Abstract
The androgen receptor (AR) plays an essential role in the development of prostate cancer, and androgen-deprivation therapy is used as a first-line treatment for prostate cancer. However, under androgen-deprivation therapy, castration-resistant prostate cancer inevitably arises, suggesting that the interacting transcriptional coregulators of AR are promising targets for developing novel therapeutics. In this study, we used novel proteomic techniques to evaluate the AR interactome, including biochemically labile binding proteins, which might go undetected by conventional purification methods. Using rapid immunoprecipitation mass spectrometry of endogenous proteins, we identified enhanced at puberty 1 (EAP1) as a novel AR coregulator, whereas its interaction with AR could not be detected under standard biochemical conditions. EAP1 enhanced the transcriptional activity of AR via the E3 ubiquitin ligase activity, and its ubiquitination substrate proteins included AR and HDAC1. Furthermore, in prostate cancer specimens, EAP1 expression was significantly correlated with AR expression as well as a poor prognosis of prostate cancer. Together, these results suggest that EAP1 is a novel AR coregulator that promotes AR activity and potentially plays a role in prostate cancer progression.
Keywords: AR, androgen, nuclear receptor, coactivator, EAP1, IRF2BPL
The androgen receptor (AR) is a prime transcription factor involved in the development and progression of prostate cancer [1-4]. Activation of AR via its cognate ligand, androgen, leads to its nuclear translocation and, as a chromatin-binding nuclear receptor (NR), binding to DNA elements known as AR response elements [5, 6]. On chromatin binding, AR recruits a series of proteins known as NR coregulators that reorganize chromatin structure to enable AR transcriptional regulation of its target genes [7-9]. Furthermore, biochemical approaches have shown that NR coregulators often act as multisubunit complexes possessing distinct enzymatic activities such as histone modification, chromatin remodeling, and histone binding [10]. Sequential interactions of various NR coregulator complexes with NRs are assumed to promote dynamic changes in chromatin structure that facilitate appropriate transcriptional control [11].
The current therapies for advanced prostate cancer were designed to target AR activation processes such as androgen production, androgen binding to AR, and AR nuclear localization [1, 12]. However, despite these therapies, progression to castration-resistant prostate cancer, which exhibits resistance to AR-targeted therapies and loss of androgen dependency, is inevitable [13]. From this viewpoint, AR coregulators are attracting attention as novel and promising therapeutic targets for prostate cancer [14, 15].
To date, hundreds of AR coregulators have been identified via various methodologies such as yeast-two hybrid assays, glutathione S-transferase pull-down assays, and Tag-based purification [7, 16-18]. Most of these methods are based on detection of biochemically stable protein-protein interactions, and therefore coregulators that associate with the target protein via labile, transient, or indirect interactions are likely not detected by these techniques.
Materials and Methods
In the present study, we used a recently developed proteomic method, rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) [19, 20], to evaluate the AR interactome. We identified enhanced at puberty 1 (EAP1/IRF2BPL) as a novel AR coregulator that promotes AR activity and might play a role in prostate cancer progression.
Cell Culture and Transfection
LNCaP cells were obtained from the American Type Culture Collection and maintained in Roswell Park Memorial Institute (RPMI) 1640 medium (Fujifilm Wako Pure Chemical) supplemented with 10% fetal bovine serum (FBS; Nichirei Biosciences), 100-U/mL penicillin G, and 100-μg/mL streptomycin (Fujifilm Wako Pure Chemical) at 37 °C in 5% CO2. We maintained 293F cells in Dulbecco’s modified Eagle’s medium (DMEM, Nissui Pharmaceutical) supplemented with 10% FBS and antibiotics. To establish short hairpin RNA (shRNA)-expressing LNCaP cells, cells were infected with retrovirus carrying an shRNA-expressing cassette and then selected with RPMI 1640 medium containing 1-μg/mL puromycin. For transfection, we used the Lipofectamine2000 (Invitrogen) according to the manufacturer’s guidance. shRNA-expressing retroviruses were generated using PLAT-A cells. The PLAT-A cells were kindly provided by Dr Toshio Kitamura (University of Tokyo). For dihydrotestosterone (DHT) treatment, the culture medium was replaced by phenol red-free RPMI-1640 medium containing 5% charcoal-stripped FBS. The cells were maintained for about 48 hours in the medium and subsequently exposed to DHT (10 nM) or vehicle (methanol) for indicated time.
Plasmids, Antibodies, and Short Hairpin RNAs
Full-length human AR and EAP1 complementary DNA (cDNA) were amplified by polymerase chain reaction (PCR) from an LNCaP cell cDNA library and were cloned in-frame into the pcDNA3.1-FLAG vector and pcDNA3.1-hemagglutinin (HA) vector, respectively. HA-tagged lysine-specific demethylase 1 (LSD1) was described previously [26, 43]. The EAP1 C715A point mutation were introduced using the PrimeSTAR Mutagenesis Basal Kit (Takara Bio) according to the manufacturer’s instructions. To generate the shRNA expression plasmid, the following shRNA sequences were inserted into the pSUPER.retro.puro vector (Oligoengine): human EAP1_1, 5′-GCCTATCCTCGGGTTTC-3′; human EAP1_2, 5′-GGAGGATACGCATTTCG-3′; and Escherichia coli LacZ shRNA, 5′-gcccatctacaccaacgtaac-3′ [26]. For trypsin-resistant tandem-repeat ubiquitin-binding entity (TR-TUBE)-fused EAP1, FLAG TR-TUBE was amplified from pcDNA-FLAG TR-TUBE (a gift from Dr Yukiko Yoshida, Tokyo Metropolitan Institute of Medical Science [49]) and cloned in-frame into pQCXIP vector with EAP1. For negative control of TR-TUBE-EAP1, FLAG-TR-TUBE was cloned into pRetroX-Tight-Pur vector. The antibodies used for immunoblot analysis and RIME were anti-AR (sc-816 (N-20, RRID:AB_1563391), sc-13062 (H-280, RRID:AB_633881); Santa Cruz Biotechnology), anti-FLAG (F1804; Sigma-Aldrich, RRID:AB_262044), and anti-HA tag (RHGT-45A-Z; ICL, RRID:AB_2861135). The antibodies used for immunohistochemistry and immunofluorescence were anti-AR (GTX29474; GeneTex, RRID:AB_367520) and anti-IRF2BPL (HPA050862; Atlas, RRID:AB_2681258). The antibodies used for PLA were anti-AR (sc-109500 (G-13), Santa Cruz Biotechnology, RRID:AB_1563387) and anti-IRF2BPL (Atlas, RRID:AB_2681258).
Rapid Immunoprecipitation Mass Spectrometry of Endogenous Proteins
The RIME experiments were performed as reported previously [50]. Briefly, 5 × 107 cells were stimulated with DHT for 8 hours and then fixed in 1% formalin for 8 minutes at 37 °C. Cell lysates were mixed with an anti-AR antibody (N-20 or H-280) or rabbit immunoglobulin G (IgG)-coated Dynabeads and incubated at 4 °C overnight. Antibody-Dynabead complexes were then washed and mixed with 10 µL 10 ng/µL Trypsin Gold (Promega) and incubated at 37 °C overnight. Digested peptides were cleaned up using the Pierce Detergent Removal Spin Column (Thermo Fisher Scientific) and analyzed using the LTQ-Orbitrap Velos (Thermo Fisher Scientific) [43, 51]. For protein identification, spectra were processed using Proteome Discoverer, version 1.3 (Thermo Fisher Scientific) with the Mascot algorithm against the human protein database from SwissProt. Peptide data were filtered using a Mascot significance threshold of less than .05. Raw data were deposited in the jPOST Repository (JPST001080) [52].
Immunoprecipitation
293F cell lysates transfected with FLAG-AR and/or HA-tagged proteins were prepared in TNE buffer (20-mM Tris-HCl pH 7.9, 150-mM NaCl, 2-mM EDTA, 1% NP-40, and protease inhibitor). The extracts were incubated with FLAG M2 beads (Sigma-Aldrich) at 4 °C. Immunoprecipitants were washed and boiled with Laemmli sample buffer, and then subjected to sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis and Western blotting with indicated antibodies.
Proximity Ligation Assay
Cells were fixed in 10% formalin and permeabilized with 0.1% Triton X-100. Then, proximity ligation assay (PLA) was performed according to the manufacturer’s instructions (Sigma-Aldrich) using the same antibodies as those used for immunofluorescence staining.
RNA Isolation, Complementary DNA Synthesis, and Quantitative Reverse Transcriptase–Polymerase Chain Reaction
Total RNA was extracted by Sepasol RNA I Super G (Nacalai Tesque) and cDNA synthesized using ReverTra AceR quantitative polymerase chain reaction (qPCR) reverse transcriptase (RT) Master Mix (TOYOBO). qPCR was performed with the Thermal Cycler Dice Real Time System II (Takara Bio) according to the manufacturer’s instructions. The primer sequences for each gene were as follows: EAP1 (forward, 5′-tcgcttcaagaaggaccact-3′ and reverse, 5′-ccgtggggtactcaatgaac-3′); PSA (forward, 5′-ggcagcattgaaccagaggag-3′ and reverse, 5′-gcatgaacttggtcaccttctg-3′); and KLK2 (forward, 5′-tcatccagtctcggattgtg-3′ and reverse, 5′-cttctttaggcaatgggcag-3′); and Nkx3.1 (forward, 5′-gccaagaacctcaagctcac-3′ and reverse, 5′-agaaggcctcctctttcagg-3′); and Rplp0 (forward, 5′-tcgacaatggcagcatctac-3′ and reverse, 5′-tgatgcaacagttgggtagc-3′). RNA levels were normalized using the Rplp0 gene as an internal control.
Chromatin Immunoprecipitation
LNCaP cells were cross-linked for 10 minutes at room temperature with 0.75% formaldehyde-containing phosphate-buffered saline; cross-linking was stopped with phosphate-buffered saline–glycine (0.3 M final). Cells were resuspended in cell lysis buffer (50-mM Tris, pH 8.1, 1% SDS, 10-mM EDTA) and incubated for 10 minutes on ice. Lysates were then sonicated to obtain DNA fragments averaging 200 to 500 bp in length. Sonicated lysates were cleared by centrifugation and diluted in dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2-mM EDTA, 20 mM Tris-HCl, pH 8.1, 167-mM NaCl) and immunoprecipitated overnight with 2 μg of indicated antibodies. Immunoprecipitated DNA was analyzed by qPCR (KAPA SYBR FAST Universal 2X qPCR Master Mix) together with 1% of the input chromatin. Specific primer pairs were designed to amplify the promoter region of the human androgen response element of the PSA gene: (5’-AACAGACCTACTCTGGAGGAACA -3’ and 5’-TCCAGGCTTGCTTACTGTCC-3’), and Nkx3.1 gene: (5’-ATCTGGGAGACTGGCAAAGA-3’ and 5’-GGCACTTCCTGAGCAAACTT-3’), which were designed to amplify one of the AR peak regions [53].
Detection of Ubiquitination Substrates of Enhanced at Puberty 1
LNCaP cells were infected with retrovirus carrying FLAG-TR-TUBE-fused EAP1 gene and selected in a medium containing 1-μg/mL puromycin. As a control, LNCaP cells were infected with retrovirus carrying Dox-inducible FLAG-TR-TUBE. Established cells (1.0 × 106 cells) were lysed and ubiquitinated peptides were purified as previously described [36]. Briefly, the cell lysates were incubated with anti-DYKDDDDK tag antibody agarose (Fujifilm Wako Pure Chemical) for 2 hours at 4 ºC. The resin was washed and subjected to Western blot.
Preparation of Clinical Prostate Cancer Samples
Human prostate cancer samples (needle biopsy samples collected from patients who provided informed consent) were obtained from the tumor tissue bank of Miyagi Cancer Center. The study was reviewed and approved by the ethical committee of Tohoku University School of Medicine (2019-1-311) and Miyagi Cancer Center (2018-031).
Immunohistochemistry
Formalin-fixed paraffin-embedded blocks from prostate cancer tissues were sectioned at 3-μm thickness and mounted onto Superfrost Plus slides. Immunohistochemistry was performed using the BenchMark ULTRA IHC/ISH staining system (Roche Diagnostics). Sections underwent dewaxing, heat-incubated antigen retrieval, and primary and secondary antibody incubations using the aforementioned antibodies. The sections were counterstained with hematoxylin.
Immunohistochemistry Scoring and Statistical Analyses
The Gleason score was categorized into 3 groups (score of 6, 7, and 8 + 9) according to a previous report [54]. EAP1 and AR immunoreactivities were detected in the nucleus of carcinoma cells and were semiquantified using a modified labeling index system according to Mehta et al [55] with some modifications. In this system, immunoreactivity was categorized as 0 (no expression), 10 (0%-10%), 20 (11%-20%)…100 (91%-100%) according to the percentage of positive immunoreactivity [Labeling index (LI)]. Statistical analysis was performed using regression analysis to calculate correlation coefficients. All other values were statistically evaluated using one-way analysis of variance and Fisher protected least significant difference. P less than .05 was considered to indicate significance.
Results
Rapid Immunoprecipitation Mass Spectrometry of Endogenous Protein Purification of Coregulators Associated With Androgen Receptors
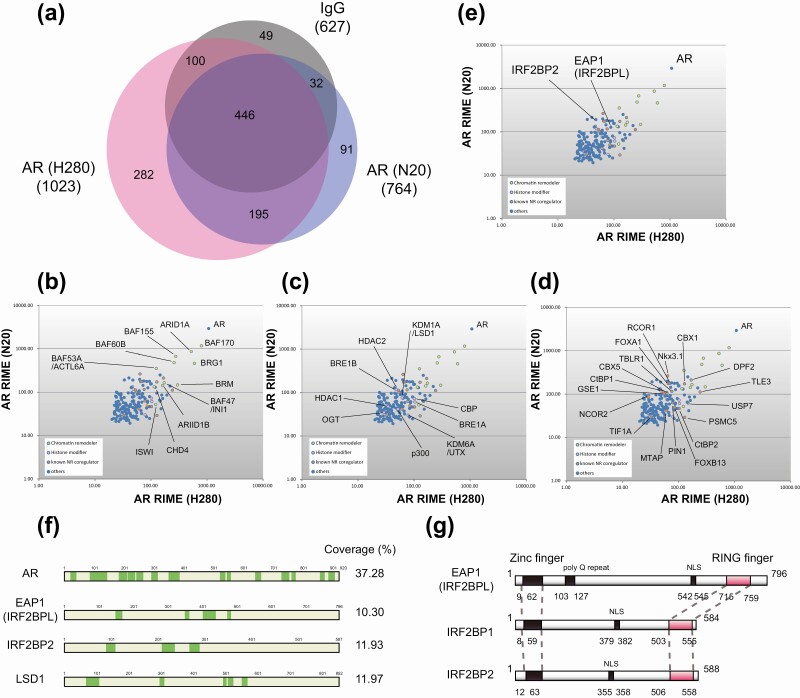
Endogenous AR was purified from 6-hour DHT-treated LNCaP prostate cancer cells using RIME technology. To exclude antibody-specific background proteins, we used 2 distinct antibodies to purify AR (N20 and H280). We considered only the proteins identified by both antibodies and excluded any proteins that appeared in IgG control (Fig. 1A). The 195 extracted proteins were plotted according to their Mascot score and functionally classified into 4 groups: chromatin remodelers, histone modifiers, known nonenzymatic NR coregulators, and others (Fig. 1B-1D, Supplementary Table 1 [21]). AR was the top protein identified by both antibodies, corroborating the validity of our approach. Both antibodies also copurified chromatin remodelers such as BRG1 and BAFs [22] (Fig. 1B), histone modifiers including p300, CBP, and LSD1/KDM1A, which are well-characterized AR coregulators [23-26] (Fig. 1C), as well as nonenzymatic NR coregulators such as FOXA1, TIF1A, and NCOR2 [27-29] (Fig. 1D). These results indicated that AR-interacting coregulators were successfully isolated in our RIME experiment. Among the identified proteins, more than 25% (49 proteins) have been previously reported to interact with NRs. Among the 75% of newly identified proteins, EAP1 was chosen for further analysis because of its uncharacterized function in transcriptional regulation [7-9] (Fig. 1E and 1F). EAP1 belongs to the IRF2BP protein family, which comprises IRF2BP1, IRF2BP2, and EAP1 [30, 31] (Fig. 1G). All of these proteins harbor a RING finger motif and are thought to be E3 ubiquitin ligases, although their substrates remain elusive.
Figure 1.
Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) purification of androgen receptor (AR)-interacting proteins in the LNCaP cells. A, AR RIME was performed using 2 distinct anti-AR antibodies (N-20 and H-280). Only proteins identified by both antibodies (195 proteins) were considered, and any protein that was identified in the immunoglobulin G (IgG) control was excluded. B, Two distinct RIME purifications were conducted, and the proteins identified in both were plotted and categorized as chromatin remodelers; C, histone modifiers; and D, known nonenzymatic nuclear receptor (NR) coregulators. The y-axis represents the log10 Mascot score of each replicate. E, Enhanced at puberty 1 (EAP1; IRF2BPL) and its family member IRF2BP2 were identified by RIME and indicated in the scatter plot. F, Total peptide coverage of specific identified proteins. Green highlighting indicates peptides identified with high confidence (false discovery rate < 0.01). G, Schematic of the protein domain structure of IRF2BP family members. All members harbor a zinc finger domain, nuclear localization signal (NLS), and RING finger domain. EAP1 contains only a poly glutamine (Q) repeat motif.
Colocalization of Androgen Receptors and Enhanced at Puberty 1 in the Nucleus
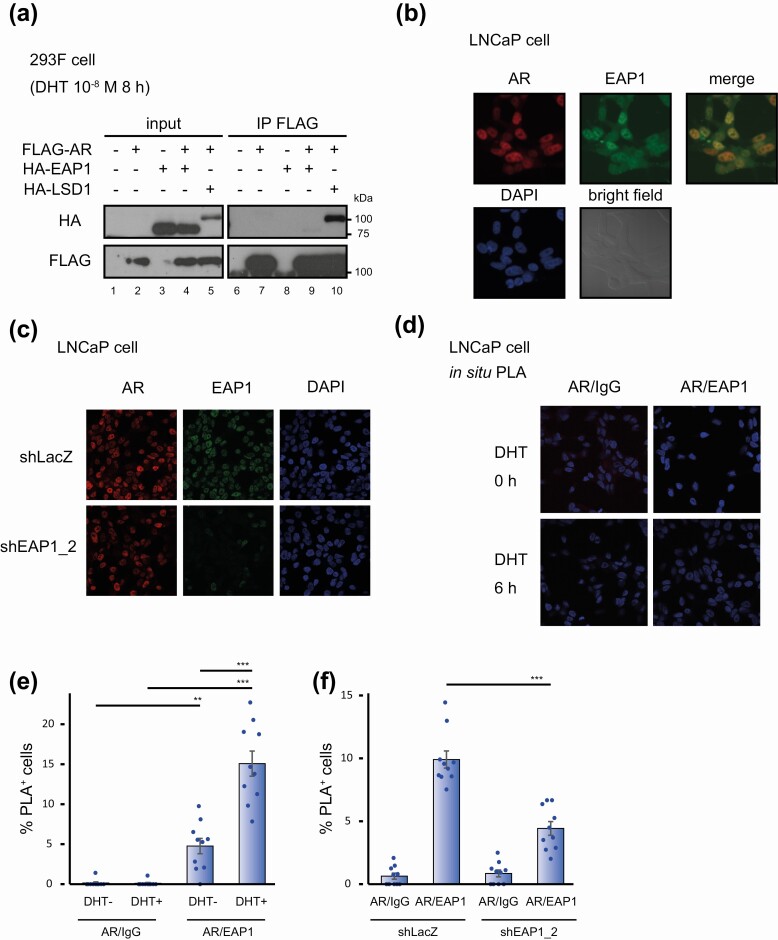
To examine the biochemical interaction between AR and EAP1, we conducted an immunoprecipitation experiment using lysates from DHT-treated 293F cells ectopically expressing FLAG-tagged AR and/or HA-tagged EAP1. Immunoblot analysis revealed that AR did not coimmunoprecipitate with EAP1, whereas LSD1, a known AR-interacting protein used as the positive control [24], did coimmunoprecipitate with AR (Fig. 2A). This suggests that the interaction between AR and EAP1 could not be detected by a conventional biochemical method, presumably because of their biochemically labile interaction.
Figure 2.
Androgen receptor (AR) interacts with enhanced at puberty 1 (EAP1) in the cells to a degree that it could not be detected under the standard biochemical condition. A, 293F cell lysates exogenously expressing FLAG-tagged AR and/or hemagglutinin (HA)-tagged EAP1 and stimulated with 10-nM dihydrotestosterone (DHT) for 8 hours were subjected to immunoprecipitation followed by immunoblot analysis using anti-FLAG and anti-HA antibodies. HA-tagged lysine-specific demethylase 1 (LSD1) was used as a positive control for coimmunoprecipitation. The molecular weights of the marker proteins are indicated on the right. B, LNCaP cells treated with 10-nM DHT for 6 hours were stained with anti-AR and anti-EAP1 antibodies and 4′,6-diamidino-2-phenylindole (DAPI). Both antigens were colocalized in the nucleus of the cells. Bright field images are also shown. C, LNCaP cells that expressed short hairpin RNA (shRNA) for EAP1 were treated with 10-nM DHT for 6 hours and stained with anti-AR and anti-EAP1. LNCaP cells expressing shRNA for LacZ were used as a control. As for EAP1 expression levels in these cells, see Fig. 3C and 3D. D, LNCaP cells were treated for 6 hours with vehicle (ethanol) or with 10-nM DHT and were analyzed by proximity ligation assay (PLA) to validate protein interactions between AR and EAP1. Images were acquired at 40× magnification. E, Quantification of the number of PLA dot–positive cells in Fig. 1D. Data were analyzed by one-way analysis of variance (ANOVA) with a post hoc Tukey-Kramer test (n = 10) Error bars indicate SDs. *P less than .05; **P less than .01; ***P less than .001. F, PLA was performed using 10-nM DHT–treated LNCaP cells (6 hours) that expressed either shLacZ or shEAP1. Quantification of the number of PLA dot–positive cells are shown. Data were analyzed by one-way ANOVA, with a post hoc Tukey-Kramer test (n = 10) Error bars indicate SDs. ***P less than .001.
To observe intracellular localization of AR and EAP1 in LNCaP cells, we performed immunofluorescence staining and found that the signals of both proteins overlapped in the nucleus in DHT-treated cells, implying that AR and EAP1 interact in the nucleus (Fig. 2B). This EAP1 staining was abolished in EAP1 knocked-down cells (shEAP1_2; see also Fig. 3C and 3D), indicating specificity of the antibody used here (Fig. 2C). The interaction between EAP1 and AR observed in the RIME experiment was further confirmed by the PLA, which is used for in situ detection of protein-protein interactions [32]. As shown in Fig. 2D and 2E, the PLA-positive cells were detected in the nucleus of LNCaP cells. Furthermore, PLA signals were significantly increased in a DHT-dependent manner (Fig. 2E), suggesting DHT-dependent complex formation of an EAP1-AR complex. Finally, this PLA signals were decreased in EAP1 knocked-down cells, supporting the validity of this assay (Fig. 2F).
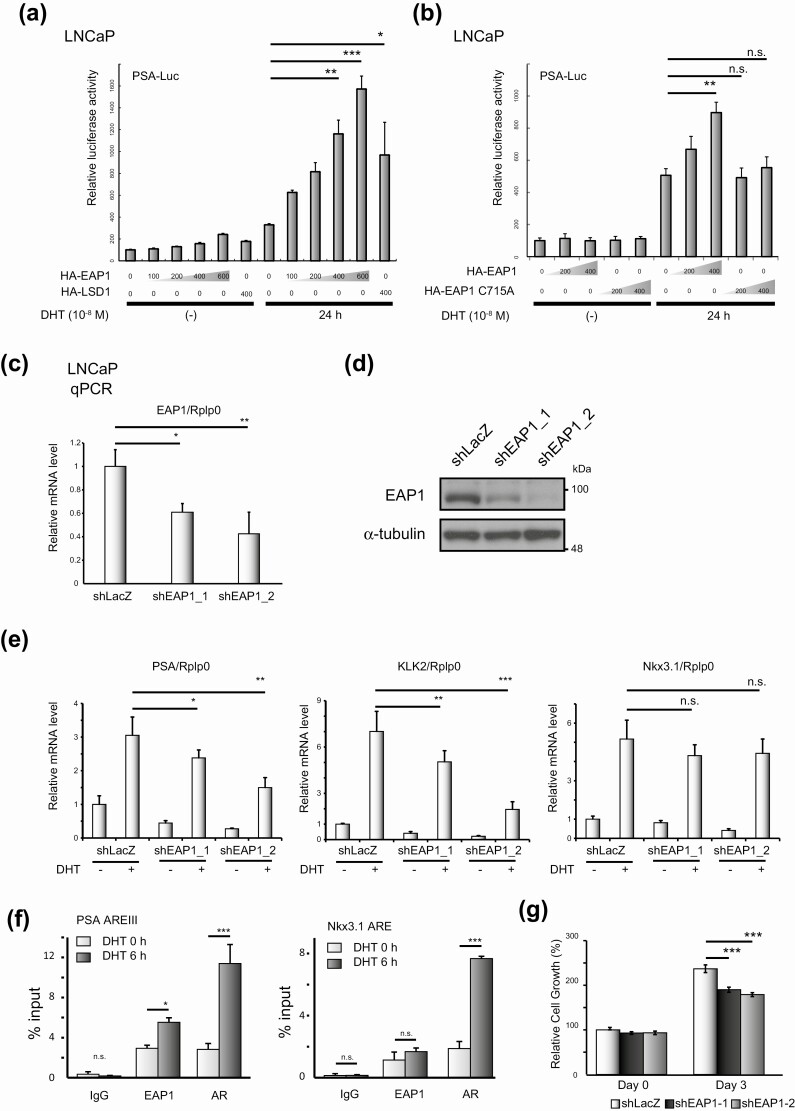
Figure 3.
Enhanced at puberty 1 (EAP1) is an androgen receptor (AR) coactivator. A, LNCaP cells were transiently transfected with the pGL4.1-PSA-Luc reporter (500 ng) and pcDNA3.1-hemagglutinin (HA)-EAP1 expression vectors at the indicated amounts (ng). After 24 hours of incubation, the cells were stimulated with vehicle (ethanol) or 10-nM dihydrotestosterone (DHT) for 24 hours. Data were analyzed by one-way analysis of variance (ANOVA) with a post hoc Tukey-Kramer test (n = 4) Error bars indicate SDs. *P less than .05; **P less than .01; ***P less than .001. HA-tagged lysine-specific demethylase 1 (LSD1), as an AR coactivator, was used as the positive control. B, LNCaP cells were transiently transfected with the pGL4.1-PSA-Luc reporter vector (500 ng) and pcDNA3.1 expressing either HA-tagged wild-type EAP1 or HA-tagged EAP1 C715A mutant at the indicated amounts. After 24 hours’ incubation, cells were stimulated with vehicle (ethanol) or 10-nM DHT for 24 hours. Data were analyzed by one-way ANOVA with a post hoc Tukey-Kramer test (n = 4) Error bars indicate SDs. **P less than .01; n.s., not significant. C, Knockdown of EAP1 by short hairpin RNA (shRNA). Quantitative polymerase chain reaction (qPCR) analysis for EAP1 was performed. LNCaP cells expressing shLacZ were used as a negative control. Gene expression was normalized to that of Rplp0 and presented as the fold change in expression relative to the expression level in cells not treated with DHT. Data were analyzed by one-way ANOVA with a post hoc Tukey-Kramer test (n = 4) Error bars indicate SDs. *P less than .05; **P less than .01. E, Knockdown of EAP1 by shRNA and its effect on the transcriptional activity of AR. Cells were treated with 10-nM DHT for 12 hours, and qPCR analysis of AR target genes was performed. Gene expression was normalized to that of Rplp0 and presented as the fold change in expression relative to the expression level in cells not treated with DHT. Data were analyzed by one-way ANOVA with a post hoc Tukey-Kramer test (n = 4) Error bars indicate SDs. *P less than .05; **P less than .01; ***P less than .001; n.s., not significant. F, Chromatin immunoprecipitation analysis of EAP1 and AR at the indicated gene enhancers. DNA fragments in LNCaP cells were precipitated with anti-EAP1 and anti-AR; rabbit immunoglobulin G (IgG) was used as a negative control for the immunoprecipitation. Precipitated DNA fragments were assessed by qPCR. The error bars indicate SDs. Data were analyzed by one-way ANOVA with a post hoc Tukey-Kramer test (n = 3) Error bars indicate SDs. *P less than .05; ***P less than .001; n.s., not significant. G, Cell proliferation assays were performed using the WST-8 cell proliferation assay. The relative absorbance of each cell at the indicated time points is shown. Data were analyzed by one-way ANOVA with a post hoc Tukey-Kramer test (n = 5) Error bars indicate SDs. ***P less than .001.
Enhanced at Puberty 1 Acts as a Coactivator of Androgen Receptor–mediated Transcription
Next, we investigated the biological significance of the interaction between AR and EAP1. Reporter analysis using the PSA-Luc construct containing the full-length 6-kb upstream regulatory region of PSA gene reporter plasmid was performed [33, 34]. As shown in Fig. 3A, EAP1 increased the DHT-induced transcriptional activity of AR dose dependently. However, when a construct containing EAP1 with the C715A mutation, in which the conserved cysteine critical for E3 ubiquitin ligase activity [35] was mutated to alanine, was used in the analysis, EAP1 showed loss of AR coactivator activity (Fig. 3B), suggesting that EAP1 promotes the transcriptional activity of AR via its E3 ubiquitin ligase activity. Furthermore, we used quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR) analysis to verify that EAP1 regulated the messenger RNA expression of AR target genes in LNCaP cells. Consistent with the aforementioned findings, the expression of DHT-induced AR target genes, such as PSA and KLK2, was downregulated when EAP1 was knocked down by shRNA specific for EAP1, but not for LacZ (Fig. 3C-3E). On the other hand, the AR target gene Nkx3.1 was not downregulated, suggesting that EAP1 might regulate a specific subset of AR target genes. Supporting this finding, chromatin immunoprecipitation assay revealed that EAP1 was recruited to the androgen response element of the PSA gene promoter but not to Nkx3.1 gene promoter in a DHT-dependent manner (Fig. 3F). Furthermore, the EAP1-specific shRNA significantly inhibited LNCaP cell proliferation (Fig. 3G). Together, these data suggest that EAP1 is a novel AR coactivator.
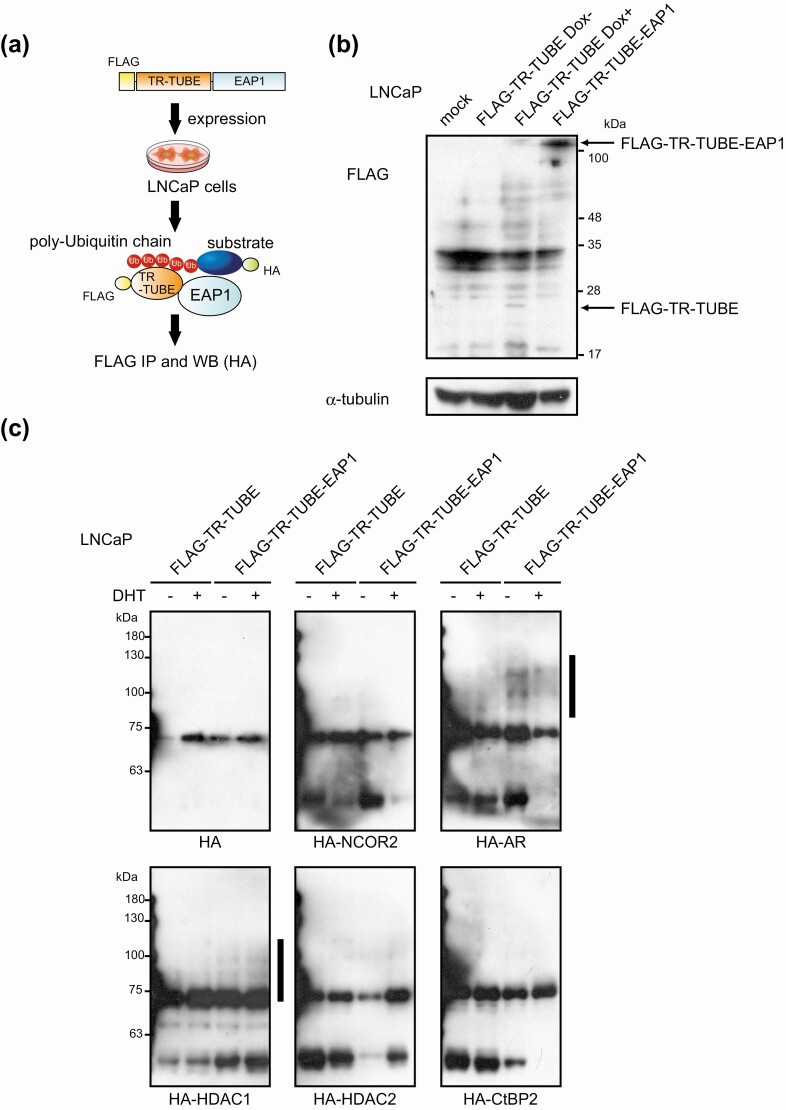
Next, we further examined the mechanism underlying E3 ubiquitin-ligase activity-dependent AR coactivation function of EAP1. To address ubiquitination substrates of EAP1, we applied a novel E3 ligase substrate-trapping strategy by fusing a trypsin-resistant–tandem ubiquitin-binding entity (TR-TUBE) with EAP1. In this method, polyubiquitinated substrates can be captured by TR-TUBE and protected from degradation or deubiquitination and, therefore, substrates can be detected with high sensitivity [36] (Fig. 4A). Because EAP1 acted as an AR coactivator, we reasoned that EAP1 might target corepressor proteins for ubiquitination. To evaluate this hypothesis, we tested a series of transcriptional corepressors identified in our RIME experiments (see Fig. 1) as candidate EAP1 substrates.
Figure 4.
Detection of ubiquitination substrates of enhanced at puberty 1 (EAP1) using trypsin-resistant–tandem ubiquitin-binding entity (TR-TUBE). A, Schematic diagrams of workflow for purifying ubiquitination substrates of EAP1 using the TR-TUBE strategy. B, LNCaP cells stably expressing FLAG-TR-TUBE-fused EAP and doxycycline (Dox)-inducible FLAG-TR-TUBE were established. For Dox treatment, 1-μg/mL Dox was added and incubated for 24 hours. Cell lysates were subjected to immunoblot analysis using indicated antibodies. The molecular weights of the marker proteins are indicated on the right. C, Cells were transfected with HA-tagged indicated proteins. For negative control, Dox-inducible FLAG-TR-TUBE–bearing cells were treated with 1-μg/mL Dox for 48 hours. After 48 hours, TR-TUBE–captured proteins were immunoprecipitated with anti-FLAG antibody and assessed by Western blot using anti-HA antibody. The molecular weights of the marker proteins are indicated on the left side. Vertical bars denote the positions of polyubiquitinated substrates.
First, we established LNCaP cells stably expressing FLAG-TR-TUBE–fused EAP1 and doxycycline (Dox)-inducible FLAG-TR-TUBE as a negative control (Fig. 4B). Using anti-FLAG immunoprecipitation product from these cells, we could detect self-ubiquitination of EAP1 by mass spectrometry (data not shown), showing E3 ubiquitin ligase activity of EAP1 and validating this experimental system. Next, cells were transfected with HA-tagged indicated proteins, and then proteins were extracted and immunoprecipitated with anti-FLAG antibody. Purified proteins were assessed by Western blot. As shown in Fig. 4C, among selected proteins, AR and HDAC1 were polyubiquitinated by EAP1, suggesting that EAP1 is an E3 ubiquitin ligase for AR and HDAC1 protein. This result implies that EAP1 regulates AR-HDAC1 complex levels via ubiquitin-proteasome pathways to activate transcriptional activity of AR.
Enhanced at Puberty 1 Is Overexpressed, and Correlated With Poor Outcome, in Prostate Cancer
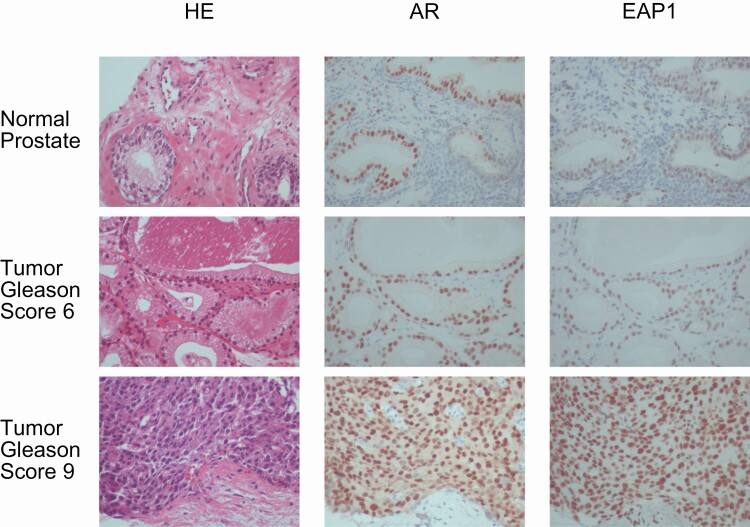
Finally, to assess the clinical relevance of EAP1 in prostate cancer, we performed immunohistochemical analyses to examine the localization of EAP1 in human prostate tumor tissues obtained by needle biopsy. As shown in representative tissue sections with varying Gleason scores, EAP1 was expressed in the nucleus both of normal prostate and prostate tumor luminal epithelial cells (Fig. 5). Importantly, these cells also expressed AR in the nucleus, supporting the formation of an AR-EAP1 complex in human prostate cancer cells. Furthermore, the EAP1 expression level was significantly correlated with the AR expression level, and also with Gleason scores and TMN classification (Table 1), indicating that EAP1 is a key AR coactivator in prostate cancer progression.
Figure 5.
Enhanced at puberty 1 (EAP1) and androgen receptor (AR) colocalized in the nuclei of human prostate cells in vivo. Immunohistochemical staining of AR and EAP1 in human normal and tumor prostate specimens with different Gleason scores. AR (middle column) and EAP1 (right column) immunoreactivities were detected in the same cell nuclei. Hematoxylin and eosin–stained images are shown in the left column. All sections were taken from the same continuous specimen, and images were acquired at 40× magnification.
Table 1.
Association between enhanced at puberty 1 (EAP1) immunohistochemical expression and clinicopathological parameters in 24 prostate cancer specimens. Association between EAP1 LI and clinicopathological parameters in 24 prostate cancer specimens
| Value | No. | EAP1 LI, % | P |
|---|---|---|---|
| Agea, y | 24 | .52 (r = –0.14) | |
| pT | |||
| pT1 | 6 | 68.3 ± 7.9 | |
| pT2 | 13 | 93.8 ± 2.4 | < .001 (pT1 vs pT2) |
| pT3 | 5 | 96.0 ± 2.5 | < .001 (pT1 vs pT3) |
| Stage | |||
| I | 13 | 93.1 ± 5.5 | |
| II | 6 | 97.7 ± 4.0 | |
| III + IV | 5 | 96.0 ± 2.5 | .26 |
| Gleason score | |||
| 6 | 5 | 78.0 ± 7.3 | |
| 7 | 10 | 84.0 ± 6.0 | |
| 8 + 9 | 9 | 97.8 ± 1.5 | .02 (6 vs 8 + 9) |
| AR LIa(%) | 24 | .002 (r = 0.61) |
Data are presented as mean ± SEM. P less than .05 is considered significant and is shown in bold.
Abbreviation: AR, androgen receptor.
a Statistical analysis was performed by correlation coefficient (r) and regression test. All other values were statistically evaluated using one-way analysis of variance and Fisher protected least significant difference.
Discussion
By applying RIME to isolate AR coregulators, we have unveiled protein-protein interaction networks involving this major driving transcription factor in a prostate cancer cell line. Of note, the identified proteins included labile binding coregulators such as EAP1, which presumably go undetected by conventional biochemical identification methods. Furthermore, using 2 distinct antibodies against AR, as well as rabbit IgG as a negative control to exclude nonspecific binding proteins, we were able to distinguish true interactions with AR from antibody-specific background interactions. Thus, the RIME data provided here contain more confident and reproducible proteins compared with experiments using a single antibody. This strategy can be applied to identify proteins associated with any transcription factor, with less risk of detecting nonspecific proteins.
Among the identified AR-associated factors, we focused on EAP1 for further study because of its uncharacterized function in transcriptional regulation, although a role of this protein in neural development has been reported [37]. We observed colocalization of AR and EAP1 in the nucleus of LNCaP cells and prostate cancer/normal prostate specimens. EAP1 enhanced the transcriptional activity of AR, which was dependent on the E3 ubiquitin ligase activity of EAP1. EAP1 belongs to the IRF2BP family, and another member of this family, IRF2BP1, has been reported to function as a transcriptional corepressor [30, 38, 39]. Recently, Lempiäinen et al [40, 41] reported that IRF2BP2 acts as a coactivator of NRs such as the glucocorticoid receptor and AR. This suggests that IRF2BP2 family proteins have dichotomous functions in gene regulation depending on chromatin context or their binding partner, similar to the dual function of LSD1 in transcriptional regulation [24, 26, 42, 43]. Furthermore, CG11138, the Drosophila counterpart of EAP1, colocalized with the Drosophila ecdysone receptor, an NR, in ecdysone-induced puffs (euchromatin region) within the larval salivary gland (data not shown, personal communication from Dr Sawatsubashi, Tokushima University [44], 2017), suggesting that EAP1 might be a highly conserved NR coregulator from Drosophila to humans.
The precise mechanism of EAP1-mediated coactivation of AR transcription still remains unclear. From the data provided here, the function of EAP1 as a coactivator requires its ubiquitin ligase activity, implying that EAP1 coactivates AR transcriptional activity via ubiquitin-proteasomal degradation of certain proteins. Several ubiquitin ligases have been reported to regulate AR transcriptional activity via distinct mechanisms, including regulation of local turnover of AR chromatin complexes, recruitment of AR coactivators, and global AR stability [45-48]. Taking the role of EAP1 as an AR coactivator into account, AR corepressor proteins including AR and HDAC1 are candidate targets of EAP1-mediated ubiquitination to be tested in the future. Furthermore, target gene preference of EAP1 also remains elusive and chromatin immunoprecipitation sequencing using next-generation sequencing might solve this limitation.
Given the key role of AR in prostate cancer progression, the findings from the present study provide new insight into the transcriptional machinery underlying AR-mediated gene regulation. We found that EAP1 expression was significantly correlated with AR expression, the Gleason score, and TMN classification. Although it is unclear why EAP1 is upregulated during cancer progression, as supported by the finding that EAP1 contributed to LNCaP cell proliferation, this protein is a potentially promising target for the treatment of prostate cancers.
Taken together, our work identifies EAP1 as an important coregulator of AR in prostate cancer cells. EAP1 coactivated AR via E3 ubiquitin ligase activity though overview of the targeted proteins of the ubiquitination is to be identified. If transcription factors have labile interacting coregulators like EAP1 adding to their conventional coregulators, there might be a requirement for reconsideration of transcriptional interactomes, leading to identification of novel mechanisms for transcriptional regulation. And from those processes, important drug target proteins might be discovered in the future.
Acknowledgements
We appreciate the technical assistance provided by Dr Saya Ito-Ueda (Kyoto Prefectural University of Medicine) and Dr Daiki Umetsu (Tohoku University).
Financial Support: This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (grant Nos. 16K08606 to A.Y., 16H03252 to A.S., and 16K15492 to A.S.), the Yamaguchi Endocrine Research Foundation (to A.Y.), and the Organization of the Smoking Research Foundation (to A.S.).
Glossary
Abbreviations
- AR
androgen receptor
- cDNA
complementary DNA
- DHT
dihydrotestosterone
- Dox
doxycycline
- EAP1
enhanced at puberty 1
- FBS
fetal bovine serum
- HA
hemagglutinin
- IgG
immunoglobulin G
- LSD1
lysine-specific demethylase 1
- NR
nuclear receptor
- PCR
polymerase chain reaction
- PLA
proximity ligation assay
- qRT-PCR
quantitative reverse transcriptase–polymerase chain reaction
- RIME
rapid immunoprecipitation mass spectrometry of endogenous proteins
- RPMI
Roswell Park Memorial Institute
- shRNA
short hairpin RNA;
- TR-TUBE
trypsin-resistant–tandem ubiquitin-binding entity.
Additional Information
Disclosures : The authors have nothing to disclose.
Data Availability
Some or all data generated or analyzed during this study are included in this published article or in the data repositories listed in “References.”
References
- 1. Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer. 2015;15(12):701-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Matsumoto T, Sakari M, Okada M, et al. The androgen receptor in health and disease. Annu Rev Physiol. 2013;75:201-224. [DOI] [PubMed] [Google Scholar]
- 3. Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24(18):1967-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lempiäinen JK, Manjur ABMK, Malinen M, Ketola K, Niskanen EA, Palvimo JJ. BCOR-coupled H2A monoubiquitination represses a subset of androgen receptor target genes regulating prostate cancer proliferation. Oncogene. 2020;39(11):2391-2407. [DOI] [PubMed] [Google Scholar]
- 5. Lubahn DB, Joseph DR, Sullivan PM, Willard HF, French FS, Wilson EM. Cloning of human androgen receptor complementary DNA and localization to the X chromosome. Science. 1988;240(4850):327-330. [DOI] [PubMed] [Google Scholar]
- 6. Mangelsdorf DJ, Thummel C, Beato M, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83(6):835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Heemers HV, Tindall DJ. Androgen receptor (AR) coregulators: a diversity of functions converging on and regulating the AR transcriptional complex. Endocr Rev. 2007;28(7):778-808. [DOI] [PubMed] [Google Scholar]
- 8. Kato S, Yokoyama A, Fujiki R. Nuclear receptor coregulators merge transcriptional coregulation with epigenetic regulation. Trends Biochem Sci. 2011;36(5):272-281. [DOI] [PubMed] [Google Scholar]
- 9. Dasgupta S, Lonard DM, O’Malley BW. Nuclear receptor coactivators: master regulators of human health and disease. Annu Rev Med. 2014;65:279-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20(11):1405-1428. [DOI] [PubMed] [Google Scholar]
- 11. Yokoyama A, Fujiki R, Ohtake F, Kato S. Regulated histone methyltransferase and demethylase complexes in the control of genes by nuclear receptors. Cold Spring Harb Symp Quant Biol. 2011;76:165-173. [DOI] [PubMed] [Google Scholar]
- 12. Tran C, Ouk S, Clegg NJ, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324(5928):787-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Crawford ED, Schellhammer PF, McLeod DG, et al. Androgen receptor targeted treatments of prostate cancer: 35 years of progress with antiandrogens. J Urol. 2018;200(5):956-966. [DOI] [PubMed] [Google Scholar]
- 14. Skowron KJ, Booker K, Cheng C, et al. Steroid receptor/coactivator binding inhibitors: an update. Mol Cell Endocrinol. 2019;493:110471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tice CM, Zheng YJ. Non-canonical modulators of nuclear receptors. Bioorg Med Chem Lett. 2016;26(17):4157-4164. [DOI] [PubMed] [Google Scholar]
- 16. Hsiao JJ, Smits MM, Ng BH, Lee J, Wright ME. Discovery proteomics identifies a molecular link between the coatomer protein complex i and androgen receptor-dependent transcription. J Biol Chem. 2016;291(36):18818-18842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stelloo S, Nevedomskaya E, Kim Y, et al. Endogenous androgen receptor proteomic profiling reveals genomic subcomplex involved in prostate tumorigenesis. Oncogene. 2018;37(3):313-322. [DOI] [PubMed] [Google Scholar]
- 18. Paltoglou S, Das R, Townley SL, et al. Novel androgen receptor coregulator GRHL2 exerts both oncogenic and antimetastatic functions in prostate cancer. Cancer Res. 2017;77(13):3417-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mohammed H, D’Santos C, Serandour AA, et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 2013;3(2):342-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mohammed H, Taylor C, Brown GD, Papachristou EK, Carroll JS, D’Santos CS. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat Protoc. 2016;11(2):316-326. [DOI] [PubMed] [Google Scholar]
- 21. Yokoyama A. September 10, 2021. Androgen receptor RIME using LNCaP cells, jPOST ID: JPST001080. https://repository.jpostdb.org/
- 22. Inoue H, Furukawa T, Giannakopoulos S, Zhou S, King DS, Tanese N. Largest subunits of the human SWI/SNF chromatin-remodeling complex promote transcriptional activation by steroid hormone receptors. J Biol Chem. 2002;277(44):41674-41685. [DOI] [PubMed] [Google Scholar]
- 23. Chen H, Lin RJ, Schiltz RL, et al. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90(3):569-580. [DOI] [PubMed] [Google Scholar]
- 24. Metzger E, Wissmann M, Yin N, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437(7057):436-439. [DOI] [PubMed] [Google Scholar]
- 25. Chikanishi T, Fujiki R, Hashiba W, Sekine H, Yokoyama A, Kato S. Glucose-induced expression of MIP-1 genes requires O-GlcNAc transferase in monocytes. Biochem Biophys Res Commun. 2010;394(4):865-870. [DOI] [PubMed] [Google Scholar]
- 26. Yokoyama A, Takezawa S, Schüle R, Kitagawa H, Kato S. Transrepressive function of TLX requires the histone demethylase LSD1. Mol Cell Biol. 2008;28(12):3995-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lupien M, Eeckhoute J, Meyer CA, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132(6):958-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Teyssier C, Ou CY, Khetchoumian K, Losson R, Stallcup MR. Transcriptional intermediary factor 1alpha mediates physical interaction and functional synergy between the coactivator-associated arginine methyltransferase 1 and glucocorticoid receptor-interacting protein 1 nuclear receptor coactivators. Mol Endocrinol. 2006;20(6):1276-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ye X, Han SJ, Tsai SY, et al. Roles of steroid receptor coactivator (SRC)-1 and transcriptional intermediary factor (TIF) 2 in androgen receptor activity in mice. Proc Natl Acad Sci U S A. 2005;102(27):9487-9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Childs KS, Goodbourn S. Identification of novel co-repressor molecules for interferon regulatory factor-2. Nucleic Acids Res. 2003;31(12):3016-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heger S, Mastronardi C, Dissen GA, et al. Enhanced at puberty 1 (EAP1) is a new transcriptional regulator of the female neuroendocrine reproductive axis. J Clin Invest. 2007;117(8):2145-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Söderberg O, Gullberg M, Jarvius M, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3(12):995-1000. [DOI] [PubMed] [Google Scholar]
- 33. Cleutjens KB, van der Korput HA, van Eekelen CC, van Rooij HC, Faber PW, Trapman J. An androgen response element in a far upstream enhancer region is essential for high, androgen-regulated activity of the prostate-specific antigen promoter. Mol Endocrinol. 1997;11(2):148-161. [DOI] [PubMed] [Google Scholar]
- 34. Shang Y, Myers M, Brown M. Formation of the androgen receptor transcription complex. Mol Cell. 2002;9(3):601-610. [DOI] [PubMed] [Google Scholar]
- 35. Freemont PS. RING for destruction? Curr Biol. 2000;10(2):R84-R87. [DOI] [PubMed] [Google Scholar]
- 36. Watanabe M, Saeki Y, Takahashi H, et al. A substrate-trapping strategy to find E3 ubiquitin ligase substrates identifies Parkin and TRIM28 targets. Commun Biol. 2020;3(1):592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marcogliese PC, Shashi V, Spillmann RC, et al. IRF2BPL is associated with neurological phenotypes. Am J Hum Genet. 2018;103(2):245-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li D, Das S, Yamada T, Samuels HH. The NRIF3 family of transcriptional coregulators induces rapid and profound apoptosis in breast cancer cells. Mol Cell Biol. 2004;24(9):3838-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yeung KT, Das S, Zhang J, et al. A novel transcription complex that selectively modulates apoptosis of breast cancer cells through regulation of FASTKD2. Mol Cell Biol. 2011;31(11):2287-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lempiäinen JK, Niskanen EA, Vuoti KM, et al. Agonist-specific protein interactomes of glucocorticoid and androgen receptor as revealed by proximity mapping. Mol Cell Proteomics. 2017;16(8):1462-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Manjur ABMK, Lempiäinen JK, Malinen M, Palvimo JJ, Niskanen EA. IRF2BP2 modulates the crosstalk between glucocorticoid and TNF signaling. J Steroid Biochem Mol Biol. 2019;192:105382. [DOI] [PubMed] [Google Scholar]
- 42. Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437(7057):432-435. [DOI] [PubMed] [Google Scholar]
- 43. Yokoyama A, Igarashi K, Sato T, et al. Identification of myelin transcription factor 1 (MyT1) as a subunit of the neural cell type-specific lysine-specific demethylase 1 (LSD1) complex. J Biol Chem. 2014;289(26):18152-18162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sawatsubashi S, Murata T, Lim J, et al. A histone chaperone, DEK, transcriptionally coactivates a nuclear receptor. Genes Dev. 2010;24(2):159-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Geng C, Rajapakshe K, Shah SS, et al. Androgen receptor is the key transcriptional mediator of the tumor suppressor SPOP in prostate cancer. Cancer Res. 2014;74(19):5631-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Qi J, Fan L, Hussain A. Implications of ubiquitin ligases in castration-resistant prostate cancer. Curr Opin Oncol. 2015;27(3):172-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Qi J, Tripathi M, Mishra R, et al. The E3 ubiquitin ligase Siah2 contributes to castration-resistant prostate cancer by regulation of androgen receptor transcriptional activity. Cancer Cell. 2013;23(3):332-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xu K, Shimelis H, Linn DE, et al. Regulation of androgen receptor transcriptional activity and specificity by RNF6-induced ubiquitination. Cancer Cell. 2009;15(4):270-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yoshida Y, Saeki Y, Murakami A, et al. A comprehensive method for detecting ubiquitinated substrates using TR-TUBE. Proc Natl Acad Sci U S A. 2015;112(15):4630-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Noro E, Yokoyama A, Kobayashi M, et al. Endogenous purification of NR4A2 (Nurr1) identified poly(ADP-Ribose) polymerase 1 as a prime coregulator in human adrenocortical H295R cells. Int J Mol Sci. 2018;19(5):1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yokoyama A, Okuno Y, Chikanishi T, et al. KIAA1718 is a histone demethylase that erases repressive histone methyl marks. Genes Cells. 2010;15(8):867-873. [DOI] [PubMed] [Google Scholar]
- 52. Okuda S, Watanabe Y, Moriya Y, et al. jPOSTrepo: an international standard data repository for proteomes. Nucleic Acids Res. 2017;45(D1):D1107-D1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xu B, Song B, Lu X, et al. Altered chromatin recruitment by FOXA1 mutations promotes androgen independence and prostate cancer progression. Cell Res. 2019;29(9):773-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Takayama KI, Suzuki T, Fujimura T, Takahashi S, Inoue S. Association of USP10 with G3BP2 inhibits p53 signaling and contributes to poor outcome in prostate cancer. Mol Cancer Res. 2018;16(5):846-856. [DOI] [PubMed] [Google Scholar]
- 55. Mehta RJ, Jain RK, Leung S, et al. FOXA1 is an independent prognostic marker for ER-positive breast cancer. Breast Cancer Res Treat. 2012;131(3):881-890. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Some or all data generated or analyzed during this study are included in this published article or in the data repositories listed in “References.”