNRNPG-mediated alternative splicing [43, 44].
m6A modulates RNA subcellular localization
Mature RNAs modified by m6A in the nucleus are recognized by readers, which subsequently mediate subcellular localization. In general, nuclear readers (e.g., YTHDC1 and HNRNPA2B1) can identify m6A-RNAs (e.g., mRNAs and circRNAs), and accelerate their exportation from the nucleus to the cytoplasm [34, 45, 60]. However, for some RNAs, m6A modification may detain them within the nucleus. For example, m6A modification of lncRNA RP11 can increase its accumulation in the nucleus and on chromatin, which may be due to its interaction with HNRNPA2B1 [61]. Interestingly, several RNAs without m6A modification can still be exported from the nucleus, indicating that the m6A is a facilitator but not an indispensable factor for translocation [14].
m6A modulates RNA stability, translation, and binding capacity
RNA exported to the cytoplasm may exert their biological functions or be degraded, and m6A modification can impact these processes via multiple cytoplasmic readers. Readers mediate the degradation of m6A-mRNAs, including YTHDF1, YTHDF2, YTHDF3, and YTHDC2, and readers enhance m6A-mRNA stability, including IGF2BPs, PRRC2A, and SND1 (Table 1) [29–32, 39–41, 47, 52, 53]. Moreover, these readers may regulate RNA stability through diverse mechanisms. For example, the carboxy-terminal domain of YTHDF2 selectively interacts with m6A-mRNAs, whereas the amino-terminal domain mediates the transposition of the YTHDF2-mRNA complex from the translatable pool to mRNA decay sites (e.g., processing bodies) [62]. IGF2BPs probably recruit RNA stabilizers, such as ELAV like RNA binding protein 1 (ELAVL1 or HuR), matrin 3 (MATR3), and poly(A) binding protein cytoplasmic 1 (PABPC1), to maintain the stability of their target m6A-RNAs [47]. Numerous studies have demonstrated that m6A can regulate translation with the assistance of readers, including YTHDF1, YTHDF2, YTHDF3, YTHDC2, IGF2BPs, and EIF3 [29–32, 39–41, 47–49], which involves several distinct mechanisms. For example, YTHDF1 can facilitate the ribosome assembly of m6A-mRNAs and interact with the initiation factor to promote translation [29]. In the absence of the cap-binding factor eIF4E, EIF3 can directly bind to the m6A in the 5′ untranslated region (UTR) and recruit the 43S complex to initiate translation [50]. METTL3 directly binds to the eukaryotic translation initiation factor 3 subunit h (eIF3h) and presumably promotes translation through ribosome recycling [63]. Promoter-bound METTL3 induces m6A in the coding region of mRNA to enhance translation by relieving ribosome stalling [64]. Moreover, m6A on 18S and 28S ribosomal RNA also play critical roles in the maintenance of ribosomal translation dynamics [20, 22]. Apart from the influence of the RNA-RBP interaction described above, m6A may also be indispensable for some RNA-RNA interactions. For example, the sufficient enrichment of the m6A modification on linc1281 is required for the interaction between linc1281 with miRNAs [65], and the m6A modified 353–357 region in the YAP 3’UTR was found to be critical for miR-582-3p targeting [66].
CircRNAs
In contrast to other RNA molecules, circRNAs have a unique circular structure, which requires a unique biogenesis process. Moreover, this stable structure may endow them with distinctive cellular functions, as well as unique approach to degradation. The associated details are described below and a summary is presented in Fig. 2.
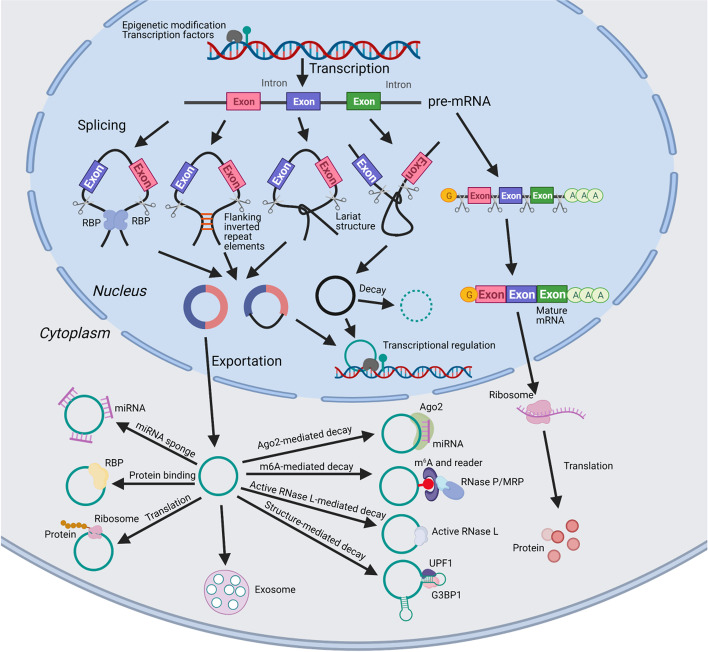
Fig. 2.
Summary of circRNA biology. Both circRNAs and mRNAs are originated from pre-mRNAs transcribed from genomic DNA which are regulated by transcriptional regulators. Distinguished from mRNA maturation (5′-capping, 3′-polyadenylation and introns removing), circRNA maturation go through various back-splicing processes which competes with the traditional mRNA splicing. CircRNAs matured in the nucleus may stay in the nucleus (EIcRNAs and ciRNAs with intron elements) or export to the cytoplasm (EcRNAs without introns). CircRNAs stay in the nucleus may participate in nuclear processes such as transcriptional regulation. CircRNAs export to the cytoplasm are involved in cytoplasmic processes such as miRNA sponging, RBP binding and translation. Some circRNAs are also enriched and stable in exosomes and secreted to extracellular components. All the circRNAs will eventually be degraded via a variety of mechanisms
Biogenesis of circRNAs
Similar to mRNA maturation, the biogenesis of circRNAs should also undergo processes, including the transcription and splicing of pre-mRNAs [7, 8, 11]. Therefore, the factors that regulate transcription (e.g., epigenetic modifications and transcription factors) may influence the generation of circRNAs [7, 8, 11]. Distinguished from the canonical splicing of mRNAs, the alternative splicing of circRNAs represents a unique mode that competes with mRNA splicing. Instead of the 5′-capping, 3′-polyadenylation and introns removing events of mRNA maturation, a downstream 5′-splice site and a 3′-splice site are connected to form a covalently closed single-stranded structure in circRNA splicing [7, 8, 11–13]. There are several acknowledged mechanisms that can be used to interpret the circularization of circRNAs [7, 13, 67]. The first is intron pairing-driven circularization, in which the flanking of inverted repeat elements form RNA double strands through base-pairing. Another model is RBP-mediated circularization, in which the RBPs bind to the upstream and downstream flanking introns to form dimers. The third is lariat-driven circularization, which is mediated by the lariat structures that form in the exon-skipping events during linear splicing or intronic lariats that escape from the debranching of canonical linear splicing. These regulatory modes serve to bring the downstream splice-donor sites into close proximity with the upstream splice-acceptor site subject to alternative splicing. Based on the involved splicing and the genomic elements, three types of circRNAs are generated: 1) exonic circRNAs (EcRNAs), which are composed by one or more exons; 2) exon-intron circRNAs (EIcRNAs) that contain both exon and intron components; and 3) intronic circRNAs (ciRNAs), which consist of only introns [11, 68, 69].
Exportation and distribution of circRNAs
As described above, the biogenesis of circRNAs occurs in the nucleus, and are then exported to the cytoplasm or detained in the nucleus. In general, most circRNAs are exported into the cytoplasm and the vast majority of cytoplasmic circRNAs are EcRNAs without introns [7, 12, 13]. Similar to many linear RNAs, circRNAs involving intron elements (e.g., EIcRNAs and ciRNAs) are usually sequestered in the nucleus [7, 12, 13]. There are some underlying mechanisms may interpret the exportation and distribution of circRNAs. The first and the most recently concerned is m6A-mediated circRNA translocation, which will be discussed in detail below. Another mechanism is the length-dependent evolutionarily conserved pathway which involves the association of circRNA lengths with the conserved proteins, UAP56 and URH49 [70]. In addition, since circRNAs are also enriched and stable in exosomes, they also widely exist in extracellular components [71, 72].
Biological functions of circRNAs
Based on the unique characteristics and distribution, circRNAs may exert various biological functions. CircRNAs enriched in the nucleus are more likely to modulate transcription and splicing and several underlying mechanisms have been reported. For instance, circSCMH1 may interact with transcription factor methyl CpG binding protein 2 (MeCP2) to restrain its transcriptional activity [73]. circMRPS35 can recruit the histone acetyltransferase, KAT7, to elicit the acetylation of H4K5 in the promoters and directly bind to the promoters of FOXO1 and FOXO3a genes to activate the transcription [74]. CircRNAs derived from exon 6 of the SEP3 gene in Arabidopsis can bind to its cognate DNA locus to form an RNA:DNA hybrid, pausing transcription and exon 6 skipping in the alternative splicing of SEP3 pre-mRNA [75].
Compared with nuclear circRNAs, cytoplasmic circRNAs are better acquainted. The most frequently reported function of circRNAs is their capacity to act as miRNA sponges. Such sponging refers to the manner by which circRNAs impair miRNA activity through sequestration in a competing endogenous RNA (ceRNA) manner, thereby raising the expression of miRNA target genes [76, 77]. Compared with this explicit inhibitory role on miRNA, circRNAs exhibit diverse binding effects on various proteins [78–81]. For example, circRNAs may not only recruit RBPs to stabilize and translate mRNAs, but also competitively bind to these RBPs to inhibit translation and degradation [62, 82, 83]. In addition, the interaction between RBPs and circRNAs may also influence the functionality and induce degradation through the ubiquitination of RBPs [84, 85].
CircRNAs are once considered as non-coding RNAs, however, recent studies have demonstrated that some cytoplasmic circRNAs carrying an initiation codon and putative open reading frames can be translated into peptides. Although lacking the traditional initiation elements (e.g., 5′ and 3’untranslated regions), circRNAs carrying an internal ribosome entry site (IRES) may undergo translation in a cap-independent manner [9, 86]. In addition, some circRNAs possess m6A and translation initiation sites may also go through m6A-driven translation with the assistance of the initiation factor, eIF4G2, and m6A reader, YTHDF3 [87]. Due to the same ORF components, several peptides translated by circRNAs are closely related to the proteins translated by their corresponding mRNAs. These peptides may act as substitutes to protect intact proteins from degradation or function as competitors to compete for regulators with intact proteins [88–90]. As proteins, while they may also play other functions [91–93], there are few relevant published studies, and further explorations remain to be performed.
Based on abundance and stability, circRNAs located in exosomes can be detected in the circulation and urine. Accumulating studies have confirmed that the circRNA content in the exosomes of some diseases is anomalous, indicating that they are promising diagnostic molecular markers [71, 72]. It is also feasible that cells transfer circRNAs to other cells or even throughout the body via excretion in exosomes. Therefore, they may act as mediators to ensure natural cell-to-cell communications. Besides, exosomes may bring abnormal amount of circRNAs to target cells, which is also an important source of various pathophysiological processes [71, 72].
Degradation of circRNAs
Although the structure is highly stable and are resistant to exonucleases [7, 8, 11], they will eventually be degraded through the involvement of several unique and diverse degradation pathways. The binding of miRNAs to circRNAs can initiate the Argonaute 2 (Ago2)-mediated RNA decay, which is executed by the RNA-induced silencing complex (RISC) [94]. However, this phenomenon may not be as common as expected since similar to linear RNAs, the overwhelming majority of circRNAs bear sequences that are only partially complementary to miRNAs [13]. CircRNAs modified by m6A may be decayed by the ribonuclease complex RNase P/MRP, which will be discussed in detail below. In addition, there is a structure-mediated RNA decay model (e.g., high overall 3′ UTR structure) formed by base pairing in circRNAs that can be targeted and degraded by UPF1 and G3BP1 [95]. Upon viral infection, circRNAs can also be globally degraded by activated RNase L, which is required for PKR activation [96]. Actually, the above-mentioned degradation pathways are also suitable for some other RNA molecules and not unique to circRNAs.
Crosstalk between m6A and circular RNAs
Through the above summary, we can find that there are many intersections between the regulatory pathway of m6A and the life cycle of circRNAs. Indeed, there have been many studies focusing on the crosstalk between m6A and circRNAs. Details are described below and a summary is shown in Table 2 and Fig. 3.
Table 2.
Crosstalk between m6A and circRNAs
| Crosstalk | circRNA | Roles | Refs |
|---|---|---|---|
| m6A regulates circRNAs expression | circMETTL3 | METTL3 facilitates circMETTL3 expression in an m6A-dependent manner | [97] |
| circ1662 | METTL3 induced circ1662 generation by binding its flanking sequences and installing m6A modifications | [98] | |
| circCUX1 | METTL3 mediates the m6A methylation of circCUX1 and stabilizes circCUX1 | [99] | |
| circRNA-SORE | m6A modification raises circRNA-SORE level by increasing RNA stability | [100] | |
| circRNAs | m6A modification cause circRNAs selectively degraded by RNase P/MRP complex | [101] | |
| m6A regulates circRNAs distribution | circGFRα1 | METTL14 promotes cytoplasmic export of m6A-modified circGFRα1 through the GGACU motif | [102] |
| circNSUN2 | m6A modification of circNSUN2 facilitates cytoplasmic export | [60] | |
| m6A regulates circRNAs function | circRNAs | Extensive m6A modifications in circRNAs drives protein translation in a cap-independent fashion | [87] |
| circRNAs | m6A modification controls circRNA immunity | [103] | |
| circRNAs regulate m6A | hsa_circ_0072309 | hsa_circ_0072309 upregulates the expression of m6A demethylase FTO by targeting miR-607 | [104] |
| circMAP2K4 | circMAP2K4 promote YTHDF1 expression by binding with hsa-miR-139-5p | [105] | |
| circRAB11FIP1 | circRAB11FIP1 regulated the m6A methylation of ATG5 and ATG7 mRNA via upregulating FTO | [106] | |
| circMEG3 | circMEG3 inhibits the expression of METTL3 dependent on HULC | [107] | |
| circNOTCH1 | circNOTCH1 regulates the m6A modification on Nothch1 mRNA by binding to METTL14. | [108] | |
| circZbtb20 | circZbtb20 enhances the interaction of ALKBH5 with Nr4a1 mRNA, leading to ablation of the m6A on Nr4a1 mRNA | [109] | |
| circSTAG1 | circSTAG1 regulates m6A modification on FAAH by mediating ALKBH5 translocation | [110] |
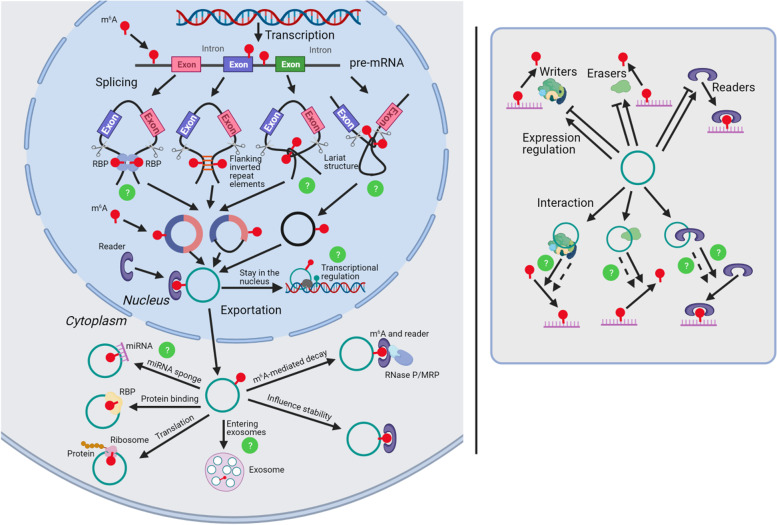
Fig. 3.
Crosstalk of m6A with circular RNAs. m6A modification is involved in the life cycle of circRNAs. First, m6A modification on pre-mRNAs may influence the splicing and generation of circRNAs. Second, m6A modification on mature circRNAs could affect the nuclear exportation of circRNAs. Third, m6A modification on circRNAs may influence the molecular functions of circRNAs, including transcription regulation, miRNA sponging, RBP binding and translation. Fourth, m6A modification may influence circRNA degradation or even entering the exosomes. Also, circRNAs can regulate m6A by affecting the expression or the functions of the m6A writers, erasers and readers. There are some predicted interactions between m6A and circRNAs have not been validated yet, which are marked with “?” in the figure
m6A modulates the expression of circRNAs
Similar to other RNA molecules, m6A can also modulate the expression of circRNAs through regulating their generation, stability, or degradation. A recent study confirmed that METTL3 can install m6A in the reverse complementary sequences of flanking introns of circ1662, as well as facilitate the generation of circ1662 based on the intron pairing-driven circularization pattern [98]. Moreover, circRNAs modified by m6A can be recognized by readers and exhibit changes in stability, resulting in altered expression [99, 100]. A subset of m6A-containing circRNAs may be endoribonucleolyticly degraded by the RNase P/MRP complex, which depends on the cooperative binding of HRSP12 and YTHDF2 [101]. The dynamic balance or the imbalance of circRNA expression is a consequence of the combined effect of these regulatory factors.
m6A modulates the distribution of circRNAs
Some studies have demonstrated that the m6A modification on circRNAs may modulate their nuclear exportation [60, 102]. This process may depend on the recognition and mediation of m6A readers [60]. Despite these initial findings, the mechanisms underlying the subcellular trafficking of m6A-circRNAs remains largely unknown.
m6A modulates the function of circRNAs
As described above, although considered to be non-coding RNAs, some circRNAs have the potential to encode proteins. Due to their unique structure, the translation of m6A modified circRNAs also differs from that of the linear RNAs. In this process, m6A-circRNAs are identified by the reader, YTHDF3, which recruits the translation initiation factors, eIF4G2 and eIF3A, to initiate translation in a cap-independent manner [87]. The m6A modification also influences the function of circRNAs in the regulation of innate immunity. Unmodified foreign circRNAs can directly trigger RIG-I signaling to promote immune activation; however, m6A-circRNAs may recruit YTHDF2 to form a complex with RIG-I and suppress the RIG-I immune signaling [103].
circRNAs modulates m6A
Conversely, circRNAs can also regulate m6A modification. Some circRNAs can regulate m6A by affecting the expression of the m6A writers, erasers, and readers [104–107]. Other circRNAs may influence the functions of m6A writers, erasers, and readers [108–110]. For example, circRNAs can competitively bind to writers and contend modifications, thereby hindering the modification of other RNAs by writers [108]. In addition, circRNAs can not only recruit ALKBH5 to ablate the m6A modification of target mRNA, but also capture ALKBH5 to suppress its translocation into the nucleus and impede its role of ablating m6A [109, 110]. Generally, circRNAs can only mediately modulate m6A modification by regulating m6A writers, erasers or readers instead of directly affect m6A modification by themselves.
Conclusions and prospectives
m6A modification and circRNA biology are undoubtedly current research hotspots, and the crosstalk between the two has attracted increasing attention from the researchers. In this review, we describe the complexity of m6A modification and circRNA biology, and present the identified crosstalk between them. Although some efforts have been devoted in this field, the study of correlation between m6A and circRNAs remains in the initial stages. In consideration of the current research realities, lots of questions remain to be addressed. In reference to the crosstalk mentioned in the previous section, we will present some perspectives which may represent potential future hot topics. These perspectives are also displayed in Fig. 3.
First, previous studies have only reported that m6A in the reverse complementary sequences of flanking introns facilitates the generation of circRNA through the intron pairing-driven circularization pattern [98]. However, other studies have verified that the m6A modification in pre-mRNA may regulate RNA-protein interactions and pre-mRNA processing [42–44]. This regulatory mode has been investigated in mRNA maturation but not in circRNAs biogenesis. Here, we propose that the m6A modification in pre-mRNA may affect the binding of some RBPs and regulate the generation of circRNA via RBP-mediated circularization. Whether m6A modification can modulate the lariat-driven circularization of circRNAs is also a topic worth examining.
Second, although it has been confirmed that m6A can regulate the stability of circRNAs [99, 100], in consideration of the specific circular structure and degradation pathway of circRNAs, is there any difference between circRNAs and linear RNAs in the mechanism of m6A regulating stability? Similarly, does the regulation of m6A on circRNAs exportation differ from its regulation on linear RNAs? In addition, studies have revealed that m6A may modulate the degradation of circRNAs [101]; however, whether this mode can participate in the normal life process or disease development has not yet been explored.
Third, while m6A are important initiators in the translation of circRNAs [88], it is unknown whether they are translation sustainers or terminators. Since m6A is able to affect RNA-RBP interactions [47], it may also influence the binding of RBPs to circRNAs. Similarly, the m6A modification on lncRNA is important for the binding of miRNAs [66], and there is an excellent probability that the m6A modification on circRNAs affects miRNA binding. Due to the recent research focus on miRNA sponging, potential m6A-mediated miRNA-circRNA interactions may represent another area of research interest.
Fourth, circRNAs can contend the modification of m6A writers [108], which may also apply to erasers and readers. In addition, while CircRNAs can recruit m6A erasers to ablate m6A modifications, it remains unknown whether they can also recruit m6A writers and readers to install and identify the m6A modifications.
Acknowledgments
Figures in this manuscript were created with BioRender.com.
Abbreviations
- m6A
N6-methyladenosine
- circRNAs
Circular RNAs
- MTC
Methyltransferases complex
- METTL3
Methyltransferase-like 3
- METTL14
Methyltransferase-like 14
- METTL5
Methyltransferase-like 5
- METTL16
Methyltransferase-like 16
- HAKAI
Cbl proto-oncogene-like 1
- WTAP
Wilms’ tumor 1-associating protein
- VIRMA
Vir Like M6A Methyltransferase Associated
- RBM15/15B
RNA Binding Motif Protein 15/15B;
- ZCCHC4
Zinc Finger CCCH-Type Containing 4
- ZC3H13
Zinc Finger CCCH-Type Containing 13
- FTO
Fat mass and obesity-associated protein
- ALKBH5
AlkB homolog 5
- YTH
YT521-B homology
- HNRNP
Heterogeneous nuclear ribonucleoprotein
- IGF2BPs
Insulin-like growth factor 2 mRNA-binding proteins
- EIF3
Eukaryotic translation initiation factor 3
- PRRC2A
Proline rich coiled-coil 2A
- SND1
Staphylococcal nuclease and tudor domain containing 1
- RBPs
RNA binding proteins
- carRNAs
Chromosome-associated regulatory RNAs
- pri-miRNAs
primary miRNAs
- ELAVL1
ELAV like RNA binding protein 1
- MATR3
Matrin 3
- PABPC1
Poly(A) binding protein cytoplasmic 1
- UTR
Untranslated region
- eIF3h
Eukaryotic translation initiation factor 3 subunit h
- EcRNAs
Exonic circRNAs
- EIcRNAs
Exon-intron circRNAs
- ciRNAs
Intronic circRNAs
- MeCP2
Methyl CpG binding protein 2
- ceRNA
Competing endogenous RNA
- IRES
Internal ribosome entry site
- Ago2
Argonaute 2
- RISC
RNA-induced silencing complex
Authors’ contributions
X.W. and R.M. wrote the manuscript; X.Z., L.C., Y.D. and W.S. retrieved literature; Y.S., C.G. and X.Z. critically revised the manuscript. All authors have read and approved the final manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (Grant no. 81673050, 81872522, 81900612, 82073429), Innovation Program of Shanghai Municipal Education Commission (No.2019-01-07-00-07-E00046), the Program of Science and Technology Commission of Shanghai Municipality (No. 18140901800), Excellent Subject Leader Program of Shanghai Municipal Commission of Health and Family Planning (No. 2018BR30), Clinical Research Program of Shanghai Hospital Development Center (No. SHDC2020CR1014B, SHDC12018X06), Program of Shanghai Academic Research Leader (No. 20XD1403300) and Research Program of Shanghai Skin Disease Hosptial (No. 2019KYQD08).
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xin Wang, Rui Ma and Xilin Zhang contributed equally to this work.
Contributor Information
Chunyuan Guo, Email: chunyuanguo@aliyun.com.
Yuling Shi, Email: shiyuling1973@126.com.
References
- 1.Frye M, Harada BT, Behm M, He C. RNA modifications modulate gene expression during development. Science. 2018;361:1346–1349. doi: 10.1126/science.aau1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbieri I, Kouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer. 2020;20:303–322. doi: 10.1038/s41568-020-0253-2. [DOI] [PubMed] [Google Scholar]
- 3.Huang H, Weng H, Chen J. m(6)A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell. 2020;37:270–288. doi: 10.1016/j.ccell.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang T, Kong S, Tao M, Ju S. The potential role of RNA N6-methyladenosine in cancer progression. Mol Cancer. 2020;19:88. doi: 10.1186/s12943-020-01204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiener D, Schwartz S. The epitranscriptome beyond m(6)A. Nat Rev Genet. 2021;22:119–131. doi: 10.1038/s41576-020-00295-8. [DOI] [PubMed] [Google Scholar]
- 6.Shulman Z, Stern-Ginossar N. The RNA modification N(6)-methyladenosine as a novel regulator of the immune system. Nat Immunol. 2020;21:501–512. doi: 10.1038/s41590-020-0650-4. [DOI] [PubMed] [Google Scholar]
- 7.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 8.Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021;21:22–36. doi: 10.1038/s41568-020-00306-0. [DOI] [PubMed] [Google Scholar]
- 9.Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19:30. doi: 10.1186/s12943-020-1135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das A, Gorospe M, Panda AC. The coding potential of circRNAs. Aging (Albany NY) 2018;10:2228–2229. doi: 10.18632/aging.101554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Ma R, Shi W, Wu Z, Shi Y. Emerging roles of circular RNAs in systemic lupus erythematosus. Mol Ther Nucleic Acids. 2021;24:212–222. doi: 10.1016/j.omtn.2021.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao MS, Ai Y, Wilusz JE. Biogenesis and functions of circular RNAs come into focus. Trends Cell Biol. 2020;30:226–240. doi: 10.1016/j.tcb.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21:475–490. doi: 10.1038/s41580-020-0243-y. [DOI] [PubMed] [Google Scholar]
- 14.Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20:608–624. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 15.Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28:616–624. doi: 10.1038/s41422-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vu LP, Cheng Y, Kharas MG. The biology of m(6)A RNA methylation in Normal and malignant hematopoiesis. Cancer Discov. 2019;9:25–33. doi: 10.1158/2159-8290.CD-18-0959. [DOI] [PubMed] [Google Scholar]
- 17.Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 2019;74:640–650. doi: 10.1016/j.molcel.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021;6:74. doi: 10.1038/s41392-020-00450-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruszkowska A. METTL16, methyltransferase-like protein 16: current insights into structure and function. Int J Mol Sci. 2021;22:2176. doi: 10.3390/ijms22042176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rong B, Zhang Q, Wan J, Xing S, Dai R, Li Y, et al. Ribosome 18S m(6)A methyltransferase METTL5 promotes translation initiation and breast cancer cell growth. Cell Rep. 2020;33:108544. doi: 10.1016/j.celrep.2020.108544. [DOI] [PubMed] [Google Scholar]
- 21.van Tran N, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P, et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019;47:7719–7733. doi: 10.1093/nar/gkz619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinto R, Vagbo CB, Jakobsson ME, Kim Y, Baltissen MP, O'Donohue MF, et al. The human methyltransferase ZCCHC4 catalyses N6-methyladenosine modification of 28S ribosomal RNA. Nucleic Acids Res. 2020;48:830–846. doi: 10.1093/nar/gkz1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren W, Lu J, Huang M, Gao L, Li D, Wang GG, et al. Structure and regulation of ZCCHC4 in m(6)A-methylation of 28S rRNA. Nat Commun. 2019;10:5042. doi: 10.1038/s41467-019-12923-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruzicka K, Zhang M, Campilho A, Bodi Z, Kashif M, Saleh M, et al. Identification of factors required for m(6)A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 2017;215:157–172. doi: 10.1111/nph.14586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, et al. VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10. doi: 10.1038/s41421-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wen J, Lv R, Ma H, Shen H, He C, Wang J, et al. Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol Cell. 2018;69:1028–38.e6. doi: 10.1016/j.molcel.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaccara S, Jaffrey SR. A unified model for the function of YTHDF proteins in regulating m(6)A-modified mRNA. Cell. 2020;181:1582–95.e18. doi: 10.1016/j.cell.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dixit D, Prager BC, Gimple RC, Poh HX, Wang Y, Wu Q, et al. The RNA m6A reader YTHDF2 maintains oncogene expression and is a targetable dependency in Glioblastoma stem cells. Cancer Discov. 2021;11:480–499. doi: 10.1158/2159-8290.CD-20-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27:315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol Cell. 2016;61:507–519. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 34.Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife. 2017;6:e31311. doi: 10.7554/eLife.31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu W, Li J, He C, Wen J, Ma H, Rong B, et al. METTL3 regulates heterochromatin in mouse embryonic stem cells. Nature. 2021;591:317–321. doi: 10.1038/s41586-021-03210-1. [DOI] [PubMed] [Google Scholar]
- 36.Liu J, Dou X, Chen C, Chen C, Liu C, Xu MM, et al. N (6)-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science. 2020;367:580–586. doi: 10.1126/science.aay6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, Gao M, He J, Wu K, Lin S, Jin L, et al. The RNA m(6)A reader YTHDC1 silences retrotransposons and guards ES cell identity. Nature. 2021;591:322–326. doi: 10.1038/s41586-021-03313-9. [DOI] [PubMed] [Google Scholar]
- 38.Li Y, Xia L, Tan K, Ye X, Zuo Z, Li M, et al. N(6)-Methyladenosine co-transcriptionally directs the demethylation of histone H3K9me2. Nat Genet. 2020;52:870–877. doi: 10.1038/s41588-020-0677-3. [DOI] [PubMed] [Google Scholar]
- 39.Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27:1115–1127. doi: 10.1038/cr.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou B, Liu C, Xu L, Yuan Y, Zhao J, Zhao W, et al. N(6) -Methyladenosine reader protein YT521-B homology domain-containing 2 suppresses liver Steatosis by regulation of mRNA stability of Lipogenic genes. Hepatology. 2021;73:91–103. doi: 10.1002/hep.31220. [DOI] [PubMed] [Google Scholar]
- 41.Wojtas MN, Pandey RR, Mendel M, Homolka D, Sachidanandam R, Pillai RS. Regulation of m(6)A transcripts by the 3′-->5′ RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian Germline. Mol Cell. 2017;68:374–87.e12. doi: 10.1016/j.molcel.2017.09.021. [DOI] [PubMed] [Google Scholar]
- 42.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017;45:6051–6063. doi: 10.1093/nar/gkx141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou KI, Shi H, Lyu R, Wylder AC, Matuszek Z, Pan JN, et al. Regulation of co-transcriptional pre-mRNA splicing by m(6)A through the low-complexity protein hnRNPG. Mol Cell. 2019;76:70–81.e9. doi: 10.1016/j.molcel.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alarcon CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015;162:1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang L, Wen M, Cao X. Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science. 2019;365:eaav0758. doi: 10.1126/science.aav0758. [DOI] [PubMed] [Google Scholar]
- 47.Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muller S, Bley N, Busch B, Glass M, Lederer M, Misiak C, et al. The oncofetal RNA-binding protein IGF2BP1 is a druggable, post-transcriptional super-enhancer of E2F-driven gene expression in cancer. Nucleic Acids Res. 2020;48:8576–8590. doi: 10.1093/nar/gkaa653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu Z, Shi Y, Lu M, Song M, Yu Z, Wang J, et al. METTL3 counteracts premature aging via m6A-dependent stabilization of MIS12 mRNA. Nucleic Acids Res. 2020;48:11083–11096. doi: 10.1093/nar/gkaa816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, et al. 5′ UTR m(6)A promotes cap-independent translation. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hwang SY, Jung H, Mun S, Lee S, Park K, Baek SC, et al. L1 retrotransposons exploit RNA m(6)A modification as an evolutionary driving force. Nat Commun. 2021;12:880. doi: 10.1038/s41467-021-21197-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu R, Li A, Sun B, Sun JG, Zhang J, Zhang T, et al. A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 2019;29:23–41. doi: 10.1038/s41422-018-0113-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baquero-Perez B, Antanaviciute A, Yonchev ID, Carr IM, Wilson SA, Whitehouse A. The Tudor SND1 protein is an m(6)A RNA reader essential for replication of Kaposi's sarcoma-associated herpesvirus. Elife. 2019;8:e47261. doi: 10.7554/eLife.47261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, et al. Differential m(6)A, m(6)Am, and m(1)a Demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol Cell. 2018;71:973–85.e5. doi: 10.1016/j.molcel.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang X, Liu QL, Xu W, Zhang YC, Yang Y, Ju LF, et al. m(6)A promotes R-loop formation to facilitate transcription termination. Cell Res. 2019;29:1035–1038. doi: 10.1038/s41422-019-0235-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, et al. m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540:301–304. doi: 10.1038/nature20577. [DOI] [PubMed] [Google Scholar]
- 58.Goh YT, Koh CWQ, Sim DY, Roca X, Goh WSS. METTL4 catalyzes m6Am methylation in U2 snRNA to regulate pre-mRNA splicing. Nucleic Acids Res. 2020;48:9250–9261. doi: 10.1093/nar/gkaa684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, et al. The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM Synthetase intron retention. Cell. 2017;169:824–35.e14. doi: 10.1016/j.cell.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10:4695. doi: 10.1038/s41467-019-12651-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu Y, Yang X, Chen Z, Tian L, Jiang G, Chen F, et al. m(6)A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer. 2019;18:87. doi: 10.1186/s12943-019-1014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561:556–560. doi: 10.1038/s41586-018-0538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millan-Zambrano G, Robson SC, et al. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature. 2017;552:126–131. doi: 10.1038/nature24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang D, Qiao J, Wang G, Lan Y, Li G, Guo X, et al. N6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 2018;46:3906–3920. doi: 10.1093/nar/gky130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang X, Xu Y, Qian Z, Zheng W, Wu Q, Chen Y, et al. circRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis. 2018;9:1091. doi: 10.1038/s41419-018-1132-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, Xu RH. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19:172. doi: 10.1186/s12943-020-01286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ma S, Kong S, Wang F, Ju S. CircRNAs: biogenesis, functions, and role in drug-resistant Tumours. Mol Cancer. 2020;19:119. doi: 10.1186/s12943-020-01231-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li J, Sun D, Pu W, Wang J, Peng Y. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer. 2020;6:319–336. doi: 10.1016/j.trecan.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 70.Huang C, Liang D, Tatomer DC, Wilusz JE. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018;32:639–644. doi: 10.1101/gad.314856.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang S, Zhang K, Tan S, Xin J, Yuan Q, Xu H, et al. Circular RNAs in body fluids as cancer biomarkers: the new frontier of liquid biopsies. Mol Cancer. 2021;20:13. doi: 10.1186/s12943-020-01298-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang L, Han B, Zhang Z, Wang S, Bai Y, Zhang Y, et al. Extracellular vesicle-mediated delivery of circular RNA SCMH1 promotes functional recovery in rodent and nonhuman primate ischemic stroke models. Circulation. 2020;142:556–574. doi: 10.1161/CIRCULATIONAHA.120.045765. [DOI] [PubMed] [Google Scholar]
- 74.Jie M, Wu Y, Gao M, Li X, Liu C, Ouyang Q, et al. CircMRPS35 suppresses gastric cancer progression via recruiting KAT7 to govern histone modification. Mol Cancer. 2020;19:56. doi: 10.1186/s12943-020-01160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Conn VM, Hugouvieux V, Nayak A, Conos SA, Capovilla G, Cildir G, et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants. 2017;3:17053. doi: 10.1038/nplants.2017.53. [DOI] [PubMed] [Google Scholar]
- 76.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 77.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 78.Jiang MP, Xu WX, Hou JC, Xu Q, Wang DD, Tang JH. The emerging role of the interactions between circular RNAs and RNA-binding proteins in common human cancers. J Cancer. 2021;12:5206–5219. doi: 10.7150/jca.58182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zheng S, Zhang X, Odame E, Xu X, Chen Y, Ye J, et al. CircRNA-protein interactions in muscle development and diseases. Int J Mol Sci. 2021;22:3262. doi: 10.3390/ijms22063262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10:3503–3517. doi: 10.7150/thno.42174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zang J, Lu D, Xu A. The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function. J Neurosci Res. 2020;98:87–97. doi: 10.1002/jnr.24356. [DOI] [PubMed] [Google Scholar]
- 82.Tsitsipatis D, Grammatikakis I, Driscoll RK, Yang X, Abdelmohsen K, Harris SC, et al. AUF1 ligand circPCNX reduces cell proliferation by competing with p21 mRNA to increase p21 production. Nucleic Acids Res. 2021;49:1631–1646. doi: 10.1093/nar/gkaa1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lou J, Hao Y, Lin K, Lyu Y, Chen M, Wang H, et al. Circular RNA CDR1as disrupts the p53/MDM2 complex to inhibit Gliomagenesis. Mol Cancer. 2020;19:138. doi: 10.1186/s12943-020-01253-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu F, Zhang Y, Wang Z, Gong W, Zhang C. Hsa_circ_0030042 regulates abnormal autophagy and protects atherosclerotic plaque stability by targeting eIF4A3. Theranostics. 2021;11:5404–5417. doi: 10.7150/thno.48389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li B, Zhu L, Lu C, Wang C, Wang H, Jin H, et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat Commun. 2021;12:295. doi: 10.1038/s41467-020-20527-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu P, Mo Y, Peng M, Tang T, Zhong Y, Deng X, et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol Cancer. 2020;19:22. doi: 10.1186/s12943-020-1147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liang WC, Wong CW, Liang PP, Shi M, Cao Y, Rao ST, et al. Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20:84. doi: 10.1186/s13059-019-1685-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- 90.Xia X, Li X, Li F, Wu X, Zhang M, Zhou H, et al. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol Cancer. 2019;18:131. doi: 10.1186/s12943-019-1056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z, Xu B, et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via hippo-YAP signaling. Mol Cancer. 2019;18:47. doi: 10.1186/s12943-019-1010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun. 2018;9:4475. doi: 10.1038/s41467-018-06862-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kleaveland B, Shi CY, Stefano J, Bartel DP. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell. 2018;174:350–62.e17. doi: 10.1016/j.cell.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fischer JW, Busa VF, Shao Y, Leung AKL. Structure-mediated RNA decay by UPF1 and G3BP1. Mol Cell. 2020;78:70–84.e6. doi: 10.1016/j.molcel.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu CX, Li X, Nan F, Jiang S, Gao X, Guo SK, et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177:865–80.e21. doi: 10.1016/j.cell.2019.03.046. [DOI] [PubMed] [Google Scholar]
- 97.Li Z, Yang HY, Dai XY, Zhang X, Huang YZ, Shi L, et al. CircMETTL3, upregulated in a m6A-dependent manner, promotes breast cancer progression. Int J Biol Sci. 2021;17:1178–1190. doi: 10.7150/ijbs.57783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen C, Yuan W, Zhou Q, Shao B, Guo Y, Wang W, et al. N6-methyladenosine-induced circ1662 promotes metastasis of colorectal cancer by accelerating YAP1 nuclear localization. Theranostics. 2021;11:4298–4315. doi: 10.7150/thno.51342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu P, Fang X, Liu Y, Tang Y, Wang W, Li X, et al. N6-methyladenosine modification of circCUX1 confers radioresistance of hypopharyngeal squamous cell carcinoma through caspase1 pathway. Cell Death Dis. 2021;12:298. doi: 10.1038/s41419-021-03558-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xu J, Wan Z, Tang M, Lin Z, Jiang S, Ji L, et al. N(6)-methyladenosine-modified CircRNA-SORE sustains sorafenib resistance in hepatocellular carcinoma by regulating beta-catenin signaling. Mol Cancer. 2020;19:163. doi: 10.1186/s12943-020-01281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, et al. Endoribonucleolytic cleavage of m(6)A-containing RNAs by RNase P/MRP Complex. Mol Cell. 2019;74:494–507.e8. doi: 10.1016/j.molcel.2019.02.034. [DOI] [PubMed] [Google Scholar]
- 102.Li X, Tian G, Wu J. Novel circGFRalpha1 promotes self-renewal of female Germline stem cells mediated by m(6)A writer METTL14. Front Cell Dev Biol. 2021;9:640402. doi: 10.3389/fcell.2021.640402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L, et al. N6-Methyladenosine modification controls circular RNA immunity. Mol Cell. 2019;76:96–109.e9. doi: 10.1016/j.molcel.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mo WL, Deng LJ, Cheng Y, Yu WJ, Yang YH, Gu WD. Circular RNA hsa_circ_0072309 promotes tumorigenesis and invasion by regulating the miR-607/FTO axis in non-small cell lung carcinoma. Aging (Albany NY) 2021;13:11629–11645. doi: 10.18632/aging.202856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chi F, Cao Y, Chen Y. Analysis and validation of circRNA-miRNA network in regulating m(6)A RNA methylation modulators reveals CircMAP2K4/miR-139-5p/YTHDF1 Axis involving the proliferation of hepatocellular carcinoma. Front Oncol. 2021;11:560506. doi: 10.3389/fonc.2021.560506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang Z, Zhu H, Hu J. CircRAB11FIP1 promoted autophagy flux of ovarian cancer through DSC1 and miR-129. Cell Death Dis. 2021;12:219. doi: 10.1038/s41419-021-03486-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jiang X, Xing L, Chen Y, Qin R, Song S, Lu Y, et al. CircMEG3 inhibits telomerase activity by reducing Cbf5 in human liver cancer stem cells. Mol Ther Nucleic Acids. 2021;23:310–323. doi: 10.1016/j.omtn.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shen Y, Li C, Zhou L, Huang JA. G protein-coupled oestrogen receptor promotes cell growth of non-small cell lung cancer cells via YAP1/QKI/circNOTCH1/m6A methylated NOTCH1 signalling. J Cell Mol Med. 2021;25:284–296. doi: 10.1111/jcmm.15997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu B, Liu N, Zhu X, Yang L, Ye B, Li H, et al. Circular RNA circZbtb20 maintains ILC3 homeostasis and function via Alkbh5-dependent m(6)A demethylation of Nr4a1 mRNA. Cell Mol Immunol. 2021;18:1412–1424. doi: 10.1038/s41423-021-00680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Huang R, Zhang Y, Bai Y, Han B, Ju M, Chen B, et al. N(6)-Methyladenosine modification of fatty acid amide hydrolase messenger RNA in circular RNA STAG1-regulated astrocyte dysfunction and depressive-like behaviors. Biol Psychiatry. 2020;88:392–404. doi: 10.1016/j.biopsych.2020.02.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.