Abstract
Objectives
To determine the time to reverse transcription-polymerase chain reaction (RT-PCR) negativity after the first positive RT-PCR test, factors associated with longer time to RT-PCR negativity, proportion of children seroconverting after proven severe acute respiratory syndrome coronavirus 2 infection, and factors associated with the lack of seroconversion.
Study design
The Epidemiological Study of Coronavirus in Children of the Spanish Society of Pediatrics is a multicenter study conducted in Spanish children to assess the characteristics of coronavirus disease 2019. In a subset of patients, 3 serial RT-PCR tests on nasopharyngeal swab specimens were performed after the first RT-PCR test, and immunoglobulin G serology for severe acute respiratory syndrome coronavirus 2 antibodies was performed in the acute and follow-up (<14 and ≥14 days after diagnosis) phase.
Results
In total, 324 patients were included in the study. The median time to RT-PCR negativity was 17 days (IQR, 8-29 days), and 35% of patients remained positive more than 4 weeks after the first RT-PCR test. The probability of RT-PCR negativity did not differ across groups defined by sex, disease severity, immunosuppressive drugs, or clinical phenotype. Globally, 24% of children failed to seroconvert after infection. Seroconversion was associated with hospitalization, persistence of RT-PCR positivity, and days of fever.
Conclusions
Time to RT-PCR negativity was long, regardless of the severity of symptoms or other patient features. This finding should be considered when interpreting RT-PCR results in a child with symptoms, especially those with mild symptoms. Seroprevalence and postimmunization studies should consider that 11 in 4 infected children fail to seroconvert.
Keywords: COVID-19, SARS-CoV-2, children, serology, RT-PCR, dynamics, seroconversion
Abbreviations: COVID-19, Coronavirus disease 2019; Ct, Cycle threshold; EPICO-AEP, Epidemiological Study of Coronavirus in Children of the Spanish Society of Pediatrics; IgG, immunoglobulin G; MIS-C, Multisystem inflammatory syndrome in children; RT-PCR, Reverse transcription-polymerase chain reaction; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2
Studies of serial respiratory viral load and serum antibody responses from children infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are limited.1 SARS-CoV-2 detection by reverse transcription-polymerase chain reaction (RT-PCR) testing of respiratory samples is used widely to diagnose and monitor infection. A better understanding of the dynamics of RT-PCR and serological testing in children with SARS-CoV-2 infection (coronavirus disease 2019 [COVID-19]) may help to inform healthcare decisions, including quarantine strategies and duration.
The severity and progression of SARS-CoV-2 infection has multifactorial associations, including host immunity, and an adequate antibody response might confer protection to future SARS-CoV-2 infection. The dynamics of the antibody response has been well-described in adults.2 Studies of multisystem inflammatory syndrome in children (MIS-C) suggest that not all patients develop detectable antibodies despite a strong dysregulated immune response.3 Knowing the proportion of children who do not develop a detectable antibody response and the factors related to the absence of seroconversion might be useful for the diagnosis of post-COVID-19 conditions and reinfections.
The aims of the present study were to determine the time to RT-PCR negativity after the first positive RT-PCR test result, to investigate factors associated with a delayed RT-PCR negative result, to assess the rate of seroconversion after proven SARS-CoV-2 infection, and to examine the factors associated with the absence of seroconversion.
Methods
The Epidemiological Study of Coronavirus in Children of the Spanish Society of Pediatrics (EPICO-AEP) is a multicenter cohort study to assess the characteristics of COVID-19 in Spanish children. The registry includes patients aged 0-18 years evaluated in any 1 of the 76 hospitals of the network, with positive SARS-CoV-2 RT-PCR test or a diagnosis of MIS-C following the World Health Organization criteria.3 Children were tested for SARS-CoV-2 by RT-PCR if they presented with symptoms consistent with SARS-CoV-2 infection. The study was approved by the Ethics Committee of Hospital 12 de Octubre, Madrid (code 20/101), and the other participating hospitals. Patients were enrolled after signed or verbal consent from parents or guardians and patients older than 12 years.
For the present substudy, patients with SARS-CoV-2 infection confirmed by RT-PCR were offered serial follow-up visits with 3 associated RT-PCR testing episodes if the hospital and the patient's situation permitted. Additionally, the interval recommended between serial RT-PCR testing episodes was 1 week, but the timing depended on logistics, the situation at the center, and the participant. Serology was performed in the acute phase (<14 days after first positive RT-PCR test result) and in the follow-up phase (≥14 days after first positive RT-PCR test result).
Eligible participants for this study were those patients with a positive RT-PCR at diagnosis from March 1, 2020, to March 18, 2021, who had at least 2 PCR tests in follow-up. Patients with a diagnosis of MIS-C were excluded because the interval between the SARS-CoV-2 infection and the diagnosis of MIS-C usually is 4-6 weeks and most children with MIS-C are not diagnosed with acute infection.
Laboratory Methods
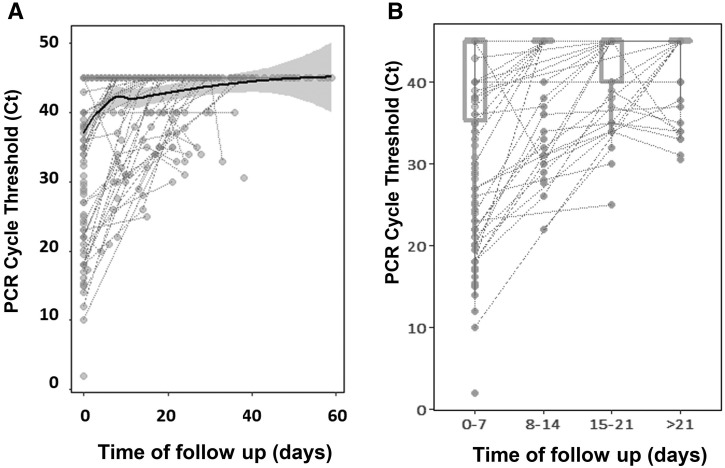
Respiratory samples were obtained from nasopharyngeal swabs. RT-PCR was performed according to standardized techniques at each center. RT-PCR was considered positive according to the limits provided by the manufacturers. For analysis purposes (Figure 1 ), negative was considered as 45 cycle thresholds (Ct). In all centers, immunoglobulin G (IgG) antibodies against the SARS-CoV-2 surface S1 domain of the spike protein or against the internal nucleocapsid protein were measured in ethylenediaminetetraacetic acid plasma samples using standardized commercial techniques.
Figure 1.
A, Ct over time. The line represents an inference of the trajectory of Cts until negativity. Negative values are presented as a Ct of 45. Negative values beyond 60 days are not presented. B, Box-and-whiskers distributions of Ct across weeks. Most of the samples had a high Ct value. At diagnosis, the median Ct was 38.5 (IQR, 35.5-40.0).
Data Management and Statistical Analyses
An electronic case report form was built, and the responsible physician of each center completed the form with clinical data and laboratory test results. Data included age, sex, comorbidities, immunosuppressant medication, date of diagnosis, syndromic diagnosis, and serial RT-PCR and serology test results.
Baseline characteristics were described using frequencies for categorical variables and medians (IQR) for continuous variables, both in the total population and after stratification based on RT-PCR test result. The χ2 or Fisher exact test (<5 frequencies) was applied to assess differences across groups for categorical variables, and the Mann-Whitney test was used for continuous variables.
For the time-to-event analysis, the time to RT-PCR negativity was analyzed using Kaplan-Meier estimates according to sociodemographic and clinical variables including sex, age, days of fever, diagnostic syndrome and severity, and the log-rank test was used for comparison of curves. The time to RT-PCR negativity and association with potential risk factors was assessed using a generalized multivariable linear model. Variable selection for the model was performed through the stepwise Akaike Information Criterion. Two-tailed P values were considered significant when the P values was less than .05.
Locally estimated scatterplot smoothing splines were fitted to the Ct curves to plot the dynamics of extinct viral load along time to follow-up. For this plot, a Ct of 45 was considered as negative or undetectable. R software was used for all analyses (R Development Core Team, Vienna, Austria, 2009).
Results
RT-PCR Test Results
A total of 324 children from 50 centers were included in the present study and followed for a median of 20 days (IQR, 13-32; range, 0-120 days). The median age was 4.5 years (IQR, 0.6-11.6 years), 188 (58%) children were male, and the median days of fever at diagnosis was 2 (IQR, 0-4). All patients had 2 (100%), 3 (162/324 [50%]), or 4 (86/324 [26.5%]) RT-PCR tests performed during follow-up. The median time between serial RT-PCR tests was 11 days (IQR, 7.0-17.0 days).
A total of 248 of the 324 patients (76.5%) had a negative RT-PCR test result in 1 or more of the serial samples after the first positive RT-PCR; 27 of the 324 (8.3%) did not have a negative result in any of 3 serial samples after the first positive RT-PCR; 49 of the 324 (15.1%) did not complete the protocol of 3 serial samples and were still positive when they were lost to follow-up. Among 27 children who completed 3 sampling episodes but did not achieve RT-PCR negativity, the median time of follow-up was 27 days (IQR, 22-34 days). Among children who did not have 3 tests performed and did not achieve negativity and were lost to follow-up, the median time to follow-up was 14 days (IQR, 9-16 days).
Overall, the median time to RT-PCR negativity was 17 days (IQR, 8.0-29.2). A total of 89 of 248 participants (35.8%) had persistent positive RT-PCR 4 weeks after the first RT-PCR. Notably, 6 children remained positive 90 days after diagnosis. The Ct values increased with time since first positive RT-PCR test.
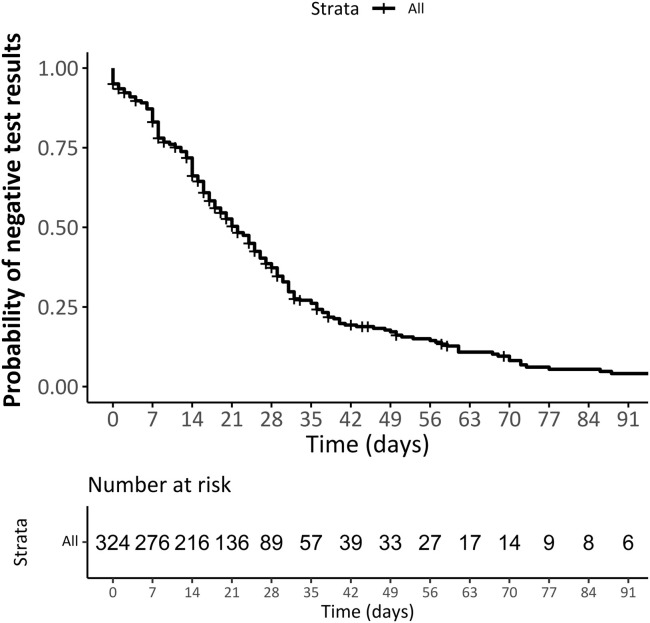
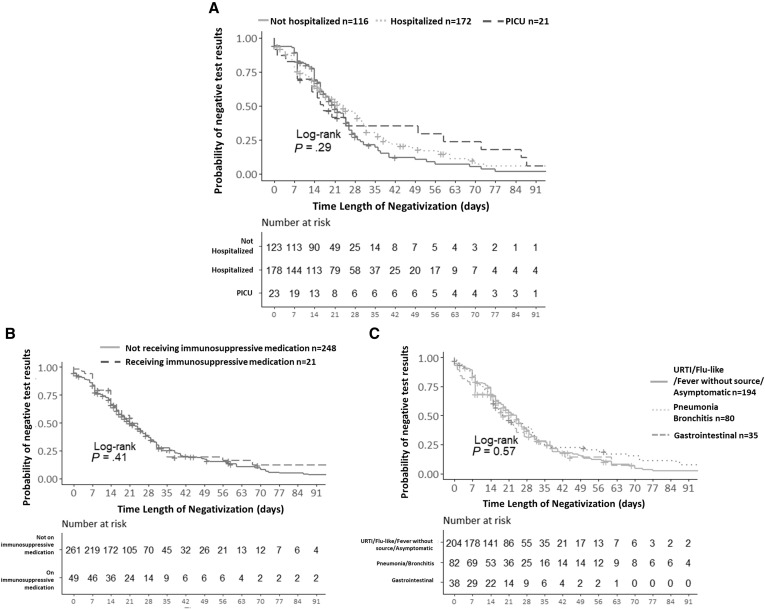
According to the Kaplan-Meier function estimate, the median time from first positive to first negative test result was 22 days (95% CI, 19-25). The probability of remaining positive at 14 days was 0.66 (95% CI, 0.61-0.71), at 21 days was 0.5 (95% CI, 0.45-0.56), and at 28 days 0.37 (95% CI, 0.31-0.43) (Figure 2; available at www.jpeds.com). The probability of a RT-PCR negative result did not differ between groups with different severity (hospitalized vs not hospitalized), receiving or not receiving immunosuppressive medication, or across different clinical phenotypes at presentation (Figure 3 ).
Figure 2.
Probability of RT-PCR negativity by A, severity, B, immunosuppressive medication or C, phenotype. PICU, pediatric intensive care unit; URTI, upper respiratory tract infection.
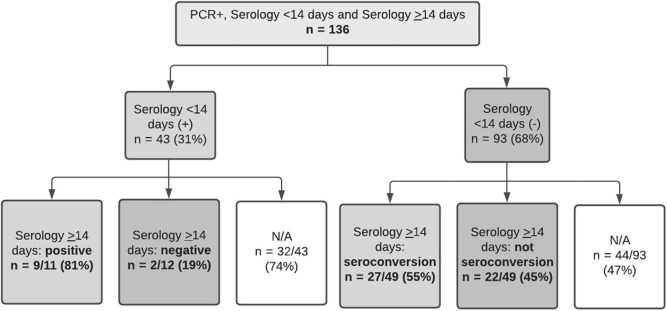
Figure 3.
Flowchart of patients with serologic follow-up. Overall, at least 22/93 (23.6%) children with a recent RT-PCR positive test and available serum specimens did not develop detectable IgG SARS-CoV-2 antibodies. N/A, not applicable.
A Cox multivariable analysis was performed to investigate whether confounding might have attenuated the suggested association with severity of disease. The covariates included in the raw model were sex, phenotype, age, days of fever, and severity. However, according to the backward stepwise selection method, only sex was selected to be included in the final best fitting model (coefficient, 0.85; 95% CI, 0.66-1.1; P = .229). We did not find any factor influencing the probability of achieving a negative RT-PCR test result. There were no differences between patients who achieved or did not achieve negativity during the follow-up period by sex, age, days of fever at diagnosis, clinical phenotype at presentation, immunosuppressive medication, hospitalization, or need for pediatric intensive care unit admission or high-flow oxygen therapy.
The Ct values at diagnosis were available for 84 of 275 patients (30%). Most samples had a high Ct value. At diagnosis, the median Ct was 38.5 (IQR, 35.5-40.0). The Ct information of all the serial RT-PCRs was available for 46 of 84 patients (54%). The trajectory line of Cts until negativity was reached quickly (around day 5) (value of 45) (Figure 1). After day 8, the median Ct value of the remaining positive patients was above the threshold of positive Ct. We observed a weak, but significant, positive correlation between initial Ct value and time to negativity (R = −0.35; P = .03)—the higher the Ct, the longer the time to negativity.
Serology
Among 324 children, a total of 136 participated in serology protocol: a first IgG serology test performed within 14 days from the first positive RT-PCR, and a follow-up IgG serology test 14 or more days after the first positive RT-PCR test result. Only 59 of 136 patients (43.3%) completed the protocol with both tests. The time between the first and second IgG test was 28.0 days (IQR, 25.5-34.5; range, 14-45 days).
The first SARS-CoV-2 IgG test was positive within the first 14 days in 43 of 136 children (31.6%) and was negative in 93 of the 136 children (68.4%). Of those with the first negative IgG test result, the follow-up IgG was available for 49 of 93 children (52.7%). Follow-up IgG was positive in 27 of 49 children (55.1%); therefore, 22 of these 49 (44.9%) did not seroconvert over the period of observation (Figure 4 ). For those who seroconverted, median time from first IgG negative to second IgG positive test was 18 days (IQR, 16.5-31.5). Overall, at least 22 of 93 children (23.6%) with a recent RT-PCR positive test result and available serologies did not develop detectable IgG antibodies against SARS-CoV-2.
Figure 4.
Probability of RT-PCR test negativity in all 324 participants.
We compared the 27 children who seroconverted with the 22 who did not (Table ). We excluded from this analysis those patients with a positive IgG less than 14 days after the positive RT-PCR to exclude the possibility of a positive IgG from a prior infection 14 days previously. Absence of seroconversion was observed in 10 of 32 hospitalized patients (31.2%) and in 12 of 17 nonhospitalized patients (70.6%; P = .02). Similarly, among patients who never achieved a negative RT-PCR test result during the study follow-up, all patients seroconverted, whereas among patients who achieved a negative RT-PCR test result during the follow-up, 22 of the 40 (55%) did not seroconvert (P = .02). Patients who seroconverted had more days of fever (median, 3; IQR 1-4) than patients who did not seroconvert (median, 0; IQR, 0-3; P = .017).
Table.
Characteristics of initially IgG-negative patients who had follow-up IgG serology
| Characteristics | Total |
Not seroconverted |
Seroconverted |
P value | OR |
|---|---|---|---|---|---|
| (n = 49) | (n = 22) | (n = 27) | [95% CI] | ||
| Sex | .827 | ||||
| Male | 27 (55.1%) | 13 (59.1%) | 14 (51.9%) | Ref. 1.00 | |
| Female | 22 (44.9%) | 9 (40.9%) | 13 (48.1%) | 1.33 [0.42-4.30] | |
| Age (months) | NA = 1 (2.0%) | NA = 0 (0.0%) | NA = 1 (3.7%) | .041 | |
| Median [IQR] | 38.3 [1.83-130] | 93.7 [13.9-163] | 10.1 [1.23-78.4] | 0.99 [0.98-1.00] | |
| Fever days | NA = 9 (18.4%) | NA = 6 (27.3%) | NA = 3 (11.1%) | .017 | |
| Median [IQR] | 2.50 [0.00-4.00] | 0.00 [0.00-3.00] | 3.00 [1.00-4.00] | 1.56 [1.04-2.35] | |
| Immunosuppressive medication | 1.000 | ||||
| No | 45 (91.8%) | 20 (90.9%) | 25 (92.6%) | Ref. 1.00 | |
| Yes | 4 (8.16%) | 2 (9.09%) | 2 (7.41%) | 0.80 [0.08-8.29] | |
| Severity | .020 | ||||
| Not hospitalized | 17 (34.7%) | 12 (54.5%) | 5 (18.5%) | Ref. 1.00 | |
| Hospitalized (PICU and normal wards) | 32 (65.3%) | 10 (45.5%) | 22 (81.5%) | 5.01 [1.43-20.1] | |
| Cycle threshold of RT-PCR at diagnosis | NA = 31 (63.3%) | NA = 10 (45.5%) | NA = 21 (77.8%) | .589 | |
| Median [IQR] | 38.5 [35.5-40.0] | 38.0 [36.5-40.0] | 40.0 [11.5-40.0] | 0.96 [0.90-1.03] | |
| PICU or high-flow oxygen necessity | NA = 1 (3.1%) | 1.000 | |||
| No | 44 (91.7%) | 20 (90.9%) | 24 (92.3%) | Ref. 1.00 | |
| Yes | 4 (8.33%) | 2 (9.09%) | 2 (7.69%) | 0.84 [0.08-8.64] | |
| Time between first (−) and second serology (+) | - | ||||
| Median [IQR] Median [Range] |
- | - | 28.0 [25.5-34.5] 28.0 (14.0-45.0) |
- | |
| RT-PCR negativity | .022 | ||||
| No | 6 (13.0%) | 0 (0.00%) | 6 (25.0%) | ||
| Yes | 40 (87.0%) | 22 (100%) | 18 (75.0%) | - | |
| Clinical phenotype | .744 | ||||
| Asymptomatic | 5 (10.2%) | 3 (13.6%) | 2 (7.41%) | Ref. 1.00 | |
| URTI/flu-like/fever without source | 30 (61.2%) | 12 (54.5%) | 18 (66.7%) | 2.15 [0.29-20.8] | |
| Pneumonia/bronchitis | 7 (14.3%) | 4 (18.2%) | 3 (11.1%) | 1.11 [0.09-14.8] | |
| Gastrointestinal | 7 (14.3%) | 3 (13.6%) | 4 (14.8%) | 1.86 [0.17;25.4] | |
PICU, pediatric intensive care unit; NA, not available; PICU, pediatric intensive care unit; URTI, upper respiratory tract infection. Values in bold are statistically significant (P-value <.05).
Discussion
In our study, after diagnosis with clinical symptoms, the median time to RT-PCR test negativity in children with SARS-Cov-2 infection was 17 days, irrespective of the type or severity of presenting features, and 35% of children remained RT-PCR positive more for more than 1 month after the first positive RT-PCR. We also showed that at least 25% of patients did not develop SARS-CoV-2 IgG antibodies after confirmed infection, particularly those not hospitalized and those with early SARS-CoV-2 RT-PCR clearance.
Time to RT-PCT negativity did not differ by sex, disease severity, receipt of immunosuppressive drugs, or clinical phenotype. Kaplan-Meier analysis indicated a trend for patients admitted to the pediatric intensive care unit remain positive for a slightly longer time than others, but the CIs overlapped, and the P value was not significant.
In adults with SARS-CoV-2 infection, RT-PCR positivity can persist for weeks after the resolution of symptoms; however, the median time to RT-PCR negativity is unclear. Some studies have reported a median time from 12 to 35 days, but there is large variability depending on age, the severity of the disease and the sample type.4 , 5 In a study of 68 children, patients in the 0- through 5-year age group had a median of 22 days to RT-PCR negativity, and patients aged 6 through 15 years had a median of 32 days, compared with 18 days for patients 16-22 years of age.6
In our study, children seemed to have more rapid clearance than adults, which might be related to the high median Ct observed (38; IQR, 35.5-40.0), indicating a low viral load. In adults, the median initial Ct value has been reported to be 28 (95% CI, 27-28).7 Our results are consistent with similar studies in children conducted with smaller sample sizes. A Korean study reporting 71 symptomatic children showed a median time to negativity of 17.6 days, a study from Singapore observed a median time of 16 days, and a study of 68 children from the US reported median duration of viral shedding (RT-PCR positivity) of 19.5 days (IQR, 12-39).1 , 6 , 8 However, it is important to consider that a proportion of patients in our study remained positive for more than 21 days after diagnosis. Consistent with our findings, it has been described that 19% of people continue to show nasopharyngeal RT-PCR positivity 2 or more weeks after the resolution of symptoms.9 Likewise, in an Italian study, 28 of 191 adults (14.7%) with a mean age of 51 years (IQR, 41-59), were persistently positive between days 41 and 60 after discharge.5 Prolonged or recurrent viral RNA detection from the nasopharynx does not necessarily indicate prolonged viral infectiousness. Isolation of infectious viruses from upper respiratory specimens more than 10 days after the onset of illness has only rarely been documented in patients who had a nonsevere infection and whose symptoms had resolved.10 Rather, our findings should be used to better understand the significance of a positive RT-PCR in children in different settings. A positive RT-PCR should not exclude other possible infections. Also, our findings can help to further our understanding of postinfectious complications such as MIS-C. In children with MIS-C, almost one-half of all patients have positive RT-PCR test results at presentation.3 Possibly, the persistence of the antigenic stimulus may stimulate immune activation, leading to the development of late complications.
Serologic tests play an important role in the clinical diagnosis of postinfectious patients or patients with late-stage SARS-CoV-2-related conditions. Our data suggest that negative SARS-CoV-2 serologic test results alone do not exclude previous SARS-CoV-2 in children. In a previous study, only one-half of the patients demonstrated adequate neutralizing antibodies during the time frame of specimen collection.6 This finding is important when considering postinfectious complications such as MIS-C. Interestingly, patients with mild disease and a shorter time to RT-PCR negativity in our study seroconverted less often than patients with more severe disease and who had more prolonged RT-PCR positivity, suggesting that the level and persistence of viral material may be key to developing antibodies. These findings also suggest that detectable antibodies by routine techniques are not a useful marker for clinical SARS-CoV-2 infection. In contrast, the absence of detectable antibodies may be related to early clearance, mild disease, and possibly a good innate response. We also should consider that some patients may not have initiated seroconversion at the time of the second test. The median time from RT-PCR positivity as studied by to seropositivity, by chemiluminescent testing, has been reported to be 18 days in children.6 Our data also should be considered for interpretation of seroprevalence studies, especially in children, who usually have mild disease seem to achieve PCR negativity earlier than adults and may remain seronegative.
Almost all adults seroconvert after confirmed COVID-19.9 However, the sensitivity of enzyme-linked immunosorbent assay (ELISA) tests targeting the nucleocapsid or spike protein, or both (combined) antigens is 0.79 (95% CI, 0.68-0.88), 0.80 (95% CI, 0.62-0.92), and 0.86 (95% CI, 0.68-0.91), respectively.4 From our data, we cannot state definitively whether the proportion of children who did not seroconvert did not develop a humoral response, or if the ELISA techniques were unable to detect the antibodies.
The strengths of our study include a large sample size, serial testing, and a multicenter approach. Our study has several limitations. As a multicenter study, the techniques used for virus detection and antibody assays were not uniform. Also, we did not analyze neutralizing antibodies or the cross-reactivity of serology tests for SARS-CoV-2, although the specificity of ELISA and chemiluminescent immuno assay ranges from 92% to 100%.4 Owing to the exceptional situation, mainly during the first months of the pandemic, several patients were lost to follow-up, and there was some variability in the timing between tests. Ct information was available for a proportion of participants only, so the analysis related to Ct should be interpreted with caution. Reporting Ct values as a tool for assessing with exactitude the viral load has been challenged.11 , 12 However, other reports have reported a good correlation between Ct value and viral loads in samples obtained from children with SARS-CoV-2 infection (r = −1), with lower values of viral loads in asymptomatic children and children aged less than 1 year.13 Some centers are reporting Ct values after normalization or in a semiquantitative format and, in practice, most centers use the Ct as a proxy for a high or low viral load. Regarding serologic results, because there was appreciable dropout from the 136 included to the 59 who completed the protocol, a selection bias should be considered.
Acknowledgments
We thank all the patients and families for their participation in the study, and the laboratory staff and clinical staff members who cared for them. We thank Kenneth McCreath for the meaningful comments and suggestions on English writing. K.M. is a medical English consultant funded by Universidad Europea de Madrid.
We appreciate the support of Instituto de Salud Carlos III (Ministry of Economy, Industry and Competitiveness), European Regional Development Fund, to the Spanish Society of Pediatrics (Asociación Española de Pediatría), to the Fundación para la Investigación Biomédica del Hospital Universitario Infanta Sofía y del Hospital del Henares (FIB 03/2020) and to Cantera Santander and Universidad Europea de Madrid Grant.
Footnotes
Supported by Project PI20/00095, from the Instituto de Salud Carlos III (Ministry of Economy, Industry and Competitiveness) and cofounded by the European Regional Development Fund. E.C. is supported by a specific Research Project of the Spanish Society of Pediatrics (Asociación Española de Pediatría). J.M. is supported by Fundación para la Investigación Biomédica del Hospital Universitario Infanta Sofía y del Hospital del Henares (FIB 03/2020). M.B. is supported by a Cantera Santander and Universidad Europea de Madrid Grant. C.G. is funded by the Spanish Ministry of Science and Innovation—Instituto de Salud Carlos III and Fondos FEDER (Contrato Río Hortega CM19/00015). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The other authors declare no conflicts of interest.
Appendix. Additional members of the EPICO-AEP Working Group
Francisco José Sanz, María Isabel Iglesias-Bouzas, Jose Antonio Alonso Cadenas, Blanca Herrero (Hospital Infantil Universitario Niño Jesús, Spain), Teresa del Rosal, Ana Méndez-Echevarría, Talía Sainz, Clara Udaondo, Fernando Baquero, Cristina Calvo, Carlos Grasa, Paula R Molino, María José Mellado, María Ceano, Victor Galán, Paula Garcia Sanchez, Sonsoles San Roman (Hospital Universitario La Paz, Spain), Alicia Hernanz Lobo, Mar Santos, Marisa Navarro, Elena Rincón, David Aguilera, Begoña Santiago, Jorge Huerta, Eduardo Bardón, Jorge Lorente (Hospital Universitario Gregorio Marañón, Spain), Pablo Rojo, Daniel Blázquez, Luis Prieto, Elisa Fernández-Cooke, David Torres-Fernández, Ángela Manzanares, Jaime Carrasco, Cristina Epalza, Jesús Contreras, Sara Domínguez, Sara Villanueva, Arantxa Gonzalez (Hospital Universitario 12 de Octubre, Spain), Cinta Moraleda, Alfredo Tagarro, Elena Cobos, Álvaro Ballesteros, Sara Domínguez-Rodriguez (Instituto de Investigación 12 de Octubre, Spain), Gemma Pons, Silvia Simó, Miguel Lanaspa, Victoria Fumadó, Rosa María Pino, María Melé (Hospital Sant Joan de Déu, Spain), María Espiau, Jacques G. Rivière, Pere Soler-Palacín, Antonio Soriano Arandes, Natalia Mendoza (Hospital Universitari Vall d’Hebron, Spain), Mercedes Herranz, María Urretavizcaya Martínez (Complejo Hospitalario de Navarra, Spain), Fernando Cabañas, Fátima Ara, Marta Baragaño (Hospital Universitario Quirónsalud Madrid, Spain), Rut del Valle, Ana González-de-Zárate, Mónica Pacheco, María Luisa Herreros, Julia Yebra, Beatriz Pérez-Seoane, María Fernández, Teresa Raga, María de la Serna, Ane Plazaola, Juan Miguel Mesa, Rosa Batista, Ana Barrios, Ignacio Navarro, Jana Rizo, Teresa Reinoso, Alfonso Cañete (Hospital Universitario Infanta Sofía, Spain), María Dolores Martín, Elena Sáez, Olga Nerea Coya, Fernando Cava (BR Salud, Spain), Enrique Otheo, Juan Carlos Galán, José Luis Vázquez, Carmen Vázquez, Victor Quintero (Hospital Universitario Ramón y Cajal, Spain), Lola Falcón, Olaf Neth, Peter Olbrich, Walter Goicoechea, Cati Márquez, Marisol Camacho, Inés Marín Cruz (Hospital Universitario Virgen del Rocío, Spain), Laura Martín (Hospital Universitario Regional de Málaga, Spain), Lucía Figueroa (Hospital de Villalba, Spain), María Llorente (Hospital Universitario del Sureste, Spain), María Penin, Claudia García, María García, Teresa Alvaredo (Hospital Universitario Príncipe de Asturias, Spain), Ma Inmaculada Olmedo, Agustín López, María Jose Pérez (Hospital Universitario Puerta de Hierro, Spain), Elvira Cobo (Hospital Fundación Alcorcón, Spain), Mariann Tovizi (Hospital del Tajo, Spain), Pilar Galán (Hospital Universitario de Fuenlabrada, Spain), Beatriz Soto, Sara Guillén (Hospital de Getafe, Spain), Adriana Navas (Hospital Universitario Infanta Leonor, Spain) M. Luz García (Hospital de Leganés, Spain), Sara Pérez (Hospital de Torrejón, Spain), Amanda Bermejo, Pablo Mendoza (Hospital de Móstoles, Spain), Gema Sabrido (Hospital Rey Juan Carlos, Spain), María José Hernández (Hospital Central de la Defensa, Spain), Ana Belén Jiménez (Fundación Jiménez Díaz, Spain), Arantxa Berzosa, José Tomás Ramos, Marta Illán (Hospital Clínico San Carlos, Spain), Ana López, Nerea Gallego (Hospital Universitari Son Espases, Spain), Beatriz Ruiz (Hospital Universitario Reina Sofía, Spain), Santiago Alfayate, Ana Menasalvas, Eloísa Cervantes (Hospital Clínico Universitario Virgen de la Arrixaca, Spain), María Méndez (Institut d'Investigació en Ciències de la Salut Germans Trias i Pujol, Spain), Ángela Hurtado (Instituto Hispalense de Pediatría, Spain), Cristina García, Inés Amich, Yolanda Ruiz (Hospital San Pedro, Spain), Manuel Oltra, Álvaro Villarroya-Villalba, Adela Cañete, Bienvenida Argiles (Hospital Universitari i Politècnic La Fe, Spain), Angustias Ocaña (Hospital La Moraleja, Spain), Isabel Romero, María Fernanda Guzmán (Hospitales Madrid, Spain), M.J. Pascual (Hospital Nisa, Spain), María Sánchez-Códez (Hospital Universitario Puerta del Mar, Spain), Elena Montesinos (Consorci Hospital General Universitari de València, Spain), Julia Jensen, María Rodríguez (Hospital Universitario Infanta Cristina, Spain), Gloria Caro (Hospital Universitario Infanta Elena, Spain), Neus Rius, Alba Gómez (Hospital Universitari Sant Joan de Reus, Spain), Rafael Bretón (Hospital Clínico Universitario de Valencia, Spain), Margarita Rodríguez, Julio Romero, Juan Francisco Pascual Gazquez (Hospital Universitario Virgen de las Nieves, Spain), Ana Campos (Hospital Universitario Sanitas La Zarzuela, Spain), Mercedes García (Hospital de Mérida, Spain), Rosa María Velasco (Complejo Hospitalario de Toledo, Spain), Zulema Lobato (Althaia, Xarxa Assistencial Universitària de Manresa, Spain), Fernando Centeno, Elena Pérez, Alfredo Cano (Hospital Universitario Río Hortega, Spain), Paula Vidal (Hospital Clínico Universitario Lozano Blesa, Spain), Corsino Rey, Ana Vivanco, Maruchi Alonso (Hospital Universitario Central de Asturias, Spain), Pedro Alcalá, Javier González de Dios, Laura Ureña Horno (Hospital General Universitario de Alicante, Spain), Eduard Solé, Laura Minguell (Hospital Universitari Arnau de Vilanova, Spain), Itziar Astigarraga, Olatz Villate, Susana García Obregón (Hospital Universitario de Cruces, Spain), Ma Ángeles Vázquez, Miguel Sánchez, Leticia Martínez Campos (Hospital Universitario Torrecárdenas, Spain), Elena Díaz (Hospital Virgen de la Luz, Spain), Eduardo Consuegra, Susana Riesco, Almudena González, Maika Mendoza (Hospital Universitario de Salamanca, Spain), María Cabanillas (Complejo Asistencial Universitario de Palencia, Spain), Yeray Novoa-Medina, Elena Colino-Gil, Ana Reyes-Domínguez, Luis Peña-Quintana (Hospital Universitario Materno Infantil de las Palmas, Spain), Elisa Garrote, Maite Goicoechea (Hospital Universitario de Basurto, Spain), Irene Centelles (Hospital General Universitari de Castelló, Spain), Santiago Lapeña, Sara Gutiérrez, Soraya Gutiérrez (Complejo Asistencial Universitario de León, Spain), Amparo Cavalle (PIUS Hospital de Valls, Spain), José María Olmos (Hospital Mare de Déu dels Lliris, Spain), Alejandro Cobo, Sara Díaz, Cristina Martinez Faci, Macarena Gonzalez Cruz, Beatriz Castro (Hospital Universitario de Canarias, Spain), Beatriz Jiménez, Cristina Alvarez Alvarez (Hospital Universitario Marqués de Valdecilla, Spain), Raúl González (Hospital Sant Joan d'Alacant, Spain), Miguel Lafuente, Matilde Bustillo (Hospital Infantil de Zaragoza, Spain), Natividad Pons, Julia Morata (Hospital Lluís Alcanyis, Spain), Elsa Segura (Hospital Universitario Son Llatzer de Palma de Mallorca, Spain), María Bernardino (Universidad Europea de Madrid, Spain), Marta Pareja León (Complejo Hospitalario Universitario de Albacete, Spain), Ana Domingo Ruiz (Hospital de Manacor, Spain), Eider Oñate, Nagore Garcia de Andoin Baran (Hospital Universitario Donostia, Spain), Nerea Domínguez-Pinilla (Hospital Virgen de la Salud, Spain), María Teresa Coll Sibina (Hospital General de Granollers, Spain), María Jesús García García (Hospital Universitario de Cáceres, Spain), Marta Osuna (Hospital HM Montepríncipe, Spain), Raquel Portugal (Hospital Universitario de Burgos, Spain), Leonor García Maset (Hospital de Sagunto, Spain), Belén Sevilla (Hospital Universitario San Cecilio Granada, Spain), Noelia Berciano Jiménez (Hospital Virgen Macarena Sevilla, Spain).
Appendix
References
- 1.Han M.S., Choi E.H., Chang S.H., Jin B.-L., Lee E.J., Kim B.N., et al. Clinical characteristics and viral RNA detection in children with coronavirus disease 2019 in the Republic of Korea. JAMA Pediatr. 2021;175:73. doi: 10.1001/jamapediatrics.2020.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Long Q.-X., Tang X.-J., Shi Q.-L., Li Q., Deng H.-J., Yuan J., et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 3.Moraleda C., Serna-Pascual M., Soriano-Arandes A., Simó S., Epalza C., Santos M., et al. Multi-inflammatory syndrome in children related to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in Spain. Clin Infect Dis. 2021;72:e397–e401. doi: 10.1093/cid/ciaa1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H., Ai J., Loeffelholz M.J., Tang Y.-W., Zhang W. Meta-analysis of diagnostic performance of serology tests for COVID-19: impact of assay design and post-symptom-onset intervals. Emerg Microbes Infect. 2020;9:2200–2211. doi: 10.1080/22221751.2020.1826362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cento V., Colagrossi L., Nava A., Lamberti A., Senatore S., Travi G., et al. Persistent positivity and fluctuations of SARS-CoV-2 RNA in clinically-recovered COVID-19 patients. J Infect. 2020;81:e90–e92. doi: 10.1016/j.jinf.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bahar B., Jacquot C., Mo Y.D., DeBiasi R.L., Campos J., Delaney M. Kinetics of viral clearance and antibody production across age groups in children with severe acute respiratory syndrome coronavirus 2 infection. J Pediatr. 2020;227:31–37.e1. doi: 10.1016/j.jpeds.2020.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singanayagam A., Patel M., Charlett A., Lopez Bernal J., Saliba V., Ellis J., et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kam K.Q., Thoon K.C., Maiwald M., Chong C.Y., Soong H.Y., Loo L.H., et al. SARS-CoV-2 viral RNA load dynamics in the nasopharynx of infected children. Epidemiol Infect. 2021;149:e18. doi: 10.1017/S095026882100008X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wajnberg A., Mansour M., Leven E., Bouvier N.M., Patel G., Firpo-Betancourt A., et al. Humoral response and PCR positivity in patients with COVID-19 in the New York City region, USA: an observational study. Lancet Microbe. 2020;1:e283–e289. doi: 10.1016/S2666-5247(20)30120-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bullard J., Dust K., Funk D., Strong J.E., Alexander D., Garnett L., et al. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis. 2020;71:2663–2666. doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahdouh E., Lázaro-Perona F., Romero-Gómez M.P., Mingorance J., García-Rodriguez J. Ct values from SARS-CoV-2 diagnostic PCR assays should not be used as direct estimates of viral load. J Infect. 2021;82:414–451. doi: 10.1016/j.jinf.2020.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Binnicker M.J. Challenges and controversies to testing for COVID-19. J Clin Microbiol. 2020;58:e01695–e016720. doi: 10.1128/JCM.01695-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pinninti S.G., Pati S., Poole C., Latting M., Seleme M.C., Yarbrough A., et al. Virological characteristics of hospitalized children with SARS-CoV-2 infection. Pediatrics. 2021;147 doi: 10.1542/peds.2020-037812. e2020037812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.