UF-MRE11 recruits PP1-a to dephosphorylate NBS1 regulating Apollo recruitment and preserving telomere leading strand.
Abstract
Ubiquitin-fold modifier 1 (UFM1) is involved in neural and erythroid development, yet its biological roles in these processes are unknown. Here, we generated zebrafish models deficient in Ufm1 and Ufl1 that exhibited telomere shortening associated with developmental delay, impaired hematopoiesis and premature aging. We further report that HeLa cells lacking UFL1 have instability of telomeres replicated by leading-strand synthesis. We uncover that MRE11 UFMylation is necessary for the recruitment of the phosphatase PP1-α leading to dephosphorylation of NBS1. In the absence of UFMylation, NBS1 remains phosphorylated, thereby reducing MRN recruitment to telomeres. The absence of MRN at telomeres favors the formation of the TRF2-Apollo/SNM1 complex consistent with the loss of leading telomeres. These results suggest that MRE11-UFMylation may serve as module to recruit PP1-α. Last, zebrafish expressing Mre11 that cannot be UFMylated phenocopy Ufm1-deficient zebrafish, demonstrating that UFMylation of MRE11 is a previously undescribed evolutionarily conserved mechanisms regulating telomere length.
INTRODUCTION
The ubiquitin family of proteins consists of Ubiquitin (Ub), and the other ubiquitin-like (Ubl) proteins, all of which share the same β-grasp fold, are posttranslational modifiers that play a central role in a variety of cellular processes, including cell cycle progression, DNA damage response, protein translation and stability, signal transduction, intracellular trafficking, and antiviral response (1). Ubiquitin-fold modifier 1 (UFM1) is one of the most recently identified Ubls. It is evolutionarily conserved, and its orthologs are found in Metazoa and plants but not in yeast (2). The process of protein modification by UFM1 involves an enzymatic cascade similar to that described for the other ubiquitin family members. Briefly, after cleavage of the last two amino acids of pro-UFM1 by the peptidase UFSP2, UFM1 is activated by the UFM1-specific E1 enzyme UBA5, transferred to the E2-conjugating enzyme UFC1 before being ligated onto substrates by the E3 ligase UFL1. UFMylation is reversible as it is cleaved from its targets by the UFM1-specific protease, UFSP2 [reviewed in (3)].
Genetic studies using knockout (KO) mouse models and the identification of patients with mutations in the UFM1 pathway provided strong evidence for the indispensable role of this posttranslational modification in animal development and homeostasis. It has been shown that UFM1 is crucial for hematopoiesis (4–6), liver development (7), brain development (8), heart failure protection (9), maintenance of intestinal homeostasis, and protection from inflammatory diseases (10).
The nuclear receptor coactivator ASC1 (Activating signal cointegrator 1) was one of the first UFMylated proteins to be identified. UFMylated ASC1 enhances the recruitment of transcription cofactors to promoters of estrogen receptor α (ERα) target genes to up-regulate their expression (11). More recently, it has been suggested that the ribosomal subunit RPL26 is the primary target of UFMylation with its UFMylation suggested to mediate ER homeostasis by regulating biogenesis of secretory proteins (12). In addition, up-regulation of protein levels of p53 and its targets and increased level of H2AX phosphorylation were recently reported in Ufl1-deficient bone marrow cells (5). This suggests that the UFM1 pathway could contribute to genome stability, although the exact mechanism whereby UFL1 exerts this function remains unknown. Very recently, it was reported that UFM1 promotes ataxia-telangiectasia mutated (ATM) activation and mono-UFMylates histone H4 in response to double-strand breaks, suggesting that H4 UFMylation contributes to the amplification of ATM activation (13).
In this study, we analyzed the functions of UFMylation in hematopoiesis by generating Ufm1- and Ufl1-deficient zebrafish. Our work reveals a hitherto unanticipated role for UFMylation in maintaining telomere length. We further define the underlying mechanism by identifying that UFMylated MRE11 recruits PP1-α that dephosphorylates NBS1 to allow the recruitment of the MRN complex to telomeres. Consistent with this result, loss of UFMylation resulted in telomere leading-strand shortening, marked shortening of telomeres during hematopoiesis, and premature aging. These findings provide a likely explanation for the observed anemia in mice lacking components of the UFM1 pathway.
RESULTS
Inactivation of the Ufm1 pathway leads to hematopoietic stem and progenitor cell death and premature aging in zebrafish
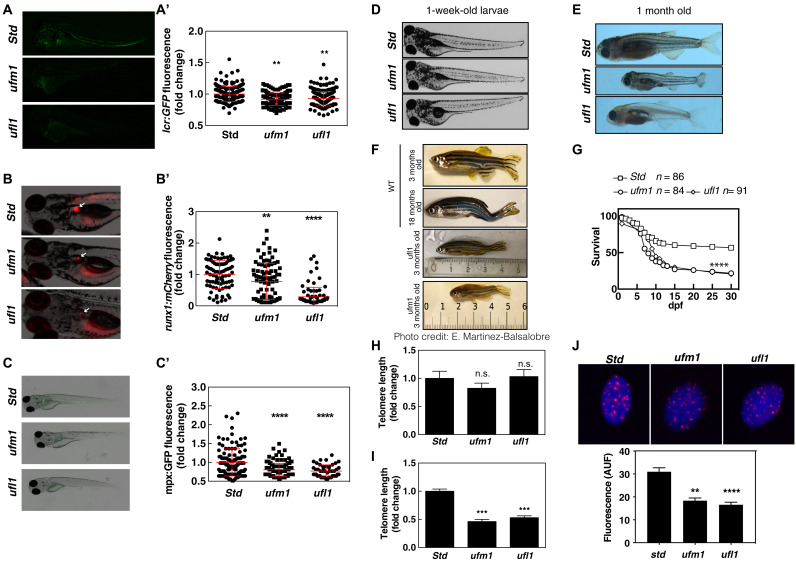
As UFM1 has been shown to be crucial for hematopoiesis in mouse models (4–6), we investigated the relevance of UFMylation in zebrafish using the unique advantages of this model for genetic analysis of hematopoiesis. As in mammals, zebrafish hematopoiesis occurs in two waves: The primitive wave generates a transient population of blood cells, while the second wave generates hematopoietic stem cells (HSCs) about 30 hours after fertilization (14). In the zebrafish line Tg(lcr:eGFP) that expresses green fluorescent protein (GFP) in erythroid cells (15), genetic inactivation of either ufm1 or ufl1 with CRISPR-Cas9 gene editing (fig. S1, A and B) resulted in reduced erythrocyte numbers in larvae at 4 days post-fertilization (dpf) (Fig. 1A, A′).
Fig. 1. UFM1 pathway prevents premature aging and telomere shortening.
Tg(lcr:eGFP) (A), Tg(runx1:GAL4; UAS:nfsb-mCherry) (B), and Tg(mpx:eGFP) (C) one-cell embryos were injected with standard control (Std), ufm1, or ufl1 sgRNA and recombinant Cas9. Representative images of green and red channel of whole larvae for the different treatments (C′) and quantitation of erythroid cells at 4 dpf (A′), HSPCs (B′) and neutrophils cells at 5 dpf are shown. White arrows indicated HSPCs in the kidney marrow of 5-dpf larvae (B). Each dot represents normalized fluorescence from a single larva, while the mean ± SEM for each group is also shown. A representative image of control and deficient ufm1 or ufl1 larval phenotype at 1-week- and 1-month-old fish is shown (D and E). Phenotypic characteristics of 3-month-old fish deficient in Ufm1 and Ufl1 are displayed with an old and young zebrafish. Larval survival curve (Kaplan-Meier representation) of genotype Ufm1- and Ufl1-deficient zebrafish compared with std. (F) Photo credit: Elena Martinez Balsalobre. Survival curve of fish deficient in Ufm1 and Ufl1. (G). Telomere length was determined by quantitative polymerase chain reaction (qPCR) in sorted erythroid cells (GFP−) (H) or (GFP+) (I) of 6-dpf larvae. Telomere was determined by quantitative fluorescence in situ hybridization (qFISH) in GFP+ cells (J). The data are shown as the means ± SEM of two independent experiments. **P < 0.01; ***P < 0.001; ****P < 0.0001 according to Student’s t test and analysis of variance followed by Tukey multiple range test. n.s., not significant.
We then analyzed the emergence, maintenance, and differentiation of HSCs using Tg(runx1:nfsB-mCherry) and Tg(mpx:eGFP) lines that fluorescently mark hematopoietic stem and progenitor cells (HSPCs) (16) and neutrophils (17), respectively. Using these models, we found that HSPC survival was impaired in Ufm1- and Ufl1-deficient F0 larvae, since a decreased number of HSPCs (Fig. 1B, B′) and neutrophils (Fig. 1C, C′) was observed at 5 dpf in the kidney marrow, the definitive hematopoietic tissue in zebrafish. However, the emergence of HSPCs (fig. S1C) and primitive myelopoiesis (fig. S1D) were both unaffected.
Although Ufm1- and Ufl1-deficient F0 embryo/larvae did not show any obvious defect during the first week of life (Fig. 1D), 40% of them displayed a broad collection of phenotypes 1 month after birth, including reduced size, curved tail, and cachexia (Fig. 1E). These phenotypes were observed in both males and females. Overall, the alterations observed in 3-month-old Ufm1- or Ufl1-deficient zebrafish resemble those normally observed in an aged 18-month-old fish (Fig. 1F). In addition, Ufm1- and Ufl1-deficient zebrafish had a shortened life span, with approximately 60% dying before 13 dpf and only 10 to 20% reaching 3 months of age (Fig. 1G).
The UFM1 pathway is required to maintain telomere length in zebrafish HSPCs
Telomeres are essential for genomic stability as they protect chromosomes ends. In the absence of special telomere maintenance mechanisms, linear chromosomes shorten with every round of DNA replication, leading to replicative senescence, making telomere shortening one of the hallmarks of cell aging (18, 19). Intriguingly, despite the premature aging of Ufm1- and Ufl1-deficient zebrafish, we were not able to detect significant changes in telomere length by quantitative polymerase chain reaction (qPCR) analysis of whole zebrafish larvae at 6 dpf (Fig. 1H). Since this analysis is on a mixture of cell types, we next sought to determine whether telomere length was impaired in HSPCs. As larvae have low numbers of HSPCs, it is technically challenging to analyze telomere length in these cells. We therefore measured telomere length in erythrocytes, which, unlike human and mice, are nucleated, and this revealed that telomere length is reduced (Fig. 1, I and J). The telomere shortening observed in Ufm1- and Ufl1-deficient HSPCs is similar to the shortening in HSPCs of telomerase (tert and terc)–deficient zebrafish larvae (20). We therefore measured telomere length in erythrocytes as a surrogate of HSPCs. When telomere length was assayed by qPCR or quantitative fluorescence in situ hybridization (qFISH), we find robust telomere shortening in both Ufm1- and Ufl1-deficient erythrocytes compared to wild type (WT) (Fig. 1, I and J).
To determine whether premature aging observed in Ufl1- and Ufm1-deficient zebrafish was due to telomere attrition, we analyzed genetic epistasis between ufl1 (or ufm1) and tert. Notably, genetic inactivation of ufl1 or ufm1 Tert-deficient G1 (obtained from tert−/− G0) zebrafish larvae resulted in impaired development and drastic reduction of their life span (fig. S1, E and F). This cumulative effect suggests that the premature aging phenotype observed in Ufm1- and Ufl1-deficient zebrafish resulted from telomere attrition but was not due to telomerase inactivation.
Loss of UFL1 results in sister chromatid fusions
To get better insights into the underlying mechanism by which loss of UFMylation leads to telomere attrition and whether this is conserved in human cells, we used CRISPR-Cas9–mediated genome editing to disrupt the UFL1 gene in HeLa cells (fig. S2, A to C). Analysis of genomic DNA from clonal populations of HeLa cells obtained using a guide RNA (gRNA) targeting exon 2 revealed a deletion and frameshift in both alleles confirming KO (fig. S2D). Consistent with published results (5), we noticed that UFL1 KO cells have a reduced growth rate [23.1 hours/per division (PD)] compared to parental HeLa cells (26.6 hours/PD) (fig. S2E). However, we did not observe any significant differences in cell cycle in the UFL1 KO cells (fig. S2F). Because telomere shortening can lead to chromosomal fusions, we next investigated whether UFL1 KO cells exhibited chromosome abnormalities. Metaphase chromosome spread experiments revealed that UFL1 KO cells displayed a threefold increase in levels of spontaneous chromosomal fusions compared to WT HeLa cells (Fig. 2, A and B). We then assessed the effect of UFL1 inactivation on telomere fusions by using FISH on metaphase chromosome spreads. This approach revealed that most of the chromosomal fusions appear to be sister chromatid fusions (Fig. 2C). We also detected four times more chromatid ends with no detectable telomeric signal in UFL1 KO cells compared to WT HeLa cells (Fig. 2, D and E). Notably, most of these losses in telomeric signal appeared to be oriented, suggesting loss of either lagging or leading telomere strands (Fig. 2D, arrow).
Fig. 2. UFL1 KO cells have defect in telomere length.
(A) Representative image of chromosome metaphase spreads from WT and UFL1−/− cells. Black arrow: telomere fusion. (B) Quantification of the chromosomes breaks and fusions. (C) Quantification of the types of fusions. (D) Representative images of telomere FISH on metaphase spreads in HeLa and HeLa UFL1−/− cells. Blue, 4′,6-diamidino-2-phenylindole(DAPI)–stained chromosomes. Red dots, telomeres; white arrows, telomere loss. (E) Quantification of (D). (F) Representative image of CO-FISH labeling. (G) Quantification of the leading and lagging telomere losses. (H) Southern blot analysis of the terminal restriction fragment (TRF) lengths in indicated cells. Red lines indicate the peak signal intensity of the TRF smears. (I) Analysis of the individual telomere length distribution by telomere shortest length assay (TeSLA). Nine TeSLA PCRs were performed for each DNA sample. (J) Quantification of the TeSLA band distributions. *P < 0.1; **P < 0.01; ***P < 0.001; ****P < 0.0001 according to Student’s t test and analysis of variance followed by Tukey multiple range test. n.s., not significant.
UFL1 KO cells predominantly lose the leading-strand telomere
To next visualize telomeres produced by the lagging- and leading-strand synthesis, we used Chromosome Orientation (CO)–FISH. Using this approach, we detected that the loss of the leading telomere signal was roughly three times more frequent than the loss of the lagging telomeric signal in UFL1 KO cells (Fig. 2, F and G). To determine whether UFL1 inactivation affected overall telomere length, we performed Southern blot analysis of terminal restriction fragments (TRFs) using two independently derived UFL1 KO clones. Both clones showed around 30% decrease of the mean telomere length compared to the WT controls (Fig. 2H). Together, these results suggest that UFL1 is required to maintain telomere length. To further characterize the changes in telomere length, we used the telomere shortest length assay (TeSLA) that reveals the length of individual telomeres from all chromosome ends (21). These analyses showed that the fraction of very short telomeres (<1 kb) is increased more than twofold in UFL1 KO compared to WT HeLa cells (Fig. 2, I and J, and fig. S3, A and B). The TeSLA analysis also revealed up to 40% reduction in the average length of amplified telomeres in UFL1 KO clones, further confirming an unappreciated role for UFL1 in telomere length regulation.
To rule out that the telomere shortening observed in UFL1 KO cells was due to loss of telomerase, we measured telomerase activity in HeLa extracts using the Telomere Repeated Amplification Protocol (TRAP) assay. No significant differences in telomerase activity between WT and UFL1 KO cells were detected (fig. S3C), indicating that loss of UFL1 does not appear to impair telomerase activity per se. Last, the labeling of γH2AX at telomeres did not show any differences in the frequency of TIFs (Telomere dysfunction Induced Foci) between WT and KO (fig. S3D). In summary, these experiments suggest a role for UFMylation in telomere maintenance.
UFL1 localizes to telomeres
A role of UFL1 and UFMylation in maintaining telomere length suggests nuclear localization of the proteins. While UFM1 was initially detected within the nucleus (2), UFL1 has been described as a protein localized in the endoplasmic reticulum (ER) via its interaction with the ER-resident protein UFBP1 (22). We therefore investigated the localization of endogenous UFL1 and UFM1 using cell fractionation of HeLa cells. In these assays, we detected UFL1 in the cytoplasmic (CET), nuclear (NEB), and chromatin (NEB+) fractions. In contrast, free (unconjugated) UFM1 appeared to be predominantly cytoplasmic, while UFMylated proteins were detected in the nucleus and chromatin (fig. S4A). We also analyzed the localization of UFM1 and UFL1 by expressing GFP-tagged forms of these proteins, which revealed both proteins to localize in the cytoplasm and the nucleus. When we use conditions that remove the proteins not bound to chromatin proteins (CSK buffer), we find UFM1 and UFL1 to form nuclear foci (fig. S4B). Monitoring colocalization between endogenous UFL1 and telomeres revealed that a fraction of UFL1 foci colocalized with telomeres (fig. S4C), suggesting that the telomeric phenotypes of UFL1 KO cells may be a direct consequence of the lack of UFMylation at telomeres.
MRE11 interacts with the UFM1 pathway members
To understand how UFL1 regulates telomere length, we sought to identify interactors of UFL1. Identification of enzyme partners has often been problematic because of the transient nature of the interaction, making it difficult to purify complexes by conventional protein coimmunoprecipitation methods. To overcome this issue, we used proximity labeling using the BioID method that biotinylates proteins in close vicinity (23). Biotinylated proteins can then be purified using streptavidin affinity capture and identified by mass spectrometry. To identify substrates and interactors of the UFM1 E3 ligase UFL1, we fused BirA (R118G) biotin ligase domain to the N terminus of UFL1 (Fig. 3A) (23). Expression of the BirA-UFL1 fusion protein can be induced in HeLa FRT-TREx cells. We first verified that the fusion of BirA on UFL1 did not alter the interaction of UFL1 with its known partner UFSP2. As expected, UFSP2 is only detected in the biotin pull-down from BirA-UFL1–expressing cells (Fig. 3B). After this validation, we performed mass spectrometry to identify proteins that were biotinylated by the BirA-UFL1 fusion. Proteins unique to the BirA-UFL1 pull-down and not detected in the BirA-GFP pull-down were considered for further analysis (table S1). Our mass spectrometry identified known partners of UFL1 such as UFBP1 and UFSP2, giving us confidence in the approach (Fig. 3C).
Fig. 3. UFL1 interacts with MRE11.
(A) Strategy for the identification of UFL1 binding proteins. (B) Biotinylated proteins are purified from cells expressing BirA-GFP or BirA-UFL1. The samples were then subjected to immunoblot with indicated antibodies. (C) List of the candidate target proteins identified by mass spectrometry. (D) Biotinylated proteins are purified from cells expressing BirA-GFP or BirA-UFL1. The samples were then subjected to immunoblot with indicated antibodies. (E) Biotinylated proteins are purified from cells expressing BirA-GFP or BirA-UFM1. The samples were then subjected to immunoblot with indicated antibodies.
Using this approach, we identified MRE11 as a high confidence protein biotinylated by BirA-UFL1 (Fig. 3C). To validate this observation, we performed biotinylation experiments using cells expressing BirA-UFM1 or BirA-UFC1, which reveals that MRE11 is also an interactor of UFM1 and UFC1 (fig. S5, A, B, D, and E). We further confirmed the presence of MRE11 in biotin pull-down from BirA-UFL1 (Fig. 3D)– and BirA-UFM1–expressing cells by immunoblotting (Fig. 3E).
MRE11 is a substrate of UFL1
MRE11 together with RAD50 and Nijmegen breakage syndrome 1 (NBS1; also known as nibrin) forms the multifunctional protein complex MRN. Besides its role in the maintenance of genome stability (24), MRN promotes C-NHEJ at dysfunctional telomeres through the activation of ATM (25–27). Last, the MRN complex has been proposed to regulate telomere length (6, 28). Given the biotinylation of MRE11 by BirA fusions of UFL1, UFM1, and UFC1, we therefore hypothesized that the UFM1 pathway may regulate telomere length via its interaction with the MRN complex.
We first investigated whether MRE11 could be UFMylated by UFL1 in an in vitro UFMylation reaction. When recombinant MRE11 was added into the reaction, we detected a band shift of ~10 kDa corresponding to the addition of a single UFM1 (Fig. 4A). This modification is dependent on the presence of UFL1, suggesting that MRE11 is a substrate of UFL1 (Fig. 4A). To test whether MRE11 is also UFMylated in vivo, we analyzed cell extracts from HeLa cells with an Mre11 antibody. In accordance with the in vitro reconstitution results, a similarly shifted band of MRE11 was detected in extracts prepared from WT cells but not UFL1-deficient cells (Fig. 4B). While UFL1 is known to only mediate the attachment of UFM1 to proteins, we cannot rule out that the observed loss of modified MRE11 in UFL1 KO cells could be an indirect effect of MRE11 being modified by a different PTM. To confirm that the higher molecular weight band of MRE11 detected is indeed UFMylated MRE11, we incubated the protein extracts from WT cells with recombinant UFSP2. Incubation with UFSP2 results in the disappearance of the slower-migrating band, confirming that the observed band is UFMylated MRE11 (Fig. 4B). Together, these results demonstrate that UFL1 UFMylates MRE11 both in vitro and in cells.
Fig. 4. MRE11 is UFMylated on K281-282.
(A) Recombinant His6-MRE11 were incubated with UFMylation enzymes and UFM1 in the presence of adenosine triphosphate and MgCl2 for 1 hour at 37°C. The reaction was stopped by the addition of 3× SDS loading buffer, and the reaction products were separated on a 4 to 12% NuPAGE SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gel. Top: Western blot analysis of UFMylation of MRE11 WT and mutants using MRE11-specific antibody. Bottom: Ponceau-stained nitrocellulose blot as loading control. (B) Cells (106) are lysed in SDS, and after treatment or not with recombinant UFSP2, samples were subjected to immunoblot with indicated antibodies. (C) Protein alignment of ASC1 UFMylated site with MRE11 sequence. (D) Same as (A) with MRE11 fragment mutated in indicated residues. (E) GFP-tagged MRE11 WT and mutant were expressed in cells with 6HIS-UFM1 as indicated. Cell lysates were subjected to pull-down with NTA resins followed by immunoblot with the indicated antibodies.
We next sought to identify lysine residues in MRE11 that are UFMylated. Because MRE11 contains 45 lysine residues, we wondered whether we could define a consensus UFMylation site based on homology with identified UFMylation sites on ASC1 (Fig. 4C). Using this approach, we identified four lysine candidates within two stretches of MRE11 (K196-K197 and K281-K282). To test whether these lysines in MRE11 are the preferred UFMylation sites, we mutated each pair to arginine. Whereas mutation of the K196-K197 or K281-K282 lysines into arginines did not affect the UFMylation of MRE11 in vitro (Fig. 4D), mutating K281R-K282R led to a strong reduction of UFMylation in HeLa cells (Fig. 4E). This result suggests that MRE11 is mainly UFMylated on K281-K282.
UFL1 does not regulate DSB repair
MRE11 is part of the MRN complex, a sensor of DSBs that controls the DNA damage response (DDR) by governing the activation of the central transducing kinase ATM. In addition, MRN regulates DSB repair through the homology directed repair (HDR) pathway (29). We thus investigated whether UFL1 could control DSB repair by monitoring the activation of ATM and ATR as well as the phosphorylation of H2AX upon DNA damage. Compared to WT cells, phosphorylation of ATR, ATM (fig. S6A), and H2AX (fig. S6B) was slightly enhanced in UFL1 KO cells exposed to various DNA damaging agents. However, contrary to recently published results (30), we do not find the sensitivity of UFL1 KO cells to x-rays to be significantly increased compared to control cells (fig. S6C). Moreover, a GFP reporter assay of DNA repair indicated that HDR was not compromised in UFL1 KO cells (fig. S6D). In summary, these results demonstrate that UFL1 does not play a critical role in DSB repair and HDR.
To further characterize the regulation of MRN by UFL1 upon IR stress, we performed MRE11 immunoprecipitation from chromatin fractions of cells exposed to IR. These experiments revealed that the MRN complex formation is not affected in UFL1 KO cells (fig. S6E). In addition, we did not observe differences in the amount of UFL1 associated with MRE11 after induction of DSB by irradiation (fig. S5E). These results are in agreement with our mass spectrometry analysis of the proteins interacting with UFM1 (fig. S5, A, D, and E), UFC1 (fig. S5, B, D, and E), and UFL1 (fig. S5, C to E). Exposure of cells to IR did not increase the biotinylation of MRE11. This indicates that the interaction between MRE11 and components of the UFM1 pathway is not increased upon DNA damage.
UFL1 regulates the interaction of MRE11 and NBS1 with telomeres
It has been shown that the recruitment of MRN to telomeres is mediated by the direct interaction between NBS1 and the shelterin protein TRF2 (31). To evaluate whether the recruitment of MRN to telomeres was impaired in UFL1 KO cells, we quantified the colocalization between endogenous MRE11 and telomeres by fluorescence microscopy. This approach revealed a reduction in colocalization of MRE11 with the telomeric probe in UFL1 KO cells (Fig. 5, A and B). To test whether the association between MRE11 and TRF2 is regulated by UFMylation, we performed immunoprecipitation of MRE11 from WT and UFL1 KO cells. While similar amounts of RAD50 coprecipitated with MRE11 in UFL1 KO and control cells, MRE11 association with TRF2 was strongly reduced in the absence of UFL1 (Fig. 5C). Collectively, these results indicate that the recruitment of MRE11 to telomeres is markedly reduced when UFL1 is inactivated, suggesting that UFMylation of MRE11 regulates its localization to telomere. To confirm that UFMylation of MRE11 is necessary for its interaction with telomeres, we expressed GFP-tagged MRE11 and mutants that cannot be UFMylated in HeLa cells and tested their association with endogenous TRF2. Whereas TRF2 coimmunoprecipitated with GFP-MRE11 and GFP-MRE11-K196-7R, it was barely coimmunoprecipitated with the UFMylation defective GFP-MRE11 K281-2R mutant. This mutant retained the ability to bind RAD50, confirming that mutating these lysines does not disrupt MRE11 folding (Fig. 5D). Together, these results suggest that UFL1 promotes localization of MRE11 at telomeres.
Fig. 5. UFL1 regulates the interaction of MRE11 and NBS1 with telomeres.
(A) Colocalization of MRE11 (MRE11 antibodies) and telomere (telomere probe) in the indicated cells. (B) Quantification of (A). (C) Protein lysates from the indicated cells were subjected to MRE11 immunoprecipitation. Immunoprecipitation with immunoglobulin G (IgG) was used as a negative control. After SDS-PAGE, samples were analyzed with the indicated antibodies. (D) GFP-tagged MRE11 WT and mutant were expressed in cells as indicated. Cell lysates were subjected to GFP TRAP followed by immunoblot with the indicated antibodies. (E) Protein lysates from the indicated cells were subjected to TRF2 or NBS1 immunoprecipitation. Immunoprecipitation with IgG was used as a negative control. After SDS-PAGE, samples were analyzed with the indicated antibodies. (F) NBS1 immunoprecipitation from WT or UFL1−/− cells were incubated with phosphatase or heat-inactivated phosphatase before Western blot analysis of NBS1. Intensities of the band along the indicated red line were quantified using Fiji and are represented on the graphs. (G) Same as (D). (H) Protein lysates from the indicated cells overexpressing Flag-Apollo were subjected to Flag or TRF2 immunoprecipitation. Immunoprecipitation with IgG was used as a negative control. After SDS-PAGE, samples were analyzed with the indicated antibodies.
Since MRE11 interacts with NBS1, which, in turn, interacts with TRF2 (31), we performed NBS1 and TRF2 coimmunoprecipitation to determine whether the interaction between NBS1 and TRF2 was impaired in UFL1 KO cells. As shown in Fig. 5E, the interaction between NBS1 and TRF2 was reduced in UFL1 KO cells, implying that the impaired recruitment of MRE11 to telomeres in UFL1 KO cells is a consequence of the reduced interaction between NBS1 and TRF2.
Dephosphorylation of NBS1 by PP1-α is regulated by UFL1
In these experiments, we noticed that the NBS1 protein from UFL1 KO cells appears to migrate as a slightly higher molecular weight form compared to parental cells (Fig. 5E). Such a size shift is reminiscent of phosphorylation. To test whether the size shift of NBS1 in UFL1 KO cells was due to phosphorylation, we incubated the NBS1 immunoprecipitation with alkaline phosphatase, which revealed that treatment with active phosphatase and not heat-inactivated phosphatase resulted in the disappearance of the slower-migrating form (Fig. 5F). Furthermore, the level of phospho-NBS1 is increased in UFL1 KO cells, suggesting that UFL1 might indirectly regulate NBS1 phosphorylation. Phosphorylation inhibits NBS1 binding to TRF2, and the phosphatase PP1-α was reported to dephosphorylate NBS1, thereby promoting NBS1-TRF2 interaction (31). PP1-α was among the high-confidence proteins biotinylated by BirA-UFL1 identified by mass spectrometry (table S1). We therefore hypothesized that the activity of PP1-α might be modulated by UFL1. We first examined the interaction of PP1-α with MRE11 by performing purification of GFP-MRE11. We find that the association of PP1-α with GFP-MRE11 appeared to be mildly reduced when the lysines that undergo UFMylation are mutated in MRE11 (Fig. 5G). We infer from this result that MRE11 UFMylation promotes the interaction between PP1-α and the MRN complex, resulting in the dephosphorylation of NBS1 to promote NBS1 interaction with TRF2.
The Apollo/SNM1B nuclease functions together with TRF2 to protect telomeres (32) and phosphorylation of NBS1 dissociates NBS1 from TRF2 to promote TRF2-Apollo/SNM1B interaction (31). Since NBS1 phosphorylation is increased in UFL1 KO cells, we asked whether recruitment of Apollo/SNM1B to telomeres was impaired in UFL1 KO cells. We find that the amount of Flag-Apollo/SNM1B coimmunoprecipitated with TRF2 was increased in UFL1 KO cells. This result suggests that the balance between dephosphorylated NBS1 and Apollo/SNM1B is regulated by UFL1 (Fig. 5H).
UFMylation of MRE11 prevents premature aging and telomere shortening in zebrafish
On the basis of the results obtained in human cells, and in zebrafish, we tested whether mre11a and its UFMylation is also critical for telomere maintenance and hematopoiesis in zebrafish. We first demonstrated that genetic inactivation of mre11a by CRISPR-Cas9 resulted in decreased erythrocyte numbers in 4-dpf larvae (Fig. 6A, A′) and telomere shortening in erythrocytes (Fig. 6, B, B′, and C). Notably, anemia (Fig. 6A, A′) and telomere attrition (Fig. 6, B, B′, and C) of Mre11a-deficient larvae were both rescued by the forced expression of WT Mre11a but not of the K283R mutant (which is equivalent to K282 of human MRE11). Like Ufm1- and Ufl1-deficient zebrafish, Mre11a-deficient larvae showed reduced life span (Fig. 6D) and impaired development observed from 1 month onward (Fig. 6E). Furthermore, genetic inactivation of mre11a in Tert-deficient larvae resulted in marked impaired development and further reduced life span (Fig. 6, F and G). Together, these results suggest that disrupting the UFMylation of MRE11 in a vertebrate animal model results in telomere attrition in vivo, with severe consequences for development likely probably due to telomere shortening in stem cells.
Fig. 6. UFMylation of MRE11 prevents premature aging and telomeres shortening in zebrafish.
(A and A′) Tg(lcr:eGFP) one-cell embryos were injected with standard control (Std) or mre11a sgRNA recombinant Cas9 and complemented with the indicated mRNA. Representative images of green channel of whole larvae for the different treatments and quantitation of erythroid cells at 4 dpf. Each dot represents normalized fluorescence from a single larva, while the mean ± SEM for each group is also shown. (B) Telomere length was determined in GFP+ cells by qFISH (B and B′) or qPCR (C). Larval survival curve (Kaplan-Meier representation) of genotype mre11a-deficient zebrafish compared with std (D). Representative images of control and deficient mre11a larval phenotype at 1-week- and 1-month-old fish are shown (E). Representative images of Tert-deficient larvae injected with mre11a phenotype at 8 dpf (F). Larval survival curve (Kaplan-Meier representation) of Tert-deficient one-cell embryos injected with mre11a gRNA (G).
DISCUSSION
Our work reveals a new role for the UFM1 pathway in maintaining telomere length and preventing premature aging in zebrafish. In human cells, loss of UFL1 results in sister chromatid fusion and the preferential loss of the leading-strand telomere. The fact that UFL1 can be localized at telomeres prompted us to focus on interactors of UFM1 E3 ligase UFL1 related to telomere length maintenance. We uncover that MRE11 is UFMylated and that this modification regulates MRE11 association with telomeres, which have previously been shown to interact with TRF2 (33). The question that arises is whether the lack of MRE11 UFMylation is the cause of the telomeric phenotypes observed in UFL1 KO cells.
The role of MRE11 at telomeres was mainly demonstrated at uncapped telomeres (25–27, 34). In cells lacking TRF2, MRE11 removes the 3′ telomeric overhang to promote chromosome fusions. At normal telomeres in telomerase-positive cells, the MRN complex was shown to play a role in the generation of G-overhangs at human telomeres (35). More recently, CDK2-mediated phosphorylation of NBS1 was shown to regulate the interaction between TRF2 and NBS1 (31) and dephosphorylation of NBS1 by PP1-α promotes its dissociation from TRF2, thereby facilitating TRF2-Apollo/SNM1B complex formation (31). Nucleolytic processing of telomeres and generation of 3′ overhangs are regulated by the concerted action of Apollo and Exo1 nucleases, and CST fill in (36, 37). We discovered that (i) PP1-α is biotinylated by BirA-UFL1, (ii) PP1 interacts with MRE11 in a UFMylation-dependent manner, (iii) inactivation of UFL1 leads to the hyper-phosphorylation NBS1, and (iv) Apollo association to telomeres is increased in UFL1-KO cells. Collectively, these findings suggest that UFMylation favors the dephosphorylation of NBS1 by PP1-α, thereby promoting the release of the MRN complex from telomeres and the association of Apollo to TRF2. The fact that the UFMylation defective MRE11-K281-2R mutant does not interact with PP1-α suggests that MRE11 UFMylation promotes the recruitment of PP1-α to the MRN complex. The loss of PP1-α recruitment by MRE11 stabilizes the phosphorylation of NBS1 and facilitates the recruitment of Apollo to telomeres (fig. S7).
Leading- and lagging-strand telomeres of mammalian cells behave differently. TRF2-deficient cells exhibit telomere-telomere fusions specifically between leading-strand telomeres (38). In addition, inactivation of TRF2 combined with deletion of Mre11 biases fusions to leading- rather than lagging-strand telomeres (26). A similar phenotype was also observed in cells expressing the hypomorphic Mre11 ATLD1 (25). Consistent with the model proposed in our study, we found that UFL1 inactivation leads to the preferential loss of the leading-strand telomere that may reflect the increased accessibility of Apollo in particular at the leading telomere (36, 37). In yeast, Mre11 was shown to be specifically recruited at the leading telomere, thereby allowing the regeneration of the 3′ overhang (39). It is tempting to speculate that MRE11 UFMylation may control the telomere processing by regulating the access of Apollo at the leading telomere.
In light of the presented results, we propose a model where UFL1 regulates telomere length in human cells by modifying MRE11. While we cannot rule out that UFL1 regulates other mechanisms involved in telomere maintenance, the reduction of telomere length observed in zebrafish deficient in MRE11 UFMylation supports a role of MRE11 and its UFMylation in telomere maintenance. The fact that MRE11 and its UFMylation affect telomere length in zebrafish may be in apparent contradiction with previous findings showing that no telomere shortening was observed in mouse embryonic fibroblasts carrying the hypomorphic Mre11(ATLD1/ATLD1) mutation, even after extensive passage through culture (25). This paradox may be explained by the fact that the lack of MRE11 UFMylation creates a gain of function by promoting a cell cycle deregulation of NBS1 phosphorylation whose effects would be distinct from a loss of function of MRE11. To our knowledge, telomere shortening in MRN-deficient vertebrates has not been reported yet. Here, we report an early premature aging phenotype that could be a consequence of a defect in telomere length maintenance in pluripotent cells. Notably, telomere length analysis in zebrafish was carried out on erythrocyte cells as a read out for telomeres in pluripotent cells. Mouse pluripotent cells display a TRF2-independent mechanism of telomere protection (40, 41). Our results in zebrafish suggest that the UFM1 pathway could contribute to telomere maintenance essentially in stem cells during development. This could explain why humans carrying point mutations in genes of the UFM1 pathway display developmental defects (8).
Our work reveals the UFM1 pathway as a new regulator involved in telomere maintenance in zebrafish and humans. Patients mutated in UFM1 display brain development defects. Whether the function of UFM1 in telomere maintenance plays a role in preventing these defects remains to be investigated.
MATERIALS AND METHODS
Cell culture and transfection
293T and HeLa FRT-T-Rex (Invitrogen) were maintained at 37°C in DMEM (Dulbecco’s modified Eagle’s medium; Gibco) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (Gibco). For transfection, each dish of adherent cells was transfected with 5 to 10 μg of plasmid DNA using Lipofectamine 2000 (Thermo Fisher Scientific). Flag Apollo was a gift of P. Revy (42).
CRISPR gene editing
CRISPR-Cas9 px335 and pBabe-U6 have been used to clone gRNA sense and antisense, respectively. Cloning has been made according to Zhang laboratory protocols. Cells have been transfected with 1 μg of combined plasmids and cell-sorted into 96-well plates. Clones have been screened using immunodetection of targeted proteins, and the mutation has been sequenced as described before (43).
Stable cell line establishment
Stable cells expressing BirA-GFP or BirA-tagged proteins were generated according to the manufacturer’s instructions. Briefly, pcDNA5 FRT BirA plasmids were cotransfected with POG44 plasmids (ratio 1/9) with Lipofectamine 2000 (Invitrogen). Cells were then selected with hygromycin and blasticidin.
Cloning
Restriction enzyme digestions, DNA ligations, and other recombinant DNA procedures were performed using standard protocols. All mutagenesis was performed using the QuikChange site-directed mutagenesis method (Stratagene) with KOD polymerase (Novagen). All DNA constructs were verified by DNA sequencing, which was performed by GATC (Eurofins). DNA for mammalian cell transfection was amplified in Escherichia coli DH5α strain, and plasmid preparation was done using QIAGEN Maxi prep kit according to the manufacturer’s protocol.
Biotin ligase assay
Expression of BirA proteins was induced for 24 hours by adding 1 μM doxycycline (Sigma-Aldrich). For purification of biotinylated substrates, 10 cm by 10 cm dishes were treated for 8 hours with 50 μM biotin. Cells were then collected, washed with phosphate-buffered saline (PBS), and resuspended in lysis buffer [0.5 ml/10 cm dish; 8 M urea, 1% SDS, 50 mM N-ethylmaleimide, 1× protease inhibitor cocktail (Roche) in PBS]. Sonication was performed as needed to reduce sample viscosity. To reduce urea concentration, the samples were diluted by adding binding buffer (3 M urea, 1 M NaCl, and 0.25% SDS; 0.5 volume). Incubation was done using 100-μl suspension of high-capacity Streptavidin-Sepharose beads (GE Healthcare) overnight at room temperature. Washes have been done with 2× WB1, 3× WB2, 1× WB3, 3× WB4, 1× WB1, 1× WB5, and 3× WB6 [WB1: 8 M urea, 0.25% SDS in PBS; WB2: 6 M guanidine hydrochloride in PBS; WB3: 6.4 M urea, 1 M NaCl, 0.2% SDS in PBS (prewarmed to 37°C); WB4: 4 M urea, 1 M NaCl, 10% isopropanol, 10% ethanol, 0.2% SDS in PBS; WB5: 8 M urea, 1% SDS in PBS; WB6: 2% SDS in PBS]. Samples were eluted in 100 μl of 4× Laemmli sample buffer with 100 mM dithiothreitol (DTT) by two cycles of heating (5 min; 99°C), with vortexing in between. For mass spectrometry analysis, the bead slurry was transferred to a Vivaclear Mini 0.8-μm PES filter (Sartorius) and spun to recover bead-free eluate.
Mass spectrometry
Pull-down elutions were separated by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Brilliant Blue G-Colloidal Concentrate (Sigma-Aldrich) according to the manufacturer’s instructions. Gel bands were excised from the whole gel lane and destained, and proteins were in-gel digested with trypsin (sequencing grade, Promega) overnight. The resulting peptide mixtures were analyzed by liquid chromatography (LC)–tandem mass spectrometry using Orbitrap Fusion Lumos Tribrid Mass Spectrometer (Thermo Electron, Bremen, Germany) online with a nanoLC Ultimate 3000 chromatography system (Dionex, Sunnyvale, CA) through an LC EASY-Spray C18 column from Dionex. All raw LC–mass spectrometry files were processed with MaxQuant software (version 1.5.3.8, www.maxquant.org) and searched against species-specific UniProt protein sequence databases and common contaminants using the Andromeda peptide search engine with a false discovery rate of 0.01 at both peptide and protein levels.
The lists of proteins identified by mass spectrometry were analyzed as follows. First, contaminants and proteins identified by only one peptide were eliminated. Then, only those proteins with at least 10-fold higher peak surface in the experiment samples versus the controls BirA-GFP were considered as positive hits.
Protein extraction
Ten-centimeter dishes were resuspended with 300 μl of lysis buffer [50 mM tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% (w/v) Triton, 1 mM sodium orthovanadate, 10 mM sodium glycerophosphate, 50 mM sodium fluoride, 10 mM sodium pyrophosphate, 0.27 M sucrose, 0.1% (v/v) 2-mercaptoethanol, 1 mM benzamidine, and 0.1 mM phenylmethylsulfonyl fluoride (PMSF)]. After a sonication of 2 min (Bioblock Vibracell) extracts were incubated with 250 U of nuclease (Universal Nuclease Pierce) at 4°C for 30 min, centrifuged at full speed, and quantified (BCA Protein Assay Kit, Pierce).
Immunobloting
For immunobloting, 30 μg of total protein extracted or 25 μl of protein fraction (subcellular protein fractionation kit, Thermo Fisher Scientific) has been separated by SDS-PAGE 4 to 12% (bis-tris Bolt, Thermo Fisher Scientific) and transferred on nitrocellulose (Turbo transfer Bio-Rad). The signal was detected with ChemiDoc (Bio-Rad). Larvae of 6 dpf were lysed in lysis buffer [50 mM tris-HCl (pH 7.5), 1% IGEPAL, 150 mM NaCl, and a 1:20 dilution of the protease inhibitor cocktail P8340 from Sigma-Aldrich]. Samples were centrifuged (13,000g, 10 min) and resolved on 4 to 12% SDS-PAGE and transferred to polyvinylidene difluoride membranes. Membranes were blocked for 1 hour at room temperature and incubated overnight at 4°C. For detection, corresponding horseradish peroxidase–conjugated secondary antibodies (1:5000 dilution) were used. After repeated washes, the signal was detected with the enhanced chemiluminescence reagent and ChemiDoc XRS Bio-Rad.
Immunoprecipitation
For immunoprecipitation of GFP, TRAP-GFP beads were used. Lysates (0.5 to 5 mg) were incubated with 10 to 20 μl of resin for 2 hours at 4°C under gentle agitation, and the immunoprecipitates were washed three times with lysis buffer containing 0.15 M NaCl and then twice with lysis buffer. Proteins were eluted by resuspending washed immunoprecipitates in 30 μl of 1× SDS sample buffer.
Immunofluorescence
Cells were washed with PBS and fixed for 10 min with 4% paraformaldehyde in PBS. Cells were permeabilized for 20 min with PBS/0.2% Triton X-100 and blocked with PBS/0.2% Tween 20 (PBS-T) containing 5% bovine serum albumin (BSA). Coverslips were incubated for 1 hour with primary antibodies and for 1 hour with appropriate secondary antibodies coupled to Alexa Fluor 488 or 594 fluorophores (Life Technologies), before being incubated with 4′,6-diamidino-2-phenylindole (DAPI; 2 μg/ml). Pictures were acquired with Z1 (Zeiss). For high-resolution imaging, z-stacks were acquired with a Z1 (z-stack of 0.2-μm interval) equipped with a 63× oil objective (ZEISS) and controlled with Zen. Deconvolutions were then performed in conservative mode. The different channels were acquired sequentially.
Bacterial protein purification
Recombinant His6-UBA5, His6-UFM1, His6-MRE11 WT, and mutants were expressed in E. coli BL21 and purified using Ni2+–nitrilotriacetic acid (NTA) affinity chromatography. Briefly, E. coli BL21 cultures expressing His6-tagged proteins were grown in 2xTY medium at 37°C until OD 0.6. Protein expression was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and the cultures were incubated at 18°C for 16 hours. Cells were lysed in lysis buffer [25 mM tris 7.5, 300 mM NaCl, 10% glycerol, 2 mM DTT, 1 mM benzamidine, 1 mM AEBSF, and protease inhibitor cocktails (Roche)]. Lysed cells were then clarified by centrifugation at 30,000g for 30 min at 4°C. The clarified lysate was then incubated with Ni2+-NTA Agarose beads for 2 hours in binding buffer (25 mM tris 7.5, 300 mM NaCl, 10% glycerol, and 10 mM imidazole), washed extensively, and eluted using binding buffer containing 300 mM imidazole. His6-tags were then cleaved off by incubating tagged proteins with 3C protease at 4°C overnight. E. coli BL21 culture was grown and induced using IPTG as described above. A two-step affinity enrichment process was used to purify the UFL1 ligase complex. First, the clarified cell lysate was incubated with Ni2+-NTA Agarose beads for 2 hours at 4°C in binding buffer containing 25 mM tris (pH 8.0), 300 mM NaCl, 2 mM DTT, and 10 mM imidazole. After washing the beads using binding buffer containing 20 mM imidazole, the protein was eluted using binding buffer containing 300 mM imidazole. In the second step, the eluted protein was passed through a StrepTrap (GE Healthcare Life Sciences) column and eluted using binding buffer containing 2.5 mM desthiobiotin. Last, the purified complex was passed through a Superdex 200 16/60 column (GE Healthcare Life Sciences) using buffer containing 25 mM tris (pH 8.0), 300 mM NaCl, and 2 mM DTT. The purified UFL1 ligase was stored in the same buffer at −80°C. Glutathione S-transferase (GST)–tagged Ufc1 was expressed in E. coli BL21 as described above. Cells were lysed in lysis buffer containing 25 mM tris (pH 7.5), 300 mM NaCl, 10% glycerol, and 2 mM DTT using ultrasonication. Glutathione B Sepharose beads were incubated with clarified lysate for 2 hours. The beads were then washed with high salt buffer containing 25 mM tris-HCl (pH 7.5), 500 mM NaCl, 10% glycerol, and 1 mM DTT followed by low-salt buffer containing 25 mM tris-HCl (pH 7.5), 150 mM NaCl, 10% glycerol, and 1 mM DTT. The GST tag was then cleaved off by incubation with C3 protease at 4°C overnight. All proteins were further purified by size exclusion chromatography using Superdex 75 16/60 and Superdex 200 16/60 columns (GE Healthcare Life Sciences). The purified proteins were then concentrated and stored at −80°C.
In vitro UFMylation assay
Recombinant fragments of MRE11 and mutants were incubated with 0.25 μM UBA5, 5 μM UFC1, 2 μM UFL1 ligase, and 30 μM UFM1 in reaction buffer containing 50 mM Hepes (pH 7.5), 10 mM MgCl2, and 5 mM adenosine triphosphate for 1 hour at 37°C. The reaction was stopped by the addition of 3× SDS loading buffer containing 10% mercaptoethanol. The reaction products were separated on a 4 to 12% NuPAGE SDS-PAGE gel and analyzed by Immunoblotting using MRE11 antibody.
Cell sensitivity assay
For sensitivity assay to irradiation (IR), 1000 cells were plated in three replicates onto 10-cm plates in complete growth medium. After cells attached, they were treated with the indicated dose of IR. The number of colonies with >100 cells was counted. For each genotype, cell viability of untreated cells was defined as 100%. Data are represented as means ± SD from three independent experiments. For the other cytotoxicity assays, 1000 cells were plated in 96-well dishes. The next day, indicated drugs were added to the wells and plates were transferred into an IncuCyte microscope (Essen BioScience). Phase contrast pictures were acquired every 3 hours over 48 hours. Percentage of cell confluence was calculated by the Cell Player integrated software (Essen BioScience) and analyzed with GraphPad Prism software.
Homologous recombination assay
HeLa WT and HeLa UFL1 KO were transfected with 5 μg of pCBA-I-SceI and 5 μg of DR-GFP. Twenty-four hours later, cells were harvested and analyzed by fluorescence-activated flow cytometry (FACS) to examine GFP-positive cells. Cells were gated to exclude cellular aggregates, debris, and GFP-negative cells in the FSC/FSC dot plot. Gates of positive cells were set and compared with a control sample (without pCBA-I-SceI). Results were normalized with transfection efficiency using mCherry plasmids.
Zebrafish experiments
For CRISPR experiments, zebrafish lines used were Tg(mpx:eGFP)i114 (17), Tg(lcr:eGFP)cz3325 (14), Tg(runx1:GAL4)utn6 (16), and Tg(UAS:nfsB-mCherry)c264 (15). The single guide RNA (sgRNAs) were obtained from IDT and prepared using the manufacturer’s manual with a concentration of resulting duplex of ~1715 ng/μl (50 μM). After assembling the ribonucleoprotein complex, 1 nl of the mix was injected into the yolk of 1-cell stage zebrafish embryos. The sequences of the guides are ufm1 5′-TGAGAGCACACCATTCACAG CGG-3′, ufl1 5′-CCC AGAGCACTTGGGTTGAGTCG-3′, mre11a 5′-GGCAACCATGATGACCCAAC TGG-3′.
WT (NM_001001407) and K283R Mre11a were synthesized by GenScript. In vitro–transcribed RNA was obtained using mMESSAGE mMACHINE kit (Ambion) following the manufacturer’s instructions and 100 pg per egg was microinjected in combination with Cas9 ribonucleoprotein complex as indicated above.
Images were acquired at 3, 4, and 5 dpf using a Leica M205 FA fluorescence stereo microscope equipped with a DFC365FX camera (Leica) and processed using ImageJ software (http://rsb.info.nih.gov/ij/) and Photoshop CS.
Approximately 300 to 500 larvae were anesthetized in tricaine, minced with a razor blade, and incubated at 28°C for 30 min with Liberase (0.077 mg/ml; Roche), and the resulting cell suspension was passed through a 40-μm cell strainer. Cell sorting was performed on a SH800Z Cell Sorter (Sony). The experiments performed comply with the Guidelines of the European Union Council (Directive 2010/63/EU) and the Spanish RD 53/2013. Experiments and procedures were performed as approved by the Bioethical Committee of the University of Murcia (approval numbers 75/2014, 216/2014, and 395/2017).
Telomere measurement by qPCR
Genomic DNA was extracted from the cells using the ChargeSwitch Genomic DNA Micro Tissue Kit (Invitrogen). The telomeric sequences were detected through real-time PCR using 16 ng of gDNA as a template. Ribosomal protein S11 (rps11) content in each sample was used for normalization of zebrafish mRNA expression, using the comparative Ct method (2-DCt). Actin and 36b4 were used as a standard value for lymphocytes and HeLa cell line, respectively. Reaction mixtures were incubated for 15 min at 95°C, followed by 40 cycles of 15 s at 95°C, 2 min at 54°C, and lastly 15 s at 95°C, 1 min at 60°C, and 15 s at 95°C. For the standard genes, reaction mixtures were incubated for 10 min at 95°C, followed by 40 cycles of 15 s at 95°C, 1 min at 60°C, and lastly 15 s at 95°C, 1 min at 60°C, and 15 s at 95°C.
The primers used were as follows: Telom F: 5′-TTTTTGAGGGTGAGGGTGAGGGTGAGGGTGAGGGT-3′ and Telom R: 5′-TCCCGACTATCCCTATCCCTATCCCTATCCCTATCCCTA-3′; rps11 F: 5′-ACAGAAATGCCCCTTCACTG-3′ and rps11 R: 5′-GCCTCTTCTCAAAACGGTTG-3′; 36b4 F: 5′-CAGCAAGTGGGAAGGTGTAATCC-3′ and 36b4 R: 5′-CCCATTCTATCATCAACGGGTACAA-3′; Actin F: 5′-GGCACCACACCTTCTACAATG-3′ and R: 5′-GTGGTGGTGAAGCTGTAGCC-3′.
The results were normalized with the control. In all cases, each PCR was performed with triplicate samples and repeated at least with two independent samples. The differences between two samples were analyzed by the Student’s t test and between three samples by one-way analysis of variance.
Metaphase spread analysis
Cells were initially plated in DMEM containing 10% FBS until cells reached 60 to 70% confluence. All cultures were harvested following the conventional cytogenetic protocol. Briefly, the cell cultures were treated with Colcemid (0.1 μg/ml; Irvine Scientific) approximately 30 min before the initiation of harvest. For chromosome preparations, the cells were harvested following the conventional cytogenetic protocol of hypotonic treatment (75 mM KCl) and freshly prepared chilled 3:1 methanol:acetic acid fixation. This was followed by four additional fixation cycles and air-dried slide preparation. The slides were “aged” in a hot oven at 60°C over 16 hours, followed by Giemsa staining (Invitrogen). A total of 25 metaphases were scored for each culture.
Analysis of cell cycle by flow cytometry
Cells were analyzed for their respective cell cycle phase distribution using flow cytometry. Cells were trypsinized, washed with PBS + 0.2% (w/v) BSA, and resuspended in flow cytometry tubes. Cells were then fixed by 70% (v/v) ice-cold ethanol and stored at −20°C until analysis. After washing fixed cells once with PBS, ribonuclease (RNase) A (50 μg/ml) and propidium iodide (PI) (50 μg/ml) were added to the cells and incubated in the dark at room temperature (25°C) for 20 min. The live cell populations were then subjected to quantitative measurement of DNA content by flow cytometry using an LSRFortessa (BD Biosciences), the cell cycle distribution, and the percentage of G2-S-G1 cells determined by the Watson (pragmatic) modeling algorithm using FlowJo software (Tree Star).
Telomere FISH
Telomere FISH on metaphase chromosome spreads was performed according to standard protocol (44). Briefly, a hybridization mix containing 50 nM Cy3-labeled PNA telomere probe (Cy3-OO-TTAGGGTTAGGGTTAGGG 3′) in 70% formamide was spotted on the slides with metaphase spreads prepared as described above. The slides were covered with coverslips, preheated to 80°C to facilitate DNA denaturation, and incubated at room temperature in the dark for a minimum of 2 to 3 hours to allow PNA probe annealing to telomeres. Following hybridization, the slides were washed two times, 15 min each, with the solution containing 10 mM tris-HCl (pH 7.2), 70% formamide, and 0.1% BSA, and then three times, 5 min each, with the solution containing 0.1 M tris-HCl (pH 7.2), 0.15 M NaCl, and 0.08% Tween 20. After washes, the chromosome spreads were dehydrated in ethanol series: 5 min in 70, 95, and 100% ethanol, air-dried, and sealed using a coverslip and a small volume of an antifade solution containing DAPI as a DNA counterstain. The images were acquired using Zeiss fluorescence microscope and the Metamorph software as described above. The PNA probe was ordered from PNA Bio Inc. (Thousand Oaks, CA, USA), dissolved at 50 μM in formamide, and stored at −80°C.
Telomere CO-FISH
Cells were incubated with 10 μM 5-bromo-2′-deoxyuridine for 12 hours, treated with Colcemid (0.5 μg/ml) for 4 hours, and harvested. Formalin-fixed metaphase spreads were stained with Hoechst 33258 (0.5 μg/ml; Sigma-Aldrich) in PBS for 15 min at room temperature before being exposed to ultraviolet (UV) light equivalent to 5.4 × 103 J/m2. After digestion with 150 U of Exonuclease III (NEB), samples were denatured at 85°C for 3 min, and incubated sequentially with LNA G–rich (FAM) and PNA C-rich telomeric probe (Exiqon).
Telomere repeat amplification protocol
To measure telomerase activity in the cellular extracts, TRAP was performed according to the original protocol (45). Briefly, the cells were extracted on ice using standard CHAPS buffer (60 μl per 106 cells) supplemented with a cocktail of protease inhibitors (Roche) and 20 U of RNase inhibitor (Applied Biosystems) followed by centrifugation at 18,000g for 30 min at 4°C. The protein concentration in the supernatant was measured using Pierce 660-nm Protein Assay, and the aliquots equivalent to 960, 480, 240, and 120 ng of protein were used to extend 50 ng of the FAM-labeled telomerase substrate (FAM-TS oligo) in the 25-μl reaction volume. The telomerase extension products were then amplified by PCR in the same tube using the standard ACX and NT primers (45). The TSNT oligo was spiked into the reactions to serve as an internal competitor of the telomerase extension products during PCR. The TRAP product was separated in the 10% nondenaturing polyacrylamide gel at 240 V for 2 hours in 1× TRis-Borate EDTA (TBE) buffer, and the FAM fluorescence was captured using ChemiDoc MP Imaging System (Bio-Rad).
Southern blot analysis of TRFs
The cells were lysed in the TNES buffer [10 mM tris-HCl (pH 7.5), 400 mM NaCl, 100 mM EDTA, and 0.6% SDS], supplemented with Proteinase K (100 μg/ml), and incubated at 55°C for 2 hours. Genomic DNA was extracted using a phenol:chloroform (1:1) mixture, precipitated with 100% ethanol-acetate, washed with 70% ethanol, dissolved in the 10 mM tris-HCl (pH 8.0), and quantified using NanoDrop spectrophotometer. The purified DNA (12 μg of each sample) was digested with Hin fI and Rsa I (1.5 U each per microgram of DNA) at 37°C overnight and separated on 0.8% agarose gel in tris-borate buffer. The DNA was transferred from the gel on the Hybond N+ membrane using the standard Southern blotting procedure in alkaline solution (0.4 N NaOH and 0.5 M NaCl). The DNA was UV–cross-linked onto the nylon membrane and then blocked and hybridized with 32P-labeled (CCCTAA)3 probe in the Church and Gilbert buffer overnight at 42°C. After washing in the Na-phosphate buffer, the membrane was exposed to PhosphorImager screen for image acquisition. Telomere/TRF length was quantified using TeloTool (46) using manual background adjustment and the correction method for probe intensity that is appropriate for standard Southern blotting where DNA is denatured.
Telomere shortest length assay
TeSLA was performed according to the protocol described by Lai et al. (17). Briefly, 50 ng of undigested genomic DNA was ligated with an equimolar mixture (50 pM each) of the six TeSLA-T oligonucleotides containing seven nucleotides of telomeric C-rich repeats at the 3′ end and 22 nucleotides of the unique sequence at the 5′ end. After overnight ligation at 35°C, genomic DNA was digested with Cvi AII, Bfa I, Nde I, and Mse I, the restriction enzymes that create short AT or TA overhangs. Digested DNA was then treated with Shrimp Alkaline Phosphatase to remove 5′ phosphate from each DNA fragment to avoid their ligation to each other during the subsequent step of adapter ligation. Upon heat inactivation of phosphatase, partially double-stranded AT and TA adapters were added (final concentration 1 μM each) and ligated to the dephosphorylated fragments of genomic DNA at 16°C overnight. Following ligation of the adapters, genomic DNA was diluted to a final concentration of 20 pg/μl, and 2 to 4 μl of it was used in a 25-μl PCR reaction to amplify terminal fragments using primers complementary to the unique sequences at the 5′ ends of the TeSLA-T oligonucleotides and the AT/TA adapters. FailSafe polymerase mix (Epicenter) with 1× FailSafe buffer H was used to efficiently amplify G-rich telomeric sequences. Entire PCR reactions were then loaded onto the 0.85% agarose gel for separation of the amplified fragments. To specifically visualize telomeric fragments, the DNA was transferred from the gel onto the nylon membrane by Southern blotting procedure and hybridized with the 32P-labeled (CCCTAA)3 probe essentially as described above for the Southern blot analysis of TRFs. The sizes of the telomeric fragments were quantified using TeSLA Quant software (21).
Acknowledgments
We acknowledge J.-H. Guervilly and M. Modesti for discussion, advice, and comment; P. Revy for the gift of Flag-Apollo plasmid; the two students in the laboratory who helped with this work, L. Verrier and L. Ameur; the CRCM facilities including TrGET, microcopy, and FACS; and I. Fuentes and P. Martínez for excellent technical assistance and zebrafish maintenance. Proteomics analyses were supported by the Institut Paoli-Calmettes and the Centre de Recherche en Cancérologie de Marseille. Proteomic analyses were done using the mass spectrometry facility of Marseille Proteomics (marseille-proteomique.univ-amu.fr) supported by IBISA (Infrastructures Biologie Santé et Agronomie), the Canceropôle PACA, the Provence-Alpes-Côte d’Azur Région, the Institut Paoli-Calmettes, and the Fonds Européen de Développement Régional (FEDER). Funding: This work was supported by the ATIP AVENIR, plan cancer (C17004AS), Canceropôle Provence Alpes Côte d’Azur, Institut National du Cancer and Région Sud (ERC booster, LCLIC number 2018_12981), the Spanish Ministry of Science, Universities and Innovation (grant BIO2017-84702-R to V.M., PID2019-105565RJ-100 to A.B.P.O., and PI13/0234 to M.L.C., cofunded with FEDER), and the University of Murcia (PhD fellowship to E.M.-B.). Work in V.G.’s laboratory is supported by the “Ligue Nationale Contre le Cancer,” Equipe labellisée. Author contributions: L.L. designed and conducted most of the experiments in human cells. A.B.P.O. and E.M.-B. made most of the zebrafish experiments. J.P. made in vitro UFMylation assay. G.A. quantified cell sensitivity. V.A. performed FACS analysis. D.R. performed the TRF2 immunoprecipitations. D.C. made TESLA and telomere Southern blot analyses. S.A. and L.C. performed mass spectrometry experiments and data analysis. C.L. designed and supervised the study with A.B.P.O., V.G., Y.K., V.M., and M.L.C. C.L. and V.G. wrote the manuscript with input from A.B.P.O., V.M., Y.K., and M.L.C. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Table S1
REFERENCES AND NOTES
- 1.Hochstrasser M., Origin and function of ubiquitin-like proteins. Nature 458, 422–429 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Komatsu M., Chiba T., Tatsumi K., Iemura S. I., Tanida I., Okazaki N., Ueno T., Kominami E., Natsume T., Tanaka K., A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. EMBO J. 23, 1977–1986 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daniel J., Liebau E., The ufm1 cascade. Cell 3, 627–638 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tatsumi K., Yamamoto-Mukai H., Shimizu R., Waguri S., Sou Y. S., Sakamoto A., Taya C., Shitara H., Hara T., Chung C. H., Tanaka K., Yamamoto M., Komatsu M., The Ufm1-activating enzyme Uba5 is indispensable for erythroid differentiation in mice. Nat. Commun. 2, 181 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang M., Zhu X., Zhang Y., Cai Y., Chen J., Sivaprakasam S., Gurav A., Pi W., Makala L., Wu J., Pace B., Tuan-Lo D., Ganapathy V., Singh N., Li H., RCAD/Ufl1, a Ufm1 E3 ligase, is essential for hematopoietic stem cell function and murine hematopoiesis. Cell Death Differ. 22, 1922–1934 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai Y., Pi W., Sivaprakasam S., Zhu X., Zhang M., Chen J., Makala L., Lu C., Wu J., Teng Y., Pace B., Tuan D., Singh N., Li H., UFBP1, a key component of the Ufm1 conjugation system, is essential for ufmylation-mediated regulation of erythroid development. PLOS GENET. 11, e1005643 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang R., Wang H., Kang B., Chen B., Shi Y., Yang S., Sun L., Liu Y., Xiao W., Zhang T., Yang J., Zhang Y., Zhu M., Xu P., Chang Y., Jia Y., Huang Y., CDK5RAP3, a UFL1 substrate adaptor, is crucial for liver development. Development 146, dev169235 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Nahorski M. S., Maddirevula S., Ishimura R., Alsahli S., Brady A. F., Begemann A., Mizushima T., Guzmán-Vega F. J., Obata M., Ichimura Y., Alsaif H. S., Anazi S., Ibrahim N., Abdulwahab F., Hashem M., Monies D., Abouelhoda M., Meyer B. F., Alfadhel M., Eyaid W., Zweier M., Steindl K., Rauch A., Arold S. T., Woods C. G., Komatsu M., Alkuraya F. S., Biallelic UFM1 and UFC1 mutations expand the essential role of ufmylation in brain development. Brain 141, 1934–1945 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J., Yue G., Ma W., Yue G., Ma W., Zhang A., Zou J., Cai Y., Tang X., Wang J., Liu J., Li H., Su H., Ufm1-specific ligase Ufl1 regulates endoplasmic reticulum homeostasis and protects against heart failure. Circ. Heart Fail. 11, e004917 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai Y., Zhu G., Liu S., Pan Z., Quintero M., Poole C. J., Lu C., Zhu H., Islam B., Riggelen J., Browning D., Liu K., Blumberg R., Singh N., Li H., Indispensable role of the Ubiquitin-fold modifier 1-specific E3 ligase in maintaining intestinal homeostasis and controlling gut inflammation. Cell Discov. 5, 7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoo H. M., Kang S. H., Kim J. Y., Lee J. E., Seong M. W., Lee S. W., Ka S. H., Sou Y. S., Komatsu M., Tanaka K., Lee S. T., Noh D. Y., Baek S. H., Jeon Y. J., Chung C. H., Modification of ASC1 by UFM1 is crucial for ERα transactivation and breast cancer development. Mol. Cell 56, 261–274 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Walczak C. P., Leto D. E., Zhang L., Riepe C., Muller R. Y., DaRosa P. A., Ingolia N. T., Elias J. E., Kopito R. R., Ribosomal protein RPL26 is the principal target of UFMylation. Proc. Natl. Acad. Sci. U.S.A. 116, 1299–1308 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin B., Yu J., Nowsheen S., Wang M., Tu X., Liu T., Li H., Wang L., Lou Z., UFL1 promotes histone H4 ufmylation and ATM activation. Nat. Commun. 10, 1242–1213 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganis J. J., Hsia N., Trompouki E., de Jong J. L. O., DiBiase A., Lambert J. S., Jia Z., Sabo P. J., Weaver M., Sandstrom R., Stamatoyannopoulos J. A., Zhou Y., Zon L. I., Zebrafish globin switching occurs in two developmental stages and is controlled by the LCR. Dev. Biol. 366, 185–194 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davison J. M., Akitake C. M., Goll M. G., Rhee J. M., Gosse N., Baier H., Halpern M. E., Leach S. D., Parsons M. J., Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev. Biol. 304, 811–824 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamplin O. J., Durand E. M., Carr L. A., Childs S. J., Hagedorn E. J., Li P., Yzaguirre A. D., Speck N. A., Zon L. I., Hematopoietic stem cell arrival triggers dynamic remodeling of the perivascular niche. Cell 160, 241–252 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Renshaw S. A., Loynes C. A., Trushell D. M. I., Elworthy S., Ingham P. W., Whyte M. K. B., A transgenic zebrafish model of neutrophilic inflammation. Blood 108, 3976–3978 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Shay J. W., Role of telomeres and telomerase in aging and cancer. Cancer Discov. 6, 584–593 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cawthon R. M., Telomere measurement by quantitative PCR. Nucleic Acids Res. 30, e47 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alcaraz-Pérez F., García-Castillo J., García-Moreno D., López-Muñoz A., Anchelin M., Angosto D., Zon L. I., Mulero V., Cayuela M. L., A non-canonical function of telomerase RNA in the regulation of developmental myelopoiesis in zebrafish. Nat. Commun. 5, 3228 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Lai T.-P., Zhang N., Noh J., Mender I., Tedone E., Huang E., Wright W. E., Danuser G., Shay J. W., A method for measuring the distribution of the shortest telomeres in cells and tissues. Nat. Commun. 8, 1356–1314 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu H., Bhatt B., Sivaprakasam S., Cai Y., Liu S., Kodeboyina S. K., Patel N., Savage N. M., Sharma A., Kaufman R. J., Li H., Singh N., Ufbp1 promotes plasma cell development and ER expansion by modulating distinct branches of UPR. Nat. Commun. 10, 1084–1015 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwon K., Beckett D., Function of a conserved sequence motif in biotin holoenzyme synthetases. Protein Sci. 9, 1530–1539 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stracker T. H., Petrini J. H. J., The MRE11 complex: Starting from the ends. Nat. Rev. Mol. Cell Biol. 12, 90–103 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Attwooll C. L., Akpinar M., Petrini J. H. J., The mre11 complex and the response to dysfunctional telomeres. Mol. Cell. Biol. 29, 5540–5551 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng Y., Guo X., Ferguson D. O., Chang S., Multiple roles for MRE11 at uncapped telomeres. Nature 460, 914–918 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dimitrova N., de Lange T., Cell cycle-dependent role of MRN at dysfunctional telomeres: ATM signaling-dependent induction of nonhomologous end joining (NHEJ) in G1 and resection-mediated inhibition of NHEJ in G2. Mol. Cell. Biol. 29, 5552–5563 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Y., Xiao S., Zhu X.-D., MRE11-RAD50-NBS1 and ATM function as co-mediators of TRF1 in telomere length control. Nat. Struct. Mol. Biol. 14, 832–840 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Oh J., Symington L. S., Role of the Mre11 complex in preserving genome integrity. Genes (Basel) 9, 589 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z., Gong Y., Peng B., Shi R., Fan D., Zhao H., Zhu M., Zhang H., Lou Z., Zhou J., Zhu W. G., Cong Y. S., Xu X., MRE11 UFMylation promotes ATM activation. Nucleic Acids Res. 47, 4124–4135 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rai R., Hu C., Broton C., Chen Y., Lei M., Chang S., NBS1 phosphorylation status dictates repair choice of dysfunctional telomeres. Mol Cell. 65, 801–817.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lenain C., Bauwens S., Amiard S., Brunori M., Giraud-Panis M.-J., Gilson E., The Apollo 5′ exonuclease functions together with TRF2 to protect telomeres from DNA repair. Curr. Biol. 16, 1303–1310 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Zhu X. D., Küster B., Mann M., Petrini J. H., de Lange T., Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 25, 347–352 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Porro A., Feuerhahn S., Lingner J., TERRA-reinforced association of LSD1 with MRE11 promotes processing of uncapped telomeres. Cell Rep. 6, 765–776 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Chai W., Du Q., Shay J. W., Wright W. E., Human telomeres have different overhang sizes at leading versus lagging strands. Mol. Cell 21, 427–435 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Lam Y. C., Akhter S., Gu P., Ye J., Poulet A., Giraud-Panis M. J., Bailey S. M., Gilson E., Legerski R. J., Chang S., SNMIB/Apollo protects leading-strand telomeres against NHEJ-mediated repair. EMBO J. 29, 2230–2241 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu P., van Overbeek M., Rooney S., de Lange T., Apollo contributes to G overhang maintenance and protects leading-end telomeres. Mol. Cell 39, 606–617 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bailey S. M., Cornforth M. N., Kurimasa A., Chen D. J., Goodwin E. H., Strand-specific postreplicative processing of mammalian telomeres. Science 293, 2462–2465 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Faure V., Coulon S., Hardy J., Geli V., Cdc13 and telomerase bind through different mechanisms at the lagging- and leading-strand telomeres. Mol. Cell 38, 842–852 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Markiewicz-Potoczny M., Lobanova A., Loeb A. M., Kirak O., Olbrich T., Ruiz S., Lazzerini Denchi E., TRF2-mediated telomere protection is dispensable in pluripotent stem cells. Nature 589, 110–115 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruis P., Van Ly D., Borel V., Kafer G. R., Carthy A. M., Howell S., Blassberg R., Snijders A. P., Briscoe J., Niakan K. K., Marzec P., Cesare A. J., Boulton S. J., TRF2-independent chromosome end protection during pluripotency. Nature 589, 103–109 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Touzot F., Callebaut I., Soulier J., Gaillard L., Azerrad C., Durandy A., Fischer A., de Villartay J. P., Revy P., Function of Apollo (SNM1B) at telomere highlighted by a splice variant identified in a patient with Hoyeraal-Hreidarsson syndrome. Proc. Natl. Acad. Sci. U.S.A. 107, 10097–10102 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munoz I. M., Szyniarowski P., Toth R., Rouse J., Lachaud C., Improved genome editing in human cell lines using the CRISPR method. PLOS ONE 9, e109752 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lansdorp P. M., Verwoerd N. P., van de Rijke F. M., Dragowska V., Little M. T., Dirks R. W., Raap A. K., Tanke H. J., Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet. 5, 685–691 (1996). [DOI] [PubMed] [Google Scholar]
- 45.Kim N. W., Wu F., Advances in quantification and characterization of telomerase activity by the telomeric repeat amplification protocol (TRAP). Nucleic Acids Res. 25, 2595–2597 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Göhring J., Fulcher N., Jacak J., Riha K., TeloTool: A new tool for telomere length measurement from terminal restriction fragment analysis with improved probe intensity correction. Nucleic Acids Res. 42, e21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Table S1