Abstract
Bone is an active organ that is continuously remodeled throughout life via formation and resorption; therefore, a fine-tuned bone (re)modeling is crucial for bone homeostasis and is closely connected with energy metabolism. Amino acids are essential for various cellular functions as well as an energy source, and their synthesis and catabolism (e.g., metabolism of carbohydrates and fatty acids) are regulated through numerous enzymatic cascades. In addition, the intracellular levels of amino acids are maintained by autophagy, a cellular recycling system for proteins and organelles; under nutrient deprivation conditions, autophagy is strongly induced to compensate for cellular demands and to restore the amino acid pool. Metabolites derived from amino acids are known to be precursors of bioactive molecules such as second messengers and neurotransmitters, which control various cellular processes, including cell proliferation, differentiation, and homeostasis. Thus, amino acid metabolism and autophagy are tightly and reciprocally regulated in our bodies. This review discusses the current knowledge and potential links between bone diseases and deficiencies in amino acid metabolism and autophagy.
Keywords: Autophagy, Amino acid metabolism, Bone, Bone formation, Bone homeostasis
1. Introduction
Continuous bone formation and resorption are critical processes for the maintenance of healthy bones throughout life and are closely intertwined with energy metabolism, which comprises a series of metabolic pathways that generate energy in the form of adenosine triphosphate (ATP) from nutrients such as carbohydrates, fats, and proteins. Both the anabolic and catabolic metabolic pathways are catalyzed by numerous enzymes that require co-factors and ATP itself for their own activation [1]. In addition to enzymatic activities, proteins, which are combinations of > 20 amino acids, serve as functional molecules (e.g., cellular components, receptors, cytoskeleton, and growth factors) in the cell, extracellular matrix, and circulation systems. The amino acids for their production are supplied through the degradation of dietary and/or cellular proteins, as well as synthesis via metabolic pathways such as glycogenesis and the tricarboxylic acid (TCA) cycle (a.k.a. citric acid cycle); the TCA cycle is dependent on the carbohydrate and fatty acid metabolic pathways, which are important for bone homeostasis [2,3]. Amino acids also act as precursors of bioactive molecules such as neurotransmitters, second messengers, and cytokines. Therefore, dysregulation of amino acid metabolism may result in various pathologies, including those affecting bone tissue and the skeleton [4–6].
Amino acid levels can also be sustained through autophagy, a cellular system for the degradation and recycling of intracellular proteins and organelles [7]. The ULK1 complex (a.k.a. ATG13-ULK1/2-FIP200-ATG101 complex) acts as a pre-initiation complex in the autophagy pathway, which is activated through AMP-dependent protein kinase (AMPK) and inactivated through the core protein of the mammalian target of rapamycin (mTOR) complex 1 (mTORC1). Under amino acid/nutrient starvation conditions, AMPK dephosphorylates ULK1 and activates the ULK1 complex. Beclin-1 (BECN1), a homolog of yeast Atg6 (autophagy-related 6) that is involved in autophagy, endocytosis, and apoptosis [8,9], interacts with Barkor/ATG14, PI3K/VPS34, VPS15, and AMBRA at the endoplasmic reticulum (ER) membrane, and forms the class III phosphoinositide 3 kinase (PI3K) class III complex I (the PI3KC3-C1 complex), which initiates formation of the isolation membrane (a.k.a. phagophore). An active ULK1 complex is known to phosphorylate BECN1 and PI3K/VPS34 in PI3KC3-C1. Conversely, interaction with B-cell lymphoma 2 (BCL2) inhibits the formation of the PI3KC3-C1 complex, while nutrient starvation induces dissociation of the Beclin-1–BCL2 complex, initiating autophagy [8,9]. At the ER, phosphorylated PI3K/VPS34 catalyzes the conversion of phosphatidylinositol (PI) to phosphatidylinositol-3-phosphate [PI(3)P] (the structure called omegasome). The accumulation of PI(3)P then promotes nucleation of the omegasome from the ER.
There are two ubiquitin-like conjugation pathways, ATG12–5 and ATG8, crucial for the autophagy regulation in autophagosome formation. ATG7 acts as an E1-like enzyme to activate both ATG12–5 and ATG8 pathways during initiation and elongation of the autophagosome membrane. ATG3 and ATG10 then act as E2-like enzymes in the ATG8 and ATG12–5 pathways, respectively. The ATG12–5 complex conjugates with ATG16L (the ATG12–5:ATG16L complex) and acts as an E3-like ligase to catalyze phosphatidylethanolamine (PE) to ATG8, transforming the inactivated cytosolic form (type I) into the activated membrane-bounded form (type II). There are three homologs of yeast Atg8 in mammals: LC3 (microtube-associated protein 1 light chain 3), GABARAP (γ-aminobutyric acid receptor-associated protein), and GATE16 (a.k.a. GABARAPL2; Golgi-associated ATPase enhancer of 16 KDa). Among them, LC3 is the best characterized in autophagy. The ATG12–5:ATG16L complex binds to the outer membrane only; by contrast, LC3-II binds to both outer and inner membranes of the isolation membrane in order to promote the elongation of the autophagic membrane (capturing unnecessary proteins/organelles) for autophagosome formation. These autophagosomes fuse with lysosomes (called autolysosomes) and degrade/recycle unnecessary proteins/organelles for supplying amino acid, lipids, and ATP [10–12].
The steady-state level of autophagy is involved in clearance and turnover of both organelles and proteins; however, in case of nutrient starvation, autophagy can be greatly induced to generate amino acids from proteins in order to meet the cellular needs [7,13]. Amino acid metabolism, its metabolites, and intracellular amino acid levels, are all involved in the regulation of autophagy, and vice versa, under physiological and pathological conditions [14–18]. In addition, recent studies show that autophagy is involved in antioxidant protection [19,20]. Under oxidative stress, reactive oxygen species (ROS) oxidize the cysteine residues in SQSTM1/p62 and ATG4 and promotes degradation of ubiquitinated proteins and lipidation of ATG8, respectively; thus, oxidative stress induces autophagic activity [21–23]. The KEAP1–NRF2 system plays a role in cellular defense against ROS, nitric oxide, and electrophilic stresses [20,24]. KEAP1 (Kelch-like ECH associated protein 1), an adaptor protein of Cullin-3 E3-like ligase, degrades NRF2 (erythroid 2-related factor 2), a transcription factor regulating the expression of anti-stress genes [25,26]. Under stress conditions, KEAP1 is inactivated, which allows NRF2 to translocate into the nuclei. Interestingly, SQSTM1/p62 can bind to KEAP1 by competing with NRF2, resulting in the stabilization and consequent translocation of NRF2 to the nuclei [24,26,27]. Thus, autophagy and KEAP1–NRF2 system are closely associated each other. On the other hand, hypoxia conditions activate both hypoxia-inducible factor 1-alpha (HIF-1α)-dependent anti-oxidative activity and ATG5-dependent mitophagy, the selective degradation of mitochondria by autophagy [28]. In this review, we discuss how amino acid metabolic aberrations, including those due to deficiencies in the autophagic machinery, lead to bone disease.
2. Role of Autophagy in Bone Development and Homeostasis
A growing number of studies suggest that autophagy is associated with bone and cartilage development and homeostasis [29,30]. For instance, mice with an osteoblast-specific deletion of the gene coding for autophagy-related 5 (Col1a1-Cre;Atg5F/F mice), a protein crucial for the formation of autophagosomes, exhibit reduced bone formation and mineralization [31]. Moreover, mice deficient for Atg7 in osteoblasts (Osx1-Cre;Atg7F/F mice) exhibit low bone mass and spontaneous fractures through the suppression of bone remodeling due to disrupted osteocyte survival and maturation [32]. Similarly, mice with a deletion of Atg7 in osteocytes (Dmp1-Cre;Atg7F/F mice) exhibit low bone mass and reduced bone remodeling due to increased oxidative stress [33]. In cartilage, deletion of the Atg7 genes results in shorter bone length and growth retardation; for example, Prx1-Cre;Atg7F/F and TamCol2a1-Cre;Atg7F/F mice show accumulation of type II procollagen in the ER of chondrocytes, resulting in ER stress mediated by fibroblast growth factor 18 [34,35]. Chondrocytes in either Col2-Cre;Atg7F/F, Col2-Cre;Atg5F/F, or TamCol2a1-Cre;Atg7F/F mutant mice show increased apoptotic cell death in the growth plate [35,36]. By contrast, Col11a2-Cre;Atg7F/Fmice exhibit severe growth retardation, due to failure in the transition of mesenchymal cells to chondrocytes, with accumulation of glycogen granules, but not elevated ER stress, in chondrocytes of the growth plate [37].
In cathepsin K-expressing osteoclasts, mice with loss of the beclin1 gene (CstK-Cre;Becn1F/F mice), which codes for a critical factor in the regulation of autophagy and cell death, show impaired osteoclast function and bone resorption as well as dysregulated chondrogenesis [38]. In addition, knockdown of Becn1 in mouse bone marrow-derived macrophages suppresses osteoclastogenesis [39]. Osteoclasts from mice with a monocyte-specific deletion of Atg5 (Lyz2-Cre;Atg5F/F mice) also show impaired bone resorption, caused by reduced lysosome secretion from the ruffled border of osteoclasts, which results in increased trabecular bone volume in long bones [40]. Moreover, mutations in SQSTM1/p62, a known substrate of selective autophagy, cause Paget’s disease of bone (PDB), which is characterized by accelerated bone remodeling leading to dysregulated bone formation and weaker bones [41–43]. Sqstmt1 null mice (a.k.a. p62−/− mice) and mice with a point mutation that substitutes proline with leucine at codon 394 of Sqstm1 (p.P394L), equivalent to the p.P392L SQSTM1 mutation in humans [44], also show accelerated osteoclastogenesis, which recapitulates PDB in humans [45,46].
mTOR, a core protein of the mTOR complex 1 (mTORC1) and complex 2 (mTORC2), acts as a serine/threonine kinase. mTORC1, which is activated by nutrients or growth factors, inhibits autophagy [10,47], and the amino acid–mTORC1–autophagy axis is closely associated with amino acid/protein intake into the cells [14,48]. Recent studies have demonstrated that mTOR plays key roles in amino acid-induced signaling via the Rag guanosine triphosphatases (GTPases) (a.k.a. Rags) [49–53]. Under amino acid starvation conditions, SQSTM1/p62, which is separated from the lysosomal membrane, binds to LC3-II at the isolation membrane, leading to degradation of SQSTM1/p62by the autophagosome. On the other hand, under amino acid-rich conditions, mTORC1 binds to SQSTM1/p62 on the lysosomal membrane and induces PI(3)P degradation. Active mTORC1 phosphorylates ULK1/2 and ATG13, leading to inactivation of the ULK1 complex that suppresses autophagic pathway.
Supplementation of L-glutamine, which is uptaken through the SLC1A5 transporter, promotes mTORC1 pathway and inhibits autophagic activation. On the other hand, inhibition of SLC1A5 suppresses mTORC1 activation and activates autophagy. L-type amino acid exchanger SLC7A5 requires mTORC1 signaling activation through exchanging intracellular L-glutamine to extracellular essential amino acid [54]. Thus, increase of intracellular L-glutamine level promotes exchanging L-glutamine to other amino acids, leading to the activation of mTORC1 and inhibition of the autophagic activity.
Leucine, a branched-chain amino acid, is catabolized by methylcrotonoyl-CoA carboxylase 1 (MCCC1) to acetyl-CoA. Inhibition of MCCC1 suppresses mTORC1 activity and activates autophagy through reduction of acetyl-CoA levels via acetylation of RAPTOR by an acetyltransferase EP300 [54].
An increasing number of studies show that mTOR plays crucial roles in osteoblasts and chondrocytes [55–58]. During endochondral ossification, mTORC1 signaling is detectable in pre-hypertrophic/hypertrophic chondrocytes, pre-osteoblasts, and osteoblasts in the primary spongiosa [59]. Mice with a conditional deletion of Mtor in skeletal mesenchymal cells (Prx1-Cre;MtorF/F) show diminished mTORC1 activity and die at birth with short limbs and exencephaly due to ossification defects in the long bones, calvaria, and sternum [59]. Mice with a pre-osteoblast specific deletion of Mtor (Osx1-Cre;MtorF/F) show defects in osteoblast differentiation through suppression of RUNX2 expression. Finally, mice deficient for Mtor in Osterix-positive osteoblasts exhibit low trabecular bone mass with ossification defects that resemble cleidocranial dysplasia in humans [60]. Regulatory-associated protein of mTOR (RAPTOR), a component of mTORC1, binds to mTOR [61], and mice with deficient Raptor in skeletal mesenchymal cells (Prx1-Cre;RaptorF/F) exhibit bone phenotypes similar to Prx1-Cre;MtorF/F mice [59]. In addition, Osx1-Cre;RaptorF/F mice exhibit a bone phenotype similar to Osx1-Cre;MtorF/F mice, with suppressed differentiation in pre-osteoblasts and chondrocytes in the growth plate [56,60,62]. Interestingly, in Raptor mutant osteoblasts the mTORC1 pathway is suppressed without affecting autophagic activity, while the mTORC2 pathway is activated [56]. The suppression of mTORC1 in osteoblasts also affects B-lymphopoiesis by reduced expression of Cxcl12 and Il7, which are key cytokines for B-lymphocyte differentiation [63]. Mice with a deletion of Raptor in osteocytes (Dmp1-Cre;RaptorF/F) show normal bone development but slightly inhibited bone resorption, resulting in an increase of trabecular bone mass [64]. In addition, mice with a mesenchymal cell-specific deletion of Rictor (Prx1-Cre;RictorF/F) exhibit short and thinner long bones due to a delay in chondrocyte differentiation in the growth plate, as well as calvarial defects due to suppression of osteoblastic differentiation [65]. Thus, mTOR signaling is indispensable for chondrocyte and osteoblast differentiation.
Mice with an osteoclast-specific deletion of Raptor (Lyz2-Cre;RaptorF/F) exhibit osteopenia due to accelerated osteoclastogenesis through the suppression of mTORC1 signaling [66,67]. By contrast, Ctsk-Cre;RaptorF/F mice present increased bone mass due to suppression of osteoclastogenesis [68]. Therefore, the osteoclast phenotype in these mice is controversial for unknown reasons, and Raptor may have different functions at each specific stage of osteoclast differentiation. On the other hand, mice with a conditional deletion of mTORC1 inhibitor Tsc in osteoclasts (Lyz2-Cre;Tsc1F/F mice and Ctsk-Cre;Tsc1F/F mice) show increased bone mass and reduced osteoclast activity [66,69]; bone marrow mesenchymal cells isolated from Tsc1 mutant mice show suppression of osteoclastogenesis [67]. On the other hand, mice with a deletion of Tsc2, which forms a heterodimer with TSC1 to regulate the mTORC1 pathway, in osteoblasts (OC-Cre;Tsc2F/F) exhibit an increase of cortical and trabecular bone thickness and bone mass. Because TSC2 regulates the insulin signaling pathway via AKT (a.k.a. protein kinase B), a loss of Tsc2 in osteoblasts lead to insulin resistance, resulting in the suppression of osteoblast proliferation and differentiation and disorganized bone formation. The normalization of mTORC1 signaling by haploinsufficiency of Mtor in Tsc mutant mice (OC-Cre;Tsc2F/F;MtorF/+ mice) can partially rescue the bone phenotype [58]. These genetic mouse models for autophagy and their bone phenotypes are summarized in Table 1.
Table 1.
Skeletal defects associated with deficiencies in autophagy
| Gene | Targeted cell type | Genotype | Phenotype | References |
|---|---|---|---|---|
| Atg5 | Osteoblast | Col1a1-Cre;Atg5 F/F | Reduced bone formation and mineralization | [16] |
| Atg7 | Osteoblast | Osx1-Cre;Atg7 F/F | Low bone mass and spontaneous fractures Suppressed bone remodeling Disrupted osteocyte survival and maturation |
[17] |
| Slc7a5 | Osteoblasts | Osx-Cre;Slc7a5 F/F | No bone phenotype | [35] |
| Atg7 | Osteocytes | Dmp1-Cre;Atg7 F/F | Low bone mass and reduced bone remodeling Increased oxidative stress |
[18] |
| Atg7 | Skeletal mesenchymal cells |
Prx1-Cre;Atg7 F/F | Accumulated type II procollagen in chondrocytes ER stress | [19, 20] |
| Atg7 | Chondrocytes | Tam Col2a1-Cre;Atg7 F/F | Accumulated type II procollagen in chondrocytes ER stress | [19, 20] |
| Col2a1-Cre;Atg7 F/F | Increased apoptotic cell death in the growth plate | [19, 20] | ||
| Col2a1-Cre;Atg5 F/F | Increased apoptotic cell death in the growth plate | [20, 21] | ||
| Col2a1-Cre;Atg7 F/F | Increased apoptotic cell death in the growth plate | [20, 21] | ||
| Col11a2-Cre;Atg7 F/F | Severe growth retardation | [20, 21] | ||
| Becn | Osteoclasts | CstK-Cre;Becn1 F/F | Impaired osteoclast function and bone resorption Dysregulated chondrogenesis |
[23] |
| Atg5 | Osteoclasts | Lyz2-Cre;Atg5 F/F | Impaired bone resorption Reduced lysosome secretion from osteoclasts Increased trabecular bone volume of long bones |
[25] |
| Slc7a5 | Osteoclasts | Tnfrsf11a-Cre;Slc7a5 F/F | Low bone mass and increased bone resorption Accelerated osteoclastogenesis Suppression of mTORC1 pathway |
[35] |
| Slc7a5 | Osteoclasts | Lyz2-Cre;Slc7a5 F/F | Low bone mass and increased bone resorption Accelerated osteoclastogenesis Suppression of mTORC1 pathway |
[35] |
| Raptor | Osteoclasts | Lyz2-Cre;Raptor F/F | Osteopenia Accelerated osteoclastogenesis Suppressed mTORC1 signaling |
[37] |
| Raptor | Osteoclasts | Ctsk-Cre;Raptor F/F | Increased bone mass Suppressed osteoclastogenesis |
[38] |
| Tsc1 | Osteoclasts | Lyz2-Cre;Tsc1 F/F | Increased bone mass and reduced osteoclast activity | [37] |
| Tsc1 | Osteoclasts | Ctsk-Cre;Tsc1 F/F | Increased bone mess and reduced osteoclast activity | [39] |
| P62 | all cell types | P62 −/− | Accelerated osteoclastogenesis | [30] |
| P62 | all cell types | P62 P394L/P394L | Accelerated osteoclastogenesis | [31] |
3. Role of Amino Acids in Bone Development and Homeostasis
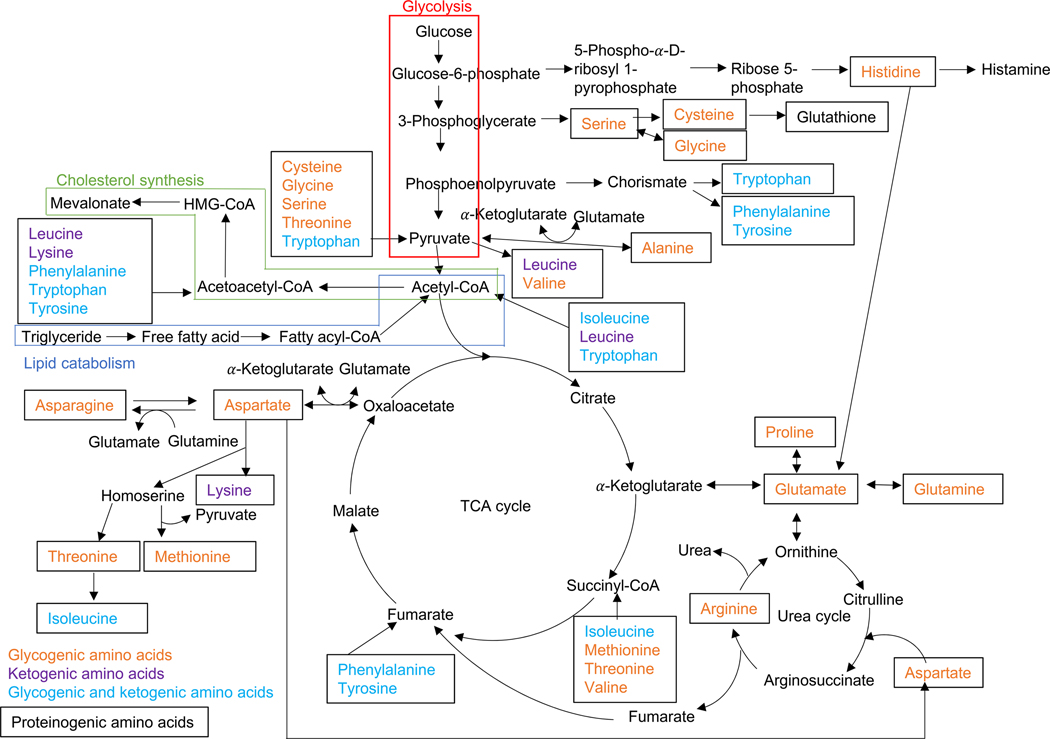
There are 20 proteogenic amino acids and numerous non-proteogenic amino acids in humans. The specific combination of amino acid determines the characteristics and functions of proteins in eukaryotes and microorganisms. Nine of the amino acids (phenylalanine, valine, threonine, tryptophan, methionine, leucine, isoleucine, lysine, and histidine) are categorized as essential amino acids and cannot be synthesized in the human body. For those amino acids that can be produced by cells, their precursors are intermediates or metabolites generated during glycolysis and the TCA cycle. As shown in Figure 1, phenylalanine, tyrosine, and tryptophan arise from chorismate, which derives from phosphoenolpyruvate, a glycolysis metabolite. Isoleucine, valine, leucine, and alanine are converted from pyruvate, whereas methionine, threonine, lysine, and asparagine are synthesized from oxaloacetate. Glutamine, proline, and arginine arise from α-ketoglutarate, which is an intermediate of the TCA cycle. Glucose 6-phosphate is converted to ribose-5-phosphate, and then eventually to histidine, whereas cysteine, glycine, and serine arise from 3-phosphoglycerate, an intermediate of glycolysis. Beyond the abovementioned metabolic pathways, metabolites generated during degradation of amino acids give rise to precursors for other biosynthesis pathways or bioactive molecules. From the perspective of the degradation pathway, amino acids are also classified, based on their catabolites, as glycogenic or ketogenic: 14 glycogenic amino acids are catabolized to either pyruvate or TCA cycle metabolites, two ketogenic amino acids are catabolized to either acetyl-CoA or acetoacetyl-CoA, and five amino acids are both glycogenic and ketogenic (Figure 1). The metabolic reactions can be converted bidirectionally. For example, around 40% of glutamate in bone marrow stromal cells are derived from glutamine [70]. Human bone diseases associated with a failure in amino acid metabolism are summarized in Table 2 (see sections below for details). Mice with deficient amino acid metabolism and their bone phenotypes are summarized in Table 3.
Figure 1.
Amino acid metabolic pathway. This figure was drawn based on ARGININE BIOSYNTHESIS (mmu00220), ALANINE, ASPARATE AND GLUTAMATE METABOLISM (mmu00250), GLYCINE, SERINE AND THREONINE METABOLISM (mmu00260), CYSTEINE AND METHIONINE METABOLISM (mmu00270), VALINE, LEUCINE AND ISOLEUCINE DEGRADATION (mmu00280), LYSINE DEGRADATION (mmu00310), ARGININE BIOSYNTHESIS (mmu00220), ARGININE AND PROLINE METABOLISM (mmu00330), HISTIDINE METABOLISM (mmu00340), TYROSINE METABOLISM (mmu0350), PHENYLALANINE METABOLISM (mmu00360), and TRYPTOPHAN METABOLISM (mmu00380) pathway maps obtained from the KEGG website.
Table 2.
Human bone diseases associated with a failure in amino acid metabolism
| Amino acid name | Genes | Disease name | Bone defects | References |
|---|---|---|---|---|
| Alanine | GPT | Nonalcoholic fatty liver disease | Lower bone mineral density | [40] |
| Proline | PYCR2 | Microcephaly, facial dysmorphism, and developmental delay | [42] | |
| Asparagine | ASNS | Epilepsy, developmental delay, and progressive microcephaly | [46] | |
| Phenylalanine | PAH | Phenylketonuria | Osteopenia | [85–87] |
| Tyrosine | Dopamine-related diseases (e.g., Parkinson’s disease, Schizophrenia) |
Osteoporosis | [93–95] |
Table 3.
Mouse bone phenotypes associated with amino acid metabolic defects
| Amino acid | Targeted genes | Bone defects | References |
|---|---|---|---|
| Alanine | Gpt −/− | Increased bone mineral contents | IMPC |
| Proline | Pycr2 −/− | Low bone mineral density | IMPC |
| Cysteine | Cod1 −/− | Short snout, facial asymmetry, irregular calvaria shape, and kyphosis | [48] |
| Histidine | Hdc −/− | Increased bone mineral contents and increased cortical bone thickness Accelerated bone formation and suppressed osteoclastogenesis |
[71] |
| Methionine | Cbs −/− | Pointed snout, osteoporosis, delayed endochondral ossification, and small body | [76–79] |
| Phenylalanine | Pah enu2/enu2 | Low mineral density, low trabecular volume, imbalance of bone homeostasis Suppressed osteogenic differentiation | [88] |
| Tyrosine | Slc6a3 −/− | Reduced bone mass and strength | [99] |
| Tyrosine | Slc6a4 −/− | Reduced bone mass and strength | [100] |
| Tryptophan | Ido −/− | Osteopenia | [112] |
3.1. Alanine (L-alanine)
The (inter)conversion between pyruvate and alanine is catalyzed by glutamate dehydrogenase and alanine aminotransferase 1 (ALT1, a.k.a. glutamic-pyruvic transaminase), which are encoded by the GPT gene. ALT1 acts on the reversible transamination between alanine/α-ketoglutarate and pyruvate/glutamate. Gpt−/− female mice, but not male mice, exhibit increased bone mineral content [reported by the International Mouse Phenotyping Consortium (IMPC), accessible at https://www.mousephenotype.org]. In addition, patients with nonalcoholic fatty liver disease and increased serum levels of ALT1 show lower bone mineral density compared with healthy individuals or patients with normal ALT1 levels [71].
In addition, alanine-glyoxylate aminotransferase 2 (AGXT2) catalyzes the conversion between alanine/glyoxylate and pyruvate/glycine. Mice with a deficiency for the Agxt2 gene (Agxt2−/− mice) exhibit increased vasodilation in skeletal and cardiac muscles [72].
3.2. Arginine (L-arginine) and Proline
In the TCA cycle, alpha-ketoglutarate is converted into glutamic acid by glutamate dehydrogenase. Glutamic acid is further converted to either glutamine by glutamine synthase, to proline by pyrroline 5-carboxylate reductase 1 and 2 (PYCR1 and 2), or to citrulline (via ornithine), and then arginine, in the urea cycle, with formation of urea from toxic catabolite ammonia in the liver. Thus, arginine, ornithine, and citrulline are all substrates of the urea cycle.
Proline is catabolized into ornithine by proline dehydrogenase 1 (PRODH) and ornithine aminotransferase (OAT), or into glutamic acid by delta-1-pyrroline-5-carboxylate dehydrogenase (P5CDH, a.k.a. ALDH4A); ornithine is further converted into glutamic acid by OAT.
Arginine enhances osteogenesis in human mesenchymal stem cells through upregulation of expression of osteogenic transcription factors: RUNX2, DLX5, and OSX (Osterix) [73]. Patients with autosomal recessive mutations in the PYCR2 gene display microcephaly, seizures, facial dysmorphism, developmental delay, and cerebral atrophy [74], and Pycr2−/− mice exhibit low bone mineral density and decreased grip strength (reported by the IMPC). The catabolism of arginine involves its conversion to glutamic acid by OAT or cycling through the urea cycle. Mutations in OAT cause gyrate atrophy, characterized by progressive retinal atrophy and cataracts, which may cause blindness [75,76].
3.3. Asparagine and Aspartic Acid and Lysine
Oxaloacetate, an intermediate in the TCA cycle, is converted into aspartic acid by transaminase, then to asparagine, a substrate of the TCA cycle, by asparagine synthetase (ASNS). Alternatively, aspartic acid can be converted into methionine by methionine synthase via homocysteine, with co-factor cobalamin (vitamin B12), into threonine by threonine synthase, and into lysine by diaminopimelate decarboxylase. Patients with ASNS mutations display epilepsy, developmental delay, progressive microcephaly, and reduced brain volume [77]. Alpha aminoadipic semialdehyde synthase (AASS) converts L-lysine to saccharopine in the mitochondria. The final metabolite of lysine is acetyl-CoA, which enters the TCA cycle. A known mutation in AASS causes hyperlysinemia, which typically results in no health issues [78]. Patients with lysinuric protein intolerance (LPI), an autosomal recessive disorder of cationic amino acid (arginine, lysine, and ornithine) transport caused by mutations in the SLC7A7 (solute carrier family 7 member 7) gene present with low bone mass, delayed bone development and osteoporosis, muscle weakness, pulmonary alveolar proteinosis, and kidney dysfunction [79–81]. Slc7a7 null mice exhibit bone mineralization defects and delayed development in the kidneys, lungs and liver, as seen in LPI patients [82].
3.4. Cysteine
Cysteine is catabolized to taurine and pyruvate, which are in turn catalyzed by cysteine dioxygenase (CDO), aspartate aminotransferase, cysteine sulfonic acid decarboxylase (CSAD), 3-mercaptopyruvate sulfurtransferase (MPST), and hypotaurine dehydrogenase.Cdo1−/− mice exhibit short snout, facial asymmetry, irregular calvaria shape, kyphosis, and hyperlaxity of the paws with low levels of taurine and excessive cysteine sulfate in adults [83]. Mpst−/− mice show anxiety-like behavior without any morphological abnormality in the brain [84], but no bone-related defects. Studies with animal models also suggest that taurine supplementation can improve bone formation [85–89]. Moreover, exogenous taurine accelerates osteogenic differentiation in human mesenchymal stem cells [90]. Taken together, taurine and taurine synthesis seem to be related to bone development and homeostasis. N-acetyl-L-cysteine (NAC) is an amino acid derivative that plays a role in the cellular antioxidant capabilities [91]. NAC, which is incorporated into cells through cysteine transporter SLC3A1 or the NAC carrier AE1 (anion exchange protein 1), is deacetylated into L-cysteine, which is transformed into glutathione, an antioxidative molecule inhibiting intracellular ROS activity, by a glutamate cysteine ligase and glutathione synthetase in the cytosol [92,93]. Pre-treatment with NAC renders the cells resistant to oxidative stress in cell viability, cell proliferation, and osteogenic differentiation in rat femur bone marrow-derived osteoblast-like cells [94]. Moreover, in vivo transplantation of the pre-treated bone marrow-derived osteoblast-like cells with NAC to bone defects in rat long bone accelerates bone regeneration through suppression of ROS [94,95]. On the other hand, a treatment with NAC in bone marrow-derived monocytes suppresses osteoclastogenesis [96].
The members of the Forkhead box class O (FoxO) family of transcription factors play roles in various cellular functions (e.g. cell proliferation, differentiation, apoptosis, resistance for oxidative stress) in a variety of cells and tissues. FoxO1 promotes osteogenic differentiation of mesenchymal stem cells through RUNX expression. Therefore, mice with a deficiency for FoxO1 in Osteocalcin (a.k.a. BGLAP)-expressing osteoblasts (Bglap-Cre;FoxO1F/F) exhibit low bone mass due to differentiation defects and increased apoptosis in osteoblasts through increased intracellular ROS [98]. As expected, treatment with NAC normalizes intracellular ROS levels and osteogenic differentiation in Bglap-Cre;FoxO1F/F mice [98]. By contrast, overexpression of Foxo1 suppresses pre-osteoblast proliferation, suggesting that FoxO1 acts bi-directionally in osteogenesis [97].
3.5. Glutamine and L-Glutamic acid (Glutamate)
In the TCA cycle, alpha-ketoglutarate is converted into glutamic acid by glutamate dehydrogenase [(GLUD1 and GLUD2 (only in humans)]. Glutamic acid is further converted into glutamine by glutamine synthase.
Glutamic acid is known a neurotransmitter in the central nervous system and a precursor of gamma-aminobutyric acid (GABA); its conversion to GABA is catalyzed by glutamate decarboxylase 1 (GAD1; a.k.a. GAD67) with co-factor vitamin B6.Glutamic acid receptors are expressed in osteoblasts and osteoclasts [99–102], and intracellular glutamic acid metabolism plays crucial roles in various aspects of metabolic homeostasis in bone [103]. For instance, the inhibition of glutamate transporter as well as the blockage of the receptor increases the number of bone resorption pits, while glutamic acid has no effect on survival or activity of mature osteoclasts [103]. In addition, glutamic acid can significantly increase osteoblast differentiation and mineralization, while inhibition of the transport shows no effect on osteoblasts [103]. Blockage of the receptors inhibits osteoblast maturation and the mineralization activity, while stimulation of the receptors accelerates bone mineralization in vitro and in vivo [101]. These evidences suggest that glutamic acid may be more important in osteoclasts rather than in osteoblasts. Moreover, patients with autosomal recessive GAD1 mutations exhibit neuronal developmental defects such as early-onset epilepsy and intellectual disability, cleft palate, and scoliosis (a sideways curvature or twist of the spine) [104]. While scoliosis can be caused by conditions such as cerebral palsy and muscular dystrophy, a failure of spine formation and osteoporosis with fractures in the spine can cause scoliosis. The cause of most scoliosis remains unclear. Gad1 null mice exhibit cleft palate and immature lung and die at birth likely due to a respiratory failure [105,106].
Glutamine metabolism also plays roles in several cellular processes in bone cells. For instance, WNT7B-mediated β-catenin independent WNT signaling activates osteoblast function through activation of mTORC1 signaling [107]. The accelerated bone formation in mice with constitutively activated Wnt7b in osteoblasts (Osx-Cre;R26R-Wnt7b) is restored by deletion of Raptor (Osx-Cre;R26R-Wnt7b-RaptorF/F) [107]. The glutamine-dependent anaplerosis is catalyzed by glutaminase (GLS; kidney type) and glutaminase 2 (GLS2; liver type) in the mitochondria, converting glutamine into glutamate. Interestingly, WNT3A-mediated β-catenin-independent, but mTORC1-dependent, signaling accelerates osteoblast differentiation through increased GLS-dependent glutamine catabolism in cultured osteoblast progenitors [108]. Mice with a deficiency for Gls in skeletal mesenchymal cells (Prx1-Cre;GlsF/F) exhibit a decrease in the osteoblast number and bone formation rate without affecting osteoclast number and bone resorption, resulting in reduction of bone mass in long bones [70]. In addition, deletion of Gls in skeletal stem cells (Lepr-Cre;GlsF/F) show reduced bone mass similar to Prx1-Cre;GlsF/F mice; however, mice with an osteoblast-specific deletion of Gls (Bglap-Cre;GlsF/F) show normal bone formation and resorption [70]. These results suggest that GLS is required for the fate determination of skeletal stem cells to osteoblasts, but not for cellular functions in osteoblasts. Osteoclastogenesis of bone marrow macrophages is accelerated by glutamine in a dose-dependent manner [109]. Mice with chondrocyte-specific deletion of Gls (Col2a1-Cre;GlsF/F) exhibit a growth arrest in the growth plate after birth [110]. These chondrocyte’s phenotypes are restored by supplementation of α-ketoglutarate, which is a metabolite in the TCA cycle [110]. On the other hand, the enzymatic activity of glutamine synthetase (GS) is suppressed by canonical WNT/β-catenin signaling induced by either WNT3A or lithium chloride (LiCl) in MG-63 cells, a human osteoblast like-osteosarcoma cell line, and rat bone marrow mesenchymal stem cells [111].
In addition, glutamine influences bone healing and regeneration. An intravenous injection of glutamine/alanine accelerates formation of fibrocartilaginous callus and hyalin cartilage in fractured bone in rats [112]. Aging of mesenchymal stem cells (MSCs) leads to decrease in the osteogenic differentiation capacity through suppression of estrogen-related receptor alpha (ERRα) and glutaminases (GLS), resulting in age-associated osteoporosis. In the mitochondria, glutamine is catabolized to glutamic acid by GLS in the anaplerotic pathway. While it has been known that ERRα, an orphan nuclear receptor, regulates target genes, a recent study suggests that ERRα directly regulates Gls expression via mTROC1 pathway [113]. Interestingly, overexpression of Erra and Gls in aged MSCs can restore decreased osteogenic differentiation and glutamine consumption, indicating that glutamine anaplerosis in the mitochondria is crucial for osteogenic differentiation in MSCs [113].
3.6. Glycine
The conversion reactions between glycine and serine are reversibly catalyzed by serine hydroxymethyltransferase 1 and 2 (SHMT1 and SHMT2), with co-factor vitamin B6. In the liver, glycine is degraded to CO2 and NH3 with co-conversion of tetrahydrofolate to 5,10-) methylenetetrahydrofolate by the glycine cleavage system. While Shmt1 or Shmt2 null mice have been generated, detailed bone analyses have not been performed yet.
3.7. Histidine
Histidine is converted to histamine, a biological amine that regulates immune response and neurotransmission, by histidine decarboxylase (HDC), with co-factor vitamin B6. Hdc−/− mice exhibit abnormal neurological responses, a suppressed immune system, increased bone mineral contents, and increased cortical bone thickness due to accelerated bone formation and suppressed osteoclastogenesis [114]. The upregulated expression of HDC in monocytes from patients with osteoporosis is negatively correlated with bone mineral density [115], suggesting that histamine may activate osteoclasts for bone resorption. T-cell ubiquitin ligand-2 (TULA-2), a histidine phosphatase that is expressed in pre- and mature-osteoclasts, suppresses osteoclast differentiation [116]. Mice with a double knockout of the Tula and Tula2 genes exhibit low bone mass due to an increase of the osteoclast number and activity [116].
3.8. Isoleucine, Leucine, and Valine
The degradation of isoleucine, leucine, and valine is catabolized by branched-chain amino acid transaminase 1 and 2 [BCAT1 (cytosolic) and BCAT2 (mitochondrial)] and branched-chain α-keto acid dehydrogenase [BCKDH; homodimer of alpha and beta (BCKDHA and BCKDHB)]. Leucine and isoleucine are eventually converted to acetyl-CoA, while isoleucine and valine are converted to succinyl-CoA; both acetyl-CoA and succinyl-CoA are transported into the TCA cycle. Mutations in either BCAT1 or BCAT2 cause hypervalinemia and hyperleucin-isoleucinemia. Although Bcat1−/− mice have been generated, their phenotype still needs to be analyzed. However, it is known that mice with a point mutation in Bcat2 or with a deficiency of Bcat2 (Bcat2−/−mice) exhibit decreased body weight [117,118]. In humans, mutations in either BCKDHA or BCKDHB cause maple syrup urine disease, which is characterized by neurodegeneration but no bone defect [119].Bckdha−/− and Bckdhb−/−null mice have been generated, but no phenotypic data have been reported. A recent study shows that MC3T3-E1 cells, an osteoblast cell line, generate adenosine triphosphate (ATP) through glycolysis and dependent upon branched chain amino acids [120]. Exogenous supplementation of leucine suppresses cell proliferation in MC3T3-E1 pre-osteoblast cells through cell senescence and DNA damage [121].
3.9. Methionine
Methionine is catabolized to S-adenosylmethionine (SAM) by methionine adenosyltransferase, and then to homocysteine; SAM acts as a substrate for methylation of multiple macromolecules such as DNA, RNA, and proteins. Homocysteine is then converted into cysteine, together with serine and with vitamin B6 as co-factor, by cystathionine beta-synthase (CBS) and cystathionin-γ-lyase (CTH, a.k.a. CSE) through reverse-transulfurylation. Cbs null mice exhibit pointed snout, osteoporosis, delayed endochondral ossification, hepatic steatosis, microphthalmia, small body, cardiovascular defects, and hyperhomocysteinemia/homocystinuria, which are also seen in patients with classical homocystinuria [122–125]. Cth null mice exhibit myofiber atrophy when cysteine is not supplemented from diets [126], and autosomal recessive CTH mutations cause primary cystathioninuria in humans [127]. Methionine-restricted diets increase longevity and lifespan by improving metabolism with reduced body size in mice and rats. Mice fed with methionine-restricted diets show weaker bones, lower bone density, and thinner trabecular bones compared to a normal diets group due to suppression of osteoblast differentiation, by suppressing Runx2, Opg and Dmp1, without affecting the number of osteoblasts and osteoclasts [128,129]. Interestingly, several microRNAs (small non-coding RNAs containing ~22 nucleotides), which alter Runx2 expression, are elevated in osteoblasts from mice fed with methionine-restricted diets [129].
Homocysteine can also be transformed back to methionine by methionine synthase (MTR; 5-methyltetrahydrofolate-homocysteine methyltransferase), with vitamin B12 as co-factor. In this conversion, 5’-methyltetrahydrofolate is transformed into tetrahydrofolate, a component of folic acid metabolism. Single-nucleotide polymorphisms (SNPs) in the MTR gene are associated with non-syndromic cleft lip with/without cleft palate [130].
3.10. Phenylalanine and Tyrosine
L-phenylalanine is converted into L-tyrosine by phenylalanine hydroxylase (PAH), or into phenethylamine, a neuromodulator, by DOPA decarboxylase (DDC). L-tyrosine is then converted by DDC into p-tyramine, also a neuromodulator, by tyrosine hydroxylase (TH) into L-DOPA, or by tyrosine aminotransferase (TAT) and fumarylacetoacetate hydrolase (FAH) into fumarate. L-DOPA can be further converted into dopamine by DDC. In the end, dopamine can be transformed into several catecholamines that act as neurotransmitters in the central and peripheral nervous systems.
Mice with a point mutation in Pah (phenylalanine hydroxylase) exhibit phenylketonuria, microcephaly, and impaired learning and motor coordination that recapitulates phenylketonuria in humans [131]. Osteopenia, risk of fracture, and low bone mineral density have also been reported in some patients with phenylketonuria [132–134]. Pah mutant (ENU-induced point mutation) mice show low mineral density, low trabecular volume, an imbalance in bone homeostasis through upregulation of parathyroid hormone PTH, and suppression of osteogenic differentiation [135].
DDC is a multifunctional enzyme that acts with co-factor vitamin B6, converting L-DOPA to dopamine, L-phenylalanine to phenethylamine, L-tyrosine to tyramine, L-histidine to histamine, L-tryptophan to tryptamine, and 5-hydroxytryptophan (5-HTP) to serotonin. All of its metabolites act as neuromodulators or neurotransmitters.
The majority of mice deficient for tyrosine hydroxylase (Th−/− mice) die at the embryonic stage, while some that survive exhibit impaired locomotor activity and growth retardation [136,137]. Mice with a knock-in mutation in the human gene (ThR233H/R233H), which results in its deficiency, survive but exhibit hypotension, hypokinesia, and impaired motor coordination, similar to what is observed in human patients [138].
Dopamine-related diseases (e.g., Parkinson’s disease, schizophrenia) pose a risk of osteoporosis [139–141], and previous studies show that dopamine can induce osteogenic differentiation via dopamine receptors [142–144]. Moreover, mice with a deficiency of either dopamine transporter (Slc6a3−/− mice) or serotonin transporter (Slc6a4−/−mice) show reduced bone mass and strength [145,146]. The skeletal phenotype has not yet been studied in Ddc or Th mutant mice.
Mutations in TAT cause tyrosinemia type II (a.k.a. Richner-Hanhart syndrome), which is characterized by photophobia, neurological dysfunction, and hyperkeratosis [147,148]. Tat−/− mice have been generated and still need to be analyzed. Finally, mutations in FAH cause tyrosinemia type I, with symptoms ranging from neonatal death to survival beyond 20 years old. Milder cases show hepatocellular carcinoma, rickets, and renal failure [149,150], which are also seen in Fah mutant mice [151,152].
3.11. Serine
Metabolite 3-phosphoglycerate is converted into 3-phosphoserine by 3-phospho-hydroxypyruvate-glutamate transaminase (PSAT), with co-conversion of glutamate to α-ketoglutarate, and then to L-serine by phosphoserine-phosphatase (PSPH). L-serine is further converted into glycine by serine hydroxymethyltransferase 1 and 2 (SHMT1 and SHMT2) with co-factor vitamin B6, or into cysteine by cystathionine beta-synthase (CBS) and cystathionin-γ-lyase (CTH), also with co-factor vitamin B6, which is called as transsulfuration pathway. During the conversion of serine to glycine, tetrahydrofolate is transformed to 5,10-methylenetetrahydrofolate as part of the folic acid metabolism. Degradation of serine is a reverse reaction to glycine or conversion to pyruvate by serine dehydratase (SDS, a.k.a. SDH), with co-factor vitamin B6.
3.12. Threonine
Threonine is converted into pyruvate by threonine dehydrogenase (TDH) or acetyl-CoA and glycine by thiolysis. In humans, TDH is inactive. Instead, threonine is converted into α-ketobutylate by serine dehydratase (SDS), with co-factor vitamin B6. Both Tdh and Sds mutant mice have been generated but not analyzed yet (reported by the IMPC).
3.13. Tryptophan
Tryptophan is a precursor of bioactive molecules, particularly those that act in the brain: 1) serotonin (5-HT), which derives from 5-HTP; 2) melatonin, a derivative of 5-HT; and 3) niacin, which results from quinolinic acid (QA), kynurenic acid (KYNA), and picolinic acid [153–155]. Tryptophan and its metabolites have been suggested to be associated with osteoporosis [156,157]. Mice with suppressed degradation of tryptophan (Ido−/− mice) exhibit osteopenia with an imbalance in the number of osteoblasts and osteoclasts [158]. Exogenous picolinic acid induces osteoblastogenesis in human mesenchymal stem cells [158]; however, excessive kynurenine induces osteoclastogenesis and bone loss in vivo [159].
Serotonin is produced at either the central nervous system or peripheral organs independently. Brain-derived neurotransmitter serotonin accelerates bone formation through inhibition of bone resorption; however, gastrointestinal-derived hormonal serotonin inhibits bone formation through suppression of osteoblast proliferation [160,161]. A human cohort study showed that long-term daily niacin intake is negatively associated with hip bone mineral density and risk of hip fracture [162]. The intestinal enterobacteria catabolize tryptophan to indole, 3-indolepropionic acid (IPA), or indole-3-carboxaldehyde (I3A), and these molecules are transfused into or taken up by the intestinal epithelium to enter the bloodstream and then transfer to the brain and liver.
3.14. Amino acid transporter and receptors
Amino acid-mediated signaling is induced by transporters and receptors. In bones, expression of amino acid transporters and receptors varies by cell type and developmental stage. For instance, glutamine transporters GLT-1 (a.k.a. EAAT2) and EAAT4 are expressed in differentiated mouse osteoclasts. By contrast, the cysteine-glutamate antiporter SLC7A11 is expressed in pre-osteoclasts although it disappears in mature osteoclasts [163]. Interestingly, exogenous supplementation of glutamate in RAW264.7 cells, an osteoclast cell line, suppresses osteoclastogenesis [163]. Inhibition of SLC7A11 by sulfasalazine enhances osteogenic differentiation in human bone marrow MSCs and suppresses bone loss in ovariectomized mice [164]. In mouse osteoblasts, glutamate transporters (SLC1A1 and SLC1A3) and glutamate receptor (GLUR2/−3/−4) are expressed [165]. Inhibition of glutamate transporters accelerates osteoblast differentiation with increased extracellular glutamate level. Importantly, a treatment with either glutamine or a glutamate receptor agonist accelerates osteoblast differentiation [165]. The glutamate-aspartate transporter GLAST1 (a.k.a. EAAT1) is expressed in osteocytes and osteoblasts in an extracellular glutamate’s dose-dependent manner [166,167]. However, Glast null mice show normal bone formation/resorption and tooth formation [168]. Mice with an osteoclast-specific deletion of the L-type amino acid transporter 1 gene (Tnfrsf11a-Cre;Slc7a5F/F and Lyz2-Cre;Slc7a5F/F mice) exhibit low bone mass and increased bone resorption due to accelerated osteoclastogenesis through suppression of the mTORC1 pathway [169]. By contrast, mice with osteoblast-specific deletion of Slc7a5 (Osx-Cre;Slc7a5F/F mice) show normal bone formation and resorption [169].
N-methyl-D-aspartate (NMDA) receptor, a glutamate receptor, is expressed in both neuronal and non-neuronal tissues including bones and cartilages. NMDA-mediated glutamatergic signaling plays crucial roles in the differentiation of chondroblasts, osteoblasts, and osteoclasts [170,171]. Eukaryotic translation initiation factor 2 alpha kinase 4 (EIF2AK4), a member of EIF2α (eukaryotic initiation factor 2 alpha), responds to amino acid deprivation and acts as an amino acid sensor. Under ER stress, phosphorylated EIF2α activates ATF4 (activating transcription factor 4) that regulates target gene’s expression [172]. Elf2ak4 null (Elf2ak4−/−) mice exhibit low bone mass, osteoblast proliferation defects, and reduction of bone turnover without any effects on osteogenic differentiation [173]. In addition, gene expression of amino acid transporters (such as Slc1a1, Slc1a5, Slc7a5, and Slc38a2) is suppressed in bone marrow MSCs from Elf2ak4−/−mice, resulting in reduced cell proliferation [173]. ELF2α phosphorylated by either amino acid deprivation, oxidative stress, or ER stresses upregulates expression of ATF4 [174], a CREB family transcription factor that plays roles in osteoblast differentiation, bone matrix synthesis, and osteoclast differentiation. The EIF2α–ATF4 signaling pathway is also induced by parathyroid hormone (PTH) or bone morphogenetic protein 2 (BMP2), and stimulates osteogenic differentiation and proliferation in osteoblasts [175,176]. In addition, a variety of signaling pathways are involved in the regulation of ATF4 through Ras, which binds small molecules GTP and GDP interchangeably and can hydrolyze GTP to GDP. For instance, mice with a conditional deficiency for Neurofibromin 1 (NF1), a GTPase-activating protein known as a negative regulator of Ras signaling,in osteoblasts (Col1a(2.3kb)-Cre;Nf1F/F mice; hereafter Nf1 cKO mice) exhibit increases of bone volume, bone formation, and bone resorption through upregulation of ATF4 phosphorylation and Osteocalcin expression. As a line of this evidence, mice with overexpression ofAtf4 under the type I collagen promotor driver control [Col1a1(2.3kb)-Atf4] display a bone phenotype similar to Nf1 cKO mice [177]. By contrast, Atf4−/−mice display decreased bone volume, bone formation, and bone resorption [177]. Consistent with these evidences, low-protein diets in Nf1 cKO mice and a high-protein diet in Atf4−/− mice can rescue the bone phenotypes [177].
4. Conclusion
An accumulating number of studies indicate that amino acid metabolism is crucial for bone development and homeostasis. Recent genetic studies highlight the link between bone diseases and metabolic disorders affected by abnormal amino acid metabolism. The molecular mechanisms and interactions between bone and non-bone cells in these networks remain to be determined. In this review, we focused on recent findings related to amino acid metabolism in bone homeostasis and diseases. As suggested by several studies, nutritional and pharmacological approaches targeting amino acid metabolism and related pathways may be suitable novel targets for the prevention and treatment of bone diseases.
Highlights.
Recent genetic studies highlight the link between bone diseases and metabolic disorders affected by abnormal amino acid metabolism.
Autophagy plays crucial roles in bone development and homeostasis
Amino acid metabolism is crucial for bone development and homeostasis.
As suggested by several studies, nutritional and pharmacological approaches targeting amino acid metabolism and related pathways may be suitable novel targets for the prevention and treatment of bone diseases.
Acknowledgments
This study was supported by grants from the National Institute of Dental and Craniofacial Research, National Institutes of Health (DE026767, DE028340, and DE029818), to JI, and UTHealth School of Dentistry faculty funds to JI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wilson MP, Plecko B, Mills PB, Clayton PT (2019) Disorders affecting vitamin B6 metabolism. J Inherit Metab Dis 42: 629–646. [DOI] [PubMed] [Google Scholar]
- 2.Montalvany-Antonucci CC, Duffles LF, de Arruda JAA, Zicker MC, de Oliveira S, et al. (2019) Short-chain fatty acids and FFAR2 as suppressors of bone resorption. Bone 125: 112–121. [DOI] [PubMed] [Google Scholar]
- 3.Lucas S, Omata Y, Hofmann J, Bottcher M, Iljazovic A, et al. (2018) Short-chain fatty acids regulate systemic bone mass and protect from pathological bone loss. Nat Commun 9: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding KH, Cain M, Davis M, Bergson C, McGee-Lawrence M, et al. (2018) Amino acids as signalling molecules modulating bone turnover. Bone 115: 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long F (2018) Energy Metabolism and Bone. Bone 115: 1. [DOI] [PubMed] [Google Scholar]
- 6.Dirckx N, Moorer MC, Clemens TL, Riddle RC (2019) The role of osteoblasts in energy homeostasis. Nat Rev Endocrinol 15: 651–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizushima N (2009) Physiological functions of autophagy. Curr Top Microbiol Immunol 335: 71–84. [DOI] [PubMed] [Google Scholar]
- 8.Levine B, Liu R, Dong X, Zhong Q (2015) Beclin orthologs: integrative hubs of cell signaling, membrane trafficking, and physiology. Trends Cell Biol 25: 533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wirawan E, Lippens S, Vanden Berghe T, Romagnoli A, Fimia GM, et al. (2012) Beclin1: a role in membrane dynamics and beyond. Autophagy 8: 6–17. [DOI] [PubMed] [Google Scholar]
- 10.Jung CH, Ro SH, Cao J, Otto NM, Kim DH (2010) mTOR regulation of autophagy. FEBS Lett 584: 1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green DR, Levine B (2014) To be or not to be? How selective autophagy and cell death govern cell fate. Cell 157: 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunlop EA, Tee AR (2014) mTOR and autophagy: a dynamic relationship governed by nutrients and energy. Semin Cell Dev Biol 36: 121–129. [DOI] [PubMed] [Google Scholar]
- 13.Kuma A, Komatsu M, Mizushima N (2017) Autophagy-monitoring and autophagy-deficient mice. Autophagy 13: 1619–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan HWS, Sim AYL, Long YC (2017) Glutamine metabolism regulates autophagy-dependent mTORC1 reactivation during amino acid starvation. Nat Commun 8: 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang N, Yang X, Yuan F, Zhang L, Wang Y, et al. (2018) Increased Amino Acid Uptake Supports Autophagy-Deficient Cell Survival upon Glutamine Deprivation. Cell Rep 23: 3006–3020. [DOI] [PubMed] [Google Scholar]
- 16.Baracco EE, Castoldi F, Durand S, Enot DP, Tadic J, et al. (2019) alpha-Ketoglutarate inhibits autophagy. Aging (Albany NY) 11: 3418–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaneko H, Kobayashi M, Mizunoe Y, Yoshida M, Yasukawa H, et al. (2018) Taurine is an amino acid with the ability to activate autophagy in adipocytes. Amino Acids 50: 527–535. [DOI] [PubMed] [Google Scholar]
- 18.Yanagisawa H, Ishii T, Endo K, Kawakami E, Nagao K, et al. (2017) L-leucine and SPNS1 coordinately ameliorate dysfunction of autophagy in mouse and human Niemann-Pick type C disease. Sci Rep 7: 15944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanchez-Martin P, Komatsu M (2020) Physiological Stress Response by Selective Autophagy. J Mol Biol 432: 53–62. [DOI] [PubMed] [Google Scholar]
- 20.Scherz-Shouval R, Elazar Z (2011) Regulation of autophagy by ROS: physiology and pathology. Trends Biochem Sci 36: 30–38. [DOI] [PubMed] [Google Scholar]
- 21.Filomeni G, Desideri E, Cardaci S, Rotilio G, Ciriolo MR (2010) Under the ROS...thiol network is the principal suspect for autophagy commitment. Autophagy 6: 999–1005. [DOI] [PubMed] [Google Scholar]
- 22.Scherz-Shouval R, Shvets E, Elazar Z (2007) Oxidation as a post-translational modification that regulates autophagy. Autophagy 3: 371–373. [DOI] [PubMed] [Google Scholar]
- 23.Carroll B, Otten EG, Manni D, Stefanatos R, Menzies FM, et al. (2018) Oxidation of SQSTM1/p62 mediates the link between redox state and protein homeostasis. Nat Commun 9: 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ichimura Y, Komatsu M (2018) Activation of p62/SQSTM1-Keap1-Nuclear Factor Erythroid 2-Related Factor 2 Pathway in Cancer. Front Oncol 8: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baird L, Dinkova-Kostova AT (2011) The cytoprotective role of the Keap1-Nrf2 pathway. Arch Toxicol 85: 241–272. [DOI] [PubMed] [Google Scholar]
- 26.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, et al. (2010) The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 12: 213–223. [DOI] [PubMed] [Google Scholar]
- 27.Copple IM, Lister A, Obeng AD, Kitteringham NR, Jenkins RE, et al. (2010) Physical and functional interaction of sequestosome 1 with Keap1 regulates the Keap1-Nrf2 cell defense pathway. J Biol Chem 285: 16782–16788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, et al. (2008) Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem 283: 10892–10903. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Yin X, Zhou C, Li J, Liu R, Shi B, et al. (2019) Autophagy in bone homeostasis and the onset of osteoporosis. Bone Res 7: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaber FA, Khan NM, Ansari MY, Al-Adlaan AA, Hussein NJ, et al. (2019) Autophagy plays an essential role in bone homeostasis. J Cell Physiol 234: 12105–12115. [DOI] [PubMed] [Google Scholar]
- 31.Nollet M, Santucci-Darmanin S, Breuil V, Al-Sahlanee R, Cros C, et al. (2014) Autophagy in osteoblasts is involved in mineralization and bone homeostasis. Autophagy 10: 1965–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piemontese M, Onal M, Xiong J, Han L, Thostenson JD, et al. (2016) Low bone mass and changes in the osteocyte network in mice lacking autophagy in the osteoblast lineage. Sci Rep 6: 24262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Onal M, Piemontese M, Xiong J, Wang Y, Han L, et al. (2013) Suppression of autophagy in osteocytes mimics skeletal aging. J Biol Chem 288: 17432–17440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cinque L, Forrester A, Bartolomeo R, Svelto M, Venditti R, et al. (2015) FGF signalling regulates bone growth through autophagy. Nature 528: 272–275. [DOI] [PubMed] [Google Scholar]
- 35.Kang X, Yang W, Feng D, Jin X, Ma Z, et al. (2017) Cartilage-Specific Autophagy Deficiency Promotes ER Stress and Impairs Chondrogenesis in PERK-ATF4-CHOP-Dependent Manner. J Bone Miner Res 32: 2128–2141. [DOI] [PubMed] [Google Scholar]
- 36.Vuppalapati KK, Bouderlique T, Newton PT, Kaminskyy VO, Wehtje H, et al. (2015) Targeted Deletion of Autophagy Genes Atg5 or Atg7 in the Chondrocytes Promotes Caspase-Dependent Cell Death and Leads to Mild Growth Retardation. J Bone Miner Res 30: 2249–2261. [DOI] [PubMed] [Google Scholar]
- 37.Horigome Y, Ida-Yonemochi H, Waguri S, Shibata S, Endo N, et al. (2020) Loss of autophagy in chondrocytes causes severe growth retardation. Autophagy 16: 501–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arai A, Kim S, Goldshteyn V, Kim T, Park NH, et al. (2019) Beclin1 Modulates Bone Homeostasis by Regulating Osteoclast and Chondrocyte Differentiation. J Bone Miner Res 34: 1753–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chung YH, Jang Y, Choi B, Song DH, Lee EJ, et al. (2014) Beclin-1 is required for RANKL-induced osteoclast differentiation. J Cell Physiol 229: 1963–1971. [DOI] [PubMed] [Google Scholar]
- 40.DeSelm CJ, Miller BC, Zou W, Beatty WL, van Meel E, et al. (2011) Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev Cell 21: 966–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Usategui-Martin R, Gestoso-Uzal N, Calero-Paniagua I, De Pereda JM, Del Pino-Montes J, et al. (2020) A mutation in p62 protein (p. R321C), associated to Paget’s disease of bone, causes a blockade of autophagy and an activation of NF-kB pathway. Bone 133: 115265. [DOI] [PubMed] [Google Scholar]
- 42.Guay-Belanger S, Picard S, Gagnon E, Morissette J, Siris ES, et al. (2015) Detection of SQSTM1/P392Lpost-zygotic mutations in Paget’s disease of bone. Hum Genet 134: 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wright T, Rea SL, Goode A, Bennett AJ, Ratajczak T, et al. (2013) The S349T mutation of SQSTM1 links Keap1/Nrf2 signalling to Paget’s disease of bone. Bone 52: 699–706. [DOI] [PubMed] [Google Scholar]
- 44.Klinck R, Laberge G, Bisson M, McManus S, Michou L, et al. (2014) Alternative splicing in osteoclasts and Paget’s disease of bone. BMC Med Genet 15: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zach F, Polzer F, Mueller A, Gessner A (2018) p62/sequestosome 1 deficiency accelerates osteoclastogenesis in vitro and leads to Paget’s disease-like bone phenotypes in mice. J Biol Chem 293: 9530–9541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daroszewska A, van ‘t Hof RJ, Rojas JA, Layfield R, Landao-Basonga E, et al. (2011) A point mutation in the ubiquitin-associated domain of SQSMT1 is sufficient to cause a Paget’s disease-like disorder in mice. Hum Mol Genet 20: 2734–2744. [DOI] [PubMed] [Google Scholar]
- 47.Rabanal-Ruiz Y, Otten EG, Korolchuk VI (2017) mTORC1 as the main gateway to autophagy. Essays Biochem 61: 565–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russell RC, Yuan HX, Guan KL (2014) Autophagy regulation by nutrient signaling. Cell Res 24: 42–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim J, Guan KL (2019) mTOR as a central hub of nutrient signalling and cell growth. Nat Cell Biol 21: 63–71. [DOI] [PubMed] [Google Scholar]
- 50.Shaw RJ (2008) mTOR signaling: RAG GTPases transmit the amino acid signal. Trends Biochem Sci 33: 565–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tokunaga C, Yoshino K, Yonezawa K (2004) mTOR integrates amino acid- and energy-sensing pathways. Biochem Biophys Res Commun 313: 443–446. [DOI] [PubMed] [Google Scholar]
- 52.Takahara T, Amemiya Y, Sugiyama R, Maki M, Shibata H (2020) Amino acid-dependent control of mTORC1 signaling: a variety of regulatory modes. J Biomed Sci 27: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Condon KJ, Sabatini DM (2019) Nutrient regulation of mTORC1 at a glance. J Cell Sci 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, et al. (2009) Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 136: 521–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Esen E, Chen J, Karner CM, Okunade AL, Patterson BW, et al. (2013) WNT-LRP5 signaling induces Warburg effect through mTORC2 activation during osteoblast differentiation. Cell Metab 17: 745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen J, Long F (2015) mTORC1 Signaling Promotes Osteoblast Differentiation from Preosteoblasts. PLoS One 10: e0130627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen J, Long F (2018) mTOR signaling in skeletal development and disease. Bone Res 6: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Riddle RC, Frey JL, Tomlinson RE, Ferron M, Li Y, et al. (2014) Tsc2 is a molecular checkpoint controlling osteoblast development and glucose homeostasis. Mol Cell Biol 34: 1850–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen J, Long F (2014) mTORC1 signaling controls mammalian skeletal growth through stimulation of protein synthesis. Development 141: 2848–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dai Q, Xu Z, Ma X, Niu N, Zhou S, et al. (2017) mTOR/Raptor signaling is critical for skeletogenesis in mice through the regulation of Runx2 expression. Cell Death Differ 24: 1886–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nojima H, Tokunaga C, Eguchi S, Oshiro N, Hidayat S, et al. (2003) The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J Biol Chem 278: 15461–15464. [DOI] [PubMed] [Google Scholar]
- 62.Fitter S, Matthews MP, Martin SK, Xie J, Ooi SS, et al. (2017) mTORC1 Plays an Important Role in Skeletal Development by Controlling Preosteoblast Differentiation. Mol Cell Biol 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martin SK, Fitter S, El Khawanky N, Grose RH, Walkley CR, et al. (2018) mTORC1 plays an important role in osteoblastic regulation of B-lymphopoiesis. Sci Rep 8: 14501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Q, Liu C, Yang Y, Yang H, Chen J (2018) Osteocyte-intrinsic mTORC1 signaling restrains trabecular bone accrual in mice. J Cell Biochem 119: 8743–8749. [DOI] [PubMed] [Google Scholar]
- 65.Chen J, Holguin N, Shi Y, Silva MJ, Long F (2015) mTORC2 signaling promotes skeletal growth and bone formation in mice. J Bone Miner Res 30: 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y, Xu S, Li K, Tan K, Liang K, et al. (2017) mTORC1 Inhibits NF-kappaB/NFATc1 Signaling and Prevents Osteoclast Precursor Differentiation, In Vitro and In Mice. J Bone Miner Res 32: 1829–1840. [DOI] [PubMed] [Google Scholar]
- 67.Hiraiwa M, Ozaki K, Yamada T, Iezaki T, Park G, et al. (2019) mTORC1 Activation in Osteoclasts Prevents Bone Loss in a Mouse Model of Osteoporosis. Front Pharmacol 10: 684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dai Q, Xie F, Han Y, Ma X, Zhou S, et al. (2017) Inactivation of Regulatory-associated Protein of mTOR (Raptor)/Mammalian Target of Rapamycin Complex 1 (mTORC1) Signaling in Osteoclasts Increases Bone Mass by Inhibiting Osteoclast Differentiation in Mice. J Biol Chem 292: 196–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu S, Zhang Y, Wang J, Li K, Tan K, et al. (2018) TSC1 regulates osteoclast podosome organization and bone resorption through mTORC1 and Rac1/Cdc42. Cell Death Differ 25: 1549–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu Y, Newman H, Shen L, Sharma D, Hu G, et al. (2019) Glutamine Metabolism Regulates Proliferation and Lineage Allocation in Skeletal Stem Cells. Cell Metab 29: 966–978 e964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Umehara T (2018) Nonalcoholic fatty liver disease with elevated alanine aminotransferase levels is negatively associated with bone mineral density: Cross-sectional study in U.S. adults. PLoS One 13: e0197900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Caplin B, Wang Z, Slaviero A, Tomlinson J, Dowsett L, et al. (2012) Alanine-glyoxylate aminotransferase-2 metabolizes endogenous methylarginines, regulates NO, and controls blood pressure. Arterioscler Thromb Vasc Biol 32: 2892–2900. [DOI] [PubMed] [Google Scholar]
- 73.Huh JE, Choi JY, Shin YO, Park DS, Kang JW, et al. (2014) Arginine enhances osteoblastogenesis and inhibits adipogenesis through the regulation of Wnt and NFATc signaling in human mesenchymal stem cells. Int J Mol Sci 15: 13010–13029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nakayama T, Al-Maawali A, El-Quessny M, Rajab A, Khalil S, et al. (2015) Mutations in PYCR2, Encoding Pyrroline-5-Carboxylate Reductase 2, Cause Microcephaly and Hypomyelination. Am J Hum Genet 96: 709–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ramesh V, Gusella JF, Shih VE (1991) Molecular pathology of gyrate atrophy of the choroid and retina due to ornithine aminotransferase deficiency. Mol Biol Med 8: 81–93. [PubMed] [Google Scholar]
- 76.Ginguay A, Cynober L, Curis E, Nicolis I (2017) Ornithine Aminotransferase, an Important Glutamate-Metabolizing Enzyme at the Crossroads of Multiple Metabolic Pathways. Biology (Basel) 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ruzzo EK, Capo-Chichi JM, Ben-Zeev B, Chitayat D, Mao H, et al. (2013) Deficiency of asparagines synthetase causes congenital microcephaly and a progressive form of encephalopathy. Neuron 80: 429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sacksteder KA, Biery BJ, Morrell JC, Goodman BK, Geisbrecht BV, et al. (2000) Identification of the alpha-aminoadipic semialdehyde synthase gene, which is defective in familial hyperlysinemia. Am J Hum Genet 66: 1736–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Posey JE, Burrage LC, Miller MJ, Liu P, Hardison MT, et al. (2014) Lysinuric Protein Intolerance Presenting with Multiple Fractures. Mol Genet Metab Rep 1: 176–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Borsani G, Bassi MT, Sperandeo MP, De Grandi A, Buoninconti A, et al. (1999) SLC7A7, encoding a putative permease-related protein, is mutated in patients with lysinuric protein intolerance. Nat Genet 21: 297–301. [DOI] [PubMed] [Google Scholar]
- 81.Sebastio G, Sperandeo MP, Andria G (2011) Lysinuric protein intolerance: reviewing concepts on a multisystem disease. Am J Med Genet C Semin Med Genet 157C: 54–62. [DOI] [PubMed] [Google Scholar]
- 82.Stroup BM, Marom R, Li X, Hsu CW, Chang CY, et al. (2020) A global Slc7a7 knockout mouse model demonstrates characteristic phenotypes of human lysinuric protein intolerance. Hum Mol Genet 29: 2171–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ueki I, Roman HB, Valli A, Fieselmann K, Lam J, et al. (2011) Knockout of the murine cysteine dioxygenase gene results in severe impairment in ability to synthesize taurine and an increased catabolism of cysteine to hydrogen sulfide. Am J Physiol Endocrinol Metab 301: E668–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nagahara N, Nagano M, Ito T, Shimamura K, Akimoto T, et al. (2013) Antioxidant enzyme, 3-mercaptopyruvate sulfurtransferase-knockout mice exhibit increased anxiety-like behaviors: a model for human mercaptolactate-cysteine disulfiduria. Sci Rep 3: 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Roman-Garcia P, Quiros-Gonzalez I, Mottram L, Lieben L, Sharan K, et al. (2014) Vitamin B(1)(2)-dependent taurine synthesis regulates growth and bone mass. J Clin Invest 124: 2988–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Choi MJ (2017) Taurine May Modulate Bone in Cholesterol Fed Estrogen Deficiency-Induced Rats. Adv Exp Med Biol 975 Pt 2: 1093–1102. [DOI] [PubMed] [Google Scholar]
- 87.Choi MJ, Chang KJ, Lee JW, Jung YJ (2017) Beneficial Function of Taurine on Bone Metabolism in Alcohol-Fed OVX Rat Model. Adv Exp Med Biol 975 Pt 2: 1059–1069. [DOI] [PubMed] [Google Scholar]
- 88.Moon PD, Kim MH, Lim HS, Oh HA, Nam SY, et al. (2015) Taurine, a major amino acid of oyster, enhances linear bone growth in a mouse model of protein malnutrition. Biofactors 41: 190–197. [DOI] [PubMed] [Google Scholar]
- 89.Choi MJ, Seo JN (2013) Effect of taurine feeding on bone mineral density and bone markers in rats. Adv Exp Med Biol 776: 51–58. [DOI] [PubMed] [Google Scholar]
- 90.Zhou C, Zhang X, Xu L, Wu T, Cui L, et al. (2014) Taurine promotes human mesenchymal stem cells to differentiate into osteoblast through the ERK pathway. Amino Acids 46: 1673–1680. [DOI] [PubMed] [Google Scholar]
- 91.Zafarullah M, Li WQ, Sylvester J, Ahmad M (2003) Molecular mechanisms of N-acetylcysteine actions. Cell Mol Life Sci 60: 6–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jiang Y, Cao Y, Wang Y, Li W, Liu X, et al. (2017) Cysteine transporter SLC3A1 promotes breast cancer tumorigenesis. Theranostics 7: 1036–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Raftos JE, Whillier S, Chapman BE, Kuchel PW (2007) Kinetics of uptake and deacetylation of N-acetylcysteine by human erythrocytes. Int J Biochem Cell Biol 39: 1698–1706. [DOI] [PubMed] [Google Scholar]
- 94.Yamada M, Watanabe J, Ueno T, Ogawa T, Egusa H (2019) Cytoprotective Preconditioning of Osteoblast-Like Cells with N-Acetyl-L-Cysteine for Bone Regeneration in Cell Therapy. Int J Mol Sci 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Watanabe J, Yamada M, Niibe K, Zhang M, Kondo T, et al. (2018) Preconditioning of bone marrow-derived mesenchymal stem cells with N-acetyl-L-cysteine enhances bone regeneration via reinforced resistance to oxidative stress. Biomaterials 185: 25–38. [DOI] [PubMed] [Google Scholar]
- 96.Soares MPR, Silva DP, Uehara IA, Ramos ES Jr., Alabarse PVG, et al. (2019) The use of apocynin inhibits osteoclastogenesis. Cell Biol Int 43: 466–475. [DOI] [PubMed] [Google Scholar]
- 97.Ma X, Su P, Yin C, Lin X, Wang X, et al. (2020) The Roles of FoxO Transcription Factors in Regulation of Bone Cells Function. Int J Mol Sci 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang Y, Xiong Y, Zhou J, Xin N, Zhu Z, et al. (2018) FoxO1 expression in osteoblasts modulates bone formation through resistance to oxidative stress in mice. Biochem Biophys Res Commun 503: 1401–1408. [DOI] [PubMed] [Google Scholar]
- 99.Hinoi E, Takarada T, Yoneda Y (2004) Glutamate signaling system in bone. J Pharmacol Sci 94: 215–220. [DOI] [PubMed] [Google Scholar]
- 100.Kalariti N, Koutsilieris M (2004) Glutamatergic system in bone physiology. In Vivo 18: 621–628. [PubMed] [Google Scholar]
- 101.Lin TH, Yang RS, Tang CH, Wu MY, Fu WM (2008) Regulation of the maturation of osteoblasts and osteoclastogenesis by glutamate. Eur J Pharmacol 589: 37–44. [DOI] [PubMed] [Google Scholar]
- 102.Skerry TM, Taylor AF (2001) Glutamate signalling in bone. Curr Pharm Des 7: 737–750. [DOI] [PubMed] [Google Scholar]
- 103.Seidlitz EP, Sharma MK, Singh G (2010) Extracellular glutamate alters mature osteoclast and osteoblast functions. Can J Physiol Pharmacol 88: 929–936. [DOI] [PubMed] [Google Scholar]
- 104.Chatron N, Becker F, Morsy H, Schmidts M, Hardies K, et al. (2020) Bi-allelic GAD1 variants cause a neonatal onset syndromic developmental and epileptic encephalopathy. Brain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Oh WJ, Westmoreland JJ, Summers R, Condie BG (2010) Cleft palate is caused by CNS dysfunction in Gad1 and Viaat knockout mice. PLoS One 5: e9758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Asada H, Kawamura Y, Maruyama K, Kume H, Ding RG, et al. (1997) Cleft palate and decreased brain gamma-aminobutyric acid in mice lacking the 67-kDa isoform of glutamic acid decarboxylase. Proc Natl Acad Sci U S A 94: 6496–6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen J, Tu X, Esen E, Joeng KS, Lin C, et al. (2014) WNT7B promotes bone formation in part through mTORC1. PLoS Genet 10: e1004145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Karner CM, Esen E, Okunade AL, Patterson BW, Long F (2015) Increased glutamine catabolism mediates bone anabolism in response to WNT signaling. J Clin Invest 125: 551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Indo Y, Takeshita S, Ishii KA, Hoshii T, Aburatani H, et al. (2013) Metabolic regulation of osteoclast differentiation and function. J Bone Miner Res 28: 2392–2399. [DOI] [PubMed] [Google Scholar]
- 110.Stegen S, Rinaldi G, Loopmans S, Stockmans I, Moermans K, et al. (2020) Glutamine Metabolism Controls Chondrocyte Identity and Function. Dev Cell 53: 530–544 e538. [DOI] [PubMed] [Google Scholar]
- 111.Olkku A, Mahonen A (2008) Wnt and steroid pathways control glutamate signalling by regulating glutamine synthetase activity in osteoblastic cells. Bone 43: 483–493. [DOI] [PubMed] [Google Scholar]
- 112.Polat O, Kilicoglu SS, Erdemli E (2007) A controlled trial of glutamine effects on bone healing. Adv Ther 24: 154–160. [DOI] [PubMed] [Google Scholar]
- 113.Huang T, Liu R, Fu X, Yao D, Yang M, et al. (2017) Aging Reduces an ERRalpha-Directed Mitochondrial Glutaminase Expression Suppressing Glutamine Anaplerosis and Osteogenic Differentiation of Mesenchymal Stem Cells. Stem Cells 35: 411–424. [DOI] [PubMed] [Google Scholar]
- 114.Fitzpatrick LA, Buzas E, Gagne TJ, Nagy A, Horvath C, et al. (2003) Targeted deletion of histidine decarboxylase gene in mice increases bone formation and protects against ovariectomy-induced bone loss. Proc Natl Acad Sci U S A 100: 6027–6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu YZ, Dvornyk V, Lu Y, Shen H, Lappe JM, et al. (2005) A novel pathophysiological mechanism for osteoporosis suggested by an in vivo gene expression study of circulating monocytes. J Biol Chem 280: 29011–29016. [DOI] [PubMed] [Google Scholar]
- 116.Back SH, Adapala NS, Barbe MF, Carpino NC, Tsygankov AY, et al. (2013) TULA-2, a novel histidine phosphatase, regulates bone remodeling by modulating osteoclast function. Cell Mol Life Sci 70: 1269–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wu JY, Kao HJ, Li SC, Stevens R, Hillman S, et al. (2004) ENU mutagenesis identifies mice with mitochondrial branched-chain aminotransferase deficiency resembling human maple syrup urine disease. J Clin Invest 113: 434–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.She P, Reid TM, Bronson SK, Vary TC, Hajnal A, et al. (2007) Disruption of BCATm in mice leads to increased energy expenditure associated with the activation of a futile protein turnover cycle. Cell Metab 6: 181–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Blackburn PR, Gass JM, Vairo FPE, Farnham KM, Atwal HK, et al. (2017) Maple syrup urine disease: mechanisms and management. Appl Clin Genet 10: 57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Guntur AR, Gerencser AA, Le PT, DeMambro VE, Bornstein SA, et al. (2018) Osteoblast-like MC3T3-E1 Cells Prefer Glycolysis for ATP Production but Adipocyte-like 3T3-L1 Cells Prefer Oxidative Phosphorylation. J Bone Miner Res 33: 1052–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.da Luz Dias R, Basso B, Donadio MVF, Pujol FV, Bartrons R, et al. (2018) Leucine reduces the proliferation of MC3T3-E1 cells through DNA damage and cell senescence. Toxicol In Vitro 48: 1–10. [DOI] [PubMed] [Google Scholar]
- 122.Watanabe M, Osada J, Aratani Y, Kluckman K, Reddick R, et al. (1995) Mice deficient in cystathionine beta-synthase: animal models for mild and severe homocyst(e)inemia. Proc Natl Acad Sci U S A 92: 1585–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Majtan T, Hulkova H, Park I, Krijt J, Kozich V, et al. (2017) Enzyme replacement prevents neonatal death, liver damage, and osteoporosis in murine homocystinuria. FASEB J 31: 5495–5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Robert K, Maurin N, Vayssettes C, Siauve N, Janel N (2005) Cystathionine beta synthase deficiency affects mouse endochondral ossification. Anat Rec A Discov Mol Cell Evol Biol 282: 1–7. [DOI] [PubMed] [Google Scholar]
- 125.Mudd SH, Skovby F, Levy HL, Pettigrew KD, Wilcken B, et al. (1985) The natural history of homocystinuria due to cystathionine beta-synthase deficiency. Am J Hum Genet 37: 1–31. [PMC free article] [PubMed] [Google Scholar]
- 126.Ishii I, Akahoshi N, Yamada H, Nakano S, Izumi T, et al. (2010) Cystathionine gamma-Lyase-deficient mice require dietary cysteine to protect against acute lethal myopathy and oxidative injury. J Biol Chem 285: 26358–26368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang J, Hegele RA (2003) Genomic basis of cystathioninuria (MIM 219500) revealed by multiple mutations in cystathionine gamma-lyase (CTH). Hum Genet 112: 404–408. [DOI] [PubMed] [Google Scholar]
- 128.Ouattara A, Cooke D, Gopalakrishnan R, Huang TH, Ables GP (2016) Methionine restriction alters bone morphology and affects osteoblast differentiation. Bone Rep 5: 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Plummer J, Park M, Perodin F, Horowitz MC, Hens JR (2017) Methionine-Restricted Diet Increases miRNAs That Can Target RUNX2 Expression and Alters Bone Structure in Young Mice. J Cell Biochem 118: 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang W, Jiao XH, Wang XP, Sun XY, Dong C (2016) MTR, MTRR, and MTHFR Gene Polymorphisms and Susceptibility to Nonsyndromic Cleft Lip With or Without Cleft Palate. Genet Test Mol Biomarkers 20: 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shedlovsky A, McDonald JD, Symula D, Dove WF (1993) Mouse models of human phenylketonuria. Genetics 134: 1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schwahn B, Mokov E, Scheidhauer K, Lettgen B, Schonau E (1998) Decreased trabecular bone mineral density in patients with phenylketonuria measured by peripheral quantitative computed tomography. Acta Paediatr 87: 61–63. [DOI] [PubMed] [Google Scholar]
- 133.Allen JR, Humphries IR, Waters DL, Roberts DC, Lipson AH, et al. (1994) Decreased bone mineral density in children with phenylketonuria. Am J Clin Nutr 59: 419–422. [DOI] [PubMed] [Google Scholar]
- 134.Modan-Moses D, Vered I, Schwartz G, Anikster Y, Abraham S, et al. (2007) Peak bone mass in patients with phenylketonuria. J Inherit Metab Dis 30: 202–208. [DOI] [PubMed] [Google Scholar]
- 135.Dobrowolski SF, Tourkova IL, Robinson LJ, Secunda C, Spridik K, et al. (2018) A bone mineralization defect in the Pah(enu2) model of classical phenylketonuria involves compromised mesenchymal stem cell differentiation. Mol Genet Metab 125: 193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhou QY, Quaife CJ, Palmiter RD (1995) Targeted disruption of the tyrosine hydroxylase gene reveals that catecholamines are required for mouse fetal development. Nature 374: 640–643. [DOI] [PubMed] [Google Scholar]
- 137.Szczypka MS, Rainey MA, Kim DS, Alaynick WA, Marck BT, et al. (1999) Feeding behavior in dopamine-deficient mice. Proc Natl Acad Sci U S A 96: 12138–12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Korner G, Noain D, Ying M, Hole M, Flydal MI, et al. (2015) Brain catecholamine depletion and motor impairment in a Th knock-in mouse with type B tyrosine hydroxylase deficiency. Brain 138: 2948–2963. [DOI] [PubMed] [Google Scholar]
- 139.Malochet-Guinamand S, Durif F, Thomas T (2015) Parkinson’s disease: A risk factor for osteoporosis. Joint Bone Spine 82: 406–410. [DOI] [PubMed] [Google Scholar]
- 140.Zhao Y, Shen L, Ji HF (2013) Osteoporosis risk and bone mineral density levels in patients with Parkinson’s disease: a meta-analysis. Bone 52: 498–505. [DOI] [PubMed] [Google Scholar]
- 141.Tseng PT, Chen YW, Yeh PY, Tu KY, Cheng YS, et al. (2015) Bone Mineral Density in Schizophrenia: An Update of Current Meta-Analysis and Literature Review Under Guideline of PRISMA. Medicine (Baltimore) 94: e1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Motyl KJ, Beauchemin M, Barlow D, Le PT, Nagano K, et al. (2017) A novel role for dopamine signaling in the pathogenesis of bone loss from the atypical antipsychotic drug risperidone in female mice. Bone 103: 168–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lee DJ, Tseng HC, Wong SW, Wang Z, Deng M, et al. (2015) Dopaminergic effects on in vitro osteogenesis. Bone Res 3: 15020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang CX, Ge XY, Wang MY, Ma T, Zhang Y, et al. (2020) Dopamine D1 receptor-mediated activation of the ERK signaling pathway is involved in the osteogenic differentiation of bone mesenchymal stem cells. Stem Cell Res Ther 11: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bliziotes M, McLoughlin S, Gunness M, Fumagalli F, Jones SR, et al. (2000) Bone histomorphometric and biomechanical abnormalities in mice homozygous for deletion of the dopamine transporter gene. Bone 26: 15–19. [DOI] [PubMed] [Google Scholar]
- 146.Bliziotes M, Gunness M, Eshleman A, Wiren K (2002) The role of dopamine and serotonin in regulating bone mass and strength: studies on dopamine and serotonin transporter null mice. J Musculoskelet Neuronal Interact 2: 291–295. [PubMed] [Google Scholar]
- 147.Culic V, Betz RC, Refke M, Fumic K, Pavelic J (2011) Tyrosinemia type II (Richner-Hanhart syndrome): a new mutation in the TAT gene. Eur J Med Genet 54: 205–208. [DOI] [PubMed] [Google Scholar]
- 148.Goldsmith LA (1978) Molecular biology and molecular pathology of a newly described molecular disease--tyrosinemia II (the Richner-Hanhart syndrome). Exp Cell Biol 46: 96–113. [DOI] [PubMed] [Google Scholar]
- 149.Holme E, Lindstedt S (1995) Diagnosis and management of tyrosinemia type I. Curr Opin Pediatr 7: 726–732. [DOI] [PubMed] [Google Scholar]
- 150.Chinsky JM, Singh R, Ficicioglu C, van Karnebeek CDM, Grompe M, et al. (2017) Diagnosis and treatment of tyrosinemia type I: a US and Canadian consensus group review and recommendations. Genet Med 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Grompe M, al-Dhalimy M, Finegold M, Ou CN, Burlingame T, et al. (1993) Loss of fumarylacetoacetate hydrolase is responsible for the neonatal hepatic dysfunction phenotype of lethal albino mice. Genes Dev 7: 2298–2307. [DOI] [PubMed] [Google Scholar]
- 152.Al-Dhalimy M, Overturf K, Finegold M, Grompe M (2002) Long-term therapy with NTBC and tyrosine-restricted diet in a murine model of hereditary tyrosinemia type I. Mol Genet Metab 75: 38–45. [DOI] [PubMed] [Google Scholar]
- 153.Stone TW, Stoy N, Darlington LG (2013) An expanding range of targets for kynurenine metabolites of tryptophan. Trends Pharmacol Sci 34: 136–143. [DOI] [PubMed] [Google Scholar]
- 154.Platten M, Nollen EAA, Rohrig UF, Fallarino F, Opitz CA (2019) Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat Rev Drug Discov 18: 379–401. [DOI] [PubMed] [Google Scholar]
- 155.Comai S, Bertazzo A, Brughera M, Crotti S (2020) Tryptophan in health and disease. Adv Clin Chem 95: 165–218. [DOI] [PubMed] [Google Scholar]
- 156.Al Saedi A, Sharma S, Summers MA, Nurgali K, Duque G (2020) The multiple faces of tryptophan in bone biology. Exp Gerontol 129: 110778. [DOI] [PubMed] [Google Scholar]
- 157.Michalowska M, Znorko B, Kaminski T, Oksztulska-Kolanek E, Pawlak D (2015) New insights into tryptophan and its metabolites in the regulation of bone metabolism. J Physiol Pharmacol 66: 779–791. [PubMed] [Google Scholar]
- 158.Vidal C, Li W, Santner-Nanan B, Lim CK, Guillemin GJ, et al. (2015) The kynurenine pathway of tryptophan degradation is activated during osteoblastogenesis. Stem Cells 33: 111–121. [DOI] [PubMed] [Google Scholar]
- 159.Refaey ME, McGee-Lawrence ME, Fulzele S, Kennedy EJ, Bollag WB, et al. (2017) Kynurenine, a Tryptophan Metabolite That Accumulates With Age, Induces Bone Loss. J Bone Miner Res 32: 2182–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ducy P, Karsenty G (2010) The two faces of serotonin in bone biology. J Cell Biol 191: 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yadav VK, Ryu JH, Suda N, Tanaka KF, Gingrich JA, et al. (2008) Lrp5 controls bone formation by inhibiting serotonin synthesis in the duodenum. Cell 135: 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Carbone LD, Buzkova P, Fink HA, Raiford M, Le B, et al. (2019) Association of Dietary Niacin Intake With Incident Hip Fracture, BMD, and Body Composition: The Cardiovascular Health Study. J Bone Miner Res 34: 643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hinoi E, Takarada T, Uno K, Inoue M, Murafuji Y, et al. (2007) Glutamate suppresses osteoclastogenesis through the cystine/glutamate antiporter. Am J Pathol 170: 1277–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Jin C, Zhang P, Zhang M, Zhang X, Lv L, et al. (2017) Inhibition of SLC7A11 by Sulfasalazine Enhances Osteogenic Differentiation of Mesenchymal Stem Cells by Modulating BMP2/4 Expression and Suppresses Bone Loss in Ovariectomized Mice. J Bone Miner Res 32: 508–521. [DOI] [PubMed] [Google Scholar]
- 165.Xie W, Dolder S, Siegrist M, Wetterwald A, Hofstetter W (2016) Glutamate Receptor Agonists and Glutamate Transporter Antagonists Regulate Differentiation of Osteoblast Lineage Cells. Calcif Tissue Int 99: 142–154. [DOI] [PubMed] [Google Scholar]
- 166.Huggett JF, Mustafa A, O’Neal L, Mason DJ (2002) The glutamate transporter GLAST-1 (EAAT-1) is expressed in the plasma membrane of osteocytes and is responsive to extracellular glutamate concentration. Biochem Soc Trans 30: 890–893. [DOI] [PubMed] [Google Scholar]
- 167.Mason DJ, Suva LJ, Genever PG, Patton AJ, Steuckle S, et al. (1997) Mechanically regulated expression of a neural glutamate transporter in bone: a role for excitatory amino acids as osteotropic agents? Bone 20: 199–205. [DOI] [PubMed] [Google Scholar]
- 168.Gray C, Marie H, Arora M, Tanaka K, Boyde A, et al. (2001) Glutamate does not play a major role in controlling bone growth. J Bone Miner Res 16: 742–749. [DOI] [PubMed] [Google Scholar]
- 169.Ozaki K, Yamada T, Horie T, Ishizaki A, Hiraiwa M, et al. (2019) The L-type amino acid transporter LAT1 inhibits osteoclastogenesis and maintains bone homeostasis through the mTORC1 pathway. Sci Signal 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Itzstein C, Cheynel H, Burt-Pichat B, Merle B, Espinosa L, et al. (2001) Molecular identification of NMDA glutamate receptors expressed in bone cells. J Cell Biochem 82: 134–144. [DOI] [PubMed] [Google Scholar]
- 171.Matta C, Juhasz T, Fodor J, Hajdu T, Katona E, et al. (2019) N-methyl-D-aspartate (NMDA) receptor expression and function is required for early chondrogenesis. Cell Commun Signal 17: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Grootjans J, Kaser A, Kaufman RJ, Blumberg RS (2016) The unfolded protein response in immunity and inflammation. Nat Rev Immunol 16: 469–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hu G, Yu Y, Tang YJ, Wu C, Long F, et al. (2020) The Amino Acid Sensor Eif2ak4/GCN2 Is Required for Proliferation of Osteoblast Progenitors in Mice. J Bone Miner Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, et al. (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11: 619–633. [DOI] [PubMed] [Google Scholar]
- 175.Zhang K, Wang M, Li Y, Li C, Tang S, et al. (2019) The PERK-EIF2alpha-ATF4 signaling branch regulates osteoblast differentiation and proliferation by PTH. Am J Physiol Endocrinol Metab 316: E590–E604. [DOI] [PubMed] [Google Scholar]
- 176.Saito A, Ochiai K, Kondo S, Tsumagari K, Murakami T, et al. (2011) Endoplasmic reticulum stress response mediated by the PERK-eIF2(alpha)-ATF4 pathway is involved in osteoblast differentiation induced by BMP2. J Biol Chem 286: 4809–4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Elefteriou F, Benson MD, Sowa H, Starbuck M, Liu X, et al. (2006) ATF4 mediation of NF1 functions in osteoblast reveals a nutritional basis for congenital skeletal dysplasiae. Cell Metab 4: 441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]