Abstract
The CCAAT sequence in the amdS promoter of Aspergillus nidulans is recognized by AnCF, a complex consisting of the three evolutionary conserved subunits HapB, HapC, and HapE. In this study we have investigated the effect of AnCF on the chromatin structure of the amdS gene. The AnCF complex and the CCAAT sequence were found to be necessary for the formation of a nucleosome-free, DNase I-hypersensitive region in the 5′ region of the amdS gene. Deletion of the hapE gene results in loss of the DNase I-hypersensitive site, and the positioning of nucleosomes over the transcriptional start point is lost. Likewise, a point mutation in the CCAAT motif, as well as a 530-bp deletion which removes the CCAAT box, results in the loss of the DNase I-hypersensitive region. The DNase I-hypersensitive region and the nucleosome positioning can be restored by insertion of a 35-bp oligonucleotide carrying the CCAAT motif. A DNase I-hypersensitive region has been found in the CCAAT-containing fmdS gene and was also hapE dependent. These data indicate a critical role for the AnCF complex in establishing an open chromatin structure in A. nidulans.
Most of the genomic DNA in a eucaryotic cell is packed into nucleosomes representing a potentially repressive chromatin structure. In such a chromatin environment the regulatory regions of genes must be accessible to specific transcription factors and the components of the general transcriptional machinery. Such sites, often characterized by their increased sensitivity to nucleases, have been identified in many genes and appear to be generated by the displacement or disruption of nucleosomes within the promoter region (9, 34). DNase I-hypersensitive sites often coincide with the binding sites for transcription factors (1, 51). Hypersensitive sites can be generated by a variety of different mechanisms. In some cases replication is required in order to disrupt a nucleosome, e.g., to activate promoters silenced by their proximity to telomeric sequences (4). Replication-independent pathways have been described for a number of inducible promoters (59, 61). The precise positioning of nucleosomes, which leave some transcription factor binding sites exposed, could create a hypersensitive site (2, 17, 73). Increased DNase I susceptibility could also result from the binding of a transcription factor which locally distorts the DNA within or adjacent to this site (62).
Recently, a variety of complexes have been identified that assist transcription factors to reconfigure chromatin. An ATP-dependent multiprotein complex, the SWI-SNF complex, capable of altering the chromatin structure and facilitating binding of TFIIA-TBP and activators to nucleosome templates is required for activation of certain genes (10, 72). Another class of transcriptional coactivators are the histone acetyltransferases (HAT), whose enzymatic activity may contribute to chromatin disruption and transcription. For example, the yeast ADA complex contains GCN5, a subunit with intrinsic HAT activity, that is necessary for transcriptional activation, and this complex is known both to modify histones locally in the vicinity of the regulated promoter and to facilitate chromatin disruption (11, 69, 70). Additional coactivators in yeasts and higher eukaryotes, including TATA-binding protein (TBP)-associated factor TAFII 250, p300/CBP and P/CAF, have been identified to be HATs (50, 56, 77).
The sequence CCAAT is found in the 5′ region of approximately 30% of eukaryotic genes (46). In Saccharomyces cerevisiae a heteromeric complex of proteins encoded by HAP2, HAP3, HAP4, and HAP5 genes binds to sequences containing a core CCAAT element upstream of genes involved in respiration (18, 24, 48). In vertebrates the NF-Y complex containing NF-YA, NF-YB, and NF-YC homologs of Hap2p, Hap3p, and Hap5p, respectively, has been found to bind to CCAAT containing sequences (39, 54). NF-YA and NF-YC contain a histone fold motif, a structural feature of histones suggesting that NF-Y might be involved in the organization of the chromatin structure (40).
The amdS gene of Aspergillus nidulans encodes an acetamidase required for the utilization of acetamide (28). The 5′ region contains a CCAAT sequence that is required for setting the basal level of expression (43). The AnCF complex, comprising the HapB, -C, and -E homologs of the Hap2p, Hap3p, and Hap5p subunits, respectively, has been shown to bind to the CCAAT sequence (57, 63, 67). Disruption of each of the hapB, hapC, and hapE genes has been shown to affect amdS expression to the same extent as mutations of the CCAAT sequence (57, 63). So far evidence for a HAP4 homologue is lacking.
We have investigated the role of AnCF in influencing the chromatin structure and found that AnCF is necessary for the establishment of a nucleosome-free, DNase I-hypersensitive site in the 5′ region of the amdS gene. Deletion or point mutation of the CCAAT box greatly decreases gene expression and loss of DNase I hypersensitivity, as well as the loss of positioned nucleosomes upstream of the core promoter and over the transcriptional unit. Disruption of the hapB, hapC, and hapE genes also results in loss of the DNase I-hypersensitive site. In addition, CCAAT sequences in the 5′ region of another gene in A. nidulans have been found to result in AnCF-dependent DNase I hypersensitivity. We have also found that AnCF function is likely to be dependent on other promoter elements since insertion of a functional CCAAT-containing motif outside a promoter context did not result in DNase I hypersensitivity. Our data implicate AnCF as a critical determinant of DNase I-hypersensitive regions in A. nidulans, a function that may be central to its role as a transcription activator.
While this manuscript was in preparation, it has been shown that a CCAAT sequence bound by NF-Y is essential for the formation of a DNase I-hypersensitive site and defines the acetylation responsiveness of the Xenopus laevis hsp70 promoter via interaction with p300/CBP in vivo (38). Further, the association of the acetyltransferase P/CAF with the mammalian CCAAT binding factor NF-Y in vitro has been reported (13, 31). Therefore, AnCF involvement in chromatin organization may occur via acetylation of components of the chromatin.
MATERIALS AND METHODS
Medium.
The minimal medium used was that of Cove (12), with 10 mM ammonium tartrate as the sole nitrogen source.
Construction of strains containing amdS 5′ mutations.
The construction of plasmids pLIT1, pLIT23, pLIT1011, and pLIT14 have been described (43). These plasmids were used to generate the strains MH 3408, MH5103, MH5095, and MH5788 by two-step gene replacement at the amdS locus (14). Point mutations of the CCAAT sequence of the amdS 5′ region were introduced by site-directed mutagenesis by the method of Kunkel (33) with an oligonucleotide of sequence 5′-TAGCTGGAGATCTGCTGGCT-3′. The resulting plasmid containing an amdS-lacZ fusion (pMH3352) was used to generate strain MH5733 by gene replacement at the amdS locus. The PCR method of Higuchi (26) was used to introduce mutations into the putative TATA box by using the oligonucleotide pair 5′-GGCATGAGAGCTCTGTAGGC-3′ and 5′-GCCTACAGAGCTCTCTCATGCC-3′. The resulting mutated fragment was cloned upstream of the amdS-lacZ fusion in pMH3779 containing a mutated argB gene, and strain MH8709 was made by insertion of a single copy of the plasmid at the argB locus by the method of Punt et al. (58). Strain MH8907 was generated by insertion of a single copy of plasmid pMH3776 containing an amdS-lacZ fusion and the mutated argB gene at the argB locus by the method of Punt et al. (58).
Chromatin analysis.
Micrococcal nuclease and DNase I-based mapping of chromatin organisation was carried out as described by Gonzales and Scazzocchio (22). Strains grown for 16 h in 0.1% glucose plus 10 mM ammonium tartrate were transferred for 2 h either to repressing conditions (1% glucose) or derepressing conditions (glucose free). The mycelia were harvested by filtration through a nylon mesh, pressed dry with a paper towel, and frozen in liquid nitrogen. Then, 200-mg portions of mycelia were ground under liquid nitrogen and resuspended in 1 ml of nuclease digestion buffer (250 mM sucrose, 60 mM KCl, 15 mM NaCl, 1 mM CaCl2, 3 mM MgCl2, 0.5 mM dithiothreitol, 15 mM Tris-Cl [pH 7.5]). Digestion mixtures containing 200 μl of crude nuclei were incubated with micrococcal nuclease (50 to 150 U/ml) or DNase I (5 to 15 U/ml) at 30°C for 5 min. The reaction was terminated with 1% sodium dodecyl sulfate–12.5 mM EDTA (final concentration). DNA was purified by two rounds of phenol-chloroform extraction and ethanol precipitation, and RNA was removed by treatment with 50 μg of RNase A at 37°C for 30 min.
Indirect-end-labeling analysis.
Indirect end labeling was carried out as described by Wu (75). After secondary digestion with the appropriate restriction enzyme, the samples were electrophoresed in 1.7% agarose gels in 1× TAE, transferred onto Hybond N+ nylon membrane (Amersham), and hybridized according to standard protocols. Labeling of specific probes was done by random oligonucleotide priming.
RESULTS
The promoter region of amdS is organized in strictly positioned nucleosomes and a DNase I-hypersensitive region.
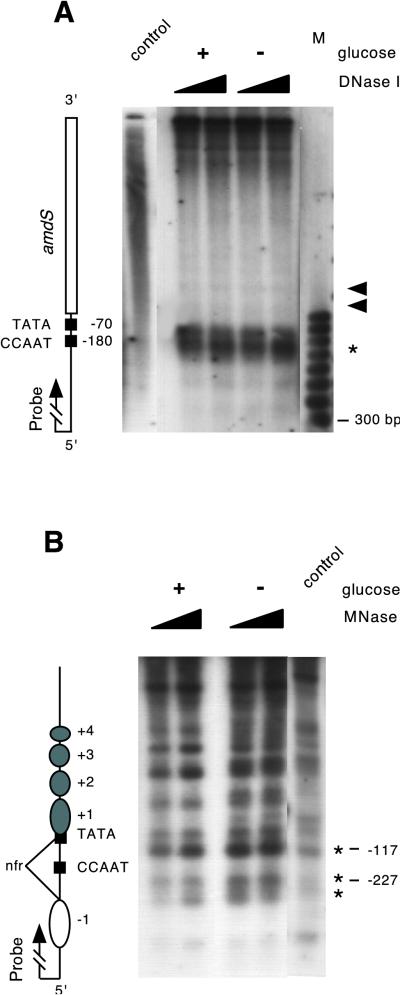
We investigated the chromatin organization of the amdS gene by using micrococcal nuclease (MNase) and DNase I. Strain MH1 (wild type) was grown in 0.1% glucose for 15 h, harvested, and transferred to either repressing conditions (1% glucose) or derepressing conditions (C-free) for 2 h. Nuclease-treated DNA was then analyzed by the indirect end-labeling technique. The DNase I experiments clearly showed a strong DNase I-hypersensitive site in the promoter region, while the transcriptional unit was resistant to DNase I treatment (Fig. 1A). Regions of increased DNase I sensitivity have been proposed to be free of nucleosomes or altered in DNA-histone interaction, leading to an open chromatin structure more accessible to transcription factors (9, 34). The hypersensitive site in the amdS promoter corresponded to positions −250 to −70 (±20 bp) relative to the translational start point and overlapped the binding sites for known transcription factors in the amdS promoter (29). This DNase I-hypersensitive site is interrupted by a region of lower sensitivity at approximately position −180, which coincides with the binding site for AnCF. The faint bands over the transcriptional unit indicate the presence of nucleosomes.
FIG. 1.
Chromatin organization of the amdS gene under repressed (+glucose) and derepressed (−glucose) conditions. Mycelia grown for 16 h in 0.1% glucose plus 10 mM ammonium tartrate were transferred for 2 h either to repressing conditions (1% glucose) or derepressing conditions (carbon free). (A) Crude nuclei were treated for 5 min with 5 and 15 U of DNase I per ml at 30°C. DNA was digested with SpeI and subjected to indirect end labeling with a SpeI (−1008)/XbaI (−650) fragment of the amdS promoter region as a probe. The control is naked genomic DNA treated with DNase I and processed similarly to chromatin samples. The arrowheads indicate internucleosomal cutting of DNase I. Lane M contains a 100-bp ladder (Bio-Rad). The vertical map at the left indicates the relative positions of TATA and CCAAT sequences and the amdS coding region. (B) Crude nuclei were treated for 5 min with 50 and 150 U of MNase per ml. DNA was digested with SacI and subjected to indirect end labeling with a SacI (−751)/SacII (−227) fragment of the amdS promoter region as a probe. The control is naked genomic DNA treated with MNase and is processed similarly to chromatin samples. Asterisks indicate nuclease-hypersensitive sites. The positions of nucleosomes are pictured at the right as ellipses and are numbered divergently from the nucleosome-free region (nfr). The closed ellipses denote nucleosomes remodeled upon derepression. The vertical map at the left indicates the relative positions of TATA and CCAAT sequences and the amdS coding region. Numbers on the right refer to the position of restriction sites in the amdS promoter that were determined by double digests of genomic DNA with SacI and SmaI (−117) and Sac and SacII (−227).
To address more directly whether the amdS promoter is free of positioned nucleosomes, we repeated the indirect end-labeling experiments with MNase. Under repressing conditions MNase generated a ladder of nucleosome-specific bands, exhibiting a repeat length of 170 bp (Fig. 1B) over the transcriptional unit (nucleosomes +1 to +4). However, the promoter region was cleaved in a pattern similar to that of naked control DNA, indicating the absence of nucleosomes. The size of the nucleosome-free region determined by use of MNase corresponded to the DNase I-hypersensitive site. Note that the sequence-specific bands in the chromatin samples were hypersensitive compared to the naked control DNA, suggesting that DNA in this region is more sensitive to MNase in a chromatin environment (Fig. 1B and Fig. 4). This result supported the DNase I experiments in indicating the absence of positioned nucleosomes over the amdS promoter. Another nucleosome is positioned upstream of this nuclease-hypersensitive site (nucleosome −1). After derepression, nucleosomal bands became more diffuse and additional, sequence-specific cutting sites appeared over the transcriptional unit in the chromatin samples, indicating that the strict positioning of the nucleosomes over the transcriptional unit was lost. Nucleosomes further upstream were unaffected by this rearrangement (data not shown).
FIG. 4.
Chromatin organization of the amdS::lacZ fusion gene in a CCAAT and hapE mutant strain. MH1 corresponds to the wild-type strain. In strain MH9206, hapE is deleted. In strain MH5733, the CCAAT motif is mutated to CAGAT (Fig. 2). Mycelia were grown for 16 h on 1% glucose plus 10 mM ammonium tartrate. Chromatin analysis was performed as indicated in the legend of Fig. 1B. Nucleosomes are pictured at the right as ellipses. The dashed ellipses indicate a less strict positioning of nucleosomes. The vertical map at the left indicates the relative positions of TATA box, CCAAT sequence, and amdS::lacZ coding region.
These data showed that the promoter region of amdS is preset in a nucleosome-free region, whereas an array of nucleosomes is positioned over the coding region and upstream of the promoter. After relief of carbon catabolite repression the chromatin structure in the amdS gene undergoes rearrangement.
DNase I hypersensitivity over the promoter region is dependent on the CCAAT sequence and functional AnCF.
The CCAAT site in the promoter region of the amdS gene is located 180 bp upstream of the translational startpoint and shows significant homology to the HAP2/3/4/5 consensus binding site (46). Gel mobility shift assays have shown that AnCF binds to this sequence in vitro and is required for setting the level of the amdS expression (43, 57, 63).
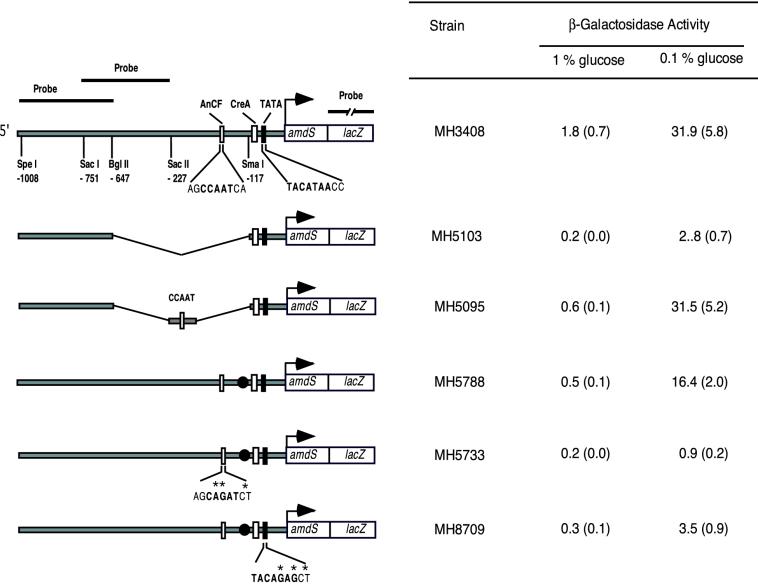
We examined the effect of mutations in the CCAAT sequence on the activity and chromatin architecture of the amdS promoter. Expression of the amdS promoter was assessed by using an amdS-lacZ fusion reporter replaced at the amdS locus (14, 33). A 530-bp deletion (−117 to −647 relative to the translational startpoint) in MH5103 resulted in greatly reduced expression of an amdS-lacZ reporter (Fig. 2). Insertion of a 35-bp sequence representing the region −185 to −151, including the amdS CCAAT sequence, in strain MH5095 restored expression (Fig. 2). We mutated this element by site-directed mutagenesis and subsequent gene replacement (32). In strain MH5733 the alteration of two bases of the CCAAT sequence resulted in a reduction in amdS-lacZ expression by an order of magnitude (Fig. 2). The level of expression was similar to the previously described affect of disruption of the hapB, hapC, and hapE genes (57, 63). These data confirm previous results (43) showing that the CCAAT motif is required for high levels of amdS expression.
FIG. 2.
Effect of promoter mutation on amdS-lacZ expression. The arrow indicates the translational startpoint. The relevant restriction sites and cis-acting sites are shown. The sequences for the CCAAT and TATA box are given in detail. Mutated nucleotides within these sequences are indicated by asterisks. β-Galactosidase activity is shown with standard errors in brackets. Repressed conditions were growth for 16 h in minimal medium with 1% glucose plus 10 mM ammonium tartrate; derepressed conditions were growth for 19 h in minimal medium with 0.1% glucose plus 10 mM ammonium tartrate. The amdS-lacZ construct in strain MH8709 is integrated into the argB locus. A corresponding control amdS-lacZ reporter strain (MH8907) showed comparable values to MH5788. The closed circle indicates a 10-bp insertion (CGGGATCCCG [43]). Probes used in indirect-end-labeling experiments are shown.
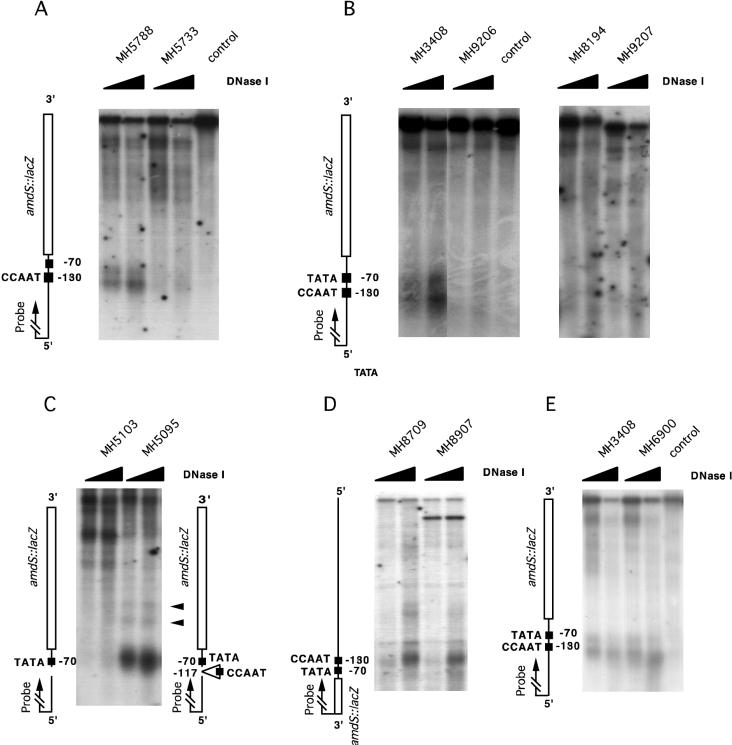
We examined the effect of mutations in the CCAAT motif on the chromatin architecture of the amdS promoter. Point mutations in the CCAAT sequence completely eliminated the DNase I hypersensitivity in the promoter region (Fig. 3A), indicating a dramatic change in the chromatin structure.
FIG. 3.
Effect of mutations on the DNase I sensitivity of the amdS promoter. Mycelia were grown for 16 h on 1% glucose plus 10 mM ammonium tartrate. DNA was digested with SacI, and a SacI (−751)/SacII (−227) fragment of the amdS promoter region was used as a probe except as noted otherwise. (A) Strains MH5788 and MH5733 carry a 10-bp oligonucleotide containing a BamHI site inserted into the SmaI (−117) site in the amdS promoter. In strain MH5733 the CCAAT motif is mutated to CAGAT (see Fig. 2). (B) Effect of the hapBCE gene deletions on the DNase I sensitivity of the amdS promoter. In MH9207 hapB is deleted, in MH8194 hapC is deleted, and in MH9206 hapE is deleted. MH3408 is wild type at all hap loci. All strains carry an amdS::lacZ fusion. (C) MH5103 carries a 530-bp BglII (−647)/SmaI (−117) promoter deletion which removes most of the regulatory sites including the CCAAT sequence. In strain MH5095 a 35-bp oligonucleotide carrying the CCAAT motif is inserted into the deletion site (see Fig. 2). DNA was digested with SpeI, and a SpeI (−1008)/XbaI (−650) fragment of the amdS promoter region was used as a probe. The arrowheads indicate internucleosomal cutting of DNase I. (D) In MH8709 the TATA box is mutated as indicated in Fig. 2. Both strains carry an amdS::lacZ fusion gene integrated into the argB locus. DNA was digested with ClaI and a HindIII (+113)/ClaI (+907) fragment of the lacZ coding region was used as a probe. (E) Strain MH6900 carries a 29-bp deletion which removes the two CreA-binding sites.
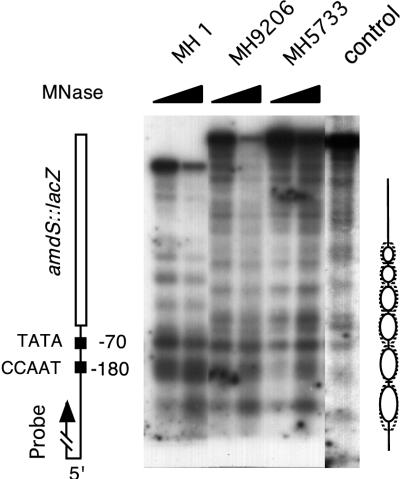
To determine whether the observed effect results from the binding of the AnCF complex, we analyzed the chromatin structure of amdS in ΔhapB, ΔhapC, and ΔhapE deletion strains where AnCF is not functional (57, 63). Strains MH9207 (ΔhapB), MH8194 (ΔhapC), and MH9206 (ΔhapE) showed no DNase I hypersensitivity in the amdS promoter (Fig. 3B). These results revealed a crucial role for AnCF in the formation of a defined chromatin structure in the amdS promoter.
To test whether the positioning of the nucleosomes is affected by these mutations, strain MH5733 and MH9206 were analyzed by MNase treatment under repressing conditions (Fig. 4). The nucleosomal organization in the MH5733 strain carrying the mutated CCAAT sequence was changed compared with the wild-type situation. The strict positioning of the nucleosomes over the transcriptional unit was lost. The appearance of sequence-specific cutting sites in addition to weaker nucleosomal bands indicated a changed organization of the nucleosomes. The same result was obtained with the hapE deletion strain (MH9206). We conclude that AnCF not only is responsible for the formation of a DNase I-hypersensitive site in the amdS 5′ region but also influences the positioning of the adjacent nucleosomes.
To determine whether the CCAAT sequence alone is sufficient to generate DNase I hypersensitivity in the amdS promoter, we carried out DNase I analysis with strains MH5103 and MH5095 (Fig. 3C). The DNase I-hypersensitive site was absent in strain MH5103 which contains the deletion from −117 to −650 but was restored in strain MH5095 in which the deleted region is replaced by a 35-bp oligonucleotide containing the amdS CCAAT sequence. However, the architecture of this DNase I-hypersensitive site was clearly different from that in the wild type. Whereas the wild-type promoter showed a clear double band (Fig. 1A), the DNase I-sensitive site in MH5095 was a more diffuse region lacking the intervening protected area.
In addition to the DNase I-hypersensitive site recreated after insertion of the oligonucleotide containing the CCAAT sequence, two positioned nucleosomes were observed downstream of the DNase I-sensitive site (indicated by the arrows in Fig. 3C). This finding strongly supports the idea that the CCAAT-mediated formation of the DNase I-sensitive site directly or indirectly determines the position of the adjacent nucleosomes, possibly by defining the position of the first nucleosome. Thus, AnCF plays a crucial role in the organization of the regulatory chromatin structure of the amdS gene. The experiments with strains MH5103 and MH5095 also showed that the regulatory elements for AreA mediated nitrogen derepression, FacB mediated acetate induction, and AmdR mediated omega amino acid induction, which are deleted in these strains, were not necessary for the formation of the DNase I-hypersensitive site.
We further tested the ability of the CCAAT sequence to generate a DNase I-hypersensitive site outside a promoter. For this purpose the 35-bp oligonucleotide containing the amdS CCAAT sequence was inserted 788 bp 3′ to the stop codon of the argB gene, and this construct was transformed into an argB mutant strain. DNase I experiments were carried out on five independent argB+ transformants, but none showed a nuclease-sensitive site in the 3′ region of the argB gene (data not shown). Therefore, additional elements may be necessary for the CCAAT-mediated formation of the DNase I-sensitive site, and these elements must be present in the truncated amdS promoter of MH5095.
Presetting the chromatin structure of the amdS promoter is not dependent on the TATA box or a CreA binding site.
The truncated amdS promoter in MH5095 still carries a potential TATA box. Mutation of this sequence in MH8709 resulted in greatly reduced amdS-lacZ expression compared to the control strain MH5788 (Fig. 2), indicating a functional role for this sequence. As the yeast homologue of AnCF, the HAP2/3/4/5 complex interacts with the ADA2/ADA3/GCN5 complex, and ADA2 is associated with TBP, we supposed that TBP could be the component acting together with AnCF (5, 20, 47, 64). However, the DNase I-hypersensitive site was still present in the mutated promoter (Fig. 3D).
Two sequences present between positions −90 and −107 have been found to be binding sites for CreA, a C2H2 finger protein responsible for carbon catabolite repression, as well as for binding by the AmdA and AmdX C2H2 finger activating proteins (3, 41, 53). Deletion of these sequences (in plasmid p23 [41]), followed by gene replacement at the amdS locus, resulted in strain MH6900. This deletion was not found to affect the formation of a DNase I-sensitive site (Fig. 3E).
The promoter regions of fmdS are organized in DNase I-sensitive sites which are dependent on AnCF.
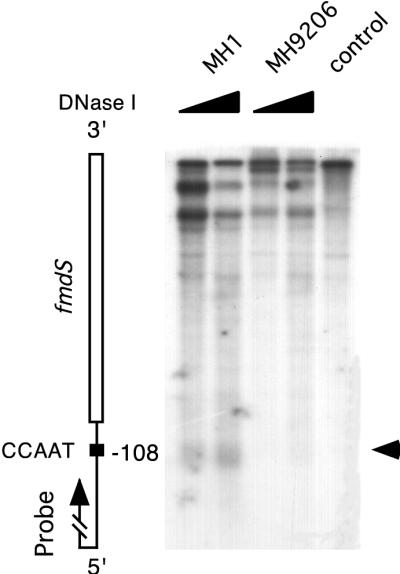
We investigated whether other genes containing CCAAT sequences showed a similar chromatin organization in their promoter regions. The promoter region of fmdS, a gene encoding a formamidase, contains a CCAAT sequence (position −108) which exhibits significant homology to the CCAAT element consensus sequence (46) and binds AnCF (19). We investigated the promoter regions of this gene by using DNase I and indirect end labeling. A DNase I-sensitive site was found which coincides with the CCAAT motif (Fig. 5). The hapE deletion present in strain MH 9206 abolished the formation of the DNase I-hypersensitive site in the promoter region of fmdS. These data indicate that the mechanism by which AnCF presets the chromatin structure in a promoter region may be the same in the two genes investigated in this work.
FIG. 5.
DNase I analysis of the fmdS gene in a wild-type strain (MH1) and a hapE deletion strain (MH9602). Mycelia were grown for 16 h on 1% glucose plus 10 mM ammonium tartrate. Chromatin analysis was performed as indicated in the legend to Fig. 1A except that DNA was digested with XhoI, and a XhoI (−599)/PstI (−90) fragment of the fmdS promoter region was used as probe. The vertical map on the left indicates the relative position of the CCAAT sequence in the fmdS promoter. The arrow indicates the DNase I-hypersensitive site.
DISCUSSION
In the present study we found that the formation of a DNase I-hypersensitive site in the amdS promoter region is strictly dependent on the presence of a functional CCAAT box and the AnCF complex. Further, the positioning of the adjacent nucleosomes is dependent on the presence of the DNase I-hypersensitive site. Our results indicate that AnCF is crucial for presetting the amdS promoter in an open chromatin structure.
The amdS locus is organized in a well-defined chromatin structure in the repressed state, with an array of positioned nucleosomes over parts of the promoter and the coding region. In both the repressed and derepressed states a constitutive DNase I-hypersensitive site exists from positions −250 to −70 relative to the translational startpoint. Adjacent to the DNase I-hypersensitive site an ordered nucleosome array covering the coding region and sequences upstream of the promoter exists under repressing conditions. Upon derepression, the discrete MNase bands become diffuse, indicating that the exact positioning of the nucleosomes is lost and that they can slide along the DNA, a change also observed in other genes and characteristic of the active state (15, 36, 68, 76).
The constitutive DNase I-hypersensitive site observed in the promoter region covers all previously identified cis-acting sequences (for a review, see reference 29), suggesting that the transcription factors required for regulation of the amdS gene may have permanent access to their binding sites. This stretch of DNA (ca. 180 bp) is nucleosome-free under the growth conditions tested. Thus, the transcription factors acting at the amdS promoter may be able to bind to their sites without first disrupting nucleosomes. This differs from the remodelling of the PHO5 promoter by Pho4p, in which transcription factor binding and nucleosome disruption seem to be linked (2, 66).
The DNase I-hypersensitive site is interrupted by a region of decreased nuclease sensitivity, a structural feature observed in DNase I-sensitive regions of several other promoters (6, 23). This protection may be caused by the binding of protein(s) to the hypersensitive region that protects DNA from DNase I. This idea is supported by the fact that some MNase cleavage sites within the promoter show hypersensitivity, indicating a structural change causing increased accessibility of the DNA in the chromatin samples compared with naked DNA. It is known that bending of DNA by DNA binding proteins can cause an increased accessibility to nucleases (25), and such a bending of DNA has been shown for the CCAAT binding complex PENR1 (which is probably identical to AnCF) (44) and NF-Y (60). Alternatively, the protection in the DNase I-hypersensitive region may reflect a conformational change of the DNA structure that prevents DNase I from cutting in both strands.
The TATA box of amdS is located at the border of the first downstream nucleosome (nucleosome +1) and the DNase I-hypersensitive site. A similar situation is found in the yeast HSP82 and the Drosphila hsp26 genes (23, 42). This is in contrast to a number of other genes where the incorporation of the TATA box into a nucleosome severely inhibits the binding of TBP (21, 30) and greatly reduces transcription initiation in vitro (32, 35, 74) and in vivo (37, 65).
Deletion or mutation of the CCAAT motif, the DNA-binding motif for AnCF in the amdS promoter, or deletion of the hapB, hapC, and hapE genes, which results in a nonfunctional AnCF complex, reduces amdS expression by an order of magnitude (Fig. 2) (43, 57, 63). Moreover, such mutations result in a distinct rearrangement of the chromatin structure of amdS. The DNase I-hypersensitive site in the promoter region and the positioning of the nucleosomes is lost. Importantly, this effect on chromatin structure is not a consequence of transcriptional inactivation, since point mutations in the TATA box which greatly reduce amdS expression (Fig. 2) retain the DNase I-hypersensitive site in the promoter region (Fig. 3D). The importance of the CCAAT box is further demonstrated by the restoration of both the DNase I sensitivity and amdS expression when a CCAAT containing sequence is inserted into a truncated amdS promoter (Fig. 3C and Fig. 2).
However, the CCAAT sequence itself is not sufficient to initiate an open chromatin structure since the CCAAT-containing oligonucleotide cloned 3′ to the argB gene failed to create such a nuclease-sensitive site. Thus, the remaining part of the promoter must carry an additional feature necessary to generate the DNase I-hypersensitive site which acts in concert with AnCF. Two known cis-acting sites in the truncated promoter are the binding site for the CreA carbon catabolite repressor with homology to Mig1p (16) and the TATA box. The lack of effect of the TATA box mutation on DNase I sensitivity (Fig. 3D) is consistent with data from the yeast HSP82 gene, where it has been shown that TBP has no influence on the organization of the DNase I-hypersensitive region (23), although in vitro data suggest that TBP may prevent the assembly of nucleosomes in a core promoter region (7, 49, 74). A strain carrying a deletion of the CreA-binding sites showed a chromatin structure corresponding to the carbon-derepressed phenotype (data not shown) but was unaffected in DNase I hypersensitivity (Fig. 3E).
Of particular interest is the fact that the insertion of the CCAAT sequence into a truncated amdS promoter not only restores the DNase I-hypersensitive site but also reassembles the downstream region into positioned nucleosomes (Fig. 3C). Together with the observation that the strict positioning of the nucleosomes over the coding region is lost in the CCAAT sequence mutant, this suggests that the AnCF-mediated assembly of the DNase I-hypersensitive site directly or indirectly determines the position of the adjacent nucleosomes, possibly by defining the position of the first nucleosome. Widlund et al. (71) found that particular sequences, e.g., A runs, extended repeats of CA, or tetramers of TATAA, are responsible for the positioning of nucleosomes. However, the amdS promoter does not contain such strong nucleosome-positioning motifs, indicating that the underlying DNA sequence is not involved in the positioning of nucleosomes. Mutation of the major HSF binding site in the yeast HSP82 promoter leads to the displacement of the DNase I-hypersensitive site by two strictly positioned nucleosomes (23). In contrast, a loss of nucleosomal positioning results from mutation of the amdS CCAAT or from inactivation of AnCF.
The amdS gene is subjected to multiple regulatory controls (29). Mutations affecting the CCAAT sequence do not eliminate responses to amdA-, areA-, facB-, or creA-mediated regulatory circuits, although overall levels of expression are greatly reduced (27, 63). Deletion of the areA gene, a major determinant of amdS expression under carbon sufficient but nitrogen limiting conditions, does not affect the DNase I-hypersensitive site (55). This indicates that the products of these genes can bind in a nucleosomal environment. A similar effect is seen in the niiA-niaD promoter, where areA-dependent chromatin remodelling still occurs in a mutant where only AreA binding sites outside the nucleosome-free region are intact (45). However, it has been shown that regulation by AmdR which binds to a sequence partially overlapping the CCAAT sequence is abolished in mutants lacking AnCF. For this transcription factor, therefore, it is probable that the AnCF-mediated chromatin structure is necessary for binding to DNA (63).
An AnCF-dependent DNase I-hypersensitive site corresponding to a CCAAT sequence has been found in the promoter region of the fmdS gene (Fig. 5). This indicates that the mechanism by which AnCF is acting at the amdS promoter could also apply to other promoters in A. nidulans.
Our results are consistent with the finding that Y boxes, bound by the AnCF homologue NF-Y, serve a similar role in the X. laevis hsp70 promoter (34, 38). The biochemical mechanism which finally prevents a tight DNA-histone interaction in a DNase I-hypersensitive site remains unclear. The NF-Y subunits B and C (corresponding to HapB and HapC in AnCF) carry a histone fold motif, showing similarity to histone H2B and H2A (40). It has been shown that NF-Y can bind to a Y box even in the presence of reconstituted nucleosomes (52). The action of acetyltransferases may play a role in the local disruption of nucleosomes since an association of GATA-1 and NF-Y with acetyltransferases p300/CBP has been shown (8, 38). However, Trichostatin A, an inhibitor of deacetylases which clearly activates the p300-triggered transcription of the X. laevis hsp70 gene, has no influence on the formation of a DNase I-sensitive site in this promoter (38).
Our data support the idea that CCAAT sequences could play a conserved role in the generation of an open chromatin structure necessary for full transcriptional activation in eukaryotic promoters.
ACKNOWLEDGMENTS
This work was supported by the Australian Research Council. F.M.N. was supported by the Austrian Science Foundation (J 1518-GEN).
Advice and suggestions from Alex Andrianopoulos, construction of the TATA mutation by Chris Stemple, and assistance by Julie Sharp are appreciated.
REFERENCES
- 1.Almer A, Horz W. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J. 1986;5:2681–2687. doi: 10.1002/j.1460-2075.1986.tb04551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almer A, Rudolph H, Hinnen A, Horz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrianopoulos A, Brons J, Davis M A, Hynes M J. The amdA regulatory gene of Aspergillus nidulans: characterization of gain-of-function mutations and identification of binding sites for the gene product. Fungal Genet Biol. 1996;21:50–63. doi: 10.1006/fgbi.1997.0968. [DOI] [PubMed] [Google Scholar]
- 4.Aparicio O M, Billington B L, Gottschling D E. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- 5.Barlev N A, Candau R, Wang L, Darpino P, Silverman N, Berger S L. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J Biol Chem. 1995;270:19337–19344. doi: 10.1074/jbc.270.33.19337. [DOI] [PubMed] [Google Scholar]
- 6.Baum J A, Giles N H. DNase I hypersensitive sites within the inducible qa gene cluster of Neurospora crassa. Proc Natl Acad Sci USA. 1986;83:6533–6537. doi: 10.1073/pnas.83.17.6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker P B, Rabindran S K, Wu C. Heat shock-regulated transcription in vitro from a reconstituted chromatin template. Proc Natl Acad Sci USA. 1991;88:4109–4113. doi: 10.1073/pnas.88.10.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 9.Bresnick E H, Bustin M, Marsaud V, Richard-Foy H, Hager G L. The transcriptionally-active MMTV promoter is depleted of histone H1. Nucleic Acids Res. 1992;20:273–278. doi: 10.1093/nar/20.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cairns B R. Chromatin remodeling machines: similar motors, ulterior motives. Trends Biochem Sci. 1998;23:20–25. doi: 10.1016/s0968-0004(97)01160-2. [DOI] [PubMed] [Google Scholar]
- 11.Candau R, Zhou J X, Allis C D, Berger S L. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 1997;16:555–565. doi: 10.1093/emboj/16.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cove D J. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim Biophys Acta. 1966;113:51–56. doi: 10.1016/s0926-6593(66)80120-0. [DOI] [PubMed] [Google Scholar]
- 13.Currie R A. NF-Y is associated with the histone acetyltransferases GCN5 and P/CAF. J Biol Chem. 1998;273:1430–1434. doi: 10.1074/jbc.273.3.1430. [DOI] [PubMed] [Google Scholar]
- 14.Davis M A, Cobbett C S, Hynes M J. An amdS-lacZ fusion for studying gene regulation in Aspergillus. Gene. 1988;63:199–212. doi: 10.1016/0378-1119(88)90525-2. [DOI] [PubMed] [Google Scholar]
- 15.del Olmo M L, Sogo J M, Franco L, Perez-Ortin J E. Chromatin structure of the yeast FBP1 gene: transcription-dependent changes in the regulatory and coding regions. Yeast. 1993;9:1229–1240. doi: 10.1002/yea.320091110. [DOI] [PubMed] [Google Scholar]
- 16.Dowzer C E, Kelly J M. Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol Cell Biol. 1991;11:5701–5709. doi: 10.1128/mcb.11.11.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992;355:219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- 18.Forsburg S L, Guarente L. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989;3:1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- 19.Fraser, J. Personal communication.
- 20.Georgakopoulos T, Thireos G. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J. 1992;11:4145–4152. doi: 10.1002/j.1460-2075.1992.tb05507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godde J S, Nakatani Y, Wolffe A P. The amino-terminal tails of the core histones and the translational position of the TATA box determine TBP/TFIIA association with nucleosomal DNA. Nucleic Acids Res. 1995;23:4557–4564. doi: 10.1093/nar/23.22.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez R, Scazzocchio C. A rapid method for chromatin structure analysis in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 1997;25:3955–3956. doi: 10.1093/nar/25.19.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gross D S, Adams C C, Lee S, Stentz B. A critical role for heat shock transcription factor in establishing a nucleosome-free region over the TATA-initiation site of the yeast HSP82 heat shock gene. EMBO J. 1993;12:3931–3945. doi: 10.1002/j.1460-2075.1993.tb06071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hahn S, Guarente L. Yeast HAP2 and HAP3: transcriptional activators in a heteromeric complex. Science. 1988;240:317–321. doi: 10.1126/science.2832951. [DOI] [PubMed] [Google Scholar]
- 25.Higgins N P, Collier D A, Kilpatrick M W, Krause H M. Supercoiling and integration host factor change the DNA conformation and alter the flow of convergent transcription in phage Mu. J Biol Chem. 1989;264:3035–3042. [PubMed] [Google Scholar]
- 26.Higuchi R. Recombinant PCR. In: Innis M A, Gelfand D H, Sninsky J S, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 177–183. [Google Scholar]
- 27.Hynes, M. J. Personal communication.
- 28.Hynes M J, Corrick C M, King J A. Isolation of genomic clones containing the amdS gene of Aspergillus nidulans and their use in the analysis of structural and regulatory mutations. Mol Cell Biol. 1983;3:1430–1439. doi: 10.1128/mcb.3.8.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hynes M J, Davis M A. Regulation of acetamide catabolism. In: Brambl R, Marzluf G, editors. The mycota III: biochemistry and molecular biology. Berlin, Germany: Springer-Verlag; 1996. pp. 381–393. [Google Scholar]
- 30.Imbalzano A N, Kwon H, Green M R, Kingston R E. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 31.Jin S, Scotto K W. Transcriptional regulation of the MDR1 gene by histone acetyltransferase and deacetylase is mediated by NF-Y. Mol Cell Biol. 1998;18:4377–4384. doi: 10.1128/mcb.18.7.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knezetic J A, Luse D S. The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell. 1986;45:95–104. doi: 10.1016/0092-8674(86)90541-6. [DOI] [PubMed] [Google Scholar]
- 33.Kunkel T A. Rapid and efficient site-directed mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landsberger N, Wolffe A P. Role of chromatin and Xenopus laevis heat shock transcription factor in regulation of transcription from the X. laevis hsp70 promoter in vivo. Mol Cell Biol. 1995;15:6013–6024. doi: 10.1128/mcb.15.11.6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laybourn P J, Kadonaga J T. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 1991;254:238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- 36.Lee M S, Garrard W T. Transcription-induced nucleosome “splitting”: an underlying structure for DNase I sensitive chromatin. EMBO J. 1991;10:607–615. doi: 10.1002/j.1460-2075.1991.tb07988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li G, Chandler S P, Wolffe A P, Hall T C. Architectural specificity in chromatin structure at the TATA box in vivo: nucleosome displacement upon beta-phaseolin gene activation. Proc Natl Acad Sci USA. 1998;95:4772–4777. doi: 10.1073/pnas.95.8.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Q, Herrler M, Landsberger N, Kaludov N, Ogryzko V V, Nakatani Y, Wolffe A P. Xenopus NF-Y pre-sets chromatin to potentiate p300 and acetylation-responsive transcription from the Xenopus hsp70 promoter in vivo. EMBO J. 1998;17:6300–6315. doi: 10.1093/emboj/17.21.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liberati C, di Silvio A, Ottolenghi S, Mantovani R. NF-Y binding to twin CCAAT boxes: role of Q-rich domains and histone fold helices. J Mol Biol. 1999;285:1441–1455. doi: 10.1006/jmbi.1998.2384. [DOI] [PubMed] [Google Scholar]
- 40.Linhoff M W, Wright K L, Ting J P. CCAAT-binding factor NF-Y and RFX are required for in vivo assembly of a nucleoprotein complex that spans 250 base pairs: the invariant chain promoter as a model. Mol Cell Biol. 1997;17:4589–4596. doi: 10.1128/mcb.17.8.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lints R, Davis M A, Hynes M J. The positively acting amdA gene of Aspergillus nidulans encodes a protein with two C2H2 zinc-finger motifs. Mol Microbiol. 1995;15:965–975. doi: 10.1111/j.1365-2958.1995.tb02365.x. [DOI] [PubMed] [Google Scholar]
- 42.Lis J T, Wu C. Transcriptional regulation of heat shock genes. In: Cronaway R C, Cronaway J W, editors. Transcription: mechanisms and regulation. New York, N.Y: Raven Press; 1994. pp. 459–475. [Google Scholar]
- 43.Littlejohn T G, Hynes M J. Analysis of the site of action of the amdR product for regulation of the amdS gene of Aspergillus nidulans. Mol Gen Genet. 1992;235:81–88. doi: 10.1007/BF00286184. [DOI] [PubMed] [Google Scholar]
- 44.Litzka O, Papagiannopolous P, Davis M A, Hynes M J, Brakhage A A. The penicillin regulator PENR1 of Aspergillus nidulans is a HAP-like transcriptional complex. Eur J Biochem. 1998;251:758–767. doi: 10.1046/j.1432-1327.1998.2510758.x. [DOI] [PubMed] [Google Scholar]
- 45.Muro-Pastor M I, Gonzales R, Strauss J, Narendja F M, Scazzocchio C. The GATA factor AreA is essential for chromatin remodelling in a eukaryotic bidirectional promoter. EMBO J. 1999;18:1584–1597. doi: 10.1093/emboj/18.6.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mantovani R. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 1998;26:1135–1143. doi: 10.1093/nar/26.5.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marcus G A, Silverman N, Berger S L, Horiuchi J, Guarente L. Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J. 1994;13:4807–4815. doi: 10.1002/j.1460-2075.1994.tb06806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McNabb D S, Xing Y, Guarente L. Cloning of yeast HAP5: a novel subunit of a heterotrimeric complex required for CCAAT binding. Genes Dev. 1995;9:47–58. doi: 10.1101/gad.9.1.47. [DOI] [PubMed] [Google Scholar]
- 49.Meisterernst M, Horikoshi M, Roeder R G. Recombinant yeast TFIID, a general transcription factor, mediates activation by the gene-specific factor USF in a chromatin assembly assay. Proc Natl Acad Sci USA. 1990;87:9153–9157. doi: 10.1073/pnas.87.23.9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 51.Moreira J M, Holmberg S. Nucleosome structure of the yeast CHA1 promoter: analysis of activation-dependent chromatin remodeling of an RNA-polymerase-II-transcribed gene in TBP and RNA pol II mutants defective in vivo in response to acidic activators. EMBO J. 1998;17:6028–6038. doi: 10.1093/emboj/17.20.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Motta M C, Caretti G, Badaracco G F, Mantovani R. Interactions of the CCAAT-binding trimer NF-Y with nucleosomes. J Biol Chem. 1999;274:1326–1333. doi: 10.1074/jbc.274.3.1326. [DOI] [PubMed] [Google Scholar]
- 53.Murphy R L, Andrianopoulos A, Davis M A, Hynes M J. Identification of amdX, a new Cys-2-His-2 (C2H2) zinc-finger gene involved in the regulation of the amdS gene of Aspergillus nidulans. Mol Microbiol. 1997;23:591–602. doi: 10.1046/j.1365-2958.1997.d01-1872.x. [DOI] [PubMed] [Google Scholar]
- 54.Nakshatri H, Bhat-Nakshatri P, Currie R A. Subunit association and DNA binding activity of the heterotrimeric transcription factor NF-Y is regulated by cellular redox. J Biol Chem. 1996;271:28784–28791. doi: 10.1074/jbc.271.46.28784. [DOI] [PubMed] [Google Scholar]
- 55.Narendja, F. M. Unpublished data.
- 56.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 57.Papagiannopoulos P, Andrianopoulos A, Sharp J A, Davis M A, Hynes M J. The hapC gene of Aspergillus nidulans is involved in the expression of CCAAT-containing promoters. Mol Gen Genet. 1996;251:412–421. doi: 10.1007/BF02172369. [DOI] [PubMed] [Google Scholar]
- 58.Punt P J, Dingemanse M A, Kuyvenhoven A, Soede R D, Pouwels P H, van den Hondel C A. Functional elements in the promoter region of the Aspergillus nidulans gpdA gene encoding glyceraldehyde-3-phosphate dehydrogenase. Gene. 1990;93:101–109. doi: 10.1016/0378-1119(90)90142-e. [DOI] [PubMed] [Google Scholar]
- 59.Reik A, Schutz G, Stewart A F. Glucocorticoids are required for establishment and maintenance of an alteration in chromatin structure: induction leads to a reversible disruption of nucleosomes over an enhancer. EMBO J. 1991;10:2569–2576. doi: 10.1002/j.1460-2075.1991.tb07797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ronchi A, Bellorini M, Mongelli N, Mantovani R. CCAAT-box binding protein NF-Y (CBF, CP1) recognizes the minor groove and distorts DNA. Nucleic Acids Res. 1995;23:4565–4572. doi: 10.1093/nar/23.22.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmid A, Fascher K D, Horz W. Nucleosome disruption at the yeast PHO5 promoter upon PHO5 induction occurs in the absence of DNA replication. Cell. 1992;71:853–864. doi: 10.1016/0092-8674(92)90560-y. [DOI] [PubMed] [Google Scholar]
- 62.Stamatoyannopoulos J A, Goodwin A, Joyce T, Lowrey C H. NF-E2 and GATA binding motifs are required for the formation of DNase I hypersensitive site 4 of the human beta-globin locus control region. EMBO J. 1995;14:106–116. doi: 10.1002/j.1460-2075.1995.tb06980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Steidl S, Papagiannopoulos P, Litzka O, Andrianopoulos A, Davis M A, Brakhage A A, Hynes M J. AnCF, the CCAAT binding complex of Aspergillus nidulans, contains products of the hapB, hapC, and hapE genes and is required for activation by the pathway-specific regulatory gene amdR. Mol Cell Biol. 1999;19:99–106. doi: 10.1128/mcb.19.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sterner D E, Grant P A, Roberts S M, Duggan L J, Belotserkovskaya R, Pacella L A, Winston F, Workman J L, Berger S L. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Straka C, Horz W. A functional role for nucleosomes in the repression of a yeast promoter. EMBO J. 1991;10:361–368. doi: 10.1002/j.1460-2075.1991.tb07957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Svaren J, Horz W. Transcription factors vs. nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem Sci. 1997;22:93–97. doi: 10.1016/s0968-0004(97)01001-3. [DOI] [PubMed] [Google Scholar]
- 67.van Heeswijck R, Hynes M J. The amdR product and a CCAAT-binding factor bind to adjacent, possibly overlapping DNA sequences in the promoter region of the Aspergillus nidulans amdS gene. Nucleic Acids Res. 1991;19:2655–2660. doi: 10.1093/nar/19.10.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vincenz C, Fronk J, Tank G A, Langmore J P. Nucleoprotein hybridization: a method for isolating active and inactive genes as chromatin. Nucleic Acids Res. 1991;19:1325–1336. doi: 10.1093/nar/19.6.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang L, Liu L, Berger S L. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 1998;12:640–653. doi: 10.1101/gad.12.5.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang L, Mizzen C, Ying C, Candau R, Barlev N, Brownell J, Allis C D, Berger S L. Histone acetyltransferase activity is conserved between yeast and human GCN5 and is required for complementation of growth and transcriptional activation. Mol Cell Biol. 1997;17:519–527. doi: 10.1128/mcb.17.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Widlund H R, Cao H, Simonsson S, Magnusson E, Simonsson T, Nielsen P E, Kahn J D, Crothers D M, Kubista M. Identification and characterization of genomic nucleosome-positioning sequences. J Mol Biol. 1997;267:807–817. doi: 10.1006/jmbi.1997.0916. [DOI] [PubMed] [Google Scholar]
- 72.Winston F, Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 73.Wolffe A P. Nucleosome positioning and modification: chromatin structures that potentiate transcription. Trends Biochem Sci. 1994;19:240–244. doi: 10.1016/0968-0004(94)90148-1. [DOI] [PubMed] [Google Scholar]
- 74.Workman J L, Roeder R G. Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell. 1987;51:613–622. doi: 10.1016/0092-8674(87)90130-9. [DOI] [PubMed] [Google Scholar]
- 75.Wu C. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- 76.Wu C, Wong Y C, Elgin S C. The chromatin structure of specific genes. II. Disruption of chromatin structure during gene activity. Cell. 1979;16:807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]
- 77.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]