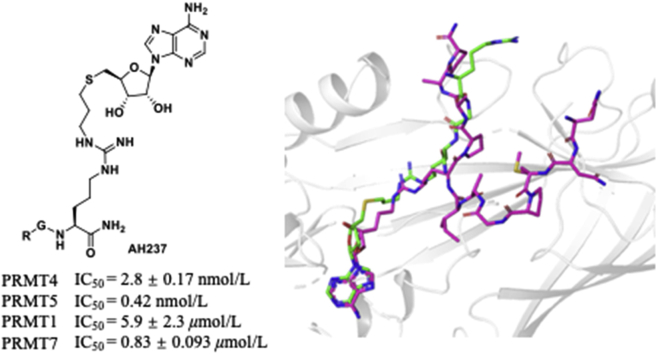
Abstract
Protein arginine methyltransferases (PRMTs) have been implicated in the progression of many diseases. Understanding substrate recognition and specificity of individual PRMT would facilitate the discovery of selective inhibitors towards future drug discovery. Herein, we reported the design and synthesis of bisubstrate analogues for PRMTs that incorporate a S-adenosylmethionine (SAM) analogue moiety and a tripeptide through an alkyl substituted guanidino group. Compound AH237 is a potent and selective inhibitor for PRMT4 and PRMT5 with a half-maximal inhibition concentration (IC50) of 2.8 and 0.42 nmol/L, respectively. Computational studies provided a plausible explanation for the high potency and selectivity of AH237 for PRMT4/5 over other 40 methyltransferases. This proof-of-principle study outlines an applicable strategy to develop potent and selective bisubstrate inhibitors for PRMTs, providing valuable probes for future structural studies.
KEY WORDS: Protein arginine methyltransferase 5, Protein arginine methyltransferase 4, Bisubstrate analogue, Protein arginine methyltransferase, Bisubstrate inhibitor
Abbreviations: GAR, glycine and arginine; IC50, half-maximal inhibition concentration; NTMTs, N-terminal methyltransferases; PKMTs, protein lysine methyltransferases; PRMTs, protein arginine methyltransferases; SAH, S-(5′-adenosyl)-l-homocysteine; SAM, S-adenosylmethionine; SNF, sinofungin; TLC, thin-layer chromatography
Graphical abstract
A potent bisubstrate PRMT inhibitor AH237 was developed, displaying an exceptional selectivity for PRMT4 and PRMT5 with IC50 of 2.8 and 0.42 nmol/L, respectively. This would facilitate the understanding of the biochemical properties for PRMT4/5 and pave the avenue to develop selective and cell-potent PRMT inhibitors.
1. Introduction
Arginine methylation is a prominent post-translational modification that regulates various physiological processes including cell differentiation, RNA splicing, and DNA damage repair1. It is catalyzed by protein arginine methyltransferases (PRMTs) that transfer the methyl group from the cofactor S-adenosylmethionine (SAM) to the guanidino group of the arginine residue in an “SN2-like” fashion. There are nine members in the PRMT family, which are categorized into three types based on the product types1,2. Type I PRMTs (PRMT1, 2, 3, 4, 6, and 8) generate asymmetric dimethylated arginine. Type II PRMTs (PRMT5 and 9) produce symmetric dimethylated arginine. Type III contains only PRMT7 that yields monomethylated arginine. Besides a typical Rossmann-fold domain for interaction with the cofactor SAM, most of PRMTs share the glycine and arginine (GAR) substrate motif3. The only exception is PRMT9 which displays the preference for FKRKY motif4. Although PRMT4 predominantly recognize arginine residues in proline-rich context5, it is able to methylate arginine residues next to proline, glycine, and methionine rich motifs in vitro. Therefore, it remains ambiguous about the physiological substrate preference for each PRMT.
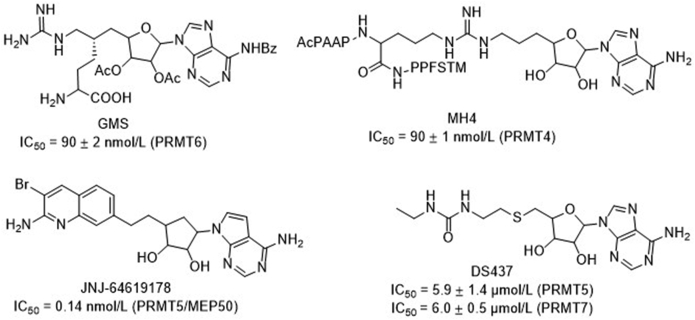
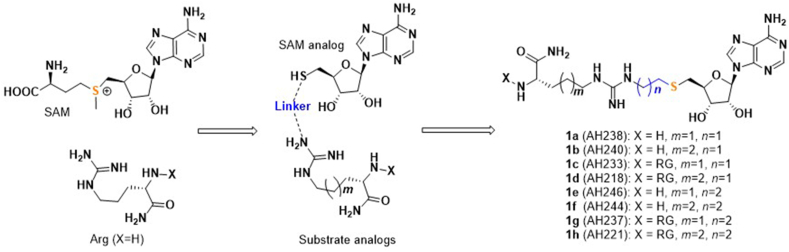
Abnormal expression or activity of PRMTs has been associated with a variety of diseases, including cancers, cardiovascular diseases, inflammatory diseases, and diabetes6, 7, 8, 9, 10. Thus, PRMTs have attracted emerging attentions to develop specific and potent inhibitors as potential therapeutic agents. Although many potent small molecule inhibitors have been reported for PRMTs to date, selectivity remains a challenge for individual PRMT isoform because of a conserved SAM binding site and similar substrate recognition motif9,11, 12, 13, 14. Bisubstrate analogue has demonstrated its potential to offer potent and specific inhibitors for several PRMTs including PRMT1, PRMT4, and PRMT6, as well as facilitate the formation of co-crystal structures to offer valuable insights into the structural basis for PRMT specificity15, 16, 17. For instance, the bisubstrate inhibitor GMS that links a guanidine moiety to a sinofungin (SNF) analogue is 17-fold more potent than SNF for PRMT6 (Fig. 1)15. Similarly, bisubstrate analogue MH4 that tethers an adenosine moiety with the PRMT4 substrate peptide PAAPRPPFSTM displays potent inhibition of PRMT4 with IC50 value of 90 nmol/L and 250-fold selectivity over PRMT117. JNJ-64619178 is a highly selective PRMT5 inhibitor in phase I clinical trial for advanced solid tumors, non-Hodgkin lymphoma, and myolodysplastic syndromes18. In this study, we designed a general platform to develop potent and selective inhibitors for PRMTs. We adopted both rational and structure-based drug design strategies to produce a series of new bisubstrate analogues that covalently connect a SAM analogue with a single amino acid (Lys or Arg) or a tripeptide (RGR or RGK) through a guanidine group (Fig. 2), which is the methyl acceptor of arginine. Among them, AH237 showed a superior selectivity for PRMT4 and PRMT5 with IC50 values in a low nanomolar range.
Figure 1.
Reported bisubstrate inhibitors for PRMTs.
Figure 2.
The designed PRMT inhibitors.
2. Results and discussion
2.1. Design
In the active site of PRMTs, two adjacent binding pockets are occupied by both S-(5′-Adenosyl)-l-homocysteine (SAH) and the Arg residue of the substrate peptide. The distance between the SAH sulfur atom and the α-nitrogen atom of the guanidinum group ranges from 3.5 to 5.5 Å15, 16, 17. Therefore, we hypothesized that covalently linking a SAM analogue moiety with a guanidinum group through a 2-C or 3-C atom linker would mimic the transition state to offer potent inhibitors as general probes for PRMTs (Fig. 2). A 2-C or 3-C atom linker length has demonstrated feasibility for various protein methyltransferases in previous studies15, 16, 17,19, 20, 21, 22. For the SAM cofactor analogue, we chose a thioadenosine as inspired by a dual PRMT5/7 inhibitor DS43723. For the substrate portion, we focused on a general peptide motif RGR since it can be essentially recognized by all PRMTs except PRMT9. To achieve the specificity for PRMTs, the methylation acceptor guanidium group was retained. We also investigated an RGK peptide, as well as a single amino acid (Arg or Lys), to explore the effect of substrate peptide moiety on the inhibition (Table 1 and Fig. 2).
Table 1.
The inhibitory IC50 of the synthesized compounds.
| Compd. | R1 | n | IC50 (μmol/L) |
||||
|---|---|---|---|---|---|---|---|
| PRMT1a | TbPRMT7a | G9ab | NTMT1b | SETD7b | |||
| 1a (AH238) | Orn | 1 | 16 ± 8 | 2.8 ± 0.2 | >100 | >100 | >100 |
| 1b (AH240) | Lys | 1 | 8.3 ± 5.4 | 3.3 ± 2.2 | >100 | >100 | >100 |
| 1c (AH233) | Arg-Gly-Orn | 1 | 1.7 ± 0.1 | 0.59 ± 0.05 | >100 | >100 | >100 |
| 1d (AH218) | Arg-Gly-Lys | 1 | 8.3 ± 0.1 | 0.91 ± 0.07 | ∼10 | >100 | ∼100 |
| 1e (AH246) | Orn | 2 | 6.5 ± 2.8 | 7.6 ± 1.0 | >100 | >100 | >100 |
| 1f (AH244) | Lys | 2 | 3.0 ± 1.1 | 20 ± 6 | >100 | >100 | >100 |
| 1g (AH237) | Arg-Gly-Orn | 2 | 15 ± 0.2 | 0.29 ± 0.03 | >100 | >100 | >100 |
| 1h (AH221) | Arg-Gly-Lys | 2 | 5.6 ± 0.1 | 3.4 ± 0.3 | >100 | >100 | >100 |
IC50 values were determined in triplicate (n = 3) and presented as mean ± standard deviation (SD).
IC50 values were estimated from three-dose screening in duplicate (n = 2).
2.2. Synthesis
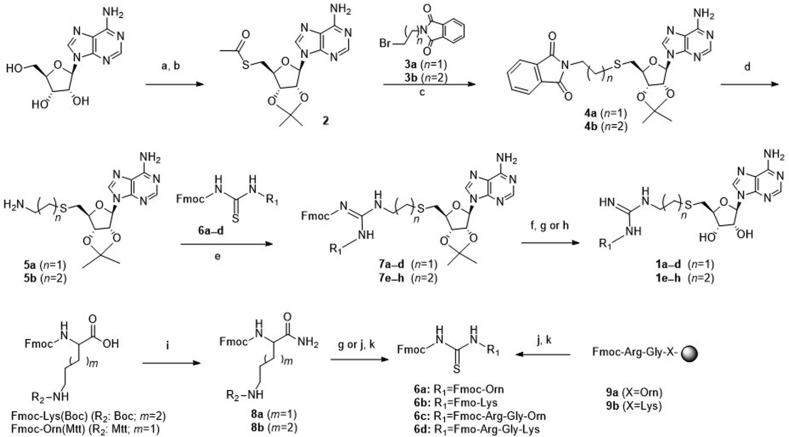
Commercially available adenosine was first subjected to a diol protection and followed by a subsequent Mistunobu reaction to produce thioester 2 (Scheme 1)19,24. Then 2 was hydrolyzed by sodium methoxide and followed by thiol alkylation with phthalimide alkyl bromides 3a and b to provide phthalimides 4a and b25. Deprotection of 4a and b by hydrazine afforded the key amine intermediates 5a and b, which were then reacted with various thioureas 6a‒d to provide 7a‒h in a convergent manner26,27. Removal of Fmoc protection group followed by acidic deprotection or cleavage offered final compounds 1a‒h28. For 1a, 1b, 1e and 1f, Fmoc-Orn(Mtt) and Fmoc-Lys (Boc) were first amidated using ammonium chloride to produce 9a and b, followed by deprotection with TFA to yield free amines to react with Fmoc-isothiocyanate at 0 °C to yield 6a and b27. To prepare peptide conjugates 1c, 1d, 1g and 1h, short peptides Fmoc-Arg-Gly-Orn/Lys (9a and b) were synthesized on solid phase and followed by removal of Mtt protection group, which were then reacted with Fmoc-isothiocyanate to yield 6c and d27.
Scheme 1.
Synthesis of compounds 1a‒h. Reagents and conditions: (a) CH(OEt)3, p-TsOH, acetone, 85%. (b) Thioacetic acid, PPh3, DIAD, THF, 94%. (c) NaOCH3, MeOH, 59%–70%. (d) Hydrazine, MeOH, 76%–92%; (e) EDC, DIPEA, CH2Cl2. (f) Piperidine, HOBt, DMF. (g) 20% TFA in CH2Cl2, 0 °C. (h) TFA:DODT:TIPS:H2O (94:2.5:1:2.5, v/v), rt, 5 h. (i) NH4Cl, HBTU, NMM, CH3CN, 70%–90%. (j) 1% TFA in CH2Cl2; (k) Fmoc-isothiocyanate, CH2Cl2, 0 °C.
2.3. Biochemical characterization
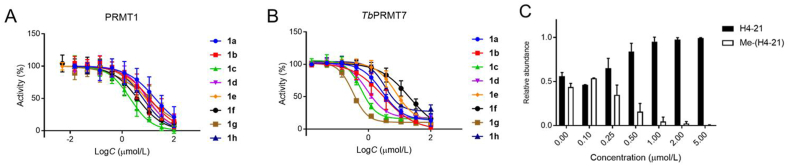
All synthesized bisubstrate analogues were first evaluated in a SAH hydrolase (SAHH)-coupled fluorescence assay under the condition of the Km values of both SAM and the respective peptide substrate for two representative PRMTs (PRMT1 and TbPRMT7)21,29. As shown in Table 1 and Fig. 3A and B, all bisubstrate analogues exhibited inhibition against PRMT1 and TbPRMT7 with IC50 values ranging from 1.7 to 14.6 μmol/L and 0.29–19.5 μmol/L, respectively. The majority of bisubstrate analogues displayed improved or comparable potency to TbPRMT7 than PRMT1 except AH244, which showed 6-fold increased potency to PRMT1. According to results from this series, PRMT1 demonstrated its preference to a 3-C atom linker while TbPRMT7 showed its preference for a 2-C atom linker. For instance, AH244 and AH246 that contained a 3-C atom linker were around 3- and 14-fold more potent for PRMT1 than AH240 and AH238 that contained a 2-C atom linker, respectively. However, AH233 and AH237 were exceptions because AH237 containing a 3-C atom linker was the most potent inhibitor (IC50 = 0.29 μmol/L) for TbPRMT7 and AH233 containing a 2-C atom linker was the most potent inhibitor (IC50 = 0.59 μmol/L) for PRMT1 in this series. Furthermore, AH237 displayed over 50-fold selectivity for TbPRMT7 over PRMT1. In terms of the effect of peptide length on inhibition, bisubstrate analogues containing a tripeptide substrate showed about 4-fold improved potency for TbPRMT7 in comparison with their respective bisubstrate analogues containing a single amino acid, while marginal effects were observed for PRMT1 except for AH233. Meanwhile, the linker length has minimal effect on the bisubstrate analogues that contain the tripeptide moiety like Arg-Gly-Arg for TbPRMT7 as shown in AH233 and AH237, but about 8-fold difference for PRMT1.
Figure 3.
(A) IC50 curves of bisubstrate compounds for PRMT1. (B) IC50 curves of bisubstrate compounds for TbPRMT7. (C) The methylation assay of AH237 for TbPRMT7 (n = 2).
Our hypothesis is that designed PRMT inhibitors would be selective for PRMTs against other protein methyltransferases such as protein lysine methyltransferases (PKMTs) and N-terminal methyltransferases (NTMTs), because all designed compounds contain a guandinium function group that is a unique methylation acceptor for PRMTs. To test this hypothesis, we chose two representative PKMTs (G9a and SETD7) and NTMT1 to examine their activities in the SAHH-coupled fluorescence assay29. Not surprisingly, all eight bisubstrate analogues did not display any significant inhibition against G9a, NTMT1, and SETD7 up to 100 μmol/L, except that AH218 inhibited 50% of G9a activity at 10 μmol/L. AH237 that incorporated a tripeptide of Arg-Gly-Arg and a 3-C atom linker was the most potent and selective inhibitor for TbPRMT7 in our series (IC50 = 0.29 ± 0.03 μmol/L). When we carried out this study, no potent and selective PRMT7 inhibitor was available except a dual PRMT5 and 7 inhibitor DS437 (IC50 = 6 μmol/L)23. Therefore, we focused on AH237 in subsequent inhibition mechanism and the comprehensive selectivity study.
2.4. MALDI-MS methylation inhibition assay
The MALDI-MS methylation assay was performed to validate the inhibitory activity of AH237 on TbPRMT730,31. The results indicated that at 0.5 μmol/L of AH237 the methylation level of H4-21 was reduced by more than 50%, while the methylated product was abolished with 5 μmol/L compound (Fig. 3C).
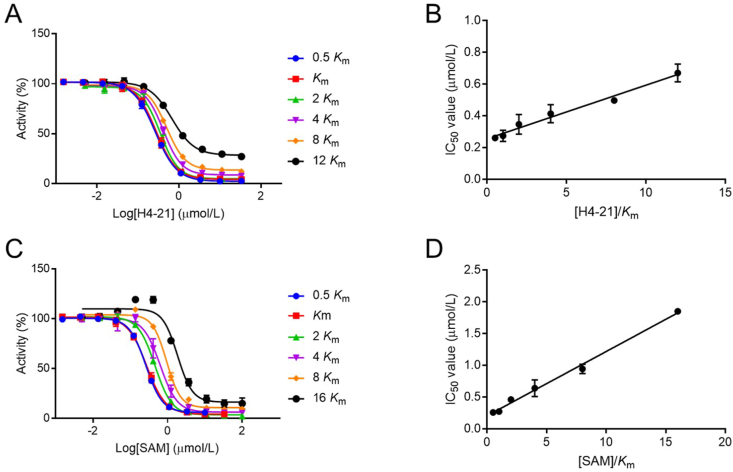
2.5. Inhibition mechanism
To examine the inhibition mechanism of AH237, a kinetic analysis was performed using the SAHH-coupled fluorescence-based assay with TbPRMT7. AH237 showed an unambiguous pattern of competitive inhibition for both the peptide substrate and SAM, as demonstrated by the linear ascending of the IC50 values depending on either the peptide substrate or SAM concentration (Fig. 4). The result indicated that compound AH237 occupied both cofactor and peptide substrate binding sites of TbPRMT7 as a bisbustrate inhibitor, supporting its feature as the bisubstrate analogue.
Figure 4.
Inhibition mechanism studies of AH237 for TbPRMT7. (A) IC50 curves of AH237 at varying concentrations of H4-21 peptide with fixed concentration of SAM. (B) Linear regression plot IC50 values with corresponding concentrations of H4-21. (C) IC50 curves of AH237 at varying concentrations of SAM with fixed concentration of H4-21. (D) Linear regression plot IC50 values with corresponding concentrations of SAM.
2.6. Selectivity studies
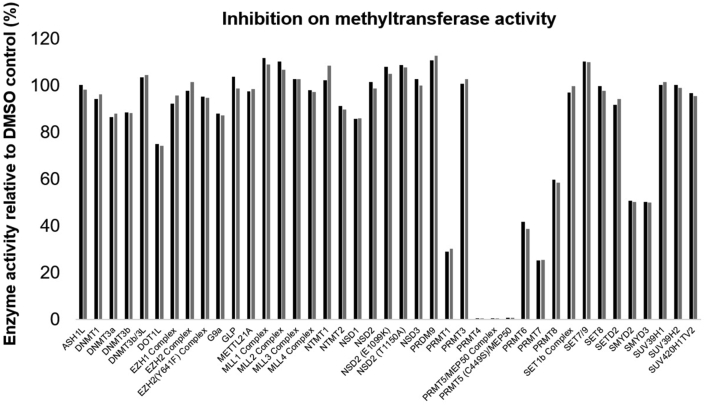
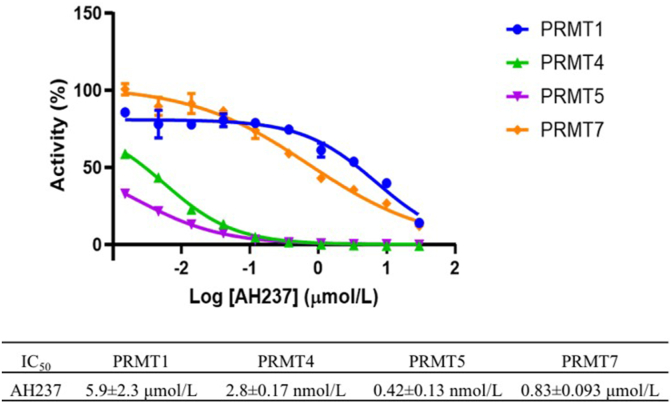
To further understand the selectivity profile of AH237, its inhibitory activity was examined for a panel of 41 MTases such as PRMTs (PRMT1, 3–8), PKMTs (ASH1L, EZHs, G9a, GLP, METTL21A, MLL complexes, NSDs, PRDM9, SETs, SMYDs, DOT1L, and SUV39Hs), NTMT1/2, and DNA methyltransferases (DNMTs) at a single dose (10 μmol/L) of AH237 (Reaction Biology Inc., Malvern, PA, USA). The results indicated that AH237 selectively reduced the activity for most PRMTs at 10 μmol/L except PRMT3. Importantly, AH237 did not show any inhibition for all PKMTs, NTMTs, and DNMTs except for SMYD2 and 3 (Fig. 5 and Supporting Information Table S1). To our surprise, AH237 completely abolished the activities of PRMT4 and 5 at the concentration of 10 μmol/L. Subsequent dose–response analysis indicated that AH237 is highly selective for PRMT4 and PRMT5 with IC50s of 2.8 ± 0.17 and 0.42 ± 0.13 nmol/L, respectively. Its potency was also confirmed for PRMT1 (IC50 = 5.9 ± 2.3 μmol/L) and PRMT7 (IC50 = 831 ± 93 nmol/L), which are comparable to the values in Table 1 (Fig. 6).
Figure 5.
HotSpot methyltransferases inhibition profile of AH237 at 10 μmol/L on various methyltransferase enzymes in duplicates (n = 2). The results indicated that PRMT enzymes, including PRMT4 and PRMT5, were selectively inhibited by AH237 except PRMT3.
Figure 6.
IC50 determination of AH237 against human PRMT1/4/5/7 (n = 2).
2.7. Docking studies
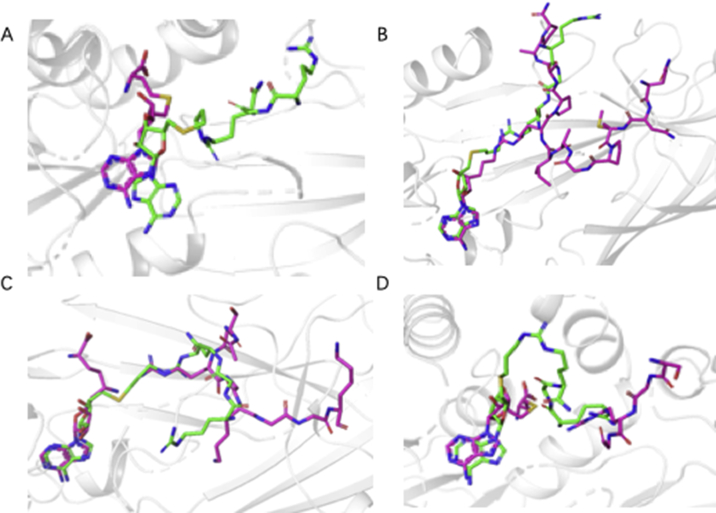
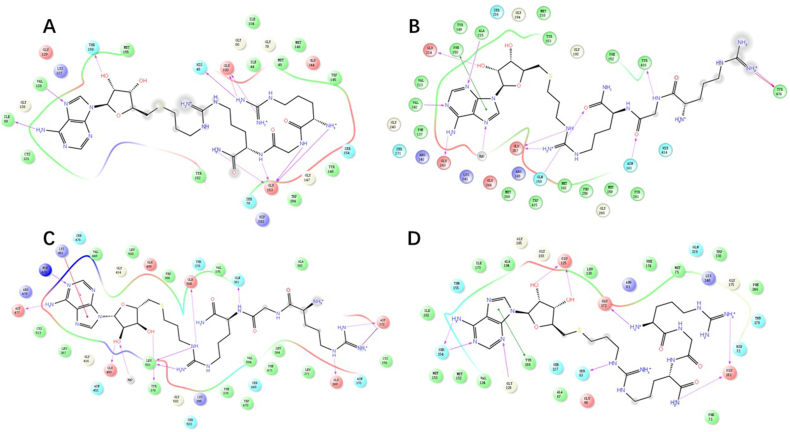
In an attempt to rationalize the observed high potency and selectivity of AH237 for PRMT4/5, we performed computational studies for PRMT1, 4, 5, and 7. Interactions of thioadenosine moiety of AH237 with those four PRMTs offered plausible explanation for its different inhibitory activities towards PRMT1/4/5/7. As shown in Fig. 7, the thioadenosine moiety of AH237 overlaid very well with SAH or SAH mimic moiety in PRMT4 and 517,32. For PRMT7, most interactions were retained except a slight shift, which possibly resulted in the loss of interaction with Glu 12533. Whereas, the interaction patterns of AH237 were quite different for PRMT1 compared to the interaction of SAH with PRMT1, yielding loss of the interactions with both Cys 101 and Glu 100 of PRMT134. Together, these results provided insight into the preferences of AH237 binding to PRMT4/5 over PRMT1/7. In addition to the adenosine moiety, the tripeptide portion of AH237 demonstrated very similar binding orientation as the peptide substrate of PRMT4/5. Furthermore, the linker guanidine group formed more interactions with PRMT4/5 than PRMT7 (Fig. 8).
Figure 7.
Predicted binding modes of AH237 in the PRMTs active site. AH237 was shown in green stick model, while peptides, SAM or SAH were displayed with magenta stick model. Residues in PRMTs were hidden for a better view. (A) PRMT1 (PDB, 1OR8). (B) PRMT4 (PDB, 6S79). (C) PRMT5 (PDB, 3UA3). (D) PRMT7 (PDB, 4M37).
Figure 8.
The estimated 2D interactions of AH237 in the pocket of PRMTs. The peptides, SAM, or SAH in PRMTs were hidden for a better view. AH237 was shown in black line model, while residues around the protein pocket were presented as colored circles. (A) PRMT1 (PDB, 1OR8). (B) PRMT4 (PDB, 6S79). (C) PRMT5 (PDB, 3UA3). (D) PRMT7 (PDB, 4M37).
3. Conclusions
In summary, we have designed and synthesized a series of new bisubstrate inhibitors for PRMTs that covalently link either a single amino or peptide with a thioadenosine through a guanidino group. All of the synthesized inhibitors showed selectivity for PRMTs over two representative PKMTs (G9a and SETD7) and NTMT1. In general, PRMT1 showed less sensitivity to bisubstrate analogues with variable linkers and substrate sequence length because there is less than a 10-fold difference among eight bisubstrate analogues. This tolerance supports a broad substrate spectrum of PRMT1 as it mediates over 85% of the reported arginine methylation events10. On the contrary, PRMT7 exhibited higher stringency towards substrate moiety as bisubstrate analogues containing a tripeptide portion were more potent for TbPRMT7 than their respective pair with a single amino acid35. Among them, AH237 showed high potency for PRMT4 and 5 with an IC50 value of 2.8 and 0.42 nmol/L, respectively. Moreover, it displayed almost 1000-fold selectivity over PRMT1 and 7, and over 10,000-fold selectivity for the other MTases. This profound selectivity corroborates the benefits of bisubstrate analogues that are able to differentiate their potency even among PRMTs family that share a similar substrate recognition preference. In summary, our study offers a glimpse of a delicate difference of transition state for PRMTs, which shed lights on the specificity of PRMT4/5 over the other PRMTs. Additionally, our study outlines a feasible and applicable synthetic strategy to develop bisubstrate inhibitors for PRMTs.
4. Experimental
4.1. Material and instruments
All chemicals and solvents were purchased from commercial suppliers and used without further purification unless stated otherwise. 1H and 13C NMR spectra were carried out on Bruker Avance 500 MHz NMR spectrometer (Billerica, MA, USA) in deuterated solvents. MALDI spectra were performed on 4800 MALDI TOF/TOF mass spectrometry (Sciex, Framingham, MA, USA) at the Mass Spectrometry and Purdue Proteomics Facility (PPF), Purdue University. Peptides were synthesized by CEM Liberty Blue peptide synthesizer (Matthews, NC, USA). Crude products were purified by chromatography using silica gel, standard grade from DAVSIL® (code number 1000179164, 35–70 micron SC). Flash chromatography was performed on Teledyne ISCO CombiFlash Companion chromatography system (Chicago, IL, USA) on RediSep prepacked silica cartridges. Thin-layer chromatography (TLC) plates (20 cm × 20 cm) were purchased from Merck KGaA (Darmstadt, Germany). All the purity of target compounds showed >95%.
4.2. Synthesis
4.2.1. Synthesis S-(((3aS,4S,6R,6aR)-6-(6-amino-9H-purin-9-yl)-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)methyl) ethanethioate (2)
To a solution of the adenosine (0.8 g, 3 mmol, 1 eq.) in acetone (100 mL) was added triethylorthoformate (2.9 g, 19.5 mmol, 6.5 eq.), and p-toluenesulfonic acid (2.55 g, 15 mmol, 5 eq.). The reaction mixture was stirred at room temperature overnight, then was quenched with sat. NaHCO3 (50 mL) and extracted with ethylacetate (3 × 40 mL). The combined organic extract was washed with brine, dried over anhydrous sodium sulfate, and the solvent was evaporated under vacuo to get (2.5 mmol) protected adenosine. Then the product was dissolved in tetrahydrofuran (40 mL) and added triphenyl phosphine (2 g, 7.5 mmol, 3 eq.), DIAD (1 g, 5 mmol, 2 eq.), and thioacetic acid (395 mg, 5 mmol, 2 eq.). The reaction mixture was stirred at room temperature for 2 h. It was concentrated in vacuo and purified by flash chromatography on silica gel in 60%–90% acetone/hexanes to get 2 as a fluffy white solid (875 mg, 80%, in two steps). 1H NMR (500 MHz, CDCl3) δ 8.36 (s, 1H), 8.17 (s, 1H), 6.16 (d, J = 2.4 Hz, 1H), 5.31–5.26 (m, 1H), 4.94 (dd, J = 6.3, 3.2 Hz, 1H), 4.58 (dt, J = 5.5, 3.7 Hz, 1H), 4.35–4.25 (m, 2H), 1.99 (s, 3H), 1.66–1.63 (s, 3H), 1.42–1.39 (s, 3H). 13C NMR (126 MHz, CDCl3) δ 194.58, 155.72, 152.77, 149.09, 140.02, 120.24, 114.52, 90.90, 86.13, 84.20, 83.68, 31.26, 30.57, 27.07, 25.35.
4.2.2. General procedure for synthesis of phthalidomides 4a and b
To a solution of 2 (730 mg, 2 mmol, 1 eq.) and bromoalkyl phthalidomides 3a and b (4 mmol, 2 eq.) in methanol (50 mL) at −20 °C was gradually added sodium methoxide (30% w/w in methanol) (0.55 mL, 3 mmol, 1.5 eq.). The reaction mixture was stirred at −20 °C for 3 h and then at room temperature overnight. The reaction was quenched with addition of dH2O (20 mL) and extracted with ethylacetate (3 × 50 mL). The combined organic extract was washed with brine, dried over anhydrous sodium sulfate, and the solvent was evaporated under vacuo and purified by flash chromatography on silica gel in 60%–90% acetone/hexanes.
4.2.2.1. 2-(2-((((3aS,4S,6R,6aR)-6-(6-Amino-9H-purin-9-yl)-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)methyl)thio)ethyl)isoindoline-1,3-dione (4a)
1H NMR (500 MHz, CDCl3) δ 8.36 (s, 1H), 7.97 (s, 1H), 7.82 (dd, J = 5.4, 3.0 Hz, 2H), 7.69 (dd, J = 5.5, 3.0 Hz, 2H), 6.31 (s, 2H), 6.08 (d, J = 2.3 Hz, 1H), 5.45 (dd, J = 6.4, 2.3 Hz, 1H), 5.05 (dd, J = 6.4, 3.3 Hz, 1H), 4.40 (td, J = 6.7, 3.4 Hz, 1H), 3.80 (t, J = 7.3 Hz, 2H), 2.96–2.86 (m, 2H), 2.81 (t, J = 7.0 Hz, 2H), 1.59 (s, 3H), 1.37 (s, 3H).
4.2.2.2. 2-(3-((((3aS,4S,6R,6aR)-6-(6-Amino-9H-purin-9-yl)-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)methyl)thio)propyl)isoindoline-1,3-dione (4b)
1H NMR (500 MHz, CDCl3) δ 8.33 (s, 1H), 7.96 (s, 1H), 7.85–7.78 (m, 2H), 7.72–7.66 (m, 2H), 6.25 (s, 2H), 6.07 (d, J = 2.2 Hz, 1H), 5.46 (dd, J = 6.4, 2.3 Hz, 1H), 5.03 (dd, J = 6.4, 3.2 Hz, 1H), 4.42–4.34 (m, 1H), 3.72 (t, J = 7.4 Hz, 3H), 2.90–2.72 (m, 2H), 2.55 (t, J = 6.6 Hz, 2H), 2.18–2.13 (m, 2H), 1.89 (p, J = 6.3 Hz, 2H), 1.59 (s, 3H), 1.38 (s, 3H).
4.2.3. General procedure for phthalidomides deprotection 5a and b
To a solution of 4a and b (1 mmol, 1 eq.) in methanol (30 mL) was added hydrazine (0.1 mL, 3 mmol, 3 eq.). The reaction mixture was stirred at room temperature overnight. 10% acetic acid (3 mL) was added and extracted with ethylacetate (5 mL). 2 mol/L sodium hydroxide (3 mL) was added and extracted with ethylacetate (3 × 5 mL). The combined organic extract was washed with brine, dried over anhydrous sodium sulfate, and the solvent was evaporated under vacuo. The residue was used in the next step without further purification.
4.2.4. General procedure for amidation
To a solution of amino acid Fmoc-Orn(Mtt) or Fmoc-Lys (Boc) (0.3 mmol, 1 eq.) in acetonitrile (3 mL) was added ammonium chloride (53 mg, 0.6 mmol, 2 eq.), HBTU (379 mg, 0.45 mmol, 1.5 eq.), and N-methyl morpholine (105 mg, 0.6 mmol, 2 eq.). The reaction mixture was stirred at room temperature overnight. The reaction was quenched with addition of dH2O (2 mL) and extracted with ethylacetate (3 × 7 mL). The combined organic extract was washed with brine, dried over anhydrous sodium sulfate, and the solvent was evaporated under vacuo and purified by flash chromatography on silica gel in 70%–90% ethylacetate/hexanes to obtain 8a and b.
4.2.5. General procedure for peptide synthesis
The Mtt-protected ornithine/lysine-containing peptides 9a and b were synthesized using a Liberty Blue™ peptide synthesizer (Matthews, NC, USA) on Rink amide MBHA resin (138 mg with a resin loading of 0.1 mmol/g, Chem-Impex, Wood Dale, IL, USA) using standard protocols with no final Fmoc deprotection. Each peptide was synthesized of 0.10 mmol and peptide couplings were performed by using 0.2 mol/L of Fmoc-amino acid (Mtt-Orn or Mtt-Lys 2 eq., Gly 6 eq., Arg 12 eq.), activators of DIC reagent (4 mL of 0.5 mol/L), and Oxyma (2 mL of 1 mol/L) in DMF, 20% piperidine in DMF (13 mL) was used for deprotection. The resin was washed with DMF (3 × 3 mL) and DCM (2 × 2 mL). Peptides were not cleaved from the resin and used directly in the next step.
4.2.6. General procedure for Mtt deprotection
To 8a, 9a and b (0.1 mmol, 1 eq.) was added 1% trifluoroacetic acid in dichloromethane (1 mL). The reaction mixture was stirred at room temperature for 2 h. The mixture was either evaporated under vacuo or washed with dichloromethane. The residue was neutralized using DIPEA (15 mg, 0.12 mmol, 1.2 eq.), which were used in the next step without further purification.
4.2.7. Compound 8b Fmoc deprotection
To 8b (0.3 mmol, 1 eq.) was added 20% trifluoroacetic acid in dichloromethane (3 mL). The reaction mixture was stirred at room temperature for 1 h. The solvent was evaporated under vacuo and the residue was neutralized using DIPEA (45 mg, 0.36 mmol, 1.2 eq.) and the obtained residue was used in the next step without further purification.
4.2.8. General procedure for synthesis of thiourea
To a solution of residues that obtained from Mtt and Fmoc deprotection in dichloromethane (3 mL) at 0 °C was added Fmoc-isothiocyanate (42 mg, 0.15 mmol, 1.5 eq.). The reaction mixture was stirred at 0 °C for 30–60 min. The mixture was evaporated under vacuo or washed. The evaporated residue was crystalized using dichloromethane:hexanes 1:1 (2 mL) to produce 6a‒d.
4.2.9. General procedure for synthesis of guanidine moiety
To a solution of amine 5a and b (0.05 mmol, 1 eq.) in dichloromethane (2 mL) was added thiourea 6a‒d (0.75 mmol, 1.5 eq.), EDC (20 mg, 0.1 mmol, 2 eq.), and DIPEA (13 mg, 0.1 mmol, 2 eq.). The reaction mixture was stirred at room temperature overnight. The mixture were washed with dichloromethane (3 × 2 mL) to produce 7a‒h.
4.2.10. General procedure for final compounds synthesis
The mixture 7a‒h were treated with 20% piperidine in DMF with 0.1 mol/L HOBt (3 × 2 mL) for 10 min each. Then it was flushed with dichloromethane (3 × 2 mL) and used for next step. Then it was dissolved in dichloromethane (2 mL) followed by adding a cleavage cocktail (TFA:TIPS:DODT:H2O = 9.4 mL:0.1 mL:0.25 mL:0.25 mL). The reaction mixture was stirred at room temperature for 5 h. The mixture was evaporated by passing N2 gas, washed with dry ether. The residue was used for semi-preparatory HPLC separation using MeOH/H2O 10%–40% to obtain final compounds 1a‒h. The purity of final compounds was confirmed through Agilent 1260 Series HPLC system (Wilmington, DE, USA) by running with 5%–30% methanol/water gradient with 0.1% TFA. All the purity of target compounds showed >95%.
4.2.10.1. AH240 1a (Lys-C2-AD)
C19H32N10O4S, MALDI, Calcd. for [M+H]+ 497.2407; Found 497.3052.
4.2.10.2. AH244 1b (Lys-C3-AD)
C20H34N10O4S, MALDI, Calcd. for [M+H]+ 511.2563; Found 511.2719.
4.2.10.3. AH238 1c (Arg-C2-AD)
C18H30N10O4S, MALDI, Calcd. for [M+H]+ 483.2172; Found 483.2628.
4.2.10.4. AH246 1d (Arg-C3-AD)
C19H32N10O4S, MALDI, Calcd. for [M+H]+ 497.2407; Found 497.2405.
4.2.10.5. AH218 1e (RGK-C2-Ad)
C26H45N15O6S, MALDI, Calcd. for [M+H]+ 710.3554; Found 710.6207.
4.2.10.6. AH221 1f (RGK-C3-Ad)
C28H49N15O6S, MALDI, Calcd. for [M+H]+ 724.3711; Found 724.5140.
4.2.10.7. AH233 1g (RGR-C2-Ad)
C26H45N15O6S, MALDI, Calcd. for [M+H]+ 696.3398; Found 696.4561.
4.2.10.8. AH237 1h (RGR-C3-Ad)
C27H47N15O6S, MALDI, Calcd. for [M+H]+ 710.3554; Found 710.4666.
4.3. The enzyme inhibition assay and selectivity studies
A fluorescence-based SAHH-coupled assay was employed to calculate their IC50 values and to study the effect of the synthesized compound on methyltransferase activity of PRMT1, G9a, SETD7, NTMT1, and TbPRMT721,22. For PRMT1, the assay was carried out in a final well volume of 40 μL: 25 mmol/L HEPES buffer (pH = 8), 25 μmol/L EDTA, 25 mmol/L NaCl, 0.01% Triton X-100, 50 μmol/L TCEP, 5 μmol/L SAHH, 0.1 μmol/L PRMT1, 5 μmol/L SAM, and 15 μmol/L ThioGlo1. The inhibitor was added at nine compound concentrations: 0.005, 0.015, 0.046, 0.14, 0.41, 1.23, 3.7, 11, and 33 μmol/L. After 10 min incubation with the inhibitor, reactions were initiated by the addition of 4 μmol/L H4-21 peptide. For G9a, the assay was performed in a final well volume of 40 μL: 25 mmol/L potassium phosphate buffer (pH = 7.6), 1 mmol/L EDTA, 2 mmol/L MgCl2, 0.01% Triton X-100, 5 μmol/L SAHH, 0.1 μmol/L G9a, 10 μmol/L SAM, and 15 μmol/L ThioGlo1. The inhibitor was added at a single compound concentration of 100 μmol/L. After 10 min incubation with the inhibitor, reactions were initiated by the addition of 4 μmol/L H3-21 peptide. For SETD7, the assay was performed in a final well volume of 40 μL: 25 mmol/L potassium phosphate buffer (pH = 7.6), 0.01% Triton X-100, 5 μmol/L SAHH, 1 μmol/L SETD7, 2 μmol/L AdoMet, and 15 μmol/L ThioGlo1. The inhibitors were added at a single compound concentration of 100 μmol/L. After 10 min incubation with inhibitors, reactions were initiated by the addition of 90 μmol/L H3-21 peptide. For NTMT1, the assay was exerted in a final well volume of 40 μL: 25 mmol/L Tris (pH = 7.5), 50 mmol/L KCl, 0.01% Triton X-100, 5 μmol/L SAHH, 0.2 μmol/L NTMT1, 3 μmol/L SAM, and 15 μmol/L ThioGlo1. The inhibitor was added at a single compound concentration of 100 μmol/L. After 10 min incubation with the inhibitor, reactions were initiated by the addition of 3 μmol/L SPKRIA peptide. For TbPRMT7, the assay was performed in a final well volume of 40 μL: 25 mmol/L Tris (pH = 7.5), 50 mmol/L KCl, 0.01% Triton X-100, 5 μmol/L SAHH, 0.2 μmol/L TbPRMT7, 3 μmol/L SAM, and 15 μmol/L ThioGlo1. After 10 min incubation with inhibitors, reactions were initiated by the addition of 60 μmol/L H4-21 peptide. Fluorescence was monitored on a BMG CLARIOstar microplate reader with excitation 400 nm and emission 465 nm. Data were processed by using GraphPad Prism software 7.0.
The HotSpot methyltransferases profiling (radioisotope filter binding platform) was used to evaluate the selectivity of compound AH237 against 41 methyltransferases (Reaction Biology Corp.). The compound was tested in single dose (10 μmol/L) with duplicate. Control compounds, SAH (S-(5′-adenosyl)-l-homocysteine), chaetocin, LLY 507, or ryuvidine were tested in 10-dose IC50 mode with 3-fold serial dilution starting at 100 or 200 μmol/L. All the reactions were carried out at 1 μmol/L SAM.
4.4. The enzyme inhibition study
For the enzyme kinetic study, the fluorescence-based SAHH-coupled assay was used. Varying concentrations of SAM (from 0.5 to 16 Km) with 60 μmol/L fixed concentration of H4-21 or varying concentration of H4-21 (from 0.5 to 12 Km) with 3 μmol/L fixed concentration of SAM were included in reactions with a series concentration of compounds. All the IC50 values were determined in triplicate. Fluorescence was monitored on a BMG CLARIOstar microplate reader (Cary, NC, USA) with excitation 380 nm and emission 505 nm. Data were processed by using GraphPad Prism software 7.0 (San Diego, CA, USA).
4.5. MS methylation inhibition analysis
MALDI-MS methylation inhibition assay was performed and analyzed via a Sciex 4800 MALDI TOF/TOF MS (MA, USA). 0.2 μmol/L TbPRMT7, 20 mmol/L Tris (pH = 7.5), 50 mmol/L KCl, 5 μmol/L AdoMet and various concentrations of compounds at 37 °C for 10 min before the addition of 2.5 μmol/L H4-21 peptide to initiate the reaction. After incubation overnight, the samples were quenched in a 1:1 ratio with a quenching solution (20 mmol/L NH4H2PO4, 0.4% (v/v) TFA in 1:1 acetonitrile/water). Samples were analyzed by MALDI-MS with 2,5-dihydroxybenzoic acid matrix solution. Duplicate was performed for all experiments. Data were processed in Data Explorer.
4.6. Molecular modeling
Glide of Schrödinger Maestro, NY, USA (Version 10.1) was used to predict the binding of AH237 against the active site of PRMT 1, 4, 5, and 7. The chemical structure of AH237 was drawn with Ligprep module and ionization state was generated at pH 7.0 ± 2.0 using Epik module. PRMT1 (1OR8), PRMT4 (6S79), PRMT5 (3UA3), and PRMT7 (4M37) were obtained from the Protein Data Bank (www.rcsb.org). Structures were prepared and refined with the protein preparation module and the energy was minimized using OPLS_2005 force field. Grids were generated with Glide by adopting the default parameters. A cubic box of specific dimensions centered around the active site residues was generated for the receptors. The bounding box was set to 15 Å × 15 Å × 15 Å. Flexible ligand docking was performed. Glide extra precision docking was performed by keeping all docking parameters as default. Ligand poses were generated for AH237 by using Monte Carlo random search algorithm, and its binding affinities to PRMTs were predicted with Glide docking score. Post-docking minimizations were taken under OPLS_2005 force field, and 30 poses per ligand were finally saved.
Acknowledgments
The authors acknowledge the support from National Institute of Health (NIH) grants R01GM117275 (RH) and P30 CA023168 (Purdue University Center for Cancer Research, West Lafayette, IN, USA). We also thank supports from the Department of Medicinal Chemistry and Molecular Pharmacology (RH) at Purdue University, West Lafayette, IN, USA.
Author contributions
Rong Huang devised the project and conceived the main conceptual ideas. Rong Huang and Ayad A. Al-Hamashi designed the compounds. Ayad A. Al-Hamashi synthesized, purified, and characterized all compounds. Ayad A. Al-Hamashi, Dongxing Chen, Guangping Dong, and Youchao Deng performed inhibition and selectivity study. Dongxing Chen characterized the inhibition mechanism and Youchao Deng carried out docking studies. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Conflicts of interest
The authors declare no competing financial interest.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting information to this article can be found online at https://doi.org/10.1016/j.apsb.2020.10.013.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Jahan S., Davie J.R. Protein arginine methyltransferases (PRMTs): role in chromatin organization. Adv Biol Regul. 2015;57:173–184. doi: 10.1016/j.jbior.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Gayatri S., Bedford M.T. Readers of histone methylarginine marks. Biochim Biophys Acta-Gene Regul Mech. 2014;1839:702–710. doi: 10.1016/j.bbagrm.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schapira M., de Freitasa R.F. Structural biology and chemistry of protein arginine methyltransferases. Med Chem Commun. 2014;5:1779–1788. doi: 10.1039/c4md00269e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Y., Hadjikyriacou A., Xia Z., Gayatri S., Kim D., Zurita-Lopez C. PRMT9 is a type II methyltransferase that methylates the splicing factor SAP145. Nat Commun. 2015;6:6428. doi: 10.1038/ncomms7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng D., Côté J., Shaaban S., Bedford M.T. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol Cell. 2007;25:71–83. doi: 10.1016/j.molcel.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 6.Yang Y., Bedford M.T. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013;13:37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 7.Krause C.D., Yang Z.H., Kim Y.S., Lee J.H., Cook J.R., Pestka S. Protein arginine methyltransferases: evolution and assessment of their pharmacological and therapeutic potential. Pharmacol Ther. 2007;113:50–87. doi: 10.1016/j.pharmthera.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 8.Bedford M.T., Clarke S.G. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanc R.S., Richard S. Arginine methylation: the coming of age. Mol Cell. 2017;65:8–24. doi: 10.1016/j.molcel.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Al-Hamashi A.A., Diaz K., Hung R. Non-histone arginine methylation by protein arginine methyltransferases. Curr Protein Pept Sci. 2020;21:699–712. doi: 10.2174/1389203721666200507091952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li A.S.M., Li F., Eram M.S., Bolotokova A., dela Seña C.C., Vedadi M. Chemical probes for protein arginine methyltransferases. Methods. 2020;175:30–43. doi: 10.1016/j.ymeth.2019.11.017. [DOI] [PubMed] [Google Scholar]
- 12.Kaniskan H.Ü., Eram M.S., Zhao K., Szewczyk M.M., Yang X., Schmidt K. Discovery of potent and selective allosteric inhibitors of protein arginine methyltransferase 3 (PRMT3) J Med Chem. 2018;61:1204–1217. doi: 10.1021/acs.jmedchem.7b01674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan-Penebre E., Kuplast K.G., Majer C.R., Boriack-Sjodin P.A., Wigle T.J., Johnston L.D. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat Chem Biol. 2015;11:432–437. doi: 10.1038/nchembio.1810. [DOI] [PubMed] [Google Scholar]
- 14.Mitchell L.H., Drew A.E., Ribich S.A., Rioux N., Swinger K.K., Jacques S.L. Aryl pyrazoles as potent inhibitors of arginine methyltransferases: identification of the first PRMT6 tool compound. ACS Med Chem Lett. 2015;6:655–659. doi: 10.1021/acsmedchemlett.5b00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu H., Zheng W., Eram M.S., Vhuiyan M., Dong A., Zeng H. Structural basis of arginine asymmetrical dimethylation by PRMT6. Biochem J. 2016;473:3049–3063. doi: 10.1042/BCJ20160537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunnell E.A., Al-Noori A., Muhsen U., Davies C.C., Dowden J., Dreveny I. Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4) Biochem J. 2020;477:787–800. doi: 10.1042/BCJ20190826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Haren M.J., Marechal N., Troffer-Charlier N., Cianciulli A., Sbardella G., Cavarelli J. Transition state mimics are valuable mechanistic probes for structural studies with the arginine methyltransferase CARM1. Proc Natl Acad Sci U S A. 2017;114:3625–3630. doi: 10.1073/pnas.1618401114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janssen Research & Development, LLC A study of JNJ-64619178, an nnhibitor of PRMT5 in participants with advanced solid tumors, NHL, and lower risk MDS. https://clinicaltrials.gov/ct2/show/NCT03573310 ClinicalTrials.gov, identifier: NCT03573310. June 29, 2018. Available from:
- 19.Zhang G., Richardson S.L., Mao Y., Huang R. Design, synthesis, and kinetic analysis of potent protein N-terminal methyltransferase 1 inhibitors. Org Biomol Chem. 2015;13:4149–4154. doi: 10.1039/c5ob00120j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen D., Li L., Diaz K., Iyamu I.D., Yadav R., Noinaj N. Novel propargyl-linked bisubstrate analogues as tight-binding inhibitors for nicotinamide N-methyltransferase. J Med Chem. 2019;62:10783–10797. doi: 10.1021/acs.jmedchem.9b01255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen D., Dong G., Noinaj N., Huang R. Discovery of bisubstrate inhibitors for protein N-terminal methyltransferase 1. J Med Chem. 2019;62:3773–3779. doi: 10.1021/acs.jmedchem.9b00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen D., Dong C., Dong G., Srinivasan K., Min J., Noinaj N. Probing the plasticity in the active site of protein N-terminal methyltransferase 1 using bisubstrate analogs. J Med Chem. 2020;63:8419–8431. doi: 10.1021/acs.jmedchem.0c00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smil D., Eram M.S., Li F., Kennedy S., Szewczyk M.M., Brown P.J. Discovery of a dual PRMT5-PRMT7 inhibitor. ACS Med Chem Lett. 2015;6:408–412. doi: 10.1021/ml500467h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Volante R.P. A new, highly efficient method for the conversion of alcohols to thiolesters and thiols. Tetrahedron Lett. 1981;22:3119–3122. [Google Scholar]
- 25.van Haren M., van Ufford L.Q., Moret E.E., Martin N.I. Synthesis and evaluation of protein arginine N-methyltransferase inhibitors designed to simultaneously occupy both substrate binding sites. Org Biomol Chem. 2015;13:549–560. doi: 10.1039/c4ob01734j. [DOI] [PubMed] [Google Scholar]
- 26.Khan M.N. Kinetic evidence for the occurrence of a stepwise mechanism in hydrazinolysis of phthalimide. J Org Chem. 1995;60:4536–4541. [Google Scholar]
- 27.Martin N.I., Beeson W.T., Woodward J.J., Marletta M.A. NG-aminoguanidines from primary amines and the preparation of nitric oxide synthase inhibitors. J Med Chem. 2008;51:924–931. doi: 10.1021/jm701119v. [DOI] [PubMed] [Google Scholar]
- 28.Zhang G., Huang R. Facile synthesis of SAM-peptide conjugates through alkyl linkers targeting protein N-terminal methyltransferase 1. RSC Adv. 2016;6:6768–6771. doi: 10.1039/C5RA20625A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richardson S.L., Mao Y., Zhang G., Hanjra P., Peterson D.L., Huang R. Kinetic mechanism of protein N-terminal methyltransferase 1. J Biol Chem. 2015;290:11601–11610. doi: 10.1074/jbc.M114.626846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blair L.P., Avaritt N.L., Huang R., Cole P.A., Taverna S.D., Tackett A.J. MassSQUIRM: an assay for quantitative measurement of lysine demethylase activity. Epigenetics. 2011;6:490–499. doi: 10.4161/epi.6.4.14531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richardson S.L., Hanjra P., Zhang G., Mackie B.D., Peterson D.L., Huang R. A direct, ratiometric, and quantitative MALDI-MS assay for protein methyltransferases and acetyltransferases. Anal Biochem. 2015;478:59–64. doi: 10.1016/j.ab.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Antonysamy S., Bonday Z., Campbell R.M., Doyle B., Druzina Z., Gheyi T. Crystal structure of the human PRMT5 : MEP50 complex. Proc Natl Acad Sci U S A. 2012;109:17960–17965. doi: 10.1073/pnas.1209814109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang C., Zhu Y., Caceres T.B., Liu L., Peng J., Wang J. Structural determinants for the strict monomethylation activity by trypanosoma brucei protein arginine methyltransferase 7. Structure. 2014;22:756–768. doi: 10.1016/j.str.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 34.Zhang X., Cheng X. Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure. 2003;11:509–520. doi: 10.1016/s0969-2126(03)00071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng Y., Hadjikyriacou A., Clarke S.G. Substrate specificity of human protein arginine methyltransferase 7 (PRMT7): the importance of acidic residues in the double e loop. J Biol Chem. 2014;289:32604–32616. doi: 10.1074/jbc.M114.609271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.