Abstract
PURPOSE
Early identification of childhood cancer survivors at high risk for treatment-related cardiomyopathy may improve outcomes by enabling intervention before development of heart failure. We implemented artificial intelligence (AI) methods using the Children's Oncology Group guideline–recommended baseline ECG to predict cardiomyopathy.
MATERIAL AND METHODS
Seven AI and signal processing methods were applied to 10-second 12-lead ECGs obtained on 1,217 adult survivors of childhood cancer prospectively followed in the St Jude Lifetime Cohort (SJLIFE) study. Clinical and echocardiographic assessment of cardiac function was performed at initial and follow-up SJLIFE visits. Cardiomyopathy was defined as an ejection fraction < 50% or an absolute drop from baseline ≥ 10%. Genetic algorithm was used for feature selection, and extreme gradient boosting was applied to predict cardiomyopathy during the follow-up period. Model performance was evaluated by five-fold stratified cross-validation.
RESULTS
The median age at baseline SJLIFE evaluation was 31.7 years (range 18.4-66.4), and the time between baseline and follow-up evaluations was 5.2 years (0.5-9.5). Two thirds (67.1%) of patients were exposed to chest radiation, and 76.6% to anthracycline chemotherapy. One hundred seventeen (9.6%) patients developed cardiomyopathy during follow-up. In the model based solely on ECG features, the cross-validation area under the curve (AUC) was 0.87 (95% CI, 0.83 to 0.90), whereas the model based on clinical features had an AUC of 0.69 (95% CI, 0.64 to 0.74). In the model based on ECG and clinical features, the cross-validation AUC was 0.89 (95% CI, 0.86 to 0.91), with a sensitivity of 78% and a specificity of 81%.
CONCLUSION
AI using ECG data may assist in the identification of childhood cancer survivors at increased risk for developing future cardiomyopathy.
INTRODUCTION
Because of improved treatment and supportive care, 5-year survival rates for childhood cancer now exceed 85% with more than 500,000 childhood cancer survivors alive in the United States today.1 Patients treated with anthracycline chemotherapy or cardiac-directed radiation are at increased risk for adverse cardiovascular sequelae,2 including cardiomyopathy, coronary artery disease, and valvular heart disease.
CONTEXT
Key Objective
To investigate whether artificial intelligence (AI) applied to electrocardiograms can predict cardiomyopathy in adult survivors of childhood cancer exposed to cardiotoxic therapies.
Knowledge Generated
Clinical models predicted cardiomyopathy with a sensitivity of 62%, a specificity of 66%, and an area under the curve (AUC) of 0.69 (95% CI, 0.64 to 0.74). An AI model based on 86 signal processing–based ECG features was 76% and 79% sensitive and specific, with an AUC of 0.87 (95% CI, 0.83 to 0.90). Combining clinical and ECG features correctly predicted 78% of patients who developed cardiomyopathy and 81% who did not, with positive and negative predictive values of 30% and 97%, respectively, and an AUC of 0.89 (95% CI, 0.86 to 0.91).
Relevance
AI may help classify adult survivors of childhood cancer before clinical recognition. Enhanced characterization of this at-risk population may help separate those in need of greater surveillance from those for whom a lower degree of monitoring may suffice.
Surveillance guidelines have been developed by a variety of oncology groups to guide long-term monitoring with the goal of identifying those who might benefit from early medical interventions.3 While offering an opportunity for early detection of myocardial dysfunction, screening guidelines do not identify patients with preserved function who may yet develop cardiomyopathy.
We investigated whether childhood cancer survivors at risk of developing late-onset cardiomyopathy could be accurately identified before the development of echocardiographic evidence of dysfunction using machine learning algorithms directed at ECG waveforms.
MATERIALS AND METHODS
Study Population and Design
The St Jude Lifetime Study (SJLIFE) cohort is an ongoing study with prospective clinical follow-up of survivors of childhood cancer treated at St Jude Children's Research Hospital, Memphis, TN.4,5 Participants in this analysis were at least 10 years from cancer diagnosis and ≥ 18 years of age at cohort entry and had completed at least two comprehensive clinical assessments on the St Jude Children's Research Hospital campus including physical examination, a core laboratory battery, 12-lead ECG (GE MAC 1200 machine), and two-dimensional Doppler ultrasound echocardiography (GE Medical Systems, Milwaukee, WI) with three-dimensional imaging for left ventricular volumes. Medical record abstraction was performed to determine cumulative anthracycline and cardiac radiation exposures. For survivors treated with radiation, cumulative dose, orientation, beam energy, field size, weighting, blocking, and anatomical position were abstracted. For each individual, all radiation therapy (RT) fields were reconstructed on computational phantoms scaled to their age at RT. To be eligible for our study, participants had to have an ejection fraction ≥ 50% at the time of their baseline SJLIFE assessment.
As of December 31, 2018, 1,326 participants had > 1 on-campus clinical assessment including ECHO and ECG screening. Our final study cohort included 1,217 of 1,326 with no cardiomyopathy at their first visit with ECHO screening (Data Supplement).
Outcome
Cardiomyopathy was defined as an ejection fraction < 50% or an absolute drop from the baseline ≥ 10% obtained by echocardiogram during any of the follow-up visits.
Treatment exposures
Four treatment exposures were included in the analysis. Cumulative anthracycline dose (milligram per square meter) was calculated by summing adriamycin, daunorubicin, epirubicin, idarubicin, and mitoxantrone in adriamycin equivalent doses.6 Radiation to the chest was used in two different forms as a yes or no variable and maximum total dose. Mean heart doses (stray dose from leakage and scatter radiation from nonchest-directed radiation) were calculated by radiation physicists at MD Anderson Cancer Center, Houston, TX.7
Additional covariates
A total of 12 demographic and clinical variables were used in the analysis: age at diagnosis and at time of echocardiography (years), race, sex, body surface area (per square meter), primary cancer diagnosis, heart rate (beats/min), respiratory rate (respirations/min), systolic and diastolic blood pressures (mm Hg), smoking (yes or no), and the presence of clinically assessed cardiovascular risk factors (diabetes [treated by diet and/or medication], hypertriglyceridemia [fasting triglycerides ≥ 150 mg/dL and/or on a lipid lowering agent], hypertension [systolic blood pressure > 140 mm Hg and/or diastolic blood pressure > 90 mm Hg and/or on antihypertensive agents], or hypercholesterolemia [fasting total cholesterol > 200 mg/dL and/or on a lipid lowering agent]). We also included respiratory rate (respiration/min) as subtle changes in respiratory rate may reflect underlying heart disease even in the early stages of disease. In addition to these, we extracted features (described in detail below) from 12-lead ECGs for use in the models.
Statistical Methods
Overview
The study was approved by the Institutional Review Board, and all survivors provided informed consent before participation. Characteristics of the survivors at baseline assessment were expressed as median and range for continuous variables and as count and percentage for categorical variables. They were compared between those who developed cardiomyopathy and those who did not. Categorical variables were compared using Pearson chi-square test and Fisher's exact test, and numerical variables were compared using Student's t-test. The results of statistical tests were considered to be significant if the two-sided P value was < .05.
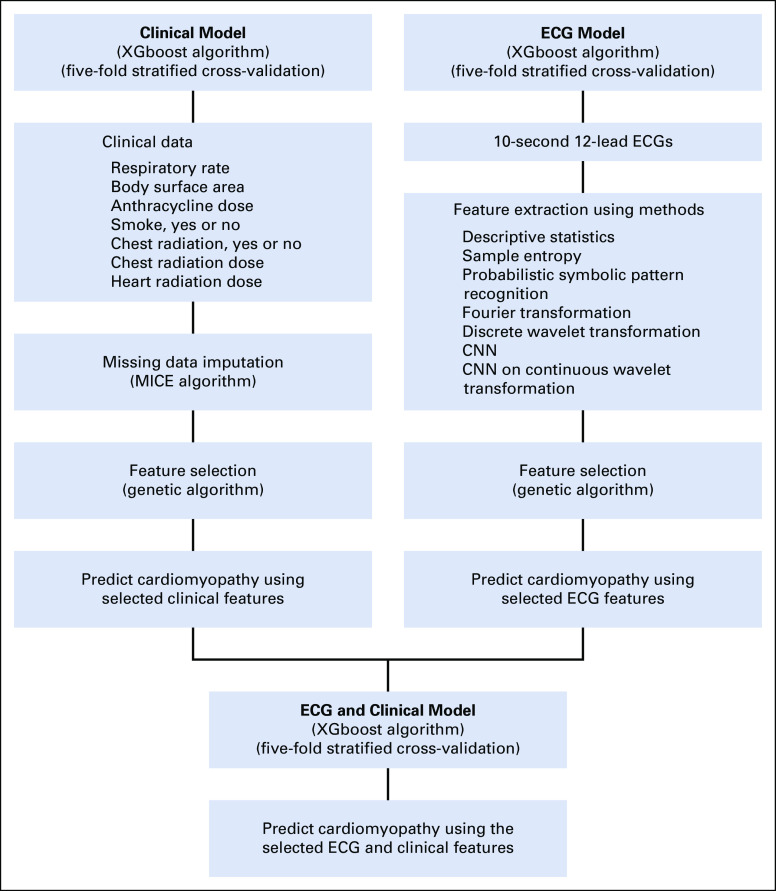
We used waveform data from 12-lead ECG recordings sampled at 500 Hz. Features were extracted using both signal processing and deep learning methods, and those features most discriminating for cardiomyopathy (diagnosed during follow-up) were selected using a stochastic optimization method, genetic algorithm (GA).8 Features were also selected from clinical data using GA (Fig 1). Extreme Gradient Boosting (XGboost)9 method was applied to the selected features of ECG from each method separately and then combined with clinical data to predict cardiomyopathy.
FIG 1.
First column shows the analytic flow for developments of the clinical model (light blue), including the missing data imputation, determination of important clinical features, and clinical model building. The second column shows the analytic flow for development of the ECG model (light blue), including the feature extraction through signal processing and deep learning methods, determination of important extracted ECG features of each method, and ECG model building. The bottom row shows the combination of selected ECG and clinical features and final model building (dark blue). CNN, convolutional neural network; XGboost, extreme gradient boosting.
Feature extraction
Using baseline ECG data, we generated measures from each lead, including mean, median, kurtosis, skewness, variance, and other general features including root mean square and mean crossing rate of observed amplitude values. Sample entropy was calculated for each ECG lead as a measure of signal regularity of the signal. We segmented each ECG recording to 1 second epochs and computed sample entropy for each epoch sequentially. For probabilistic symbolic pattern recognition features, dissimilarities were calculated with reference to 1,100 healthy ECG recordings.10 We decomposed ECG signals using a Fourier transform into basic sinusoidal waves at different frequencies and used amplitudes and phases of the waves as a feature. Both continuous and discrete wavelet transformations were used to extract features from the frequency spectrum. The discrete wavelet transformation was implemented by calculating descriptive statistics from a five-level wavelet decomposition. The continuous wavelet transformation was completed by calculating the similarity between signal and wavelets at each frequency and time points by shifting wavelets through signals to obtain a two-dimensional scaleogram. We then trained a convolutional neural network to extract features from the scaleograms. Finally, features were extracted using waveform data from 12-lead ECG recordings. Each 12-lead ECG signal was overlaid on top of each other to form one signal with 12 channels. Residual neural networks were used to extract features. We designed our neural network architectures in Python using the Keras library with TensorFlow backend (see the Data Supplement for full listing of all feature extraction approaches).
Feature selection
We implemented a GA algorithm to select the best subset of all ECG features and clinical variables for the predictive model, which provided the minimum classification error. The GA was started with a random subset of features (different variable combinations) and iteratively mixed subsets (to obtain different feature combinations) based on their performance (area under the curve [AUC] value in predicting cardiomyopathy) to obtain feature combinations yielding better performance. XGboost was used as a fitness function on the iterated GA processes from 40 generations, with a crossover probability of .5 and a mutation probability of .05. We implemented GA separately on features extracted from each feature extraction method and then combined selected features for the final prediction models.
Missing data imputation
Of the 20 clinical variables, all except two had < 1% missingness. Dose to the heart from RT had the highest missing rate (2.8%), followed by chest radiation dose (0.9%), and chest radiation (yes or no) (0.8%). We imputed missing clinical data using the multivariate imputation by chained equation method in the MICE package in R.11
Predictive modeling
Although there are many machine learning algorithms for classification tasks, XGBoost has shown superior performance in several complex problems and data science competitions such as Kaggle challenges.9,12 For the computationally intensive classification at multiple stages (classification models based on features from individual feature extraction methods, ECG features selected from feature extraction methods and clinical data, and a combination of ECG features and clinical data), we used the XGboost algorithm for all classification tasks. We used Bayesian hyperparameter optimization for tuning hyperparameters in the XGBoost model. For the XGBoost model (Data Supplement), optimization occurred when the learning rate was 0.16 and the total number of trees was 1,869. Following five-fold stratified cross-validation, model performance was assessed using metrics including sensitivity, specificity, and AUC.
RESULTS
Study Population Characteristics
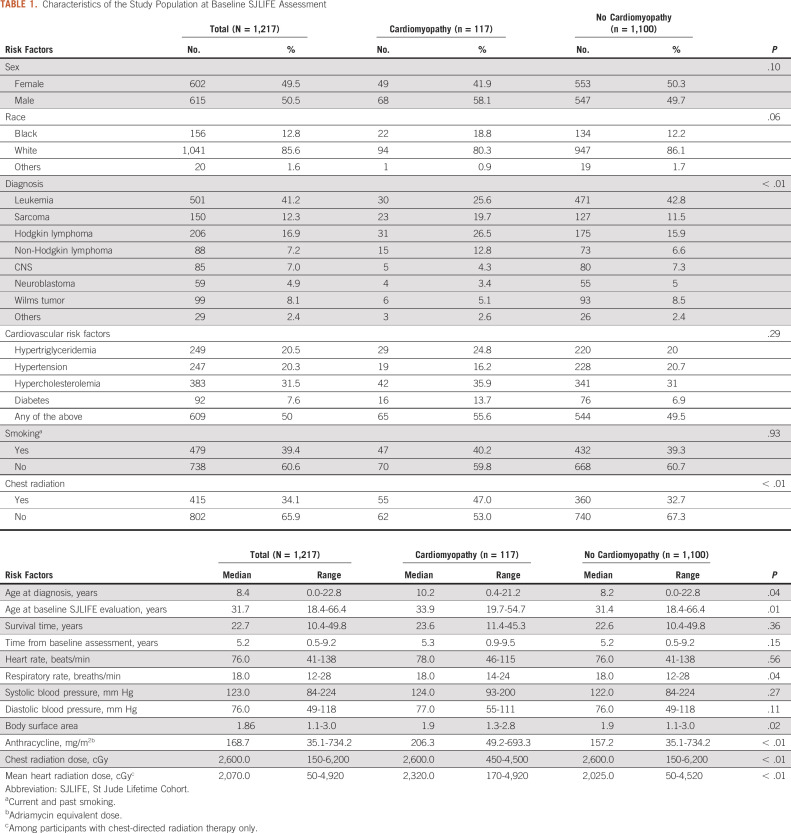
Among 1,217 eligible survivors (51% male and 86% White), the median time between baseline and subsequent visits was 5.2 (range 0.5-9.2) years. The median age at cancer diagnosis was 8.4 (0.0-22.7) years and 31.7 (18.4-66.4) years at baseline SJLIFE assessment. Eight hundred seventeen (67.1%) were exposed to chest radiation (median dose 2,600 cGy, 150-6,200), and 932 (76.6%) to anthracycline therapy (median cumulative dose 168.7 mg/m2, 35.1-734.2). Half (n = 609) had at least one cardiovascular risk factor, including 92 (7.6%) with diabetes, 249 (20.5%) with hypertriglyceridemia, 247 (20.3%) with hypertension, and 383 (31.5%) with hypercholesterolemia. Demographic and clinical characteristics of the study participants at the time of baseline visit are shown in Table 1.
TABLE 1.
Characteristics of the Study Population at Baseline SJLIFE Assessment
One hundred seventeen (9.6%) survivors developed cardiomyopathy between the baseline and subsequent follow-up visit. Survivors who developed cardiomyopathy were significantly older at cancer diagnosis (10.2 v 8.2; P < .04) and baseline SJLIFE assessment (33.9 v 31.4; P < .01). Those with cardiomyopathy had a higher median anthracycline (206.3 mg/m2 v 157.2 mg/m2; P < .001) and heart radiation (2,320 cGy v 2,025 cGy; P < .001) dose.
Cardiomyopathy Prediction
Feature extraction
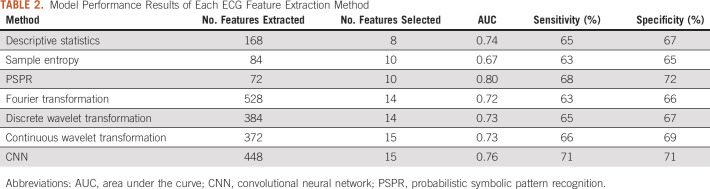
A total of 168 ECG features (such as mean, median, skewness, etc) were extracted from 12 leads (14 features per lead). Among these features, eight were selected by GA and found by XGBoost to predict cardiomyopathy with a sensitivity of 65%, a specificity of 67%, and a five-fold cross-validated AUC of 0.74. In a similar manner, 72 features reflecting sample entropy were extracted and, of these, 10 variables were selected by GA. With XGboost, these 10 features predicted cardiomyopathy with a sensitivity of 63%, a specificity of 65%, and a five-fold cross-validated AUC of 0.67. The list of all feature extraction methods considered and their associated five-fold cross-validated model performance metrics are shown in Table 2. Detailed information about the predictors is presented in the Data Supplement.
TABLE 2.
Model Performance Results of Each ECG Feature Extraction Method
Model performance of ECG data
Overall model performance was calculated using the 86 selected ECG features extracted from the combination of methods listed in the Data Supplement and applying XGBoost. Cardiomyopathy was predicted with a five-fold cross-validated sensitivity of 76%, a specificity of 79%, and an AUC of 0.87 (95% CI, 0.83 to 0.90) using these 86 ECG features.
Model performance of clinical data
Of the 20 clinical features that were evaluated, seven were selected for the clinical data model: respiratory rate, body surface area, cumulative anthracycline dose, smoking, chest radiation (both as a dichotomous variable [yes or no] and cumulative dose), and mean heart dose from RT. The model constructed with these variables had a five-fold cross-validated sensitivity of 62%, a specificity of 66%, and an AUC of 0.69 (95% CI, 0.64 to 0.74).
Model performance of ECG plus clinical data
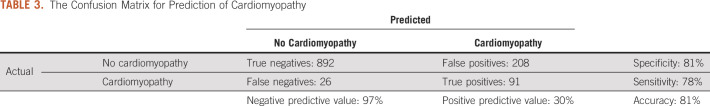
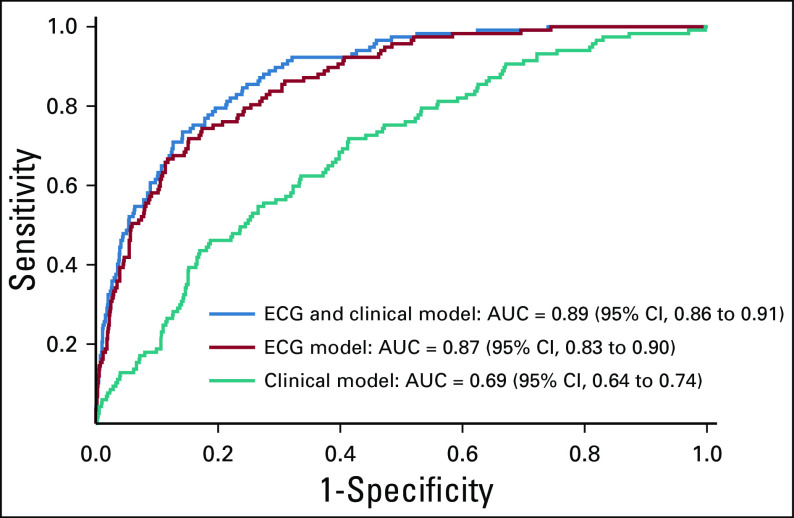
The 86 selected ECG features were combined with the seven clinical features to build a final model. The confusion matrix for prediction of cardiomyopathy is provided in Table 3. The model correctly predicted 78% of patients who developed cardiomyopathy, with a positive predictive value of 30%. The model correctly predicted 81% of patients who did not develop cardiomyopathy, with a negative predictive value of 97%. Overall, 81% of the model predictions were correct, with an AUC of 0.89 (95% CI, 0.86 to 0.91). Receiver operating characteristic curves of these models are shown in Figure 2.
TABLE 3.
The Confusion Matrix for Prediction of Cardiomyopathy
FIG 2.
Receiver operating characteristic curves of the models based on ECG features, clinical features and ECG, and clinical features. AUC, area under the curve.
Subgroup analysis
A subgroup analysis was conducted to assess model performance for predicting cardiomyopathy at 0.5-5 years and 5-9 years following the baseline SJLIFE assessment. At 0.5-5 years, the sensitivity was 79%, specificity 81%, and AUC 0.89 (95% CI, 0.85 to 0.93), and 78%, 81%, and 0.88 (95% CI, 0.84 to 0.92), respectively, 5-9 years after the baseline visit (Data Supplement).
DISCUSSION
Machine learning techniques applied to ECG data collected among a large clinically assessed cohort of adult survivors of childhood cancer (22 years from cancer diagnosis) were able to accurately classify the risk for late-onset cardiomyopathy (AUC 0.89 for the model incorporating ECG and clinical data). A model using only ECG features was similar (AUC 0.87, sensitivity 76%, and specificity 79%), and both outperformed the clinical model (AUC 0.69 sensitivity 62%, and specificity 66%). Our model performance predicting cardiomyopathy in 0.5-5 years or 5-9.2 years was similar, suggesting time invariant cardiomyopathy risk up to 9.2 from the baseline ECG. Artificial intelligence (AI) may help classify high-risk cancer survivors before clinical detection could occur and in doing so identify survivors in need of enhanced surveillance and/or early intervention to ameliorate long-term morbidity. If these findings hold, AI may also help limit subsequent costly evaluations and procedures in patients who are less likely to develop events such as cardiomyopathy.
Predictive models for heart disease are increasingly common. Our model performance exceeded that reported among survivors participating in the well-characterized Childhood Cancer Survivor Study. Using self-reported data from more than 13,000 five-year cancer survivors (median age at follow-up 32 years and range 6-59 years; median survival time 19 years and range 0-34 years) in a model focused primarily on chemotherapy and radiation exposures yielded an AUC ranging from 0.68-0.82 for predicting heart failure at age 40 years13 Using similar models, the same investigators reported AUCs ranging from 0.66-0.67 for ischemic heart disease and 0.68-0.72 for stroke by age 50 years in this population.14 Chen et al15 incorporated traditional cardiovascular risk factors (hypertension, dyslipidemia, and diabetes) reported by Childhood Cancer Survivor Study participants to predict risk for heart failure at 50 years and obtained AUCs between 0.69 and 0.77 across age categories 20-35 years. Additionally, applying convolutional neural networks in a large (n = 180,922) noncancer population evaluated over 24 years, Attia et al16 recently reported an AUC of 0.87 for identifying atrial fibrillation based on a single ECG. Our model, which applies AI to ECG data, predicts cardiomyopathy as well as, or better than, these previous studies.
AI has also shown promise for the early detection of cardiac dysfunction in other populations. Cardiac sympathetic denervation has been recognized as an early feature in Parkinson's Disease (PD),17-20 and reduced heart rate variability has been associated with increased risk of PD.21 Work from our group has shown that classification of PD is possible using ECGs collected in the presymptomatic state.22 Attia et al23 have also used neural networks on 12-lead ECGs to identify asymptomatic left ventricular dysfunction, suggesting that early disease might subtly affect cardiac conduction.
The use of a relatively inexpensive and widely available test such as an ECG to generate risk stratification for childhood cancer survivors holds appeal. Currently recommended as a baseline assessment by the Children's Oncology Group's Long-Term Follow-up Guidelines for Survivors of Childhood, Adolescent, and Young Adult Cancers, the focus has largely been to monitor QT intervals (25). Our study suggests additional value from ECGs obtained on cancer survivors. The addition of seven clinical variables to the ECG data yielded only a marginal improvement in AUC, from 0.87 to 0.89, highlighting the informative value of the digital ECG data. The performance of such predictive models suggests the possibility for remote monitoring via mobile or wearable devices with ECG recording functionality. With such an approach, at-risk patients can potentially be identified for heightened surveillance that could potentially be facilitated by remote applications.
Understandably, the features identified in our models are not immediately interpretable and bear little relationship to classical ECG characteristics such as p-waves, QRS complexes, and t-waves typically used for diagnosis of chamber dysfunction. Clinical use of our model will require its execution on the digital data underlying standard 12-lead ECGs—a computationally simple task—to generate a risk score for future cardiomyopathy. The development of such predictive models might assist clinicians by identifying those who could benefit from early initiation of preventive therapeutics to reduce or minimize the risk of future cardiomyopathy and associated heart failure.
Despite the large volume of clinically assessed data available in the SJLIFE cohort, there are a number of limitations to acknowledge. Cohort participants comprised childhood cancer survivors evaluated at a single institution, and although we performed five-fold cross-validation for model training and testing, external validation is warranted in other cancer cohort data sets where clinically assessed long-term follow-up is available. However, it should be noted that there is no other comparable childhood cancer survivor cohort to SJLIFE in the United States with similarly clinically assessed data for external validation. Therefore, future effort for external validation will require international collaboration with cohorts with sufficient long-term follow-up ECHO and ECG screening of childhood cancer survivors cohorts. Second, although the cohort size was relatively large (1,217 patients), the small number of patients who developed cardiomyopathy precluded our ability to test whether the model performed differently by sex, age, race, or other characteristics. Additional follow-up as the SJLIFE cohort ages may permit theses analyses. Finally, we did not evaluate whether deep learning applied to other tools, such as echocardiograms, could also predict cardiomyopathy, add to, or outperform the predictions made from the ECG. Future work on these topics is needed.
In conclusion, we developed an AI-assisted model based on ECG features predictive of cardiomyopathy with a sensitivity of 76%, a specificity of 79%, and an AUC of 0.87, suggesting that machine learning may play a role in the identification of childhood cancer survivors at risk for developing cardiomyopathy. Future investigation will focus on validation in other cancer survivor populations and the application of remote monitoring techniques to screen and monitor long-term survivors of childhood cancer.
Rebecca M. Howell
Research Funding: MD Anderson Cancer Center
John L. Jefferies
Honoraria: Genzyme, Amicus Therapeutics, Pfizer, Abbott Diagnostics, Stealth Biotherapeutics, Novartis, Chiesi
Consulting or Advisory Role: Abbott Diagnostics, Amicus Therapeutics, Chiesi, Medtronic, CHF Solutions, Stealth Biotherapeutics, Pfizer, Novartis
Speakers' Bureau: Genzyme, Pfizer
Research Funding: Medtronic, Myokardia, Sanofi, Innolife, Novartis, Lilly, CHF Solutions, Regeneron
Travel, Accommodations, Expenses: Genzyme, Abbott Diagnostics, Amicus Therapeutics, Novartis, Medtronic, Chiesi, PQBypass
Deo Kumar Srivastava
Consulting or Advisory Role: GENERAL DYNAMICS Information Technology Peer Review and Science Management
Melissa M. Hudson
Consulting or Advisory Role: Oncology Research Information Exchange Network, Princess Máxima Center
No other potential conflicts of interest were reported.
PRIOR PRESENTATION
Presented in part at ASCO Annual Meeting (virtual), May 29-June 2, 2020 and its abstract was published at JCO Clinical Cancer Informatics (38:10545 [15_suppl]).
SUPPORT
Supported by the National Cancer Institute (U01 CA195547, M.M.H. and L.L.R., Principal Investigators). Support to St Jude Children's Research Hospital was also provided by the Cancer Center Support (CORE) grant (CA21765, C. Roberts, Principal Investigator) and the American Lebanese Syrian Associated Charities (ALSAC).
AUTHOR CONTRIBUTIONS
Conception and design: Fatma Güntürkün, Oguz Akbilgic, Robert L. Davis, Gregory T. Armstrong, Rebecca M. Howell, John L. Jefferies, Ibrahim Karabayir, Deo Kumar Srivastava, Melissa M. Hudson, Daniel A. Mulrooney
Financial support: Gregory T. Armstrong, Melissa M. Hudson, Leslie L. Robison
Administrative support: Robert L. Davis, Gregory T. Armstrong, Kirsten K. Ness, Melissa M. Hudson
Provision of study materials or patients: Gregory T. Armstrong, Melissa M. Hudson, Leslie L. Robison, Elsayed Z. Soliman
Collection and assembly of data: Robert L. Davis, Gregory T. Armstrong, Kirsten K. Ness, Melissa M. Hudson, Leslie L. Robison, Daniel A. Mulrooney
Data analysis and interpretation: Fatma Güntürkün, Oguz Akbilgic, Robert L. Davis, Gregory T. Armstrong, Rebecca M. Howell, John L. Jefferies, Kirsten K. Ness, John T. Lucas, Deo Kumar Srivastava, Melissa M. Hudson, Elsayed Z. Soliman, Daniel A. Mulrooney
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Rebecca M. Howell
Research Funding: MD Anderson Cancer Center
John L. Jefferies
Honoraria: Genzyme, Amicus Therapeutics, Pfizer, Abbott Diagnostics, Stealth Biotherapeutics, Novartis, Chiesi
Consulting or Advisory Role: Abbott Diagnostics, Amicus Therapeutics, Chiesi, Medtronic, CHF Solutions, Stealth Biotherapeutics, Pfizer, Novartis
Speakers' Bureau: Genzyme, Pfizer
Research Funding: Medtronic, Myokardia, Sanofi, Innolife, Novartis, Lilly, CHF Solutions, Regeneron
Travel, Accommodations, Expenses: Genzyme, Abbott Diagnostics, Amicus Therapeutics, Novartis, Medtronic, Chiesi, PQBypass
Deo Kumar Srivastava
Consulting or Advisory Role: GENERAL DYNAMICS Information Technology Peer Review and Science Management
Melissa M. Hudson
Consulting or Advisory Role: Oncology Research Information Exchange Network, Princess Máxima Center
No other potential conflicts of interest were reported.
REFERENCES
- 1.Howlader N, Noone AM, Krapcho M, et al. SEER Cancer Statistics Review, 1975-2017. Bethesda, MD: National Cancer Institute; 2020. [Google Scholar]
- 2.Mulrooney DA, Armstrong GT, Huang S, et al. Cardiac outcomes in adult survivors of childhood cancer exposed to cardiotoxic therapy: A cross-sectional study Ann Intern Med 16493–1012016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armenian SH, Hudson MM, Mulder RL, et al. Recommendations for cardiomyopathy surveillance for survivors of childhood cancer: A report from the International Late Effects of Childhood Cancer Guideline Harmonization Group Lancet Oncol 16e123–e1362015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hudson MM, Ness KK, Nolan VG, et al. Prospective medical assessment of adults surviving childhood cancer: Study design, cohort characteristics, and feasibility of the St. Jude Lifetime cohort study Pediatr Blood Cancer 56825–8362011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hudson MM, Ehrhardt MJ, Bhakta N, et al. Approach for classification and severity grading of long-term and late-onset health events among childhood cancer survivors in the St. Jude Lifetime Cohort Cancer Epidemiol Biomarkers Prev 26666–6742017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feijen EAM, Leisenring WM, Stratton KL, et al. Derivation of anthracycline and anthraquinone equivalence ratios to doxorubicin for late-onset cardiotoxicity JAMA Oncol 5864–8712019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howell RM, Smith SA, Weathers RE, et al. Adaptations to a generalized radiation dose reconstruction methodology for use in epidemiologic studies: An update from the MD Anderson Late Effect Group Radiat Res 192169–1882019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holland JH. Adaptation in Natural and Artificial Systems: An Introductory Analysis with Applications to Biology, Control and Artificial Intelligence. Cambridge, MA, MIT Press: 1992. [Google Scholar]
- 9.Chen T, Guestrin C. XGBoost: A scalable tree boosting system. Presented at the Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, California, USA, August, 2016.
- 10.Akbilgic O, Howe JA.Symbolic pattern recognition for sequential data Sequential Anal 36528–5402017 [Google Scholar]
- 11.van Buuren S, Groothuis-Oudshoorn K. mice: Multivariate imputation by chained equations in R. J Stat Softw. 2011;45:67. [Google Scholar]
- 12.Pafka S. Benchmarking Machine Learning Tools for Scalability, Speed and Accuracy, Epoch H2O World Conference. Mountain View: 2015. [Google Scholar]
- 13.Chow EJ, Chen Y, Kremer LC, et al. Individual prediction of heart failure among childhood cancer survivors J Clin Oncol 33394–4022015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chow EJ, Chen Y, Hudson MM, et al. Prediction of ischemic heart disease and stroke in survivors of childhood cancer J Clin Oncol 3644–522018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Chow EJ, Oeffinger KC, et al. Traditional cardiovascular risk factors and individual prediction of cardiovascular events in childhood cancer survivors J Natl Cancer Inst 112256–2652020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Attia ZI, Noseworthy PA, Lopez-Jimenez F, et al. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: A retrospective analysis of outcome prediction Lancet 394861–8672019 [DOI] [PubMed] [Google Scholar]
- 17.Goldman S, Kim A, Meng C, et al. Heart rate variability (HRV) from a 10-second electrocardiogram (ECG) in Parkinson's disease and control. Neurology. 2018;90 [Google Scholar]
- 18.Singleton A, Gwinn-Hardy K, Sharabi Y, et al. Association between cardiac denervation and parkinsonism caused by alpha-synuclein gene triplication Brain 127768–7722004 [DOI] [PubMed] [Google Scholar]
- 19.Goldstein DS, Holmes C, Lopez GJ, et al. Cardiac sympathetic denervation predicts PD in at-risk individuals Parkinsonism Relat Disord 5290–932018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pellicano C, Benincasa D, Pisani V, et al. Prodromal non-motor symptoms of Parkinson's disease Neuropsychiatr Dis Treat 3145–1522007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alonso A, Huang X, Mosley TH, et al. Heart rate variability and the risk of Parkinson disease: The Atherosclerosis Risk in Communities study Ann Neurol 77877–8832015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akbilgic O, Kamaleswaran R, Mohammed A, et al. Electrocardiographic changes predate Parkinson's disease onset. Scientific Rep. 2020;10:11319. doi: 10.1038/s41598-020-68241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Attia ZI, Kapa S, Lopez-Jimenez F, et al. Screening for cardiac contractile dysfunction using an artificial intelligence-enabled electrocardiogram Nat Med 2570–742019 [DOI] [PubMed] [Google Scholar]