Abstract
Nitrate is both an important nutrient and a critical signaling molecule that regulates plant metabolism, growth, and development. Although several components of the nitrate signaling pathway have been identified, the molecular mechanism of nitrate signaling remains unclear. Here, we showed that the growth-related transcription factors HOMOLOG OF BRASSINOSTEROID ENHANCED EXPRESSION2 INTERACTING WITH IBH1 (HBI1) and its three closest homologs (HBIs) positively regulate nitrate signaling in Arabidopsis thaliana. HBI1 is rapidly induced by nitrate through NLP6 and NLP7, which are master regulators of nitrate signaling. Mutations in HBIs result in the reduced effects of nitrate on plant growth and ∼22% nitrate-responsive genes no longer to be regulated by nitrate. HBIs increase the expression levels of a set of antioxidant genes to reduce the accumulation of reactive oxygen species (ROS) in plants. Nitrate treatment induces the nuclear localization of NLP7, whereas such promoting effects of nitrate are significantly impaired in the hbi-q and cat2 cat3 mutants, which accumulate high levels of H2O2. These results demonstrate that HBI-mediated ROS homeostasis regulates nitrate signal transduction through modulating the nucleocytoplasmic shuttling of NLP7. Overall, our findings reveal that nitrate treatment reduces the accumulation of H2O2, and H2O2 inhibits nitrate signaling, thereby forming a feedback regulatory loop to regulate plant growth and development.
Transcriptional regulation of ROS by HBI family proteins contributes to nitrate signal transduction through regulating the nucleocytoplasmic shuttling of NLP7.
Introduction
Nitrogen is an essential macronutrient for plants, and its availability is a key determinant of plant growth and development and crop yield (Xu et al., 2012; O'Brien et al., 2016; Wang et al., 2018b; Fredes et al., 2019; Vidal et al., 2020). Hence, nitrogen fertilizers have been used as the major driving force for the improvement of crop yield worldwide (Stitt, 1999; Crawford and Forde, 2002; Wang et al., 2018b; Vidal et al., 2020). Nitrate is the main form of nitrogen available in the soil for plant uptake. Moreover, nitrate serves as the nitrogen signal for the rapid induction of a transcriptional response, called the “primary nitrate response” (PNR), which regulates a wide range of developmental processes including seed germination, root architecture, shoot development, and flowering (Forde, 2014; Yan et al., 2016; Lin and Tsay, 2017; Gras et al., 2018; Landrein et al., 2018). Nitrate is sensed by the dual-affinity transceptor (transporter/receptor) NITRATE TRANSPORTER 1.1 (NRT1.1/CHL1/NPF6.3), which is a dual-affinity transporter and can switch between high and low affinities through phosphorylation and dephosphorylation at the Thr101 site in response to different nitrate concentrations (Ho et al., 2009). At low concentrations of nitrate, NRT1.1 is phosphorylated by CIPK23–CBL9 complex (CIPK, CALCINEURIN B-LIKE (CBL)-INTERACTION PROTEIN KINASE; CBL, CALCINEURIN B-LIKE PROTEIN), shifting into a high-affinity nitrate transporter. In contrast, at high nitrate levels, NRT1.1 is dephosphorylated and turns into a low-affinity transporter (Ho et al., 2009; Hu et al., 2009). Nitrate triggers the accumulation of calcium ions (Ca2+) in the cytoplasm and nucleus, thereby activating three Ca2+-SENSOR PROTEIN KINASES (CPKs), namely CPK10, CPK30, and CPK32, which phosphorylate the NIN-like protein Nin-Like Protein (NLP)7 to promote its nuclear localization, where it acts as a master transcription factor to regulate the expression of a subset of nitrate-responsive genes in Arabidopsis thaliana (Marchive et al., 2013; Liu et al., 2017; Alvarez et al., 2020).
Reactive oxygen species (ROS), as the by-products of aerobic metabolism, initially studied in relation to their damaging effects, have emerged as major regulators of normal plant growth and development, as well as plant responses to stress (Mittler, 2017; Waszczak et al., 2018). Under nitrogen deficiency conditions, ROS concentration increases in Arabidopsis roots, suggesting that root hair cells may contain a sensing system for nitrogen deprivation (Shin et al., 2005). Recent studies in Arabidopsis uncovered an important role of ROS in nitrogen demand and nitrogen-mediated signal transduction (Jung et al., 2018; Safi et al., 2018). Deprivation of nitrogen triggers the accumulation of hydrogen peroxide (H2O2), which functions as a versatile signaling modulator in various regulatory pathways. The inhibition of H2O2 accumulation by chemical agents, such as potassium iodide or diphenyleniodonium NADPH-oxidase inhibitor), blocked the induction of gene expression associated with the nitrogen-starvation response (Safi et al., 2018). In maize (Zea mays), nitrogen depletion or supply affects the balance between superoxide () and H2O2 in the root apex transition zone through the regulation of Zmupb1 and Zmprx112 transcription (Trevisan et al., 2019). These findings indicated that ROS plays critical roles in nitrogen signal transduction pathway. However, the interplay between ROS and nitrogen signaling in plant development remains unclear.
HOMOLOG OF BRASSINOSTEROID-ENHANCED EXPRESSION2 INTERACTING WITH IBH1 (HBI1), a basic helix–loop–helix (bHLH) transcription factor, has been shown to function as a major hub in the central growth circuit of plants to regulate plant growth and immunity (Bai et al., 2012; Fan et al., 2014; Malinovsky et al., 2014; Wang et al., 2014, 2018a; Neuser et al., 2019; Cai et al., 2020). Growth-promoting signals, including dark, shade, high temperature, auxin, gibberellin, and brassinosteroids activate HBI1 at the post-transcriptional level to promote cell elongation through the BZR-ARF-PIF/DELLA (BAP/D) module coupled with a tripartite module of HLH and bHLH factors (Chaiwanon et al., 2016). In contrast, the pathogen Pst DC3000 and flagellin inhibit the activity of HBI1 at transcriptional level to repress plant immunity (Fan et al., 2014). A recent study reported that HBI1 regulates the tradeoff between growth and immunity by controlling apoplastic ROS homeostasis through the different regulation of the genes encoding NADPH oxidase and peroxidase. HBI1 directly induces the expression of RESPIRATORY BURST OXIDASE HOMOLOG C (RbohC), but represses the transcript accumulation of RbohA to promote cell expansion during leaf growth (Neuser et al., 2019). In addition, HBI1 interacts with the transcription factor TCP20 to positively regulate the C‐terminally Encoded Peptides (CEP)-mediated systemic nitrate acquisition (Chu et al., 2020).
In this study, we show that HBI1 positively regulates nitrate signal transduction through controlling ROS homeostasis in plants. Nitrate supply significantly induces HBI1 and BEE2 expressions through NLP6 and NLP7. HBI1 promotes ROS scavenging by directly regulating the expression of CAT2. Loss of functions of HBI1, BEE2, and another two closet homolog genes lead to ROS accumulation, the reduced promoting effects of nitrate on shoot growth and root development, and ∼22% of nitrate-responsive genes were no longer regulated by nitrate. Nitrate treatment induced the nuclear localization of NLP7, but such effects of nitrates were impaired in hbi-q and cat2 cat3 mutants, which accumulated high levels of H2O2 in plants. Together, our study reveals that HBI-mediated ROS homeostasis regulates nitrate signaling by modulating the nucleocytoplasmic shuttling of NLP7.
Results
HBI1 and BEE2 are induced by nitrate
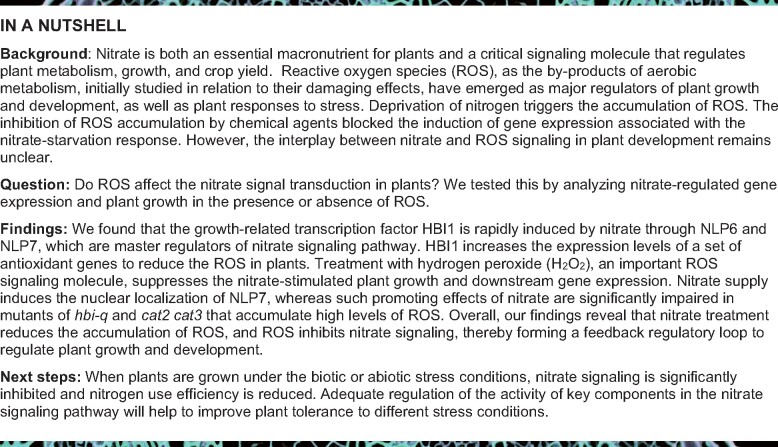
We have previously reported that HBI1 integrates a wide range of environmental signals and endogenous programs to regulate plant growth and immunity (Bai et al., 2012; Fan et al., 2014; Chu et al., 2020). To further understand the function of HBI1 in plant growth and development, we performed gene-expression analysis of HBI1 in Arabidopsis transcriptomic data and found that the expression of HBI1 and its homolog gene BEE2 was rapidly induced by nitrate. Integrative network bioinformatics identified HBI1 as a potential regulatory transcription factor in the nitrate response, ranking among the top five candidate regulatory genes in Arabidopsis (Alvarez et al., 2014). To test whether HBI1 and BEE2 are regulated by nitrate, we analyzed the expression levels of HBI1 and BEE2 upon nitrate treatment at different times or different concentrations. The transcript levels of HBI1 and BEE2 significantly increased as early as 15 min after nitrate treatment and increased over time (Figure 1A;Supplemental Figure S1A). When grown under different concentrations of nitrate, HBI1 and BEE2 expression increased at more than 300-µM nitrate concentration and became more prominent with increasing nitrate concentrations, indicating that nitrate induced the expression of HBI1 and BEE2 in a dose-dependent pattern (Figure 1B). The levels of HBI1-YFP protein expressed from the constitutive 35S promoter did not change dramatically after nitrate treatment, whereas the level of HBI1-YFP protein derived from its endogenous HBI1 promoter significantly increased upon nitrate treatment, confirming that nitrate regulated HBI1 at the transcriptional level (Supplemental Figure S1, B and C). These results strongly indicated that HBI1 and BEE2 are the early responsive genes of nitrate signaling.
Figure 1.
Nitrate induces the expression of HBI1 and BEE2. A, HBI1 and BEE2 are the early responsive genes of nitrate. Seedlings of Col-0 grown on medium with 7-mM KNO3 for 7 days were transferred to nitrogen-free medium for 2 days, and then treated with 10-mM KNO3 for different times. B, Nitrate induces the expression of HBI1 and BEE2 in a dose-dependent manner. Wild-type (Col-0) plants were grown on medium containing different concentrations of KNO3 for 14 days. The transcript levels of HBI1 and BEE2 were quantified by real-time PCR. PP2A gene was used as an internal control. Error bars are sd of three biological replicates. C, NLP6 and NLP7 play critical roles in the nitrate-induced expression of HBI1 and BEE2. Seedlings of Col-0, nlp7-1, and nlp6-1 nlp7-1 grown on medium with 7-mM KNO3 for 7 days were transferred to nitrogen-free medium for 2 days, and then treated with 10-mM KNO3 for 3 h. The transcript levels of HBI1 and BEE2 were quantified by real-time PCR. PP2A gene was used as an internal control. Error bars are sd of three biological replicates. Different letters above bars indicate statistically significant difference between samples (two-way ANOVA analysis followed by Tukey’s test, P < 0.05). D, E, Transient gene expression assay showed NLP7 promoted the expression of HBI1 and BEE2. The promoters of HBI1 or BEE2 fused to the luciferase reporter gene were co-transfected with 35Spro:GFP or 35Spro:NLP7-GFP into mesophyll protoplast of wild-type plants with or without nitrate treatment. The luciferase activities were normalized by Renilla luciferase as an internal control. Different letters above bars indicate statistically significant difference between samples (two-way ANOVA analysis followed by Tukey’s test, P < 0.05). F, G, ChIP-qPCR analysis of NLP7 binding to the promoters of HBI1 and BEE2. The 35Spro:NLP7-YFP and 35Spro:YFP transgenic plants growing in half-strength Murashige and Skoog (MS) liquid medium for 12 days were used to perform the ChIP analysis. Schematic illustration of different regions in the promoters of HBI1 and BEE2. The levels of NLP7 binding were calculated as the ratio between 35Spro:NLP7-YFP and 35Spro:YFP control, and then normalized to that of the control gene PP2A. Error bars indicate sd of three independent experiments. Asterisks between bars indicate statistically significant differences between samples (Student’s t test, *P < 0.05).
Due to the rapid increase in the transcription levels of HBI1 and BEE2 upon nitrate treatment, we speculated that the core transcription factors involved in nitrate signaling contribute to the transcriptional regulation of HBI1 and BEE2. NLP6 and NLP7 have been reported to be the master transcription factors in the nitrate signaling pathway (Marchive et al., 2013; Liu et al., 2017; Alvarez et al., 2020). To test whether the regulation of HBI1 and BEE2 by nitrate is dependent on NLP6 and NLP7, we performed reverse transcription quantitative polymerase chain reaction (RT-qPCR) experiments using wild-type plants, nlp7-1 and nlp6-1 nlp7-1 with or without nitrate treatment. Nitrate significantly induced the expression of HBI1 and BEE2 in wild-type plants, while such effects of nitrate were dramatically reduced in nlp7-1 and nlp6-1 nlp7-1 mutants (Figure 1C). The expression levels of HBI1 homologous genes, including bHLH44, bHLH50, bHLH137, and bHLH62, were significantly induced by nitrate supply in wild-type plants, but weakly induced in nlp6-1 nlp7-1 mutants (Supplemental Figure S2, A–F). Protoplast transient gene-expression assays showed that co-expression of NLP7 increased the expression of the luciferase reporter driven by the HBI1 or BEE2 promoters (Figure 1, D and E). The recent published chromatin immunoprecipitation sequencing data of NLP7 showed HBI1 is the target gene of NLP7 (Alvarez et al., 2020). Consistent with this, our chromatin immunoprecipitation–quantitative PCR (ChIP–qPCR) analysis confirmed that NLP7 directly bound to the promoters of HBI1 and BEE2 (Figure 1, F and G). These results indicated NLP6 and NLP7 play critical roles in the nitrate-induced expression of HBI1 and BEE2.
HBI and its close homologs (HBIs) positively regulate nitrate-mediated plant growth
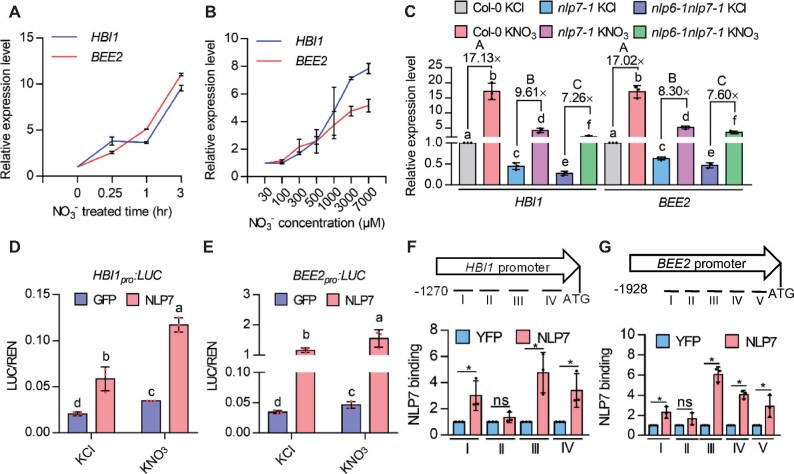
Because HBI1 and BEE2 are early nitrate-responsive genes, we wanted to determine whether HBI1 and BEE2 are involved in nitrate-regulated plant growth and development. Nitrate has been reported to stimulate shoot growth and regulate the elongation of primary and lateral root (Araya et al., 2014; Moreno et al., 2020; Swift et al., 2020). We first analyzed the nitrate response in the loss-of-function mutants of HBI1 and BEE2. The results showed hbi1 and bee2 single mutants displayed slightly decreased cotyledon size and fresh weight, and shorter primary and total lateral root lengths compared to wild-type plants in the presence of nitrate (Supplemental Figure S3, A–F). Considering the functional redundancy of HBI1 family, three closest genes of HBI1 including BEE2, bHLH31, and bHLH79 were selected to generate higher-order HBI mutants. The hbi1 bee2 double mutants showed a significant reduction in cotyledon size, fresh weight, and root growth; the addition of mutations in bHLH31 or bHLH79 enhanced the phenotype of hbi1 bee2; and simultaneous mutation of hbi1, bee2, bhlh31, and bhlh79 (hbi-q) resulted in significantly reducing the nitrate-promoting effects on plant growth. Overexpression of HBI1 (HBI1-Ox) led to an enlarged cotyledon in all concentrations of nitrate and hypersensitivity to nitrate in terms of the promotion of plant fresh weight (Figure 2, A–C; Supplemental Figure S4, A and B). These results indicated that HBI1 and its homologs, which we termed HBIs, play important roles in nitrate-mediated plant growth and development.
Figure 2.
HBIs positively regulate nitrate-mediated plant growth. A, B, Quantification of HBI-stimulated shoot growth in the presence of different concentrations of nitrate. Seedlings of the wild-type, HBI1-Ox, and hbi-q were grown on the medium with different concentrations of nitrate for 7 days. Scale bar represents 5 mm. Error bars indicate sd (n = 16–30). Asterisks indicate statistically significant differences among Col-0, HBI1-OX, and hbi-q at individual nitrate concentration (Student’s t test, *P < 0.05). C, Quantification of the fresh weight of the wild type, HBI1-Ox, and hbi-q that were grown on the medium with different concentrations of nitrate under 16-h light/8-h dark for 4 weeks. Error bars indicate sd (n = 15–26). Asterisks indicate statistically significant differences among Col-0, HBI1-OX, and hbi-q at individual nitrate concentration (Student’s t test, *P < 0.05). D, E, Quantification of the promoting effects of HBIs on the shoot growth in the presence of different nitrogen sources. Seedlings of the wild-type, HBI1-Ox, and hbi-q were grown on the medium containing 7 mM different nitrogen sources for 7 days. Scale bar represents 5 mm. Error bars indicate sd (n = 20–36). Different letters above bars indicate statistically significant differences between samples (two-way ANOVA analysis followed by Tukey’s test, P < 0.05). F, Quantification of the fresh weight of the wild-type, HBI1-Ox, and hbi-q that were grown on the medium with different nitrogen sources for 12 days. Every three plants is one group. Error bars indicate sd (n = 4). Different letters above bars indicate statistically significant differences between samples (two-way ANOVA analysis followed by Tukey’s test, P < 0.05).
To examine the functions of HBI1 in the nitrate-mediated plant growth and development, we analyzed the growth phenotypes of the wild-type Col-0, hbi-q, and HBI1-Ox that were grown in the medium containing the 7 mM different nitrogen sources under 16-h light/8-h dark photoperiod for 7 days. HBI1-Ox showed increased shoot growth and root development phenotype in all growth conditions (Figure 2, D–F). However, when plants were grown on medium containing KCl or , the hbi-q mutant displayed a similar growth phenotype to wild-type plants; and when plants were grown on the medium with KNO3, nitrate significantly stimulated shoot growth in wild-type plants, but such effects of nitrate were significantly reduced in hbi-q (Figure 2, D–F). The hbi-q mutant exhibits deficiency in nitrate-regulated shoot growth and root development, indicating HBIs play important roles in the nitrate-mediated plant growth and development.
To further determine the functions of HBI1 in the nitrate signaling, we analyzed the shoot-growth phenotypes of the wild-type, HBI1-Ox, chl1-5, and HBI1-Ox/chl1-5. The results showed that chl1-5 partially suppressed the promoting effects of HBI1 on shoot growth in the presence of nitrate (Supplemental Figure S5, A and B). These results suggested the promoting effects of HBI1 on plant growth are partial dependent on the nitrate transceptor CHL1/NRT1.1. Together, these results strongly demonstrated that HBIs play important roles in nitrate-promoted plant growth and development.
HBIs are required for nitrate-regulated gene expression
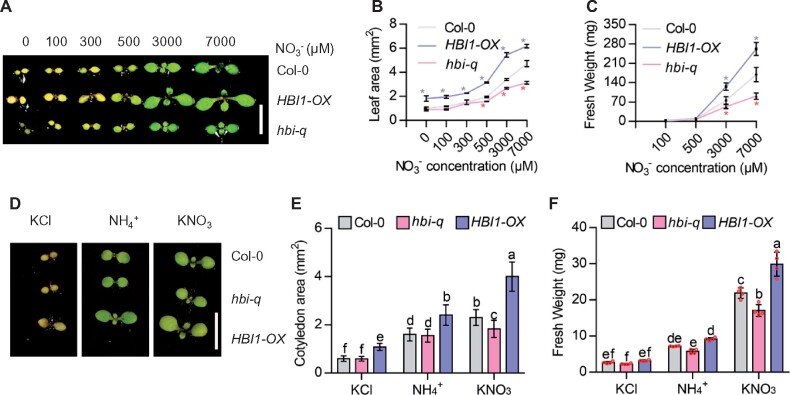
Given the important roles of HBIs in nitrate-mediated plant growth and the rapid induction of HBIs by nitrate, we speculated that HBIs are involved in the transcriptomic response to nitrate. To test this hypothesis, we carried out RNA sequencing (RNA-Seq) experiments with the wild-type Col-0, hbi-q, and nlp7-1 plants that were grown on MGRL medium (7-mM nitrate) for 7 days, subjected to nitrate starvation 2 days, and then treated with 10-mM KCl or 10-mM KNO3 for 3 h. RNA-Seq analysis identified 3,431, 3,703, and 3,541 differentially expressed genes (DEGs) that responded to nitrate in wild-type plants, hbi-q, and nlp7-1 mutants, respectively (Figure 3, A and B; Supplemental Data Sets S1–S4), and they significantly overlapped with each other. Among the 3,431 genes regulated by nitrate in wild-type plants, 764 genes (22.3%) were no longer responsive to nitrate in hbi-q mutant, and 1,447 (42.2%) genes were not regulated by nitrate in the nlp7-1 mutant. Of 764 HBI-affected nitrate-responsive genes, 604 genes (79.1%) also depended on NLP7 to respond to nitrate, indicating HBIs and NLP7 regulate the expression of nitrate-responsive genes through a common signaling pathway (Figure 3, A and B). Gene ontology (GO) analysis revealed that genes involved in nitrate and organonitrogen, transmembrane transporter, gibberellin metabolic, root development, and starch metabolism are the most significantly enriched functional classes of the HBI-affected nitrate-regulated genes and NLP7-affected nitrate-regulated genes, suggesting that HBIs, similar to NLP7, play important roles in diverse nitrate-mediated cellular and metabolic processes (Figure 3C).
Figure 3.
HBIs are required for nitrate-regulated gene expression. A, Venn diagram showing the overlap between sets of genes significantly regulated by nitrate in the wild-type, nlp7-1, and hbi-q mutants. B, Hierarchical cluster analysis of the expression data of 3,431 genes regulated by nitrate in the wild-type, nlp7-1, and hbi-q. The numerical values for the blue-to-red gradient bar represent log2 of the ratio. C, GO analysis of nitrate-regulated HBI1-affected, NLP7-affected, or HBI1- and NLP7 co-affected genes. Numbers indicate the percentages of genes belonging to each GO category. Genomic denotes the random genomic control. D, E, HBIs are required for nitrate-induced expression of NRT1.1 and NRT2.1. Seedlings of Col-0 and hbi-q were grown on medium with 7-mM KNO3 for 5 days, transferred to nitrogen-free medium for 2 days, and then treated with 10-mM KNO3 for 30 min. The transcript levels of NRT1.1 and NRT2.1 were quantified by real-time PCR. PP2A gene was used as an internal control. Error bars are sd of three biological replicates. Different letters above bars indicate statistically significant differences between samples (two-way ANOVA analysis followed by Tukey’s test, P < 0.05). Asterisks between bars indicate statistically significant different effects of nitrate on the expression of NRT1.1 and NRT2.1 in Col-0 and hbi-q (Student’s t test, *P < 0.05). F, G, HBI1 directly binds to the promoters of NRT1.1 and NRT2.1. The 35Spro:HBI1-YFP and 35Spro:YFP transgenic plants growing in half-strength MS liquid medium for 12 days were used to perform the ChIP analysis. The levels of HBI1 binding were calculated as the ratio between 35Spro:HBI1-YFP and 35Spro:YFP control, and then normalized to that of the control gene PP2A. Error bars indicate sd of three independent experiments. Asterisks between bars indicate statistically significant differences between samples (Student’s t test, *P < 0.05). H, I, Transient assays showed that HBI1 induces the expression of NRT1.1 and NRT2.1. The promoters of NRT1.1 or NRT2.1 fused to the luciferase reporter gene were co-transfected with 35Spro:GFP or 35Spro:HBI-GFP into mesophyll protoplasts of wild-type plants with or without nitrate treatment. The luciferase activities were normalized by Renilla luciferase as an internal control. Asterisks between bars indicate statistically significant differences between samples (Student’s t test, *P < 0.05).
To further determine the function of HBIs in nitrate-regulated gene expression, we performed the RT-qPCR to compare the expression of nitrate-responsive genes NRT1.1 and NRT2.1 in Col-0 and hbi-q in response to nitrate. When seedlings were treated with nitrate for 3 h, like in the RNA-Seq experiments, we found that nitrate had similar inducing effects on the expression of NRT1.1 and NRT2.1 in wild-type plants and hbi-q mutants. However, when nitrate treatment was for 30 min, the nitrate-increased expression of NRT1.1 and NRT2.1 was significantly reduced in hbi-q mutant, suggesting that HBIs might play an important role in the early induction of NRT1.1 and NRT2.1 by nitrate (Figure 3, D and E). We validated these results using another internal control, ACT2 (Supplemental Figure S6A). ChIP–qPCR analysis revealed that HBI1 directly binds to the promoters of NRT1.1 and NRT2.1 (Figure 3, F and G). Furthermore, protoplast transient gene-expression assays showed that HBI1 significantly induced the expression of NRT1.1 and NRT2.1 (Figure 3, H and I). These results indicated that HBIs play important roles in the nitrate regulation of downstream gene expression.
HBIs contribute to the nitrate-triggered ROS scavenging
A recent study reported that HBI1 controls the tradeoff between plant growth and immunity through the transcriptional regulation of ROS homeostasis (Neuser et al., 2019). The significant enrichment of ROS-related genes among HBI-affected nitrate-regulated genes suggested that ROS might play an important function in the HBI-mediated nitrate response (Supplemental Figure S6B). Peroxidases play critical roles in the ROS homeostasis of plants. Our transcriptomic analysis showed the expression of 14 peroxidases was regulated by nitrate treatment, of which 8 were upregulated and 6 were downregulated by nitrate. Mutation of HBIs significantly reduced the promoting effects of nitrate on the most of upregulated peroxidases, but only weak effects on the nitrate-repressed peroxidases (Supplemental Figure S6C). RT-qPCR analysis further confirmed that nitrate significantly induced the expression of peroxidase genes including PRX5, PRX27, PRX28, PRX52, and PRX62, and such promoting effects of nitrate were impaired in hbi-q mutants (Supplemental Figure S6D).
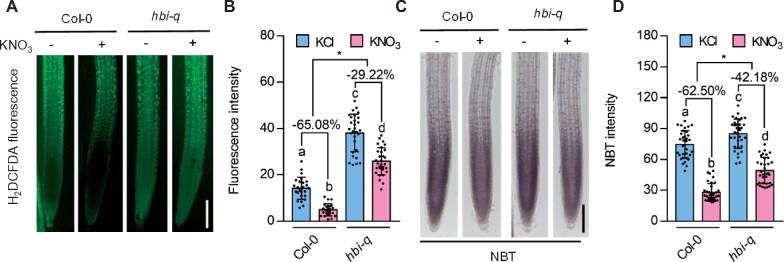
To determine whether nitrate regulates ROS levels in plants, we measured ROS content in wild-type plants and hbi-q mutants with or without nitrate treatment using the fluorescent dye 2′, 7′-dichlorofluorescin diacetate (H2DCFDA) and nitroblue tetrazolium (NBT; Bielski et al., 1980; Yao and Greenberg, 2006). The nitrate treatment significantly reduced the ROS content in wild-type seedlings. Loss of functions of HBIs resulted in the high accumulation of ROS in the absence of nitrate and failure to effectively scavenge ROS after nitrate treatment (Figure 4, A to D; Supplemental Figure S7, A and B). These results indicate that nitrate induces scavenging of ROS through HBI-mediated increases in the expression levels of a set of antioxidant genes.
Figure 4.
HBIs contribute to the nitrate-triggered ROS scavenging. A, H2DCFDA staining for H2O2 in the primary root tips of the wild-type and hbi-q treated with KCl or KNO3. Bar = 100 μm. Wild-type plants and hbi-q mutants growing on medium with 10-mM KNO3 for 5 days were transferred to nitrogen-free medium for 2 days and then treated with 10-mM KCl or KNO3 for 1 h. B, Quantification of H2DCFDA fluorescence intensities in the images of (A). Error bars, sd (n = 22–29 images). C, NBT staining showed that nitrate treatment significantly reduced the contents of ROS in the primary roots of wild-type plants (Col-0), but had weak effects in those of hbi-q. Bar = 100 μm. Wild-type plants and hbi-q mutants growing on medium with 10-mM KNO3 for 5 days were transferred to nitrogen-free medium for 2 days then treated with 10-mM KCl or KNO3 for 1 h. D, Quantification of NBT intensities in the images of (C). Error bars, sd (n = 34 images). For (B) and (D), different letters above bars indicate statistically significant difference between samples (two-way ANOVA analysis followed by Tukey’s test, P < 0.05). Asterisks between bars indicate statistically significant different effects of nitrate on the scavenging ROS in Col-0 and hbi-q (Student’s t test, *P < 0.05).
Catalase plays a critical role in HBI1-mediated plant growth
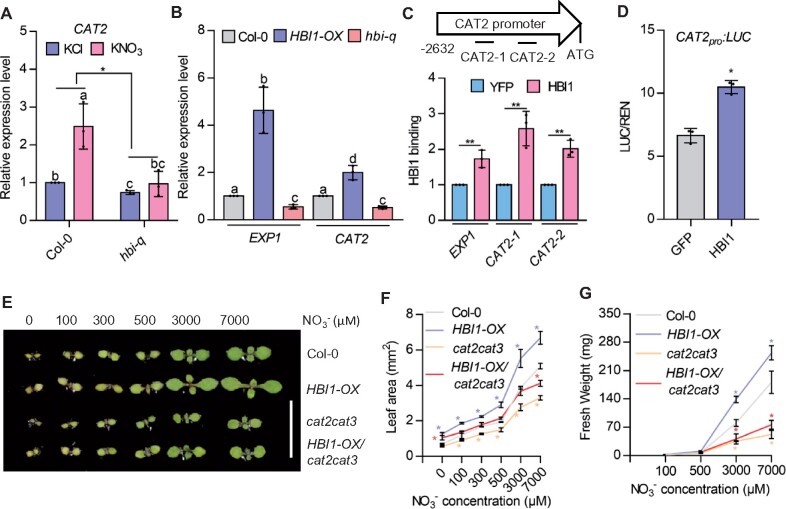
Catalase is a key H2O2 scavenging enzyme in plants (Su et al., 2018). Our RNA-Seq and RT-qPCR results revealed that nitrate induced the expression of CAT2 through HBIs (Figure 5A;Supplemental Data Set S1). We selected CAT2 to determine whether the promoting effects of HBIs on plant growth are dependent on ROS scavenging. We observed that the overexpression of HBI1 induced CAT2 expression, and mutation in HBIs led to a decreased transcript of CAT2 (Figure 5B). ChIP–qPCR analysis showed that HBI1 directly binds to the promoter of CAT2 (Figure 5C). The analysis of the protoplast transient assays showed that HBI1 promoted the expression of CAT2 (Figure 5D). These results indicated HBI1 directly promoted CAT2 expression.
Figure 5.
Catalase plays critical roles in HBI-promoted nitrate responses. A, HBIs are required for nitrate-induced the expression of CAT2. Seedlings of Col-0 and hbi-q grown on medium with 7-mM KNO3 for 5 days were transferred to nitrogen-free medium for 2 days, and then treated with 10-mM KNO3 for 3 h. The transcript levels of CAT2 were quantified by real-time PCR. PP2A was used as an internal control. Error bars are sd of three biological replicates. Different letters above bars indicate statistically significant difference between samples (two-way ANOVA analysis followed by Tukey’s test, P < 0.05). Asterisks between bars indicate statistically significant different effects of nitrate on the CAT2 expression in the wild-type and hbi-q mutant (Student’s t test, *P < 0.05). B, RT-qPCR analysis of the expression of CAT2 and EXP1 in the wild-type, HBI1-Ox, and hbi-q that were grown on half-strength MS medium under 16-h light/8-h dark photoperiod for 7 days. PP2A was used as an internal control and EXP1 was used as a positive control. Error bars indicate sd of three independent experiments. Different letters above bars indicate statistically significant differences between samples (one-way ANOVA analysis followed by Tukey’s test, P < 0.05). C, HBI1 directly binds to the promoter of CAT2. The 35Spro:HBI1-YFP and 35Spro:YFP transgenic plants growing in half-strength MS liquid medium for 12 days were used to perform the ChIP analysis. Illustration of different regions in the promoter of CAT2. The levels of HBI1 binding were calculated as the ratio between 35Spro:HBI1-YFP and 35Spro:YFP control, and then normalized to that of the control gene PP2A. EXP1 was used as a positive control. Error bars indicate sd of three independent experiments. Asterisks between bars indicate statistically significant differences between samples (Student’s t test, *P < 0.05). D, Transient assays showed HBI1 promotes the expression of CAT2. The CAT2 promoter fused to the luciferase reporter gene was co-transfected with 35Spro:GFP or 35Spro:HBI1-GFP into mesophyll protoplasts of wild-type plants. The luciferase activities were normalized by Renilla luciferase as an internal control. Error bars indicate sd of three independent experiments. Asterisks between bars indicate statistically significant differences between samples (Student’s t test, *P < 0.05). E, F, Mutations of CAT2 and CAT3 suppressed the HBI1-promoted shoot growth. Seedlings of Col-0, cat2cat3, HBI1-Ox, and HBI1-Ox/cat2cat3 were grown on medium with different concentrations of nitrate for 7 days. Scale bar represents 10 mm. Error bars indicate sd (n = 24–36). Asterisks indicate statistically significant differences between Col-0, cat2 cat3, HBI1-Ox, and HBI1-Ox/cat2 cat3 (Student’s t test, *P < 0.05). G, Quantification of the fresh weight of the wild-type, Col-0, cat2 cat3, HBI1-Ox, and HBI1-Ox/cat2 cat3 that were grown on the medium with different concentrations of nitrate under 16h light/8h dark for 4 weeks. Error bars indicate sd (n = 15–26). Asterisks indicate statistically significant differences between, cat2 cat3, HBI1-Ox, and HBI1-Ox/cat2 cat3 (Student’s t test, *P < 0.05).
Next, we generated the HBI1-Ox/cat2 cat3 plants by crossing HBI1-Ox transgenic plants with cat2 cat3 mutants. The analysis of ROS content showed that nitrate treatment significantly reduced the ROS levels in the wild-type and HBI1-Ox plants, but had weaker effects in the cat2 cat3 and HBI1-Ox/cat2 cat3 (Supplemental Figure S8, A to D). In the wild-type background, overexpression of HBI1 led to larger cotyledons and higher fresh weight compared to wild-type plants. However, mutation in CAT2 and CAT3 significantly reduced the promoting effects of HBI1 on plant growth (Figure 5, E to G; Supplemental Figure S9, A to E). These results indicated that HBI1 might promote plant growth by promoting ROS scavenging through catalase.
H2O2 treatment inhibits the nitrate-mediated plant growth
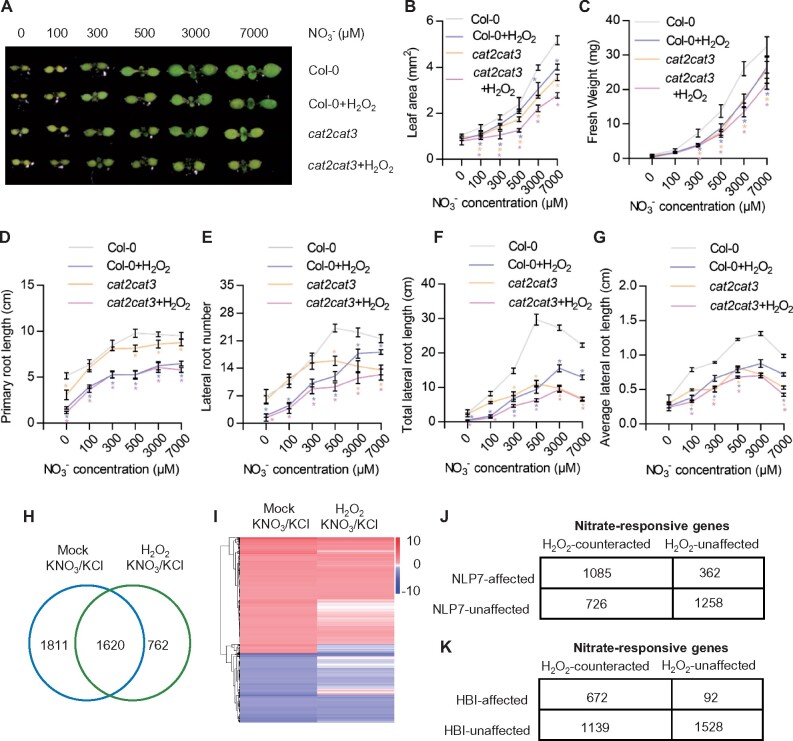
To investigate whether ROS are involved in nitrate-mediated plant growth and development, we analyzed the growth phenotypes of wild-type plants and cat2 cat3 mutants that were grown on medium with different nitrogen sources. In the presence of KCl, , or Gln, cat2 cat3 and wild-type plants showed a similar growth phenotype. However, when grown on the medium with KNO3, nitrate supply significantly stimulated shoot growth of wild-type plants, but had weaker effects on the growth of cat2 cat3, suggesting CAT2 and CAT3 are required for nitrate-promoted shoot growth (Supplemental Figure S9, D and E). To further examine the effects of ROS on nitrate-stimulated plant growth and development, we analyzed the growth phenotypes of wild-type plants and cat2 cat3 mutants in the presence of different concentrations of nitrate and/or H2O2. Nitrate treatment promoted cotyledon expansion, stimulated shoot growth, and increased primary root length, total lateral root length and lateral root number in wild-type plants. However, mutations of CAT2 and CAT3 or co-treatment with H2O2 strongly counteracted the effects of nitrate on plant growth and development (Figure 6, A to G). Additionally, we analyzed the effects of different concentrations of H2O2 on plant growth at the same nitrate concentration and found that the length of primary roots and the number of lateral roots were significantly reduced with increasing H2O2 concentration, indicating that H2O2 inhibited root growth and development in a dose-dependent manner (Supplemental Figure S10, A–D). These results suggested that nitrate-mediated removal of ROS contributes to nitrate-induced plant growth and development.
Figure 6.
H2O2 negatively regulates the nitrate-mediated plant growth and gene expression. A, Seedlings of wild-type plants and cat2 cat3 mutants grown on medium containing different concentrations of nitrate with or without 0.5-mM H2O2 for 7 days. Scale bar represents 10 mm. B, Measurement of the cotyledon area of plants in (A). Error bars indicate sd (n = 16–28). Asterisks indicate statistically significant differences between Col-0 and cat2 cat3 at individual concentration (Student’s t test, *P < 0.05). C, Measurement of the fresh weight of the wild-type and cat2 cat3 grown on medium containing different concentrations of nitrate with or without 0.5-mM H2O2 for 15 days. Error bars indicate sd (n = 21–30). Asterisks indicate statistically significant differences between Col-0 and cat2 cat3 at individual concentration (Student’s t test, *P < 0.05). D–G, H2O2 inhibited the nitrate-regulated root development. Wild-type and cat2cat3 seedlings were grown on medium with or without 0.5-mM H2O2 and different concentrations KNO3 for 12 days. Primary root length (D), lateral root number (E), total lateral root length (F), and average lateral root length (G) were quantified. Error bars indicate sd (n = 30). Asterisks indicate statistically significant differences between Col-0 and cat2 cat3 at individual concentration (Student’s t test, *P < 0.05). H, Venn diagram showing the overlap between sets of genes significantly regulated by nitrate in the wild-type with or without H2O2 treatment. I, Hierarchical cluster analysis of the expression data of 3,431 genes regulated by nitrate in wild-type plants with or without H2O2 treatment. The numerical values for the blue-to-red gradient bar represent log2 of the ratio. J, K, Comparison of the HBI- or NLP7-affected nitrate-responsive genes with H2O2-counteracted nitrate-responsive genes.
H2O2 treatment inhibits the regulation of gene expression by nitrate
To determine the effects of ROS on the nitrate-regulated gene expression, we performed RT-qPCR in the wild-type and cat2 cat3 with or without nitrate treatment. RT-qPCR analysis showed that mutations of CAT2 and CAT3 significantly inhibited the nitrate-induced expression of NRT1.1 and NRT2.1 (Supplemental Figure S11, A and B). NRP-YFP is a nitrate-responsive marker line (Wang et al., 2009). Nitrate treatment significantly increased the fluorescence intensity of NRP-YFP in the wild-type background, but such effects of nitrate were impaired in cat2 cat3 mutants (Supplemental Figure S11, C and D). These results indicated that CAT-mediated scavenging of ROS plays an important role in the nitrate signaling pathway.
To further determine the roles of ROS in nitrate-regulated gene expression at the genomic level, we performed the RNA-Seq experiments with the wild-type Col-0 grown on MGRL medium (7-mM nitrate) for 7 days. After this period, nitrate starvation was imposed for 2 days, and then plants were treated with 10-mM KCl, 10-mM and/or 0.5-mM H2O2 for 3 h. There were 3,431 genes affected with more than two-fold changes by only nitrate treatment, 1,810 genes regulated by only H2O2 treatment, and 2,382 genes regulated by nitrate and H2O2 co-treatment (Figure 6H;Supplemental Data Sets S1, S5, and S6). The genes regulated by only nitrate or only H2O2 treatment did not display a clear regulation pattern, which may be due to the experimental conditions we used to carry out RNA-Seq (Supplemental Figure S12, A and B). Before the plants were treated with nitrate or H2O2, the plants were grown on nitrate-free medium for 2 days, which resulted in high accumulation of ROS in plants. However, among 3,431 genes regulated by nitrate under normal conditions, 1,811 genes (52.8%) were no longer responsive to nitrate when co-treated with H2O2; we termed these H2O2-counteracted nitrate-responsive genes, and the other 1,620 genes were termed H2O2-unaffected nitrate-responsive genes (Figure 6, H and I). GO analysis showed that terms related to nitrate-regulated genes, response to nitrogen compound, starvation, nutrient levels, oxidoreductase activity, anion transport, starch metabolism, and gibberellin metabolic processes were highly enriched in the H2O2-counteracted nitrate-regulated genes (Supplemental Figure S13). These results demonstrated that H2O2 plays a negative role in nitrate signal transduction.
To further determine whether NLP7 and HBIs are involved in the H2O2–counteracted gene expression in response to nitrate, we compared the NLP7- or HBI-affected nitrate-responsive genes with H2O2-counteracted nitrate-responsive genes. The results showed that most of H2O2-counteracted nitrate-responsive genes are not regulated by nitrate in nlp7-1 mutants, and most NLP7-unaffected nitrate-responsive genes are also not affected by H2O2 treatment (Figure 6J), suggesting NLP7 plays critical roles in the H2O2 inhibition of nitrate-regulated downstream gene expression. Among the 764 HBI-affected nitrate-responsive genes, 672 genes (88%) were regulated by nitrate through the H2O2-counteracted pathway, whereas among H2O2-counteracted nitrate-responsive genes, HBI-dependent nitrate-responsive genes were not enriched (Figure 6K), indicating that HBIs are not the targets of H2O2 in inhibiting nitrate-regulated gene expression. These transcriptomic analyses, together with genetic data showing the catalase is required for the functions of HBIs, indicated that HBIs regulate plant growth and gene expression in response to nitrate by modulating ROS homeostasis in plants.
H2O2 inhibits the nitrate-triggered nuclear localization of NLP7
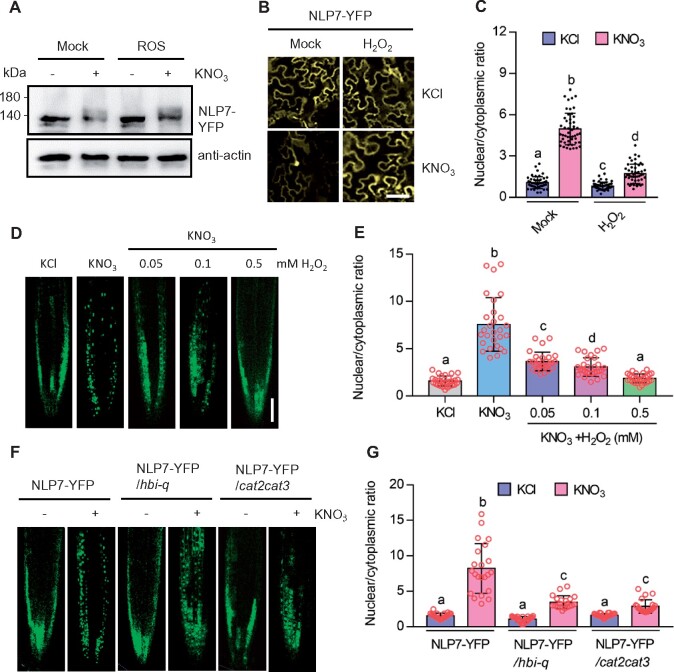
Considering that >50% of nitrate-responsive genes are counteracted by H2O2 supply, we speculated that the key components of nitrate signaling pathways were the targets of H2O2. To determine which key components of nitrate signal transduction are regulated by H2O2, we compared the difference in nitrate-induced phosphorylation of NLP7 proteins with or without H2O2 treatment. We reasoned that if H2O2 could alter the phosphorylation levels of NLP7, H2O2 may target upstream of NLP7, possibly the transceptor NRT1.1 or kinase CPKs; conversely, if H2O2 had no significant effect on the phosphorylation of NLP7, H2O2 may act on NLP7 itself or downstream of NLP7. Immunoblot analysis showed that nitrate treatment caused the phosphorylation of NLP7, but co-treatment with H2O2 had no obvious effect on the levels of phosphorylated NLP7 (Figure 7A;Supplemental Figure S14), thus indicating that H2O2 probably regulates nitrate signal transduction through the transcription factor NLP7 or downstream of NLP7.
Figure 7.
H2O2 attenuates the nitrate-induced nuclear localization of NLP7-YFP. A, H2O2 treatment had no significant effects on nitrate-induced phosphorylation of NLP7. The immunoblots were probed using anti-YFP and anti-actin antibodies. Actin bands showed protein loading. Transgenic plant 35Spro:NLP7-YFP growing on medium with 10-mM KNO3 for 5 days were transferred to nitrogen-free medium for 2 days, and then treated with 10-mM KCl, KNO3, and/or 0.5-mM H2O2 for 0.5 h. B, C, H2O2 inhibited the nitrate-induced nuclear localization NLP7-YFP in the epidermal cells of N. Benthamiana. Plants that were scooped out from soil and grown on the nitrate-free medium for 2 days were transformed with Agrobacterium containing 35Spro:NLP7-YFP construct. After 36 h, these plants were treated with 10-mM KCl, 10-mM KNO3, and/or 0.5-mM H2O2 for 1 h. Scale bar: 10 μm. Different letters above the bars indicate statistically significant differences between the samples (two-way ANOVA analysis followed by Tukey’s test, P < 0.05). D, E, H2O2 antagonizes nitrate in the regulation of the nuclear localization of NLP7-YFP in plants. Seedlings of 35Spro:NLP7-YFP transgenic plants were grown on the medium with or without 10-mM KCl, 10-mM KNO3, and/or different concentrations of H2O2 for 7 days. Scale bar, 50 μm. Different letters above the bars indicate statistically significant differences between the samples (one-way ANOVA analysis followed by Tukey’s test, P < 0.05). F, G, Nitrate-induced nuclear localization of NLP7-YFP was impaired in hbi-q and cat2 cat3 mutants. Seedlings of 35Spro:NLP7-YFP, 35Spro:NLP7-YFP/hbi-q, and 35Spro:NLP7-YFP/cat2 cat3 were grown on medium containing 10-mM KCl or 10-mM KNO3 for 7 days. Scale bar: 50 μm. Fluorescent signals were visualized by using the LSM-700 laser scanning confocal microscope (Zeiss) and the signal intensities of YFP were determined by ImageJ software. Error bars, sd (n = 28 images). Different letters above the bars indicate statistically significant differences between the samples (two-way ANOVA analysis followed by Tukey’s test, P < 0.05).
To understand how H2O2 modulates the function of NLP7 in plants, we examined whether H2O2 affects the subcellular localization of NLP7 by analyzing NLP7-YFP proteins in the epidermal cells of Nicotiana benthamiana leaves. NLP7 was fused to YFP and then transiently expressed in the leaves of N. benthamiana, which were first grown in the soil to the 3- to 4-leaf stage, then washed with water to clean off soil particles, and transferred to a nitrogen-deficient hydroponic solution for about 48 h. Under nitrogen starvation, a few fluorescent signals of NLP7-YFP were found in the nucleus, and most fluorescent signals were found outside the nucleus. Nitrate treatment induced the nuclear localization of NLP7-YFP and the ratio of nuclear to cytoplasmic NLP7-YFP was significantly increased. However, co-treatment with nitrate and H2O2 significantly attenuated the nitrate-induced transport of NLP7-YFP from the cytoplasm to the nucleus (Figure 7, B and C). These findings were validated in the primary roots of the 35Spro:NLP7-YFP and NLP7pro:NLP7-YFP transgenic plants. Nitrate supply significantly induced the nuclear localization of NLP7-YFP, while in the presence of 0.05-mM H2O2, such effects of nitrate on the subcellular location of NLP7-YFP were slightly reduced. As the concentration of H2O2 increased, a growing amount of NLP7-YFP was localized in the cytoplasm, indicating H2O2 inhibits the nitrate-induced nuclear localization of NLP7 in a dose-dependent manner (Figure 7, D and E; Supplemental Figure S15, A and B). Leptomycin B (LMB) is a streptomycin metabolite that inhibits the export of nuclear proteins (Haasen et al., 1999). LMB treatment abolished the inhibiting effects of H2O2 on the nuclear localization of NLP7 (Supplemental Figure S16, A and B). To further determine the specific effects of H2O2 on the subcellular location of HBI1, we analyzed the subcellular localization of HBI1-YFP in the presence of nitrate and/or H2O2. The results showed that nitrate and H2O2 had no significant effects on the subcellular localization of HBI1-YFP (Supplemental Figure S17, A and B). These results indicated that H2O2 treatment reduces the nitrate-triggered nuclear localization of NLP7.
To corroborate these pharmacologic data, we analyzed the effects of H2O2 on the subcellular location of NLP7 in the cat2 cat3 and hbi-q mutants, which accumulate high levels of H2O2. The results showed that nitrate treatment significantly induces the nuclear localization of NLP7-YFP, but such effects of nitrate were reduced in hbi-q and cat2 cat3 mutants (Figure 7, F and G). Furthermore, we found overexpression of HBI1 promoted the nuclear localization of NLP7 in response to nitrate (Supplemental Figure S18, A and B). NLP6, the homolog of NLP7, also showed nitrate-induced nuclear localization, whereas in the presence of H2O2, NLP6 was retained in the cytoplasm (Supplemental Figure S19, A–D). These results indicated that H2O2 inhibited the nitrate-induced nuclear localization of NLP6 and NLP7.
Discussion
Nitrate is the main form of nitrogen that affects plant growth and crop yield and also an important molecular signal for plant adaptation to changing soil conditions (Xu et al., 2012; O'Brien et al., 2016; Wang et al., 2018b). Nitrate binding to the dual-affinity transceptor NRT1.1 triggers the accumulation of calcium in plants, which in turn activates three CPKs to phosphorylate and promote the nuclear location of NLP7 (Liu et al., 20017). In this study, we further showed that the bHLH transcription factors HBI1 and BEE2 are targets of NLP7. Nitrate-activated NLP7 directly induces the expression of HBI1 and BEE2, which then promotes ROS scavenging in plants by directly regulating the expression of CAT2. Mutation in HBI genes resulted in ROS accumulation, and about 22% of nitrate-responsive genes were no longer regulated by nitrate. Exogenous H2O2 treatment repressed the nitrate-stimulated plant growth and downstream gene expression by inhibiting the nuclear localization of NLP7. Nitrate treatment reduced the accumulation of H2O2, and H2O2 inhibits nitrate signaling, thereby forming a feedback regulatory loop to regulate plant growth and development (Figure 8).
Figure 8.
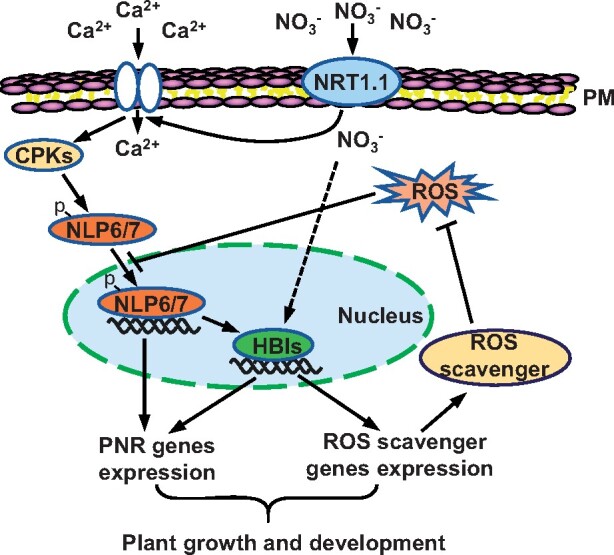
A model for the contribution of HBIs in nitrate-mediated gene expression and plant growth. Perception of nitrate signals by the transceptor NRT1.1 triggers the phosphorylation and nuclear localization of NLP6 and NLP7 to induce the expression of HBIs. HBIs not only regulate some of nitrate-responsive gene expression, but also increase the expression levels of a set of antioxidant genes to reduce the accumulation of H2O2, which inhibits the nitrate-induced nuclear localization of NLP6 and NLP7, thereby forming a feedback regulatory loop to enhance the nitrate signaling in plants. P, phosphorylation; PM, plasma membrane.
HBI1 has been reported to integrate diverse environmental, phytohormone, and plant pathogen signals to regulate plant growth and immunity (Bai et al., 2012; Fan et al., 2014). Here, we further demonstrate through several lines of evidence that HBI1 and its homologs participate in nitrate signaling. First, HBI1 and BEE2 are the early nitrate-responsive genes. Nitrate treatment induces HBI1 and BEE2 expression in as little as 15 min. Second, HBIs play critical roles in the nitrate-regulated gene expression. Transcriptomic analysis showed that ∼22% of the genes were no longer responsive to nitrate in hbi-q. Nitrate supply significantly induces the expression of NRT1.1 and NRT2.1, whereas such effects of nitrate were reduced in hbi-q mutants. HBI1 could directly bind to the promoters of NRT1.1 and NRT2.1 to induce their expression. Third, HBIs are involved in nitrate-stimulated plant growth and development. The plants lacking HBI1 and its three homologs displayed similar growth phenotypes to that of wild-type plants when grown on the medium with KCl or , but exhibited deficiency in nitrate promotion of plant growth and development, suggesting that HBIs play important roles in nitrate-mediated plant growth. Overexpression of HBI1 led to the increased shoot growth and root development phenotype in all growth conditions, which may be due to dual functions of HBI1 in plant growth. On the one side, HBI1 could induce the expression of ROS scavenging genes to reduce the ROS content in the plant and then activate NLP7 to regulate downstream gene expression. On the other side, HBI1 could directly bind to the promoters of growth-related genes to induce the expression of these genes and promote plant growth. Furthermore, the chl1-5 partially suppressed the increased growth phenotypes of HBI1-Ox plants. Together, these results revealed that HBIs positively regulate nitrate signaling to control plant growth and development.
ROS are involved in numbers of signaling pathways and their homeostasis affects many developmental processes and responses to diverse biotic and abiotic stresses (Mittler, 2017; Waszczak et al., 2018). Previous studies reported that nitrate starvation induces the accumulation of ROS (Safi et al., 2018). Here, we showed that nitrate supply reduces the ROS levels by inducing the expression of antioxidant genes such as peroxidases, glutaredoxin, and catalase. HBIs play an important role in the transcriptional induction of this detoxification program. HBI1 directly binds to the promoter of CAT2 to induce its expression and reduce the ROS content in plants. However, nitrate had weak but still significant effects on the induction of antioxidant gene expression in hbi-q mutants, suggesting that other regulatory factors are involved in the nitrate-mediated expression of these antioxidant genes. A recent study showed that the chromatin factor HIGH NITROGEN INSENSITIVE9 (HNI9) and transcription factor ELONGATED HYPOCOTYL5 (HY5) regulate the ROS homeostasis of plants in response to high nitrate provision through the induction of a subset of antioxidant genes (Bellegarde et al., 2019). Mutations of HNI9 or HY5 lead to the highly accumulated ROS under high nitrate provision. Transcriptomic analysis showed that HNI9 and HY5 induced the expression of numbers of genes involved in ROS scavenging in response to high nitrate provision. HNI9 has been reported to interact with BES1, a master transcription factor of brassinosteroid (BR) signaling, to promote downstream gene expression (Li et al., 2010). BR and BES1 increase the activity of HBI1 by activating an HLH/bHLH tripartite growth regulatory cascade (Bai et al., 2012). Thus, HBIs, HNI9, and HY5 form a transcriptional regulator network that modulates the expression of ROS-related genes to fine-tune the ROS homeostasis in response to fluctuating external nitrate supply.
H2O2 is an important redox signaling molecule that is involved in plant developmental processes and stress responses, due to its remarkable universality within cells and the reversible oxidation of target proteins (García-Santamarina et al., 2014; Waszczak et al., 2018). In the present study, we demonstrated that H2O2 negatively regulates nitrate signal transduction through modulating the subcellular location of NLP7. Under nitrate deprivation, the expression of HBIs is not induced, which leads to the low expression of antioxidant genes and H2O2 accumulation, thereby inhibiting plant growth. Constitutive expression of HBI1 reduced the H2O2 levels and partially rescued the growth defect in the low-nitrate conditions. The hbi-q and cat2 cat3 mutants, which accumulate high levels of H2O2, had impaired nitrate-regulated downstream gene expression. Transcriptomic analysis showed that more than half of the nitrate-regulated genes were no longer responsive to nitrate in the presence of H2O2, and most H2O2-counteracted nitrate-responsive genes were regulated by nitrate through the NLP7-dependent pathway, indicating that H2O2 inhibits the regulatory effects of nitrate on downstream gene expression through modulating the activity of NLP7. Furthermore, nitrate treatment induced the nuclear localization of NLP7, whereas such promoting effects of nitrate were significantly reduced by co-treatment with H2O2 or mutation of HBI or CAT genes. The overexpression of HBI1 slightly increased the nuclear localization of NLP7 in the absence of nitrate, suggesting that other signals might regulate the nuclear location of NLP7, and HBI1 may have a function in this process. These results demonstrated that H2O2 contributes to nitrate signal transduction partially through regulating the nucleocytoplasmic shuttling of NLP7.
As a molecular signal, H2O2 generally regulates the activity of target proteins through cysteine oxidative modification of the target proteins. For example, H2O2 has been reported to induce the oxidation of BZR1, a master regulator in the signaling pathway for the plant hormone brassinosteroid. Oxidative modification enhanced the transcription activity of BZR1 through increasing the binding affinity of BZR1 to its partners PIF4 and ARF6 (Tian et al., 2018). Here, we showed that H2O2 regulates the subcellular location of NLP7 to negatively modulate the nitrate signal transduction, but its molecular mechanism remains unclear. One possibility is that H2O2 directly induces the oxidative modification of NLP7. NLP7 contains 25 cysteine residues, which may be the putative target sites of oxidation. Two of them, Cys273 and Cys296, are located in the NLP7 GAF-like domain that is the nitrate-responsive domain; two of them, Cys615 and Cys623, are located in the RWP-PK domain that is the DNA binding domain; two of them, Cys919 and Cys926, are located in the PB1 domain that is protein–protein interaction domain. Site-directed mutagenesis of these cysteines to analyze their effects on the subcellular location of NLP7 will help to reveal whether H2O2 modulates the subcellular location of NLP7 through oxidative modification of these cysteines. Another possibility is that H2O2 induces the oxidative modification of the partners of NLP7. OsNLP3 is the closest homolog of NLP7 in rice. Nitrate treatment significantly induced the nuclear location of OsNLP3, and mutation in OsNLP3 caused a severe defect in nitrate-induced gene expression, indicating OsNLP3 plays an important role in the rice nitrate signaling pathway (Hu et al., 2019). A recent study showed that OsNLP3 physically interacts with OsSPX4 and OsPHR2, which are the key transcription factors of the phosphate signaling pathway. These interactions increase the cytoplasmic retention of OsNLP3 and OsPHR2, and repress the downstream phosphate and nitrate responses (Hu et al., 2019). H2O2 might induce the oxidation of OsSPX4 to enhance the interaction between OsSPX4 and OsNLP3, thereby increasing the cytoplasmic retention of OsNLP3.
ROS and calcium (Ca2+), acting as ubiquitous secondary messengers in the cell, exhibit crosstalk to regulate many cellular processes. Both ROS and Ca2+ are involved in nitrate signaling. Ca2+ positively regulates nitrate signal transduction in plants, whereas ROS negatively regulates it. Using the ultrasensitive Ca2+ biosensors GCaMP6s and a nuclear mCherry, time-lapse recordings in single cells revealed that nitrate specifically stimulates a unique and dynamic Ca2+ signature in the nucleus and cytosol. Ca2+-activated protein kinases CPK10, CPK30, and CPK32 phosphorylate NLP7 to promote its nuclear localization and regulate the nitrate-responsive gene expression (Liu et al., 2017). Here, we showed that nitrate supply reduces the ROS accumulation by promoting the expression of ROS-scavenging genes. ROS treatment inhibits the nitrate-triggered nuclear localization of NLP7 to repress the nitrate signaling. ROS and Ca2+ antagonize to regulate nitrate signal transduction by modulating the nuclear localization of NLP7. Ca2+ has been reported to regulate the ROS level by binding directly to the EF-hand motif in the N-terminus of RBOH protein (Ogasawara et al., 2008), and/or by regulating the activities of CPK5 and CIPK26, which induce the phosphorylation of RBOH proteins (Kobayashi et al., 2007; Drerup et al., 2013). ROS regulate the change of Ca2+ by directly modulating the Ca2+ channel or pump, such as ROS-activating Ca2+-permeable channels in root cells and Ca2+-influx channels in stomates. However, how the production of ROS and Ca2+ are coordinated in the process of nitrate signaling remains to be further studied.
When plants are grown under the biotic or abiotic stress conditions, the nitrate content significantly decreases in both the leaves and roots (Song et al., 2019), compared to that under normal conditions, but the molecular mechanism remains unclear. Given that ROS generation is induced by diverse biotic and abiotic stresses, it would be expected that the biotic and abiotic stresses promote the cytoplasmic retention of NLP7 by inducing the accumulation of ROS in plants, thereby inhibiting the nitrate signal transduction and affecting the nitrate utilization efficiency of plants. Taken together, redox regulation of NLP7 sets a molecular framework for H2O2 crosstalk between nitrate signaling and plant stress responses. This mechanism of crosstalk is likely to provide insight into the molecular mechanisms involved in stress inhibition of nitrate utilization efficiency. Further, adequate regulation of the activity of key components in the nitrogen signaling pathway should improve plant tolerance to different stress conditions.
Materials and methods
Plant materials and growth conditions
Arabidopsis ecotype Columbia (Col-0) was used as the wild-type. Mutants hbi1 (CS870840), bee2 (Salk_205833) bhlh031 (CS879790), and bhlh079 (Salk_142716) were obtained from the Arabidopsis Biological Resource Centre (ABRC). Homozygous T-DNA lines were identified using HBI gene-specific primers and T-DNA left-border primers. The gene specific primers used are listed in Supplemental Data Set S7. The higher-order mutants were obtained by genetic crosses between hbi1, bee2, bhlh031, and bhlh079, and confirmed by PCR (Chu et al., 2020). The 35Spro:HBI1-YFP (HBI1-Ox), 35Spro:NLP7-YFP (NLP7-Ox), NLP7pro:NLP7-YFP, nlp7-1, nlp6-1 nlp7-1, and cat2 cat3 have been described previously (Huang et al., 1996; Bai et al., 2012; Li et al., 2015; Xu et al., 2016; Guan et al., 2017; Su et al., 2018; Chu et al., 2020). Seeds were either surface sterilized and plated on half-strength MS basal salt medium (Phyto-Technology Laboratories) or grown directly in soil. Arabidopsis thaliana plants were grown in a greenhouse or in a growth chamber under a 16-h/8-h light/dark cycle (white fluorescent lamp [intensity of 100–120 μmol m−2 s−1]) at 22–24°C for general growth and seed harvesting. For short-term (1–3 weeks) hydroponic culture (for nitrate-induction assays and gene expression analyses), wild-type Col-0 seedlings and different mutants were cultivated in a growth chamber under a 16-h/8-h light/dark cycle at 22–24°C. In order to analyze the effect of different nitrate concentrations on plant growth, seeds were germinated and grown on MGRL medium consisting in 1.5-mM KH2PO4, 2-mM K2HPO4, 1.5-mM MgSO4, 2-mM Ca(NO3)2, 3-mM KNO3, 67-μM Na2EDTA, 8.6-μM FeSO4, 10.3-μM MnSO4, 30-μM H3BO3, 1-μM ZnSO4, 24-nM Na2MoO4, 130-nM CoCl2, 1-μM CuSO4, 0.05% MES, 1% sucrose, and 1% agar, among which Ca(NO3)2 and KNO3 are replaced by CaCl2 and KCl, depending on nitrate concentration. For root length measurement, seedlings were photocopied, and their primary roots and lateral roots were measured using Image J software (http://rsb.info.nih.gov/ij). Statistical analysis was performed with one-way or two-way ANOVA analysis followed by Tukey’s test, P < 0.05, or Student’s t test, *P < 0.05 (Supplemental Data Set S8).
Plasmid constructs and transgenic plants
Full-length cDNA of HBI1, NLP7 without stop codon were amplified by PCR and cloned into pENTRTM/SD/D-TOPOTM vectors (Thermo Fisher), and then recombined with destination vector pGAL4BDGW (N-GAL4BD), pGAL4ADGW (N-GAL4AD), pX-nYFP (35Spro:C-nYFP), pX-cYFP (35Spro:C-cYFP), pDEST15 (N-GST), and pMAL2CGW (N-MBP). The ∼2.1-kb promoters of NRT1.1 and ∼2-kb promoter of CAT2 were amplified from Arabidopsis genomic DNA and cloned into the pENTRTM/SD/D-TOPOTM vectors (Thermo Fisher), and then recombined with destination vector pGREEN-GW (Promoter: LUC). The ∼1.2-kb promoter of HBI1 was amplified and constructed into a vector to drive the expression of the luciferase gene. Then HBI1pro:LUC transgenic plants were generated by Agrobacterium (GV3101)-mediated transformation by floral dip. Oligo primers used for cloning are listed in Supplemental Data Set S7.
Transient gene expression assays
Protoplast isolation and PEG transformation was carried out as described previously (Yoo et al., 2007; Wu et al., 2009). Plasmid DNAs were extracted using the Qiagen Plasmid Maxi Kit according to manufacturer instructions. Aliquots of 5 × 104 isolated mesophyll protoplasts were transfected with a mixture of 20 μg of DNA and incubated overnight. Protoplasts were harvested by centrifugation and lysed in 100 µL of passive lysis buffer (Promega). Firefly and Renilla (as internal standard) luciferase activities were measured using a dual-luciferase reporter kit (Promega).
RNA isolation, RT-PCR, and RT-qPCR
Wild-type plants were grown on nitrate concentration gradient medium for 14 days; wild-type and hbi-q growing on medium with 7-mM KNO3 for 7 days were transferred to nitrogen-free medium for 2 days and then treated with 10-mM KNO3 at different times, or treated with 10-mM KNO3, 0.5-mM H2O2 for 3 h, respectively. Whole plants were harvested and rapidly frozen in liquid nitrogen. Total RNA was extracted using the Trizaol RNA extraction kit (Transgene). The first-strand cDNAs were synthesized by RevertAid reverse transcriptase (Thermo) and used as RT-PCR templates. Q-PCR analyses using a SYBR green reagent (Roche) was carried on a CFX connect real-time PCR detection system (Bio-Rad) with gene-specific primers (see Supplemental Data Set S7).
ChIP-qPCR assay
ChIP assays were performed as described previously using 35Spro:YFP, 35SproNLP7-YFP, and 35Spro:HBI1-YFP transgenic plants (Bai et al., 2012). Briefly, plants were grown in liquid half-strength MS medium containing 1% sucrose under a 16-h/8-h light/dark cycle for 12 days. The seedlings were crosslinked for 10 min in 1% formaldehyde and quenched by 0.25 M glycerine. Total chromatin was extracted and sonicated into fragments with sizes below 200 bp by Bioruptor (Diagenode), immunoprecipitation was performed using YFP-Trap bound to Protein A agarose. Immunoprecipitated protein and DNA were eluted with 1% SDS and 0.1 M NaHCO3, and the crosslink was reversed by incubation at 95°C 1 h in the presence of 250-mM NaCl. DNA was extracted by a PCR purification kit (Fermentas) and analyzed by qPCR using the oligonucleotide primers listed in Supplemental Data Set S7. The enrichment was calculated as the ratio between 35Spro:NLP7-YFP and the 35Spro:YFP or 35Spro:HBI1-YFP and the 35Spro:YFP, and then normalized to that of the control gene PP2A. Three biological repeats were conducted.
Short-term nitrate induction assay of HBI1 expression
The HBI1pro:LUC transgenic line was grown on nitrogen-free medium for 5 days and then transferred to 10-mM KNO3 plates containing 1-mM D-luciferin at different times; subsequently, fluorescence images were detected by Tanon 5200 fully automatic chemiluminescence image analysis system.
Measurements of ROS production
ROS was detected by H2DCFDA, NBT, and 3,3'-diaminobenzidine tetrahydrochloride (DAB) staining. For H2DCFDA staining, seedlings were incubated with 50-μM H2DCFDA solution for 10 min in the dark and then washed with distilled water to remove excess dye. Examinations of fluorescence intensity were performed using a laser scanning confocal microscope (excitation, 488 nm; emission, 505–550 nm; Zeiss, LSM700). For NBT and DAB staining, seedlings were incubated with 1-mg·mL-1 NBT solution or 1-mg·mL-1 DAB solution for 2 h at room temperature. Samples were then immersed in 80% (v/v) ethanol for 10 min to terminate the staining before being photographed.
Subcellular localization assay
To analyze NLP7-YFP nuclear retention triggered by nitrate and H2O2 in Arabidopsis, seedlings of 35Spro:NLP7-YFP transgenic plants were grown on the medium with or without 10-mM KCl, 10-mM KNO3, and/or different concentrations of H2O2 for 7 days. To analyze NLP6-YFP and NLP7-YFP nuclear retention triggered by nitrate and H2O2 in N. benthamiana, 35Spro:NLP6-YFP and 35Spro:NLP7-YFP construct were transformed into Agrobacterium strain GV3101, agrobacterium was diluted to a final concentration of OD600 = 0.4, and then injected into N. benthamiana leaves with a needle tube. The plants were scooped out from soil and grown on the nitrate-free medium for 2 days before transformation with Agrobacterium containing the 35Spro:NLP7-YFP construct. After 36 h, these plants were treated with 10-mM KCl, 10-mM KNO3, and/or 0.5-mM H2O2 for 1 h. To analyze nitrate-induced nuclear localization of NLP7-YFP in hbi-q and cat2 cat3 mutants. Seedlings of 35Spro:NLP7-YFP, 35Spro:NLP7-YFP/hbi-q, 35Spro:NLP7-YFP/cat2 cat3, NLP7pro:NLP7-YFP, 35Spro:NLP7-YFP/35Spro:HBI1-Myc were grown on medium containing 10-mM KCl or 10-mM KNO3 for 7 days. The fluorescence of YFP in the leaves was imaged using a confocal laser scanning microscope (Zeiss, LSM700); excitation was set to 488 nm (YFP) and images at emissions 505–550 nm (YFP) were collected. Image J was used to quantitatively analyze the data; florescence intensity from at least 50 pavement cells was recorded.
Phos-tag sodium dodecyl sulphate–polyacrylamide gel electrophoresis assay
Transgenic plant 35Spro:NLP7-YFP growing on medium with 10 mM KNO3 for 5 days was transferred to nitrogen-free medium for 2 days, and then treated with 10-mM KCl, KNO3 and/or 0.5-mM H2O2 for 0.5 h. Harvested seedlings were ground in liquid nitrogen, powder was suspended in 1 X sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) sample buffer, and centrifuged at 13,523g for 10 min and the supernatant was used for Phos-tag SDS–PAGE analyses. Phos-Tag was purchased from FUJIFILM Wako Pure Chemical Corporation and used according to the manufacturer’s protocol. Gels for Phos-tag SDS–PAGE consisted of a separating gel added 25-μM Phos-tag acrylamide, 100-μM ZnCl2. The anti-YFP (TransGen Biotech, Cat: HT801, 1:5,000 dilution) and anti-actin (Invitrogen, Cat: MA1-744, 1:5,000 dilution) antibodies were used to detect NLP7-YFP and actin, respectively.
RNA-Seq
Wild-type Col-0 seedlings, nlp7-1 and hbi-q mutants growing on medium with 7-mM KNO3 for 7 days were transferred to nitrogen-free medium for 2 days then treated with 10-mM KNO3 for 3 h. To analyze the effects of H2O2 on nitrate regulated gene expression, Col-0 seedlings growing on medium with 7-mM KNO3 for 7 days were transferred to nitrogen-free medium for 2 days then treated with 10-mM KNO3 with or without 0.5-mM H2O2 for 3 h or treated with H2O2 for 3 h. Total RNA was extracted with Trizaol RNA extraction kit (Transgene), and mRNA sequencing libraries construction and sequencing on the BGISEQ-500 platform were performed at Beijing Genomics Institute. The sequence reads were mapped to the Arabidopsis genome using HISAT and Bowtie2 software. The gene read counts were calculated by htseq-count with default parameters (Anders et al., 2015), and differential gene expression was analyzed using DESeq2 tool with the generalized linear model (Love et al., 2014). DEGs were defined by two-fold expression difference with false discovery rate <0.05.
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AT2G18300 (HBI1), AT4G36540 (BEE2), At1g59640 (bHLH031), At5g62610 (bHLH079), At1g18400 (BEE1), At4g34530 (CIB1), AT2G43060 (IBH1), AT4G24020 (NLP7), AT1G12110 (NRT1.1), AT1G08090 (NRT2.1), AT1G08100 (NRT2.2), AT1G77760 (NIA1), AT4G35090 (CAT2), AT1G20620 (CAT3), AT3G01190 (PRX27), AT1G14550 (PRX5), AT5G05340 (PRX52), AT3G03670 (PRX28), AT5G39580 (PRX62), AT1G03850 (GRXS13), PP2A (AT1G13320), and AT5G40850 (UPM1). All sequencing data that support the finding of this study have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) and are accessible through the GEO series accession number GSE174025.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nitrate increases the activity of HBI1 at the transcriptional level (supports Figure 1).
Supplemental Figure S2. Nitrate induces the expression of several members of HBI1 gene family (supports Figure 1).
Supplemental Figure S3. Mutation of HBIs reduces the nitrate-mediated plant growth and development (supports Figure 2).
Supplemental Figure S4. Mutations of HBIs reduces the nitrate sensitivity of shoot growth (supports Figure 2).
Supplemental Figure S5. NRT1.1 is required for HBI1-promoted shoot growth (supports Figure 2).
Supplemental Figure S6. HBI1 participates in the nitrate-regulated expression of some ROS-related Genes (supports Figure 3).
Supplemental Figure S7. Mutation in HBIs reduces the nitrate-promoted scavenging of ROS in plants (supports Figure 4).
Supplemental Figure S8. Catalase plays important roles in HBI1-induced ROS scavenging (supports Figure 5).
Supplemental Figure S9. CATs are required for HBI1-stimulated plant growth (supports Figure 5).
Supplemental Figure S10. H2O2 inhibits the nitrate-induce root growth in a dose-dependent manner (supports Figure 6).
Supplemental Figure S11. CATs are required for the nitrate-induced expression of downstream genes (supports Figure 6).
Supplemental Figure S12. RNA-Seq analyzed the effects of H2O2 and nitrate on the gene expression (supports Figure 6).
Supplemental Figure S13. GO analysis the H2O2 and nitrate co-regulated genes (supports Figure 6).
Supplemental Figure S14. H2O2 has no significant effects on the nitrate-induced phosphorylation of NLP7 (supports Figure 7).
Supplemental Figure S15. H2O2 reduces the nitrate-induced nuclear localization of NLP7-YFP in the NLP7pro:NLP7-YFP transgenic plants (supports Figure 7).
Supplemental Figure S16. LMB attenuates the H2O2-inhibited nuclear localization of NLP7-YFP (Supports Figure 7).
Supplemental Figure S17. H2O2 has no significant effects on the subcellular localization of HBI1-YFP (supports Figure 7).
Supplemental Figure S18. HBI1 Enhances the nitrate-induced nuclear localization of NLP7-YFP (supports Figure 7).
Supplemental Figure S19. H2O2 attenuates the nitrate-induced nuclear localization of NLP6 (supports Figure 7).
Supplemental Data Set S1. Nitrate-regulated genes in wild-type plants.
Supplemental Data Set S2. Nitrate-regulated genes in hbi-q mutant.
Supplemental Data Set S3. Nitrate-regulated genes in nlp7-1 mutant.
Supplemental Data Set S4. Nitrate-regulated overlap genes in Col-0, hbi-q, and nlp7-1 mutants.
Supplemental Data Set S5. Genes regulated by nitrate in the presence of H2O2.
Supplemental Data Set S6. H2O2-regulated genes in wild-type plants.
Supplemental Data Set S7. Oligo used in this study.
Supplemental Data Set S8. ANOVA analysis in this study.
Supplemental Data Set S9. Amino acid sequences used for generating phylogenetic tree.
Supplementary Material
Acknowledgments
We thank Haiyan Yu and Xiaomin Zhao from the Analysis and Testing Center of SKLMT (State Key Laboratory of Microbial Technology, Shandong University) for assistance with the laser scanning confocal microscopy.
Funding
This work was funded by the National Natural Science Foundation of China (grant no. 31970306, 32070210, and 31870262), by the Shandong Province Natural Science Foundation (grant no. ZR2019ZD16 and 2019LZGC-015), by the China Postdoctoral Science Foundation (grant no. 2017M612259, 2018T110684 and 2020M672047), and by the Shandong Province Postdoctoral Science Foundation (grant no. 11200078311023 and 61200070311209).
Conflict of interest statement. The authors declare no conflict of interest.
These authors contributed equally to this work (X.C., J-G.W).
X.C., J.W., and M.B. together designed the experiments. X.C. performed the statistical analysis of plant growth, transient expression. X.C., J.W., and X.Y. performed the RNA-Seq and GO analysis. J.W. performed the western blot, subcellular location analysis, and ChIP–qPCR. M.L., C.H., M.F., S.Z., Y.G., X.C., and J.W. generated HBI1-related mutants, HBI1-Ox/cat2cat3 and NLP7-YFP/cat2cat3 transgenic plants. G.L., F.X., Y.W., and C.X. provided the critical discussion on the work. X.C. and J.W. performed all other experiments. X.C., J.W., and M. B. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Ming-Yi Bai (baimingyi@sdu.edu.cn).
References
- Alvarez JM, Riveras E, Vidal EA, Gras DE, Contreras‐López O, Tamayo KP, Aceituno F, Gómez I, Ruffel S, Lejay L (2014) Systems approach identifies TGA 1 and TGA 4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J 80: 1–13 [DOI] [PubMed] [Google Scholar]
- Alvarez JM, Schinke A-L, Brooks MD, Pasquino A, Leonelli L, Varala K, Safi A, Krouk G, Krapp A, Coruzzi GM (2020) Transient genome-wide interactions of the master transcription factor NLP7 initiate a rapid nitrogen-response cascade. Nat Commun 11: 1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya T, Miyamoto M, Wibowo J, Suzuki A, Kojima S, Tsuchiya YN, Sawa S, Fukuda H, von Wirén N, Takahashi H (2014) CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc Natl Acad Sci USA 111: 2029–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M-Y, Fan M, Oh E, Wang Z-Y (2012) A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 24: 4917–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellegarde F, Maghiaoui A, Boucherez J, Krouk G, Lejay L, Bach L, Gojon A, Martin A (2019) The chromatin factor HNI9 and ELONGATED HYPOCOTYL5 maintain ROS homeostasis under high nitrogen provision. Plant Physiol 180: 582–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielski BHJ, Shiue GG, Bajuk S (1980) Reduction of nitro blue tetrazolium by and radicals. J Phys Chem 84: 830–833 [Google Scholar]
- Cai H, Chai M, Chen F, Huang Y, Zhang M, He Q, Liu L, Yan M, Qin Y (2020) HBI1 acts downstream of ERECTA and SWR1 in regulating inflorescence architecture through the activation of the brassinosteroid and auxin signaling pathways. New Phytol 229: 414–428 [DOI] [PubMed] [Google Scholar]
- Chaiwanon J, Wang W, Zhu J-Y, Oh E, Wang Z-Y (2016) Information integration and communication in plant growth regulation. Cell 164: 1257–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu X, Li M, Zhang S, Fan M, Han C, Xiang F, Li G, Wang Y, Xiang C-b, Wang J-G, et al. (2020). HBI1-TCP20 interaction positively regulates the CEPs-mediated systemic nitrate acquisition. J Integr Plant Biol. DOI: 10.1111/jipb.13035. [DOI] [PubMed] [Google Scholar]
- Crawford NM, Forde BG (2002) Molecular and developmental biology of inorganic nitrogen nutrition. Arabidopsis Book 1: e0011–e0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drerup MM, Schlücking K, Hashimoto K, Manishankar P, Steinhorst L, Kuchitsu K, Kudla J (2013) The calcineurin B-like calcium sensors CBL1 and CBL9 together with their interacting protein kinase CIPK26 regulate the Arabidopsis NADPH oxidase RBOHF. Mol Plant 6: 559–569 [DOI] [PubMed] [Google Scholar]
- Fan M, Bai M-Y, Kim J-G, Wang T, Oh E, Chen L, Park CH, Son S-H, Kim S-K, Mudgett MB, et al. (2014). The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern–triggered immunity in Arabidopsis. Plant Cell 26: 828–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG (2014) Nitrogen signalling pathways shaping root system architecture: an update. Curr Opin Plant Biol 21: 30–36 [DOI] [PubMed] [Google Scholar]
- Fredes I, Moreno S, Díaz FP, Gutiérrez RA (2019) Nitrate signaling and the control of Arabidopsis growth and development. Curr Opin Plant Biol 47: 112–118 [DOI] [PubMed] [Google Scholar]
- García-Santamarina S, Boronat S, Hidalgo E (2014) Reversible cysteine oxidation in hydrogen peroxide sensing and signal transduction. Biochemistry 53: 2560–2580 [DOI] [PubMed] [Google Scholar]
- Gras DE, Vidal EA, Undurraga SF, Riveras E, Moreno S, Dominguez-Figueroa J, Alabadi D, Blázquez MA, Medina J, Gutiérrez RA (2018) SMZ/SNZ and gibberellin signaling are required for nitrate-elicited delay of flowering time in Arabidopsis thaliana. J Exp Bot 69: 619–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan P, Ripoll J-J, Wang R, Vuong L, Bailey-Steinitz LJ, Ye D, Crawford NM (2017) Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc Natl Acad Sci USA 114: 2419–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasen D, Köhler C, Neuhaus G, Merkle T (1999) Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine-rich nuclear export signals in Arabidopsis thaliana. Plant J 20: 695–705 [DOI] [PubMed] [Google Scholar]
- Ho C-H, Lin S-H, Hu H-C, Tsay Y-F (2009) CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Hu B, Jiang Z, Wang W, Qiu Y, Zhang Z, Liu Y, Li A, Gao X, Liu L, Qian Y (2019) Nitrate–NRT1. 1B–SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat Plants 5: 401. [DOI] [PubMed] [Google Scholar]
- Hu HC, Wang YY, Tsay YF (2009) AtCIPK8, a CBL‐interacting protein kinase, regulates the low‐affinity phase of the primary nitrate response. Plant J 57: 264–278 [DOI] [PubMed] [Google Scholar]
- Huang NC, Chiang CS, Crawford NM, Tsay YF (1996) CHL1 encodes a component of the low-affinity nitrate uptake system in Arabidopsis and shows cell type-specific expression in roots. Plant Cell 8: 2183–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J-Y, Ahn JH, Schachtman DP (2018) CC-type glutaredoxins mediate plant response and signaling under nitrate starvation in Arabidopsis. BMC Plant Biol 18: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Ohura I, Kawakita K, Yokota N, Fujiwara M, Shimamoto K, Doke N, Yoshioka H (2007) Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 19: 1065–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrein B, Formosa-Jordan P, Malivert A, Schuster C, Melnyk CW, Yang W, Turnbull C, Meyerowitz EM, Locke JCW, Jönsson H (2018) Nitrate modulates stem cell dynamics in Arabidopsis shoot meristems through cytokinins. Proc Natl Acad Sci USA 115: 1382–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu J, Wang G, Cha J-Y, Li G, Chen S, Li Z, Guo J, Zhang C, Yang Y, et al. (2015). A chaperone function of NO CATALASE ACTIVITY1 is required to maintain catalase activity and for multiple stress responses in Arabidopsis. Plant Cell 27: 908–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ye H, Guo H, Yin Y (2010) Arabidopsis IWS1 interacts with transcription factor BES1 and is involved in plant steroid hormone brassinosteroid regulated gene expression. Proc Natl Acad Sci USA 107: 3918–3923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-L, Tsay Y-F (2017) Influence of differing nitrate and nitrogen availability on flowering control in Arabidopsis. J Exp Bot 68: 2603–2609 [DOI] [PubMed] [Google Scholar]
- Liu K-h, Niu Y, Konishi M, Wu Y, Du H, Sun Chung H, Li L, Boudsocq M, McCormack M, Maekawa S, et al. (2017). Discovery of nitrate–CPK–NLP signalling in central nutrient–growth networks. Nature 545: 311–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovsky FG, Batoux M, Schwessinger B, Youn JH, Stransfeld L, Win J, Kim S-K, Zipfel C (2014) Antagonistic regulation of growth and immunity by the Arabidopsis basic helix-loop-helix transcription factor homolog of brassinosteroid enhanced expression2 interacting with increased leaf inclination1 binding bHLH1. Plant Physiol 164: 1443–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchive C, Roudier F, Castaings L, Bréhaut V, Blondet E, Colot V, Meyer C, Krapp A (2013) Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat Commun 4: 1713. [DOI] [PubMed] [Google Scholar]
- Mittler R (2017) ROS are good. Trends Plant Sci 22: 11–19 [DOI] [PubMed] [Google Scholar]
- Moreno S, Canales J, Hong L, Robinson D, Roeder AHK, Gutiérrez RA (2020) Nitrate defines shoot size through compensatory roles for endoreplication and cell division in Arabidopsis thaliana. Curr Biol 30: 1988–2000 [DOI] [PubMed] [Google Scholar]
- Neuser J, Metzen CC, Dreyer BH, Feulner C, van Dongen JT, Schmidt RR, Schippers JHM (2019) HBI1 mediates the trade-off between growth and immunity through its impact on apoplastic ROS homeostasis. Cell Reports 28: 1670–1678 [DOI] [PubMed] [Google Scholar]
- O'Brien JA, Vega A, Bouguyon E, Krouk G, Gojon A, Coruzzi G, Gutiérrez RA (2016) Nitrate transport, sensing, and responses in plants. Mol Plant 9: 837–856 [DOI] [PubMed] [Google Scholar]
- Ogasawara Y, Kaya H, Hiraoka G, Yumoto F, Kimura S, Kadota Y, Hishinuma H, Senzaki E, Yamagoe S, Nagata K, et al. (2008). Synergistic activation of the Arabidopsis NADPH oxidase AtrbohD by and phosphorylation. J Biol Chem 283: 8885–8892 [DOI] [PubMed] [Google Scholar]
- Safi A, Medici A, Szponarski W, Marshall-Colon A, Ruffel S, Gaymard F, Coruzzi G, Lacombe B, Krouk G (2018) HRS1/HHOs GARP transcription factors and reactive oxygen species are regulators of Arabidopsis nitrogen starvation response. bioRxiv, 164277. [Google Scholar]
- Shin R, Berg RH, Schachtman DP (2005) Reactive oxygen species and root hairs in Arabidopsis root response to nitrogen, phosphorus and potassium deficiency. Plant Cell Physiol 46: 1350–1357 [DOI] [PubMed] [Google Scholar]
- Song Y, Li J, Liu M, Meng Z, Liu K, Sui N (2019) Nitrogen increases drought tolerance in maize seedlings. Funct Plant Biol 46: 350–359 [DOI] [PubMed] [Google Scholar]
- Stitt M (1999). Nitrate regulation of metabolism and growth. Curr Opin Plant Biol 2: 178–186 [DOI] [PubMed] [Google Scholar]
- Su T, Wang P, Li H, Zhao Y, Lu Y, Dai P, Ren T, Wang X, Li X, Shao Q, et al. (2018). The Arabidopsis catalase triple mutant reveals important roles of catalases and peroxisome-derived signaling in plant development. J Integr Plant Biol 60: 591–607 [DOI] [PubMed] [Google Scholar]
- Swift J, Alvarez JM, Araus V, Gutiérrez RA, Coruzzi GM (2020) Nutrient dose-responsive transcriptome changes driven by Michaelis–Menten kinetics underlie plant growth rates. Proc Natl Acad Sci USA 117: 12531–12540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Fan M, Qin Z, Lv H, Wang M, Zhang Z, Zhou W, Zhao N, Li X, Han C, et al. (2018). Hydrogen peroxide positively regulates brassinosteroid signaling through oxidation of the BRASSINAZOLE-RESISTANT1 transcription factor. Nat Commun 9: 1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevisan S, Trentin AR, Ghisi R, Masi A, Quaggiotti S (2019) Nitrate affects transcriptional regulation of UPBEAT1 and ROS localisation in roots of Zea mays L. Physiol Plant 166: 794–811 [DOI] [PubMed] [Google Scholar]
- Vidal EA, Alvarez JM, Araus V, Riveras E, Brooks MD, Krouk G, Ruffel S, Lejay L, Crawford NM, Coruzzi GM, et al. (2020). Nitrate 2020: thirty years from transport to signaling networks. Plant Cell 32: 2094–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Xing X, Wang Y, Tran A, Crawford NM (2009) A genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1. Plant Physiol 151: 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Li L, Xu P, Lian H, Wang W, Xu F, Mao Z, Zhang T, Yang H (2018a) CRY1 interacts directly with HBI1 to regulate its transcriptional activity and photomorphogenesis in Arabidopsis. J Exp Bot 69: 3867–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Bai M-Y, Wang Z-Y (2014) The brassinosteroid signaling network—a paradigm of signal integration. Curr Opin Plant Biol 21: 147–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-Y, Cheng Y-H, Chen K-E, Tsay Y-F (2018b) Nitrate transport, signaling, and use efficiency. Annu Rev Plant Biol 69: 85–122 [DOI] [PubMed] [Google Scholar]
- Waszczak C, Carmody M, Kangasjärvi J (2018) Reactive oxygen species in plant signaling. Annu Rev Plant Biol 69: 209–236 [DOI] [PubMed] [Google Scholar]
- Wu F.-H., Shen S.-C., Lee L.-Y., Lee S.-H., Chan M.-T., Lin C.-S. (2009). Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Fan X, Miller AJ (2012) Plant nitrogen assimilation and use efficiency. Annu Rev Plant Biol 63: 153–182 [DOI] [PubMed] [Google Scholar]
- Xu N, Wang R, Zhao L, Zhang C, Li Z, Lei Z, Liu F, Guan P, Chu Z, Crawford NM, Wang Y (2016) The Arabidopsis NRG2 protein mediates nitrate signaling and interacts with and regulates key nitrate regulators. Plant Cell 28: 485–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Easwaran V, Chau V, Okamoto M, Ierullo M, Kimura M, Endo A, Yano R, Pasha A, Gong Y, et al. (2016). NIN-like protein 8 is a master regulator of nitrate-promoted seed germination in Arabidopsis. Nat Commun 7: 13179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao N, Greenberg JT (2006) Arabidopsis ACCELERATED CELL DEATH2 modulates programmed cell death. Plant Cell 18: 397–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S-D, Cho Y-H, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.