BET inhibitor (BETi) response is governed by leukemia differentiation state, with monocytic AMLs exhibiting intrinsic sensitivity due to expression of transcription factors that recruit histone acetylation machinery–BETi targets.
Abstract
To understand mechanisms of response to BET inhibitors (BETi), we mined the Beat AML functional genomic data set and performed genome-wide CRISPR screens on BETi-sensitive and BETi-resistant acute myeloid leukemia (AML) cells. Both strategies revealed regulators of monocytic differentiation—SPI1, JUNB, FOS, and aryl-hydrocarbon receptor signaling (AHR/ARNT)—as determinants of BETi response. AHR activation synergized with BETi, whereas inhibition antagonized BETi-mediated cytotoxicity. Consistent with BETi sensitivity dependence on monocytic differentiation, ex vivo sensitivity to BETi in primary AML patient samples correlated with higher expression of the monocytic markers CSF1R, LILRs, and VCAN. In addition, HL-60 cell line differentiation enhanced its sensitivity to BETi. Further, screens to rescue BETi sensitivity identified BCL2 and CDK6 as druggable vulnerabilities. Finally, monocytic AML patient samples refractory to venetoclax ex vivo were significantly more sensitive to combined BETi + venetoclax. Together, our work highlights mechanisms that could predict BETi response and identifies combination strategies to overcome resistance.
Significance:
Drug resistance remains a challenge for AML, and new therapies, such as BETi, will require combination approaches to boost single-agent responses. We conducted genome-wide CRISPR screens and functional genomics on AML patient samples to identify leukemic differentiation state and AHR signaling as primary mediators of BETi response.
Introduction
Acute myeloid leukemia (AML) is an aggressive hematologic malignancy, diagnosed primarily in elderly patients. Many patients cannot tolerate the intensive 7 + 3 chemotherapy regimen (cytarabine + anthracycline), which has been a standard of care for >40 years (1). These patients rely on alternative treatment strategies, such as targeted small-molecule inhibitors. Recently, combinations of the BCL2 inhibitor venetoclax with hypomethylating agents were approved for treatment of patients unfit for chemotherapy, but not all patients respond to this therapeutic regimen (2). Most patients, particularly those treated with monotherapy regimens, will develop resistance and relapse. Therefore, understanding molecular mechanisms driving drug resistance is critical for the development of drug combinations that yield durable remissions and extend survival.
AML is a heterogeneous cancer that is primarily driven by four classes of mutations: (i) activation of proliferative and antiapoptotic genes, (ii) block of differentiation, (iii) epigenetic regulators, and (iv) splicing machinery (3,4,5). As such, we have seen the implementation of many small-molecule inhibitors targeting these pathways (6).
The bromodomain and extra-terminal domain (BET) protein family consists of bromodomain containing the proteins BRD2, BRD3, BRD4, and BRDT, which interact with acetylated histone tails to facilitate many downstream functions, such as chromatin remodeling and transcriptional regulation. Epigenetic inhibitors targeting BET family proteins (BETi) have recently come to the forefront of development due to evident cytotoxicity in hematologic settings (7,8,9,10,11). BRD4 binds acetylated histone tail and recruits positive transcription elongation factor b (P-TEFb) to enhancer regions to mediate the phosphorylation of the c-terminal domain of RNA pol II, required for elongation of the nascent mRNA (12, 13). BRD4 also acts as a histone acetyltransferase (HAT; ref. 14), an atypical kinase (15), and interacts with splicing machinery (16). Previous RNAi studies identified BRD4 loss as a potent inhibitor of leukemic growth (17). BET proteins have also been linked to driving leukemia disease by recruiting transcription machinery to MYC and BCL2 promoters (18, 19). BETi treatment in leukemia cells has been shown to dramatically reduce transcription of these oncogenes and induce cell death (8, 18, 20). Clinically, the BETi OTX-015 achieved complete remissions in a small subset of patients who failed alternative therapies. However, many patients were unresponsive to OTX-015, prompting the investigation of potential intrinsic resistance mechanisms to BETi (21).
Several studies have reported on a wide scope of genetic resistance mechanisms to BETi involving autophagy (22), WNT signaling driven by leukemia stem cells and transcriptional plasticity (23, 24), and PP2A (25). Further, Bell and colleagues have shown that nongenetic resistance can arise upon BETi exposure (26). These studies prompted us to mine the Beat AML functional genomic data set and also utilize genome-wide CRISPR screens to further understand drivers of resistance to BETi in AML and identify therapeutically druggable dependencies.
Results
Monocytic Markers Correlate with BETi Sensitivity in AML Patient Samples
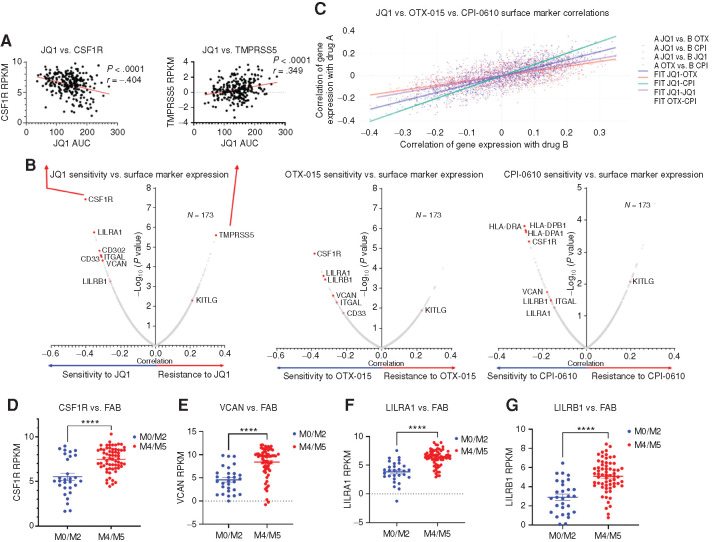
To identify potential genetically driven BETi resistance mechanisms, we correlated ex vivo drug-sensitivity data for three BETi (JQ1, OTX-015, and CPI-610) against recurrent genetic mutations using the Beat AML cohort, an integrative data set containing ex vivo drug-sensitivity analyses, exome sequencing, and RNA sequencing for over 500 primary AML patient specimens (3). Of the entire database, we were able to obtain BETi ex vivo data for 173 unique patient samples. Interestingly, we found no genetic mutations or cytogenetic patterns that significantly correlated with resistance or sensitivity to any of the three BETi (Supplementary Fig. S1A–S1F). In recent work by our lab and others, similar explorations of response to venetoclax identified leukemic differentiation state as a primary determinant of sensitivity and resistance (27,28,29,30). Accordingly, because no mutations correlated with BETi response, we next asked whether BETi sensitivity and resistance correlated with expression of cell-surface markers that are known indicators of cell differentiation state. Indeed, within the Beat AML database, ex vivo responses to JQ1, OTX-015, and CPI-0610 in 173 primary human patient samples were significantly correlated with expression of monocytic markers such as CSF1R, VCAN, CD33, ITGAL, and LILRA1 (Fig. 1A and B). In addition, we found high congruence of drug-sensitivity versus surface marker expression correlations across all three BETi (Fig. 1C). Historically, AML cases have been classified based on the French–American–British (FAB) M0 to M7 classification system, where M0, M1, and M2 represent tumors comprised of minimally or undifferentiated cells, and M4 and M5 represent tumors of a myelomonocytic or monocytic cell state. As expected, we also found that CSF1R, VCAN, LILRA1, and LILRB1 were highly expressed in monocytic leukemia FAB subtypes (M4–M5) compared with undifferentiated cases (M0–M2; Fig. 1D–G). Together, these findings indicate that BETi may be more efficacious in differentiated leukemias and highlight a novel vulnerability for these leukemias.
Figure 1.
Identification of correlates of BETi resistance and sensitivity in AML patient samples. A, Correlations between ex vivo sensitivity to JQ1 in 173 patient samples and expression of monocytic differentiation markers with the Beat AML data set. Each dot represents a unique patient sample ex vivo response to a BETi [area under the curve (AUC), determined by MTS viability assay] plotted against mRNA level of the surface marker denoted (CSF1R, TMPRSS5). A simple linear regression model was fit separately for each inhibitor and gene, with the inhibitor AUC values as the outcome. The t-statistic and P value test whether the slope is nonzero and corrected for multiple comparisons. Corresponding correlation coefficients were computed using the relationship to the linear model: slope*(sd(expr)/sd(auc)), where sd indicates standard deviation. RPKM, reads per kilobase per million mapped. B, Pearson correlation coefficients and significance, corrected for multiple comparisons, are calculated for each surface marker mRNA expression and ex vivo sensitivity to a BETi in 173 Beat AML patient sample data set. Each dot represents a correlation coefficient and corresponding P value for individual surface marker mRNA versus BETi AUC (as shown in A, extended to all known surface markers). Points of interest are highlighted in red. C, Pearson correlations for sensitivity to each individual BETi (JQ1, OTX-015, and CPI-0610) versus surface marker expression calculated previously in B, plotted against each other with a line of best fit highlighting their congruence. Each point thus represents the XY coordinate location of correlations for a surface marker between two different BETi, for example, a single point will be X = JQ1 versus CSF1R correlation, Y = OTX-015 versus CSF1R correlation, or X = OTX-015 versus ITGAL correlation versus Y = CPI-0610 versus ITGAL correlation. Dark purple dots represent comparisons between JQ1 and OTX-015, orange represents comparisons between JQ1 and CPI-0610, green represents comparisons between JQ1 against itself, and light purple represents comparisons between OTX-015 and CPI-0610. Corresponding lines of best fit match the above comparisons. D–G, Comparison of RNA expression levels (RPKM) of CSF1R (D), VCAN (E), LILRA1 (F), and LILRB1(G) across FAB groups M0 to M2 (29 samples), undifferentiated AML, and M4 and M5 (34 samples), myelomonocytic–monocytic AML (83) within the Beat AML patient data set (3). P values determined by Mann–Whitney t tests. The x-axis denotes FAB subtype; the y-axis denotes RPKM expression of a given gene.
Genome-wide CRISPR Screen Identifies Monocytic Differentiation Regulators of BETi Resistance
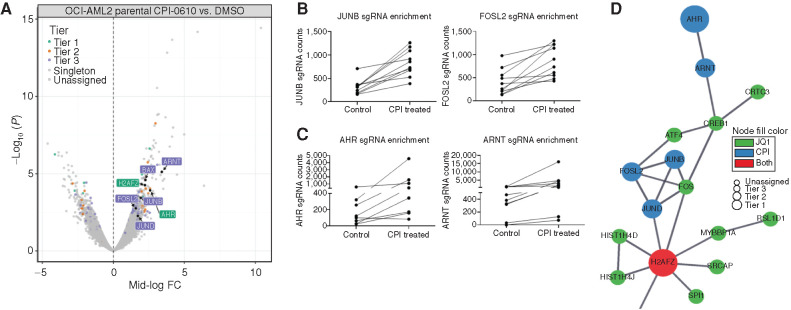
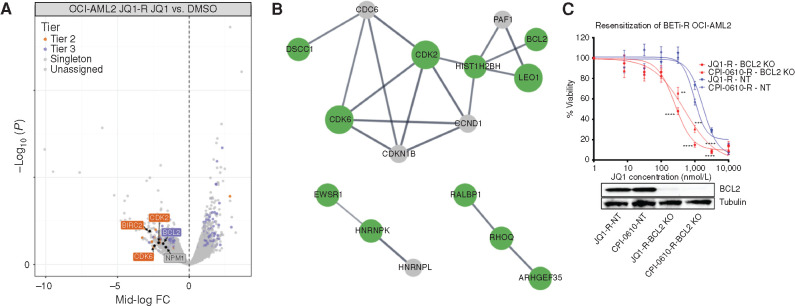
To further study mechanisms of resistance to BETi in AML, we performed a genome-wide CRISPR resistance screen in OCI-AML2 cells under selection of the BETi CPI-0610. Distribution of single-guide RNAs (sgRNA) in deep-sequenced libraries from drug-treated cells was compared with DMSO-treated controls using edgeR (31). To prioritize hits, we used a tiering structure that we previously developed for CRISPR screens (32). This scheme organizes top candidates into three tiers based on evidence (determined by the number of sgRNA guide hits per gene), concordance (indicated by the agreement across the set of guides for a given gene), and discovery (based on expanding effect size threshold). Using this prioritization scheme and focusing on tiered genes with a mid-log fold change > 1.5, CPI-0610 (Fig. 2A) selected cells showed enrichment for guides targeting hematopoietic transcription factors (TF; e.g., FOSL2, JUNB; Fig. 2B) and aryl-hydrocarbon signaling (e.g., AHR, ARNT; Fig. 2C) compared with DMSO. We ran a parallel screen using overlay of a related BETi, JQ1, instead of CPI-0610. Although the results of this screen did not yield hits that were as statistically significant as the CPI-0610 screen with no genes assigned to a tier, we focused attention to genes with a mid-log fold change > 2. Using this threshold, we also identified resistance-enriched guides that targeted genes in the same category of TFs known to regulate myelopoiesis (SPI1, FOS, CREB1; Supplementary Fig. S2A and S2B). To prioritize hits for follow-up analysis, we assessed enrichment of hits from the two screens in a pathway context. We took the combined list of gene hits from both screens and seeded a STRING (Search Tool for Retrieval of Interacting Genes/Proteins; ref. 33) network to investigate associations between this union of candidate genes from both screens (Fig. 2D; Supplementary Fig. S2C). In addition, we took these same hits and analyzed them via Gene Ontology Cellular Component Ontology and found high concordance between genes from both screens under shared ontological components (Supplementary Fig. S2D). Using both methods, a core set of gene hits—SPI1, FOS, JUNB, AHR, and ARNT—were mapped in close proximity across multiple pathway annotations, and these genes were chosen for downstream validation.
Figure 2.
Whole-genome CRISPR screen identifies hematopoietic TFs and AHR signaling as drivers of BETi resistance. Cas9-expressing OCI-AML2 were lentivirally transduced with a genome-wide CRISPR library containing an average of five guides per gene, collectively targeting 18,010 human genes (32, 77). Cells were treated with 500 nmol/L CPI-0610 or vehicle (DMSO) for 21 days, followed by DNA harvest and PCR amplification of sgRNA guide sequences. A, Volcano plot comparing enrichment of sgRNAs relative to DMSO control in CPI-0610-treated OCI-AML2 cells versus –log10-transformed median P value after 21 days, with corresponding significance tiers. Each dot represents a combined sgRNA knockout (KO) enrichment score (combining approximately five to six unique sgRNAs targeting the same gene) relative to control versus corresponding P value. Significance of each sgRNA was determined via edgeR (RRID: SCR_012802) after trimmed mean of M values (TMM; ref. 78) normalization. Briefly, considering only significant sgRNAs (FDR < 0.05), genes were classified into five ordered groups. Tier 1 genes had more than one significant sgRNA, a minimum log2 fold change ≥2, 75% of sgRNAs per gene present, and concordance among sgRNAs per gene ≥75%; tier 2 hits had log2 fold change ≥2 and 100% concordance among sgRNAs per gene; and tier 3 hits had log2 fold change ≥1 and 100% concordance among sgRNAs per gene. Singleton hits represent significantly enriched genes with log2 fold change ≥2 but only a single significant sgRNA. Enriched hits not satisfying these criteria were classified into the unassigned group. The x-axis corresponds to median log fold change (CPI treated/control treated) sgRNA counts. The y-axis corresponds to –log10 median P value. FC, fold change. B, Enrichment of cells containing the relative sgRNA KO relative to DMSO control (roughly five to six unique sgRNAs per gene KO for Yusa library) targeting hematopoietic TFs JUNB (left) and FOSL2 (right) in CPI-0610–treated OCI-AML2. Each line represents the guide counts of the relevant unique sgRNA KOs in control- or CPI-0610-treated cells. C, Enrichment of cells containing the relative sgRNA KO targeting aryl-hydrocarbon receptor signaling components AHR (left) and ARNT (right) in CPI-0610–treated OCI-AML2. Each line represents the guide counts of the relevant sgRNA KOs in control or CPI-0610–treated cells. D, A STRING (33) network, which is used to model known or predicted protein–protein interactions from custom gene lists, was seeded with significant hits from both the CPI-0610 and JQ1 CRISPR screens, and identifies a concordant network of enriched hits of interest across both JQ1 and CPI-0610 CRISPR screens centered on hematopoietic TFs, AHR, and histone-modifying machinery. Size of node denotes tier (largest = tier 1, smallest = unassigned); color denotes gene screen of origin.
Screen Validation
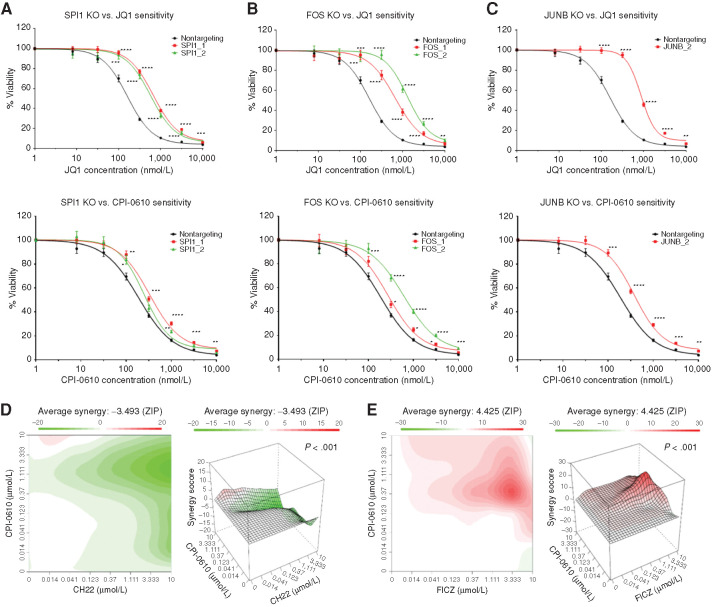
Because both the JQ1 and CPI screens pointed to the same pathways but yielded less agreement at the level of individual genes, we wanted to determine whether these specific gene knockout events were truly causing differential resistance to the two drugs or whether targeting of these genes yields more of a pan-BETi resistance phenotype. Accordingly, we designed two sgRNAs per gene to target SPI1, FOS, or JUNB or nontargeting. Knockouts were validated by Tracking of Indels by DEcomposition (TIDE) analysis or Western blot (Supplementary Tables S1 and S2; Supplementary Fig. S3A). Sensitivity to BETi (JQ1 and CPI-0610) was determined by MTS assay after selection of OCI-AML2 cells with control or gene-targeting sgRNAs. With this approach, we found that loss of SPI1, FOS, and JUNB resulted in pan-BETi resistance (Fig. 3A–C). Small-molecule AHR inhibitors and activators are readily available and, thus, to investigate altered AHR signaling in mediating BETi resistance, we tested the combination of CPI-0610 with either an AHR inhibitor (AHRi), CH-223191, or an AHR agonist, FICZ. As expected, the AHRi antagonized CPI-0610 cytotoxicity in OCI-AML2s (Fig. 3D), with a zero interaction potency (ZIP) synergy score of −3.493, whereas FICZ enhanced these effects, with a ZIP synergy score of 4.425 (Fig. 3E). We validated these findings further by knocking out AHR and ARNT with single sgRNAs and observed multi-BETi resistance (Supplementary Fig. S3A and S3B; Supplementary Table S3). Convergent lines of evidence suggest that hematopoietic TFs (34, 35) and AHR (36, 37) recruit histone-modifying machinery to active transcription sites and interact with the P-TEFb complex (38, 39). The subsequent hyperacetylation at these regions increases dependency on BRD4 and induces monocytic differentiation. Therefore, we hypothesized that monocytic leukemias, which characteristically have high expression of these genes, may exhibit greater sensitivity to BETi, and that undifferentiated blasts will be intrinsically resistant to BETi due to decreased expression of these genes.
Figure 3.
Validation of whole-genome CRISPR screen hits SPI1, FOS, JUNB, and AHR signaling. A–C, OCI-AML2 cells were transduced with lentiviruses carrying custom-designed single sgRNA/Cas9 constructs (two different sgRNAs/gene) targeting SPI1, FOS, or JUNB or nontargeting. After puromycin selection and 10 days of transduction, sensitivity to JQ1 (top row), and CPI-0610 (bottom row) was measured in replicates of six via colorimetric MTS-based viability assay. Plots represent sensitivity to BETi in SPI1 KO (A; left), FOS KO (B; middle), and JUNB KO (C; right) in single sgRNA KOs (colored) versus nontargeting control (black) in OCI-AML2 cells. Error bars represent the SE margin between the six replicates. Drug curve consists of eight concentrations ranging from 0 μmol/L and 0.0137 to 10 μmol/L in log increments. P values for individual sgRNAs versus nontargeting were calculated using a two-stage linear step-up procedure of Benjamini–Krieger–Yekutieli. The x-axis represents BETi concentration; the y-axis represents OCI-AML2 viability. KO, knockout. D and E, OCI-AML2 cells were subjected to dilutions of CPI-0610 in combination with titrations of either the AHRi CH-223191 (D) or the AHR ligand FICZ (E) and evaluated for viability via colorimetric MTS-based assay. The ZIP synergy score was calculated at each drug combination dose pair as previously described (81, 82). The ZIP score average was computed across the dose matrix to provide an overall index of drug interaction. A positive average ZIP score indicates the combination was synergistic (red), whereas a negative average ZIP score indicates antagonism (green). ZIP scores were −3.493 and 4.425 for CH-223191 and FICZ, respectively. Significance of synergy determined by one-sample Wilcoxon signed rank test comparing individual dose–pair combinations across the entire 7 × 7 matrix. The x-axis represents increasing concentrations of the AHRi CH-223191 (left) or the AHR agonist FICZ (right). the y-axis represents increasing concentrations of CPI-0610.
BETi-Resistant Cells Exhibit Decreased Markers of Leukemic Differentiation, and Forced Myeloid Differentiation Increases Sensitivity to BETi
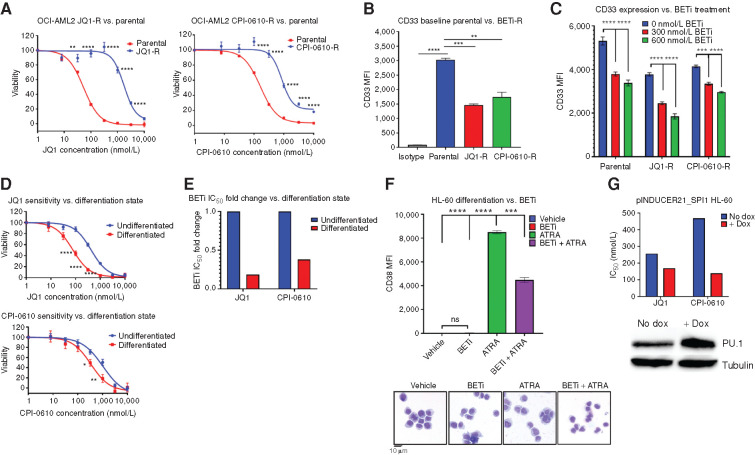
Given our proposed mechanism, which posited that BETi sensitivity is driven by monocytic differentiation, we asked whether acquired BETi-resistant (BETi-R) cells had reduced marks of myeloid differentiation. We generated JQ1- and CPI-0610–resistant (JQ1-R and CPI-0610-R) OCI-AML2 cell models by serially passaging them under increasing selective pressure of BETi and reached IC50 values 5-fold higher than parental OCI-AML2 cells (JQ1: 52 nmol/L parental, 1.8 μmol/L resistant; CPI-0610: 170 nmol/L parental, 1.1 μmol/L resistant; Fig. 4A). Immunophenotyping of OCI-AML2 parental and BETi-R cells revealed a significant reduction in myeloid differentiation marker CD33 expression in BETi-R cells at baseline (Fig. 4B). To test whether BETi treatment selects for less differentiated clones, we treated both BETi-naïve and BETi-R OCI-AML2 and observed decreased CD33 expression in both BETi-naïve and BETi-R cells in a dose-dependent manner, further suggesting that BETi may select for less differentiated clones (Fig. 4C). HL-60 is an undifferentiated M2 FAB leukemia cell line that has been used as a model of myeloid differentiation due to the ability of HL-60s to differentiate in response to all-trans retinoic acid (ATRA), marked by a dramatic increase in expression of CD38. Upon exposure to ATRA, HL-60s rapidly upregulate myeloid differentiation programs and dramatically increase expression of many genes such as SPI1, CDKN1A, AHR, FOS, JUN, CD38, and others (40). Given our previous findings that showed that BETi-R cells exhibit decreased marks of differentiation, and that loss of these differentiation regulators drives resistance, we directly asked whether inducing myeloid differentiation increases BETi sensitivity. Indeed, HL-60s, differentiated with 1 μmol/L ATRA for 72 hours prior to exposing to BETi, showed dramatically enhanced sensitivity to BETi as compared with undifferentiated HL-60s (Fig. 4D and E). In addition, doxycycline-inducible PU.1 overexpression in HL-60s sensitized to multiple BETi (Fig. 4F). Lastly, to answer whether BETi directly target differentiation programs and, thus, support the notion that BETi may modulate the process of differentiation, we added BETi concomitantly with ATRA in HL-60s and found that differentiation was significantly attenuated, as seen by a significant reduction in CD38 expression and morphologic changes consistent with myeloid differentiation after 72 hours (Fig. 4G). Taken together, these findings validate that BETi treatment selects for less differentiated clones and that forced differentiation can enhance sensitivity to BETi.
Figure 4.
BETi-R OCI-AML2 exhibit hallmarks of decreased differentiation, and forced myeloid differentiation of HL-60s increases sensitivity to BETi. A–C, BETi-R OCI-AML2 cells were generated via incubation with serially increasing concentration of JQ1 or CPI-0610 over a several month period. A, MTS viability assays were used to assess BETi dose responses in JQ1-R (left) and CPI-R (right) OCI-AML2 cells. Error bars represent the SE margin between the six replicates. P values were calculated using a two-stage linear step-up procedure of Benjamini–Krieger–Yekutieli comparing parental to BETi-R OCI-AML2 cells. The x-axis represents BETi concentration; the y-axis represents OCI-AML2 percentage viability. B, Median fluorescence intensity (MFI) of myeloid differentiation marker CD33, as determined by flow cytometry, on untreated BETi-naïve OCI-AM2 cells and BETi-R OCI-AML2 cells withdrawn from drug for 1 week. Significance determined by ordinary one-way ANOVA with multiple comparison corrections from three replicates. C, CD33 MFI, as determined by flow cytometry, of BETi-naïve or BETi-R OCI-AML2 cells treated with vehicle, 300 nmol/L JQ1, or 600 nmol/L JQ1 for 72 hours. Significance determined by two-way ANOVA with multiple comparison corrections from three replicates. D, HL-60 cells were differentiated with 1 μmol/L ATRA for 72 hours or left undifferentiated in vehicle for 72 hours. The cells were then washed in PBS, and viability was assessed (>95%) by guava easycyte. Drug dose–response curves, as determined by MTS viability assay, were then assessed for JQ1 (top) and CPI-0610 (bottom) comparing undifferentiated (vehicle) and ATRA-differentiated HL-60 cells. Error bars represent the standard error margin between the six replicates. P values were calculated using a two-stage linear step-up procedure of Benjamini–Krieger–Yekutieli comparing undifferentiated (vehicle-treated cells) to differentiated (ATRA-treated cells). The x-axis represents BETi concentration; the y-axis represents HL-60 viability. E, Fold change IC50 values for JQ1 and CPI-0610 in undifferentiated HL-60s versus ATRA-differentiated HL-60s from D. F, Top, flow cytometry measurement of CD38 MFI, which marks HL-60 differentiation state, in naïve HL-60s treated for 72 hours with vehicle (undifferentiated), BETi, ATRA (differentiated), or simultaneously added BETi and ATRA. Significance determined by one-way ANOVA from three replicates. Bottom, Giemsa stain of HL-60 cells that were previously subjected to vehicle, BETi, ATRA, or BETi + ATRA for 72 hours. G, HEK 293T cells were used to generate inducible PU.1 virus (pINDUCER21-SPI1) with packaging vectors psPAX2 and VSVG. HL-60 cells were then lentivirally infected with the inducible virus or empty vector and GFP sorted. HL-60 cells were then induced with doxycycline (dox) for 5 days, and JQ1 and CPI-0610 IC50 values were determined by the MTS assay. Corresponding Western blot validating PU.1 overexpression in HL-60 cells at day 5 (bottom). The x-axis denotes control versus pINDUCER21-SPI1 cells; the y-axis denotes IC50 for JQ1 or CPI-0610.
Genome-Wide CRISPR Screening in BETi-R Cells Identifies BCL2 and CDK2/6 as Resensitizing to BETi
To determine acquired vulnerabilities in BETi-R cells, we performed a genome-wide CRISPR dropout screen on JQ1-R OCI-AML2 cells to generate knockout events that resensitize JQ1-R cells to BETi. Hits were identified by comparing depleted sgRNAs from JQ1-treated cells to DMSO. Using STRING analyses, we identified a subnetwork consisting of cell-cycle genes (CDK2 and CDK6) and antiapoptotic genes (BCL2, ROCK1, and BIRC2; Fig. 5A and B). Independently derived sgRNA guides targeting BCL2 resensitized both JQ1-R and CPI-0610-R OCI-AML2 cells to BETi (Fig. 5C). Data collected from the Beat AML biorepository, bloodspot.eu, and data deposited by Pei and colleagues (29) show that SPI1, FOS, FOSL2, JUNB, and AHR are highly expressed in monocytic leukemias, whereas BCL2 and CDK2/4/6 are enriched in undifferentiated blasts (Supplementary Fig. S4A–S4F). Collectively, these data further support our hypothesis that BETi sensitivity and resistance are tethered to differentiation state and identify potentially targetable vulnerabilities BCL2 and CDK2/6.
Figure 5.
Whole-genome CRISPR screening of BETi-R OCI-AML2 cells identifies targetable vulnerabilities to CDK4/6 and BCL2. Cas9-expressing BETi-R OCI-AML2 cells were generated using Cas9Blst. Loss-of-function screens were performed as described (9), using a human genome-wide sgRNA library (77). 100e6 cells were used for viral transduction at MOI (multiplicity of infection) 0.3, selected with puromycin and then subjected to DMSO or 200 nmol/L JQ1 for 14 days. A, Volcano plot comparing enrichment of sgRNAs relative to DMSO control in JQ1-treated JQ1-R OCI-AML2 cells (generated in Fig. 4A) versus –log10-transformed median P value. Hits of interest are highlighted and identified. Significance and prioritization of hits performed as previously performed on JQ1 and CPI-0610 screens (Fig. 2A). The x-axis denotes median log fold change (FC) of guides (treated control). The y-axis denotes –log10 P value. B, STRING analysis performed as previously in Fig. 2D from hits (defined as P < 0.05, median log fold change ≤1.5×) identified in the JQ1-R OCI-AML2 CRISPR screen identifies a network of targetable genes, BCL2, CDK2, and CDK6. C, JQ1-R and CPI-0610-R OCI-AML2 cells (generated in Fig. 4A) were infected with either nontargeting (NT) or BCL2-targeting lentiviruses and selected for puromycin. Top, MTS viability assay was then used to compare JQ1 sensitivity in single sgRNA KO of BCL2 or nontargeting in JQ1-R or CPI-0610-R OCI-AML2 cells. Bottom, whole-cell extracts from JQ1-R, CPI-0610-R, and nontargeting cells were evaluated by Western blot showing BCL2 KO. Error bars represent the standard error margin between the six replicates. P values were calculated using a two-stage linear step-up procedure of Benjamini–Krieger–Yekutieli comparing nontargeting BETi-R cells to BCL2 KO BETi-R OCI-AML2 cells. The x-axis represents BETi concentration; the y-axis represents BETi-R OCI-AML2 viability. KO, knockout.
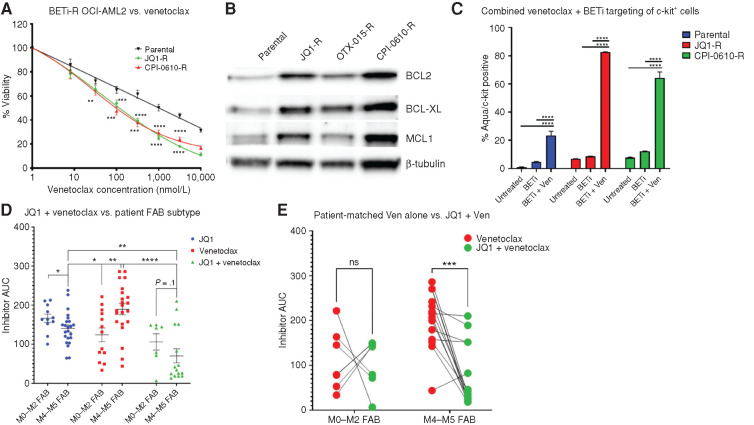
BETi-R AML Cells Are Sensitive to BCL2 Inhibitors and the Combination of BETi + BCL2 Inhibitor Specifically Rescues Intrinsically BCL2 Inhibitor–Resistant Monocytic AMLs
We and others have shown that BCL2 inhibitors (BCL2i) are more effective in undifferentiated blasts, and monocytic differentiation is a driver of BCL2i resistance in AML (27,28,29,30)—the inverse correlation that we have seen here where BETi are more effective on AML cells of monocytic differentiation state and an undifferentiated state promotes BETi resistance. Thus, we tested the efficacy of the BCL2i venetoclax in BETi-naïve and BETi-R cells and found increased sensitivity in BETi-R cells (Fig. 6A). This correlated with increased expression of BCL2, MCL1, and BCL2L1 (BCL-XL) in the BETi-R cells (Fig. 6B). In addition, the combination of venetoclax and BETi significantly increased cytotoxicity in undifferentiated c-kit+ cells in BETi-naïve and more so in BETi-R cells (Fig. 6C). Finally, AML patient samples treated ex vivo with JQ1 showed enhanced sensitivity in M4/M5 FABs but were intrinsically resistant to venetoclax, as previously published (28,29,30). However, the combination of JQ1 + venetoclax showed significantly enhanced sensitivity compared with venetoclax alone, specifically in matched monocytic leukemias, rescuing intrinsic BCL2i resistance due to monocytic differentiation (Fig. 6D and E).
Figure 6.
BETi-R cells have increased BCL family member expression and concomitant enhanced sensitivity to BCL2i. A, BETi-naïve and BETi-R OCI-AML2 cells were subjected to titrations of the BCL2i venetoclax (Ven) for 72 hours and then assessed for viability by MTS assay. Error bars represent the standard error margin between the six replicates. P values were calculated using a two-stage linear step-up procedure of Benjamini–Krieger–Yekutieli comparing parental to BETi-R OCI-AML2 cells. B, Whole-cell extracts from naïve or BETi-R OCI-AML2 cells were subjected to immunoblot analysis using antibodies specific for BCL2L1 (BCL-XL), MCL1, and BCL2. C, Flow-cytometric analysis of treatment of BETi-naïve or BETi-R OCI-AML2 cells with vehicle, 600 nmol/L JQ1 48 hours, or 600 nmol/L JQ1 + 100 nmol/L venetoclax 48 hours versus viability by Zombie Aqua. Significance determined by two-way ANOVA with multiple comparison corrections from three replicates. OTX-015-R, OTX-015 resistant. D, Ex vivo drug sensitivities, as determined by MTS assay, in Beat AML patient samples derived from the Beat AML biorepository (3), comparing responses to JQ1, venetoclax, or JQ1 + venetoclax combined and stratified by FAB subtype (M0–M2, undifferentiated AML; M4–M5, myelomonocytic-monocytic AML). Significance determined by two-way ANOVA. N = 11 JQ1 M0 to M2, 23 M4 and M5; N = 13 venetoclax M0 to M2, 22 M4 and M5; N = 7 JQ1 + venetoclax M0 to M2, 15 M4 and M5. AUC, area under the curve. E, Sensitivity to venetoclax alone and, separately, JQ1 in combination with venetoclax, as determined by MTS assay in matched Beat AML patient samples stratified by FAB subtype. Lines connect individual patient sample responses to single-agent venetoclax (red) compared with response to JQ1 + venetoclax (green). N = 7 M0 to M2, 14 M4 and M5. Significance determined by two-way ANOVA.
As discussed previously, our data indicate that BETi selects for less differentiated clones, which correlates with increased CDK2/6 expression, and was resensitized by sgRNAs targeting these genes. Thus, we tested the combination of the CDK4/6 inhibitors (CDK4/6i) palbociclib or abemaciclib with JQ1, OTX-015, or CPI-203 in BETi-naïve OCI-AML2 and OCI-AML3 cells and observed strong synergy (Supplementary Fig. S5A). In concordance with our proposed mechanism, we found that palbociclib resistance correlates with increased monocytic markers, inverse of BETi (Supplementary Fig. S5B), and that forced myeloid differentiation of HL-60s drove strong resistance to palbociclib (59.45 nmol/L IC50 undifferentiated, 3766 nmol/L IC50 differentiated; Supplementary Fig. S5C). In addition, AML patient samples treated with palbociclib or JQ1 + palbociclib recapitulated our findings with venetoclax, with M4/M5 FAB patient samples exhibiting resistance to single-agent palbociclib treatment but enhanced sensitivity with BETi combination treatment (Supplementary Fig. S5D). Collectively, these data suggest that the synergy shown here and by others previously between BCL2i or CDK4/6i and BETi is driven by differentiation state, and that myeloid differentiation can drive resistance to CDK4/6i and BCL2i.
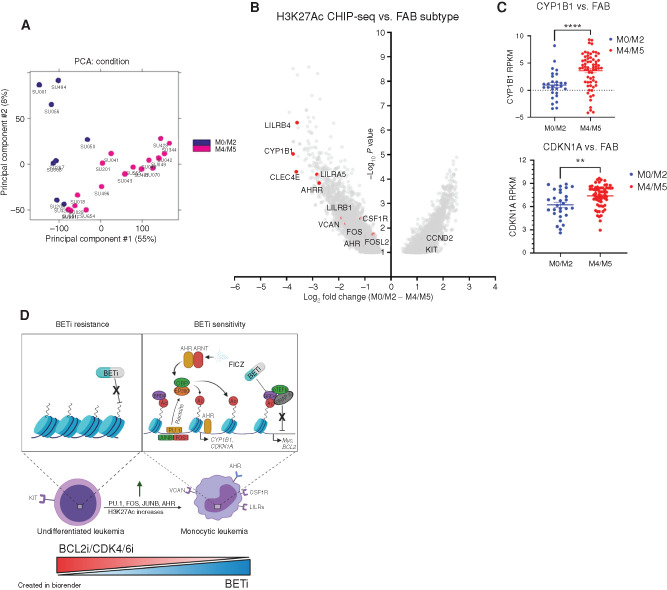
H3K27Ac Chromatin Immunoprecipitation Sequencing Stratified by FAB Subtype Reveals Enrichment of AHR Signaling and Hematopoietic TFs in Monocytic Leukemias
Finally, to determine whether undifferentiated blasts (FAB subtype: M0–M2) have differentially acetylated histone residue profiles in comparison with monocytic leukemias (FAB subtype: M4–M5), as a consequence of differential expression of the monocytic differentiation regulators (e.g., SPI1, FOS, AHR), which recruit histone-modifying machinery, we analyzed H3K27Ac chromatin immunoprecipitation sequencing (ChIP-seq) data deposited by McKeown and colleagues (41) and called differentially acetylated regions by FAB subtype. The undifferentiated M0 to M2 FAB subtype samples separated distinctly by principal component analysis (PCA) from the differentiated M4 and M5 subtypes (Fig. 7A). A total of 6,076 differential affinity peaks were identified between M0 to M2 and M4 and M5 samples. As expected, we observed increased acetylation at monocytic surface markers such as VCAN/LILRs and most significantly increased acetylation at the canonical AHR transcriptional target CYP1B1 (42, 43) and its repressor AHRR, which is induced during constitutive AHR signaling (refs. 44, 45; Fig. 7B). This suggests increased AHR signaling in monocytic AMLs. To further validate that enhanced AHR signaling is found in monocytic leukemias, we asked whether M4 and M5 patient samples had increased expression of canonical AHR-regulated genes CYP1B1 and CDKN1A. Indeed, both were found to be significantly increased in M4 and M5 AML (Fig. 7C). In addition, we evaluated genes coexpressed with CYP1B1 and AHR within AML patient samples in the Beat AML database and found significant positive correlations with monocytic surface markers, hematopoietic TFs previously identified within the Beat AML database, and BETi-naïve whole-genome CRISPR screens as well as negative correlations with CDKs and BCL2 (Supplementary Fig. S6A and S6B). In conclusion, these data show that monocytic AML patient samples have increased binding at BRD4 targets as well as AHR. Further, we show that AHR signaling is enhanced in monocytic leukemias, validating our findings in the initial whole-genome CRISPR screens and subsequent validation with AHR agonists and antagonists.
Figure 7.
H3K27Ac ChIP-seq of AML patient samples stratified by FAB subtype identifies enrichment of AHR signaling components and hematopoietic TFs in monocytic leukemias. A and B, Analysis of H3K27Ac ChIP-Seq AML samples (41) with known FAB designations (n = 8 M0–M2 FAB, 17 M4–M5). A, PCA. B, Volcano plots representing the 6,076 differentially H3K27-acetylated genes between the undifferentiated M0 to M2 FAB subtype and M4 and M5 FAB monocytic subtype. Each point represents a single gene enrichment versus –log10 P value. C, Relative expression levels (reads per kilobase per million mapped, RPKM) of canonical AHR genes across FAB groups within the Beat AML patient RNA-sequencing data set (all samples with both RNA-sequencing data and FAB subtype data, 29 M0–M2 samples and 34 M4–M5 samples). D, Cartoon schematic detailing findings and hypothesis. In brief, we hypothesize that BETi sensitivity and resistance are governed by leukemic differentiation state. Monocytic AMLs have higher expression of TFs (SPI1, FOS, JUNB, etc.) and increased AHR signaling, which recruit histone acetylation machinery to acetylate histone residues that BRD4 binds to mediate downstream affects. Thus, monocytic AMLs have more targetable sites for BETi and increased sensitivity. Conversely, BCL2i/CDK4/6i are more efficacious in undifferentiated leukemias, which characteristically have higher expression of these genes, and the combination with BETi may help overcome drug resistance development as a consequence of altered maturation state. Left, decreased BRD4 dependency due to decreased recruitment of histone-modifying machinery and AHR signaling. Right, increased BRD4 dependency due to increased histone acetylation driven by hematopoietic TFs and AHR recruitment.
Discussion
BETi have shown clinical promise in a small subset of patients, but most are intrinsically resistant to BET inhibition as a monotherapy (46), which is consistent with Beat AML biorepository data, with 48 of 287 (17%) patients exhibitingex vivo sensitivity (<100 nmol/L IC50, average 886 nmol/L) to the BETi JQ1 (3). Although we found no association of BETi sensitivity with known mutations, our data correlate dependency of intrinsic resistance to BETi on leukemic differentiation state. Our study indicates that primary patient samples with higher expression of genes associated with a more differentiated, monocytic phenotype exhibit enhanced sensitivity to BETi, and reveals a potential therapeutic strategy for monocytic leukemias.
Consistent with the observed patient sample ex vivo BETi responses correlating with monocytic surface markers, our CRISPR BETi-resistance screen identified SPI1, FOS, JUNB, and AHR—genes with high expression in monocytic leukemias and that regulate monocytic differentiation—as primary drivers of BETi resistance (47,48,49,50). Our CRISPR resensitization screen of BETi-R cells revealed CDK4/6, BCL2, and NPM1 as top hits in resensitizing to BETi. Thus, our study identified both resistance and resensitization components of BETi sensitivities, tethered to blast differentiation state. Loss of function of genes expression enriched in monocytic AML, such as hematopoietic TFs SPI1, FOS, and JUNB as well as AHR signaling, is revealed as a primary driver of resistance to BET inhibition, whereas inactivation of genes that are expression enriched in primitive AML, such as CDK6 and BCL2, resensitizes resistant cells to BETi.
Dysregulation of Hematopoietic TFs in AML
Hematopoietic TFs such as SPI1 and JUNB suppress myeloid leukemias by enabling proper differentiation (51,52,53). Not surprisingly, mutations that cause reduced function in these TFs are frequently found in AML and generally associate with a poor prognosis (3,54,55,56). Despite the large body of evidence surrounding the biology of TFs in hematologic malignancies, the manner by which they can affect drug sensitivity is largely unknown.
In normal hematopoiesis, SPI1 drives monocytic differentiation by positively regulating the AP-1 TFs, JUN/FOS, and, as such, their expression highly correlates with AML M4 and M5 subtypes (51). Interestingly, SPI1 expression in AML has demonstrated the ability to bidirectionally transition between differentiated and undifferentiated states (57), which supports the notion that leukemia cells could acquire BETi resistance by selecting for cells in a repressed SPI1 state without acquiring de novo mutations. Both the AP-1 complex and AHR signaling regulate cellular proliferation by directly (indirectly for AHR via upregulation of CDKN1A/p21) repressing Cyclin D1 and regulating expression of CDK2 and CDK4 (52, 58). Together, these data may explain our findings that CDK4/6i and BETi synergize. We also noted increased sensitivity to BCL2i and resensitization to genetic knockdown in acquired BETi-R cells that overexpressed BCL family member proteins. Previous work has shown that sustained expression of NFkB-regulated antiapoptotic genes occurs in SPI1-depleted AML cells (59).
Monocytic Surface Markers as Correlates of BETi Efficacy
We identified monocytic markers CSF1R, VCAN, and LILRA1 as strongly correlating with BETi sensitivity in AML patient samples. Of particular interest is CSF1R, as there are highly specific flow-cytometry antibodies that could be integrated into clinical immunophenotyping panels. CSF1R is a surface receptor that binds CSF1 or IL34 to promote proliferation, survival, and differentiation of monocytes and macrophages. Upon activation, CSF1R activates proliferation and survival pathways that upregulate the SPI1 (60). VCAN is an extracellular matrix proteoglycan expressed across many human tissues. It increases activation and adhesion of monocytes through interactions with CD44. Interestingly, monocytes highly upregulate VCAN, allowing a stronger self-promoting response to inflammation (61,62,63). LILRs are highly expressed in monocytes/macrophages, neutrophils, and B cells, and interact with HLA class I molecules, which have both activating and inhibitory functions (64). Together, our findings suggest a possibility of predicting BETi sensitivity based on CSF1R/VCAN/LILR expression and further suggest the impetus for investigating the efficacy of combining CSF1R and BET inhibitors in monocytic leukemias.
Combined BETi-R and BETi-Naïve Genome-Wide Screening to Identify Novel Combinatorial Treatment Strategies to Overcome BETi Resistance
Our study highlights the power of performing genome-wide CRISPR drug screens on both treatment-naïve and treatment-resistant AML cells to identify rational combinatorial treatment strategies to overcome resistance. We identified in both settings that targeting BCL2 or CDK4/6 may sensitize BETi-R AML cells. Several groups have published in vitro and in vivo efficacy of targeting BCL2 in combination with BETi, lending credence to these approaches (35,65,66,67,68). However, we believe we are the first to rationally describe this phenomenon in the context of differentiation state and highlight a novel therapeutic vulnerability in differentiated AMLs. In addition, we describe a novel role for AHR signaling in mediating BETi response in AML and as a measure of leukemic differentiation state, a topic that we will be exploring further. Further, in silico predictions based on the Beat AML data set identified BETi and CDK4/6i as a synergistic combination strategy, indicating a potentially viable combination strategy for both treatment-naïve and BETi-R settings (69, 70). Future studies will investigate unreported combination strategies identified in the screens, such as AHR agonists with BETi.
BETi Vulnerability as a Consequence of Leukemic Differentiation State
Hematopoietic differentiation is accompanied by large changes to the chromatin landscape and requires HATs, methyltransferases, and others (71, 72). Monocytic differentiation in particular is associated with stark increases in histone acetylation and chromatin accessibility at hematopoietic TF binding sites (73). Hematopoietic TFs recruit BRD4 by directly stimulating activity of HATs, such as EP300, which is corroborated by studies demonstrating that EP300 inhibition phenocopies BRD4 inhibition (7, 13, 34, 74). In this context, BRD4 acts as a cofactor for hematopoietic TFs, offering a likely explanation of increased sensitivity to BETi in hematologic malignancies.
Our findings suggest that BETi resistance is driven by differentiation state as a consequence of evolving histone acetylation status. This is consistent with studies demonstrating WNT-driven transcriptional plasticity as a driver for BETi resistance in AML (23, 26). Further, Sheng and colleagues have recently described a mechanism for WNT signaling blocking monocytic differentiation through inhibition of SPI1 (75). Intriguingly, studies have also shown that AHR activation induces SPI1 production and monocytic markers in human AML (40, 76). Thus, we propose a unified mechanism of resistance involving decreased expression of hematopoietic TFs as a consequence of differentiation state, which can be driven by loss of WNT signaling, that consequently decreases BRD4 dependence and BETi sensitivity. Differentiated leukemia cells upregulate hematopoietic TFs, which recruit histone-modifying machinery and serve to attract BRD4 to target sites, resulting in increased BETi sensitivity (Fig. 7D). Conversely, less differentiated cells have lower expression of these TFs and thus have reduced BRD4 recruitment and, subsequently, reduced sensitivity to a BETi. Further complexity to the proposed mechanisms could result from the transient and often unpredictable nature of differentiation/epigenetic dysregulation in leukemia, driven by differences in patient-specific mutations. These are important topics for investigation moving forward. However, we believe these data support more targeted clinical exploration of BETi in AML, such as an emphasis on recruiting M4 and M5 FAB subtypes or patients with high expression of CSF1R. In addition, our findings provide a further mechanistic rationale for potentially effective drug combination strategies bridging compounds targeting more undifferentiated AML cells (e.g., BCL2i or CDK4/6i) with therapies that are more effective against AML cells further differentiated along the monocytic lineage (e.g., BETi). This strategy may mitigate relapse due to changed maturation state of the tumor.
Methods
Cell Lines
OCI-AML2 (DSMZ cat. #AC-99, RRID: CVCL_1619), OCI-AML3 (DSMZ cat. #AC-582, RRID: CVCL_1844), and HL-60 (CLS cat. #300209/p671_HL-60, RRID: CVCL_0002) were obtained from ATCC. Cell lines were authenticated using the Oregon Health and Science DNA Serviced Core Facility and tested biweekly for Mycoplasma. All cell lines were maintained in RPMI, 20% FBS, L-Glutamine, penicillin/streptomycin, amphotericin-B, and normocin. BETi-R cells were generated by incubating OCI-AML2s at respective JQ1 IC90 or CPI-0610 IC90 concentration twice weekly, and viability was monitored three times per week by guava easyCyte. Once viability returned to ∼80%, cell sensitivity was remeasured and cells were incubated at their increased IC90 concentration. This was repeated until a 5× IC50 increase was achieved. Cells were maintained resistant by treating with 800 nmol/L JQ1 or 800 nmol/L CPI-0610 weekly.
CRISPR/Cas9 Library Screen and CRISPR/Cas9 Gene Inactivation by Individual sgRNA
Cas9-expressing cells were generated using Cas9Blst (Addgene, #52962). Loss-of-function screens were performed as described (9), using a human genome-wide sgRNA library (77), purchased from Addgene (#67989). High-titer lentivirus was generated using calcium phosphate precipitation procedures in HEK 293T cells (NCBI cat. #C498, RRID: CVCL_0063). Viral supernatant was concentrated and titered using a viral titration kit (ABMgood). Cells (100e6) were used for viral transduction at MOI (multiplicity of infection) 0.3 and selected with puromycin for 5 to 7 days. Cells were screened with 200 nmol/L JQ1, 500 nmol/L CPI-0610, or DMSO. DNA was harvested after puromycin selection on day 14 (JQ1) and day 21 (CPI-0610). PCR-amplified barcode libraries were generated as previously described (77) and deep sequenced using Illumina platform.
Single sgRNA Knockouts
Single sgRNA sequences were designed using Synthego design tool (https://design.synthego.com/#/) and converted into DNA sequences (Supplementary Table S2). Individual sgRNAs were cloned into plentiCRISPRV2 (Addgene, #52961, RRID: Addgene_127644). Phosphorylated complementary oligonucleotides were annealed and ligated into BsmbI-digested pLentiCRISPRV2 backbone, which contains sequences for Cas9 and puromycin resistance, and then validated by Sanger sequencing. Lipofectamine 2000 (Invitrogen, #11668019) was used to transfect HEK 293T cells with single transfer vectors with packaging plasmids psPax2 (Addgene, #12260, RRID: Addgene_12260) and VSVG (Invitrogen) to generate virus. Viral supernatants were collected, filtered through 0.45-μm filters, and used for transduction of AML cells using spinoculation method as described (77). Cells were selected with 2 μg/mL of puromycin for 5 to 7 days and outgrown for 14 days in culture before testing for BETi resistance.
Biostatistical Analysis of CRISPR Screens
Pipeline for executing analyses of CRISPR library sequences was performed using modified Mageck analyses. Adaptor sequences were removed using cutadapt, and reads were aligned to the K. Yusa library using bowtie2 (RRID: SCR_016368). Sequences that aligned to more than one region were discarded. An overall alignment rate of 92% was achieved. The overall set of sgRNAs was first filtered to remove any that had zero reads in all samples or in the plasmid. For each contrast, sgRNAs that did not achieve 100 reads (adjusting for library size) in at least half the samples were also removed. Significance of each sgRNA was determined via edgeR (RRID: SCR_012802) after trimmed mean of M values (TMM; ref. 78) normalization for the following contrasts: JQ1 screen at day 14 to DMSO screen at day 14, and CPI-0610 screen at day 21 to DMSO screen day 21. Sequencing data are deposited at Gene Expression Omnibus (GEO; GSE159689). Hits were prioritized according to a previously described tiering structure (32). Briefly, considering only significant sgRNAs (FDR < 0.05), genes were classified into five ordered groups. Tier 1 genes had more than one significant sgRNA, a minimum log2 fold change ≥2, 75% of sgRNAs per gene present, and concordance among sgRNAs per gene ≥75%; tier 2 hits had log2 fold change ≥2 and 100% concordance among sgRNAs per gene; and tier 3 hits had log2 fold change ≥1 and 100% concordance among sgRNAs per gene. Singleton hits represent significantly enriched genes with log2 fold change ≥2 but only a single significant sgRNA. Enriched hits not satisfying these criteria were classified into the unassigned group.
Biostatistical Analysis of H3K27Ac ChIP-seq Data Set
Processing was performed using ENCODE's protocol for unreplicated H3K27Ac ChIP-seq experiments. Differential binding was determined using DIffBind (RRID: SCR_012918) from Bioconductor (RRID: SCR_006442; ref. 79). The Annotatr Package from Bioconductor (RRID: SCR_006442) was then used to annotate regions 1 to 5 kb from gene (80). Volcano plots were generated by plotting −log10 transformed P value and log2-transformed fold changes (M0–M2 and M4–M5).
Drug Viability Testing and ZIP Synergy Scores
Drug viability testing was performed as previously described (3, 69) using MTS-based assays. Absorbance values were normalized to a kill control (10 μmol/L FSV) and media/cells only. AML cells were plated for 72 hours in replicates of six at 1,250 cells/well in R20 and titrated using a log scale (0–10 μmol/L) against JQ1 and CPI-0610. Viability of the cells was measured using Guava easyCyte prior to plating to ensure >90% viability. P values for individual sgRNAs versus nontargeting were calculated using a two-stage linear step-up procedure of Benjamini–Krieger–Yekutieli. Combinations of CPI-0610 with CH-223191 or FICZ were tested for efficacy using the MTS assay and a matrix dose layout wherein each drug/ligand in the combination was titrated over eight concentrations. Dose-specific normalized cell viability percentages were averaged across replicates, and ZIP synergy scores were calculated as previously described (81, 82) using the “synergyfinder” R package.
Beat AML Patient Sample Surface Marker Analyses
A simple linear regression model was fit separately for each inhibitor and gene with the inhibitor area under the curve (AUC) values as the outcome. The T-statistic and P value test whether the slope is nonzero and corrected for multiple comparisons. Corresponding correlation coefficients were computed using the relationship to the linear model: slope*(sd(expr)/sd(auc)), where sd indicates standard deviation. Log2 reads per kilobase per million mapped (RPKM) values are available in Tyner and colleagues (3).
HL-60 Differentiation Assays and Drug Sensitivity
In triplicate, 250,000 HL-60 cells (>95% viability) were plated in a 6-well dish in 3 mL R20 and treated with either vehicle (100% EtOH), 300 nmol/L JQ1, 1 μmol/L ATRA (Sigma, #R2625) suspended in 100% EtOH, or 1 μmol/L ATRA and 300 nmol/L BETi. After 72 hours, cells were resuspended in fresh R20. A portion of the vehicle- and ATRA-alone–treated cells were taken and assessed for BETi sensitivity by MTS as previously described. The rest of the vehicle, BETi alone, ATRA alone, or BETi + ATRA cells were stained for viability and differentiation marker CD38 as described below.
HL-60 Morphologic Assessment
HL-60 cells were subjected to vehicle, 300 nmol/L JQ1, 1 μmol/L ATRA, or 300 nmol/L JQ1 + 1 μmol/L ATRA for 72 hours as described previously for flow-cytometric assessment. Cells were then spun onto glass slides via cytospin (800 rpm/3 min), fixed in methanol, and stained with 1:20 Giemsa (Sigma-Aldrich) for 20 minutes prior to brightfield imaging (Leica).
Doxycycline-Inducible SPI1-Expressing HL-60s
Lipofectamine 2000 was used to transfect HEK 293T cells with pINDUCER21-SPI1 (Addgene #97039) with packaging plasmids psPax2 (Addgene, #12260, RRID: Addgene_12260) and VSVG (Invitrogen) to generate virus. Viral supernatants were collected, filtered through 0.45-μm filters and used for transduction of AML cells using the spinoculation method as described (77). GFP+ HL-60 cells were then sorted via FACSARIAIII.
HL-60s were then induced with vehicle control or 1 μg/mL doxycycline for 5 days, washed with media, and then tested for BETi sensitivity using MTS assay as previously described. A portion of cells was taken for Western blotting prior to plating for MTS.
Flow-Cytometry Staining
A total of 500,000 OCI-AML2 parental, JQ1-R, or CPI-0610-R cells were treated with BETi, BETi combined with venetoclax, or left untreated and washed in PBS and resuspended in 100 μL PBS. One microliter Zombie Aqua (BioLegend, #423101) was added to the cells and incubated for 15 minutes at room temperature in the dark. Cells were then washed with FACS buffer (PBS, 2% bovine calf serum, 0.005% sodium azide) and resuspended in 20 μL/sample 1:50 Human Fc block (BioLegend, #422302) in FACS buffer and incubated for 5 minutes on ice. Without washing, a 20 μL/sample cocktail in FACS buffer of 1:20 human antibodies, which included anti-CD33 BV711 (BioLegend, #303424, RRID: AB_2565775), 1:20 human anti–c-kit (BioLegend, #313228, RRID: AB_2566215), and 1:20 anti-CD38 A647 (BioLegend, #303514, RRID: AB_493090) for a final staining concentration of 1:100 Fc block and 1:40 antibodies, was added. The cells were then incubated in the dark, on ice, for 30 minutes, at which point they were washed, fixed for 20 minutes in PFA, and resuspended in FACS buffer prior to running on a BD Fortessa. Data were analyzed using FlowJo Software.
Western Blotting
Briefly, lysates were generated using cell lysis buffer (Cell Signaling Technologies, #9803S), 1 mmol/L complete mini protease inhibitor (Roche), 1 mmol/L phosphatase inhibitor (Sigma), and 1 mmol/L PMSF. Protein concentration was quantified and normalized via BCA. Lysates were given a solution of 6% SDS, 150 mmol/L Tris (pH 6.8), 30% Glycerol, 0.1% bromophenol blue, and beta-mercaptoethanol (BME) and boiled at 95°C for 5 minutes before gel separation. Proteins were transferred to a PVDF membrane using an iblot2 dry transfer system, blocked with BSA, and incubated with 1:1,000 in TBST primary antibody overnight at 4°C. The membrane was then washed with TBST before incubation in appropriate HRP-conjugated secondary for 3 hours and activated for imaging. Antibodies used were BCL2 (Cell Signaling Technologies, #4223S), JUNB (Cell Signaling Technologies, #3746S), Tubulin (Cell Signaling Technologies, #2146S), PU.1 (Cell Signaling Technologies, #2258S), MCL1 (Cell Signaling Technologies, #94269S), and BCL-XL (Cell Signaling Technologies, #2764S).
Statistical Analyses
Specific statistical analyses are described in figure legends. In all figures, “ns” denotes not significant (P > 0.05), and *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, respectively.
Supplementary Material
Acknowledgments
The authors thank all of the patients who provided specimens for donating precious time and tissue. This work was supported by the NCI/NIH (1U01CA217862, 1U54CA224019, R01CA245002, and R01CA183947). T. Nechiporuk is supported by a Research Specialist award from the NIH/NCI (1R50CA251708). J.W. Tyner was supported by the V Foundation for Cancer Research, the Gabrielle's Angel Foundation for Cancer Research, the Anna Fuller Fund, the Mark Foundation for Cancer Research, and the Silver Family Foundation. R. Majeti is a Scholar of the Leukemia & Lymphoma Society.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Blood Cancer Discov 2021;2:518–31
Authors' Disclosures
R. Majeti is on the board of directors for BeyondSpring Inc.; on the scientific advisory boards for Kodikaz Therapeutic Solutions Inc. and Coherus Biosciences; a consultant for Acuta Capital Partners; and an inventor on a number of patents related to CD47 cancer immunotherapy licensed to Gilead Sciences, Inc. J.W. Tyner reports other support from Constellation during the conduct of the study, as well as other support from Agios, Aptose, Array, AstraZeneca, Genentech, Gilead, Incyte, Janssen, Petra, Seattle Genetics, Syros, Takeda, and Tolero outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
K.A. Romine: Conceptualization, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. T. Nechiporuk: Conceptualization, methodology. D. Bottomly: Conceptualization, formal analysis, visualization, methodology. S. Jeng: Formal analysis, visualization. S.K. McWeeney: Conceptualization, resources, formal analysis, visualization. A. Kaempf: Formal analysis. M.R. Corces: Resources, data curation. R. Majeti: Resources, data curation. J.W. Tyner: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1.Dombret H, Gardin C.An update of current treatments for adult acute myeloid leukemia. Blood 2016;127:53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DiNardo CD, Jonas BA, Pullarkat V, Thirman MJ, Garcia JS, Wei AH, et al. Azacitidine and venetoclax in previously untreated acute myeloid leukemia. N Engl J Med 2020;383:617–29. [DOI] [PubMed] [Google Scholar]
- 3.Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018;562:526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DiNardo CD, Cortes JE.Mutations in AML: prognostic and therapeutic implications. Hematology Am Soc Hematol Educ Program 2016;2016:348–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lagunas-Rangel FA, Chavez-Valencia V, Gomez-Guijosa MA, Cortes-Penagos C.Acute myeloid leukemia-genetic alterations and their clinical prognosis. Int J Hematol Oncol Stem Cell Res 2017;11:328–39. [PMC free article] [PubMed] [Google Scholar]
- 6.Kim TK, Gore SD, Zeidan AM.Epigenetic therapy in acute myeloid leukemia: current and future directions. Semin Hematol 2015;52:172–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature 2010;468:1067–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albrecht BK, Gehling VS, Hewitt MC, Vaswani RG, Cote A, Leblanc Y, et al. Identification of a benzoisoxazoloazepine inhibitor (CPI-0610) of the bromodomain and extra-terminal (BET) family as a candidate for human clinical trials. J Med Chem 2016;59:1330–9. [DOI] [PubMed] [Google Scholar]
- 9.Shi X, Liu C, Liu B, Chen J, Wu X, Gong W.JQ1: a novel potential therapeutic target. Pharmazie 2018;73:491–3. [DOI] [PubMed] [Google Scholar]
- 10.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 2011;478:529–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawson MA, Gudgin EJ, Horton SJ, Giotopoulos G, Meduri E, Robson S, et al. Recurrent mutations, including NPM1c, activate a BRD4-dependent core transcriptional program in acute myeloid leukemia. Leukemia 2014;28:311–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 2005;19:535–45. [DOI] [PubMed] [Google Scholar]
- 13.Shi J, Vakoc CR.The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell 2014;54:728–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devaiah BN, Case-Borden C, Gegonne A, Hsu CH, Chen Q, Meerzaman D, et al. BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat Struct Mol Biol 2016;23:540–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devaiah BN, Lewis BA, Cherman N, Hewitt MC, Albrecht BK, Robey PG, et al. BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc Natl Acad Sci U S A 2012;109:6927–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uppal S, Gegonne A, Chen Q, Thompson PS, Cheng D, Mu J, et al. The bromodomain protein 4 contributes to the regulation of alternative splicing. Cell Rep 2019;29:2450–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 2011;478:524–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A 2011;108:16669–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roe JS, Vakoc CR.The essential transcriptional function of BRD4 in acute myeloid leukemia. Cold Spring Harb Symp Quant Biol 2016;81:61–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coude MM, Braun T, Berrou J, Dupont M, Bertrand S, Masse A, et al. BET inhibitor OTX015 targets BRD2 and BRD4 and decreases c-MYC in acute leukemia cells. Oncotarget 2015;6:17698–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berthon C, Raffoux E, Thomas X, Vey N, Gomez-Roca C, Yee K, et al. Bromodomain inhibitor OTX015 in patients with acute leukaemia: a dose-escalation, phase 1 study. Lancet Haematol 2016;3:e186–95. [DOI] [PubMed] [Google Scholar]
- 22.Jang JE, Eom JI, Jeung HK, Cheong JW, Lee JY, Kim JS, et al. AMPK-ULK1-mediated autophagy confers resistance to BET inhibitor JQ1 in acute myeloid leukemia stem cells. Clin Cancer Res 2017;23:2781–94. [DOI] [PubMed] [Google Scholar]
- 23.Rathert P, Roth M, Neumann T, Muerdter F, Roe JS, Muhar M, et al. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature 2015;525:543–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fong CY, Gilan O, Lam EY, Rubin AF, Ftouni S, Tyler D, et al. BET inhibitor resistance emerges from leukaemia stem cells. Nature 2015;525:538–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shu S, Lin CY, He HH, Witwicki RM, Tabassum DP, Roberts JM, et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature 2016;529:413–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bell CC, Fennell KA, Chan YC, Rambow F, Yeung MM, Vassiliadis D, et al. Targeting enhancer switching overcomes non-genetic drug resistance in acute myeloid leukaemia. Nat Commun 2019;10:2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Majumder MM, Leppa AM, Hellesoy M, Dowling P, Malyutina A, Kopperud R, et al. Multi-parametric single cell evaluation defines distinct drug responses in healthy hematological cells that are retained in corresponding malignant cell types. Haematologica 2020;105:1527–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuusanmaki H, Leppa AM, Polonen P, Kontro M, Dufva O, Deb D, et al. Phenotype-based drug screening reveals association between venetoclax response and differentiation stage in acute myeloid leukemia. Haematologica 2020;105:708–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pei S, Pollyea DA, Gustafson A, Stevens BM, Minhajuddin M, Fu R, et al. Monocytic subclones confer resistance to venetoclax-based therapy in patients with acute myeloid leukemia. Cancer Discov 2020;10:536–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H, Nakauchi Y, Koehnke T, Stafford M, Bottomly D, Thomas R, et al. Integrated analysis of patient samples identifies biomarkers for venetoclax efficacy and combination strategies in acute myeloid leukemia. Nat Cancer 2020;1:826–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCarthy DJ, Chen Y, Smyth GK.Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res 2012;40:4288–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nechiporuk T, Kurtz SE, Nikolova O, Liu T, Jones CL, D'Alessandro A, et al. The TP53 apoptotic network is a primary mediator of resistance to BCL2 inhibition in AML cells. Cancer Discov 2019;9:910–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607–D13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roe JS, Mercan F, Rivera K, Pappin DJ, Vakoc CR.BET bromodomain inhibition suppresses the function of hematopoietic transcription factors in acute myeloid leukemia. Mol Cell 2015;58:1028–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fiskus W, Sharma S, Qi J, Valenta JA, Schaub LJ, Shah B, et al. Highly active combination of BRD4 antagonist and histone deacetylase inhibitor against human acute myelogenous leukemia cells. Mol Cancer Ther 2014;13:1142–54. [DOI] [PubMed] [Google Scholar]
- 36.Bunaciu RP, MacDonald RJ, Jensen HA, Gao F, Wang X, Johnson L, et al. Retinoic acid and 6-formylindolo(3,2-b)carbazole (FICZ) combination therapy reveals putative targets for enhancing response in non-APL AML. Leuk Lymphoma 2019;60:1697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hestermann EV, Brown M.Agonist and chemopreventative ligands induce differential transcriptional cofactor recruitment by aryl hydrocarbon receptor. Mol Cell Biol 2003;23:7920–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian Y, Ke S, Chen M, Sheng T.Interactions between the aryl hydrocarbon receptor and P-TEFb. Sequential recruitment of transcription factors and differential phosphorylation of C-terminal domain of RNA polymerase II at cyp1a1 promoter. J Biol Chem 2003;278:44041–8. [DOI] [PubMed] [Google Scholar]
- 39.Wang S, Ge K, Roeder RG, Hankinson O.Role of mediator in transcriptional activation by the aryl hydrocarbon receptor. J Biol Chem 2004;279:13593–600. [DOI] [PubMed] [Google Scholar]
- 40.Tasseff R, Jensen HA, Congleton J, Dai D, Rogers KV, Sagar A, et al. An effective model of the retinoic acid induced HL-60 differentiation program. Sci Rep 2017;7:14327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKeown MR, Corces MR, Eaton ML, Fiore C, Lee E, Lopez JT, et al. Superenhancer analysis defines novel epigenomic subtypes of non-APL AML, including an RARalpha dependency targetable by SY-1425, a potent and selective RARalpha agonist. Cancer Discov 2017;7:1136–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang X, Solomon S, Fraser LR, Trombino AF, Liu D, Sonenshein GE, et al. Constitutive regulation of CYP1B1 by the aryl hydrocarbon receptor (AhR) in pre-malignant and malignant mammary tissue.J Cell Biochem 2008;104:402–17. [DOI] [PubMed] [Google Scholar]
- 43.Chang JT, Chang H, Chen PH, Lin SL, Lin P.Requirement of aryl hydrocarbon receptor overexpression for CYP1B1 up-regulation and cell growth in human lung adenocarcinomas. Clin Cancer Res 2007;13:38–45. [DOI] [PubMed] [Google Scholar]
- 44.Mimura J, Ema M, Sogawa K, Fujii-Kuriyama Y.Identification of a novel mechanism of regulation of Ah (dioxin) receptor function. Genes Dev 1999;13:20–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hahn ME, Allan LL, Sherr DH.Regulation of constitutive and inducible AHR signaling: complex interactions involving the AHR repressor. Biochem Pharmacol 2009;77:485–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Braun T, Gardin C.Investigational BET bromodomain protein inhibitors in early stage clinical trials for acute myelogenous leukemia (AML). Expert Opin Investig Drugs 2017;26:803–11. [DOI] [PubMed] [Google Scholar]
- 47.Rojo R, Pridans C, Langlais D, Hume DA.Transcriptional mechanisms that control expression of the macrophage colony-stimulating factor receptor locus. Clin Sci 2017;131:2161–82. [DOI] [PubMed] [Google Scholar]
- 48.Fontana MF, Baccarella A, Pancholi N, Pufall MA, Herbert DR, Kim CC.JUNB is a key transcriptional modulator of macrophage activation. J Immunol 2015;194:177–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Platzer B, Richter S, Kneidinger D, Waltenberger D, Woisetschlager M, Strobl H.Aryl hydrocarbon receptor activation inhibits in vitro differentiation of human monocytes and Langerhans dendritic cells. J Immunol 2009;183:66–74. [DOI] [PubMed] [Google Scholar]
- 50.Lord KA, Abdollahi A, Hoffman-Liebermann B, Liebermann DA.Proto-oncogenes of the fos/jun family of transcription factors are positive regulators of myeloid differentiation. Mol Cell Biol 1993;13:841–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steidl U, Rosenbauer F, Verhaak RG, Gu X, Ebralidze A, Otu HH, et al. Essential role of Jun family transcription factors in PU.1 knockdown-induced leukemic stem cells. Nat Genet 2006;38:1269–77. [DOI] [PubMed] [Google Scholar]
- 52.Somervaille TC, Cleary ML.PU.1 and Junb: suppressing the formation of acute myeloid leukemia stem cells. Cancer Cell 2006;10:456–7. [DOI] [PubMed] [Google Scholar]
- 53.Cook WD, McCaw BJ, Herring C, John DL, Foote SJ, Nutt SL, et al. PU.1 is a suppressor of myeloid leukemia, inactivated in mice by gene deletion and mutation of its DNA binding domain. Blood 2004;104:3437–44. [DOI] [PubMed] [Google Scholar]
- 54.Mueller BU, Pabst T, Osato M, Asou N, Johansen LM, Minden MD, et al. Heterozygous PU.1 mutations are associated with acute myeloid leukemia. Blood 2002;100:998–1007. [DOI] [PubMed] [Google Scholar]
- 55.Schnittger S, Dicker F, Kern W, Wendland N, Sundermann J, Alpermann T, et al. RUNX1 mutations are frequent in de novo AML with noncomplex karyotype and confer an unfavorable prognosis. Blood 2011;117:2348–57. [DOI] [PubMed] [Google Scholar]
- 56.Lin LI, Chen CY, Lin DT, Tsay W, Tang JL, Yeh YC, et al. Characterization of CEBPA mutations in acute myeloid leukemia: most patients with CEBPA mutations have biallelic mutations and show a distinct immunophenotype of the leukemic cells. Clin Cancer Res 2005;11:1372–9. [DOI] [PubMed] [Google Scholar]
- 57.McKenzie MD, Ghisi M, Oxley EP, Ngo S, Cimmino L, Esnault C, et al. Interconversion between tumorigenic and differentiated states in acute myeloid leukemia. Cell Stem Cell 2019;25:258–72. [DOI] [PubMed] [Google Scholar]
- 58.Mechta-Grigoriou F, Gerald D, Yaniv M.The mammalian Jun proteins: redundancy and specificity. Oncogene 2001;20:2378–89. [DOI] [PubMed] [Google Scholar]
- 59.Haimovici A, Humbert M, Federzoni EA, Shan-Krauer D, Brunner T, Frese S, et al. PU.1 supports TRAIL-induced cell death by inhibiting NF-kappaB-mediated cell survival and inducing DR5 expression. Cell Death Differ 2017;24:866–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stanley ER, Chitu V.CSF-1 receptor signaling in myeloid cells. Cold Spring Harb Perspect Biol 2014;6:a021857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Asplund A, Stillemark-Billton P, Larsson E, Rydberg EK, Moses J, Hulten LM, et al. Hypoxic regulation of secreted proteoglycans in macrophages. Glycobiology 2010;20:33–40. [DOI] [PubMed] [Google Scholar]
- 62.Wu YJ, La Pierre DP, Wu J, Yee AJ, Yang BB.The interaction of versican with its binding partners. Cell Res 2005;15:483–94. [DOI] [PubMed] [Google Scholar]
- 63.Masuda A, Yasuoka H, Satoh T, Okazaki Y, Yamaguchi Y, Kuwana M.Versican is upregulated in circulating monocytes in patients with systemic sclerosis and amplifies a CCL2-mediated pathogenic loop. Arthritis Res Ther 2013;15:R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hirayasu K, Arase H.Functional and genetic diversity of leukocyte immunoglobulin-like receptor and implication for disease associations. J Hum Genet 2015;60:703–8. [DOI] [PubMed] [Google Scholar]
- 65.Lin KH, Rutter JC, Xie A, Pardieu B, Winn ET, Bello RD, et al. Using antagonistic pleiotropy to design a chemotherapy-induced evolutionary trap to target drug resistance in cancer. Nat Genet 2020;52:408–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li W, Gupta SK, Han W, Kundson RA, Nelson S, Knutson D, et al. Targeting MYC activity in double-hit lymphoma with MYC and BCL2 and/or BCL6 rearrangements with epigenetic bromodomain inhibitors. J Hematol Oncol 2019;12:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramsey HE, Greenwood D, Zhang S, Childress M, Arrate MP, Gorska AE, et al. BET inhibition enhances the antileukemic activity of low-dose venetoclax in acute myeloid leukemia. Clin Cancer Res 2021;27:598–607. [DOI] [PubMed] [Google Scholar]
- 68.Zhang S, Zhao Y, Heaster TM, Fischer MA, Stengel KR, Zhou X, et al. BET inhibitors reduce cell size and induce reversible cell cycle arrest in AML. J Cell Biochem 2018Nov 11 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kurtz SE, Eide CA, Kaempf A, Khanna V, Savage SL, Rofelty A, et al. Molecularly targeted drug combinations demonstrate selective effectiveness for myeloid- and lymphoid-derived hematologic malignancies. Proc Natl Acad Sci U S A 2017;114:E7554–E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Drusbosky LM, Vidva R, Gera S, Lakshminarayana AV, Shyamasundar VP, Agrawal AK, et al. Predicting response to BET inhibitors using computational modeling: a BEAT AML project study. Leuk Res 2019;77:42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sharma S, Gurudutta G.Epigenetic regulation of hematopoietic stem cells. Int J Stem Cells 2016;9:36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu X, Wu C, Bhavanasi D, Wang H, Gregory BD, Huang J.Chromatin dynamics during the differentiation of long-term hematopoietic stem cells to multipotent progenitors. Blood Adv 2017;1:887–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen S, Yang J, Wei Y, Wei X.Epigenetic regulation of macrophages: from homeostasis maintenance to host defense. Cell Mol Immunol 2020;17:36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Donati B, Lorenzini E, Ciarrocchi A.BRD4 and cancer: going beyond transcriptional regulation. Mol Cancer 2018;17:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sheng Y, Ju W, Huang Y, Li J, Ozer H, Qiao X, et al. Activation of wnt/beta-catenin signaling blocks monocyte-macrophage differentiation through antagonizing PU.1-targeted gene transcription. Leukemia 2016;30:2106–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bunaciu RP, Jensen HA, MacDonald RJ, LaTocha DH, Varner JD, Yen A.6-Formylindolo(3,2-b)carbazole (FICZ) modulates the signalsome responsible for RA-induced differentiation of HL-60 myeloblastic leukemia cells. PLoS One 2015;10:e0135668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tzelepis K, Koike-Yusa H, De Braekeleer E, Li Y, Metzakopian E, Dovey OM, et al. A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep 2016;17:1193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Robinson MD, Oshlack A.A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 2010;11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 2012;481:389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cavalcante RG, Sartor MA.annotatr: genomic regions in context. Bioinformatics 2017;33:2381–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.He L, Kulesskiy E, Saarela J, Turunen L, Wennerberg K, Aittokallio T, et al. Methods for high-throughput drug combination screening and synergy scoring. Methods Mol Biol 2018;1711:351–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yadav B, Wennerberg K, Aittokallio T, Tang J.Searching for drug synergy in complex dose-response landscapes using an interaction potency model. Comput Struct Biotechnol J 2015;13:504–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Arber DA.The 2016 WHO classification of acute myeloid leukemia: what the practicing clinician needs to know. Semin Hematol 2019;56:90–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.