Abstract
We have isolated the RPN9 gene by two-hybrid screening with, as bait, RPN10 (formerly SUN1), which encodes a multiubiquitin chain receptor residing in the regulatory particle of the 26S proteasome. Rpn9 is a nonessential subunit of the regulatory particle of the 26S proteasome, but the deletion of this gene results in temperature-sensitive growth. At the restrictive temperature, the Δrpn9 strain accumulated multiubiquitinated proteins, indicating that the RPN9 function is needed for the 26S proteasome activity at a higher temperature. We analyzed the proteasome fractions separated by glycerol density gradient centrifugation by native polyacrylamide gel electrophoresis and found that a smaller amount of the 26S proteasome was produced in the Δrpn9 cells and that the 26S proteasome was shifted to lighter fractions than expected. The incomplete proteasome complexes were found to accumulate in the Δrpn9 cells. Furthermore, Rpn10 was not detected in the fractions containing proteasomes of the Δrpn9 cells. These results indicate that Rpn9 is needed for incorporating Rpn10 into the 26S proteasome and that Rpn9 participates in the assembly and/or stability of the 26S proteasome.
The ubiquitin-proteasome pathway is a major proteolytic system acting in various cellular processes (15). Proteins to be degraded by this pathway are first tagged with ubiquitins, except in one case so far (14), and multiubiquitin chains attached to the proteins are recognized by the 26S proteasome, which degrades the target proteins in an ATP-dependent manner and releases ubiquitins for repeated use. The ubiquitination machinery is a multicomponent system in which ubiquitin is activated by E1 enzyme (ubiquitin-activating enzyme), transferred to E2 enzymes (ubiquitin-conjugating enzymes) and then finally transferred to the target proteins (15). Depending upon the proteins to be ubiquitinated, E3 enzyme (ubiquitin ligase) is needed for the final step of ubiquitination. Since this proteolysis system resides intracellularly, the proteolytic activity must be strictly controlled, otherwise nonspecific degradation of cellular proteins may be hazardous to the cells. The selectivity of proteolysis and the temporal control of its execution are important for its proper function. How are the selectivity and timing of proteolysis controlled? One level of control is obviously at the step of ubiquitination, because the presence of multiple E2 and E3 enzymes and combinations of them contribute to the selection of a protein to be degraded.
As mentioned above, the ubiquitination step has been emphasized in the regulation of ubiquitin-mediated proteolysis whereas the 26S proteasome is believed to be constitutively active and has not attracted much attention as a regulatory molecule. However, according to recent progress in the structural analyses of the 26S proteasome (4, 11, 12), the specificity of proteolysis by the ubiquitin-proteasome pathway may well be modulated by the 26S proteasome. The 26S proteasome is a multicatalytic protease of about 2,000 kDa, and its structure is well conserved throughout eukaryotes (2, 25, 33, 39). It consists of two subcomplexes, the 20S proteasome and the 19S regulatory particle, attached to one or both ends of the 20S proteasome. In yeast, 14 genes encoding subunits, seven α and seven β subunits, of the 20S proteasome have been elucidated (4, 13). The structure of the yeast 20S proteasome was analyzed by X-ray crystallography (13), and it was found that the protease activity is sequestered inside the β ring and there is no opening on the α ring for protein substrates to get into the lumen of the 20S proteasome. For the activity of the 26S proteasome, the 19S regulatory particle plays crucial roles at the ends of the 20S proteasome; it binds a multiubiquitin chain to select the substrate and unfolds substrates so that they become accessible to the lumen of the 20S proteasome. According to the biochemical analyses of the regulatory component of the yeast 26S proteasome by Glickman et al. (12), the 19S regulatory particle is composed of 6 ATPases and 11 or more non-ATPase subunits. Furthermore, Glickman et al. (11) demonstrated that the 19S regulatory particle can be subdivided into two components, the lid and the base; the former consists of non-ATPases, and the latter consists of six ATPases and two non-ATPase subunits, Rpn1 and Rpn2.
In cell extracts, the 26S proteasome and subcomplexes of it are likely to be in an equilibrium. However, it is only poorly understood how subunits assemble into each subcomplex. Recently, Ramos et al. (26) found the UMP1 gene, which encodes a protein that is needed for proper assembly of the 20S proteasome. Interestingly, Ump1p becomes a substrate of the proteasomes when assembly of the 20S proteasome is completed. It can be assumed that there are proteins functioning as a chaperone to stimulate assembly of the lid or the base or both. Finding and analyzing such a protein may well provide clues to understand the regulation of the 26S proteasome-mediated proteolysis. Fortunately, the high conservation of components of the 26S proteasome throughout eukaryotes (5, 9, 10, 34, 38) enables us to exploit the yeast genetic system. In this study, we attempted two-hybrid screening by using the RPN10 gene encoding a yeast multiubiquitin receptor (21, 35) as bait to identify the protein(s) which interacts with Rpn10. Here we describe the isolation and characterization of the RPN9 gene, which encodes a nonessential component of the 26S proteasome. We found that Rpn9 exerts a novel function in the assembly or stability of the 26S proteasome and allows Rpn10 to be incorporated into the 26S proteasome.
MATERIALS AND METHODS
Strains and microbiological methods.
The principal Saccharomyces cerevisiae strains and plasmids used in this study are listed in Table 1. To obtain J106 and J107, we first integrated pJUN315 (see below) linearized with AflII at the RPT1 locus of W303-1A to replace the resident RPT1 gene with 6xHis-RPT1-URA3. The resulting transformant was crossed with J44 (MATα Δrpn9::LEU2), and the heterozygous diploid was dissected. Among the progeny, a Ura+ segregant (J106; 6xHis-RPT1-URA3) and a Ura+ Leu+ segregant (J107; 6xHis-RPT1-URA3 Δrpn9::LEU2) were saved for further study. Escherichia coli DH5α [endA1 gyrA96 hsdR17(rk− mk+) recA1 relA1 supE44 thi-1 deoR Δ(lacZYA-argF)U169 φ80lacZΔM15 F− λ−] was used for the propagation and construction of plasmids. YPD contained 2% glucose, 2% polypepton (Daigo Eiyo, Tokyo, Japan), and 1% yeast extract (Difco Laboratories, Detroit, Mich.). Synthetic medium (SD) was prepared by the recipe described by Sherman (30). SC is fully supplemented SD medium. Omission media were prepared by removing an appropriate nutrient from SC medium and designated, for example, SC − Ura for synthetic medium lacking uracil. Sporulation medium contained 1% potassium acetate. The permissive and restrictive temperatures for the temperature-sensitive mutants were 25 and 35 to 37°C, respectively. Yeast transformations were done by the method described by Ito et al. (19) and Schiestl and Gietz (29). Luria broth LB (pH 7.0) consisting of 1% Bacto Tryptone (Difco Laboratories), 0.5% Bacto Yeast Extract (Difco Laboratories), and 0.5% NaCl was used for growing E. coli cells. Ampicillin (50 μg/ml) was added as appropriate. Competent cells for bacterial transformations were prepared as described by Inoue et al. (18). Two-hybrid screening was carried out as described by Fields and Sternglanz (7).
TABLE 1.
Yeast strains and plasmids
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| L40 | MATa his3 leu2 trp1 URA3::lexA-lacZ LYS2::lexA-HIS3 gal80 ade | 36 |
| YK109 | MATa rpn12-1 leu2 his3 trp1 ura3 ade1 | 20 |
| KA31α | MATα leu2 his3 trp1 ura3 | Our stock |
| KA31D | MATa/MATα leu2/leu2 his3/his3 trp1/trp1 ura3/ura3 | Our stock |
| W303D | MATa/MATα leu2/leu2 his3/his3 trp1/trp1 ura3/ura3 ade2/ade2 can1/can1 | Our stock |
| W303-1A | MATa leu2 his3 trp1 ura3 ade2 can1 | Our stock |
| J33 | MATα Δrpn9::LEU2 leu2 his3 trp1 ura3 | This study |
| J38 | MATα Δrpn10::HIS3 leu2 his3 trp1 ura3 | This study |
| J44 | MATα Δrpn9::LEU2 leu2 his3 trp1 ura3 ade2 can1 | This study |
| J106 | MATa 6xHis-RPT1-URA3 leu2 his3 trp1 ura3 ade2 can1 | This study |
| J107 | MATa Δrpn9::LEU2 6xHis-RPT1-URA3 leu2 his3 trp1 ura3 ade2 can1 | This study |
| Plasmids | ||
| pACTII | PADH AD 2μm ori TRP1 | 7 |
| pBTM116 | PADH lexA 2μm ori LEU2 | 7 |
| pBTM-RPN10 | pBTM116-RPN10 | This study |
| pRS306 | URA3 | 31a |
| pKT10 | PTDH3 2μm ori URA3 | Our stock |
| TOp59 | PTDH3 2μm ori URA3 | This study |
| YCUp4 | CEN4 URA3 ARS1 | A. Fujita |
| pJUN180 | KS−-Δrpn9::LEU2 | This study |
| pJUN197 | YCUp4-RPN9 | This study |
| pJUN217 | GST-RPN9 fusion | This study |
| pJUN227 | pBluescript SK−-116a10 | This study |
| pJUN228 | pBluescript SK−-242c5 | This study |
| pJUN235 | pBluescript SK−-TO6D8.8 ORF | This study |
| pJUN238 | PTDH3-TO6D8.8 ORF 2μm ori URA3 | This study |
| pJUN315 | 6xHis-RPT1 (3′ truncated)-URA3 | This study |
| DP1 | 6xHis-RPT1 TRP1 CEN4 | D. Finley |
DNA manipulation.
The methods adopted in this study for engineering DNA were those described by Sambrook et al. (28). Yeast genomic DNA was isolated by the glass bead method described by Hoffman and Winston (16). Restriction endonucleases and DNA-modifying enzymes were purchased from Takara Shuzo (Kyoto, Japan) and Toyobo Biochemicals (Kyoto, Japan). Gene Clean II was from Bio 101, Inc. (Vista, Calif.).
Construction of plasmids.
The RPN9 gene was disrupted by inserting the LEU2 gene at the NcoI site encompassing codons 122 and 123 of the RPN9 open reading frame (ORF) (pJUN180), and one of the chromosomal RPN9 alleles of the diploid KA31D was replaced with the disrupted rpn9::LEU2 gene by one-step gene replacement (27). The correct disruption was confirmed by Southern hybridization. Tetrad dissection of this diploid gave rise to four viable spore clones, and the Leu+ phenotype segregated 2+:2− in every ascus, indicating that the RPN9 gene is not an essential gene. However, the disruptants showed temperature-sensitive growth. The disruptant with a complete deletion of the ORF of YDR427w was reported to be temperature sensitive for its growth (12, 17).
pJUN217, containing the GST-RPN9 fusion gene, was constructed and used for production of glutathione S-transferase (GST)-Rpn9 fusion protein, which was used as the antigen to immunize rabbits. The RPN9 ORF (codons 1 to 393) was amplified by PCR with a pair of primers, 5′-gggggggatccaccacattatatttcgc-3′ (a forward primer) and 5′-gggggagatctacaacccagatggattggc-3′ (a reverse primer). Amplified DNA was cut with BamHI and BglII and ligated at the BamHI site of pBluescript KS−, and a plasmid whose BglII-BamHI junction is situated nearer to the EcoRI site was selected. The BamHI-EcoRI fragment containing the RPN9 ORF was excised from this plasmid and ligated into the gap of BamHI-EcoRI of pGEX-5x-3 (Pharmacia Biotech, Uppsala, Sweden), resulting in pJUN217, containing the GST-RPN9 gene.
pJUN238 expressing the Caenorhabditis elegans TO6D8.8 ORF in yeast was constructed as follows. Two λ cosmid clones containing cDNA of C. elegans, yk116a10 and yk242c5, were donated by Y. Kohara (National Institute of Genetics, Mishima, Japan). SK plasmid clones, SK-116a10 (pJUN227) and SK-242c5 (pJUN228), were recovered from each of the cosmids. The complete ORF was successfully reconstructed from these two incomplete but complementary clones. In brief, the 5′ portion of the ORF was excised from pJUN228 as the XbaI (in SK sequence)-ClaI (in the ORF) fragment and inserted into the XbaI-ClaI gap of pJUN227, resulting in pJUN235, which contains the full length of the TO6D8.8 ORF. The SmaI-XhoI fragment excised from pJUN235, which contains the full ORF, was inserted in the PvuII-XhoI gap of vector TOp59 to be expressed under the TDH3 promoter.
To construct pJUN315, 3′-terminally truncated 6xHis-RPT1 excised as a HindIII-BglII fragment from DP1 was inserted at the HindIII-BamHI gap of pRS306.
Detection of multiubiquitinated proteins.
The heat block method was used to prepare yeast lysate (21). In brief, cells were cultured in YPD medium at 25°C to an optical density at 600 nm (OD600) of 1.0, and then transferred to 37°C. Cells corresponding to 5.0 OD600 units were periodically harvested and washed with deionized water once. The pellet was suspended in 100 μl of lysis buffer A (phosphate-buffered saline with 1 μg each of leupeptin, pepstatin A, antipain, and aprotinin per ml and 1 mM phenylmethylsulfonyl fluoride), heated at 97°C for 10 min, and then vortexed and heated for 30 s each. The vortexing and heating were repeated six times. A 25-μl volume of 5x Laemmli sampling buffer (22) was added to the lysate, and the resultant mixture was heated for 10 min at 97°C and centrifuged for 15 min at 15,000 rpm (TOMY MR-150 centrifuge) at 4°C. A 15-μl volume of supernatant was loaded onto a sodium dodecyl sulfate (SDS)–7.5% polyacrylamide gel. FK1 monoclonal antibody against multiubiquitin (8) was used as the primary antibody.
Density gradient centrifugation.
Cells harvested from a 1-liter culture when it attained an OD600 of 1.0 were washed with deionized water and resuspended in 1.0 ml of lysis buffer B (25 mM Tris-HCl [pH 7.5], 2 mM ATP, 1 mM dithiothreitol [DTT], 1 μg each of leupeptin, pepstatin A, antipain, and aprotinin per ml, 1 mM phenylmethylsulfonyl fluoride). The cells were disrupted by vortexing with glass beads for 10 min at 4°C. Insoluble material was removed by centrifugation at 8,000 rpm for 30 min at 4°C. Supernatant (5 mg of protein/ml of extract) was divided in half; one half was incubated with 5 mM MgCl2 and the other half was incubated without MgCl2 for 30 min at the indicated temperature. Then, 1 ml of extract was loaded on 35 ml of 10 to 40% glycerol density gradient that had been made by Gradient Mate (Towakagaku, Tokyo, Japan) and centrifuged at 25,000 rpm in a SW28 rotor with the L8-55 Ultracentrifuge (Beckman) for 22 h at 4°C. Fractions (1 ml) were collected by puncturing the bottom of the tube. In some experiments, cell extract was prepared with buffer C (buffer B containing 5 mM MgCl2) and preincubation of extract before glycerol density gradient was omitted.
Biochemical methods.
The protein concentration was determined by the method described by Bradford (3). Peptidase activity was assayed by using fluorogenic succinyl-Leu-Leu-Val-Tyr-4-methyl-coumaryl-7-amide (Suc-LLVY-MCA) as a substrate. Suc-LLVY-MCA (0.1 mM) was incubated with an enzyme source for 60 min at 37°C in the presence or absence of 0.05% SDS in 100 mM Tris-HCl (pH 8.0) as described previously (32). The reaction was stopped by adding 100 μl of 10% SDS and 2 ml of 100 mM Tris-HCl (pH 9.0), and the fluorescence at 460 nm of the reaction products was measured with excitation at 380 nm.
Pull-down experiments with the 6xHis-RPT1 strains.
6xHis-Rpt1 was pulled down from high-speed supernatant (see below) with Ni-nitrilotriacetic acid (NTA) agarose beads (Qiagen, Valencia, Calif.) as specified by the manufacturer.
Immunological methods.
Anti-Rpn9 antibody was raised as follows. GST-Rpn9 fusion protein was induced in E. coli DH5α (pJUN217) by incubation with 2 mM isopropylthiogalactoside for 4 h at 37°C. Gst-Rpn9 fusion protein produced as insoluble protein was separated from soluble proteins and purified by SDS-polyacrylamide gel electrophoresis (PAGE). The band containing GST-Rpn9 fusion protein was excised, and the fusion protein was eluted by electrophoresis. Purified GST-Rpn9 fusion protein was injected into rabbits to raise anti-GST-Rpn9 antibody. Antiserum containing anti-Rpn9 antibody was passed through a GST-Sepharose column to remove anti-GST antibody. Anti-Rpn9 antibody in the pass-through fraction was further purified on a protein A column (Pharmacia Biotech). The following antibodies were described previously: anti-Rpn12 antibody (20), anti-Rpn10 antibody (21), anti-20S proteasome antibody (32), anti-Rpt1 peptide antibody (31), anti-rabbit immunoglobulin G (IgG) goat antibody conjugated with alkaline phosphatase or horseradish peroxidase (Promega Corp., Madison, Wis.), anti-actin C4 monoclonal antibody (Boehringer Mannheim), anti-mouse IgG goat antibody conjugated with horseradish peroxidase (Promega) and anti-multiubiquitin chain monoclonal antibody (FK1) (8). The chemiluminescence reagent for Western blot was from DuPont NEN (Boston, Mass.).
Immunoprecipitation experiments.
Polyclonal antibody against the 20S proteasome and nonimmune rabbit IgG (60 μg each in 40 μl of buffer H containing 100 mM Tris-HCl [pH 7.6], 2 mM ATP, 2 mM MgCl2, 0.5 mM EDTA, and 2% glycerol) were mixed with protein A-Sepharose beads and mixtures were rotated at 4°C for 2 h. The beads were then treated with skim milk in buffer H, washed three times with buffer H, and added to the indicated sample. After the mixtures were rotated at 4°C for 2 h, supernatant (40 μl) was recovered by centrifugation and mixed with sample buffer (20 μl) for SDS-PAGE, while the resulting beads were washed three times with buffer H and suspended in sample buffer (60 μl). A 20-μl volume each of supernatant and bead suspension were subjected to SDS-PAGE in a slab gel containing 12.5% polyacrylamide followed by Western analysis. For Western blot analysis, the separated proteins were electrically transferred to a polyvinylidene difluoride filter (Millipore, Bedford, Mass.). Then, the filter was processed for Western blotting as recommended by the manufacturer.
Native PAGE.
The procedures for preparation of a native acrylamide gel and for electrophoresis were described by Glickman et al. (12). All procedures were performed at 4°C. A native gel contained 0.18 M Tris-borate (pH 8.3), 5 mM MgCl2, 1 mM ATP, 1 mM DTT, and 4% acrylamide-bisacrylamide (at a ratio of 37.5:1) polymerized with 0.1% N,N,N′,N′-tetramethylethylenediamine (TEMED)–0.2% ammonium persulfate. Running buffer contained 0.18 M Tris-borate (pH 8.3), 5 mM MgCl2, 1 mM ATP, and 1 mM DTT. A 20-μl volume of alternate fraction from fractions 15 to 25 was loaded on the native gel. To carry out Western blotting analysis, 100 μl of sample was concentrated with a Millipore spin column (Ultrafree-MC) to one-fifth of the original volume and loaded. Electrophoresis was performed at 15 mA for 3.6 h. The overlay assay of peptidase activity of proteasomes was described by Glickman et al. (12). In brief, a native polyacrylamide gel, after electrophoresis, was overlaid with 2 ml of reaction mixture containing Suc-LLVY-MCA with or without 0.05% SDS and incubated at room temperature for 10 min. Peptidase activity was visualized by irradiating the gel with 380-nm UV light.
RESULTS
Isolation and characterization of the RPN9 gene.
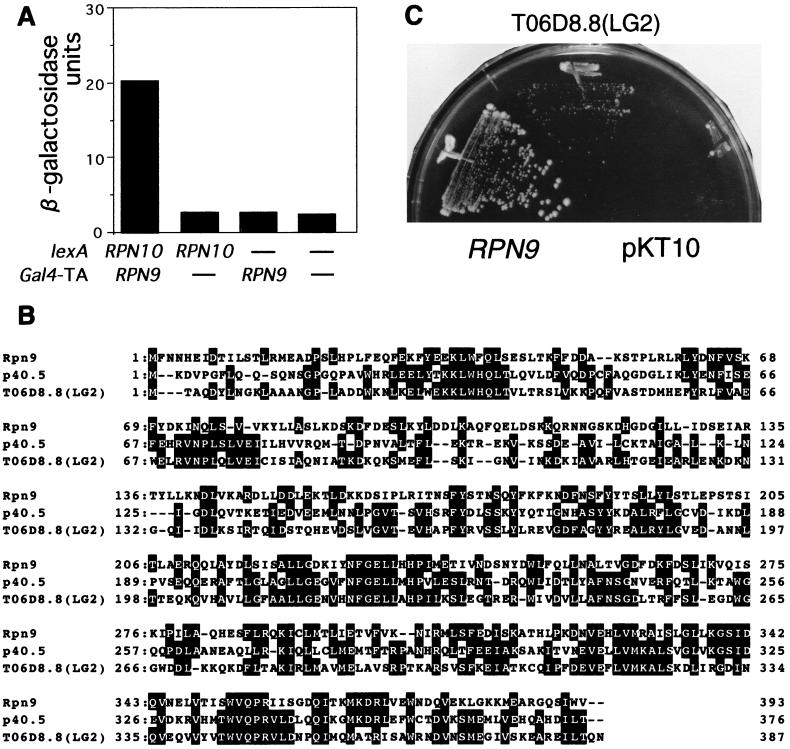
To identify protein interacting with a component of the 26S proteasome, we have been attempting two-hybrid screening with a lexA-RPN gene fusion as bait. Here, we performed two-hybrid screening with the lexA-RPN10 gene as bait. By surveying approximately 30,000 colonies, each carrying the bait pBTM-RPN10 and plasmid of a library, we found one positive clone, pACTII-RPN9 (Fig. 1A). Partial sequencing at the cloning junction revealed the RPN9 gene, which encodes a protein consisting of 393 amino acid residues and shows homology to the C. elegans gene encoding the hypothetical protein TO6D8.8 (Fig. 1B) and to a gene, p40.5, encoding a subunit of the human 26S proteasome (17) (Fig. 1B). We confirmed that the RPN9 gene is a nonessential gene and that the null mutant showed temperature-sensitive growth. To examine whether the homology between Rpn9 and TO6D8.8 is biologically significant, the complete ORF encoding TO6D8.8 was regenerated by using two cDNA clones, yk116a10 and yk242c5, and expressed in the Δrpn9 cells (see below) under a strong promoter of yeast, the TDH3 promoter. Expression of the C. elegans gene partially complemented temperature-sensitive growth of the Δrpn9 strain (Fig. 1C), indicating that the C. elegans ORF TO6D8.8 encodes a functionally homologous gene to the RPN9 gene.
FIG. 1.
Characterization of the RPN9 gene. (A) Two-hybrid interaction. The extent of the two-hybrid interaction between the bait (pBTM-RPN10) and the fish (pACTII-RPN9) was estimated by assaying β-galactosidase activity, which was expressed as Miller units (24). Each transformant was grown in SC − (Trp + Leu) medium at 25°C overnight, and cells were harvested at OD600 = 1 and subjected to an enzyme assay as described by Miller (24). Dashes indicate the empty vectors. (B) Alignment of the amino acid sequences of Rpn9, the p40.5 subunit of the human 26S proteasome, and the C. elegans ORF TO6D8.8 (LG2). Identical amino acids are highlighted. Gaps were introduced to attain the highest matching. (C) Complementation. The C. elegans ORF TO6D8.8 was reconstructed from two cDNAs (yk116a10 and yk242c5) as described in Materials and Methods and was fused to the TDH3 promoter (pJUN238). pJUN238 containing PTDH3-TO6D8.8 ORF [a section labeled TO6D8.8(LG2)], pJUN197 containing the RPN9 gene (a section labeled RPN9), and the vector (a section labeled pKT10) were separately introduced into the Δrpn9 strain (J33). One representative transformant from each transformation experiment was streaked across a YPD plate. The plate was incubated at 35.5°C for 7 days.
Rpn9 is a component of the yeast 26S proteasome.
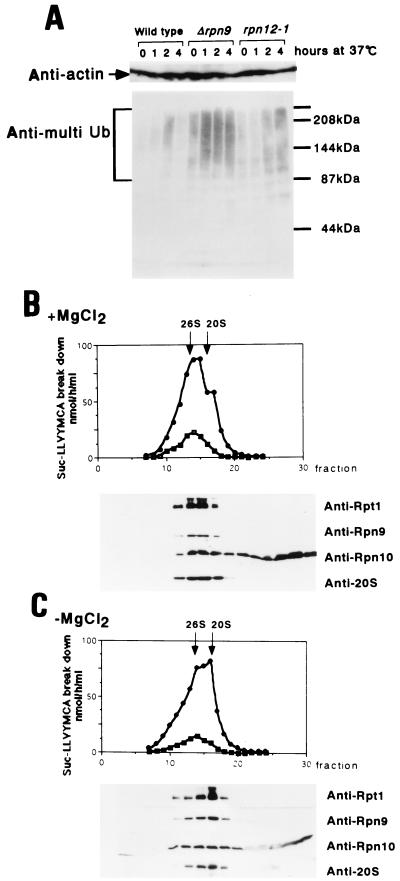
Since RPN9 was identified as a gene whose product interacts with Rpn10, a component of the 26S proteasome, we anticipated some roles of Rpn9 in ubiquitin-mediated proteolysis. This consideration prompted us to examine whether Rpn9 participates in degradation of multiubiquitinated proteins. Wild-type strain KA31α, Δrpn9 strain J33, and the rpn12-1 (formerly nin1-1) strain YK109 (20) were grown at 25°C to the mid-logarithmic phase and then shifted to 37°C. At the indicated time points, a portion of each culture was harvested and subjected to detection of multiubiquitinated proteins as described previously (20). As shown in Fig. 2A, the Δrpn9 strain accumulated a large amount of multiubiquitinated proteins at the restrictive temperature, as did the rpn12-1 strain, indicating that the 26S proteasome function is defective in the Δrpn9 cells at the restrictive temperature. Accumulation of a small amount of multiubiquitinated proteins was seen in the wild-type strain after 2 h of incubation at 37°C; however, these proteins disappeared during further incubation.
FIG. 2.
Rpn9 is a component of the 26S proteasome. (A) Accumulation of multiubiquitinated proteins. KA31α (wild type), J33 (Δrpn9), and YK109 (rpn12-1) cells were grown in YPD at 25°C to the mid-logarithmic phase and then shifted to 37°C. At the indicated time after the shift, cells corresponding to 5 OD600 units were harvested and disintegrated by vortexing with glass beads (20). Proteins were separated by electrophoresis in an SDS–7.5% polyacrylamide gel, and the multiubiquitin chain was detected by Western blotting with a anti-multiubiquitin chain (Anti-multi Ub)-specific monoclonal antibody, FK1 (8). Actin was detected with C4 monoclonal antibody as an internal reference. The positions of the size markers are shown on the right. (B and C) Glycerol density gradient centrifugation. The wild-type yeast KA31α cells grown exponentially in YPD at 25°C were collected from a 1-liter culture. Extract was prepared as described by Kominami et al. (20) and preincubated at 30°C for 30 min with (B) and without (C) ATP-MgCl2. Peptidase activity assayed in the presence (circles) or absence (squares) of 0.05% SDS is shown at the top; Western blotting with anti-Rpt1, anti-Rpn10, anti-Rpn9, and anti-20S proteasome antibodies is shown at the bottom. Fractions are numbered from the bottom to the top.
Next, we examined whether Rpn9 is a component of the 26S proteasome by glycerol density gradient centrifugation followed by Western blotting with antibodies against several components of the 26S proteasome. Extract prepared from the wild-type strain was preincubated at 30°C for 30 min with or without ATP-Mg2+, under conditions which should promote either assembly or disassembly, respectively, of the 26S proteasome, followed by glycerol density gradient centrifugation. Preincubation without ATP-Mg2+ promotes dissociation of the 26S proteasome into the 20S proteasome and the 19S regulatory particle in vitro (Fig. 2C). Under these conditions, Rpn9 cosedimented around the 20S region with Rpt1, an authentic subunit of the base component of the 19S regulatory particle. On the other hand, when extract was preincubated with ATP-Mg2+ (Fig. 2B), Rpn9 and Rpt1 moved to the 26S proteasome fractions. The behavior of Rpn10 in the gradient was quite different from that of other subunits, in that some Rpn10 did exist in the 19S regulatory complex (Fig. 2C) and in the 26S proteasome (Fig. 2B) but the majority was detected in lighter fractions, as had been described by van Nocker et al. (35).
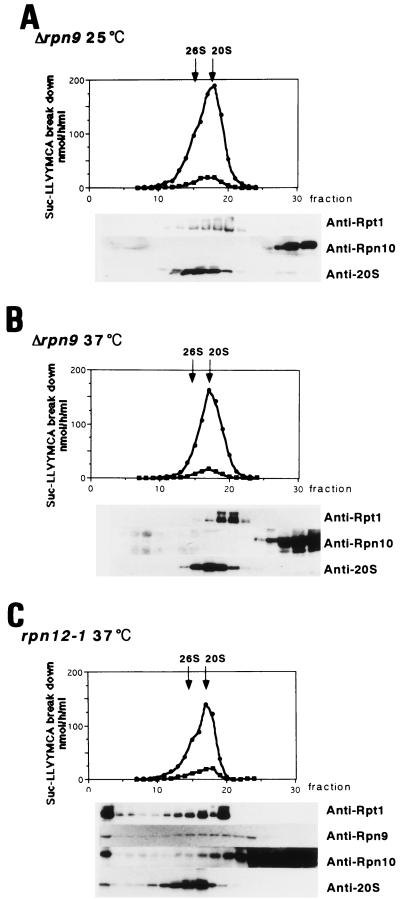
The 26S proteasome in the Δrpn9 cells.
As mentioned above, Δrpn9 cells show temperature-sensitive growth. The logarithmic-phase culture of the Δrpn9 cells stopped growing 4 h after a shift to 37°C. Since it is well established that the 26S proteasome function is indispensable for yeast growth, the functional 26S proteasome is likely to be produced in the absence of Rpn9. To test this possibility, extracts were prepared from the Δrpn9 cells grown at 25 and 37°C, preincubated with ATP-Mg2+ for 30 min at 30°C to stimulate reconstruction of the 26S proteasome, and then subjected to analysis by glycerol density gradient centrifugation (Fig. 3A and B). Unexpectedly, the profiles of proteasomes and of peptidase activity in the gradients were similar irrespective of the growth temperature. The 26S proteasome peak was not clearly seen in either centrifugation profile. Furthermore, Rpt1 was found in a large protein complex, and the peak of the 20S proteasome was distributed in the gradient at a denser position than that of Rpt1. This profile is in clear contrast to that shown in Fig. 2B, where peaks of Rpt1 and the 20S proteasome are superimposed. Furthermore, Western blot analysis with anti-Rpn10 antibody gave rise to a surprising result; Rpn10 was not detected in the proteasome fractions, whereas Rpt1 was. In contrast, extract prepared from the rpn12-1 cells grown at 37°C for 4 h did have Rpn10 in the regulatory complex and the 26S proteasome (Fig. 3C).
FIG. 3.
The proteasomes in the Δrpn9 cells. J33 (Δrpn9) cells were grown at 25°C in 1 liter of YPD to mid-logarithmic phase, and half of the culture was shifted to 37°C (B) and the other half was incubated at 25°C (A) for 4 h. Cell extracts prepared with buffer B from each culture were incubated at 30°C for 30 min in the presence of 5 mM MgCl2–2 mM ATP and analyzed by glycerol density gradient centrifugation as described in the text. Peptidase assay and immunoblotting were done as described in the legend to Fig. 2. (C) The rpn12-1 strain grown at 25°C was shifted to 37°C and incubated for 4 h. Then, extract prepared as described above was treated with 5 mM MgCl2–2 mM ATP at 30°C for 30 min and subjected to glycerol density gradient centrifugation.
To further examine whether the 26S proteasome is present in the Δrpn9 cells, extracts were prepared from wild-type cells and Δrpn9 cells grown in YPD at 25°C for 24 h. Each extract was treated with anti-20S proteasome antibody, and the resulting immunoprecipitates were analyzed by Western blotting with anti-Rpn10 and anti-Rpn12 antibodies. As shown in Fig. 4, a smaller amount of Rpn12 was detected in the immunoprecipitates from the Δrpn9 cells than from the wild-type cells whereas comparable amounts of the 20S proteasome were detected in the two extracts. Furthermore, Rpn10 was not detected in the immunoprecipitates of the Δrpn9 cells.
FIG. 4.
Estimation of the quantity of the 26S proteasome in cell extract. The KA31α (wild type) and J33 (Δrpn9) strains were each grown in 40 ml of YPD at 25°C for 24 h with shaking. Cells were harvested, resuspended in homogenization buffer H (100 mM Tris-HCl [pH 7.6], 2 mM ATP, 2 mM MgCl2, 0.5 mM EDTA, 100 mM NaCl, 2% glycerol, 0.5 mM diisoprppylfluorophosphate, 1.5 μg of pepstatin A per ml), and disrupted with glass beads for 15 min at 4°C. Extract was centrifuged at 20,000 × g for 15 min at 4°C followed by high-speed centrifugation at 100,000 × g for 20 min at 4°C. The resulting supernatant, equivalent to 5 mg of protein, was immunoprecipitated with 20 μl (6.5 mg of IgG/ml) of anti-20S proteasome antibody conjugated to protein A-Sepharose beads (designated 20S) or with nonimmune rabbit IgG conjugated to protein A-Sepharose beads as control (designated C at the description of the immunoprecipitation experiments). The resulting immunoprecipitates were subjected to SDS-PAGE followed by Western blot analysis with anti-Rpn12 antibody (A), anti-Rpn10 antibody (B), or anti-20S proteasome antibody (C). IP, immunoprecipitate; Blot, Western blotting. The positions of the size markers are shown on the right.
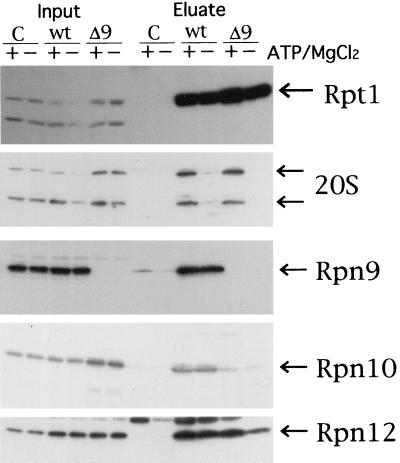
To confirm the above result that Rpn10 is missing from proteasomes produced in the Δrpn9 cells, we examined whether Rpn10 is coprecipitated with 6xHis-Rpt1 by Ni-NTA agarose beads. High-speed supernatant was prepared from a log-phase culture of each of J106, J107, and W303-1A. The same amount of high-speed supernatant (4 mg of protein) was subjected to the pull-down experiment with Ni-NTA agarose as described in Materials and Methods. High-speed supernatant and eluate from Ni-NTA agarose was analyzed by SDS-PAGE followed by Western blotting with anti-Rpt1, anti-Rpn10, anti-20S proteasome, anti-Rpn9, and anti-Rpn12 antibodies (Fig. 5). All the subunits detected in this experiment were present in similar amounts in extract (lanes labeled Input). A comparable amount of the 20S proteasome was coprecipitated with 6xHis-Rpt1 from J106 and J107 extracts prepared in the presence of ATP plus MgCl2 but not from extracts prepared in the absence of ATP plus MgCl2. On the other hand, comparable amounts of components of the regulatory particle, such as Rpn9, Rpn10, and Rpn12, were coprecipitated with the His tag irrespective of the presence of ATP and MgCl2 in the extract of J106. Rpn10 was not detected in the eluate from Ni-NTA agarose beads incubated with J107 extract. The results shown in Fig. 4 and 5 are consistent with those obtained by the glycerol density gradient centrifugation experiments (Fig. 3).
FIG. 5.
Pull-down experiment with 6xHis-Rpt1. J106 (6xHis-RPT1-URA3), J107 (6xHis-RPT1-URA3 Δrpn9::LEU2), and W303-1A (no His tag) were grown in 100 ml of YPD at 25°C. Cells were harvested from a 50-ml culture at OD600 = 1.0, resuspended in 300 μl of buffer D (100 mM Tris [pH 7.5], 10% glycerol, 1 mM DTT, 5 mM MgCl2, 2 mM ATP) or buffer E (buffer D without both MgCl2 and ATP) and disrupted by vortexing with glass beads for 10 min at 4°C. Homogenates were centrifuged at 15,000 rpm for 15 min (TOMY MR-150). Supernatant was collected and centrifuged at 100,000 × g for 20 min in a Beckman TLA-100.2 rotor. Then 4 mg of protein from each sample was gently mixed with 60 μl of slurry of 50% Ni-NTA agarose beads at 4°C. After 2 h of mixing, Ni-NTA agarose beads incubated with high-speed supernatants prepared in the presence (+) or absence (−) of ATP and MgCl2 were washed with buffer F (buffer D containing 100 mM NaCl and 10 mM imidazole) or buffer G (buffer E containing 100 mM NaCl and 10 mM imidazole), respectively. Then 40 μl of phosphate-buffered saline containing 1 M imidazole was added to elute proteins bound with the Ni-NTA agarose beads. The resulting eluate (Eluate), along with high-speed supernatant (Input), was analyzed by SDS-PAGE followed by Western blotting with antibody against the subunit shown on the right side. Lanes: C, W303-1A; wt, J106; Δ9, J107.
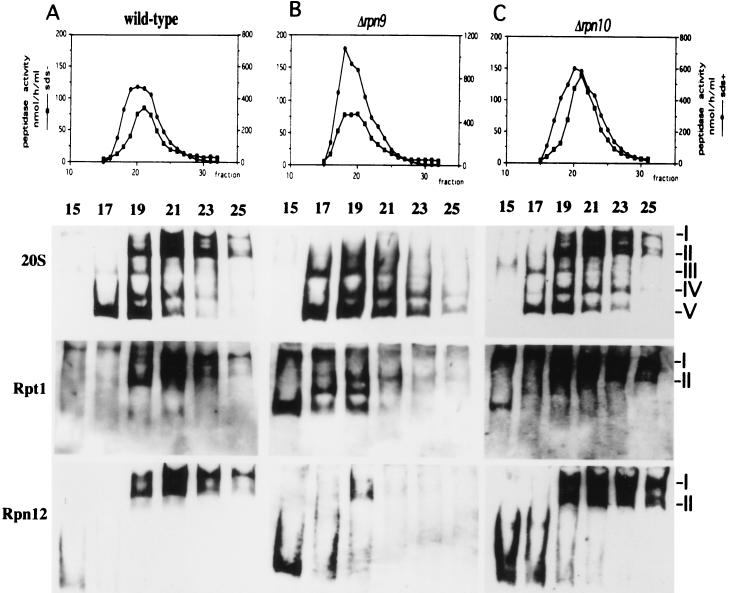
Proteasome species separated by native PAGE.
Intracellular proteasomes most probably exist in a mixture of molecular species including the 20S proteasome, the 26S proteasome, and proteasomes with intermediate sizes, which can hardly be separated by glycerol density gradient centrifugation. To separate molecular species of proteasomes, we adopted nondenaturing PAGE (native PAGE) to analyze fractions separated by glycerol density gradient centrifugation (Fig. 6). In the following experiments, the preincubation of extracts before glycerol density gradient centrifugation was omitted to avoid possible artifacts which might be caused by preincubation at 30°C. Separated proteins were blotted to a nitrocellulose filter and then subjected to Western blotting with anti-20S proteasome, anti-Rpn12, and anti-Rpt1 antibodies. When the sample of the wild-type cell lysate (Fig. 6A) which was prepared with buffer containing ATP and MgCl2 was analyzed with anti-20S proteasome antibody, five major bands were obtained. The band with the highest mobility (band V) corresponds to the 20S proteasome, and the two bands with the lowest mobilities, I and II, correspond to the symmetric and asymmetric forms of the 26S proteasome, respectively, because they were detected by anti-Rpt1 antibody as well as by anti-Rpn12 antibody. Two bands, III and IV, which react with anti-20S proteasome and anti-Rpt1 antibodies appeared between the bands of 20S and the 26S proteasomes. Because these two molecular species were not detected by anti-Rpn12 antibody, it is likely that they do not contain the lid and that they are asymmetric (band IV) and symmetric (band III) forms of the 20S proteasome with one base and two bases, respectively. Extract prepared from the wild-type cells in buffer without MgCl2 buffer gave rise to a similar result described above (data not shown). We used these five bands, which were detected in the wild-type sample, as references of molecular species of the proteasomes. It should be noted that there is no band which reacts with both anti-Rpt1 antibody and anti-Rpn12 antibody but does not react with anti-20S proteasome antibody.
FIG. 6.
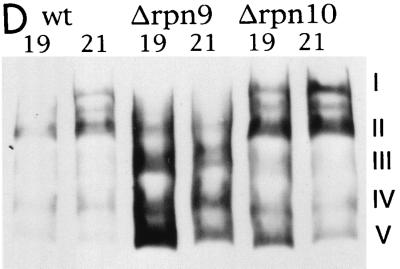
Molecular species of the proteasomes separated by native PAGE. (A to C) Cell extracts were prepared in buffer C from the wild-type strain (KA31α) (A), the Δrpn9 strain (J33) (B), and the Δrpn10 strain (J38) (C), as described in Materials and Methods. Each extract was fractionated by glycerol density gradient centrifugation, and fractions 15 to 25 were loaded onto native PAGE and analyzed by immunoblotting with anti-20S proteasome antibody (top row), anti-Rpt1 antibody (middle row), or anti-Rpn12 antibody (bottom row). (D) Comparison of proteasome species in peak fractions separated by glycerol density gradient centrifugation. Fractions 19 and 21 from each gradient shown in panels A to C were separated by native PAGE followed by Western blotting with anti-20S proteasome antibody. Fractions are numbered from the top to the bottom of the gradient. The categories of molecular species of proteasomes are shown on the right (see the text for details).
To analyze the proteasome species produced in the Δrpn9 cells, extract was prepared from the Δrpn9 cells and fractionated without preincubation by glycerol density gradient centrifugation. Fractions containing the proteasomes were subjected to native PAGE followed by Western blotting with anti-20S proteasome antibody. As in the previous experiment with wild-type extract, five major bands were detected; however, the band pattern was quite different from that seen in Fig. 6A. In the Δrpn9 extract (Fig. 6B), the amount of a symmetric form containing the base (band III) increased (see the blot with anti-20S proteasome antibody and the blot with anti-Rpt1 antibody). Figure 6B also shows that the amount of the 26S proteasome decreased in the Δrpn9 extract. The change in the 26S proteasome of the Δrpn9 cells became more evident when the blotting was performed with anti-Rpn12 antibody; the top two bands corresponding to the 26S proteasome were detected weakly, and they shifted to slightly lighter fraction (fraction 19), whereas the 26S proteasome of the wild-type cells was found in fractions 19, 21, 23, and 25. These are the reasons why the 26S proteasome peak was not seen at 26S in glycerol density gradient centrifugation of the Δrpn9 extract. In Fig. 6B, a new complex which reacts with anti-Rpt1 antibody but not with anti-Rpn12 or anti-20S proteasome antibody was detected in fraction 15. This complex migrates to a similar position to band II but is clearly different from it, because this complex does not react with anti-20S proteasome antibody but band II does. Fraction 15 in Fig. 6A to C also contains a protein complex containing Rpn12 but not Rpt1, most probably the lid.
The fact that the proteasomes produced in the Δrpn9 cells are missing Rpn10 prompted us to examine molecular species of the proteasomes in the Δrpn10 strain. Extract was prepared from the Δrpn10 cells and fractionated by glycerol density gradient centrifugation, and then fractions containing the proteasomes were subjected to native PAGE. As shown in Fig. 6C, the molecular species of the proteasomes in the Δrpn10 cells are similar to those seen in the wild-type cells.
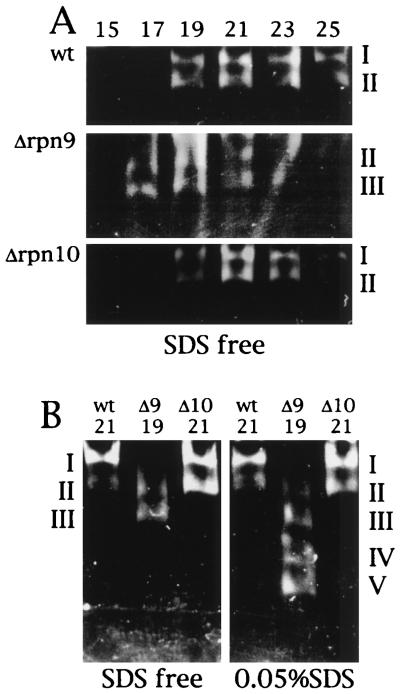
To provide a better comparison between patterns of separation of proteasome species on native PAGE, peak fractions of each extract separated by glycerol density gradient centrifugation were analyzed in the same gel followed by Western blotting with anti-20S proteasome antibody (Fig. 6D). Molecular species of proteasomes produced by the wild-type strain were identical to those produced by the Δrpn10 strain, whereas fast-moving species were accumulated in the Δrpn9 strain. Differences in proteasome species between the wild-type and Δrpn9 strains were also demonstrated by an in-gel assay of peptidase (Fig. 7). The indicated fractions of the glycerol density gradient centrifugation were separated by native PAGE, and peptidase activity was assayed by the gel overlay method with a reaction mixture without 0.05% SDS (Fig. 7A). The peptidase activities of the wild-type and Δrpn10 strains were seen at the position of the 26S proteasome, peaking at fraction 21, whereas the peak displayed by the Δrpn9 strain moved to a lighter fraction, peaking at fraction 19. The peak fractions were loaded on two native polyacrylamide gels. After electrophoresis, peptidase activity was assayed in the reaction mixture with or without 0.05% SDS. In the reaction mixture without SDS, the peptidase activity of the wild-type and Δrpn10 strains was seen at the position of the 26S proteasome whereas the peptidase activity of the Δrpn9 strain was seen at the position of a fast-moving species of proteasomes. When peptidase was assayed in the presence of SDS, peptidase activity of the wild-type and Δrpn10 strains was again seen at the position of the 26S proteasome but peptidase was activated at the fast-moving species of proteasomes in the sample of the Δrpn9 strain.
FIG. 7.
Gel overlay assay of peptidase activity of proteasomes separated by native PAGE. (A) Fractions 15 to 25 from the experiment in Fig. 6 were separated by native PAGE. Peptidase activity was assayed by the gel overlay method with the reaction mixture without 0.05% SDS. Activity was visualized by irradiating the gel with UV light (380 nm). (B) Peak fractions of peptidase activity in the gradient (Fig. 6) were separated by native PAGE. Peptidase activity was assayed by the gel overlay method with the reaction mixture with 0.05% SDS or without SDS (denoted SDS free). wt, KA31α (wild type); Δ9, J33 (Δrpn9); Δ10, J38 (Δrpn10).
Altogether, there are four remarkable features of proteasomes produced in the Δrpn9 cells: (i) proteasomes with an intermediate size and the 20S proteasome are accumulated, (ii) the amount of the 26S proteasome was decreased, (iii) the 26S proteasome migrated slightly slowly in glycerol density gradient centrifugation, and (iv) Rpn10 was not incorporated into the 26S proteasome.
DISCUSSION
The YDR427w ORF was found to encode a component of the yeast 26S proteasome by two groups independently (12, 17). We obtained the YDR427w gene, now designated RPN9, by two-hybrid screening with the RPN10 gene as bait and found that the Δrpn9 mutant accumulated multiubiquitinated proteins at a restrictive temperature. Since it is well established that the 26S proteasome is necessary for yeast growth, we expected that the 26S proteasome would be present in the Δrpn9 cell extract. To test this idea, Δrpn9 extract was analyzed by glycerol density gradient centrifugation, and it was found that the 26S proteasome was not clearly seen in the gradient (Fig. 3). Since glycerol density gradient analysis may not be sensitive enough to detect a small amount of the 26S proteasome, we used the immunological method to detect the 26S proteasome in the Δrpn9 extract. As shown in Fig. 4, a component of the lid, Rpn12, was coprecipitated with the 20S proteasome from the Δrpn9 extract, although a smaller amount of Rpn12 was precipitated from Δrpn9 cell extract than that from wild-type cell extract, implying that the Δrpn9 cells possess the 26S proteasome in a reduced amount.
Since the peak of the 26S proteasome of the Δrpn9 cells was not well separated in glycerol density gradient centrifugation, it was necessary to characterize the molecular species of the proteasomes in a more sensitive way. We analyzed the fractions obtained by glycerol density gradient centrifugation by native PAGE followed by immunoblotting with three different antibodies, i.e., anti-20S proteasome antibody, anti-Rpt1 antibody, and anti-Rpn12 antibody, to detect the 20S proteasome, the base, and the lid, respectively. Proteasomes with an intermediate size, which correspond to the 20S proteasome with two bases, are abundant in the Δrpn9 cells (Fig. 6A and B). From this result, we suggest that Rpn9 is an important subunit to connect the lid with the base in vivo. Our claim at this point contradicts that made by Glickman et al. (11), in that they interpreted Rpn10 as a protein linking the lid and the base. Furthermore, immunoblotting of the native PAGE gel with anti-Rpn12 antibody revealed that the Δrpn9 cells do have the 26S proteasome, although in a reduced amount, and that the top two bands corresponding to the 26S proteasome were detected in slightly lighter fractions in the Δrpn9 extract. The fact that the 26S proteasome produced in the Δrpn9 cells lacks Rpn9 and Rpn10 may explain the reduction of the molecular weight of the 26S proteasome.
Glycerol density gradient centrifugation analysis demonstrated, to our surprise, that Rpn10 was not incorporated into the 19S regulatory particle and the 26S proteasome in the Δrpn9 cells (Fig. 3A and B). This result was reinforced by the experiment in Fig. 5. 6xHis-Rpt1 in high-speed supernatant from the Δrpn9 cells coprecipitated with the lid component Rpn12, whereas Rpn10 was not coprecipitated with 6xHis-Rpt1 from the same high-speed supernatant. This result indicates that the Rpt1 and Rpn12 form a complex, probably the regulatory particle, in the Δrpn9 extract. However, Rpn10 is not contained in the regulatory particle of the Δrpn9 cells although it is present in the extract. This result suggests that Rpn9 is necessary for Rpn10 to be incorporated into the 19S regulatory particle whereas other subunits such as Rpt1 are successfully accommodated in the regulatory particle without Rpn9. It should be noted that the protein complex containing Rpt1 in Δrpn9 cell extracts seems smaller than that in wild-type extracts (Fig. 2 and 3).
A difference in the spectrum of the molecular species of proteasomes among the wild-type, Δrpn9, and Δrpn10 strains is also evident by the gel overlay assay of peptidase of proteasomal fractions (Fig. 7). When the peak fractions of the 26S proteasome were compared, peptidase activity in the Δrpn9 sample was detected at fast-moving bands whereas in the wild-type and Δrpn10 samples, peptidase activities were found at the position of the 26S proteasome, suggesting that proteasomes are unstable in the absence of Rpn9.
The absence of Rpn10 in proteasomes produced in the Δrpn9 strain is consistent with the fact that RPN9 was isolated by a two-hybrid screening with RPN10 as bait. However, incorporation of Rpn10 into the 26S proteasome is not likely to be a sole function of Rpn9, because Δrpn10 cells grow like the wild-type cells do whereas Δrpn9 cells are temperature sensitive and because the profile of the proteasomes of Δrpn10 extract in glycerol density gradient centrifugation was similar to that of wild-type extract. This fact suggests that Rpn9 is an important subunit of the regulatory particle or that there is a subunit(s) of the 26S proteasome other than Rpn10 to be accommodated in the regulatory particle with the aid of Rpn9. Alternatively, since the 26S proteasome produced in Δrpn9 cells always misses Rpn9 and Rpn10, the simultaneous loss of these two subunits may not permit the 26S proteasome to be active at a higher temperature. To examine this possibility, the 26S proteasome missing only Rpn9 must be produced, but this approach is not possible at present.
The 26S proteasome produced in Δrpn9 cells is clearly different in size and subunit composition from that produced in the wild-type cells. In spite of such differences, the 26S proteasome in Δrpn9 cells retains its protease activity. For example, Sic1p was degraded efficiently in the Δrpn9 cells (our unpublished observation).
Assembly and disassembly of the 26S proteasome are promising targets of regulation of the 26S proteasome functions. The mammalian regulatory particle, PA700, was described as a 700-kDa multisubunit ATP-dependent proteasome activator (23), which corresponds to the 19S regulatory particle. It forms a complex with the 20S proteasome to produce a larger proteasome complex resembling the purified 26S proteasome in vitro. Furthermore, DeMartino et al. (6) found a new protein complex, a modulator (1), that functions as a PA700-dependent activator of the 20S proteasome. They found that the modulator was effective only in the presence of PA700 and the 20S proteasome and that a larger complex, probably the 26S proteasome, was produced only in the presence of three components, PA700, the 20S proteasome, and the modulator. The modulator consists of three subunits, p27, p42, and p50, the last two of which are ATPase components of PA700, and their yeast counterparts are known as Rpt4 and Rpt5, respectively. Recently, a yeast homologue of p27 was reported as Nas2p (37). However, a protein complex corresponding to such a modulator has not been described in yeast.
In vitro reconstruction of the 26S proteasome occurs in yeast extract in an ATP-dependent manner (20). This fact strongly suggests that yeast proteasomes undergo assembly and disassembly as in animal cells. The facts that the quantity of the 26S proteasome in the Δrpn9 cells is reduced and that a larger amount of proteasome species with an intermediate size was found in the Δrpn9 cells led us to believe that Rpn9 plays a key role in facilitating the assembly of the 26S proteasome or in stabilizing the structure of the 26S proteasome.
ACKNOWLEDGMENTS
We thank Y. Kohara (National Institute of Genetics, Mishima, Japan) for cDNA clones of C. elegans and D. Finley for the plasmid carrying the 6xHis-RPT1 gene.
This study was supported in part by the grants for scientific research from Monbusho and CREST (Core Research for Evolutional Science and Technology) of Japan Science and Technology Corporation. J.T. is a recipient of Fellowship of JSPS for Japanese Junior Scientists.
REFERENCES
- 1.Adams G M, Falke S, Goldberg A L, Slaughter C A, DeMartino G N, Gogol E P. Structural and functional effects of PA700 and modulator protein on proteasomes. J Mol Biol. 1997;273:646–657. doi: 10.1006/jmbi.1997.1334. [DOI] [PubMed] [Google Scholar]
- 2.Baumeister W, Walz J, Zuhl F, Seemiller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Coux O, Tanaka K, Goldberg A L. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 5.DeMarini D J, Papa F R, Swaminathan S, Ursic D, Rasmussen T P, Culbertson M R, Hochstrasser M. The yeast SEN3 gene encodes a regulatory subunit of the 26S proteasome complex required for ubiquitin-dependent pathway degradation in vivo. Mol Cell Biol. 1995;15:6311–6321. doi: 10.1128/mcb.15.11.6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeMartino G N, Proske R J, Moomaw C R, Strong A A, Song X, Hisamatsu H, Tanaka K, Slaughter C A. Identification, purification, and characterization of a PA700-dependent activator of the proteasome. J Biol Chem. 1996;271:3112–3118. doi: 10.1074/jbc.271.6.3112. [DOI] [PubMed] [Google Scholar]
- 7.Fields S, Sternglanz R. The two-hybrid system: an assay for protein-protein interactions. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- 8.Fujimuro M, Sawada H, Yokosawa H. Production and characterization of monoclonal antibodies specific to multiubiquitinated chains of polyubiquitinated proteins. FEBS Lett. 1994;349:173–180. doi: 10.1016/0014-5793(94)00647-4. [DOI] [PubMed] [Google Scholar]
- 9.Fujimuro M, Tanaka K, Yokosawa H, Toh-e A. Son1p is a component of the 26S proteasome of the yeast Saccharomyces cerevisiae. FEBS Lett. 1998;423:149–154. doi: 10.1016/s0014-5793(98)00084-2. [DOI] [PubMed] [Google Scholar]
- 10.Ghislain M, Udvardy A, Mann C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature (London) 1993;366:358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- 11.Glickman M H, Rubin D M, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried V A, Finley D. A subcomple of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 12.Glickman M H, Rubin D M, Fried V A, Finley D. The regulatory particle of the S. cerevisiae proteasome. Mol Cell Biol. 1998;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik H D, Huber R. Structure of 20S proteasome from yeast at 2.4A resolution. Nature (London) 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi S, Murakami Y, Matsufuji S. Ornithine decarboxylase antizyme: a novel type of regulatory protein. Trends Biochem Sci. 1996;21:27–30. [PubMed] [Google Scholar]
- 15.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 16.Hoffman C S, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 17.Hori T, Kata S, Saeki M, DeMartino G N, Slaughter C A, Takeuchi J, Toh-e A, Tanaka K. cDNA cloning and functional analysis of p28 (Nas6p) and p40.5 (Nas7p), two novel regulatory subunits, of the 26S proteasome. Gene. 1998;216:113–122. doi: 10.1016/s0378-1119(98)00309-6. [DOI] [PubMed] [Google Scholar]
- 18.Inoue H, Nojima H, Okayama H. High frequency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 19.Ito H, Fukuda K, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1993;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kominami K, DeMartino G N, Moomaw C R, Slaughter C A, Shimbara N, Fujimuro M, Yokosawa H, Hisamatsu H, Tanahashi N, Shimizu Y, Tanaka K, Toh-e A. Nin1p, a regulatory subunit of the 26S proteasome, is necessary for activation of Cdc28p kinase of Saccharomyces cerevisiae. EMBO J. 1995;14:3105–3115. doi: 10.1002/j.1460-2075.1995.tb07313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kominami K, Okura N, Kawamura M, DeMartino G N, Slaughter C A, Shimbara N, Chung C H, Fujimuro M, Yokosawa H, Shimizu Y, Tanahashi N, Tanaka K, Toh-e A. Yeast counterparts of subunits S5a and p58 (S3) of the human 26S proteasome are encoded by two multicopy suppressors of nin1-1. Mol Biol Cell. 1997;8:171–187. doi: 10.1091/mbc.8.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assemble of the head of bacteriphage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Ma C-P, Vu J H, Proske R J, Slaughter C A, DeMartino G N. Identification, purification, and characterization of a high molecular weight, ATP-dependent activator (PA700) of the 26S proteasome. J Biol Chem. 1994;269:3539–3547. [PubMed] [Google Scholar]
- 24.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 25.Peters J-M. Proteasomes: protein degradation machineries of the cells. Trends Biochem Sci. 1994;19:377–382. doi: 10.1016/0968-0004(94)90115-5. [DOI] [PubMed] [Google Scholar]
- 26.Ramos P C, Hockendorft J, Johnson E S, Varshavsky A, Dohmen R J. Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell. 1998;92:489–499. doi: 10.1016/s0092-8674(00)80942-3. [DOI] [PubMed] [Google Scholar]
- 27.Rothstein R. One step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Schiestl R H, Gietz R D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 30.Sherman F. Getting started with yeast. Method Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 31.Shibuya H, Irie K, Ninomiya-Tsuji J, Goebl M, Taniguchi T, Matsumoto K. New human gene encoding a positive modulator of HIV tat-mediated transactivation. Nature (London) 1992;357:700–702. doi: 10.1038/357700a0. [DOI] [PubMed] [Google Scholar]
- 31a.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka K, Yoshimura T, Kumatori A, Ichihara A, Ikai A, Nisihigai M, Kameyama K, Takagi T. Proteasomes (multi-protease complexes) as 20S ring-shaped particles in a variety of eukaryotic cells. J Biol Chem. 1988;263:16209–19217. [PubMed] [Google Scholar]
- 33.Tanaka K, Tanahashi N, Tsurumi C, Yokota K, Shimbara N. Proteasomes and antigen processing. Adv Immunol. 1997;64:1–38. doi: 10.1016/s0065-2776(08)60885-8. [DOI] [PubMed] [Google Scholar]
- 34.Tsurumi C, Shimizu Y, Saeki M, Kato S, DeMartino G N, Slaughter C A, Fujimuro M, Yokosawa H, Yamasaki M, Hendil K B, Toh-e A, Tanahashi N, Tanaka K. cDNA cloning and functional analysis of the p97 subunit of the 26S proteasome, a polypeptides identical to the type-1 tumor-necrosis-factor-receptor associated protein-2/55-11. Eur J Biochem. 1996;239:912–921. doi: 10.1111/j.1432-1033.1996.0912u.x. [DOI] [PubMed] [Google Scholar]
- 35.van Nocker S, Sadis S, Rubin D M, Glickman M, Fu H, Coux O, Wefes I, Finley D, Viestra R D. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol Cell Biol. 1996;16:6020–6028. doi: 10.1128/mcb.16.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe T K, Saito A, Suzuki M, Fujiwara T, Takahashi E, Slaughter C A, DeMartino G N, Hendi K B, Chung C H, Tanahashi N, Tanaka K. cDNA cloning and characterization of a human proteasomal modulator subunit p27. Genomics. 1998;50:241–250. doi: 10.1006/geno.1998.5301. [DOI] [PubMed] [Google Scholar]
- 38.Yokota K, Kagawa S, Shimizu Y, Akioka H, Tsurumi C, Noda C, Fujimuro M, Yokosawa H, Fujiwara T, Takahashi E, Ohba M, Yamasaki M, DeMartino G N, Slaughter C A, Toh-e A, Tanaka K. cDNA cloning of p112, the largest regulatory subunit of the human 26S proteasome, and functional analysis of its yeast homolog, Sen3p. Mol Biol Cell. 1996;7:853–870. doi: 10.1091/mbc.7.6.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshimura T, Kameyama K, Takagi T, Kai A, Tokunaga F, Koide T, Tanahashi N, Tamura T, Cejka Z, Baumeister W, Tanaka K, Ichihara A. Molecular characterization of the “26S” proteasome complex from rat liver. J Struct Biol. 1993;111:200–211. doi: 10.1006/jsbi.1993.1050. [DOI] [PubMed] [Google Scholar]