Abstract
The association of Sm proteins with U small nuclear RNA (snRNA) requires the single-stranded Sm site (PuAU4–6GPu) but also is influenced by nonconserved flanking RNA structural elements. Here we demonstrate that a nonameric Sm site RNA oligonucleotide sufficed for sequence-specific assembly of a minimal core ribonucleoprotein (RNP), which contained all seven Sm proteins. The minimal core RNP displayed several conserved biochemical features of native U snRNP core particles, including a similar morphology in electron micrographs. This minimal system allowed us to study in detail the RNA requirements for Sm protein-Sm site interactions as well as the kinetics of core RNP assembly. In addition to the uridine bases, the 2′ hydroxyl moieties were important for stable RNP formation, indicating that both the sugar backbone and the bases are intimately involved in RNA-protein interactions. Moreover, our data imply that an initial phase of core RNP assembly is mediated by a high affinity of the Sm proteins for the single-stranded uridine tract but that the presence of the conserved adenosine (PuAU…) is essential to commit the RNP particle to thermodynamic stability. Comparison of intact U4 and U5 snRNAs with the Sm site oligonucleotide in core RNP assembly revealed that the regions flanking the Sm site within the U snRNAs facilitate the kinetics of core RNP assembly by increasing the rate of Sm protein association and by decreasing the activation energy.
Small nuclear ribonucleoproteins (snRNPs) mediate essential RNA processing events, including pre-mRNA splicing (reviewed in references 43 and 45). The major spliceosomal snRNP particles consist of snRNA (U1, U2, U4/U6, or U5) and numerous proteins, which can be classified as either snRNP specific or common to each snRNP particle. The common proteins are collectively referred to as Sm proteins and were originally identified due to their antigenicity in the autoimmune disease systemic lupus erythematosus (26). The best characterized of these are the seven HeLa Sm proteins, which are named B/B′, D1, D2, D3, E, F, and G and range in size from 241 (for B′) to 76 (for G) amino acids (reference 16 and references therein). B and B′ are alternatively spliced products of the same gene, differing only in 11 amino acids at their C termini (42), and are referred to here as B/B′.
Association of the Sm proteins with U snRNA plays a pivotal role in the further biogenesis of U snRNP particles. The metabolic stability of the U snRNP particles is dependent on the Sm protein association (10, 20, 37). Following export from the nucleus, each U snRNA (with the exception of U6) assembles cytoplasmically with a set of Sm proteins, resulting in its core RNP particle. Hypermethylation of the U snRNA 5′ cap structure from monomethyl guanosine (m7G) to trimethylguanosine (m3G) is dependent upon formation of the core RNP domain, and in particular requires the presence of the B/B′ and D3 Sm proteins (30, 34, 36). These Sm proteins are the last to assemble (36, 38), linking cap hypermethylation to the completion of core RNP assembly. Cytoplasmic 3′-end processing trims various numbers of nucleotides from several U snRNAs, and a final nuclear 3′ end trimming to the mature U snRNA form is dependent on the presence of the m3G cap (29, 33, 44). Nuclear import of the U snRNP particles is mediated by a composite nuclear localization signal (NLS) composed of the core RNP domain and the m3G cap structure (4, 5, 11, 31). The import receptor for the m3G cap-mediated but not the core-mediated NLS is Snurportin 1, which is an importin α-like adapter (18). The nature of the core-mediated NLS and the identity of its import receptor are still unknown. Finally, the core RNP domain contributes to the integration of at least some of the specific proteins into their respective U snRNP particles (12, 32).
Each Sm protein interacts in vitro with one or more of the other Sm proteins, giving rise to three stable, RNA-free heteromeric complexes of EFG, D1D2, and B/B′D3 (16, 25, 36). An Sm sequence motif present in each Sm protein (2, 16, 40) mediates the protein-protein interactions (16, 21). Crystallization of two Sm protein complexes, BD3 and D1D2, demonstrated a common folding pattern of the Sm proteins, with an N-terminal helix followed by a strongly bent five-stranded antiparallel β-sheet, as well as a conserved interaction surface between the Sm proteins within each dimer (21). None of the Sm proteins contains known RNA-binding domain(s) (16), and no single Sm protein or heteromeric complex can directly interact with the U snRNA in a stable manner in vitro (36), suggesting that interactions among the Sm protein complexes define their RNA-interacting surface and specificity. A model based on the Sm protein crystal structures predicts that within the core RNP domain, seven Sm proteins (with either B or B′) form a doughnut-like structure, with each protein contributing to an RNA-binding surface surrounding the central hole of the doughnut (21). This correlates well with the ultrastructure observed for the HeLa U snRNP core particle in negatively stained electron micrographs: the core RNP domain is characterized by a round area approximately 8 nm in diameter, with a central accumulation of stain that could be accounted for by a reduction in density (22). Sm protein complexes assemble onto U snRNA in a stepwise and ordered manner. An initial and cooperative association of both the EFG and the D1D2 complexes with the U snRNA forms the U snRNP subcore particle (8, 36). The U snRNP subcore is the only stable RNP intermediate formed during core RNP assembly and provides a unique binding site for the B/B′D3 complex, whose association completes the core RNP particle (36, 38). Recently, a cytoplasmic protein complex of ca. 300 kDa that contains, among others, several Sm proteins and the survival-of-motor-neurons (SMN) protein has been demonstrated to play an essential role in U snRNP core assembly in vivo (3, 28). It is not yet clear at which step(s) in the assembly this complex is involved.
The RNA determinants guiding core RNP assembly appear highly complex. One prerequisite for Sm protein binding is the presence of the Sm site element, a short, single-stranded region usually flanked by stem-loop structures, with the consensus sequence of PuAU4–6GPu (1, 27, 31). The Sm site element is conserved among the U2- and U12-type Sm-spliceosomal U snRNAs (i.e., U1, U2, U4, U5, U11, U12, and U4atac) and the histone-processing U7 snRNA (for a review, see reference 45). In vivo selection for Sm protein binding and nucleocytoplasmic transport of a U1-like RNA containing a short combinatorial library selected an optimal sequence that perfectly matched the consensus Sm site element (9). UV cross-linking of HeLa U1 snRNPs revealed a direct protein-RNA interaction between the 5′ end of the Sm site (AAU) and the Sm protein G (15). A much more extensive Sm protein-U snRNA interaction surface is predicted by experimental data, since both micrococcal nuclease digestion (27) and hydroxyl radical probing (14) of HeLa core U snRNP particles demonstrated that a continuous, 20- to 25-nucleotide (nt) RNA region (including the Sm site element) was protected within U snRNP core particles. Mutagenesis of the Sm site in yeast U5 snRNA revealed that core RNP assembly in vivo was highly tolerant to point mutations in the Sm site (20), suggesting that contacts with Sm site bases important for RNA recognition are few or redundant. In contrast, point mutations of the Sm site in the yeast U4 snRNA were deleterious (17). Additional U snRNA elements were found to contribute to the assembly of U1 and U5 snRNP core particles in Xenopus oocytes, which were, importantly, neither conserved nor mutually exchangeable between U snRNA molecules (19). Distinctions in binding determinants on the U snRNAs could reflect differences in the Sm proteins which bind per se; however, the Sm proteins appear to be shared and not distinct for each type of U snRNA (39). Thus, the RNA-binding determinants for the Sm proteins appear to be composed of the conserved Sm site element as well as the specific regions of each U snRNA.
Here we address the role played by the Sm site element in the RNA-protein interactions leading to core RNP assembly. To eliminate influences of other RNA elements or RNA tertiary structure on Sm protein binding, a nonameric Sm site RNA oligonucleotide was analyzed in an in vitro reconstitution assay with HeLa Sm proteins. Strikingly, all seven Sm proteins associated with the Sm site oligonucleotide in a sequence-specific manner, forming a minimal core RNP particle that displayed properties characteristic of native U snRNP core particles. Core RNP assembly was found to be promoted by a high affinity of the Sm proteins for uracil-rich RNA, but it required the presence of the conserved adenosine base (PuAU…) for the formation of a thermodynamically stable RNP. Kinetic differences between assembly of the Sm proteins onto the Sm site oligonucleotide and either U4 or U5 snRNA revealed that the nonconserved regions of the U snRNA increase the rate of assembly and decrease the activation energy.
MATERIALS AND METHODS
Preparation of RNA-free, native snRNP TPs.
Native snRNP total proteins (TPs) were prepared as described previously (41) from a preparation of anti-cap immunaffinity-purified HeLa U snRNPs which contained predominantly U1 snRNP and small amounts of U2 snRNP. Briefly, proteins were dissociated from the U snRNPs in the presence of DEAE-cellulose (DE 53; Whatman) and EDTA. After the DEAE-cellulose had been removed by centrifugation, the supernatant containing the RNA-free proteins was dialyzed at 4°C against reconstitution buffer (20 mM HEPES-KOH [pH 7.9], 50 mM KCl, 5 mM MgCl2, 5% [vol/vol] glycerol, 0.2 mM EDTA). To concentrate the samples, dialysis against reconstitution buffer containing 30% (wt/vol) polyethylene glycol 6000 (Merck) was carried out, followed by a final dialysis against reconstitution buffer. Protein concentrations were approximately 0.3 mg/ml, as determined by the Bradford assay (Sigma). Proteins were analyzed on high-N,N,N′,N′-tetramethylethylenediamine (TEMED) 12.5% polyacrylamide–sodium dodecyl sulfate (SDS) gels (24) and visualized by Coomassie staining with brilliant blue G-colloidal (Sigma). The approximate protein molarities were estimated from the relative intensities of the protein bands on the Coomassie-stained gels.
RNA preparation.
RNA oligonucleotides were prepared by phosphoramidite chemistry on an Applied Biosystems 394 DNA/RNA synthesizer. Native U4 and U5 snRNAs were prepared from partially purified snRNPs by phenol-chloroform extraction and gel fractionation. Oligonucleotides and U snRNA were 5′-end radiolabelled with T4 polynucleotide kinase (New England Biolabs) and a twofold molar excess of [γ-32P]ATP (5000 Ci/mmol; Amersham), to ensure quantitative labelling and thus allow more accurate measurement of the RNA concentration. The oligonucleotides used for the chemical probing assays were 3′-end radiolabelled with [5′-32P]pCp (3,000 Ci/mmol; Amersham) and T4 RNA ligase (New England Biolabs). RNA was purified prior to use on a denaturing 12% (for U snRNA) or 22% (for oligonucleotides) polyacrylamide (acrylamide/bisacrylamide ratio, 20:1)–8 M urea gel. UV shadowing was used to visualize nonradiolabelled RNA for gel excision. RNA was eluted from the gel overnight with 400 μl of TNES buffer (20 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA, 0.01% [wt/vol] SDS). Eluted RNA was precipitated directly from the buffer with 3 volumes of ethanol in the presence of 10 μg of glycogen (Boehringer Mannheim Biochemicals), washed once with 70% ethanol, and dried under vacuum.
In vitro reconstitution.
For in vitro reconstitution, radiolabelled RNA (∼5 nM) was mixed with TPs (∼100 nM, unless stated otherwise), 10 μM Escherichia coli tRNA, 0.01% Nonidet P-40 (NP-40) and, when appropriate, competitor RNA oligonucleotide, and the volume was adjusted to 10 μl with reconstitution buffer. The mixtures were incubated at 30°C for 45 min, except when the rate of assembly was determined, in which case they were incubated for the time stated. Reconstitution mixtures for streptavidin precipitation contained 200 nM RNA oligonucleotide and 400 nM TPs in a final volume of 60 μl. Samples for gradient sedimentation contained 400 nM RNA and/or 400 nM TPs in a final volume of 150 μl, and the assays were performed in the absence of tRNA and NP-40 to allow subsequent analysis by electron microscopy. To determine the thermodynamic stability, reconstitution assays were started at various times, so that all the assays were completed simultaneously following the different time intervals (0 to 120 min) of incubation at 30°C with competitor RNA (5 μM Sm site oligonucleotide).
Streptavidin-agarose precipitation.
Reconstitution assay mixtures containing biotinylated RNA oligonucleotide or mock assay mixtures without RNA oligonucleotide were incubated with 10 μg of prewashed streptavidin-agarose beads (Boehringer Mannheim) in 400 μl of phosphate-buffered saline (pH 8.0). Samples were incubated for 90 min at 4°C with gentle, end-over-end rotation. The beads were washed five times with 1 ml of IP buffer (10 mM Tris-HCl [pH 8.0], 0.1% NP-40, 0.15 to 2 M KCl, as indicated). Since free Sm proteins bind avidly to microtube walls, the slurry was transferred to new tubes after the second wash. The beads were vacuum dried and then boiled in SDS sample buffer (50 mM Tris-HCl [pH 6.8], 2% [wt/vol] SDS, 50 mM dithiothreitol, 10% [vol/vol] glycerol, 0.01% bromophenol blue). Following brief centrifugation, an aliquot of the supernatant was loaded onto a high-TEMED, 12.5% polyacrylamide–SDS gel for protein analysis. The gels were stained with Coomassie blue and (for the experiment in Fig. 1B) subsequently with silver for better visualization.
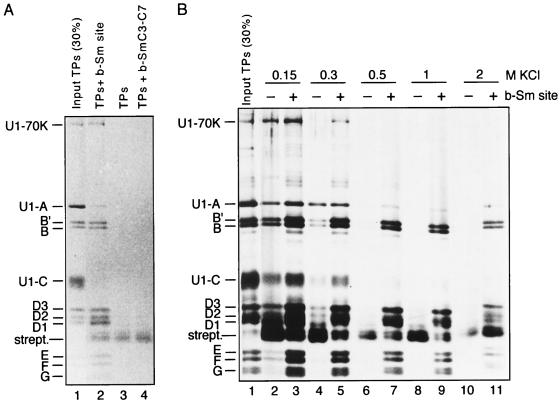
FIG. 1.
All of the Sm proteins associate in a specific and stable manner with a biotinylated Sm site oligonucleotide. (A) Reconstitution with TPs was carried out with biotinylated Sm site oligonucleotide (AAUUUUUGA [lane 2]) or, as negative controls, in the absence of biotinylated RNA (lane 3) or with biotinylated SmC3-C7 (AACCCCCGA) oligonucleotide (lane 4). Following streptavidin-agarose precipitation, samples were washed extensively with buffer containing 150 mM KCl. Bound proteins were analyzed by SDS-polyacrylamide gel electrophoresis and stained with Coomassie blue. (B) Reconstitution with TPs was performed in either the presence or absence of biotinylated Sm site oligonucleotide (b-Sm site), as indicated above each lane. The salt stability of the coprecipitation of U1-specific and Sm proteins was analyzed by extensive washing with buffers containing various concentrations of salt (0.15 to 2 M KCl, as indicated above the lanes). Following SDS-polyacrylamide gel electrophoresis, proteins were visualized by staining first with Coomassie blue and then with silver.
Sucrose gradient fractionation and electron microscopy.
Sample volumes were adjusted to 200 μl with reconstitution buffer prior to application to 10 to 30% sucrose gradients containing 20 mM KPO4 (pH 8.0) and 400 mM KCl. The gradients were centrifuged at 4°C in a TLS55 rotor (Beckman) for 10 h at 173,500 × g. Fractions of 75 μl were removed manually from top to bottom of the gradient, and those containing 32P-labelled RNA were analyzed by scintillation counting. For the gradients containing either proteins or RNA-protein complexes, half of each gradient fraction was removed and the proteins were separated from RNA by phenol-chloroform extraction (for the samples containing RNA-protein), precipitated with 5 volumes of acetone, and analyzed by high-TEMED SDS-polyacrylamide gel electrophoresis. Sedimentation standards used were cytochrome c (2.3S), creatine kinase (4.8S), and aldolase (7.8S).
For electron microscopy (EM) analysis, the remaining aliquot of each gradient fraction was used directly following fractionation for EM sample preparations. Negative staining with 2.5% (wt/vol) uranyl formate was performed by the double-carbon-film method as described previously (22). Preparations were examined with a Philips CM120 Biotwin electron microscope operating at 120 kV. Electron micrographs were obtained at a magnification of ×105,000.
Modification with DMS.
Dimethyl sulfate (DMS) modification was performed essentially as previously described (14). Briefly, modification of the 3′-end radiolabelled Sm site oligonucleotide, within the reconstituted core RNP or in the absence of Sm proteins, was carried out in 200 μl of DMS buffer (50 mM cacodylic acid-KOH [pH 7.0], 50 mM KCl, 10 mM MgCl2). The reaction was started by the addition of 1 μl of DMS (Fluka), the mixture was incubated for 10 min on ice, and then the reaction was stopped by addition of 50 μl of DMS stop buffer (1 M Tris-HCl [pH 4.6], 2 M β-mercaptoethanol, 12.5 mM EDTA). RNA was ethanol precipitated, resuspended, and reprecipitated with ethanol. For the NaBH4 reaction, RNA was precipitated once more in the presence of 13 μg of DMS-modified carrier tRNA. NaBH4 reaction conditions, aniline treatment, RNA recovery, and the diethylpyrocarbonate (DEPC) and Fe(II)-EDTA reactions were performed as described elsewhere (14).
Electrophoretic mobility shift assays (EMSA).
Aliquots of the reconstitution assay mixtures (5 μl) were mixed with 5 μl of loading buffer (16% glycerol, 4 M urea, 10 mg of heparin per ml) and loaded directly onto a native gel containing 4% glycerol, 0.5× Tris-borate-EDTA (TBE) buffer, and either 6 or 8% polyacrylamide (acrylamide/bisacrylamide ratio, 80:1), to analyze assay mixtures containing either U snRNA or oligonucleotides, respectively. Prior to loading, the gels were prerun for 1 h at 4°C. To determine the rate of assembly, aliquots from a single reconstitution mixture were removed following the indicated incubation times, mixed with loading buffer, and applied directly onto a gel during electrophoresis. The gels were run for approximately 4 h in 0.5× TBE buffer at 4°C with a constant current of 35 mA. The gels were dried, exposed to film, and quantitated with a Molecular Dynamics PhosphorImager.
RESULTS
A complete set of Sm proteins interacts with an Sm site oligonucleotide in a salt-resistant manner.
To elucidate whether the Sm site sequence can associate with one or more Sm proteins in the absence of the flanking stem-loop structures, we chemically synthesized an RNA oligonucleotide with the human U4 Sm site sequence (Sm site oligonucleotide; AAUUUUUGA) and a 5′-biotin label. This oligonucleotide was incubated with RNA-free, native HeLa snRNP proteins (TPs), prepared from U snRNPs consisting of predominantly U1 and some U2 snRNP particles, under standard in vitro reconstitution conditions (41), and subsequently precipitated with streptavidin-agarose. To minimize nonspecific interactions, detergent (0.01% NP-40) and a 50-fold molar excess of E. coli tRNA over Sm site oligonucleotide were added to the reconstitution mixture. All Sm proteins were efficiently coprecipitated with the biotinylated Sm site oligonucleotide (Fig. 1A, lane 2, compared to 30% input TPs [lane 1]) to a level significantly higher than that observed for the control assays with either no biotinylated RNA oligonucleotide (lane 3) or an Sm site-derived, biotinylated RNA oligonucleotide (b-SmC3-C7) in which the uridines had been replaced with cytidines (AACCCCCGA) (lane 4). Thus, coprecipitation of the Sm proteins with the biotinylated Sm site oligonucleotide was sequence specific and was not due to an affinity for either the biotin label or single-stranded RNA as such. To determine whether the interaction of Sm proteins with the biotinylated Sm site oligonucleotide was stable at high ionic strength, the agarose beads were extensively washed with buffers containing different salt concentrations. Increasing the ionic strength of the wash buffer from 150 mM to 300 mM KCl had little effect on the Sm protein association with the Sm site oligonucleotide (Fig. 1B; compare lanes 3 and 5). Indeed, efficient coprecipitation of the Sm proteins was still observed after washing with either 500 mM (lane 7) or 1 M (lane 9) salt. While increasing the salt concentration of the wash buffer to 2 M greatly reduced the amount of Sm protein coprecipitation, a significant amount still remained stably associated with the Sm site oligonucleotide (lane 11). We thus conclude that all of the Sm proteins associate with the Sm site oligonucleotide, forming a “minimal” core RNP that, like native U snRNP core particles (19, 20), resists dissociation at high salt concentrations.
Interestingly, the U1-specific proteins U1-70K and, to a lesser degree, U1-A and U1-C were efficiently coprecipitated at both low (150 mM) and moderate (300 mM) salt concentrations (Fig. 1A, lane 2; Fig. 1B, lanes 3 and 5). In contrast to the Sm proteins, the specific proteins completely dissociated following 500 mM salt washes (Fig. 1B, lane 7). We conclude that the U1-70K, U1-A, and U1-C proteins associate with the minimal core RNP via salt-labile interactions with the Sm proteins, as has been demonstrated within HeLa U1 snRNP particles for the U1-70K and U1-C proteins (32).
Sedimentation behavior of the minimal core RNP.
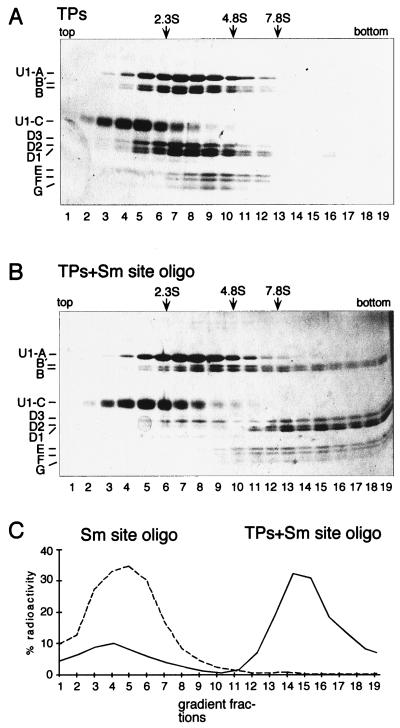
To independently characterize the minimal core RNP, sedimentation analysis on sucrose gradients was performed (Fig. 2). A reconstitution mixture containing equal concentrations of TPs and nonbiotinylated Sm site oligonucleotide (with a 1:10 ratio of radiolabelled to unlabelled) was fractionated on a 10 to 30% sucrose gradient containing 400 mM KCl (Fig. 2B). Mock reconstitution mixtures containing only TPs (Fig. 2A) or only Sm site oligonucleotide (depicted schematically in Fig. 2C) were fractionated in parallel. A dramatic shift in the sedimentation behavior of both the Sm proteins and Sm site oligonucleotide was observed upon core RNP reconstitution: whereas the Sm protein complexes or the Sm site oligonucleotide alone peaked in approximately the top third of the gradient (fractions 6 to 9 and 4 to 6, respectively [Fig. 2A and C]), the majority of the Sm site oligonucleotide and Sm proteins cosedimented in the bottom third of the gradient following reconstitution together (Fig. 2B and C). In contrast, no shift in sedimentation of the Sm proteins was observed when the reconstitution was performed with the SmC3-C7 oligonucleotide (data not shown). Consistent with the fact that the gradients contained a high-salt buffer, the sedimentation behavior of the U1-specific A and C proteins did not change in the presence of the Sm site oligonucleotide (compare Fig. 2A and B; see also Fig. 1). These results thus provide direct evidence that the minimal core RNP contains all of the Sm proteins.
FIG. 2.
Analysis of minimal core RNP particle sedimentation in sucrose gradients. (A) Sedimentation behavior of TPs incubated under reconstitution conditions in the absence of Sm site oligonucleotide. The reconstitution assays were fractionated on a 10 to 30% sucrose gradient containing 400 mM KCl. Fractionation was from top to bottom (corresponding to left to right). Proteins were analyzed by SDS-polyacrylamide gel electrophoresis and visualized by silver staining. (B) Sedimentation behavior of TPs after reconstitution with the Sm site oligonucleotide performed as described for panel A. Note that the apparent overabundance of nonshifted B and B′ protein following reconstitution can be attributed to the U2-specific A′ and B" proteins, which cosedimented with a peak in lanes 9 and 10 of both TP-containing gradients (A). Some B, B′, and D3 cosedimented as a free complex following reconstitution, most probably due to their slight overabundance. (C) Sedimentation behavior of the Sm site oligonucleotide. The amount of radioactivity in each fraction was determined by scintillation counting and expressed as the percentage of the input cpm. The dashed line indicates Sm site oligonucleotide in a mock reconstitution, and the solid line indicates Sm site oligonucleotide reconstituted with TPs. The protein sedimentation profile of the latter is shown in panel B.
Structural and conformational characteristics of native U snRNP core particles are displayed by the minimal core RNP.
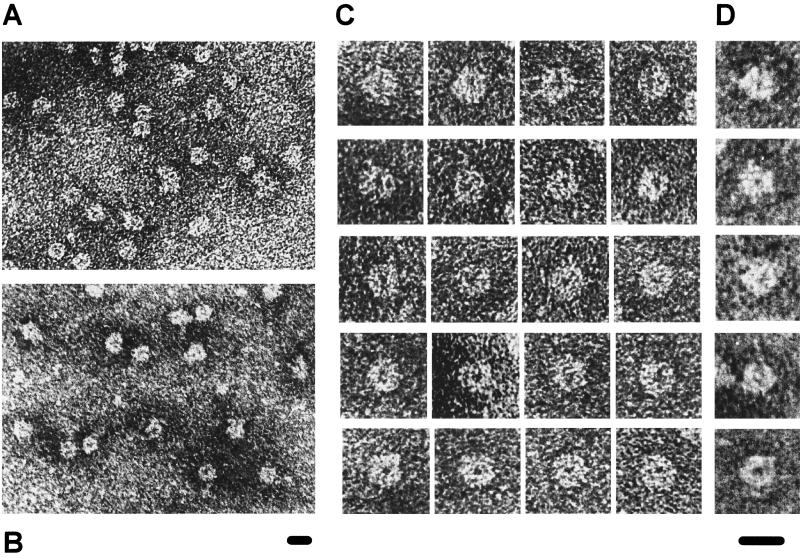
The ultrastructure of the core RNP domain, visible by negative-staining EM, is a highly conserved feature of native HeLa U snRNPs (22). To test whether the association of the Sm proteins with the Sm site oligonucleotide generates a similar morphology, a fraction containing minimal core RNPs purified by sucrose gradient sedimentation (equivalent to fraction 15 from Fig. 2B) was analyzed by negative-staining EM (Fig. 3). The EM overview reveals numerous particles with a morphology similar to that of native U snRNP core RNPs, displaying in particular smooth, round outlines with a diameter of approximately 8 nm (Fig. 3A; compare with an overview of native HeLa core U5 snRNP particles, prepared as described previously [22], in Fig. 3B). Moreover, detailed structural similarities between the minimal core RNP (Fig. 3C) and native U5 snRNP core particles (Fig. 3D) were also evident, as depicted by each row of images (see the figure legend for image descriptions). No typical core structures were observed in the fraction taken from the top third of the RNP gradient (fraction 7 in Fig. 2B) or from the counterpart fractions of the TP gradient (fractions 7 and 15 in Fig. 2A and data not shown). Significantly, the structure of the minimal core RNP exhibits a stronger resemblance to fully assembled U snRNP core particles than to either U snRNP subcore particles or EFG protein complexes (described in reference 35). We conclude that the minimal core RNP has the typical morphological structure of native U snRNP core particles.
FIG. 3.
Electron micrographs of negatively stained minimal core RNP particles purified by sucrose gradient sedimentation. (A) Overview micrograph of the minimal core RNP sample from fraction 15 of the RNP gradient (Fig. 2B). (B) Overview micrograph of native 10S core U5 snRNP particles, which contains only U5 snRNA and Sm proteins, isolated by an independent procedure and shown for comparison. (C and D) Typical views of the minimal core RNP (C) and the U5 core snRNP (D). Particles with similar structural details are arranged in rows (C) and each row corresponds to those described previously for U5 core snRNP particles (22) (D). Briefly, images in the first and second rows show forms with a line of stain that roughly bisects the core RNP domain, with additional short extensions in the second row; images in the third row show forms with a wedge-shaped structure; and images in the fourth and fifth rows show forms with a light or dark central dot. The bar at the bottom of each picture represents 10 nm with respect to the images depicted above it.
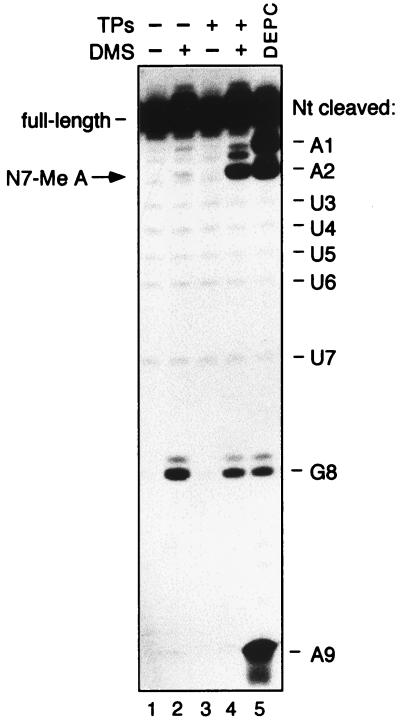
A further characteristic of properly assembled U snRNP core particles is their sensitivity to N7 methylation at the conserved adenosine at position +2 of the Sm site (PuAU…) following treatment with DMS (14). Notably, in naked U snRNA, DMS modifies the N7 position of guanosine but not adenosine residues, demonstrating that this particular Sm site adenosine takes on an unusual conformation when incorporated into core RNP particles. The acquisition of the RNA conformation necessary for this modification has been linked to the functionality of U snRNPs with respect to their nuclear import (14). To test for this chemical sensitivity, minimal core RNP particles were reconstituted with 3′-end-radiolabelled Sm site oligonucleotide and modified with DMS. The RNA was subsequently purified, reduced with NaBH4, and treated with aniline to obtain cleavage 3′ of any modification sites. Indeed, the conserved adenosine at position +2 (AAUUUUUGA) was methylated when reconstitution was performed in the presence of TPs (Fig. 4, lane 4; the relevant cleavage product is indicated by an arrow) but not in the control reactions performed in the absence of either TPs (lane 2), DMS (lane 3), or both DMS and TPs (lane 1). Thus, similar to the situation observed for native U snRNA, N7 methylation sensitivity of the adenosine at position +2 of the Sm site oligonucleotide is dependent on the interactions with the Sm proteins. This underscores the structural similarity between the minimal core RNP and native U snRNP core particles and corroborates the above findings that the minimal core RNP is a properly assembled core RNP particle.
FIG. 4.
Formation of the minimal core RNP imparts N7 methylation sensitivity to the conserved A2 adenosine. The Sm site oligonucleotide was 3′-end labelled with [32P]pCp and incubated in the presence of TPs (lanes 3 and 4) or, as mock reconstitution assays, in the absence of TPs (lanes 1 and 2). RNA in each assay was then treated with NaBH4-aniline following incubation with DMS (lanes 2 and 4) or, as controls for nonspecific RNA cleavage by NaBH4-aniline, without DMS (lanes 1 and 3). The position of the N7-methylated adenosine is indicated by an arrow on the left. The weaker band approximately halfway between those marked A1 and A2 is probably due to N3 methylation of the cytidine of the 3′-pCp label. To verify the positions of the adenosines, 32P-labelled Sm site oligonucleotide was treated with DEPC (lane 5). A base ladder marker generated by hydroxyl radical cleavage of the 32P-labelled Sm site oligonucleotide was performed in parallel and is depicted schematically at the right, with the 5′-most nucleotide (Nt) of the cleaved RNA oligonucleotide indicated.
Differences in assembly kinetics between the Sm site oligonucleotide and native U4 and U5 snRNAs.
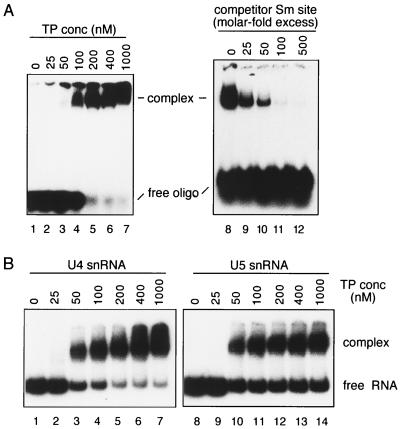
Our results demonstrate that the Sm site element suffices for sequence-dependent assembly of a stable core RNP particle. However, the specific RNA structural elements flanking the Sm site element contribute to U snRNP core assembly in vivo (19). To determine in which way the specific RNA elements affect assembly, we compared the kinetics of in vitro core RNP assembly of the Sm site oligonucleotide with that of the full-length, native HeLa U snRNAs by using EMSA. We chose to use U4 and U5 snRNAs since the TP preparation originated mainly from U1 and U2 snRNPs and contained, in addition to Sm proteins, U1- and U2-specific proteins but little or no U4- or U5-specific proteins. To eliminate nonspecific interactions, a vast excess of tRNA and 0.01% NP-40 were added to the reconstitution assay mixtures. A high concentration of urea (2 M) was included in the loading buffer to select against nonspecific or weak RNA-protein interactions; note that native U snRNP core particles are stable under these conditions (data not shown). Sm site oligonucleotide (∼5 nM) was effectively shifted into a single lower-mobility complex when incubated with TP concentrations of 100 nM or higher (Fig. 5A, lanes 4 to 7). The specificity of this shift was demonstrated by adding an excess of unlabelled Sm site oligonucleotide (25-, 50-, 100-, or 500-molar excess over labelled oligonucleotide) to the reconstitution mixture containing 100 nM TPs (lanes 9 to 12): complex formation was completely inhibited by a 100-fold molar excess (500 nM) of cold oligonucleotide (lane 11). Thus, the lower-mobility complex can be attributed to the stable association of the Sm proteins with the Sm site oligonucleotide. Similar to the Sm site oligonucleotide, each of the native HeLa U4 and U5 snRNAs (∼5 nM) was shifted into a predominantly single band by its association with Sm proteins (Fig. 5B, lanes 3 to 7 for U4 and lanes 10 to 14 for U5). Previous studies have shown that the only stable RNP intermediate of core RNP assembly is the U snRNP subcore particle, which lacks the B/B′ and D3 Sm proteins (36). However, reconstitution of a TP mixture depleted of the B/B′ and D3 proteins resulted in faster-migrating complexes than those formed from the full TP mixture (data not shown). This strongly suggests that the complexes observed in Fig. 5 correspond to the fully assembled core RNP complex for each RNA. At the highest concentration of TPs, the majority of the Sm site oligonucleotide and the U4 snRNA was shifted but a large amount of the U5 snRNA remained free; this apparent block in U5 snRNP assembly of the remaining RNA was consistently observed with the native U5 snRNA and remains to be clarified. Importantly, an RNP complex shift was first observed at 100 nM TPs for the Sm site oligonucleotide (Fig. 5A, lane 4) and at 50 nM for both U4 and U5 snRNAs (Fig. 5B, lanes 3 and 10, respectively), representing only a slight (twofold) reduction in apparent Sm protein affinity for the Sm site oligonucleotide compared to the native U snRNAs. We conclude that the high affinity of the Sm proteins for the single-stranded Sm site element provides a major driving force during U snRNP core particle assembly.
FIG. 5.
Comparison of core RNP assembly kinetics for the Sm site oligonucleotide and U4 and U5 snRNAs, as measured by EMSA. (A) Radiolabelled Sm site oligonucleotide (∼5 nM) was incubated with TP concentrations between 0 and 1,000 nM, as indicated (lanes 1 to 7). Competition of the Sm site oligonucleotide shift was performed by adding an excess of unlabelled Sm site oligonucleotide at the onset of the reconstitution in a 25-, 50-, 100-, or 500-molar excess over the labelled Sm site oligonucleotide; the TP concentration for these assays was 100 nM (lanes 8 to 12). Native gels contained either 8% (left) or 6% (right) acrylamide. (B) Radiolabelled, native HeLa U4 (lanes 1 to 7) or U5 (lanes 8 to 12) snRNA (∼5 nM) was incubated with various TP concentrations, as indicated above each lane. Complexes were analyzed as above, on native gels containing 6% acrylamide.
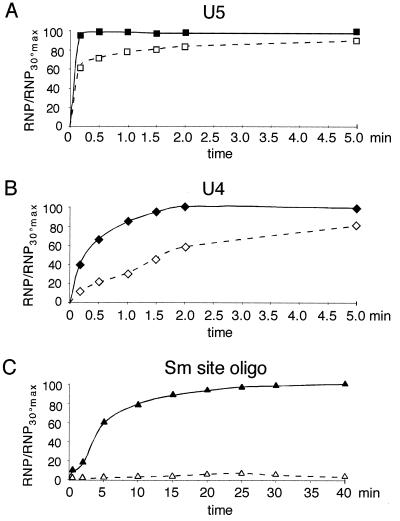
Next, we investigated whether the rates of core RNP assembly differed for U4 snRNA, U5 snRNA, and the Sm site oligonucleotide. For this, each RNA was incubated at 30°C with a sufficient concentration of TPs to give an efficient shift (50 nM for U4 and U5 and 100 nM for the Sm site oligonucleotide) and aliquots of the assay mixtures were applied to a native gel at various time intervals for EMSA analysis. Here it is important to note that once the samples are applied to the gel, additional complex formation is not probable (7). The results of these experiments are represented graphically as the percentage of complex formed at each time point with respect to the total complex at the end time point (Fig. 6). The U4 and U5 snRNA were shifted rapidly into complexes (Fig. 6A and B, solid lines), with >40% of the complex being formed after 10 s. Assembly of U5 snRNP was completed after 30 s, but that of U4 snRNP was completed only after 2 min. This could indicate that the U4 snRNP core assembly is slightly less efficient than that for U5, or it could be due to the inability of some of the U5 snRNA to be incorporated into complexes (Fig. 5B). In contrast, the assembly of the Sm site oligonucleotide into an RNP complex was significantly slower than that for either full-length U snRNA: efficient complex formation (>50%) was first observed after 5 min, with completion of the assembly after >20 min (Fig. 6C, solid line). Thus, the nonconserved, specific regions of the U snRNA increase the rate of core RNP assembly.
FIG. 6.
Rate of association of Sm proteins with U5 snRNA (A), U4 snRNA (B), or the Sm site oligonucleotide (C), as measured by EMSA and represented graphically. Radiolabelled RNA (∼5 nM) was incubated at 30°C (solid line) or 0°C (dashed line) in the presence of a TP concentration of approximately 100 nM for the Sm site oligonucleotide and 50 nM for the U4 and U5 snRNAs. Aliquots from each assay were withdrawn at time intervals ranging from 10 s to 5 min for the U4 and U5 snRNAs and 10 s to 40 min for the Sm site oligonucleotide, mixed with loading buffer, and applied to a running native polyacrylamide gel. The shifted complex for each RNA was measured densitometrically. The percentage of total complex, relative to the maximum concentration of complex formed at 30°C, is plotted on the y axis, and the time intervals are plotted on the x axis. The standard error was less than 7%.
Finally, we determined the temperature dependency of the assembly reaction by comparing assembly at 30°C (Fig. 6, solid lines) with that at 0°C (dashed lines). No drastic shift in the assembly rate was observed for U5, although the overall complex formation was less efficient (Fig. 6A). U4 snRNP assembly was slower at 0°C, requiring approximately 2 min for 50% assembly (Fig. 6B). Strikingly, however, the Sm site oligonucleotide was no longer assembled into complexes at 0°C (Fig. 6C), even after a 40-min time interval. Performing the reconstitution with higher concentrations of TPs (up to 2 μM) did not lead to significant RNP complex formation (data not shown), demonstrating that the 0°C block in assembly could not be overcome by an approximately 20-fold increase in Sm proteins. This suggests that the activation energy necessary for core RNP assembly onto the Sm site oligonucleotide is greater than that for full-length U snRNAs.
Sm site determinants for assembly of the minimal core RNP.
An important conclusion of our results is that the Sm proteins can recognize and stably associate with the single-stranded Sm site element directly, without the contribution of flanking U snRNA regions. To investigate which Sm site RNA constituents act as recognition determinants for these stable interactions, we used EMSA to test a series of Sm site-derived oligonucleotides, with either ribose or base modifications, for in vitro assembly with Sm proteins. The apparent affinity of the Sm proteins for the modified oligonucleotides was determined relative to that for the wild-type Sm site oligonucleotide, which was tested in parallel in each experiment.
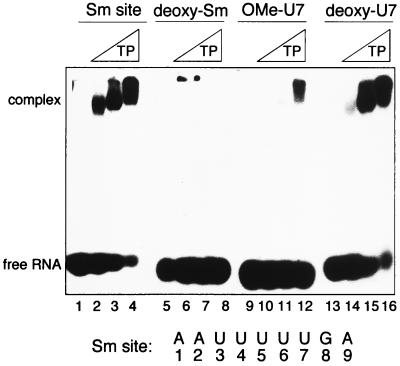
Modifications of the 2′-OH groups were made to determine whether any 2′-OH had a position-specific (in relation to the base sequence) effect on Sm protein binding. Changing all nine positions in the Sm site oligonucleotide to either 2′-deoxymethoxy (2′-OMe [data not shown]) or 2′-deoxy (Fig. 7, lanes 5 to 8) completely disrupted Sm protein binding, demonstrating that the 2′-OH positions in general are important for Sm protein association. The introduction of 2′-OMe at any single position (data not shown) drastically reduced the affinity of the Sm proteins for the oligonucleotide (>50-fold); an example of this is shown with an oligonucleotide modified at the uridine in position 7 (OMe-U7) (lanes 9 to 12). 2′-O-methylation could interfere with Sm protein association by preventing the modified ribose 2′-groups from acting as a hydrogen bond donor and/or by exerting steric effects due to the bulky methyl group. To distinguish between these possibilities, 2′-deoxy substitutions were made at single positions, since replacement of -OH with -H prevents the 2′ groups from acting as a hydrogen bond donor or acceptor but should have no steric effects. In contrast to the drastic reduction observed by the methoxyoligonucleotides, each single deoxy substitution resulted in an only slightly reduced (∼threefold) binding affinity of the Sm proteins, as exemplified by an oligonucleotide modified at U7 (Fig. 7, lanes 13 to 16). This implies first that steric hindrance at the riboses disrupts the association of the Sm proteins with the Sm site RNA and second that while no specific position is critical, each 2′-hydroxyl moiety contributes to the recognition of the Sm site element by the Sm proteins.
FIG. 7.
Effects of substitutions of the 2′-hydroxyl groups of the Sm site oligonucleotide on the binding specificity of the Sm proteins. EMSA analysis of reconstitution mixtures containing the indicated radiolabelled oligonucleotide (5 nM) with no TPs (first lane of each group) or increasing concentrations (∼100, 300, and 600 nM) of TPs is shown. The oligonucleotides used are indicated above the gel. Sm site, wild-type Sm site oligonucleotide; deoxy-Sm; 2′-H at all positions; OMe-U7, 2′-OMe at the uridine at position 7; deoxy-U7, 2′-H at the uridine at position 7.
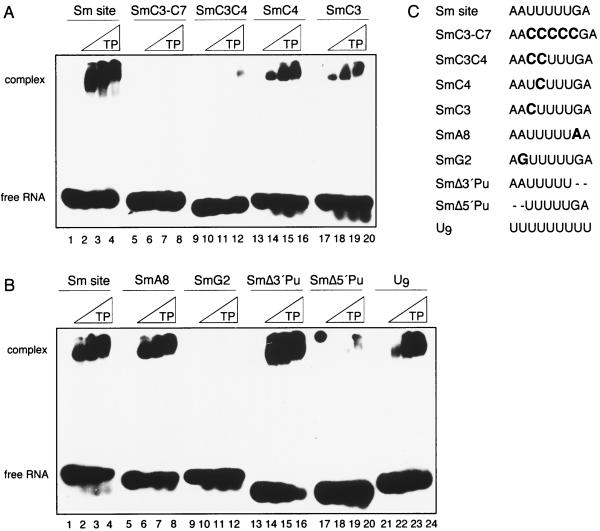
The base identities of the conserved positions (the uridine tract and the 5′ A and 3′ G flanking the U tract) were next analyzed for their potential roles in Sm site recognition. Exchanging all uridines with cytidines (SmC3-C7) blocked complex formation (Fig. 8A, lanes 5 to 8); this is in agreement with the inability of the biotinylated SmC3-C7 to coprecipitate Sm proteins (Fig. 1A). Indeed, a double change of U to C at any two positions within the U tract was sufficient to abolish assembly, as exemplified by the SmC3C4 oligonucleotide (Fig. 8A, lanes 9 to 12). Since this could be due to the requirement for a minimum length of the U tract as well as to the site-specific recognition of a certain uridine, oligonucleotides with single changes of U to C were tested. Those with substitutions at any one of the last four uridine positions (AAUUUUUGA) assembled almost as efficiently as the wild-type oligonucleotide did, with only slight (threefold) reductions in affinity (exemplified by SmC4 [lanes 13 to 16]). A more significant decrease (sixfold) in RNP assembly was observed for an oligonucleotide in which the first U had been changed to C (SmC3 [lanes 17 to 20]). This indicates that, while not essential, the uridine base identity at the 5′ border of the U tract plays an important role in the assembly process. Furthermore, similar to the 2′-hydroxyl moieties, the uridine bases appear to collectively provide a recognition determinant for core RNP assembly.
FIG. 8.
Effects of base modifications or deletions on oligonucleotide binding specificity of the Sm proteins. (A and B) EMSA analysis of reconstitution mixtures containing the indicated radiolabelled oligonucleotide (5 nM) with no TPs (first lane of each group) or increasing concentrations (∼100, 300, and 600 nM) of TPs is shown. The RNA oligonucleotides used are shown above the gel. Note that the dot at the top of lane 17 of panel B is due to a nonspecific contamination on the dried gel. Panels A and B represent results from two experiments with different RNAs. (C) Sequences of RNA oligonucleotides used in the assays in panels A and B.
Single substitutions of the highly conserved bases A2 and G8 (AAUUUUUGA) were likewise tested. Changing G8 to A had no effect on the assembly process (SmA8 [Fig. 8B, lanes 5 to 8]). In contrast, exchanging A2 with G (SmG2) blocked assembly (lanes 9 to 12). Interestingly, the 5′-terminal A in the SmG2 oligonucleotide did not compensate for the A-to-G substitution at position 2 (AGUUUUUGA). Since this could be due to sterical hindrance by the guanosine residue, we tested an RNA oligonucleotide in which both 5′ adenosines had been deleted (i.e., UUUUUGA [SmΔ5′Pu]). Similar to the SmG2 oligonucleotide, the SmΔ5′Pu oligonucleotide did not support assembly (Fig. 8B, lanes 17 to 20). While this suggests that the adenosine at position 2 is essential for core RNP assembly, the Δ5′ oligonucleotide could also be inefficient due to its shorter length (7 versus 9 nt). That the latter is not the case was demonstrated by deleting 2 nt from the 3′ end of the Sm site (AAUUUUU); the SmΔ3′Pu oligonucleotide was slightly more efficient in RNP assembly than was the wild-type Sm site oligonucleotide (∼threefold [Fig. 8B, lanes 13 to 16]). Thus, while the identity of the conserved guanosine at position 8 was not important for core RNP assembly, that of the conserved adenosine at position 2 was crucial. In sum, the length of the U tract (requiring at least three uridines), the adenosine 5′ to the U tract, and the accessibility of the ribose 2′-hydroxyl groups are crucial requirements for the association of the Sm site RNA with the Sm proteins. Position-specific binding determinants are provided by the 5′ adenosine as well as, to a lesser degree, the uridine adjacent to it (AAUUUUUGA).
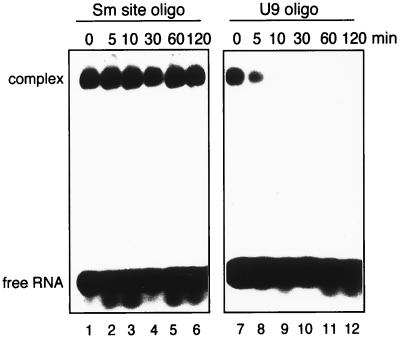
Lastly, we investigated the extent to which the Sm proteins would assemble onto an oligonucleotide that lacked the sequence-specific binding determinants, by testing for Sm protein binding onto a 9-meric uridine (U9) oligonucleotide. Intriguingly, the U9 oligonucleotide was shifted only slightly (threefold) less efficiently than the Sm site oligonucleotide was (Fig. 8B, lanes 21 to 24). This implies that the Sm proteins have a sufficiently high affinity for uridine-rich RNA to stably associate with it, even in the absence of bordering purines. These results, at first sight, suggested that a long U tract could alleviate the observed requirement for an upstream adenosine. We were therefore interested to determine whether the U9-RNP and the minimal core RNP (with the Sm site oligonucleotide) displayed equivalent thermodynamic stability. Following reconstitution, a 1,000-fold molar excess of unlabelled Sm site oligonucleotide over labelled RNA was added to each mixture, and the samples were further incubated at 30°C for 0 to 2 h and analyzed by EMSA (Fig. 9). Strikingly, no significant dissociation of the minimal core RNP (containing the Sm site oligonucleotide) was observed, even after 2 h of incubation (lane 6). This amount of competitor is more than sufficient to prevent association when added at the onset of the Sm site oligonucleotide-TP reconstitution assay (Fig. 5A), ensuring that any complexes assembled after the addition of competitor would contain unlabelled Sm site oligonucleotide and hence not be detectable. This low dissociation rate therefore indicates that the minimal core RNP has a high degree of thermodynamic stability. In sharp contrast, the U9-RNP completely dissociated after 10 min (Fig. 9, lane 9). Thus, although the U9-RNP was stable enough to withstand EMSA in the presence of 2 M urea, it was not thermodynamically stable. Collectively, these data suggest that an initial phase of core RNP assembly is mediated by a high affinity of the Sm proteins for uridine-rich, single-stranded RNA but that the presence of the 5′ adenosine in the Sm site is essential to “lock in” the RNA-protein interactions, thereby committing the RNP particle to thermodynamic stability.
FIG. 9.
Comparison of the thermodynamic stability of the RNP containing either the U9 or the Sm site oligonucleotide. Radiolabelled oligonucleotide (5 nM) was incubated for 45 min at 30°C with approximately 100 nM TPs. Following reconstitution, a 1,000-fold excess of cold Sm site oligonucleotide (5 μM) was added to each assay mixture. Incubation at 30°C was continued for 0 to 120 min, as indicated. Reconstitution assays were started at different times so that all sample incubations were completed simultaneously.
DISCUSSION
We demonstrate here that a nonameric Sm site RNA oligonucleotide suffices for assembly of a minimal core RNP that has acquired several characteristic features of U snRNP core particles. This minimal system allowed us to study in detail the kinetics of core RNP assembly. Specific regions of the U snRNA were found to facilitate the assembly kinetics by accelerating the rate of Sm protein association and reducing the activation energy of core RNP assembly. Formation of a thermodynamically stable minimal core RNP required the presence of the conserved adenosine base. Furthermore, recognition determinants for the Sm proteins were found to be provided collectively by the 2′ hydroxyl moieties and the uridine bases, as well as specifically by the conserved adenosine base, which was essential for assembly commitment.
Sm site RNA determinants of core RNP assembly.
Several Sm site RNA constituents were shown to be important determinants for the stable association of Sm proteins onto the Sm site element. While the presence of the 2′ hydroxyl groups is collectively essential for the interactions, no position-specific group is critical (Fig. 7). This suggests that an intimate association with the ribose backbone is pivotal for the Sm site RNA-Sm protein interactions during the assembly process. Consistent with this idea, hydroxyl radical probing of in vitro-reconstituted HeLa U1 snRNP particles revealed a protection of the ribose backbone throughout the Sm site element (14).
The Sm site consensus sequence (PuAU4–6GPu) can be considered to be two entities: the central, uridine-rich tract, and the flanking purines. We demonstrate that the U tract provides one of the main recognition determinants for the Sm protein interactions. Shortening it to three or fewer uridines, by either deletions (data not shown) or base exchanges, abolished RNP assembly, while a single base change of U to C at any one position led to a three- to sixfold decrease in affinity (Fig. 8). This suggests that, similar to the 2′-hydroxyl groups, the uridines collectively provide a specific interaction surface for the Sm proteins during assembly. Accordingly, in native HeLa U2 snRNPs, the N3 position of each uridine within the Sm site was protected from chemical modification (14). In vivo mutational analysis of the yeast U4 Sm site (AAU5GG) revealed that each of four different point mutations within the U tract either was lethal or led to a growth defect (17). In the yeast U5 Sm site (UAU6GG), three of the six uridine positions were sensitive to point mutations (20). The yeast U5 Sm site might be more tolerant to mutations due to the longer length of the U tract, or, alternatively, the yeast U4 Sm site might be less tolerant due to the lack of a 3′-flanking stem-loop structure. In any case, both studies corroborate the general importance of the U tract for the interactions with the Sm proteins. However, a U-rich stretch is not sufficient to induce a thermodynamically stable RNP: a 9-meric uridine RNA oligonucleotide was efficiently shifted into an RNP particle that was stable enough to withstand the 2 M urea included in the loading buffer (Fig. 8), but it displayed rapid dissociation (<10 min) in comparison to the minimal core RNP (>2 h) (Fig. 9).
For the assembly of a thermodynamically stable core RNP, the presence of the conserved upstream adenosine (PuAU4–6GPu) appears to be essential. No complex formation was observed when the A2 adenosine was removed either by replacement with a G or by deletion of both 5′ purines (Fig. 8). The inability of the 5′-most adenosine to functionally replace the substituted adenosine in the SmA2-G oligonucleotide (AGUUUUUGA) suggests that the presence of a uridine (or pyrimidine) 3′ to the adenosine is important. Consistent with this idea, the uridine at this position within the Sm site oligonucleotide was the most sensitive to mutation; replacement of this uridine with C led to a sixfold decrease in Sm protein affinity (Fig. 8). Interestingly, this region of the Sm site (AAU) was previously identified, by UV cross-linking experiments with HeLa U1 snRNPs, to be a site of interaction with the Sm G protein (15). Our finding that the A2 adenosine plays a crucial role in core RNP assembly appears to contradict results from an in vivo mutagenesis study of the Sm site in yeast U5 snRNA (20), which did not reveal a critical function for the conserved adenosine site. One possible explanation for this is that when the conserved adenosine base in the yeast U5 snRNA was mutated, an upstream adenosine was used instead: in yeast U5, the Sm site is preceded directly by an adenosine followed by a uridine (caUAUU…; the position corresponding to A2 is underlined). Alternatively, core RNP assembly in yeast may differ from the mammalian system. For example, specific elements may play a more dominant role in determining the specificity.
In contrast to the A2 position, the identity of the highly conserved G8 guanosine (PuAU4–6GPu) was not essential for Sm protein interactions (Fig. 8). In vivo selection for Sm protein binding and nuclear import of a U1-like RNA containing a randomized stretch resulted in a sequence that matched the Sm site consensus and contained both conserved purines (AAUUUUUGG) (9), suggesting that this guanosine has a critical function in vivo which has yet to be determined. An important role as specificity determinants of the 3′ purines per se is suggested by our observation that the Sm proteins did not bind the SmΔ5′ oligonucleotide (UUUUUGA) but did bind to uridine RNA oligonucleotides with a length of either 9 nt (Fig. 8) or 5 nt (data not shown). This suggests that the 3′ purines impede Sm protein binding. In agreement with this, we observed that removal of the 3′ purines (in the SmΔ3′ oligonucleotide) led to an apparent increase in binding (∼threefold [Fig. 8]). One could imagine that the 3′ purines reduce the affinity of the Sm proteins for the uridine-rich RNA, such that the presence of an adenosine 5′ to the U tract is necessary for the molecular stabilization of the RNA-protein interactions; this would lead to an increase in specificity.
Recently, the crystal structure of the Sex-lethal (Slx) protein bound to a 12-nt, single-stranded RNA derived from its cognate binding partner, tra mRNA, has been reported (13). Significantly, this structure revealed that a uridine-rich region of 9 nt is continuously involved in interactions with the Slx protein, with an extensive recognition of the phosphate-sugar backbone and uridine bases. This study thus provides the first molecular explanation of how an RNA element which lacks intramolecular base pairing can be specifically recognized by a protein. The resemblance of these recognition determinants with those elucidated here biochemically for the Sm protein-U snRNA interactions is interesting; the sugar backbone and uridine bases of the single-stranded Sm site element collectively provide the basic recognition determinants.
Involvement of specific U snRNA regions in core RNP assembly.
Our results suggest that, to a large degree, the stable nature of U snRNP particles can be attributed to the RNA-protein interactions which occur directly at the single-stranded Sm site element. Since previous studies have revealed that specific structural elements of U snRNAs can strongly influence the assembly of core U snRNP particles in vivo (19), the kinetics of core assembly in vitro onto either the Sm site oligonucleotide or native U4 and U5 snRNAs was compared. Despite the drastic reduction in RNA length (from approximately 150 to 9 nt), the relative binding efficiency of the Sm proteins was similar for the Sm site oligonucleotide and for the full-length U snRNAs (with only an apparent twofold reduction in affinity for the Sm site oligonucleotide). However, the core RNP assembly onto U4 and U5 snRNAs was >100 times faster than that onto the Sm site oligonucleotide (Fig. 6). In addition, assembly of the Sm proteins onto the Sm site oligonucleotide showed a stronger temperature dependency; performing the reconstitution at 0°C rather than 30°C slightly decreased the efficiency of U4 and U5 snRNP assembly but completely blocked assembly onto the Sm site oligonucleotide (Fig. 6). Taken together, these results suggest that a general role for the specific elements of U snRNA is to act as assembly facilitators, by reducing the activation energy of core RNP assembly and accelerating the assembly process.
Reduction of the U snRNA to solely the Sm site element should have eliminated any stable higher-order RNA structures, yet it resulted in a slower, rather than faster, assembly. This implies that extensive protein rearrangements are required for stable core RNP formation and that these rearrangements are rate limiting during core assembly. The stepwise assembly of the Sm protein complexes onto U snRNA (8, 36) is likely to be an important regulatory control for U snRNP core assembly and suggests two ways, which are not mutually exclusive, in which RNA-protein interactions may be necessary to trigger protein rearrangements prior to stable association with U snRNA. The first possibility is that the protein interaction surfaces of the EFG (and perhaps also of the D1D2 and B/B′D3) complex are blocked for association with the other Sm complexes until an initial RNA-protein contact(s) exposes them. This could explain why no stable higher-order protein complex containing all seven Sm proteins exists in the absence of U snRNA, either in vitro or in vivo (6, 8, 36). The RNA-free HeLa EFG complex apparently consists of two copies of each protein and displays a ring-shaped morphology in negatively stained electron micrographs (35, 36); such a structure could prevent further complex formation with D1D2 and B/B′D3. Destabilization of the EFG hexameric ring could be induced by contacts with the Sm site element (or with structural elements of the U snRNA), thus allowing a single trimeric EFG complex to form protein-protein contacts with D1D2 and to bind subsequently to the Sm site element. A second possibility is that RNA-induced rearrangements among the D1D2 and EFG complexes are required to create an intermolecular RNA-binding domain, allowing the D1D2-EFG complex to bind stably and specifically to the Sm site element. It is likely that the specific elements of U snRNA assist in these protein rearrangements.
Our observation that the minimal core RNP can interact with the U1-70K protein (Fig. 1) suggests that an additional role of the specific U snRNA regions is to prevent the binding of noncognate specific proteins to the core RNP surface. Specifically, a previous study found that a truncated U1-70K protein, lacking its RNA-binding domain, was able to interact with the Sm B/B′ and D2 proteins within a U1 snRNP core particle but not within U2 or U5 core particles (32). This binding discrimination could be explained if differences in the Sm site sequences (in which the uridine tract of the U1 Sm site is interrupted by a G) cause variations in the core RNP surface. However, our Sm site oligonucleotide has a U4 Sm site sequence. It is therefore more probable that the core RNP has a high degree of plasticity in its interaction surface, which is regulated by RNA-protein interactions between the specific U snRNA regions and the Sm proteins.
Recent findings demonstrate a critical role in spliceosomal U snRNP assembly in vivo for the SMN protein (3, 28), which is responsible for the genetic disease spinal muscular atrophy (23). Our findings reveal that core RNP formation can proceed by self-assembly in vitro and provide a basis for understanding how such non-snRNP factors might affect assembly in vivo. For example, they might act as fidelity factors, by increasing the requirement for the structural RNA regions flanking the Sm site element. The in vitro core RNP assembly system presented here is well suited for further elucidation of the role of SMN in U snRNP assembly.
ACKNOWLEDGMENTS
We thank P. Kempkes for HeLa cell preparation; I. Öchsner, W. Lorenz, and A. Badouin for U snRNP preparation; B. Rückert for EM technical assistance; and M. Krause for RNA oligonucleotide synthesis. We are grateful to K. Nagai, C. L. Will, and L. Di Croce for their helpful comments and discussions.
This work was supported by the Gottfried Wilhelm Leibniz Program, Fonds der Chemischen Industrie, and Deutsche Forschungsgemeinschaft grants SFB 286 to R.L. and Ka 805/2 to B.K.
REFERENCES
- 1.Branlant C, Krol A, Ebel J P, Lazar E, Haendler B, Jacob M. U2 RNA shares a structural domain with U1, U4, and U5 RNAs. EMBO J. 1982;1:1259–1265. doi: 10.1002/j.1460-2075.1982.tb00022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cooper M, Johnston L H, Beggs J D. Identification and characterization of Uss1p (Sdb23p): a novel U6 snRNA-associated protein with significant similarity to core proteins of small nuclear ribonucleoproteins. EMBO J. 1995;14:2066–2075. doi: 10.1002/j.1460-2075.1995.tb07198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 4.Fischer U, Lührmann R. An essential signaling role for the m3G cap in the transport of U1 snRNP to the nucleus. Science. 1990;249:786–790. doi: 10.1126/science.2143847. [DOI] [PubMed] [Google Scholar]
- 5.Fischer U, Sumpter V, Sekine M, Satoh T, Lührmann R. Nucleocytoplasmic transport of U snRNPs: definition of a nuclear location signal in the Sm core domain that binds a transport receptor independently of the m3G cap. EMBO J. 1993;12:573–583. doi: 10.1002/j.1460-2075.1993.tb05689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisher D E, Conner G E, Reeves W H, Wisniewolski R, Blobel G. Small nuclear ribonucleoprotein particle assembly in vivo: demonstration of a 6S RNA-free core precursor and posttranslational modification. Cell. 1985;42:751–758. doi: 10.1016/0092-8674(85)90271-5. [DOI] [PubMed] [Google Scholar]
- 7.Fried M, Crothers D M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fury M, Andersen J, Ponda P, Aimes R, Zieve G W. Thirteen anti-Sm monoclonal antibodies immunoprecipitate the three cytoplasmic snRNP core protein precursors in six distinct subsets. J Autoimmunity. 1999;12:91–100. doi: 10.1006/jaut.1998.0266. [DOI] [PubMed] [Google Scholar]
- 9.Grimm C, Lund E, Dahlberg J E. In vivo selection of RNAs that localize in the nucleus. EMBO J. 1997;16:793–806. doi: 10.1093/emboj/16.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grimm C, Stefanovic B, Schümperli D. The low abundance of U7 snRNA is partly determined by its Sm binding site. EMBO J. 1993;12:1229–1238. doi: 10.1002/j.1460-2075.1993.tb05764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamm J, Darzynkiewicz E, Tahara S M, Mattaj I W. The trimethylguanosine cap structure of U1 snRNA is a component of a bipartite nuclear targeting signal. Cell. 1990;62:569–577. doi: 10.1016/0092-8674(90)90021-6. [DOI] [PubMed] [Google Scholar]
- 12.Hamm J, Kazmaier M, Mattaj I W. In vitro assembly of U1 snRNPs. EMBO J. 1987;6:3479–3485. doi: 10.1002/j.1460-2075.1987.tb02672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Handa N, Nureki O, Kurimoto K, Kim I, Sakamoto H, Shimura Y, Muto Y, Yokoyama S. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature. 1999;398:579–585. doi: 10.1038/19242. [DOI] [PubMed] [Google Scholar]
- 14.Hartmuth K, Raker V A, Huber J, Branlant C, Lührmann R. An unusual chemical reactivity of Sm site adenosines strongly correlates with proper assembly of core U snRNP particles. J Mol Biol. 1999;285:133–147. doi: 10.1006/jmbi.1998.2300. [DOI] [PubMed] [Google Scholar]
- 15.Heinrichs V, Hackl W, Lührmann R. Direct binding of small nuclear ribonucleoprotein G to the Sm site of small nuclear RNA. Ultraviolet light cross-linking of protein G to the AAU stretch within the Sm site (AAUUUGUGG) of U1 small nuclear ribonucleoprotein reconstituted in vitro. J Mol Biol. 1992;227:15–28. doi: 10.1016/0022-2836(92)90678-d. [DOI] [PubMed] [Google Scholar]
- 16.Hermann H, Fabrizio P, Raker V A, Foulaki K, Hornig H, Brahms H, Lührmann R. snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein-protein interactions. EMBO J. 1995;14:2076–2088. doi: 10.1002/j.1460-2075.1995.tb07199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu J, Xu D, Schappert K, Xu Y, Friesen J D. Mutational analysis of Saccharomyces cerevisiae U4 small nuclear RNA identifies functionally important domains. Mol Cell Biol. 1995;15:1274–1285. doi: 10.1128/mcb.15.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huber J, Cronshagen U, Kadokura M, Marshallsay C, Wada T, Sekine M, Lührmann R. Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J. 1998;17:4114–4126. doi: 10.1093/emboj/17.14.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarmolowski A, Mattaj I W. The determinants for Sm protein binding to Xenopus U1 and U5 snRNAs are complex and non-identical. EMBO J. 1993;12:223–232. doi: 10.1002/j.1460-2075.1993.tb05648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones M H, Guthrie C. Unexpected flexibility in an evolutionarily conserved protein-RNA interaction: genetic analysis of the Sm binding site. EMBO J. 1990;9:2555–2561. doi: 10.1002/j.1460-2075.1990.tb07436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kambach C, Walke S, Young R, Avis J M, de la Fortelle E, Raker V A, Lührmann R, Li J, Nagai K. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999;96:375–387. doi: 10.1016/s0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- 22.Kastner B, Bach M, Lührmann R. Electron microscopy of small nuclear ribonucleoprotein (snRNP) particles U2 and U5: evidence for a common structure-determining principle in the major U snRNP family. Proc Natl Acad Sci USA. 1990;87:1710–1714. doi: 10.1073/pnas.87.5.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lefebvre S, Burglen L, Reboullet S C, Burlet O, P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 24.Lehmeier T, Foulaki K, Lührmann R. Evidence for three distinct D proteins, which react differentially with anti-Sm autoantibodies, in the cores of the major snRNPs U1, U2, U4/U6 and U5. Nucleic Acids Res. 1990;18:6475–6484. doi: 10.1093/nar/18.22.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lehmeier T, Raker V A, Hermann H, Lührmann R. cDNA cloning of the Sm proteins D2 and D3 from human small nuclear ribonucleoproteins: evidence for a direct D1-D2 interaction. Proc Natl Acad Sci USA. 1994;91:12317–12321. doi: 10.1073/pnas.91.25.12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lerner M R, Steitz J A. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc Natl Acad Sci USA. 1979;76:5495–5499. doi: 10.1073/pnas.76.11.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liautard J P, Sri-Widada J, Brunel C, Jeanteur P. Structural organization of ribonucleoproteins containing small nuclear RNAs from HeLa cells. Proteins interact closely with a similar structural domain of U1, U2, U4 and U5 small nuclear RNAs. J Mol Biol. 1982;162:623–643. doi: 10.1016/0022-2836(82)90392-8. [DOI] [PubMed] [Google Scholar]
- 28.Liu Q, Fischer U, Wang F, Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- 29.Madore S J, Wieben E D, Pederson T. Intracellular site of U1 small nuclear RNA processing and ribonucleoprotein assembly. J Cell Biol. 1984;98:188–192. doi: 10.1083/jcb.98.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mattaj I W. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell. 1986;46:905–911. doi: 10.1016/0092-8674(86)90072-3. [DOI] [PubMed] [Google Scholar]
- 31.Mattaj I W, De Robertis E M. Nuclear segregation of U2 snRNA requires binding of specific snRNP proteins. Cell. 1985;40:111–118. doi: 10.1016/0092-8674(85)90314-9. [DOI] [PubMed] [Google Scholar]
- 32.Nelissen R L, Will C L, van Venrooij W J, Lührmann R. The association of the U1-specific 70K and C proteins with U1 snRNPs is mediated in part by common U snRNP proteins. EMBO J. 1994;13:4113–4125. doi: 10.1002/j.1460-2075.1994.tb06729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neuman de Vegvar H E, Dahlberg J E. Nucleocytoplasmic transport and processing of small nuclear RNA precursors. Mol Cell Biol. 1990;10:3365–3375. doi: 10.1128/mcb.10.7.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plessel G, Fischer U, Lührmann R. m3G cap hypermethylation of U1 small nuclear ribonucleoprotein (snRNP) in vitro: Evidence that the U1 small nuclear RNA-(guanosine-N2)-methyltransferase is a non-snRNP cytoplasmic protein that requires a binding site on the Sm core domain. Mol Cell Biol. 1994;14:4160–4172. doi: 10.1128/mcb.14.6.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plessel G, Lührmann R, Kastner B. Electron microscopy of assembly intermediates of the snRNP core: morphological similarities between the RNA-free (E.F.G) protein heteromer and the intact snRNP core. J Mol Biol. 1997;265:87–94. doi: 10.1006/jmbi.1996.0713. [DOI] [PubMed] [Google Scholar]
- 36.Raker V A, Plessel G, Lührmann R. The snRNP core assembly pathway: identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J. 1996;15:2256–2269. [PMC free article] [PubMed] [Google Scholar]
- 37.Reddy R, Bush H. Small nuclear RNAs and RNA processing. Prog Nucleic Acid Res Mol Biol. 1983;30:127–162. doi: 10.1016/s0079-6603(08)60685-6. [DOI] [PubMed] [Google Scholar]
- 38.Sauterer R A, Goyal A, Zieve G W. Cytoplasmic assembly of small nuclear ribonucleoprotein particles from 6 S and 20 S RNA-free intermediates in L929 mouse fibroblasts. J Biol Chem. 1990;265:1048–1058. [PubMed] [Google Scholar]
- 39.Ségault V, Will C L, Sproat B S, Lührmann R. In vitro reconstitution of mammalian U2 and U5 snRNPs active in splicing: Sm proteins are functionally interchangeable and are essential for the formation of functional U2 and U5 snRNPs. EMBO J. 1995;14:4010–4021. doi: 10.1002/j.1460-2075.1995.tb00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Séraphin B. Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J. 1995;14:2089–2098. doi: 10.1002/j.1460-2075.1995.tb07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sumpter V, Kahrs A, Fischer U, Kornstädt U, Lührmann R. In vitro reconstitution of U1 and U2 snRNPs from isolated proteins and snRNA. Mol Biol Rep. 1992;16:229–240. doi: 10.1007/BF00419662. [DOI] [PubMed] [Google Scholar]
- 42.Van Dam A, Winkel I, Zijlstra-Baalbergen J, Smeenk R, Cuypers H T. Cloned human snRNP proteins B and B′ differ only in their carboxyl-terminal part. EMBO J. 1989;8:3853–3860. doi: 10.1002/j.1460-2075.1989.tb08563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Will C L, Lührmann R. Protein functions in pre-mRNA splicing. Curr Opin Cell Biol. 1997;9:320–328. doi: 10.1016/s0955-0674(97)80003-8. [DOI] [PubMed] [Google Scholar]
- 44.Yang H, Moss M L, Lund E, Dahlberg J E. Nuclear processing of the 3′-terminal nucleotides of pre-U1 RNA in Xenopus laevis oocytes. Mol Cell Biol. 1992;12:1553–1560. doi: 10.1128/mcb.12.4.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu Y-T, Scharl E C, Smith C M, Steitz J A. The growing world of small nuclear ribonucleoproteins. In: Gesteland R F, Cech T R, Aktins J F, editors. The RNA World. 2nd ed. New York: Cold Spring Harbor; 1999. pp. 487–524. [Google Scholar]