Abstract
The microbiota—the diverse set of commensal microbes that normally colonize humans—represents the first line of defense against infectious diseases. In this review, we summarize the direct and indirect mechanisms by which the microbiota modulates susceptibility to—and severity of—infections, with a focus on immunological mechanisms. Moreover, we highlight some of the ways that modern-world lifestyles have influenced the structure–function relationship between the microbiota and infectious diseases. Ultimately, understanding how the microbiota influences infectious risks will facilitate development of microbiota-derived therapeutics that bolster host defenses.
The incidence of infectious diseases decreased dramatically throughout the 20th century owing to improvements in sanitation, advances in vaccinology, and the development of antimicrobials and other infectious disease control measures (1); however, infections still accounted for 3 of the top 10 causes of death worldwide in 2019 (2). Because of this importance of infections to public health, there has been intense investigation over the past century to better understand how pathogens cause disease and what can be done to stop this process. Ever since Pasteur published his germ theory in the mid-19th century, there has been an almost singular focus on studying pathogens. This view was further reinforced over the next ~125 years with the introduction of Koch’s postulates and Falkow’s molecular postulates (3, 4), which focused on identifying the microbe and/or microbial gene(s) that caused infectious diseases and provided the conceptual backdrop for most microbial pathogenesis research in the 20th century.
Even in Pasteur’s time, though, there was disagreement regarding the relative importance of the pathogen and the context of the host–pathogen interaction. Antoine Béchamp, a contemporary of Pasteur, argued that the “terrain” and “internal milieu” of the body was critical for the development of disease after infection by the pathogen (5). Although Pasteur’s germ theory carried the day, Pasteur, shortly before his death, recognized the strength of Béchamp’s argument by stating: “Béchamp was right—the microbe is nothing. The terrain is everything” (6). Although Béchamp was incorrect to ignore the importance of the pathogen itself, there is now growing appreciation for his central tenet that the general landscape in which the infection occurs is also important. This concept has more recently been expanded upon with the damage response framework and the idea of disease tolerance (7, 8). Both of these paradigms invoke the notion that the host response—along with attributes of the pathogen itself—is necessary to determine whether, and how severely, disease may develop or whether the host is able to tolerate the infectious challenge. This idea of a context-dependent nature to infection that started with Béchamp and continues to evolve into the 21st century must now also integrate the microbiota, the diverse collection of host-associated microorganisms that regulates host physiology, into the concept of “terrain”.
The intricate interactions between these three factors—the host, pathogen, and microbiota—determine the outcome of an infection. However, the microbiota itself is incredibly dynamic. In addition to intra- and inter-individual variability (9–12), there are notable changes to the microbiota associated with modernization, whether it be related to dietary shifts (13, 14), technological and medical advancements (15, 16), or increases in lifestyle-associated diseases (17, 18). Given that the microbiota, virulence strategies of pathogens, and host-specific defense mechanisms have individually been reviewed extensively (19–22), this review focuses on the mechanisms underlying how the microbiota modulates infectious diseases (Figure 1). Given the numerous studies in this area, it is challenging to be truly comprehensive. As such, we highlight exemplar vignettes of how the microbiota regulates infections, with a general focus on immune mechanisms.
Figure 1. The microbiota impacts infectious diseases via multiple mechanisms.
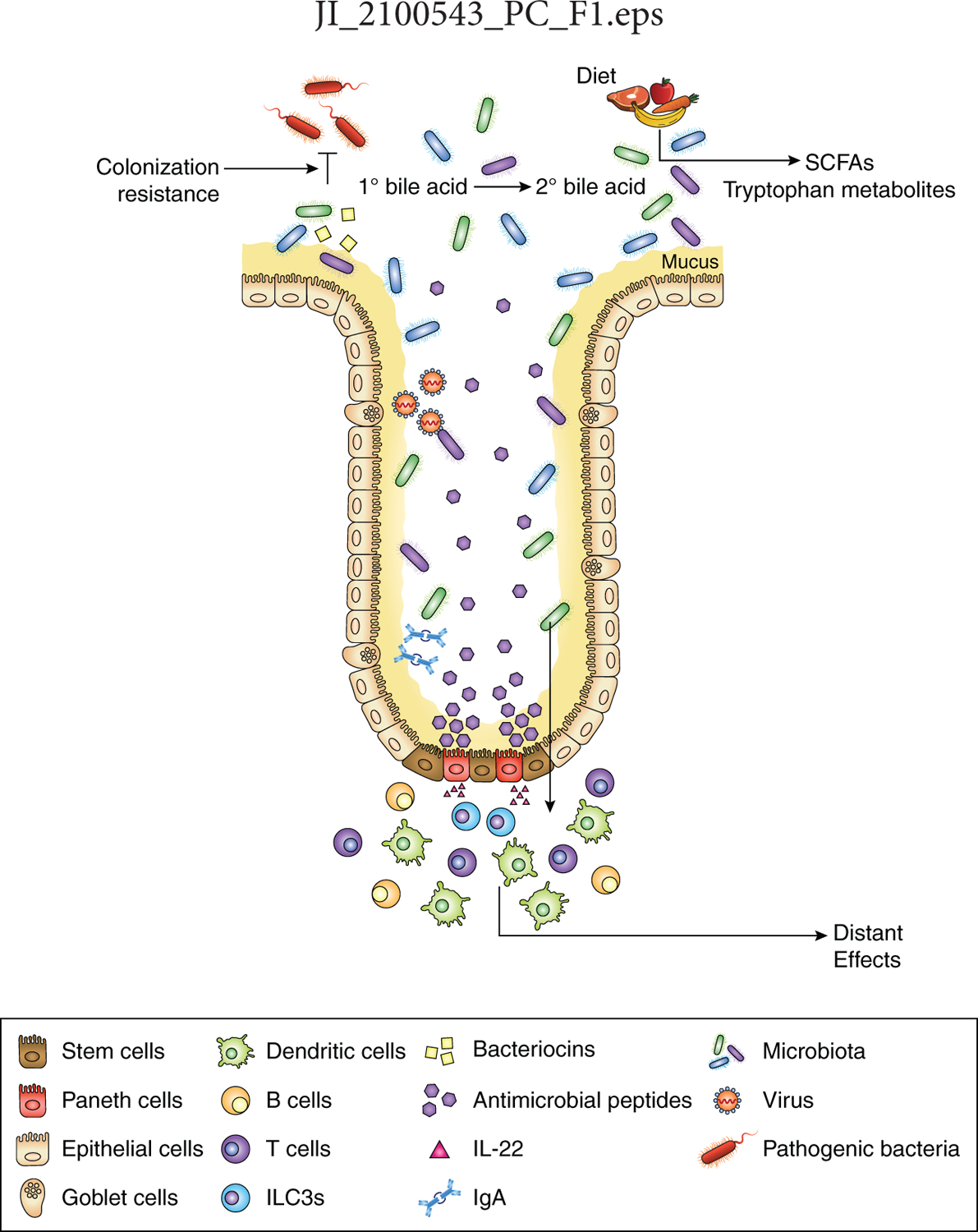
The microbiota directly affects the ability of pathogens to colonize the host by niche exclusion (colonization resistance) and producing molecules (e.g., bacteriocins) that target pathogens. Bacterial products can both influence the infectivity of pathogens and also modulate the immune system. The microbiota indirectly alters the course of infection by modulating epithelial cell responses [e.g., mucus production, expression of antimicrobial peptides (AMPs), stem cell regeneration) and through the production of various bacterial metabolites. These various mechanisms can alter the course of infectious diseases locally and also at distant sites. Although these processes occur at all mucosal surfaces, the small-intestinal epithelium is depicted with enterocytes in yellow, goblet cells in purple, stem cells in brown, and Paneth cells in red.
Microbe–microbe interactions
The members of the microbiota exist in a delicate ecological balance with one another. Within these networks, numerous microbe–microbe interactions occur that enable persistence of some species (i.e., cross-feeding interactions) and exclusion of others (23). One major mechanism by which the microbiota protects against infections is by simply occupying the same niche that the pathogen would normally colonize. Seminal studies performed in the 1950’s and 1960’s demonstrated that the intestinal microbiota protected animals against infection (24–27). This manner of protection, later coined “colonization resistance” (28), could be abrogated by antibiotic treatment and restored with supplementation of commensal microbes. It has been known since the 1970’s that this protection is largely reliant on anaerobic organisms (28, 29), and the subsequent half-century has been spent trying to identify the specific microbe(s) involved (30). In addition to niche competition, the microbiota can also produce molecules, such as bacteriocins, that directly antagonize would-be pathogens (discussed in more detail below). Moreover, there is precedence for microbiota-derived quorum-sensing molecules to impact infection: expression of luxS [autoinducer 2 (AI-2) synthase] by Ruminococcus obeum induces repression of several of Vibrio cholerae colonization factors (31). Although the precise mechanism for this repression is unknown, it does not require the V. cholerae AI-2 sensor, luxP. Importantly, though, this example highlights that commensal bacteria can “talk” to pathogens as a means to protect against infection. Although the host and microbiota normally exist in a mutualistic state of détente, slight perturbations to this equilibrium result in inflammation, which subtly changes the redox state of the intestine in a way that favors pathogens and results in a bloom of Enterobacteriaceae (32, 33). The inability of the microbiota to adequately deal with these increases in reactive oxygen and nitrogen species predisposes the host to enteric infection.
Beyond these interbacterial interactions, bacteria–virus interactions have recently been demonstrated to be important as well. Intriguingly, some enteric viruses (e.g., poliovirus, reovirus, mouse mammary tumor virus) require the presence of commensal bacteria for pathogenicity (34, 35). In these cases, the viruses bind bacterial surface molecules (e.g., lipopolysaccharide, peptidoglycan), thereby triggering Toll-like receptor (TLR) responses that promote viral infectivity and enhancing receptor binding (34, 35). Interestingly, the specific bacterial requirements may vary for different viruses, even those that are closely related (36). In a more complex interaction, the microbiota differentially influences the pathogenicity of norovirus infections in a region-specific manner (37, 38). Biotransformation of bile acids in the proximal small intestine primes the type III interferon response and inhibits norovirus infection; in contrast, the microbiota stimulates norovirus infection in the distal gut (37). Finally, bacteriophages—viruses that are specific for bacteria—can also influence infections. While there is less robust data regarding the role of endogenous bacteriophages in preventing infection, there has recently been resurging interest in the clinical use of bacteriophages for treatment of infections that are otherwise medically recalcitrant (39, 40). Although these reports are still at the case report level, it underscores the important role that bacteriophages may play in modulating susceptibility to various infections.
Microbe-host interactions
Timing and memory of microbial exposures
The first glimpse of the microbial world begins in utero. Although the existence of a placental microbiome remains controversial (41), it is widely established that maternal transfer of antibodies and microbe-derived metabolites—both of which occur prenatally and via breast milk—provides passive immunity against neonatal infections and augments neonatal immune development (42–45). In some cases, these maternal antibodies are induced by the microbiota, cross-react with pathogens, and confer protection against infection to the offspring (46). Neonatal acquisition of specific commensal bacterial taxa, such as clostridial species, is necessary for protection against infection (47). Conversely, the neonatal microbiota is required for the development of immunosuppressive CD71+ erythroid cells, which protect against excessive commensal microbe-induced inflammation but also leaves neonates more susceptible to infections (48). Moreover, some of the immunoregulatory changes induced in early postnatal life (e.g., invariant NK T cells, regulatory T cells, lymphoid tissue inducer-like cells) have life-long consequences that may similarly impact susceptibility to infection later in life (49–51). Considered together, the microbiota orchestrates host defenses against infections beginning in the prenatal period and provides multiple layers of transient protection during neonatal windows of vulnerability, which may also have lasting effects. It is likely that this process is disrupted due to peri- and postpartum antibiotic exposure and may contribute to elevated rates of sepsis observed in antibiotic-exposed infants (52, 53).
These microbial exposures are important not only in shaping the acute response against infection, but they are also critical for the development of immunological memory after an infection. For example, the microbiota is required for the generation of virus-specific CD8+ memory T cells (54, 55). Moreover, gastrointestinal infection with Toxoplasma gondii causes a loss of tolerance to the microbiota, with microbiota-specific CD4+ T cells forming memory cells that are phenotypically similar to pathogen-specific T cells (56). Notably, these studies did not directly demonstrate that these microbiota-induced memory T cells actually confer protection against a secondary infectious challenge. More recent work has demonstrated that enteric infection causes an expansion of taurine-metabolizing bacterial species in the microbiota that confers greater resistance to subsequent infection (57).
Conceptually analogous to this idea that the microbiota influences the immune response after an active infection, it can also modulate the response to vaccines, which attempt to mimic immunologically key aspects of infection. Indeed, murine experiments have demonstrated that microbiota–TLR 5 interactions are required for a robust vaccine response (58, 59), though this appears to be more apparent for non-adjuvanted viral subunit vaccines (e.g., inactivated influenza, inactivated polio) and not live or adjuvanted vaccines (e.g., live attenuated yellow fever, Tdap/alum). In humans, disruption of the microbiota with antibiotics resulted in attenuated responses to influenza vaccination (60), findings that indicate the microbiota also potentiates vaccine responsiveness in humans. However, in some cases, the gut microbiota has been observed to distract vaccine-induced immunity, thereby leading to a poorer response (61). Furthermore, it has long been noted that many oral vaccines (e.g., polio, cholera, rotavirus) have lower immunogenicity in low- and middle-income countries as compared to high-income countries (62, 63). Although the exact reasons for this disparity remain unknown, it is thought to relate to differences in the intestinal milieu, including dysbiosis of the microbiota and higher incidence of environmental enteric dysfunction (63, 64). Taken together, these data suggest that the microbiota may serve as an adjuvant for certain vaccine types and could lead to novel microbiome-based methods for improving vaccine efficacy.
Local versus distant effects
In addition to the timing of microbial exposure, the location of these host–microbe interactions may also be important. In thinking about location, one must consider not only the macroenvironment (e.g., the organ or even the specific region of a given organ) but also the microenvironment (e.g., intestinal crypt or villus). This is true in all anatomic sites, as it is known that microbial communities in adjacent anatomic regions may be vastly different from one another (10, 65). These spatial differences, which reflect variability in physicochemical characteristics and immune effectors among other things, result in microbes selectively occupying locations suited to their adaptations (65–67).
Intuitively, it makes sense that spatial proximity may be required for the commensal microbiota to influence susceptibility to infectious diseases. For example, the gastric microbiota regulates clearance of Helicobacter pylori from the stomach by inducing local production of immunoglobulin A (68). However, it is now clear that the microbiota can also exert its effects across long distances. For example, segmented filamentous bacteria (SFB), which is most abundant in the distal ileum and largely absent from the colon, is known to induce ileal Th17 cells; however, it also protects against colonic infection with Citrobacter rodentium (69). That said, SFB also induces colonic expression of IL-17 (70), which may explain this apparent dichotomy between the site of a commensal bacteria and location of its effects. Furthermore, intestinal SFB protects against pneumonia due to Staphylococcus aureus (71), a finding that further highlights the distant effects conferred by the microbiota. Indeed, this notion of the gut microbiota impacting infections in distant organs has now been demonstrated for multiple sites, including the liver, lungs, and brain among others (71–75). However, virtually nothing is known about how microbial signals from the intestines are transmitted to these distant sites. Possibilities include that bacteria translocate across the intestinal epithelium and travel to these remote sites, that immune cells educated in the intestines travel to other organs (76, 77), that bacterial factors and/or metabolites produced in the gut are somehow ferried to distant areas (42, 78), or that there is a direct hard-wired connection (e.g., the vagus nerve for the gut–brain axis) (79). None of these possibilities has conclusively been demonstrated as necessary or sufficient for modulating extra-intestinal infectious diseases.
Modulation of epithelial cell physiology
Epithelial cells are a critical partner to the microbiota in regulating susceptibility to infection as they can sense and quickly respond to changes in microbial signals delivered by commensal organisms. Two key defense mechanisms that provide direct protection against pathogens include establishing and maintaining a mucus barrier as well as expressing a variety of antimicrobial peptides (e.g., lysozyme, Reg3 proteins, defensins). Both of these features are regulated by the microbiota (80, 81). The importance of mucus in protecting the host against infection has been shown in the lungs and gut (82–86). In the large intestine, each crypt opening is guarded by a sentinel goblet cell, which is able to detect TLR ligands only when their concentrations are high as occurs during breaches of the mucus layer (87). In addition to increasing their own mucus secretion, these sentinel goblet cells generate an intercellular gap junction signal that activates adjacent goblet cells to also increase mucus secretion (87). Antimicrobial peptides help maintain a bacteria-free zone adjacent to epithelial cells and have critical roles in protecting against infection due to a variety of pathogens (88–91). The mechanism for microbiota-induced antimicrobial peptide expression has been best studied for Reg3γ, where the working model holds that two independent pathways are simultaneously required (92): luminal microbe-associated molecular patterns (e.g., lipopolysaccharide) signal through intestinal epithelial cell TLRs and a MyD88-dependent pathway to increase Reg3γ expression while group 3 innate lymphoid cell secretion of IL-22 in the lamina propria is also required for Reg3γ expression (93–96). However, while MyD88 signaling is required for Reg3γ expression in the colon, it is not required in the ileum (97). Moreover, it is less likely that TLR agonists are generically able to stimulate Reg3γ expression in the small intestine given that very few bacterial species have this capacity (80, 98). Finally, mice deficient in IL-22 still express Reg3γ (99), an observation that indicates that IL-22 is not absolutely required for Reg3γ expression. Although exogenously administered IL-22 leads to increased Reg3γ expression (100), this could be through a pathway distinct from commensal-induced Reg3γ. Taken together, while Reg3γ expression—and antimicrobial peptide expression more generally—is dependent on the microbiota, the specific mechanisms underlying induction may differ among commensal microbes.
In addition to these mechanisms that directly protect against infection, the microbiota also helps regulate stem cell activity (101–103), which is crucial for maintaining barrier integrity in the setting of infection (104, 105). Finally, epithelial cells are also able to process signals from commensal bacteria to modulate the underlying immune system. For example, commensal bacteria regulate MHCII expression on intestinal epithelial cells such that they can present antigens to immune cells in the lamina propria (106). SFB represents a specific illustration of this, where SFB proteins are endocytosed, processed, and presented by intestinal epithelial cells to induce Th17 cells (107). Moreover, commensal organisms can alter cytokine production by epithelial cells, which results in a different immune microenvironment that is less permissive to invading microbes (108, 109). A conceptually similar example involves clostridial regulation of retinoic acid levels in intestinal epithelial cells, which results in regulating IL-22 responses and colonization by pathogens (110). While the vast majority of studies examining how the microbiota regulates host epithelial cell biology focus on commensal bacteria, there is an increasing appreciation for the importance of other microbial classes. For example, Tritrichomonas musculis, a murine commensal protist, has been shown to protect against enteric infection by inducing IL-18 in epithelial cells (111) Taken together, these examples highlight how microbiota–epithelial cell interactions help modulate susceptibility to infections through both direct and indirect mechanisms.
Microbiota-derived molecules that modulate infectious diseases
Commensal microbes produce chemicals at a diversity that rivals that of any other microbial ecosystem (112, 113), a feature that attests to their vast potential in modulating host physiology, including susceptibility to infection. The molecules can either be produced directly by commensal microbes (i.e., microbial products) or result from microbe-mediated modification of host compounds (i.e., microbial metabolites). Regardless of how they are produced, these microbiota-derived molecules can influence infections by either modulating the host response or directly impacting pathogens.
Microbial products
The first century of microbiology largely focused on pathogens, with attempts to clarify the molecular basis for how they cause disease. A wide variety of virulence factors have been identified, and one major class includes pattern recognition receptor (PRR) ligands, such as lipopolysaccharide, lipoteichoic acid, and peptidoglycan (114). Just as these molecules help pathogens shape the host response to infection, the host immune system is similarly guided by the TLR agonists expressed by commensal bacteria. NOD1 recognition of microbiota-derived peptidoglycan enhances neutrophil function to help protect against pneumococcal sepsis (115), and glycolipids from various Bacteroides species induce interferon β that leads to protection against infection with either influenza virus or vesicular stomatitis virus (116). It is not clear what drives these divergent responses across different bacteria, though it may relate to subtle—yet important—structural variations in the bacterial products. Moreover, the context of how these molecules are delivered is also critical. Bacteroides fragilis polysaccharide A, the archetypal immunomodulatory molecule from a commensal bacterium, is required for abdominal abscess formation but also promotes immunoregulatory changes in other disease settings (117). In contrast to direct production of PRR ligands impacting infection severity, Enterococcus faecium produces a peptidoglycan hydrolase, SagA, that protects against Salmonella and Clostridiodes difficile infection in a MyD88- and NOD2-dependent fashion (118, 119).
Beyond expression of PRR ligands that modulate host immunity, commensal microbes also produce molecules that can directly antagonize pathogens, a function that helps them maintain their niche in an ecologically dense environment. One manner in which bacteria accomplish this is through the secretion of bacteriocins, which are peptide-based toxins that target and kill other bacteria. In many cases, these bacteriocins specifically target similar or closely related species, such as observed with microcin-producing Escherichia coli protecting against infection with adherent-invasive E. coli and Salmonella (120). In some cases, the bacteriocin confers protection against more distantly related species: a Blautia producta-produced lantibiotic reduces colonic colonization of vancomycin-resistant enterococci (121). Finally, gassericin A, a bacteriocin expressed by Lactobacillus gasseri that targets various food-borne pathogens, confers resistance to diarrhea by binding to keratin 19 in the plasma membrane of intestinal epithelial cells and enhancing fluid absorption (122), a finding that demonstrates bacteriocins can also directly affect the host.
Microbial Metabolites
Some of the best studied microbiota-derived metabolites include short chain fatty acids (SCFAs), bile acids, and tryptophan breakdown products. There are a growing number of examples where each of these classes of molecules help protect against infection. Commensal bacteria differ in their capacity and propensity to produce SCFAs, with differences also present in which specific SCFAs are made; however, multiple SCFAs have been shown to reduce infections. Microbiota-derived butyrate, particularly from clostridial organisms, helps maintain the intestinal barrier integrity and protects against Salmonella infection (123, 124); acetate produced by Bifidobacterium longum can improve intestinal defense and protect against enteropathogenic infection (125); and succinate generated by Tritrichomonas, a protozoan, induces small-intestinal remodeling and limits infection by other helminths (126). Furthermore, SCFAs help acidify the proximal colon and directly facilitate clearance of a variety of aerobic pathogens by eliminating the benefit of their O2 and NO3 respiration (127). More broadly, SCFAs offer a dual-pronged defense against infection by also promoting host antimicrobial immune responses (126, 128–131). In the case of bile acids, many commensal bacteria are able to hydrolyze primary bile acids into free bile acids, but only a small subset are able to dehydroxylate them into secondary bile acids (132). Generation of secondary bile acids by Clostridium scindens has been shown to protect against C. difficile and Entamoeba histolytica (133, 134). Moreover, colonization resistance to Vibrio cholerae is mediated via microbiome-derived bile salt hydrolase activity, and the abundance of this enzyme in the fecal microbiota of humans correlates with infection (135). Similar to SCFAs, microbiota-derived tryptophan metabolites (e.g., indole and its derivatives) can act directly on bacterial and fungal pathogens, in this case by inhibiting virulence pathways and/or pathogen growth (136–140). Moreover, tryptophan metabolites also modulate the mucosal barrier function and the host immune response (most notably, levels of IL-22 and group 3 innate lymphoid cells) to impact bacterial and fungal infections (141–144). Intriguingly, indole-3-carbinol, a specific tryptophan metabolite that regulates the number of intestinal group 3 innate lymphoid cells, is vertically transmitted from mother to infant via breast milk (42), thereby providing a mechanism for microbiome-mediated multigenerational control of immune responses that can impact severity of infections (145, 146).
In addition to these widely studied metabolites, there is growing evidence of other bacterial metabolites that impact infection outcomes. Microbiome-mediated increases in taurine levels enhances NLRP6 inflammasome-induced colonic IL-18 secretion and downstream antimicrobial peptide expression, while histamine, spermine, and putrescine suppresses IL-18 secretion and antimicrobial peptide expression (147). While this study did not directly link these changes to infection severity, it is well established that antimicrobial peptides constitute a key host defense against infection. Finally, Clostridium orbiscindens metabolizes dietary flavonoids to produce desaminotyrosine, which induces type I interferon and protects against influenza virus (148). These examples, which highlight the range of potential microbiome-derived molecules and their effects, underscore how much more there is to be learned regarding bacterial metabolites. Efforts are ongoing to exhaustively catalog the genes and phenotypes of commensal microbes, which will undoubtedly unveil broader classes of metabolites that impact infection severity.
Modern world influences on the intestinal microbiota
As highlighted by the examples above, it is evident that the microbiota plays a critical role in modulating susceptibility to infection. It is important to note that medical and technological advancements over the past century have had profound impacts on the structure of the microbiota and risk of infections (Figure 2). For example, the advent of antibiotics has drastically reduced morbidity and mortality due to infectious diseases (1); however, these treatments have profound and lasting effects on the microbiome (15, 149). Even a quarter of drugs that are not conventional antimicrobials influence the growth of commensal microbes (150). While antibiotic treatment has clearly been linked to increased risk of subsequent infections, it is likely—but as of yet unproven—that some of these non-antimicrobial therapies similarly modify the microbiota in ways that alter susceptibility to infection. This concept of collateral damage to the microbiota has been incorporated into antimicrobial stewardship strategies, and it is likely that a similar consideration may be needed for other drugs as well. Other changes in medical practice beyond therapeutics also alter the microbiota and can impact infectious risk. Indeed, childbirth via Cesarean section, which has become much more common in modern times, is associated with a small, but statistically significant, increase in rates of enteric infections in children less than 5 years of age (151). This association between Cesarean sections and an altered microbiota in early life has given rise to attempts to normalize this dysbiotic microbial community (152), though it remains to be seen whether this sort of intervention alters health outcomes. A large-scale randomized clinical trial, however, has shown that treatment of high-risk infants with a combination of prebiotics and probiotics resulted in a decreased incidence of sepsis (153), a finding that demonstrates modulation of the microbiota in early life can impact rates of infection.
Figure 2. Modernization of lifestyles has altered the microbiota and infectious risks.
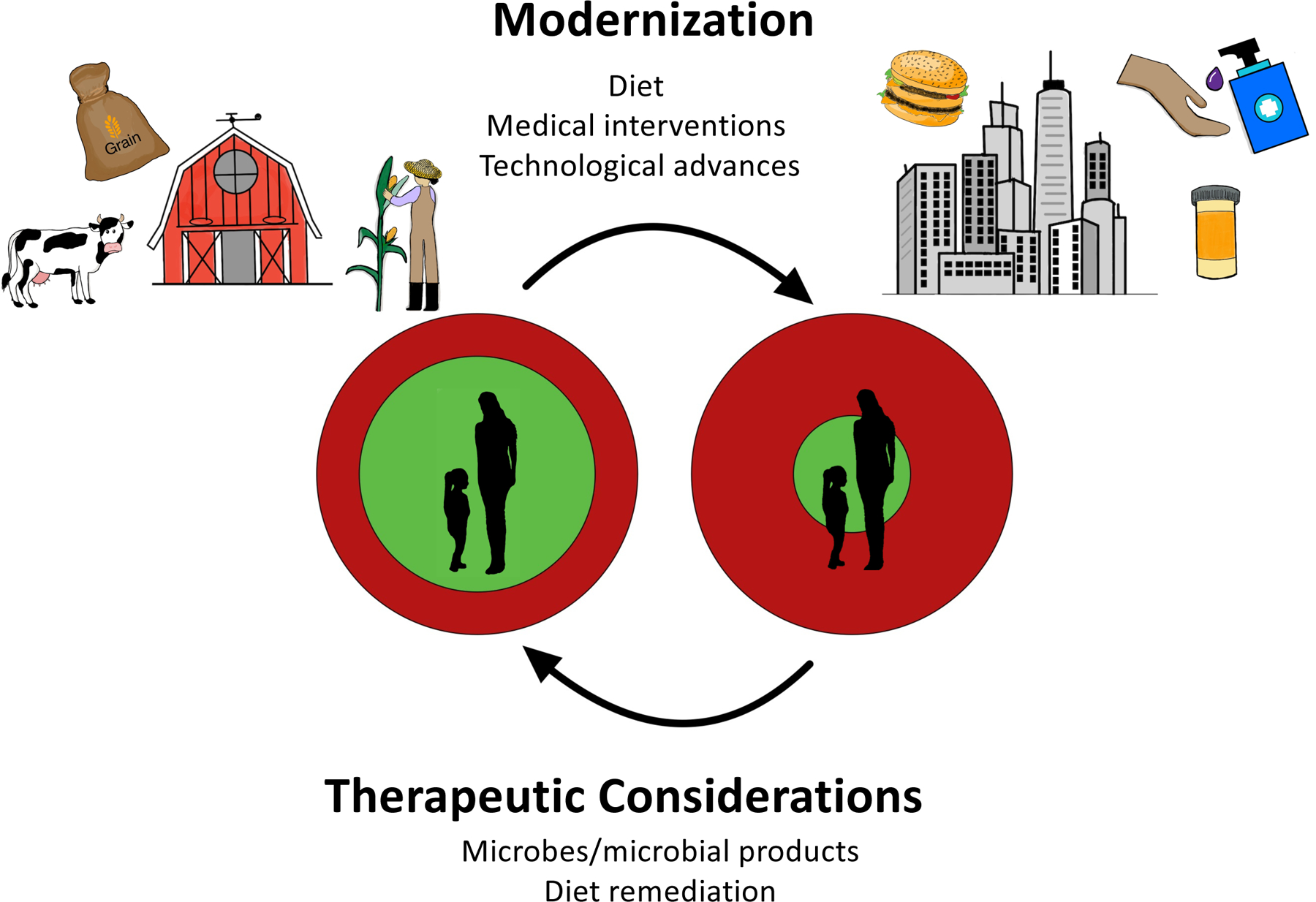
Although individuals benefit from the many conveniences of urbanization and technological developments, these same factors have altered the microbiota. In doing so, the protective “shroud” offered by the commensal microbiota (green circle) is insufficient to defend against pathogenic exposures (red circle; right side). With ongoing development of microbiome-based therapeutics—including prebiotics to alter the structure–function of the microbiome—there is an opportunity to restore the microbiome such that its protective shroud begins to cover the host again and provides balance to host–microbiota–pathogen interactions (left side).
Beyond changing medical practices, technological advancements have completely reshaped dietary habits such that the typical Western diet bears little resemblance to that of a hunter–gatherer (154). These changes have had a drastic impact on the microbiota as evidenced in both controlled and cross-sectional studies (12, 13, 155). Diets that contain low amounts of fiber—a hallmark of Western diets—are linked with thinner mucus layers, disrupted spatial localization of the microbiota, reduced SCFA production, and altered susceptibility to infections (85, 156–158). In addition, the increased protein content of Western diets leads to greater tryptophan intake, which may lead to functionally important differences in tryptophan metabolites and infection severity (144, 159). Of note, microbiota changes induced by dietary shifts impact many non-immune facets of host physiology, which may also impact the development of infectious diseases (160). These dietary effects on the microbiota have increased interest in developing dietary interventions and rational prebiotics that help shape the microbiota into beneficial states (161), though the specific intervention may need to be individualized (162).
Conclusions
Although the microbiota has long been known to provide colonization resistance against infections, it is now clear that it also modulates susceptibility to infectious diseases through a variety of mechanisms. Alterations in the microbiota, particularly those induced by modern world changes, influence and—in many cases—increase susceptibility to infections; however, this should not be seen as a call to revert back to a “simpler” lifestyle. Rather, careful consideration of how these changes impact microbiome-dependent infectious risks will highlight high-priority areas—some of which are detailed in this review—that can be exploited as therapeutic strategies. Ultimately, understanding the molecular mechanisms underlying how the microbiota impacts infectious risks and severity of infections will empower development of therapies that can shift this host–microbiota–pathogen balance back to a healthier state.
Grant Support:
C.Y.T. is supported by a National University of Singapore Development Grant. Z.E.R. is supported by the National Science Foundation grant DGE 1545220. N.K.S. receives support from The Hartwell Foundation, as a Translating Duke Health Scholar, and as a Whitehead Scholar.
References
- 1.Centers for Disease, C., and Prevention. 1999. Control of infectious diseases. MMWR. Morb. Mortal. Wkly. Rep 48: 621–629. [PubMed] [Google Scholar]
- 2.WHO. 2020. The top 10 causes of death.
- 3.Falkow S 1988. Molecular Koch’s postulates applied to microbial pathogenicity. Rev. Infect. Dis 10Suppl 2: S274–276. [DOI] [PubMed] [Google Scholar]
- 4.Koch R 1876. Untersuchungen über Bakterien: V. Die Ätiologie der Milzbrand-Krankheit, begründet auf die Entwicklungsgeschicte des Bacillus anthracis. Cohns Beitrage zur Biologie der Pflanzen 2: 277–310. [Google Scholar]
- 5.James DG 1982. The Hunterian oration on Louis Pasteur’s final judgement. Host reaction, soil or terrain. Trans. Med. Soc. Lond 99–100: 131–147. [PubMed] [Google Scholar]
- 6.Zumla A, Maeurer M, and C. Host-Directed Therapies Network. 2015. Host-Directed Therapies for Tackling Multi-Drug Resistant Tuberculosis: Learning From the Pasteur-Bechamp Debates. Clin. Infect. Dis 61: 1432–1438. [DOI] [PubMed] [Google Scholar]
- 7.Casadevall A, and Pirofski L.-a.. 2003. The damage-response framework of microbial pathogenesis. Nature Reviews Microbiology 1: 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider DS, and Ayres JS. 2008. Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nature Reviews Immunology 8: 889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.David LA, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, Perrotta A, Erdman SE, and Alm EJ. 2014. Host lifestyle affects human microbiota on daily timescales. Genome Biol 15: R89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.C. Human Microbiome Project 2012. Structure, function and diversity of the healthy human microbiome. Nature 486: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, and Ley RE. 2011. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. U. S. A 108Suppl 1: 4578–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, and Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486: 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, and Turnbaugh PJ. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505: 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smits SA, Leach J, Sonnenburg ED, Gonzalez CG, Lichtman JS, Reid G, Knight R, Manjurano A, Changalucha J, Elias JE, Dominguez-Bello MG, and Sonnenburg JL. 2017. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 357: 802–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dethlefsen L, and Relman DA. 2011. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. U. S. A 108Suppl 1: 4554–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shao Y, Forster SC, Tsaliki E, Vervier K, Strang A, Simpson N, Kumar N, Stares MD, Rodger A, Brocklehurst P, Field N, and Lawley TD. 2019. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 574: 117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shelton CD, and Byndloss MX. 2020. Gut Epithelial Metabolism as a Key Driver of Intestinal Dysbiosis Associated with Noncommunicable Diseases. Infect. Immun 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, and Gordon JI. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027–1031. [DOI] [PubMed] [Google Scholar]
- 19.Finlay BB, and Falkow S. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev 61: 136–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martens EC, Neumann M, and Desai MS. 2018. Interactions of commensal and pathogenic microorganisms with the intestinal mucosal barrier. Nat. Rev. Microbiol 16: 457–470. [DOI] [PubMed] [Google Scholar]
- 21.Nish S, and Medzhitov R. 2011. Host defense pathways: role of redundancy and compensation in infectious disease phenotypes. Immunity 34: 629–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Surana NK, and Kasper DL. 2014. Deciphering the tete-a-tete between the microbiota and the immune system. J. Clin. Invest 124: 4197–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rakoff-Nahoum S, Foster KR, and Comstock LE. 2016. The evolution of cooperation within the gut microbiota. Nature 533: 255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bohnhoff M, Drake BL, and Miller CP. 1954. Effect of streptomycin on susceptibility of intestinal tract to experimental Salmonella infection. Proc. Soc. Exp. Biol. Med 86: 132–137. [DOI] [PubMed] [Google Scholar]
- 25.Bohnhoff M, Miller CP, and Martin WR. 1964. Resistance of the Mouse’s Intestinal Tract to Experimental Salmonella Infection. I. Factors Which Interfere with the Initiation of Infection by Oral Inoculation. J Exp Med 120: 805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freter R 1956. Coproantibody and bacterial antagonism as protective factors in experimental enteric cholera. J. Exp. Med 104: 419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freter R 1956. Experimental enteric Shigella and Vibrio infections in mice and guinea pigs. J. Exp. Med 104: 411–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Waaij D, Berghuis-de Vries JM, and Lekkerkerk L.-v.. 1971. Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J. Hyg. (Lond) 69: 405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Waaij D, Berghuis JM, and Lekkerkerk JE. 1972. Colonization resistance of the digestive tract of mice during systemic antibiotic treatment. J. Hyg. (Lond) 70: 605–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ratner M 2016. Seres’s pioneering microbiome drug fails mid-stage trial. Nat. Biotechnol 34: 1004–1005. [DOI] [PubMed] [Google Scholar]
- 31.Hsiao A, Ahmed AM, Subramanian S, Griffin NW, Drewry LL, Petri WA Jr., Haque R, Ahmed T, and Gordon JI. 2014. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature 515: 423–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, and Baumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467: 426–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD, Popova IE, Parikh SJ, Adams LG, Tsolis RM, Stewart VJ, and Baumler AJ. 2013. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339: 708–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kane M, Case LK, Kopaskie K, Kozlova A, MacDearmid C, Chervonsky AV, and Golovkina TV. 2011. Successful transmission of a retrovirus depends on the commensal microbiota. Science 334: 245–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, Hooper LV, Dermody TS, and Pfeiffer JK. 2011. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science 334: 249–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson CM, Woods Acevedo MA, McCune BT, and Pfeiffer JK. 2019. Related Enteric Viruses Have Different Requirements for Host Microbiota in Mice. J. Virol 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grau KR, Zhu S, Peterson ST, Helm EW, Philip D, Phillips M, Hernandez A, Turula H, Frasse P, Graziano VR, Wilen CB, Wobus CE, Baldridge MT, and Karst SM. 2020. The intestinal regionalization of acute norovirus infection is regulated by the microbiota via bile acid-mediated priming of type III interferon. Nat Microbiol 5: 84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR, Gonzalez-Hernandez MB, Iovine NM, Wobus CE, Vinje J, Tibbetts SA, Wallet SM, and Karst SM. 2014. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 346: 755–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dedrick RM, Guerrero-Bustamante CA, Garlena RA, Russell DA, Ford K, Harris K, Gilmour KC, Soothill J, Jacobs-Sera D, Schooley RT, Hatfull GF, and Spencer H. 2019. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med 25: 730–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.El Haddad L, Harb CP, Gebara MA, Stibich MA, and Chemaly RF. 2019. A Systematic and Critical Review of Bacteriophage Therapy Against Multidrug-resistant ESKAPE Organisms in Humans. Clin. Infect. Dis 69: 167–178. [DOI] [PubMed] [Google Scholar]
- 41.Blaser MJ, Devkota S, McCoy KD, Relman DA, Yassour M, and Young VB. 2021. Lessons learned from the prenatal microbiome controversy. Microbiome 9: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gomez de Agüero M, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, Steinert A, Heikenwalder M, Hapfelmeier S, Sauer U, McCoy KD, and Macpherson AJ. 2016. The maternal microbiota drives early postnatal innate immune development. Science 351: 1296. [DOI] [PubMed] [Google Scholar]
- 43.Macpherson AJ, de Aguero MG, and Ganal-Vonarburg SC. 2017. How nutrition and the maternal microbiota shape the neonatal immune system. Nat. Rev. Immunol 17: 508–517. [DOI] [PubMed] [Google Scholar]
- 44.Koch MA, Reiner GL, Lugo KA, Kreuk LS, Stanbery AG, Ansaldo E, Seher TD, Ludington WB, and Barton GM. 2016. Maternal IgG and IgA Antibodies Dampen Mucosal T Helper Cell Responses in Early Life. Cell 165: 827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zinkernagel RM 2001. Maternal antibodies, childhood infections, and autoimmune diseases. N. Engl. J. Med 345: 1331–1335. [DOI] [PubMed] [Google Scholar]
- 46.Zheng W, Zhao W, Wu M, Song X, Caro F, Sun X, Gazzaniga F, Stefanetti G, Oh S, Mekalanos JJ, and Kasper DL. 2020. Microbiota-targeted maternal antibodies protect neonates from enteric infection. Nature 577: 543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim Y-G, Sakamoto K, Seo S-U, Pickard JM, Gillilland MG, Pudlo NA, Hoostal M, Li X, Wang TD, Feehley T, Stefka AT, Schmidt TM, Martens EC, Fukuda S, Inohara N, Nagler CR, and Núñez G. 2017. Neonatal acquisition of Clostridia species protects against colonization by bacterial pathogens. Science 356: 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elahi S, Ertelt JM, Kinder JM, Jiang TT, Zhang X, Xin L, Chaturvedi V, Strong BS, Qualls JE, Steinbrecher KA, Kalfa TA, Shaaban AF, and Way SS. 2013. Immunosuppressive CD71+ erythroid cells compromise neonatal host defence against infection. Nature 504: 158–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen VL, Surana NK, Duan J, and Kasper DL. 2013. Role of murine intestinal interleukin-1 receptor 1-expressing lymphoid tissue inducer-like cells in salmonella infection. PLoS One 8: e65405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, and Blumberg RS. 2012. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336: 489–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramanan D, Sefik E, Galvan-Pena S, Wu M, Yang L, Yang Z, Kostic A, Golovkina TV, Kasper DL, Mathis D, and Benoist C. 2020. An Immunologic Mode of Multigenerational Transmission Governs a Gut Treg Setpoint. Cell 181: 1276–1290 e1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singer JR, Blosser EG, Zindl CL, Silberger DJ, Conlan S, Laufer VA, DiToro D, Deming C, Kumar R, Morrow CD, Segre JA, Gray MJ, Randolph DA, and Weaver CT. 2019. Preventing dysbiosis of the neonatal mouse intestinal microbiome protects against late-onset sepsis. Nature Medicine 25: 1772–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou P, Zhou Y, Liu B, Jin Z, Zhuang X, Dai W, Yang Z, Feng X, Zhou Q, Liu Y, Xu X, and Zhang L. 2020. Perinatal Antibiotic Exposure Affects the Transmission between Maternal and Neonatal Microbiota and Is Associated with Early-Onset Sepsis. mSphere 5: e00984–00919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bachem A, Makhlouf C, Binger KJ, de Souza DP, Tull D, Hochheiser K, Whitney PG, Fernandez-Ruiz D, Dähling S, Kastenmüller W, Jönsson J, Gressier E, Lew AM, Perdomo C, Kupz A, Figgett W, Mackay F, Oleshansky M, Russ BE, Parish IA, Kallies A, McConville MJ, Turner SJ, Gebhardt T, and Bedoui S. 2019. Microbiota-Derived Short-Chain Fatty Acids Promote the Memory Potential of Antigen-Activated CD8+ T Cells. Immunity 51: 285–297.e285. [DOI] [PubMed] [Google Scholar]
- 55.Tanaka K, Sawamura S, Satoh T, Kobayashi K, and Noda S. 2007. Role of the Indigenous Microbiota in Maintaining the Virus-Specific CD8 Memory T Cells in the Lung of Mice Infected with Murine Cytomegalovirus. The Journal of Immunology 178: 5209. [DOI] [PubMed] [Google Scholar]
- 56.Hand TW, Dos Santos LM, Bouladoux N, Molloy MJ, Pagán AJ, Pepper M, Maynard CL, Elson CO, and Belkaid Y. 2012. Acute Gastrointestinal Infection Induces Long-Lived Microbiota-Specific T Cell Responses. Science 337: 1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stacy A, Andrade-Oliveira V, McCulloch JA, Hild B, Oh JH, Perez-Chaparro PJ, Sim CK, Lim AI, Link VM, Enamorado M, Trinchieri G, Segre JA, Rehermann B, and Belkaid Y. 2021. Infection trains the host for microbiota-enhanced resistance to pathogens. Cell 184: 615–627 e617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lynn MA, Tumes DJ, Choo JM, Sribnaia A, Blake SJ, Leong LEX, Young GP, Marshall HS, Wesselingh SL, Rogers GB, and Lynn DJ. 2018. Early-Life Antibiotic-Driven Dysbiosis Leads to Dysregulated Vaccine Immune Responses in Mice. Cell Host Microbe 23: 653–660 e655. [DOI] [PubMed] [Google Scholar]
- 59.Oh JZ, Ravindran R, Chassaing B, Carvalho FA, Maddur MS, Bower M, Hakimpour P, Gill KP, Nakaya HI, Yarovinsky F, Sartor RB, Gewirtz AT, and Pulendran B. 2014. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity 41: 478–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hagan T, Cortese M, Rouphael N, Boudreau C, Linde C, Maddur MS, Das J, Wang H, Guthmiller J, Zheng N-Y, Huang M, Uphadhyay AA, Gardinassi L, Petitdemange C, McCullough MP, Johnson SJ, Gill K, Cervasi B, Zou J, Bretin A, Hahn M, Gewirtz AT, Bosinger SE, Wilson PC, Li S, Alter G, Khurana S, Golding H, and Pulendran B. 2019. Antibiotics-Driven Gut Microbiome Perturbation Alters Immunity to Vaccines in Humans. Cell 178: 1313–1328.e1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams WB, Liao H-X, Moody MA, Kepler TB, Alam SM, Gao F, Wiehe K, Trama AM, Jones K, Zhang R, Song H, Marshall DJ, Whitesides JF, Sawatzki K, Hua A, Liu P, Tay MZ, Seaton KE, Shen X, Foulger A, Lloyd KE, Parks R, Pollara J, Ferrari G, Yu J-S, Vandergrift N, Montefiori DC, Sobieszczyk ME, Hammer S, Karuna S, Gilbert P, Grove D, Grunenberg N, McElrath MJ, Mascola JR, Koup RA, Corey L, Nabel GJ, Morgan C, Churchyard G, Maenza J, Keefer M, Graham BS, Baden LR, Tomaras GD, and Haynes BF. 2015. Diversion of HIV-1 vaccine–induced immunity by gp41-microbiota cross-reactive antibodies. Science 349: aab1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Plotkin SA, Lebrun A, and Koprowski H. 1960. Vaccination with the CHAT strain of type 1 attenuated poliomyelitis virus in Leopoldville. Belgian Congo. 2. Studies of the safety and efficacy of vaccination. Bull. World Health Organ 22: 215–234. [PMC free article] [PubMed] [Google Scholar]
- 63.Church JA, Parker EP, Kosek MN, Kang G, Grassly NC, Kelly P, and Prendergast AJ. 2018. Exploring the relationship between environmental enteric dysfunction and oral vaccine responses. Future Microbiol 13: 1055–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harris VC, Haak BW, Handley SA, Jiang B, Velasquez DE, Hykes BL Jr., Droit L, Berbers GAM, Kemper EM, van Leeuwen EMM, Boele van Hensbroek M, and Wiersinga WJ. 2018. Effect of Antibiotic-Mediated Microbiome Modulation on Rotavirus Vaccine Immunogenicity: A Human, Randomized-Control Proof-of-Concept Trial. Cell Host Microbe 24: 197–207 e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nava GM, Friedrichsen HJ, and Stappenbeck TS. 2011. Spatial organization of intestinal microbiota in the mouse ascending colon. The ISME Journal 5: 627–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stacy A, McNally L, Darch SE, Brown SP, and Whiteley M. 2016. The biogeography of polymicrobial infection. Nature Reviews Microbiology 14: 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Caballero S, Carter R, Ke X, Susac B, Leiner IM, Kim GJ, Miller L, Ling L, Manova K, and Pamer EG. 2015. Distinct but Spatially Overlapping Intestinal Niches for Vancomycin-Resistant Enterococcus faecium and Carbapenem-Resistant Klebsiella pneumoniae. PLoS Pathog 11: e1005132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Satoh-Takayama N, Kato T, Motomura Y, Kageyama T, Taguchi-Atarashi N, Kinoshita-Daitoku R, Kuroda E, Di Santo JP, Mimuro H, Moro K, and Ohno H. 2020. Bacteria-Induced Group 2 Innate Lymphoid Cells in the Stomach Provide Immune Protection through Induction of IgA. Immunity 52: 635–649 e634. [DOI] [PubMed] [Google Scholar]
- 69.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, and Littman DR. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139: 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, Eberl G, Snel J, Kelly D, and Cerf-Bensussan N. 2009. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31: 677–689. [DOI] [PubMed] [Google Scholar]
- 71.Gauguet S, D’Ortona S, Ahnger-Pier K, Duan B, Surana NK, Lu R, Cywes-Bentley C, Gadjeva M, Shan Q, Priebe GP, and Pier GB. 2015. Intestinal microbiota of mice influences resistance to Staphylococcus aureus pneumonia. Infect. Immun 83: 4003–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McAleer JP, and Kolls JK. 2018. Contributions of the intestinal microbiome in lung immunity. Eur. J. Immunol 48: 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brown DG, Soto R, Yandamuri S, Stone C, Dickey L, Gomes-Neto JC, Pastuzyn ED, Bell R, Petersen C, Buhrke K, Fujinami RS, O’Connell RM, Stephens WZ, Shepherd JD, Lane TE, and Round JL. 2019. The microbiota protects from viral-induced neurologic damage through microglia-intrinsic TLR signaling. eLife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Erny D, Hrabě de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, Keren-Shaul H, Mahlakoiv T, Jakobshagen K, Buch T, Schwierzeck V, Utermöhlen O, Chun E, Garrett WS, McCoy KD, Diefenbach A, Staeheli P, Stecher B, Amit I, and Prinz M. 2015. Host microbiota constantly control maturation and function of microglia in the CNS. Nature Neuroscience 18: 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McDonald B, Zucoloto AZ, Yu IL, Burkhard R, Brown K, Geuking MB, and McCoy KD. 2020. Programing of an Intravascular Immune Firewall by the Gut Microbiota Protects against Pathogen Dissemination during Infection. Cell Host Microbe 28: 660–668 e664. [DOI] [PubMed] [Google Scholar]
- 76.Morton AM, Sefik E, Upadhyay R, Weissleder R, Benoist C, and Mathis D. 2014. Endoscopic photoconversion reveals unexpectedly broad leukocyte trafficking to and from the gut. Proc. Natl. Acad. Sci. U. S. A 111: 6696–6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ochoa-Reparaz J, Mielcarz DW, Wang Y, Begum-Haque S, Dasgupta S, Kasper DL, and Kasper LH. 2010. A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal. Immunol 3: 487–495. [DOI] [PubMed] [Google Scholar]
- 78.Mazmanian SK, Liu CH, Tzianabos AO, and Kasper DL. 2005. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122: 107–118. [DOI] [PubMed] [Google Scholar]
- 79.Kaelberer MM, Buchanan KL, Klein ME, Barth BB, Montoya MM, Shen X, and Bohorquez DV. 2018. A gut-brain neural circuit for nutrient sensory transduction. Science 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Geva-Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz-Lopez A, Yanortsang TB, Yang L, Jupp R, Mathis D, Benoist C, and Kasper DL. 2017. Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell 168: 928–943 e911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Johansson ME, Jakobsson HE, Holmen-Larsson J, Schutte A, Ermund A, Rodriguez-Pineiro AM, Arike L, Wising C, Svensson F, Backhed F, and Hansson GC. 2015. Normalization of Host Intestinal Mucus Layers Requires Long-Term Microbial Colonization. Cell Host Microbe 18: 582–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ehre C, Worthington EN, Liesman RM, Grubb BR, Barbier D, O’Neal WK, Sallenave JM, Pickles RJ, and Boucher RC. 2012. Overexpressing mouse model demonstrates the protective role of Muc5ac in the lungs. Proc. Natl. Acad. Sci. U. S. A 109: 16528–16533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Johansson ME, Larsson JM, and Hansson GC. 2011. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl. Acad. Sci. U. S. A 108Suppl 1: 4659–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Roy MG, Livraghi-Butrico A, Fletcher AA, McElwee MM, Evans SE, Boerner RM, Alexander SN, Bellinghausen LK, Song AS, Petrova YM, Tuvim MJ, Adachi R, Romo I, Bordt AS, Bowden MG, Sisson JH, Woodruff PG, Thornton DJ, Rousseau K, De la Garza MM, Moghaddam SJ, Karmouty-Quintana H, Blackburn MR, Drouin SM, Davis CW, Terrell KA, Grubb BR, O’Neal WK, Flores SC, Cota-Gomez A, Lozupone CA, Donnelly JM, Watson AM, Hennessy CE, Keith RC, Yang IV, Barthel L, Henson PM, Janssen WJ, Schwartz DA, Boucher RC, Dickey BF, and Evans CM. 2014. Muc5b is required for airway defence. Nature 505: 412–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, Young VB, Henrissat B, Wilmes P, Stappenbeck TS, Nunez G, and Martens EC. 2016. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 167: 1339–1353 e1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bergstrom KS, Kissoon-Singh V, Gibson DL, Ma C, Montero M, Sham HP, Ryz N, Huang T, Velcich A, Finlay BB, Chadee K, and Vallance BA. 2010. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog 6: e1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Birchenough GM, Nystrom EE, Johansson ME, and Hansson GC. 2016. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science 352: 1535–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, Schnabl B, DeMatteo RP, and Pamer EG. 2008. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 455: 804–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Choi SM, McAleer JP, Zheng M, Pociask DA, Kaplan MH, Qin S, Reinhart TA, and Kolls JK. 2013. Innate Stat3-mediated induction of the antimicrobial protein Reg3gamma is required for host defense against MRSA pneumonia. J. Exp. Med 210: 551–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.van Ampting MT, Loonen LM, Schonewille AJ, Konings I, Vink C, Iovanna J, Chamaillard M, Dekker J, van der Meer R, Wells JM, and Bovee-Oudenhoven IM. 2012. Intestinally secreted C-type lectin Reg3b attenuates salmonellosis but not listeriosis in mice. Infect. Immun 80: 1115–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bel S, Pendse M, Wang Y, Li Y, Ruhn KA, Hassell B, Leal T, Winter SE, Xavier RJ, and Hooper LV. 2017. Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science 357: 1047–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mukherjee S, and Hooper LV. 2015. Antimicrobial defense of the intestine. Immunity 42: 28–39. [DOI] [PubMed] [Google Scholar]
- 93.Brandl K, Plitas G, Schnabl B, DeMatteo RP, and Pamer EG. 2007. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. J. Exp. Med 204: 1891–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C, and Diefenbach A. 2009. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat. Immunol 10: 83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, and Hooper LV. 2008. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc. Natl. Acad. Sci. U. S. A 105: 20858–20863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, and Hooper LV. 2011. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 334: 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Larsson E, Tremaroli V, Lee YS, Koren O, Nookaew I, Fricker A, Nielsen J, Ley RE, and Backhed F. 2012. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut 61: 1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Surana NK, and Kasper DL. 2017. Moving beyond microbiome-wide associations to causal microbe identification. Nature 552: 244–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McAleer JP, Nguyen NL, Chen K, Kumar P, Ricks DM, Binnie M, Armentrout RA, Pociask DA, Hein A, Yu A, Vikram A, Bibby K, Umesaki Y, Rivera A, Sheppard D, Ouyang W, Hooper LV, and Kolls JK. 2016. Pulmonary Th17 Antifungal Immunity Is Regulated by the Gut Microbiome. J. Immunol 197: 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhao D, Kim YH, Jeong S, Greenson JK, Chaudhry MS, Hoepting M, Anderson ER, van den Brink MR, Peled JU, Gomes AL, Slingerland AE, Donovan MJ, Harris AC, Levine JE, Ozbek U, Hooper LV, Stappenbeck TS, Ver Heul A, Liu TC, Reddy P, and Ferrara JL. 2018. Survival signal REG3alpha prevents crypt apoptosis to control acute gastrointestinal graft-versus-host disease. J. Clin. Invest 128: 4970–4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lesher S, Walburg HE Jr., and Sacher GA Jr. 1964. Generation Cycle in the Duodenal Crypt Cells of Germ-Free and Conventional Mice. Nature 202: 884–886. [DOI] [PubMed] [Google Scholar]
- 102.Peck BC, Mah AT, Pitman WA, Ding S, Lund PK, and Sethupathy P. 2017. Functional Transcriptomics in Diverse Intestinal Epithelial Cell Types Reveals Robust MicroRNA Sensitivity in Intestinal Stem Cells to Microbial Status. J. Biol. Chem 292: 2586–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, and Medzhitov R. 2004. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118: 229–241. [DOI] [PubMed] [Google Scholar]
- 104.Hefele M, Stolzer I, Ruder B, He GW, Mahapatro M, Wirtz S, Neurath MF, and Gunther C. 2018. Intestinal epithelial Caspase-8 signaling is essential to prevent necroptosis during Salmonella Typhimurium induced enteritis. Mucosal. Immunol 11: 1191–1202. [DOI] [PubMed] [Google Scholar]
- 105.Mileto SJ, Jarde T, Childress KO, Jensen JL, Rogers AP, Kerr G, Hutton ML, Sheedlo MJ, Bloch SC, Shupe JA, Horvay K, Flores T, Engel R, Wilkins S, McMurrick PJ, Lacy DB, Abud HE, and Lyras D. 2020. Clostridioides difficile infection damages colonic stem cells via TcdB, impairing epithelial repair and recovery from disease. Proc. Natl. Acad. Sci. U. S. A 117: 8064–8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Koyama M, Mukhopadhyay P, Schuster IS, Henden AS, Hulsdunker J, Varelias A, Vetizou M, Kuns RD, Robb RJ, Zhang P, Blazar BR, Thomas R, Begun J, Waddell N, Trinchieri G, Zeiser R, Clouston AD, Degli-Esposti MA, and Hill GR. 2019. MHC Class II Antigen Presentation by the Intestinal Epithelium Initiates Graft-versus-Host Disease and Is Influenced by the Microbiota. Immunity 51: 885–898 e887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ladinsky MS, Araujo LP, Zhang X, Veltri J, Galan-Diez M, Soualhi S, Lee C, Irie K, Pinker EY, Narushima S, Bandyopadhyay S, Nagayama M, Elhenawy W, Coombes BK, Ferraris RP, Honda K, Iliev ID, Gao N, Bjorkman PJ, and Ivanov II. 2019. Endocytosis of commensal antigens by intestinal epithelial cells regulates mucosal T cell homeostasis. Science 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kobayashi T, Voisin B, Kim DY, Kennedy EA, Jo JH, Shih HY, Truong A, Doebel T, Sakamoto K, Cui CY, Schlessinger D, Moro K, Nakae S, Horiuchi K, Zhu J, Leonard WJ, Kong HH, and Nagao K. 2019. Homeostatic Control of Sebaceous Glands by Innate Lymphoid Cells Regulates Commensal Bacteria Equilibrium. Cell 176: 982–997 e916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Scharschmidt TC, Vasquez KS, Pauli ML, Leitner EG, Chu K, Truong HA, Lowe MM, Sanchez Rodriguez R, Ali N, Laszik ZG, Sonnenburg JL, Millar SE, and Rosenblum MD. 2017. Commensal Microbes and Hair Follicle Morphogenesis Coordinately Drive Treg Migration into Neonatal Skin. Cell Host Microbe 21: 467–477 e465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Grizotte-Lake M, Zhong G, Duncan K, Kirkwood J, Iyer N, Smolenski I, Isoherranen N, and Vaishnava S. 2018. Commensals Suppress Intestinal Epithelial Cell Retinoic Acid Synthesis to Regulate Interleukin-22 Activity and Prevent Microbial Dysbiosis. Immunity 49: 1103–1115 e1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chudnovskiy A, Mortha A, Kana V, Kennard A, Ramirez JD, Rahman A, Remark R, Mogno I, Ng R, Gnjatic S, Amir ED, Solovyov A, Greenbaum B, Clemente J, Faith J, Belkaid Y, Grigg ME, and Merad M. 2016. Host-Protozoan Interactions Protect from Mucosal Infections through Activation of the Inflammasome. Cell 167: 444–456 e414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Donia MS, and Fischbach MA. 2015. Small molecules from the human microbiota. Science 349: 1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tan TG, Lim YS, Tan A, Leong R, and Pavelka N. 2019. Fungal Symbionts Produce Prostaglandin E2 to Promote Their Intestinal Colonization. Front Cell Infect Microbiol 9: 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Philpott DJ, and Girardin SE. 2004. The role of Toll-like receptors and Nod proteins in bacterial infection. Mol. Immunol 41: 1099–1108. [DOI] [PubMed] [Google Scholar]
- 115.Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, and Weiser JN. 2010. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nature Medicine 16: 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Stefan KL, Kim MV, Iwasaki A, and Kasper DL. 2020. Commensal Microbiota Modulation of Natural Resistance to Virus Infection. Cell 183: 1312–1324.e1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Surana NK, and Kasper DL. 2012. The yin yang of bacterial polysaccharides: lessons learned from B. fragilis PSA. Immunol. Rev 245: 13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pedicord VA, Lockhart AAK, Rangan KJ, Craig JW, Loschko J, Rogoz A, Hang HC, and Mucida D. 2016. Exploiting a host-commensal interaction to promote intestinal barrier function and enteric pathogen tolerance. Sci Immunol 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rangan KJ, Pedicord VA, Wang YC, Kim B, Lu Y, Shaham S, Mucida D, and Hang HC. 2016. A secreted bacterial peptidoglycan hydrolase enhances tolerance to enteric pathogens. Science 353: 1434–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sassone-Corsi M, Nuccio S-P, Liu H, Hernandez D, Vu CT, Takahashi AA, Edwards RA, and Raffatellu M. 2016. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 540: 280–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim SG, Becattini S, Moody TU, Shliaha PV, Littmann ER, Seok R, Gjonbalaj M, Eaton V, Fontana E, Amoretti L, Wright R, Caballero S, Wang Z-MX, Jung H-J, Morjaria SM, Leiner IM, Qin W, Ramos RJJF, Cross JR, Narushima S, Honda K, Peled JU, Hendrickson RC, Taur Y, van den Brink MRM, and Pamer EG. 2019. Microbiota-derived lantibiotic restores resistance against vancomycin-resistant Enterococcus. Nature 572: 665–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hu J, Ma L, Nie Y, Chen J, Zheng W, Wang X, Xie C, Zheng Z, Wang Z, Yang T, Shi M, Chen L, Hou Q, Niu Y, Xu X, Zhu Y, Zhang Y, Wei H, and Yan X. 2018. A Microbiota-Derived Bacteriocin Targets the Host to Confer Diarrhea Resistance in Early-Weaned Piglets. Cell Host & Microbe 24: 817–832.e818. [DOI] [PubMed] [Google Scholar]
- 123.Kelly Caleb J., Zheng L, Campbell Eric L., Saeedi B, Scholz Carsten C., Bayless Amanda J., Wilson Kelly E., Glover Louise E., Kominsky Douglas J., Magnuson A, Weir Tiffany L., Ehrentraut Stefan F., Pickel C, Kuhn Kristine A., Lanis Jordi M., Nguyen V, Taylor Cormac T., and Colgan Sean P.. 2015. Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host & Microbe 17: 662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rivera-Chávez F, Zhang Lillian F., Faber F, Lopez Christopher A., Byndloss Mariana X., Olsan Erin E., Xu G, Velazquez Eric M., Lebrilla Carlito B., Winter Sebastian E., and Bäumler Andreas J.. 2016. Depletion of Butyrate-Producing Clostridia from the Gut Microbiota Drives an Aerobic Luminal Expansion of Salmonella. Cell Host & Microbe 19: 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, Taylor TD, Itoh K, Kikuchi J, Morita H, Hattori M, and Ohno H. 2011. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469: 543–547. [DOI] [PubMed] [Google Scholar]
- 126.Schneider C, O’Leary CE, von Moltke J, Liang H-E, Ang QY, Turnbaugh PJ, Radhakrishnan S, Pellizzon M, Ma A, and Locksley RM. 2018. A Metabolite-Triggered Tuft Cell-ILC2 Circuit Drives Small Intestinal Remodeling. Cell 174: 271–284.e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sorbara MT, Dubin K, Littmann ER, Moody TU, Fontana E, Seok R, Leiner IM, Taur Y, Peled JU, van den Brink MRM, Litvak Y, Bäumler AJ, Chaubard J-L, Pickard AJ, Cross JR, and Pamer EG. 2018. Inhibiting antibiotic-resistant Enterobacteriaceae by microbiota-mediated intracellular acidification. Journal of Experimental Medicine 216: 84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schulthess J, Pandey S, Capitani M, Rue-Albrecht KC, Arnold I, Franchini F, Chomka A, Ilott NE, Johnston DGW, Pires E, McCullagh J, Sansom SN, Arancibia-Cárcamo CV, Uhlig HH, and Powrie F. 2019. The Short Chain Fatty Acid Butyrate Imprints an Antimicrobial Program in Macrophages. Immunity 50: 432–445.e437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sencio V, Barthelemy A, Tavares LP, Machado MG, Soulard D, Cuinat C, Queiroz-Junior CM, Noordine M-L, Salomé-Desnoulez S, Deryuter L, Foligné B, Wahl C, Frisch B, Vieira AT, Paget C, Milligan G, Ulven T, Wolowczuk I, Faveeuw C, Le Goffic R, Thomas M, Ferreira S, Teixeira MM, and Trottein F. 2020. Gut Dysbiosis during Influenza Contributes to Pulmonary Pneumococcal Superinfection through Altered Short-Chain Fatty Acid Production. Cell Reports 30: 2934–2947.e2936. [DOI] [PubMed] [Google Scholar]
- 130.Wu T, Li H, Su C, Xu F, Yang G, Sun K, Xu M, Lv N, Meng B, Liu Y, Hu L, Liu Y, Gao Y, Wang H, Lan Y, Xu D, and Li J. 2020. Microbiota-Derived Short-Chain Fatty Acids Promote LAMTOR2-Mediated Immune Responses in Macrophages. mSystems 5: e00587–00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Morita N, Umemoto E, Fujita S, Hayashi A, Kikuta J, Kimura I, Haneda T, Imai T, Inoue A, Mimuro H, Maeda Y, Kayama H, Okumura R, Aoki J, Okada N, Kida T, Ishii M, Nabeshima R, and Takeda K. 2019. GPR31-dependent dendrite protrusion of intestinal CX3CR1+ cells by bacterial metabolites. Nature 566: 110–114. [DOI] [PubMed] [Google Scholar]
- 132.Winston JA, and Theriot CM. 2020. Diversification of host bile acids by members of the gut microbiota. Gut Microbes 11: 158–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Burgess SL, Leslie JL, Uddin J, Oakland DN, Gilchrist C, Moreau GB, Watanabe K, Saleh M, Simpson M, Thompson BA, Auble DT, Turner SD, Giallourou N, Swann J, Pu Z, Ma JZ, Haque R, and Petri WA Jr. 2020. Gut microbiome communication with bone marrow regulates susceptibility to amebiasis. The Journal of clinical investigation 130: 4019–4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MR, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, and Pamer EG. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517: 205–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Alavi S, Mitchell JD, Cho JY, Liu R, Macbeth JC, and Hsiao A. 2020. Interpersonal Gut Microbiome Variation Drives Susceptibility and Resistance to Cholera Infection. Cell 181: 1533–1546.e1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bommarius B, Anyanful A, Izrayelit Y, Bhatt S, Cartwright E, Wang W, Swimm AI, Benian GM, Schroeder FC, and Kalman D. 2013. A Family of Indoles Regulate Virulence and Shiga Toxin Production in Pathogenic E. coli. PloS one 8: e54456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Negatu DA, Yamada Y, Xi Y, Go ML, Zimmerman M, Ganapathy U, Dartois V, Gengenbacher M, and Dick T. 2019. Gut Microbiota Metabolite Indole Propionic Acid Targets Tryptophan Biosynthesis in Mycobacterium tuberculosis. mBio 10: e02781–02718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kumar A, and Sperandio V. 2019. Indole Signaling at the Host-Microbiota-Pathogen Interface. mBio 10: e01031–01019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lee J-H, Cho HS, Kim Y, Kim J-A, Banskota S, Cho MH, and Lee J. 2013. Indole and 7-benzyloxyindole attenuate the virulence of Staphylococcus aureus. Applied Microbiology and Biotechnology 97: 4543–4552. [DOI] [PubMed] [Google Scholar]
- 140.Oh S, Go GW, Mylonakis E, and Kim Y. 2012. The bacterial signalling molecule indole attenuates the virulence of the fungal pathogen Candida albicans. Journal of Applied Microbiology 113: 622–628. [DOI] [PubMed] [Google Scholar]
- 141.Bansal T, Alaniz RC, Wood TK, and Jayaraman A. 2010. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proceedings of the National Academy of Sciences 107: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet Alexandre P., Qiu Z, Maher L, Redinbo Matthew R., Phillips Robert S., Fleet James C., Kortagere S, Mukherjee P, Fasano A, Le Ven J, Nicholson Jeremy K., Dumas Marc E., Khanna Kamal M., and Mani S. 2014. Symbiotic Bacterial Metabolites Regulate Gastrointestinal Barrier Function via the Xenobiotic Sensor PXR and Toll-like Receptor 4. Immunity 41: 296–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zelante T, Iannitti Rossana G., Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D’Angelo C, Massi-Benedetti C, Fallarino F, Carvalho A, Puccetti P, and Romani L. 2013. Tryptophan Catabolites from Microbiota Engage Aryl Hydrocarbon Receptor and Balance Mucosal Reactivity via Interleukin-22. Immunity 39: 372–385. [DOI] [PubMed] [Google Scholar]
- 144.Schiering C, Wincent E, Metidji A, Iseppon A, Li Y, Potocnik AJ, Omenetti S, Henderson CJ, Wolf CR, Nebert DW, and Stockinger B. 2017. Feedback control of AHR signalling regulates intestinal immunity. Nature 542: 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Song C, Lee JS, Gilfillan S, Robinette ML, Newberry RD, Stappenbeck TS, Mack M, Cella M, and Colonna M. 2015. Unique and redundant functions of NKp46+ ILC3s in models of intestinal inflammation. J. Exp. Med 212: 1869–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Satoh-Takayama N, Vosshenrich CAJ, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, Mention J-J, Thiam K, Cerf-Bensussan N, Mandelboim O, Eberl G, and Di Santo JP. 2008. Microbial Flora Drives Interleukin 22 Production in Intestinal NKp46+ Cells that Provide Innate Mucosal Immune Defense. Immunity 29: 958–970. [DOI] [PubMed] [Google Scholar]
- 147.Levy M, Thaiss CA, Zeevi D, Dohnalova L, Zilberman-Schapira G, Mahdi JA, David E, Savidor A, Korem T, Herzig Y, Pevsner-Fischer M, Shapiro H, Christ A, Harmelin A, Halpern Z, Latz E, Flavell RA, Amit I, Segal E, and Elinav E. 2015. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell 163: 1428–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Steed AL, Christophi GP, Kaiko GE, Sun L, Goodwin VM, Jain U, Esaulova E, Artyomov MN, Morales DJ, Holtzman MJ, Boon ACM, Lenschow DJ, and Stappenbeck TS. 2017. The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science 357: 498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Dethlefsen L, Huse S, Sogin ML, and Relman DA. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6: e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, Brochado AR, Fernandez KC, Dose H, Mori H, Patil KR, Bork P, and Typas A. 2018. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555: 623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bager P, Simonsen J, Ethelberg S, and Frisch M. 2010. Cesarean Delivery and Risk of Intestinal Bacterial Infection. The Journal of Infectious Diseases 201: 898–902. [DOI] [PubMed] [Google Scholar]
- 152.Dominguez-Bello MG, De Jesus-Laboy KM, Shen N, Cox LM, Amir A, Gonzalez A, Bokulich NA, Song SJ, Hoashi M, Rivera-Vinas JI, Mendez K, Knight R, and Clemente JC. 2016. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat. Med 22: 250–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Panigrahi P, Parida S, Nanda NC, Satpathy R, Pradhan L, Chandel DS, Baccaglini L, Mohapatra A, Mohapatra SS, Misra PR, Chaudhry R, Chen HH, Johnson JA, Morris JG, Paneth N, and Gewolb IH. 2017. A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature 548: 407–412. [DOI] [PubMed] [Google Scholar]
- 154.Schnorr SL, Candela M, Rampelli S, Centanni M, Consolandi C, Basaglia G, Turroni S, Biagi E, Peano C, Severgnini M, Fiori J, Gotti R, De Bellis G, Luiselli D, Brigidi P, Mabulla A, Marlowe F, Henry AG, and Crittenden AN. 2014. Gut microbiome of the Hadza hunter-gatherers. Nat Commun 5: 3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Brito IL, Yilmaz S, Huang K, Xu L, Jupiter SD, Jenkins AP, Naisilisili W, Tamminen M, Smillie CS, Wortman JR, Birren BW, Xavier RJ, Blainey PC, Singh AK, Gevers D, and Alm EJ. 2016. Mobile genes in the human microbiome are structured from global to individual scales. Nature 535: 435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Riva A, Kuzyk O, Forsberg E, Siuzdak G, Pfann C, Herbold C, Daims H, Loy A, Warth B, and Berry D. 2019. A fiber-deprived diet disturbs the fine-scale spatial architecture of the murine colon microbiome. Nature Communications 10: 4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hryckowian AJ, Van Treuren W, Smits SA, Davis NM, Gardner JO, Bouley DM, and Sonnenburg JL. 2018. Microbiota-accessible carbohydrates suppress Clostridium difficile infection in a murine model. Nat Microbiol 3: 662–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Oliveira BCM, Bresciani KDS, and Widmer G. 2019. Deprivation of dietary fiber enhances susceptibility of mice to cryptosporidiosis. PLoS Negl Trop Dis 13: e0007411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D’Angelo C, Massi-Benedetti C, Fallarino F, Carvalho A, Puccetti P, and Romani L. 2013. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39: 372–385. [DOI] [PubMed] [Google Scholar]
- 160.Kolodziejczyk AA, Zheng D, and Elinav E. 2019. Diet-microbiota interactions and personalized nutrition. Nat Rev Microbiol 17: 742–753. [DOI] [PubMed] [Google Scholar]
- 161.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen Y-Y, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, and Lewis JD. 2011. Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes. Science 334: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A, Ben-Yacov O, Lador D, Avnit-Sagi T, Lotan-Pompan M, Suez J, Mahdi JA, Matot E, Malka G, Kosower N, Rein M, Zilberman-Schapira G, Dohnalova L, Pevsner-Fischer M, Bikovsky R, Halpern Z, Elinav E, and Segal E. 2015. Personalized Nutrition by Prediction of Glycemic Responses. Cell 163: 1079–1094. [DOI] [PubMed] [Google Scholar]


