Abstract
Photocaged cell-permeable ubiquitin probe holds promise in profiling the activity of cellular deubiquitinating enzymes (DUBs) with the much needed temporal control. Here we report a new photocaged cell-permeable ubiquitin probe that undergoes photoactivation upon 365 nm UV treatment and enables intracellular deubiquitinating enzyme profiling. We used a semisynthetic approach to generate modular ubiquitin-based probe containing a tetrazole-derived warhead at the C-terminus of ubiquitin and employed a cyclic polyarginine cell-penetrating peptide (cR10) conjugated to the N-terminus of ubiquitin via a disulfide linkage to deliver the probe into live cells. Upon 365 nm UV irradiation, the tetrazole group is converted to a nitrilimine intermediate in situ, which reacts with nearby nucleophilic cysteine residue from the DUB active site. The new photocaged cell-permeable probe showed good reactivity toward purified DUBs, including USP2, UCHL1, and UCHL3, upon photoirradiation. The Ub-tetrazole probe was also assessed in HeLa cell lysate and showed robust labeling only upon photoactivation. We further carried out protein profiling in intact HeLa cells using the new photocaged cell-permeable ubiquitin probe and identified DUBs captured by the probe using label-free quantitative (LFQ) mass spectrometry. Importantly, the photocaged cell-permeable ubiquitin probe captured DUBs specifically in respective G1/S and G2/M phases in synchronized HeLa cells. Moreover, using this probe DUBs were profiled at different time points following the release of HeLa cells from G1/S phase. Our results showed that photocaged cell-permeable probe represents a valuable new tool for achieving a better understanding of the cellular functions of DUBs.
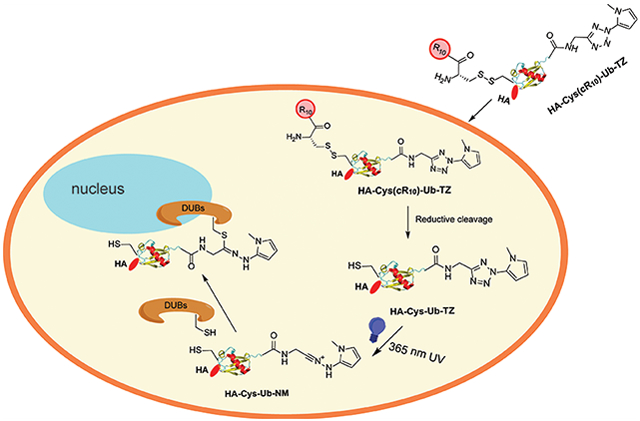
Graphical Abstract
INTRODUCTION
Ubiquitination, a reversible post-translational modification (PTM), is important for the normal cellular function of eukaryotes. Ubiquitin (Ub), a 76-amino acid protein, can be covalently attached to the ε-amino group of a lysine residue in the acceptor protein via an isopeptide bond by a three-enzyme cascade (E1–E2–E3).1 Since its discovery, protein ubiquitination has emerged as a crucial regulatory mechanism in almost every aspect of eukaryotic biological processes, including cell signaling, protein degradation, trafficking.2-4 Ubiquitination is also known to modulate enzyme catalytic activity and protein–protein interaction.5 One of the best-studied ubiquitination pathways is proteasomal protein degradation in eukaryotes.6 Recently, by exploiting the ubiquitin–proteasome system, a PROTAC (proteolysis targeting chimera) strategy was developed to degrade targeted cellular proteins by hijacking the activities of E3 ubiquitin ligases and the 26S proteasome.7-9
Deubiquitinating enzymes (DUBs) oppose ubiquitination by removing ubiquitin or polyubiquitin chains from proteins in various cellular processes. Over the past decades, DUBs have attracted increasing attention as promising new targets for treating cancer, neurodegeneration, and other human diseases.10 Activity-based probes (ABPs) are instrumental in the development of DUB inhibitors by allowing profiling of DUB activities in cell lysates.11 A comprehensive understanding of the activities and functions of DUBs in intact cells is required for the development of new therapeutics for treating a wide range of human diseases. We and others recently reported cell-permeable activity-based ubiquitin probes that enabled intracellular profiling of human DUBs.12-14 To obtain a deeper understanding of the dynamic control of cellular DUB activities with desired temporal precision, we set out to develop a photocaged cell-permeable ubiquitin probe. Precise control of probe activity with high specificity and temporal resolution through photoirradiation provides an attractive route to profile cellular protein activity under different biological contexts, providing information that cannot be obtained by gene knockout or knockdown.15-20
Huisgen and co-workers reported a photoactivated cycloaddition reaction between 2,5-diphenyltetrazole and methyl crotonate more than 50 years ago.21 A reaction mechanism was proposed. Upon photoirradiation, the diaryl tetrazole can release N2 and generate a nitrilimine dipole in situ, which undergoes a facile cyclization reaction with an alkene dipolarophile to afford a pyrazoline cycloadduct. The nitrilimine–alkene cycloaddition is an ultra rapid reaction and has been used in photoinduced protein labeling.22-29 Moreover, Huisgen et al. also reported that the nitrilimine dipole can react with thiophenol through nucleophilic addition.30 Recently, several studies reported that the nitrilimine intermediate generated from UV irradiation of tetrazole can react with nearby nucleophilic groups.31-37 Inspired by these findings, we decided to develop a Ub probe containing a C-terminal tetrazole as a photocaged warhead. Upon 365 nm UV irradiation, the probe can be activated to trap nearby nucleophilic residues in the DUB active site. We demonstrated that the DUB catalytic cysteine indeed forms a covalent adduct with the nitrilimine group in the probe and the labeling of DUBs can be triggered efficiently by 365 nm UV irradiation. With the cR10 conjugated to the N-terminal region of the Ub through a disulfide bond,12 we were able to carry out light-induced profiling of DUBs in live HeLa cells. As a proof of concept, we showed that the new photocaged cell-permeable ubiquitin probe can capture specific DUBs in G1/S and G2/M phases when synchronized HeLa cells were treated with the probe. Moreover, the probe allowed intracellular temporal profiling of DUBs at different time points (0 h, 1 h, 4 h) following the release of HeLa cells from the G1/S phase.
RESULTS AND DISCUSSION
Generation of Photocaged Cell-Permeable Ubiquitin Probe.
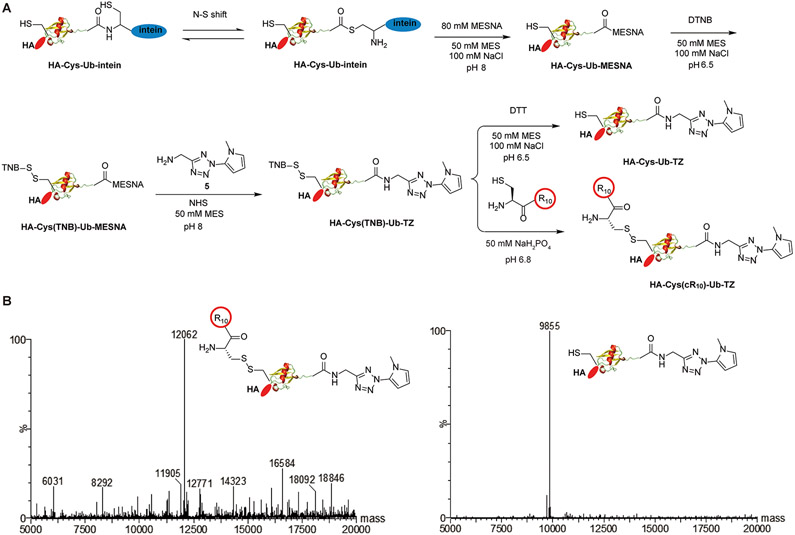
In order to introduce the tetrazole functional group at the C-terminus of ubiquitin, we designed and synthesized molecule 5 (2-(1-methyl-1H-pyrrol-2-yl)-2H-tetrazol-5-yl)-methanamine (Figure 1A, Scheme S1). The tetrazole-containing molecule 5 was generated starting from 1, which was synthesized from ethyl 2-(1-methyl-1H-pyrrol-2-yl)-2H-tetrazole-5-carboxylate following a previous publication.37 Then, LiAlH4 was used to reduce the ester in 1 to hydroxyl group to generate 2. CBr4 and PPh3 were next used to substitute the hydroxyl group in 2 with bromide to obtain molecule 3. The sodium azide was used to substitute the bromide in 3 with azide, leading to 4. Finally, 5 was obtained by traceless Staudinger reduction to reduce the azide in 4 to amine. All synthesized compounds were characterized by NMR and mass spectrometry (ESI).
Figure 1.
Generation and characterization of HA-Cys(cR10)-Ub-TZ and HA-Cys-Ub-TZ probes: (A) chemical steps of generating photocaged cell-permeable HA-Cys(cR10)-Ub-TZ and HA-Cys-Ub-TZ probes; (B) ESI-MS characterization of HA-Cys(cR10)-Ub-TZ and HA-Cys-Ub-TZ probes.
To generate photocaged cell-permeable probe HA-Cys-(cR10)-Ub-TZ (Figure 1A), site-directed mutagenesis was first used to introduce a cysteine residue between the HA-tag and the N-terminus of Ub1–75, which we named as HA-Cys-Ub. By use of an intein-based expression strategy, HA-Cys-Ub-MESNA was obtained. 12,38-41 The cysteine residue was then reacted with 5,5′- dithiobis(2-nitrobenzoic acid) (DTNB) to generate HA-Cys(TNB)-Ub-MESNA. Excess amount of 5 was then reacted with HA-Cys(TNB)-Ub-MESNA to generate HA-Cys(TNB)-Ub-TZ. HA-Cys(TNB)-Ub-TZ was subsequently reacted with a thiol-containing cR10 through disulfide bond exchange reaction to yield HA-Cys(cR10)-Ub-TZ as the final product. cR10 was generated using solid phase peptide synthesis according to a previous publication.42 Following HPLC purification, lyophilization, and refolding in MES buffer (50 mM MES, 100 mM NaCl, pH 6.5), the identity of HA-Cys(cR10)-Ub-TZ was confirmed by ESI-MS. The observed mass of 12 062 Da agrees well with the expected molecular weight of 12 063 Da (Figure 1B). cR10 will be released from ubiquitin probe upon entry into cells through reductive cleavage facilitated by the reducing environment in cells to generate HA-Cys-Ub-TZ. We thus generated HA-Cys-Ub-TZ by treating HA-Cys(TNB)-Ub-TZ with 100 mM DTT. The identity of HA-Cys-Ub-TZ was confirmed by ESI-MS. The observed mass of 9855 Da agrees well with the expected molecular weight of 9855 Da (Figure 1B).
UV Activation of Photocaged Probe.
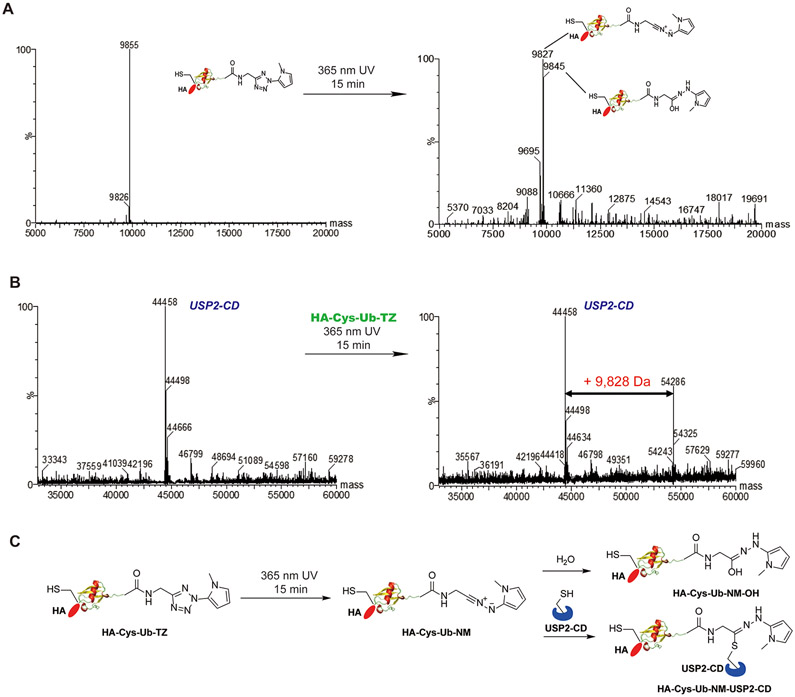
With the probes in hand, we assessed the decaging of HA-Cys-Ub-TZ upon UV irradiation using ESI-MS. First, we carried out a time-dependent UV irradiation of 15 μM HA-Cys-Ub-TZ probe for 1, 3, 5, 10, 15 min using 365 nm UV light. The ESI-MS result showed that upon irradiation of HA-Cys-Ub-TZ at 365 nm for 15 min, the tetrazole can be fully decaged as the peak of HA-Cys-Ub-TZ disappeared in mass spectrometry (Figures 2A and S1). One major peak of 9827 Da, with a decrease of 28 Da from the original Ub probe, was detected. This agrees with the formation of a nitrilimine intermediate HA-Cys-Ub-NM, as the expected mass is identical to the detected mass. Two other peaks at 9845 (or 9846) Da and 9695 Da were also detected in the mass spectrometry. We assigned 9845 (or 9846) Da as the hydroxylation product of nitrilimine (HA-Cys-Ub-NM-OH) as it was previously found to slowly react with H2O.27 The 9695 Da corresponds to the Ub species HA-Cys-Ub-COOH that was carried over from the previous ligation reaction of HA-Cys(TNB)-Ub-MESNA with 5.
Figure 2.
Photoactivation of HA-Cys-Ub-TZ probe and labeling of deubiquitinating enzyme: (A) ESI-MS characterization of HA-Cys-Ub-TZ probe with 365 nm UV treatment for 15 min; (B) ESI-MS characterization of USP2-CD incubated with HA-Cys-Ub-TZ under 365 nm UV treatment for 15 min; (C) proposed reaction mechanism of HA-Cys-Ub-TZ with deubiquitinating enzyme USP2-CD under 365 nm UV treatment.
We next assessed the reactivity of HA-Cys-Ub-TZ toward recombinantly purified USP2 catalytic domain (USP2-CD) upon 365 nm UV irradiation using ESI-MS (Figure 2B). A major peak of 44 458 Da and a minor peak of 44 498 Da were detected for the untreated USP2-CD. We then incubated 15 μM USP2-CD with 15 μM HA-Cys-Ub-TZ in MES buffer (pH 6.5). In the absence of UV irradiation, no USP2-CD-Ub adduct was detected in the ESI-MS (Figure S2). Upon 365 nm UV irradiation for 15 min, we detected two new peaks at 54 286 Da and 54 325 Da, respectively. Notably, the molecular weight of 54 286 Da represents a 9828 Da increase of 44 458 Da while 54 325 Da represents a 9827 Da increase of 44 498 Da. The expected molecular weight of HA-Cys-Ub-NM is 9827 Da and agrees well with the detected molecular weight increase of USP2-CD. Thus, the HA-Cys-Ub-TZ probe forms a covalent adduct with USP2-CD upon 365 nm UV irradiation through the generation of nitrilimine intermediate. To identify the residue that reacts with HA-Cys-Ub-NM, we subjected the labeling band of USP2-CD by HA-Cys-Ub-TZ probe to in-gel trypsin digestion followed by LC–MS/MS analysis. We were able to identify the active site cysteine-containing peptide fragment with the modification by the nitrilimine warhead as shown in Figure S3. We propose a reaction mechanism of HA-Cys-Ub-TZ with USP2-CD under 365 nm UV irradiation as shown in Figure 2C. Upon irradiation of HA-Cys-Ub-TZ with 365 nm UV light for 15 min, the nitrilimine intermediate HA-Cys-Ub-NM is formed, which reacts with the active site cysteine of USP2-CD and forms a stable covalent adduct. At the same time, nitrilimine intermediate can also undergo hydroxylation and form HA-Cys-Ub-NM-OH.27
In Vitro Labeling of Purified DUBs Using Photocaged Cell-Permeable Ubiquitin Probe.
Next, we assessed the probe activity toward USP2-CD using a 20% SDS–PAGE gel. As shown in Figure 3, with 365 nm UV irradiation for 15 min, the USP2-CD can be efficiently labeled by tetrazole probes (HA-Cys-Ub-TZ and HA-Cys(cR10)-Ub-TZ). Without UV irradiation, no discernible labeling band was detected for HA-Cys-Ub-TZ or HA-Cys(cR10)-Ub-TZ. HA-Ub-VME and HA-Ub-PA were used as positive control. Structures of the ubiquitin probes used are shown in Figure S4. By comparison of the labeling efficiency of the Ub-TZ probes with HA-Ub-VME and HA-Ub-PA, the tetrazole warhead-containing probes showed lower labeling efficiency toward USP2-CD. We also observed a slight smearing of USP2-CD labeling band by HA-Cys(cR10)-Ub-TZ upon 365 nm UV irradiation for 15 min. We attributed this to partial DTT-induced cleavage of the cR10, due to DTT carried over from the USP2-CD stock solution.
Figure 3.
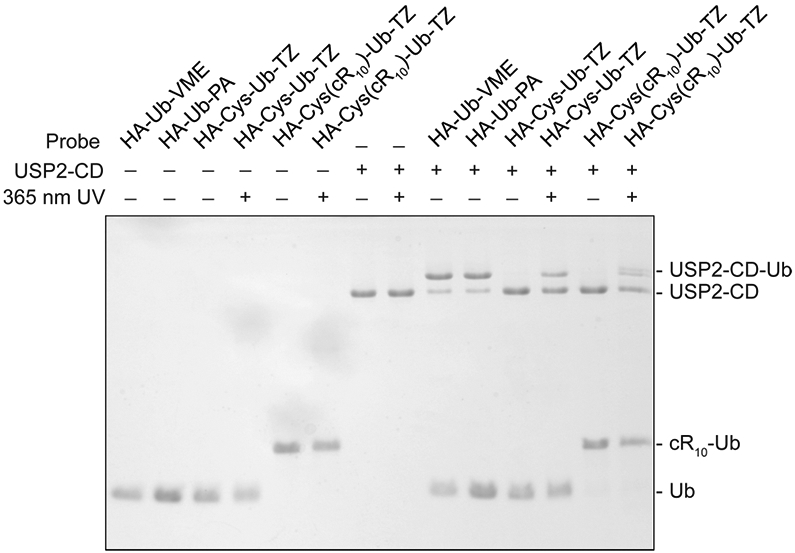
Labeling of USP2-CD by photocaged tetrazole probes. HA-Ub-VME and HA-Ub-PA were used as comparison. The labeling reaction products were resolved on a 20% SDS–PAGE gel and stained using Coomassie brilliant blue.
Moreover, we assessed the reactivity of HA-Cys-Ub-TZ probe toward USP2-CD active site cysteine mutants (USP2-CD C276S, USP2-CD C276A).43,44 While HA-Cys-Ub-TZ can label wild type USP2-CD efficiently upon 365 nm UV irradiation for 15 min, a very faint labeling band was detected for active site cysteine mutant USP2-CD C276S and USP2-CD C276A as shown in Figure S5. We speculate that the faint labeling band may be due to the labeling of nucleophilic residue close to the catalytic cysteine of USP2-CD,45 as previous studies showed that nitrilimine can react with aspartate and histidine.35-37 This result indicates that the active site cysteine of USP2-CD contributes to the labeling by tetrazole probes. The reactivity of the tetrazole probes was also evaluated using ubiquitin C-terminal hydrolase 3 (UCHL3) and ubiquitin C-terminal hydrolase 1 (UCHL1) that belong to the ubiquitin C-terminal hydrolase DUB family, different from USP2.46 The tetrazole probes showed good labeling reactivity toward UCHL3 (Figure S6) and UCHL1 (Figure S7) only upon 365 nm UV irradiation. Interestingly, compared with HA-Ub-VME and HA-Ub-PA probe, Ub-TZ probes showed varied reactivity toward DUBs. While Ub-TZ probes showed decreased reactivity toward USP2-CD than HA-Ub-VME and HA-Ub-PA probes, similar reactivity toward UCHL3 was observed for all probes. Moreover, Ub-TZ and HA-Ub-VME probes showed higher reactivity toward UCHL1 than HA-Ub-PA.
To rule out that the labeling bands of DUBs by Ub-TZ probes are due to nonspecific cross-linking, BSA was used in a similar labeling reaction. With 365 nm UV irradiation for 15 min, no discernible labeling band of BSA was detected (Figure S8). Next, we tested the reactivity of HA-Cys-Ub-TZ toward thiol-containing small molecules, glutathione and DTT, upon 365 nm UV irradiation. No glutathione adduct was detected with or without UV irradiation according to the ESI-MS results (Figure S9). However, the more reactive thiol containing reagent DTT can form covalent adduct with the HA-Cys-Ub-TZ probe upon 365 nm UV treatment for 15 min as shown in Figure S10. Since 5 mM DTT can react with HA-Cys-Ub-TZ upon 365 nm UV irradiation according to the ESI-MS result, we tested whether DTT can interfere with the labeling activity of tetrazole probe toward DUBs. Different concentrations of DTT were incubated with the mixture of USP2-CD and the probe. After 365 nm UV irradiation for 15 min, a 20% SDS–PAGE gel was used to evaluate the labeling efficiency of USP2-CD by HA-Cys-Ub-TZ probe. As high as 100 mM DTT did not prevent the labeling of USP2-CD by HA-Cys-Ub-TZ upon 365 nm UV irradiation for 15 min as shown in Figure S11. Also, we used different concentrations of probe HA-Cys-Ub-TZ (5 μM, 15 μM, 30 μM, 45 μM, 60 μM) to label USP2-CD and UCHL3. As shown in Figure S12, 15 μM or higher concentrations of the probe showed similar labeling efficiency. We thus chose 15 μM probe concentration in the following studies.
Cell Lysate and In-Cell Labeling Using Photocaged Cell-Permeable Ubiquitin Probe.
We next carried out HeLa cell lysate labeling using the photocaged HA-Cys-Ub-TZ and HA-Cys(cR10)-Ub-TZ probe detected by immunoblotting using an anti-HA antibody. We observed a number of labeling bands by HA-Cys-Ub-TZ and HA-Cys(cR10)-Ub-TZ probe upon 365 nm UV irradiation and similar labeling efficiency comparing the HA-Cys-Ub-TZ with HA-Cys(cR10)-Ub-TZ as shown in Figure 4A, indicating that the addition of cR10 did not affect the labeling efficiency in HeLa cell lysate. As expected, without UV irradiation few labeling bands were detected for cell lysate labeling using tetrazole probes HA-Cys-Ub-TZ and HA-Cys(cR10)-Ub-TZ. Notably, we observed a quite different labeling profile by Ub-TZ probes compared to those using HA-Ub-VME and HA-Ub-PA probes, suggesting different reactivities of these probes toward the cellular DUBs.
Figure 4.
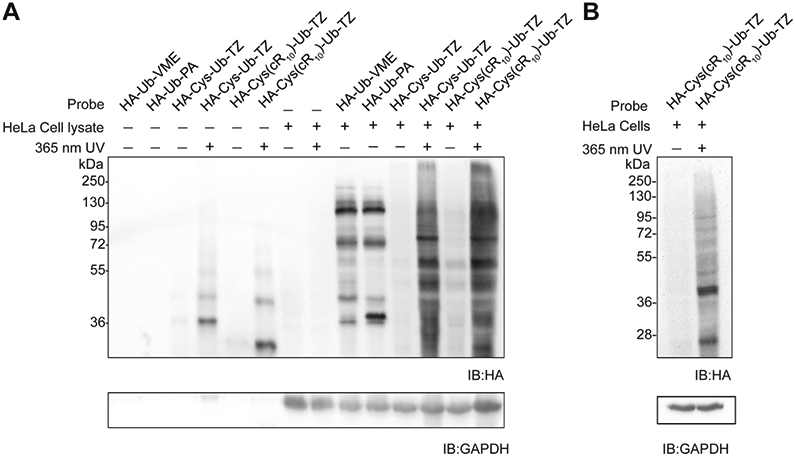
Lysate and cell-based profiling using HA-Cys(cR10)-Ub-TZ. (A) HeLa cell lysate-based profiling using 15 μM probes. Labeled proteins were resolved using 12% SDS–PAGE gel and analyzed by immunoblotting using an anti-HA antibody. GAPDH was utilized as a loading control. (B) HeLa cell-based profiling using 15 μM HA-Cys(cR10)-Ub-TZ. Labeled proteins were resolved using 12% SDS–PAGE gel and analyzed by immunoblotting using an anti-HA antibody. GAPDH was utilized as a loading control.
Next, we investigated intracellular protein profiling using the photocaged cell-permeable probe HA-Cys(cR10)-Ub-TZ detected by immunoblotting using an anti-HA antibody. Previous studies demonstrated efficient cellular uptake of cR10 cell-permeable ubiquitin probes.12,14 HeLa cells were treated with 15 μM HA-Cys(cR10)-Ub-TZ for 4 h at 37 °C with 5% CO2. After probe treatment, cells were rinsed with a trypsin–EDTA solution followed by several cold DPBS washes to remove residual probe not taken up by the cells. Then HeLa cells were irradiated with 365 nm UV for 15 min in cold DPBS buffer or without UV treatment. Cell pellet was harvested, and cells were lysed by RIPA lysis buffer. Immunoblotting using an anti-HA antibody revealed a number of labeling bands upon cellular treatment of HA-Cys(cR10)-Ub-TZ with 365 nm UV irradiation for 15 min (Figure 4B). No discernible band was observed in the cellular treatment of HA-Cys(cR10)-Ub-TZ without UV irradiation.
Proteome-Wide DUB Profiling Using Photocaged Cell-Permeable Ubiquitin Probe.
To identify the proteins captured by the photocaged cell-permeable probe, we performed intracellular proteome-wide protein profiling using label-free quantitative (LFQ) mass spectrometry. HeLa cells were incubated with 15 μM HA-Cys(cR10)-Ub-TZ probe at 37 °C with 5% CO2 for 4 h. Cells were then rinsed with a trypsin–EDTA solution followed by several cold DPBS washes to remove untransduced probe. Then HeLa cells were irradiated with 365 nm UV for 15 min in cold DPBS buffer or without any UV treatment. Cell pellet was harvested and cells were lysed by RIPA lysis buffer. Anti-HA beads were used to enrich the proteins captured by the photocaged cell-permeable probe. After elution with a 50 mM NaOH solution and neutralization with 1 M NH4HCO3 buffer, proteins were reduced by DTT and alkylated by iodoacetamide. The samples were then digested by trypsin and cleaned with C18 ZipTip and subjected to LC–MS/MS Orbitrap analysis.
Raw MS data sets were processed with MaxQuant to search against the human proteome using the built-in Andromeda peptide search engine.47-50 The MaxLFQ module within MaxQuant was used for label-free quantification of protein intensities comparing the pulldown experiments, i.e., cellular treatment of HA-Cys(cR10)-Ub-TZ probe in the presence or absence of 365 nm UV irradiation, respectively. Identified DUBs and their corresponding LFQ intensity scores are listed in Table S1. Twenty-three DUBs were identified at least once in any of the pulldowns, while others are largely housekeeping proteins.
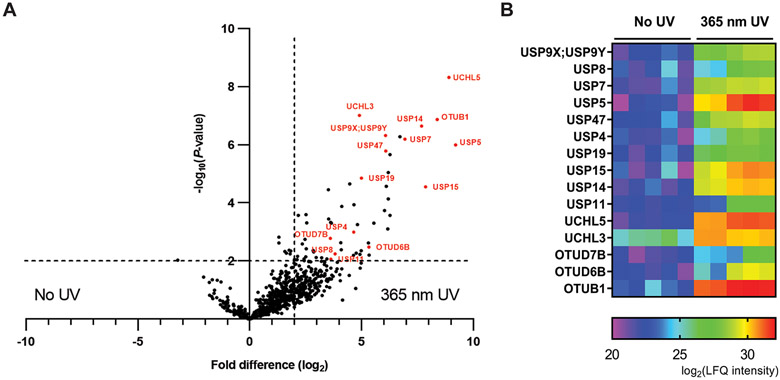
A two-sample t test was next performed in Perseus to obtain P-values of fold difference comparing LFQ intensity scores of two paired pulldown experiments. A volcano plot was generated by plotting the −log10(P-value) against log2(fold difference) of the LFQ intensities of the paired samples. Protein groups with a fold difference of 4 or greater and a P-value of <0.01 are considered significant in this experiment. These cutoffs were commonly used in similar studies.51-53 Comparison of HeLa cell pulldown samples by HA-Cys(cR10)-Ub-TZ treated with 365 nm UV to samples without UV treatment revealed 15 DUBs, labeled and colored as red in Figure 5A (USP5, UCHL5, OTUB1, USP15, USP14, USP7, USP47, USP9X;USP9Y, OTUD6B, USP19, UCHL3, USP4, USP8, USP11, OTUD7B), as significantly enriched. Notably, 39 protein groups other than DUBs were also enriched. The LFQ intensities of these proteins are listed in Table S2. Among the 39 protein groups, many are proteasome related proteins (PSMD1, PSMD11, PSMD12, PSMD2, PSMD3, PSMD5, PSMC1, PSMC2, PSMC3, PSMC4, PSMC5, PSMC6). This is likely due to co-pulldown with the proteasome DUB USP14 by the HA-Cys(cR10)-Ub-TZ probe. The MaxQuant output was also visualized using a heat map. Significantly enriched DUBs are shown as Figure 5B. A heat map of other significantly enriched protein groups is shown similarly in Figure S13. Red indicates that the protein group is highly enriched (higher LFQ intensity), while purple indicates a weak enrichment (lower LFQ intensity). Also, in a comparison of HeLa cells pulldown sample by HA-Cys(cR10)-Ub-TZ without UV irradiation to the control pulldown (no probe and no UV treatment), only UCHL3 was significantly enriched by HA-Cys(cR10)-Ub-TZ (Figure S14). This demonstrated the lack of activity of the photocaged probe in the absence of photoirradiation.
Figure 5.
Comparison of HeLa cell-based pulldown by 15 μM HA-Cys(cR10)-Ub-TZ in the presence and absence of 365 nm UV irradiation analyzed using MS-based LFQ. (A) Volcano plot of pairwise comparison of protein groups pulled down by HA-Cys(cR10)-Ub-TZ with 365 nm UV irradiation relative to no UV irradiation. Significantly enriched DUBs are colored as red and labeled. (B) Heat map representing the LFQ intensity scores of protein groups significantly enriched by HA-Cys(cR10)-Ub-TZ with 365 nm UV irradiation. Red and purple represent high and low enrichment, respectively.
Proteome-Wide DUB Profiling in G1/S and G2/M Phase Cells Using Photocaged Cell-Permeable Ubiquitin Probe.
Lastly, as a proof of concept, we carried out cell cycle dependent (G1/S phase and G2/M phase) DUB profiling using the photocaged cell-permeable ubiquitin probe HA-Cys(cR10)-Ub-TZ. We used double thymidine treatment to arrest the HeLa cells in the G1/S phase. The cyclin A level as detected by anti-cyclin A antibody upon release confirmed the effective synchronization of HeLa cells at the G1/S phase (Figure S15A). Then, double thymidine block synchronized HeLa cells were incubated with 15 μM HA-Cys(cR10)-Ub-TZ probe in cell culture media containing 2 mM thymidine at 37 °C and 5% CO2 for 4 h. Cells were then rinsed with a trypsin–EDTA solution followed by several cold DPBS wash to remove untransduced probe. Then HeLa cells were irradiated with 365 nm UV for 15 min in cold DPBS buffer or without UV treatment. The HeLa cells were next harvested, and pulldown was carried out using anti-HA magnetic beads as described above. A total of 22 DUBs were significanly enriched (USP9X, USP8, USP7, USP5, USP47, USP4, USP19, USP15, USP14, USP11, UCHL5, UCHL3, OTUD7B, OTUD6B, OTUB1, USP48, USP3, USP24, USP10, BAP1, USP36, USP16) by HA-Cys(cR10)-Ub-TZ with 365 nm UV irradiation at the G1/S phase (Figure S16). The heat map of significantly enriched DUBs at the G1/S phase is shown in Figure S17.
In another experiment, 0.5 μg/mL nocodazole was used to arrest HeLa cells at the G2/M phase, as confirmed by the cyclin B1 level of the released cells (Figure S15B). The G2/M phase HeLa cells were incubated with 15 μM HA-Cys(cR10)-Ub-TZ probe in media containing 0.5 μg/mL nocodazole at 37 °C and 5% CO2 for 4 h. Cells were then rinsed with a trypsin–EDTA solution followed by several cold DPBS washes to remove untransduced probe. Then HeLa cells were irradiated with 365 nm UV for 15 min in cold DPBS buffer or without any UV treatment. The HeLa cells were harvested, and pulldown was carried out using anti-HA magnetic beads. As shown in the volcano plot (Figure S18) and heat map (Figure S19), 18 DUBs were significantly enriched by HA-Cys(cR10)-Ub-TZ with 365 nm UV irradiation at the G2/M phase, including USP9X, USP8, USP7, USP5, USP47, USP4, USP36, USP19, USP16, USP15, USP14, USP11, UCHL5, UCHL3, OTUD7B, OTUD6B, OTUD5, OTUB1. A comparison of DUBs significantly enriched at the respective G1/S and G2/M phases and the unsynchronized HeLa cells revealed 5 DUBs (BAP1, USP48, USP3, USP24, USP10) were exclusively enriched at the G1/S phase, 1 DUB (OTUD5) was exclusively enriched at the G2/M phase, and 2 DUBs (USP16, USP36) were enriched at both G1/S and G2/M phases but not in unsynchronized HeLa cells (Figure S20). Notably, previous studies showed that BAP1 regulates G1/S transition by inhibition of HCF-1 ubiquitination,54 and USP36 stabilizes c-Myc in G1/S phase.55 Moreover, USP16 regulates kinetochore localization of Plk1 in G2/M phase to promote proper chromosome alignment and segregation.56
We also carried out intracellular DUBs profiling at different time points (0 h, 1 h, 4 h) controlled by UV-induced activation of the probe following the release of HeLa cells from G1/S phase. Double thymidine block synchronized HeLa cells were incubated with 15 μM HA-Cys(cR10)-Ub-TZ probe in media containing 2 mM thymidine at 37 °C and 5% CO2 for 4 h. Cells were then rinsed with a trypsin–EDTA solution followed by several cold DPBS washes to remove untransduced probe. For the 0 h time point treatment, the HeLa cells were irradiated immediately with 365 nm UV for 15 min in cold DPBS buffer. For the 1 and 4 h treatments, the HeLa cells were incubated with fresh media without thymidine for 1 and 4 h, respectively. Then the cells were rinsed with cold DPBS buffer before irradiation with 365 nm UV for 15 min in cold DPBS buffer. The HeLa cells were next harvested, and pulldown was carried out using anti-HA magnetic beads as described above. Our proteomic data revealed 24 DUBs were detected in total considering all three time points. The LFQ intensities of the DUBs were determined from five repeats for each time point. We observed DUBs being specifically detected at different time points, particularly USP16 was detected in 0 h and 1 h but not in 4 h. Through a pairwise comparison of DUBs captured at different time points following the release from G1/S phase using MaxQuant analysis (Table S3), we also identified several other DUBs with reliable fold difference in LFQ intensity. These include USP19 with a respective 4.2- and 2.4-fold higher activity-based capture at 1 and 4 h than 0 h and USP10 with a 3.0- and 3.7-fold lower capture at 4 h than that of 0 h and 1 h, respectively. This result supports the feasibility of temporally profiling DUB activities in live cells.
CONCLUSIONS
A better understanding of DUBs’ cellular function and regulation requires profiling of their activities in a temporally controlled manner. We herein report the development and implementation of a tetrazole photocaged cell-permeable ubiquitin probe to profile DUB activities in live cells. This was made feasible through the usage of a cyclic polyarginine cell penetrating peptide, linked to the N-terminus of Ub via a disulfide bond, released upon cellular entry due to the reducing environment. We chose tetrazole as a photocaged warhead for DUBs. The tetrazole warhead can be activated through the generation of an active nitriliamine intermediate upon 365 nm UV irradiation. Immunoblotting analysis using an anti-HA antibody demonstrated the photocaged tetrazole-warhead DUB probe can label proteins in live HeLa cells upon 365 nm UV irradiation. Using MS-based LFQ proteome-wide analysis, we identified DUBs from unsynchronized cells and cells synchronized to either G1/S phase or G2/M phase. In addition to some common DUBs, specific DUBs were identified using the photoactivated cell-permeable probe at G1/S or G2/M phase, respectively. Moreover, we profiled cellular DUB activities at different time points following the release of HeLa cells from G1/S phase.
The photocaged cell-permeable probe HA-Cys(cR10)-Ub-TZ described in this work allows the interrogation of DUB activities in live cells with temporal control. During the revision of this manuscript, Taylor et al. reported a photoinitiated ubiquitin probe allowing the labeling of DUBs in cell lysate through thiol–ene chemistry.57 Cellular pathways such as cell cycle progression, DNA damage repair and tolerance, and antiviral immune response can be conviniently studied following the treatment of human cells by extracellular stimuli. As a result, important information on the dynamic regulation of DUB activities can be garnered in live cells. This strategy can also be adapted to cellular profiling of deISGylases, deNeddylases, deSUMOylases, and other ubiquitin pathway enzymes. Our photocaged cell-permeable Ub-based DUB ABPs will enable future studies into the diverse cellular roles of human DUBs and facilitate the DUB inhibitor development.
Supplementary Material
ACKNOWLEDGMENTS
We dedicate this manuscript to the memory of Huib Ovaa, who has made tremendous contribution to the ubiquitin field, especially in the development of chemical probes and tools for investigating protein ubiquitination and deubiquitination. This work was supported in part by U.S. National Institutes of Health Grants R21 NS085509 and R01 GM129468 to Z.Z. Special thanks go to PapaNii Asare-Okai of the Mass Spectrometry Facility at the University of Delaware for the help in the proteomics data collection and valuable insights offered for data processing. Additionally, this work was made possible by the Delaware COBRE program, supported by grants from the National Institute of General Medical Sciences, NIGMS (Grants P30 GM110758, P20 GM104316), from the National Institutes of Health.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.9b12426.
Figures, tables, and methods; synthetic schemes; characterization of new compounds (PDF)
The authors declare no competing financial interest.
Contributor Information
Weijun Gui, Department of Chemistry and Biochemistry, University of Delaware, Newark, Delaware 19716, United States.
Siqi Shen, Department of Chemistry and Biochemistry, University of Delaware, Newark, Delaware 19716, United States.
Zhihao Zhuang, Department of Chemistry and Biochemistry, University of Delaware, Newark, Delaware 19716, United States.
REFERENCES
- (1).Ikeda F; Crosetto N; Dikic I What determines the specificity and outcomes of ubiquitin signaling? Cell 2010, 143 (5), 677–81. [DOI] [PubMed] [Google Scholar]
- (2).Komander D; Rape M The ubiquitin code. Annu. Rev. Biochem 2012, 81, 203–29. [DOI] [PubMed] [Google Scholar]
- (3).Clague MJ; Heride C; Urbé S The demographics of the ubiquitin system. Trends Cell Biol. 2015, 25 (7), 417–26. [DOI] [PubMed] [Google Scholar]
- (4).Yau R; Rape M The increasing complexity of the ubiquitin code. Nat. Cell Biol 2016, 18 (6), 579–86. [DOI] [PubMed] [Google Scholar]
- (5).Swatek KN; Komander D Ubiquitin modifications. Cell Res. 2016, 26 (4), 399–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Schwartz AL; Ciechanover A Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annu. Rev. Pharmacol. Toxicol 2009, 49, 73–96. [DOI] [PubMed] [Google Scholar]
- (7).Lai AC; Crews CM Induced protein degradation: an emerging drug discovery paradigm. Nat. Rev. Drug Discovery 2017, 16 (2), 101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Cromm PM; Crews CM Targeted Protein Degradation: from Chemical Biology to Drug Discovery. Cell Chem. Biol 2017, 24 (9), 1181–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Paiva SL; Crews CM Targeted protein degradation: elements of PROTAC design. Curr. Opin. Chem. Biol 2019, 50, 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Harrigan JA; Jacq X; Martin NM; Jackson SP Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat. Rev. Drug Discovery 2018, 17 (1), 57–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Gopinath P; Ohayon S; Nawatha M; Brik A Chemical and semisynthetic approaches to study and target deubiquitinases. Chem. Soc. Rev 2016, 45 (15), 4171–98. [DOI] [PubMed] [Google Scholar]
- (12).Gui W; Ott CA; Yang K; Chung JS; Shen S; Zhuang Z Cell-Permeable Activity-Based Ubiquitin Probes Enable Intracellular Profiling of Human Deubiquitinases. J. Am. Chem. Soc 2018, 140 (39), 12424–12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Hameed DS; Sapmaz A; Gjonaj L; Merkx R; Ovaa H Enhanced Delivery of Synthetic Labelled Ubiquitin into Live Cells by Using Next-Generation Ub-TAT Conjugates. ChemBioChem 2018, 19 (24), 2553–2557. [DOI] [PubMed] [Google Scholar]
- (14).Mann G; Satish G; Meledin R; Vamisetti GB; Brik A Palladium-Mediated Cleavage of Proteins with Thiazolidine-Modified Backbone in Live Cells. Angew. Chem., Int. Ed 2019, 58 (38), 13540–13549. [DOI] [PubMed] [Google Scholar]
- (15).Bardhan A; Deiters A Development of photolabile protecting groups and their application to the optochemical control of cell signaling. Curr. Opin. Struct. Biol 2019, 57, 164–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Nguyen DP; Mahesh M; Elsässer SJ; Hancock SM; Uttamapinant C; Chin JW Genetic encoding of photocaged cysteine allows photoactivation of TEV protease in live mammalian cells. J. Am. Chem. Soc 2014, 136 (6), 2240–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Abo M; Weerapana E A Caged Electrophilic Probe for Global Analysis of Cysteine Reactivity in Living Cells. J. Am. Chem. Soc 2015, 137 (22), 7087–90. [DOI] [PubMed] [Google Scholar]
- (18).Liu J; Li S; Aslam NA; Zheng F; Yang B; Cheng R; Wang N; Rozovsky S; Wang PG; Wang Q; Wang L Genetically Encoding Photocaged Quinone Methide to Multitarget Protein Residues Covalently in Vivo. J. Am. Chem. Soc 2019, 141 (24), 9458–9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wang J; Liu Y; Liu Y; Zheng S; Wang X; Zhao J; Yang F; Zhang G; Wang C; Chen PR Time-resolved protein activation by proximal decaging in living systems. Nature 2019, 569 (7757), 509–513. [DOI] [PubMed] [Google Scholar]
- (20).Courtney TM; Deiters A Optical control of protein phosphatase function. Nat. Commun 2019, 10 (1), 4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Clovis JS; Eckell A; Huisgen R; Sustmann R 1.3-Dipolare Cycloadditionen, XXV. Der Nachweis des freien Diphenylnitrilimins als Zwischenstufe bei Cycloadditionen. Chem. Ber 1967, 100, 60–70. [Google Scholar]
- (22).Ramil CP; Lin Q Photoclick chemistry: a fluorogenic light-triggered in vivo ligation reaction. Curr. Opin. Chem. Biol 2014, 21, 89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Wang Y; Rivera Vera CI; Lin Q Convenient synthesis of highly functionalized pyrazolines via mild, photoactivated 1,3-dipolar cycloaddition. Org. Lett 2007, 9 (21), 4155–8. [DOI] [PubMed] [Google Scholar]
- (24).Song W; Wang Y; Qu J; Madden MM; Lin Q A photoinducible 1,3-dipolar cycloaddition reaction for rapid, selective modification of tetrazole-containing proteins. Angew. Chem., Int. Ed 2008, 47 (15), 2832–5. [DOI] [PubMed] [Google Scholar]
- (25).Song W; Wang Y; Qu J; Lin Q Selective functionalization of a genetically encoded alkene-containing protein via “photoclick chemistry” in bacterial cells. J. Am. Chem. Soc 2008, 130 (30), 9654–5. [DOI] [PubMed] [Google Scholar]
- (26).Wang Y; Song W; Hu WJ; Lin Q Fast alkene functionalization in vivo by Photoclick chemistry: HOMO lifting of nitrile imine dipoles. Angew. Chem., Int. Ed 2009, 48 (29), 5330–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Wang XS; Lee YJ; Liu WR The nitrilimine-alkene cycloaddition is an ultra rapid click reaction. Chem. Commun. (Cambridge, U. K.) 2014, 50 (24), 3176–9. [DOI] [PubMed] [Google Scholar]
- (28).Zengeya TT; Garlick JM; Kulkarni RA; Miley M; Roberts AM; Yang Y; Crooks DR; Sourbier C; Linehan WM; Meier JL Co-opting a Bioorthogonal Reaction for Oncometabolite Detection. J. Am. Chem. Soc 2016, 138 (49), 15813–15816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Kulkarni RA; Briney CA; Crooks DR; Bergholtz SE; Mushti C; Lockett SJ; Lane AN; Fan TW; Swenson RE; Marston Linehan W; Meier JL Photoinducible Oncometabolite Detection. ChemBioChem 2019, 20 (3), 360–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Huisgen R; Sauer J; Seidel M Ringöffnungen der Azole, VI. Die Thermolyse 2.5-disubstituierter Tetrazole zu Nitriliminen. Chem. Ber 1961, 94, 2503–2509. [Google Scholar]
- (31).Tian Y; Lin Q Recent Development of Photo-Cross-Linkers as Tools for Biomedical Research. Chimia 2018, 72 (11), 758–763. [DOI] [PubMed] [Google Scholar]
- (32).Zhang Y; Liu W; Zhao ZK Nucleophilic trapping nitrilimine generated by photolysis of diaryltetrazole in aqueous phase. Molecules 2014, 19 (1), 306–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Feng W; Li L; Yang C; Welle A; Trapp O; Levkin PA UV-Induced Tetrazole-Thiol Reaction for Polymer Conjugation and Surface Functionalization. Angew. Chem., Int. Ed 2015, 54 (30), 8732–5. [DOI] [PubMed] [Google Scholar]
- (34).Li Z; Qian L; Li L; Bernhammer JC; Huynh HV; Lee JS; Yao SQ Tetrazole Photoclick Chemistry: Reinvestigating Its Suitability as a Bioorthogonal Reaction and Potential Applications. Angew. Chem., Int. Ed 2016, 55 (6), 2002–6. [DOI] [PubMed] [Google Scholar]
- (35).Herner A; Marjanovic J; Lewandowski TM; Marin V; Patterson M; Miesbauer L; Ready D; Williams J; Vasudevan A; Lin Q 2-Aryl-5-carboxytetrazole as a New Photoaffinity Label for Drug Target Identification. J. Am. Chem. Soc 2016, 138 (44), 14609–14615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Cheng K; Lee JS; Hao P; Yao SQ; Ding K; Li Z Tetrazole-Based Probes for Integrated Phenotypic Screening, Affinity-Based Proteome Profiling, and Sensitive Detection of a Cancer Biomarker. Angew. Chem., Int. Ed 2017, 56 (47), 15044–15048. [DOI] [PubMed] [Google Scholar]
- (37).Tian Y; Jacinto MP; Zeng Y; Yu Z; Qu J; Liu WR; Lin Q Genetically Encoded 2-Aryl-5-carboxytetrazoles for Site-Selective Protein Photo-Cross-Linking. J. Am. Chem. Soc 2017, 139 (17), 6078–6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Chen J; Ai Y; Wang J; Haracska L; Zhuang Z Chemically ubiquitylated PCNA as a probe for eukaryotic translesion DNA synthesis. Nat. Chem. Biol 2010, 6 (4), 270–2. [DOI] [PubMed] [Google Scholar]
- (39).Li G; Liang Q; Gong P; Tencer AH; Zhuang Z Activity-based diubiquitin probes for elucidating the linkage specificity of deubiquitinating enzymes. Chem. Commun. (Cambridge, U. K.) 2014, 50 (2), 216–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Yang K; Li G; Gong P; Gui W; Yuan L; Zhuang Z Chemical Protein Ubiquitylation with Preservation of the Native Cysteine Residues. ChemBioChem 2016, 17 (11), 995–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Gong P; Davidson GA; Gui W; Yang K; Bozza WP; Zhuang Z Activity-based ubiquitin-protein probes reveal target protein specificity of deubiquitinating enzymes. Chem. Sci 2018, 9 (40), 7859–7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Herce HD; Schumacher D; Schneider AFL; Ludwig AK; Mann FA; Fillies M; Kasper MA; Reinke S; Krause E; Leonhardt H; Cardoso MC; Hackenberger CPR Cell-permeable nanobodies for targeted immunolabelling and antigen manipulation in living cells. Nat. Chem 2017, 9 (8), 762–771. [DOI] [PubMed] [Google Scholar]
- (43).Bozza WP; Liang Q; Gong P; Zhuang Z Transient kinetic analysis of USP2-catalyzed deubiquitination reveals a conformational rearrangement in the K48-linked diubiquitin substrate. Biochemistry 2012, 51 (50), 10075–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Tencer AH; Liang Q; Zhuang Z Divergence in Ubiquitin Interaction and Catalysis among the Ubiquitin-Specific Protease Family Deubiquitinating Enzymes. Biochemistry 2016, 55 (33), 4708–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Zhang W; Sulea T; Tao L; Cui Q; Purisima EO; Vongsamphanh R; Lachance P; Lytvyn V; Qi H; Li Y; Ménard R Contribution of active site residues to substrate hydrolysis by USP2: insights into catalysis by ubiquitin specific proteases. Biochemistry 2011, 50 (21), 4775–85. [DOI] [PubMed] [Google Scholar]
- (46).Mevissen TET; Komander D Mechanisms of Deubiquitinase Specificity and Regulation. Annu. Rev. Biochem 2017, 86, 159–192. [DOI] [PubMed] [Google Scholar]
- (47).Cox J; Mann M MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 2008, 26 (12), 1367–72. [DOI] [PubMed] [Google Scholar]
- (48).Cox J; Neuhauser N; Michalski A; Scheltema RA; Olsen JV; Mann M Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res 2011, 10 (4), 1794–805. [DOI] [PubMed] [Google Scholar]
- (49).Cox J; Hein MY; Luber CA; Paron I; Nagaraj N; Mann M Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 2014, 13 (9), 2513–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Tyanova S; Temu T; Cox J The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc 2016, 11 (12), 2301–2319. [DOI] [PubMed] [Google Scholar]
- (51).Rudolph JD; Cox J A Network Module for the Perseus Software for Computational Proteomics Facilitates Proteome Interaction Graph Analysis. J. Proteome Res 2019, 18 (5), 2052–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Zhang X; Smits AH; van Tilburg GB; Ovaa H; Huber W; Vermeulen M Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat. Protoc 2018, 13 (3), 530–550. [DOI] [PubMed] [Google Scholar]
- (53).Jiang B; Wang ES; Donovan KA; Liang Y; Fischer ES; Zhang T; Gray NS Development of Dual and Selective Degraders of Cyclin-Dependent Kinases 4 and 6. Angew. Chem., Int. Ed 2019, 58 (19), 6321–6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Machida YJ; Machida Y; Vashisht AA; Wohlschlegel JA; Dutta A The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J. Biol. Chem 2009, 284 (49), 34179–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Sun XX; He X; Yin L; Komada M; Sears RC; Dai MS The nucleolar ubiquitin-specific protease USP36 deubiquitinates and stabilizes c-Myc. Proc. Natl. Acad. Sci U. S. A 2015, 112 (12), 3734–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Zhuo X; Guo X; Zhang X; Jing G; Wang Y; Chen Q; Jiang Q; Liu J; Zhang C Usp16 regulates kinetochore localization of Plk1 to promote proper chromosome alignment in mitosis. J. Cell Biol 2015, 210 (5), 727–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Taylor NC; Hessman G; Kramer HB; McGouran JF Probing enzymatic activity – a radical approach. Chemical Science 2020, 11 (11), 2967–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.