Abstract
PURPOSE
Nucleophosmin 1 (NPM1) mutations are associated with a favorable prognosis in acute myeloid leukemia (AML) when an internal tandem duplication (ITD) in the fms-related tyrosine kinase 3 gene (FLT3) is absent (FLT3-ITDneg) or present with a low allelic ratio (FLT3-ITDlow). The 2017 European LeukemiaNet guidelines assume this is true regardless of accompanying cytogenetic abnormalities. We investigated the validity of this assumption.
METHODS
We analyzed associations between karyotype and outcome in intensively treated patients with NPM1mut/FLT3-ITDneg/low AML who were prospectively enrolled in registry databases from nine international study groups or treatment centers.
RESULTS
Among 2,426 patients with NPM1mut/FLT3-ITDneg/low AML, 2,000 (82.4%) had a normal and 426 (17.6%) had an abnormal karyotype, including 329 patients (13.6%) with intermediate and 83 patients (3.4%) with adverse-risk chromosomal abnormalities. In patients with NPM1mut/FLT3-ITDneg/low AML, adverse cytogenetics were associated with lower complete remission rates (87.7%, 86.0%, and 66.3% for normal, aberrant intermediate, and adverse karyotype, respectively; P < .001), inferior 5-year overall (52.4%, 44.8%, 19.5%, respectively; P < .001) and event-free survival (40.6%, 36.0%, 18.1%, respectively; P < .001), and a higher 5-year cumulative incidence of relapse (43.6%, 44.2%, 51.9%, respectively; P = .0012). These associations remained in multivariable mixed-effects regression analyses adjusted for known clinicopathologic risk factors (P < .001 for all end points). In patients with adverse-risk chromosomal aberrations, we found no significant influence of the NPM1 mutational status on outcome.
CONCLUSION
Karyotype abnormalities are significantly associated with outcome in NPM1mut/FLT3-ITDneg/low AML. When adverse-risk cytogenetics are present, patients with NPM1mut share the same unfavorable prognosis as patients with NPM1 wild type and should be classified and treated accordingly. Thus, cytogenetic risk predominates over molecular risk in NPM1mut/FLT3-ITDneg/low AML.
INTRODUCTION
Mutations in the nucleophosmin 1 (NPM1) gene have been associated with a favorable prognosis in the absence of concomitant internal tandem duplications (ITD) of the fms-related tyrosine kinase 3 (FLT3) gene in cytogenetically normal acute myeloid leukemia (AML).1-6 It was later observed that patients with an NPM1 mutation (NPM1mut) and a low allelic ratio FLT3-ITD (FLT3-ITDlow) had similar outcomes to patients with an NPM1 mutation but no FLT3-ITD.7,8 Thus, both groups are now being classified as favorable risk in the 2017 update of the European LeukemiaNet recommendations on genetic risk classification (ELN 2017).9
Another key change in ELN 2017 has been to consider the NPM1mut/FLT3-ITDneg/low status favorable regardless of coexisting chromosomal abnormalities, as in the case of core-binding factor AML.9 However, the evidence supporting this modification is limited, and previous studies have so far only compared the impact of abnormal versus normal cytogenetics, without specific focus on the rare co-occurrence of NPM1 mutations and karyotype abnormalities that are classically associated with an adverse risk, such as deletions or monosomies of chromosomes 5 or 7, abnormalities of chromosome 17p, or complex or monosomal karyotypes, among others.10,11 Even though the combination of an NPM1 mutation with these abnormalities is rare, the prognostic effect of adverse cytogenetics in NPM1mut AML has important implications for postremission treatment decisions, in particular, the current recommendation that patients who are NPM1mut/FLT3-ITDneg/low not receive allogeneic hematopoietic stem cell transplantation (HSCT), given their presumed low risk of relapse might be altered if the adverse karyotype increased the risk. We evaluated the potential prognostic impact of karyotype in 2,426 intensively treated patients with NPM1mut/FLT3-ITDneg/low AML in a pooled analysis of individual patient data from nine study group registries or treatment centers worldwide.
METHODS
Patients
Individual patient data were collected from nine international AML study group registries or treatment centers: Study Alliance Leukemia, Programa Español para el Tratamiento de las Hemopatías Malignas, Acute Leukemia French Association, Toulouse-Bordeaux AML database, MD Anderson Cancer Center, Swedish AML Registry, Czech Leukemia Study Group for Life, Australasian Leukemia and Lymphoma Group, and Fred Hutchinson Cancer Research Center. Only intensively treated patients with AML 18 years of age or older with a known karyotype and who carried an NPM1 mutation in the absence of an FLT3-ITD mutation with a high (≥ 0.5)9 allelic ratio were included. From each cohort, data on intensively treated patients with NPM1 wild type (NPM1wt) and adverse cytogenetics as defined in the ELN 2017 guidelines were used as an adverse-risk reference cohort. For each patient, a predefined minimal data set was collected, including the variables age, sex, date of AML diagnosis, type of AML (de novo or secondary), bone marrow (BM) blast count, WBC count, karyotype, NPM1/FLT3-ITD mutational status including mutant/wild-type ratio, type of and response to induction chemotherapy, date of allogeneic HSCT in first complete remission (CR1), date of allogeneic HSCT beyond CR1, events (induction failure, relapse, death), and date of last contact. Patients with myelodysplastic syndromes or acute promyelocytic leukemia were excluded. This study was performed in accordance with the Declaration of Helsinki, all registries were approved by the local institutional review boards, and written informed consent was obtained from all patients through the participating centers.
Central Cytogenetic Review and Genetic Analyses
All karyotypes were centrally reviewed by a hematologist and a geneticist, L.A. and R.E., following International System for Human Cytogenomic Nomenclature 2016 guidelines.12 Cytogenetic abnormalities were classified as favorable, intermediate, or adverse as defined in ELN 2017.9 Among patients with FLT3-ITD-positive disease, only those with an allelic ratio of less than 0.5 (FLT3-ITDlow) were included.9
Statistical Analyses
Baseline variables were compared using χ2 or Fisher`s exact test for categorical and Mann-Whitney or Kruskal-Wallis test for continuous variables. Time-to-event variables and complete remission (CR) were defined as described in ELN 2017,9 with the exception that overall survival (OS) and event-free survival (EFS) were measured from initial AML diagnosis. Furthermore, because induction strategies and time points of response evaluation differed between cohorts, for reasons of comparability, response to induction was based on a patient’s status at day 90, counting induction failure as an event at day 90 for EFS.13 Survival probabilities were determined with the Kaplan-Meier and the Aalen-Johansen estimator and compared using log-rank and Gray’s test, respectively; all survival probabilities are given at 5 years. Follow-up time was calculated by the reverse Kaplan-Meier method. Multivariable mixed-effects Cox proportional hazard, cause-specific hazard, and logistic regression models were generated to assess statistical significance of prognostic factors with respect to OS, EFS, cumulative incidence of relapse (CIR), and CR. Cytogenetic risk was adjusted for age, type of AML (de novo v secondary), WBC count, and FLT3-ITD mutational status (present with low allelic ratio v absent). A registry center–specific random effects term was included in the multivariable models to account for potential heterogeneity in baseline risk among centers. The proportional hazards assumption was verified for each variable individually by inspection of scaled Schoenfeld residuals. The consistency of association with outcomes across subgroups was examined with Cox proportional hazard models and test for interaction. To evaluate associations of allogeneic HSCT as postremission therapy with prognosis, time-to-event variables were measured from CR1, and allogeneic HSCT was introduced as a time-dependent covariable. The time-dependent association was plotted according to the method of Simon and Makuch.14 Missing data were not imputed. Two-sided P values < .05 were considered significant. All analyses were performed using R (www.r-project.org), version 3.5.1.
RESULTS
Association With Clinical and Molecular Features
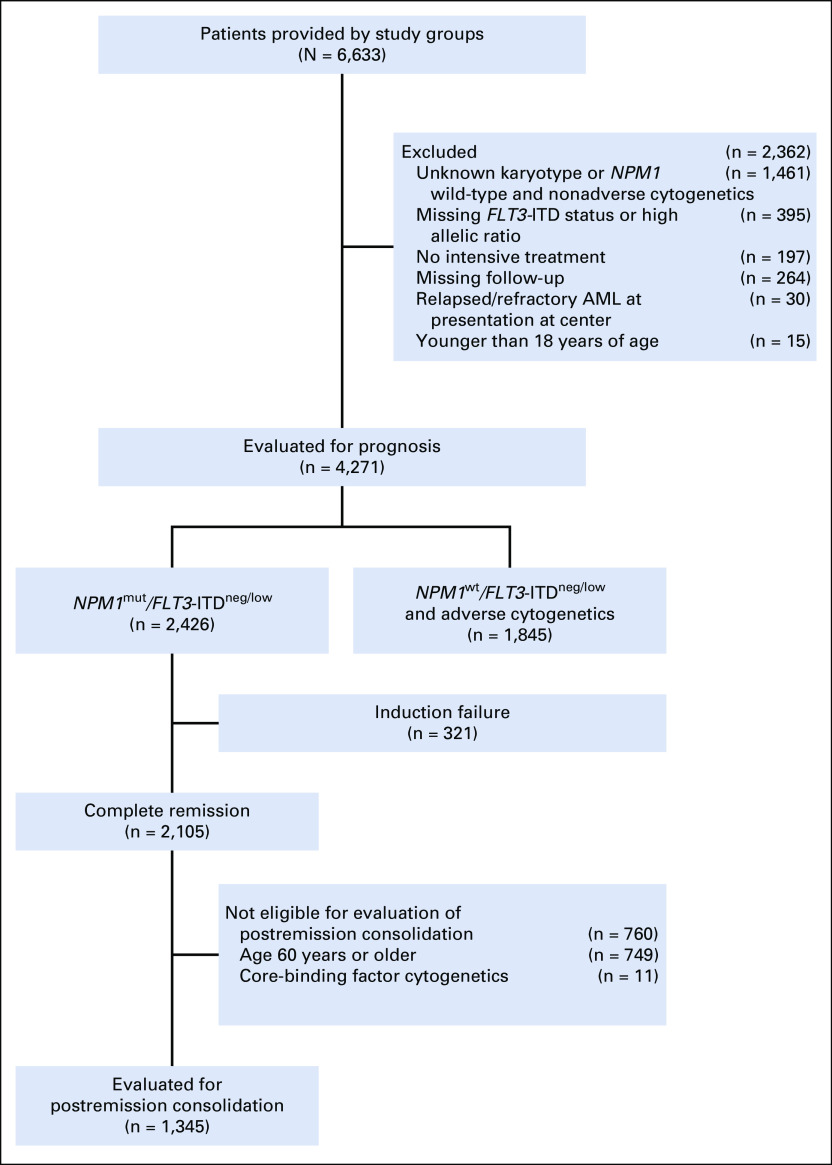
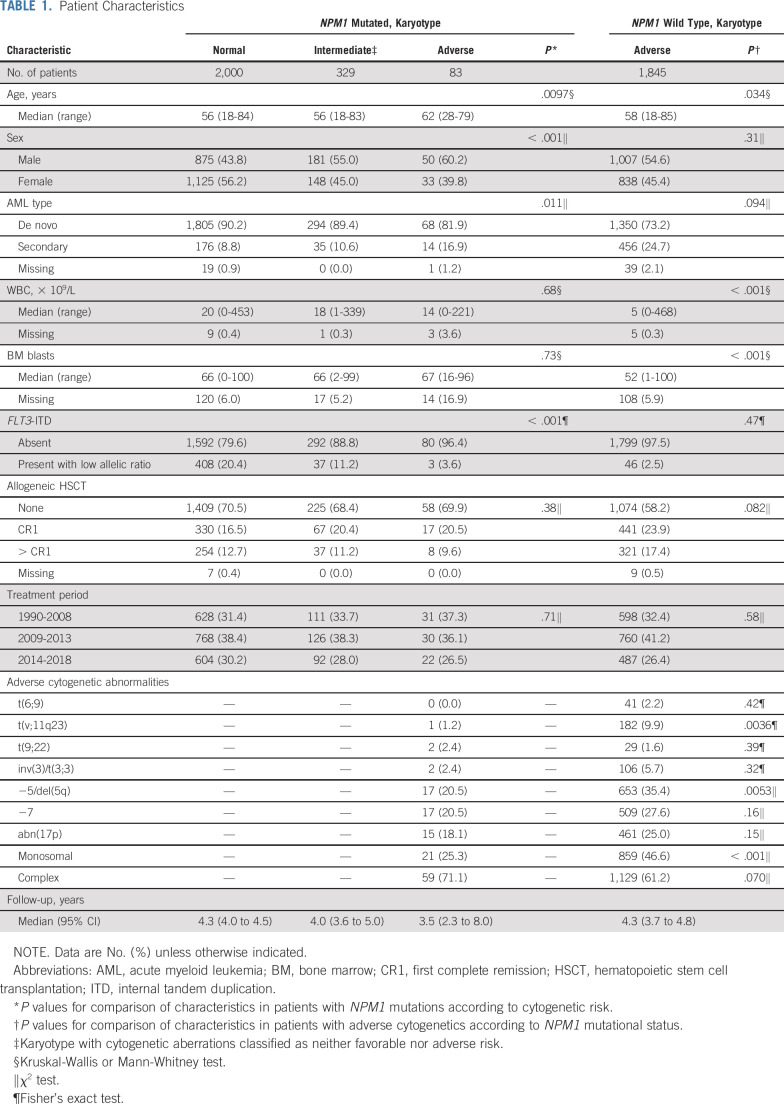
A total of 2,426 patients with NPM1mut/FLT3-ITDneg/low AML were identified (Fig 1). Of these, 2,000 (82.4%) had a normal karyotype and 426 (17.6%) had an abnormal karyotype, including 329 patients (13.6%) with chromosomal abnormalities of intermediate risk (ie, with aberrations not classified as adverse or favorable according to ELN 2017), 83 (3.4%) with chromosomal abnormalities of adverse risk, and 14 (0.6%) with favorable core-binding factor cytogenetics. A total of 1,845 patients with NPM1wt/FLT3-ITDneg/low and adverse-risk cytogenetics were identified. Baseline characteristics are listed in Table 1. In patients with NPM1mut/FLT3-ITDneg/low AML, the presence of adverse-risk cytogenetics was associated with older age (P = .0097), male sex (P < .001), secondary AML (P = .032), and negative FLT3-ITD status (P < .001). There were no significant differences in rates of allogeneic HSCT between cytogenetic risk groups in patients with NPM1mut/FLT3-ITDneg/low (P = .40). Compared with patients with NPM1wt/FLT3-ITDneg/low with adverse cytogenetics, patients with NPM1mut/FLT3-ITDneg/low with adverse cytogenetics were older, had higher BM blast counts, and had higher WBC counts. Rearrangements of 11q23, -5/del(5q), and a monosomal karyotype were under-represented in patients with NPM1mut/FLT3-ITDneg/low with adverse-risk karyotype compared with the NPM1wt reference group. Rates of allogeneic HSCT were higher in patients with adverse-risk cytogenetics with NPM1wt than in those with NPM1mut (41.5% v 30.1%; P = .082).
FIG 1.
Study profile. AML, acute myeloid leukemia.
TABLE 1.
Patient Characteristics
Karyotype and Outcome in NPM1mut/FLT3-ITDneg/low AML
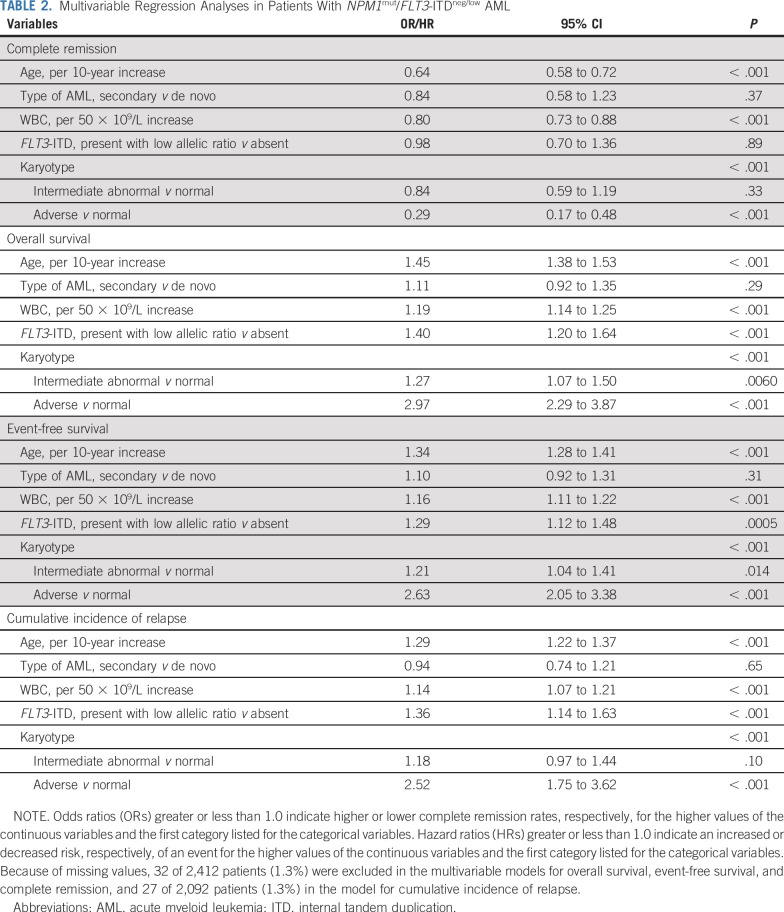
The 83 patients with NPM1mut/FLT3-ITDneg/low AML and adverse cytogenetics had a lower CR rate (66.3%) than patients with NPM1mut/FLT3-ITDneg/low AML and a normal karyotype (87.7%) or intermediate-risk cytogenetic abnormalities (86.0%; P < .001), but a similar CR rate to patients with NPM1wt/FLT3-ITDneg/low carrying adverse cytogenetic abnormalities (57.5%; P = .11). Median follow-up time for all patients was 4.23 years (95% CI, 4.05 to 4.46 years). Among patients with NPM1mut/FLT3-ITDneg/low, 5-year EFS rates were lower in the presence of adverse cytogenetics (18.1%) than with aberrant intermediate (36.0%) or normal karyotypes (40.6%; P < .001; Fig 2). The same applied considering OS: 19.5% for adverse, 44.8% for intermediate, and 52.4% for normal cytogenetics (P < .001; Fig 2). Likewise, CIR was 43.6%, 44.2%, and 51.9%, respectively (P = .0012; Fig 2). Median EFS was 0.43, 1.45, and 2.13 years, and median OS was 0.63, 2.99, and 6.62 years for adverse, intermediate, and normal cytogenetics, respectively. When adjusting for age, AML type, WBC, and FLT3-ITD status in multivariable analyses (Table 2), patients with NPM1mut/FLT3-ITDneg/low with adverse cytogenetics had a substantially reduced likelihood of achieving a CR (odds ratio [OR], 0.29; 95% CI, 0.17 to 0.48; P < .001) and an increased risk of death (hazard ratio [HR], 2.97; 95% CI, 2.29 to 3.87; P < .001) and experiencing an event (HR, 2.63; 95% CI, 2.05 to 3.38; P < .001) or relapse (HR, 2.52; 95% CI, 1.75 to 3.62; P < .001) compared with patients with a normal karyotype. Most importantly, we found no significant hazard risk of death (P = .77), experiencing an event (P = .68), or relapse (P = .53) when comparing patients with NPM1wt/FLT3-ITDneg/low and NPM1mut/FLT3-ITDneg/low with adverse-risk cytogenetic abnormalities. When comparing intermediate and normal cytogenetics in patients with NPM1mut/FLT3-ITDneg/low, intermediate cytogenetics remained associated with inferior OS (HR, 1.27; 95% CI, 1.07 to 1.50; P = .0060) and EFS (HR, 1.21; 95% CI, 1.04 to 1.41; P = .014), but not with a higher CIR (HR, 1.18; 95% CI, 0.97 to 1.44; P = .10) after adjustment for age, AML type, WBC, and FLT3-ITD status. We found no significant difference in outcome between normal karyotype and favorable cytogenetic abnormalities among patients with NPM1mut/FLT3-ITDneg/low genotype (Data Supplement).
FIG 2.
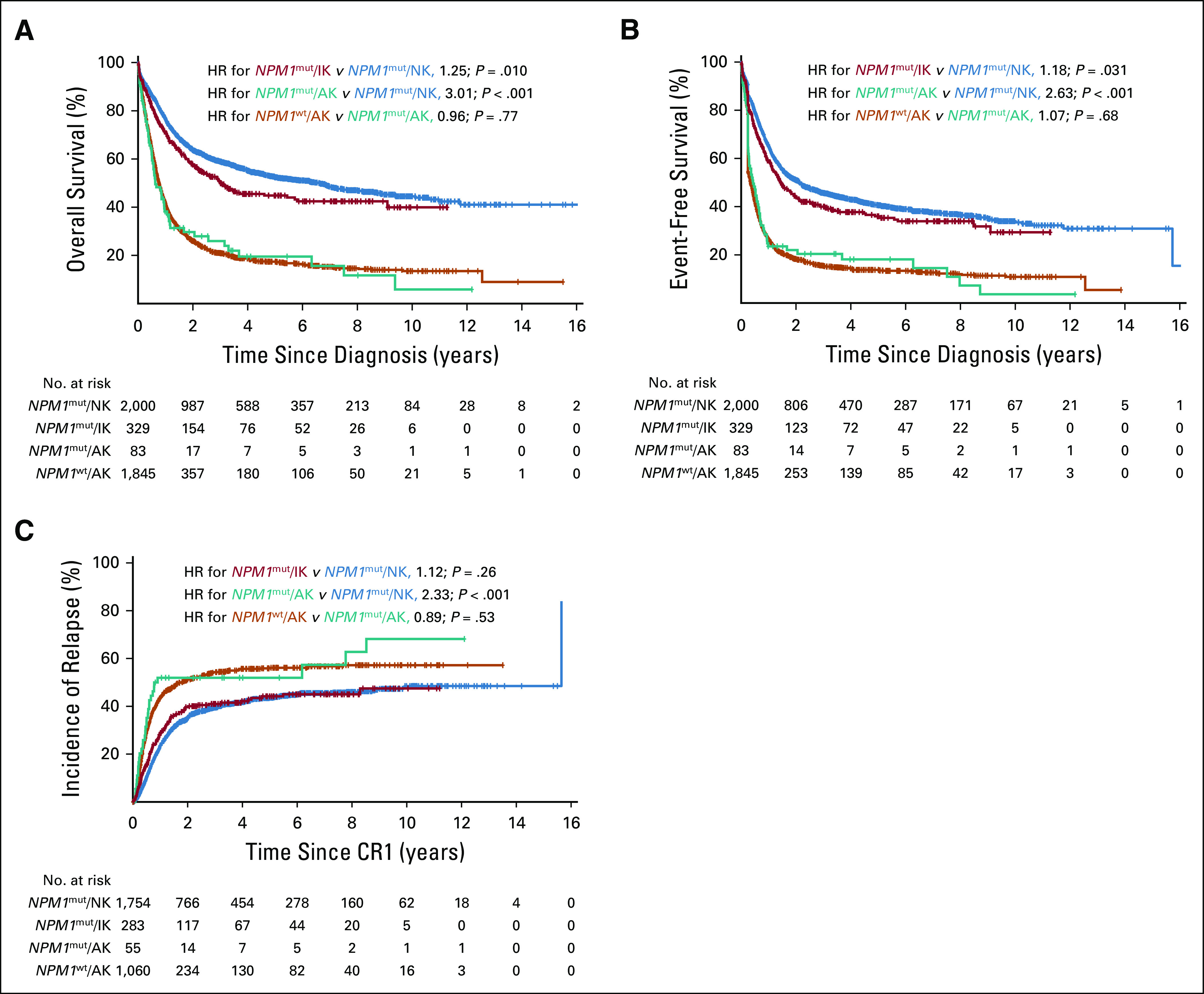
Cytogenetic abnormalities and survival in patients with NPM1mut/FLT3- ITDneg/low acute myeloid leukemia from nine international cohorts. (A) Overall survival, (B) event-free survival, and (C) cumulative incidence of relapse, according to cytogenetic risk. NPM1wt/FLT3-ITDneg/low patients with adverse-risk cytogenetics from the same cohorts served as the adverse-risk comparator arm. AK, adverse karyotype; CR1, first complete remission; HR, hazard ratio; IK, intermediate abnormal karyotype; NK, normal karyotype; wt, wild type.
TABLE 2.
Multivariable Regression Analyses in Patients With NPM1mut/FLT3-ITDneg/low AML
Recently, the categorization of NPM1mut AML with an FLT3-ITDlow mutation as favorable risk has been questioned.15 Thus, we reanalyzed the data excluding the 448 patients with an FLT3-ITDlow mutation. Again, adverse cytogenetic abnormalities remained associated with an unfavorable outcome in patients with NPM1mut/FLT3-ITDneg disease after adjustment for age, AML type, and WBC (HR for death, 2.94; 95% CI, 2.25 to 3.84; HR for event, 2.62; 95% CI, 2.03 to 3.38; HR for relapse, 2.45; 95% CI, 1.68 to 3.57; all P < .001; Data Supplement). Of note, the FLT3-ITDlow genotype was virtually absent in the adverse karyotype group regardless of NPM1 mutational status (Table 1).
Cytogenetics and Prognosis by Subgroup
We found a significant interaction of age with OS according to karyotype abnormalities in patients with NPM1mut/FLT3-ITDneg/low disease (Pinteraction = .0096; Fig 3) such that the risk of death associated with adverse cytogenetics was 3.97 in younger patients (< 60 years) but only 1.95 in older patients (≥ 60 years) with NPM1mut/FLT3-ITDneg/low AML. Similar results were obtained for EFS (HR, 3.07 and 1.88, respectively; Pinteraction = .086; Data Supplement) and CIR (HR, 3.59 and 1.25, respectively; Pinteraction = .0046; Data Supplement). Younger patients with adverse cytogenetics had a two-thirds reduced likelihood of achieving a CR (OR, 0.29; 95% CI, 0.12 to 0.71; P = .0062) and an increased risk of death (HR, 4.71; 95% CI, 3.11 to 7.14; P < .001), experiencing an event (HR, 3.56; 95% CI, 2.38 to 5.31; P < .001) or relapse (HR, 4.62; 95% CI, 2.81 to 7.60; P < .001) after adjustment for age, AML type, WBC, and FLT3-ITD status compared with patients with normal cytogenetics (Data Supplement). Also in older patients with NPM1mut/FLT3-ITDneg/low, adverse cytogenetics remained associated with inferior CR (OR, 0.28; 95% CI, 0.15 to 0.52; P < .001), OS (HR, 2.16; 95% CI, 1.53 to 3.05; P < .001), and EFS (HR, 2.07; 95% CI, 1.50 to 2.87; P < .001), but not with CIR (HR, 1.43; 95% CI, 0.83 to 2.46; P = .20) in multivariable analyses (Data Supplement). We found no significant heterogeneity of associations between cytogenetic risk and OS by sex, type of AML, FLT3-ITD status, WBC, BM blast counts, treatment period, or cohort (Fig 3).
FIG 3.
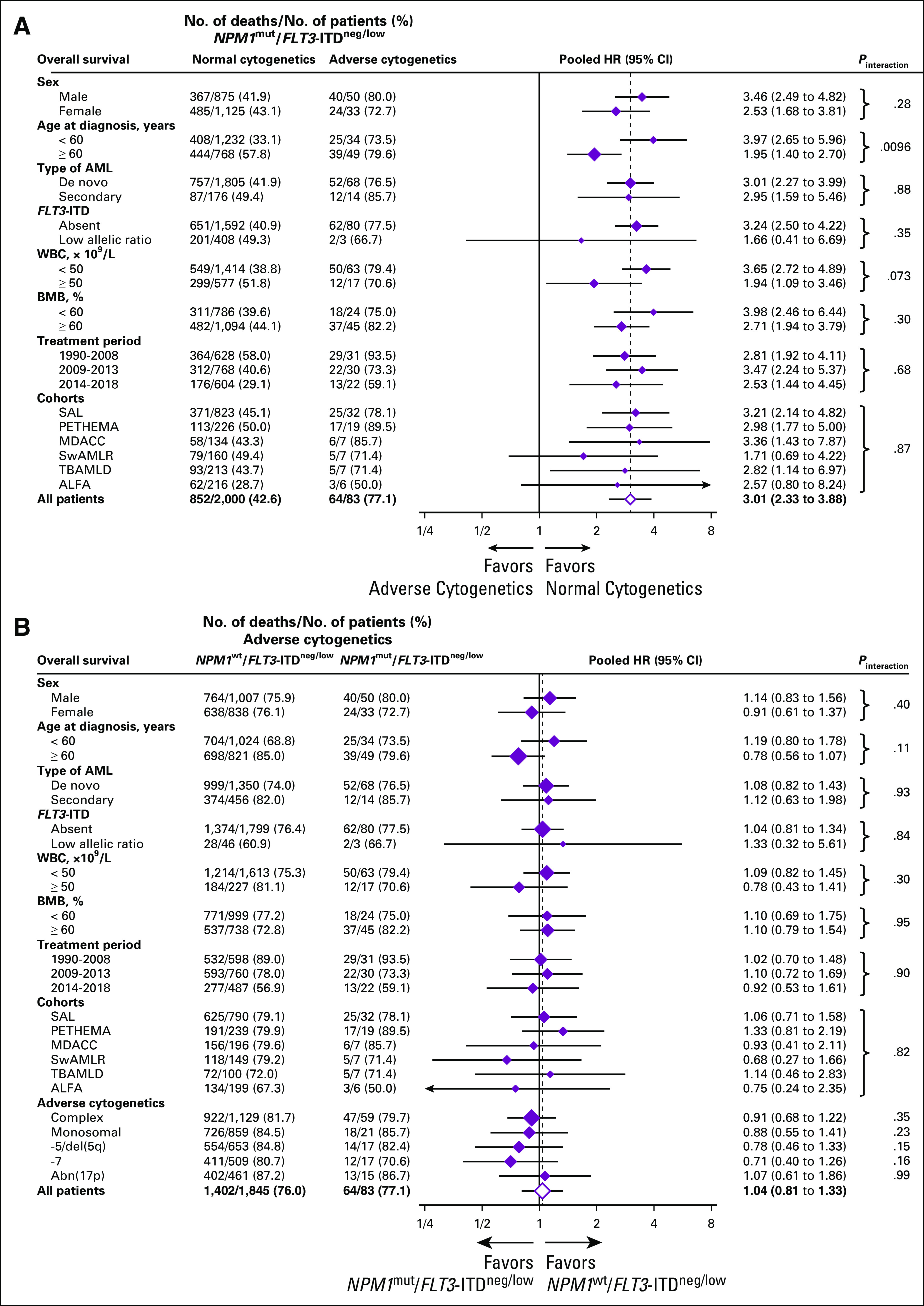
Overall survival according to cytogenetics and NPM1 mutational status by selected baseline categories. (A) Normal versus adverse cytogenetics in NPM1mut/FLT3- ITDneg/low acute myeloid leukemia (AML). (B) NPM1mut/FLT3-ITDneg/low versus NPM1wt/FLT3-ITDneg/low status in patients with AML with adverse-risk cytogenetics. Diamonds represent the pooled unadjusted hazard ratios (HRs). Horizontal lines represent the 95% CIs. The dotted vertical line represents the HR from the complete cohort. The P values are for interaction of unadjusted HRs by subgroups and represent heterogeneity. For analysis of heterogeneity among cohorts, only those with more than five patients with NPM1mut/FLT3-ITDneg/low/adverse karyotype were included. ALFA, Acute Leukemia French Association; BMB, bone marrow blasts; ITD, internal tandem duplication; MDACC, MD Anderson Cancer Center; PETHEMA, Programa Español para el Tratamiento de las Hematopatías Malignas; SAL, Study Alliance Leukemia; SwAMLR, Swedish AML Registry; TBAMLD, Toulouse-Bordeaux AML database; wt, wild type.
Individual Adverse-Risk Cytogenetic Abnormalities
There were no obvious differences in the association of individual adverse abnormalities with risk of death when comparing patients with NPM1wt/FLT3-ITDneg/low with patients with NPM1mut/FLT3-ITDneg/low in the five largest adverse-risk cytogenetic subgroups, but the number of deaths was small [-5/del(5q): 14; -7: 12; abn(17p): 13; complex karyotype: 47; monosomal karyotype: 18; Fig 3; Data Supplement].
Allogeneic HSCT in CR1 in Patients With NPM1mut/FLT3-ITDneg/low
To evaluate the association of postremission allogeneic HSCT with survival, time-to-event variables were measured from CR1. Because of the low frequency of allogeneic HSCT in CR1 in patients 60 years of age or older (9.2%), only younger patients (< 60 years) were included in these analyses. Of 1,466 younger patients with NPM1mut/FLT3-ITDneg/low, 1,345 (91.7%) achieved a CR and 345 of these (25.7%) subsequently received allogeneic HSCT. Baseline characteristics of patients in CR1 stratified by postremission therapy are shown in the Data Supplement. Patients undergoing transplantation in CR1 more frequently had secondary AML (P = .0079) or an FLT3-ITD mutation (P < .001). Survival for all patients with NPM1mut/FLT3-ITDneg/low according to postremission therapy is shown in Figures 4A and 4B. Patients who underwent transplantation had a lower CIR (22.6% v 41.4%; P < .001) but a higher nonrelapse mortality (NRM, 18.3% v 7.4%; P < .001) compared with patients who did not undergo transplantation in CR1. There were no differences in OS between patients who underwent transplantation and those who did not (65.6% v 67.2%; P = .34).
FIG 4.
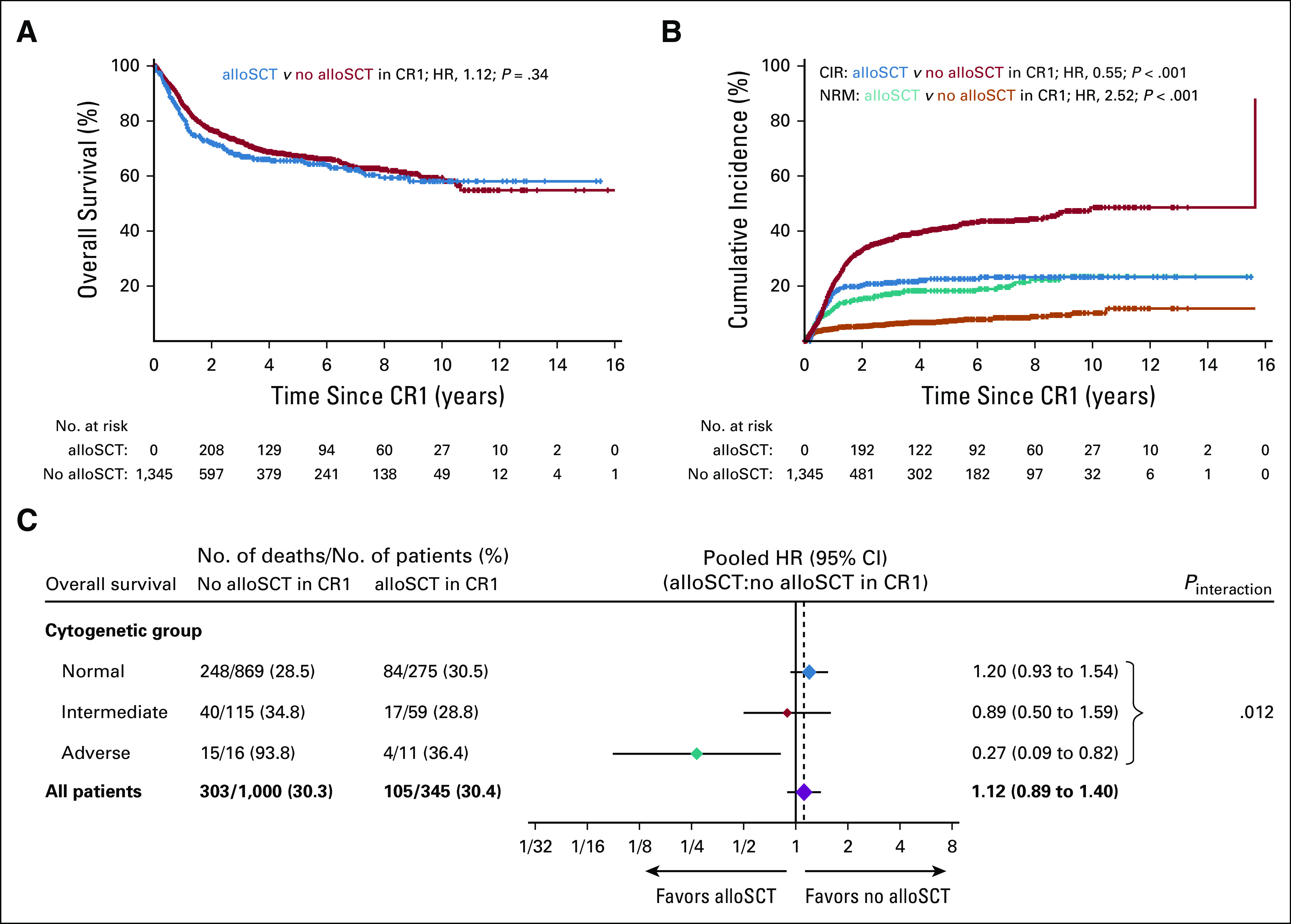
Survival by transplantation status in NPM1mut/FLT3ITDneg/low acute myeloid leukemia. (A) Overall survival and (B) cumulative incidence of relapse (CIR) and nonrelapse mortality (NRM) according to postremission therapy in younger patients in first complete remission (CR1). (C) Overall survival according to allogeneic hematopoietic stem cell transplantation (alloSCT) status in subgroups by cytogenetic risk. Diamonds represent the pooled unadjusted hazard ratios (HRs). Horizontal lines represent the 95% CIs. The dotted vertical line represents the HR from the complete cohort. The P value for interaction is from the χ2 test comparing the interaction HRs across subgroups and represents heterogeneity. AlloSCT was included as a time-dependent covariable.
However, among patients with NPM1mut/FLT3-ITDneg/low with adverse cytogenetics, those who received allogeneic HSCT in CR1 had a four-fold lower risk of death than patients who did not receive allogeneic HSCT in CR1 (HR, 0.27; 95% CI, 0.09 to 0.82; Pinteraction = .012; Fig 4C). We found no significant interaction of cytogenetics and postremission treatment (transplantation v no transplantation) with CIR (Pinteraction = .41) or NRM (Pinteraction = .53). Among patients with adverse cytogenetics, the association of allogeneic HSCT in CR1 with better OS was more pronounced in patients with NPM1mut (HR for death, 0.27; 95% CI, 0.09 to 0.82) than in patients with NPM1wt (HR for death, 0.71; 95% CI, 0.58 to 0.87; Pinteraction = .073).
DISCUSSION
Pretreatment cytogenetic and molecular abnormalities are routinely used to assess risk of relapse in AML and thus to guide treatment strategies in AML, in particular, use of allogeneic HSCT in CR1.9 In the absence of FLT3-ITD with a high allelic ratio, NPM1 mutations have been considered to confer a favorable prognosis regardless of concomitant cytogenetic abnormalities, most influentially in the most recent ELN 2017 genetic risk classification.9 This view was based on two reports investigating the prognostic effects of an abnormal karyotype in patients with NPM1mut/FLT3-ITDneg AML. In the larger cohort (n = 355, derived from two independent cohorts),10 no significant impact of cytogenetic abnormalities on survival for NPM1mut/FLT3-ITDneg AML was found, whereas in the smaller cohort (n = 95),11 a significantly inferior EFS, but not OS, was reported for patients with abnormal cytogenetics. In both studies, however, the individual cytogenetic abnormalities were not reported for patients with the NPM1mut/FLT3-ITDneg genotype, and no distinction was made between chromosomal abnormalities of intermediate and adverse risk, owing to the rarity of the latter in NPM1mut AML.
In contrast, in this study, we were able to distinguish between intermediate and adverse cytogenetics consequent to the inclusion of 2,426 patients with NPM1mut/FLT3-ITDneg/low AML from sites in Europe, Australia, and the United States. As expected, the majority of these patients had a normal karyotype, whereas cytogenetic abnormalities were found in 17.6% of patients, including 329 patients with intermediate and 83 with adverse-risk aberrations. However, it has to be noted that our analysis was performed retrospectively in patients who were, in part, not originally classified according to ELN 2017 guidelines. Thus, NPM1/FLT3 genetic analyses might have been underperformed in patients with an aberrant karyotype, and patients with an abnormal karyotype might have been under-represented in our study.
We found that concomitant chromosomal abnormalities were significantly associated with prognosis in patients with NPM1mut/FLT3-ITDneg/low AML. Whereas the difference in 5-year OS and EFS between normal and intermediate aberrant karyotypes was modest (OS, 52.4% and 44.8%, respectively; EFS, 40.6% and 36.0%, respectively), patients with NPM1mut/FLT3-ITDneg/low with adverse chromosomal aberrations had a considerably poorer prognosis (OS, 19.5%; EFS, 18.1%). In fact, patients carrying adverse-risk cytogenetics shared a virtually identical unfavorable outcome, regardless of whether the otherwise beneficial NPM1mut/FLT3-ITDneg/low status was present or not. The type of the adverse chromosomal abnormality did not seem to influence this effect, although low numbers might obscure detection of heterogeneity among individual aberrations. Furthermore, we found similar results when repeating the analyses excluding patients with an FLT3-ITDlow mutation. Thus, it is unlikely that varying assays for FLT3-ITD mutational status and allelic ratio significantly affected our analyses. Given that only three patients with NPM1mut AML and adverse cytogenetics also had an FLT3-ITDlow mutation, the impact of adverse cytogenetics in this particular genotype remains unclear.
Chromosome aberrations in NPM1mut AML are considered secondary genetic events,16 and the NPM1 mutation is stable during the course of the disease.16 In the latest update of WHO classification of myeloid neoplasms, AML with mutated NPM1 has become a distinct entity.17 Our data, however, suggest that adverse karyotype abnormalities seem to predominate over the favorable biologic impact of the NPM1 founder mutation and dictate the course of the disease. However, because of the methodology of detection without use of subclone analysis, we were unable to dissect whether NPM1 mutations and concurrent genetic abnormalities occurred in the same or in different leukemic cell populations. We also did not have access to relapse samples, lacking the possibility to test whether the NPM1 mutation, the cytogenetic abnormalities, or both are retained during the course of the disease.
Allogeneic HSCT is widely accepted as the best postremission treatment of transplant-eligible patients with adverse cytogenetics.18 The results of our analysis support previous data from smaller cohorts that allogeneic HSCT in CR1 is also associated with a reduction in risk of relapse in the total cohort of patients with NPM1mut/FLT3-ITDneg/low, including those with normal cytogenetics.19 However, the reduced risk of relapse did not translate into a superior OS. We found no excessive NRM (18.3%) in our cohort. It is likely that effective salvage treatments in relapsing patients, who did not receive allogeneic HSCT as primary postremission consolidation, offset the potential beneficial effect of transplantation in CR1. Minimal residual disease–guided treatment decisions might further reduce the effect of primary allogeneic HSCT in patients with NPM1mut/FLT3-ITDneg/low, especially those with intermediate-risk cytogenetics.20,21 Given the retrospective nature of our study, these results demand additional validation within prospective trials. In contrast, in patients with NPM1mut/FLT3-ITDneg/low with adverse cytogenetics, allogeneic HSCT in CR1 was associated with a significantly improved survival compared with consolidation chemotherapy, further emphasizing that these patients should be classified as high risk and managed as such.
In summary, this international collaborative study clearly shows that cytogenetic abnormalities are important determinants of outcome in NPM1mut/FLT3-ITDneg/low AML. Most importantly, patients with NPM1 mutations with the FLT3-ITDneg/low genotype and adverse-risk cytogenetics share the same unfavorable prognosis as their counterparts with NPM1wt and should be classified accordingly.
ACKNOWLEDGMENT
We thank all participating patients, hospitals, and investigators contributing data to the Acute Leukemia French Association, Australasian Leukemia and Lymphoma Group, Czech Leukemia Study Group for Life, Fred Hutchinson Cancer Research Center, MD Anderson Cancer Center, Programa Español para el Tratamiento de las Hemopatías Malignas, Study Alliance Leukemia, Swedish AML Registry, and Toulouse-Bordeaux AML database trials or registries.
L.A. is supported by the Innovative Medical Research Fund of the University of Münster Medical School (AN111813). G.L. and W.E.B. are supported by the German Research Foundation (DFG EXC 1003, Cluster of Excellence Cells in Motion). C. Schliemann is supported by the Innovative Medical Research Fund of the University of Münster Medical School (SC211008). The Toulouse-Bordeaux acute myeloid leukemia database was supported by grants from the French government (ANR-11-PHUC-001) and by the Groupement Interrégional de Recherche Clinique et d’Innovation-Sud Ouest Outre Mer (APITHEM 2014). Z.R. and P.Z. are supported by Ministry of Health of the Czech Republic (Grant No. 15-25809A). A.H.W. is supported by the National Health and Medical Research Council of Australia, Medical Research Future Fund, and the Victorian Cancer Agency.
U.K. and C. Schliemann share senior authorship.
AUTHOR CONTRIBUTIONS
Conception and design: Linus Angenendt, Christoph Röllig, Elihu Estey, Jan-Henrik Mikesch, Gerhard Ehninger, Wolfgang E. Berdel, Utz Krug, Christoph Schliemann
Administrative support: Christoph Röllig
Provision of study materials or patients: Linus Angenendt, Christoph Röllig, Pau Montesinos, David Martínez-Cuadrón, Eva Barragan, Pilar Martínez, Farhad Ravandi, Tapan Kadia, Jorge Cortes, Gunnar Juliusson, Vladimir Lazarevic, Christian Recher, Arnaud Pigneux, Pierre-Yves Dumas, Hervé Dombret, Claude Preudhomme, Andrew H. Wei, Jan-Henrik Mikesch, Hubert Serve, Wolfgang E. Berdel
Collection and assembly of data: Linus Angenendt, Christoph Röllig, Pau Montesinos, David Martínez-Cuadrón, Eva Barragan, Raimundo García, Carmen Botella, Pilar Martínez, Farhad Ravandi, Tapan Kadia, Jorge Cortes, Gunnar Juliusson, Sören Lehmann, Christian Recher, Sarah Bertoli, Pierre-Yves Dumas, Hervé Dombret, Claude Preudhomme, Jean-Baptiste Micol, Christine Terré, Zdeněk Ráčil, Jan Novák, Pavel Žák, Andrew H. Wei, Ing S. Tiong, Meaghan Wall, Elihu Estey, Carole Shaw, Rita Exeler, Lisa Wagenführ, Friedrich Stölzel, Christian Thiede, Wolfgang E. Berdel, Michael Kramer, Utz Krug, Christoph Schliemann
Data analysis and interpretation: Linus Angenendt, Christoph Röllig, Pau Montesinos, Eva Barragan, Tapan Kadia, Hagop M. Kantarjian, Gunnar Juliusson, Vladimir Lazarevic, Martin Höglund, Arnaud Pigneux, Jan Novák, Andrew H. Wei, Ing S. Tiong, Meaghan Wall, Elihu Estey, Rita Exeler, Matthias Stelljes, Georg Lenz, Jan-Henrik Mikesch, Hubert Serve, Gerhard Ehninger, Wolfgang E. Berdel, Michael Kramer, Utz Krug, Christoph Schliemann
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Chromosomal Abnormalities and Prognosis in NPM1-Mutated Acute Myeloid Leukemia: A Pooled Analysis of Individual Patient Data From Nine International Cohorts
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Christoph Röllig
Consulting or Advisory Role: AbbVie/Genentech, Amgen, Bristol-Myers Squibb, Celgene, Daiichi Sankyo, Janssen-Cilag, Jazz Pharmaceuticals, Novartis, Pfizer, Roche
Research Funding: Bayer Health (Inst), AbbVie (Inst), Novartis (Inst), Pfizer (Inst), Janssen-Cilag (Inst), Celgene (Inst)
Pau Montesinos
Consulting or Advisory Role: AbbVie, Pfizer, Shire, Daiichi Sankyo, Novartis, Celgene, Jazz Pharmaceuticals
Speakers' Bureau: Otsuka, Celgene, Daiichi Sankyo
Research Funding: Celgene (Inst), Janssen-Cilag (Inst), Pfizer (Inst)
Travel, Accommodations, Expenses: Amgen
David Martínez-Cuadrón
Consulting or Advisory Role: Jazz Pharmaceuticals, Novartis, Pfizer, Teva
Speakers' Bureau: Pfizer, Janssen-Cilag, Amgen
Travel, Accommodations, Expenses: Jazz Pharmaceuticals, Novartis, Pfizer
Eva Barragan
Consulting or Advisory Role: Novartis
Speakers' Bureau: Novartis, Janssen-Cilag
Research Funding: Incyte
Travel, Accommodations, Expenses: Novartis
Raimundo García
Research Funding: Alexion Pharmaceuticals, Merck Sharp & Dohme, Janssen
Farhad Ravandi
Honoraria: Sunesis Pharmaceuticals, Amgen, Seattle Genetics, Pfizer, Astellas Pharma, Orsenix, Celgene, Agios, AbbVie/Genentech
Consulting or Advisory Role: Seattle Genetics, Sunesis Pharmaceuticals, Amgen, Astellas Pharma, Orsenix, Celgene, Jazz Pharmaceuticals, Agios, AbbVie/Genentech, Bristol-Myers Squibb, Sunesis Pharmaceuticals, Amgen, Seattle Genetics, Merck, Macrogenix, Xencor, Selvita, Cellerant
Tapan Kadia
Consulting or Advisory Role: Novartis, Jazz Pharmaceuticals, Pfizer, AbbVie/Genentech, Bristol-Myers Squibb, Celgene, Sanofi, Amgen, BiolineRx, Incyte, Genentech/AbbVie, Pfizer, Jazz Pharmaceuticals
Hagop M. Kantarjian
Honoraria: AbbVie, Amgen, Ariad, Bristol-Myers Squibb, Immunogen, Orsenix, Pfizer
Research Funding: Pfizer (Inst), Amgen (Inst), Bristol-Myers Squibb (Inst), Novartis (Inst), Ariad (Inst), Astex Pharmaceuticals (Inst)
Jorge Cortes
Consulting or Advisory Role: Bristol-Myers Squibb, BiolineRx, Novartis, Pfizer, Amphivena Therapeutics, Daiichi Sankyo, Bio-Path Holdings, Astellas Pharma, Takeda, Jazz Pharmaceuticals
Research Funding: Bristol-Myers Squibb (Inst), Novartis (Inst), Pfizer (Inst), Celgene (Inst), Arog (Inst), Astellas Pharma (Inst), Celator (Inst), Immunogen (Inst), Sun Pharma (Inst), Takeda (Inst), Merus (Inst), Daiichi Sankyo (Inst), Tolero Pharmaceuticals (Inst), Trovagene (Inst)
Gunnar Juliusson
Honoraria: Daiichi Sankyo
Consulting or Advisory Role: Astellas Pharma, AbbVie, Novartis
Christian Recher
Consulting or Advisory Role: Celgene, Amgen, Novartis, Jazz Pharmaceuticals, AbbVie, Janssen, Astellas Pharma, Macrogenics, Daiichi Sankyo
Research Funding: Celgene (Inst), Amgen (Inst), Novartis (Inst), Jazz Pharmaceuticals (Inst), Astellas Pharma (Inst), Chugai Pharma (Inst), Agios (Inst), Daiichi Sankyo (Inst), MaatPharma (Inst)
Travel, Accommodations, Expenses: Incyte, Celgene, Sanofi, Amgen, Novartis, Daiichi Sankyo
Arnaud Pigneux
Honoraria: Novartis
Consulting or Advisory Role: Roche, AbbVie, Astellas Pharma, Amgen, Daichi, Jazz Pharmaceuticals, Pfizer
Travel, Accommodations, Expenses: Sanofi
Sarah Bertoli
Honoraria: Daiichi Sankyo, Astellas Pharma, Jazz Pharmaceuticals, Sanofi
Consulting or Advisory Role: Daiichi Sankyo, Astellas Pharma
Travel, Accommodations, Expenses: Pfizer, Janssen-Cilag, Celgene, Astellas Pharma
Hervé Dombret
Honoraria: Amgen, Celgene, Pfizer, Novartis, Incyte, Jazz Pharmaceuticals, Cellectis, Immunogen, Agios, Sunesis Pharmaceuticals, Daiichi Sankyo, Astellas Pharma, Janssen, Servier, Shire, AbbVie, Otsuka, Menarini
Research Funding: Amgen (Inst), Novartis (Inst), Pfizer (Inst), Jazz Pharmaceuticals (Inst), Incyte (Inst), Servier (Inst)
Jean-Baptiste Micol
Honoraria: Jazz Pharmaceuticals
Consulting or Advisory Role: AbbVie
Travel, Accommodations, Expenses: Jazz Pharmaceuticals
Zdeněk Ráčil
Consulting or Advisory Role: Pfizer, Novartis, MSD
Speakers' Bureau: Novartis
Expert Testimony: Novartis
Travel, Accommodations, Expenses: Novartis
Jan Novák
Consulting or Advisory Role: Pfizer, Amgen
Andrew H. Wei
Honoraria: Amgen, Servier, Novartis, Celgene, AbbVie/Genentech, Roche, Pfizer, Janssen Oncology
Consulting or Advisory Role: Servier, Novartis, Amgen, AbbVie/Genentech
Speakers' Bureau: AbbVie/Genentech, Novartis
Research Funding: Novartis (Inst), Celgene (Inst)
Patents, Royalties, Other Intellectual Property: A.H.W. is a former employee of the Walter and Eliza Hall Institute, which receives milestone and royalty payments related to venetoclax and is eligible for benefits related to these payments. A.H.W. receives payments from the Walter and Eliza Hall Institute related to venetoclax.
Expert Testimony: Amgen
Travel, Accommodations, Expenses: AbbVie, Amgen, Novartis, Celgene
Friedrich Stölzel
Honoraria: Jazz Pharmaceuticals, Shire
Research Funding: Astellas Pharma
Travel, Accommodations, Expenses: Neovii Biotech
Christian Thiede
Employment: AgenDix
Leadership: AgenDix
Stock and Other Ownership Interests: AgenDix
Honoraria: Novartis, GWT
Consulting or Advisory Role: Astellas Pharma
Research Funding: Bayer Schering Pharma (Inst)
Matthias Stelljes
Consulting or Advisory Role: Pfizer, Jazz Pharmaceuticals, Gilead Sciences, MSD, Amgen
Speakers' Bureau: Pfizer, Medac, MSD, Incyte
Research Funding: Pfizer (Inst)
Travel, Accommodations, Expenses: Medac, Neovii Biotech
Georg Lenz
Honoraria: Janssen-Cilag, Bayer, Celgene, Genentech, AstraZeneca, Bristol-Myers Squibb, MorphoSys, Gilead Sciences, Novartis
Consulting or Advisory Role: Genentech, Janssen-Cilag, Bristol-Myers Squibb, Bayer, Novartis, Celgene, MorphoSys, AstraZeneca, Gilead Sciences
Research Funding: Janssen-Cilag, Genentech, AstraZeneca, Aquinox Pharmaceuticals, Bayer, Gilead Sciences, MorphoSys, Verastem
Expert Testimony: MorphoSys
Travel, Accommodations, Expenses: Janssen-Cilag, Celgene, AstraZeneca
Jan-Henrik Mikesch
Consulting or Advisory Role: Pfizer Deutschland, Daiichi Sankyo Deutschland
Travel, Accommodations, Expenses: Daiichi Sankyo Deutschland, Celgene, Kite Pharma
Hubert Serve
Honoraria: Novartis, Robert-Bosch-Gesellschaft für Medizinische Forschung, Gilead Sciences
Consulting or Advisory Role: Gilead Sciences, IKP Stuttgart (Robert-Bosch-Gesellschaft für Medizinische Forschung)
Patents, Royalties, Other Intellectual Property: Patent on Samhd1 modulation for treating resistance to cancer therapy (Inst), patent on oncogene redirection, companion diagnostics for leukemia treatment (Inst), markers for responsiveness to an inhibitor of FLT3
Gerhard Ehninger
Employment: Rhoen Klinikum
Leadership: Rhoen Klinikum
Stock and Other Ownership Interests: Fresenius Healthcare, Celgene
Honoraria: Janssen-Cilag
Wolfgang E. Berdel
Consulting or Advisory Role: Philogen
Patents, Royalties, Other Intellectual Property: I hold international patent rights for vascular targeting of tissue factor. So far this not connected with any return of money
Expert Testimony: Philogen
Travel, Accommodations, Expenses: Pfizer, Philogen
Michael Kramer
Consulting or Advisory Role: Cellex patient treatment, GEMoaB
Utz Krug
Honoraria: Celgene, Daiichi Sankyo, Janssen, Jazz Pharmaceuticals, AbbVie, Roche, Boehringer Ingelheim, Genesis Pharmaceuticals, Sirtex, Chugai Pharma
Consulting or Advisory Role: Janssen, Celgene, Daiichi Sankyo, Jazz Pharmaceuticals, Pfizer
Travel, Accommodations, Expenses: Celgene, Janssen, Merck Serono, Daiichi Sankyo
Christoph Schliemann
Consulting or Advisory Role: AbbVie, Jazz Pharmaceuticals, Pfizer, Novartis, Takeda
Travel, Accommodations, Expenses: Celgene, PharmaMar, Pfizer
No other potential conflicts of interest were reported.
REFERENCES
- 1.Falini B, Mecucci C, Tiacci E, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype N Engl J Med 352254–2662005 [DOI] [PubMed] [Google Scholar]
- 2.Suzuki T, Kiyoi H, Ozeki K, et al. Clinical characteristics and prognostic implications of NPM1 mutations in acute myeloid leukemia Blood 1062854–28612005 [DOI] [PubMed] [Google Scholar]
- 3.Döhner K, Schlenk RF, Habdank M, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: Interaction with other gene mutations Blood 1063740–37462005 [DOI] [PubMed] [Google Scholar]
- 4.Thiede C, Koch S, Creutzig E, et al. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML) Blood 1074011–40202006 [DOI] [PubMed] [Google Scholar]
- 5.Verhaak RG, Goudswaard CS, van Putten W, et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): Association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance Blood 1063747–37542005 [DOI] [PubMed] [Google Scholar]
- 6.Schnittger S, Schoch C, Kern W, et al. Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype Blood 1063733–37392005 [DOI] [PubMed] [Google Scholar]
- 7.Gale RE, Green C, Allen C, et al. The impact of FLT3 internal tandem duplication mutant level, number, size, and interaction with NPM1 mutations in a large cohort of young adult patients with acute myeloid leukemia Blood 1112776–27842008 [DOI] [PubMed] [Google Scholar]
- 8.Pratcorona M, Brunet S, Nomdedéu J, et al. Favorable outcome of patients with acute myeloid leukemia harboring a low-allelic burden FLT3-ITD mutation and concomitant NPM1 mutation: Relevance to post-remission therapy Blood 1212734–27382013 [DOI] [PubMed] [Google Scholar]
- 9.Döhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel Blood 129424–4472017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haferlach C, Mecucci C, Schnittger S, et al. AML with mutated NPM1 carrying a normal or aberrant karyotype show overlapping biologic, pathologic, immunophenotypic, and prognostic features Blood 1143024–30322009 [DOI] [PubMed] [Google Scholar]
- 11.Micol JB, Boissel N, Renneville A, et al. The role of cytogenetic abnormalities in acute myeloid leukemia with NPM1 mutations and no FLT3 internal tandem duplication Blood 1144601–46022009 [DOI] [PubMed] [Google Scholar]
- 12.McGowan-Jordan J, Simons A, Schmid M. ISCN 2016: An International System for Human Cytogenomic Nomenclature. Basel, Switzerland, Karger,: 2016. [Google Scholar]
- 13.Büchner T, Schlenk RF, Schaich M, et al. Acute myeloid leukemia (AML): Different treatment strategies versus a common standard arm--combined prospective analysis by the German AML Intergroup J Clin Oncol 303604–36102012 [DOI] [PubMed] [Google Scholar]
- 14.Simon R, Makuch RW.A non-parametric graphical representation of the relationship between survival and the occurrence of an event: Application to responder versus non-responder bias Stat Med 335–441984 [DOI] [PubMed] [Google Scholar]
- 15.Straube J, Ling VY, Hill GR, et al. The impact of age, NPM1mut, and FLT3ITD allelic ratio in patients with acute myeloid leukemia Blood 1311148–11532018 [DOI] [PubMed] [Google Scholar]
- 16.Heath EM, Chan SM, Minden MD, et al. Biological and clinical consequences of NPM1 mutations in AML Leukemia 31798–8072017 [DOI] [PubMed] [Google Scholar]
- 17.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia Blood 1272391–24052016 [DOI] [PubMed] [Google Scholar]
- 18.Stelljes M, Krug U, Beelen DW, et al. Allogeneic transplantation versus chemotherapy as postremission therapy for acute myeloid leukemia: A prospective matched pairs analysis J Clin Oncol 32288–2962014 [DOI] [PubMed] [Google Scholar]
- 19.Röllig C, Bornhäuser M, Kramer M, et al. Allogeneic stem-cell transplantation in patients with NPM1-mutated acute myeloid leukemia: Results from a prospective donor versus no-donor analysis of patients after upfront HLA typing within the SAL-AML 2003 trial J Clin Oncol 33403–4102015 [DOI] [PubMed] [Google Scholar]
- 20.Ivey A, Hills RK, Simpson MA, et al. Assessment of minimal residual disease in standard-risk AML N Engl J Med 374422–4332016 [DOI] [PubMed] [Google Scholar]
- 21.Jongen-Lavrencic M, Grob T, Hanekamp D, et al. Molecular minimal residual disease in acute myeloid leukemia N Engl J Med 3781189–11992018 [DOI] [PubMed] [Google Scholar]