A pectate lyase-like gene influences wall mechanics and cell pressurization in guard cells, thereby contributing to stomatal dynamics and plant growth.
Abstract
Plant cell deformations are driven by cell pressurization and mechanical constraints imposed by the nanoscale architecture of the cell wall, but how these factors are controlled at the genetic and molecular levels to achieve different types of cell deformation is unclear. Here, we used stomatal guard cells to investigate the influences of wall mechanics and turgor pressure on cell deformation and demonstrate that the expression of the pectin-modifying gene PECTATE LYASE LIKE12 (PLL12) is required for normal stomatal dynamics in Arabidopsis thaliana. Using nanoindentation and finite element modeling to simultaneously measure wall modulus and turgor pressure, we found that both values undergo dynamic changes during induced stomatal opening and closure. PLL12 is required for guard cells to maintain normal wall modulus and turgor pressure during stomatal responses to light and to tune the levels of calcium crosslinked pectin in guard cell walls. Guard cell-specific knockdown of PLL12 caused defects in stomatal responses and reduced leaf growth, which were associated with lower cell proliferation but normal cell expansion. Together, these results force us to revise our view of how wall-modifying genes modulate wall mechanics and cell pressurization to accomplish the dynamic cellular deformations that underlie stomatal function and tissue growth in plants.
Introduction
Stomatal dynamics regulate CO2 and water flux in plants to enable photosynthesis and transpiration. Inflation or deflation of guard cells results in the opening or closure of stomata, respectively. Guard cell deformations are thought to be driven by turgor pressure changes and constrained by the cell wall (Aylor et al., 1973; DeMichele and Sharpe, 1973). However, the influence of cell wall architecture and metabolism on cell biomechanics and pressurization during deformation is poorly understood. Improving our understanding of stomatal biomechanics and how cell wall-related genes impinge on those biomechanics has the potential to open new opportunities to engineer stomatal activity for optimal plant growth under challenging conditions such as drought.
The primary wall of growing plant cells is a composite material, with cellulose microfibrils embedded in a pectin-containing matrix (Cosgrove, 2018). Pectins appear to be particularly important for stomatal function, since they are present in guard cells of plant species that contain very little overall pectin (Jones et al., 2005). Stomatal responses to environmental stimuli are influenced by pectin methylesterases (PMEs) and polygalacturonases (PGs), which respectively modulate the methylation state of pectic homogalacturonan (HG) and hydrolyze demethylated HG (pectate) (Jones et al., 2003, 2005; Amsbury et al., 2016; Huang et al., 2017; Rui et al., 2017; Yi et al., 2018). Pectate lyases (PLs) cleave pectate by β-elimination, but their functions in stomatal dynamics are unclear. We hypothesize that PLs contribute to stomatal dynamics in a manner different from PGs because even though both classes of enzymes cleave the HG backbone, they have differing mechanisms of action and have independent evolutionary histories (McCarthy et al., 2014).
Because pectins are negatively charged and form hydrated gels and thus have tunable biochemical and biomechanical properties, pectin modification is thought to facilitate cell expansion (Xiao et al., 2014) or cell separation (Babu and Bayer, 2014) in different developmental contexts. Pectate can also be crosslinked by calcium, which increases the elastic modulus of pectin gels in vitro (Ström et al., 2007). However, in plant cell walls, both higher (Daher et al., 2018) and lower (Peaucelle et al., 2011) wall stiffness are associated with more calcium crosslinking, and the functions of pectin and its metabolism, configurations, and crosslinking in plant cell deformation remain unclear.
In the context of stomatal dynamics, mathematical models (Woolfenden et al., 2017; Yi et al., 2018) and atomic force microscopy (AFM) of plasmolyzed guard cells (Carter et al., 2017) have begun to reveal the contributions of HG to the mechanics of guard cell walls, but exactly how HG and other wall components, plus their interactions, influence the extent and kinetics of guard cell deformation during stomatal responses to stimuli are not known. Because pectins function in wall integrity signaling (Kohorn et al., 2009; Feng et al., 2018), pectinases might act not only directly on cell wall mechanics but also via intracellular signaling pathways to influence cell pressurization, which ultimately drives plant cell expansion (Ortega, 1985). Although a pressure probe can be used to measure turgor pressure in species with large guard cells (Franks et al., 1995), turgor pressure has been more difficult to quantify in organisms with smaller guard cells such as Arabidopsis thaliana, which has a multitude of genetic resources for investigating the cell wall. Although AFM has been used for dynamic measurement of cell wall mechanics (Milani et al., 2014; Yakubov et al., 2016) and for measurement of turgor pressure (Beauzamy et al., 2015), simultaneous tracking of wall modulus and turgor pressure during stomatal movements requires a nondisruptive method that is beyond the capabilities of the pressure probe or AFM methods in isolation. Nanoindentation was recently combined with finite element modeling (FEM) to simultaneously estimate wall mechanics and turgor pressure values in living, pressurized pavement cells of Arabidopsis (Routier-Kierzkowska et al., 2012; Forouzesh et al., 2013; Weber et al., 2015; Li et al., 2021), opening the possibility to quantify time-resolved mechanical dynamics for Arabidopsis guard cells in motion.
Here, we integrated biomechanics, genetic, and physiological approaches to study the influence of pectin on guard cell walls and stomatal dynamics. Our nanoindentation–FEM analyses revealed unexpected dynamic changes in both wall modulus and turgor pressure during stomatal responses to changing light conditions in wild-type plants. We demonstrated that the A. thaliana PL gene PL LIKE12 (PLL12) is required for normal stomatal function and used nanoindentation-FEM to show that PLL12 is required to build guard cells with normal biomechanical properties including directional wall modulus and turgor pressure, likely due to its influence on HG abundance and crosslinking in the guard cell wall. The phenotypes of guard cell-specific knockdown lines for PLL12 indicate that the guard cell-specific functions of this gene are required for cell proliferation and plant growth.
Results
PLL12 encodes a putative pectate lyase and is widely expressed in Arabidopsis plants
To investigate the functions of PLs in stomatal biology, we first mined Arabidopsis transcriptome data (Hachez et al., 2011) gathered after the induced expression of FAMA, a transcription factor that drives stomatal differentiation (Ohashi-Ito and Bergmann, 2006), for PL genes. PLL12 expression is upregulated 48 h after FAMA induction (Hachez et al., 2011), suggesting a role in guard cell development and function. Both splice variants encoded by PLL12 have a PL-C domain and a signal peptide; for transgenic analyses, we used splice variant 1 (Supplemental Figure S1). PLL12 shows high sequence similarity with PLs from multiple plant species and contains conserved residues involved in Ca2+ binding, substrate binding, and catalytic activity (Yoder and Jurnak, 1995; Scavetta et al., 1999; Supplemental Figure S1).
To analyze PLL12 expression patterns, we transformed Arabidopsis plants of the Columbia-0 (Col-0) ecotype with a construct containing the 2-kb region upstream of the PLL12 start codon fused to the β-glucuronidase (GUS) gene. GUS activity varied across three independent transformant lines (Supplemental Figure S2), as previously reported (Sun and van Nocker, 2010). Nevertheless, GUS activity was commonly higher in older rosette leaves than in younger leaves (Supplemental Figure 2A). In young seedlings, roots and cotyledons showed GUS activity (Supplemental Figure S2B). In the leaf epidermis, GUS activity was detected in both guard cells and surrounding pavement cells (Supplemental Figure S2C). These data and previous Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR) results (Palusa et al., 2007; Sun and van Nocker, 2010) demonstrate that PLL12 is widely expressed in roots, leaves, stems, and inflorescences of Arabidopsis.
To analyze the function of PLL12, we isolated a T-DNA knockout mutant that we named pll12-1 (Supplemental Figure S2D). No PLL12 RT-PCR product was detected in this mutant, suggesting that it is a null mutation (Supplemental Figure S2D). Complementation lines (PLL12comp) were generated by transforming a construct containing a PLL12 promoter:PLL12 CDS fusion into the pll12-1 mutant background. RT-PCR showed restored PLL12 expression in three independent transformant lines (Supplemental Figure S2E), and PLL12comp-1 was named PLL12comp and used for further analyses. A PLL12 overexpression line was generated by transforming a construct containing a 35S pro:PLL12 CDS fragment into the Col background, and given the similarly elevated levels of PLL12 expression in three independent transformant lines (Supplemental Figure S2E), PLL12OE-1 was chosen for further analysis and named PLL12OE.
Given the broad expression of PLL12, we also constructed guard cell-specific knockdown lines for this gene. Three different sets of transgenic lines were generated by transforming Col plants with constructs containing the guard cell-specific promoter pGC1 (Yang et al., 2008) fused with artificial microRNA (amiRNA) sequences targeting one of three different sites in PLL12 (Supplemental Figure S2G), and were designated PLL12kd1 to 3. As controls, we constructed GFPkd1 to 3 transgenic plants targeting GFP, which is not present in Arabidopsis.
We examined the expression levels of PLL12 in Col and transgenic plants using qPCR (Supplemental Figure S2F). Given the age-dependent expression of PLL12 in leaves (Supplemental Figure S2A), only leaves 5–8 from 21-day-old rosettes were used for qPCR and subsequent assays (Supplemental Figure S2F). Using a (log2) difference of two as a cutoff, pll12-1 and all PLL12kd leaves had lower PLL12 transcript levels, PLL12OE had higher transcript levels, and GFPkd lines had transcript levels similar to those of the Col controls (Supplemental Figure S2F). PLL12comp leaves had significantly higher PLL12 expression levels than Col (Supplemental Figure S2F). Comparing PLL12kd to GFPkd lines, PLL12 transcript levels in whole leaves of PLL12kd2 and 3 were significantly lower than those of GFPkd1 and 3 but not GFPkd2, and PLL12 expression levels in PLL12kd1 leaves were not significantly different from those of any GFPkd line (Supplemental Figure S2F). These results indicate that PLL12 transcripts were more efficiently silenced in guard cells of PLL12kd2 and 3 plants than in PLL12kd1 plants.
PLL12 functions in stomatal dynamics
To determine the role of PLL12 in stomatal function, we assayed stomatal responses to various stimuli in plants of the genotypes described above. The hormone abscisic acid (ABA) or darkness was applied to excised leaves to induce stomatal closure; Fusicoccin (FC), a proton pump activator, or light was used to induce stomatal opening. Every 30 min after treatment, a leaf epidermis was peeled and imaged to track stomatal dynamics for each genotype. In addition to stomatal pore area, pore area: stomatal complex area ratios were calculated for each stomatal complex to quantify the degree of stomatal opening/closing in a manner that accounted for variation in stomatal complex size across different genotypes (see below).
In pll12-1 knockout leaves, stomata responded more slowly to closure and opening stimuli than Col stomata (Figure 1; Supplemental Figure S3, A–F). pll12-1 stomata also closed further than Col stomata in response to ABA or darkness (Figure 1A; Supplemental Figure S3, A, C, and E) and did not open as widely as Col stomata after FC or light induction (Figure 1C; Supplemental Figure S3, B, D, and F). PLL12OE stomata closed slightly more slowly in response to ABA or dark than Col stomata (Figure 1, A–D; Supplemental Figure S3, A, C, and E). In the opening assays, although PLL12OE stomata were slightly less responsive to FC than Col stomata, they opened as rapidly as Col stomata in response to light, and they opened to the same degree as Col stomata (Figure 1C; Supplemental Figure S3, B, D, and F). Pore areas in PLL12OE stomata were similar to Col at the beginning of FC or light treatment experiments, indicating that PLL12OE stomata can close to the same degree as Col stomata (Figure 1C; Supplemental Figure S3, B, D, and F). Together, these data suggest that the loss of PLL12 expression results in stiffer and/or less readily pressurized guard cells that cannot respond efficiently to environmental stimuli, whereas overexpression of PLL12 results in guard cells that are as deformable as Col cells, but contract more slowly in response to closure stimuli, implying that they have a defect in the kinetics of wall contractibility and/or guard cell depressurization.
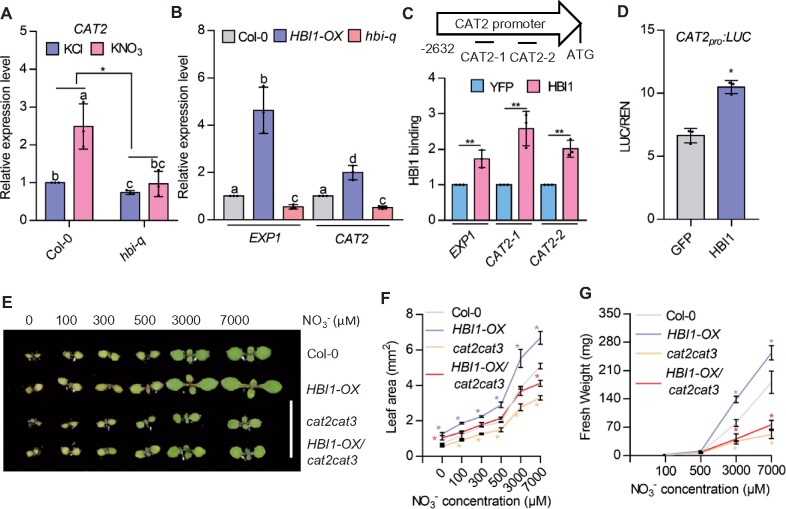
Figure 1.
Expression of PLL12 affects stomatal function. A, Stomatal responses to 50 µM ABA in Col (black), pll12-1 (magenta), and PLL12OE (green) genotypes. Error bars indicate standard error of the mean (sem); n ≥ 121 stomata per genotype per time point from three independent experiments. Different letters at each timepoint indicate P < 0.05 across genotypes for that timepoint, one-way analysis of variance (ANOVA) and Tukey’s test (dashed gray box indicates statistical comparison group). B, Representative images of stomata in Col, pll12-1, and PLL12OE plants before and after 2.5 h ABA treatment. Stomatal complexes and pores are outlined by yellow dashed lines. Bar = 5 µm. C–D, As in (A–B) but treated with 1 µM FC and n ≥ 122 stomata per genotype per timepoint. Bar = 5 µm. E, Comparison of stomatal responses to 50 µM ABA between GFP kd-2 (black) and PLL12 kd-2 (blue) plants. Error bars indicate SEM; asterisks indicate significant difference between genotypes at each time point examined: ****P < 0.0001, Student’s t test; n ≥ 102 stomata per genotype per time point from three independent experiments and six biological replicates (individual leaves from different plants) in total. F, As in (B) but for GFP kd-2 and PLL12 kd-2. Images without outlines of PLL12 kd2 are also shown. Bar = 5 µm. G–H, As in (E–F) but treated with 1 µM FC and n ≥ 112 stomata. Bar = 5 µm.
Although we observed altered stomatal dynamics in pll12-1 plants, the broad expression of PLL12 (Supplemental Figure S2, A–C) raises uncertainty as to whether PLL12 regulates stomatal function directly via its expression in guard cells. To address this uncertainty, we measured stomatal responses to ABA and FC in PLL12kd lines where PLL12 expression was specifically knocked down in guard cells, as well as in GFPkd controls. In these experiments, although PLL12kd guard cells did not differ in size from the controls (see below), PLL12kd stomata showed defective responses to some stimuli: for example, PLL12kd2 stomata closed and opened more slowly in response to ABA or FC, respectively, than GFPkd2 stomata (Figure 1, E–H). We measured stomatal pore widths after 2.5 h ABA or FC treatment for all three PLL12kd lines. Abnormal stomatal responses were seen in PLL2kd2 and PLL12kd3 plants (Supplemental Figure S3, I–L), but not in PLL12kd1 plants (Supplemental Figure S3, G–H), which did not show as extensive a reduction in PLL12 transcript levels (Supplemental Figure S2F). Measurements made 2.5 h after FC treatment and at the beginning of ABA treatments (after leaves were pre-incubated in the light for 2.5 h) indicated that PLL12kd stomata are unable to open as widely as the controls (Figure 1, E–H; Supplemental Figure S3, I–L). In most cases after the induction of stomatal closure, PLL12kd stomata were closed to a higher degree than the GFPkd controls (Figure 1G, time 0; Supplemental Figure S3, I, J, and L), although in other experiments measuring stomatal responses to ABA, PLL12kd stomata ultimately closed to a similar degree to GFPkd stomata (Figure 1E; Supplemental Figure S3K). Together, these data support a specific function of PLL12 in guard cells in facilitating normal stomatal dynamics.
PLL12 balances turgor pressure and wall mechanics in guard cells
After establishing that PLL12 is required for normal stomatal dynamics, we next investigated the underlying physical mechanism(s) by which PLL12 affects guard cell behavior. Previous studies of plant cell mechanics propose that cell wall modifications alter wall mechanics, which in combination with water uptake and cell pressurization determine the rate and extent of cell expansion during diffuse, irreversible growth (Cosgrove, 2016, 2018). One hypothesis to explain the observed defects in the rates and ranges of stomatal opening and closure in PLL12 mutant plants is that in Col plants, PLL12 cleaves HG in guard cell walls to prevent extensive pectin crosslinking, reducing wall modulus to facilitate stomatal opening and closure. Alternatively, PLL12 might influence guard cell pressurization without changing wall modulus, or it might influence both properties. To resolve these hypotheses, real-time, simultaneous measurements of wall modulus and turgor pressure in guard cells during responses to physiological stimuli are necessary. However, such measurements are challenging to achieve in guard cells, which are small and undergo large changes in turgor pressure (Franks et al., 1998).
Advances in nanoindentation combined with FEM, which have been employed to investigate the mechanics of epidermal cells of Arabidopsis and other systems (Forouzesh et al., 2013; Bidhendi and Geitmann, 2019; Li et al., 2021), now enable us to probe the mechanics of functioning guard cells. Nanoindentation directly and rapidly measures the force exerted between a probe tip and a cell indented sequentially at precise depths. With these data, local stiffness (force/length) can be quantified at specific depths. The local stiffness is governed by a combination of cell morphology, wall modulus, and turgor pressure. Thus, both wall modulus and turgor pressure can be estimated using a computational model (FEM) of the measurements. This model includes the measured shape of a given guard cell, with the model being used to characterize the relationship between the local stiffness and the probe indentation depth, wall modulus, and turgor pressure. This approach is effective because the local stiffness is depth-dependent, with shallow indentations (less than the thickness of the wall) influenced more by wall modulus and deeper indentations influenced more by turgor pressure. By measuring stiffness values at different depths in the same location and using FEM, the wall modulus and turgor pressure at a given time point can be estimated, and these measurements can be made repeatedly over the course of a physiological response experiment without killing the cell.
In our experiments, plants that had been kept in the dark overnight to induce stomatal closure were placed in a nanoindenter, and their stomata were induced to open and then to close by turning a light on and then off (Figure 2). Individual guard cells from attached leaves were indented every 10 min or less (Figure 2, A and B), and apparent stiffness at each specific depth was quantified from the unloading curve (Figure 2B). It should be noted that apparent stiffness increased soon after the light was turned on and dropped after the light was turned off for all the genotypes (Supplemental Figure S4).
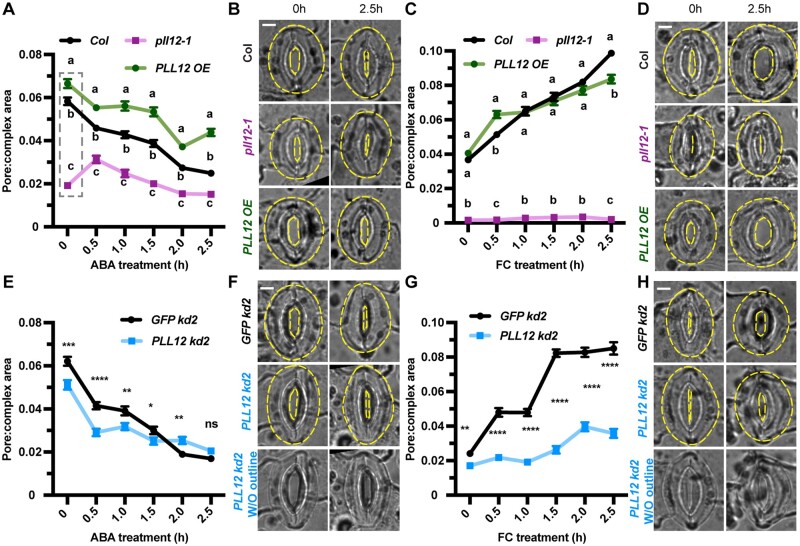
Figure 2.
Altered PLL12 expression affects turgor pressure and wall modulus in guard cells. A, Illustration of stiffness measurements by nanoindentation and experimental set up. Plants were kept in the dark overnight to ensure stomatal closure, and nanoindentation was performed at least every 10 min during the experiment. At 20 min after the first measurement (T0), light was turned on for 60 min to induce stomatal opening, then the light was turned off and measurements were collected for 30 min. The abaxial side of leaf seven was gently twisted to face upward, a target guard cell was located under the microscope, then the nanoindentation probe (tip radius = 2–3 μm) was moved to the same location. At each time point, stiffness in the same guard cell was measured at six different depths (B). B, Diagram of indentation at different depths, representative force–depth curve for one guard cell, and definition of stiffness. Loading–unloading curves of all six different depths are shown by blue numbers. A zoomed in view of a loading (orange) and unloading (blue) curve at the fifth depth is shown in the blue box. Stiffness is defined as the slope of the unloading curve. C, Simulation of calculated stiffness to six different depths. Blue dots represent the calculated stiffness from nanoindentation measurements; dashed line represents the results of the simulation. Bars = SD. D, Representative FEM models. Upper and lower images represent modeled guard cell shapes before (dark) and after pressurization (light), respectively. Longitudinal modulus E1 was set to be the same as radial modulus E3, and both are 4 times smaller than circumferential modulus E2. E, Illustration of the iteration process to derive turgor pressure and wall modulus from nanoindentation simulation and FEM. F–G, Wall modulus (F) and turgor pressure (G) derived for each genotype. n = 9 guard cells from at least five plants per genotype. Different letters at each timepoint indicate P < 0.05 across genotypes for that timepoint, two-way ANOVA and Tukey’s test. Bars = sem; ns, no significant difference.
To disentangle the contributions of wall modulus and turgor pressure to changes in apparent stiffness (Figure 2B) and stomatal aperture, each measurement for every cell was modeled using FEM. The model was constructed using measured cell size for each indented cell and wall thickness for each genotype (Supplemental Figure S5, B–C). The cell wall was modeled as an anisotropic elastic material with circumferential (E2), longitudinal (E1), and radial (E3) moduli (Figure 2D). The E2 direction is aligned with the orientation of cellulose microfibrils and was assumed to remain constant during the light on/off stages because guard cells undergo much less circumferential deformation than elongation during stomatal opening (Meckel et al., 2007). E1 and E3 moduli, which represent potential mechanical contributions from cellulose and wall matrix polymers, were assumed to be equal, were defined to be 4 times lower than E2 for the initial dark condition (see “Materials and Methods”; Marom et al., 2017; Yi et al., 2018), and were allowed to change during stomatal opening/closing. Simulations of nanoindentation measurements (Figure 2C) were performed iteratively (Figure 2E) to match each measured apparent stiffness as a function of indentation depth in order to estimate the E1 and E3 moduli, turgor pressure, and geometrical deformation (Supplemental Figure S6A).
This analysis revealed that in Col guard cells, wall modulus increased significantly within 5 min after light stimulation, then diminished slowly during the light-on phase; after light-off, the wall modulus in Col guard cells dropped suddenly then slowly recovered (Figure 2F; Supplemental Figure S6C). Turgor pressure in Col guard cells also increased rapidly when the light was turned on and continued to increase more slowly over the course of light stimulation; when the light was turned off, turgor pressure dropped promptly within the first 5 min and then continued to decrease (Figure 2G; Supplemental Figure S6E).
In pll12-1 guard cells, wall modulus also rose immediately, then gradually increased upon light stimulation, with a sudden drop and gradual recovery after the light was turned off. However, E1 and E2 moduli in pll12-1 guard cells were higher than in Col cells, both when stomata were closed in the first 20 min of the experiment and after the light was turned on (Figure 2F; Supplemental Figure S6B). Conversely, turgor pressure in pll12-1 guard cells was lower in the initial closed state, failed to increase as much within the first 5 min after light stimulation, and remained lower throughout the experiment than in Col cells (Figure 2G; Supplemental Figure S6F). In PLL12OE guard cells, no significant difference in wall modulus was detected compared with Col cells. However, turgor pressure initially dropped, then plateaued in PLL12OE guard cells after the light was turned off, a pattern that differed slightly from that in Col cells (Figure 2G). These abnormalities in wall modulus and turgor pressure dynamics in both genotypes were consistent with the results of the stomatal function assays, where pll12-1 stomatal opening was reduced and the stomata opened more slowly than Col stomata in response to light (Supplemental Figure S3, D and F) or FC (Figure 1C), and PLL12OE stomata opened normally, but stomatal closing was reduced compared to Col in response to dark (Supplemental Figure S3, C and E) or ABA (Figure 1A).
For comparison, we also measured turgor pressure in guard cells using incipient plasmolysis (Weber et al., 2015). Turgor pressure in guard cells was estimated in leaves exposed to light with fully open stomata and in leaves treated with ABA and darkness for 2.5 h to induce stomatal closure. Using incipient plasmolysis, turgor pressure values in guard cells of open and closed stomata were estimated to be 1.67 ± 0.46 MPa and 0.65 ± 0.02 MPa, respectively, in Col leaves; 4.58 ± 1.13 and 3.30 ± 0.81 MPa in pll12-1 leaves; and 1.48 ± 0.02 MPa and 0.90 ± 0.08 MPa in PLL12OE leaves. These values for Col and PLL12OE guard cells were comparable to the maximal and minimal turgor pressure values derived from nanoindentation–FEM analyses (Figure 2G). However, turgor pressure values estimated by incipient plasmolysis for pll12-1 guard cells were ∼3 times those from nanoindentation–FEM analyses. The majority of the incipient plasmolysis results were consistent with the nanoindentation–FEM results with the exception of pll12-1, where changes in wall structure might inhibit osmolyte diffusion and/or water transport, complicating the estimation of turgor pressure by incipient plasmolysis, which is known to be time- and condition-dependent (Willmer and Beattie, 1978).
We tested the sensitivity of FEM models to different E1:E2 modulus ratios for single Col and pll12-1 guard cells (Supplemental Figure S6G). Changing the E1:E2 ratio from 1:4 to 1:2 led to 3% and 25% increases in estimated turgor pressure, whereas changing the ratio to 1:8 resulted in 18% and 12% decreases in estimated turgor pressure, respectively. These changes are much smaller than the approximately two-fold difference in estimated turgor pressure between Col and pll12-1 guard cells (Figure 2G). Examining E1 and E2 values, we observed increases in E1 of 18% and 27% upon changing the E1:E2 ratio from 1:4 to 1:2, and decreases in E1 of 20% and 16% upon changing the E1:E2 ratio from 1:4 to 1:8 in Col and pll12-1 guard cells, respectively. In contrast, E2 decreased by 41% and 36% upon changing the E1:E2 ratio from 1:4 to 1:2, whereas E2 increased by 59% and 68% upon changing the E1:E2 ratio from 1:4 to 1:8 in Col and pll12-1 guard cells, respectively. As for turgor pressure values, these changes are smaller than the difference in estimated E1 modulus between Col and pll12-1 guard cells (Figure 2F).
To further examine the guard cell-specific function of PLL12 in stomatal dynamics, we performed nanoindentation–FEM analyses on PLL12kd2 and GFPkd2 plants that were grown in the dark overnight. Estimated turgor pressure was significantly lower in PLL12kd2 guard cells than in GFPkd2 guard cells (Supplemental Figure S6H), whereas estimated wall modulus in PLL12kd2 guard cells was similar to that in GFPkd2 guard cells (Supplemental Figure S6H). Together, these data imply that PLL12 is required to establish normal cell pressurization and wall mechanics in guard cells to facilitate dynamic stomatal responses, and that normal cell pressurization depends on the expression of PLL12 specifically in guard cells.
Changes in PLL12 expression alter HG composition in guard cell walls
The above data reveal a physical mechanism by which PLL12 modulates guard cell mechanics. The next step was to determine whether the molecular status of the cell wall correlated with these physical and functional changes in PLL12 mutants. To test the hypothesis that PLL12 cleaves HG in the guard cell wall to alter the status of the pectin network and influence wall mechanics, we labeled guard cells of wild-type and mutant genotypes with dyes and antibodies that recognize different forms of HG (Figure 3). Chitosan oligosaccharide-Alexa 488 (COS488) interacts with low methyl-esterified (low DM) HG (Mravec et al., 2014), and propidium iodide (PI) binds to negatively charged uronic acids (UAs) in HG (Rounds et al., 2011; Figure 3A). The antibody 2F4 interacts with calcium crosslinked HGs (Powell et al., 1982; Liners, 1989), and LM19 and LM20 recognize low and high DM HG, respectively (Verhertbruggen et al., 2009; Figure 3A). Negative controls for immunolabeling did not show specific signals (Supplemental Figure S7). We also measured total UA content in leaves to estimate the abundance of HG.
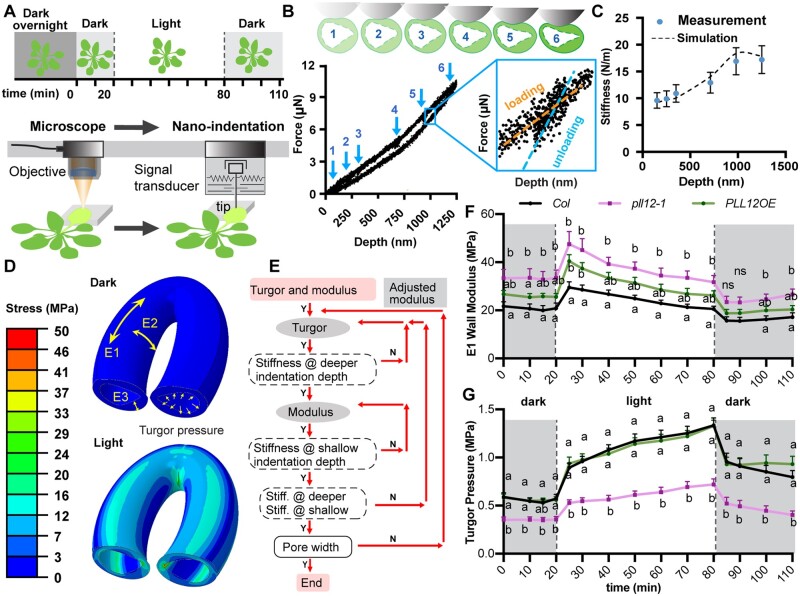
Figure 3.
PLL12 influences HG labeling in guard cell walls. A, Diagram showing different epitopes of HG that are recognized by the dyes and antibodies. Blue pentagon represents GalA, with or without red circle on Gal represents methylated or unmethylated GalA, respectively. Yellow diamond represents PI molecule. B, Representative XY (upper) and XZ (lower) maximum projection images of COS488 labeling. Images are displayed with a fire look-up table, XZ projections were made from the midline in the Y direction. Bar = 5 µm. C, Quantification of relative intensity of COS488 labeling in guard cells from 3- to 4-week-old Col, pll12-1, and PLL12 OE plants. Whiskers extend to min and max, box boundaries indicate first and third quartiles of datasets, and horizontal lines inside boxes represent medians; n ≥ 153 guard cells per genotype from two independent experiments, six plants in total. Different letters indicate P < 0.05, one-way ANOVA and Tukey’s test. D and E, as in (B and C) but stained with PI and n ≥ 131 guard cells per genotype. F, Representative images of cross-sections of guard cells from Col, pll12-1, and PLL12 OE plants, immunolabeled with 2F4 (green) and counterstained with S4B (magenta). Bars = 5 µm. G, Quantification of 2F4 labeling intensity in cross-sections of guard cells from 3- to 4-week-old Col, pll12-1 and PLL12 OE plants. Error bars indicate SEM; n ≥ 112 guard cells per genotype from at least two independent experiments, three different leaves. Different letters indicate P < 0.05, or no significant differences across genotypes, one-way ANOVA and Tukey’s test. H–K, as in (F and G) but labeled with LM19 (H and I) or LM20 (J and K). L, UA content, and PL, PG, and PME enzymatic activity in Col, pll12-1, and PLL12 OE rosettes. For UA content, PG, and PME assays, n = three plants per genotype assayed independently, each with five technical replicates per plant. For the PL assay, n = three plants per genotype assayed independently, each with three technical replicates per plant. Values are mean ± SD. Different letters indicate P < 0.05, one-way ANOVA and Tukey’s test.
In pll12-1 guard cells, COS488 labeling intensity was lower and 2F4 and PI labeling intensities were slightly but significantly higher than in the Col controls, but LM19 and LM20 labeling intensity did not differ from the Col controls (Figure 3, A–K). Total UA content appeared to be slightly lower in pll12-1 rosettes than in Col, but this difference was not statistically significant (Figure 3L). These results suggest that the walls of pll12-1 guard cells contain less de-methylesterified HG that is available for COS488 binding, but higher amounts of calcium crosslinked HG, than Col. In guard cell walls of PLL12 OE plants, PI staining intensity was higher than in the Col controls (Figure 3, D and E), but labeling intensity with other probes and UA content in leaves did not differ from Col (Figure 3, B–C and F–L), suggesting that HG in guard cell walls of PLL12OE plants might contain more de-methylesterified UAs. UA content was also higher in PLL12OE leaves than in pll12-1 leaves (Figure 3L).
The altered wall composition in pll12-1 and PLL12OE guard cells supports the influence of PLL12 on HG abundance and modification status. To probe pectin metabolism further, we measured total PL, PG, and PME activities in protein extracts from rosettes. No significant differences in the activities of these enzymes were detected across genotypes (Figure 3L), although these assays with total protein from leaves might obscure more specific changes in pectin metabolism in guard cells in relation to altered PLL12 expression. Overall, the data in Figures 1–3 indicate that excessive HG crosslinking might account for the higher wall modulus observed in pll12-1 guard cells that accompany their defective stomatal opening and closure. In PLL12OE guard cells, slower stomatal closure and opening are accompanied by more subtle changes in cell biomechanics and wall composition.
Normal PLL12 expression in guard cells is required for cell proliferation but not cell expansion in growing rosette leaves
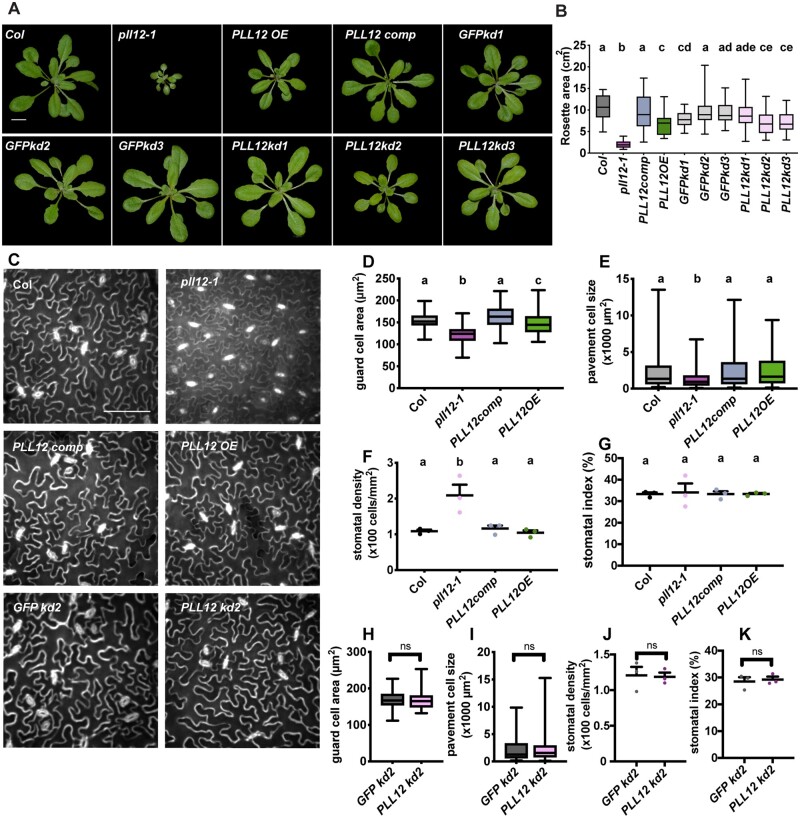
We investigated the physiological function of PLL12 by examining the sizes of 21-day-old plants of different genotypes. Average rosette area in pll12-1 plants was approximately one-fifth of that in Col plants, and complementation with PLL12 fully rescued the pll12-1 dwarf phenotype (Figure 4). PLL12OE plants also had smaller rosettes than Col (Figure 4, B and C). GFPkd1 control plants had smaller rosettes than Col, and both PLL12kd2 and 3 plants, but not PLL12kd1 plants, had smaller rosettes than Col and GFPkd2 and 3, in keeping with the qPCR data, where PLL12kd2 and 3 but not PLL12kd1 plants had significantly reduced PLL12 transcript levels in rosette leaves (Supplemental Figure S2F).
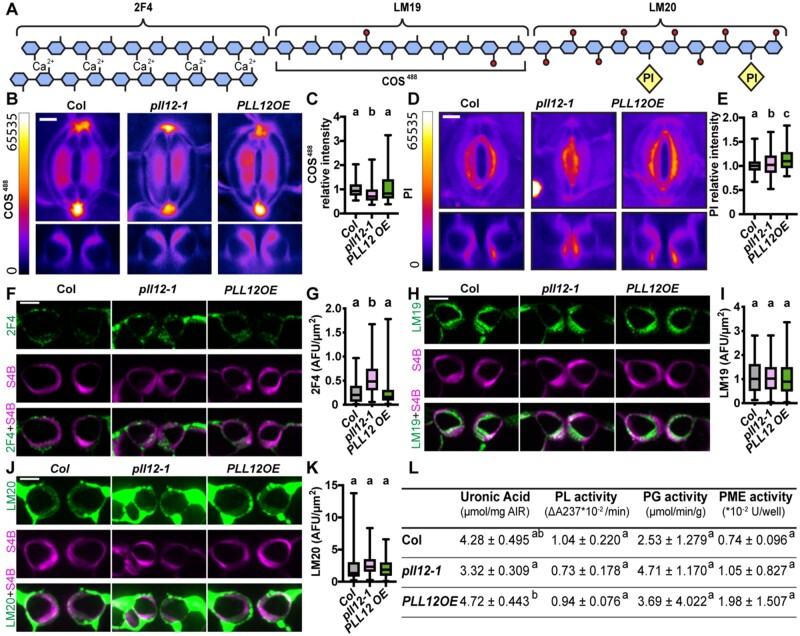
Figure 4.
PLL12 expression affects plant growth, epidermal cell expansion, and proliferation. A, Representative rosette images of 21-day-old Col, pll12-1, PLL12 overexpression (PLL12 OE), PLL12 complementation (PLL12 comp), and guard cell-specific knockdown pGC1:GFP kd-1 to -3 and pGC1:PLL12 kd-1 to -3 lines. Bar = 1 cm. B, Rosette areas of 21-day-old Col and PLL12 transgenic plants; n ≥ 35 plants per genotype from three independent experiments. C, Representative images of epidermal cells stained with PI in 21-day-old Col, pll12-1, PLL12 OE, PLL12 comp, GFP kd-2, and PLL12 kd-2 plants. Enhance contrast was performed on the maximum projection of z-stack images. Bar = 100 µm. D–G, Quantification of guard cell area (D), pavement cell size (E), stomatal density (F), and stomatal index (G) in 21-day-old Col, pll12-1, PLL12 OE, and PLL12 comp plants. n ≥ 74 (D) and n ≥ 99 (H) guard cells from three individual plants per genotype. n ≥ 223 (E) and n ≥ 105 (I) pavement cells from at least three individual plants per genotype. n = three individual plants (F, G, J, and K) with five fields of each were imaged and quantified for density and index analysis. Stomatal index = number of stomata (ns) divided by the sum of stomata number and pavement cell number (np) per field (ns/(ns + np)). Different letters denote P < 0.05, one-way ANOVA and Tukey’s test. F–I, as in (B–E) but for GFP kd-2 and PLL12 kd-2 plants. Dark gray dots or box represent Col, magenta for pll12-1, blue for PLL12comp, green for PLL12OE, light gray for GFPkd, and pink for PLL12kd. Error bars indicate sd. Whiskers extend to min and max, box boundaries indicate first and third quartiles of datasets, and horizontal lines inside boxes represent medians.
We next sought to determine how PLL12 affects rosette growth at the cellular level. Compared to the Col controls, pll12-1 plants showed ∼20% and ∼50% reductions in guard cell and pavement cell area, respectively, with an approximately two-fold increase in stomatal density (stomata/area, Figure 4, D–F) but no change in stomatal index (stomata/(stomata + pavement cells)). This smaller cell size alone does not account for the 80% reduction in rosette area in the pll12-1 mutant (Figure 4, Aand B), suggesting that both cell expansion and proliferation might be affected. Restoration of PLL12 rescued these phenotypes in PLL12comp plants (Figure 4, C–G). Despite their reduced leaf size, cell size and stomatal patterning were unaltered in PLL12OE plants (Figure 4, C–G). Likewise, PLL12kd2 plants did not show changes in cell size or stomatal patterning (Figure 4, C, and H–K), implying that the reduced rosette area in these plants (Figure 4, A and B) arises mainly from reduced cell proliferation.
Discussion
Here we report that in addition to PG and PME genes (Amsbury et al., 2016; Huang et al., 2017; Rui et al., 2017), the PL gene PLL12 also influences stomatal function. Although both PL and PG genes encode enzymes that degrade demethylated HGs in the cell wall, loss of PLL12 expression alters stomatal dynamics differently from the loss of the guard cell-expressed PG gene PGX3 (Rui et al., 2017). Whereas pll12-1 stomata open and close more slowly over a smaller dynamic range than Col stomata, pgx3-1 knockout stomata open normally but close with a slightly smaller dynamic range and in a step-wise fashion, potentially reflecting differences in substrate specificity between the PL and PG. The higher amount of calcium crosslinked HG in pll12-1 cell walls, as detected by 2F4 immunolabeling, is consistent with the canonical function of PLs in degrading pectate. Similarly, pgx3-1 walls show increased 2F4 labeling, suggesting that calcium crosslinked HG might be a common substrate for both PL and PG enzymes (Rui et al., 2017). However, the findings that LM19 labeling of low-methylesterified HG is increased in pgx3-1 guard cells but similar to the controls in pll12-1 mutants suggests that PLL12 might target a type of HG that is distinct from that targeted by PGX3.
PME6 is also required for stomatal function (Amsbury et al., 2016). A common phenotype of pme6-1 and pll12-1 guard cells is that the stomata in both mutants open and close within a smaller dynamic range, but whereas pll12-1 stomata could reach a more closed state than WT, pme6-1 stomata could not close (Amsbury et al., 2016). In addition to having different enzymatic effects on the cell wall, turgor pressure dynamics might be differentially altered in these mutants. Further empirical and modeling analyses of additional genetic and/or biochemical perturbations of guard cell walls will be required to better understand how different classes of pectin-modifying enzymes influence guard cell mechanics and function (Jones et al., 2003; Amsbury et al., 2016).
The nanoindentation–FEM approach described here can be used to simultaneously track wall modulus and turgor pressure in living guard cells undergoing physiological responses. For Col guard cells, E1 and E3 modulus values in the dark were estimated to be around 20 MPa (Figure 2F), which is very close to previous AFM measurements of apparent modulus in plasmolyzed guard cells (Carter et al., 2017). Upon light stimulus, we observed an immediate initial increase in directional wall modulus that slowly diminished over the course of light stimulation (Figure 2F). A study in tobacco (Nicotiana tabacum) stomata combining the use of a pressure probe to inflate guard cells with calculations of volumetric elastic modulus found that the bulk modulus of guard cells increases as stomata open wider (Zhang et al., 2011), and a theoretical analysis of wall mechanics predicted an increase in E2 in the guard cell wall during stomatal opening that was proposed to arise from strain-stiffening (Wu and Sharpe, 1979). Therefore, we suspect that the rapid initial increase in modulus we observed (Figure 2F) might be caused by strain stiffening of the wall (Cosgrove, 1993; Kierzkowski et al., 2012), whereas the subsequent slow reduction in modulus might represent time-dependent wall relaxation accomplished by wall-loosening proteins such as expansins (Zhang et al., 2011; Cosgrove, 2016). Together, our findings, along with previous studies, support the existence of dynamic changes in directional wall modulus during guard cell deformation, opening avenues for studying the interplay between wall mechanics and cell wall remodeling during stomatal dynamics.
In Col plants, our nanoindentation–FEM analysis revealed an initially sharp, then gradual increase in turgor pressure from ∼0.5 MPa to ∼1.3 MPa over 60 min of light exposure that accompanied an increase in stomatal pore width of ∼0.8 µm (Figure 2G; Supplemental Figure S6A). These dynamic changes in turgor pressure are potentially driven by initial rapid ion flux into guard cells upon light-induced membrane depolarization and a corresponding drop in osmotic potential that causes water influx: both fluxes slow but do not cease as guard cells continue to respond to the light stimulus (Jezek and Blatt, 2017). Due to their small size, it is quite difficult to perform turgor pressure measurements for Arabidopsis guard cells. However, for species where guard cells have been probed, turgor pressure increases by ∼0.25 to 1 MPa along with each ∼1 µm increase in stomatal pore width (Franks et al., 1998), which is within the same order of magnitude as the turgor pressure pore width correlation we report here.
The ability to capture dynamic changes in turgor pressure during stomatal opening and closure, especially the rapid increase and decrease in turgor pressure we observed upon switching the light stimulus on or off, paves the way for investigating the functional connections between rapid signaling events and turgor pressure in guard cells. The asymmetry, we observed in both wall modulus and turgor pressure changes is further evidence for the cryptic hysteresis (apparently symmetrical behavior that belies a mechanistic dependence on the direction of the process) that we hypothesize to be a feature of guard cell biomechanics, in which biophysical hysteresis (dependence on history) underlies the apparently symmetrical opening and closure behaviors of guard cells (Rui et al., 2017).
In pll12-1 knockout plants, we found that although wall modulus and turgor pressure values in guard cells derived from nanoindentation–FEM analyses showed similar dynamic trends as in Col plants, wall moduli remained consistently higher, whereas turgor pressures were consistently lower than in Col guard cells. The nanoindentation–FEM results for PLL12OE plants were more complex, with wall modulus values lying in between values for Col and pll12-1 but not differing significantly from either genotype at most timepoints, whereas turgor pressure in PLL12OE stomata was estimated to be nearly indistinguishable from Col, except for showing a slower reduction after lights off. We conclude that the observed dysfunctional stomatal dynamics in pll12-1 plants (Figure 1) arise from the combined effects of stiffer cell walls and a smaller initial jump and slower increase in turgor pressure (Figure 2), while the observed slower stomatal closure in PLL12OE plants (Figure 1) arises from the slower decrease in turgor pressure (Figure 2). The more closed stomata in PLL12kd lines than GFPkd lines after 2.5-h ABA treatment or before FC treatment (Figure 1G; Supplemental Figure S3, I–L) are attributable to a significant lower turgor pressure, but not wall modulus, in PLL12kd lines (Supplemental Figure S6H).
The application of exogenous PG to guard cells reduces their apparent elastic modulus, as measured by AFM (Carter et al., 2017), underscoring the importance of pectin modification in determining wall mechanics in guard cells. A higher proportion of cross-linkable HG in pll12-1 (Figure 3) likely accounts for the observed increase in wall stiffness, as previous findings demonstrate that a higher proportion of calcium-crosslinked pectins in pectin gels increases their elastic modulus in vitro (Ström et al., 2007). An AFM study of Arabidopsis hypocotyl cells also revealed a correlation between more abundant calcium-crosslinking pectin and higher wall modulus (Daher et al., 2018). The finding that wall modulus in guard cells is significantly increased in pll12-1 plants but not in PLL12kd2 plants might be attributable to remaining PLL12 activity in PLL12kd plants.
Our study showed that knocking out PLL12 inhibits guard cell pressurization after light stimulus, whereas overexpressing PLL12 prevents continued guard cell depressurization upon light removal (Figure 2G). Knocking down PLL12 specifically in guard cells also reduces turgor pressure in guard cells (Supplemental Figure S6H), suggesting that maintaining stomatal pore aperture, which depends on guard cell pressurization, requires PLL12 expression specifically in guard cells. These findings were unexpected and highlight a potential connection between HG degradation by PLs and signaling networks, such as cell wall integrity sensing (Bai et al., 2009; Ringli, 2010; Ma et al., 2019), that include stomatal responses to pectin oligosaccharides (Lee et al., 1999). These results suggest that wall-modifying enzymes might affect cell mechanics and dynamic behaviors through a signaling-mediated influence on turgor pressure in addition to their direct effects on wall modulus. They also demonstrate the need to measure both turgor pressure and wall modulus simultaneously in studies of stomatal responses and to apply genetic and molecular tools to investigate how feedback from wall integrity signaling might influence wall mechanics.
Because pectins are thought to exist in complex three-dimensional networks, whereas cellulose orientation is constrained within the plane of the cell wall, it is possible that altering PLL12 expression changes directional wall moduli differently, a subject for further refinement and testing of the nanoindentation-FEM approach described here. In our FEM analysis, the ratio of E1:E2 wall modulus was assumed to be 1:4 for the closed state given the estimated proportions, physical properties, and orientations of cellulose and matrix polysaccharides in the guard cell wall (Marom et al., 2017; Yi et al., 2018). However, if pectin is the only polysaccharide affected in pll12-1 plants, and more extensive pectin crosslinking in the guard cell wall gives rise to higher moduli in all dimensions, E1 would be expected to increase relative to E2, moving the E1:E2 ratio closer to or past 1:2. If the ratio were 1:2, turgor pressure in pll12-1 guard cells would be 25% higher than our current estimation (Supplemental Figure S6G) but would still be lower than in Col guard cells (Figure 2G).
We also employed incipient plasmolysis for comparison with our nanoindentation–FEM analyses. Turgor pressure estimates obtained for Col and PLL12OE were similar between the two methods. Additionally, the difference in turgor pressure between open and closed stomata, as estimated by incipient plasmolysis, was smaller in PLL12OE guard cells (0.58 MPa) than in Col guard cells (1.02 MPa). This result is consistent with the slower decrease in turgor pressure in PLL12OE guard cells than Col after the light was turned off, as suggested by nanoindentation–FEM analyses, although that difference was not statistically significant (Figure 2G). However, the turgor pressure values estimated in guard cells of pll12-1 knockout plants with open and closed stomata were ∼3 times the values obtained by nanoindentation–FEM analyses. This discrepancy raises the possibility that the nanoindentation–FEM approach does not accurately derive turgor pressure in the pll12-1 mutant in contrast to the highly correlated results for other genotypes as well as pavement cells (Li et al., 2021). Instead, we think it is more likely that turgor pressure in the pll12-1 mutant is less able to be accurately measured by incipient plasmolysis, because this method depends on factors such as plasmolysis time, solute identity, and the diffusion rates of water and solutes (Willmer and Beattie, 1978). The altered stomatal behavior and wall composition observed in pll12-1 further complicate interpretations of incipient plasmolysis data. In contrast, nanoindentation is rapid, and FEM takes into account differences in cell geometry between genotypes, highlighting its utility for estimating turgor pressure in guard cells.
We found that PLL12 affects rosette size by affecting both cell expansion and proliferation (Figure 4), which is consistent with the function of the rice (Oryza sativa) PLL gene DEL1 in cell cycle progression and leaf growth (Leng et al., 2017). However, another Arabidopsis PL gene, PMR6, affects cell expansion but not proliferation (Vogel, 2002). PLs might potentiate cell expansion by loosening the pectin network in the wall via a mechanism comparable to that of PGs (Xiao et al., 2014; Rui et al., 2017). Alternatively, PLL12 might function in cell expansion by helping to establish the molecular architecture required to maintain proper wall integrity (Anderson, 2016; Leng et al., 2017). The defects of the PLL12kd line in cell proliferation but not expansion suggest that PLL12 influences cell proliferation in a guard cell-specific manner to influence rosette growth, potentially by simply enabling sufficient CO2 capture through stomata to drive photosynthesis and provide energy for leaf cell proliferation. Our findings suggest a dual role for PLL12 in rosette leaf growth, in that it might facilitate guard and pavement cell expansion by degrading pectin and enable cell proliferation by potentiating stomatal dynamics.
In summary, we found that the putative PL, PLL12 reduces the levels of cross-linkable HG in the guard cell wall and is required for the ability of guard cells to maintain sufficient turgor pressure for driving guard cell expansion in response to light. By enabling normal stomatal function, guard cell-expressed PLL12 influences cell proliferation and leaf growth. The ability to measure stomatal biomechanics in real time and combine realistic models of guard cells with sophisticated material simulations has allowed us to shed light on the unexpected biophysical mechanisms by which stomatal guard cells respond to external stimuli and revealed how a cell wall-modifying gene influences guard cell biomechanics and stomatal dynamics. Further dissection of these mechanisms should enable plant improvement, leading to the development of resilient and sustainable crops that will benefit human societies.
Materials and methods
Generation of transgenic plants
Seeds of wild-type A. thaliana Col-0 and T-DNA insertion mutant pll12-1 (CS878465) were obtained from the Arabidopsis Biological Resource Center (ABRC). To constitutively overexpress PLL12, the PLL12 coding sequence (generated by PCR using primer PLL12F and primer PLL12R) was cloned into entry vector pCR8/GW/TOPO using a TA Cloning Kit (Invitrogen, Waltham, MA, USA); then the coding sequence was inserted into destination vector pEarleyGate 101 (Earley et al., 2006) using Gateway LR Clonase II (Invitrogen, Waltham, MA, USA); the overexpression construct was then transformed into the Col-0 background and homozygous plants were selected using 5-μM methionine sulfoximine. For PLL12 expression pattern analysis, a 2-kb fragment upstream of the PLL12 start codon (PCR using primer pPLL12 F and primer pPLL12 R) was TA cloned into the pCR8/GW/TOPO entry vector and then LR cloned into pMDC162 (Curtis and Grossniklaus, 2003), which contains a GUS coding sequence; the plasmid was then transformed into Col-0, and transgenic plants were selected on 25-μg/mL hygromycin to obtain homozygous lines. To generate the PLL12 complementation line, the 2 kb PLL12 native promoter and PLL12 coding sequence described above were ligated (primer PLL12 p/CDS overlap) and cloned into the pCR8/GW/TOPO entry vector, then LR cloned into vector pMDC110 (Curtis and Grossniklaus, 2003); the plasmid was transformed into pll12-1 heterozygous plants and selected using 5-μM methionine sulfoximine and 25-μg/mL hygromycin to obtain homozygous lines for both alleles. To obtain guard cell-specific knockdown lines and technical controls, amiRNAs targeting three different sites of PLL12 and GFP were designed (see Supplemental Table S1) and inserted into vector pMDC32B-AtMIR390a-B/c (Carbonell et al., 2014) separately. The 35S promoter in the vector was replaced by the guard cell-specific promoter pGC1 (PCR using primers pGC1 D1 F and pGC1 D1 R and restriction enzymes PstI and KpnI, T4 DNA ligase [NEB]) (Yang et al., 2008) to achieve cell-specific expression of amiRNAs. Plant transformation was performed using an Agrobacterium (GV3101)-based floral dip method.
Plant growth conditions
Surface sterilized (20 min in 30% bleach + 0.1% SDS) A. thaliana seeds were stratified at 4°C for 3–10 days before being plated on Murashige and Skoog (MS) plates containing 2.2 g/L MS salts (Caisson Laboratories, North Logan, UT, USA), 0.6 g/L MES, 1% (w/v) Suc, and 0.8% (w/v) agar (Sigma, St Louis, MO, USA), pH 5.6. Seedlings were grown at 22°C under 24 h of illumination (4100K Fluorescent Lamp, 800-900 PPFD) for 10 days before being transferred to soil supplemented with Miracle-Gro (The Scotts Company, Marysville, OH, USA). Plants were grown in a chamber under 16-h light/8-h dark light conditions at 22°C.
Gene expression analysis
For GUS staining, 6-day-old seedlings, epidermal peels, or rosettes of 3- to 4-week-old PLL12 pro:GUS lines were soaked in GUS staining solution (50 mM sodium phosphate, pH 7.2, 0.2% (v/v) Triton X-100, and 2 mM X-Gluc) in the dark for 3–16 h before de-staining with 70% ethanol. A Zeiss Discovery V12 fluorescence dissecting microscope was used to collect images of seedlings; a Zeiss Axio Observer microscope attached to a Nikon D5100 DSLR camera was used for the epidermis, and a Scanjet 8300 scanner (HP) at 600 dpi was used for rosette imaging.
For qPCR, total RNA was extracted from 21-day-old rosette leaves 5–8 using a NucleoSpin RNA Plant kit (Machery-Nagel, Düren, Germany), and cDNA was synthesized using Quanta qScript cDNA Supermix (Quantabio, Beverly, MA, USA). The cDNA and PLL12 qPCR primers (PLL12qF and PLL12qR) were mixed with Quanta PerfeCTa SYBR Green Fastmix ROX (Quantabio; catalog no. 95073-250). Reactions and quantification were performed on a StepOne Plus Real-Time PCR machine (Applied Biosystems, Waltham, MA, USA) using the following program: 2 min at 95°C; 10 s at 95°C, then 30 s at 60°C for 40 cycles. To calculate the relative expression of PLL12 across different transgenic plants, ACTIN2 (ACT2) (ACT2-qF and ACT2-qR) and Col-0 were used as controls.
Plant growth analysis
Twenty-one-day-old plants were imaged with a Nikon D5100 DSLR camera. Images were segmented based on color threshold: images were opened in ImageJ and based on the HSV (Hue, Saturation, and value) color space, green regions were selected to separate rosettes from the background. The background was then removed and the wand tool was used to select each rosette to measure its area. For the epidermal cell dimension and patterning study, leaves 5–8 from 21-day-old plants were excised and imaged. To measure guard cell size, the epidermis of a leaf was peeled and soaked in 100 μg/mL PI for 5 min. For pavement cell size and stomatal density and index, intact leaves were used. Images were collected on a Zeiss Axio Observer microscope with a Yokogawa CSU-X1 spinning disk head using a 63X 1.4 NA oil immersion objective for guard cell size and a 20X 0.5 NA air objective for other measurements. A 561 nm excitation laser and a 617/73-nm emission filter were used to image PI. Five fields of three plants per genotype were imaged and quantified using ImageJ.
Stomatal function assays
Fully expanded mature leaves (leaves 5–8 from 3- to 4-week-old plants) were excised and used for stomatal function assays (Rui and Anderson, 2016). To record stomatal opening in response to FC, excised leaves were acclimated in dark solution (20 mM KCl, 1 mM CaCl2, and 5 mM MES-KOH, pH 6.15) for 2.5 h, and the leaves were then incubated in 1 mM FC in dark solution in the dark for another 2.5 h. To record stomatal closure in response to ABA, excised leaves were acclimated in light solution (containing 50 mM KCl, 0.1 mM CaCl2, and 10 mM MES-KOH, pH 6.15) for 2.5 h, and the leaves were then incubated in light solution containing 50 mM ABA in the light (4100K Fluorescent Lamp, 800-900 PPFD) for another 2.5 h. To track stomatal opening in response to light, excised leaves were acclimated in light solution (containing 50 mM KCl, 0.1 mM CaCl2, and 10 mM MES-KOH, pH 6.15) for 2.5 h in the dark, and then transferred to the light for another 2.5 h. To record stomatal closure responses to dark, excised leaves were acclimated in dark solution (20 mM KCl, 1 mM CaCl2, and 5 mM MES-KOH, pH 6.15) for 2.5 h in the light and then transferred to the dark for another 2.5 h. For Col, pll12-1, PLL12OE, GFPkd2, and PLL12kd2, epidermal peels from two leaves of each genotype were collected every 30 min and imaged; to compare pore width in GFPkd1-3 and PLL12kd1-3, epidermal peels were collected 0 and 150 min after incubation in ABA or FC. Samples were imaged on a Zeiss Axio Observer microscope; 10 fields per epidermis were imaged. Each assay was repeated at least 3 times, and in each experiment, epidermal peels were collected from two leaves from two individual plants. Stomatal pore area, complex area, and pore width were measured using ImageJ. To account for different guard cell sizes, especially in pll12-1, area ratio (pore area/complex area) was calculated and displayed to reflect the degree of stomatal opening.
HG labeling of intact guard cells
Fully expanded mature leaves (leaves 5–8 from 3- to 4-week-old plants) were used for both COS488 and PI labeling. For COS488 staining (Mravec et al., 2014), epidermal peels were collected and stained in a 1:1,000 diluted solution for 20 min, and after rinsing, z-stack images (0.5 μm z distance) were taken using a 488-nm excitation laser and a 525/50-nm emission filter. For PI staining, epidermal peels were stained in 100 μg/ml PI for 5 min, and after rinsing away excess dye, z-stack images (0.5 μm z distance) were taken using a 561-nm excitation laser, and a 617/73-nm emission filter. Images were collected with a Zeiss Axio Observer microscope with a 63X 1.4 NA oil immersion objective. To quantify fluorescence intensity, z-stack images were projected using the SUM algorithm in ImageJ, then areas and raw integrated density of entire guard cell regions for COS488 labeling and guard cell regions without phenolic rings for PI staining were measured. Relative fluorescence intensity was calculated by dividing raw integrated density by traced area.
Immunolabeling and dye staining of guard cell cross-sections
For section preparation, square 3-mm leaf patches cut from leaves were vacuum infiltrated in 4% (w/v) formaldehyde in PEM buffer (0.1 M PIPES, 2 mM EGTA, 1 mM MgSO4, pH 7) and incubated for 1 h (Rui et al., 2017). The leaf patches were then dehydrated in an ethanol series (30 min each in 30%, 50%, 70%, and 100% ethanol) and infiltrated with LR White Resin (Electron Microscopy Science, Hatfield, PA, USA) diluted in ethanol (30 min each in 10%, 20%, 30%, 50%, 70%, 90%, and 100%); 100% LR White Resin was replaced two more times with at least 8 h incubations. Samples were placed vertically in gelatin capsules (Ted Pella) filled with resin for 7 days at 37°C. Sections of 2 µm thickness were cut using a Leica UC6 ultramicrotome with a glass knife and collected on positively charged glass slides. For immunolabeling, sections were blocked in 3% (w/v) BSA in KPBS (0.01 M K3PO4 and 0.5 M NaCl, pH 7.1) for LM19 and LM20 labeling, or in TCaS buffer (20 mM Tris–HCl, 0.5 mM CaCl2, and 150 mM NaCl, pH 8.2) for 2F4 labeling, for 4 h. Sections were then incubated with a ten-fold dilution of primary antibodies in 3% (w/v) BSA in KPBS or TCaS for 24 h at room temperature. After rinsing 3 times with KPBS or TCaS solution, sections were incubated in secondary antibody (100-fold dilution in 3% (w/v) BSA in KPBS or TCaS) for 16 h. For LM19 and LM20, the secondary antibody Alexa Fluor 488-conjugated goat anti-rat IgG (H + L), was used; for 2F4, Alexa Fluor 488-conjugated goat anti-mouse IgG (H + L) was used. The samples were rinsed again 3 times before being counterstained with 0.1% (w/v) S4B in (KPBS or TCaS) for 30 min. After rinsing with KPBS or TCaS, the samples were imaged under a Zeiss Axio Observer microscope with a 100X 1.4 NA oil objective. A 488-nm excitation laser and a 525/50-nm emission filter were used for Alexa Fluor 488 signals, and a 561-nm excitation laser and a 617/73-nm emission filter were used for S4B signals. To quantify the arbitrary fluorescence units (AFUs), the area of the guard cell wall was traced using the S4B staining image, and raw integrated density from the same region was measured. To account for background noise, the AFU of samples that were stained with only secondary antibody was also calculated and subtracted from the AFU of samples with both primary and secondary antibody. All antibodies are described in Supplemental Table S2.
Cell wall thickness was measured by staining sections prepared as for immunolabeling with 0.05% (w/v) toluidine blue for 10–30 s and rinsing with water. Samples were imaged using a Zeiss Axio Observer microscope with a 100X 1.4 NA oil objective and a Nikon D5100 DSLR camera. Thicknesses of guard cell walls at different positions were measured using ImageJ. Sections were prepared from three leaves from three individual plants.
Enzymatic assays
Total protein extraction for PL activity assays was adapted from (Silva-Sanzana et al., 2019). Rosettes of 4–5-week-old plants were ground in extraction buffer (1 M NaCl, 0.2 M Na2HPO4, 0.1 M citric acid, pH 6.5) and incubated at 4°C for 1.5 h. The homogenate was centrifuged at 15,000 g for 10 min at 4°C, and the supernatant was transferred to a new tube and centrifuged again, and the supernatant was used for the PL activity assay. A Bradford assay was used to measure protein concentration. Total protein was incubated with 0.12% (w/v) polygalacturonic acid (Sigma, St Louis, MO, USA) in a solution containing 30 mM Tris–HCl pH 8.5 and 0.15 mM CaCl2 at room temperature, and absorbance at 237 nm was measured every minute for 10 min. PL activity was defined as the increase in 237 nm absorbance per min per amount of total protein.
Total protein extraction for PG activity assays was performed as in (Xiao et al., 2014). Rosettes of 4—5-week-old plants were ground in liquid N2, and the powder was incubated in protein extraction buffer (50 mM Tris–HCl, 1 M NaCl, 3 mM EDTA, 2.5 mM 1,4-dithiothreitol (Sigma-Aldrich, St. Louis, MO, USA), 2 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich), and 10% (v/v) glycerol, pH 7.5) for 1 h. The mixture was dialyzed in 50 mM sodium acetate buffer (pH 5.0) at 4°C for 16 h. Protein concentration was measured using a Bradford assay. PG activity was quantified by measuring the release of reducing ends. The dialyzed total protein was incubated with 0.5% (w/v) polygalacturonic acid in 37.5 mM NaOAc (pH 4.4) at 30°C for 3 h, and 100 mM sodium tetraborate buffer (pH 9.0) and 200 µL 1% (w/v) 2-cyanoacetamide were added to label reducing ends. D-galacturonic acid (Sigma) was used as a standard. PG activity was defined as the amount of reducing ends produced per minute per amount of total protein.
A PECTOPLATE assay was used to measure PME activity (Lionetti, 2015). Total protein was extracted as for the PG activity assay. PECTOPLATEs contained 0.1% (w/v) apple pectin (Sigma), 1% (w/v) SeaKem LE Agarose, 12.5 mM citric acid, 50 mM Na2HPO4 at pH 6.5. Twenty microliters of 25 µg/mL protein samples were loaded in wells made by punching the PECTOPLATE with a cork borer with a 5-mm diameter. After incubation at 30°C for 16 h, the plates were stained with 0.05% (w/v) Ruthenium Red (Sigma) for 30 min, then rinsed with water at least 3 times until halos were clear for imaging. Photographs were taken using a Scanjet 8300 scanner (HP), and halo area was measured using ImageJ. A standard curve of PME activity and halo area was made using commercial PME (Prozomix PO, Haltwhistle, UK).
Nanoindentation
A Hysitron Triboscan (Ti950, USA) was used to conduct nanoindentation experiments. The machine was equipped with a 50X objective so that guard cells could be easily identified. The diameter of the conical-type tip of the probe is 2–3 μm. The tip was scanned using a confocal microscope and its geometry rendered for the computational model. A set force of 2–5 μN was used to engage each targeted cell. Displacement control was set for the input load function (Forouzesh et al., 2013), and the loading rate was 100 nm/s. As the indentation depth increased from 150 to 1250 nm, the contact area was estimated to increase from ∼1.77 μm2 (with a diameter of ∼1.5 µm) to ∼11.34 μm2 (with a diameter of ∼3.8 μm). The corresponding change to the internal cell volume caused by the indentations increased from ∼1% to ∼6.5% of the total cell volume. For the maximum indentation depth of 1,250 nm, the effect of the neighboring cells in the local cell wall mechanical response was not substantial (Mosca et al., 2017; Li et al., 2021). Two blue bulbs (40 W each) were positioned in front of the probe at a distance of 14–15 inches to provide 200–250 μmol m2·s−1 light intensity. The door of the instrument was covered with foil to ensure dark conditions. To provide the blue light needed to activate guard cells, the front door of the machine was opened. Before testing, the leaf was mounted on a support using epoxy and then was placed in a dark growth chamber for >12 h. The sample was settled for 1–2 h on the stage of the nanoindenter before testing. The lighting illumination of the microscope was set to a minimum level to reduce its effect on the guard cells. The middle of each guard cell from the top view was set as the indentation position. Indentation was performed every 5–10 min. For each genotype, experiments were performed using nine guard cells from five plants.
FEM
Mechanical analysis of nanoindentation experiments was conducted using commercial finite element software (Abaqus, 2019) to estimate the wall modulus and turgor pressure of the cell. A structural model of each guard cell was constructed using the lofting method in Abaqus using the polar length, complex width, and guard cell width evaluated from the optical image of the nanoindenter microscope. The thickness distribution of the cross-section of the cell was set based on previous measurements (Supplemental Figure S5B). Similar to previous studies (Marom et al., 2017; Yi et al., 2018), a linear anisotropic elastic model (transverse isotropy) was assigned uniformly across the whole cell, and based on the estimated proportions, physical properties, and orientations of cellulose and matrix polysaccharides in the guard cell wall, the anisotropic modulus was assumed to have a relation E1:E2:E3 = 1:4:1 for the closed state. E2 defined the wall modulus along the circumferential direction of the cell. Poisson’s ratios were set to ν12 = ν23 = 0.3, and ν13 = 0.47. Shear modulus was assumed to have a relation G12 = G23 = E1, and G13 can be determined by G13 = E1/(0.5 + ν13). As a result, only two unknowns, turgor pressure and modulus E2, need to be determined. For boundary conditions, the materials at the polar positions were confined, ventral edges were free of constraint, and dorsal edges were constrained in the vertical direction to represent constraints from adjacent pavement cells. The analysis was conducted in two steps: cell pressurization and nanoindentation. The pore width at the end of the pressurization and the stiffness at shallow and deep indentation depths were used to compare with experimental measurements iteratively. Once the optical and mechanical measurements were matched, the turgor pressure and the cell wall modulus were estimated.
To measurements of pore width (Supplemental Figure S5C) used in the FE analyses, pore width in the first dark phase was obtained by subtracting the pore width of stomata that were plasmolyzed by incubation in 1–3 M sorbitol for 1 h (turgor pressure is ∼0 MPa) (see source data) from the stomata that were left under dark overnight (dark phase). Pore width in the light phase and the second dark phase were estimated using the stomatal opening and closure rate as determined using the stomatal function assay results for light and dark treatments (Supplemental Figure S5C).
Incipient plasmolysis
Leaves were excised directly from 3- to 4-week-old plants that were growing in the light (3000K Fluorescent Bulb, 150-250 PPFD) to constitute an open stomata group. To induce stomatal closure, excised leaves were treated with 50 mM ABA in the dark for 2.5 h. Leaves with open or closed stomata were soaked in 0.1 mM FM1–43 in sorbitol solutions of differing concentrations (0, 0.2, 0.4, 0.6, 0.8, 1 M for Col and PLL12OE, 0, 1, 2, 2.5, 3, 4 M for pll12-1) for 40 min before imaging under a Zeiss Axio Observer microscope with a 63X 1.4 NA oil immersion objective. Z-stack images (0.5 μm z interval) were collected using a 488 nm excitation laser and a 525/50 nm emission filter. ImageJ was used to quantify and calculate the ratio of plasmolyzed: total guard cells. A function of the ratio to sorbitol concentration was plotted using DESMOS and fitted to an S curve, and the concentration of sorbitol at the point where 50% of guard cells were plasmolyzed was estimated and used to calculate turgor pressure as the osmotic potential at incipient plasmolysis according to the equation , where c is the concentration of sorbitol, R is the ideal gas constant (8.314 kPa·L/mol·K), and T is the temperature in Kelvin (298 K). For both open stomata and closed stomata groups, nine leaves were imaged and quantified for each sorbitol concentration.
Quantification and statistical analysis
Statistical analysis in this study was conducted with GraphPad. Protein alignment was performed using the MAFFT plugin implemented in Geneious. DESMOS was used to derive the sorbitol concentration at which guard cells are incipiently plasmolyzed. Details of statistical tests are listed in Supplemental File S1.
Accession numbers
Genes analyzed in this article have the following accession numbers: PLL12 (At5g04310), ACT2 (At3g18780), Cryj1 (BAA05542), PMR6 (At3g54920), DEL1 (LOC_Os10g31910), ZePel (Y09541), MaPel1 (AAF19195), GhPel (ADB90478), PtxtPL1-27 (EU379971), NJJS25 (AFF339024). Further information and requests for vectors, transgenic plants constructed in Supplemental Data. PLL12 patterns the guard cell wall to coordinate turgor pressure and wall mechanics for proper stomatal function in Arabidopsis Plant Cell. Further information and requests for vectors and transgenic lines constructed in this study will be fulfilled by the corresponding author. Modeling and analysis code will be provided upon request.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . PLL12 encodes a putative PL in A. thaliana.
Supplemental Figure S2 . PLL12 is widely expressed in A. thaliana.
Supplemental Figure S3 . PLL12 also functions in stomatal responses to light conditions, and its roles in stomatal responses to ABA and FC are partially dependent on its expression in guard cells.
Supplemental Figure S4 . Apparent stiffness values of Col, pll12-1, and PLL12OE guard cells at all depths.
Supplemental Figure S5 . Guard cell wall thickness used for FE analysis.
Supplemental Figure S6 . Additional FE analysis for guard cells in Col, pll12-1, and PLL12OE.
Supplemental Figure S7 . Negative controls of immunolabeling in guard cell walls.
Supplemental Table S1 . Primers and oligonucleotides used in this study
Supplemental Table S2 . Antibodies used in this study
Supplemental File S1 . ANOVA and T-test results.
Supplementary Material
Acknowledgments
Thanks to Silu Shen, Rayna Marshall, and Katya Iatsenko for help with data analysis, Dr Jozef Mravec for the gift of COS488, Missy Hazen and Gang Ning in the Huck Microscopy Core Facility at Penn State for technical support, and Dr Yue Rui, Dr Daniel Cosgrove, Dr Hojae Yi, and members of the Anderson lab for helpful discussions.
Funding
This work was supported by the National Science Foundation under Grant MCB-1616316 awarded to C.T.A., James Z. Wang, and Virendra M. Puri. Nanoindentation measurements were performed at the UNL Nano-Engineering Research Core Facility, which is partially funded by the Nebraska Research Initiative.
Conflict of interest statement. The authors declare no conflict of interest.
Contributor Information
Yintong Chen, Department of Biology, The Pennsylvania State University, University Park, Pennsylvania, 16802 USA.
Wenlong Li, Department of Mechanical and Materials Engineering, University of Nebraska-Lincoln, Lincoln, Nebraska, 68588 USA.
Joseph A. Turner, Department of Mechanical and Materials Engineering, University of Nebraska-Lincoln, Lincoln, Nebraska, 68588 USA
Charles T. Anderson, Department of Biology, The Pennsylvania State University, University Park, Pennsylvania, 16802 USA.
Y.C. and C.T.A. designed the project; Y.C. generated plant material and characterized plant growth, stomatal responses, and cell wall modifications; W.L. and J.A.T. designed and developed the iterative experimental–computational method to analyze nanoindentation data; W.L. performed nanoindentation experiments and FE analysis; Y.C. and C.T.A. wrote the manuscript; Y.C., W.L., J.A.T., and C.T.A. edited the manuscript.
The author responsible for distribution of materials integral to the findings in this article in accordance with the policy described in the Instruction for Authors (https://academic.oup.com/plcell) is: Charles T. Anderson (cta3@psu.edu).
References
- Amsbury S, Hunt L, Elhaddad N, Baillie A, Lundgren M, Verhertbruggen Y, Scheller HV, Knox JP, Fleming AJ, Gray JE (2016) Stomatal function requires pectin de-methyl-esterification of the guard cell wall. Curr Biol 26: 2899–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CT (2016) We be jammin’: an update on pectin biosynthesis, trafficking and dynamics. J Exp Bot 67: 495–502 [DOI] [PubMed] [Google Scholar]
- Aylor DE, Parlange JY, Krikorian AD (1973) STOMATAL MECHANICS. Am J Bot 60: 163–171 [Google Scholar]
- Babu Y, Bayer M (2014) Plant Polygalacturonases involved in cell elongation and separation—the same but different? Plants 3: 613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Zhang G, Zhou Y, Zhang Z, Wang W, Du Y, Wu Z, Song CP (2009) Plasma membrane‐associated proline‐rich extensin‐like receptor kinase 4, a novel regulator of Ca2+ signalling, is required for abscisic acid responses in Arabidopsis thaliana. Plant J 60: 314–327 [DOI] [PubMed] [Google Scholar]
- Beauzamy L, Derr J, Boudaoud A (2015) Quantifying hydrostatic pressure in plant cells by using indentation with an atomic force microscope. Biophys J 108: 2448–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidhendi AJ, Geitmann A (2019) Methods to quantify primary plant cell wall mechanics. J Exp Bot 70: 3615–3648 [DOI] [PubMed] [Google Scholar]
- Carbonell A, Takeda A, Fahlgren N, Johnson SC, Cuperus JT, Carrington JC (2014) New generation of artificial MicroRNA and synthetic trans-acting small interfering RNA vectors for efficient gene silencing in Arabidopsis. Plant Physiol 165: 15–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter R, Woolfenden H, Baillie A, Amsbury S, Carroll S, Healicon E, Sovatzoglou S, Braybrook S, Gray JE, Hobbs J, et al. (2017) Stomatal opening involves polar, not radial, stiffening of guard cells. Curr Biol 27: 2974–2983 e2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ (1993) Wall extensibility: its nature, measurement and relationship to plant cell growth. New Phytol 124: 1–23 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ (2016) Plant cell wall extensibility: connecting plant cell growth with cell wall structure, mechanics, and the action of wall-modifying enzymes. J Exp Bot 67: 463–476 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ (2018) Diffuse growth of plant cell walls. Plant Physiol 176: 16–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daher FB, Chen Y, Bozorg B, Clough J, Jönsson H, Braybrook SA (2018) Anisotropic growth is achieved through the additive mechanical effect of material anisotropy and elastic asymmetry. Elife 7: e38161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMichele DW, Sharpe PJH (1973) An analysis of the mechanics of guard cell motion. J Theor Biol 41: 77–96 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway‐compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Feng W, Kita D, Peaucelle A, Cartwright HN, Doan V, Duan Q, Liu MC, Maman J, Steinhorst L, Schmitz-Thom I, et al. (2018) The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca(2+) signaling. Curr Biol 28: 666–675 e665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forouzesh E, Goel A, Mackenzie SA, Turner JA (2013) In vivoextraction of Arabidopsis cell turgor pressure using nanoindentation in conjunction with finite element modeling. Plant J 73: 509–520 [DOI] [PubMed] [Google Scholar]
- Franks P, Cowan I, Tyerman S, Cleary A, Lloyd J, Farquhar G (1995) Guard cell pressure/aperture characteristics measured with the pressure probe. Plant Cell Environ 18: 795–800 [Google Scholar]
- Franks PJ, Cowan 2 I. R, Farquhar GD (1998) A study of stomatal mechanics using the cell pressure probe. Plant Cell Environ 21: 94–100 [Google Scholar]
- Hachez C, Ohashi-Ito K, Dong J, Bergmann DC (2011) Differentiation of Arabidopsis guard cells: analysis of the networks incorporating the basic helix-loop-helix transcription factor, FAMA. Plant Physiol 155: 1458–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YC, Wu HC, Wang YD, Liu CH, Lin CC, Luo DL, Jinn TL (2017) PECTIN METHYLESTERASE34 contributes to heat tolerance through its role in promoting stomatal movement. Plant Physiol 174: 748–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezek M, Blatt MR (2017) The membrane transport system of the guard cell and its integration for stomatal dynamics. Plant Physiol 174: 487–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L, Milne JL, Ashford D, McCann MC, McQueen-Mason SJ (2005) A conserved functional role of pectic polymers in stomatal guard cells from a range of plant species. Planta 221: 255–264 [DOI] [PubMed] [Google Scholar]
- Jones L, Milne JL, Ashford D, McQueen-Mason SJ (2003) Cell wall arabinan is essential for guard cell function. Proc Natl Acad Sci U S A 100: 11783–11788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierzkowski D, Nakayama N, Routier-Kierzkowska AL, Weber A, Bayer E, Schorderet M, Reinhardt D, Kuhlemeier C, Smith RS (2012) Elastic domains regulate growth and organogenesis in the plant shoot apical meristem. Science 335: 1096–1099 [DOI] [PubMed] [Google Scholar]
- Kohorn BD, Johansen S, Shishido A, Todorova T, Martinez R, Defeo E, Obregon P (2009) Pectin activation of MAP kinase and gene expression is WAK2 dependent. Plant J 60: 974–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Choi H, Suh S, Doo IS, Oh KY, Choi EJ, Taylor ATS, Low PS, Lee Y (1999) Oligogalacturonic acid and chitosan reduce stomatal aperture by inducing the evolution of reactive oxygen species from guard cells of tomato and Commelina communis. Plant Physiol 121: 147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng Y, Yang Y, Ren D, Huang L, Dai L, Wang Y, Chen L, Tu Z, Gao Y, Li X et al. (2017) A rice PECTATE LYASE-LIKE gene is required for plant growth and leaf senescence. Plant Physiol 174: 1151–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Keynia S, Belteton SA, Afshar-Hatam F, Szymanski DB, Turner JA (2021) Protocol for mapping the spatial variability in cell wall mechanical bending behavior in living leaf pavement cells. bioRxiv, 2021.2002.2023.432478; 10.1101/2021.02.23.432478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liners F, Letesson J, Didembourg C, Cutsem PV (1989) Monoclonal antibodies against pectins: recognition of a conforma- tion induced by calcium. Plant Physiol 91: 1419–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti V (2015) PECTOPLATE: the simultaneous phenotyping of pectin methylesterases, pectinases, and oligogalacturonides in plants during biotic stresses. Front Plant Sci 6: 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zhang X, Yang L, Tang M, Wang K, Wang L, Bai L, Song C (2019) Hydrogen peroxide plays an important role in PERK4-mediated abscisic acid-regulated root growth in Arabidopsis. Funct Plant Biol 46: 165–174 [DOI] [PubMed] [Google Scholar]
- Marom Z, Shtein I, Bar-On B (2017) Stomatal opening: the role of cell-wall mechanical anisotropy and its analytical relations to the bio-composite characteristics. Front Plant Sci 8: 2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy TW, Der JP, Honaas LA, dePamphilis CW, Anderson CT (2014) Phylogenetic analysis of pectin-related gene families in Physcomitrella patensand nine other plant species yields evolutionary insights into cell walls. BMC Plant Biol 14: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckel T, Gall L, Semrau S, Homann U, Thiel G (2007) Guard cells elongate: relationship of volume and surface area during stomatal movement. Biophys J 92: 1072–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani P, Mirabet V, Cellier C, Rozier F, Hamant O, Das P, Boudaoud A (2014) Matching patterns of gene expression to mechanical stiffness at cell resolution through quantitative tandem epifluorescence and nanoindentation. Plant Physiol 165: 1399–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravec J, Kracun SK, Rydahl MG, Westereng B, Miart F, Clausen MH, Fangel JU, Daugaard M, Van Cutsem P, De Fine Licht HH, et al. (2014) Tracking developmentally regulated post-synthetic processing of homogalacturonan and chitin using reciprocal oligosaccharide probes. Development 141: 4841–4850 [DOI] [PubMed] [Google Scholar]
- Mosca G, Sapala A, Strauss S, Routier-Kierzkowska AL, Smith RS (2017) On the micro-indentation of plant cells in a tissue context. Phys Biol 14:015003. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Bergmann DC (2006) Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18: 2493–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega JK (1985) Augmented growth equation for cell wall expansion. Plant Physiol 79: 318–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palusa SG, Golovkin M, Shin SB, Richardson DN, Reddy AS (2007) Organ‐specific, developmental, hormonal and stress regulation of expression of putative pectate lyase genes in Arabidopsis. New Phytol 174: 537. [DOI] [PubMed] [Google Scholar]
- Peaucelle A, Braybrook SA, Le Guillou L, Bron E, Kuhlemeier C, Hofte H (2011) Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr Biol 21: 1720–1726 [DOI] [PubMed] [Google Scholar]
- Powell D, Morris E, Gidley M, Rees D (1982) Conformations and interactions of pectins: II. Influence of residue sequence on chain association in calcium pectate gels. J Mol Biol 155: 517–531 [DOI] [PubMed] [Google Scholar]
- Ringli C (2010) Monitoring the outside: cell wall-sensing mechanisms. Plant Physiol 153: 1445–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounds CM, Lubeck E, Hepler PK, Winship LJ (2011) Propidium iodide competes with Ca(2+) to label pectin in pollen tubes and Arabidopsis root hairs. Plant Physiol 157: 175–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routier-Kierzkowska AL, Weber A, Kochova P, Felekis D, Nelson BJ, Kuhlemeier C, Smith RS (2012) Cellular force microscopy for in vivo measurements of plant tissue mechanics. Plant Physiol 158: 1514–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui Y, Anderson CT (2016) Functional analysis of cellulose and xyloglucan in the walls of stomatal guard cells of Arabidopsis. Plant Physiol 170: 1398–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui Y, Xiao C, Yi H, Kandemir B, Wang JZ, Puri VM, Anderson CT (2017) POLYGALACTURONASE INVOLVED IN EXPANSION3 functions in seedling development, rosette growth, and stomatal dynamics in Arabidopsis thaliana. Plant Cell 29: 2413–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scavetta RD, Herron SR, Hotchkiss AT, Kita N, Keen NT, Benen JA, Kester HC, Visser J, Jurnak F (1999) Structure of a plant cell wall fragment complexed to pectate lyase C. Plant Cell 11: 1081–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Sanzana C, Celiz-Balboa J, Garzo E, Marcus SE, Parra-Rojas JP, Rojas B, Olmedo P, Rubilar MA, Rios I, Chorbadjian RA (2019) Pectin methylesterases modulate plant homogalacturonan status in defenses against the aphid Myzus persicae. Plant Cell 31: 1913–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ström A, Ribelles P, Lundin L, Norton I, Morris ER, Williams MA (2007) Influence of pectin fine structure on the mechanical properties of calcium− pectin and acid− pectin gels. Biomacromolecules 8: 2668–2674 [DOI] [PubMed] [Google Scholar]
- Sun L, van Nocker S (2010) Analysis of promoter activity of members of the PECTATE LYASE-LIKE (PLL) gene family in cell separation in Arabidopsis. BMC Plant Biol 10: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhertbruggen Y, Marcus SE, Haeger A, Ordaz-Ortiz JJ, Knox JP (2009) An extended set of monoclonal antibodies to pectic homogalacturonan. Carbohydr Res 344: 1858–1862 [DOI] [PubMed] [Google Scholar]
- Vogel JP (2002) PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell Online 14: 2095–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A, Braybrook S, Huflejt M, Mosca G, Routier-Kierzkowska AL, Smith RS (2015) Measuring the mechanical properties of plant cells by combining micro-indentation with osmotic treatments. J Exp Bot 66: 3229–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmer C, Beattie LN (1978) Cellular osmotic phenomena during stomatal movements ofCommelina communis. Protoplasma 95: 321–332 [Google Scholar]
- Woolfenden HC, Bourdais G, Kopischke M, Miedes E, Molina A, Robatzek S, Morris RJ (2017) A computational approach for inferring the cell wall properties that govern guard cell dynamics. Plant J 92: 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HI, Sharpe PJ (1979) Stomatal mechanics II: material properties of guard cell walls. Plant Cell Environ 2: 235–244 [Google Scholar]
- Xiao C, Somerville C, Anderson CT (2014) POLYGALACTURONASE INVOLVED IN EXPANSION1 functions in cell elongation and flower development in Arabidopsis. Plant Cell 26: 1018–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakubov GE, Bonilla MR, Chen H, Doblin MS, Bacic A, Gidley MJ, Stokes JR (2016) Mapping nano-scale mechanical heterogeneity of primary plant cell walls. J Exp Bot 67: 2799–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Costa A, Leonhardt N, Siegel RS, Schroeder JI (2008) Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods 4: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H, Rui Y, Kandemir B, Wang JZ, Anderson CT, Puri VM (2018) Mechanical effects of cellulose, xyloglucan, and pectins on stomatal guard cells of Arabidopsis thaliana. Front Plant Sci 9: 1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder MD, Jurnak F (1995) The refined three-dimensional structure of pectate lyase C from Erwinia chrysanthemi at 2.2 angstrom resolution (implications for an enzymatic mechanism). Plant Physiol 107: 349–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XQ, Wei PC, Xiong YM, Yang Y, Chen J, Wang XC (2011) Overexpression of the Arabidopsis α-expansin gene AtEXPA1 accelerates stomatal opening by decreasing the volumetric elastic modulus. Plant Cell Rep 30: 27–36 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.