Abstract
Nucleotides are an important class of odorants for aquatic vertebrates such as frogs and fishes, but also have manifold signaling roles in other cellular processes. Recently, an adenosine receptor believed to belong to the adora2 clade has been identified as an olfactory receptor in zebrafish. Here, we set out to elucidate the evolutionary history of both this gene and its olfactory function. We have performed a thorough phylogenetic study in vertebrates, chordates and their sister group, ambulacraria, and show that the origin of the zebrafish olfactory receptor gene can be traced back to the most recent common ancestor of all three groups as a segregate sister clade (adorb) to the adora gene family. Eel, carp, and clawed frog all express adorb in a sparse and distributed pattern within their olfactory epithelium very similar to the pattern observed for zebrafish that is, consistent with a function as olfactory receptor. In sharp contrast, lamprey adorb-expressing cells are absent from the sensory region of the lamprey nose, but form a contiguous domain directly adjacent to the sensory region. Double-labeling experiments confirmed the expression of lamprey adorb in nonneuronal cells and are consistent with an expression in neuronal progenitor cells. Thus, adorb may have undergone a switch of function in the jawed lineage of vertebrates towards a role as olfactory receptor.
Keywords: adenosine receptor, lamprey, olfaction
Significance
The adenosine receptor A2c/adorb is known as olfactory receptor in zebrafish, but the origin of this gene and of its olfactory function were unknown. We have identified the origin of adorb in the most recent common ancestor of deuterostomes, much earlier than other olfactory receptor families. We show in representative species that jawed vertebrates express adorb in sparsely distributed olfactory receptor neurons. In sharp contrast, the jawless lamprey expresses adorb in a contiguous region of nonneuronal cells, which could be neuronal progenitor cells. Thus the adorb receptor appears to have acquired an olfactory function in bony vertebrates, a long time after the birth of this gene. These findings constitute an important step in understanding the evolution of olfactory receptor function.
Introduction
The sense of smell is both evolutionary ancient and of primary importance in many different animal species, who employ it to detect prey or food, evade predators, guide mating behavior, and sculpt manifold social interactions. In vertebrates perception of odors is mediated by four main families of olfactory receptors, the odorant receptors (ORs) proper, the trace-amine-associated receptors (TAAR and TAAR-like), and the vomeronasal receptors types I and II—V1R/ORA and V2R/OlfC, respectively (Korsching 2016). The evolutionary origin of these four families differs, with ORs already observed in chordates (Churcher and Taylor 2009) but see (Nordström et al. 2008), V1R and TAAR-like receptors first reported in vertebrates, and V2Rs and TAARs only found in jawed vertebrates (Korsching 2016). Additionally, several smaller receptor families or even single genes have been repurposed for an olfactory function at differing evolutionary stages, in some cases rather recently, for example, some formyl peptide receptors in rodents, but not in other mammals (Dietschi et al. 2017).
For most of the main odor classes, we have a good understanding of their respective receptors. The OR families of tetrapods often recognize volatile that is, hydrophobic ligands, whereas the TAAR families of both tetrapods and teleost fish appear to be specialized for detection of amines (Korsching 2020). Tetrapod and teleost V1R/ORA are assumed to detect pheromones, whereas the V2R/OlfC family appears to detect peptide pheromones in tetrapods, but amino acids (food odors) in teleosts and amphibians (Korsching 2020). Another important component of food odors in aquatic animals are nucleotides/nucleosides, but for a long time the molecular nature of their olfactory receptors was unknown. There are three well-described purinergic receptor families (metabotropic P1 and P2Y, ionotropic P2X; Fredholm et al. 2011), but none of their genes seems involved in olfactory detection of nucleotides or nucleosides in zebrafish (Wakisaka et al. 2017). However, the same study identified a novel gene as an olfactory adenosine receptor. This gene was found to be related to adenosine receptors type A, adora genes, and the name A2C, for adora2c, was suggested (Wakisaka et al. 2017). A preliminary phylogenetic analysis has detected the presence of this gene in many other vertebrate species (Wakisaka et al. 2017), but its evolutionary origin is currently unknown, and it is also unclear at what point in its evolutionary lineage it acquired an olfactory function.
Here, we report that this novel gene is already present as a sister clade to adora genes in the common ancestor of chordates and echinoderms that is, close to the origin of deuterostomes (one of two superphyla in bilateral animals), which prompted us to (re)name the gene clade adorb (adenosine receptors B), in close analogy to the accepted naming of the adenosine receptor A genes (adora). Interestingly, a role as olfactory receptor seems to have been acquired only much later in evolution: we observed an olfactory receptor-like expression pattern for the adorb gene in several teleost and one amphibian species, but not in lamprey, consistent with adorb first becoming an olfactory receptor in jawed vertebrates. The expression pattern of adorb in lamprey is consistent with a role in differentiation of olfactory neuron progenitor cells.
Results
The Adenosine Receptor adorb (Synonym A2c) Is Already Present in the Common Ancestor of Chordates and Echinoderms, That Is, Close to the Origin of Deuterostomes
We have performed an in-depth phylogenetic analysis of the evolutionary origin of the adorb gene, making use of the recent availability of genomes representing many early-derived lineages within vertebrates (three jawless and three cartilaginous fish), chordates (five urochordates and four cephalochordates) and even the sister clades of chordates (10 echinoderms and two hemichordates). We report a single clade with very high branch support comprising orthologs for the zebrafish gene named A2c (Wakisaka et al. 2017) in reptiles, amphibians, teleost fish, ray-finned and lobe-finned fish, cartilaginous and jawless fish, cephalochordates, hemichordates, and echinoderms (figs. 1 and 2). At all evolutionary levels analyzed these zebrafish A2c-related genes form a separate sister clade to the adora clade encompassing the well-known adora1, adora2a, adora2b, and adora3 genes. This result differs from that of a preliminary study that was restricted to vertebrates, used neighbor-joining algorithm for construction of the gene tree, and a less diverse outgroup (Wakisaka et al. 2017). We have employed well over 100 genes representing all major subgroups of rhodopsin class G protein-coupled receptors (GPCRs) and even rhodopsin-associated genes including many genes from early-diverging species as outgroup (see supplementary files 1–3, Supplementary Material online). In our experience, it is essential to use large and diverse outgroups in the phylogenetic analysis to obtain a robust tree topology. We consequently suggest renaming the adora2c (A2C) gene as adorb for adenosine receptor B to accurately reflect the phylogenetic position and distance to the adenosine receptor A clade (adora). An analysis of splice sites confirms the inferred segregation of adora and adorb genes: all vertebrate adora genes exhibit the same, conserved splice site, whereas adorb genes are mostly monoexonic, with occcasional intron gains at positions differing from that of adora genes (figs. 1–3, and supplementary file 2, Supplementary Material online).
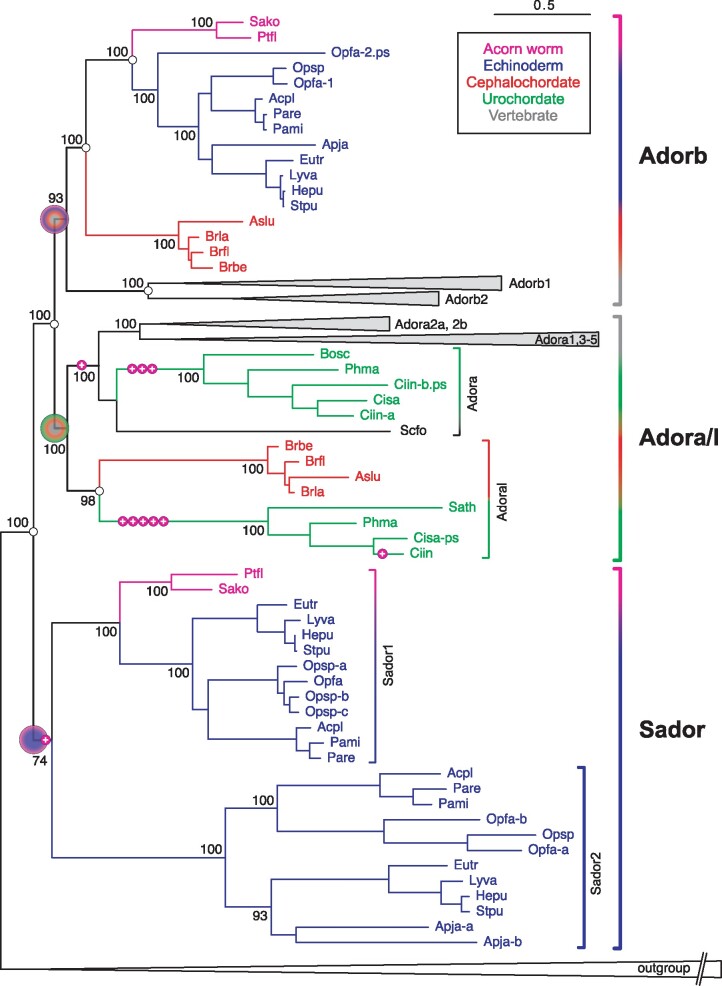
Fig. 1.
The evolutionary origin of the nucleotide receptor adorb in the MRCA of deuterostomes. Maximum likelihood tree for adora, adorb, and related receptors. Branch support is given in %, scale bar denotes number of amino acid substitutions per site. Species groups are color-coded as indicated, all vertebrate clades are collapsed. Magenta circles with plus sign indicate intron gains, small empty circles denote absence of introns. Gene clades are visualized by angular parentheses, color-coded according to the species represented in that clade. Species are indicated by four letter abbreviation, for full names see supplementary file 3, Supplementary Material online. The root of the adora, adorb, and sador clade is denoted by large circles, color-coded according to species represented within that clade. Adora, adoral, adorb, and sador clades are well separated with high to maximum branch support. Adorb is present in all deuterostome groups analyzed except urochordates. The outgroup (collapsed) consists of over 100 genes encompassing all subgroups of rhodopsin class GPCRs as well as rhodopsin-associated class genes (see supplementary file 3, Supplementary Material online).
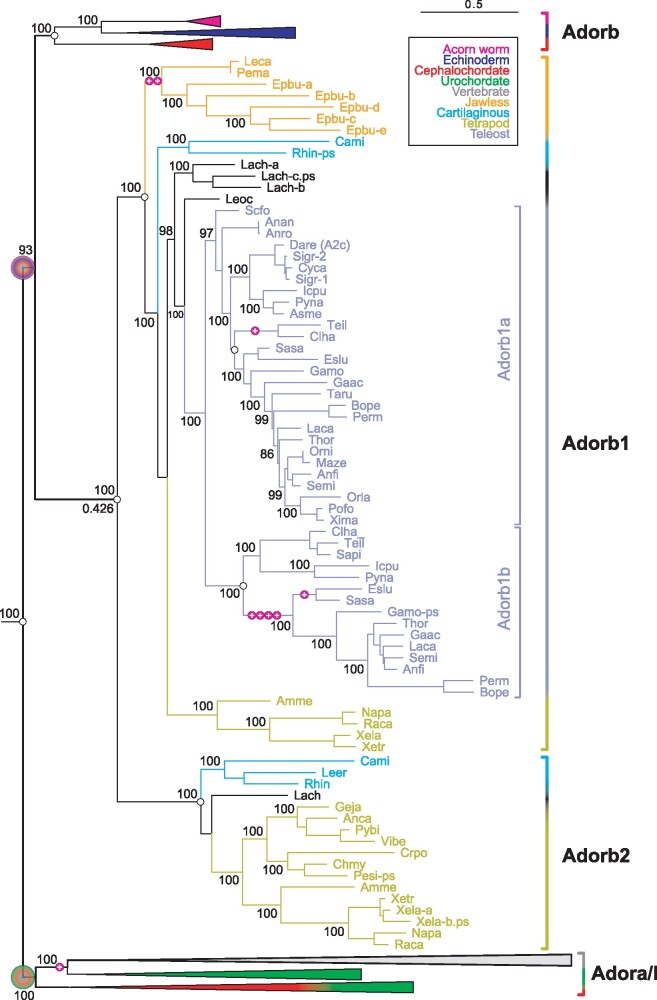
Fig. 2.
Evolution of the adorb gene clade in vertebrates. Maximum likelihood tree for adora, adorb, and related receptors, same tree file as figure 1, only vertebrate adorb clade expanded here. Branch support is given in %, for the root of the adorb clade also the common branch length is given (0.426); scale bar denotes number of amino acid substitutions per site. Species groups are color-coded as indicated, all nonvertebrate nodes and the vertebrate adora node are collapsed. Magenta circles with plus sign indicate intron gains; small empty circles denote absence of introns. Gene clades are visualized by angular parantheses, color-coded according to the species represented in that clade. Species are indicated by four letter abbreviation, for full names see supplementary file 3, Supplementary Material online. The outgroup (not shown) consists of over 100 genes encompassing all subgroups of rhodopsin class GPCRs as well as rhodopsin-associated class genes (see supplementary file 3, Supplementary Material online). The root of the adora and adorb clade is denoted by large circles, color-coded according to species represented within that clade. The vertebrate adorb clade is well separated with maximum branch support. Coelacanth (Lach) and hagfish (Epbu) show small gene expansions, the remaining increases in gene number result from two whole genome duplications or polyploidy (Sigr, Xela).
The tree topology of the adorb clade generally reproduces the known species relationships faithfully, with hemichordate (acorn worms) and echinoderm (sea stars and sea urchins) adorb genes forming a common sister clade to chordates. Within chordates, cephalochordates (lancelets) form a sister clade to vertebrates, and within vertebrates, jawless fish adorb genes are basal to those of cartilaginous fish, which themselves are basal to bony fish orthologs. Thus, the origin of the zebrafish adenosine receptor adorb (formerly A2C, Wakisaka et al. 2017) can be traced back to the MRCA of deuterostomes that is, well beyond the chordate branch of the tree of life.
Rare Gene Duplications and Several Losses Characterize the Evolution of the adorb Gene
To better understand the evolution of the adorb gene, we have also searched for its closest homologs outside of the adora/adorb clade. The adorb gene is distinct from adora as far back as the MRCA of deuterostomes (see above), but both genes share a common ancestor, which may be named ador as it is the ancestor of both adora and adorb genes; fig. 1 and supplementary files 1–4, Supplementary Material online). Our search for the origin of this deduced ador gene has identified a sister gene, sador (sister of ador) in all 12 ambulacraria genomes analyzed (two hemichordates and ten echinoderms), which together form the sister group of chordates. Despite extensive searches, no chordate orthologs could be identified, suggesting a loss of sador early in the chordate lineage. Sador itself has undergone an early duplication event in ambulacraria resulting in sador1 and sador2, the latter not found in either of the two acorn worm genomes available that is, presumably lost. sador1 and sador2 mostly have remained singletons in ambulacraria, except two gene duplications in a starfish sador1 (Ophiothrix spiculata) and one gene duplication each in sador2 from another starfish and a sea cucumber (Ophionereis fasciata and Apostichopus japonicus, respectively). Ligands for sador genes are unknown.
The sador/adora/adorb monophyletic clade is supported with maximal branch support and a common branch of considerable length (0.34 amino acid substitutions/site), segregating it clearly from other rhodopsin alpha group genes including aminergic neurotransmitter receptors, as well as from the other rhodopsin family subgroups (beta, gamma, and delta, Fredriksson et al. 2003), which were used as further outgroup (fig. 1 and supplementary files 1–3, Supplementary Material online).
Within the adorb node, we identified echinoderm, ambulacraria, cephalochordate, and vertebrate representatives, but never observed any urochordate genes (urochordate adora genes were localized easily). We conclude that adorb has been lost in urochordates, which are closer related to chordates than cephalochordates (Delsuc et al. 2006). Within vertebrates adorb has duplicated to adorb1 and adorb2, presumably as a consequence of a whole genome duplication in early vertebrates (Nakatani et al. 2007), since in species with chromosome annotation these two genes lie on different chromosomes (data not shown). Adorb1 and adorb2 show very diverging evolutionary histories. Adorb1 is present in jawless fish (three species), cartilaginous fish (two species), coelacanth, a ray-finned fish, all representative teleost fish species analyzed (28 species), and in amphibians (five species), but absent in reptiles, suggesting widespread conservation, but a loss in terrestrial species. Adorb2 is not found in jawless fish, absent in all ray-finned fish and teleosts analyzed, but present in cartilaginous fish (three species), coelacanth, amphibians (five species), and reptiles (seven species), suggesting losses in jawless and in ray-finned fish, and a somewhat later loss inside the terrestrial lineage (no avian or mammalian orthologs were found). The whole genome duplication in the MRCA of teleost fish (Nakatani et al. 2007) led to another duplication of adorb1 into adorb1a and adorb1b, which again have a very different evolutionary history. Although adorb1a has been retained in all but one teleost species (and the absence in the sardine genome may well have technical reasons), adorb1b has been lost repeatedly in different sublineages. Interestingly, the adorb1b gene has gained four introns in the MRCA of Euteleosteomorpha and a fifth in salmonids, but has remained intronless in Otomorpha (herring, sardine, catfish, and piranha), whereas herring/sardine adorb1a is the only adorb1a gene with an intron gain (fig. 2). Two unrelated intron gains have occurred in the MRCA of jawless fish in adorb1 (fig. 2).
A Thorough Delineation of the adora Family Reveals Two Novel Clades
Unexpectedly, our control searches for adora genes revealed the presence of two novel clades, adora4 and adora5 (fig. 3). To substantiate our results for the adorb clade being separate from all adora genes, we considered it essential to perform a thorough delineation of the adora clade in all basal and some of the derived species we had examined for the presence of adorb orthologs. Adora1, 2a, 2b, and 3 were identified in a ray-finned and a lobe-finned fish (spotted gar and coelacanth [minus adora3], respectively) as well as in early-derived (dragonfish, eel, and zebrafish) and later-derived (salmon, medaka, and stickleback) teleost fish (fig. 3). Some species-specific, late gene births were observed, as well as partial retention of genes duplicated in the whole genome duplication occurring in the MRCA of teleost fish (Nakatani et al. 2007), in particular for adora1 and adora2a (fig. 3). Additionally, the polyploidization of salmon increased the total number of adora1–3 genes to 14. Adora1, -2a, -2b, and -3 genes were present in an amphibian (clawed frog), a reptile (python), and a bird (chicken), consistent with earlier reports (Wakisaka et al. 2017).
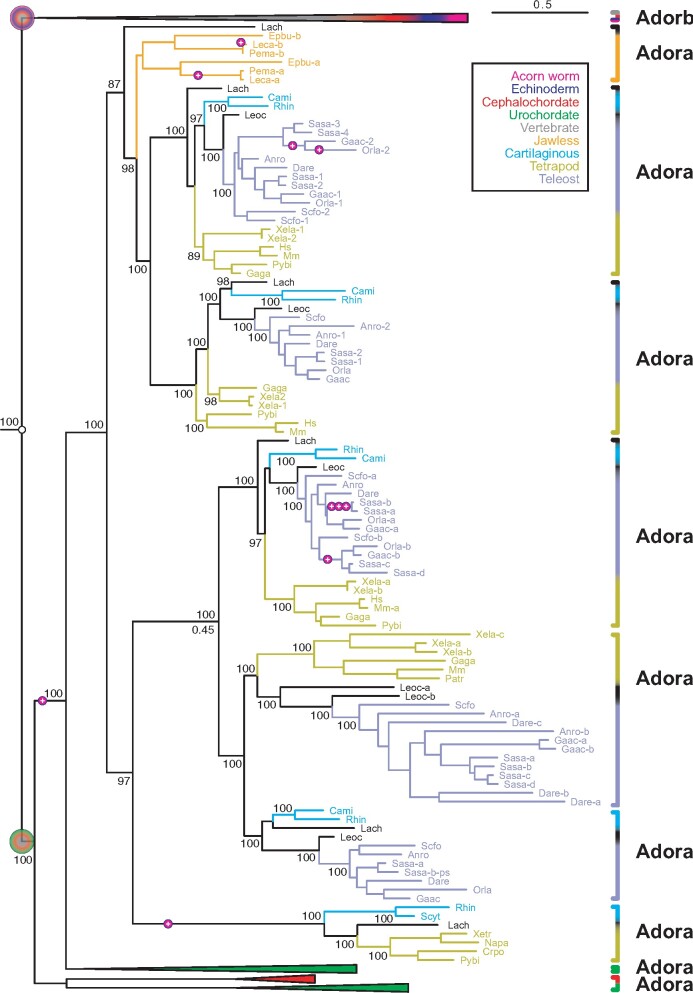
Fig. 3.
Two novel adora genes in vertebrates. Maximum likelihood tree for adora, adorb and related receptors, same tree file as figure 1, only vertebrate adora clade expanded here. Branch support is given in %, scale bar denotes number of amino acid substitutions per site. Species groups are color-coded as indicated; all nonvertebrate nodes and the vertebrate adorb node are collapsed. Magenta circles with plus sign indicate intron gains; small empty circles denote absence of introns. Gene clades are visualized by angular parantheses, color-coded according to the species represented in that clade. Species are indicated by four letter abbreviation, for full names see supplementary file 3, Supplementary Material online. The outgroup (not shown) consists of over 100 genes encompassing all subgroups of rhodopsin class GPCRs as well as rhodopsin-associated class genes (see supplementary file 3, Supplementary Material online). The root of the adora and adorb clade is denoted by large circles, color-coded according to species represented within that clade. The vertebrate adora clade is well separated with maximum branch support. Orthologs for mammalian genes adora1, 2a, 2b, and 3 are present in teleosts, for the first three also in cartilaginous fish. Two novel clades, adora4 and 5, are present in cartilaginous fish, lobe-finned fish, and ray-finned fish, respectively amphibians. Remnants of the teleost-specific whole genome duplication are visible, some other increases in gene number result from polyploidy, for example, salmon.
A fifth clade is apparent in the phylogenetic tree as sister clade to adora3 and was named adora4. This clade contains shark, coelacanth, spotted gar, and teleost fish orthologs, but seems to have been lost in tetrapods, as no amphibian, reptilian, or avian representative could be uncovered (fig. 3). Finally, we describe another novel clade basal to adora1, 3, 4, which we named adora5. This clade contains shark, coelacanth, amphibian, and reptilian representatives, but seems to have been lost in ray-finned fish, birds, and mammals (fig. 3). Ligands for adora4 and adora5 are unknown but may be expected to include adenosine, which is the physiological ligand both for the four previously described adora genes (Fredholm et al. 2011) and the recently described zebrafish adorb gene (Wakisaka et al. 2017, here named A2c).
All gene birth events leading to adora1–5 have taken place within the vertebrate lineage, but the ancestral adora gene can be followed back further to the MRCA of urochordates and vertebrates (figs. 1 and 3). A sister gene (adora-like, adoral) is present in urochordates as well as cephalochordates, but not in vertebrates, consistent with a loss of adora in cephalochordates and a loss of adoral in vertebrates (figs. 1 and 3). Ligands for adoral genes are unknown, but could include adenosine, the ligand for adora and adorb genes (Fredholm et al. 2011; Wakisaka et al. 2017). No ambulacraria adora genes were detected despite intensive searches, consistent with a loss of adora in ambulacraria or, alternatively, with a later origin of adora as gene duplication of an earlier adorb gene within the chordate lineage.
An Olfactory Receptor-like Expression Pattern of adorb1 in Representative Teleost and Amphibian Species
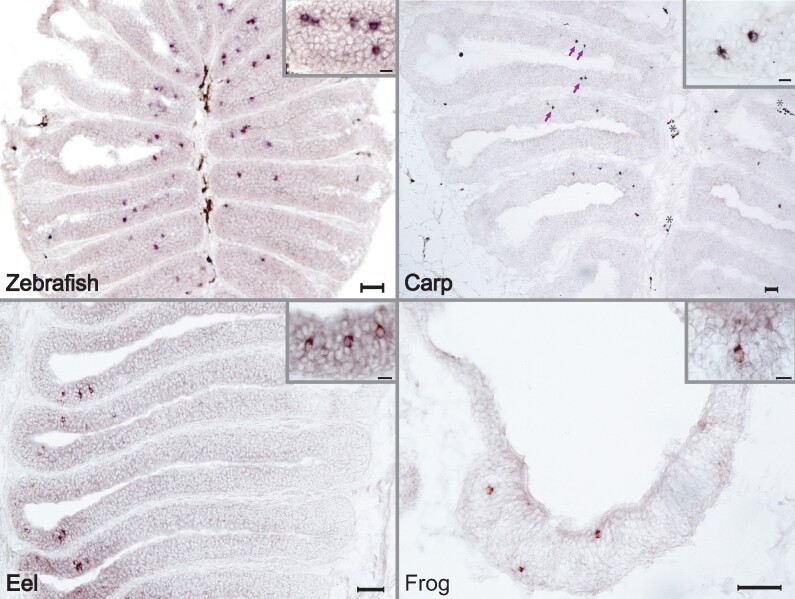
Next, we wished to analyze the expression pattern of adorb in representative species. The zebrafish adorb1a gene has been shown to be expressed in sparsely distributed olfactory receptor neurons (Wakisaka et al. 2017), the characteristic spatial expression pattern of an olfactory receptor (Korsching 2020). We chose common carp as a close relative to zebrafish, European eel as a rather distantly related and much earlier-diverged teleost, and the African clawed frog as an amphibian representative. Zebrafish was included as control. For all four species, we performed in situ hybridization (ISH) on cryostat sections of the olfactory organ. We observe sparsely distributed labeled cells in each of the olfactory organs (fig. 4), in the typical pattern observed for olfactory receptor genes (Weth et al. 1996; Syed et al. 2013; Korsching 2020), suggesting that the adorb1 gene has served an olfactory function throughout the evolution of bony fish, from the MRCA of the ray-finned (carp, eel, and zebrafish) and the lobe-finned (frog) lineage, about 430 Ma (Kumar et al. 2017), up to its loss in nonamphibian tetrapods. A quantitative analysis of the spatial distribution of adorb1-expressing cells in zebrafish shows the expression domain of adorb1 to be of comparable width with those of OR genes, with a center of gravity rather close to the center of the olfactory epithelium, and within the range covered by OR genes (supplementary files 1 and 6, Supplementary Material online). Thus, zebrafish adorb1 expression conforms to the spatial pattern observed for and characteristic for other olfactory receptor gene families (Weth et al. 1996; Ahuja et al. 2018).
Fig. 4.
Across a wide evolutionary range adorb is expressed in sparsely distributed cells within the olfactory epithelium. Species-specific adorb1 probes were used to perform chromogenic ISH of horizontal cryostat sections of olfactory epithelia for three fish (zebrafish, top left panel; carp, top right panel; eel, bottom left panel) and one amphibian species (clawed frog, bottom right panel). Insets at higher magnification; scale bars in main panels 50 µm, in insets 10 µm.
Lamprey adorb1 Is Expressed in a Nonsensory Region of the Olfactory Organ, but Absent in Olfactory Sensory Neurons
We then went on to investigate whether the lamprey adorb1 gene would also show an olfactory expression pattern. Much to our surprise, we found a completely different situation than that observed for bony fish and frog. The lamprey adorb1 gene is indeed expressed in the nose, but as one contiguous region within a lamellae, which appeared to be adjacent to the sensory area (fig. 5A and B). Together, these regions form an outer “ring.” In some cases, a second zone of expression is observed close to the inner tip of lamella (fig. 5C), and in rare cases additional intermediate zones are seen (fig. 5D). In all cases, expression is in contiguous areas, not in isolated cells. For comparison, a lamprey olfactory receptor (Berghard and Dryer 1998) shows the sparsely distributed expression pattern typical for olfactory receptor genes (fig. 5E). The accessory olfactory system (Ren et al. 2009, Green et al. 2017) does not express adorb1 (supplementary file 5, Supplementary Material online).
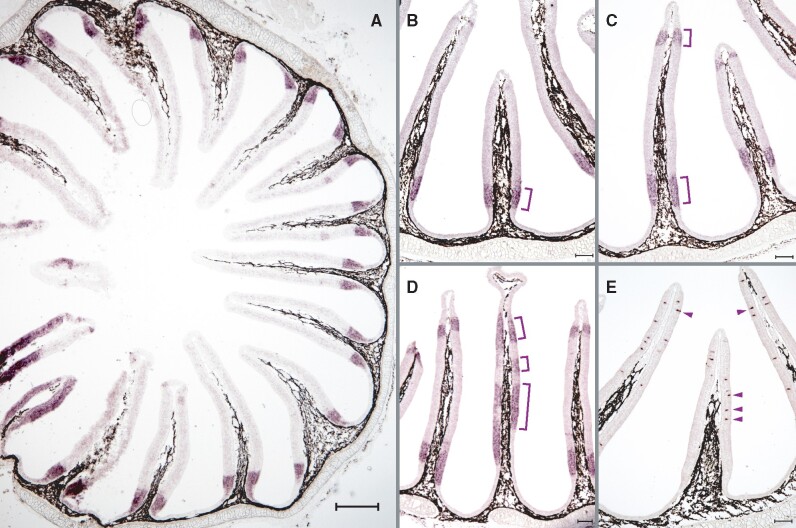
Fig. 5.
Expression of lamprey adorb in contiguous segments of the nose is distinctly different from that observed in jawed vertebrates. ISH was performed in horizontal cross-sections of the olfactory organ of L. fluviatilis using NBT/BCIP for visualizing adorb1 (A–D) and tarl7a (aka Lor3) expression (E). Adorb1 is expressed in a contiguous region adjacent to the olfactory sensory area (A, B), in some cases close to the inner tip (C) and rarely in intermediate zones (D) of the OE. In contrast, a known olfactory receptor (Lor3, Berghard and Dryer 1998), is sparsely distributed in the olfactory sensory area (E). Scale bar (A) 300 µm, (B–E) 100 µm.
The unusual expression pattern of lamprey adorb1 suggests that it is not expressed in olfactory sensory neurons. To substantiate this hypothesis, we performed double labeling with the panneuronal marker HuC (Kim et al. 1996) and established markers for ciliated and microvillous neurons, the two main subpopulations of olfactory sensory neurons in vertebrates, including lamprey (Laframboise et al. 2007; Korsching 2020). The HuC antibody staining delineates the known sensory region of the olfactory lamella, all of which is negative for adorb1 expression (fig. 6). Even at high magnification no coexpression is seen (fig. 6). We then proceeded to perform double ISH for adorb1 and golf, a marker for ciliated neurons (fig. 7). golf has been shown to label a subpopulation of OSNs also in lamprey and thus demarkates the sensory area also in this species (Frontini et al. 2003). The expression zones for golf and adorb1 are adjacent, but clearly segregated, and no overlap is seen. As a control, double-labeling was performed with golf and HuC. As expected, there was extensive overlap of staining observed (fig. 7), confirming the suitability of Golf to delineate the sensory area. Next, we performed double ISH for adorb1 and trpc2, an accepted marker for microvillous neurons of bony vertebrates (fig. 7). Unexpectedly, trpc2 expression was not restricted to the sensory segment as defined by HuC labeling and thus does not seem to be a specific marker for microvillous olfactory neurons in lamprey, in contrast to bony vertebrates (Sato et al. 2005). Nevertheless, we observe no overlap of trpc2 with adorb1 expression at the cellular level (fig. 7).
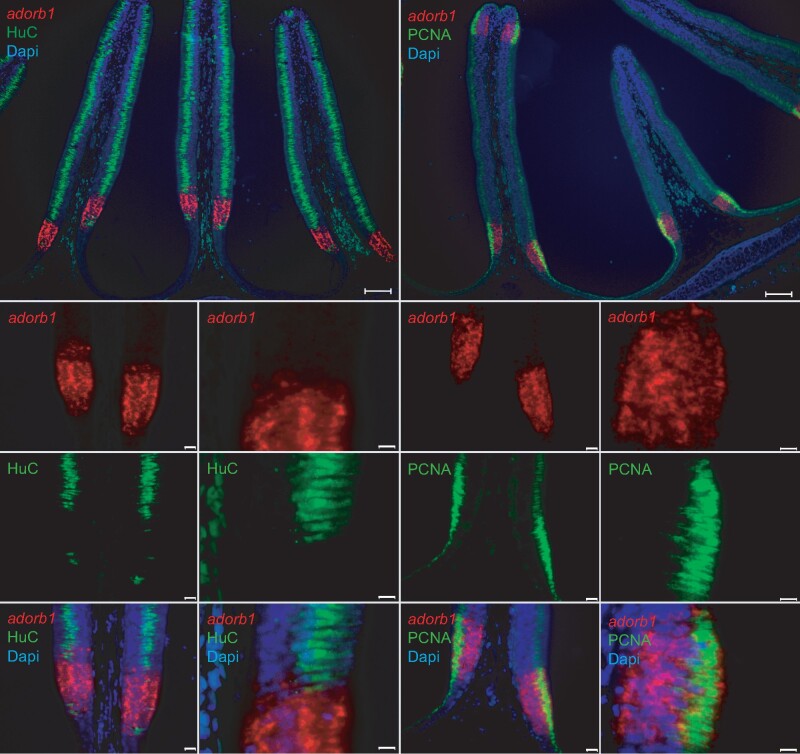
Fig. 6.
Lamprey adorb1 expression zone borders the sensory area of the olfactory organ. Double labeling of horizontal cross-sections of L. fluviatilis olfactory epithelium with HuC antibody and PCNA antibody confirms the expression of adorb1 (visualized by ISH) in a nonsensory region. Genes and color as indicated on the respective panels, nuclei are visualized by DAPI staining (blue). The neuronal marker HuC (left columns, green) labels the olfactory sensory area of the lamella, whereas PCNA (right columns, green) labels the adjacent mitotic regions. Adorb1 (red) is expressed in a contiguous manner in the PCNA-positive region, but absent from the sensory region. Note that PCNA labeling (green) is restricted to the apical layer, whereas adorb1(red) staining is distributed more broadly. Top panels show an overview (scale bars 100 µm); the smaller bottom panels are enlarged (scale bars 50 and 10 µm, respectively).
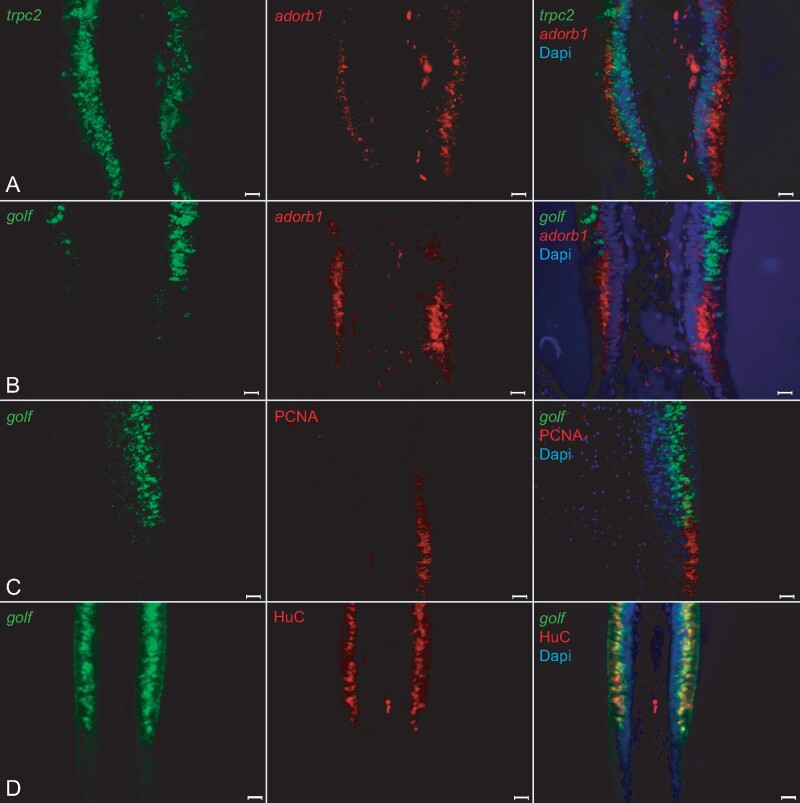
Fig. 7.
Adorb1-expressing cells in lamprey are negative for several neuronal markers. Horizontal cross-sections of L. fluviatilis olfactory epithelium were double labeled with OSN markers in several combinations; nuclei are visualized with DAPI. Left and middle columns, single label; right column, merged pictures. Orientation of panels: the adorb1-positive region is always below the adjacent sensory region. All scale bars are 20 µm. (A, B) Double ISH with neuronal markers for microvillous and ciliated OSNs (trpc2, and golf, respectively) and adorb1 shows no coexpression with either marker. (A) Unexpectedly, trpc2 (green) is expressed both in the sensory region and the adorb1-positive (red) segment. However, no double-labeled cells are seen. The nature of the trpc2-positive cells in the nonsensory region is unclear. (B) golf (green) is expressed in the sensory area, and exclusive of adorb1 expression (red). (C, D) Double labeling of golf with HuC and PCNA antibodies shows extensive overlap of golf and HuC at the cellular level, and complete absence of overlap for golf and PCNA, as expected.
In summary, adorb1-positive cells express neither a general neuronal marker nor markers characteristic for ciliated and microvillous neurons, and thus are clearly nonneuronal, in sharp contrast to the expression patterns observed in bony fish (figs. 4–7 andsupplementary files 1 and 6, Supplementary Material online).
Lamprey adorb Expression Is Consistent with Expression in Neural Progenitor Cells
So, what are these adorb1-expressing cells? Interestingly they are present in a region directly adjacent to the neuronal segment of the olfactory lamellae, suggesting this region to be a potential source of newly born olfactory receptor neurons. Similar mitotic zones have been shown in the zebrafish olfactory organ and migration from this zone into the sensory region of the nose has been demonstrated (Oehlmann et al. 2004; Bayramli et al. 2017). The morphology of the lamprey olfactory organ is somewhat different from that of jawed fish, as it consists of a single, medially fused structure. However, olfactory lamella that are fused to outer nonsensory structures and merge in the center of the organ occur in lamprey as well as jawed fish, and topology-wise the outer and inner adorb1-expressing regions would correspond to the outer and inner “ring” of mitotically active cells in the zebrafish nose (Oehlmann et al. 2004). In zebrafish, these rings express proliferating cell nuclear antigen (PCNA), and no overlap with the adorb1-expressing sensory region is seen (supplementary files 1 and 7, Supplementary Material online). In contrast, in lamprey double labeling with PCNA stains exactly those segments of the olfactory lamella, which are expressing adorb1 (fig. 6 and supplementary files 1 and 7, Supplementary Material online). Nevertheless, at the cellular level, there is little overlap, with PCNA-expressing cells forming a contiguous apical layer, whereas the adorb1 expression is centered in more basal layers of the lamella (fig. 6). PCNA is essential for replication and an established marker for neuroepithelial cells (Ino and Chiba 2000; Bayramli et al. 2017), that is, a marker of stem cells in the neuronal lineage. Adorb1-expressing cells might represent a different (later) stage in the neuronal lineage, but our attempts at identifying suitable markers for such stages in lamprey have so far not been successful. However, the close association with PCNA-expressing cells and the absence of neuronal markers in adorb1-expressing cells is consistent with these cells representing an intermediate stage in the differentiation from neuroepithelial stem cells to mature neurons.
Discussion
Nucleotides and nucleosides constitute important food-derived odors for aquatic organisms including fishes. The recent description of the first olfactory receptor for a nucleoside (Wakisaka et al. 2017) prompted us to investigate the evolution of this receptor and its olfactory function as estimated by the presence or absence of the characteristic expression pattern—sparsely distributed cells within the sensory region of the olfactory epithelium. We report that the receptor itself is very ancient and already present in the common ancestor of deuterostomes, since it is found in hemichordates, echinoderms, cephalochordates, and vertebrates, in the latter as a sister clade to the adora gene family (fig. 8). In contrast, the adora genes are only observed in vertebrates and their closest relatives, the urochordates, consistent with either a loss in hemichordates, echinoderms, and cephalochordates, or a later birth of adora in the MRCA of vertebrates and urochordates. Thus, we suggest to replace the initial designation (adora2c; Wakisaka et al. 2017) with adorb (adenosine receptor b), which appears better suited to the phylogenetic position as sister clade to adora (adenosine receptor a).
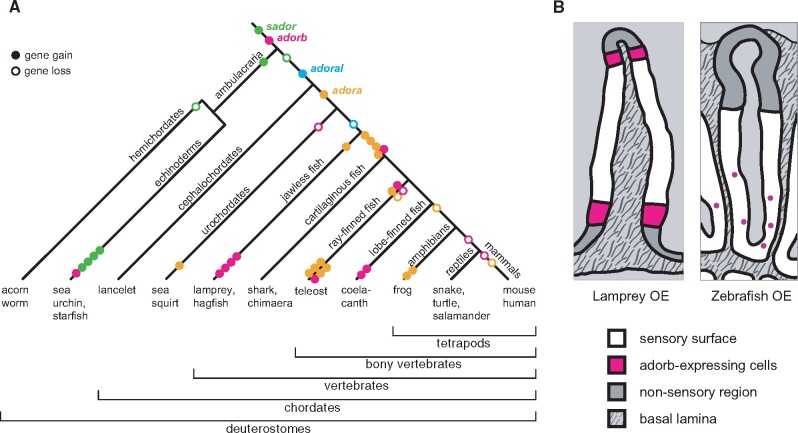
Fig. 8.
Evolutionary dynamics of adorb and related clades and transition in adorb expression patterns between jawless and jawed fish. (A) Dynamic evolution in the adora, adorb and related clades inferred from the phylogenetic analysis. The evolutionary history is depicted using maximum parsimony. Genes are color-coded as indicated and gene name is given at the birth of the respective clade. Each gene duplication event is shown as filled circle, an empty circle denotes a gene loss in the respective species group. Late gene duplication events are shown close to the tips, species-specific gene losses are not shown to keep the figure legible. Gene duplications resulting from a whole genome duplication are counted as one event. (B) Schematic representation of adorb expression in lamprey and zebrafish. Cross sections of a lamella are shown for lamprey (left panel) and zebrafish (right panel). Magenta, cells expressing adorb; white, sensory surface; light gray, lumen; medium gray, nonsensory region; stippled gray, basal lamina.
We identified several related genes in the nonvertebrate clades analyzed, which we named according to their phylogenetic position, adoral for adora-like gene, and sador for sister of ador, ador being the deduced ancestral gene for adora and adorb. The sador clade is present in ambulacraria (both echinoderms and hemichordates), but absent from chordates, and the adoral clade is present inside the chordate lineage (both cephalochordates and urochordates), but absent from vertebrates, consistent with losses of these genes in the respective lineages (fig. 8). Such an evolutionary dynamic parallels findings for other receptors when analyzing the lineage of a known vertebrate receptor family outside of vertebrates (see e.g., Ramos-Vicente et al. 2018).
As a proxy for a function as olfactory receptor gene, we examined the spatial expression of adorb genes in an array of vertebrate species. Most olfactory receptors, including the zebrafish adorb1a gene (Wakisaka et al. 2017) have a highly characteristic expression pattern consisting of isolated cells dispersed between cells expressing other olfactory receptors (Korsching 2020)—for lamprey see Berghard and Dryer (1998). We find such sparsely dispersed neurons expressing adorb in fishes closely (common carp) and not so closely (eel) related to zebrafish, as well as in a tetrapod (frog). This suggests that the function of the adorb gene as an olfactory receptor is at least as old as the MRCA of the ray-finned and the lobe-finned lineage, about 430 Ma (Kumar et al. 2017). In contrast, the contiguous expression of the lamprey adorb gene in a region adjacent to, but clearly different from the sensory surface, argues strongly against a function as an olfactory receptor (fig. 8). The regions expressing adorb in lamprey are colabeled with a marker for proliferating cells (PCNA) and appear to be identical to the proliferative regions of the lamprey nose described in an earlier study (Thornhill 1970). Olfactory sensory neurons have a finite life time of a few weeks and have to be replenished regularly from dividing precursor cells throughout adult life in both mammals and fishes (Mackay-Sim and Kittel 1991; Ferrando et al. 2010). In zebrafish, it has been shown that immature neurons migrate from sensory-adjacent regions into the sensory surface (Oehlmann et al. 2004; Bayramli et al. 2017), so it is conceivable that the proliferative regions in the lamprey nose might serve the same purpose. Thus, the adorb expression pattern is consistent with a role in the development of olfactory sensory neurons. In this context, it is worth noting that purines (ATP and ADP, adenosine triphosphate and adenosine diphosphate, respectively) are in fact mitotic signals for olfactory stem cells in frog and fish (Hassenklöver et al. 2009; Demirler et al. 2020).
It is tempting to suggest that lamprey might represent the ancestral situation, in which the adorb gene may have served a function in proliferating precursor cells of the olfactory organ, and that at some time point after the divergence of jawed fish and before the emergence of ray- and lobe-finned fish a function as olfactory receptor may have been acquired. In another case of neofunctionalization—immune system peptide receptors of the formyl peptide receptor family became olfactory receptors in rodents—it seems to have been sufficient to come under control of transcription factors regulating olfactory receptor-like expression patterns (Dietschi et al. 2017). Even the beta adrenergic receptor, expressed as a knock-in under olfactory receptor promoter control, was able to generate a specific target glomerulus like proper OR genes (Feinstein et al. 2004).
Alternatively, the nonsensory expression of lamprey adorb could constitute the derived pattern, in analogy to what has been observed for several OR genes, whose coding regions came under regulatory control of nonolfactory genes, resulting in extra-nasal expression (Feingold et al. 1999). However, this appears less likely for several reasons, among them the function of the presumptive ligands ATP and adenosine as mitotic signals acting on proliferating progenitor cells (Hassenklöver et al. 2009; Demirler et al. 2020). A final answer will require expression analysis in a larger array of chordate, hemichordate, echinoderm, and additional early-derived vertebrate species.
In an initial attempt to shed light on the change in adorb function between jawless and bony fish, we analyzed the genomic surrounding of the adorb1 gene in bony, cartilaginous, and jawless fish. The adorb1 gene is situated inside the large fourth intron of the parp1 gene for spotted gar, zebrafish, medaka, and Western clawed frog (Wakisaka et al. 2017)—incidentally this has led to many misannotations of the adorb gene as parp1 by automatted gene prediction methods. We observed the same genomic location inside the fourth parp1 intron in whale shark, elephant shark, sea lamprey, and arctic lamprey (data not shown), the latter being a very close relative of the lamprey species we used for expression studies (Lampetra fluviatilis). However, chordate and ambulacraria adorb genes do not exhibit this association with parp1, which often only possesses small introns in these species (data not shown). Thus the genomic surrounding of adorb changed in vertebrates compared with chordates, but not between jawless and jawed vertebrates. In other words, the association with parp1 predates the olfactory function of the adorb gene, and consequently cannot explain the aquisition of olfactory function in jawed vertebrates.
Taken together, we have shown the presence of the adorb gene in the MRCA of ambulacraria and chordates, which makes this gene more ancient than even the OR genes, previously known as the most ancient olfactory receptor genes, which are limited to chordates (Churcher and Taylor 2009). However, the olfactory function itself appears to have been acquired much later in jawed vertebrates, because the jawless vertebrate lamprey expresses adorb in nonolfactory cells, albeit directly adjacent to the olfactory region (fig. 8). The evolutionary process converting the olfactory-adjacent contiguous expression in lamprey to the expression in scattered olfactory sensory neurons in jawed vertebrates remains to be elucidated.
Materials and Methods
Animal Handling
Experiments were performed with tissue from adult river lampreys (L.fluviatilis) in the anadromous stage, African clawed frog (Xenopus laevis) in the larval stage 50–54, juvenile Common carp (Cyprinus carpio), juvenile European eel (Anguilla anguilla), and adult zebrafish (Danio rerio) of either sex. Lampreys were kept at 4 °C and zebrafish at 28 °C both with a 12 h/12 h light/dark cycle. Animal housing and maintenance was licensed by the office for environment and consumer protection of the city of Cologne, Germany. Eel and carp were anesthetized and dissected direct after delivery. The experimental procedures were approved by the Federal ministry for nature, environment and consumer protection of Nordrhine-Westfalia, Germany, and the local ethics committee in Stockholm (Northern Stockholm Animal Review Board), Sweden, and were in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (1996 revision).
Dissection of Olfactory Tissue
Lamprey
Lampreys were handled according to (von Twickel et al. 2019). In short, they were anesthetized with MS-222 (ethyl 3-minobenzoate methanesulfonate; 100 mg/l water; Sigma, St Louis, MO) dissolved in water from the aquarium at 4 °C. Animals were then decapitated with a scalpel caudal to the gills and moved to a preparation tray on ice (containing Sylgard gel; Dow Corning, Midland, MI) filled with 4 °C artificial cerebral spinal fluid, with the following composition (in mM): NaCl 125, KCl 2.6, MgCl2 1, CaCl2 2, NaHCO3 25, and Glucose10. After dissection of an olfactory epithelium, it was placed for 17–40 h in 4% paraformaldehyde (PFA), then 3 h in 20% sucrose, and 15 min in Tissue-Tek O.C.T. Compound before freezing.
Zebrafisch, Carp, and Eel
Before the preparation of olfactory epithelia, the fish was anesthetized with the appropriate amounts of MS-222 (300 mg/l for zebrafish and carp, double the amount for eel). After this the fish was decapitated with a sharp razor blade and the head was fixed from the top with a sterile forceps in a petridish with prechilled phosphate-buffered saline (PBS) ph 7.4. To simplify the removal of the olfactory epithelia, both eyes were taken out and the gills were pulled into an anterior direction. Next, the skin at the top of the head and around the nose holes was removed to expose the olfactory epithelia. Then as much cartilage as possible was removed around the olfactory epithelia before final removal. For ISH, the complete epithelia were stored in Tissue-Tek O.C.T. Compound at −80 °C. In case of immunohistochemistry (IHC), the epithelia were fixed in 4% PFA in PBS pH 7.4 for 5 min and then stored in tissue tec-OCT Compound at −80 °C.
Frog
Frog olfactory tissue was prepared as published in Syed et al. (2017).
mRNA Isolation, cDNA Synthesis, and ISH Probes
After dissection, noses were immediately placed in RNALater (Sigma-Aldrich). Total mRNA was extracted using the easy-spin Total RNA Extraction Kit (iNtRON Biotechnology) resulting in highly concentrated mRNA (OD260 > 0.9). Complementary DNA (cDNA) was synthesized using SuperScript II Reverse Transcriptase (Invitrogen, No. 18064022), with forward (FW) and reverse (RV) primers for probe synthesis as listed (table 1). The T3 promoter sequence (ATTAACCCTCACTAAAGG) was added 5′ to the RV primer for generation of RNA probes. The lamprey primers were originally developed based on a cDNA of Petromyzon marinus and tested in L.fluviatilis. For tarl7a (aka Lor3) a previously published primer-pair was used (Berghard and Dryer 1998). All PCR products were confirmed by sequencing.
Table 1.
Primer Sequences
| Species | Gene | Forward 5′–3′ primer | Reverse 5′–3′ Primer | No. of bp |
|---|---|---|---|---|
| Carp | adorb1 | TCTACGCGCAGATCTTCGTC | TTGTTCATCGCCATCACCGA | 362 |
| Eel | adorb1 | AACAGGAAGATGCACACCGT | ATGAGGAAGGACAGGACCCA | 308 |
| Frog | adorb1 | TTAACGTCTCTGTCCGTGGC | AGAGGGGTTTTATGCCAGCC | 314 |
| Zebrafish | adorb1 | TTCGTCACGGTTAAGCGTCA | ATTCCCCTGCAGCACATGAA | 329 |
| Lamprey | adorb1 | TCTACGCGCAGATCTTCGTC | TGTTCATCGCCATCACCGA | 530 |
| Lamprey | Golf | ATTTTTCGACCACGTGAAGG | AGGATGATCGAGATGGTTCG | 432 |
| Lamprey | trpc2 | GAGTGCAAGGAGGTGTGGAT | ATGAGCATGTTGAGCAGCAC | 599 |
| Lamprey | tarl7a | CCGCAACGCGTGGTCCTGAT | TCCTAAAGTTGAATAGATCCGTC | 961 |
In table 1, species, genes, and primer direction as indicated. Number of bp denotes the length of the PCR product used for generation of the RNA probes.
ISH and ISH Combined with IHC
Ten-micrometer cryosections of olfactory epithelia were placed onto Superfrost Plus slides. ISH and IHC were performed as described (von Twickel et al. 2019). Primary antibodies used in the IHC are mouse anti-PCNA (1:200, Merck) and mouse anti-HuC (1:200, Invitrogen). A strong PCNA signal in proliferating neuronal precursor cells is easily distinguished from very faint labeling of neuronal cells, similar to observations by (Ino and Chiba 2000). The HuC signal is highly specific for the neuronal region of the nose, whereas acetylated tubulin antibody (1:200, Sigma) stained both sensory and nonsensory regions of the adult lamprey nose (data not shown), in contrast to more specific staining of the olfactory sensory surface of larval lamprey (Frontini et al. 2003). 4′,6-diamidino-2-phenylindole (DAPI) was used as counterstain for fluorescent detection. Micrographs were taken using a Keyence BZ-9000 fluorescence microscope, and absence of crosstalk between channels was confirmed.
Data Mining and Phylogenetic Trees
Initial searches using zebrafish adorb and adora amino acid sequences as queries and tBLASTn (Gertz et al. 2006) in Whole Genome Shotgun (WGS) databases available at NCBI showed widespread presence of adorb in fish, amphibians, and reptiles. Representative species for detailed analysis of adorb evolution were selected as follows (numbers for adora after the slash): 3/3 jawless fish, 3/2 cartilaginous fish, 1/1 lobe-finned fish (Latimeriachalumnae), 1/1 nonteleost ray-finned fish (Lepisosteusoculatus), 28/6 early- and later-derived teleost fish, 5/1 amphibians, and 7/1 reptiles (supplementary files 1 and 3, Supplementary Material online). However, in case of a predicted gene loss all available genomes from the respective phylogenetic group were analyzed. To investigate the evolutionary origin of adorb and adora genes all nonvertebrate deuterostome genomes available at NCBI were examined—ten echinoderm, two hemichordate, four cephalochordate, and five urochordate genomes (supplementary files 1 and 3, Supplementary Material online). Blast hit regions were extracted with 0.5–1 kb flanking sequence. For monoexonic genes Expasy (Artimo et al. 2012) was then used to identify the appropriate open reading frame, for multiexonic genes genewise (Madeira et al. 2019) was used with close homologues as template. Most candidates could be obtained as full length, but all candidates down to 100 amino acid length were evaluated further. In few cases, gene predictions were made by splicing several contigs together. This was only done, when a template with high homology was available, and when no ambiguity was possible. Searches were performed recursively using more closely related genes as query once they had been validated in phylogenetic trees with reference and outgroup genes (see below). BLAST hits were ordered by decreasing % identity, and evaluated until several consecutive candidates fell into the outgroup. Candidate genes were validated in phylogenetic trees with known reference genes and a large outgroup consisting of representative members from all subgroups of rhodopsin (alpha, beta, gamma, and delta) and rhodopsin-associated receptors including genes from early-diverging species (Fredriksson et al. 2003; Nordström et al. 2011; supplementary files 1 and 3, Supplementary Material online). Phylogenetic tree construction was performed as described (Dieris et al. 2021). In short, MAFFT version 7 with E-INSI setting (Katoh and Standley 2013; Katoh et al. 2019) was used to generate the multiple sequence alignment, which was then degapped at 90% threshold. The tree was constructed using the maximum likelihood algorithm PhyML-aLRT with smart model selection (resulting in substitution model JTT+G + F), subtree grafting and chi square-based aLRT to estimate branch support (Dereeper et al. 2008; Guindon et al. 2010; Lefort et al. 2017).
Naming of Genes
Adora genes newly identified in this study were named according to phylogenetic relationship with already named genes, whenever possible. Adora genes outside the known clades of adora1, 2a, 2b, and 3 fell into two clades segregating with maximal branch support, which were named adora4 and adora5. An urochordate sister group of vertebrate+urochordate adora genes was named adoral, for adora-like. The zebrafish gene initially named a2c (adora2c; Wakisaka et al. 2017) belongs to a sister clade of [adora (adenosine receptor a) + adoral genes] and was therefore renamed as adorb (adenosine receptor b). Adora and adorb genes together possess a sister clade, which was named sador (sister of ador). Subdivisions of adorb, adoral and sador were named first by number and then by letter.
Inference of Gene Birth and Death Events
The topology of the gene tree was generally similar to that of the taxonomic tree, for example, within ambulacraria the hemichordate genes always formed a sister group to the echinoderms, and within vertebrates the jawless fish were more basal than cartilaginous than bony fish. Thus, it was mostly straight-forward to infer gene birth and death events by eye, and no program was used. In a few exceptions, we considered maximum parsimony to infer gene birth/death events, not the tree topology, since in our experience a small number of genes in a particular clade can distort its position in the tree (Dieris et al. 2021).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Mehmet Saltürk for taking good care of the fish. We are grateful to Katharina Dittrich, Arndt von Twickel, and Hans-Peter Bollhagen for tissue samples (frog and lamprey, respectively). Adnan Syed was involved in an early stage of the study.
Data Availability
The data underlying this article are available in the article and in its Supplementary Material online.
Literature Cited
- Ahuja G, et al. 2018. Overlapping but distinct topology for zebrafish V2R-like olfactory receptors reminiscent of odorant receptor spatial expression zones. BMC Genomics. 19(1):383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artimo P, et al. 2012. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40(Web Server issue):W597–W603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayramli X, Kocagöz Y, Sakizli U, Fuss SH.. 2017. Patterned arrangements of olfactory receptor gene expression in zebrafish are established by radial movement of specified olfactory sensory neurons. Sci Rep. 7(1):5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghard A, Dryer L.. 1998. A novel family of ancient vertebrate odorant receptors. J Neurobiol. 37(3):383–392. [PubMed] [Google Scholar]
- Churcher AM, Taylor JS.. 2009. Amphioxus (Branchiostoma Floridae) has orthologs of vertebrate odorant receptors. BMC Evol Biol. 9:242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H.. 2006. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 439(7079):965–968. [DOI] [PubMed] [Google Scholar]
- Demirler MC, et al. 2020. Purinergic signalling selectively modulates maintenance but not repair neurogenesis in the zebrafish olfactory epithelium. FEBS J. 287(13):2699–2722. [DOI] [PubMed] [Google Scholar]
- Dereeper A, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36(Web Server issue):W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieris M, Kowatschew D, Korsching SI.. 2021. Olfactory function in the trace amine-associated receptor family (TAARs) evolved twice independently. Sci Rep. 11(1):7807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietschi Q, et al. 2017. Evolution of immune chemoreceptors into sensors of the outside world. Proc Natl Acad Sci U S A. 114(28):7397–7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feingold EA, Penny LA, Nienhuis AW, Forget BG.. 1999. An olfactory receptor gene is located in the extended human beta-globin gene cluster and is expressed in erythroid cells. Genomics 61(1):15–23. [DOI] [PubMed] [Google Scholar]
- Feinstein P, Bozza T, Rodriguez I, Vassalli A, Mombaerts P.. 2004. Axon guidance of mouse olfactory sensory neurons by odorant receptors and the beta2 adrenergic receptor. Cell 117(6):833–846. [DOI] [PubMed] [Google Scholar]
- Ferrando S, et al. 2010. Cell proliferation and apoptosis in the olfactory epithelium of the shark Scyliorhinus canicula. J Chem Neuroanat. 40(4):293–300. [DOI] [PubMed] [Google Scholar]
- Fredholm BB, Ijzerman AP, Jacobson KA, Linden J, Müller CE.. 2011. International union of basic and clinical pharmacology. LXXXI. Nomenclature and classification of adenosine receptors—an update. Pharmacol Rev. 63(1):1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson R, Lagerström MC, Lundin LG, Schiöth HB.. 2003. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 63(6):1256–1272. [DOI] [PubMed] [Google Scholar]
- Frontini A, et al. 2003. Glomerular territories in the olfactory bulb from the larval stage of the sea lamprey Petromyzon marinus. J Comp Neurol. 465(1):27–37. [DOI] [PubMed] [Google Scholar]
- Gertz EM, Yu Y-K, Agarwala R, Schäffer AA, Altschul SF.. 2006. Composition-based statistics and translated nucleotide searches: improving the TBLASTN module of BLAST. BMC Biol. 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green WW, et al. 2017. Odorant organization in the olfactory bulb of the sea lamprey. J Exp Biol. 220(Pt 7):1350–1359. [DOI] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. [DOI] [PubMed] [Google Scholar]
- Hassenklöver T, Schwartz P, Schild D, Manzini I.. 2009. Purinergic signaling regulates cell proliferation of olfactory epithelium progenitors. Stem Cells. 27(8):2022–2031. [DOI] [PubMed] [Google Scholar]
- Ino H, Chiba T.. 2000. Expression of proliferating cell nuclear antigen (PCNA) in the adult and developing mouse nervous system. Brain Res Mol Brain Res. 78(1-2):163–174. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD.. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CH, et al. 1996. Zebrafish elav/HuC homologue as a very early neuronal marker. Neurosci Lett. 216(2):109–112. [DOI] [PubMed] [Google Scholar]
- Korsching SI.2020. Olfaction. In: Currie S, Evans DH, editors. The physiology of fishes. 5th ed. Bd. Chapter 14. Boca Raton (FL: ): CRC Press, Taylor & Francis Group. [Google Scholar]
- Korsching SI.2016. Aquatic olfaction. In: Zufall F, Munger SD, editors. Chemosensory transduction. New York, NY: Academic Press. p. 81–100. [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB.. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 34(7):1812–1819. [DOI] [PubMed] [Google Scholar]
- Laframboise AJ, Ren X, Chang S, Dubuc R, Zielinski BS.. 2007. Olfactory sensory neurons in the sea lamprey display polymorphisms. Neurosci Lett. 414(3):277–281. [DOI] [PubMed] [Google Scholar]
- Lefort V, Longueville J-E, Gascuel O.. 2017. SMS: smart model selection in PhyML. Mol Biol Evol. 34(9):2422–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay-Sim A, Kittel PW.. 1991. On the life span of olfactory receptor neurons. Eur J Neurosci. 3(3):209–215. [DOI] [PubMed] [Google Scholar]
- Madeira F, et al. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47(W1):W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, Takeda H, Kohara Y, Morishita S.. 2007. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 17(9):1254–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström KJV, Fredriksson R, Schiöth HB.. 2008. The Amphioxus (Branchiostoma Floridae) genome contains a highly diversified set of G protein-coupled receptors. BMC Evol Biol. 8:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström KJV, Sallman Almen M, Edstam MM, Fredriksson R, Schioth HB.. 2011. Independent HHsearch, Needleman–Wunsch-based, and motif analyses reveal the overall hierarchy for most of the G protein-coupled receptor families. Mol Biol Evol. 28(9):2471–2480. [DOI] [PubMed] [Google Scholar]
- Oehlmann VD, Berger S, Sterner C, Korsching SI.. 2004. Zebrafish beta tubulin 1 expression is limited to the nervous system throughout development, and in the adult brain is restricted to a subset of proliferative regions. Gene Expr Patterns. 4(2):191–198. [DOI] [PubMed] [Google Scholar]
- Ramos-Vicente D, et al. 2018. Metazoan evolution of glutamate receptors reveals unreported phylogenetic groups and divergent lineage-specific events. Elife 7:e35774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, et al. 2009. Projections from the accessory olfactory organ into the medial region of the olfactory bulb in the sea lamprey (Petromyzon marinus): a novel vertebrate sensory structure? J Comp Neurol. 516(2):105–116. [DOI] [PubMed] [Google Scholar]
- Sato Y, Miyasaka N, Yoshihara Y.. 2005. Mutually exclusive glomerular innervation by two distinct types of olfactory sensory neurons revealed in transgenic zebrafish. J Neurosci. 25(20):4889–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed AS, Sansone A, Nadler W, Manzini I, Korsching SI.. 2013. Ancestral amphibian V2rs are expressed in the main olfactory epithelium. Proc Natl Acad Sci U S A. 110(19):7714–7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed AS, Sansone A, Hassenklöver T, Manzini I, Korsching SI.. 2017. Coordinated shift of olfactory amino acid responses and V2R expression to an amphibian water nose during metamorphosis. Cell Mol Life Sci. 74(9):1711–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornhill RA.1970. Cell division in the olfactory epithelium of the lamprey, Lampetra fluviatilis. Z Zellforsch Mikrosk Anat. 109(2):147–157. [DOI] [PubMed] [Google Scholar]
- von Twickel A, et al. 2019. Individual dopaminergic neurons of lamprey SNc/VTA project to both the striatum and optic tectum but restrict co-release of glutamate to striatum only. Curr Biol. 29(4):677–685.e6. [DOI] [PubMed] [Google Scholar]
- Wakisaka N, et al. 2017. An adenosine receptor for olfaction in fish. Curr Biol. 27(10):1437–1447.e4. [DOI] [PubMed] [Google Scholar]
- Weth F, Nadler W, Korsching SI.. 1996. Nested expression domains for odorant receptors in zebrafish olfactory epithelium. Proc Natl Acad Sci U S A. 93(23):13321–13326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its Supplementary Material online.