Abstract
The use of enzymes in industrial processes is often limited by the unavailability of biocatalysts with prolonged stability. Thermostable enzymes allow increased process temperature and thus higher substrate and product solubility, reuse of expensive biocatalysts, resistance against organic solvents, and better “evolvability” of enzymes. In this work, we have used an activity-independent method for the selection of thermostable variants of any protein in Thermus thermophilus through folding interference at high temperature of a thermostable antibiotic reporter protein at the C-terminus of a fusion protein. To generate a monomeric folding reporter, we have increased the thermostability of the moderately thermostable Hph5 variant of the hygromycin B phosphotransferase from Escherichia coli to meet the method requirements. The final Hph17 variant showed 1.5 °C higher melting temperature (Tm) and 3-fold longer half-life at 65 °C compared to parental Hph5, with no changes in the steady-state kinetic parameters. Additionally, we demonstrate the validity of the reporter by stabilizing the 2-keto-3-deoxy-l-rhamnonate aldolase from E. coli (YfaU). The most thermostable multiple-mutated variants thus obtained, YfaU99 and YfaU103, showed increases of 2 and 2.9 °C in Tm compared to the wild-type enzyme but severely lower retro-aldol activities (150- and 120-fold, respectively). After segregation of the mutations, the most thermostable single variant, Q107R, showed a Tm 8.9 °C higher, a 16-fold improvement in half-life at 60 °C and higher operational stability than the wild-type, without substantial modification of the kinetic parameters.
Keywords: aldolase, directed evolution, hygromycin B phosphotransferase, in vivo selection, thermostability, Thermus thermophilus
Short abstract
This work demonstrates a method for selecting thermostable variants of any protein of interest to be used in biocatalysis, increasing the sustainability of industrial chemical transformations.
Introduction
Reaction conditions of enzymes in industrial biocatalysis are usually far from those in nature: non-natural substrates are used in high concentrations while higher temperatures and organic cosolvents are needed to promote substrate and product solubility. In this context, enzyme engineering constitutes an efficient methodology to tailor enzyme activity, substrate selectivity, or stability under operational conditions to each industrial process.1
The rational prediction of thermostability is a complex task because methods are based on different structure–function hypotheses, leading to different solutions, which in many cases do not result in direct increases in stability.2 Therefore, directed evolution is preferred, since it allows exploration of a large sequence space (in the range of 106 to 109 individuals),3 albeit at the cost of increasing the screening effort to cover a meaningful fraction of this man-made diversity.
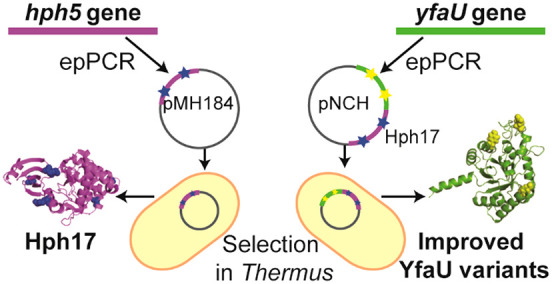
Screening for thermostable enzyme variants in large libraries can be carried out in a thermophile, provided its growth is coupled to the stability of the target protein.4 In 2007, we reported a procedure for the in vivo selection of thermostable variants of any protein independently of its activity using Thermus thermophilus as a host.5 The method was based on the folding interference phenomenon that occurs in a protein fusion between a thermosensitive target protein in the N-terminus and a thermostable kanamycin nucleotidyl transferase4 (Kat) in the C-terminus (Figure S1). This method has proven useful for the isolation of thermostable variants of human interferons and enzymes for biocatalysis, such as lipase A from Bacillus subtilis, formate dehydrogenase from Pseudomonas sp. 101,5 and more recently, the esterase I from Pseudomonas fluorescens.6
In the course of generating thermostable variants of the latter enzyme, we encountered a large number of false positives that we attributed to having used a dimeric folding interference reporter, such as Kat. Therefore, we evolved the monomeric, moderately thermostable hygromycin B phosphotransferase variant (Hph5) from Escherichia coli reported by Nakamura et al.7 Hph5 accumulated five amino acid substitutions that allowed T. thermophilus to grow at temperatures up to 67 °C. However, lower transformation efficiency of this marker in Thermus had been reported at that temperature,7 compromising the throughput of our selection method as well as limiting the selection pressure, i.e. temperature, that can be applied.
Consequently, in this work we engineered a bespoke, highly thermostable, monomeric folding reporter (Hph17) and used it to stabilize the E. coli 2-keto-3-deoxy-l-rhamnonate aldolase (YfaU). YfaU is a class II pyruvate aldolase that accepts a wide range of electrophiles, and even though the natural nucleophilic substrate is pyruvate, it can also use homologous ketoacids. The aldol addition of pyruvate or homologues to a wide variety of N-carboxybenzyl-amino aldehydes are especially relevant since the resulting aldol adducts are intermediates of new proline, pyrrolizidine-3-carboxylic acid, pipecolic acid, and β-hydroxy-γ-amino acid derivatives.8,9 Moreover, YfaU plays an important role in the biocatalytic cascade for the synthesis of the noncanonical amino acid (S)-2-amino-4-hydroxybutanoic acid (l-homoserine). YfaU can synthesize (S)- or (R)-2-amino-4-hydroxybutanoic acid with ee values of >99% using pyruvate and formaldehyde as substrates, and a transaminase provides pyruvate from alanine, thus l-homoserine is produced using formaldehyde and alanine as sole and inexpensive starting materials10
Results and Discussion
Library Generation and Selection of Hph Variants
In order to improve the stability of Hph5 for its use as a folding interference reporter, the hph5 gene was randomized by error-prone PCR (epPCR) in the presence of 0.2 mM Mn2+ to introduce 3–6 nucleotide replacements per gene, which represent between 2 and 5 amino acid changes, in good agreement with most directed evolution studies.11 The epPCR Hph5 library was generated in E. coli and then transformed in T. thermophilus for selection. The generated E. coli library of 4.5 × 104 individuals was selected at 70 °C and 100 μg/mL of hygromycin B (HygB), at which transformants expressing parental Hph5 could not grow (Figure S2). Under permissive conditions (60 °C and 100 μg/mL of HygB), 9961 CFU/ng plasmid were obtained, while under selection pressure (70 °C and 100 μg/mL of HygB) only 32 CFU/ng of plasmid were selected, which represents a selection factor of 0.32%. Because of the high number of transformants obtained under those conditions, the temperature had to be subsequently increased to 71 °C, leading to 2 CFU/ng plasmid and a selection factor of 0.02%.
Twenty randomly selected clones were verified for HygB resistance using a serial dilution assay at 71 °C (Figure S3, A). A particular variant (Hph17) harboring five changes (R61H, S86G, Q96P, A185V, and V322E) was found four times in the pool and enabled growth of Thermus even at 74 °C (Figure S3B). It seems unlikely that all of these four individuals originated independently during epPCR, but their recurrence is likely a natural consequence of library construction in E. coli prior to selection in Thermus. Most importantly, unlike the in vivo mutagenesis used by Nakamura to generate Hph5,7in vitro mutagenesis by epPCR likely enabled the creation of a larger and more diverse sequence space, from which a fitter variant can be selected. In fact, it took a combination of natural evolution, DNA shuffling, and random amino acid duplications to confer a similar degree of thermostability to a hygromycin phosphotransferase from Streptomyces hygroscopicus (Hyg10).12
Kinetic, Thermodynamic, and Structural Characterization of Hph17
The mutations of the moderately thermostable Hph5 were mostly situated in the hydrophobic core.13 In contrast, three out of the four thermostabilizing positions mutated in the Hph17 variant (R61H, S86G, Q96P, and A185V) are found on the protein surface, except A185V, which is located in a hydrophobic core (Figure 1), reducing the distance between the adjacent β-strands and contributing toward compactness.14 On the other hand, residue 96 is placed in a loop and substitution of Gln to Pro in a loop diminishes the RMSD of that region, contributing to the overall stabilization of the enzyme. Finally, A185V strengthens the hydrophobic interactions and increases the protein packing3 since Val is bulkier than Ala.
Figure 1.
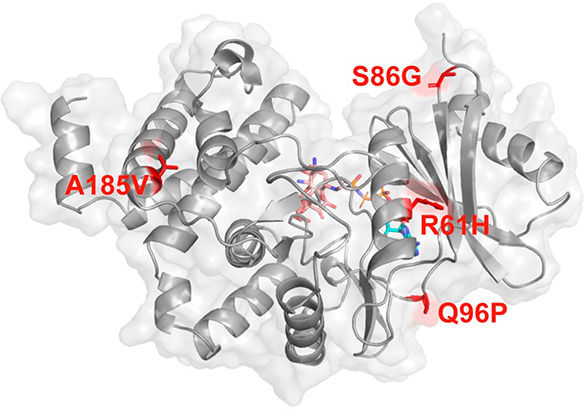
Location of stabilizing mutations in Hph17 variant. Amino acid substitutions are shown as red sticks. Substrates hygromycin B (HygB) and phosphoaminophosphonic acid-adenylate ester (ANP) are depicted as sticks in CPK colors with carbon atoms in salmon and cyan, respectively.
As shown in Table 1, both the catalytic constant, kcat, and the efficiency for ATP, KM,ATP, remained unaltered in Hph17 compared with the parental enzyme. However, KM,HygB, was 2.4-fold higher for Hph17 respect to that of Hph5, with a consequent reduction in the catalytic efficiency. Regarding thermostability, the melting temperature of Hph17 was 1.5 °C higher than that of Hph5, while its half-life at 65 °C doubled, with the main contributions toward this enhancement originating from replacements S86G and Q96P. Increases in kinetic stability usually suggest that these mutations may interfere with an initial step on the path toward the irreversible unfolded state, thus avoiding further global unfolding.15,16 Therefore, we used constraint network analysis (CNA), to simulate protein unfolding.17 As shown in Figure 2, positions Ser86 and Gln96 were some of the most flexible loci in the protein (highest ri values), congruently with the postulated “hinge” function of neighboring Val98.13 Thus, replacing Ser86 and Gln96 would restrict local movements leading to unfolded states by irreversible denaturation, which might explain the increase in half-life of variants S86G and Q96P (Table 1).
Table 1. Kinetica and Stability Parameters for Hph5, Hph17, and the Segregated Variants Containing the Amino Acid Replacements of Hph17b.
| Hph variant | KM,HygB (mM) | KM,ATP (mM) | kcat (min–1) | kcat/KM,HygB (min–1 mM–1) | kcat/KM,ATP (min–1 mM–1) | Tm (°C) | half-life (min) | kd (min–1) |
|---|---|---|---|---|---|---|---|---|
| Hph5 | 0.29 ± 0.03 | 0.37 ± 0.02 | 2984 ± 72 | 10290 | 8065 | 58.2 ± 0.1 | 2.2 ± 0.4 | 0.33 ± 0.06 |
| Hph17 | 0.7 ± 0.10 | 0.39 ± 0.03 | 2847 ± 153 | 4067 | 7300 | 59.7 ± 0.1 | 7 ± 3 | 0.12 ± 0.05 |
| R61H | 0.84 ± 0.09 | 0.46 ± 0.04 | 3578 ± 133 | 4260 | 7778 | 59.5 ± 0.1 | 2.2 ± 0.2 | 0.31 ± 0.03 |
| S86G | 0.50 ± 0.07 | 0.55 ± 0.03 | 3202 ± 44 | 6404 | 5822 | 59.6 ± 0.3 | 4.7 ± 0.9 | 0.15 ± 0.04 |
| Q96P | 0.41 ± 0.03 | 0.31 ± 0.03 | 3555 ± 76 | 8671 | 11468 | 59.5 ± 0.2 | 3.0 ± 0.1 | 0.23 ± 0.01 |
| A185V | 0.39 ± 0.06 | 0.32 ± 0.02 | 2672 ± 108 | 6851 | 8350 | 57.9 ± 0.1 | 1.6 ± 0.2 | 0.43 ± 0.06 |
| V322E | 0.57 ± 0.09 | 0.39 ± 0.02 | 3196 ± 145 | 5607 | 8195 | 59.1 ± 0.4 | 1.9 ± 0.4 | 0.38 ± 0.08 |
Steady-state kinetic constants were determined at 60 °C.
Half-lives and deactivation constants (Kd) were determined at 65 °C. Values represent the mean ± standard deviation of three independent determinations.
Figure 2.
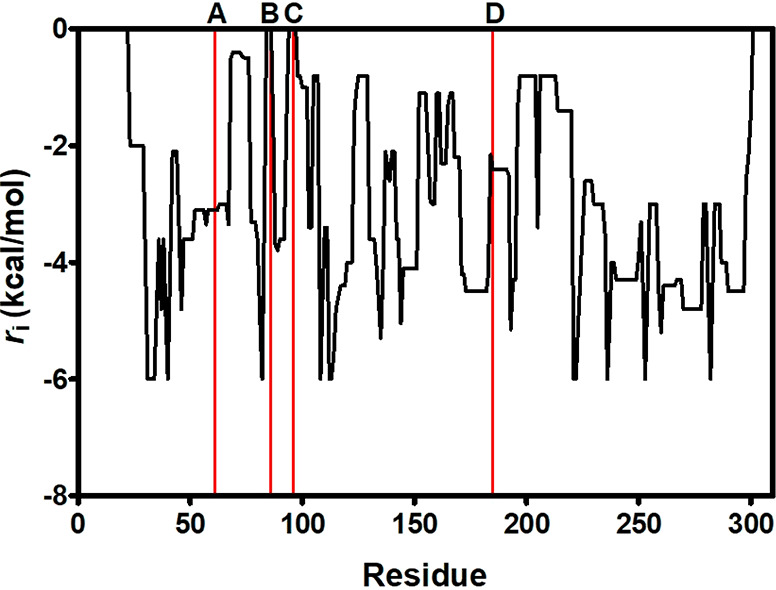
Constraint network analysis of Hph5. Lines indicate the four amino acid replacements in Hph17 (line A corresponds to position Arg61; line B, Ser86; line C, Gln96; and line D, Ala185) that can be mapped in the homology model.
When Hph5 was evolved from Hph, a marked increase in thermodynamic stability was observed, despite the lack of a clear structural explanation.13,18 However, neither Hph17 nor the individual variants showed an increase in melting temperature (Tm) over the parental Hph5, suggesting that a further increase of protein rigidity significant enough to gain thermodynamic stability could be detrimental for the enzyme activity. This result is not incompatible with the putative higher protein stability in vivo, which could be enhanced by factors such as the molecular crowding and compatible solutes of the Thermus cytoplasm19 whereas Tm determinations are carried out with the protein in buffer. In contrast, different scaffolds, such as Hyg10, have been evolved to both higher Tm (12.2 °C) and specific activity (2-fold) at the optimum activity temperature.12 However, the sequence identity of the Hyg10 and Hph proteins is approximately 30%, and their activity is not identical, as Hyg10 phosphorylates HygB in a different hydroxyl group.
The folding free energy of the mutants (ΔΔG) was estimated using FoldX. The ΔΔG values obtained were 0.78, 0.50, −0.92, 0.21, and 0.58 kcal/mol for the single variants R61H, S86G, Q96P, A185V, and the multiple mutant containing R61H, S86G, Q96P, and A185V, respectively. In this case, FoldX cannot predict correctly the found mutants given the very low differences in Tm between variants, and the standard deviation of predicted values (between 1.0 and 1.7 kcal/mol).2
Library Generation and Selection of Thermostable YfaU Variants
Hph17 was used as a folding interference reporter to engineer higher stability in YfaU (Figures 3 and S1). The class II pyruvate aldolase, YfaU, was mutagenized by epPCR in the presence of 0.3 mM Mn2+. The library sequences analyzed contained between 1 and 8 nucleotide replacements, i.e., 1–6 amino acid substitutions. The generated library of 1.5 × 105 individuals was selected in T. thermophilus at 67 °C and 100 μg/mL of HygB, conditions under which the transformants expressing the wild-type YfaU (YfaU-wt) could not grow (Figure S4).
Figure 3.
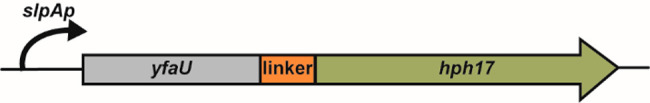
Gene fusion of yfaU to hph17, expressed under the promoter slpA (slpAp).
Due to the large number of variants selected, 54 unique clones were randomly picked to perform a dilution assay on plate at 67 °C (Figure S5). The 12 variants with the highest growth (variants 2, 8, 14, 15, 48, 50, 63, 66, 70, 99, 103, and 105) were chosen for subsequent sequencing and characterization.
Characterization of Thermostable YfaU Variants
The 12 selected YfaU variants and YfaU-wt were cloned into pET28b, transformed in E. coli BL21, and expressed using autoinduction medium at 20 °C. The solubility of these variants was checked by SDS-PAGE; supernatant and pellet were run separately (Figure S6). Only four of the variants showed the presence of the protein in the supernatant fraction. The lack of solubility of these putative thermostable YfaU variants could arise from differences between the context in which they were selected and produced,27 i.e., a fusion protein in a thermophile host vs a standalone protein in a mesophile. Also, the low solubility of YfaU has been previously described, requiring expression in fusion with either dihydrofolate reductase (DHFR) or maltose binding protein (MBP) at the N-terminus.10
These four YfaU variants and YfaU-wt were purified by immobilized metal affinity chromatography (IMAC), and their Tm values were measured. Variants 2 (H49Q and G118D) and 14 (G39D and I73F) showed Tms 8.5 and 5.5 °C lower than YfaU-wt, while variants 99 (L4F, G90S, Q107R, Q141L, F215L, A252E, F254I, and I263 K) and 103 (V122F, P187T, and P261Q) increased their Tms by 2.0 and 2.9 °C, respectively, compared with YfaU-wt. However, the assessment of variants 99 and 103 using a straightforward retro-aldol reaction showed a 150- and 120-fold reduction in activity, respectively (Table 2). These results agree with previous studies of randomized libraries, in which an increase in thermal stability resulted in lower activity,20,21 due to a gradual loss of flexibility as the number of mutations increases.22 Furthermore, selection by folding interference is an activity-independent process, which may be convenient in cases where a functional selection is either complex or impossible5 but, in this case, led to lower activity values due to lack of selective pressure towards function.
Table 2. Specific Retro-Aldol Activity,aTm, and Half-Lifeb of YfaU Variants.
| YfaU variant | specific retro-aldol activity (U/mg) | Tm (°C) | half-life (min) |
|---|---|---|---|
| wild-type | 60 ± 1 | 60.5 ± 0.0 | 0.9 ± 0.1 |
| YfaU99 | 0.4 ± 0.2 | 62.5 ± 1.0 | nm |
| YfaU103 | 0.5 ± 0.3 | 63.4 ± 0.2 | nm |
| Q107R | 48 ± 7 | 69.4 ± 0.2 | 14 ± 1 |
| Q141L | 62 ± 6 | 62.7 ± 0.2 | 3.0 ± 0.3 |
Specific activity was determined at 25 °C.
Half-life was measured at 60 °C. Values represent the mean and the standard deviation of three independent determinations. nm: nonmeasurable.
To remediate the observed activity–stability trade-off, the amino acid replacements of these two variants were segregated and their Tms and specific activities were measured individually (Table 2). Only variants Q107R and Q141L (both derived from variant 99) increased their Tms by 8.9 and 2.2 °C, respectively, compared to YfaU-wt while increasing or maintaining the retro-aldol activities of the wild-type enzyme. In addition, the half-lives of Q107R and Q141L were 16- and 3.3-fold higher compared to YfaU-wt, respectively. Considering that only 0.01–0.5% of random mutations are beneficial,23 the increase in thermostability of Q107R seems to arise from a truly beneficial mutation, and the rest of the mutations in variant 99 have a deleterious or neutral effect on enzyme stability.
Performance of YfaU Q107R and Q141L in the Aldol Addition of Pyruvate to Formaldehyde
To test the proficiency of the best YfaU variants in a biocatalytically relevant reaction, the aldol addition of pyruvate to formaldehyde was assayed, modeled and the steady-state kinetic parameters were calculated for YfaU-wt, Q107R, and Q141L (Table 3 and Figure S7).
Table 3. Steady-State Kinetic Parameters for the Aldol Addition of Pyruvate to Formaldehyde Catalyzed by YfaU-wt, Q107R, and Q141La.
| YfaU variant | kcat (min–1) | KM,formaldehyde (mM) | kcat/KM,formaldehyde (min–1 mM–1) | Ki,formadehyde (mM) | KM,pyruvate (mM) | kcat/KM,pyruvate (min–1 mM–1) | Ki,pyruvate (mM) |
|---|---|---|---|---|---|---|---|
| wild-type | 113 ± 40 | 24 ± 4 | 4.71 | 95 ± 14 | 209 ± 83 | 0.54 | 47 ± 18 |
| Q107R | 180 ± 53 | 26 ± 6 | 6.92 | 75 ± 16 | 61 ± 20 | 2.95 | 46 ± 15 |
| Q141L | 242 ± 28 | 17 ± 2 | 14.2 | 113 ± 12 | 66 ± 10 | 3.67 | 151 ± 22 |
Values represent the mean and the standard deviation of three independent determinations.
The ca. 2-fold increase in kcat and decrease of KM for both substrates for variant Q141L resulted in a 3.1- and 6.8-fold increase in catalytic efficiency (kcat/KM) for formaldehyde and pyruvate, respectively. Moreover, Ki for both substrates increased. Variant Q107R showed better turnover and similar KM values compared to YfaU-wt, while Ki for formaldehyde decreased.
The operational stability of the Q107R and Q141L variants in this reaction was evaluated in a batch reactor. Assuming that the decay in operational stability can be described by first order kinetics (Figure S8), the calculated deactivation constants (kd) for variants Q107R and Q141L are approximately 2-fold lower than for YfaU-wt (Table 4). Both variants showed similar values in terms of operational stability (kd and half-life), which contrasts with their differences in kinetic thermostability, where Q107R showed a half-life at 60 °C almost 5-fold higher than Q141L (Table 2). These differences between kinetic and operational stability could be explained by the fact that half-life at high temperature considers only the stability of the protein molecule in buffer, while operational stability considers enzyme activity in the reactor in the presence of substrate, cofactor, and products.24
Table 4. Estimated Values of Operational Stability Decay Rate Constants (kd) and the Corresponding Half-Life Times in a Batch Reactor at 25 °Ca.
| YfaU variant | kd (h–1) | half-life (h) |
|---|---|---|
| wild-type | 0.144 ± 0.013 | 4.8 ± 0.4 |
| Q107R | 0.068 ± 0.006 | 10.2 ± 0.9 |
| Q141L | 0.061 ± 0.006 | 11 ± 1 |
Values represent the mean and the standard deviation of three independent determinations.
Structure–Function Analysis of YfaU Q107R and Q141L
To investigate the reason why both mutants were more thermostable, homology models of Q107R and Q141L were built using the crystal structures of YfaU-wt (PDB: 2VWS and 2VWT). YfaU presents a hexameric assembly composed by a trimer (3-fold axis) of (β/α)8 barrel dimers (2-fold axis). Since the 2-fold related subunits superpose with an RMSD of 0.25 Å25 and residues Gln107 and Gln141 are not involved in the interaction between subunits, only the 3-fold related subunits were considered for the analysis (Figure S9). The replacement Q107R decreased the number of H-bonds with the replaced residue or with other amino acids in its hydrogen bond network. Similar results were found for the mutant Q141L (Figure 4).
Figure 4.
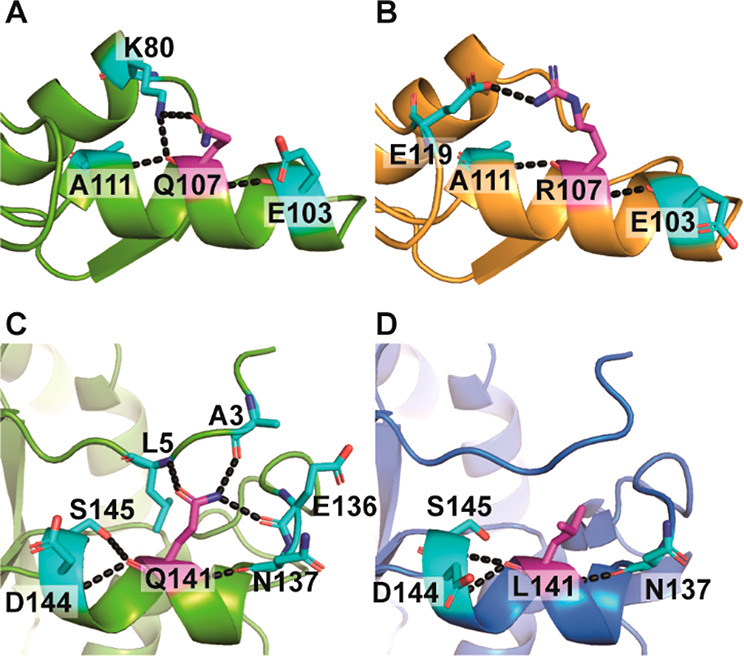
Hydrogen bonds formed between residues 107 and 141 with their surrounding residues, respectively. A. Residue Gln107 of YfaU-wt. B. Residue Arg107 of variant Q107R. C. Residue Gln141 of YfaU-wt. D. Residue Leu141 of variant Q141L. Target residues are depicted in magenta sticks in CPK colors, while residues which form hydrogen bonds are illustrated as cyan sticks.
Rigidity index (ri) from the CNA algorithm was also used to monitor the degree of rigidity of the residues from YfaU-wt (Figure 5). As previously described, only the trimeric assembly was considered for structural analysis. Considering this and since CNA does not relate residues from different chains, ri has been averaged from the three different chains. According to CNA, with a ri value of −2.8 kcal/mol, residue Gln107 is not in a flexible region of the protein. However, residue Gln141 has a ri value of −0.84 kcal/mol, which implies a certain degree of flexibility in this region.
Figure 5.
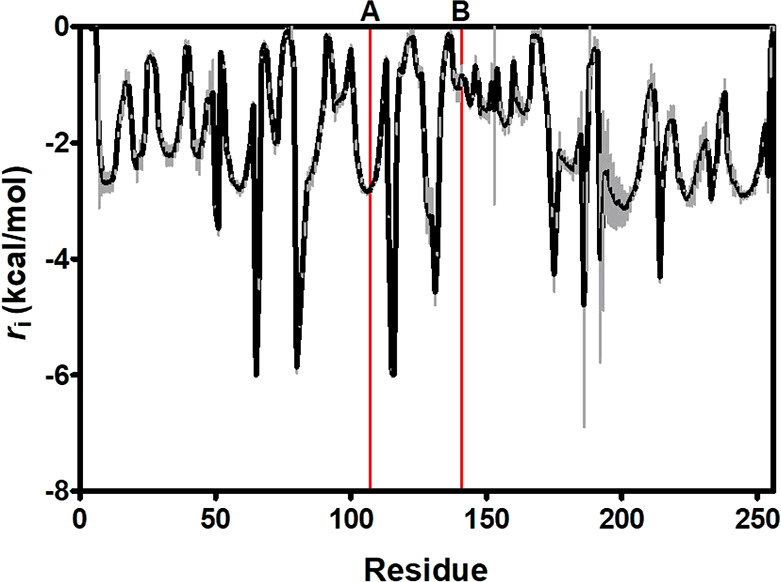
Rigidity index (ri) for YfaU-wt calculated by CNA. Lines indicate the amino acid replacements (line A corresponds to position 107 and line B, position 141). Since YfaU is a homotrimer, ri has been averaged for the same residue of each chain. The mean value is shown in black, and the standard deviations are in gray.
Finally, FoldX calculations were carried out to estimate the folding free energy of the mutants (ΔΔG). Q107R caused a ΔΔG of −2.55 kcal/mol. Considering ΔΔG from FoldX and the general rule that correlates ΔGunfold and ΔTm,26 the corresponding empirical ΔTm would be 9.2 °C, which is similar to the experimental ΔTm, 8.9 °C. By contrast, ΔΔG of variant Q141L was 0.08 kcal/mol, which would represent a ΔTm of −0.3 °C, while the experimental ΔTm was 2.2 °C.
Considering the output of the chosen methods and algorithms used for structure–function analysis, we could identify beneficial mutations using our screening system, which would not be made easily evident by bioinformatics tools. However, the folding interference principle in T. thermophilus allowed the identification of these stabilizing positions, in consonance with a recent study in which stabilizing positions were identified in the esterase I from Pseudomonas fluorescens also by folding interference, using the kanamycin nucleotidyl tranferase gene as folding reporter instead.6
Conclusions
The improvement of hygromycin B phosphotransferase (Hph17) enabled the thermal stabilization of the pyruvate aldolase from E. coli YfaU. The only two selected variants that were expressed in soluble form, YfaU99 and 103, showed higher Tm than the wild-type protein, 2.0 and 2.9 °C, respectively, at the cost of a lower specific activity. However, the low solubility issue can be solved using complementary rational design strategies, such as specific solubility-enhancing algorithms or back-to-consensus mutations that restore conserved amino acids, which usually yield active and more soluble proteins.24,26
With the aim of knowing the effect of individual mutations both in enzyme activity and stability, all mutations were segregated and characterized individually. The Q107R and Q141L replacements conferred higher kinetic and thermodynamic stability. Especially interesting is the case of variant Q107R, with an increase of 8.9 °C in Tm, 16-fold longer half-life, and similar kinetic constants than YfaU-wt. Regarding variant Q141L, the improvement in stability was much more modest, but this variant had better turnover, affinity, and lower substrate inhibition compared to the wild-type.
YfaU is a relevant enzyme for biocatalysis, allowing for instance the synthesis of l-homoserine using alanine and up to 3 M formaldehyde, when coupled with a transaminase.10 Our highly active and thermostable Q107R and Q141L variants have twice the operational stability of YfaU-wt in the synthesis of 4-hydroxy-2-oxobutanoate, which would allow a longer-term usage in this cascade reaction, with the consequent reduction in the cost of the process.
Acknowledgments
This work has been funded through the European Union’s Research and Innovation program Horizon 2020 through grant agreement no. 635595 (CarbaZymes) and by the Spanish Ministry of Economy and Competitiveness through grant BIO-2013-44963R. Institutional grants from the Fundación Ramón Areces and Banco Santander to the CBMSO are also acknowledged. S.B. is the recipient of a Ph.D. fellowship from UAM. D.M.M. was supported by a Research Talent Attraction contract from the Community of Madrid. A generous allocation of computing time at the Scientific Computation Center of the UAM (CCC-UAM) is also acknowledged.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssuschemeng.1c00699.
Tables, figures, and experimental procedures (PDF)
Author Present Address
⊥ E.S.-F.: Francisco de Vitoria University, Carretera de Pozuelo a Majadahonda Km 1,800, 28223 Madrid, Spain. Email: esther.sanchez@ufv.es.
Author Present Address
○ M.L.d.P.: Institute of Organic Chemistry. Spanish National Research Council (IQAC-CSIC), Juan de la Cierva 3, 28006 Madrid, Spain. Email: mldelpozo@mncn.csic.es.
Author Present Address
■ J.Q.: Section for GeoGenetics, Globe Institute, University of Copenhagen, Øster Voldgade 5-7, Copenhagen, Denmark. Email: jwm781@alumni.ku.dk.
Author Present Address
# M.C̆.: Ekonerg d.o.o., Koranska ul. 5, 10000, Zagreb. Email: morana.cesnik@ekonerg.hr.
Author Contributions
S.B. performed the experiments on library generation, selection, and characterization of Hph and YfaU variants, compiled and analyzed all data, and wrote the first version of the manuscript. J.Q. did the characterization of the Hph single variants. M.L.d.P. contributed to the experiments on library generation and selection of YfaU. E.S.-F. contributed to the characterization of the YfaU variants. M.C. performed the modeling of the steady-state kinetic parameters and operational stability of YfaU. Y.Q. did the production of pyruvate kinase. S.B. and D.M.M. contributed to data analysis, figure design, writing, and editing of the manuscript. K.H., P.C., D.V.-R. Z.F.B., and J.B. contributed to manuscript writing. A.H. conceived and supervised the project and wrote the final version of the manuscript. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Woodley J. M. Protein Engineering of Enzymes for Process Applications. Curr. Opin. Chem. Biol. 2013, 17 (2), 310–316. 10.1016/j.cbpa.2013.03.017. [DOI] [PubMed] [Google Scholar]
- Buß O.; Rudat J.; Ochsenreither K. FoldX as Protein Engineering Tool: Better Than Random Based Approaches?. Comput. Struct. Biotechnol. J. 2018, 16, 25–33. 10.1016/j.csbj.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mate D. M.; Gonzalez-Perez D.; Mateljak I.; Gomez de Santos P.; Vicente A. I.; Alcalde M. The Pocket Manual of Directed Evolution: Tips and Tricks. In Biotechnology of Microbial Enzymes: Production, Biocatalysis and Industrial Applications 2017, 185–213. 10.1016/B978-0-12-803725-6.00008-X. [DOI] [Google Scholar]
- Matsumura M.; Aiba S. Screening for Thermostable Mutant of Kanamycin Nucleotidyltransferase by the Use of a Transformation System for a Thermophile, Bacillus stearothermophilus. J. Biol. Chem. 1985, 260 (28), 15298–15303. 10.1016/S0021-9258(18)95735-2. [DOI] [PubMed] [Google Scholar]
- Chautard H.; Blas-Galindo E.; Menguy T.; Grand’Moursel L.; Cava F.; Berenguer J.; Delcourt M. An Activity-Independent Selection System of Thermostable Protein Variants. Nat. Methods 2007, 4 (11), 919–921. 10.1038/nmeth1090. [DOI] [PubMed] [Google Scholar]
- Mate D. M.; Rivera N. R.; Sanchez-Freire E.; Ayala J. A.; Berenguer J.; Hidalgo A. Thermostability Enhancement of the Pseudomonas fluorescens Esterase I by in Vivo Folding Selection in Thermus thermophilus. Biotechnol. Bioeng. 2020, 117 (1), 30–38. 10.1002/bit.27170. [DOI] [PubMed] [Google Scholar]
- Nakamura A.; Takakura Y.; Kobayashi H.; Hoshino T. In Vivo Directed Evolution for Thermostabilization of Escherichia Coli Hygromycin B Phosphotransferase and the Use of the Gene as a Selection Marker in the Host-Vector System of Thermus thermophilus. J. Biosci. Bioeng. 2005, 100 (2), 158–163. 10.1263/jbb.100.158. [DOI] [PubMed] [Google Scholar]
- Hernández K.; Gómez A.; Joglar J.; Bujons J.; Parella T.; Clapés P. 2-Keto-3-Deoxy-l-Rhamnonate Aldolase (YfaU) as Catalyst in Aldol Additions of Pyruvate to Amino Aldehyde Derivatives. Adv. Synth. Catal. 2017, 359 (12), 2090–2100. 10.1002/adsc.201700360. [DOI] [Google Scholar]
- Hernandez K.; Joglar J.; Bujons J.; Parella T.; Clapes P. Nucleophile Promiscuity of Engineered Class II Pyruvate Aldolase YfaU from E. Coli. Angew. Chem., Int. Ed. 2018, 57 (14), 3583–3587. 10.1002/anie.201711289. [DOI] [PubMed] [Google Scholar]
- Hernandez K.; Bujons J.; Joglar J.; Charnock S. J.; Domínguez De María P.; Fessner W. D.; Clapés P. Combining Aldolases and Transaminases for the Synthesis of 2-Amino-4-Hydroxybutanoic Acid. ACS Catal. 2017, 7 (3), 1707–1711. 10.1021/acscatal.6b03181. [DOI] [Google Scholar]
- Wong T.; Zhurina D.; Schwaneberg U. The Diversity Challenge in Directed Protein Evolution. Comb. Chem. High Throughput Screening 2006, 9 (4), 271–288. 10.2174/138620706776843192. [DOI] [PubMed] [Google Scholar]
- Sugimoto N.; Takakura Y.; Shiraki K.; Honda S.; Takaya N.; Hoshino T.; Nakamura A. Directed Evolution for Thermostabilization of a Hygromycin B Phosphotransferase from Streptomyces hygroscopicus. Biosci., Biotechnol., Biochem. 2013, 77 (11), 2234–2241. 10.1271/bbb.130486. [DOI] [PubMed] [Google Scholar]
- Iino D.; Takakura Y.; Fukano K.; Sasaki Y.; Hoshino T.; Ohsawa K.; Nakamura A.; Yajima S. Crystal Structures of the Ternary Complex of APH(4)-Ia/Hph with Hygromycin B and an ATP Analog Using a Thermostable Mutant. J. Struct. Biol. 2013, 183 (1), 76–85. 10.1016/j.jsb.2013.05.023. [DOI] [PubMed] [Google Scholar]
- Bednar D.; Beerens K.; Sebestova E.; Bendl J.; Khare S.; Chaloupkova R.; Prokop Z.; Brezovsky J.; Baker D.; Damborsky J. FireProt: Energy- and Evolution-Based Computational Design of Thermostable Multiple-Point Mutants. PLoS Comput. Biol. 2015, 11 (11), e1004556. 10.1371/journal.pcbi.1004556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommarius A. S.; Paye M. F. Stabilizing Biocatalysts. Chem. Soc. Rev. 2013, 42 (15), 6534–6565. 10.1039/c3cs60137d. [DOI] [PubMed] [Google Scholar]
- Brouns S. J. J.; Wu H.; Akerboom J.; Turnbull A. P.; De Vos W. M.; Van Der Oost J. Engineering a Selectable Marker for Hyperthermophiles. J. Biol. Chem. 2005, 280 (12), 11422–11431. 10.1074/jbc.M413623200. [DOI] [PubMed] [Google Scholar]
- Krüger D. M.; Rathi P. C.; Pfleger C.; Gohlke H. CNA Web Server: Rigidity Theory-Based Thermal Unfolding Simulations of Proteins for Linking Structure, (Thermo-)Stability, and Function. Nucleic Acids Res. 2013, 41, 340–348. 10.1093/nar/gkt292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A.; Takakura Y.; Sugimoto N.; Takaya N.; Shiraki K.; Hoshino T. Enzymatic Analysis of a Thermostabilized Mutant of an Escherichia coli Hygromycin B Phosphotransferase. Biosci., Biotechnol., Biochem. 2008, 72 (9), 2467–2471. 10.1271/bbb.80285. [DOI] [PubMed] [Google Scholar]
- Borges N.; Ramos A.; Raven N. D. H.; Sharp R. J.; Santos H. Comparative Study of the Thermostabilizing Properties of Mannosylglycerate and Other Compatible Solutes on Model Enzymes. Extremophiles 2002, 6 (3), 209–216. 10.1007/s007920100236. [DOI] [PubMed] [Google Scholar]
- Mayer S.; Rüdiger S.; Ang H. C.; Joerger A. C.; Fersht A. R. Correlation of Levels of Folded Recombinant P53 in Escherichia coli with Thermodynamic Stability in Vitro. J. Mol. Biol. 2007, 372 (1), 268–276. 10.1016/j.jmb.2007.06.044. [DOI] [PubMed] [Google Scholar]
- Kurahashi R.; Tanaka S. i.; Takano K. Activity-Stability Trade-off in Random Mutant Proteins. J. Biosci. Bioeng. 2019, 128 (4), 405–409. 10.1016/j.jbiosc.2019.03.017. [DOI] [PubMed] [Google Scholar]
- Tokuriki N.; Stricher F.; Serrano L.; Tawfik D. S. How Protein Stability and New Functions Trade Off. PLoS Comput. Biol. 2008, 4 (2), 35–37. 10.1371/journal.pcbi.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modarres H. P.; Mofrad M. R.; Sanati-Nezhad A. Protein Thermostability Engineering. RSC Adv. 2016, 6 (116), 115252–115270. 10.1039/C6RA16992A. [DOI] [Google Scholar]
- Bloom J. D.; Arnold F. H. In the Light of Directed Evolution: Pathways of Adaptive Protein Evolution. Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (Supplement 1), 9995–10000. 10.1073/pnas.0901522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommarius A. S.; Drauz K.; Klenk H.; Wandrey C. Operational Stability of Enzymes. Acylase-Catalyzed Resolution of N-Acetyl Amino Acids to Enantiomerically Pure L-Amino Acids. Ann. N. Y. Acad. Sci. 1992, 672, 126–136. 10.1111/j.1749-6632.1992.tb32669.x. [DOI] [PubMed] [Google Scholar]
- Rea D.; Hovington R.; Rakus J. F.; Gerlt J. A.; Fülöp V.; Bugg T. D. H.; Roper D. I. Crystal Structure and Functional Assignment of YfaU, a Metal Ion Dependent Class II Aldolase from Escherichia coli K12. Biochemistry 2008, 47 (38), 9955–9965. 10.1021/bi800943g. [DOI] [PubMed] [Google Scholar]
- Kazlauskas R. Engineering More Stable Proteins. Chem. Soc. Rev. 2018, 47 (24), 9026–9045. 10.1039/C8CS00014J. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


