Abstract
The machinery that inserts mitochondrially encoded proteins into the inner membrane and translocates their hydrophilic domains through the membrane is poorly understood. We have developed a genetic screen for Saccharomyces cerevisiae mutants defective in this export process. The screen is based on the fact that the hydrophilic polypeptide Arg8mp is exported from the matrix if it is synthesized within mitochondria as a bifunctional Cox2p-Arg8mp fusion protein. Since export of Arg8mp causes an Arg− phenotype, defective mutants can be selected as Arg+. Here we show that mutations in the nuclear gene PNT1 block the translocation of mitochondrially encoded fusion proteins across the inner membrane. Pnt1p is a mitochondrial integral inner membrane protein that appears to have two hydrophilic domains in the matrix, flanking a central hydrophobic hairpin-like anchor. While an S. cerevisiae pnt1 deletion mutant was more sensitive to H2O2 than the wild type was, it was respiration competent and able to export wild-type Cox2p. However, deletion of the PNT1 orthologue from Kluyveromyces lactis, KlPNT1, caused a clear nonrespiratory phenotype, absence of cytochrome oxidase activity, and a defect in the assembly of KlCox2p that appears to be due to a block of C-tail export. Since PNT1 was previously described as a gene affecting resistance to the antibiotic pentamidine, our data support a mitochondrial target for this drug.
While the majority of mitochondrial proteins are encoded in the nucleus and imported into the organelle from the cytoplasm, a few important subunits of respiratory complexes are encoded in mitochondrial DNA (mtDNA), synthesized in the matrix, and inserted into the inner membrane from within (3, 47). Each of the mitochondrially encoded respiratory-chain subunits of Saccharomyces cerevisiae contains at least one hydrophilic tail or loop that is exported to the outside surface of the inner membrane by translocation across the lipid bilayer (48). In addition to their important role in assembly of the respiratory complexes, mitochondrial export translocation systems might play a role in transporting mitochondrially encoded polypeptides out of the organelle in animal (9) and plant (1) cells. Export systems also could be involved in intracellular signaling by mitochondria (31) and possibly, by analogy to whole cells (4, 50), in removal of toxic components from the organelle.
The translocation machinery that carries out import of proteins from the cytoplasm to mitochondria has been extensively studied (43, 45, 51). However, little is known about the inner membrane proteins responsible for export of mitochondrially coded polypeptide domains. While the topology of mitochondrial export resembles that of bacterial secretion, the yeast genome does not encode detectable homologues of the bacterial Sec translocase that function in mitochondria (18). The single protein known to be involved in export is Oxa1p, a conserved nuclearly encoded integral inner membrane protein that is required for the export of the hydrophilic N and C tails of the mitochondrially encoded precursor to cytochrome c oxidase subunit II (Cox2p) and also plays a role in ATP synthase formation (5, 7, 8, 20, 22, 25, 28, 37). While Oxa1p interacts directly with nascent mitochondrially synthesized polypeptides (23), its precise role in membrane insertion is not clear. Surprisingly, oxa1 null mutations can be suppressed by mutations in the nuclear gene encoding the cytochrome c1 subunit of the bc1 complex (19). This suppression indicates that the conserved Oxa1p function can be bypassed in the membrane insertion process, although the mechanism of suppression has not been established.
To study the mitochondrial export machinery in vivo, we have adapted the gene fusion approach to the study of topogenesis of proteins encoded in S. cerevisiae mtDNA (20). As a passenger protein-coding sequence, we used the synthetic mitochondrial gene ARG8m, which directs the synthesis of a soluble polypeptide within the organelle and complements nuclear arg8 mutations (53). ARG8m specifies, in the yeast mitochondrial genetic code, the biosynthetic enzyme normally encoded by the nuclear gene ARG8, which in wild-type cells is synthesized in the cytoplasm and imported into the mitochondrial matrix. Expression of a chimeric gene that encodes a fusion protein with the Arg8mp moiety attached to the C terminus of Cox2p results in export of the hydrophilic passenger protein through the inner membrane to the intermembrane space (IMS) (20).
Here we report the successful development of a genetic selection for yeast mutants defective in the export of mitochondrially coded proteins. The strategy is based on the fact that when Arg8mp is exported from the matrix, it can no longer participate in arginine biosynthesis. While novel in its application to mitochondria, the rationale for our selection is based on schemes used successfully to identify genes in yeast (10) and Escherichia coli (44) that encode components of cellular secretion machineries. The efficacy of this selection is demonstrated by the fact that it identified a recessive mutation that prevents the export of passenger protein moieties attached to Cox2p. The mutated gene, PNT1, encodes a mitochondrial integral inner membrane protein. While deletion of PNT1 in an otherwise wild-type S. cerevisiae strain has a surprisingly modest phenotype, we found that deletion of the Kluyveromyces lactis functional homologue, KlPNT1, caused a tight nonrespiratory phenotype and prevented normal assembly of KlCox2p into cytochrome oxidase, suggesting that it caused defective export of wild-type KlCox2p in that species.
MATERIALS AND METHODS
Plasmids and DNA manipulation.
Construction of the gene fusions COX2(UTL)::ARG8m and COX2::ARG8m and their subsequent integration into ρ+ mtDNA were as previously described (20). To construct a nuclear ARG8 plasmid (pSH303) that encodes the same truncated Arg8p as the above ARG8m fragment, ARG8 was amplified from genomic DNA by PCR, and ligated to the NotI site on pDB20, a 2μm expression vector (13). To create BLEm, another artificial gene for mitochondrial expression, four oligonucleotides of 100 bases each (data not shown) were designed according to the Ble protein sequence encoded by the bacterial Tn5 transposon (36), with consensus yeast mitochondrial codon usage (11). The four oligonucleotides sequentially overlapped for 20 to 25 bp and were used to synthesize the BLEm gene by PCR. To construct the pnt1Δ::LEU2 deletion, a SpeI-NcoI fragment encompassing the PNT1 locus was first cloned to Bluescript. PNT1 codons 3 to 348 were then replaced by a PCR-generated LEU2 gene. The resulting plasmid, pSH401, was linearized before transformation into S. cerevisiae cells. Deletion of the chromosomal PNT1 was confirmed by PCR analysis and genetic complementation. To tag the wild-type chromosomal PNT1 with three HA epitopes, an HA-URA3-HA cassette (52) was PCR amplified with the primers TATCATTGTTAAAATATAATTCAGAATTGGTAGCTACGAAAGATATTCAAAGGGAACAAAAGCTGG and GTTTTAAACAATATGTACAGCATAGATAATGTCATGAATATTTACATCTACTATAGGGCGAATTGG, each tailed at the 5′ end with a 50-bp PNT1 sequence for in-frame insertion before the PNT1 stop codon. The resulting PCR fragment was used to transform S. cerevisiae cells. PNT1-tagged transformants were identified by PCR analysis and were then plated on 5-fluoroorotic acid medium to isolate PNT1::HA strain that had lost URA3 through homologous recombination.
To construct the Klpnt1 disruption plasmid, pSH502, a 550-bp upstream fragment and a 830-bp downstream fragment immediately flanking the KlPNT1 coding sequence (see below) were PCR amplified and ligated to the BglII-BamHI HisG-URA3-HisG cassette released from pNKY51 (2) and the resulting fusion was inserted into a pBluescript vector. To disrupt KlPNT1 in K. lactis, pSH502 was linearized with XbaI and XhoI and transformed into strain KB101 by the same procedure as that for S. cerevisiae. A strain with expected KlPNT1 deletion, as verified by PCR analysis, was subjected to 5-fluoroorotic acid selection, resulting in strain SH702. To clone KlPNT1 into a K. lactis expression vector, KlPNT1 was PCR amplified from a genomic subclone (data not shown) by using a primer pair that also introduced a His6 epitope tag at the C terminus of KlPnt1p. The gene fusion was then ligated to the EcoRI-HindIII sites of the K. lactis/E. coli shuttle vector pROCS2 (6), resulting in pSH505.
Strains, growth conditions, and genetic and general methods.
Yeast strains used in this study are listed in Table 1. S. cerevisiae JKR102, DFS188, SH39, SH40, SH404, and SH412 are isogenic or congenic to D273-10B, and DFS160, SH45B, SH45B-41, SH401, SH349, SH402, SH135, SH405, and SH407 are isogenic or congenic to DBY947. SH45B was constructed by transferring the ρ+ mtDNA of SH39, including the gene fusion COX2::ARG8m, to the ρ0 recipient DFS160 by cytoduction. K. lactis KB101 was obtained from the American Type Culture Collection (ATCC 96265). Standard genetic methods were as described previously (14, 49). Nuclear transformation, mitochondrial transformation, and gene replacement were as described previously (20). To stabilize K. lactis transformants containing pROCS2 or pSH505, K. lactis plasmid pKD1 (6) was reconstituted from the pSPH1 clone (obtained from H. Fukuhara via N. A. Da Silva), and cotransformed with the shuttle vectors. Yeast growth was monitored with either a Klett-Summerson photoelectric colorimeter or a spectrophotometer.
TABLE 1.
Strains used in this study
| Strain | Nuclear [mitochondrial] genotype | Reference or source |
|---|---|---|
| JKR102 | MATa leu2-3,112 his4-519 ura3Δ [rho+] | 53 |
| DFS160 | MATα ade2-101 ura3-52 leu2Δ arg8Δ::URA3 kar1-1 [rho0] | 53 |
| DFS188 | MATa ura3-52 leu2-3,112 lys2 his3Δ arg8::hisG [rho+] | 53 |
| SH39 | MATa ura3-52 leu2-3,112 lys2 his3Δ arg8::hisG [rho+COX2::ARG8m] | 20 |
| SH40 | MATa ura3-52 leu2-3,112 lys2 his3Δ arg8::hisG [rho+COX2(UTL)::ARG8m] | 20 |
| SH45B | MATα ade2-101 ura3-52 leu2Δ arg8Δ::URA3 kar1-1 [rho+COX2::ARG8m] | This study |
| SH45B-41 | MATα ade2-101 ura3-52 leu2Δ arg8Δ::URA3 pnt1-41 kar1-1 [rho+COX2::ARG8m] | This study |
| SH404 | MATα ade2-101 ura3Δ PNT1::HA [rho+] | This study |
| SH401 | MATα ade2-101 ura3-52 leu2Δ arg8Δ::URA3 kar1-1 pnt1Δ::LEU2 [rho+COX2::ARG8m] | This study |
| SH349 | MATa ade2-101 leu2Δ arg8Δ::URA3 [rho+] | This study |
| SH402 | MATa ade2-101 leu2Δ arg8Δ::URA3 pnt1Δ::LEU2 [rho+] | This study |
| SH135 | MATα ade2-101 ura3-52 leu2Δ arg8Δ::URA3 kar1-1 [rho+COX2::BLEm] | This study |
| SH405 | MATα ade2-101 ura3-52 leu2Δ arg8Δ::URA3 kar1-1 pnt1Δ::LEU2 [rho+COX2::BLEm] | This study |
| SH412 | MATa ura3-52 leu2-3,112 lys2 his3Δ arg8::hisG pnt1Δ::LEU2 [rho+] | This study |
| SH407 | MATa ade2-101 ura3-52 pnt1Δ::LEU2 [rho+COX2::ARG8m] | This study |
| KB101 | MATa ade trp1 ura3 gal80-1 [rho+] | ATCC 96265 |
| SH702 | MATa ade trp1 ura3 Klpnt1Δ::hisG gal80-1 [rho+] | This study |
Genetic screening and gene cloning.
Independent SH45B cultures (1 ml each) were grown to stationary phase in yeast extract-peptone-dextrose (YPD). Each culture was washed and spread on minimal glucose plates lacking Arg to identify spontaneous mutants. After incubation at 30°C for 7 days, Arg+ colonies were replica plated onto nonfermentable glycerol-ethanol medium (YPEG). About 10% of the arginine-prototrophs were respiration defective. These colonies were streak purified and characterized. One to three colonies from each plate that appeared phenotypically stable and relatively tight for respiratory deficiency were selected as putative mutants defective in export for further study.
The gene mutated in the tight Pet− mutant SH45B-41 was cloned by complementation with yeast genomic DNA libraries. Transformants were first selected on synthetic medium and then replica plated to YPEG to identify respiring colonies (Pet+). Total genomic DNA was extracted from each colony and used to transform E. coli to purify the plasmid, which was then reintroduced to SH45B-41. Clones that restored the Pet+ phenotype were selected for restriction mapping and subcloning.
To clone KlPNT1, a 2 μm plasmid-based K. lactis genomic DNA library obtained from L. A. Grivell (39) was screened for plasmids complementing the Pet− phenotype of S. cerevisiae SH407 by using the same procedure as for the cloning of PNT1. The DNA sequence of a 2,600-bp fragment encompassing KlPNT1 was determined.
Mitochondrial isolation, subfractionation, and protein analyses.
Mitochondrial isolation and purification, mitochondrial membrane fractionation, alkaline extraction, mitoplasting, proteinase K protection assay, and Western blot analysis were as previously described (15, 17, 20), except that K. lactis strains were grown in YPD. Cytochrome c oxidase activity was determined spectrophotometrically as previously described (35), except that mitochondria were purified as above.
Nucleotide sequence accession number.
The sequence of the 2,600-bp fragment encompassing KlPNT1 has been deposited in GenBank under accession no. AF157501.
RESULTS
Genetic screen for mutants defective in mitochondrial protein export.
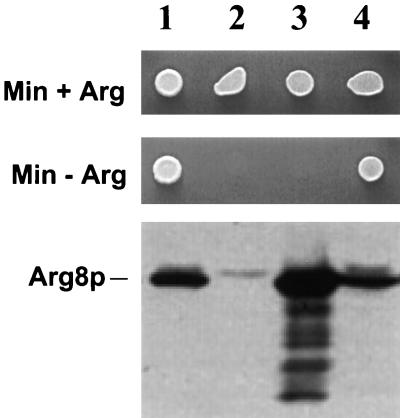
The wild-type nuclear ARG8 gene specifies a precursor protein that is imported into mitochondria under the direction of an amino-terminal targeting signal (53). Within the matrix, Arg8p functions as acetylornithine aminotransferase in arginine biosynthesis (21, 27). We have found that this enzyme must be localized in the mitochondrial matrix to participate in arginine synthesis. A strain whose nuclear ARG8 gene was truncated at the 5′ end, such that its product lacked the amino-terminal 21 residues, exhibited an Arg− growth phenotype despite the presence of abundant Arg8p in the cell, as revealed by Western analysis of total protein (Fig. 1, lane 3). The inability of such cells to make arginine must be due to mislocalization of the truncated protein, since expression of the identical truncated protein within mitochondria, from a synthetic mitochondrial structural gene inserted at the COX2 locus, leads to robust Arg+ growth despite deletion of the nuclear arg8 gene (lane 4) (20). Thus, Arg8p located outside of the mitochondrial inner membrane cannot participate in arginine biosynthesis.
FIG. 1.
Arg8p must be present in the mitochondrial matrix to participate in arginine biosynthesis. The top two panels show the growth of yeast strains on glucose medium (SD) that either contains Arg (Min + Arg) or lacks Arg (Min − Arg). The bottom panel is a Western blot of total-cell extracts probed with anti-Arg8p antibody. The position of Arg8p in the gel is indicated. Lanes: 1, wild-type ARG8 strain with wild-type mtDNA (JKR102); 2, arg8::hisG strain with wild-type mtDNA (DFS188); 3, arg8::hisG strain with wild-type mtDNA (DFS188) transformed with a nuclear 2 μm expression plasmid bearing a truncated ARG8 gene that lacks the N-terminal 21 codons and thus the mitochondrial targeting sequence (pSH303; see Materials and Methods); 4, arg8::hisG strain whose mtDNA contains a truncated ARG8m gene in place of the COX2 coding sequence (SH40). Mitochondrial expression of this gene produces the same protein as in lane 3 but within mitochondria.
We have previously found that a 402-amino-acid residue Arg8mp moiety, synthesized within mitochondria, can be exported in vivo through the inner membrane to the IMS when fused to the C terminus of Cox2p (20), which is normally located in the IMS (54). If this export process sufficiently depletes the matrix of mitochondrially synthesized Arg8mp, cells expressing the fusion protein should be Arg−. (The outer membrane is porous to small molecules such as acetylornithine [42], while the inner membrane is not, making the IMS topologically equivalent to the cytoplasm for this purpose.) In this case, a collection of Arg+ mutants selected from a parent expressing the Cox2p-Arg8mp fusion protein might include strains with defects in the protein export process that allow the Arg8mp moiety of the fusion protein to remain in the matrix.
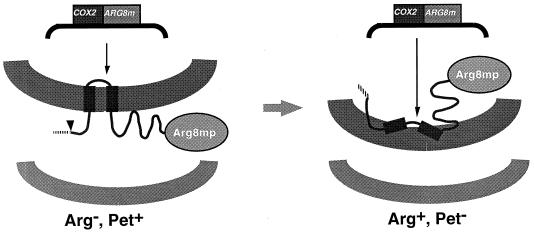
Mitochondrial expression of the Cox2p-Arg8mp fusion protein supports robust respiratory growth (20). This fact demonstrates that the N and C tails of the Cox2p moiety of the fusion protein are translocated normally through the inner membrane. Proteolytic removal of the Arg8mp moiety in the IMS yields a Cox2p-cross-reacting species comparable in size to wild-type Cox2p (unpublished results), which presumably functions in cytochrome oxidase. We would expect that an Arg+ mutant selected as described above, which was deficient in the export translocation process, would not be able to translocate the C tail of Cox2p normally and would therefore also exhibit a nonrespiratory (Pet−) growth phenotype (Fig. 2).
FIG. 2.
Rationale for a selection scheme to identify mutants defective in protein export from the mitochondrial matrix. An arg8 strain containing the COX2::ARG8m chimeric gene in its mtDNA (SH45B) exhibits an Arg− growth phenotype due to export of the Arg8mp moiety, which can function only in the matrix (Fig. 1). The strain can respire (Pet+) due to the normal translocation of Cox2p through the inner membrane followed by proteolytic removal of the Arg8mp moiety (not shown). An Arg+ growth phenotype could result from a defect in export that results in accumulation of the Arg8mp moiety in the matrix. If the defect is severe enough, it will also prevent the export of Cox2p domains to the IMS and the assembly of cytochrome oxidase, producing a Pet− phenotype.
Our previous study was performed with strains in the D273-10B (ATCC 25657) genetic background, which exhibits vigorous respiratory growth and mitochondrial gene expression. While the bulk of Arg8mp was exported, we were also able to detect a low level of unexported, matrix-localized Arg8mp in the mitochondria of such a strain expressing the Cox2p-Arg8mp fusion protein. Consistent with this observation, that strain (SH39) had an Arg+ growth phenotype (20). Thus, our previously described strain was unsuitable for the selection scheme. We reasoned that lowering the rate of mitochondrial gene expression might lower the level of residual unexported Arg8mp sufficiently to produce an Arg− phenotype. This speculation was supported by the fact that a D273-10B-derived diploid, heterozygous for a deletion of the limiting (18a) COX2-mRNA-specific translational activator gene PET111 (40), was Arg−.
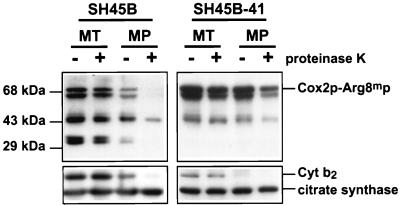
To construct an Arg− haploid strain for our selection scheme, we transferred the mitochondrial genome encoding the Cox2p-Arg8mp fusion protein into a strain whose nuclear genetic background was derived from DBY947 (41), a strain closely related to S288C (ATCC 26108). The respiration rate of this strain background was roughly half that of D273-10B related strains, when cells were grown in YPEG (unpublished data). The resulting strain, SH45B, was indeed Arg−, and its steady-state cellular level of Arg8mp was lower than that of the corresponding strain in the D273-10B background (unpublished results). However, it is important to note that mitochondrial expression of unexported forms of Arg8mp in strains isogenic to SH45B does produce an Arg+ phenotype, suggesting that it is export of the SH45B fusion protein that depletes the matrix of active enzyme. Western blot analysis of protease-treated mitochondria and mitoplasts derived from SH45B confirmed that the Arg8mp moiety of the full-length Cox2p-Arg8mp fusion protein was largely exported through the inner membrane to the IMS, as were two small Arg8mp-cross-reacting fragments (Fig. 3, left). Furthermore, SH45B exhibited a Pet+ growth phenotype, demonstrating normal export and assembly of the Cox2p moiety. Thus, strain SH45B appears to be a useful parent for the isolation of export mutants by selecting for an Arg+ phenotype and screening among those mutants for clones that also exhibit a respiratory defect (to eliminate background mutations that, for example, simply increase mitochondrial gene expression).
FIG. 3.
Export defect of the mitochondrially synthesized Cox2p-Arg8mp fusion protein from mitochondria of the mutant SH45B-41. Mitochondria (MT) expressing Cox2p-Arg8mp were isolated from the parental strain SH45B (left panel) and the mutant strain SH45B-41 (right panel) and were converted to mitoplasts (MP) in the absence or presence of proteinase K (100 μg/ml), as indicated above each lane. Samples were run on sodium dodecyl sulfate (SDS)–10% polyacrylamide gels, immunoblotted, and probed with anti-Arg8p (top panels). The position of the full-length Cox2p-Arg8mp fusion protein is indicated. Two Arg8mp fragments, of roughly 33 kDa, known to be located in the IMS (20), are present in the mitochondria of SH45B but not SH45B-41. The same blots were stripped and reprobed with anti-cytochrome b2 (Cyt b2) (an IMS marker) and anti-citrate synthase (a matrix marker) (bottom).
Isolation and characterization of an export-defective nuclear mutant.
SH45B spontaneously gives rise to Arg+ clones, and roughly 10% of these also exhibit at least a partial Pet− phenotype. To test whether this screen might be effective in identifying strains with mitochondrial export defects, we chose one such mutant with a tight Pet− phenotype, SH45B-41, for detailed analysis. SH45B-41 contains a single recessive nuclear mutation: when mated with a ρ0 tester strain, it yielded respiring diploids. When these diploids were sporulated, the Pet− phenotype segregated 2:2.
To determine whether the Arg+ Pet− phenotype of the SH45B-41 mutant was due to defective export of the Cox2p-Arg8mp fusion protein, we isolated its mitochondria and subjected them to mitoplasting (disruption of the outer membrane) and protease digestion (Fig. 3, right). These experiments revealed that the full-length fusion protein of the mutant was largely protected from external protease by mitoplasts. Furthermore, the mitochondria from the mutant lacked the two small Arg8mp-cross-reacting fragments that are present in the parental strain (Fig. 3, right). As shown previously (20) and confirmed by the experiment of Fig. 3, these fragments of the mitochondrially synthesized fusion protein are localized in the IMS. Thus, in contrast to the parental strain, the Arg8mp moiety of the mutant had remained inside the inner membrane. (The partial protease sensitivity of the full-length fusion protein in mutant mitoplasts apparently reflects some leakage of proteinase K across the inner membrane.) These results strongly suggest that the Arg+ Pet− phenotype of SH45B-41 is due to failure of translocation of the C-terminal portion of the Cox2p-Arg8mp fusion protein across the inner membrane.
The export defect is caused by mutation of the PNT1 gene.
To identify the nuclear gene mutated in SH45B-41, two genomic libraries of S. cerevisiae DNA were screened for clones that restored a Pet+ phenotype to the mutant. Nine overlapping clones were obtained, and the complementing gene, PNT1, was identified by subcloning. PNT1 had been previously identified as a gene that caused resistance to the drug pentamidine when present on a high-copy-number vector in S. cerevisiae (34). It is a unique gene that encodes a protein of 423 amino acids. The protein, Pnt1p, has no significant sequence similarity to any other known or predicted protein. Sequence analysis of the gene isolated from SH45B-41 revealed that the mutation, pnt1-41, caused a missense substitution, W382R, in the C-terminal portion of Pnt1p.
We disrupted the chromosomal copy of PNT1 in the parental strain SH45B by replacing codons 3 to 348 with LEU2 (see Materials and Methods). The resulting pnt1Δ::LEU2 strain (SH401) exhibited the same Arg+ Pet− phenotype as the original mutant SH45B-41 (see below), and it was complemented by PNT1 on a plasmid. Moreover, a strain bearing the pnt1Δ::LEU2 allele failed to complement SH45B-41 for respiratory growth.
Pnt1p is a mitochondrial inner membrane protein.
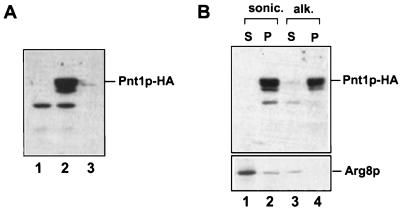
The protein product of PNT1 has not been previously characterized. To allow the detection of Pnt1p, it was tagged at its C terminus with three HA epitopes by addition of the appropriate sequence (52) to the 3′ end of the chromosomal PNT1 coding sequence (see Materials and Methods). This alteration did not interfere with Pnt1p function as judged by the ability of PNT1-HA to fully complement the Pet− phenotype of pnt1-41. To determine the subcellular location of Pnt1p, we constructed an otherwise wild-type strain, SH404, in which PNT1-HA replaced PNT1. Highly purified mitochondria from SH404 contained a protein of the expected size that reacted with anti-HA antibody (Fig. 4A, lane 2). This protein was barely detectable in the cytosolic fraction from SH404 (lane 3). The anti-HA antibody failed to react with any protein of this size in mitochondria from a wild-type strain lacking the epitope (lane 1), confirming that the species detected in SH404 was Pnt1p-HA. We conclude that Pnt1p is a nuclearly encoded mitochondrial protein.
FIG. 4.
Pnt1p is an integral mitochondrial membrane protein. (A) Epitope-tagged Pnt1p copurified with mitochondria. Mitochondria from wild-type strain DFS188 (lane 1) and from the PNT1-HA strain SH404 (lane 2) were purified on Nycodenz gradients (17). These mitochondria (20 μg) and the cytosolic fraction from SH404 (20 μg) (lane 3) were run on an SDS–10% polyacrylamide gel, immunoblotted, and probed with anti-HA monoclonal antibody (see Materials and Methods). (B) Pnt1p was not extractable from membranes by alkaline carbonate. Mitochondria purified from SH404 expressing Pnt1p-HA were sonicated and separated into a membrane pellet (lane 2) and soluble supernatant by centrifugation (lane 1). An aliquot of the membrane fraction was extracted with sodium carbonate (pH 11.5), followed by centrifugation to separate pelleted integral membrane proteins (lane 4) and solubilized proteins in the supernatant (lane 3). The sample were resolved by SDS-polyacrylamide gel electrophoresis (PAGE), immunoblotted, and probed with anti-HA antibody (top). The same blot was then stripped and reprobed with antiserum against Arg8p, a known mitochondrial matrix protein (bottom).
To determine whether Pnt1p-HA is soluble or membrane bound, the purified mitochondria from SH404 were sonicated and centrifuged. Pnt1p-HA was present in the membrane pellet (Fig. 4B, lane 2) but absent from the soluble supernatant (lane 1). Extraction of the pelleted membranes with alkaline sodium carbonate (15) failed to solubilize Pnt1p-HA (lanes 3 and 4). These results indicate that Pnt1p is an integral mitochondrial membrane protein.
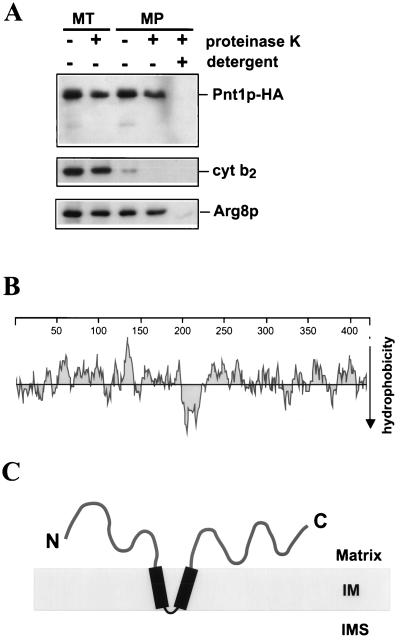
To examine the submitochondrial location and topology of Pnt1p-HA, purified mitochondria from SH404 were subjected to mitoplasting and protease digestion prior to Western analysis (Fig. 5A). Proteinase K treatment of detergent-solubilized mitochondria eliminated the Pnt1p-HA signal, demonstrating that the epitope is labile to protease. However, Pnt1p-HA was protected from digestion by proteinase K in both whole mitochondria and mitoplasts: the C-terminal epitope remained detectable on the full-length protein, and no shorter immunoreactive species were generated. Similar results were obtained in experiments where Pnt1p was tagged at its C terminus with a hexahistidine epitope (unpublished results). Taken together with the data in Fig. 4B, these results strongly suggest that Pnt1p is an integral inner membrane protein that is not accessible to protease digestion from the outside.
FIG. 5.
Pnt1p is an inner membrane protein that is inaccessible to protease added to mitoplasts, leading to a proposed topology. (A) Mitochondria (MT) were purified from strain SH404 and converted to mitoplasts (MP) by osmotic shock (20) in the absence or presence of proteinase K (100 μg/ml), as indicated for each lane. Mitoplasts were also treated with proteinase K in the presence of 1% octylglucoside (right lane) to solubilize the inner membrane. Treated samples were resolved by SDS-PAGE, immunoblotted, and probed with anti-HA (top), with anti-cytochrome b2 (cyt b2) as an IMS marker (middle), and with anti-Arg8p as a matrix marker (bottom). (B) Pnt1p hydrophobicity plot (30) showing a distinct hydrophobic domain of about 31 residues near the middle of the protein. (C) Proposed topology for Pnt1p in the mitochondrial inner membrane based on protease protection assays, hydrophobicity analysis, and the positive inside rule (16). IM, inner membrane.
A hydrophobicity plot of the Pnt1p sequence (Fig. 5B) reveals a highly hydrophobic region in the center of the protein between residues 198 and 228. Residues 209 and 210 are both acidic (E), suggesting that this region could form a hairpin-like membrane anchor. Both domains flanking the hydrophobic region are largely hydrophilic and positively charged. Taking these observations together, we propose (Fig. 5C) that Pnt1p is anchored to the inner membrane by a hairpin-like hydrophobic domain, with both the N- and the C-terminal domains facing the matrix. Virtually no residues would be exposed on the outer surface of the inner membrane.
The absence of Pnt1p has only modest effects on otherwise wild-type S. cerevisiae cells.
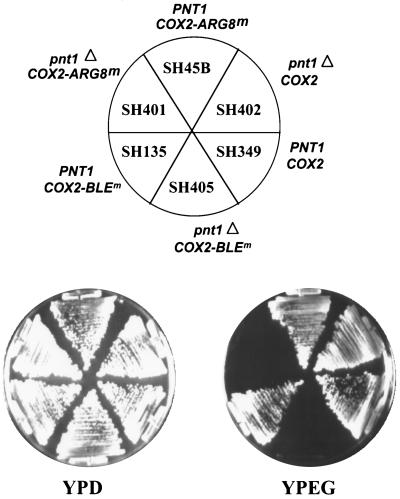
Both the pnt1-41 and pnt1Δ::LEU2 mutations prevent respiratory growth of cells whose mtDNA encodes the Cox2p-Arg8mp fusion protein. We also examined the effect of the pnt1Δ::LEU2 deletion on the growth of strains whose mtDNA encoded either another fusion protein, Cox2p-Blemp, or wild-type Cox2p (Fig. 6). The synthetic gene BLEm specifies, in the yeast mitochondrial genetic code, the 126-residue bleomycin-resistant protein (36) of the bacterial transposon Tn5 (unpublished results). Fusion of this relatively short passenger moiety to the C terminus of Cox2p did not affect the respiratory growth of an otherwise wild-type strain. However, expression of the Cox2p-Blemp fusion protein in a pnt1Δ::LEU2 nuclear mutant resulted in a Pet− phenotype (Fig. 6), suggesting that membrane translocation of this fusion protein was also dependent on Pnt1p function. Consistent with this interpretation, pnt1Δ::LEU2 cells expressing either mitochondrially encoded fusion protein lacked cytochrome oxidase activity (data not shown).
FIG. 6.
Deletion of PNT1 prevents respiratory growth of strains expressing Cox2p fusion proteins but not wild-type Cox2p. Each strain was streaked on YPD and YPEG, and the plates were incubated at 30°C for 4 or 5 days, respectively. The relevant nuclear genotypes (PNT1 or pnt1Δ) and mitochondrial genotypes (COX2, COX2::ARG8m or COX2::BLEm) are indicated for each sector.
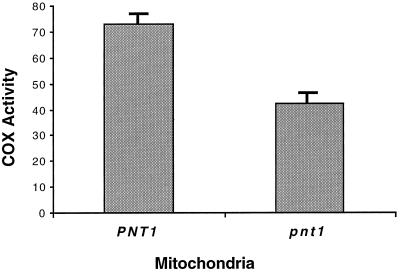
Interestingly, the pnt1Δ::LEU2 mutation did not appear to affect the respiratory growth of a strain containing wild-type mtDNA on solid nonfermentable medium (Fig. 6), although in liquid medium with poor aeration the pnt1Δ::LEU2 mutant grew slower than the wild type did. To determine whether the absence of Pnt1p had any effect on the biogenesis of cytochrome oxidase, we assayed this enzyme in mitochondria purified from isogenic PNT1 and pnt1Δ::LEU2 strains growing on nonfermentable medium (Fig. 7). In the absence of Pnt1p, cytochrome oxidase specific activity was reduced to 58% of the wild-type level. This suggests that Pnt1p plays a role in the efficient biogenesis of cytochrome oxidase. However, export of the wild-type Cox2p C-tail cannot be completely dependent upon Pnt1p function.
FIG. 7.
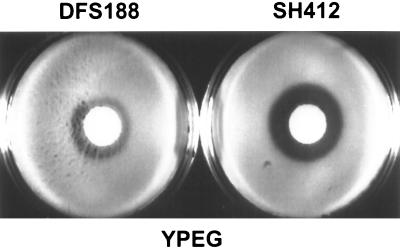
The absence of Pnt1p reduces cytochrome c oxidase activity in mitochondria from an S. cerevisiae strain containing wild-type mtDNA. Mitochondria were isolated from mid-log-phase cells of two isogenic strains, DFS188 (PNT1) and SH412 (pnt1Δ::LEU2), that were grown in YPEG at 30°C. The mitochondria were purified on Nycodenz gradients (17) prior to the assay. Specific activity was determined spectrophotometrically as the rate of oxidation of reduced horse heart cytochrome c and is expressed as the change in optical density at 550 nm per minute per milligram of mitochondrial proteins (35). The error bars show the standard deviation of four independent measurements.
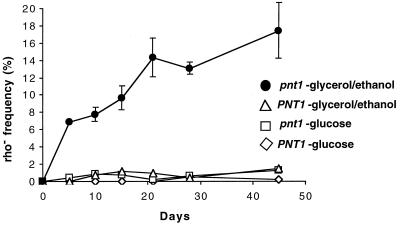
We also examined the frequency of ρ− (cytoplasmic petite) mtDNA deletion mutants in cultures of isogenic wild-type and pnt1Δ::LEU2 mutant strains, both of which contained wild-type mtDNA. Cultures of each strain were grown in complete medium containing either fermentable (glucose [YPD]) or nonfermentable (ethanol plus glycerol [YPEG]) carbon sources and sampled during logarithmic growth as well as during the stationary phase for a period of several weeks. mtDNA of the wild-type strain was very stable under all these conditions (Fig. 8). During logarithmic growth on glucose and subsequent incubation of the cultures, the absence of Pnt1p had no effect on the stable maintenance of ρ+ mtDNA. However, the pnt1Δ::LEU2 culture grown in YPEG accumulated increased numbers of ρ− cells, which rose dramatically over a period of days and weeks (Fig. 8). After 45 days, roughly 17% of the viable pnt1Δ::LEU2 cells had lost the ability to respire. These nonrespiring cells were ρ− mutants with deletions of various regions of mtDNA, as judged by their ability to recombine with mit− testers (data not shown). The fact that ρ− pnt1Δ::LEU2 cells accumulated during growth on nonfermentable carbon sources, which selects against nonrespiring mutants, suggests that the absence of Pnt1p has a more deleterious effect in cells actively engaged in respiration. Consistent with this idea, we found that the pnt1Δ::LEU2 mutation increased the sensitivity of otherwise wild-type cells to growth inhibition by H2O2 on medium containing nonfermentable carbon sources (Fig. 9).
FIG. 8.
Absence of Pnt1p causes increased ρ− accumulation when grown in nonfermentable carbon sources. Single colonies of the isogenic strains DFS188 (PNT1) and SH412 (pnt1Δ::LEU2) grown on nonfermentable medium (YPEG) were inoculated to liquid YPD and allowed to grow to saturation. Aliquots of cells of each strain were then inoculated into fresh liquid YPD or YPEG and incubated at 30°C for the times indicated. Samples were taken periodically from each culture, diluted, and plated onto YPD. These YPD plates were replica-plated to YPEG, and colonies that failed to grow on YPEG were scored as ρ−.
FIG. 9.
Deletion of PNT1 causes increased sensitivity to oxidative damage by H2O2. Equal amounts of cells of strains DFS188 (PNT1) and SH412 (pnt1Δ::LEU2), grown to early stationary phase, were evenly spread on plates (35 by 10 mm) containing 3 ml of YPEG, and 3 μl of 30% H2O2 was pipetted onto filter disks in the center of the plates. The plates were incubated for 2 days at 30°C and photographed.
In K. lactis, absence of the homologous KlPnt1p prevents respiratory growth, assembly of cytochrome oxidase, and, apparently, export of the Cox2p C tail.
We isolated a functional homologue of PNT1 from K. lactis, KlPNT1, by its ability to complement the Pet− phenotype of a S. cerevisiae pnt1 deletion strain that carried COX2::ARG8m in its mtDNA (see Materials and Methods). KlPnt1p and Pnt1p have only about 27% identity and 48% similarity in amino acid sequence. However, the hydrophobicity plot of the KlPnt1p sequence (not shown) resembles that of Pnt1p (Fig. 5B), and the central hydrophobic region is also interrupted by two acidic E residues.
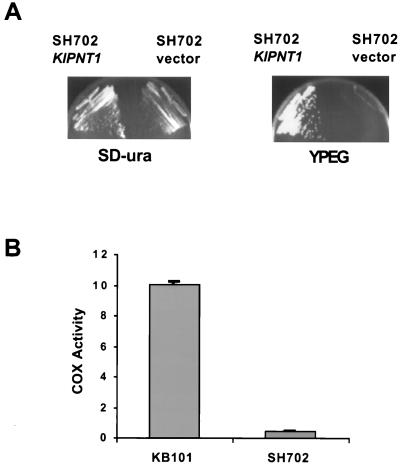
We deleted the KlPNT1 gene from the nuclear genome of a K. lactis strain with normal mtDNA (see Materials and Methods). The resulting mutant was unable to grow on nonfermentable medium (Fig. 10A), indicating that respiratory growth is more dependent on KlPNT1 in K. lactis than on PNT1 in S. cerevisiae. To determine whether this defect could be due to diminished cytochrome c oxidase, we assayed the enzyme in mitochondria isolated from the Klpnt1 deletion strain as well as the parental wild-type strain. In K. lactis, the deletion essentially eliminated cytochrome oxidase activity (Fig. 10B), in contrast to the modest effect of the corresponding mutation in S. cerevisiae (Fig. 7). Western blot analysis of these mitochondria with anti-Cox2p revealed that the Klpnt1 deletion did not block the accumulation of Cox2p, although it did reduce the steady-state level relative to the wild type (Fig. 11 and data not shown).
FIG. 10.
Deletion of the K. lactis gene KlPNT1 causes a nonrespiratory phenotype and the absence of cytochrome c oxidase activity. (A) The Klpnt1 deletion strain (SH702) was transformed with either a KlPNT1 plasmid (pSH505) or the empty vector (pROCS2). The transformants were streaked on glucose minimal medium (SD-ura) selecting for the plasmids or on YPEG and incubated at 30°C for 2 days before being photographed. (B) Crude mitochondria were isolated from KB101 (KlPNT1) and SH702 (Klpnt1Δ::hisG) as described in Materials and Methods, and cytochrome oxidase was assayed by the same procedure as in the experiment Fig. 7. The error bars show the standard deviation of four independent measurements.
FIG. 11.
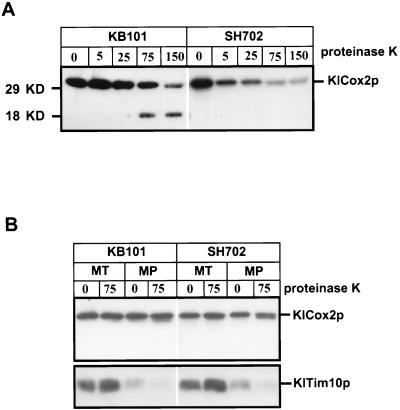
Absence of KlPnt1p causes a defect in KlCox2p assembly that is apparently due to a block of C-tail export. (A) The absence of KlPnt1p blocks the assembly of KlCox2p into the cytochrome oxidase complex. Mitochondria were isolated and purified from KB101 (KlPNT1) and SH702 (Klpnt1Δ::hisG). Purified mitochondria (75 μg for each sample) were solubilized with 1% octylglucoside, mixed with the indicated amounts of proteinase K (0 to 150 μg/ml) in a total volume of 200 μl, and incubated on ice for 20 min. Proteinase K was inactivated by the addition of 2 mM phenylmethylsulfonyl fluoride followed by incubation with 10% trichloroacetic acid at 65°C for 10 min. Samples were chilled on ice for 5 min and centrifuged. Protein samples (10 μg/lane for KB101, and 20 μg/lane for SH702) were separated on a 14% polyacrylamide–SDS gel, blotted, and probed with the anti-Cox2p monoclonal antibody CCO6. The right panel was exposed longer to achieve comparable intensities for unproteolyzed wild-type and mutant samples. (B) The purified mitochondria were subjected to mitoplasting in the absence or presence of 75 μg of proteinase K per ml, as described for S. cerevisiae (see Materials and Methods). Proteinase K inactivation and immunoblotting were carried out as described for panel A. The same gel blot was then stripped and reprobed with anti-Tim10p, which recognizes a soluble IMS protein in S. cerevisiae (29), to demonstrate successful removal of the outer membrane.
To ask whether the cytochrome oxidase deficiency of the K. lactis Klpnt1 mutant might be due to defective export of Cox2p domains, we first examined the protease K sensitivity of KlCox2p in detergent-solubilized mitochondria. Solubilized cytochrome oxidase from wild-type S. cerevisiae is highly resistant to proteolysis (12). Purified wild-type (KB101) and mutant (SH702) K. lactis mitochondria were solubilized with the mild detergent octyl glucoside, incubated with various amounts of protease K, and then analyzed by Western blotting with the anti-Cox2p monoclonal antibody CCO6 (46), which, as we have previously shown (20), recognizes an epitope in the C-tail domain (Fig. 11A). Assembled KlCox2p from solubilized wild-type mitochondria was highly resistant to proteinase K digestion, and at high concentrations of proteinase K an 18-kDa immunoreactive fragment was generated, suggesting preferential cleavage of the KlCox2p matrix loop. In contrast, solubilized KlCox2p from the Klpnt1 deletion mutant mitochondria was much more sensitive to protease K, and there was no evidence of the stable 18-kDa subfragment under any conditions, suggesting that KlPnt1p is necessary for assembly of KlCox2p into the protease-resistant cytochrome oxidase complex. In parallel experiments, KlCox2p from wild-type mitochondria was completely resistant to trypsin, while the KlCox2p from mutant mitochondria was highly sensitive (data not shown).
If the unassembled protease-sensitive KlCox2p were normally exported in the mitochondria lacking KlPnt1p, it should be sensitive to digestion by protease K in mitoplasts. On the other hand, if export were defective in the absence of KlPnt1p, this unassembled KlCox2p should be protected from protease K by the intact inner membrane of mutant (SH702) mitoplasts. As expected for an export defect, we found that KlCox2p was largely protected from protease K in mitoplasts lacking KlPnt1p (Fig. 11B, right). The assembled KlCox2p in wild-type K. lactis mitoplasts was, of course, also resistant to protease K digestion (Fig. 11B, left), as previously shown for S. cerevisiae (24). Interestingly, a fraction of KlCox2p from the mutant mitoplasts was shortened by proteinase K treatment (Fig. 11B, right, lane 4), suggesting the possibility that some N tail export occurred in the absence of KlPnt1p. These results suggest that in K. lactis, KlPnt1p is required for the export of the KlCox2p C tail and is at least partially required for the export of the KlCox2p N tail.
DISCUSSION
The machinery that inserts mitochondrially encoded proteins into the inner membrane and translocates their hydrophilic domains through the membrane is poorly understood. We have developed a genetic screen for S. cerevisiae mutants defective in the translocation of mitochondrially encoded hydrophilic protein domains through the inner membrane to the IMS. The screen is based on the fact that the hydrophilic polypeptide Arg8mp is exported from the matrix when it is synthesized within mitochondria as a bifunctional Cox2p-Arg8mp fusion protein (20). Apparently, the export system that translocates the large acidic C-tail of Cox2p through the inner membrane continues to translocate the Arg8mp fused to it (Fig. 2). However, Arg8mp participates in the arginine biosynthetic pathway only if it is located within the matrix (Fig. 1). Thus, export of Arg8mp causes an Arg− growth phenotype in the strain developed for this study. Export of the fusion protein also supports the assembly of functional cytochrome c oxidase (20), presumably after proteolytic removal of Arg8mp in the IMS. Thus, the parent strain is Pet+ (respiratory competent).
From our parent strain, we selected Arg+ mutants, with the expectation that some should be Arg+ because they fail to export Arg8mp and thus allow it to function in the matrix. We anticipated that among the Arg+ mutants isolated, those that were deficient in protein export should also be unable to assemble cytochrome c oxidase normally and therefore would exhibit an unselected Pet− (respiratory deficient) phenotype. In this study we have analyzed the gene PNT1, identified by one such recessive nuclear mutation, and the protein it encodes. In the absence of functional Pnt1p, the C-terminal Arg8mp moiety of the Cox2p-Arg8mp fusion protein is not translocated through the inner membrane and remains largely in the matrix. Furthermore, the Cox2p moiety of the fusion protein is not assembled into active cytochrome c oxidase, suggesting that its C tail is also trapped on the inside. Thus, the selection scheme appears to yield export-defective mutants.
We examined the subcellular location and topology of a functional, epitope-tagged form of Pnt1p that was expressed from the PNT1 chromosomal locus. Pnt1p-HA was specifically located in mitochondria. Submitochondrial fractionation and protease protection experiments demonstrated that Pnt1p-HA was an integral inner membrane protein, as would be expected for a protein involved in export. Proteolytic treatment of mitoplasts failed either to destroy the C-terminal HA epitope of Pnt1p-HA or to generate a shorter epitope-tagged product, suggesting that none of the polypeptide is accessible on the outside of the inner membrane.
The predicted Pnt1p sequence contains a central hydrophobic region of approximately 31 amino acids (residues 198 to 228) that is interrupted by two acidic Glu residues (at positions 209 and 210). Flanking this hydrophobic region are N- and C-terminal positively charged hydrophilic domains. We therefore propose that Pnt1p is anchored to the inner membrane by a central hairpin-like membrane anchor, with both flanking hydrophilic domains on the inside (matrix side) of the membrane (Fig. 5). Such a topology is consistent with the “positive inside rule” (55), although this “rule” is broken by several nuclearly encoded mitochondrial inner membrane proteins (16). Interestingly, our original mutation, pnt1-41, caused a missense substitution, W382R, in the C-terminal domain. However, while our data show that Pnt1p is necessary for export of the Cox2p-Arg8mp fusion protein, they do not demonstrate that it plays a direct role in translocation.
The failure of pnt1 mutants to export the C-terminal portion of the Cox2p-Arg8mp fusion protein results in a Pet− growth phenotype. We also observed the same Pet− phenotype in a pnt1 mutant whose mtDNA encoded a different fusion protein, Cox2p-Blem (Fig. 6). However, the pnt1 deletion mutation had only modest effects on S. cerevisiae strains containing wild-type mtDNA, reducing cytochrome c oxidase activity by about 40% without affect respiratory growth under normal conditions. In contrast to the modest effects of pnt1 deletion in S. cerevisiae, we found that deletion of the functionally homologous gene, KlPNT1, from the nuclear genome of a K. lactis strain containing wild-type mtDNA caused a clear nonrespiratory phenotype, the absence of cytochrome oxidase activity, and a defect in the assembly of KlCox2p that appears to be due to a C-tail export defect. Taken together, these results demonstrate an important physiological role for KlPnt1p in K. lactis and suggest that overlapping functions may exist in S. cerevisiae.
The possible redundancy of Pnt1p in S. cerevisiae is not simply due to a second homologous gene, since PNT1 is unique in the genome. However, partial functional redundancy involving nonhomologous proteins has a well-known precedent in the translocation apparatus that imports cytoplasmically synthesized precursors into the organelle. For example, the mitochondrial import system can function at least partially in the absence of either the Tom70p or Tom20p receptor proteins (26, 32, 38). Furthermore, as noted above, even the highly conserved protein Oxa1p can be rendered dispensable for inner membrane translocation in yeast by mutations affecting the membrane anchor of cytochrome c1 (19). While the mechanism of this suppression is not clear, it does support the idea that there is at least latent functional redundancy in the mitochondrial export translocation machinery in S. cerevisiae.
While Pnt1p has not been studied as intensively as Oxa1p, it appears to have a distinct function. In the absence of Pnt1p, mitochondrially synthesized Cox2p-Arg8mp cannot be exported and the accumulated enzyme in the matrix is sufficient to support arginine biosynthesis. However, while oxa1 mutations also prevent the export of Cox2p-Arg8mp, they greatly decrease the steady-state level of the accumulated fusion protein (20) to the point that cells exhibit an Arg− growth phenotype (unpublished results). Thus, Oxa1p plays a role in stabilizing mitochondrial proteins, a phenomenon also noted by others (19), that Pnt1p does not.
Pnt1p is not necessary for the maintenance of wild-type (ρ+) mtDNA. In pnt1 deletion mutant cells grown on glucose medium and incubated on the same medium for several weeks, there was no significant accumulation of ρ− mtDNA deletions. However, cultures of pnt1 mutant cells grown under respiratory conditions accumulated significantly more ρ− mutants upon prolonged incubation than did similarly incubated wild-type cultures. This result suggests that the activity of Pnt1p may play a role, direct or indirect, in protecting mitochondria from oxidative damage. Consistent with this idea, pnt1 deletion mutants exhibited increased sensitivity to the oxidant H2O2, compared to the wild type. An interesting hypothesis for the role of Pnt1p in protecting against oxidative damage is that it could promote transport out of the matrix of components damaged by reactive oxygen species. Such a “detoxification” system could be required for the export of the aberrant C-terminally modified Cox2p-fusion proteins we generated.
PNT1 was originally isolated in a screen for yeast genes that affected resistance to pentamidine, an antimicrobial drug that has been used clinically to treat pneumonia in AIDS patients infected with the fungus Pneumocystis carinii (33). As shown previously (33) and confirmed by us (data not shown), overexpression of PNT1 increases pentamidine resistance while deletion of PNT1 makes yeast more sensitive. Growth of S. cerevisiae on nonfermentable carbon sources is far more sensitive to pentamidine than is growth on fermentable sugar, suggesting that the drug interacts with mitochondrial targets (34). While the identity of these targets remains unknown, our finding that Pnt1p is a mitochondrial inner membrane protein confirms the importance of the organelle in pentamidine action. The involvement of Pnt1p in mitochondrial export suggests the possibility that drug resistance involves the removal of damaged targets, or pentamidine itself, from the matrix by an active-transport mechanism.
ACKNOWLEDGMENTS
We thank T. L. Mason for anti-Cox2p and for help in performing cytochrome oxidase assays. We also thank B. S. Glick, C. Koehler, and G. Schatz for other antisera; N. Da Silva, H. Fukuhara, and M. Bianchi for K. lactis plasmids; and L. A. Grivell for the K. lactis library.
This work was supported by the U.S. National Institutes of Health in the form of a National Research Service Award (GM18093) to S.H. and a grant (GM29362) to T.D.F.
REFERENCES
- 1.Abad A R, Mehrtens B J, Mackenzie S A. Specific expression in reproductive tissues and fate of a mitochondrial sterility-associated protein in cytoplasmic male-sterile bean. Plant Cell. 1995;7:271–285. doi: 10.1105/tpc.7.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Attardi G, Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- 4.Balzi E, Goffeau A. Yeast multidrug resistance: the PDR network. J Bioenerg Biomembr. 1995;27:71–76. doi: 10.1007/BF02110333. [DOI] [PubMed] [Google Scholar]
- 5.Bauer M, Behrens M, Esser K, Michaelis G, Pratje E. PET1402, a nuclear gene required for proteolytic processing of cytochrome oxidase subunit 2 in yeast. Mol Gen Genet. 1994;245:272–278. doi: 10.1007/BF00290106. [DOI] [PubMed] [Google Scholar]
- 6.Bianchi M M, Santarelli R, Frontali L. Plasmid functions involved in the stable propagation of the pKD1 circular plasmid in Kluyveromyces lactis. Curr Genet. 1991;19:155–161. doi: 10.1007/BF00336481. [DOI] [PubMed] [Google Scholar]
- 7.Bonnefoy N, Chalvet F, Hamel P, Slonimski P P, Dujardin G. OXA1, a Saccharomyces cerevisiae nuclear gene whose sequence is conserved from prokaryotes to eukaryotes controls cytochrome oxidase biogenesis. J Mol Biol. 1994;239:201–212. doi: 10.1006/jmbi.1994.1363. [DOI] [PubMed] [Google Scholar]
- 8.Bonnefoy N, Kermorgant M, Groudinsky O, Minet M, Slonimski P P, Dujardin G. Cloning of a human gene involved in cytochrome oxidase assembly by functional complementation of an oxa1− mutation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1994;91:11978–11982. doi: 10.1073/pnas.91.25.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dabhi V M, Lindahl K F. MtDNA-encoded histocompatibility antigens. Methods Enzymol. 1995;260:466–485. doi: 10.1016/0076-6879(95)60159-7. [DOI] [PubMed] [Google Scholar]
- 10.Deshaies R J, Schekman R. A yeast mutant defective at an early stage in import of secretory protein precursors into the endoplasmic reticulum. J Cell Biol. 1987;105:633–645. doi: 10.1083/jcb.105.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dujon B. Mitochondrial genetics and functions. In: Strathern J N, Jones E W, Broach J R, editors. The molecular biology of the yeast Saccharomyces: life cycle and inheritance. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1981. pp. 505–635. [Google Scholar]
- 12.Eytan G D, Schatz G. Cytochrome c oxidase from bakers’ yeast. V. Arrangement of the subunits in the isolated and membrane-bound enzyme. J Biol Chem. 1975;250:767–774. [PubMed] [Google Scholar]
- 13.Fikes J D, Becker D M, Winston F, Guarente L. Striking conservation of TFIID in Schizosaccharomyces pombe and Saccharomyces cerevisiae. Nature. 1990;346:291–294. doi: 10.1038/346291a0. [DOI] [PubMed] [Google Scholar]
- 14.Fox T D, Folley L S, Mulero J J, McMullin T W, Thorsness P E, Hedin L O, Costanzo M C. Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol. 1991;194:149–165. doi: 10.1016/0076-6879(91)94013-3. [DOI] [PubMed] [Google Scholar]
- 15.Fujiki Y, Hubbard A L, Fowler S, Lazarow P B. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gavel Y, Von Heijne G. The distribution of charged amino acids in mitochondrial inner-membrane proteins suggests different modes of membrane integration for nuclearly and mitochondrially encoded proteins. Eur J Biochem. 1992;205:1207–1215. doi: 10.1111/j.1432-1033.1992.tb16892.x. [DOI] [PubMed] [Google Scholar]
- 17.Glick B S, Pon L A. Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 1995;260:213–223. doi: 10.1016/0076-6879(95)60139-2. [DOI] [PubMed] [Google Scholar]
- 18.Glick B S, von Heijne G. Saccharomyces cerevisiae mitochondria lack a bacterial-type Sec machinery. Protein Sci. 1996;5:2651–2652. doi: 10.1002/pro.5560051229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Green-Willms, N. S., and T. D. Fox. Unpublished data.
- 19.Hamel P, Lemaire C, Bonnefoy N, Brivet-Chevillotte P, Dujardin G. Mutations in the membrane anchor of yeast cytochrome c1 compensate for the absence of Oxa1p and generate carbonate-extractable forms of cytochrome c1. Genetics. 1998;150:601–611. doi: 10.1093/genetics/150.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He S, Fox T D. Membrane translocation of mitochondrially coded Cox2p: distinct requirements for export of amino- and carboxy-termini, and dependence on the conserved protein Oxa1p. Mol Biol Cell. 1997;8:1449–1460. doi: 10.1091/mbc.8.8.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heimberg H, Boyen A, Crabeel M, Glansdorff N. Escherichia coli and Saccharomyces cerevisiae acetylornithine aminotransferases: evolutionary relationship with ornithine aminotransferases. Gene. 1990;90:69–78. doi: 10.1016/0378-1119(90)90440-3. [DOI] [PubMed] [Google Scholar]
- 22.Hell K, Herrmann J, Pratje E, Neupert W, Stuart R A. Oxa1p mediates the export of the N- and C-termini of pCoxII from the mitochondrial matrix to the intermembrane space. FEBS Lett. 1997;418:367–370. doi: 10.1016/s0014-5793(97)01412-9. [DOI] [PubMed] [Google Scholar]
- 23.Hell K, Herrmann J M, Pratje E, Neupert W, Stuart R A. Oxa1p, an essential component of the N-tail protein export machinery in mitochondria. Proc Natl Acad Sci USA. 1998;95:2250–2255. doi: 10.1073/pnas.95.5.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrmann J M, Koll H, Cook R A, Neupert W, Stuart R A. Topogenesis of cytochrome oxidase subunit II: mechanisms of protein export from the mitochondrial matrix. J Biol Chem. 1995;270:27079–27086. doi: 10.1074/jbc.270.45.27079. [DOI] [PubMed] [Google Scholar]
- 25.Herrmann J M, Neupert W, Stuart R A. Insertion into the mitochondrial inner membrane of a polytopic protein, the nuclear-encoded Oxa1p. EMBO J. 1997;16:2217–2226. doi: 10.1093/emboj/16.9.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hines V, Brandt A, Griffiths G, Horstmann H, Brütsch H, Schatz G. Protein import into yeast mitochondria is accelerated by the outer membrane protein MAS70. EMBO J. 1990;9:3191–3200. doi: 10.1002/j.1460-2075.1990.tb07517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jauniaux J-C, Urrestarazu L A, Wiame J-M. Arginine metabolism in Saccharomyces cerevisiae: subcellular localization of the enzymes. J Bacteriol. 1978;133:1096–1107. doi: 10.1128/jb.133.3.1096-1107.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kermorgant M, Bonnefoy N, Dujardin G. Oxa1p, which is required for cytochrome c oxidase and ATP synthase complex formation, is embedded in the mitochondrial inner membrane. Curr Genet. 1997;31:302–307. doi: 10.1007/s002940050209. [DOI] [PubMed] [Google Scholar]
- 29.Koehler C M, Jarosch E, Tokatlidis K, Schmid K, Schweyen R J, Schatz G. Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science. 1998;279:369–373. doi: 10.1126/science.279.5349.369. [DOI] [PubMed] [Google Scholar]
- 30.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 31.Liao X, Butow R A. RTG1 and RTG2: Two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell. 1993;72:61–71. doi: 10.1016/0092-8674(93)90050-z. [DOI] [PubMed] [Google Scholar]
- 32.Lithgow T, Junne T, Wachter C, Schatz G. Yeast mitochondria lacking the two import receptors Mas20p and Mas70p can efficiently and specifically import precursor proteins. J Biol Chem. 1994;269:15325–15330. [PubMed] [Google Scholar]
- 33.Ludewig G, Staben C. Characterization of the PNT1 pentamidine resistance gene of Saccharomyces cerevisiae. Antimicrob Agents Chemother. 1994;38:2850–2856. doi: 10.1128/aac.38.12.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ludewig G, Williams J M, Li Y, Staben C. Effects of pentamidine isethionate on Saccharomyces cerevisiae. Antimicrob Agents Chemother. 1994;38:1123–1128. doi: 10.1128/aac.38.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mason T L, Poyton R O, Wharton D C, Schatz G. Cytochrome c oxidase from bakers’ yeast. I. Isolation and properties. J Biol Chem. 1973;248:1346–1354. [PubMed] [Google Scholar]
- 36.Mazodier P, Cossart P, Giraud E, Gasser F. Completion of the nucleotide sequence of the central region of Tn5 confirms the presence of three resistance genes. Nucleic Acids Res. 1985;13:195–205. doi: 10.1093/nar/13.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyer W, Bauer M, Pratje E. A mutation in cytochrome oxidase subunit 2 restores respiration of the mutant pet ts1402. Curr Genet. 1997;31:401–407. doi: 10.1007/s002940050222. [DOI] [PubMed] [Google Scholar]
- 38.Moczko M, Ehmann B, Gartner F, Honlinger A, Schafer E, Pfanner N. Deletion of the receptor MOM19 strongly impairs import of cleavable preproteins into Saccharomyces cerevisiae mitochondria. J Biol Chem. 1994;269:9045–9051. [PubMed] [Google Scholar]
- 39.Mulder W, Scholten I, de Boer R, Grivell L. Sequence of the HAP3 transcription factor of Kluyveromyces lactis predicts the presence of a novel 4-cysteine zinc-finger motif. Mol Gen Genet. 1994;245:96–106. doi: 10.1007/BF00279755. [DOI] [PubMed] [Google Scholar]
- 40.Mulero J J, Fox T D. Alteration of the Saccharomyces cerevisiae COX2 5′-untranslated leader by mitochondrial gene replacement and functional interaction with the translational activator protein PET111. Mol Biol Cell. 1993;4:1327–1335. doi: 10.1091/mbc.4.12.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neff N F, Thomas J H, Grisafi P, Botstein D. Isolation of the β-tubulin gene from yeast and demonstration of its essential function in vivo. Cell. 1983;33:211–219. doi: 10.1016/0092-8674(83)90350-1. [DOI] [PubMed] [Google Scholar]
- 42.Nelson B D, Kabir F. The role of the mitochondrial outer membrane in energy metabolism of tumor cells. Biochimie. 1986;68:407–415. doi: 10.1016/s0300-9084(86)80008-6. [DOI] [PubMed] [Google Scholar]
- 43.Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 44.Oliver D B, Beckwith J. E. coli mutant pleiotropically defective in the export of secreted proteins. Cell. 1981;25:765–772. doi: 10.1016/0092-8674(81)90184-7. [DOI] [PubMed] [Google Scholar]
- 45.Pfanner N, Craig E A, Honlinger A. Mitochondrial preprotein translocase. Annu Rev Cell Dev Biol. 1997;13:25–51. doi: 10.1146/annurev.cellbio.13.1.25. [DOI] [PubMed] [Google Scholar]
- 46.Pinkham J L, Dudley A M, Mason T L. T7 RNA polymerase-dependent expression of COXII in yeast mitochondria. Mol Cell Biol. 1994;14:4643–4652. doi: 10.1128/mcb.14.7.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pon L, Schatz G. Biogenesis of yeast mitochondria. In: Broach J R, Pringle J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis and energetics. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 333–406. [Google Scholar]
- 48.Poyton R O, Duhl D M J, Clarkson G H D. Protein export from the mitochondrial matrix. Trends Cell Biol. 1992;2:369–375. doi: 10.1016/0962-8924(92)90049-s. [DOI] [PubMed] [Google Scholar]
- 49.Rose M D, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 50.Saier M H, Jr, Paulsen I T, Sliwinski M K, Pao S S, Skurray R A, Nikaido H. Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J. 1998;12:265–274. doi: 10.1096/fasebj.12.3.265. [DOI] [PubMed] [Google Scholar]
- 51.Schatz G. The protein import system of mitochondria. J Biol Chem. 1996;271:31763–31766. doi: 10.1074/jbc.271.50.31763. [DOI] [PubMed] [Google Scholar]
- 52.Schneider B L, Seufert W, Steiner B, Yang Q H, Futcher A B. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- 53.Steele D F, Butler C A, Fox T D. Expression of a recoded nuclear gene inserted into yeast mitochondrial DNA is limited by mRNA-specific translational activation. Proc Natl Acad Sci USA. 1996;93:5253–5257. doi: 10.1073/pnas.93.11.5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 Å. Science. 1995;269:1069–1074. doi: 10.1126/science.7652554. [DOI] [PubMed] [Google Scholar]
- 55.von Heijne G. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 1986;5:3021–3027. doi: 10.1002/j.1460-2075.1986.tb04601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]