Abstract
The death domain-containing receptor superfamily and their respective downstream mediators control whether or not cells initiate apoptosis or activate NF-κB, events critical for proper immune system function. A screen for upstream activators of NF-κB identified a novel serine-threonine kinase capable of activating NF-κB and inducing apoptosis. Based upon domain organization and sequence similarity, this novel kinase, named mRIP3 (mouse receptor interacting protein 3), appears to be a new RIP family member. RIP, RIP2, and mRIP3 contain an N-terminal kinase domain that share 30 to 40% homology. In contrast to the C-terminal death domain found in RIP or the C-terminal caspase-recruiting domain found in RIP2, the C-terminal tail of mRIP3 contains neither motif and is unique. Despite this feature, overexpression of the mRIP3 C terminus is sufficient to induce apoptosis, suggesting that mRIP3 uses a novel mechanism to induce death. mRIP3 also induced NF-κB activity which was inhibited by overexpression of either dominant-negative NIK or dominant-negative TRAF2. In vitro kinase assays demonstrate that mRIP3 is catalytically active and has autophosphorylation site(s) in the C-terminal domain, but the mRIP3 catalytic activity is not required for mRIP3 induced apoptosis and NF-κB activation. Unlike RIP and RIP2, mRIP3 mRNA is expressed in a subset of adult tissues and is thus likely to be a tissue-specific regulator of apoptosis and NF-κB activity. While the lack of a dominant-negative mutant precludes linking mRIP3 to a known upstream regulator, characterizing the expression pattern and the in vitro functions of mRIP3 provides insight into the mechanism(s) by which cells modulate the balance between survival and death in a cell-type-specific manner.
Apoptosis, or programmed cell death, is critical for the development of many tissue types during embryogenesis and in the adult is most clearly required for immune system homeostasis. In fact, deregulation of apoptosis has been implicated in autoimmune disorders such as rheumatoid arthritis and Crohn’s disease, as well as cancer and AIDS (46). A critical component of the apoptotic machinery is a superfamily of plasma membrane-spanning death domain-containing receptors that includes: TNFR1, Fas receptor (FasR; also called CD95 or Apo1), death receptor 3 (DR3; also called Apo3, WSL-1, TRAMP, or LARD), DR4 (also called Apo2 or TRAIL-R1), DR5 (also called TRAIL-R2, TRICK 2, or KILLER), and DR6. Ligation of death domain-containing receptors by ligands such as tumor necrosis factor (TNF), Fas, Apo3L, and TRAIL initiates immune responses and programmed cell death (1, 9, 12, 40, 41, 50). One way that death domain-containing receptors are regulated is by tissue-specific expression, as TNFR1 and DR4-6 are expressed in many normal and tumorigenic tissues, whereas a specific DR3 isoform is expressed in lymphoid tissues (3, 6, 28, 31, 38, 39, 41, 51).
The Fas, TNFR1, and DR3-6 death signaling pathways are initiated by ligand-induced receptor oligomerization, followed by binding of adapter molecules mediated through death domain interactions. The TNF receptor-associated death domain (TRADD) protein is recruited to TNFR1 (22) and DR3 (6). In turn, TRADD recruits the Fas-associated death domain (FADD) protein, whereas Fas receptor recruits FADD directly (5, 21). FADD facilitates the autocatalytic activation of caspase-8 (also known as FLICE [25, 36]). Caspase-8 activates other proteases, such as caspase-3, culminating in apoptosis (for a review, see references 13, 41, and 50).
Activation of NF-κB is also achieved through the TNFR1 and DR3-6 receptors but not through the Fas receptor. TNFR1 activation of NF-κB is mediated, in part, through the TRADD-dependent recruitment of a TNFR-associated factor (TRAF) homo- or heterodimer and a serine/threonine kinase, receptor interacting protein (RIP) (20, 21, 49). The TNFR1-TRADD-TRAF-RIP complex formation results in the activation of downstream kinases that lead to IκB phosphorylation and subsequent degradation. Once released from IκB, NF-κB translocates to the nucleus and activates target genes important for immunity and cell survival (for a review, see references 29, 30, and 32).
As already noted, one key downstream mediator of TNF-mediated apoptosis and NF-κB activation is RIP. RIP has three domains: an N-terminal kinase, an intermediate domain, and a C-terminal death domain (43). In vitro overexpression of RIP induces NF-κB-dependent transcription (20, 47). In support of the conclusion that RIP is critical to NF-κB activity, RIP−/− Abelson-transformed embryonic liver pre-B cells are unable to activate NF-κB in response to TNF. Whether other RIP−/− tissues have NF-κB activity was not reported (26). Interestingly, a RIP subdomain that lies between the kinase and the death domain is sufficient for NF-κB activation. Therefore, RIP, like the TRAF proteins, may function as an adapter in death domain-containing receptor signaling pathways. In vitro, RIP overexpression also induces apoptosis via a C-terminal death domain. Although RIP mRNA is expressed in almost all tissues except skeletal muscle and colon, the RIP−/− mice only undergo extensive apoptosis in lymphoid and adipose tissues (26). This indicates that RIP is a tissue-specific cell survival factor in vivo (26, 43). RIP2 (also known as RICK or CARDIAK), a RIP homologue, is most highly expressed in the heart, testis, placenta, spleen, and peripheral blood lymphocytes (24, 33, 45). RIP2 has an N-terminal kinase domain and a C-terminal caspase recruiting domain (CARD) (19). As with RIP, RIP2 overexpression induces NF-κB activity and apoptosis, and its functions are likely to be tissue specific. Thus, identifying RIP homologues present in restricted tissue groups is important in understanding development and how viability is regulated in various cell types.
We describe the identification of a new RIP family member, mouse RIP3 (mRIP3). mRIP3 shares a higher degree of sequence homology with RIP2 and displays functional properties similar to RIP and RIP2 in vitro, since it induces apoptosis and activates NF-κB. mRIP3 has a more restricted tissue distribution than RIP or RIP2, providing important evidence that cell viability in different tissue types is regulated by unique adapters in death signaling pathways. Since mRIP3 contains neither a CARD nor a death domain, its mechanism of action is likely to be novel.
MATERIALS AND METHODS
Library screening and Northern blots.
The mRIP3 cDNA probe was obtained in a screen of cDNAs amplified with degenerate primers to the serine/threonine consensus site and the Mg2+-ATP binding sites of the pelle, raf, and mos kinases. The degenerate primers were used to amplify related sequences from a mouse day-17 embryonic cDNA library (Clontech, Inc., Palo Alto, Calif.) by using reverse transcriptase PCR (RT-PCR) (48). The same library (3 × 105 plaques) was screened with the PCR probe containing subdomains I to VIb of the mRIP3 kinase domain radioactively labeled by random priming with [α-32P]dCTP (3,000 mCi/ml; Amersham, Arlington Heights, Ill.). Three independent clones were obtained, and both strands were sequenced (Indiana University Biotechnology Facility). All three phage clones contained an open reading frame of 1,461 nucleotides that was 99% identical from nucleotides 260 to 577 to the PCR probe sequence. For Northern analysis, the same probe (radioactively labeled as described above) was hybridized to a mouse embryo and an adult Multiple Tissue Northern Blots (Clontech) according to the manufacturer’s recommendations.
mRIP3 mutants.
An mRIP3 site-directed mutant was made by using the Altered Sites II in vitro Mutagenesis System (Promega, Madison, Wis.). The mutation changed nucleotide 631 from a G to an A, resulting in a conserved amino acid substitution of the aspartic acid (amino acid 161) with an asparagine in the putative Mg2+-ATP binding site. The mRIP3 deletion constructs were produced by PCR amplification of the indicated region (see Fig. 1C) and subsequently subcloned into a mammalian expression vector, pcDNA3.1 (Invitrogen, Inc., Carlsbad, Calif.). The kinase-only construct includes the coding region for amino acids 1 to 248, and the tail-only construct includes the coding region for amino acids 1 to 13 and 245 to 486 fused in frame. All four constructs contained a C-terminal Myc-His tag fused in frame with the mRIP3 coding sequence.
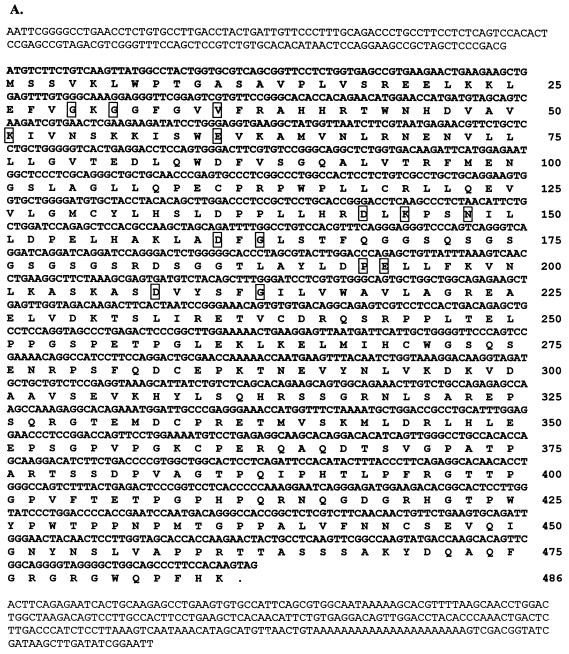
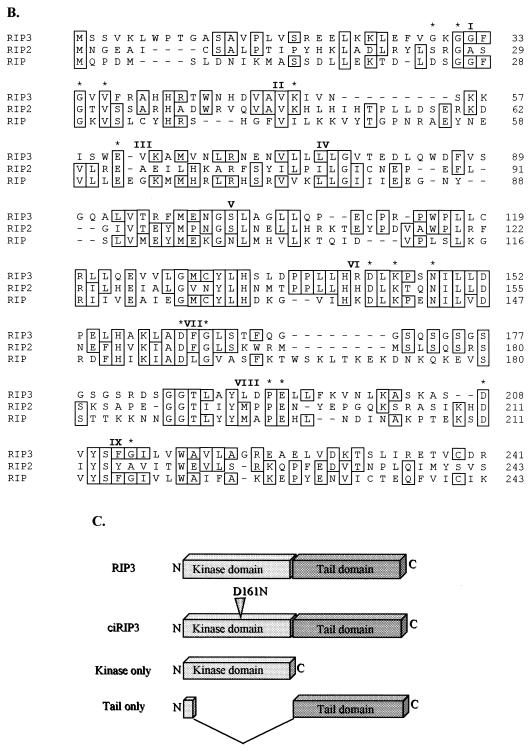
FIG. 1.
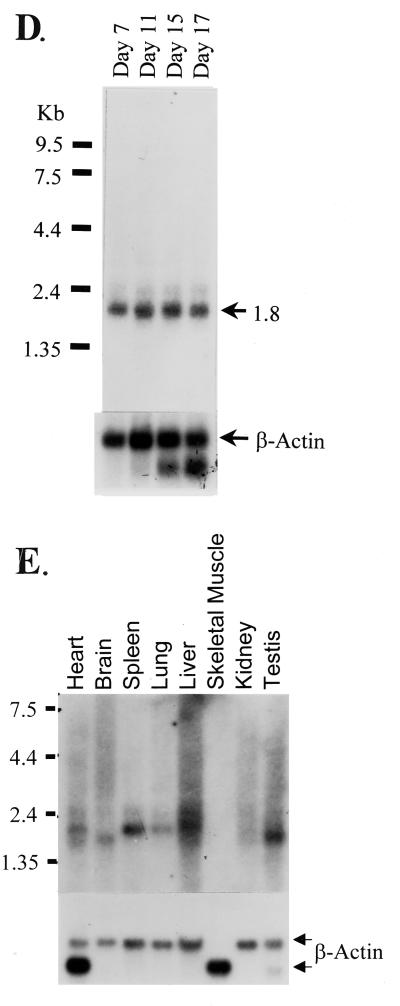
mRIP3 is a novel serine/threonine kinase related to the RIP family of kinases. (A) Nucleotide sequence of the mRIP3 cDNA and putative open reading frame with the number of amino acid residues listed at the right. Boxed residues are the conserved amino acids found in serine/threonine kinases. (B) Amino acid sequence alignments of RIP, RIP2, and mRIP3 N-terminal kinase domains. Roman numerals indicate the location of the kinase subdomains. Asterisks indicate the invariant kinase residues boxed in panel A. The analysis was performed by using the MegAlign program (DNAStar, Inc., Madison, Wis.). (C) Schematic of mRIP3 constructs. mRIP3 (wild-type protein), ciRIP3 (catalytically inactive mRIP3 in which the Mg2+-ATP binding site aspartic acid [amino acid 161] has been changed to asparagine), and tail-only and kinase-only (two deletion mutants in which each domain is expressed separately) are depicted. Kinase-only and tail-only proteins include amino acids 1 to 248 and amino acids 1 to 13 fused to 245 to 486, respectively. (D) mRIP3 is expressed in embryonic mouse tissues. A Clontech Mouse Multiple Tissue Embryo blot was probed with a radioactively labeled cDNA probe (see Materials and Methods for details). (E) mRIP3 mRNA is present in the heart, brain, spleen, lung, liver, kidney, and testis. The Clontech Adult Mouse Multiple Tissue Northern blot was probed with a radioactively labeled mRIP3 cDNA probe (see Materials and Methods for details). Size markers (in kilobases) are indicated on the left side of the panel.
Cell culture and transfections.
For transfections, human embryonic kidney cells, HEK 293 (American Type Culture Collection) plated at 106 cells/60-mm dish were maintained in Dulbecco modified Eagle medium containing 4.5 g of glucose per ml, 10% fetal calf serum, 100 IU of penicillin per ml, 100 μg of streptomycin per ml, and 2 mM glutamine. Approximately 30 h later, the cells were transfected with the indicated DNAs by using the calcium phosphate method described previously (16). The DNA amount added to each dish was equalized by the addition of pcDNA3.1 containing no insert.
DNA laddering and apoptosis assays.
Approximately 24 h after transfection, floating and adherent cells were harvested, and genomic DNA was obtained by using the Purgene DNA Isolation Kit (Gentra Systems, Minneapolis, Minn.). Fifteen micrograms of total genomic DNA from each sample was electrophoresed through 1% agarose by standard procedures.
For cellular death quantification, HEK 293 cells were cultured and transfected as described above with the addition of pCMV-β-Gal as a marker of transfection efficiency. At 24 h after transfection, adherent cells and floating cells were harvested. β-Galactosidase (β-Gal) activity was determined for each fraction by using Galacto-Light Plus (Tropix, Bedford, Mass.) according to the manufacturer’s recommendations. The percentage of the total β-Gal activity was calculated for the adherent cells in three separate experiments. The average value is reported with the standard deviation. A loss of β-Gal activity in the adherent cell fraction represents an induction of apoptosis.
Reporter gene assays.
HEK 293 cells were cultured and transfected as described above. To inhibit apoptosis, precipitates contained CrmA, which had no effect on the activation of the three promoter constructs tested (data not shown). At 15 to 17 h after transfection, adherent cells were harvested and assayed for luciferase activity by using the Luciferase Assay System (Promega). As a control for transfection efficiency, β-Gal activity was measured according to manufacturer’s recommendation (Tropix). Transfection efficiency was normalized by calculating the ratio of luciferase activity to β-Gal activity for each test. Each datum point represents the average of four independent tests, with the standard error indicated.
Immunoprecipitation.
HEK 293 cells were transfected with the indicated constructs in addition to CrmA. At 24 h after transfection, the adherent cells were harvested and lysed in 1 ml of immunoprecipitation lysis buffer (10 mM HEPES, pH 7.6; 150 mM NaCl; 5 mM EDTA; 1% Triton X-100; Complete Protease Cocktail [Roche Biochemicals, Indianapolis, Ind.]). Cell lysates were spun at 14,000 rpm at 4°C for 5 min to remove cellular debris and nuclei. Then, 1 μg of anti-c-myc antibody (mouse monoclonal clone 9E10; Roche Biochemicals) was added to the lysate and mixed gently overnight at 4°C. Next, 200 μl of protein A-Sepharose (10% [vol/vol] slurry) was added, and lysates were mixed for an additional 2 h at 4°C. Immunocomplexed material was isolated by gentle centrifugation, and the pellets were washed twice with immunoprecipitation wash buffer (10 mM HEPES, pH 7.6; 150 mM NaCl; 5 mM EDTA; 0.1% Triton X-100; Complete Protease Cocktail). Immunocomplexed material was subjected to in vitro kinase assays or analyzed by sodium dodecyl sulfate (SDS)–12% polyacrylamide gel electrophoresis (PAGE) followed by Western blotting.
In vitro kinase assays.
Immunocomplexed material was adjusted to contain 20 mM Tris-Cl (pH 7.6), 20 mM MgCl2, 20 mM β-glycerol phosphate, 1 mM sodium orthovanadate, 1 mM benzamidine, 0.4 mM phenylmethyl sulfonyl fluoride, 2 μM ATP, and 10 μCi of [γ-32P]dATP (3,000 mCi/ml). Reactions were incubated at 30°C for 30 min and stopped by the addition of 2× Laemmli buffer. Reactions were electrophoresed in a SDS–12% PAGE, the gel was stained with GelCode Blue (Pierce, Rockford, Ill.), and the gels were then dried on Whatman paper. Phosphoproteins were identified by autoradiography.
Western blots.
Immunocomplexed material was subjected to SDS–12% PAGE by using standard techniques and then transferred to polyvinylidene difluoride membranes by liquid tank transfer. The membrane was blocked in 5% milk–KPBS-T (140 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4, 0.4% Tween 20) for 1 h. The Western blots were probed with a monoclonal antibody to the myc epitope (Roche Biochemicals) for 2 h at room temperature. The blot was washed three times with KPBS-T and probed with anti-mouse F(ab)2 fragments conjugated to HRP (Jackson Immunochemicals, West Grove, Pa.) for 1 h. The blot was washed three more times in KPBS-T, and the horseradish peroxidase (HRP) tag was visualized by enhanced chemiluminescence (Amersham).
RESULTS
mRIP3 is a novel member of the RIP family of serine/threonine protein kinases.
To identify novel serine/threonine kinases involved in NF-κB regulation, conserved domains of pelle, raf, and mos were identified by sequence alignment. Pelle, a serine/threonine kinase, mediates activation of the Drosophila NF-κB homologue that controls dorsal and ventral polarity in early development and immunity to fungal pathogens in adult flies (2, 10, 34). raf and mos are mammalian serine/threonine kinases with a high degree of homology to pelle. Degenerate primers based upon the serine/threonine consensus motifs and the Mg2+-ATP binding site motifs were used to amplify novel gene fragments from an embryonic day 17 mouse cDNA library. A PCR product identified in the screen encoded a novel sequence with homology to RIP, which we named mRIP3. A day 17 mouse embryonic cDNA library was screened for a full-length mRIP3 clone. Three were identified, each containing an open reading frame of 1,461 nucleotides, which were identical to the probe sequence from nucleotides 260 to 577 (Fig. 1A). The N-terminal half of the deduced amino acid sequence has 29% identity to RIP and 36% identity to RIP2 (Fig. 1B). In addition, the mRIP3 N-terminal domain has all of the conserved hallmark features of a serine/threonine kinase (Fig. 1A and B) (14, 15, 27, 42). In vitro kinase assays demonstrate that mRIP3 is catalytically active, since immunoprecipitated mRIP3 undergoes autophosphorylation (see Fig. 3A). Sequence analysis indicates that the C terminus of mRIP3, unlike RIP and RIP2, contains neither a death domain nor a CARD and has no significant homology with any protein presently in the National Center for Biotechnology Information (NCBI) database.
FIG. 3.
mRIP3 autophosphorylates site(s) in the C-terminal domain. HEK 293 cells were transfected with the indicated constructs plus CrmA and Myc-His-LacZ (Invitrogen). Cell lysates were harvested, and fusion proteins were immunoprecipitated as described in Materials and Methods. Immunoprecipitates were divided in half and were subjected to in vitro kinase assays (A) or Western blot analysis (B) to determine the protein expression level.
To determine which mouse tissues express mRIP3, an mRIP3-specific probe was hybridized to poly(A)+ RNA isolated from developing mouse embryos and various adult tissues. One primary transcript (∼1.8 kb) of equal abundance in all stages of mouse development was detected, which suggests that mRIP3 is expressed early in development (Fig. 1D). Only certain adult tissues, the liver, lung, spleen, brain, heart, and testis, expressed the mRIP3 mRNA (Fig. 1E). The 1.8-kb transcript corresponded to the size of the cDNA obtained in the library screening; therefore, the library clone appears to contain the full-length mRIP3 transcript. In adult tissues, a second band slightly smaller than the 1.8-kb transcript was detected and may represent a splice variant.
mRIP3-induced apoptosis is FADD and caspase dependent.
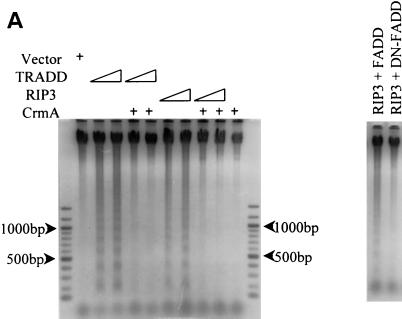
The overexpression of RIP and RIP2 induces apoptosis; therefore, the ability of mRIP3 to induce apoptosis was examined. A mammalian expression vector containing the mRIP3 open reading frame was transfected into HEK 293 cells (16). Genomic DNA was harvested and assessed for fragmentation by agarose gel electrophoresis. Genomic DNA from cells overexpressing TRADD was used as a positive control. DNA from cells transfected with TRADD, as well as with mRIP3, contained fragments that are approximately 200 bp apart, a result indicative of apoptosis (Fig. 2A). Since DNA fragmentation occurs in response to caspase activation, we determined whether mRIP3-induced fragmentation was caspase dependent. CrmA, a nonspecific caspase inhibitor from the cowpox virus (44), was coexpressed with mRIP3 in HEK 293 cells. CrmA abolished mRIP3-dependent DNA fragmentation (Fig. 2A). FADD, another inducer of apoptosis, is a component of the TNFR1, the FasR, and the DR3-5 death signal transduction pathways (4–6). Deletion of the FADD death effector domain (DED) acts in a dominant-negative manner and inhibits FADD-induced apoptosis (5, 36, 37). To determine whether mRIP3-induced apoptosis is dependent upon or requires FADD, dominant-negative FADD (DN-FADD) was cotransfected with mRIP3 into HEK 293 cells. DNA fragments were not detected in cells coexpressing mRIP3 and DN-FADD (Fig. 2A). Thus, mRIP3-induced apoptosis is FADD and caspase dependent.
FIG. 2.
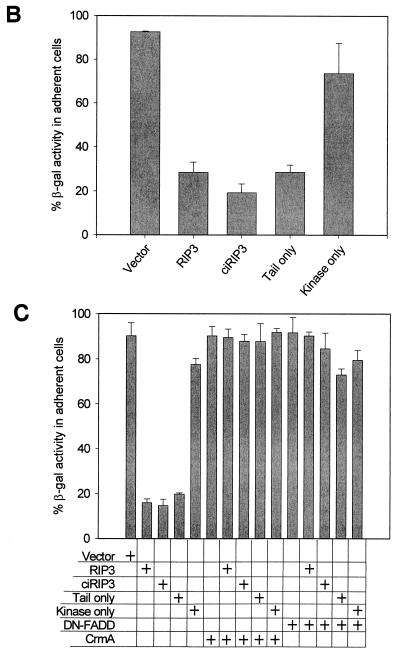
mRIP3 induces apoptosis. (A) mRIP3 overexpression induces DNA fragmentation. HEK 293 cells were transfected with 0.5 or 1.0 μg of TRADD with or without a fourfold excess of CrmA (lanes 2 to 5, respectively). In lanes 6 to 9, HEK 293 cells were transfected with 0.5 or 1.0 μg of mRIP3 with or without a fourfold excess of CrmA. In the right panel, 2 μg of mRIP3 was cotransfected with 2 μg of FADD or 10 μg of DN-FADD. At 24 h after transfection, genomic DNA was isolated and processed as described in Materials and Methods. (B) mRIP3 kinase domain does not induce apoptosis. HEK 293 cells were transfected with mRIP3, ciRIP3, and tail-only or kinase-only constructs plus 0.25 μg of pCMV-β-Gal. β-Gal activity was measured for adherent cells and floating apoptotic cells as described in Materials and Methods. The mean percentage of β-Gal in the adherent-cell fraction was calculated for three experiments and plotted with the standard deviation. (C) DN-FADD and CrmA inhibit mRIP3, ciRIP3, or mRIP3–tail-only-induced apoptosis. HEK 293 cells were transfected with the indicated mRIP3 constructs. Cell lysates were assayed as in panel B.
In addition to DNA fragmentation, apoptotic cells undergo membrane blebbing that can be visualized by the release of adherent cells from tissue culture dishes (18). To quantify the proportion of cells induced to undergo apoptosis in response to mRIP3, cotransfection assays were performed with mammalian expression vectors containing mRIP3 and β-Gal cDNAs. The total number of transfected cells was determined by measuring the amount of β-Gal activity in the floating apoptotic bodies and the adherent cells, and then the percentage of the total β-Gal activity present in each fraction was calculated. The loss of β-Gal activity in the adherent fraction represents cellular release and apoptosis. After HEK 293 cells were transfected with mRIP3, little β-Gal activity remained in the adherent cells (Fig. 2B). mRIP3-dependent apoptosis was reversed by the addition of DN-FADD or CrmA (Fig. 2A and C).
The C-terminal domain of mRIP3 induces apoptosis.
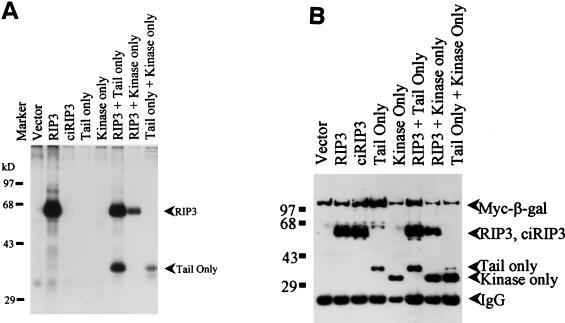
An intriguing aspect of RIP and RIP2 in vitro activity is that neither induction of apoptosis nor NF-κB activity is dependent upon kinase catalytic activity (20, 24, 26, 33, 45, 47). To determine whether mRIP3 catalytic activity was necessary for the induction of apoptosis, the conserved aspartic acid residue in the Mg2+-ATP binding site was changed to asparagine (14, 15, 27, 42). The Asp→Asn substitution had no effect on protein expression (Fig. 3B) but completely inhibited the mRIP3 autophosphorylation (Fig. 3A). To determine whether mRIP3 kinase activity was required for apoptosis, β-Gal and catalytically inactive mRIP3 (ciRIP3) cotransfection assays were performed to quantify the apoptotic membrane blebbing and cellular release. Consistent with results obtained with RIP and RIP2, ciRIP3 induced apoptosis as well as the wild-type mRIP3 (Fig. 2B). To determine the domain responsible for apoptosis, two deletion constructs were made. In the mRIP3 kinase-only mutant, the entire tail region downstream of the last kinase subdomain was deleted. The second construct, mRIP3 tail-only, deletes the entire kinase domain, leaving the first 13 amino acids of the coding region fused in frame to the C-terminal half (Fig. 1C). Both deletion proteins are expressed, although the tail-only protein migrates at a higher molecular mass than expected (34 kDa instead of 30 kDa [see Fig. 3B]). Overexpression of the kinase-only protein did not induce apoptosis, whereas overexpression of the mRIP3 tail-only protein did induce apoptosis (Fig. 2B and C). As with wild-type mRIP3, apoptosis induced by ciRIP3 or the tail-only protein was reversed by DN-FADD and CrmA. Therefore, the mRIP3 C-terminal domain induces death in a FADD- and CrmA-dependent pathway, even though it does not contain a CARD or death domain, which are present in RIP2 and RIP.
mRIP3 autophosphorylates its C-terminal domain.
To assess whether the mRIP3 kinase domain was catalytically active, in vitro kinase assays were performed with immunoprecipitated mRIP3. As seen in Fig. 3B, immunoprecipitated mRIP3 migrates slightly faster than the predicted size of full-length mRIP3 (53 kDa). In the mRIP3 immunoprecipitates, a phosphorylated protein that comigrates with mRIP3 was detected after the addition of [γ-32P]ATP, suggesting that wild-type mRIP3 undergoes autophosphorylation. In contrast, full-length mRIP3 with a mutation in the Mg2+-ATP binding site has no autocatalytic activity. The catalytic activity of the mRIP3 deletion constructs was also assessed. As expected, immunoprecipitates of the tail-only protein did not produce a phosphoprotein at 34 kDa. Surprisingly, the kinase-only construct also did not produce a phosphoprotein. The latter observation suggests either that the kinase-only protein is catalytically inactive due to disruption of tertiary structure or that the autophosphorylation site is located in the C-terminal domain. To distinguish between these possibilities, the tail-only and the kinase-only constructs (each myc-tagged) were coexpressed in HEK 293 cells. The truncated proteins were coimmunoprecipitated and subjected to in vitro kinase assays. A phosphoprotein corresponding to the tail-only protein was detected, suggesting that the truncated kinase-only mutant is catalytically active and that the tail-only mutant was a substrate for the kinase activity (Fig. 3B). These data support the conclusion that mRIP3 autophosphorylation site(s) lie in the C-terminal tail.
mRIP3 induction of NF-κB is inhibited by ciNIK or ΔTRAF2.
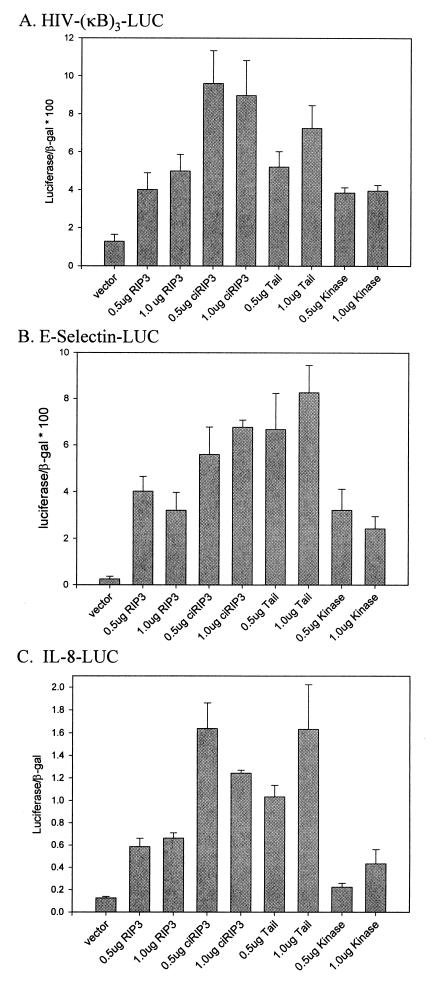
To determine whether mRIP3 could transactivate NF-κB, mRIP3 expression constructs were cotransfected with a luciferase reporter gene under the control of the interleukin-8 (IL-8) promoter (IL-8–LUC) (35), the E-selectin promoter (E-selectin–LUC) (52), or an artificial promoter containing three tandem NF-κB sites [HIV-(κB)3–LUC] (17). E-selectin and IL-8 promoters contain cis-acting elements in addition to the NF-κB binding site but are considered NF-κB dependent. CrmA was included in these transfections to inhibit mRIP3-induced apoptosis and did not independently induce transactivation of the reporter constructs (data not shown). mRIP3 overexpression induces NF-κB activity in assays with each of the reporter constructs (Fig. 4). Induction of NF-κB was approximately 2.5-fold lower in wild-type mRIP3 than in wild-type RIP transfectants, as assayed by transactivation of the HIV-(κB)3–LUC promoter (data not shown). Additionally, DN-RIP (deletion of the intermediate domain) did not block mRIP3 induced HIV-(κB)3–LUC promoter activity. Catalytically inactive mRIP3 also induced NF-κB activity. Thus, mRIP3 kinase activity is not required for NF-κB activation in vitro. The mRIP3 kinase-only and tail-only proteins have NF-κB inducing activity; therefore, segments that transactivate NF-κB may be embedded within each domain. In contrast to RIP, which transactivates both NF-κB and AP-1 in vitro, mRIP3 did not activate reporter constructs under the control of AP-1 (data not shown).
FIG. 4.
mRIP3 induces NF-κB activity. HEK 293 cells were transfected the indicated expression constructs plus HIV-(κB)3–LUC (A), E-selectin–LUC (B), or IL-8–LUC (C). Cell lysates were assayed for luciferase and β-Gal activity as described in Materials and Methods. The mean of four experiments with the standard error is shown.
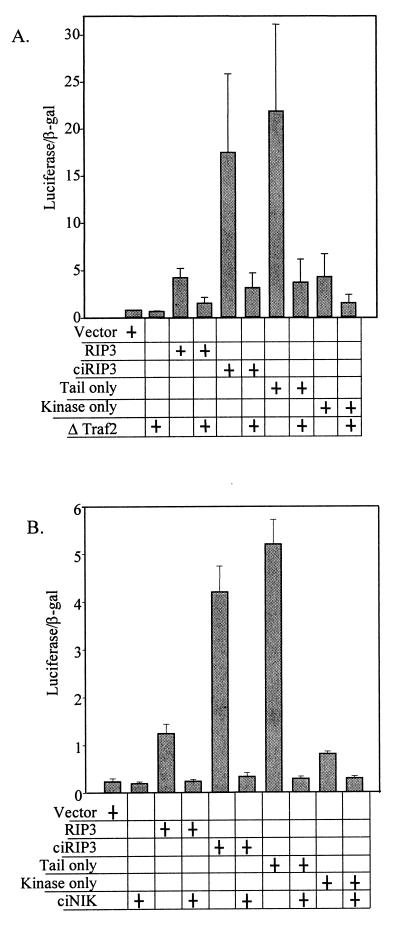
To identify other molecules involved in the mRIP3 signal transduction pathway, mRIP3-dependent NF-κB activation was assayed in cells transfected with dominant-negative TRAF2 or dominant-negative NIK. The RING finger motif of TRAF2 is critical for NF-κB activation, and without this domain TRAF2 blocks its own signal as well as a TNF-induced NF-κB activity (8). TRAF2 deleted for the RING finger (ΔTRAF2) completely inhibited IL-8–LUC activation by mRIP3 (Fig. 5A). In addition, a dominant-negative version of NIK completely blocked mRIP3-induced NF-κB activity (Fig. 5B).
FIG. 5.
mRIP3-induced NF-κB activity is blocked by ciNIK and ΔTRAF2. HEK 293 cells were transfected with the indicated constructs. Cell lysates were harvested and assayed as described in the legend to Fig. 4.
DISCUSSION
This study identifies and characterizes a novel tissue-specific kinase, mRIP3, that induces apoptosis and activates NF-κB. The method used to clone mRIP3 was previously used by our group to clone mouse pelle-like kinase (mPLK), a homologue of IL-1 receptor-associated kinase (IRAK), a TNF- and IL-1-responsive regulator of NF-κB activity (48, 49). The mRIP3 in vitro function, domain structure, and homology suggest that this protein belongs to the RIP family. Although the N-terminal kinase domain has 30 to 40% homology to RIP and RIP2, the mRIP3 C-terminal tail has no significant homology with RIP, RIP2, or any other protein presently in the NCBI database. Even though the C terminus is does not contain a CARD or death domain, overexpression of the truncated C-terminal protein induces apoptosis in a FADD- and caspase-dependent pathway. In addition to inducing apoptosis, mRIP3 induces NF-κB activity. Catalytically inactive mRIP3 retains the ability to induce apoptosis and activate NF-κB; therefore, the role of the mRIP3 kinase activity is unclear. Since a dominant-negative version of mRIP3 was not identified, the upstream regulators of mRIP3 activity remain unknown. Yet, mRIP3 is expressed in a restricted group of tissues and is, therefore, a novel cell-type-specific mediator of apoptosis and NF-κB. Also in distinction to RIP and RIP2, mRIP3 is unable to activate AP-1-dependent promoters, indicating that RIP family members have overlapping and distinct functions.
All three RIP family members have a C-terminal domain that induces death when overexpressed in vitro: RIP has a C-terminal death domain and RIP2 has a C-terminal CARD motif. Overexpression of the mRIP3 C-terminal tail can induce apoptosis, although this domain has no significant homology to any known protein or to any previously identified motif associated with cell death. Therefore, we propose that mRIP3 may induce apoptosis through a novel mechanism. Fine mapping of the C terminus is needed to determine the minimal sequence requirements for this activity. Hydropathy analysis and crystal structure analysis of CARDs, death domains, and death effector domains show high α-helical content that is postulated to mediate protein-protein interactions (7, 11, 23). Hydropathy analysis of mRIP3 C-terminal domain predicts very little α-helical secondary structure; therefore, the mechanism by which mRIP3 induces death may be due to a domain with a different secondary structure than the previously identified death-inducing domains.
The mRIP3 C terminus activates apoptosis in a FADD- and caspase-dependent pathway. FADD mediates the death signal from a variety of death receptors, including Fas receptor, TNFR1, and DR3-6 (41). Since the Fas receptor is not thought to activate NF-κB (9), mRIP3 is most likely not a component of the Fas pathway. In the absence of a dominant-negative mRIP3, we are unable to exclusively link mRIP3 to a known receptor pathway. mRIP3 may either be a downstream component of one or more of the known receptor signaling pathways and/or a component of a novel receptor signaling pathway.
mRIP3 activation of NF-κB is blocked by dominant-negative TRAF2 or dominant-negative NIK. Although NF-κB activity is not defective in TRAF2 knockout animals, the animals may be able to compensate for the loss of TRAF2 by using oligomers containing other TRAF members. Overexpression of DN-TRAF2 can disrupt TNF-induced NF-κB activity as well as mRIP3 induction of NF-κB, suggesting that TRAF2 somehow affects mRIP3 signal transduction to NF-κB. In addition, mRIP3 is unable to transactivate AP-1-dependent promoters; therefore, the TRAF2 effect is most likely due to the ability to DN-TRAF2 to disrupt activation of the NF-κB signaling pathway.
The mRIP3 kinase domain has all the conserved elements of a serine/threonine kinase and is catalytically active in vitro. Interestingly, mRIP3 catalytic activity is not required for apoptosis or NF-κB activation. Similarly, RIP kinase activity is not required for apoptosis or NF-κB induction via TNFR1. Thus, the emerging family of RIP-like kinases appears to have an unidentified use for the kinase activity. Further analysis will be necessary to determine what role mRIP3 kinase activity plays in signal transduction.
The lack of expression of mRIP3 mRNA in skeletal muscle and varied levels of mRNA in other tissues suggest that mRIP3 is a tissue-specific homologue. Since RIP and RIP2 are expressed in a wide variety of tissues, whereas mRIP3 seem to be restricted to certain tissues, this growing family of kinases may be an important point of regulation between death domain receptor-activated cell death and cell survival. Thus, mRIP3 is a novel kinase involved in the activation of NF-κB, but not AP-1, and in the induction of apoptosis. Understanding the pattern of mRIP3 expression in relation to RIP1 and RIP2 provides insight into the mechanism through which the balance between survival and death is modulated in distinct cell types.
ACKNOWLEDGMENTS
We thank J. M. Kyriakis for the RIP constructs used in this study.
This research was supported in part by National Institutes of Health grants AI42798 (M.A.H.) and CA67891 and CA73023 (D.B.D.) and in part by the Project Development Program, Research and Sponsored Programs, Indiana University at Indianapolis (M.A.H.).
ADDENDUM IN PROOF
Two independent groups (P. W. Yu et al., Curr. Biol. 9:539–542, 1999; X. Sun et al., J. Biol. Chem. 274:16871–16875, 1999) have recently reported cloning of human RIP3.
REFERENCES
- 1.Ashkenazi A, Dixit V M. Death receptors: signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 2.Belvin M P, Anderson K V. A conserved signaling pathway: the Drosophila toll-dorsal pathway. Ann Rev Cell Dev Biol. 1996;12:393–416. doi: 10.1146/annurev.cellbio.12.1.393. [DOI] [PubMed] [Google Scholar]
- 3.Bodmer J L, Burns K, Schneider P, Hofmann K, Steiner V, Thome M, Bornand T, Hahne M, Schroter M, Becker K, Wilson A, French L E, Browning J L, MacDonald H R, Tschopp J. TRAMP, a novel apoptosis-mediating receptor with sequence homology to tumor necrosis factor receptor 1 and Fas(Apo-1/CD95) Immunity. 1997;6:79–88. doi: 10.1016/s1074-7613(00)80244-7. [DOI] [PubMed] [Google Scholar]
- 4.Chaudhary P M, Eby M, Jasmin A, Bookwalter A, Murray J, Hood L. Death receptor 5, a new member of the TNFR family, and DR4 induce FADD-dependent apoptosis and activate the NF-κB pathway. Immunity. 1997;7:821–830. doi: 10.1016/s1074-7613(00)80400-8. [DOI] [PubMed] [Google Scholar]
- 5.Chinnaiyan A M, O’Rourke K, Tewari M, Dixit V M. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 6.Chinnaiyan A M, O’Rourke K, Yu G L, Lyons R H, Garg M, Duan D R, Xing L, Gentz R, Ni J, Dixit V M. Signal transduction by DR3, a death domain-containing receptor related to TNFR-1 and CD95. Science. 1996;274:990–992. doi: 10.1126/science.274.5289.990. [DOI] [PubMed] [Google Scholar]
- 7.Chou J J, Matsuo H, Duan H, Wagner G. Solution structure of the RAIDD CARD and model for CARD/CARD interaction in caspase-2 and caspase-9 recruitment. Cell. 1998;94:171–180. doi: 10.1016/s0092-8674(00)81417-8. [DOI] [PubMed] [Google Scholar]
- 8.Dadgostar H, Cheng G. An intact zinc ring finger is required for tumor necrosis factor receptor-associated factor-mediated nuclear factor-kappaB activation but is dispensable for c-Jun N-terminal kinase signaling. J Biol Chem. 1998;273:24775–24780. doi: 10.1074/jbc.273.38.24775. [DOI] [PubMed] [Google Scholar]
- 9.Depraetere V, Golstein P. Fas and other cell death signaling pathways. Semin Immunol. 1997;9:93–107. doi: 10.1006/smim.1997.0062. [DOI] [PubMed] [Google Scholar]
- 10.Dushay M S, Eldon E D. Drosophila immune responses as models for human immunity. Am J Hum Genet. 1998;62:10–14. doi: 10.1086/301694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eberstadt M, Huang B, Chen Z, Meadows R P, Ng S C, Zheng L, Lenardo M J, Fesik S W. NMR structure and mutagenesis of the FADD (Mort1) death-effector domain. Nature. 1998;392:941–945. doi: 10.1038/31972. [DOI] [PubMed] [Google Scholar]
- 12.Golstein P. Cell death: TRAIL and its receptors. Curr Biol. 1997;7:R750–R753. doi: 10.1016/s0960-9822(06)90000-1. [DOI] [PubMed] [Google Scholar]
- 13.Gravestein L A, Borst J. Tumor necrosis factor receptor family members in the immune system. Semin Immunol. 1998;10:423–434. doi: 10.1006/smim.1998.0144. [DOI] [PubMed] [Google Scholar]
- 14.Hanks S K, Quinn A M. Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Methods Enzymol. 1991;200:38–62. doi: 10.1016/0076-6879(91)00126-h. [DOI] [PubMed] [Google Scholar]
- 15.Hanks S K, Quinn A M, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 16.Harrington M A, Konicek B, Song A, Xia X L, Fredericks W J, den Rauscher F J. Inhibition of colony-stimulating factor-1 promoter activity by the product of the Wilms’ tumor locus. J Biol Chem. 1993;268:21271–21275. [PubMed] [Google Scholar]
- 17.Hedin K E, Bell M P, Kalli K R, Huntoon C J, Sharp B M, McKean D J. Delta-opioid receptors expressed by Jurkat T cells enhance IL-2 secretion by increasing AP-1 complexes and activity of the NF-AT/AP-1-binding promoter element. J Immunol. 1997;159:5431–5440. [PubMed] [Google Scholar]
- 18.Hengartner M O. Apoptosis and the shape of death. Dev Genet. 1997;21:245–248. doi: 10.1002/(SICI)1520-6408(1997)21:4<245::AID-DVG1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 19.Hofmann K, Bucher P, Tschopp J. The CARD domain: a new apoptotic signalling motif. Trends Biochem Sci. 1997;22:155–156. doi: 10.1016/s0968-0004(97)01043-8. [DOI] [PubMed] [Google Scholar]
- 20.Hsu H, Huang J, Shu H B, Baichwal V, Goeddel D V. TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity. 1996;4:387–396. doi: 10.1016/s1074-7613(00)80252-6. [DOI] [PubMed] [Google Scholar]
- 21.Hsu H, Shu H B, Pan M G, Goeddel D V. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 22.Hsu H, Xiong J, Goeddel D V. The TNF receptor 1-associated protein TRADD signals cell death and NF-κB activation. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 23.Huang B, Eberstadt M, Olejniczak E T, Meadows R P, Fesik S W. NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature. 1996;384:638–641. doi: 10.1038/384638a0. [DOI] [PubMed] [Google Scholar]
- 24.Inohara N, del Peso L, Koseki T, Chen S, Nunez G. RICK, a novel protein kinase containing a caspase recruitment domain, interacts with CLARP and regulates CD95-mediated apoptosis. J Biol Chem. 1998;273:12296–12300. doi: 10.1074/jbc.273.20.12296. [DOI] [PubMed] [Google Scholar]
- 25.Juo P, Kuo C J, Yuan J, Blenis J. Essential requirement for caspase-8/FLICE in the initiation of the Fas-induced apoptotic cascade. Curr Biol. 1998;8:1001–1008. doi: 10.1016/s0960-9822(07)00420-4. [DOI] [PubMed] [Google Scholar]
- 26.Kelliher M A, Grimm S, Ishida Y, Kuo F, Stanger B Z, Leder P. The death domain kinase RIP mediates the TNF-induced NF-κB signal. Immunity. 1998;8:297–303. doi: 10.1016/s1074-7613(00)80535-x. [DOI] [PubMed] [Google Scholar]
- 27.Kennelly P J, Krebs E G. Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J Biol Chem. 1991;266:15555–15558. [PubMed] [Google Scholar]
- 28.Kitson J, Raven T, Jiang Y P, Goeddel D V, Giles K M, Pun K T, Grinham C J, Brown R, Farrow S N. A death-domain-containing receptor that mediates apoptosis. Nature. 1996;384:372–375. doi: 10.1038/384372a0. [DOI] [PubMed] [Google Scholar]
- 29.Lee J I, Burckart G J. Nuclear factor kappa B: important transcription factor and therapeutic target. J Clin Pharmacol. 1998;38:981–993. doi: 10.1177/009127009803801101. [DOI] [PubMed] [Google Scholar]
- 30.Maniatis T. Catalysis by a multiprotein IκB kinase complex. Science. 1997;278:818–819. doi: 10.1126/science.278.5339.818. [DOI] [PubMed] [Google Scholar]
- 31.Marsters S A, Sheridan J P, Donahue C J, Pitti R M, Gray C L, Goddard A D, Bauer K D, Ashkenazi A. Apo-3, a new member of the tumor necrosis factor receptor family, contains a death domain and activates apoptosis and NF-κB. Curr Biol. 1996;6:1669–1676. doi: 10.1016/s0960-9822(02)70791-4. [DOI] [PubMed] [Google Scholar]
- 32.May M J, Ghosh S. Signal transduction through NF-kappa B. Immunol Today. 1998;19:80–88. doi: 10.1016/s0167-5699(97)01197-3. [DOI] [PubMed] [Google Scholar]
- 33.McCarthy J V, Ni J, Dixit V M. RIP2 is a novel NF-κB-activating and cell death-inducing kinase. J Biol Chem. 1998;273:16968–16975. doi: 10.1074/jbc.273.27.16968. [DOI] [PubMed] [Google Scholar]
- 34.Medzhitov R, Janeway C A., Jr An ancient system of host defense. Curr Opin Immunol. 1998;10:12–15. doi: 10.1016/s0952-7915(98)80024-1. [DOI] [PubMed] [Google Scholar]
- 35.Mukaida N, Morita M, Ishikawa Y, Rice N, Okamoto S, Kasahara T, Matsushima K. Novel mechanism of glucocorticoid-mediated gene repression. Nuclear factor-κB is target for glucocorticoid-mediated interleukin 8 gene repression. J Biol Chem. 1994;269:13289–13295. [PubMed] [Google Scholar]
- 36.Muzio M, Chinnaiyan A M, Kischkel F C, O’Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz J D, Zhang M, Gentz R, Mann M, Krammer P H, Peter M E, Dixit V M. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 37.Newton K, Harris A W, Bath M L, Smith K G C, Strasser A. A dominant interfering mutant of FADD/MORT1 enhances deletion of autoreactive thymocytes and inhibits proliferation of mature T lymphocytes. EMBO J. 1998;17:706–718. doi: 10.1093/emboj/17.3.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schall T J, Lewis M, Koller K J, Lee A, Rice G C, Wong G H, Gatanaga T, Granger G A, Lentz R, Raab H, et al. Molecular cloning and expression of a receptor for human tumor necrosis factor. Cell. 1990;61:361–370. doi: 10.1016/0092-8674(90)90816-w. [DOI] [PubMed] [Google Scholar]
- 39.Screaton G R, Xu X N, Olsen A L, Cowper A E, Tan R, McMichael A J, Bell J I. LARD: a new lymphoid-specific death domain containing receptor regulated by alternative pre-mRNA splicing. Proc Natl Acad Sci USA. 1997;94:4615–4619. doi: 10.1073/pnas.94.9.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheridan J P, Marsters S A, Pitti R M, Gurney A, Skubatch M, Baldwin D, Ramakrishnan L, Gray C L, Baker K, Wood W I, Goddard A D, Godowski P, Ashkenazi A. Control of TRAIL-induced apoptosis by a family of signaling and decoy receptors. Science. 1997;277:818–821. doi: 10.1126/science.277.5327.818. [DOI] [PubMed] [Google Scholar]
- 41.Singh A, Ni J, Aggarwal B B. Death domain receptors and their role in cell demise. J Interferon Cytokine Res. 1998;18:439–450. doi: 10.1089/jir.1998.18.439. [DOI] [PubMed] [Google Scholar]
- 42.Songyang Z, Lu K P, Kwon Y T, Tsai L H, Filhol O, Cochet C, Brickey D A, Soderling T R, Bartleson C, Graves D J, DeMaggio A J, Hoekstra M F, Blenis J, Hunter T, Cantley L C. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol Cell Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stanger B Z, Leder P, Lee T H, Kim E, Seed B. RIP: a novel protein containing a death domain that interacts with Fas/APO-1 (CD95) in yeast and causes cell death. Cell. 1995;81:513–523. doi: 10.1016/0092-8674(95)90072-1. [DOI] [PubMed] [Google Scholar]
- 44.Tewari M, Telford W G, Miller R A, Dixit V M. CrmA, a poxvirus-encoded serpin, inhibits cytotoxic T-lymphocyte-mediated apoptosis. J Biol Chem. 1995;270:22705–22708. doi: 10.1074/jbc.270.39.22705. [DOI] [PubMed] [Google Scholar]
- 45.Thome M, Hofmann K, Burns K, Martinon F, Bodmer J L, Mattmann C, Tschopp J. Identification of CARDIAK, a RIP-like kinase that associates with caspase-1. Curr Biol. 1998;8:885–888. doi: 10.1016/s0960-9822(07)00352-1. [DOI] [PubMed] [Google Scholar]
- 46.Thompson C B. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 47.Ting A T, Pimentel-Muinos F X, Seed B. RIP mediates tumor necrosis factor receptor 1 activation of NF-κB but not Fas/APO-1-initiated apoptosis. EMBO J. 1996;15:6189–6196. [PMC free article] [PubMed] [Google Scholar]
- 48.Trofimova M, Sprenkle A B, Green M, Sturgill T W, Goebl M G, Harrington M A. Developmental and tissue-specific expression of mouse pelle-like protein kinase. J Biol Chem. 1996;271:17609–17612. doi: 10.1074/jbc.271.30.17609. [DOI] [PubMed] [Google Scholar]
- 49.Vig E, Green M, Liu Y, Donner D B, Mukaida N, Goebl M G, Harrington M A. Modulation of tumor necrosis factor and interleukin-1-dependent NF-κB activity by mPLK/IRAK. J Biol Chem. 1999;274:13077–13084. doi: 10.1074/jbc.274.19.13077. [DOI] [PubMed] [Google Scholar]
- 50.Ware C F, VanArsdale S, VanArsdale T L. Apoptosis mediated by the TNF-related cytokine and receptor families. J Cell Biochem. 1996;60:47–55. doi: 10.1002/(SICI)1097-4644(19960101)60:1%3C47::AID-JCB8%3E3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 51.Warzocha K, Ribeiro P, Charlot C, Renard N, Coiffier B, Salles G. A new death receptor 3 isoform: expression in human lymphoid cell lines and non-Hodgkin’s lymphomas. Biochem Biophys Res Commun. 1998;242:376–379. doi: 10.1006/bbrc.1997.7948. [DOI] [PubMed] [Google Scholar]
- 52.Whitley M Z, Thanos D, Read M A, Maniatis T, Collins T. A striking similarity in the organization of the E-selectin and beta interferon gene promoters. Mol Cell Biol. 1994;14:6464–6475. doi: 10.1128/mcb.14.10.6464. [DOI] [PMC free article] [PubMed] [Google Scholar]