Abstract
Background
Diagnosis of active tuberculosis (ATB) currently relies on detection of Mycobacterium tuberculosis (Mtb). Identifying patients with extrapulmonary TB (EPTB) remains challenging because microbiological confirmation is often not possible. Highly accurate blood-based tests could improve diagnosis of both EPTB and pulmonary TB (PTB) and timely initiation of anti-TB therapy.
Methods
A case-control study was performed using discriminant analyses to validate an approach using Mtb-specific CD4+T-cell activation markers in blood to discriminate PTB and EPTB from latent TB infection (LTBI) as well as EPTB from PTB in 270 Brazilian individuals. We further tested the effect of human immunodeficiency virus (HIV) coinfection on diagnostic performance. Frequencies of interferon-γ +CD4+T cells expressing CD38, HLADR, and/or Ki67 were assessed by flow cytometry.
Results
EPTB and PTB were associated with higher frequencies of CD4+T cells expressing CD38, HLADR, or Ki67 compared with LTBI (all P values < .001). Moreover, frequencies of HLADR+ (P = .03) or Ki67+ (P < .001) cells accurately distinguished EPTB from PTB. HIV infection did not affect the capacity of these markers to distinguish ATB from LTBI or EPTB from PTB.
Conclusions
Cell activation markers in Mtb-specific CD4+T cells distinguished ATB from LTBI and EPTB from PTB, regardless of HIV infection status. These parameters provide an attractive approach for developing blood-based diagnostic tests for both active and latent TB.
Keywords: tuberculosis, biomarker, extrapulmonary TB, T cells, immune activation
Activation markers on Mycobacterium tuberculosis-specific CD4+ T cells distinguished active extrapulmonary and pulmonary tuberculosis from latent infection, regardless of human immunodeficiency virus status in a large case-control study, serving as an attractive approach for blood-based diagnostic tests for tuberculosis.
Diagnosis of active tuberculosis (ATB) disease currently relies on microbiologic tests such as acid-fast smear and culture and on molecular polymerase chain reaction–based assays such as GeneXpert [1] that detect Mycobacterium tuberculosis (Mtb) in patients’ sputum. However, the sensitivity of these tests can be low, particularly in extrapulmonary TB (EPTB), in which the bacillary burden is low [2]. Thus, better methods are needed to identify EPTB as well as to discriminate between EPTB, pulmonary TB (PTB), and asymptomatic latent TB infection (LTBI).
EPTB involves organs other than the lungs, such as the lymph nodes (LNs), the pleura, and meninges, and occurs with increased frequency in immunocompromised persons, including those living with human immunodeficiency virus (HIV) [3]. Diagnosis of EPTB is often more difficult than PTB because patients are more likely to have negative sputum-based tests. Indeed, radiographic-based diagnosis and empirical data on response to anti-TB therapy are commonly used to guide diagnosis of EPTB [4]. Thus, more sensitive and specific diagnostic assays for EPTB that are faster and less invasive would be a great advance for the field.
Current diagnostic tests for LTBI (eg, the tuberculin skin test and interferon gamma [(IFN)-γ] release assays) are unable to distinguish between LTBI and ATB and have decreased sensitivity in persons living with HIV. A diagnostic test that could accurately distinguish between latent and active TB, including among persons living with HIV, would substantially improve our ability to accurately diagnoses and treat these 2 disease states.
We have previously identified a blood-based assay in which Mtb-specific CD4+T cells are examined for activation and proliferation markers for diagnosis of active TB [5]. In this assay, the frequency of Mtb-specific CD4+ T cells expressing the immune activation markers CD38 and HLADR as well as the intracellular proliferation marker Ki67, can accurately identify ATB and successfully distinguish ATB from LTBI in persons from Georgia in the United States [5] and the Western Cape, South Africa [6]. Subsequently, it was demonstrated that these markers can also identify ATB in individuals living with HIV [6]. However, the performance of these diagnostic assays for identifying EPTB has not been previously explored. In the present study, we sought to extend our previously published findings on PTB to additional populations by evaluating cryopreserved peripheral blood mononuclear cell (PBMC) samples from Brazilian patients with EPTB, PTB, and LTBI. We report that CD38+ IFN-γ +, HLADR+IFN-γ +, and Ki-67+IFN-γ + CD4+ T cells successfully distinguished ATB from LTBI and EPTB from PTB, regardless of HIV status.
METHODS
Clinical Study Design
A case-control study was performed using cryopreserved PBMC samples and corresponding clinical and epidemiological data obtained from participants enrolled in a translational study performed at the Instituto Brasileiro para Investigação da Tuberculose (IBIT) and at the Hospital Especializado Octavio Mangabeira (HEOM), Salvador, Bahia, northeast Brazil, between December 2015 and January 2018. The parent study was focused on characterization of inflammatory markers in different clinical forms of TB and recruited 1792 individuals with presumptive TB at the referral primary care clinic at IBIT. These patients underwent clinical assessments and radiological (chest x-ray) examination. In addition, acid-fast bacilli (AFB) screening in sputum smears (by microscopy) and sputum cultures (Lowenstein–Jensen solid cultures) was performed in all patients. At this stage, 235 (13%) individuals were diagnosed with culture-confirmed PTB, and 215 (12%) had PTB excluded and were suspected to have EPTB. Further investigation to confirm EPTB was conducted at a TB referral site at HEOM by performing LN fine needle aspirates (with AFB screening and culture) for TB lymphadenitis and by pleural fluid drainage with lung biopsy for pleuropulmonary TB. Among the confirmed EPTB cases (n = 211, 7%), there were 102 with TB lymphadenitis, 105 with pleuropulmonary TB, 1 case of TB meningitis, 2 had spinal TB, and 1 had abdominal tuberculomas. All individuals were tested for HIV; those who tested positive had CD4+ T-cell counts and HIV viral loads (RNA copies/μL) assessed. All patients who screened positive for HIV were diagnosed at the time of study enrollment and had not been treated with antiretroviral therapy previously. The parent study also included participants who were asymptomatic contacts of TB index cases. At the time of study enrollment, individuals not living with HIV who tested positive for QuantiFERON TB Gold-in-Tube (QFT) enzyme-linked immunosorbent assay (Qiagen) were considered to have LTBI, and individuals who were QFT-negative were considered uninfected healthy controls (HCs).
At the time of study enrollment and prior to initiation of anti-TB treatment, 10 mL of venous blood was collected in sodium heparin tubes for isolation of PBMCs from a subset of participants who consented to blood collection. Cells were cryopreserved in liquid nitrogen at the biorepository of the Laboratory of Inflammation and Biomarkers, Fundação Oswaldo Cruz, Salvador, Brazil. For the immunological assays performed in the present study, selected samples from individuals with confirmed PTB and EPTB were matched on age (±5 years) and sex, with subgroups of patients living with and without HIV as well as within HCs and those with LTBI. For this study, only patients with TB lymphadenitis without pulmonary involvement were included in the EPTB group. Samples used for flow cytometry studies and characteristics of the corresponding study participants are shown in Table 1. Sample sizes were determined based on calculations of study power of 80% (alpha error, 5%) to detect differences in median frequencies of T-cell subsets >2% between active and latent TB, based on a previous study from our group [5].
Table 1.
Characteristics of the Study Participants
| Characteristic | Healthy Controls | Latent Tuberculosis Infection | PTB/HIV– | PTB/HIV+ | EPTB/HIV– | EPTB/HIV+ | P Value |
|---|---|---|---|---|---|---|---|
| N | 20 | 50 | 50 | 50 | 50 | 50 | |
| Age, median (interquartile range), y | 25 (20–32) | 27 (20–33) | 29 (19–34) | 27 (19–31) | 25 (21–29) | 26 (20–32) | >.999 |
| Male, no. (%) | 10 (50) | 25 (50) | 25 (50) | 25 (50) | 25 (50) | 25 (50) | >.999 |
| Non-white race, no. (%) | 17 (85) | 45 (90) | 45 (90) | 48 (96) | 43 (86) | 49 (98) | .198 |
| Illicit drug use, no. (%) | 2 (10) | 1 (2) | 6 (12) | 12 (14) | 3 (6) | 18 (36) | <.001 |
| Smoking, no. (%) | 1 (5) | 3 (6) | 5 (10) | 10 (20) | 3 (6) | 7 (14) | .152 |
| Alcohol abuse, no. (%) | 5 (25) | 10 (20) | 12 (14) | 22 (44) | 21 (42) | 18 (36) | .050 |
| Prior tuberculosis, no. (%) | 0 (0) | 0 (0) | 0 (0) | 1 (2) | 1 (2) | 1 (2) | NA |
| Acid-fast bacilli smear grade, no. (%) | |||||||
| 0 | 20 (100) | 50 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | <.001 |
| 1+/scanty | 0 (0) | 0 (0) | 1 (2) | 30 (60) | 15 (30) | 35 (70) | |
| 2+ | 0 (0) | 0 (0) | 24 (48) | 15 (30) | 30 (60) | 13 (26) | |
| ≥3+ | 0 (0) | 0 (0) | 25 (50) | 5 (10) | 5 (10) | 2 (4) |
The Kruskal-Wallis test was used to compare continuous variables between the groups and the distributions of age, while the Pearson χ2 test was used to compare frequencies. All tuberculosis patients had a positive culture for Mycobacterium tuberculosis. P values in bold font are statistically significant. Acid-fast bacilli smear grade was compared between PTB and EPTB groups with or without HIV using the Pearson χ2 test (the healthy control and latent tuberculosis infection groups were excluded from this analysis). Smear grade from sputum samples for PTB patients or lymph node aspirates for EPTB.
Abbreviations: EPTB, extrapulmonary tuberculosis; HIV, human immunodeficiency virus; NA, nonapplicable; PTB, pulmonary tuberculosis.
Flow Cytometry
Cryopreserved PBMCs were thawed and resuspended in 1640 Roswell Park Memorial Institute medium supplemented with 10% fetal bovine serum at 106 cells per well in 96-well plates and rested for 2 hours at 37°C in 5% CO2. Cells were washed and resuspended in complete media with Brefeldin-A (Biolegend, San Diego, CA) and Monensin (Biolegend, San Diego, CA) to block cytokine secretion and stimulated with ESAT-6 and CFP-10 peptide pools (10 μg/mL) overnight at 37°C in 5% CO2. Cells were then stained for cell surface markers with the following panel of antibodies: CD3 APC-CY7 (clone SK7), CD4 PerCp-Cy5.5 (clone L200), HLADR PE-Cy7 (clone L243), and CD38 PE (clone HB7), all from BD Biosciences. Cells were then fixed and permeabilized using the Foxp3 Fixation and Permeabilization Buffer (eBioscience). Intracellular staining was performed to detect IFN-γ Alexa Fluor 700 (clone B27) and Ki67 FITC (clone B56), all from BD Bioscience. Acquisition of stained cells was performed using a BD LSRFortessa cell analyzer (BD Bioscience, San Jose, CA) and analyzed using FlowJo software (BD Bioscience, San Jose, CA). Overall gating strategies together with representative plots are shown in Supplementary Figure 1.
Statistical Analyses
Median values with interquartile ranges were compared using the Mann-Whitney U test. Receiver operator characteristic (ROC) curve analysis was used to test the ability of frequencies of CD38+, HLADR+, and Ki67+ CD4+ T cells to distinguish ATB from LTBI and PTB from EPTB in individuals not living with HIV. The overall accuracy of the biomarkers was examined by comparing the area under the curve with C-statistics. The Fisher exact test was used to compare frequencies of virologically suppressed individuals living with HIV between PTB and EPTB groups. Spearman correlation rank analysis was performed to test the correlations between CD4+T-cell counts and frequency of IFN-γ + CD4+ T cells expressing CD38, HLADR, or Ki67 in patients living with HIV. A P value of < .05 was considered statistically significant after adjustment for multiple comparisons using the Holm-Bonferroni method. The statistical analyses were performed using GraphPad Prism 7.0 (GraphPad Software Inc) and JMP 14.0 software.
Ethics Statement
This study was conducted according to the principles expressed in the Declaration of Helsinki and approved by the Maternidade Climério de Oliveira Ethics Committee, Federal University of Bahia. Written informed consent was obtained from all participants.
RESULTS
Characteristics of the Study Population
The study groups were similar with regard to age and sex and most of the clinical and epidemiological characteristics. The highest frequency of reported illicit drug use was observed in EPTB patients living with HIV (n = 18, 36%, P < .001; Table 1). Additional analyses revealed that patients with active TB (PTB or EPTB) who had HIV coinfection exhibited lower smear grade values more frequently compared with those not living with HIV (Table 1).
Higher Frequencies of Mtb-specific CD4+ T Cells Expressing CD38, HLADR, or Ki67 in Brazilian Patients With ATB Compared With LTBI
Frequencies of IFN-γ producing Mtb-specific CD4+ T cells expressing CD38, HLADR, and Ki67 were compared between ATB and LTBI patients. We found higher frequencies of ESAT-6/CFP-10–specific IFN-γ +CD4 T-cells expressing the immune activation markers CD38 and HLADR as well as the intracellular proliferation marker Ki67 in ATB patients (Figure 1A). Next, we used cutoff values for these markers, which were established in a previous study from our group [5], and reexamined the discriminatory power in our study sample. Importantly, ROC curve analysis confirmed that each biomarker had the potential to identify ATB cases with high overall accuracy (Figure 1B). These results validated the use of the CD4+ T-cell activation markers as potential diagnostic biomarkers for ATB in this study population.
Figure 1.
Expression of CD38, HLADR, and Ki67 on IFN-γ +CD4+ T cells distinguishes active from latent tuberculosis (TB). A, Frequencies of CD38+, HLADR+, and Ki67+cells within IFN-γ + CD4+ T cells were examined in peripheral blood mononuclear cells stimulated with ESAT6-CFP10 peptide pools (10 µg/mL) from individuals with latent TB infection (LTBI; n = 50) or active TB (ATB; n = 100). Lines represent median values and interquartile ranges. Data were analyzed using the Mann-Whitney U test. B, Receiver operator characteristic (ROC) curve analyses of the frequency of CD38+, HLADR+, or Ki67+IFN-γ + CD4+ T cells were used to test accuracy to distinguish ATB from LTBI. In (A), dashed lines represent the discrimination thresholds obtained in ROC curve analysis. C, Graphs in (A) were merged. The red, dashed lines represent the discrimination threshold for each marker and show cutoff values of 18%, 60%, and 5% for CD38+IFN-γ +, HLADR+IFN-γ +, and Ki67+IFN-γ +, respectively. Such thresholds were published previously [5]. Abbreviations: AUC, the area under the curve; CI, confidence interval; IFN-γ, interferon gamma; Sens, sensitivity; Spec, specificity.
Evaluating the Predictive Value of CD38+IFN-γ+, HLADR+IFN-γ+, and Ki67+IFN-γ+CD4+ T Cells in Distinguishing EPTB from LTBI and PTB
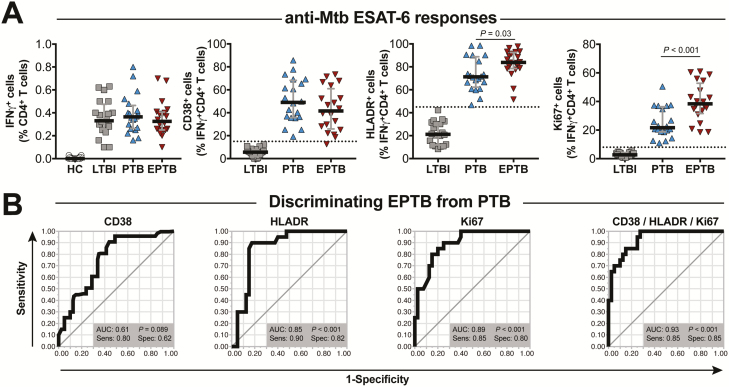
Next, we investigated whether expression of CD38, HLADR, and Ki-67 on antigen-specific IFN-γ + CD4 T cells could identify patients with EPTB. We stimulated PBMCs from individuals not living with HIV with EPTB and compared their activation profiles to those with PTB and LTBI and with HCs. CD4+ T cells from EPTB, PTB, and LTBI groups exhibited similar frequencies of IFN-γ–producing CD4+ T cells in response to in vitro stimulation with ESAT-6/CFP-10 peptides (Figure 2A). However, when the activation and proliferation markers CD38, HLADR, and Ki67 were assessed within the gate of IFN-γ + CD4+ T cells in the stimulated conditions, major differences were observed between the groups, with both disease groups displaying significantly higher frequencies of cells expressing these markers compared with the LTBI group (Figure 2A). Frequencies of CD4+ T cells expressing any of the 3 biomarkers tested were significantly higher in EPTB patients compared with those with LTBI (Figure 2A). Interestingly, the frequencies of cells expressing HLADR or Ki67, but not CD38, were higher in patients with EPTB compared with those with PTB (Figure 2A). ROC analysis confirmed that the frequency of cells expressing these markers was able to distinguish EPTB from PTB with high accuracy. The highest performance was achieved when the discriminant model was composed of data on simultaneous expression of the CD38, HLADR, and Ki67 (Figure 2B).
Figure 2.
In individuals without human immunodeficiency virus (HIV), EPTB can be distinguished from PTB based on frequencies of IFN-γ +CD4+ T-cell lymphocytes expressing HLADR+ and Ki67+. A, Frequencies of total IFN-γ +CD4+ T cells as well as of CD38+, HLADR+, and Ki67+ cells within IFN-γ + CD4+ T-lymphocytes from peripheral blood mononuclear cells stimulated with ESAT6-CFP10 peptide pools (10 µg/mL) obtained from HIV-unexposed healthy controls (n = 20), LTBI (n = 50), EPTB (n = 50), or PTB patients (n = 50). Lines represent median values and interquartile ranges. Data from EPTB and PTB were compared using the Mann-Whitney U test. B, Receiver operator characteristic curve analyses of frequencies of CD38+, HLADR+, and Ki67+ and when all parameters were considered simultaneously to distinguish EPTB and PTB patients. Abbreviations: AUC, area under the curve; EPTB, extrapulmonary tuberculosis; IFN-γ, interferon gamma; LTBI, latent tuberculosis infection; Mtb, Mycobacterium tuberculosis; PTB, pulmonary tuberculosis; Sens, sensitivity; Spec, specificity.
Frequencies of Activated Mtb-specific IFN-γ+CD4+ T Cells Expressing CD38, HLADR, and Ki67 in PTB and EPTB Are Comparable in Individuals Living With and Without HIV
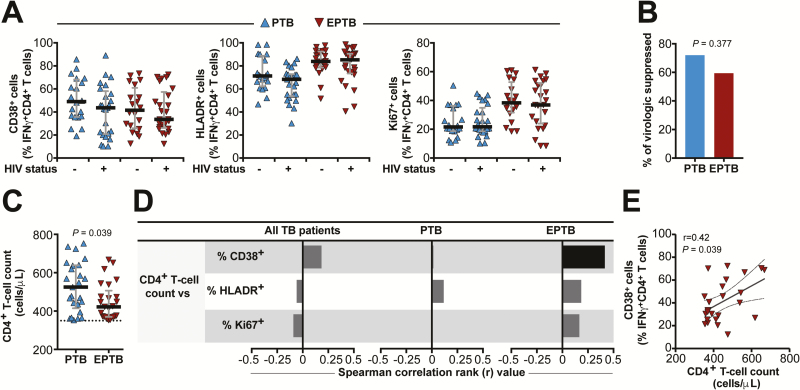
Next, we sought to determine whether the ability of Mtb-specific T cells expressing CD38, HLADR, and Ki67 to identify PTB and EPTB is altered by HIV status. We stratified the PTB and EPTB groups according to HIV status and determined the frequencies of CD38+ IFN-γ +, HLADR+ IFN-γ +, and Ki67+IFN-γ + CD4+ T cells in each group. As shown in Figure 3A, HIV status did not substantially alter the frequencies of cells expressing CD38, HLADR, or Ki67. Previous studies have suggested that progression of HIV disease gradually impairs the capacity to restrain Mtb growth, thus, favoring bacterial dissemination and extrapulmonary manifestations of TB [7, 8]. In our study, the frequency of virologically suppressed patients was just slightly lower in EPTB patients compared with those with PTB, without reaching statistical significance (Figure 3B). Nevertheless, median total CD4+ T-cell counts were lower in EPTB vs PTB (P = .039; Figure 3C), although all patients living with HIV included in the study had total CD4+ T-cell counts above 350 cells/µL and were thus not highly immunosuppressed. We further tested correlations between the total CD4+ T-cell count values and the frequencies of Mtb-specific CD4+ T cells expressing CD38, HLADR, or Ki67 in the subgroup of patients living with HIV. We found that the frequency of CD38+ Mtb-specific CD4+ T cells was positively correlated with total CD4+ T-cell counts only in EPTB patients, with all other associations not reaching statistical significance (Figure 3D). Figure 3E shows the Spearman correlation plot of such significant association (r = .42, P = .039).
Figure 3.
Frequencies of IFN-γ + CD4+ T cells expressing CD38, HLADR, or Ki67 are not substantially altered in active TB patients living with HIV. A, Frequencies of IFN-γ + CD4+ T cells expressing CD38, HLADR, or Ki67 from peripheral blood mononuclear cells stimulated with ESAT6-CFP10 peptide pools (10 µg/mL) obtained from with PTB and or EPTB stratified by HIV status (50 patients living with HIV and 50 patients not living with HIV for each disease group). Lines represent median values and interquartile ranges. Data were compared between patients living with HIV and patients not living with HIV using the Mann-Whitney U test. B, The proportion of patients living with HIV presenting with virological suppression (HIV RNA <80 RNA copies/µL) was compared between PTB and EPTB groups using the Fisher exact test. C, CD4+T-cell counts were compared between PTB and EPTB patients living with HIV using the Mann-Whitney U test. D, Spearman correlations between CD4+ T-cell counts and frequency of IFN-γ+ CD4+ T lymphocytes expressing CD38, HLADR, or Ki67 in patients living with HIV. Bars represent the strength of correlation (r values). Black bars indicate a statistically significant correlation, while gray bars were nonsignificant. E, The representative plot of correlation between CD4+T-cell counts and frequency of IFN-γ + CD4+ T cells expressing CD38 in EPTB patients living with HIV lines represent linear regression with a 95% confidence interval. Abbreviations: EPTB, extrapulmonary TB; HIV, human immunodeficiency virus; IFN-γ, interferon gamma; PTB, pulmonary TB; TB, tuberculosis.
DISCUSSION
In the present study, we explored the quantification of Mtb-specific CD4+ T cells expressing activation markers CD38 and HLADR and the intracellular proliferation marker Ki67 to diagnose ATB in a Brazilian population. Our primary results extend results from our previous studies in patients from Georgia in the United States and the Western Cape, South Africa [5], to Brazilian patients. We demonstrate that ATB patients from Brazil express higher frequencies of Mtb-specific CD4+ T cells expressing CD38, HLADR, and Ki67 when compared with individuals with LTBI. To reliably validate results from our earlier studies in our Brazilian population, we used cutoff values previously established for each marker. Using these cutoffs, we observed that quantification of cells expressing these activation markers could reliably distinguish active from latent TB in this study population with high specificity and sensitivity, as reported previously [9]. Interestingly, similar to IFN-γ release assays, IFN-γ + CD4+ T cells did not distinguish ATB from LTBI. Our findings represent an important step toward validating these 3 blood-based diagnostic biomarkers and demonstrate their high reliability when used in patient populations from different geographical locations.
Importantly, our studies also show that these biomarkers are useful for diagnosing EPTB. Extrapulmonary disease accounts for about 20%–50% of reported TB cases [10]. The most frequent locations are the pleura, LNs, bones and joints, the central nervous system, and gastrointestinal or genitourinary areas. Such anatomical sites represent a challenge for direct visualization of Mtb and therefore hinder accurate diagnosis. According to the World Health Organization, diagnosis of EPTB is currently based on a positive culture from sputum or extrapulmonary sites, positive histology, or strong clinical evidence consistent with active EPTB. Notably, 2 of these 3 criteria require invasive techniques that can delay the diagnosis of TB [11, 12]. Thus, blood-based markers are attractive for the diagnosis of EPTB and represent a desirable alternative tool for diagnosing PTB and EPTB. Several recent studies have explored nonsputum-based approaches for diagnosing ATB, including transcriptome [13], metabolome [14], proteome [15], and cellular assays [16]. We recently found that a combination of 3 plasma markers can distinguish EPTB from PTB and also from HCs in children [17]. In adults, plasma markers have also been shown to differ between PTB and EPTB [9]. However, these studies were exploratory and lacked specificity for Mtb infection. The present study significantly advances the field because it not only validates previously reported cellular biomarkers for identifying ATB in a Brazilian population [5] but also expands its potential use by accurately identifying EPTB cases. Thus, our results demonstrate that ATB patients can be identified by higher frequencies of CD38+ IFN-γ +, HLADR+ IFN-γ +, and Ki67+ IFN-γ + CD4+ and that persons with EPTB are distinguished by even higher values of such markers compared with those with either LTBI or PTB disease alone.
Diagnosis of TB in individuals living with HIV remains challenging. HIV-induced immunosuppression leads to reduced frequency of cavitation, further dampening the sensitivity of sputum-based [18] or radiographic-based assessments. Although clinical/empiric diagnoses are often used, these approaches can also be problematic since clinical manifestations of TB (pulmonary and extrapulmonary) are usually atypical. Moreover, the delayed time to diagnosis in individuals living with HIV via microbiological-based techniques directly affects clinical prognosis, with increased odds of death and treatment failure [19, 20]. Our finding that the higher frequencies of Mtb-specific CD4+ T cells expressing CD38, HLADR, and Ki67 present in ATB patients with PTB and EPTB disease compared with those with LTBI were not significantly influenced by HIV infection status is therefore of great interest. Our studies in HIV-coinfected patients corroborate previously reported observations on PTB from South Africa [21, 22] and extend the utility of these biomarkers to diagnosis of EPTB in populations living with and without HIV in Brazil. Thus, the Mtb-specific T-cell–based assay described here can be used as a blood-based diagnostic tool for both PTB and EPTB for all individuals with presumptive TB, independent of their HIV coinfection status. Although flow cytometry–based TB diagnostic tests may not be feasible in some programmatic settings, efforts are underway to develop reasonably priced commercial assays that could be used in clinical reference laboratories.
Our study has some limitations. We performed a cross-sectional investigation and examined samples obtained from a single time point, which precluded us from making conclusions about the utility of using these biomarkers for assessing response to treatment. However, we have previously reported on this in other populations [5]. The number of individuals investigated in this study was relatively small but was determined by the study power calculations, and the groups were carefully matched to reduce the influence of potential confounding factors. In addition, because we did not have drug-susceptibility test results, we were unable to investigate whether drug-susceptible vs drug-resistant Mtb strains affected immune responses; this is an important avenue for future investigation. Among the persons living with HIV who were investigated here, CD4+ T-cell counts were ≥350 cells/µL, so no highly immunosuppressed patients were examined. Future studies are necessary to test whether the performance of the biomarkers evaluated here are similar at lower CD4+ T-cell counts. All patients with PTB in our study exhibited positive AFB in sputum smears. A potential area of interest for future studies is testing the performance of these biomarkers in persons with lower mycobacterial loads (eg, AFB-negative). Despite these limitations, our study presents robust data that demonstrate that biomarkers on CD4+ T cells can distinguish EPTB from LTBI as well as from PTB cases.
In summary, we have validated a reliable, fast, noninvasive blood-based approach that accurately identified patients with active TB compared to LTBI and also distinguished PTB from EPTB—and both independent of HIV status. These observations are relevant to guide further development of point-of-care diagnosis for both active and latent TB.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Acknowledgments. The authors acknowledge study participants and also Mrs Elze Leite for logistics support.
Disclaimer. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Financial support. The study was supported by the Intramural Research Program of the Fundação Oswaldo Cruz, Departamento de Ciência e Tecnologia–Secretaria de Ciência e Tecnologia–Ministério da Saúde, Brazil (25029.000507/2013-07) and the National Institutes of Allergy and Infectious Diseases (U01-AI069923). P. S. S. M. and D. O. S. received scholarship funding from Fundação de Amparo à Pesquisa do Estado da Bahia. The work of B. B. A. was supported by grants from the National Institutes of Health (U01AI115940, U01AI069923). B. B. A. is a senior scientist from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). C. L. V. received a fellowship from CNPq. K. F. F. received a research fellowship from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (finance code 001). J. R. was supported by a Center for AIDS Research (immunology core grant P30AI050409) to Emory University and the Yerkes National Primate Research Center, US base grant through the Office of Research Infrastructure Programs (OD P51OD11132).
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1.Parrish NM, Carroll KC. Role of the clinical mycobacteriology laboratory in diagnosis and management of tuberculosis in low-prevalence settings. J Clin Microbiol 2011; 49:772–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mandal N, Anand PK, Gautam S, Das S, Hussain T. Diagnosis and treatment of paediatric tuberculosis: an insight review. Crit Rev Microbiol 2017; 43:466–80. [DOI] [PubMed] [Google Scholar]
- 3.Jones BE, Young SM, Antoniskis D, Davidson PT, Kramer F, Barnes PF. Relationship of the manifestations of tuberculosis to CD4 cell counts in patients with human immunodeficiency virus infection. Am Rev Respir Dis 1993; 148:1292–7. [DOI] [PubMed] [Google Scholar]
- 4.Purohit M, Mustafa T. Laboratory diagnosis of extra-pulmonary tuberculosis (EPTB) in resource-constrained setting: state of the art, challenges and the need. J Clin Diagn Res 2015; 9:EE01–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adekambi T, Ibegbu CC, Cagle S, et al. . Biomarkers on patient T cells diagnose active tuberculosis and monitor treatment response. J Clin Invest 2015; 125:1827–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilkinson KA, Oni T, Gideon HP, Goliath R, Wilkinson RJ, Riou C. Activation profile of Mycobacterium tuberculosis-specific CD4(+) T cells reflects disease activity irrespective of HIV status. Am J Respir Crit Care Med 2016; 193:1307–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kassu A, Mengistu G, Ayele B, et al. . Coinfection and clinical manifestations of tuberculosis in human immunodeficiency virus-infected and -uninfected adults at a teaching hospital, northwest Ethiopia. J Microbiol Immunol Infect 2007; 40:116–22. [PubMed] [Google Scholar]
- 8.Meintjes G. Management of drug-resistant TB in patients with HIV co-infection. J Int AIDS Soc 2014; 17:19508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vinhaes CL, Oliveira-de-Souza D, Silveira-Mattos PS, et al. . Changes in inflammatory protein and lipid mediator profiles persist after antitubercular treatment of pulmonary and extrapulmonary tuberculosis: a prospective cohort study. Cytokine 2019; 123:154759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Norbis L, Alagna R, Tortoli E, Codecasa LR, Migliori GB, Cirillo DM. Challenges and perspectives in the diagnosis of extrapulmonary tuberculosis. Expert Rev Anti Infect Ther 2014; 12:633–47. [DOI] [PubMed] [Google Scholar]
- 11.Hamilton CD, Swaminathan S, Christopher DJ, et al. . RePORT International: Advancing tuberculosis biomarker research through global collaboration. Clin Infect Dis 2015; 61(Suppl 3):S155–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geadas C, Stoszek SK, Sherman D, et al. . Advances in basic and translational tuberculosis research: Proceedings of the first meeting of RePORT International. Tuberculosis (Edinb) 2017; 102:55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deffur A, Wilkinson RJ, Coussens AK. Tricks to translating TB transcriptomics. Ann Transl Med 2015; 3:S43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weiner J 3rd, Parida SK, Maertzdorf J, et al. . Biomarkers of inflammation, immunosuppression and stress with active disease are revealed by metabolomic profiling of tuberculosis patients. PLoS One 2012; 7:e40221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yari S, Hadizadeh Tasbiti A, Ghanei M, Siadat SD, Yari F, Bahrmand A. Proteome-scale MDR-TB-antibody responses for identification of putative biomarkers for the diagnosis of drug-resistant Mycobacterium tuberculosis. Int J Mycobacteriol 2016; 5(Suppl 1):134–5. [DOI] [PubMed] [Google Scholar]
- 16.Qiu Z, Zhang M, Zhu Y, et al. . Multifunctional CD4 T cell responses in patients with active tuberculosis. Sci Rep 2012; 2:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albuquerque VVS, Kumar NP, Fukutani KF, et al. . Plasma levels of C-reactive protein, matrix metalloproteinase-7 and lipopolysaccharide-binding protein distinguish active pulmonary or extrapulmonary tuberculosis from uninfected controls in children. Cytokine 2019; 123:154773. [DOI] [PubMed] [Google Scholar]
- 18.Gardiner JL, Karp CL. Transformative tools for tackling tuberculosis. J Exp Med 2015; 212:1759–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das D, Dwibedi B. Delay in diagnosis among pulmonary tuberculosis patients of Rayagada District, Odisha, India. Int J Mycobacteriol 2016; 5(Suppl 1):172–3. [DOI] [PubMed] [Google Scholar]
- 20.Osei E, Akweongo P, Binka F. Factors associated with DELAY in diagnosis among tuberculosis patients in Hohoe municipality, Ghana. BMC Public Health 2015; 15:721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.du Bruyn E, Peton N, Esmail H, Howlett PJ, Coussens AK, Wilkinson RJ. Recent progress in understanding immune activation in the pathogenesis in HIV-tuberculosis co-infection. Curr Opin HIV AIDS 2018; 13:455–61. [DOI] [PubMed] [Google Scholar]
- 22.Lesosky M, Rangaka MX, Pienaar C, et al. . Plasma biomarkers to detect prevalent or predict progressive tuberculosis associated with human immunodeficiency virus-1. Clin Infect Dis 2019; 69:295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.