ABSTRACT
Triple-negative breast cancer (TNBC) is an aggressive histological subtype of breast cancer. It has been reported that the circular RNA (circRNA) circ-ERBB2 (circBase identifier hsa_circ_0007766) is mainly distributed in the cytoplasm of TNBC cells and promotes the proliferation and invasion of TNBC cells. This study aimed to explore the molecular mechanism of circ-ERBB2 regulating the progression of TNBC. The expression of circ-ERBB2 was detected by quantitative real-time PCR (qRT-PCR). Loss-of-function experiments were performed to investigate the function of circ-ERBB2 in TNBC cells in vitro and in vivo. The regulatory mechanism of circ-ERBB2 was surveyed by bioinformatics analysis and dual-luciferase reporter and RNA immunoprecipitation (RIP) or RNA pulldown assays. We observed that circ-ERBB2 was overexpressed in TNBC, and TNBC patients with high circ-ERBB2 expression levels had a poor prognosis. Functionally, circ-ERBB2 knockdown constrained TNBC growth in vivo, reduced the Warburg effect, accelerated apoptosis, and repressed the proliferation, migration, and invasion of TNBC cells in vitro. Mechanically, circ-ERBB2 sponged microRNA 136-5p (miR-136-5p) to elevate pyruvate dehydrogenase kinase 4 (PDK4) expression. In conclusion, circ-ERBB2 facilitated the Warburg effect and malignancy of TNBC cells by the miR-136-5p/PDK4 pathway, at least in part. This study supports circ-ERBB2 as a prognostic indicator for TNBC.
KEYWORDS: TNBC, circ-ERBB2, miR-136-5p, PDK4, triple-negative breast cancer
INTRODUCTION
Triple-negative breast cancer (TNBC) is an aggressive histological subtype of breast cancer (1). It is characterized by the lack of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) (2). Also, TNBC patients are not suitable for hormone or anti-HER2 therapy (3, 4). At present, cytotoxic chemotherapy and radiotherapy are still the treatment methods for TNBC patients (5). However, TNBC has a poor prognosis due to its aggressiveness and lack of effective therapeutic targets (6). Therefore, exploring the mechanism of TNBC progression is crucial for the development of new treatment strategies.
Oxidative phosphorylation (OXPHOS) and glycolysis are the two main metabolic pathways that produce ATP, which is a coenzyme responsible for the production of energy for cellular processes. The Warburg effect, also termed aerobic glycolysis, is an energy transfer from mitochondrial OXPHOS to aerobic glycolysis (7). Also, this metabolic reprogramming is a characteristic of cancer (8). Moreover, targeting the Warburg effect is a promising cancer treatment strategy (9–11).
Increasing evidence has proven that noncoding RNAs are involved in the Warburg effect of cancer cells, including circular RNAs (circRNAs) (12). circRNAs, covalently closed RNA molecules, are generated by the back-splicing of pre-mRNAs (13). Due to their special structure, most circRNAs are stable and resistant to RNase R (14). Also, circRNAs can function as competing endogenous RNAs (ceRNAs) that regulate gene expression by sponging microRNAs (miRs) (15). In recent years, circRNAs have been disclosed to be associated with the tumorigenesis of a series of cancers. For instance, circRNA circ-0008234 elevated the Warburg effect and contributed to gallbladder cancer growth via regulating pyruvate kinase L/R (PKLR) expression (16). circRNA circ-BICD2 sponged miR-107 to increase HK2 expression, resulting in facilitating the Warburg effect and tumor advancement in oral squamous cell cancer (17). circRNA circ-ERBB2 (circBase identifier hsa_circ_0007766), located on chromosome 17 (chr17) at positions 37864573 to 37866734, is produced by the back-splicing of the erb-b2 receptor tyrosine kinase 2 (ERBB2) gene, and its length is 676 bp (http://www.circbank.cn/search.html?selectValue=hsa_circ_0007766). It was reported that circ-ERBB2 acted as an oncogene in TNBC (18). Nevertheless, the regulatory mechanisms of circ-ERBB2 in TNBC have not been fully clarified.
Accordingly, this research aimed to survey the impact of circ-ERBB2 on the Warburg effect and tumor growth in TNBC. Our results showed that circ-ERBB2 had a promoting impact on the Warburg effect and tumor growth in TNBC through modulating the miR-236-5p/PDK4 (pyruvate dehydrogenase kinase 4) pathway.
RESULTS
circ-ERBB2 expression was prominently increased in TNBC.
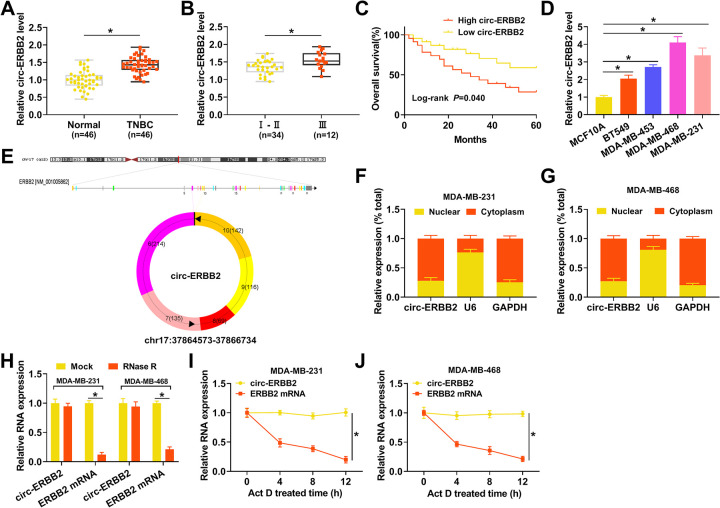
RNA sequencing (RNA-seq) uncovered that circ-ERBB2 expression was apparently increased in TNBC tissues compared to adjacent normal tissues (19). To validate this result, we conducted quantitative real-time PCR (qRT-PCR) to detect circ-ERBB2 expression in 46 paired TNBC tissues and adjacent normal tissues. The results exhibited a prominent upregulation of circ-ERBB2 in TNBC tissues in contrast to adjacent normal tissues (Fig. 1A). Moreover, TNBC patients with high circ-ERBB2 expression levels were closely associated with tumor node metastasis (TNM) stage (P = 0.033) and lymph node metastasis (P = 0.008) (Table 1). qRT-PCR showed that circ-ERBB2 expression was observably elevated in stage III TNBC tissues compared to that in stage I and II TNBC tissues (Fig. 1B). Kaplan-Meier survival analysis demonstrated that TNBC patients with high circ-ERBB2 expression levels had poor overall survival compared to those with low circ-ERBB2 expression levels (log rank P value of 0.040) (Fig. 1C). Next, we detected circ-ERBB2 expression in TNBC cells (BT549, MDA-MB-453, MDA-MB-468, and MDA-MB-231), and the results displayed that circ-ERBB2 expression was markedly higher in TNBC cells than in MCF10A cells, particularly MDA-MB-231 and MDA-MB-468 cells (Fig. 1D). circ-ERBB2 is formed by splicing the head and tail of exon 2 to exon 6 of the ERBB2 gene (Fig. 1E). Subsequently, we surveyed the distribution of circ-ERBB2 in MDA-MB-231 and MDA-MB-468 cells. A subcellular isolation assay indicated that circ-ERBB2 was preferentially distributed in the cytoplasm of MDA-MB-231 and MDA-MB-468 cells (Fig. 1F and G). Also, we observed that there was a marked downregulation of ERBB2 mRNA under RNase R treatment, while circ-ERBB2 resisted RNase R digestion (Fig. 1H). In addition, circ-ERBB2 had a longer half-life than ERBB2 mRNA in MDA-MB-231 and MDA-MB-468 cells under actinomycin D (Act D) treatment (Fig. 1I and J). These data indicated that high circ-ERBB2 expression in TNBC was associated with a poor prognosis.
FIG 1.
circ-ERBB2 expression was elevated in TNBC. (A) qRT-PCR revealed the expression of circ-ERBB2 in TNBC tissues and adjacent normal tissues. (B) qRT-PCR presented the expression of circ-ERBB2 in TNBC tissues, which were in stages I and II or stage III. (C) Kaplan-Meier and log rank tests uncovered the overall survival of TNBC patients with high or low circ-ERBB2 expression. Patients were divided into a high-circ-ERBB2-expression group and a low-circ-ERBB2-expression group according to the median level of circ-ERBB2 expression in tumor tissues. (D) qRT-PCR exhibited the expression of circ-ERBB2 in TNBC cells (BT549, MDA-MB-453, MDA-MB-468, and MDA-MB-231) and MCF10A cells. (E) Schematic diagram illustrating the formation of circ-ERBB2. (F and G) qRT-PCR revealed the subcellular distribution of circ-ERBB2 in MDA-MB-231 and MDA-MB-468 cells. (H) qRT-PCR presented the expression levels of circ-ERBB2 and ERBB2 mRNAs in RNA of MDA-MB-231 and MDA-MB-468 cells under RNase R treatment. (I and J) qRT-PCR unveiled the expression levels of circ-ERBB2 and ERBB2 mRNAs in MDA-MB-231 and MDA-MB-468 cells treated with Act D. *, P < 0.05.
TABLE 1.
Clinicopathological factors and circ-ERBB2 expression in patients with TNBCa
| Parameter | No. of patients |
P valueb | ||
|---|---|---|---|---|
| Total | circ-ERBB2 expression |
|||
| High (n = 23) | Low (n = 23) | |||
| Age (yrs) | ||||
| ≤50 | 18 | 8 | 10 | 0.546 |
| >50 | 28 | 15 | 13 | |
| Menopausal status | ||||
| Premenopausal | 20 | 12 | 8 | 0.234 |
| Postmenopausal | 26 | 11 | 15 | |
| Histological type | ||||
| Ductal | 42 | 20 | 22 | 0.295 |
| Lobular | 4 | 3 | 1 | |
| Tumor size (cm) | ||||
| ≤2 | 18 | 6 | 12 | 0.070 |
| >2 | 28 | 17 | 11 | |
| TNM stage | ||||
| I–II | 29 | 11 | 18 | 0.033* |
| III | 17 | 12 | 5 | |
| Lymph node metastasis | ||||
| No | 21 | 6 | 15 | 0.008* |
| Yes | 25 | 17 | 8 | |
Abbreviations: TNBC, triple-negative breast cancer; TNM, tumor node metastasis.
By a chi-square test. *, P < 0.05.
Inhibition of circ-ERBB2 reduced the Warburg effect and cured the malignancy of TNBC cells.
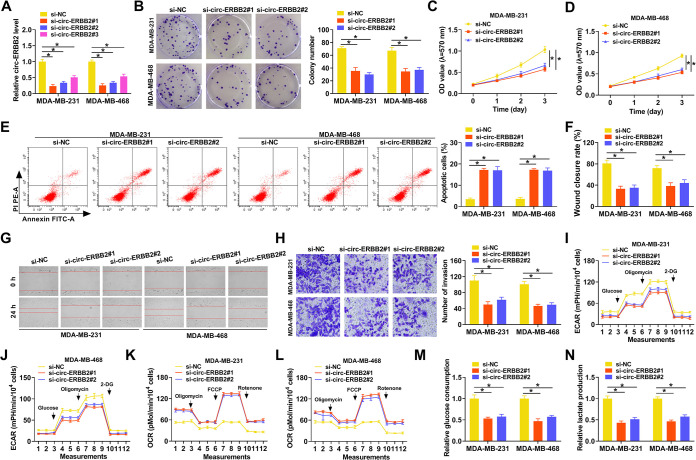
Subsequently, we performed loss-of-function experiments to investigate the function of circ-ERBB2 in TNBC. The knockdown efficiency of the circ-ERBB2 inhibitor in MDA-MB-231 and MDA-MB-468 cells was confirmed by qRT-PCR, as exhibited in Fig. 2A. Colony formation and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays displayed that the proliferation of circ-ERBB2-silenced MDA-MB-231 and MDA-MB-468 cells was curbed (Fig. 2B to D). Cell apoptosis analysis indicated that the inhibition of circ-ERBB2 elevated the apoptosis rate of MDA-MB-231 and MDA-MB-468 cells (Fig. 2E). Wound-healing and transwell assays exhibited that the migration and invasion of MDA-MB-231 and MDA-MB-468 cells were repressed after transfection with a small interfering RNA (siRNA) targeting circ-ERBB2 (si-circ-ERBB2#1 or si-circ-ERBB2#2) (Fig. 2F to H). As expected, circ-ERBB2 silencing elevated the protein level of cleaved caspase 3 and decreased the protein levels of proliferating cell nuclear antigen (PCNA) and matrix metalloproteinase 9 (MMP9) in TNBC cells (see Fig. S1 in the supplemental material). To investigate the influence of circ-ERBB2 on the Warburg effect in TNBC cells, we determined the extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) in circ-ERBB2-silenced MDA-MB-231 and MDA-MB-468 cells. As exhibited in Fig. 2I to L, circ-ERBB2 silencing reduced the ECAR, whereas it elevated the OCR, in MDA-MB-231 and MDA-MB-468 cells. In addition, the levels of glucose consumption and lactate production were decreased in circ-ERBB2-inhibited MDA-MB-231 and MDA-MB-468 cells (Fig. 2M and N). Collectively, these results indicated that circ-ERBB2 elevated the Warburg effect and promoted the malignancy of TNBC cells.
FIG 2.
Influence of circ-ERBB2 inhibition on the Warburg effect and malignancy of TNBC cells. (A) The relative expression of circ-ERBB2 in MDA-MB-231 and MDA-MB-468 cells transfected with si-circ-ERBB2#1, si-circ-ERBB2#2, or si-circ-ERBB2#3 was validated by qRT-PCR. si-NC acted as a control. (B to H) The proliferation, apoptosis, migration, and invasion of MDA-MB-231 and MDA-MB-468 cells transfected with si-circ-ERBB2#1, si-circ-ERBB2#2, or si-NC were assessed by a colony formation assay, MTT assay, flow cytometry assay, wound-healing assay, or transwell assay. (I to L) An XF96 glycolysis analyzer was utilized to analyze the ECAR and OCR in MDA-MB-231 and MDA-MB-468 cells transfected with si-circ-ERBB2#1, si-circ-ERBB2#2, or si-NC. (M and N) The levels of glucose consumption and lactate production in MDA-MB-231 and MDA-MB-468 cells transfected with si-circ-ERBB2#1, si-circ-ERBB2#2, or si-NC were detected with matching kits. 2-DG, 2-deoxy-d-glucose; PI, propidium iodide; mPH: miles per hour; FCCP, carbonyl cyanide p-(trifluoromethoxy) phenylhydrazone. In addition, PE-A in panel E comes with the apoptosis detection instrument. *, P < 0.05.
circ-ERBB2 was identified as a sponge for miR-136-5p in TNBC cells.
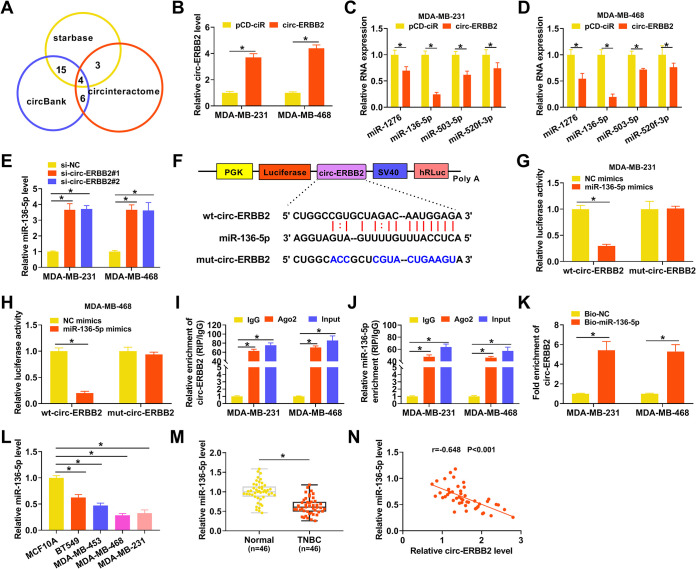
Based on the distribution of circ-ERBB2 in MDA-MB-231 and MDA-MB-468 cells, we speculated that circ-ERBB2 might function as a ceRNA. Online bioinformatics prediction (circinteractome, circBank, and starbase) revealed that 4 miRs (miR-1276, miR-136-5p, miR-503-5p, and miR-520f-3p) had complementary binding sequences with circ-ERBB2 (Fig. 3A). The overexpression efficiency of circ-ERBB2 was validated by qRT-PCR (Fig. 3B). Also, the elevation of circ-ERBB2 repressed the expression of miR-1276, miR-136-5p, miR-503-5p, and miR-520f-3p in MDA-MB-231 and MDA-MB-468 cells, especially miR-136-5p (Fig. 3C and D). Furthermore, miR-136-5p was highly expressed in circ-ERBB2-inhibited MDA-MB-231 and MDA-MB-468 cells (Fig. 3E). To verify the relationship between miR-136-5p and circ-ERBB2, we constructed a luciferase reporter containing wild-type circ-ERBB2 (wt-circ-ERBB2) or mutant circ-ERBB2 (mut-circ-ERBB2), as presented in Fig. 3F. The results of a dual-luciferase reporter assay indicated that miR-136-5p mimics reduced the luciferase activity of the luciferase reporter containing wt-circ-ERBB2 but not the luciferase reporter containing mut-circ-ERBB2 (Fig. 3G and H). An RNA immunoprecipitation (RIP) assay exhibited that miR-136-5p and circ-ERBB2 were overtly enriched in the immunoprecipitates in the Ago2 group compared to the IgG group (Fig. 3I and J). An RNA pulldown assay exhibited that circ-ERBB2 could be pulled down by the biotinylated miR-136-5p (Bio-miR-136-5p) probe but not the biotinylated negative-control (Bio-NC) probe (Fig. 3K). As expected, there was an overt downregulation of miR-136-5p in TNBC cells and tissues (Fig. 3L and M). Pearson’s correlation analysis showed that the expression of miR-136-5p and circ-ERBB2 had a negative correlation in TNBC tissues (Fig. 3N). Together, these data demonstrated that circ-ERBB2 acted as a sponge for miR-136-5p in TNBC cells.
FIG 3.
circ-ERBB2 served as a sponge for miR-136-5p in TNBC cells. (A) Venn diagram showing miRs with a complementary binding sequence to circ-ERBB2 in the circinteractome, circBank, and starbase databases. (B) The relative expression of circ-ERBB2 in MDA-MB-231 and MDA-MB-468 cells transfected with pCD-ciR or -circ-ERBB2 was measured by qRT-PCR. (C and D) The influence of circ-ERBB2 overexpression on the expression of miR-1276, miR-136-5p, miR-503-5p, and miR-520f-3p in MDA-MB-231 and MDA-MB-468 cells was determined by qRT-PCR. (E) The impact of circ-ERBB2 inhibition on the expression of miR-136-5p in MDA-MB-231 and MDA-MB-468 cells was assessed by qRT-PCR. (F) Schematic illustration of a luciferase reporter containing wt-circ-ERBB2 or mut-circ-ERBB2. SV40, simian virus 40. (G and H) MDA-MB-231 and MDA-MB-468 cells were cotransfected with the luciferase reporter containing wt-circ-ERBB2 or mut-circ-ERBB2 and miR-136-5p mimics or NC mimics, and luciferase activity was analyzed by a dual-luciferase reporter assay. (I and J) The enrichment of circ-ERBB2 and miR-136-5p in immunoprecipitates of the Ago2 and IgG groups was analyzed by qRT-PCR. (K) After an RNA pulldown assay, the abundance of circ-ERBB2 in the Bio-miR-136-5p and Bio-NC groups was analyzed by qRT-PCR. (L and M) The relative expression of miR-136-5p in TNBC cells and tissues was evaluated using qRT-PCR. (N) Pearson’s correlation analysis revealed the correlation between circ-ERBB2 and miR-136-5p in TNBC tissues. PGK, phosphoglycerate kinase; hRLuc, Renilla luciferase. *, P < 0.05.
circ-ERBB2 regulated the Warburg effect and malignancy of TNBC cells by sponging miR-136-5p.
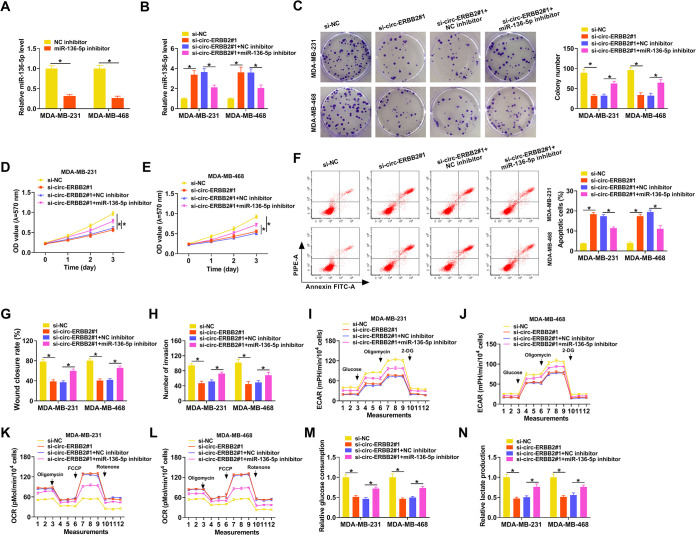
To further validate the involvement of circ-ERBB2 and miR-136-5p in TNBC progression, we performed rescue experiments. There was a marked downregulation of miR-136-5p in MDA-MB-231 and MDA-MB-468 cells after miR-136-5p inhibitor transfection relative to the negative-control (NC) inhibitor (Fig. 4A). Also, the upregulation of miR-136-5p in circ-ERBB2-inhibited MDA-MB-231 and MDA-MB-468 cells was partly neutralized after miR-136-5p downregulation (Fig. 4B). Moreover, the miR-136-5p inhibitor partially counteracted the repressive influence of circ-ERBB2 silencing on the proliferation of MDA-MB-231 and MDA-MB-468 cells (Fig. 4C to E). Furthermore, the miR-136-5p inhibitor overturned the circ-ERBB2 knockdown-mediated impacts on apoptosis, migration, and invasion of MDA-MB-231 and MDA-MB-468 cells (Fig. 4F to H). In addition, the decrease of the ECAR and the increase of the OCR in si-circ-ERBB2#1-transfected MDA-MB-231 and MDA-MB-468 cells were offset after miR-136-5p inhibition (Fig. 4I to L). Similarly, the decreases of glucose consumption and lactate production in MDA-MB-231 and MDA-MB-468 cells caused by circ-ERBB2 inhibition were partly restored after miR-136-5p silencing (Fig. 4M and N). These results demonstrated that circ-ERBB2 sponged miR-136-5p to modulate the Warburg effect and malignancy of TNBC cells.
FIG 4.
circ-ERBB2 adsorbed miR-136-5p to regulate the Warburg effect and malignancy of TNBC cells. (A) qRT-PCR uncovered the expression of miR-136-5p in MDA-MB-231 and MDA-MB-468 cells transfected with the miR-136-5p inhibitor or NC inhibitor. (B to N) MDA-MB-231 and MDA-MB-468 cells were transfected with si-NC, si-circ-ERBB2#1, si-circ-ERBB2#1 plus the NC inhibitor, or si-circ-ERBB2#1 plus the miR-136-5p inhibitor. (B) The relative expression of miR-136-5p in MDA-MB-231 and MDA-MB-468 cells was analyzed using qRT-PCR. (C to H) The influence of miR-136-5p inhibition on the proliferation, apoptosis, migration, and invasion of circ-ERBB2-silenced MDA-MB-231 and MDA-MB-468 cells was evaluated using a colony formation assay, MTT assay, flow cytometry assay, wound-healing assay, or transwell assay. (I to L) The impacts of miR-136-5p silencing on the ECAR and OCR of circ-ERBB2-repressed MDA-MB-231 and MDA-MB-468 cells were analyzed using an XF96 glycolysis analyzer. (M and N) The effects of miR-136-5p downregulation on glucose consumption and lactate production of circ-ERBB2-inhibited MDA-MB-231 and MDA-MB-468 cells were surveyed using the corresponding kits. *, P < 0.05.
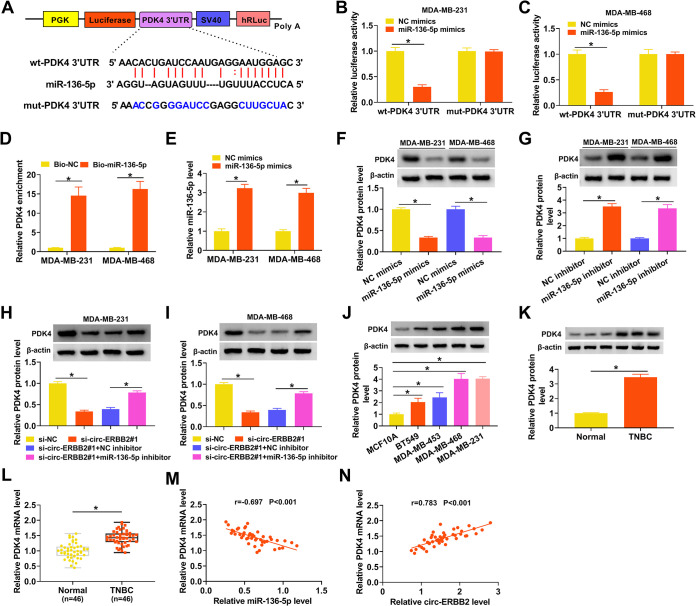
PDK4 was verified as a target for miR-136-5p in TNBC cells.
To survey the biological mechanism of miR-136-5p in TNBC, we employed starbase to predict the targets of miR-136-5p. We discovered that PDK4 might be a target of miR-136-5p and constructed a luciferase reporter containing the wt-PDK4 3′ untranslated region (UTR) or mut-PDK4 3′ UTR, as displayed in Fig. 5A. Also, the luciferase activity of the luciferase reporter containing the wt-PDK4 3′ UTR in MDA-MB-231 and MDA-MB-468 cells was curbed in the presence of miR-136-5p mimics, but there was no conspicuous change in the luciferase reporter containing the mut-PDK4 3′ UTR (Fig. 5B and C). An RNA pulldown assay showed that PDK4 could be pulled down by the Bio-miR-136-5p probe but not the Bio-NC probe (Fig. 5D). We also confirmed the transfection efficiency of miR-136-5p mimics in MDA-MB-231 and MDA-MB-468 cells, as shown in Fig. 5E. Western blotting displayed that miR-136-5p mimics had an inhibitory effect on the level of PDK4 protein in MDA-MB-231 and MDA-MB-468 cells, whereas the miR-136-5p inhibitor had an opposing impact (Fig. 5F and G). Moreover, circ-ERBB2 inhibition reduced the level of PDK4 protein in MDA-MB-231 and MDA-MB-468 cells, but this decrease was partially restored after miR-136-5p inhibition (Fig. 5H and I). We also observed an apparent elevation of the PDK4 protein level in TNBC cells (Fig. 5J). Congruously, the levels of PDK4 protein and mRNA were overtly increased in TNBC tissues (Fig. 5K and L). Additionally, the expression of PDK4 mRNA was negatively correlated with miR-136-5p and positively correlated with circ-ERBB2 in TNBC tissues (Fig. 5M and N). Collectively, these data indicated that PDK4 acted as a downstream target of miR-136-5p and was regulated by the circ-ERBB2/miR-136-5p axis.
FIG 5.
circ-ERBB2 sponged miR-136-5p to regulate PDK4 expression. (A) Schematic illustration of a luciferase reporter containing the wt-PDK4 3′ UTR or mut-PDK4 3′ UTR. (B and C) The targeting relationship between miR-136-5p and PDK4 was verified by a dual-luciferase reporter assay. (D) After an RNA pulldown assay, the enrichment of PDK4 mRNA was analyzed by qRT-PCR. (E) The transfection efficiency of miR-136-5p mimics in MDA-MB-231 and MDA-MB-468 cells was confirmed by qRT-PCR. (F and G) The influence of the miR-136-5p mimics or inhibitor on the level of PDK4 protein in MDA-MB-231 and MDA-MB-468 cells was determined by Western blotting. (H and I) The impact of the miR-136-5p inhibitor on the protein level of PDK4 in circ-ERBB2-suppressed MDA-MB-231 and MDA-MB-468 cells was assessed using Western blotting. (J and K) The relative protein level of PDK4 in TNBC cells and tissues was determined by Western blotting. (L) The relative expression of PDK4 mRNA in TNBC tissues was measured using qRT-PCR. (M and N) The correlation between PDK4 mRNA and miR-136-5p or circ-ERBB2 was determined by Pearson’s correlation analysis. *, P < 0.05.
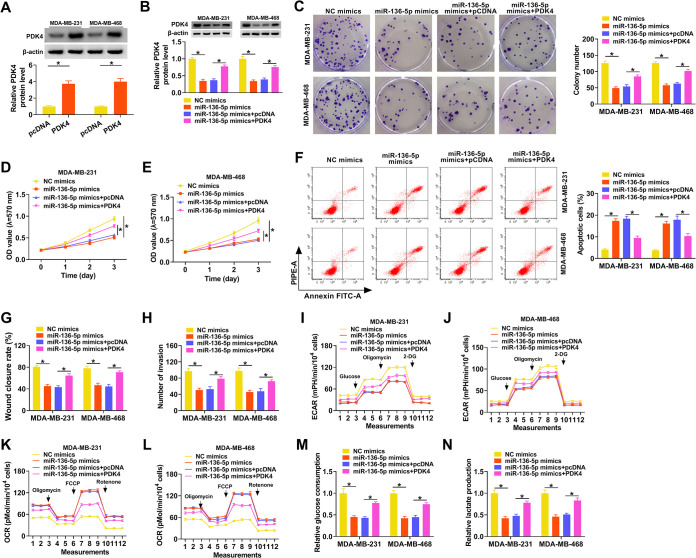
miR-136-5p targeted PDK4 to regulate the Warburg effect and malignancy of TNBC cells.
Given that miR-136-5p targeted PDK4 in TNBC cells, we further surveyed whether miR-136-5p regulated the Warburg effect and malignancy of TNBC cells by targeting PDK4. Western blotting revealed that the level of PDK4 protein was upregulated in TNBC cells after PDK4 transfection compared to the control pcDNA (Fig. 6A). Moreover, the forced expression of PDK4 partly offset the suppressive influence of miR-136-5p mimics on the protein levels of PDK4 in MDA-MB-231 and MDA-MB-468 cells (Fig. 6B). miR-136-5p mimics repressed the proliferation and accelerated the apoptosis of MDA-MB-231 and MDA-MB-468 cells, but these impacts were partly counteracted by PDK4 overexpression (Fig. 6C to F). Also, the elevation of PDK4 overturned the repressive influence of miR-136-5p mimics on the migration and invasion of MDA-MB-231 and MDA-MB-468 cells (Fig. 6G and H). Furthermore, miR-136-5p upregulation reduced the ECAR and elevated the OCR in MDA-MB-231 and MDA-MB-468 cells, but these tendencies were reversed after PDK4 elevation (Fig. 6I to L). In addition, the decreases of glucose consumption and lactate production in miR-136-5p mimic-transfected MDA-MB-231 and MDA-MB-468 cells were offset by forced PDK4 expression (Fig. 6M and N). These results indicated that miR-136-5p reduced the Warburg effect and curbed the malignancy of TNBC cells through targeting PDK4.
FIG 6.
miR-136-5p decreased the Warburg effect and repressed the malignancy of TNBC cells via targeting PDK4. (A) Western blotting revealed the protein level of PDK4 in MDA-MB-231 and MDA-MB-468 cells transfected with pcDNA or PDK4. (B to N) MDA-MB-231 and MDA-MB-468 cells were transfected with NC mimics, miR-136-5p mimics, miR-136-5p mimics plus pcDNA, or miR-136-5p mimics plus PDK4. (B) The protein level of PDK4 in MDA-MB-231 and MDA-MB-468 cells was surveyed using Western blotting. (C to H) The proliferation, apoptosis, migration, and invasion of MDA-MB-231 and MDA-MB-468 cells were evaluated using a colony formation assay, MTT assay, flow cytometry assay, wound-healing assay, or transwell assay. (I to L) The ECAR and OCR of MDA-MB-231 and MDA-MB-468 cells were measured by an XF96 glycolysis analyzer. (M and N) Glucose consumption and lactate production in MDA-MB-231 and MDA-MB-468 cells were determined using the corresponding kits. *, P < 0.05.
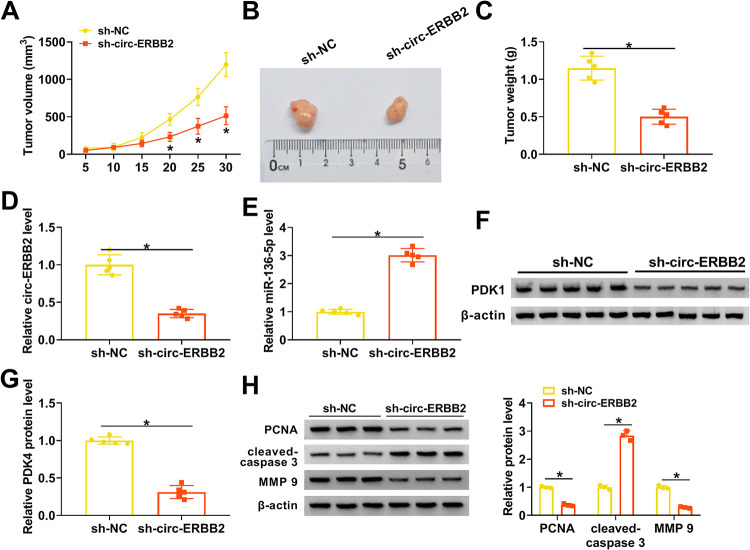
circ-ERBB2 inhibition reduced TNBC growth in vivo.
Based on the above-described findings, we further confirmed the role of circ-ERBB2 in TNBC by xenograft experiments. The results exhibited that tumor volume and weight were decreased in mice injected with MDA-MB-231 cells stably expressing short hairpin RNA (shRNA) targeting ERBB2 (sh-circ-ERBB2) compared with the sh-NC group (Fig. 7A to C). Moreover, circ-ERBB2 expression was overtly lower, whereas miR-136-5p expression was apparently higher, in mouse tumor tissues in the sh-circ-ERBB2 group than in the sh-NC group (Fig. 7D and E). Also, there was a significant downregulation of PDK4 protein in mouse tumor tissues in the sh-circ-ERBB2 group (Fig. 7F and G). In addition, protein levels of PCNA and MMP9 were downregulated in xenograft tumors of the sh-circ-ERBB2 group, but the protein level of cleaved caspase 3 had the opposite tendency (Fig. 7H). These data indicated that circ-ERBB2 knockdown decreased TNBC growth in vivo.
FIG 7.
circ-ERBB2 facilitated TNBC growth in vivo. (A) Tumor volume of mice injected with MDA-MB-231 cells carrying sh-circ-ERBB2 or sh-NC. (B) Tumor images of mice injected with MDA-MB-231 cells carrying sh-circ-ERBB2 or sh-NC. (C) Tumor weight of mice injected with MDA-MB-231 cells carrying sh-circ-ERBB2 or sh-NC. (D to G) The expression of circ-ERBB2, miR-136-5p, and PDK4 protein in mouse tumor tissues in the sh-circ-ERBB2 and sh-NC groups was assessed by qRT-PCR or Western blotting. (H) The protein levels of PCNA, cleaved caspase 3, and MMP9 in tumor tissues of the sh-circ-ERBB2 and sh-NC groups were evaluated by Western blotting. *, P < 0.05.
DISCUSSION
Growing evidence has unmasked that circRNAs exert a vital regulatory role in diverse biological processes, especially in the initiation and growth of various malignant tumors (20). Recently, many studies have disclosed that circRNAs take part in the advancement of TNBC. For instance, circRNA circ-AHNAK1 (21) and circRNA circ-TADA2As (22) played an inhibitory role in TNBC, but circRNA circ-AGFG1 (23), circRNA circ-EPSTI1 (24), and circRNA circ-SEPT9 (25) had the opposite function. Nevertheless, the specific mechanisms by which circRNAs regulate the tumorigenesis of TNBC remain unclear.
It was reported that circ-ERBB2 expression was higher in TNBC tissues (19). Li et al. revealed that high circ-ERBB2 expression was negatively correlated with the overall survival of TNBC patients, and circ-ERBB2 silencing repressed proliferation, invasion, and tumorigenesis in vitro and in vivo by inhibiting HER2-103 encoded by it (18). Here, circ-ERBB2 was highly expressed in TNBC. High circ-ERBB2 expression was related to lymph node metastasis and TNM stage. Also, elevated circ-ERBB2 expression had a negative correlation with the overall survival of TNBC patients. These data indicated that circ-ERBB2 might serve as a prognostic indicator for TNBC. Furthermore, circ-ERBB2 knockdown reduced the Warburg effect and malignancy of TNBC cells in vitro and constrained TNBC cell growth in vivo. A previous study disclosed that circ-ERBB2 acted as an unfavorable prognostic factor in gastric cancer, and circ-ERBB2 overexpression accelerated tumor growth by modulating the miR-637/MMP-19 and miR-1233-3p/GDF15 pathways (26). Huang et al. showed that the circ-ERBB2–PA2G4–TIFIA axis had a promoting impact on the growth of gallbladder cancer (27). These results revealed that circ-ERBB2 exerted a carcinogenic role in TNBC. Unfortunately, we did not explore the effect of circ-ERBB2 on MCF10A cells, which can be further studied in the future.
Accumulating evidence has demonstrated that circRNAs can function as ceRNAs and modulate gene expression through adsorbing miRs (28). Here, we observed that circ-ERBB2 was preferentially distributed in the cytoplasm of TNBC cells, suggesting that circ-ERBB2 might be a sponge for miRs. Through online bioinformatics prediction, we found that circ-ERBB2 was a sponge of miR-136-5p and confirmed this by dual-luciferase reporter and RIP assays. miR-136-5p had been found to play a tumor-inhibiting role in diverse tumors, such as retinoblastoma (29), cervical cancer (30), and gastric cancer (31). Moreover, circRNA circ-TLK1 sponged miR-136-5p to accelerate cell migration and proliferation in renal cell cancer (32). Also, long noncoding RNA (lncRNA) FAM83H-AS1 increased metadherin expression by absorbing miR-136-5p, resulting in the growth of TNBC (33). Here, miR-136-5p was lowly expressed in TNBC. Furthermore, miR-136-5p downregulation partly restored the repressive impact of circ-ERBB2 downregulation on the Warburg effect and malignancy of TNBC cells. Therefore, we inferred that circ-ERBB2 contributed to the Warburg effect and malignancy of TNBC cells, at least in part, via adsorbing miR-136-5p. In addition, circ-ERBB2 may play a regulatory role in TNBC by adsorbing other miRs or proteins, which can be further explained in the future.
Pyruvate dehydrogenase kinases (PDKs) are responsible for the phosphorylation of pyruvate dehydrogenase and the metabolic conversion from mitochondrial respiration to glycolysis (34). PDK4, a member of the PDK family, plays a critical role in several cancer types. Woolbright et al. indicated that the silencing of PDK4 could elevate cisplatin-induced bladder cancer cell death (35). Moreover, miR-16-5p targeted PDK4 to decrease glycolysis and chemoresistance in cervical cancer (36). Also, FAM210B knockdown reduced PDK4 expression, thereby reducing cancer cell glycolysis, the epithelial-mesenchymal transition (EMT), and invasion (37). Furthermore, miR-211 could target PDK4 to repress the Warburg effect and accelerate the apoptosis of breast cancer cells (38). Here, PDK4 acted as a target for miR-136-5p. The overexpression of PDK4 overturned the inhibitory impact of miR-136-5p mimics on the Warburg effect and malignancy of TNBC cells. Of note, circ-ERBB2 sponged miR-136-5p to modulate PDK4 expression. Thus, we concluded that circ-ERBB2 accelerated the Warburg effect and malignancy of TNBC cells through modulating the miR-136-5p/PDK4 axis.
In sum, circ-ERBB2 acted as an oncogene in TNBC. Moreover, circ-ERBB2 accelerated the Warburg effect and tumor growth in TNBC by the miR-136-5p/PDK4 pathway, at least in part. This study uncovered that circ-ERBB2 might be a prognostic biomarker.
MATERIALS AND METHODS
Case selection.
The protocols of the study were reviewed and approved by the Ethics Committee of Fuzhou Second Hospital affiliated with Xiaomen University. All tissue samples (TNBC tissues and adjacent normal tissues) were collected from 46 TNBC patients who underwent TNBC resection at the Fuzhou Second Hospital affiliated with Xiaomen University. The clinicopathological characteristics of TNBC patients are displayed in Table 1. All recruited patients provided written informed consent.
Cell culture.
Four TNBC cell lines (BT549, MDA-MB-453, MDA-MB-468, and MDA-MB-231) and a normal human breast epithelial cell line (MCF10A) were purchased from Procell (Wuhan, China). MCF10A cells were cultured in its special medium (catalog number CM-0525; Procell). BT549 cells were cultured in RPMI 1640 medium (Procell) comprising insulin (0.023 IU/ml; Sigma, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS) (Procell) and 1% penicillin-streptomycin (P/S) (Procell). MDA-MB-453, MDA-MB-468, and MDA-MB-231 cells were maintained in Leibovitz’s L-15 medium (Procell) supplemented with 10% FBS (Procell) and 1% P/S (Procell). These cells were incubated in a humidified chamber at 37°C with 5% CO2.
Transient transfection.
All oligonucleotides used in this study were synthesized by GenePharma (Shanghai, China), including small interfering RNA (siRNA) targeting circ-ERBB2 (si-circ-ERBB2#1 [5′-ACTGCCTGTCCCTGATATCCA-3′], si-circ-ERBB2#2 [5′-GTGACTGCCTGTCCCTGATAT-3′], and si-circ-ERBB2#3 [5′-GCCTGTCCCTGATATCCAGGA-3′]) and the negative control (NC) (si-NC), an miR-136-5p mimic (5′-ACUCCAUUUGUUUUGAUGAUGGA-3′) and the matching NC (NC mimics) (5′-UCACAACCUCCUAGAAAGAGUAGA-3′), and an miR-136-5p inhibitor (5′-TCCATCATCAAAACAAATGGAGT-3′) and the corresponding NC (NC inhibitor) (5′-TGACTGTACTGAACTCGACTG-3′). Transient transfection was carried out using the Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific, Waltham, MA, USA) or Lipofectamine 3000 reagent (Thermo Fisher Scientific). The pcDNA-PDK4 (PDK4) plasmid was constructed by subcloning the sequence of PDK4 into the pcDNA vector (Thermo Fisher Scientific) between the BamHI and XhoI sites. For the pCD-ciR-circ-ERBB2 (circ-ERBB2) plasmid, the sequence of circ-ERBB2 was subcloned into the pCD-ciR vector (Geneseed, Guangzhou, China) between the EcoRI and BamHI sites.
Quantitative real-time PCR.
Total RNA was extracted using the MagNA Pure compact RNA isolation kit (Roche, Basel, Switzerland). Subcellular fractionation of TNBC cells was conducted using the Paris kit (Ambion, Austin, TX, USA). For RNase R digestion, total RNA (2 μg) of TNBC cells was treated with RNase R (3 U/μg; Epicentre Technologies, Madison, WI, USA) at 37°C for 20 min, and diethyl pyrocarbonate (DEPC)-treated water (Thermo Fisher Scientific) was utilized as a control. To prevent the transcription of TNBC cells, these cell media were supplemented with actinomycin D (Act D) (2 mg/ml; Sigma), and dimethyl sulfoxide (2 mg/ml; Sigma) was utilized as a control. qRT-PCR was performed using the SYBR green master mix kit (Roche) on the LightCycler 480 system (Roche). The primer sequences are displayed in Table 2. Total RNA was reverse transcribed with a Transcriptor high-fidelity cDNA synthesis kit (Roche) or the miRcut Plus miRNA first-strand cDNA synthesis kit (Tiangen, Beijing, China). The results were calculated by the 2−ΔΔCT method and normalized to U6 (U6 small nuclear RNA) or β-actin.
TABLE 2.
Primers used for qRT-PCRa
| Target | Direction | Primer sequence |
|---|---|---|
| circ-ERBB2 | F | 5′-ACGTTTGAGTCCATGCCCAA-3′ |
| R | 5′-ACCTGCCTCACTTGGTTGTG-3′ | |
| ERBB2 | F | 5′-GGAAGTACACGATGCGGAGACT-3′ |
| R | 5′-ACCTTCCTCAGCTCCGTCTCTT-3′ | |
| PDK4 | F | 5′-AGGTGGAGCATTTCTCGCGCTA-3′ |
| R | 5′-GAATGTTGGCGAGTCTCACAGG-3′ | |
| miR-1276 | F | 5′-GCGCGTAAAGAGCCCTGTG-3′ |
| R | 5′-AGTGCAGGGTCCGAGGTATT-3′ | |
| miR-136-5p | F | 5′-CGCGACTCCATTTGTTTTGAT-3′ |
| R | 5′-AGTGCAGGGTCCGAGGTATT-3′ | |
| miR-503-5p | F | 5′-CGTAGCAGCGGGAACAGTT-3′ |
| R | 5′-CGTAGCAGCGGGAACAGTT-3′ | |
| miR-520f-3p | F | 5′-CGCGAAGTGCTTCCTTTTAG-3′ |
| R | 5′-CGCGAAGTGCTTCCTTTTAG-3′ | |
| U6 | F | 5′-CGCTTCGGCAGCACATATAC-3′ |
| R | 5′-TTCACGAATTTGCGTGTCATC-3′ | |
| β-Actin | F | 5′-GTCACCAACTGGGACGACAT-3′ |
| R | 5′-GAGGCGTACAGGGATAGCAC-3′ | |
| GAPDH | F | 5′-TGACTTCAACAGCGACACCCA-3′ |
| R | 5′-CACCCTGTTGCTGTAGCCAAA-3′ | |
F, forward; R, reverse; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Cell proliferation analysis.
The proliferation of TNBC cells was assessed by plate colony formation and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays. For the colony formation assays, transfected TNBC cells (5 × 102) were implanted into 6-well plates and cultured for 2 weeks. Thereafter, these plates were gently washed and stained with phosphate-buffered saline (PBS) (Thermo Fisher Scientific) and 0.1% crystal violet (Sigma), respectively. The number of colonies was counted with a Nikon microscope (Eclipse E600; Nikon Instruments, Melville, NY, USA).
The MTT assay was performed using cell proliferation kit I (Roche) according to the manufacturer’s instructions. In short, transfected TNBC cells (5 × 104) were incubated in 96-well plates and cultured for different times. Thereafter, the MTT reagent (5 mg/ml; 10 μl) was added to each well. After incubation for 4 h, the purple crystals were dissolved using a solubilization solution (100 μl). The optical density (OD) value at 570 nm was measured with an absorbance reader (ELx808; BioTek, Winooski, VT, USA).
Flow cytometry assay.
The annexin V-fluorescein isothiocyanate (FITC)-propidium iodide (PI) apoptosis detection kit (Solarbio, Beijing, China) was used to determine the apoptosis of treated TNBC cells according to the manufacturer’s procedures. In short, the cells (1 × 106) were digested and washed with trypsin (Sigma) and PBS (Thermo Fisher Scientific), respectively. After centrifugation (300 × g for 10 min), the cells were resuspended in 1× binding buffer (1 ml). Subsequently, the cells (1 × 105) were stained with annexin V (5 μl) and PI (5 μl) for 15 min in the dark. The apoptosis rate was analyzed with the FACSCalibur system (BD Biosciences, San Jose, CA, USA).
Assessment of cell migration.
For the wound-healing assay, the cells (5 × 105 cells) were inoculated into 6-well plates. The wounds were made in the cell monolayer with a sterile 200-μl pipette tip after culture overnight. Next, these plates were cultured for 24 h under the appropriate conditions. The wounded gaps were captured with a Nikon microscope (Eclipse E600; Nikon Instruments) at 0 and 24 h, and the distances were analyzed with ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Transwell assay.
The invasion of TNBC cells was assessed with transwell invasion chambers (catalog number 354480; Costar, Cambridge, MA, USA). Briefly, transfected TNBC cells (1 × 105 cells) were resuspended in 200 μl serum-free medium and then inoculated into the upper chambers. Also, the culture medium (600 μl) containing 10% FBS (Procell) was added to the lower chambers. The invasive cells were stained with 0.1% crystal violet (Sigma) after incubation for 24 h. The number of invading cells was determined using the Nikon microscope (magnification, ×100) (Eclipse E600; Nikon Instruments).
Measurements of the extracellular acidification rate and oxygen consumption rate.
The extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) of TNBC cells were analyzed using the Seahorse XF96 glycolysis analyzer (Seahorse Bioscience, North Billerica, MA, USA) with a Seahorse XF glycolysis stress test kit (Seahorse Bioscience) or the Seahorse XF cell mito stress test kit (Seahorse Bioscience) as described previously by Chen et al. (39).
Measurements of glucose consumption and lactate production.
Glucose concentrations and lactate production were calculated using a glucose assay kit (Sigma) or a colorimetric lactate assay kit (BioVision, Milpitas, CA, USA) in accordance with the procedures recommended by the manufacturer. The glucose consumption rate was the glucose concentration in fresh medium minus the glucose concentration remaining in the medium.
Bioinformatics analysis.
miRs, which could bind to circ-ERBB2, were jointly predicted by circinteractome, circBank, and starbase. The targets of miR-136-5p were predicted by starbase.
Dual-luciferase reporter assay.
The sequence of the circ-ERBB2 or PD4K 3′ untranslated region (UTR) was synthesized and inserted into the pmirGLO luciferase reporter (Promega, Madison, WI, USA) to construct the luciferase reporter carrying the wild-type circ-ERBB2 (wt-circ-ERBB2) or wt-PD4K 3′ UTR. The same method was performed to establish the luciferase reporter carrying the mutant circ-ERBB2 (mut-circ-ERBB2) or mut-PD4K 3′ UTR. TNBC cells were cotransfected with a luciferase reporter and NC mimics or miR-136-5p mimics using Lipofectamine 3000 reagent (Thermo Fisher Scientific). After 48 h, the cells were lysed, and the luciferase activities were assessed with a luciferase assay kit (BioVision) in a TD20/20 luminometer (Turner Biosystems, Sunnyvale, CA, USA).
RNA immunoprecipitation assay.
An RNA immunoprecipitation (RIP) assay was carried out using the Magna RIP RNA-binding protein immunoprecipitation kit (Millipore, Billerica, MA, USA). In short, magnetic beads were incubated with anti-Ago2 or anti-IgG antibodies to generate antibody-coated beads. TNBC cells were lysed in RIP lysis buffer and then incubated with the antibody-coated beads overnight at 4°C. RNA was extracted from immunoprecipitates with TRIzol regent (Thermo Fisher Scientific), followed by the detection of the enrichment of circ-ERBB2 and miR-136-5p by qRT-PCR.
RNA pulldown assay.
TNBC cells were transfected with biotinylated miR-136-5p (Bio-miR-136-5p) or Bio-NC. Forty-eight hours later, the lysates of TNBC cells were incubated with C-1 magnetic beads. After RNA extraction, the abundance of circ-ERBB2 or PD4K mRNA was analyzed by qRT-PCR.
Western blotting.
Radioimmunoprecipitation assay (RIPA) buffer (Thermo Fisher Scientific) was used to extract total protein from tissue samples and cells. The extracted total protein was quantified with the QuantiPro bicinchoninic acid (BCA) assay kit (Sigma) and separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Thereafter, the separated proteins were transferred onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad), followed by sealing with 5% nonfat milk. After washing with Tris-buffered saline–Tween (TBST) (Thermo Fisher Scientific), the membranes were incubated with primary antibodies against PDK4 (catalog number ab115777, 1:1,000; Abcam, Cambridge, MA, USA), proliferating cell nuclear antigen (PCNA) (catalog number ab18197, 1 μg/ml; Abcam), cleaved caspase 3 (catalog number ab2302, 1:200; Abcam), matrix metalloproteinase 9 (MMP9) (catalog number ab38898, 1:1,000; Abcam), and β-actin (catalog number ab115777, 1:200; Abcam). Next, the membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (catalog number ab205718, 1:2,000; Abcam), and the blots were detected by using the enhanced chemiluminescence (ECL) substrate (PerkinElmer, Waltham, MA, USA).
Animal experiments.
The animal experiments were ratified by the Animal Ethics Committee of Fuzhou Second Hospital affiliated with Xiaomen University. Ten female BALB/c nude mice (4 to 6 weeks old) were bought from Vital River Laboratory (Beijing, China). MDA-MB-231 cells (1 × 107) stably transduced with short hairpin RNA (shRNA) against circ-ERBB2 (sh-circ-ERBB2) or sh-NC were subcutaneously injected into 10 nude mice. The tumor volume was measured every 5 days. At the endpoint (30 days later), all mice were killed, and their tumor tissues were excised, weighed, and conserved at −80°C for subsequent analysis. The tumor volume was calculated based on the following equation: volume = (length × width2)/2.
Statistical analysis.
All experiments were repeated 3 times, and each experiment was done in triplicate. SPSS statistics version 17.0 (SPSS Inc., Chicago, IL, USA) was utilized for statistical analysis. All data are presented as means ± standard deviations. The association between circ-ERBB2 expression and clinicopathological characteristics was assessed by a chi-square test. Overall survival for TNBC patients was evaluated by Kaplan-Meier and log rank tests. The correlations were analyzed by Pearson’s correlation analysis. The differences between 2 groups or among multiple groups were determined with Student’s t test or one-way analysis of variance with Tukey’s post hoc test. A P value of <0.05 was the threshold of significance.
Data availability.
The analyzed data sets generated during the present study are available from the corresponding author upon reasonable request
ACKNOWLEDGMENTS
We declare that we have no competing interests.
This work was supported by the Chengdu Municipal Health Commission Project (number 2019107).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Lebert JM, Lester R, Powell E, Seal M, McCarthy J. 2018. Advances in the systemic treatment of triple-negative breast cancer. Curr Oncol 25:S142–S150. 10.3747/co.25.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garrido-Castro AC, Lin NU, Polyak K. 2019. Insights into molecular classifications of triple-negative breast cancer: improving patient selection for treatment. Cancer Discov 9:176–198. 10.1158/2159-8290.CD-18-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim C, Gao R, Sei E, Brandt R, Hartman J, Hatschek T, Crosetto N, Foukakis T, Navin NE. 2018. Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell 173:879–893.e13. 10.1016/j.cell.2018.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gangi A, Chung A, Mirocha J, Liou DZ, Leong T, Giuliano AE. 2014. Breast-conserving therapy for triple-negative breast cancer. JAMA Surg 149:252–258. 10.1001/jamasurg.2013.3037. [DOI] [PubMed] [Google Scholar]
- 5.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. 2016. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol 13:674–690. 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayer IA, Abramson VG, Lehmann BD, Pietenpol JA. 2014. New strategies for triple-negative breast cancer—deciphering the heterogeneity. Clin Cancer Res 20:782–790. 10.1158/1078-0432.CCR-13-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liberti MV, Locasale JW. 2016. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci 41:211–218. 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. Cell 144:646–674. 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Poff A, Koutnik AP, Egan KM, Sahebjam S, D’Agostino D, Kumar NB. 2019. Targeting the Warburg effect for cancer treatment: ketogenic diets for management of glioma. Semin Cancer Biol 56:135–148. 10.1016/j.semcancer.2017.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzgerald G, Soro-Arnaiz I, De Bock K. 2018. The Warburg effect in endothelial cells and its potential as an anti-angiogenic target in cancer. Front Cell Dev Biol 6:100. 10.3389/fcell.2018.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ponisovskiy MR. 2011. Warburg effect mechanism as the target for theoretical substantiation of a new potential cancer treatment. Crit Rev Eukaryot Gene Expr 21:13–28. 10.1615/critreveukargeneexpr.v21.i1.20. [DOI] [PubMed] [Google Scholar]
- 12.Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T, Shu Y. 2019. circRNAs in cancer metabolism: a review. J Hematol Oncol 12:90. 10.1186/s13045-019-0776-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patop IL, Wüst S, Kadener S. 2019. Past, present, and future of circRNAs. EMBO J 38:e100836. 10.15252/embj.2018100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. 2015. Circular RNA: a new star of noncoding RNAs. Cancer Lett 365:141–148. 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 15.Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, Ren D, Ye X, Li C, Wang Y, Wei F, Guo C, Wu X, Li X, Li Y, Li G, Zeng Z, Xiong W. 2018. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer 17:79. 10.1186/s12943-018-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S, Zhang Y, Cai Q, Ma M, Jin LY, Weng M, Zhou D, Tang Z, Wang JD, Quan Z. 2019. Circular RNA FOXP1 promotes tumor progression and Warburg effect in gallbladder cancer by regulating PKLR expression. Mol Cancer 18:145. 10.1186/s12943-019-1078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu L-S, Wang Y-L, Li R, Xu X-T, Li K-Y, Zuo C-R. 2020. circ_BICD2 acts as a ceRNA to promote tumor progression and Warburg effect in oral squamous cell carcinoma by sponging miR-107 to enhance HK2. Am J Transl Res 12:3489–3500. [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Ma M, Yang X, Zhang M, Luo J, Zhou H, Huang N, Xiao F, Lai B, Lv W, Zhang N. 2020. Circular HER2 RNA positive triple negative breast cancer is sensitive to pertuzumab. Mol Cancer 19:142. 10.1186/s12943-020-01259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng K, He B, Yang BB, Xu T, Chen X, Xu M, Liu X, Sun H, Pan Y, Wang S. 2018. The pro-metastasis effect of circANKS1B in breast cancer. Mol Cancer 17:160. 10.1186/s12943-018-0914-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin Y, Long J, He Q, Li Y, Liao Y, He P, Zhu W. 2019. Emerging roles of circRNA in formation and progression of cancer. J Cancer 10:5015–5021. 10.7150/jca.30828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao W, Zheng S, Zou Y, Yang A, Xie X, Tang H, Xie X. 2019. circAHNAK1 inhibits proliferation and metastasis of triple-negative breast cancer by modulating miR-421 and RASA1. Aging (Albany NY) 11:12043–12056. 10.18632/aging.102539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu J-Z, Shao C-C, Wang X-J, Zhao X, Chen J-Q, Ouyang Y-X, Feng J, Zhang F, Huang W-H, Ying Q, Chen C-F, Wei X-L, Dong H-Y, Zhang G-J, Chen M. 2019. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis 10:175. 10.1038/s41419-019-1382-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang R, Xing L, Zheng X, Sun Y, Wang X, Chen J. 2019. The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol Cancer 18:4. 10.1186/s12943-018-0933-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Chen B, Wei W, Huang X, Xie X, Kong Y, Dai D, Yang L, Wang J, Tang H, Xie X. 2018. circEPSTI1 as a prognostic marker and mediator of triple-negative breast cancer progression. Theranostics 8:4003–4015. 10.7150/thno.24106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng X, Huang M, Xing L, Yang R, Wang X, Jiang R, Zhang L, Chen J. 2020. The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple-negative breast cancer. Mol Cancer 19:73. 10.1186/s12943-020-01183-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, He M, Guo J, Cao T. 2019. Upregulation of circular RNA circ-ERBB2 predicts unfavorable prognosis and facilitates the progression of gastric cancer via miR-503/CACUL1 and miR-637/MMP-19 signaling. Biochem Biophys Res Commun 511:926–930. 10.1016/j.bbrc.2019.03.010. [DOI] [PubMed] [Google Scholar]
- 27.Huang X, He M, Huang S, Lin R, Zhan M, Yang D, Shen H, Xu S, Cheng W, Yu J, Qiu Z, Wang J. 2019. Circular RNA circERBB2 promotes gallbladder cancer progression by regulating PA2G4-dependent rDNA transcription. Mol Cancer 18:166. 10.1186/s12943-019-1098-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Fang L. 2018. Advances in circular RNAs and their roles in breast cancer. J Exp Clin Cancer Res 37:206. 10.1186/s13046-018-0870-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X-L, Hao Y-J, Wang B, Gu X-L, Wang X-X, Sun J-F. 2020. Long noncoding RNA NORAD promotes the progression of retinoblastoma by sponging miR-136-5p/PBX3 axis. Eur Rev Med Pharmacol Sci 24:1278–1287. 10.26355/eurrev_202001_20185. [DOI] [PubMed] [Google Scholar]
- 30.Zhao J, Yang T, Li L. 2020. lncRNA FOXP4-AS1 is involved in cervical cancer progression via regulating miR-136-5p/CBX4 axis. Onco Targets Ther 13:2347–2355. 10.2147/OTT.S241818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S, Zhang X, Li Z, Wang W, Li B, Huang X, Sun G, Xu J, Li Q, Xu Z, Xia Y, Wang L, Zhang Q, Li Q, Zhang L, Chen J, Wu Y, Cao J, Xu P, Zhang D, Xu H, Xu Z. 2019. Circular RNA profile identifies circOSBPL10 as an oncogenic factor and prognostic marker in gastric cancer. Oncogene 38:6985–7001. 10.1038/s41388-019-0933-0. [DOI] [PubMed] [Google Scholar]
- 32.Li J, Huang C, Zou Y, Ye J, Yu J, Gui Y. 2020. circTLK1 promotes the proliferation and metastasis of renal cell carcinoma by sponging miR-136-5p. Mol Cancer 19:103. 10.1186/s12943-020-01225-2. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Han C, Fu Y, Zeng N, Yin J, Li Q. 2020. lncRNA FAM83H-AS1 promotes triple-negative breast cancer progression by regulating the miR-136-5p/metadherin axis. Aging (Albany NY) 12:3594–3616. 10.18632/aging.102832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeoung NH. 2015. Pyruvate dehydrogenase kinases: therapeutic targets for diabetes and cancers. Diabetes Metab J 39:188–197. 10.4093/dmj.2015.39.3.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woolbright BL, Choudhary D, Mikhalyuk A, Trammel C, Shanmugam S, Abbott E, Pilbeam CC, Taylor JA. 2018. The role of pyruvate dehydrogenase kinase-4 (PDK4) in bladder cancer and chemoresistance. Mol Cancer Ther 17:2004–2012. 10.1158/1535-7163.MCT-18-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Z, Ji M, Wang Q, He N, Li Y. 2020. miR-16-5p/PDK4-mediated metabolic reprogramming is involved in chemoresistance of cervical cancer. Mol Ther Oncolytics 17:509–517. 10.1016/j.omto.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun S, Liu J, Zhao M, Han Y, Chen P, Mo Q, Wang B, Chen G, Fang Y, Tian Y, Zhou J, Ma D, Gao Q, Wu P. 2017. Loss of the novel mitochondrial protein FAM210B promotes metastasis via PDK4-dependent metabolic reprogramming. Cell Death Dis 8:e2870. 10.1038/cddis.2017.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guda MR, Asuthkar S, Labak CM, Tsung AJ, Alexandrov I, Mackenzie MJ, Prasad DVR, Velpula KK. 2018. Targeting PDK4 inhibits breast cancer metabolism. Am J Cancer Res 8:1725–1738. [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Z, Zuo X, Zhang Y, Han G, Zhang L, Wu J, Wang X. 2018. miR-3662 suppresses hepatocellular carcinoma growth through inhibition of HIF-1α-mediated Warburg effect. Cell Death Dis 9:549. 10.1038/s41419-018-0616-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Download MCB.00609-20-s0001.pdf, PDF file, 0.2 MB (167KB, pdf)
Data Availability Statement
The analyzed data sets generated during the present study are available from the corresponding author upon reasonable request