Abstract
Transcription factors, such as nuclear receptors, often exist in various forms that are generated by highly conserved splicing events. Whereas the functional significance of these splicing variants is often not known, it is known that nuclear receptors activate transcription through interaction with coactivators. The parameters, other than ligands, that might modulate those interactions, however, are not well characterized, nor is the role of splicing variants. In this study, transient transfection, yeast two-hybrid, and GST pulldown assays are used to show not only that nuclear receptor hepatocyte nuclear factor 4 α1 (HNF4α1, NR2A1) interacts with GRIP1, and other coactivators, in the absence of ligand but also that the uncommonly large F domain in the C terminus of the receptor inhibits that interaction. In vitro, the F domain was found to obscure an AF-2-independent binding site for GRIP1 that did not map to nuclear receptor boxes II or III. The results also show that a natural splicing variant containing a 10-amino-acid insert in the middle of the F domain (HNF4α2) abrogates that inhibition in vivo and in vitro. A series of protease digestion assays indicates that there may be structural differences between HNF4α1 and HNF4α2 in the F domain as well as in the ligand binding domain (LBD). The data also suggest that there is a direct physical contact between the F domain and the LBD of HNF4α1 and -α2 and that that contact is different in the HNF4α1 and HNF4α2 isoforms. Finally, we propose a model in which the F domain of HNF4α1 acts as a negative regulatory region for transactivation and in which the α2 insert ameliorates the negative effect of the F domain. A conserved repressor sequence in the F domains of HNF4α1 and -α2 suggests that this model may be relevant to other nuclear receptors as well.
Nuclear receptors comprise a large superfamily of relatively conserved transcription modulators that play a role in nearly every aspect of growth, differentiation, and development in organisms ranging from nematodes to humans (for reviews, see references 13, 58, 59, and 70). Family members are defined by the presence of two conserved functional domains. In the N-terminal portion of the protein there is a DNA binding domain (DBD) that contains two zinc fingers; in the C-terminal portion there is a large hydrophobic domain, termed the ligand binding domain (LBD), which is responsible for ligand binding, protein dimerization, and transcriptional activation. Recently, our understanding of the mechanism by which nuclear receptors modulate transcription was greatly enhanced by the finding that certain members interact in a ligand-dependent fashion with non-DNA binding coactivators from several different gene families, e.g., the p300 family, such as p300 and CBP (5, 28, 48), and the p160 family, such as GRIP1/TIF2 (38, 86) and SRC1/p160/ERAP160 (26, 48, 68) (reviewed in references 77 and 83). For several receptors, such as the thyroid hormone receptor, vitamin D receptor, retinoid (retinoic acid and retinoid X) receptors (RAR and RXR, respectively), and steroid (progesterone, estrogen, androgen, and glucocorticoid) receptors (PR, ER, AR, and GR, respectively), the binding of ligand is known to produce a conformational change in the protein which makes it more resistant to protease (1, 53, 54; see also reference 91 and references therein). Structural studies have shown that this change makes a small conserved region important for transactivation in the C-terminal end of the LBD, termed AF-2, more accessible to solvent, and therefore presumably to coactivators (3, 14, 73, 90). Coactivators in both the p300 and p160 families are known to bind receptors in a ligand-dependent fashion (5, 28, 37, 43, 60, 68, 84, 86). This interaction appears to be dependent upon nuclear receptor (NR) boxes in the coactivators which are comprised of LXXLL motifs (10, 32, 84, 85), and, at least in the p160 family, the AF-2 region of the receptor (37, 43, 60, 68, 86). Once tethered to the appropriate promoter by the receptor, the coactivators are thought to stimulate transcription by interacting with the basal transcription machinery (reviewed in reference 41) and/or by modulating the local nucleosome structure via histone acetylation (reviewed in references 39 and 92). The majority of the more than 150 different members of the nuclear receptor superfamily, however, have not yet been found to respond to ligands. The question then arises as to whether these so-called orphan receptors will also interact with coactivators and, if they do, whether the structural basis for that interaction is similar to that for receptors with known ligands.
Nuclear receptors, like many other proteins, are often subjected to alternative splicing which generates multiple isoforms of the receptors (23). Some isoforms are found more readily in cancerous than in noncancerous tissue (15, 21), while levels of others vary depending on the tissue or developmental state (34, 61, 69). Whereas the functional relevance of some of those variants is evident (e.g., producing alterations in DBDs or LBDs), the significance of other splicing variants is less clearly established, despite conservation among different species (17, 69). The question then arises as to whether some of these splicing variants differ in their interactions with different coactivators.
We wished to examine some of these issues with hepatocyte nuclear factor 4α (HNF4α, NR2A1) (8), a highly conserved member of the superfamily. Three HNF4 genes HNF4α, HNF4β, and HNF4γ (11, 35, 80), have been identified thus far in vertebrates, although most work has been done with the first cDNA cloned, HNF4α1 (80). HNF4α1 is directly linked to several human diseases: the coding region of HNF4α1 is mutated in maturity-onset diabetes of the young (MODY1) (4, 18, 27, 55, 62, 93), an HNF4α1 response element is mutated in hemophilia B Leyden (72), and HNF4α1 transcriptionally activates several hepatitis B viral genes (19, 24, 71). Deletion of the HNF4α1 gene from the mouse genome results in embryonic lethality at day 10 (7). HNF4α1 is known to activate a wide variety of genes involved in glucose, fatty acid, cholesterol, and amino acid metabolism in the liver, kidney, intestine, and pancreas (reviewed in reference 79). Whereas HNF4α1 is capable of activating transcription and binding DNA in the absence of exogenously added ligand (25, 44, 57, 80), there is a recent report of a potential ligand for HNF4α1 (33). However, many questions remain about the role of these compounds in HNF4α1 function.
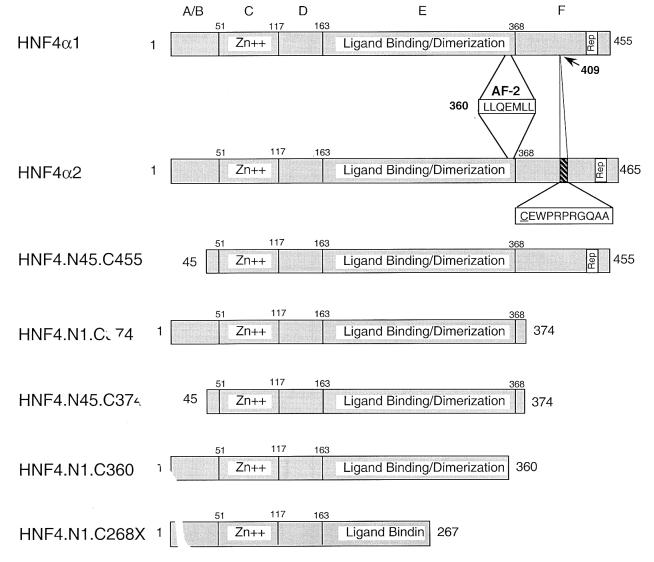
Aside from its physiological importance, HNF4α1 is of interest in that it possesses unique protein dimerization and DNA binding properties which define a distinct subfamily of nuclear receptors: HNF4α1 exists in solution and binds DNA response elements consisting of direct repeats exclusively as a homodimer (44). Furthermore, HNF4α1 is rather unique in its ability to activate transcription in the absence of exogenously added ligand in mammalian cells, in yeast cells, and in vitro (16, 25, 44, 57, 80). Finally, HNF4α1 possesses an unusually large region C terminal to the LBD (the F domain). Of the two splicing variants of HNF4α1 involving the F domain thus far identified (30, 50), one contains an additional 10 amino acids (aa) that have been inserted into the middle of the F domain (HNF4α2) (Fig. 1). Since deletion of the F domain has been shown to increase the activation of transcription by HNF4α1 in vivo (25), we wished to determine whether the F domain can modulate interaction with coactivators and whether the 10-aa splicing insert affects that modulation.
FIG. 1.
HNF4α constructs used in this study. Shown are naturally occurring rat HNF4α1 and HNF4α2 splicing variants and experimentally generated constructs containing the indicated amino acids. N1C268X is derived from a mutation found in a MODY1 patient (81, 97). The amino acid sequence of the conserved AF-2 region and the insertion in HNF4α2 are given in single-letter code. The underlined cysteine at position 409 is a serine in HNF4α1. The remaining residues shown are unique to the α2 insert; all others are identical between HNF4α1 and HNF4α2 (30). Conventional nomenclature for nuclear receptor domains (A to F) is given at the top. Numbers indicate amino acid residues. Zn++, zinc finger region; Rep, repressor region (aa 428 to 441 in HNF4α1).
In this study we used a variety of in vivo and in vitro assays, including transient transfections into mammalian cells, yeast two-hybrid and glutathione S-transferase (GST) pulldown assays, and protease digestion, to examine the interaction between HNF4α1 and HNF4α2 and coactivators GRIP1, SRC1a, p300, and CBP. The results show not only that HNF4α1 is capable of interacting physically and functionally with coactivators in vivo and in vitro in the absence of exogenously added ligand but that the F domain interferes with that interaction. We also show that the 10-aa insertion in HNF4α2 somewhat abrogates the interference by the F domain and propose a mechanism for that abrogation.
MATERIALS AND METHODS
Plasmids.
All HNF4α constructs were derived from rat HNF4α1 (80) or HNF4α2 (30) and ligated into the vector pMT7 (46) via EcoRI adapters (Promega, Madison, Wis.), unless noted otherwise, for expression in vitro and in vivo. HNF4α2 cDNA was removed from HNF-4CL4 (kindly provided by S. Hata) by digestion with BamHI/EcoRI. HNF4.N1C374 and HNF4.N45.C455 were constructed by PCR amplification of HNF4α1 with previously described primers N1 (previously Npf7) and C374 or N45 and C455 (previously Cpf7), respectively (45, 46). N1C360 was constructed in a similar fashion with primers N1 and C360 (5′-GCGCTCGAGCTACAGGTTGTCAATCTTGGCCATC-3′) but ligated into the BamHI/XhoI sites of pcDNA3.1(+) (Invitrogen, Carlsbad, Calif.). Construction of HNF4.N45.C374 and N1C268X in pMT7 has been previously described (46, 78). pSG5.GRIP1 contained full-length mouse GRIP1 coding region ligated into pSG5 (10). pRc/RSV-mCBP.HA.RK (kindly provided by R. Goodman) contained full-length mouse CBP with a C-terminal hemagglutinin (HA) tag driven by the Rous sarcoma virus promoter. pCMV.HA.p300 contained human p300 from nucleotides 1134 to 8329 with an HA tag fused to the NheI site at nucleotide 8329 driven by a cytomegalovirus promoter (12). The reporter construct pZLHIVAI-4 contained four HNF4 response elements (site A) from the human apolipoprotein AI gene (75) inserted at the BamHI site immediately upstream of positions −57 to +80 of the human immunodeficiency virus long terminal repeat (74) driving the firefly luciferase gene in pZLuc (78). Fusions of the Gal4 DBD to HNFα1 (Gal4DBD-HNF4 constructs) for the yeast two-hybrid assay were prepared by inserting the appropriate PCR-amplified HNF4α1 coding regions into plasmid pGBT9 (Clontech, Palo Alto, Calif.) at the BamHI/SalI site for constructs HNF4α1.128-455 and HNF4α1.128-415 and at the EcoRI/SalI site for construct HNF4α1.128-370. Gal4 activation domain (Gal4AD)-GRIP1 and Gal4AD-SRC1a constructs have been previously described (10). GST.127.374 was constructed by inserting a PCR product containing amino acids 127 to 374 of rat HNF4α1 into the pGEX6P-1 vector (Pharmacia, Piscataway, N.J.), using EcoRI/XhoI sites. The fusion protein was expressed in Escherichia coli BL21(DE3)(pLysS) and bound to glutathione-agarose (Sigma, St. Louis, Mo.) by using standard protocols (2). A fragment containing residues 127 to 374 plus eight residues from the vector (N-Gly-Pro-Leu-Gly-Ser-Pro-Glu-Phe-C) was released from GST by cleavage with Precision Protease as directed by the manufacturer (Pharmacia) and verified by sequencing the N terminus via Edman degradation (UC Riverside peptide sequencing facility). The sequence of all PCR-derived products were verified by dideoxy sequencing.
Transient transfection assays.
Human embryonic kidney 293T cells and COS-7 cells were maintained at 37°C under 5% CO2 in Dulbecco modified Eagle medium supplemented with penicillin-streptomycin and with 5% fetal calf and 10% bovine calf serum, respectively. Transient transfections into these cells using calcium phosphate precipitation were carried out essentially as previously described (78). All transfection results were normalized to the RSV.βgal construct level; assays were performed at least twice in triplicate. Fold inductions were calculated relative to transfections lacking expression vectors. Activation of the reporter construct by coactivators in the absence of HNF4α was minimal compared to activation in the presence of HNF4α (not shown). Production of HNF4α protein was verified by expression in COS-7 cells by Western or gel shift analysis as previously described (44, 45). Production of HNF4α1 and HNF4α2 protein in 293T cells was verified by harvesting 3.0 × 106 cells transfected with 25 μg of plasmid DNA (quantified by readings of optical density at 260 nm and analysis of ethidium bromide-stained agarose gels) 24 h after glycerol shock. The cells were lysed in 100 μl of 293T lysis buffer (50 mM HEPES [pH 7.9], 100 mM NaCl, 1.0 mM MgCl2, 1 mM EDTA, 10% glycerol, 1% Triton X-100), gently agitated for 40 min at 4°C and centrifuged for 30 min at 12,000 × g to pellet the debris. The protein concentration of the supernatant was determined by the Bio-Rad assay, and equivalent amounts of total protein were analyzed by Western blot analysis as described in the figure legends.
GST pulldown assays.
In vitro protein-protein interactions between various HNF4α constructs and GST-GRIP1 (aa 563 to 1121) were analyzed by a GST pulldown assay performed essentially as previously described (10). Briefly, 2 to 5 μl of in vitro-synthesized 35S-labeled HNF4α (TNT kit; Promega) was incubated with 10 μl of packed conjugated glutathione-agarose beads (containing approximately 1 μg of GST fusion protein per μl) in 0.01% NETN in a total of 30 to 50 μl for approximately 1 h at 4°C with gentle agitation. The beads were spun in a Sorvall MC 12-V Microfuge at 2,000 rpm for 1 min and washed three times for 30 s each with 100 to 250 μl of 0.01% NETN. Finally, the beads were boiled in sodium dodecyl sulfate (SDS) loading buffer containing 10% β-mercaptoethanol and pelleted. The resulting supernatants were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) on a 10% gel (2). The gels were either dried or transferred to polyvinylidene difluoride membrane (Millipore, Madison, Wis.) in 25 mM Tris base–0.19 M glycine and subjected to autoradiography. A PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) was used for quantification. GST-GRIP1 NRmut was made by amplifying aa 563 to 1121 from a full-length GRIP1 construct containing two mutations in both NR box II (L693A and L694A) and NR box III (L748A and L749A) (10) and ligating it into the BamHI/EcoRI sites of pGEX-2TK (Pharmacia).
Protease digestion assay.
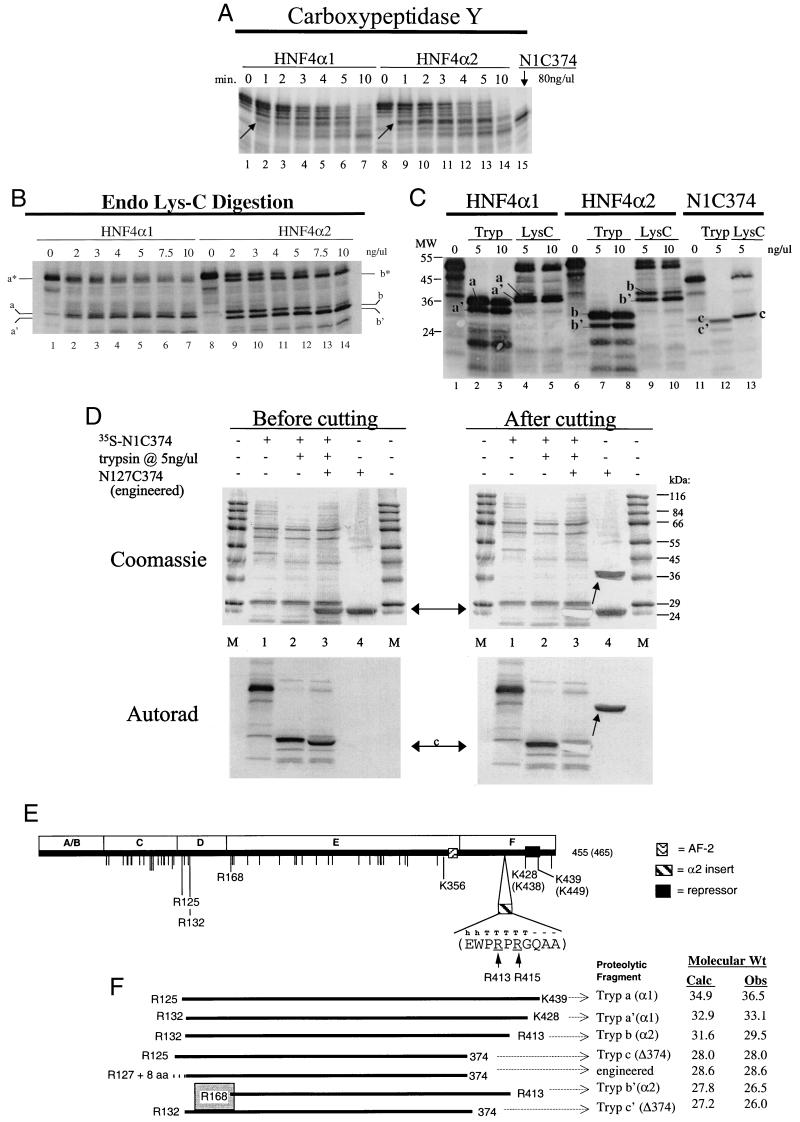
Protease digestion assays with N-tosyl-l-phenylalanine chloromethyl ketone (TPCK)-treated trypsin (Sigma) were carried out by the addition of the indicated amount of protease to in vitro-synthesized 35S-labeled HNF4α. One microliter of appropriately diluted trypsin was added to 2 μl of lysate diluted in 7 μl of 50 mM Tris (pH 7.0). Reactions were stopped after incubation at room temperature for 15 min by the addition of SDS loading buffer. Samples were subsequently analyzed as described above for the pulldown assays. Molecular weight (MW) markers (SigmaMarker, wide-MW range) included in a parallel lane in the gel were visualized by Coomassie blue staining of the membrane. Digestion with endoproteinase LysC (EndoLysC; Boehringer Mannheim, Indianapolis, Ind.) was carried out in a similar fashion except that the digestion buffer was 50 mM Tris (pH 8.6)–0.3 M NaCl–1 mM EDTA and incubation was for 1 to 2 h. Time courses of digestion with carboxypeptidase Y (Boehringer Mannheim) were carried out according to a previously published protocol (54). Briefly, lysate (10 μl) was incubated at room temperature with protease in a 210-μl reaction in 50 mM Tris-HCl (pH 6.7), and 21-μl aliquots were removed at the indicated times. Reactions were stopped by the addition of SDS loading buffer and 10 mM phenylmethylsulfonyl fluoride and placement on dry ice and were subsequently analyzed by SDS-PAGE and autoradiography. Proteolytic cleavage sites in Fig. 5E were determined by a comparison of observed MW measured from Rf values in SDS-PAGE to predicted MW based on amino acid composition, and by comparison of the bands to each other as explained in the text. When more than one residue could yield a fragment of a particular MW, the residue with the greatest surface probability according to Emini as determined by PEPTIDESTRUCTURE in the Genetics Computer Group package (20), was chosen as the potential cleavage site (e.g., R131 has a surface probability of 3.054 whereas R132 has one of 3.571). Efforts were made to minimize the number of differences between HNF4α1 and HNF4α2.
FIG. 5.
The presence of the α2 insert alters the protease sensitivity of HNF4α. (A) Autoradiograph after SDS-PAGE (10% gel) of a time course (in minutes) of proteolytic digestion of 35S-labeled HNF4α1 and HNF4α2 with carboxypeptidase Y (80 ng/μl, final concentration) which cleaves sequentially from the C terminus (see Materials and Methods for details). Uncleaved N1C374 serves as an MW marker in lane 15. Arrows point to protected fragments mentioned in the text. (B) As for panel A except that HNF4α1 and HNF4α2 were digested with increasing amounts of EndoLysC as indicated. a, a′, b, and b′, arbitrary labeling of fragments referred to in the text. a∗ and b∗ represent cleavage in the very C terminus only (most likely K439 in HNF4α1 and K449 in HNF4α2) since cleavage at the first N-terminal lysine residue, K61, would yield a band migrating much faster. (C) As for panel B except digestion of HNF4α1 and HNF4α2 was compared to digestion of N1C374. Tryp, trypsin; LysC, EndoLysC. Labeling of EndoLysC fragments is the same as in panel B. MW, MW markers (positions are indicated in kilodaltons). (D) As for panel C except that N127.374, an engineered fragment of 28.6 kDa containing residues 127 to 374 of HNF4α1 plus an additional eight residues (described in Materials and Methods), was added to the 35S-N1C374 trypsin digestion right before loading on the gel. Coomassie blue-stained blots before and after cutting out the engineered fragment (double-edged arrow) are shown on the top, and the corresponding autoradiographs (Autorad) are shown on the bottom. M, MW markers; c, same as in panel C. The arrow in the after-cutting blots shows how the band corresponding to the engineered fragment was moved. (E) Map of potential cleavage sites for trypsin (Lys-X or Arg-X) in rat HNF4α1 and HNF4α2. Tick marks indicate either a lysine (K) or an arginine (R) residue. To simplify the presentation, only those residues that are thought to be cleaved (plus K356) are indicated by a residue number. Note the complete absence of the Arg and Lys residues in the A/B domain. Also shown are the receptor domains (A to F), the AF-2 region (aa 360 to 368), a previously identified repressor region (aa 428 to 441) (40) (see Discussion), and the amino acid sequence of the insert in HNF4α2 (aa 410 to 419) and its predicted secondary structure as determined by the Chou-Fasman algorithm in PEPTIDESTRUCTURE in the Genetics Computer Group package (20). T, strong probability of a beta turn; h, possible alpha helix; −, no predicted structure. (F) Schematic representation of the trypsin digestion products shown in panels C and D and observed (Obs) and calculated MW (Calc) MW (in thousands) of each fragment in descending order. As discussed in the text, HNF4α2 appears to have a tryptic cleavage site near R168 that is not present in HNF4α1. The large number of K and R residues in domain C results in some ambiguity in the determination of the N-terminal cleavage sites. The ambiguity was resolved by relying on predictions of MW and surface probability as explained in the text. When two potential cleavage sites were close together and exhibited similar surface probabilities, such as R413 and R415 in HNF4α2, the internal most site was used (e.g., R413). Band c of N1C374 is predicted to represent cleavage only in the N terminus since carboxypeptidase Y data indicated that its C terminus is rather resistant to digestion (data not shown). α1, HNF4α1; α2, HNF4α2, Δ374, HNF4.N1C374. Numbers indicate amino acid residues.
RESULTS
The F domain of HNF4α1 negatively regulates coactivator-mediated transcription in vivo.
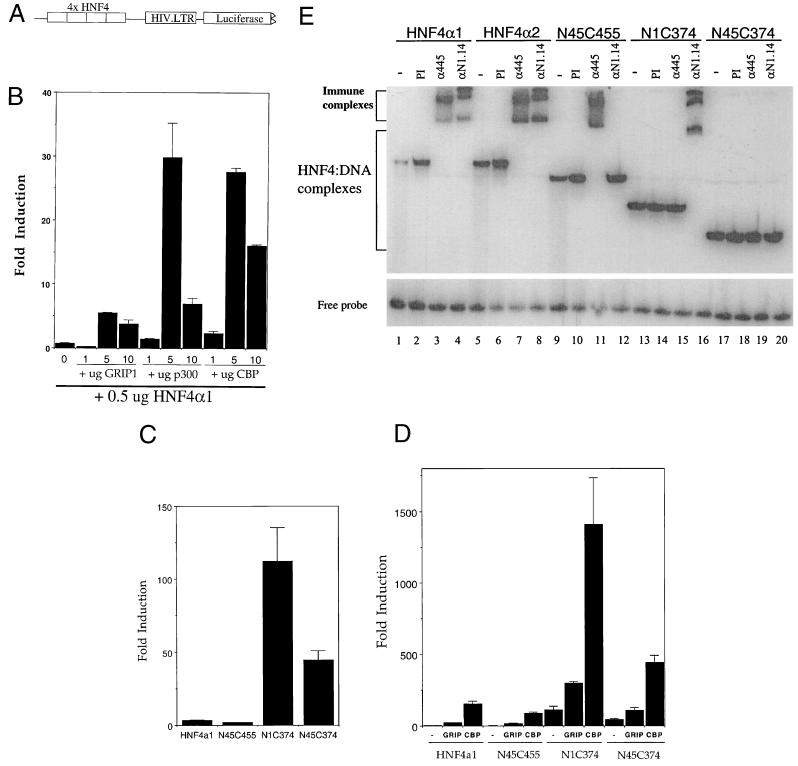
Full-length HNF4α1 and truncated forms lacking either or both of the N and C termini (HNF4.N1.C374, HNF4.N45.C455, and HNF4.N45.C374, respectively [Fig. 1]) were tested for responsiveness to coactivators GRIP1, p300, and CBP. HNF4α1 was cotransfected into 293T cells with the luciferase reporter construct pZLHIVAI-4 (Fig. 2A) and increasing amounts of GRIP1, p300, or CBP. All three coactivators were capable of enhancing transcriptional activation by full-length HNF4α1, although p300 and CBP yielded much greater enhancement than GRIP1 (Fig. 2B). The significance of the difference between the coactivators, if any, is not known. All three coactivators also enhanced HNF4α1-dependent transcription in other cell types, such as HeLa, and on other promoter constructs (data not shown).
FIG. 2.
The F domain of HNF4α1 inhibits transcriptional enhancement by coactivators. (A) Diagram of promoter region of reporter construct pZLHIVAI-4. HIV.LTR, human immunodeficiency virus long terminal repeat. (B to D) Transient cotransfections into 293T cells were performed as described in Materials and Methods with 2 μg of reporter construct, 0.5 μg of various HNF4α1 expression vectors as described in Fig. 1, and various amounts of GRIP1 (pSG5.GRIP1 full length), p300 (CMV.HA.p300 partial), or CBP (pRC.RSV.HA.CBP full length) expression vectors as indicated. Shown is the average fold induction of the relative light units normalized to a β-Gal control (0.5 to 1.0 μg of RSV.βgal) from one of several experiments. Error bars indicate the range of triplicate samples. Panel C and minus in panel D, no added coactivators. (D) One microgram GRIP1 and 5 μg of CBP expression vectors were used. Note the difference in scale of the y axes in panels C and D. (E) Electrophoretic mobility shift analysis of crude nuclear extracts from COS-7 cells transiently transfected with the various HNF4α expression vectors as indicated (HNF4α1 and HNF4α2, 25 μg of expression vector per 1.6 × 106 cells; N45C455, N1C374, and N45C374, 50 μg per 3.4 × 106 cells). DNA was introduced into the cells by standard calcium phosphate procedure, and the cells were harvested 40 h after glycerol shock. 32P-labeled APF1 oligonucleotide (0.5 ng per 7.5-μl reaction) was incubated with the protein extract (0.5 μg) in the presence of nonspecific DNA (0.5 μg of dI-dC, 0.5 μg of sonicated salmon sperm DNA) for 20 min at room temperature before the addition of antiserum (0.5 μl) as indicated. The incubation was continued for another 20 min before electrophoresis on a 6% native polyacrylamide gel. Details of procedures have been described previously (44, 78). Lanes: −, no antiserum added; PI, preimmune antiserum; α445, antiserum to the C terminus of rat HNF4α1 (80); αN1.14, antiserum raised in rabbit to a synthetic peptide corresponding to the first 14 aa of the N terminus of rat HNF4α1.
As previously seen in another system (25), the F domain of HNF4α1 inhibited transcriptional activation whereas the A/B domain in the N terminus was necessary for full activation (Fig. 2C). The same profile of activation by HNF4α1 and its truncated forms was seen in the presence of the coactivators GRIP1 and CBP (Fig. 2D). Similar results were obtained with p300 (data not shown). Appropriate expression and DNA binding ability of all HNF4α1 constructs were verified by electrophoretic mobility shift analysis (Fig. 2E). These results indicate that HNF4α1 responds to GRIP1 and CBP/p300 coactivators in vivo and that the response is inhibited by the presence of the F domain.
Physical interaction of HNF4α1 with coactivators GRIP1 and SRC1a is inhibited by the F domain.
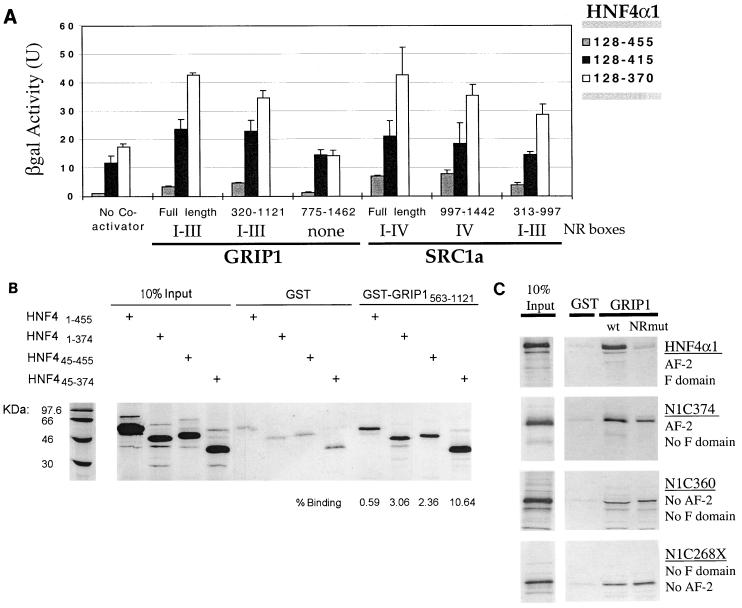
To determine whether HNF4α1 interacts physically with coactivators, a yeast two-hybrid assay was performed (Fig. 3A). When the entire hinge region plus LBD and F domain were fused to the Gal4 DBD (HNF4α1.128-455), a small but significant amount of β-galactosidase (β-Gal) activity was produced in the absence of coactivators, indicating that the HNF4α1 fragment contains a transcriptional activation domain active in yeast. Since no GRIP1 or SRC1 homologs have been found in Saccharomyces cerevisiae, the HNF4α construct must interact with either some other coactivator in yeast or the basal transcription machinery directly. Interestingly, when the Gal4-HNF4 fusion construct was truncated at amino acid 415 (HNF4α1.128-415) or amino acid 370 (HNF4α1.128-370), the basal level of β-Gal activity in the absence of any exogenously added coactivator was greatly increased, as observed in mammalian cells (Fig. 2C). The presence of the GRIP1-Gal4AD fusion protein significantly enhanced the β-Gal activity further but only with those GRIP1 constructs containing three NR boxes (full-length and 320 to 1121). The increase in β-Gal activity indicates a physical interaction in the yeast two-hybrid system. Similar results were obtained with the related coactivator SRC1a (Fig. 3A). These results indicate that, like other nuclear receptors, HNF4α1 interacts with GRIP1 and SRC1. What has not been shown for other receptors, however, is the inhibition of interaction between a nuclear receptor and coactivators by portions of the F domain.
FIG. 3.
Interaction between HNF4α1 and coactivators in vivo and in vitro is inhibited by the presence of the F domain. (A) Interactions between HNF4α1 and coactivators GRIP1 and SRC1a were examined in the yeast two-hybrid assay (Clontech) with various Gal4DBD-HNF4 (pGBT9) and Gal4AD-GRIP1 or -SRC1a (pGAD424) constructs as described in Materials and Methods and previously (10). Shown is one representative experiment of two or more independent transformations into S. cerevisiae SFY526 containing an integrated β-Gal reporter construct. Standard deviations are from four independent clones from a single transformation. Numbers indicate amino acid sequence encoded in the various constructs. NR boxes, nuclear receptor interaction motifs as previously described (10). (B) In vitro pulldown assays between the GST control and GST-GRIP1 (aa 563 to 1121) and in vitro-translated 35S-HNF4α1 constructs as indicated were performed as described in Materials and Methods. Shown is the phosphorimage after SDS-PAGE of eluted material as well as percent binding of input protein (10% input is shown). One of several experiments is shown. Positions of 14C-labeled MW markers are shown at the left. Negative controls for the pulldown assays shown in panel B, using in vitro-translated 35S-C/EBPα and 35S-luciferase, are not shown. (C) As for panel B. wt, GST-GRIP1 as in panel B; GRIP1 NRmut, as wt GRIP1 except with mutations in NR boxes II and III. The presence of the F domain and AF-2 is indicated for each HNF4α construct.
To verify the results of the yeast two-hybrid assay, in vitro pulldown assays were performed with GST-GRIP1 and 35S-labeled HNF4α (Fig. 3B). Full-length HNF4α1 interacts with GRIP1, and that interaction is enhanced severalfold when the F domain is deleted (HNF4.1-374). In contrast to the transient transfection assays, however, the interaction between HNF4α1 and GRIP1 also appears to be increased when the A/B domain in the N terminus is deleted (HNF4.45-455 and HNF4.45-374). The reason for this is not known, but it could be an indication that the A/B domain of HNF4α1 is required for transcriptional activation via a mechanism independent of GRIP1. For example, HNF4α1 has been shown to bind TFIIB in vitro in an AF-2-independent fashion (57) and numerous parts of the basal transcriptional machinery (TFIIB, TATA binding protein, TAFII31, TAFII80, and TAFIIH-p62) (22, 51). Interestingly, however, several coactivators (CBP, ADA2, and PC4) have also been shown to interact with the A/B domain of HNF4α1 (22). This suggests that GRIP1, and perhaps other members of the p160 family, activate HNF4α1-mediated transcription by a different mechanism.
The F domain of HNF4α1 obscures an AF-2-independent binding site for GRIP1.
Since the F domain begins immediately following the AF-2 region, we hypothesized that it might inhibit transcription by obscuring the AF-2 region and thereby impeding access to coactivators. To test this, we first needed to verify that the AF-2 region of HNF4α1 is required for interaction with GRIP1. Since the AF-2 regions of other receptors have been found to interact with the NR boxes of coactivators, we also examined the role of GRIP1 NR boxes II and III in HNF4α1 binding in vitro. The results (Fig. 3C) were rather surprising. As expected, full-length HNF4α1 did not significantly bind a GST-GRIP1 construct mutated in two of the three NR boxes (NRmut). However, when the F domain was deleted, there was appreciable binding to the GRIP1 mutant as well as to the wild-type (wt) GRIP1 (N1C374). Perhaps even more surprising, when HNF4α1 was further truncated to remove the AF-2 region, significant binding to both wt and mutant GRIP1 was again observed (N1C360). A similar result was observed with even further truncation of HNF4α1 (N1C268X).
These results indicate that the F domain obscures a site somewhere in the first 267 aa of HNF4α1 that binds GRIP1 in a fashion independent of at least two NR boxes and the AF-2 region. This is not to suggest, however, that binding of the NR boxes to the AF-2 region plays no role in the interaction between GRIP1 and HNF4α1. Indeed, the binding of N1C374 to the GRIP1 NR mutant was less than to wt GRIP1, suggesting that the NR boxes do play a role in binding HNF4α1. Furthermore, there was no such difference in binding of the two GRIP1 constructs to N1C360, which bound both constructs less well than N1C374. This finding suggests that the AF-2 region of HNF4α1 plays a role in binding GRIP1 but that there is also an AF-2-independent binding site in HNF4α1 which does not require NR boxes II or III. It is this site that is apparently obscured by the F domain. Finally, this result is not necessarily in conflict with the yeast two-hybrid data which showed no interaction between GRIP1 and HNF4α in the absence of three NR boxes for two reasons: the GST-GRIP1 NR mutant still contains the first NR box, and the HNF4α yeast two-hybrid construct contained only the LBD and the F domain. It is possible that there are interactions between GRIP1 and HNF4α1 involving either the first NR box and/or the other portions of HNF4α1 (i.e., the A/B domain and/or the DNA binding domain).
HNF4α2 preferentially activates transcription and responds to coactivators in vivo and in vitro.
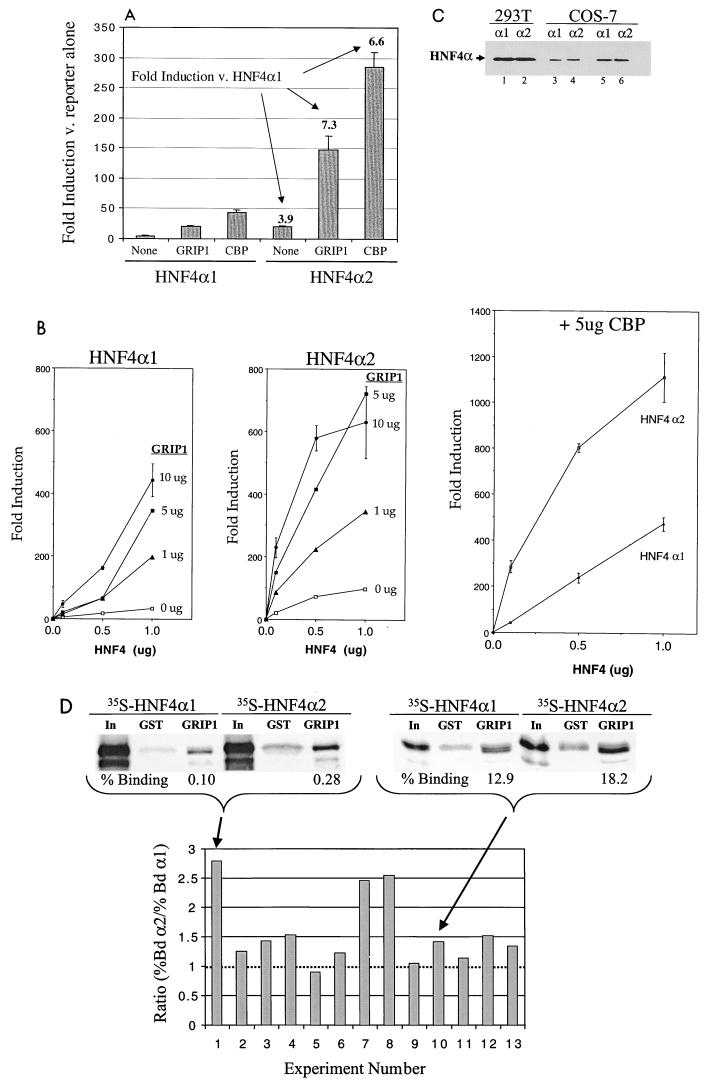
Since the results presented above indicated that the F domain of HNF4α1 modulated the interaction with coactivators and since there is a prevalent splicing variant of HNF4α1 that contains a modified F domain (HNF4α2 [Fig. 1]), we determined the effect of this splicing variant on HNF4α transactivation function. Transient transfection analysis comparing levels of activation by HNF4α1 and HNF4α2 showed not only that HNF4α2 activated transcription approximately fourfold better than HNF4α1 in the absence of added coactivator but also that GRIP1 and CBP each stimulated transcription by HNF4α2 approximately sevenfold more than they stimulated transcription by HNF4α1 (Fig. 4A). The greater effect of both coactivators on HNF4α2 was also seen over a range of DNA concentrations (Fig. 4B), although at larger amounts of DNA the effect of the coactivators was somewhat less. Western blot analysis verified that the HNF4α1 and HNF4α2 proteins were expressed to similar levels in the 293T cells used for the transfections and in COS-7 cells (Fig. 4C).
FIG. 4.
HNF4α2-mediated transcription is preferentially enhanced by coactivators GRIP1 and CBP. (A) Transient cotransfections were performed as for Fig. 2 with 0.1 μg of HNF4α1 or HNF4α2 and 5 μg of GRIP1 or CBP expression vectors as indicated. Error bars indicate range of the fold induction between triplicate samples. Plotted on the y axis is the fold induction compared to the reporter alone. Numbers in the plot represent fold induction of HNF4α2 relative to HNF4α1 under similar conditions. (B) As for panel A except with increasing amounts of expression vectors as shown. Error bars not shown for 0, 1, and 5 μg of GRIP1 were all less than 10%. Note difference in scale in y axis in between the GRIP1 and CBP panels. The difference between HNF4α1 and HNF4α2 in the presence and absence of coactivators was seen in at least three independent experiments, all done in triplicate. (C) Western blot analysis of HNF4α1 and HNF4α2 proteins transiently expressed in 293T and COS-7 cells, using chemiluminescence (Pierce, Rockford, Ill.). Twenty-five micrograms of total protein of 293T extracts (lanes 1 and 2) and 10 μg (lanes 3 and 4) or 25 μg (lanes 5 and 6) of nuclear extracts of COS cells were analyzed with a 1:5,000 dilution of α445 (see the legend to Fig. 2). Extracts from nontransfected cells showed no bands in this region of the gel (not shown). (D) GST pulldown experiments were performed as for Fig. 3B with 35S-HNF4α1 and 35S-HNF4α2 and GST control (GST) or GST-GRIP1 (aa 563 to 1121) (GRIP1). Shown are 2 of 13 representative experiments with percent binding of input (In, 10%) normalized to GST control beads and a graph of the ratio of percent bound HNF4α2 to percent bound HNF4α1 (% Bd α2/% Bd α1) (each controlled for binding GST beads) for all 13 experiments (two of which are the average of duplicate samples). The dashed line indicates a ratio of 1.0, which one would expect if there were no difference between HNF4α1 and HNF4α2. The average ratio for the 13 experiments was 1.6. A nonparametric paired t test (Wilcoxon signed-rank test) performed by the STATView program yielded a P value of 0.0024 for the 13 experiments, indicating that the difference noted between HNF4α1 and HNF4α2 is statistically significant.
The transient transfection results in Fig. 4A and B suggested that HNF4α2 may bind coactivators more efficiently than HNF4α1. To test this hypothesis directly, GST-GRIP1 pulldown assays were performed with in vitro-synthesized 35S-labeled HNF4α1 and HNF4α2. The results (Fig. 4D) indicate that HNF4α2 interacts with GRIP1 in vitro more efficiently than HNF4α1. For example, in one experiment 18.2% of the input HNF4α2 bound GRIP1, compared to 12.9% of the HNF4α1, when controlled for background binding to GST. In another experiment, the total amount bound was lower but the difference between HNF4α2 and HNF4α1 was maintained—0.28% versus 0.1%. This difference between HNF4α2 and HNF4α1 was very reproducible in that 12 of 13 independent experiments, often using different preparations of GST-GRIP1 and/or lysates, showed HNF4α2 binding GRIP1 better than HNF4α1. Furthermore, despite the variation in the absolute amount of binding as evident in the examples given above, the difference between HNF4α2 and HNF4α1 was statistically significant for the 13 experiments (P = 0.0024 for GST controlled and 0.0015 for non-GST controlled). The fact that this in vitro binding data shows somewhat less of a difference between HNF4α1 and HNF4α2 than that seen in vivo (1.6-fold versus 4- to 7-fold) could be due to the fact that in vivo the effect of the interaction between HNF4α1 and -α2 and GRIP1 is amplified. There could also be other mechanisms involved in vivo in addition to enhanced interaction with GRIP1.
The presence of the α2 insert alters the protease sensitivity of HNF4α.
To determine whether there are any structural differences between HNF4α1 and HNF4α2 that could explain the enhanced binding and responsiveness to GRIP1 (and CBP) we performed a series of protease digestion experiments using the 35S-labeled HNF4α constructs. First, a time course of digestion of HNF4α1 and HNF4α2 was performed with carboxypeptidase Y, an exopeptidase that sequentially cleaves amino acids in a C- to N-terminal fashion. The results indicate that there is indeed a difference between HNF4α1 and HNF4α2. Not only did the full-length HNF4α2 begin to disappear faster than the full-length HNF4α1 (Fig. 5A; compare lanes 8 to 10 to lanes 1 to 3), but HNF4α2 yielded a protected fragment that was significantly more pronounced than a similar fragment in HNF4α1 (compare lanes 9 to 13 to lanes 2 to 6). Since the protected fragment migrates slightly slower than uncleaved N1C374 (lane 15), this finding suggests that there might be a structural difference between HNF4α1 and HNF4α2 C terminal to aa 374 (i.e., in the F domain).
The finding that HNF4α1 and HNF4α2 differ structurally is supported by results with another protease. EndoLysC, which cleaves at internal lysine residues, yielded a pair of bands for HNF4α1 (bands a and a′) and for HNF4α2 (bands b and b′) that migrated at a molecular mass of approximately 38 kDa (Fig. 5B). Interestingly, however, even though HNF4α2 is 10 aa longer than HNF4α1, the faster-migrating band of the HNF4α2 pair (band b′) migrated slightly but reproducibly faster than the analogous band from HNF4α1 (band a′). This result suggests that HNF4α2 is cleaved by EndoLysC at a different lysine than HNF4α1 in either the C or the N terminus or both. Whereas the exact location of the EndoLysC cut sites remain to be determined, the results nonetheless support the conclusion that HNF4α2 is structurally distinct from HNF4α1.
Digestion with a third protease, trypsin, which cleaves at both arginines and lysines, provided further insight into the differences between HNF4α1 and HNF4α2. Digestion of HNF4α1 yielded a pair of bands that migrated just slightly faster than band a′ of EndoLysC (Fig. 5C; compare lanes 2 and 3 to lanes 4 and 5), whereas HNF4α2 yielded a pair of bands that migrated much faster than the EndoLysC band b′ (compare lanes 7 and 8 to lanes 9 and 10). Since there are two arginine residues in the α2 insert itself (Fig. 1), we initially thought that the increased migration was due only to cleavage in the α2 insert. However, a closer analysis of the observed and calculated molecular masses and comparison with cleavage products from N1C374 (lanes 12 and 13) suggested that tryptic bands b and b′ from HNF4α2 must represent an N-terminal cleavage distinct from that observed for HNF4α1, in addition to cleavage in the α2 insert.
Digestion of N1C374 with either trypsin or EndoLysC yielded a band c that migrates between trypsin bands b and b′ of HNF4α2 (Fig. 5C; compare lanes 12 and 13 to lanes 7 and 8). The observed molecular mass of this band c was approximately 28 kDa, based on comparison with commercial MW markers. To verify this, we spiked the N1C374 trypsin digestion with approximately 2 μg of an engineered fragment containing residues 127 to 374 of HNF4α1 (plus an additional eight residues at the N terminus from a fusion construct). After electrophoresis and transfer, the blot was stained with Coomassie blue to identify the position of the engineered fragment (Fig. 5D, top left, lanes 3 and 4). After the blot was subjected to autoradiography (Fig. 5D, bottom left), the engineered fragment, visualized by the Coomassie blue stain, was cut out and placed on another part of the blot (Fig. 5D, top right). Finally, the blot was resubjected to autoradiography (Fig. 5D, bottom right). The results show that the radiolabeled band c moved with the Coomassie blue-stained engineered fragment (Fig. 5D, right; compare lanes 3 and 4, top and bottom). This finding indicates that N1C374 trypsin band c has a molecular mass similar to that of a fragment containing residues 127 to 374 (plus eight additional residues, 28.6 kDa), thereby confirming a molecular mass of roughly 28 kDa. The fact that the migration of the 35S-labeled trypsin fragment is also altered by the presence of the engineered fragment (Fig. 5D, bottom left; compare lane 3 to lane 2) confirms the comigration, and therefore similar molecular mass of the trypsin fragment with the engineered fragment.
Using the 28-kDa size of band c as a reference point and taking into account the observed and calculated sizes of the various proteolytic products as well as the predicted surface probabilities, we attempted to determine the various trypsin cut sites in HNF4α1 and HNF4α2. The results (Fig. 5E) show that band b′ of HNF4α2 could indeed represent a fragment with an N terminus distinct from that of HNF4α1, such as R168. This is based on the finding that band b′ represents a C-terminal cut in the α2 insert and the fact that the arginine or lysine residues closest to R168 (R132 and K183) would yield fragments either too large (R132/R413, 31.6 kDa) or too small (K183/R413, 26.2 kDa; band b′ would have to migrate like band c′ in Fig. 5C, which it clearly does not). K170 and R171 would also yield fragments similar in size to R168, but their surface probabilities (1.26 and 0.86, respectively) are much less than that of R168 (2.08). Finally, in order for trypsin to also cleave HNF4α1 at R168, it would have to yield a fragment of 29.2 kDa which would migrate just slightly slower than band c (assuming a C-terminal cut at K428). However, no such fragment is evident in Fig. 5C or in any one of many other trypsin digestions performed (data not shown), supporting the notion that HNF4α1 is not as readily cleaved in the vicinity of R168 as is HNF4α2.
Potential cleavage in the vicinity of R168 in HNF4α2 but not HNF4α1 presents some interesting structural predictions. Assuming that the overall structure of the LBD of HNF4α is similar to those of the LBDs of other receptors whose structures have been solved, then R168 would be in a highly exposed, unstructured region right before the beginning of helix 3. In fact, R168 corresponds to the omega loop in RARγ and RXRα which switches position upon ligand binding and plays a role in the relative position of helix 12, which contains the activation domain AF-2 (73). The PR LBD structure also shows that the region of the omega loop is spatially close to helix 12 and AF-2 (90). Finally, the receptor-bound coactivator SRC1 may contain regions that are spatially close to the R168 region of peroxisome proliferator-activated receptor gamma (66). The possibility of enhanced cleavage of R168 in HNF4α2 suggests, therefore, that there may be some contact between the F domain and this region and that that contact may be different between HNF4α1 and HNF4α2. Considering the role of this region in other receptors, it is possible then that this region also plays a role in activation by HNF4α1 and HNF4α2.
DISCUSSION
The results of this study show that HNF4α1 responds to transcriptional coactivators GRIP1, p300, and CBP in vivo (Fig. 2 and 4). They also show a direct physical interaction between HNF4α1 and GRIP1 and SRC1a which appears to involve at least some of the NR boxes of GRIP1 and SRC1a (Fig. 3). Unlike most other nuclear receptors, however, addition of an exogenously added ligand was not required for the in vivo or in vitro effects of coactivators on HNF4α1. While this work was in progress, others observed similar interactions between HNF4α1 and CBP and between SRC1 and GRIP1 (9, 22, 64, 87, 94). Our results, however, show for the first time that the F domain of a nuclear receptor can partially block interaction with a coactivator (Fig. 2 and 3) and that the blockage is abrogated by a 10-aa insertion in the F domain generated by naturally occurring alternative splicing (HNF4α2 [Fig. 4]). They also show that the insertion induces structural changes in the F domain and elsewhere in the protein which could explain the altered blockage (Fig. 5).
There are a few reports of splicing variants of coactivators and corepressors acting differentially with nuclear receptors (31, 47, 76). There is also one report of nuclear receptor splicing variants differentially responding to corepressors (36). However, to our knowledge, this is the first published report showing that a naturally occurring splicing variant in a nuclear receptor interacts differentially with a coactivator. This is an important finding since nearly every nuclear receptor gene exhibits some degree of alternative splicing, although the functional significance of that splicing is not always known. As is seen in Fig. 4, interaction with coactivators can accentuate the difference between transcription factor isoforms that are otherwise difficult to detect, especially in transient transfection systems in which the factors tend to be expressed far beyond physiological concentrations. Furthermore, this phenomenon of splicing variants interacting differentially with coactivators might be more generally applicable to other transcription factors systems for which alternative splicing has also been shown to play a role in the control of gene expression (81).
Proposed mechanism for inhibitory action of the HNF4α F domain.
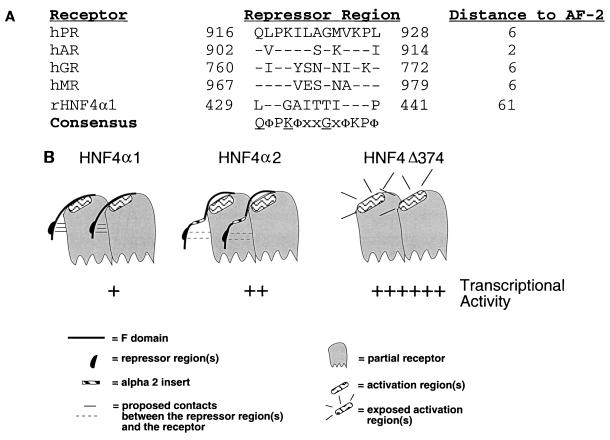
Iyemere et al. (40) recently reported a repressor region from aa 428 to 441 in rat HNF4α1 and observed that it showed a significant degree of similarity to a previously defined repressor region in the C-terminal extension of the LBD of PR (49, 91) (Fig. 6A). Shortly thereafter, the three-dimensional crystal structure of the LBD of PR was solved and showed that the repressor region forms a beta strand which is tightly fixed in position by an antiparallel beta-sheet interaction with another beta strand between helix 8 and helix 9 in the LBD (90). We have incorporated these findings and our current results into a model to explain the mechanism by which the F domain of HNF4α1 inhibits transcription (Fig. 6B). We propose that the F domain of HNF4α1 inhibits transcription by virtue of the repressor region, and possibly other regions, contacting another portion of the protein, most likely the LBD as in PR. This contact might obscure, at least partially, an activation region(s) such as AF-2 and thereby limit access to coactivators (Fig. 3 to 5). In HNF4α2, the predicted structure of the region suggests that the 10-aa insert introduces a turn in the F domain (Fig. 5E), which might cause a partial displacement of the repressor region(s), thereby exposing a protease cleavage site (Fig. 5) and the activation region(s). The net result is that the activation region(s) is somewhat more accessible to coactivators and that HNF4α2 activates transcription more efficiently than HNF4α1 (Fig. 4). In HNF4.N1.C374 and HNF4.N45.C374 (HNF4Δ374), there is no F domain to obscure the activation region(s), resulting in maximal physical contact with coactivators and hence transactivation (Fig. 2 and 3).
FIG. 6.
Model for the inhibition of transactivation and coactivator binding by the F domain of HNF4α1 and HNF4α2. (A) Shown is an alignment of the repressor regions in rat HNF4α1 (rHNF4α1) and human PR (hPR) to a region in the C-terminal extension of human GR, AR, and MR (hGR, hAR, and hMR) and the distance to the AF-2 which is located at the C-terminal end of the LBD. Residues that differ from the consensus are shown. Identical residues are indicated by dashes. Numbers indicate amino acid residues, which are given in single-letter code. φ, hydrophobic residue; x, any residue. (B) Schematic diagram (not drawn to scale) of proposed interactions between the F domain and the remainder of the receptor of HNF4α1, HNF4α2, and HNF4.N1.C374 or HNF4.N45.C374 (HNF4Δ374). Relative transcriptional activities from Fig. 2 and 4 are indicated. Specific regions shown are defined in the key at the bottom. The activation region(s) includes the AF-2 region, and possibly other regions, as discussed in the text.
Others have proposed a different model for HNF4α1 (40) and PR (91) in which the repressor region binds an unidentified corepressor molecule. There is also a report of identification of a cofactor of PR activation that is postulated to act by relieving the repression of the C-terminal extension (49). Interestingly, however, this activity had no significant effect on the ability of Xenopus HNF4α1 to activate transcription in vitro (49). All of these models, however, were proposed before the three-dimensional structure of the PR LBD showed that the repressor region contacted the LBD (90).
We favor the idea that the repressor region of HNF4α1 acts primarily by contacting another portion of HNF4α1 for an additional reason. The presence of the repressor region of HNF4α1 (aa 416 to 455) significantly decreased the ability of HNF4α1 to activate transcription in yeast (Fig. 3A). This suggests either that there is a corepressor endogenous to yeast that interacts with the C terminus of rat HNF4α1 or, more likely, that the repressor region is binding HNF4α1 itself, as in the model proposed in Fig. 6B. Furthermore, the results in Fig. 5 suggest that the F domain makes an intramolecular contact with the LBD of HNF4α1, although it remains to be shown that the contact necessarily involves the repressor region(s). Nonetheless, since the F domain consists of over 80 aa, one cannot rule out the possibility that other factors also contribute to the regulatory function of the F domain.
Finally, it is interesting that the repressor region of PR is also highly conserved in the C-terminal extension of GR, AR, and the mineralcorticoid receptor (MR) (Fig. 6A). Whereas this region has been shown to be required for ligand binding for PR, GR, and AR, and to possibly play a role in ligand specificity (42, 52, 90, 95), there are no functional data on it in the literature for MR. Furthermore, the role of the C-terminal extension in transactivation also seems to vary between different receptors as it inhibits transactivation by PR (91), whereas it is required for transactivation by GR (52, 95) and AR (42), presumably because of its requirement for ligand binding. In any case, the question then arises as to whether the analogous region in HNF4α1 is also required for ligand binding. Whereas a putative ligand for HNF4α1 has been reported (33), it has not yet been proven that the compounds proposed—fatty acyl coenzyme A thioesters—actually serve as a traditional ligand, such as by introducing a conformational change or by promoting binding to coactivators. (In fact, we have not been able to see such effects by the purported ligands [unpublished data]). Furthermore, one can imagine that the role of the repressor region in HNF4α1 might be somewhat different from that in the steroid receptors since the sequence of this region differs from the consensus sequence in at least three residues and is located in the primary amino acid sequence farther away from the C-terminal end of the LBD than in the steroid receptors, which tend to have short (<20-aa) C-terminal extensions. Nonetheless, conceptually, one possible role of a putative HNF4α ligand would be to somehow displace the F domain, thereby exposing the activation region(s) to coactivators. If this is the case, then one would expect the ligand to bind both HNF4α1 and HNF4α2 although perhaps with different affinities and/or consequences.
Several questions about the model remain. For example, whereas the AF-2 region of HNF4α is clearly critical for transactivation (9, 25), we (Fig. 3C) and others (9) have observed AF-2-independent binding of HNF4α1 to coactivators GRIP1 and CBP when the F domain is deleted. This suggests that there are regions of HNF4α in addition to AF-2 that may play a role in binding coactivators. One possible region is the AF-1 in the A/B domain which has been found by us and others to be necessary for full transactivation (Fig. 2; references 9 and 25). There are in fact recent reports of the AF-1 region of other nuclear receptors interacting with p160 family members in an NR box-independent fashion (56, 67, 88). However, whereas the AF-1 of HNF4α1 has been shown to interact directly with CBP (22), the presence of AF-1 seemed to inhibit only binding of HNF4α1 to GRIP1 (Fig. 3B). Further investigation of the mechanism of HNF4α1 binding to coactivators is clearly required.
The exact contact(s) between the F domain and the LBD must also be established. In addition to the repressor region at aa 428 to 441 identified by Iyemere et al. (40), our yeast two-hybrid data suggest that there may be another repressor region (Fig. 3A). Truncation of HNF4α1 at aa 370 increased interaction with GRIP1 and SRC1 even more than did truncation at aa 415, suggesting that residues 371 to 414 may contribute to repression by blocking interaction with coactivators. A computer analysis (PEPTIDESTRUCTURE) of HNF4α1 shows that aside from aa 428 to 441, which are predicted to form a beta strand, the only regions in the F domain that are predicted to form significant secondary structure (and hence be more likely to be involved in protein-protein contacts) are aa 383 to 389 (alpha helix) and aa 392 to 396 (beta strand). Interestingly, a mutation at residue 393 (V393I) which causes a twofold decrease in transactivation potential was recently identified in a form of inherited type II diabetes (27). It is intriguing to speculate that the mutation may cause increased contact between the F domain and the LBD and therefore result in a greater inhibitory effect of the F domain. Finally, it remains to be determined whether the F domain contacts the LBD of the same monomer or of the monomer partner. The latter would be reminiscent of the model proposed for RXR-RAR heterodimers in which the RXR AF-2 contacts the RAR partner, obscuring coactivator access (89).
A final question that arises is whether our model for HNF4α is applicable to other receptors with long F domains. RAR and ER are the two best characterized nuclear receptors with discernible AF-2 regions that have sizable F domains (>20 aa). However, whereas the F domain of human ERα is also thought to influence protein conformation and potentially protein-protein contacts, the F domain usually enhances the transcriptional activity of ERα (63). In contrast, the F domain of human RARα acts more like that of HNF4α, inhibiting transcriptional activity (82). Furthermore, we anticipate that the role of the F domain in HNF4α function will be different from that of RAR and ER since no sequence similarity could be found between the repressor region consensus noted in Fig. 6A, or any other part of the HNF4α1/α2 F domain, and the F domains of human ERα, ERβ, RARα, RARβ, or RARγ.
In conclusion, we report that the F domain of HNF4α1 acts as a negative regulatory region, impeding access of coactivators, and that a naturally occurring splicing variation in the F domain in HNF4α2 alters that function. Unfortunately, to date, none of the developmental work on HNF4α can distinguish between the two splicing variants. However, it is known that HNF4α2 mRNA is the more predominant form in several adult tissues, including liver, kidney, pancreatic islets, and enterocyte-like cells (27, 29). It is also known that the splicing variation is conserved across the three mammalian species analyzed thus far (rat, mouse, and human [6, 29]), suggesting that it is biologically important. The results presented in this report now add functional relevance to the splicing event. Finally, there are other HNF4α splicing variants, one (HNF4α3) with a completely distinct F domain (50) and two (HNF4α4 and HNF4α7) with alterations in the A/B domain (11, 18, 65). It will be of interest to determine whether these isoforms also exhibit differential interactions with coactivators and to determine the role of all the HNF4α isoforms in vivo.
ACKNOWLEDGMENTS
We thank the following colleagues for reagents: S. Hata (rat HNF4α2 cDNA), B. O’Malley (SRC1a cDNA), R. Goodman (CBP vector), and Y. Maeda (GST.127.374). We are grateful to C. Weinberger, H. Ingraham, and A. Bogan for scientific input, to V. Laudet for sharing compilations of nuclear receptors, and to D. Eastmond for advice on statistics.
This work was supported by NIH grant DK43093 to M.R.S. and American Heart Association grant-in-aid 96-267A and NIH grant DK53892 to F.M.S.
REFERENCES
- 1.Allan G F, Leng X, Tsai S Y, Weigel N L, Edwards D P, Tsai M J, O’Malley B W. Hormone and antihormone induce distinct conformational changes which are central to steroid receptor activation. J Biol Chem. 1992;267:19513–19520. [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1990. [Google Scholar]
- 3.Brzozowski A M, Pike A C W, Dauter Z, Hubbard R E, Bonn T, Engstrom O, Ohman L, Greene G L, Gustafsson J, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 4.Bulman M P, Dronsfield M J, Frayling T, Appleton M, Bain S C, Ellard S, Hattersley A T. A missense mutation in the hepatocyte nuclear factor 4 alpha gene in a UK pedigree with maturity-onset diabetes of the young. Diabetologia. 1997;40:859–862. doi: 10.1007/s001250050760. [DOI] [PubMed] [Google Scholar]
- 5.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 6.Chartier F L, Bossu J-P, Laudet V, Fruchart J-C, Laine B. Cloning and sequencing of cDNAs encoding the human hepatocyte nuclear factor 4 indicate the presence of two isoforms in human liver. Gene. 1994;147:269–272. doi: 10.1016/0378-1119(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 7.Chen W S, Manova K, Weinstein D C, Duncan S A, Plump A S, Prezioso V R, Bachvarova R F, Darnell J E., Jr Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 1994;8:2466–2477. doi: 10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
- 8.Committee, Nuclear Receptor Nomenclature. A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:161–163. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- 9.Dell H, Hadzopoulou-Cladaras M. CREB-binding protein is a transcriptional coactivator for hepatocyte nuclear factor-4 and enhances apolipoprotein gene expression. J Biol Chem. 1999;274:9013–9021. doi: 10.1074/jbc.274.13.9013. [DOI] [PubMed] [Google Scholar]
- 10.Ding X F, Anderson C M, Ma H, Hong H, Uht R M, Kushner P J, Stallcup M R. Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): multiple motifs with different binding specificities. Mol Endocrinol. 1998;12:302–313. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- 11.Drewes T, Senkel S, Holewa B, Ryffel G U. Human hepatocyte nuclear factor 4 isoforms are encoded by distinct and differentially expressed genes. Mol Cell Biol. 1996;16:925–931. doi: 10.1128/mcb.16.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 13.Escriva H, Safi R, Hanni C, Langlois M C, Saumitou L P, Stehelin D, Capron A, Pierce R, Laudet V. Ligand binding was acquired during evolution of nuclear receptors. Proc Natl Acad Sci USA. 1997;94:6803–6808. doi: 10.1073/pnas.94.13.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng W, Ribeiro R C, Wagner R L, Nguyen H, Apriletti J W, Fletterick R J, Baxter J D, Kushner P J, West B L. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science. 1998;280:1747–1749. doi: 10.1126/science.280.5370.1747. [DOI] [PubMed] [Google Scholar]
- 15.Ferguson A T, Davidson N E. Regulation of estrogen receptor alpha function in breast cancer. Crit Rev Oncog. 1997;8:29–46. doi: 10.1615/critrevoncog.v8.i1.20. [DOI] [PubMed] [Google Scholar]
- 16.Fuernkranz H A, Wang Y, Karathanasis S K, Mak P. Transcriptional regulation of the apoAI gene by hepatic nuclear factor 4 in yeast. Nucleic Acids Res. 1994;22:5665–5671. doi: 10.1093/nar/22.25.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuller P J. The steroid receptor superfamily: mechanisms of diversity. FASEB J. 1991;5:3092–3099. doi: 10.1096/fasebj.5.15.1743440. [DOI] [PubMed] [Google Scholar]
- 18.Furuta H, Iwasaki N, Oda N, Hinokio Y, Horikawa Y, Yamagata K, Yano N, Sugahiro J, Ogata M, Ohgawara H, Omori Y, Iwamoto Y, Bell G I. Organization and partial sequence of the hepatocyte nuclear factor-4 alpha/MODY1 gene and identification of a missense mutation, R127W, in a Japanese family with MODY. Diabetes. 1997;46:1652–1657. doi: 10.2337/diacare.46.10.1652. [DOI] [PubMed] [Google Scholar]
- 19.Garcia A D, Ostapchuk P, Hearing P. Functional interaction of nuclear factors EF-C, HNF-4, and RXRα with hepatitis B virus enhancer I. J Virol. 1993;67:3940–3950. doi: 10.1128/jvi.67.7.3940-3950.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Genetics Computer Group. Program manual for the Wisconsin Package, version 8. Madison, Wis: Genetics Computer Group; 1994. [Google Scholar]
- 21.Giangrande P H, Pollio G, McDonnell D P. Mapping and characterization of the functional domains responsible for the differential activity of the A and B isoforms of the human progesterone receptor. J Biol Chem. 1997;272:32889–32900. doi: 10.1074/jbc.272.52.32889. [DOI] [PubMed] [Google Scholar]
- 22.Green V J, Kokkotou E, Ladias J A A. Critical structural elements and multitarget protein interactions of the transcriptional activator AF-1 of hepatocyte nuclear factor 4. J Biol Chem. 1998;273:29950–29957. doi: 10.1074/jbc.273.45.29950. [DOI] [PubMed] [Google Scholar]
- 23.Gronemeyer H, Laudet V. Transcription factors 3: nuclear receptors. Protein Profile. 1995;2:1173–1308. [PubMed] [Google Scholar]
- 24.Guo W, Chen M, Yen T S B, Ou J-H. Hepatocyte-specific expression of the hepatitis B virus core promoter depends on both positive and negative regulation. Mol Cell Biol. 1993;13:443–448. doi: 10.1128/mcb.13.1.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hadzopoulou-Cladaras M, Kistanova E, Evagelopoulou C, Zeng S, Cladaras C, Ladias J A. Functional domains of the nuclear receptor hepatocyte nuclear factor 4. J Biol Chem. 1997;272:539–550. doi: 10.1074/jbc.272.1.539. [DOI] [PubMed] [Google Scholar]
- 26.Halachmi S, Marden E, Martin G, MacKay H, Abbondanza C, Brown M. Estrogen receptor-associated proteins: possible mediators of hormone-induced transcription. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- 27.Hani E H, Suaud L, Boutin P, Chevre J C, Durand E, Philippi A, Demenais F, Vionnet N, Furuta H, Velho G, Bell G I, Laine B, Froguel P. A missense mutation in hepatocyte nuclear factor-4 alpha, resulting in a reduced transactivation activity, in human late-onset non-insulin-dependent diabetes mellitus. J Clin Investig. 1998;101:521–526. doi: 10.1172/JCI1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanstein B, Eckner R, DiRenzo J, Halachmi S, Liu H, Searcy B, Kurokawa R, Brown M. p300 is a component of an estrogen receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hata S, Inoue T, Kosuga K, Nakashima T, Tsukamoto T, Osumi T. Identification of two splice isoforms of mRNA for mouse hepatocyte nuclear factor 4 (HNF-4) Biochim Biophys Acta. 1995;1260:55–61. doi: 10.1016/0167-4781(94)00177-5. [DOI] [PubMed] [Google Scholar]
- 30.Hata S, Tsukamoto T, Osumi T. A novel isoform of rat hepatocyte nuclear factor 4 (HNF-4) Biochim Biophys Acta. 1992;1131:211–213. doi: 10.1016/0167-4781(92)90080-j. [DOI] [PubMed] [Google Scholar]
- 31.Hayashi Y, Ohmori S, Ito T, Seo H. A splicing variant of steroid receptor coactivator-1 (SRC-1E): the major isoform of SRC-1 to mediate thyroid hormone action. Biochem Biophys Res Commun. 1997;236:83–87. doi: 10.1006/bbrc.1997.6911. [DOI] [PubMed] [Google Scholar]
- 32.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 33.Hertz R, Magenheim J, Berman I, Bar T J. Fatty acyl-CoA thioesters are ligands of hepatic nuclear factor-4alpha. Nature. 1998;392:512–516. doi: 10.1038/33185. [DOI] [PubMed] [Google Scholar]
- 34.Hodin R A, Lazar M A, Chin W W. Differential and tissue-specific regulation of the multiple rat c-erbA messenger RNA species by thyroid hormone. J Clin Investig. 1990;85:101–105. doi: 10.1172/JCI114398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holewa B, Zapp D, Drewes T, Senkel S, Ryffel G U. HNF4β, a new gene of the HNF4 family with distinct activation and expression profiles in oogenesis and embryogenesis of Xenopus laevis. Mol Cell Biol. 1997;17:687–694. doi: 10.1128/mcb.17.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hollenberg A N, Monden T, Madura J P, Lee K, Wondisford F E. Function of nuclear co-repressor protein on thyroid hormone response elements is regulated by the receptor A/B domain. J Biol Chem. 1996;271:28516–28520. doi: 10.1074/jbc.271.45.28516. [DOI] [PubMed] [Google Scholar]
- 37.Hong H, Kohli K, Garabedian M J, Stallcup M R. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong H, Kohli K, Trivedi A, Johnson D L, Stallcup M R. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imhof A, Yang X J, Ogryzko V V, Nakatani Y, Wolffe A P, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 40.Iyemere V, Davies N, Brownlee G. The activation function 2 domain of hepatic nuclear factor 4 is regulated by a short C-terminal proline-rich repressor domain. Nucleic Acids Res. 1998;26:2098–2104. doi: 10.1093/nar/26.9.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janknecht R, Hunter T. Versatile molecular glue. Transcriptional control. Curr Biol. 1996;6:951–954. doi: 10.1016/s0960-9822(02)00636-x. [DOI] [PubMed] [Google Scholar]
- 42.Jenster G, van der Korput H A G M, van Vroonhoven C, van der Kwast T H, Trapman J, Brinkmann A O. Domains of the human androgen receptor involved in steroid binding, transcriptional activation, and subcellular localization. Mol Endocrinol. 1991;5:1396–1404. doi: 10.1210/mend-5-10-1396. [DOI] [PubMed] [Google Scholar]
- 43.Jeyakumar M, Tanen M R, Bagchi M K. Analysis of the functional role of steroid receptor coactivator-1 in ligand-induced transactivation by thyroid hormone receptor. Mol Endocrinol. 1997;11:755–767. doi: 10.1210/mend.11.6.0003. [DOI] [PubMed] [Google Scholar]
- 44.Jiang G, Nepomuceno L, Hopkins K, Sladek F M. Exclusive homodimerization of orphan receptor hepatocyte nuclear factor 4 defines a new subclass of nuclear receptors. Mol Cell Biol. 1995;15:5131–5143. doi: 10.1128/mcb.15.9.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang G, Nepomuceno L, Yang Q, Sladek F M. Serine/threonine phosphorylation of orphan receptor hepatocyte nuclear factor 4. Arch Biochem Biophys. 1997;340:1–9. doi: 10.1006/abbi.1997.9914. [DOI] [PubMed] [Google Scholar]
- 46.Jiang G, Sladek F M. The DNA binding domain of hepatocyte nuclear factor 4 mediates cooperative, specific binding to DNA and heterodimerization with the retinoid X receptor α. J Biol Chem. 1997;272:1218–1225. doi: 10.1074/jbc.272.2.1218. [DOI] [PubMed] [Google Scholar]
- 47.Kalkhoven E, Valentine J E, Heery D M, Parker M G. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 1998;17:232–243. doi: 10.1093/emboj/17.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 49.Klotzbucher M, Schwerk C, Holewa B, Klein H L. Activation of transcription by progesterone receptor involves derepression of activation functions by a cofactor. Mol Endocrinol. 1997;11:768–778. doi: 10.1210/mend.11.6.0016. [DOI] [PubMed] [Google Scholar]
- 50.Kritis A A, Argyrokastritis A, Moschonas N K, Power S, Katrakili N, Zannis V I, Cereghini S, Talianidis I. Isolation and characterization of a third isoform of human hepatocyte nuclear factor 4. Gene. 1996;173:275–280. doi: 10.1016/0378-1119(96)00183-7. [DOI] [PubMed] [Google Scholar]
- 51.Ktistaki E, Talianidis I. Chicken ovalbumin upstream promoter transcription factors act as auxiliary cofactors for hepatocyte nuclear factor 4 and enhance hepatic gene expression. Mol Cell Biol. 1997;17:2790–2797. doi: 10.1128/mcb.17.5.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lanz R B, Rusconi S. A conserved carboxy-terminal subdomain is important for ligand interpretation and transactivation by nuclear receptors. Endocrinology. 1994;135:2183–2195. doi: 10.1210/endo.135.5.7956941. [DOI] [PubMed] [Google Scholar]
- 53.Lazennec G, Ediger T R, Petz L N, Nardulli A M, Katzenellenbogen B S. Mechanistic aspects of estrogen receptor activation probed with constitutively active estrogen receptors: correlations with DNA and coregulator interactions and receptor conformational changes. Mol Endocrinol. 1997;11:1375–1386. doi: 10.1210/mend.11.9.9983. [DOI] [PubMed] [Google Scholar]
- 54.Lin B C, Hong S H, Krig S, Yoh S M, Privalsky M L. A conformational switch in nuclear hormone receptors is involved in coupling hormone binding to corepressor release. Mol Cell Biol. 1997;17:6131–6138. doi: 10.1128/mcb.17.10.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lindner T, Gragnoli C, Furuta H, Cockburn B N, Petzold C, Rietzsch H, Weiss U, Schulze J, Bell G I. Hepatic function in a family with a nonsense mutation (R154X) in the hepatocyte nuclear factor-4alpha/MODY1 gene. J Clin Investig. 1997;100:1400–1405. doi: 10.1172/JCI119660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma H, Hong H, Huang S, Irvine R A, Webb P, Kushner P J, Coetzee G A, Stallcup M R. Multiple signal input and output domains of the 160-kDa nuclear receptor coactivator proteins. Mol Cell Biol. 1999;19:6164–6173. doi: 10.1128/mcb.19.9.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malik S, Karathanasis S K. TFIIB-directed transcriptional activation by the orphan nuclear receptor hepatocyte nuclear factor 4. Mol Cell Biol. 1996;16:1824–1831. doi: 10.1128/mcb.16.4.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mangelsdorf D, Evans R. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 59.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Masuyama H, Brownfield C M, St-Arnaud R, MacDonald P N. Evidence for ligand-dependent intramolecular folding of the AF-2 domain in vitamin D receptor-activated transcription and coactivator interaction. Mol Endocrinol. 1997;11:1507–1517. doi: 10.1210/mend.11.10.9990. [DOI] [PubMed] [Google Scholar]
- 61.Michael L F, Lazar M A, Mendelson C R. Peroxisome proliferator-activated receptor gammal expression is induced during cyclic adenosine monophosphate-stimulated differentiation of alveolar type II pneumonocytes. Endocrinology. 1997;138:3695–3703. doi: 10.1210/endo.138.9.5373. [DOI] [PubMed] [Google Scholar]
- 62.Moller A M, Urhammer S A, Dalgaard L T, Reneland R, Berglund L, Hansen T, Clausen J O, Lithell H, Pedersen O. Studies of the genetic variability of the coding region of the hepatocyte nuclear factor-4alpha in Caucasians with maturity onset NIDDM. Diabetologia. 1997;40:980–983. doi: 10.1007/s001250050778. [DOI] [PubMed] [Google Scholar]
- 63.Montano M M, Muller V, Trobaugh A, Katzenellenbogen B S. The carboxy-terminal F domain of the human estrogen receptor: role in the transcriptional activity of the receptor and the effectiveness of antiestrogens as estrogen antagonists. Mol Endocrinol. 1995;9:814–825. doi: 10.1210/mend.9.7.7476965. [DOI] [PubMed] [Google Scholar]
- 64.Muraoka R S, Waltz S E, Degen S J. Expression of hepatocyte growth factor-like protein is repressed by retinoic acid and enhanced by cyclic adenosine 3′,5′-monophosphate response element-binding protein (CREB)-binding protein (CBP) Endocrinology. 1999;140:187–196. doi: 10.1210/endo.140.1.6441. [DOI] [PubMed] [Google Scholar]
- 65.Nakhei H, Lingott A, Lemm I, Ryffel G U. An alternative splice variant of the tissue specific transcription factor HNF4alpha predominates in undifferentiated murine cell types. Nucleic Acids Res. 1998;26:497–504. doi: 10.1093/nar/26.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 67.Onate S A, Boonyaratanakornkit V, Spencer T E, Tsai S Y, Tsai M J, Edwards D P, O’Malley B W. The steroid receptor coactivator-1 contains multiple receptor interacting and activation domains that cooperatively enhance the activation function 1 (AF1) and AF2 domains of steroid receptors. J Biol Chem. 1998;273:12101–12108. doi: 10.1074/jbc.273.20.12101. [DOI] [PubMed] [Google Scholar]
- 68.Onate S A, Tsai S Y, Tsai M-J, O’Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 69.Pemrick S M, Lucas D A, Grippo J F. The retinoid receptors. Leukemia. 1994;8:1797–1806. [PubMed] [Google Scholar]
- 70.Perlmann T, Evans R M. Nuclear receptors in Sicily. Cell. 1997;90:391–397. doi: 10.1016/s0092-8674(00)80498-5. . (Meeting review.) [DOI] [PubMed] [Google Scholar]
- 71.Raney A K, Johnson J L, Palmer C N, McLachlan A. Members of the nuclear receptor superfamily regulate transcription from the hepatitis B virus nucleocapsid promoter. J Virol. 1997;71:1058–1071. doi: 10.1128/jvi.71.2.1058-1071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reijnen M J, Sladek F M, Bertina R M, Reitsma P H. Disruption of a binding site for hepatocyte nuclear factor 4 results in hemophilia B. Leyden. Proc Natl Acad Sci USA. 1992;89:6300–6303. doi: 10.1073/pnas.89.14.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Renaud J, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Crystal structure of the RAR-γ ligand-binding domain bound to all-trans retinoic acid. Nature. 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 74.Rosen C A, Sodroski J G, Haseltine W A. The location of cis-acting regulatory sequences in the human T cell lymphotropic virus type III (HTLV-III/LAV) long terminal repeat. Cell. 1985;41:813–823. doi: 10.1016/s0092-8674(85)80062-3. [DOI] [PubMed] [Google Scholar]
- 75.Rottman J N, Widom R L, Nadal-Ginard B, Mahdavi V, Karathanasis S K. A retinoic acid-responsive element in the apolipoprotein AI gene distinguishes between two different retinoic acid response pathways. Mol Cell Biol. 1991;11:3814–3820. doi: 10.1128/mcb.11.7.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sande S, Privalsky M L. Identification of TRACs (T3 receptor-associating cofactors), a family of cofactors that associate with, and modulate the activity of, nuclear hormone receptors. Mol Endocrinol. 1996;10:813–825. doi: 10.1210/mend.10.7.8813722. [DOI] [PubMed] [Google Scholar]
- 77.Shibata H, Spencer T E, Onate S A, Jenster G, Tsai S Y, Tsai M J, O’Malley B W. Role of co-activators and co-repressors in the mechanism of steroid/thyroid receptor action. Recent Prog Horm Res. 1997;52:141–164. [PubMed] [Google Scholar]
- 78.Sladek F, Dallas-Yang Q, Nepomuceno L. MODY1 mutation Q268X in hepatocyte nuclear factor 4α allows for dimerization in solution but causes abnormal subcellular localization. Diabetes. 1998;47:985–990. doi: 10.2337/diabetes.47.6.985. [DOI] [PubMed] [Google Scholar]
- 79.Sladek F M. Hepatocyte nuclear factor 4 (HNF-4) In: Tronche F, Yaniv M, editors. Liver gene expression. R. G. Austin, Tex: Landes Co.; 1994. pp. 207–230. [Google Scholar]
- 80.Sladek F M, Zhong W, Lai E, Darnell J E. Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 1990;4:2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- 81.Smith C W J, Patton J G, Nadal-Ginard B. Alternative splicing in the control of gene expression. Annu Rev Genet. 1989;23:527–577. doi: 10.1146/annurev.ge.23.120189.002523. [DOI] [PubMed] [Google Scholar]
- 82.Tate B F, Allenby G, Perez J R, Levin A A, Grippo J F. A systematic analysis of the AF-2 domain of human retinoic acid receptor alpha reveals amino acids critical for transcriptional activation and conformational integrity. FASEB J. 1996;10:1524–1531. doi: 10.1096/fasebj.10.13.8940298. [DOI] [PubMed] [Google Scholar]
- 83.Torchia J, Glass C, Rosenfeld M G. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 84.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 85.Voegel J J, Heine M J, Tini M, Vivat V, Chambon P, Gronemeyer H. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Voegel J J, Heine M J, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 87.Wang J C, Stafford J M, Granner D K. SRC-1 and GRIP1 coactivate transcription with hepatocyte nuclear factor 4. J Biol Chem. 1998;273:30847–30850. doi: 10.1074/jbc.273.47.30847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Webb P, Nguyen P, Shinsako J, Anderson C, Feng W, Nguyen M P, Chen D, Huang S M, Subramanian S, McKinerney E, Katzenellenbogen B S, Stallcup M R, Kushner P J. Estrogen receptor activation function 1 works by binding p160 coactivator proteins. Mol Endocrinol. 1998;12:1605–1618. doi: 10.1210/mend.12.10.0185. [DOI] [PubMed] [Google Scholar]
- 89.Westin S, Kurokawa R, Nolte R T, Wisely G B, McInerney E M, Rose D W, Milburn M V, Rosenfeld M G, Glass C K. Interactions controlling the assembly of nuclear-receptor heterodimers and co-activators. Nature. 1998;395:199–202. doi: 10.1038/26040. [DOI] [PubMed] [Google Scholar]
- 90.Williams S P, Sigler P B. Atomic structure of progesterone complexed with its receptor. Nature. 1998;393:392–396. doi: 10.1038/30775. [DOI] [PubMed] [Google Scholar]
- 91.Xu J, Nawaz Z, Tsai S Y, Tsai M J, O’Malley B W. The extreme C terminus of progesterone receptor contains a transcriptional repressor domain that functions through a putative corepressor. Proc Natl Acad Sci USA. 1996;93:12195–12199. doi: 10.1073/pnas.93.22.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu L, Glass C K, Rosenfeld M G. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 93.Yamagata K, Furuta H, Oda N, Kaisaki P J, Menzel S, Cox N J, Fajans S S, Signorini S, Stoffel M, Bell G I. Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 94.Yoshida E, Aratani S, Itou H, Miyagishi M, Takiguchi M, Osumu T, Murakami K, Fukamizu A. Functional association between CBP and HNF4 in trans-activation. Biochem Biophys Res Commun. 1997;241:664–669. doi: 10.1006/bbrc.1997.7871. [DOI] [PubMed] [Google Scholar]
- 95.Zhang S, Liang X, Danielsen M. Role of the C terminus of the glucocorticoid receptor in hormone binding and agonist/antagonist discrimination. Mol Endocrinol. 1996;10:24–34. doi: 10.1210/mend.10.1.8838142. [DOI] [PubMed] [Google Scholar]