Abstract
The Rho family of GTPases plays key roles in the regulation of cell motility and morphogenesis. They also regulate protein kinase cascades, gene expression, and cell cycle progression. This multiplicity of roles requires that the Rho GTPases interact with a wide variety of downstream effector proteins. An understanding of their functions at a molecular level therefore requires the identification of the entire set of such effectors. Towards this end, we performed a two-hybrid screen using the TC10 GTPase as bait and identified a family of putative effector proteins related to MSE55, a murine stromal and epithelial cell protein of 55 kDa. We have named this family the Borg (binder of Rho GTPases) proteins. Complete open reading frames have been obtained for Borg1 through Borg3. We renamed MSE55 as Borg5. Borg1, Borg2, Borg4, and Borg5 bind both TC10 and Cdc42 in a GTP-dependent manner. Surprisingly, Borg3 bound only to Cdc42. An intact CRIB (Cdc42, Rac interactive binding) domain was required for binding. No interaction of the Borgs with Rac1 or RhoA was detectable. Three-hemagglutinin epitope (HA3)-tagged Borg3 protein was mostly cytosolic when expressed ectopically in NIH 3T3 cells, with some accumulation in membrane ruffles. The phenotype induced by Borg3 was reminiscent of that caused by an inhibition of Rho function and was reversed by overexpression of Rho. Surprisingly, it was independent of the ability to bind Cdc42. Borg3 also inhibited Jun kinase activity by a mechanism that was independent of Cdc42 binding. HA3-Borg3 expression caused substantial delays in the spreading of cells on fibronectin surfaces after replating, and the spread cells lacked stress fibers. We propose that the Borg proteins function as negative regulators of Rho GTPase signaling.
Motility and morphogenesis are probably among the most complicated processes that a cell performs. The proteins and molecular mechanisms that regulate these processes are only now beginning to be elucidated. Adherent cells elaborate an extracellular matrix of proteins to which they bind through receptors called integrins (8). The integrins cluster at focal adhesion complexes and transmit signals through these complexes to the actin-myosin cytoskeleton (11; for reviews, see references 10 and 46). Signaling through other types of receptor, such as those that bind growth factors, can be modulated by engagement of integrins with the extracellular matrix (39). A key role in controlling focal adhesions and the actin cytoskeleton is played by the small, Rho-like GTPases (9, 19, 28, 29), of which there are at least 13 types in mammalian cells (15, 36, 54). The most intensively studied members of this family of GTPases are RhoA, Rac1A, and Cdc42. Many of these proteins induce dramatic changes in the actin cytoskeleton when expressed ectopically as gain-of-function mutants (23, 35, 40, 41; reviewed in reference 16) and can perturb cell adhesion, cell spreading, motility, and cytokinesis (11, 20, 44, 50; reviewed in reference 26 and 55). The replating of fibroblasts from suspension culture onto a fibronectin-coated surface causes dramatic membrane ruffling and the rapid production of lamellipodia and microspikes around the edges of the spreading cells. These changes are mediated by the activation of the Rac and Cdc42 GTPases (5). The RhoA GTPase is transiently inhibited after replating and then activated at a later stage of spreading, at which time actin stress fibers appear within the cytosol (11). Similar changes in the activities of these proteins may occur at the leading edge of motile cells. Additionally, the Rho GTPases can activate protein kinases cascades and transcription factors and can regulate entry into the cell cycle (2, 12, 24, 37, 55).
Given this wealth of responses, it is not surprising that each of the Rho family GTPases has been found to interact with a plethora of target proteins that likely act as downstream effectors. These proteins include a variety of types of kinase and of adapters, plus other proteins of unknown function (for reviews, see reference 51 and 55). Several of the adapter proteins interact with known components of the actin cytoskeleton such as profilin, but their roles remain unclear (49, 56).
Because the Rho family GTPases mediates changes in gene expression and cell division that are independent of the actin cytoskeleton, different subsets of effectors likely participate in distinct signal transduction pathways downstream of the GTPases (24, 52). It is only through the identification and detailed analysis of the complete set of Rho family GTPases and of their effector proteins, therefore, that we will achieve satisfactory understanding of the molecular basis for those aspects of motility and morphogenesis that are influenced by these GTPases.
Toward this end, we performed a large two-hybrid screen of a whole mouse embryo library, using the TC10 GTPase as bait. Although its cDNA was cloned almost a decade ago (14), TC10 has been characterized only recently (14, 33). Of the >250 positives clones that we isolated in the screen, many contained open reading frame fragments of previously described proteins that interact with Cdc42 (33). These proteins include the protein kinases α-PAK, β-PAK, γ-PAK, PAK4, myotonic dystrophy kinase-related Cdc42-binding kinase (MRCK)-α and -β, and mixed-lineage kinase 2 (MLK2) (1, 25, 48, 51). We also isolated a partial cDNA for a putative new MRCK protein that we called MRCK-γ. In addition, the screen identified N-WASP (30) and an uncharacterized protein called MSE55 (marrow stromal/endothelial cell protein with a molecular mass of 55,000 Da) (3). All of these clones contained a CRIB (Cdc42, Rac interactive binding) domain, which has been shown to be necessary and sufficient to permit association with the Cdc42 and/or Rac proteins when in the GTP-bound state (7). Three other downstream effectors of Cdc42 that also contain CRIB domains, namely, WASP, p120ACK, and MLK3 (51), were not identified in the two-hybrid screen. These proteins do not bind TC10 (33).
We now describe a new family of five related TC10 and Cdc42 binding proteins, which we have called Borgs (binders of Rho GTPases). Three of the cDNAs for these proteins were identified in the two-hybrid screen, and two more were uncovered by database searches. One protein of this family (Borg5) is identical to the previously reported protein MSE55 (3), which was discovered serendipitously as a protein cross-reacting with an anti-hemonectin antibody. A Northern blot detected MSE55 mRNA only in endothelial and bone marrow stromal cells (3). The cDNA encodes a protein of 391 amino acid residues with two regions of weak similarity to some calcium binding domains. More recently, a database search revealed that the protein contains a CRIB domain, and purified glutathione S-transferase (GST)–MSE55 protein was shown to bind efficiently to [γ-32P]GTP-Cdc42 and more weakly to [γ-32P]GTP-Rac (7). We demonstrate that the other Borg family members are more widely expressed, that bind TC10 and/or Cdc42, and can inhibit the Cdc42-mediated activation of the c-Jun N-terminal kinase (JNK). Ectopic expression of Borg1 or Borg3 induces distinctive changes in cell morphology and significantly delays cell spreading on fibronectin as well as cell motility in a wound test.
MATERIALS AND METHODS
Cloning of Borg cDNAs.
The two-hybrid screen has been described previously (33). The bait was TC10(Q75L)ΔC, a gain-of-function mutant of TC10 lacking the C-terminal isoprenylation signal and fused to the DNA binding domain of the yeast GAL4 protein. The library was from a mouse embryo (day 9) fused to the activating domain of VP16 and was kindly provided by Stan Hollenberg (Seattle, Wash.). Four positive clones encoded a protein sequence highly similar to that of the human protein MSE55 (amino acid residues 1 to 110) (3). Differences in 11 amino acids residues between the clones and MSE55 most likely represent interspecies differences.
A set of two identical clones and another of five clones each encoded new CRIB domains which showed closer similarity to that of MSE55 than to CRIB domains of other known Rho family GTPase binding proteins. The complete human cDNA of the first gene was obtained from expressed sequence tag (EST) plasmid R13749/26651 and was named Borg1. Both fragments found in the screen correspond to amino acids 1 to 181 of the human open reading frame of Borg1 and encompassed a complete CRIB domain. Alignments of sequences were by made using the clustal algorithm in the Megalign program from the DNAStar software package. The mouse sequence from the screen possesses four additional amino acid residues (PSPQ) between positions 143 and 144. Overall, the mouse and human sequences are 81% identical. The set of five clones encode a reading frame, which we have termed Borg4, for which no full-length ESTs were present in the database. A partial human sequence of Borg4 was obtained from EST AA013011/360199. The sequence encodes two additional amino acid residues (SS) located between positions 68 and 69 of the mouse sequence.
When the databases were searched by BLAST, using a region of the CRIB domains of Borg1 and MSE55/Borg5 as the query, we identified two new open reading frames for closely related proteins, which we named Borg2 and Borg3. EST AA046144/376758 encoded the complete human Borg2 protein, and EST W85265/408069 encoded the complete mouse Borg3 protein. In all cases, we have defined “complete” as meaning that the open reading frames begin with an initiation codon downstream of a Kozak consensus sequence (22) and that in-frame stop codons are found upstream of this sequence. Full-length Borg1, Borg2, and Borg3 cDNAs were mutagenized by PCR so as to create a BamHI (or, for Borg2, a BglII) site at the 5′ end and an EcoRI site at the 3′ end of the open reading frames. These DNA fragments were then cloned in frame into the pKH3 (6), pGEX2T, and VP16 vectors that had been cut with BamHI and EcoRI. The vectors produce fusion proteins with an N-terminal triple-HA1 (hemagglutinin epitope) tag, GST, or VP16 activation domain, respectively.
Two conserved residues within the CRIB domain of Borg3 (Ile23 and Ser24) were mutated to Ala residues by megaprimer PCR mutagenesis (4), to produce CRIB (I23A,S24A). A truncation mutant in which the C-terminal 67 amino acid residues of Borg3 were deleted was produced by PCR. Recombinant proteins were expressed and purified from Escherichia coli as previously described (6).
Yeast conjugation assay.
Conjugation assays were performed as previously described (33). Briefly, Saccharomyces cerevisiae HF7c (MATa) was transformed with VP16 vectors that express fragments or full-length sequences of Borg proteins fused to the VP16 activation domain. Strain W303 (MATα) was transformed with modified pGBT9 vectors expressing GAL4 DNA binding domain fusions with different activated mutants of Rho family GTPases lacking the C-terminal isoprenylation signal. The two mating types were mixed and incubated overnight on YPD plates to permit mating. The following day, the spots were replica plated onto selective medium (L−/W−/H− plus 10 mM 3 aminotriazole) and incubated at 30°C to permit growth of diploids containing interacting proteins. Other known target proteins that interact with Rac(G12V)ΔC and Rho(G14V)ΔC were tested in the same assay as positive controls for expression of these GTPases.
In vitro binding assays.
Recombinant GST-Borg proteins were purified, and 2 μg of each, or of GST alone, was spotted onto nitrocellulose and allowed to dry for 1 h at room temperature. The nitrocellulose was blocked in binding buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 5 mM MgCl2, 0.1 mM dithiothreitol) plus 5% dried milk for 1 h at 4°C. TC10 and Cdc42 proteins (3 μg) were loaded with [α-32P]GTP or [α-32P]GDP (3,000 Ci/mmol) as described previously (33) and separated from unincorporated nucleotide by passage over a Pharmacia PD10 size exclusion column equilibrated in binding buffer. Equal amounts of radiolabeled proteins (ca. 0.5 μCi of each) were incubated with the nitrocellulose in 10 ml of binding buffer for 1 h at 4°C. The blots were subsequently washed three times with 50 ml of binding buffer, and associated proteins were detected by fluorography.
Immunodetection, immunofluorescence, and imaging.
NIH 3T3 fibroblasts were cultured on 10-cm-diameter plates or on two-well LabTek chamber slides (Nunc) in Dulbecco modified Eagle medium (DMEM) supplemented with 5% fetal calf serum, 5% calf serum, penicillin, and streptomycin. The cells were transfected by the calcium phosphate precipitation technique as previously described (43). For each transfection experiment, 12 μg of DNA was mixed with the calcium phosphate reagents in a final volume of 1.2 ml. One-sixth of the mix was added per LabTek chamber (for immunofluorescence studies), and the remainder was added per 10-cm-diameter plate of cells (for immunoblotting). When two plasmids (12 μg of each) are mixed and cotransfected, we find that under these conditions about 90% of the transfected cells express both proteins.
Cell protein extracts were prepared from the plates 48 h posttransfection, submitted to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes as described previously. Proteins were detected with monoclonal antibody 12CA5 (anti-HA; 1/5,000) or 9E10 (anti-Myc; 1/20,000) and revealed with a horseradish peroxidase-conjugated anti-mouse secondary antibody by using chemiluminescence (Kirkegaard & Perry Laboratories, Gaithersburg, Md.). For immunofluorescence, cells were washed in phosphate-buffered saline (PBS) 40 h posttransfection and then fixed in paraformaldehyde (4% [wt/vol] in PBS) for 15 min at room temperature. Cells were washed in PBS, and residual formaldehyde was quenched with 50 mM ammonium chloride for 15 min. Cells were permeabilized with Triton X-100 (0.2% [vol/vol] in PBS) for 3 min and blocked in PBS plus 3% (wt/vol) bovine serum albumin for at least 20 min. Cells were then incubated with mouse monoclonal anti-HA antibody 12CA5 (1/500) in PBS plus 0.3% bovine serum albumin for 1 h. After being washed in the same buffer, cells were incubated for 45 min with the secondary, Texas red-coupled antibody (1/1,200) and fluorescein isothiocyanate (FITC)-phalloidin (1/1,000; Sigma). Slides were mounted in Citifluor (Ted Pella, Redding, Calif.) and imaged by using a 60× water immersion objective lens on a Nikon inverted microscope equipped with a Hamamatsu charge-coupled device camera. Data were captured and processed by using Openlab 2.0 (Improvision) and Adobe Photoshop 5.0 software.
JNK assay.
Cos or NIH 3T3 cells, grown in DMEM supplemented with 5% fetal calf serum, 5% calf serum, penicillin, and streptomycin, were cotransfected with plasmids as described above. Control vectors pKH3 and pRK7 were used as necessary to normalize the amount of DNA transfected to 9 μg per 100-mm-diameter plate. Cells were incubated overnight in serum-free DMEM 24 h posttransfection; then 100 μg of anisomycin was added (control), and cells were incubated for 20 min at 37°C. Cells were washed twice with 10 ml of ice-cold PBS and lysed in 400 μl of lysis buffer (25 mM HEPES [pH 7.4], 300 mM NaCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol, 20 mM β-glycerophosphate, 1 mM Na3VO4, 0.1% Triton X-100, 20 μg of aprotinin/ml, 10 μg of leupeptin/ml, 1 mM phenylmethylsulfonyl fluoride, 1 μM okadaic acid). Lysed cells were scraped off the plates, and the supernatant was cleared by centrifugation (5 min at 14,000 × g, 4°C). Equal volumes (100 μl) of the supernatant were removed for immunoblot analysis and separated on 12% polyacrylamide gels by SDS-PAGE. The proteins were transferred to a nitrocellulose membrane and immunoblotted. HA3-tagged proteins were immunoprecipitated from the remaining supernatant and washed three times in buffer A (2 mM Na3VO4 and 1% NP-40 in PBS), once in buffer B (100 mM morpholinepropanesulfonic acid [MOPS; pH 7.5], 0.5 M LiCl), and once in kinase buffer (12.5 mM MOPS [pH 7.5], 12.5 mM β-glycerophosphate, 7.5 mM MgCl2, 0.5 mM EGTA, 0.5 mM NaF, 0.5 mM Na3VO4). Immunoprecipitated proteins were resuspended in 30 μl of kinase buffer, and 2 μg of purified GST—c-Jun(1–79) plus 2 μCi of [γ-32P]ATP were added to each reaction tube. The reactions proceeded at 30°C for 20 min and were stopped by the addition of 10 μl of 4× Laemmli sample buffer. Kinase reaction complexes were separated on 12% polyacrylamide gels by SDS-PAGE and analyzed by fluorography.
Cell spreading and motility assays.
NIH 3T3 cells in 10-cm-diameter plates were transfected with 10 μg of DNA. Forty hours after transfection, cells were trypsinized, maintained in suspension for 15 to 20 min, and then replated on LabTek chambers that had been coated with fibronectin (0.5 ml of a 50-μg/ml solution). At different times (18, 45, or 180 min) after replating, cells were fixed with paraformaldehyde and processed for immunofluorescence as described above. Transfected cells were analyzed by using Openlab 2.0 software to measure cell surface areas in contact with the fibronectin surface. For each time point, the areas of 100 cells were counted. To compare expression levels of the Borg proteins within the transfected cells, images were captured under conditions such that no camera pixels were saturated, and the mean fluorescence of the counted cells was determined for each experiment. Differences in mean cell area at 18 min postplating were analyzed by an unpaired t test assuming equal variances (variances were calculated for each data set and found to be approximately equal). Probabilities (P values) were also calculated for the t-test values and degrees of freedom (i.e., 198).
To quantitate cell mobility, a wounding assay was used. Cells were cotransfected with the plasmid of choice plus pK7-GFP, which expresses a bright mutant of green fluorescent protein (GFP). Cells were split and replated at high density and low density onto two 35-mm-diameter plates. After 3 days, when the cells were confluent, an aspirator was drawn across the center of the high-density culture, to produce a clean 1-mm-wide wound area free of cell debris. After a further 24 h of incubation to permit migration of cells into the empty area of the plate, the numbers of green and nonfluorescent cells in the cleared area were counted and compared to the ratio of green to nonfluorescent cells in the low-density, unwounded culture. Cells cotransfected with pK7-GFP and the pRK7 empty plasmid gave a ratio close to 1.0, a value expected if the GFP itself has no effect on motility. Statistical significance of the data was assessed by a two-sample t test.
Nucleotide sequence accession numbers.
Accession numbers for the Borg open reading frame sequences are AF163840 (Borg1), AF164118 (Borg2), AF164119 (Borg3), and AF165114 (Borg4).
RESULTS
Cloning of Borg family cDNAs.
A two-hybrid screen of a 9-day mouse embryo library using an activated TC10 GTPase as bait identified numerous clones that encoded protein fragments incorporating a CRIB domain. Three groups of clones had almost identical CRIB domains and similar flanking sequences, suggesting that they belonged to a family of related proteins that interact with Rho family GTPases (Fig. 1). Database searches using the protein sequence of each group uncovered five sets of human ESTs encoding proteins with a high degree of similarity to the two hybrid clones. One of these clones was almost identical to the previously reported human MSE55 protein (3) and is most likely the mouse counterpart of MSE55. We obtained from different ESTs the complete open reading frames for three proteins of this new family but found no ESTs encompassing the 3′ end of one cDNA found in the screen.
FIG. 1.
(A) Schematic of Borg protein domain structures. Bars showing the regions of similarity within the primary sequences of the Borg proteins are aligned according to the CRIB domains. The C-terminal sequence of Borg4 is not known. Borg5 is the putative mouse homolog of human protein MSE55. (B) Alignment of BH1, BH2, and BH3 domains. The N-terminal region comprises the CRIB domain and the BH1 domain.
Because the predicted sizes of the members of this family are all lower than 55 kDa, because the tissue expression of the corresponding mRNAs is not restricted to endothelial and stromal cells (see below), and because the name MSE55 does not indicate function, we decided to rename the whole family Borg proteins. Borg1 is the complete new protein of which a part was revealed in the two-hybrid screen, Borg2 and Borg3 are the two proteins for which DNA sequences were found in the EST database, Borg4 corresponds to the incomplete cDNA identified in the screen, and Borg5 is MSE55. Two uncharacterized sequences which correspond to Borg1 have been deposited by others in the GenBank database (accession no. AF001436 and AF094521).
Surprisingly, the mouse sequence of Borg1 contains four additional amino acids after position 143 that are a repeat of the four previous amino acids (PSPQ). This discrepancy is confirmed in two other independent mouse EST clones. Additionally, the human sequence of Borg4 contains an insertion of two amino acids between positions 68 and 69 of the mouse sequence (SSSK versus SK). These differences in otherwise well-conserved sequences might reflect the presence of different splice variants of the same proteins.
The domain structures of the Borg proteins are diagrammed in Fig. 1A. Borg1, Borg2, and especially Borg3 (210 [22.5 kDa], 254 [27.5 kDa], and 150 [15.5 kDa] amino acid residues, respectively) are smaller than Borg5/MSE55 (which consists of 379 amino acid residues, for a predicted molecular mass of 39 kDa). Bahou et al. (3) described MSE55 as a 391-amino-acid protein, but the initiation codon that they indicated corresponds, at the mRNA level, to a very unlikely translation start. The Kozak context (22) for translation is unfavorable: CUGAUGC versus the best consensus described, ACCAUGG (the first adenine being present in 90% of cases) (initiation codon indicated by underlining). The context of the second in-frame Met codon (ACCAUGA) is closer to the Kozak consensus motif and aligns naturally with the initiation codon of the other Borg proteins (Fig. 1B). We suggest, therefore, that it most probably represents the true start site for translation of Borg5 and have numbered residues in Fig. 1 accordingly. The N-terminal regions of all five Borg proteins are highly similar, comprising a short basic region (from 13 amino acids for Borg1 to as short as 3 amino acids for Borg5) followed by a CRIB domain sequence, ISXPLGDFRHTXH(I/V)G. This sequence matches the consensus motif, I(S/G)XPXXFXHXXHVG (7), with an additional residue in the Borg CRIB domains between the P and F residues.
A well-conserved, short domain of about 12 amino acids that is not found in any other known protein follows the CRIB domain. We therefore call it Borg homology 1 (BH1) domain. The central and C-terminal parts of the proteins are more divergent. Two other well-conserved motifs that we call the BH2 and BH3 domains can be defined (Fig. 1). The BH2 domain is not present in Borg3; the BH3 domain has a central location in Borg5, whereas it is localized at the C-terminal parts of Borg1, Borg2, and Borg3. Neither domain is present within the N-terminal fragment of Borg4 for which we have sequence information. A proline-rich domain (about 40% of Pro in each case) can be observed in the central regions of Borg1, Borg3, and Borg5. The four amino acid residues insertion of the mouse Borg1 sequence is located within this region. Borg5 also contain eight in-tandem heptad repeats (consensus PAANPPA) located at the C-terminal side of the BH3 domain.
Borg mRNAs show distinct expression patterns.
The MSE55 gene is expressed in endothelial and bone marrow stromal cells but not in monkey liver, spleen, brain, lung, and kidney (3). To determine the tissue expression patterns of other members of the family, fragments of cDNA encoding human Borg1, Borg2, or Borg4 or mouse Borg3 were used to probe a human tissue poly(A)+ mRNA blot (Fig. 2). Borg1, Borg2, and Borg3 mRNAs were detected in all eight tissues studied, though at very different levels. For all three genes, multiple transcripts were observed. Borg1 is expressed as two transcripts of about 1.8 and 2 kb and at high levels in heart. Borg1 mRNA is less abundant in pancreas and especially liver. The Borg2 probe revealed four different transcripts of various sizes (from 2 to 5.5 kb). Borg2 is also expressed at high levels in heart but at low levels in brain, and the largest transcript was not detected in liver. The Borg3 probe was not of human origin and gave a weaker signal. However, transcripts of about 1 and 2 kb are visible (Fig. 2) and were more clearly seen in a rat cardiomyocyte mRNA extract (data not shown). In addition to these two messengers, present in all examined tissues, a more abundant transcript of about 3.5 kb is detectable specifically in skeletal muscle. The presence of multiple transcripts suggests the possible existence of differentially spliced mRNAs and of Borg protein variants. Indeed, two slightly different mRNAs of MSE55 have been described (3). No transcripts were found in any tissues probed with the Borg4 probe. The Borg4 gene might, like Borg5, be expressed in only a few, specific tissues or cell types. However, it is striking that all available ESTs are of embryonic or fetal origin. Therefore, Borg4 may not be expressed in adult tissues.
FIG. 2.
Tissue-specific expression of Borg genes. A nylon membrane containing human poly(A)+ mRNAs from different tissues (Clontech) was hybridized with α-32P-labeled human (mouse for Borg3) cDNA probes encoding the Borg proteins. Ran GTPase cDNA was used as a loading control. The same membrane was used in all four Northern blots. Molecular sizes of RNA standards are indicated (in kilobases) on the left. Arrows indicate the multiple transcripts that hybridized with each probe.
Borg3 interacts specifically with Cdc42.
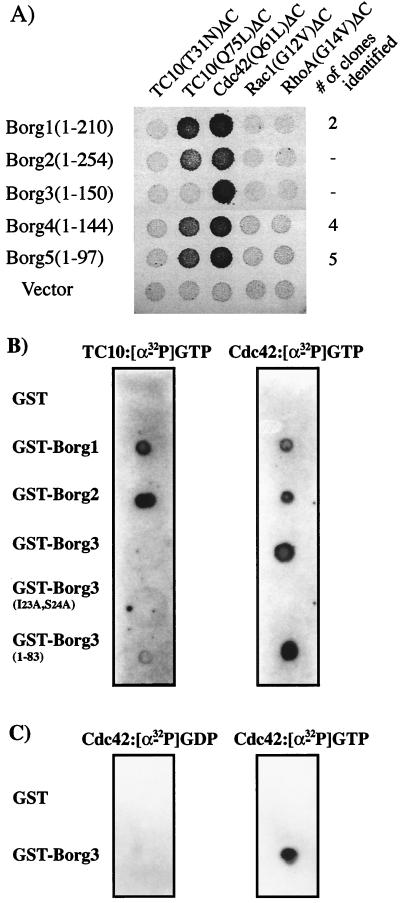
Partial cDNAs for Borg1, Borg4, and Borg5 were found in the two-hybrid screen. All clones of these cDNAs contained the 5′ coding region including the region corresponding to the CRIB and BH1 domains of the proteins. In a yeast two-hybrid conjugation assay, the expressed protein fragments of Borg1, Borg4, and Borg5 interacted specifically with the activated TC10 GTPase deleted of its isoprenylation signal [TC10(Q75L)ΔC] and not with the GDP-bound TC10 mutant [TC10(T31N)ΔC] (Fig. 3A shows data for Borg4 and Borg5). They also bound to an activated mutant of Cdc42 [Cdc42(Q61L)ΔC]. To check that the interaction was not an artifact of the use of fragments rather than full-length proteins, we repeated the conjugation assay using the complete open reading frames of Borg1 to Borg3. Surprisingly, although the Borg1 and Borg2 interacted specifically with TC10, Borg3 did not (Fig. 3A). Borg3 did, however, interact strongly with Cdc42. Burbelo et al. reported a weak binding of MSE55/Borg5 to GTP-loaded Rac1 in a dot blot assay (7), but in our hands, no member of the Borg family gave a positive response in the two-hybrid assay with activated mutants of either Rac1 or RhoA (Fig. 3A).
FIG. 3.
Binding of Borg proteins to different Rho-like GTPases. (A) Yeast two-hybrid conjugation assays. S. cerevisiae HF7c (MATa) was transformed with VP16 empty vector or with VP16 containing Borg cDNAs (the complete open reading frame of Borg1, Borg2, or Borg3 or a fragment obtained from the two-hybrid screen for Borg4 and Borg5). The transformants were then conjugated to S. cerevisiae W303 (MATα) which had been transformed with pGBT9 plasmids containing cDNA encoding different Rho-like GTPases. Each GTPase open reading frame had been truncated to remove the C-terminal isoprenylation signal. Results show the growth of diploids after replica plating onto selective medium (L−/W−/H− plus 10 mM 3-aminotriazole). For the three Borg cDNAs identified in the screen, the number of independent clones identified is noted. (B and C) Binding of GST-Borg proteins to TC10-GTP, Cdc42-GTP, or Cdc42-GDP. Purified recombinant GST or GST-Borg1, GST-Borg2, GST-Borg3, GST-Borg3(I23A,S24A), and GST-Borg3(1–83) (2 μg of each) were spotted onto a nitrocellulose membrane, which was then incubated with TC10-[α-32P]GTP, Cdc42-[α-32P]GTP, or Cdc42-[α-32P]GDP (0.5 μCi; 3,000 Ci/mmol). Interactions were revealed by fluorography after the membranes were washed in binding buffer as described in Materials and Methods.
To determine whether the observed interactions between the small GTPases and the Borgs is direct, we spotted equal amounts of recombinant GST-Borg1, GST-Borg2, and GST-Borg3 fusion proteins onto nitrocellulose and incubated them with recombinant TC10 or Cdc42 that had been loaded with [α-32P]GTP. After washing, the GST-Borg1 and GST-Borg2 proteins showed strong binding to both Cdc42 and TC10. Binding of GST-Borg3 to TC10 was undetectable, although it bound the GTP-loaded Cdc42 with high affinity (Fig. 3B). [α-32P]GDP-Cdc42 did not bind to GST-Borg3, demonstrating that, as expected, the binding of Cdc42 to Borg3 is nucleotide dependent (Fig. 3C). These results confirm the validity of the yeast two-hybrid conjugation assay and demonstrate that the original two-hybrid screen was unable to identify Borg3 not because it was absent from the library but because it does not recognize TC10. It is interesting that the Borg3 CRIB domain, though closely related to the CRIB domains of the other Borg family members, nonetheless contains determinants that can differentiate TC10 from Cdc42, despite the fact that these GTPase effector domain sequences are almost identical. Similarly, we have found that while MLK2 and N-WASP interact with TC10, MLK3 and WASP do not, though all seem to bind with similar affinity to Cdc42 (33).
To confirm that the CRIB domain constitutes the only binding site on Borg proteins for Cdc42, we created a GST-Borg3 mutant in which the two first, highly conserved amino acid residues (Ile and Ser) of the CRIB motif were converted to Ala residues [Borg3(I23A,S24A)]. This fusion protein did not detectably bind [α-32P]GTP-loaded Cdc42 (Fig. 3B). We also created a C-terminal truncation mutant of Borg3, GST-Borg3(1–83), which lacks BH3 and part of the Pro-rich domain. This mutant bound Cdc42 as well as did the wild-type protein. These results argue that the CRIB domain region of Borg3 is necessary and sufficient for interaction with the Cdc42 GTPase.
Borg3 inhibits JNK independently of Cdc42 binding.
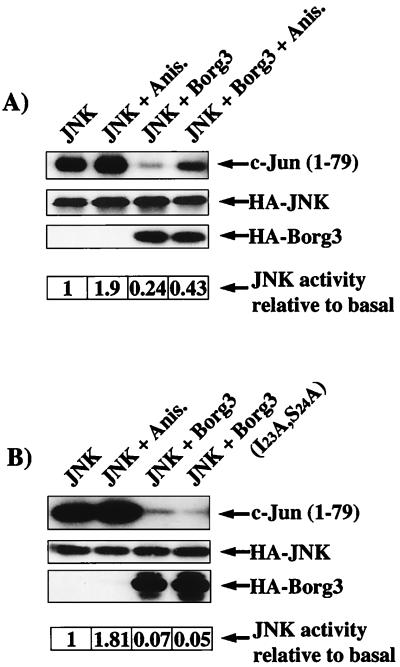
JNK is the terminal protein kinase of a cascade that can be activated by numerous stress-related signal inputs and by a variety of seven-transmembrane domain receptors that couple to heterotrimeric G proteins (31). The βγ subunits released from the G protein upon activation of these receptors are believed to activate exchange factors for Rho family GTPases (13). Cdc42 can then activate the protein kinase cascade that switches on JNK. The mixed-lineage kinase MLK2 may mediate this activation (18). However, there remains the possibility that other downstream effectors of Cdc42 participate in the pathway. We therefore tested the effects of Borg expression on the activity of JNK in transfected Cos cells.
As shown in Fig. 4, the coexpression of HA3-Borg3 suppressed the basal activity of HA-JNK. Anisomycin stimulated both the basal JNK and Borg-inhibited JNK activities by about the same degree, indicating that Borg3 does not interfere with the stress-mediated signaling pathway that regulates JNK. The inhibition by HA3-Borg3 was not a result of changes in the level of expression of the HA-JNK or of competition for the anti-HA antibody. In fact, HA3-Borg3 is not efficiently immunoprecipitated by the anti-HA3 tag antibody, perhaps because the tag is hidden by interaction of the Borg with other proteins or because it is partially buried within the folded protein (not shown). To assess whether this inhibition is dependent on the binding of Cdc42, we used in the same assay a CRIB domain mutant, Borg3(I23A,S24A), that cannot bind Cdc42. Expression of this mutant inhibited basal JNK activity to the same extent as the wild-type Borg3 (Fig. 4B). In each case, we checked the presence of all ectopically expressed proteins in the lysates by immunoblotting (Fig. 4). A similar inhibition of JNK activity by HA3-Borg3 and HA3-Borg3(I23A,S24A) was observed in transfected NIH 3T3 cells (not shown).
FIG. 4.
Inhibition of JNK by Borg3. Cos cells were transfected as described in Materials and Methods and incubated overnight in serum-free DMEM. After 24 h, 100 μg of anisomycin (Anis.) was added (control) and the cells were incubated for 20 min at 37°C. Cells were lysed in 400 μl of lysis buffer. After clearing, equal volumes (100 μl) of the supernatant were removed for immunoblot analysis and separated by SDS-PAGE. Proteins were transferred to a nitrocellulose membrane and immunoblotted as described in Materials and Methods. HA3-tagged proteins were immunoprecipitated from the remaining supernatant and resuspended in 30 μl of kinase buffer, and 2 μg of purified GST–c-Jun(1–79) as the substrate plus 2 μCi of [γ-32P] ATP were added to each reaction tube. The reactions proceeded at 30°C for 20 min and were stopped by the addition of 10 μl of 4× Laemmli sample buffer. Kinase reaction complexes were separated by SDS-PAGE and analyzed by fluorography.
These data indicate that the inhibition of the JNK pathway by Borg proteins is independent of the binding to Rho-like GTPases and therefore is not a result of competition for another downstream effector such as MLK2, which is required for JNK activation.
Effects of ectopic expression of Borg proteins on cell shape.
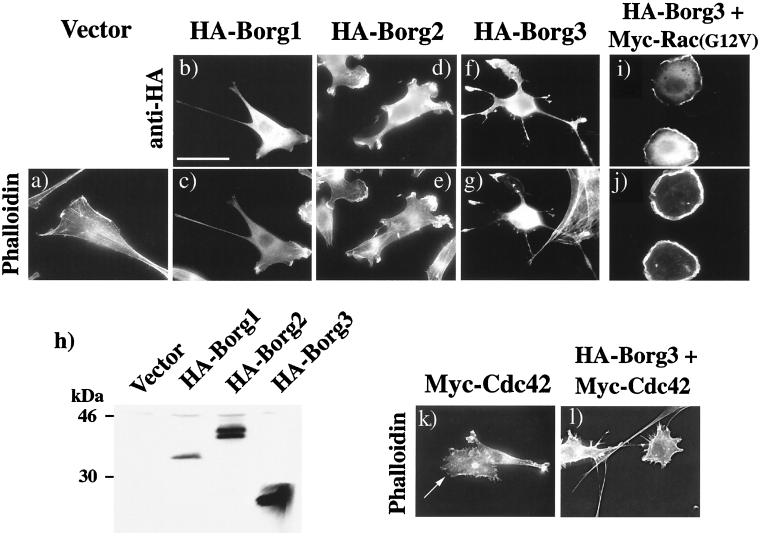
To examine whether Borgs might, like many downstream effectors of Cdc42/Rac, play a role in the elaboration or remodeling of the actin cytoskeleton, we expressed Borg1, Borg2, and Borg3 as HA3-tagged fusion proteins in NIH 3T3 fibroblasts. As a negative control, cells were transfected with the empty vector, pKH3. Expression of the Borg protein was detected with the monoclonal anti-HA antibody 12CA5 and a secondary Texas red-coupled antibody. The F-actin framework was revealed by using FITC-phalloidin. Expression of HA3-Borg1 did not cause any dramatic changes in cell shape, but there was a substantially reduced abundance of stress fibers in most of the transfected cells, and unusually long, thin extensions were common (Fig. 5b and c). Similar but more pronounced morphological changes were seen when we ectopically expressed HA3-Borg2 and especially HA3-Borg3. Most of the transfected cells displayed a disturbed shape, in which the cell bodies had pulled in, but also extended long, sometimes beaded processes and protrusive lamellipodia (Fig. 5d to g). This phenotype is reminiscent of that produced by the overexpression of RhoGDI or RhoGAPs such as p190 or by injection of C3 botulinum toxin which inhibits Rho function (40, 53). The HA3-Borgs were all localized mainly in the cytosol but also in filopodia and at the edges of lamellipodia, where they were coincident with cortical F-actin structures (Fig. 5b to g). These phenotypes were also seen in other cell types that were tested, including baby hamster kidney (BHK21) and Swiss 3T3 cells (data not shown). We confirmed by immunoblotting that Borg1 to Borg3 were expressed and were of the expected size (Fig. 5h). To verify the preferential targeting of the Borg proteins to the ruffles of lamellipodia, we coexpressed a Myc-tagged, activated Rac1 [Rac1(G12V)] with HA3-Borg3. Expression of activated Rac1 in NIH 3T3 fibroblasts induces the formation of rounded cells with large, multilayered ruffles around the entire edge of the cell (41). Ectopic expression of HA3-Borg3 did not perturb this phenotype, as shown by phalloidin staining (Fig. 5j), but the Borg protein was concentrated in the ruffles (Fig. 5i). However, the interaction of the Borg with these structures is relatively weak, because treatment with 0.008% digitonin to permeabilize the plasma membrane resulted in the loss of all detectable HA3-Borg3 from the cells, though it did not visibly perturb the actin cytoskeleton (data not shown).
FIG. 5.
Cell morphologies induced by ectopic expression of HA3-tagged Borg proteins. NIH 3T3 fibroblasts were transfected with pKH3 empty vector (a) or with pKH3 or pKMyc vector containing cDNA encoding either Borg1 (b and c), Borg2 (d and e), Borg3 (f and g), or Cdc42 (k). Some cells were cotransfected with pKH3-Borg3 plus pKMyc-Rac(G12V) (i and j) or with pKH3-Borg3 plus pKMyc-Cdc42 (l). Cells were fixed 40 h after transfection and processed for immunofluorescence using either the monoclonal mouse anti-HA antibody 12CA5 and a secondary, Texas red-coupled anti-mouse antibody to detect Borg proteins (b, d, f, and i) or with FITC-phalloidin to reveal F-actin (a, c, e, g, j, k, and l) as described in Materials and Methods. Lysates from transfected cells were analyzed by immunoblotting using anti-HA antibody 12CA5. Detection was by chemiluminescence (h). For panels k and l, expression of Myc-Cdc42 (k) and HA3-Borg3 (l) was checked by immunofluorescence using monoclonal antibodies 9E10 (anti-Myc) and 12CA5, respectively, and a secondary Texas red-coupled antibody (not shown). In panel k, arrow indicates a transfected cell. Bar = 40 μm.
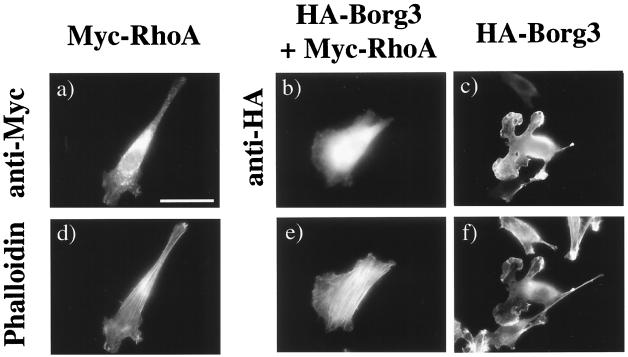
We then wanted to assess whether the ectopic expression of Borgs would inhibit the cytoskeletal remodeling induced by Cdc42. Overexpression of wild-type HA3-tagged or Myc-tagged Cdc42 induces the formation of lamellipodia bordered with microspikes around the cell edge and a partial loss of stress fibers (Fig. 5k). The coexpression of HA3-Borg3 with Myc-Cdc42 did not inhibit this phenotype but instead produced a different, “porcupine” phenotype, in which the cells radiated an abundance of actin-filled spikes that were sometimes branched and the cell bodies were devoid of stress fibers (Fig. 5l). Equivalent results were obtained when HA3-Borg1 or HA3-Borg2 was coexpressed with Myc-Cdc42 (not shown). This phenotype is not caused by a Borg-mediated enhancement of Cdc42 function, for the following reasons. First, the phenotype is not induced by expression of a gain-of-function mutant of Cdc42. Second, we observed a similar phenotype when the p190 RhoGAP was coexpressed with Myc-Cdc42 (data not shown). We conclude, therefore, that Borgs neither inhibit Cdc42 function nor act downstream of Cdc42 to trigger filopodia formation, but that the results are consistent with an inhibition of Rho function induced by Borg proteins. To test this hypothesis, we examined whether the Borg phenotype could be reversed by the coexpression of wild-type Rho. NIH 3T3 cells were transfected either with HA3-Borg3 alone, with Myc-tagged RhoA alone, or with HA3-Borg3 plus Myc-RhoA. As can be seen in Fig. 6c and f, the Borg3 produced a characteristic distortion of the normal cell morphology, with loss of stress fibers and the extension of protrusive lamellipodia. Coexpression of the Myc-RhoA dramatically increased stress fiber formation and the restoration of a more normal fibroblast appearance. This result supports the hypothesis that Borg3 inhibits Rho function and suggests that it acts upstream of or parallel to Rho rather than by inhibiting a downstream effector of Rho.
FIG. 6.
Borg phenotype is abolished by coexpression of Myc-RhoA. NIH 3T3 fibroblasts were transfected with either pKMyc containing cDNA encoding for RhoA protein (a and d) or pKH3 containing cDNA encoding Borg3 protein (c and f) or were cotransfected with plasmids encoding HA3-Borg3 and Myc-RhoA proteins (b and e). Cells were fixed and processed for immunofluorescence. Tagged proteins were detected with either monoclonal anti-HA antibody 12CA5 (b and c) or monoclonal anti-Myc antibody 9E10 (a). Both antibodies were revealed with a secondary, Texas red-coupled anti-mouse antibody. F-actin was revealed with FITC-phalloidin (d to f). Bar = 40 μm.
Finally, to determine whether the effect of the Borg proteins on the cell phenotype is dependent on binding to Cdc42, we used the CRIB (I23A,S24A) mutant of Borg3, which is defective in Cdc42 binding (Fig. 3B). Surprisingly, when transfected into NIH 3T3 fibroblasts, the CRIB-defective mutant induced the same dramatic phenotype as did wild-type HA3-Borg3, with loss of stress fibers and the extension of large, protrusive lamellipodia (Fig. 7b and e). The expression of a C-terminal deletion of Borg3 (residues 1 to 83) gave a weaker phenotype, similar to the one obtained when Borg1 is overexpressed. Few cells produced protrusive lamellipodia, but many cells extended long processes (Fig. 7c). Although the CRIB-defective mutant was expressed at levels equivalent to those of the wild-type HA3-Borg3, the Borg3(1–83) fragment did not express well, and only few cells expressed the protein at a high level. However, no degradation of the protein could be detected (Fig. 7g). The data suggest that the characteristic phenotype induced by expression of Borg3 is independent of its interaction with Cdc42.
FIG. 7.
Borg3 phenotype is independent of Cdc42 binding. NIH 3T3 fibroblasts were transfected with pKH3 vector containing cDNA encoding either Borg3 (a and d), Borg3(I23A,S24A) (b and e), or Borg3(1–83) (c and f). Cells were fixed and processed for immunofluorescence by using either monoclonal mouse anti-HA antibody 12CA5 revealed with a secondary, Texas red-coupled anti-mouse antibody to detect HA3-Borg proteins (a to c) or FITC-phalloidin to reveal F-actin (d to f). Bar = 40 μm. In parallel the expression levels of the different proteins were checked by immunoblotting with anti-HA antibody after transfer of cell lysate proteins to a nitrocellulose membrane (g). Proteins were revealed by chemiluminescence using a secondary anti-mouse antibody coupled to horseradish peroxidase. A control with cells transfected with pKH3 empty vector is also included. For cell lysates containing HA3-Borg3(1–83), a 10-fold longer exposure of the membrane is shown (×10). Arrows indicate the different Borg3 proteins.
Expression of Borg proteins delays cell spreading in a CRIB domain-dependant manner.
Recent evidence has implicated several Rho family GTPases in cell motility and in spreading on fibronectin-coated surfaces (11, 34). Engagement of integrin receptors by fibronectin results in a rapid and dramatic activation of Cdc42 and Rac, which promotes their interaction with a plethora of downstream effectors (11, 55). The function of many of these interactions in motility and spreading remains unclear. To determine whether the expression of Borg proteins would affect the ability of cells to spread, NIH 3T3 fibroblasts were transfected with either the empty pKH3 vector, pKH3-Borg1, or pKH3-Borg3. Forty hours after transfection, cells were trypsinized, maintained in suspension for 15 to 20 min, and then replated onto fibronectin-coated slides. Cells were fixed with paraformaldehyde at intervals and processed for immunofluorescence labeling. Spreading was quantitated by measuring total cell surface areas of transfected or control cells. Results are shown in Fig. 8. At early times after replating (18 min), control cells were rounded, with lamellipodia encircling the cell bodies. Cells were at this stage devoid of stress fibers (Fig. 8a, untransfected cells). On the other hand, many of the cells expressing HA3-Borg1 or HA3-Borg3 remained very small (cell area < 500 μm2), whereas almost no control cells were of that size (Fig. 8m). By 45 min, control cells were still rounded but had formed some stress fibers (Fig. 8b). In contrast, although the Borg-expressing cells began to spread, most of them were still small and completely devoid of stress fibers. Three hours after replating, control cells possessed abundant stress fibers and had attained a normal fibroblastic shape (Fig. 8c). Curiously, although the majority of Borg-expressing cells had by 3 h reached surface areas similar to those of the control (Fig. 8m), they exhibited an unusual phenotype, with a highly convoluted perimeter. These cells contained few stress fibers (Fig. 8c).
FIG. 8.
Borg3 expression delays cell spreading of NIH 3T3 cells on fibronectin-coated surface in a Cdc42-binding-dependent manner. NIH 3T3 fibroblasts were transfected with pKH3 empty vector or with pKH3 containing cDNA encoding either Borg1, Borg3, Borg3(I23A,S24A), or Borg3(1–83); 40 h after transfection, cells were trypsinized and replated on fibronectin-coated slides. Cells were fixed and processed for immunofluorescence at specific times. FITC-phalloidin was used to reveal F-actin (a to c), and anti-HA antibody 12CA5 and a secondary, Texas Red-coupled anti-mouse antibody were used to reveal Borg proteins (d to l). Arrows in panels a to c show HA3-Borg3-expressing cells. Bar = 40 μm. For quantitation of cell surface area at 18 and 180 min following replating (m and n), cells were visualized by FITC-phalloidin (Control) or anti-HA staining. Measurement were made on 100 cells for each time point, using the Openlab 2.0 software. Cells were then grouped according to size. Results of one representative experiment are shown. Two to four independent experiments were carried out. Data for the 18-min time point were analyzed by an unpaired t test. Variances were approximately equal for all data sets. Mean areas for Borg1- and Borg3-transfected cells were very highly significantly different from those of the control (P << 0.001) and Borg3(I23A,S24A)-transfected (P << 0.001) cells.
These results suggest that the ectopic expression of Borg1 or Borg3 can interfere with the normal process of spreading, both by delaying the rate of spreading and by interfering with the attainment of a normal fibroblastic shape after spreading is complete. The Rho GTPase is initially inhibited after engagement of integrin receptors by fibronectin but is activated at later stages of cell spreading. The effect of the Borg3 on cell shape is therefore consistent with the proposal that the Borgs partially inhibit Rho function.
As discussed above, the loss of stress fibers and changes in cell shape induced by Borg3 are independent of Cdc42 binding. To determine whether the reduction in spreading rate or alteration in cell shape is mediated by a Borg3-Cdc42 interaction, we assessed the effect of the mutant protein Borg3(I23A,S24A) and of Borg3(1–83). Ectopic expression of HA3-Borg3(I23A,S24A) significantly reduced the delay in cell spreading seen with the wild-type HA3-Borg3 (Fig. 8g and h). Within 18 min following replating, 48% of the cells expressing HA3-Borg3(I23A,S24A) were larger than 103 μm2 (Fig. 8n), a proportion similar to that for the control cells (54%) and substantially higher than that for the cells expressing wild-type HA3-Borg3 (13%). These differences were very highly significant, as determined by an unpaired t test (equal variances). Borg3(1–83) gave an intermediate result. The effects of the two mutants are not additive since the results obtained with a double mutant, HA3-Borg3(I23A,S24A,1–83), was similar to that of cells expressing the CRIB(I23A,S24A) mutant (not shown). The differences in spreading rate were not due to differences in expression levels of the various proteins, as judged by quantitation of the immunofluorescence of transfected cells counted in the assay (not shown).
These data indicate that the inhibition of spreading by Borg3 depends largely on its ability to bind Cdc42 and to a lesser extent on an intact BH3 and/or Pro-rich domain. The inhibition may be partially explained by a competition with other downstream effectors for Cdc42, which is rapidly activated upon integrin receptor engagement by fibronectin.
Contrary to the effects on spreading rate, however, the two Borg3 mutants remained fully capable of interfering with the attainment of a normal fibroblastic cell shape. As can be seen in Fig. 8i and l, the transfected cells after 3 h on fibronectin possess highly convoluted, lace-like peripheries with several holes near their edges. These cells also lack stress fibers (not shown). This result is consistent with the possibility that a domain near the N terminus of Borg3 (e.g., BH1) can inhibit Rho function independently of Cdc42 binding.
Dynamic regulation of Rho is also required for cell migration (34). We therefore examined whether the ectopic expression of HA3-Borg3 could inhibit the movement of cells into a cleared area of a monolayer. This wounding assay is expected to be less sensitive to small changes in signaling functions, because it occurs over a prolonged time period (24 h). Thus, cells that have a reduced motility rate might still have time to migrate into the cleared area. Nonetheless, as shown in Table 1, HA3-Borg1, HA3-Borg3, and HA3-Borg3(I23A,S24A), when coexpressed with GFP, did significantly reduce the fraction of green fluorescent cells that migrated.
TABLE 1.
Effects of ectopic expression of Borg proteins on fibroblasts migration in a wound test
| Protein expressed with GFP | Ectopic expression (% of migrating cells expressing GFP/% of cells expressing GFP in the population) | No. of experiments |
|---|---|---|
| None (Vector) | 1.21 ± 0.23 | 2 |
| HA-Borg1 | 0.77 ± 0.13a | 3 |
| HA-Borg3 | 0.77 ± 0.17b | 3 |
| HA-Borg3(I23A,S24A) | 0.88 ± 0.13b | 3 |
Value is significantly different from control, P < 0.025 (two-sample t test for independent samples).
P < 0.05.
DISCUSSION
We have described a new family of proteins, named Borg1 to Borg5, that are putative downstream effectors of Cdc42 and/or TC10. Borg1, Borg4, and Borg5 were identified from a two-hybrid screen using the TC10 GTPase as bait; Borg2 and Borg3 were discovered from database searches for Borg-related ESTs. Borg5 is most likely the human homolog of a previously reported protein called MSE55 that was shown to interact with Cdc42 and Rac through a conserved CRIB domain (3). Each member of the Borg family contains a highly related CRIB domain near the N terminus of the protein and several further regions of close similarity to one another, which we have called BH1, BH2, and BH3 domains, that were not found in any other proteins in the database. Remarkably, although Borg1, Borg2, Borg4, and Borg5 appear to bind with similar affinities to both Cdc42 and TC10, Borg3 does not interact detectably with TC10. Compared with the other members of the family, the Borg3 protein sequence lacks three amino acids immediately upstream of the CRIB domain. This deletion might account for the differential binding to the small GTPases. This result extends our previous observations that closely related pairs of effectors, such as MLK2-MLK3 and WASP–N-WASP, interact differentially with TC10 (33). TC10 has also been reported to be insensitive to the nucleotide exchange factor Dbl (17). Thus, TC10 may be designed to respond in a restricted way to only a subset of upstream signals and to activate only a specific subset of the downstream effectors that are regulated by Cdc42. Such restrictions may be essential if TC10 operates mainly in highly differentiated tissues such as muscle, where proliferative signals could be deleterious to function.
The tissue distribution of Borgs varies with the isoform. Borg1 mRNA was detected in all tissues examined, while Borg2 was expressed only at very low levels in brain and liver. Borg3 mRNA appears to be present at very low levels in most tissues, but a much longer transcript was clearly detectable in skeletal muscle. Whether the multiple Borg transcripts represent splice variants or partially processed mRNAs remains to be determined. We were unable to clone the full-length sequence of Borg4, and no mRNA was detected for this protein in any of the adult human tissues tested. It may therefore be expressed only in fetal or embryonic tissues.
We studied Borg1 and Borg3 in more detail. When expressed ectopically in NIH 3T3 cells (or in BHK cells), the proteins appeared to be predominantly cytoplasmic and induced a characteristic phenotype that included the formation of long thin extensions and protrusive lamellipodia as well as the partial loss of stress fibers. The effect may be related to the loss of stress fibers caused by inhibition of Rho function, which permits a more extensive remodeling of the F-actin into filopodia (35). The idea that there is competition between different GTPase-mediated signals for structuring the finite amount of actin present within the cell has been also proposed by others (32). We tested this hypothesis further by coexpressing Borg3 with wild-type Rho. The overexpression of Rho restored stress fibers and a more normal fibroblastic cell shape, supporting the idea that Borg3 can inhibit Rho function.
By what mechanism might the Borg proteins exert their effects on the cell? Conceivably, the Borg CRIB domain might bind to Cdc42 and/or TC10 and compete out other downstream effectors for these GTPases. We do not favor this mechanism, however, because a CRIB mutant of Borg3 that is unable to bind Cdc42 was fully capable of inducing the characteristic Borg phenotype. The CRIB mutant data also rule out the possibility that the phenotype might result from the interaction of GTP-bound Cdc42 and/or TC10 with Borg, which activates a downstream signal. Moreover, the coexpression of Borg3 with wild-type Cdc42 did not inhibit the formation of filopodia by Cdc42 but caused a dramatic increase in their length. A similar phenotype could be induced by the coexpression of the RhoGAP, p190, with Cdc42, supporting the proposal that Borgs can inhibit Rho function. Therefore, the Borg proteins most likely induce the observed effects on the cytoskeleton independently of a direct interaction with the small GTPases, although binding to Cdc42 may well modify these effects, either by changing the subcellular distribution of the Borg proteins or by inducing conformational changes which would alter their interactions with other proteins.
It is important to note that changes in stress fiber formation can occur independently of Rho activity. For example, the drug cytochalasin D activates Rho but causes the dissolution of stress fibers (38). Rho is believed to regulate stress fiber formation through its downstream effector, ROCK, which phosphorylates and inhibits the myosin light-chain (MLC) phosphatase. Phosphorylation of MLC mediates the formation of actomyosin bundles and stimulates contraction (21). A protein kinase effector of Cdc42 and Rac (PAK1) has been reported to phosphorylate and inhibit the MLC kinase, thereby reducing MLC phosphorylation and reducing actomyosin bundling (45), but these results have been disputed, and in another system PAK1 appears to be able to stimulate MLC phosphorylation (47). Our present data do not, therefore, definitively prove that Borg proteins cause disassembly of stress fibers through an inhibition of Rho function, but they are consistent with this hypothesis.
The cytoskeletal responses of cells to Rac have been shown to be independent of the effects of this GTPase on other signaling pathways such as that which activates JNK (24). A similar situation probably holds for other Rho family GTPases, including Cdc42. To determine whether Borg proteins participate in these other pathways, we examined whether the expression of Borg3 interferes with or potentiates JNK activation. We showed that wild-type HA3-tagged Borg3 inhibited the basal JNK activity. Surprisingly, like its effects on the cytoskeleton, the Borg3-mediated inhibition of JNK was independent of its ability to bind Cdc42. It remains to be determined whether the mechanism by which the Borg protein interferes with JNK is the same as that by which it inhibits Rho function.
Our observations on the effects of Borg3 on cell spreading revealed two distinct responses, only one of which was independent of Cdc42 binding. After replating onto fibronectin, the cells that expressed HA3-Borg3 spread very slowly, and the spread cells possessed an unusual cell shape and lacked stress fibers. Using the CRIB mutant which is defective in Cdc42 binding, we were able to partially restore the normal spreading rate but did not restore normal cell shape. We propose that the rate of spreading is at least partially dependent on the activation of Cdc42 and its interaction with a set of downstream effectors, which may be inhibited by competition with the Borg protein. WASP and N-WASP interact with the Arp2-Arp3 complex. This complex, which enhances actin nucleation, branching, and cross-linking of actin filaments in vitro, is involved in the formation of lamellipodia (27, 42). However, competition cannot completely account for the inhibition in cell spreading, because expression of the mutant Borg3(1–83), which can bind Cdc42, was also relatively ineffective in inhibiting spreading. Borg3(1–83) is expressed much less efficiently than wild-type protein, but even when only cells that expressed similar amounts of protein were compared, the Borg3(1–83) mutant did not show the same degree of inhibition of spreading as did the wild-type protein. It is likely, therefore, that protein-protein interactions involving the C-terminal domains of Borg are required, in addition to Cdc42 binding, to inhibit cell spreading on fibronectin. The abnormal cell shape caused by expression of either the CRIB mutant or the C-terminal deletion mutant is consistent with our other data that implicate Borgs in Rho inhibition and with evidence from other groups that Rho is switched on at a late stage in cell spreading and may be necessary for the assumption of a normal fibroblastic cell shape (5, 11).
The wounding assay for cell motility was less sensitive to ectopic expression of Borg3. Although a small inhibition was consistently observed, it was much less dramatic than the inhibition of spreading. This difference may reflect the very different times over which the two experiments were conducted. Motility was estimated after 24 h, during which time the cell could compensate for partial inhibition of endogenous functions either by changes in gene expression or by adaptation in the responsiveness to external signals.
Overall, our studies suggest that Borg proteins may interfere with the activity of the Rho GTPase and function as negative regulators in cross talk between Cdc42- and Rho-mediated signaling pathways. The mechanism by which Borg proteins inhibit actin stress fiber formation remains to be determined. They could inhibit signaling through the lysophosphatidic acid receptor or through the Gα proteins that couple to Rho-specific guanine nucleotide exchange factors, or they could inhibit the exchange factors themselves. Alternatively, they may interfere with the phosphorylation of MLC. It will be of interest, therefore, to identify binding partners for the Borg proteins other than Cdc42 and TC10 as a first step toward a complete description of their function.
ACKNOWLEDGMENTS
We thank Stan Hollenberg, Rick Cerione, Mark Rush, and Gary Bokoch for their kind gifts of reagents, and we thank Cheryl Neudauer, Nia Tatsis, and Tom Parsons for helpful discussions.
This work was supported by the Markey Center for Cell Signaling and by grant CA 56300 from the National Institutes of Health.
REFERENCES
- 1.Abo A, Qu J, Cammarano M S, Dan C, Fritsch A, Baud V, Belisle B, Minden A. PAK4, a novel effector for Cdc42Hs, is implicated in the reorganization of the actin cytoskeleton and in the formation of filopodia. EMBO J. 1998;17:6527–6540. doi: 10.1093/emboj/17.22.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagrodia S, Derijard B, Davis R J, Cerione R A. Cdc42 and PAK-mediated signaling leads to Jun kinase and p38 mitogen-activated protein kinase activation. J Biol Chem. 1995;270:27995–27998. doi: 10.1074/jbc.270.47.27995. [DOI] [PubMed] [Google Scholar]
- 3.Bahou W F, Campbell A D, Wicha M S. cDNA cloning and molecular characterization of MSE55, a novel human serum constituent protein that displays bone marrow stromal/endothelial cell-specific expression. J Biol Chem. 1992;267:13986–13992. [PubMed] [Google Scholar]
- 4.Barik S. Site-directed mutagenesis by double polymerase chain reaction: megaprimer method. In: White B A, editor. PCR protocols: current methods and applications. Totowa, N.J: Humana Press; 1993. pp. 277–286. [DOI] [PubMed] [Google Scholar]
- 5.Bourdoulous S, Orend G, MacKenna D A, Pasqualini R, Ruoslahti E. Fibronectin matrix regulates activation of Rho and Cdc42 GTPases and cell cycle progression. J Cell Biol. 1998;143:267–276. doi: 10.1083/jcb.143.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brondyk W H, McKiernan C J, Fortner K A, Stabila P, Holz R W, Macara I G. Interaction cloning of Rabin3, a novel protein that associates with the Ras-like GTPase Rab3A. Mol Cell Biol. 1995;15:1137–1143. doi: 10.1128/mcb.15.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burbelo P D, Drechsel D, Hall A. A conserved binding motif defines numerous candidate target proteins for both Cdc42 and Rac GTPases. J Biol Chem. 1995;270:29071–29074. doi: 10.1074/jbc.270.49.29071. [DOI] [PubMed] [Google Scholar]
- 8.Burridge K, Chrzanowska-Wodnicka M. Focal adhesions, contractility, and signaling. Annu Rev Cell Dev Biol. 1996;12:463–518. doi: 10.1146/annurev.cellbio.12.1.463. [DOI] [PubMed] [Google Scholar]
- 9.Chrzanowska-Wodnicka M, Burridge K. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J Cell Biol. 1996;133:1403–1415. doi: 10.1083/jcb.133.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark E A, Brugge J S. Integrins and signal transduction pathways: the road taken. Science. 1995;268:233–239. doi: 10.1126/science.7716514. [DOI] [PubMed] [Google Scholar]
- 11.Clark E A, King W G, Brugge J S, Symons M, Hynes R O. Integrin-mediated signals regulated by members of the Rho family of GTPases. J Cell Biol. 1998;142:573–586. doi: 10.1083/jcb.142.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coso O A, Chiariello M, Yu J C, Teramoto H, Crespo P, Xu N, Miki T, Gutkind J S. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 13.Coso O A, Teramoto H, Simonds W F, Gutkind J S. Signaling from G protein-coupled receptors to c-Jun kinase involves beta gamma subunits of heterotrimeric G proteins acting on a Ras and Rac1-dependent pathway. J Biol Chem. 1996;271:3963–3966. doi: 10.1074/jbc.271.8.3963. [DOI] [PubMed] [Google Scholar]
- 14.Drivas G T, Shih A, Coutavas E, Rush M G, D’Eustachio P. Characterization of four novel ras-like genes expressed in a human teratocarcinoma cell line. Mol Cell Biol. 1990;10:1793–1798. doi: 10.1128/mcb.10.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guasch R M, Scambler P, Jones G E, Ridley A J. RhoE regulates actin cytoskeleton organization and cell migration. Mol Cell Biol. 1998;18:4761–4771. doi: 10.1128/mcb.18.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 17.Hart M J, Eva A, Zangrilli D, Aaronson S A, Evans T, Cerione R A, Zheng Y. Cellular transformation and guanine nucleotide exchange activity are catalyzed by a common domain on the dbl oncogene product. J Biol Chem. 1994;269:62–65. [PubMed] [Google Scholar]
- 18.Hirai S, Katoh M, Terada M, Kyriakis J M, Zon L I, Rana A, Avruch J, Ohno S. MST/MLK2, a member of the mixed lineage kinase family, directly phosphorylates and activates SEK1, an activator of c-Jun N-terminal kinase/stress-activated protein kinase. J Biol Chem. 1997;272:15167–15173. doi: 10.1074/jbc.272.24.15167. [DOI] [PubMed] [Google Scholar]
- 19.Hotchin N A, Hall A. The assembly of integrin adhesion complexes requires both extracellular matrix and intracellular Rho/Rac GTPases. J Cell Biol. 1995;131:1857–1865. doi: 10.1083/jcb.131.6.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keely P J, Westwick J K, Whitehead I P, Der C J, Parise L V. Cdc42 and Rac1 induce integrin-mediated cell motility and invasiveness through Pi(3)K. Nature. 1997;390:632–636. doi: 10.1038/37656. [DOI] [PubMed] [Google Scholar]
- 21.Kimura K, Ito M, Amano M, Chihara K, Fukata Y, Nakafuku M, Yamamori B, Feng J, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase) Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- 22.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 23.Kozma R, Ahmed S, Best A, Lim L. The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol Cell Biol. 1995;15:1942–1952. doi: 10.1128/mcb.15.4.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamarche N, Tapon N, Stowers L, Burbelo P D, Aspenstrom P, Bridges T, Chant J, Hall A. Rac and Cdc42 induce actin polymerization and G1 cell cycle progression independently of P65(Pak) and the JNK/SAPk map kinase cascade. Cell. 1996;87:519–529. doi: 10.1016/s0092-8674(00)81371-9. [DOI] [PubMed] [Google Scholar]
- 25.Leung T, Chen X Q, Tan I, Manser E, Lim L. Myotonic dystrophy kinase-related Cdc42-binding kinase acts as a Cdc42 effector in promoting cytoskeletal reorganization. Mol Cell Biol. 1998;18:130–140. doi: 10.1128/mcb.18.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Machesky L M, Hall A. Rho—a connection between membrane receptor signalling and the cytoskeleton. Trends Cell Biol. 1996;6:304–310. doi: 10.1016/0962-8924(96)10026-x. [DOI] [PubMed] [Google Scholar]
- 27.Machesky L M, Insall R H. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr Biol. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- 28.Machesky L M, Hall A. Role of actin polymerization and adhesion to extracellular matrix in Rac- and Rho-induced cytoskeletal reorganization. J Cell Biol. 1997;138:913–926. doi: 10.1083/jcb.138.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackay D J, Esch F, Furthmayr H, Hall A. Rho- and Rac-dependent assembly of focal adhesion complexes and actin filaments in permeabilized fibroblasts: an essential role for ezrin/radixin/moesin proteins. J Cell Biol. 1997;138:927–938. doi: 10.1083/jcb.138.4.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miki H, Sasaki T, Takai Y, Takenawa T. Induction of filopodium formation by a Wasp-related actin-depolymerizing protein N-Wasp. Nature. 1998;391:93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- 31.Minden A, Lin A, Claret F X, Abo A, Karin M. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 32.Moorman J P, Luu D, Wickham J, Bobak D A, Hahn C S. A balance of signaling by Rho family small GTPases RhoA, Rac1 and Cdc42 coordinates cytoskeletal morphology but not cell survival. Oncogene. 1999;18:47–57. doi: 10.1038/sj.onc.1202262. [DOI] [PubMed] [Google Scholar]
- 33.Neudauer C L, Joberty G, Tatsis N, Macara I G. Distinct cellular effects and interactions of the Rho-family GTPase TC10. Curr Biol. 1998;8:1151–1160. doi: 10.1016/s0960-9822(07)00486-1. [DOI] [PubMed] [Google Scholar]
- 34.Nobes C, D, Hall A. Rho GTPases control polarity, protrusion, and adhesion during cell movement. J Cell Biol. 1999;144:1235–1244. doi: 10.1083/jcb.144.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nobes C D, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 36.Nobes C D, Lauritzen I, Mattei M G, Paris S, Hall A, Chardin P. A new member of the Rho family, Rnd1, promotes disassembly of actin filament structures and loss of cell adhesion. J Cell Biol. 1998;141:187–197. doi: 10.1083/jcb.141.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olson M F, Ashworth A, Hall A. An essential role for Rho, Rac, and Cdc42 GTPases in cell cycle progression through G1. Science. 1995;269:1270–1272. doi: 10.1126/science.7652575. [DOI] [PubMed] [Google Scholar]
- 38.Ren X-D, Kiosses W B, Schwartz M A. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 1999;18:578–585. doi: 10.1093/emboj/18.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Renshaw M W, Ren X-D, Schwartz M A. Growth factor activation of MAP kinase requires cell adhesion. EMBO J. 1997;16:5592–5599. doi: 10.1093/emboj/16.18.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ridley A J, Hall A. The small GTP-binding protein Rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 41.Ridley A J, Paterson H F, Johnston C L, Diekmann D, Hall A. The small GTP-binding protein Rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 42.Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kischner M W. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory Manual. Plainview, N.Y: CSH Press; 1989. pp. 16.32–16.36. [Google Scholar]
- 44.Sander E E, van Delft S, den Klooster J P, Reid T, van der Kammen R A, Michiels F, Collard J G. Matrix-dependent Tiam1/Rac signaling in epithelial cells promotes either cell-cell adhesion or cell migration and is regulated by phosphatidylinositol 3-kinase. J Cell Biol. 1998;143:1385–1398. doi: 10.1083/jcb.143.5.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanders L C, Matsumura F, Bokoch G M, de Lanerolle P. Inhibition of myosin light chain kinase by p21-activated kinase. Science. 1999;283:2083–2085. doi: 10.1126/science.283.5410.2083. [DOI] [PubMed] [Google Scholar]
- 46.Schwartz M A, Schaller M D, Ginsberg M H. Integrins: emerging paradigms of signal transduction. Annu Rev Cell Dev Biol. 1995;11:549–599. doi: 10.1146/annurev.cb.11.110195.003001. [DOI] [PubMed] [Google Scholar]
- 47.Sells M A, Boyd J T, Chernoff J. p21-Activated kinase (Pak1) regulates cell motility in mammalian fibroblasts. J Cell Biol. 1999;145:837–849. doi: 10.1083/jcb.145.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sells M A, Knaus U G, Bagrodia S, Ambrose D M, Bokoch G M, Chernoff J. Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr Biol. 1997;7:202–210. doi: 10.1016/s0960-9822(97)70091-5. [DOI] [PubMed] [Google Scholar]
- 49.Suetsugu S, Miki H, Takenawa T. The essential role of profilin in the assembly of actin for microspike formation. EMBO J. 1998;17:6516–6526. doi: 10.1093/emboj/17.22.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takaishi K, Sasaki T, Kotani H, Nishioka H, Takai Y. Regulation of cell-cell adhesion by Rac and Rho small G proteins in Mdck cells. J Cell Biol. 1997;139:1047–1059. doi: 10.1083/jcb.139.4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tapon N, Hall A. Rho, Rac and Cdc42 GTPases regulate the organization of the actin cytoskeleton. Curr Opin Cell Biol. 1997;9:86–92. doi: 10.1016/s0955-0674(97)80156-1. [DOI] [PubMed] [Google Scholar]
- 52.Tapon N, Nagata K, Lamarche N, Hall A. A new Rac target POSH is an SH3-containing scaffold protein involved in the JNK and NF-kappaB signalling pathways. EMBO J. 1998;17:1395–1404. doi: 10.1093/emboj/17.5.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tatsis N, Lannigan D A, Macara I G. The function of the p190 Rho GTPase-activating protein is controlled by its N-terminal GTP binding domain. J Biol Chem. 1998;273:34631–34638. doi: 10.1074/jbc.273.51.34631. [DOI] [PubMed] [Google Scholar]
- 54.Valencia A, Chardin P, Wittinghofer A, Sander C. The Ras protein family: evolutionary tree and role of conserved amino acids. Biochemistry. 1991;30:4637–4648. doi: 10.1021/bi00233a001. [DOI] [PubMed] [Google Scholar]
- 55.Van Aelst L, D’Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 56.Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, Saito Y, Nakao K, Jockusch B M, Narumiya S. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 1997;16:3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]