Abstract
V(D)J recombination is initiated by the specific binding of the RAG1-RAG2 (RAG1/2) complex to the heptamer-nonamer recombination signal sequences (RSS). Several steps of the V(D)J recombination reaction can be reconstituted in vitro with only RAG1/2 plus the high-mobility-group protein HMG1 or HMG2. Here we show that the RAG1 homeodomain directly interacts with both HMG boxes of HMG1 and HMG2 (HMG1,2). This interaction facilitates the binding of RAG1/2 to the RSS, mainly by promoting high-affinity binding to the nonamer motif. Using circular-permutation assays, we found that the RAG1/2 complex bends the RSS DNA between the heptamer and nonamer motifs. HMG1,2 significantly enhance the binding and bending of the 23RSS but are not essential for the formation of a bent DNA intermediate on the 12RSS. A transient increase of HMG1,2 concentration in transfected cells increases the production of the final V(D)J recombinants in vivo.
A hallmark of lymphoid differentiation is the generation of diverse antigen receptors that can recognize any given foreign antigen. Diversity of the immune repertoire is achieved by the somatic assembly of the variable antigen receptor gene segments in a process termed V(D)J site-specific recombination. Each antigen receptor segment is flanked by highly conserved recombination signal sequences (RSS) that direct the site of rearrangement. Efficient recombination occurs only between a 12RSS-23RSS pair, a restriction termed the 12/23 rule (32). V(D)J rearrangement is initiated by two key lymphoid-specific proteins, RAG1 and RAG2 (36, 45). The RAG1-RAG2 (RAG1/2) complex binds specifically to the nonamer and heptamer sequences of the RSS (13, 23, 47). This interaction is assisted by the high-mobility-group proteins HMG1 and HMG2 (collectively, HMG1,2) (44, 52). Subsequently, the 12RSS and 23RSS are bridged in a synaptic complex (14, 54) and RAG1/2 cleaves the DNA at the coding-heptamer border, producing a covalently sealed hairpin coding end and a 5′ phosphorylated blunt signal end (33, 53). The hairpin intermediate is in turn asymmetrically processed by the RAG1/2 complex, yielding nucleotide overhangs that result in P nucleotide addition at the coding ends (5, 45a). After cleavage, RAG1 and RAG2 remain stably bound to the signal ends, as well as the coding ends (24), in a complex with the ubiquitous DNA repair activities Ku70, Ku80, and DNA-PK (reviewed in references 10, 19, and 25) and with HMG1,2 (2).
The initial step of V(D)J recombination is recognition of the nonamer motif of the RSS. This binding is mediated by the homeodomain (HD) of RAG1 (13, 47), which shows structural and functional homology to the DNA binding domain of the Hin recombinase, which mediates flagellar variation in the prokaryote Salmonella typhimurium (1). The DNA binding site of Hin (hix) consists of two motifs, one of which displays striking homology to the nonamer motif of the RSS recognized by RAG1 (13, 47). Replacement of the RAG1 homeodomain with that of the Hin invertase produces a hybrid protein that is partly functional in V(D)J recombination (47). Hin-mediated recombination is strongly stimulated by HU, a nonspecific prokaryotic DNA binding and -bending protein (22) which also stimulates MuA transposase binding to its cognate DNA binding sites (30). Moreover, HU can be efficiently replaced in the Hin recombination reaction by its mammalian counterparts HMG1,2 (38), with which it has no sequence similarity (6). HMG1,2 are ubiquitous proteins that bind to the minor groove of DNA in a sequence-independent manner and bend the double helix (reviewed in references 7 and 12). They are recruited through protein-protein interactions by other DNA binding proteins to distort the DNA and facilitate the assembly of large nucleoprotein complexes.
Given the functional parallels between the RAG1 and Hin DNA binding domains on one hand and between HU and HMG1,2 on the other, we addressed the mechanisms by which HMG1,2 exert their effect on V(D)J recombination. Here we demonstrate that there is a direct interaction between the RAG1 HD and either HMG1 or HMG2 through their HMG boxes. This interaction enhances the binding of RAG1 alone and consequently of the RAG1/2 complex to the RSS, both in vitro and in vivo. We also find that RAG1/2 induces bending of the RSS DNA even in the absence of HMG1,2. Binding and bending of the 23RSS is, however, very inefficient unless assisted by HMG1,2, which suggests that the crucial contribution of HMG1,2 is the stabilization of the complex between RAG1,2 and the bent 23RSS substrate. The cooperation of RAG1/2 with HMG1,2 in the first step of the V(D)J recombination leads to the stimulation of the overall recombination reaction in vivo.
MATERIALS AND METHODS
Recombinant plasmid constructs.
For the construction of plasmids expressing glutathione S-transferase (GST) fusion proteins in mammalian cells, RAG1 and RAG2 cDNA fragments were subcloned in the pEBG vector as previously described (47). Similarly, for the eukaryotic expression of (hemagglutinin [HA]-tagged) HMG proteins, the corresponding cDNAs were subcloned in the vector pEBB as 5′-SalI-NotI-3′ fragments. For the construction of plasmids expressing His-tagged HMG proteins in bacteria, the corresponding cDNAs were subcloned as 5′-BamHI-NotI-3′ fragments in the pET28a vector (Novagen). The TCF-1b (p45-TCF-1) and HMG-I(Y) (pET25b-HMGI) cDNAs were kindly provided by H. Clevers and D. Thanos, respectively. The RAG-VP16 expression constructs (R1cVP16/pCJM199 and R2cVP16/pCJM170) and the reporter constructs (pMJD) for the in vivo one-hybrid assay were kindly provided by D. Schatz (13). For the expression of the GST-RAG1-VP16 fusions, the VP16 portion of R1cVP16/pCJM199 was subcloned into pEBG-RAG1 constructs as a 5′-MluI-NotI-3′ fragment, replacing the last 32 amino acids of RAG1 (amino acids [aa] 1008 to 1040). The HMG1 constructs were previously described (9). The basic HMG2 construct was derived from plasmid pNLVP16HMG2 (kindly provided by T. Wirth), which was cut with XhoI, treated with T4 DNA polymerase, and cut again with BamHI. The insert was ligated to pT7-7 vector cut with NdeI, treated with T4 DNA polymerase, and recut with BamHI. All other HMG2 constructs were generated by PCR of the pNLVP16HMG2 template with pairs of primers containing the ATG translation start site and a stop codon, respectively. PCR products and pT7-7 vector were digested with NdeI and BamHI and ligated. The PCR primers were oligo 1 (5′-GGAATTCCATATGGGCAAGGGTGACC-3′) and oligo 2 (5′-CGGGATCCTAGGGGTCTTTTTTCTTTCC-3′) for the M1-P92 fragment, oligo 1 and oligo 3 (5′-CGGGATCCTAAGGAACATAGTTCTTCATC-3′) for the M1-P80 fragment, oligo 4 (5′-GGAATTCCATATGCCTCCCAAAGGGGATAA-3′) and oligo 5 (5′-CGGGATCCTAGCCTGTTGGCCTACC-3′) for the P80-G180 fragment, and oligo 6 (5′-GGAATTCCATATGGCTCCGAAGAGACCA-3′) and oligo 5 for the A94-G180 fragment. The bending constructs were created by subcloning the 12RSS (AGCTTACACAGTGATACAGCCCTGAACAAAAACC) or the 23RSS (AGCTTACACAGTGATGCAGGCCAAGTGTGAAGCCATACAAAAACC) in the pBend2 vector (28) as 5′-XbaI-SalI-3′ fragments. All constructs were fully verified by sequencing.
Protein expression and purification.
GST fusion recombinant forms of RAG1 and RAG2 proteins were overexpressed in 293T cells and purified as previously described (47). The HMG proteins used in electrophoretic mobility shift assays (EMSAs) were expressed as histidine-tagged forms in Escherichia coli BL21 and purified on nickel beads (Qiagen) according to the manufacturer’s protocol. These proteins were dialyzed in cleavage buffer (25 mM Tris-HCl [pH 8.0], 150 mM KCl, 2 mM dithiothreitol [DTT], and 20% glycerol). The HMG proteins used in protein-protein interaction assays were expressed in E. coli BL21 and purified as indicated previously (9). The purified proteins were dialyzed in storage buffer (10 mM Na phosphate [pH 7.5], 100 mM NaCl, 0.5 mM DTT, and 10% glycerol). All proteins were quantified by Coomassie blue staining following sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stored at −80°C. In vitro-translated HMG1 and -2 derivatives were synthesized by transcription and translation of the corresponding plasmids as described previously (9).
In vitro RAG-HMG protein interaction assay.
Tailless HMG1 (M1-V176) and full-length HMG2 proteins (1 mg/ml of packed beads) were covalently coupled to activated CH Sepharose 4B (Pharmacia) as indicated by the manufacturer. GST-HD (25 μg/ml of packed beads) was bound to glutathione-Sepharose 4B (Pharmacia). Ten microliters of Sepharose beads bearing immobilized HMG or RAG derivatives were incubated with RAG or HMG derivatives, respectively, in a total volume of 160 μl of binding buffer (1 mM DTT, 5 mM MgCl2, 10 μg of bovine serum albumin [BSA] in phosphate-buffered saline) for 1 h at room temperature. The beads were then washed three times with 1 ml of binding buffer. The material retained on the resin, the supernatant, and an equal amount of input material were applied to an SDS-PAGE gel and transferred to Immobilon P filters (Millipore). GST-RAG derivatives were visualized with an anti-GST monoclonal antibody and the Amersham ECL kit. HMG1 and HMG2 in vitro-translated derivatives were visualized by autoradiography. To avoid protein-DNA interactions, the interaction studies were also performed in buffers containing 150 μg of ethidium bromide/ml, with identical results.
EMSA and in vitro cleavage.
The conditions for EMSAs were as previously described, with 1 mM MgCl2 and 50 ng of each RAG protein (43). Complexes were resolved on 4% native polyacrylamide gels and visualized by autoradiography. The substrates were as previously described (43). The heptamer mutants carry the mutation acgAGTG (7mer ml) or CACAtga (7mer m2), and the nonamer mutant (9mer m) carries the mutation AACAAccgCC (mutated bases are lowercased). The upper strand of the substrates was 5′ end labelled with T4 polynucleotide kinase and annealed to the unlabelled lower strand. The bending substrates were prepared by digesting the bending vector(s) with the appropriate restriction enzymes, dephosphorylation with calf intestinal phosphatase, and purification and 5′ end labelling with T4 polynucleotide kinase. In the bending studies the concentration of dimethyl sulfoxide was reduced to 5%. Cleavage reactions were as previously described (43). The products were analyzed on 16% polyacrylamide–6 M urea denaturing gels.
Circular-permutation assay.
The circular-permutation assay detects DNA deformation by measuring the electrophoretic mobility of protein-DNA complexes (59). To map the locus of protein-DNA interaction and to estimate the amount of distortion introduced in the DNA, we used a simple geometrical model previously described in detail (17). Briefly, the mobilities of protein-DNA complexes are normalized to the mobility of free DNA (Rbound/Rfree [see Fig. 6C]). The distances between the 5′ end of the probe and the center of the sequence cloned in pBend are normalized to the total length of the probe (D/L; flexure displacement [see Fig. 6C]). The experimental values for Rbound/Rfree are interpolated by using the quadratic function Rbound/Rfree = 2K(1 + cosθ) (D/L)2 − 2K(1 + cosθ) (D/L) + K, where θ is the angle between the DNA to the left and the DNA to the right of the flexure site (for no bending, θ would be equal to 180°) and K is a parameter that is chosen to maximize the fit of the parabola to the experimental points. The minimum of the parabola identifies the locus of flexure. The amplitude of θ can be readily derived from the coefficients for the second-order and first-order terms of the equation. A slightly different geometrical treatment has been described (51); however, the results obtained by both algorithms (17, 51) are numerically similar.
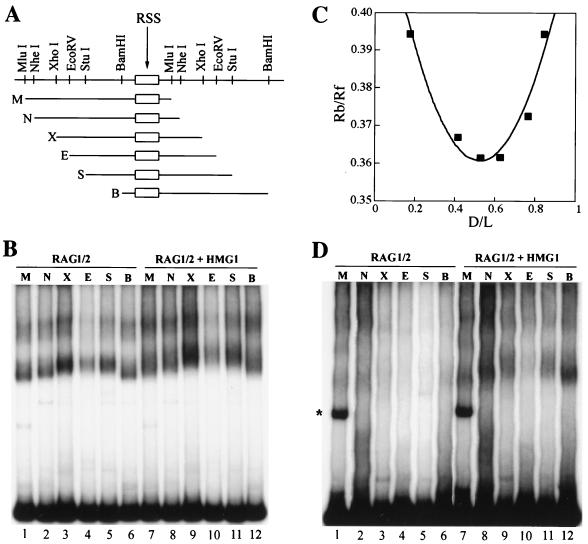
FIG. 6.
Bending of the RSS DNA by RAG1/2 and HMG1. (A) Schematic representation of the isomeric probes used in the circular-permutation analysis. (B and D) EMSAs with equimolar amounts of the isomeric probes shown in panel A carrying 12RSS (B) or 23RSS (D) sequences (the asterisk indicates a probe artifact). The probes were 5′ end labelled to the same specific activity and incubated with 50 ng each of RAG1ΔN380 and RAG2ΔC. Forty nanograms of HMG1 was added where indicated. (C) The locus and extent of bending were estimated as described in Materials and Methods. We used several gels for each estimate; shown is one example of the data obtained for 12RSS. Rb/Rf (vertical axis) is the relative mobility of bound versus free DNA. D/L (horizontal axis) is the fractional distance of the center of the RSS from the 5′ end of the probe. M, MluI; N, NheI; X, XhoI; E, EcoRV; S, StuI; B, BamHI.
One-hybrid assay.
The one-hybrid assay was performed essentially as described previously (13). Briefly, 293T (or 293 or NIH 3T3) cells were cotransfected with the RAG-VP16 expression constructs (plus or minus HMGs) and the reporter plasmid containing the luciferase gene driven by the minimal human cytomegalovirus immediate-early promoter downstream of tandem arrays of RSS sites. All luciferase values, which are expressed in arbitrary units, were normalized for transfection efficiency by cotransfection of another reporter plasmid (pRL; Promega) carrying the Renilla luciferase gene. The luciferase values were further normalized by dividing all values in any given transfection by the value obtained from a transfection of the reporter without RAG (no-RAG control) and expressed finally as fold transactivation over no-Rag control. The expression of the firefly luciferase and Renilla luciferase genes was measured with the Dual Luciferase kit (Promega).
In vivo recombination.
In vivo recombination assays were performed with 293T cells essentially as previously described (56). 293T (or 293 or NIH 3T3) cells were cotransfected with the inversional recombination substrate pJH299 (5 μg) and various combinations of GST-RAG1ΔN380 (6 μg), GST-RAG2ΔC (6 μg), HA-HMG1 (3 μg), HA-HMG2 (3 μg), HA-HMG-I(Y) (3 μg), and HA-TCF-1b (3 μg) expression constructs. The cells were harvested 48 h later, and DNA was isolated as described previously (36) and analyzed for recombination frequency by PCR analysis (20 cycles of 94°C for 30 s, 65°C for 60 s, and 74°C for 60 s). The linear range of the PCR assay was determined by serial dilutions of the rescued recombined plasmid. Oligonucleotides detect the recombined products by annealing to the joined heptamer signal (oligonucleotide RA5) and to the chloramphenicol acetyltransferase (CAT) gene present in pJH299 (oligonucleotide RA14 [56]). As a loading control, a 154-bp fragment of the CAT gene was amplified (oligonucleotides RA1 and RA14 [57]) under identical conditions. The amplified products were visualized by autoradiography following electrophoresis on a 10% polyacrylamide gel.
RESULTS
Direct interaction between the RAG1 HD and the HMG boxes of HMG1,2.
HMG1,2 are highly homologous and are functionally interchangeable in several systems (12). They have been shown to interact directly with the HDs of HOX and OCT proteins (60, 62). Therefore, we tested the ability of the RAG1 HD to interact with HMG1,2.
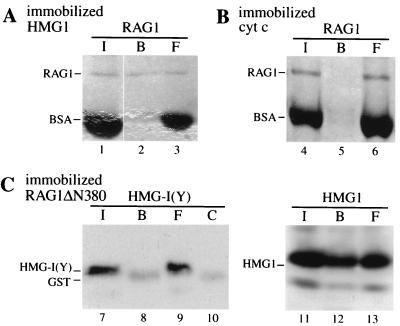
Initial experiments showed that full-length purified RAG1 associates with Sepharose beads bearing immobilized HMG1 (Fig. 1A) but not with Sepharose beads coated with BSA (not shown) or cytochrome c (Fig. 1B), which has a pI similar to that of the immobilized form of HMG1. The association was not quantitative, since about half of the input RAG1 did not bind to the beads. If all input RAG1 were active and all of the HMG1 on the beads had the same activity as native, soluble HMG1, the dissociation constant for the RAG1-HMG1 interaction would be on the order of 10−5 M. This estimate is probably conservative, but nonetheless compares favorably with the concentration of HMG1 in the cell nucleus, which is about 10−6 M (12). The in vitro interaction of HMG1 and RAG1 may thus occur in vivo as well.
FIG. 1.
RAG1 interacts with HMG1. Purified GST-RAG1 (about 0.2 μg in 160 μl) was incubated with Sepharose beads bearing immobilized, bacterially expressed tailless HMG1 (M1-V176) (A) or control beads bearing immobilized cytochrome c (B). Conversely, RAG1ΔN380, a GST fusion derivative of RAG1 that retains enzymatic activity, was bound to Sepharose-glutathione beads and used to pull down soluble HMG-I(Y), an HMG protein structurally diverse from HMG1,2, and HMG1 (C). Input (I), bound (B), and free (F) RAG1 and HMG proteins were detected by Coomassie blue staining. The protein in lane 8 is GST, as demonstrated by the appearance of the same protein in lane 10 (control [C]), where RAG1ΔN380 beads were directly boiled in loading buffer without prior exposure to HMG-I(Y). The RAG1ΔN380 protein itself migrates much higher and is not shown in the gel.
The reverse experiment was also performed (Fig. 1C): an enzymatically active fusion protein formed between GST and a truncated form of RAG1 (RAG1ΔN380), immobilized to glutathione-Sepharose beads, partially retained soluble HMG1 but did not retain HMG-I(Y), a structurally different high-mobility-group protein that facilitates the assembly of nucleoprotein complexes required for the transcription of several lymphoid cell-specific genes (16, 50). TCF-1b, another HMG box protein with a DNA binding domain structurally similar to that of HMG1 and necessary for T-cell development (55), did not interact with RAG1 either (not shown).
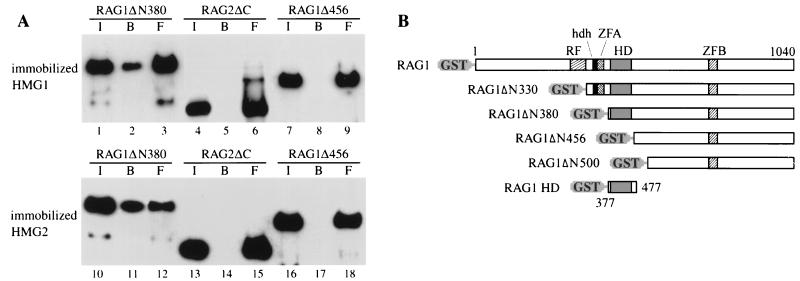
We next investigated whether the HD of RAG1 is required for the interaction with HMG1. Soluble RAG1ΔN380, which contains the HD, and RAG1Δ456, from which the HD has been deleted (Fig. 2B), were incubated with immobilized HMG1 or HMG2. Both HMG1 and HMG2 beads retained RAG1ΔN380 but not RAG1ΔN456 (Fig. 2A). In addition, a polypeptide corresponding to the RAG1 HD alone (aa 377 to 477) also interacted with HMG1,2 (data not shown). In contrast, RAG2 (active core, RAG2ΔC; aa 1 to 387) showed no obvious association with HMG1,2 (Fig. 2A, lanes 4 to 6 and 13 to 15). In controls, GST, cytochrome c, and BSA failed to retain the RAG1 protein (data not shown). Posttranslational modifications are not required for the RAG1-HMG1,2 interaction, since RAG1 expressed in either bacteria or mammalian cells interacted with HMG1 with equal efficiency (data not shown).
FIG. 2.
HMG1,2 directly interact with RAG1 through its HD. (A) Purified, eukaryotically expressed, GST-fused RAG1ΔN330, RAG1ΔN456, or RAG2ΔC (aa 1 to 388) was incubated with Sepharose beads bearing immobilized, bacterially expressed tailless HMG1 (M1-V176) or full-length HMG2, as described in Materials and Methods. The input (I), bound (B), and free (F) materials were immunoblotted with an anti-GST antibody, following SDS-PAGE. (B) Schematic representation of full-length RAG1 and derivatives. RF, ring finger (aa 288 to 330); hdh, homodimerization helices (aa 340 to 351); ZFA, zinc finger A (aa 353 to 374); HD, homeodomain (aa 389 to 446); ZFB, zinc finger B (aa 727 to 750) (3, 41).
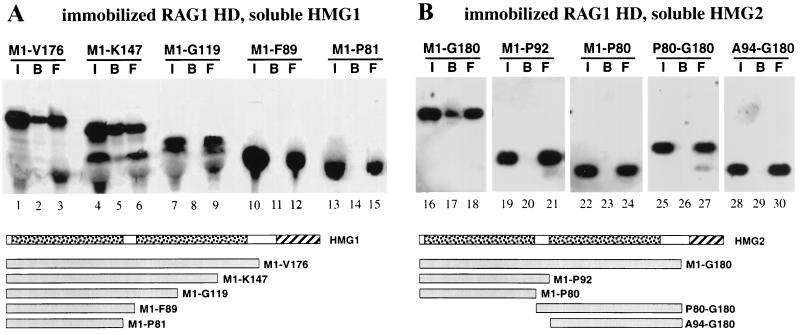
We identified the surface of interaction for RAG1 on HMGs with the reverse experiment. The immobilized HD of RAG1 was able to retain in vitro-translated tailless HMG1 and 2, respectively (Fig. 3A and B). A more extensively truncated form of HMG1 (M1-K147) was also retained, but truncated versions containing only one HMG box were not. Thus, both HMG boxes of HMG1 or -2 are necessary for interaction with the HD of RAG1.
FIG. 3.
The RAG1 HD directly interacts with HMG boxes A and B of HMG1 and -2. In vitro-translated HMG1 (A) and HMG2 (B) derivatives were incubated with Sepharose beads bearing immobilized, eukaryotically expressed RAG1 HD (Fig. 2B). The input (I), bound (B), and free (F) materials were visualized by autoradiography following SDS-PAGE. Schematic representations of full-length HMG1 and -2 are shown below panels A and B, respectively. The HMG boxes are stippled, and the acidic tails are hatched. The derivatives are identified by their first and last amino acids.
These data establish that HMG1,2 interact via their HMG boxes with the RAG1 HD even in the absence of DNA.
HMG1,2 promote the interaction of the RAG1 HD with the RSS.
We next tested whether the protein-protein interaction between RAG1 and HMG1,2 promoted the binding of the RAG1/2 complex to DNA. RAG1/2 binding to the 23RSS was examined with serially deleted forms of RAG1 (Fig. 2B).
Full-length RAG1 associated with the active core of RAG2 (RAG2ΔC) and the RSS DNA to yield a complex (Fig. 4A, lane 1) whose formation was enhanced about fivefold by HMG1 (Fig. 4A, lane 2).
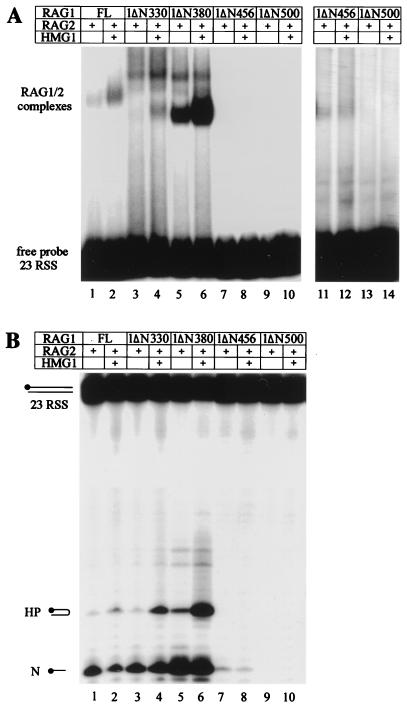
FIG. 4.
HMG1 stimulates RAG1/2 binding through the HD of RAG1. (A) EMSAs with a radiolabelled 23RSS oligonucleotide probe. RAG1 deletion derivatives and RAG2ΔC (50 ng each) and 40 ng of HMG1 were added as indicated (+). Lanes 11 to 14 represent a longer exposure of lanes 7 to 10. (B) In vitro cleavage assays. A 23RSS radiolabelled oligonucleotide probe was 5′ end labelled on the upper strand and incubated with 50 ng of each RAG1 derivative and 50 ng of RAG2ΔC; 40 ng of HMG1 was added where indicated (+). The products of the reaction, analyzed in a 16% polyacrylamide-6 M urea gel, are indicated. HP, hairpin; N, nick.
RAG1ΔN330, where aa 1 to 330 are deleted, formed two complexes with RSS DNA in the presence of RAG2ΔC, a prominent upper one and a minor lower one (Fig. 4A, lane 3). While the precise compositions of the two complexes are still to be determined, both contain RAG1 and RAG2 (43). RAG1ΔN380, which represents the active core of the protein (42, 46), lacks aa 330 to 380, which include the homodimerization helices of RAG1 (41). RAG1ΔN380/RAG2ΔC bound the RSS DNA with higher efficiency and predominantly formed the lower band (Fig. 4A, lane 5). HMG1 stimulated the formation of both complexes when RAG1ΔN330 was used and stimulated the formation of the lower one only when RAG1ΔN380 was used (Fig. 4A, lanes 4 and 6). This suggests that different homo- or heteromultimerized complexes of RAG1/2 are differently affected by HMG1.
Further deletion of RAG1 to aa 456 (which entirely removes the HD) very severely reduced the binding of RAG1ΔN456/RAG2ΔC to the RSS (Fig. 4A, lane 7); however, some residual binding is apparent after long autoradiography exposures (Fig. 4A, lanes 11 and 12), and the protein retains weak activity (43) (Fig. 4B, lane 7). Deletion to aa 500 (RAG1ΔN500) eliminated binding (Fig. 4A, lanes 9 and 13). HMG1 did not enhance RSS binding and cleavage by the RAG1 forms lacking the HD (Fig. 4A, compare lanes 11 and 12 and lanes 13 and 14; Fig. 4B, compare lanes 7 and 8).
Similar results were obtained when the 12RSS was used, but the binding and cleavage activities of the RAG1/2 complex were enhanced only twofold (data not shown). HMG2, but not HMG-I(Y) or TCF-1b, was able to stimulate RAG1/2 binding in a very similar manner (data not shown). These results were also confirmed in vivo (see below and Fig. 7).
FIG. 7.
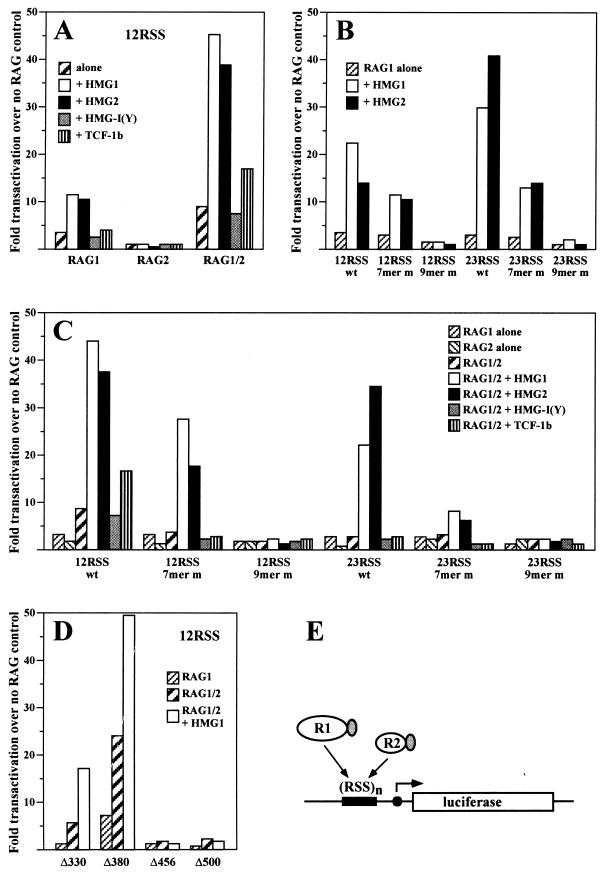
In vivo stimulation of RAG1 and RAG1/2 binding by HMG1,2. The binding of RAG proteins to a multimerized RSS in vivo can be measured by means of a mammalian one-hybrid assay (13). RAG1 and RAG2 proteins were transformed into transcriptional activators by fusing them to the VP16 transactivation domain. The occupancy of the binding site by the RAG-VP16 fusion proteins is proportional to the expression in 293 cells of a reporter luciferase gene driven by a cytomegalovirus minimal promoter. The binding site contains either 8 copies of the consensus 12RSS or 10 copies of the consensus 23RSS or mutated forms of the RSS where indicated. Comparable expression of proteins encoded by transfected plasmids was ascertained by Western blotting (see Fig. 8 for examples). The values are normalized to the expression of the reporter construct in the absence of RAG-VP16 fusion proteins. The plotted values represent the means of 8 to 10 individual experiments. The standard deviation was <8% of the mean and is not indicated. (A) Transactivation of the luciferase gene (12RSS construct) by RAG1-VP16, RAG2-VP16, or a combination of RAG1-VP16 and RAG2-VP16 as indicated below the diagram, either alone or in combination with cotransfected HMG1, HMG2, HMG-I(Y), or TCF-1b. (B) Transactivation of the reporter construct indicated below the diagram by RAG1-VP16 alone or RAG1-VP16 plus HMG1 or HMG2. The mutant 7-mer contains the sequence CGACGTC; the mutant 9-mer contains the sequence ACACTGGTA. wt, wild type. (C) Effect of HMG1, HMG2, HMG-I(Y), and TCF-1b on transactivation by RAG1/2-VP16. The transactivation by RAG1-VP16 alone or RAG2-VP16 alone is also reported for comparison. The reporter constructs are indicated below the corresponding groups of bars. (D) Transactivation of the luciferase gene (12RSS construct) by different RAG1-VP16 fusion proteins, either alone, in combination with RAG2-VP16, or in combination with RAG2-VP16 and in the presence of HMG1. (E) Schematic representation of the reporter construct. RAG1-VP16 is indicated as R1, and RAG2-VP16 is indicated as R2. The grey oval represents the VP16 transactivation domain.
HMG1,2 do not alter the sequence requirements for RSS recognition by RAG1/2.
RAG1/2 binding to the RSS is dependent on both the nonamer and heptamer motifs (13, 23, 47). In order to explore whether the stimulatory effect of HMG1 on RAG1/2 binding is also dependent on both motifs, we assayed RAG1ΔN380/RAG2ΔC binding to mutant 23RSS. HMG1 stimulated binding to the heptamer mutants, while it failed to boost binding to the nonamer mutant (Fig. 5B).
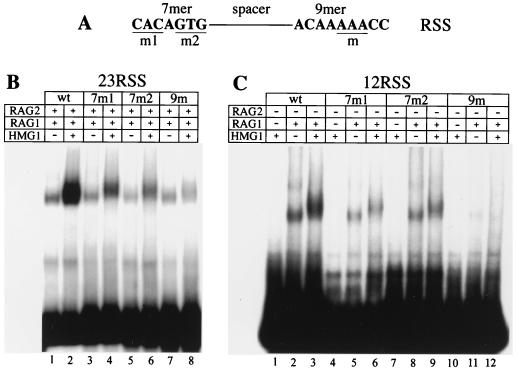
FIG. 5.
HMG1 enhances nonamer-dependent binding to the RSS. EMSAs with radiolabelled oligonucleotide probes carrying either wild-type (wt) or mutated RSS sequences. (A) Schematic representation of the RSS and the positions of the mutations employed. The mutations are further described in Materials and Methods. (B and C) RAG1ΔN380 and RAG2ΔC (50 ng each) were incubated (+) with 23RSS (B) or 12RSS (C) radiolabelled substrates carrying the indicated mutations. Forty nanograms of HMG1 was added (+) where indicated.
Using surface plasmon resonance (BIAcore), we previously showed that the RAG1 HD establishes specific interactions with the nonamer motif of the RSS, even in the absence of RAG2 (47). To study the effect of HMG1 on the binding of RAG1 alone, we assayed the binding of RAG1ΔN380 alone to 12RSS mutants in the presence or the absence of HMG1 (Fig. 5C). Mutation of the first 3 or last 3 nucleotides of the heptamer (7mers m1 and m2, respectively) reduced RAG1ΔN380 binding, but HMG1 was still incorporated in the RAG-RSS complex, as shown by the slight supershift of the band corresponding to the complex. The HMG1-dependent stimulation of RAG1 binding to the mutated heptamer was slight (less than twofold) but reproducible (Fig. 5C, lanes 4 to 9). Mutation of positions 5 to 7 of the nonamer abolished binding of RAG1ΔN380 (Fig. 5C, lane 11), and addition of HMG1 failed to rescue it (lane 12). Essentially the same results were obtained when 23RSS mutants were used, except that RAG1 binding was significantly lower, as expected (data not shown). These results were confirmed in vivo (see below and Fig. 7).
Thus, HMG1,2 stimulate RSS binding by RAG1 alone and in combination with RAG2 but do not alter the relative dependence of RAG1/2 binding on the heptamer and the nonamer.
Bending of RSS DNA by the RAG1/2-HMG1 complex.
RAG1/2 binds with greater affinity on the 12RSS than on the 23RSS (references 43, 46, and 52 and data not shown). Conversely, HMG1,2 have a more pronounced effect on the binding of RAG1/2 to the 23RSS. Based on this, it has been suggested that HMG1,2 bind and bend the spacer region of the 23RSS to bring the heptamer and nonamer motifs into close proximity (44, 52). This is in accordance with the DNA-flexing function of HMG1,2, which are known to bind to irregular or prebent DNA structures (8, 40) and to mediate bending of normal B-form DNA in ring closure assays (37, 39). Hence, we investigated by circular-permutation analysis whether HMG1 enhanced binding of RAG1/2 to the 23RSS through DNA bending. In this assay, proteins that induce DNA distortions show differential electrophoretic migration when bound to isomeric DNA probes containing their cognate DNA binding site placed at different sites along the probe (59).
The 12RSS and 23RSS motifs were subcloned into the bending vector pBend2 (28) and used as probes in EMSAs (Fig. 6A). In the absence of RAGs, HMG1 failed to interact with the pBend2-RSS probes (data not shown). RAG1/2 bound to the isomeric pBend2-RSS DNA with reduced overall efficiency compared to oligonucleotide probes. Moreover, due to the nonspecific DNA binding affinity of RAG1/2, binding to the large (150- to 161-bp) isomeric probes produced increased background levels compared to those with oligonucleotide probes (43 to 54 bp).
Unexpectedly, the RAG1/2 complex showed an intrinsic ability to bend the 12RSS DNA, even in the absence of HMG1,2 (Fig. 6B, lanes 1 to 6). The 12RSS probe was deflected by an angle that was estimated at between 43 and 49°, with the site of bending corresponding to the 12RSS itself (several gels were used to estimate the angle; Fig. 6C shows an example of the data from one such gel). Addition of HMG1 increased the deflection to between 55 and 60° without changing the site of bending (Fig. 6B; compare lanes 7 to 12 to lanes 1 to 6).
Binding and bending of the 23RSS probes by the RAG1/2 complex was almost undetectable (Fig. 6D, lanes 1 to 6), but the addition of HMG1 significantly stimulated binding (Fig. 6D, lanes 7 to 12). The RAG1/2-HMG1 complex bent the 23RSS DNA to a pattern similar to that of the 12RSS.
The DNA-bending properties of RAG1/2 on the 12RSS were also addressed by phasing analysis, where the RSS were placed at increasing distances from an intrinsic DNA bend induced by in-phase AT tracts (61). The results on the phasing DNA probes (not shown) were comparable to the circular-permutation data.
HMG1,2 stimulate specific RAG1 and RAG1/2 binding in vivo.
To explore the effect of HMG1,2 on the DNA binding activity of RAG1/2 in vivo, we utilized the previously described one-hybrid assay (13). Briefly, the RAG1 and RAG2 proteins were converted to transcriptional activators by adding the acidic domain of the herpes simplex virus protein VP16. A reporter construct provides the substrate for binding of RAG1/2 to the RSS: multiple copies of the RSS are cloned in front of a minimal promoter driving expression of the luciferase gene (Fig. 7E). Cotransfection of RAG-VP16 constructs with the reporter in mammalian cells leads to transactivation of the luciferase gene to a degree directly proportional to RAG binding. Luciferase activity was normalized for transfection efficiency and background levels as described in Materials and Methods. The expression levels of all recombinant proteins were verified by Western analysis to ensure comparability of the results.
Overexpression of HMG1 or HMG2 stimulated binding to the 12RSS of RAG1 and RAG1/2 but not of RAG2 alone (Fig. 7A). HMG-I(Y) had no major effect, but TCF-1b invariably slightly enhanced the binding of RAG1/2 to the 12RSS (Fig. 7A).
HMG1,2 increased binding of RAG1 alone to the 12RSS by 3- to 4-fold and enhanced binding to the 23RSS by about 10-fold (Fig. 7B). Mutation of the nonamer severely reduced binding of RAG1 alone and was not compensated for by addition of HMG1,2, while mutation of the heptamer allowed RAG1 binding and stimulation by HMG1,2 (Fig. 7B). These findings are in accordance with the in vitro results showing that HMG1,2 did not alter the sequence requirements for RSS recognition by RAG1 (Fig. 5).
The lack of effect of HMG1,2 on the sequence requirements of the RAG1/2 complex (as opposed to RAG1 alone) was also verified (Fig. 7C). It is worth noting that HMG2 stimulated better binding to the 23RSS than HMG1 (Fig. 7B and C).
As further controls, and to allow direct comparison with the in vitro results, GST fusion deletion mutants (homologous to the ones used for the in vitro DNA binding assays [Fig. 2B]) were produced as RAG1-VP16 fusions and analyzed in the one-hybrid assay (Fig. 7D). Only RAG1ΔN330 and -ΔN380 retained specific binding to the RSS DNA, which was stimulated by HMG1,2. Conversely, RAG1ΔN456 and -ΔN500, from which the HD has been deleted, showed no specific binding, which was unaffected by overexpression of HMG1,2.
HMG1,2 stimulate V(D)J recombination in vivo.
We tested the overall effect of HMGs on V(D)J recombination by conducting the extrachromosomal substrate recombination assay in the presence of overexpressed HMGs (Fig. 8). RAG1, RAG2, the recombination substrate pJH299, and vectors expressing the various HMGs were cotransfected in 293T cells, and recombined products (signal joints) were detected by PCR analysis (56). The amounts of PCR products were strictly proportional to the input material, as indicated by titration experiments (not shown) and by 10-fold dilution of the template (compare Fig. 8B and B′).
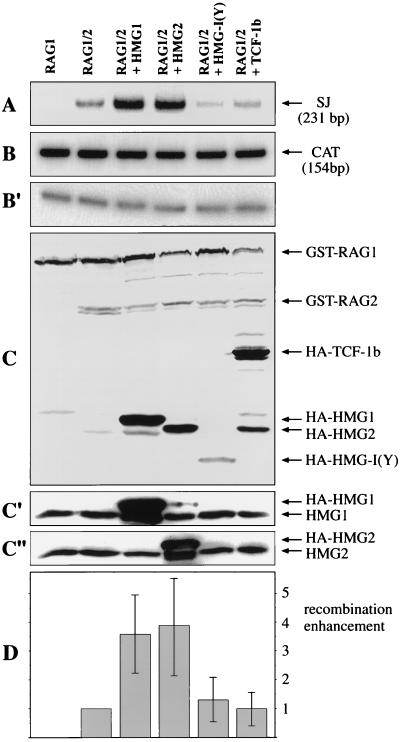
FIG. 8.
HMG1,2 increase the yield of V(D)J recombination products in vivo. 293T cells were cotransfected with the recombination substrate pJH299 and the expression constructs for the indicated proteins. The RAGs were GST tagged, while HMG1, HMG2, HMG-I(Y), and TCF-1b were HA tagged. (A) Recombination efficiency was measured by PCR analysis (20 cycles) of the recovered plasmid, using primers that amplify only the recombined sequences (signal joints [SJ]), as described in Materials and Methods. (B) Loading control of panel A. A fragment of the CAT gene, present in the pJH299 recombination substrate, was amplified under the same conditions as for panel A. (B′) Templates as in panel B were diluted 10-fold prior to PCR. The amount of PCR product obtained was proportional to the template input. (C) Expression control of panel A. Total cellular extract was immunoblotted with anti-GST and anti-HA antibodies, following SDS-PAGE. (C′) The same membrane shown in panel C was stripped and immunoblotted with anti-HMG1 antibodies. (C") The same membrane shown in panel C was stripped once more and immunoblotted with anti-HMG2 antibodies. (D) The autoradiogram shown in panel A and those from three more experiments were scanned, and the amounts of recombination products were normalized to the efficiency of V(D)J recombination with RAG1/2 alone, which was set to 1. The error bars indicate standard deviations.
We directly estimated the amounts of protein expression directed from the transfected plasmids by Western blotting with anti-GST or anti-HA antibodies and judged them to be comparable for different HMGs and very similar for RAGs in all samples (Fig. 8C). Finally, we estimated the amounts of HMG1 and HMG2 overexpression by Western blotting with anti-HMG1 (Fig. 8C′) or anti-HMG2 (Fig. 8C") antibodies. The proteins expressed from transfected plasmids contain a tag and run with slightly lower mobility than the natural HMG1,2. Our results indicate that the cellular pool of HMG1,2 can be transiently increased about threefold in 293 cells.
HMG1,2 drastically stimulated V(D)J recombination efficiency, in contrast to HMG-I(Y) and TCF-1b (Fig. 8D). We obtained identical results with assays conducted in 293 and 3T3 cells, using either the deletional substrate pJH200 or the inversional substrate pJH299 (data not shown). Thus, the ability of HMG1,2 to boost the DNA binding properties of RAG1/2 has a clear effect on the overall efficiency of V(D)J recombination in vivo.
DISCUSSION
During the initial step of V(D)J recombination, RAG1/2 establishes specific interactions with the RSS signals. The architectural chromatin components HMG1,2 were previously shown to enhance RSS recognition in vitro (29, 44, 52). In this study we showed that HMG1,2 are limiting for V(D)J recombination in transfection assays, since their transient increase leads to a higher yield of recombination products. This effect is partially or totally due to a protein-protein interaction between the major DNA binding domain of RAG1—the HD—and both HMG boxes of HMG1,2. Through this interaction, HMG1,2 enhance the binding of RAG1/2 to the 12RSS 2- to 5-fold, whereas binding to 23RSS is enhanced up to 10-fold, both in vitro and in vivo. However, and contrary to expectations, HMG1,2 do not endow RAG1/2 with DNA-flexing ability: the ability to distort the 12RSS and the 23RSS appears to be intrinsic to the RAG1/2 recombinase. Thus, the major role of HMG1,2 appears to be the stabilization of the RAG-RSS complex. The same molecular mechanism by which HMG1,2 facilitate RAG binding to RSS in naked DNA most likely underpins the facilitation of RAG binding to nucleosomal complexes (29). Moreover, it is economical to envision that HMG1,2 will likewise facilitate the interaction of the RAGs with coding hairpins and their nicking (5).
Direct interaction of HMG1,2 with the RAG1 HD.
By analogy with other transactions where HMG1,2 are involved, we suspected a direct protein-protein interaction with the major players in V(D)J recombination, RAG1 and RAG2. Using deletional analysis, we determined that the RAG1 HD directly interacts with boxes A and B of HMG1,2. Several Hox proteins directly interact with HMG1,2 through helix I of their HDs (60). The RAG1 HD contains several amino acid residues that have been highly conserved among different HDs (47), and it is therefore conceivable that HMG1,2 establish specific interactions with helix I of the RAG1 HD.
There is a single significant difference, however, between the association of HMG1,2 with RAG1 and their association with other interactors. Both boxes A and B of HMG1,2 are required for interaction with the RAG1 HD, whereas Hox proteins, Oct proteins, TBP, or steroid receptors all interact with either box A or box B of HMG1,2 (11, 48, 60, 62). The structural and functional significance of this peculiarity remains to be determined. However, given the ability of core RAG1 HD to homodimerize (41a), it is possible that HMG boxes A and B interact simultaneously with both HDs of the RAG1 homodimer.
Efforts to coimmunoprecipitate RAG1 or RAG1/2 from transfected mammalian cells with antibodies directed against HMG1 (or HMG1 tagged with the HA epitope) were unsuccessful. We had previously tried to coimmunoprecipitate HMG1 with HOXD9 and steroid hormone receptors, with negative results (6a). Apparently, the in vivo physical association between HMG1 and its partner proteins is unstable and readily reversible, whereas it is much more stable when the interactors are present in purified form. Likewise, HMG1 stably associates with purified nucleosomes but is not stably associated with interphase or metaphase chromosomes (15).
A comparison of the functions of DNA-bending proteins in prokaryotes and in eukaryotes is useful. Prokaryotic proteins, such as HU or IHF, exert their effect by direct binding to the DNA in the absence of any protein-protein interactions. No direct interaction among IHF, HU, and bacterial recombinases has been described. In Mu transposition, HU facilitates the proximity of the two transposase binding sites without establishing any direct protein-protein interactions (30). In flagellar variation, HU bends the DNA between the two hix sites and facilitates the interaction of Hin with its two cognate sites (22). Although HU is incorporated in the Hin-invertasome complex to bend the DNA, it does not establish direct interactions with Hin. Remarkably, HMG1,2 can substitute for the function of HU in the Hin-invertasome complex (38) and in Mu transposition (31). Thus, it appears that HMG proteins in eukaryotes have evolved the ability to bend the DNA through nonspecific binding as well as to enhance DNA binding through direct protein-protein interactions. In this role, HMG proteins provide a “scaffold” for the assembly of higher-order nucleoprotein complexes (21).
HMG1,2 assist RAG1 binding to the RSS.
We find that HMG1,2 facilitate the specific binding of RAG1 to RSS, as shown by both in vitro and in vivo assays. Moreover, HMG1,2 appear to be incorporated in the RAG1/2-DNA complex right from the initial RSS recognition stage (41a), and the association may persist until the formation of the stable postcleavage complex (2). Stimulation of sequence-specific target DNA recognition by HMG1,2 is a common theme. HMG1 facilitates the binding to DNA of HOX and OCT proteins (60, 62), steroid hormone receptors (11), TBP (18, 48), and p53 (26). Strikingly, these DNA binding proteins directly interact with HMG1,2 through their DNA binding domains. In so doing, these proteins increase the protein surface contacting the DNA from both the major and minor grooves to ultimately achieve high-affinity interaction with their cognate DNA sites. HMG1,2 demonstrate no inherent sequence specificity in DNA binding and very low affinity for linear, B-form DNA. Thus, they are in effect recruited to DNA by their partner to increase DNA binding affinity without altering sequence specificity. This is also true for V(D)J recombination: HMG1,2 increase the affinity of RAG1 for the nonamer without changing its sequence requirements.
Specific interactions of RAG1 with the RSS have been previously detected by surface plasmon resonance (47) and were recently observed in footprinting and gel retardation assays (35, 49). Mutations in the nonamer abolish RAG1 binding, and the binding cannot be rescued by HMG1,2. In contrast, mutations in the heptamer reduce but still allow RAG1 binding, and HMG1,2 can enhance the residual binding.
If one considers the interaction of the RAG1/2 complex with DNA, mutations in the nonamer reduce but do not abolish binding. However, HMG1 will not enhance the residual interaction of RAG1/2 with RSS containing nonamer mutations. It will nonetheless enhance the interaction of RAG1/2 with RSS containing heptamer mutations.
The same effects of HMG1,2 on the binding of RAG1 and RAG1/2 are reflected in the in vivo one-hybrid assay. Taken together, the data suggest that HMG1,2 increase the affinity of RAG1 (either alone or in complex with RAG2) for the 12RSS and the 23RSS but have no effect on the sequence requirements of the interaction.
Bending of RSS DNA by RAG1/2-HMG1,2.
The different efficiencies of RAG1 binding to the 12RSS and 23RSS and the different stimulations by HMG1,2 suggested that perhaps the 23RSS might be a worse binding substrate because the heptamer and the nonamer are separated by an additional DNA turn. To establish contacts to RAG1 similar to those of the 12RSS, the 23RSS would have to be distorted—a very likely function for DNA-flexing proteins like HMG1,2. However, we found that DNA-flexing ability is not provided uniquely by HMG1,2: surprisingly, the RAG1/2 complex by itself has an intrinsic ability to bend the 12RSS and the 23RSS DNAs. Homology between RAGs and DNA-bending proteins has been previously suggested (4).
Circular-permutation analysis indicated that the locus of the bending by the RAG1/2 complex is within the 12RSS (Fig. 6B and C). The distortion could be the direct effect of the simultaneous binding of RAG1/2 to both nonamer and heptamer motifs, which would cause the intervening sequence to bend. Alternatively, RAG1/2 may bind to the nonamer first and then bend the DNA in the immediate vicinity, thereby increasing the proximity to the heptamer. These two scenarios are not easily distinguishable, but we favor the latter for the following reasons: (i) mutations at the nonamer have a more profound effect in binding than those at the heptamer; (ii) heptamer mutations decrease binding of RAG1 and RAG1/2 (23, 43) (Fig. 5), and increasing the distance from the nonamer by one helical turn (23RSS) has the same effect; (iii) interaction of RAG1/2 with the heptamer might be transient, since protection of the heptamer is not prominent in footprinting assays (49); and (iv) the heterogeneity of the RAG1/2-23RSS complexes in EMSAs suggests that several distinct species might be in rapid equilibrium, with only a fraction of 23RSS molecules bent to optimally fit the RAG1/2 binding surfaces.
We envision the role of HMG1,2 as twofold: it would increase the effective DNA binding surface of RAG1, stabilizing its first contact with the nonamer, and it would then assist RAG1 in its intrinsic DNA-bending activity so that the heptamer would come in sufficient proximity to its cognate protein binding surface. The role of HMG1,2 would then be more significant for the 23RSS, where the heptamer is further away.
It has recently been shown that HMG1,2 are instrumental in allowing RAG1/2 to perform V(D)J recombination on nucleosomal substrates (29). HMG1,2 can interact with in vitro-reconstituted nucleosomes but do not appear to be stably bound to chromatin in the nuclei of living cells (15). Thus, it may well be that HMG1,2 associate with RAGs in the absence of DNA and then facilitate their transient association with nucleosomes. Once RAG1/2 has gained access to the DNA in nucleosomes, HMG1,2 would promote its binding to the RSS and DNA bending, as we have shown in detail.
In vivo effects of HMG1,2 in V(D)J recombination.
We have shown that a transient increase in the concentration of HMG1 or HMG2 in cells transfected with V(D)J recombination intermediates and RAG proteins results in an increased production of recombined molecules. This effect might seem surprising in view of the presence of endogenous HMG1,2 proteins, but it has already been observed in the context of interactions with Hox proteins and nuclear hormone receptors (11, 60). Thus, HMG1,2 appear to be limiting for V(D)J recombination, as well as for the other DNA transactions in which they have been implicated (reviewed in reference 7). It is perhaps surprising that mice lacking the HMG1 protein show no obvious alteration in the immune system (12a). However, HMG2 is expressed at high levels only in lymphoid cells and testes in the adult.
HMG1,2 have been implicated in the establishment of the 12/23 synaptic rule (27, 52, 58) and have been detected as part of the 12/23-dependent stable postcleavage complex formed by blunt 12- and 23RSS, RAG1/2, Ku70/80, and DNA-PK (2). In addition, HMG1,2 enhance the hairpin-nicking activity of RAG1/2 (5, 45a) and the activity of DNA ligase IV (34), an enzyme involved in the final stage of V(D)J recombination (20). The direct interaction of HMG1,2 with the RAG1 HD provides a basis for understanding these multiple roles. By binding directly to RAG1, HMG1,2 are recruited to the site of V(D)J recombination, where they can facilitate interactions among multiple proteins and DNA and possibly among the proteins themselves. By acting as integral components of the recombination machinery, HMG1,2 may enhance the kinetics of most sequential reactions in V(D)J recombination.
ACKNOWLEDGMENTS
We are grateful to H. Clevers and M. van de Wetering for the TCF-1b cDNA clones, D. Schatz and M. Difilippantonio for the VP16-RAG and luciferase plasmids, D. Thanos for the HMG-I(Y) cDNA clone and pBend2 vectors, T. Wirth for the HMG2 cDNA, and A. Hodtsev and A. Han for critical reading of the manuscript.
This work was supported by NIH grant AI40191 to E.S. and AIRC and MURST grants to M.E.B. E.S. was a Cancer Research Institute Clinical Investigator and Howard Hughes Medical Institute Assistant Investigator.
REFERENCES
- 1.Affolter M, Percival-Smith A, Muller M, Billeter M, Qian Y Q, Otting G, Wuthrich K, Gehring W J. Similarities between the homeodomain and the Hin recombinase DNA binding domain. Cell. 1991;64:879–880. doi: 10.1016/0092-8674(91)90311-l. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal A, Schatz D G. RAG1 and RAG2 form a stable post-cleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- 3.Bellon S F, Rodgers K K, Schatz D G, Coleman J E, Steitz T A. Crystal structure of the Rag1 dimerization domain reveals multiple zinc-binding motifs including a novel zinc binuclear cluster. Nat Struct Biol. 1997;4:586–591. doi: 10.1038/nsb0797-586. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein R M, Schluter S F, Bernstein H, Marchalonis J J. Primordial emergence of the recombination activating gene 1 (RAG1): sequence of the complete shark gene indicates homology to microbial integrases. Proc Natl Acad Sci USA. 1996;93:9454–9459. doi: 10.1073/pnas.93.18.9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Besmer E, Mansilla-Soto J, Cassard S, Sawchuk D J, Brown G, Sadofsky M, Lewis S M, Nussenzweig M C, Cortes P. Hairpin coding end opening is mediated by RAG1 and RAG2 proteins. Mol Cell. 1998;2:817–828. doi: 10.1016/s1097-2765(00)80296-8. [DOI] [PubMed] [Google Scholar]
- 6.Bianchi M E. Prokaryotic HU and eukaryotic HMG1: a kinked relationship. Mol Microbiol. 1994;61:1011–1051. doi: 10.1111/j.1365-2958.1994.tb01261.x. [DOI] [PubMed] [Google Scholar]
- 6a.Bianchi, M. E. Unpublished data.
- 7.Bianchi M E, Beltrame M. Flexing DNA: HMG-box proteins and their partners. Am J Hum Genet. 1998;63:1573–1577. doi: 10.1086/302170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bianchi M E, Beltrame M, Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989;243:1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- 9.Bianchi M E, Falciola L, Ferrari S, Lilley D M J. The DNA binding site of HMG1 protein is composed of two similar segments, both of which have counterparts in other eukaryotic regulatory proteins. EMBO J. 1992;11:1055–1063. doi: 10.1002/j.1460-2075.1992.tb05144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bogue M, Roth D B. Mechanism of V(D)J recombination. Curr Opin Immunol. 1996;8:175–180. doi: 10.1016/s0952-7915(96)80055-0. [DOI] [PubMed] [Google Scholar]
- 11.Boonyaratanakornkit V, Melvin V, Prendergast P, Altmann M, Ronfani L, Bianchi M E, Taraseviciene L, Nordeen S K, Allegretto E A, Edwards D P. High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol Cell Biol. 1998;18:4471–4487. doi: 10.1128/mcb.18.8.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bustin M, Reeves R. High mobility group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 12a.Calogero S, Grassi F, Aguzzi A, Voigtländer T, Ferrier P, Ferrari S, Bianchi M E. The lack of chromosomal protein HMG1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat Genet. 1999;22:276–280. doi: 10.1038/10338. [DOI] [PubMed] [Google Scholar]
- 13.Difilippantonio M, McMahan C J, Eastman Q M, Spanopoulou E, Schatz D G. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell. 1996;87:253–262. doi: 10.1016/s0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- 14.Eastman Q M, Leu T M J, Schatz D G. Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature. 1996;380:85–88. doi: 10.1038/380085a0. [DOI] [PubMed] [Google Scholar]
- 15.Falciola L, Spada F, Calogero S, Längst G, Voit R, Grummt I, Bianchi M E. High mobility group 1 (HMG1) protein is not stably associated with the chromosomes of somatic cells. J Cell Biol. 1997;137:19–26. doi: 10.1083/jcb.137.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falvo J V, Thanos D, Maniatis T. Reversal of intrinsic DNA bends in the IFN-β gene enhancer by transcription factors and the architectural protein HMG-I(Y) Cell. 1995;83:1101–1111. doi: 10.1016/0092-8674(95)90137-x. [DOI] [PubMed] [Google Scholar]
- 17.Ferrari S, Harley V R, Pontiggia A, Goodfellow P N, Lovell-Badge R, Bianchi M E. SRY, like HMG1, recognizes sharp angles in DNA. EMBO J. 1992;11:4497–4506. doi: 10.1002/j.1460-2075.1992.tb05551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ge H, Roeder R G. The high mobility group protein HMG1 can reversibly inhibit class II gene transcription by interaction with the TATA-binding protein. J Biol Chem. 1994;269:17136–17140. [PubMed] [Google Scholar]
- 19.Grawunder U, West R B, Lieber M R. Antigen receptor gene rearrangement. Curr Opin Immunol. 1998;10:172–180. doi: 10.1016/s0952-7915(98)80246-x. [DOI] [PubMed] [Google Scholar]
- 20.Grawunder U, Wilm M, Wu X, Kulesza P, Wilson T E, Mann M, Lieber M R. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 21.Grosschedl R. Higher-order nucleoprotein complexes in transcription analogies with site-specific recombination. Curr Opin Cell Biol. 1995;5:362–370. doi: 10.1016/0955-0674(95)80091-3. [DOI] [PubMed] [Google Scholar]
- 22.Haykinson M H, Johnson R C. DNA looping and the helical repeat in vitro and in vivo: effect of HU protein and enhancer location of Hin invertasome assembly. EMBO J. 1993;12:2503–2512. doi: 10.1002/j.1460-2075.1993.tb05905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiom K, Gellert M. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell. 1997;88:65–72. doi: 10.1016/s0092-8674(00)81859-0. [DOI] [PubMed] [Google Scholar]
- 24.Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol Cell. 1998;1:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 25.Jackson S P, Jeggo P A. DNA double strand break repair and V(D)J recombination: involvement of DNA-PK. Trends Biochem Sci. 1995;20:412–415. doi: 10.1016/s0968-0004(00)89090-8. [DOI] [PubMed] [Google Scholar]
- 26.Jayaraman L, Moorthy N C, Murthy K G K, Manley J L, Bustin M, Prives C. High mobility group protein-1 (HMG1) is a unique activator of p53. Genes Dev. 1998;12:462–472. doi: 10.1101/gad.12.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim D R, Oettinger M A. Functional analysis of coordinated cleavage in V(D)J recombination. Mol Cell Biol. 1998;18:4679–4688. doi: 10.1128/mcb.18.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J, Zwieb C, Wu C, Adhya S. Bending of DNA by gene-regulatory proteins: construction and use of a DNA bending vector. Gene. 1989;85:15–23. doi: 10.1016/0378-1119(89)90459-9. [DOI] [PubMed] [Google Scholar]
- 29.Kwon J, Imbalzano A N, Matthews A, Oettinger M A. Accessibility of nucleosomal DNA to V(D)J cleavage is modulated by RSS positioning and HMG1. Mol Cell. 1998;2:829–839. doi: 10.1016/s1097-2765(00)80297-x. [DOI] [PubMed] [Google Scholar]
- 30.Lavoie B D, Chaconas G. Immunoelectron microscopic analysis of the A, B and HU protein content of bacteriophage Mu transposomes. J Biol Chem. 1990;265:1623–1627. [PubMed] [Google Scholar]
- 31.Lavoie B D, Chaconas G. A second high affinity binding site in the phage Mu transposome. J Biol Chem. 1994;269:15571–15576. [PubMed] [Google Scholar]
- 32.Lewis S M. The mechanism of V(D)J joining: lessons from molecular, immunological and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 33.McBlane J F, van Gent D C, Ramsden D A, Romeo C, Cuomo C A, Gellert M, Oettinger M A. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 34.Nagaki S, Yamamoto M, Yumoto Y, Shirakawa H, Yoshida M, Teraoka H. Non-histone chromosomal proteins HMG1 and 2 enhance ligation reaction of DNA double-strand breaks. Biochem Biophys Res Commun. 1998;246:137–141. doi: 10.1006/bbrc.1998.8589. [DOI] [PubMed] [Google Scholar]
- 35.Nagawa F, Ishiguro K-I, Tsuboi A, Yoshida T, Ishikawa A, Takemori T, Otsuka A J, Sakano H. Footprint analysis of the RAG protein recombination signal sequence complex for V(D)J type recombination. Mol Cell Biol. 1998;18:655–663. doi: 10.1128/mcb.18.1.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oettinger M A, Schatz D G, Gorka C, Baltimore D. RAG1 and RAG2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 37.Oñate S A, Prendergast P, Wagner J P, Nissen M, Reeves R, Pettijohn D E, Edwards D P. The DNA bending protein HMG-1 enhances progesterone receptor binding to its target DNA sequences. Mol Cell Biol. 1994;14:3376–3391. doi: 10.1128/mcb.14.5.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paull T T, Haykinson M J, Johnson R C. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 1993;7:1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- 39.Pil P M, Chow C S, Lippard S J. High-mobility-group 1 protein mediates DNA bending as determined by ring closures. Proc Natl Acad Sci USA. 1993;90:9465–9469. doi: 10.1073/pnas.90.20.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pil P M, Lippard S J. Specific binding of chromosomal protein HMG1 to DNA damaged by the anticancer drug cisplatin. Science. 1992;256:234–236. doi: 10.1126/science.1566071. [DOI] [PubMed] [Google Scholar]
- 41.Rodgers K K, Bu Z, Fleming K G, Schatz D G, Engelman D M, Coleman J E. A zinc-binding domain involved in the dimerization of Rag1. J Mol Biol. 1996;260:70–84. doi: 10.1006/jmbi.1996.0382. [DOI] [PubMed] [Google Scholar]
- 41a.Rodgers K K, Villey I J, Ptaszek L, Corbett E, Schatz D G, Coleman J E. A dimer of the lymphoid protein RAG1 recognizes the recombination signal sequence and the complex stably incorporates the high mobility group protein HMG2. Nucleic Acids Res. 1999;27:2938–2946. doi: 10.1093/nar/27.14.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sadofsky M J, Hesse J E, McBlane J F, Gellert M. Expression and V(D)J recombination activity of mutated Rag-1 proteins. Nucleic Acids Res. 1993;21:5644–5650. doi: 10.1093/nar/21.24.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santagata S, Aidinis V, Spanopoulou E. The effect of Me2+ cofactors at the initial stages of V(D)J recombination. J Biol Chem. 1998;273:16325–16331. doi: 10.1074/jbc.273.26.16325. [DOI] [PubMed] [Google Scholar]
- 44.Sawchuk D J, Weis-Garcia F, Malik S, Besmer E, Bustin M, Nussenweig M C, Cortes P. V(D)J recombination: modulation of RAG1 and RAG2 cleavage activity on 12/23 substrates by whole cell extract and DNA bending proteins. J Exp Med. 1997;185:2025–2032. doi: 10.1084/jem.185.11.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schatz D G, Oettinger M A, Baltimore D. The V(D)J recombination activating gene RAG1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 45a.Shockett P E, Schatz D G. DNA hairpin opening mediated by the RAG1 and RAG2 proteins. Mol Cell Biol. 1999;19:3674–3683. doi: 10.1128/mcb.19.6.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silver D P, Spanopoulou E, Mulligan R C, Baltimore D. Dispensable sequence motifs in the Rag-1 and Rag-2 genes for plasmid V(D)J recombination. Proc Natl Acad Sci USA. 1993;90:6100–6104. doi: 10.1073/pnas.90.13.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spanopoulou E, Zaitseva F, Wang F-H, Santagata S, Baltimore D, Panayotou G. The homeodomain region of RAG1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell. 1996;87:263–276. doi: 10.1016/s0092-8674(00)81344-6. [DOI] [PubMed] [Google Scholar]
- 48.Sutrias-Grau M, Bianchi M E, Bernués J. HMG1 interacts with the core domain of human TBP and interferes with TFIIB within the pre-initiation complex. J Biol Chem. 1999;274:1628–1634. doi: 10.1074/jbc.274.3.1628. [DOI] [PubMed] [Google Scholar]
- 49.Swanson P, Desiderio S. V(D)J recombination signal recognition: distinct, overlapping DNA-protein contacts in complexes containing RAG1 with and without RAG2. Immunity. 1998;9:115–125. doi: 10.1016/s1074-7613(00)80593-2. [DOI] [PubMed] [Google Scholar]
- 50.Thanos D, Maniatis T. The high mobility group protein HMG-I(Y) is required for NF-κB-dependent virus induction of the human IFN-β gene. Cell. 1992;71:777–789. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- 51.Thompson J F, Landy A. Empirical estimation of protein-induced DNA bending angles: application to site-specific recombination complexes. Nucleic Acids Res. 1988;16:9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Gent D C, Hiom K, Paull T T, Gellert M. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Gent D C, McBlane J F, Ramsden D A, Sadofsky M J, Hesse J E, Gellert M. Initiation of V(D)J recombination in a cell-free system. Cell. 1995;81:925–934. doi: 10.1016/0092-8674(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 54.van Gent D C, Ramsden D A, Gellert M. The Rag-1 and Rag-2 proteins establish the 12/23 rule in V(D)J recombination. Cell. 1996;85:107–114. doi: 10.1016/s0092-8674(00)81086-7. [DOI] [PubMed] [Google Scholar]
- 55.Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, van de Wetering M, Oosterwegel M, Wilson A, MacDonald H R, Clevers H. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374:70–74. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- 56.Villa A, Santagata S, Bozzi F, Giliani S, Frattini A, Imberti L, Benerini Gatta L, Ochs H D, Schwarz K, Notarangelo L D, Vezzoni P, Spanopoulou E. Partial V(D)J recombination activity leads to Omenn syndrome. Cell. 1998;93:885–896. doi: 10.1016/s0092-8674(00)81448-8. [DOI] [PubMed] [Google Scholar]
- 57.Weis-Garcia F, Besmer E, Sawchuk D J, Yu W, Hu Y, Cassard S, Nussenweig M C, Cortes P. V(D)J recombination: in vitro coding joint formation. Mol Cell Biol. 1997;17:6379–6385. doi: 10.1128/mcb.17.11.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.West R B, Lieber M R. The RAG-HMG1 complex enforces the 12/23 rule of V(D)J recombination at the double-hairpin formation step. Mol Cell Biol. 1998;18:6408–6415. doi: 10.1128/mcb.18.11.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu H M, Crothers D M. The locus of sequence-directed and protein-induced bending. Nature. 1984;308:509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- 60.Zappavigna V, Falciola L, Helmer Citterich M, Mavilio F, Bianchi M E. HMG1 interacts with HOX proteins and enhances their DNA binding and transcriptional activation. EMBO J. 1996;15:4981–4991. [PMC free article] [PubMed] [Google Scholar]
- 61.Zinkel S S, Crothers D M. DNA bend direction by phase sensitive detection. Nature. 1987;328:178–181. doi: 10.1038/328178a0. [DOI] [PubMed] [Google Scholar]
- 62.Zwilling S, Konig H, Wirth T. High mobility group protein 2 functionally interacts with the POU domains of octamer transcription factors. EMBO J. 1995;14:1198–1208. doi: 10.1002/j.1460-2075.1995.tb07103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]